Abstract
SMNrp, also termed SPF30, has recently been identified in spliceosomes assembled in vitro. We have functionally characterized this protein and show that it is an essential splicing factor. We show that SMNrp is a 17S U2 snRNP-associated protein that appears in the pre-spliceosome (complex A) and the mature spliceosome (complex B) during splicing. Immunodepletion of SMNrp from nuclear extract inhibits the first step of pre-mRNA splicing by preventing the formation of complex B. Re-addition of recombinant SMNrp to immunodepleted extract reconstitutes both spliceosome formation and splicing. Mutations in two domains of SMNrp, although similarly deleterious for splicing, differed in their consequences on U2 snRNP binding, suggesting that SMNrp may also engage in interactions with splicing factors other than the U2 snRNP. In agreement with this, we present evidence for an additional interaction between SMNrp and the [U4/U6⋅U5] tri-snRNP. A candidate that may mediate this interaction, namely the U4/U6-90 kDa protein, has been identified. We suggest that SMNrp, as a U2 snRNP-associated protein, facilitates the recruitment of the [U4/U6⋅U5] tri-snRNP to the pre-spliceosome.
Keywords: assembly/pre-mRNA splicing/SMNrp/SPF30/spliceosome/U2 snRNP
Introduction
The spliceosome, which catalyses the removal of introns from nuclear pre-mRNA, contains the small nuclear ribonucleoprotein particles (U snRNPs) U1, U2, U4/U6 and U5, and a large number of non-snRNP splicing factors (Krämer, 1996; Burge et al., 1999; Staley and Guthrie, 1998 and references therein). U snRNPs purified from HeLa nuclear extracts under mild conditions can be distinguished by their characteristic sedimentation as 12S U1 snRNP, 17S U2 snRNP and 25S [U4/U6⋅U5] tri-snRNP. At increased salt concentrations, the [U4/U6⋅U5] tri-snRNP particle dissociates into a 20S U5 snRNP and a 12S U4/U6 snRNP, whereas several proteins dissociate from 17S U2 snRNP, giving rise to its 12S form (Behrens and Lührmann, 1991; Behrens et al., 1993a,b). Each of the aforementioned U snRNPs contains the so-called Sm proteins B/B′, D1, D2, D3, E, F and G (Hermann et al., 1995; Séraphin, 1995 and references therein). Recently, close relatives of the Sm proteins, termed Like-Sm (LSm) proteins, have been shown to associate with the U6 snRNA (Achsel et al., 1999; Salgado-Garido et al., 1999). In addition, the U snRNPs contain a characteristic set of specific proteins that are thought to contribute to the individual functions of each particle (Krämer, 1996; Will and Lührmann, 1997; Reed, 2000)
U snRNPs and non-snRNP splicing factors assemble on to pre-mRNA in a highly ordered pathway to form the mature spliceosome. In HeLa cells, pre-mRNA is first bound by an undefined number of RNA-binding proteins (complex H). Recognition of the 5′ splice site by U1 snRNP, and binding of additional non-snRNP factors to the branch-point region then commit pre-mRNA to splicing (Legrain et al., 1988; Séraphin and Rosbash, 1989; Michaud and Reed, 1991). The resulting complex E has also been shown recently to contain loosely bound 17S U2 snRNP (Das et al., 2000). In the next step, U2 snRNP stably binds to the branch-point region in an ATP-dependent fashion, forming the pre-spliceosome (complex A) (Smith et al., 1989; Fu and Maniatis, 1990; Zamore et al., 1992; Das et al., 2000). The addition of the [U4/U6⋅U5] tri-snRNP particle then marks the conversion of the pre-spliceosome into the mature spliceosome (complex B) (Lamm et al., 1991; Maroney et al., 2000). Within complex B, a series of RNA–RNA rearrangements occur, which ultimately form a catalytic RNA network of the spliceosome (Staley and Guthrie, 1998). Once formed, these RNA–RNA interactions allow the chemical reactions of splicing to occur, as monitored by the appearance of complex C. Hereafter, the spliceosome is disassembled and U snRNPs are recycled to engage in a new round of splicing.
The survival of motor neuron (SMN) protein, which is mutated in patients with spinal muscular atrophy, has recently been implicated in the assembly and regeneration of U snRNPs after the splicing reaction (Lefebvre et al., 1995, 1998; Pellizzoni et al., 1998). SMN is the only splicing factor known to date that contains a so-called Tudor domain (Ponting, 1997). This domain can be found in a large number of proteins and is anticipated to mediate protein–protein interactions (Selenko et al., 2001). Indeed, the Tudor domain of SMN functions as a protein–protein interaction surface and mediates binding to the spliceosomal Sm proteins (Bühler et al., 1999). Based on its similarity to SMN, a novel protein termed SMN-related protein (SMNrp) has been identified (Talbot et al., 1998), which likewise contains a Tudor domain. Within this region, the human proteins share 51% identity and 57% similarity, whereas flanking regions are only poorly conserved. Remarkably, in a recent study, SMNrp (here termed SPF30) has been biochemically purified along with spliceosomes assembled in vitro (Neubauer et al., 1998). This raised the intriguing possibility that SMNrp may function in pre-mRNA splicing.
In this study, we provide evidence that SMNrp is a 17S U2 snRNP-associated essential splicing factor found in complexes A and B during the splicing reaction. Immuno depletion of SMNrp from nuclear extracts stalls spliceosome assembly at complex A and consequently inhibits the first transesterification reaction of splicing. By mutational analysis, we show further that the N-terminus and the phylogenetically conserved Tudor domain of SMNrp are required for splicing, but engage in distinct interactions. Finally, evidence is provided that SMNrp interacts in vitro with the [U4/U6⋅U5] tri-snRNP, potentially via direct binding to the U4/U6-90 kDa protein. Based on these data, we propose that SMNrp enters the pre-spliceosome in association with 17S U2 snRNP and mediates the subsequent assembly of the mature spliceosome.
Results
SMNrp localizes in nuclear domains implicated in transcription and splicing
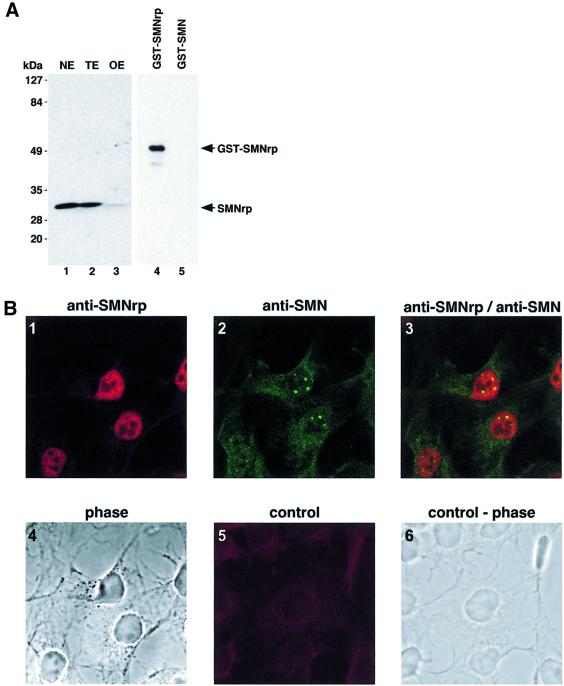
SMNrp has previously been shown to localize predominantly in the nucleus when expressed as a green fluorescent protein (GFP) fusion (Talbot et al., 1998). To examine further the subnuclear localization of endogenous SMNrp, we generated a rabbit polyclonal antiserum against the full-length protein. The specificity of this serum was tested by immunoblotting of Xenopus laevis oocyte extract, as well as nuclear and whole-cell extract from HeLa cells (Figure 1A). A single band migrating at the predicted size of SMNrp was detected in all cell fractions tested (lanes 1–3). Furthermore, the same antiserum discriminates between SMNrp and SMN in western blots (Figure 1A) and in immunoprecipitations (Figure 4C), indicating that the antiserum is monospecific.
Fig. 1. SMNrp localizes in nuclear domains implicated in transcription and splicing. (A) Detection of SMNrp in HeLa nuclear extracts (NE), whole-cell extracts (TE) and Xenopus laevis oocyte extracts (OE) by western blotting using anti-SMNrp antibodies. The anti-SMNrp antibody also efficiently detects recombinant GST-tagged SMNrp (lane 4), but fails to recognize the homologous SMN protein (lane 5). (B) Confocal immunofluorescence microscopy of COS-1 cells with anti-SMNrp antibodies (1), and with anti-SMN antibody 7B10 (2). Panel 3, superimposed images 1 and 2; panel 4, a phase contrast of the same cells; panel 5, a control immunofluorescence with anti-SMNrp antibodies pre-adsorbed with recombinant SMNrp; panel 6, the phase contrast of the same cells. (C) Immunolocalization of SMNrp in the nucleus of an ultrathin section of cryofixed HTC cells by electron microscopy. Most label is associated with perichromatin fibrils (some indicated by small arrows); interchromatin granule clusters (area surrounded and indicated by large arrows) and condensed chromatin (‘C’) remain largely unlabelled. In the lower part of the image, some protein can be seen in the cytoplasm sometimes near the nuclear membrane (NE). Bar, 0.5 µm.
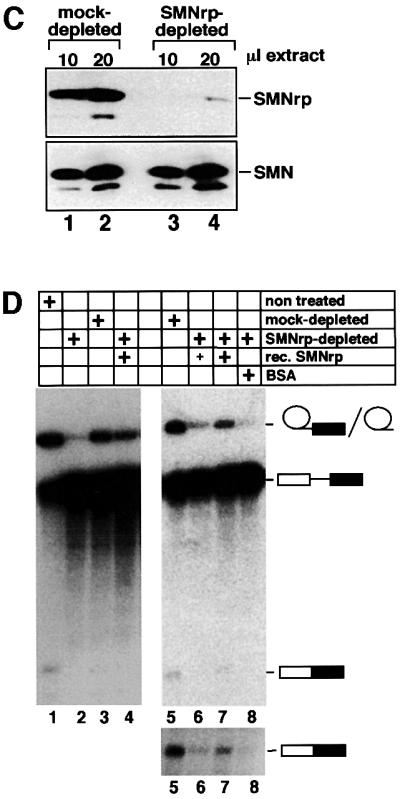
Fig. 4. SMNrp is an essential pre-mRNA splicing factor. (A) Affinity-purified anti-SMNrp antibodies inhibit pre-mRNA splicing in vitro. The splicing reactions were pre-incubated for 30 min with 5 µg of control antibody (lane 2), or 1, 3 or 5 µg of affinity-purified anti-SMNrp antibody (lanes 3–5). Thereafter, 32P-labelled pAd48 pre-mRNA was added and incubated for 1 h. RNA was separated on a denaturing RNA gel and visualized by autoradiography. Lane 6 shows the splicing reaction in the absence of any antibody; lane 1, the pre-mRNA transcript. (B) Analysis of splicing in Xenopus laevis oocytes. pAd48 pre-mRNA was micro-injected into the nucleus of oocytes that were pre-injected with either 200 ng of control antibody (lane 2) or anti-SMNrp antibody (lane 3). Nuclei from injected oocytes were isolated 60 min later and the RNA analysed as described in (A). (C) Western blot analysis of SMNrp (upper panel) and SMN (lower panel) in splicing extracts that were either mock-depleted (lanes 1 and 2) or immuno depleted with anti-SMNrp antibodies (lanes 3 and 4). (D) Recombinant SMNrp reconstitutes pre-mRNA splicing in immunodepleted nuclear extract. Nuclear extracts that had been either immunodepleted (lanes 2, 4 and 6–8), mock-depleted (lanes 3 and 5) or not treated (lane 1) were used for splicing of pAd48 pre-mRNA in vitro. Reactions were supple mented with either 1 µg (lanes 4 and 6) or 5 µg (lane 7) of recombinant SMNrp or with 5 µg of BSA (lane 8). The lower part of the autoradio graphy is also shown in a longer exposure to visualize better the spliced product of the reactions.
The intracellular localization of SMNrp was examined in COS-1 cells by indirect confocal immunofluorescence microscopy (Figure 1B). The nucleoplasm was strongly labelled when probed with anti-SMNrp antibodies (panel 1), whereas pre-absorption of the antibody with recombinant SMNrp strongly reduced the signal (panel 5). The localization of SMNrp appeared to differ from SMN, which was predominantly cytoplasmic and in nuclear gems (panels 2 and 3) (Liu and Dreyfuss, 1996). To gain further insight into the subnuclear distribution of SMNrp, we examined its localization by electron microscopy (Figure 1C). In cryofixed HTC cells, intense labelling with anti-SMNrp antiserum was obtained in the nucleus on the border of condensed chromatin areas mostly associated with perichromatin RNP fibrils (Figure 1C, dark areas, indicated by small arrows) (Fakan, 1994). However, only marginal labelling was observed in clusters of interchromatin granules (large arrows) or at sites of condensed chromatin (marked ‘C’). Thus, SMNrp predominantly localizes in perichromatin fibrils, i.e. the nuclear sites where pre-mRNA splicing takes place (Fakan, 1994).
SMNrp associates with the 17S U2 snRNP and spliceosomal complexes A and B
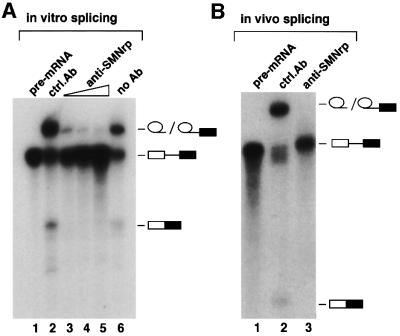
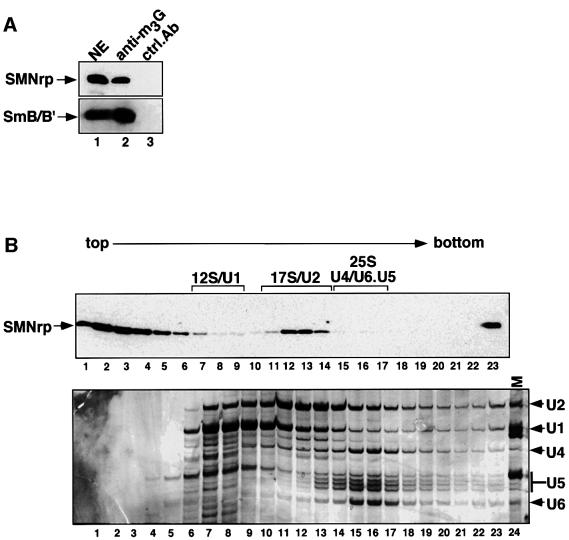
The above results, together with previous data showing that SMNrp co-purified with spliceosomes assembled in vitro (Neubauer et al., 1998), suggested that this protein may be involved in pre-mRNA splicing. To test this possibility, we initially analysed whether SMNrp could be co-eluted with U snRNPs from an anti-m3G-cap affinity column (Bringmann et al., 1983; Will et al., 1993). Components of nuclear extract that bound to the column were eluted with an excess of competing m7G-nucleoside and analysed by western blotting using anti-SMNrp and anti-Sm antibodies. SMNrp was eluted along with U snRNPs from the anti-m3G column, as indicated by the presence of spliceosomal Sm proteins B/B′ and SMNrp in the same eluate (Figure 2A, lane 2, upper and lower panels). Neither of these proteins was eluted from a control column on to which a non-related antibody had been coupled (lane 3 in both panels). This observation encouraged us to analyse further whether SMNrp could bind directly to a specific class of U snRNPs. For this purpose, we analysed the sedimentation of SMNrp in nuclear extract (Figure 2B). As shown in Figure 4B (lower panel), snRNAs were detected according to their characteristic sedimentation of the 12S U1 snRNP (lanes 7–10), 17S U2 snRNP (lanes 11–14) and 25S [U4/U6⋅U5] tri-snRNP (lanes 15–17). An immunoblot of these fractions revealed that SMNrp sediments in three major regions of the gradient corresponding to Svedberg values (S) of <6 (lanes 1–6), 17 (lanes 11–14) and >30 (the pellet fraction 23) (Figure 2B, upper panel). This sedimentation pattern is highly reproducible, although the amount of SMNrp in the 17S region varied between 20 and 80% depending on the nuclear extract prepared (compare Figures 2B, 5B and 7).
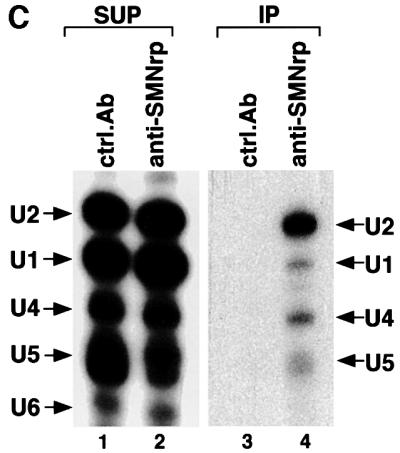
Fig. 2. SMNrp is a 17S U2 snRNP-associated protein. (A) Nuclear extract was passed over an anti-m3G/m7G-column (H-20) (lane 2) or a control column (lane 3). Bound proteins were eluted with m7G-nucleoside and analysed by western blotting. Lane 1 shows 5% of the nuclear extract applied on to the columns. SMNrp and SmB were detected using anti-SMNrp antiserum and Y12 monoclonal antibody, respectively. (B) Sedimentation analysis of SMNrp in nuclear extract by linear 15–45% sucrose gradient centrifugation. Proteins from each fraction were analysed by western blotting using anti-SMNrp antibodies (upper panel). The lower panel shows silver staining of the RNAs from the gradient fractions. (C) SMNrp is associated with 17S U2 snRNP. 17S peak fractions were immunoprecipitated with either anti-SMNrp antiserum (lane 4) or an unrelated control antibody (lane 3). RNAs from the immunoprecipitates (lanes 3 and 4) and 25% of the corresponding supernatants (lanes 1 and 2) were 3′-labelled with [32P]pCp and analysed by denaturing RNA gel electrophoresis.
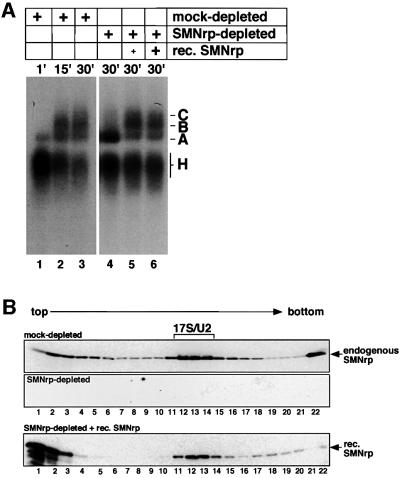
Fig. 5. Spliceosome assembly is arrested in SMNrp-depleted nuclear extract at the level of complex A. (A) pAd48 pre-mRNA was incubated with mock-depleted (lanes 1–3) or SMNrp-depleted (lanes 4–6) nuclear extract and analysed by native gel electrophoresis. Reactions shown in lanes 5 and 6 were complemented with 1, 5 and 3 µg of recombinant SMNrp, respectively. (B) Recombinant SMNrp is incorporated into 17S U2 snRNP in SMNrp-depleted nuclear extract. Nuclear extract that was either mock-depleted (upper panel), SMNrp-depleted (middle panel) or SMNrp-depleted and supplemented with recombinant His-tagged SMNrp (lower panel) was separated by gradient centrifugation and analysed by western blotting using anti-SMNrp antibody.
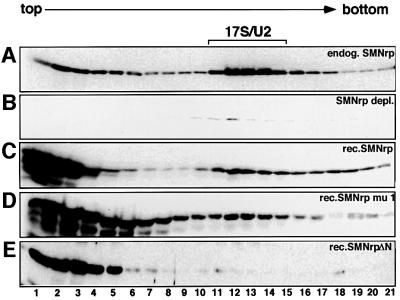
Fig. 7. Association of SMNrp mutants with 17S U2 snRNP. Sucrose gradient centrifugation of SMNrp-depleted extract supplemented with either buffer (B), 0.5 µg of recombinant proteins SMNrp (C), SMNrpmu1 (D) or SMNrpΔN (E). The mock-treated extract is shown in gradient (A). Gradient samples were separated by SDS–PAGE and SMNrp visualized by western blotting using an anti-SMNrp antibody.
The sedimentation of SMNrp in the 17S region raised the question of whether this protein associates with U2 snRNP. Indeed, as shown by radiolabelling of the RNA, the U2 snRNP could be efficiently co-immunoprecipitated with SMNrp from these gradient fractions (Figure 2C, lane 4). Consistently, antibodies directed against the SF3a subunit of the 17S U2 snRNP co-precipitated SMNrp from these fractions (data not shown). We conclude from these data that SMNrp is a 17S U2 snRNP-associated protein.
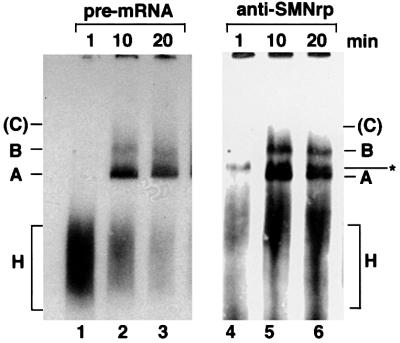
To test whether SMNrp associates with distinct spliceosomal complexes, particularly those containing the U2 snRNP (Behrens et al., 1993a), nuclear extract was incubated with a radiolabelled pre-mRNA encoded by the Adenovirus 2 major late transcription unit (pAd48; Fischer et al., 1995) and analysed by native gel electrophoresis (Figure 3). Spliceosomal complexes were subsequently transferred to a nitrocellulose membrane and visualized by autoradiography (lanes 1–3). The same blot was then probed with anti-SMNrp antibodies (lanes 4–6). SMNrp was readily detectable in splicing complexes A and B and in a band of unknown origin (lanes 5 and 6). However, SMNrp was not detected in complex H or C, although the absence in the latter may be due to the fact that this complex is unstable in native gels. These data suggest that the U2 snRNP-associated SMNrp is incorporated into spliceosomal complexes A and B.
Fig. 3. SMNrp is a component of splicing complexes A and B. Radiolabelled pAd48 pre-mRNA was incubated for the indicated time points with nuclear extract active in splicing and separated by native gel electrophoresis. The gel was subsequently blotted on to a nitrocellulose membrane and splicing complexes were visualized by autoradiography (lanes 1–3). Splicing complexes H, A, B and C are indicated. The unstable complex C is only visible after extended exposure and is hence indicated in brackets. SMNrp was visualized by probing the same blot with anti-SMNrp antibodies (lanes 4–6). The band marked with an asterisk indicates an SMNrp-specific complex of unknown identity that does not contain radiolabelled pre-mRNA.
SMNrp is an essential splicing factor
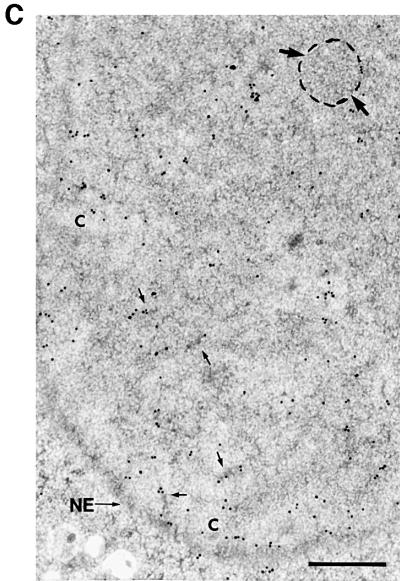
Next we analysed whether antibodies directed against SMNrp could interfere with pre-mRNA splicing in vitro and in vivo (Figure 4A and B). Radiolabelled pAd48 was efficiently spliced when incubated with nuclear extract, giving rise to the excised intron lariat and the ligated exons (Figure 4A, lane 6). As reported earlier, the lariat–exon 2 intermediate that is generated in the first step of splicing co-migrates with the excised intron lariat in this gel, and hence cannot be detected as a separate band (Fischer et al., 1995). Strikingly, addition of anti-SMNrp antibody strongly inhibited the first transesterification reaction, as indicated by the absence of products from either chemical step of splicing (lanes 3–5), whereas a control antibody did not affect splicing (lane 2). To test whether the inhibitory effect on pre-mRNA splicing could also be observed in vivo, we injected pAd48 pre-mRNA into the nucleus of oocytes that were pre-injected with either anti-SMNrp antibody or a control antibody (Figure 4B). The subsequent analysis of the RNA revealed that the anti-SMNrp antibody but not the control antibody blocked the first step of splicing (lanes 2 and 3).
To exclude the possibility that the observed inhibition of splicing by the antibody was indirect (for example, caused by steric hindrance of other proteins in the vicinity of SMNrp) we analysed splicing in nuclear extracts immunodepleted of endogenous SMNrp (Figure 4C and D). Nuclear extract was passed over a protein G–Sepharose column on to which anti-SMNrp antibodies had been coupled. As determined by immunoblotting, this procedure depleted >95% of endogenous SMNrp from the nuclear extract (Figure 4C, compare lanes 1 and 2 with 3 and 4 in the upper panel), whereas the level of the homologous SMN protein, which was monitored as a control, remained unchanged (lower panel). Splicing was severely inhibited in SMNrp-depleted extract (Figure 4D, lane 2) when compared with either untreated or mock-depleted extracts (lanes 1 and 3). Importantly, the inhibition could be reversed by the addition of purified recombinant SMNrp to the immunodepleted extract, but not by an equivalent amount of bovine serum albumin (BSA) (lanes 4 and 8). The same result was obtained from independent experiments using different batches of either recombinant SMNrp or HeLa nuclear extract (Figure 4D, lanes 5–8). Hence, our data provide direct evidence that SMNrp is an essential splicing factor required for the first transesterification reaction.
SMNrp is required for the formation of the mature spliceosome
To gain insight into the mode of action of SMNrp in splicing, radiolabelled pAd48 pre-mRNA was incubated with nuclear extract that was either mock-treated (Figure 5A, lanes 1–3) or depleted of SMNrp (lanes 4–6). As determined by native gel electrophoresis, spliceosome assembly occurred efficiently in the control reaction, as indicated by the early formation of complex H and the appearance of complexes A, B and C at later time points (lanes 2 and 3). In striking contrast, spliceosome assembly in SMNrp-depleted extract proceeded from complex H to A, but subsequent steps were almost entirely blocked (lane 4). Since it is expected that a block in complex B formation results in splicing inhibition at the first step, these results are consistent with, and in fact, further corroborate the results shown in Figure 4.
Importantly, the block in spliceosome assembly could be fully restored by re-addition of SMNrp as indicated by the formation of complexes B and C at the expense of complex A (Figure 5A, lanes 5 and 6). Moreover, when added to immunodepleted extract, recombinant SMNrp was readily incorporated into 17S U2 snRNP as shown by gradient centrifugation (Figure 5B, compare upper and lower panels). Together, these data show that SMNrp is required for the assembly of the mature spliceosome.
Two distinct domains of SMNrp mediate essential steps in spliceosome assembly
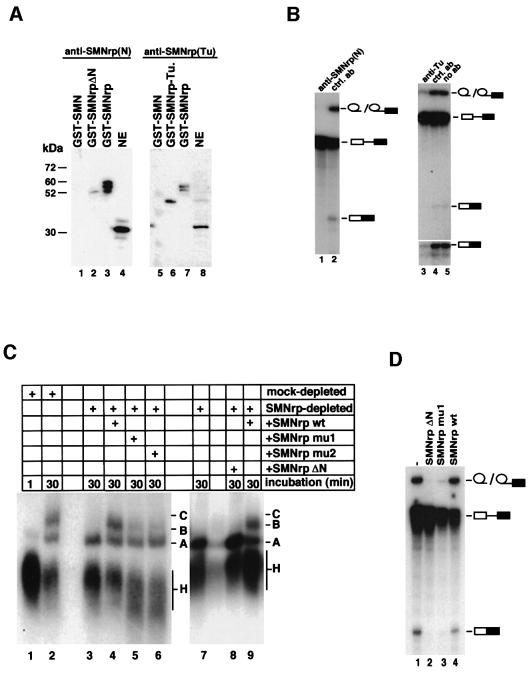
A sequence alignment of SMNrp from human and Schizo saccharomyces pombe indicated that the N-terminus and the Tudor domain have been highly conserved in evolution (Talbot et al., 1998; see also Supplementary data, available at The EMBO Journal Online, for a sequence alignment of SMNrp with a homologue in S.pombe). This raised the possibility that these domains mediate important functions in splicing. Hence, we analysed whether antibodies directed against either domain could interfere with splicing in vitro. Two anti-SMNrp antibodies were generated that specifically recognize either the N-terminus (Figure 6A, lanes 1–4) or the Tudor domain (lanes 5–8), but did not cross-react with SMN (lanes 1 and 5). As shown in Figure 6B, both antibodies inhibited splicing when added to an in vitro splicing reaction (lanes 1 and 3), whereas a non-related antibody had no effect (lanes 2 and 4). To study further the role of both domains, we generated mutants lacking either the N-terminus (SMNrpΔN) or containing amino acid substitutions in the highly conserved positions 83–87 (SMNrpmu1) or 110–115 (SMNrpmu2) of the Tudor domain (Selenko et al., 2001). Interestingly, in contrast to wild-type SMNrp (Figure 6C, lanes 4 and 9), none of the mutants supported the efficient formation of the mature spliceosome in SMNrp-depleted nuclear extract (compare lanes 1 and 2 with 4–6 and 9). Moreover, when added to nuclear extract in excess over endogenous SMNrp, SMNrpΔN and SMNrpmu1, but not the wild-type protein, exhibited a dominant-negative phenotype on pre-mRNA splicing (Figure 6D, lanes 1–4).
Fig. 6. The N-terminus and the Tudor domain of SMNrp mediate essential functions in spliceosome assembly. (A) Western blot analysis of affinity-purified antibodies directed against the N-terminus (lanes 1–4) and Tudor domain (lanes 5–8) of SMNrp. Recombinant GST-tagged SMN (lanes 1 and 5), SMNrp (lanes 3 and 7), SMNrp lacking the N-terminus (SMNrpΔN) (lane 2), Tudor domain of SMNrp (SMNrp-Tu) (lane 6) and nuclear extract (lanes 4 and 8) were tested to define the specificity of the antibodies. (B) Antibodies directed against the N-terminus and Tudor domain of SMNrp interfere with pre-mRNA splicing in vitro. Nuclear extract was incubated with either no antibody (lane 5) or antibodies directed against the N-terminus (lane 1), Tudor domain (lane 3) or an unrelated antibody (lanes 2 and 4). Splicing was analysed as described in Figure 4. The lower part of the autoradiography is also shown in a longer exposure (lanes 3–5). (C) Recombinant SMNrp harbouring amino acid substitutions in the Tudor domain or lacking the N-terminus fails to reconstitute spliceosome assembly in vitro. Mock-depleted (lanes 1 and 2) and SMNrp-depleted (lanes 3–9) nuclear extracts were incubated with 32P-labelled pAd48 pre-mRNA for the indicated time points in the presence of buffer (lanes 1–3 and 7), recombinant wild-type SMNrp (lanes 4 and 9), SMNrpmu1 (lane 5), SMNrpmu2 (lane 6) or SMNrpΔN (lane 8). The spliceosomal complexes were separated by native gel electrophoresis and visualized by autoradiography. (D) SMNrpΔN and SMNrpmu1 exhibit a trans-dominant-negative phenotype on pre-mRNA splicing. 32P-labelled pAd48 pre-mRNA was incubated for 60 min with nuclear extract in the presence of either SMNrp (lane 4), SMNrpmu1 (lane 3), SMNrpΔN (lane 2) (5 µg each) or buffer (lane 1). Splicing was analysed as described in Figure 4.
We next asked whether the aforementioned mutations interfere with the ability of SMNrp to interact with U2 snRNP. Nuclear extract immunodepleted of SMNrp was incubated with either recombinant wild-type SMNrp, SMNrpmu1 or SMNrpΔN, and was subsequently analysed by gradient centrifugation (Figure 7). SMNrp of the mock-treated extract sedimented at the top of the gradient and in the 17S range, whereas hardly any signal was detectable in the SMNrp-depleted extract (compare Figure 7A and B). Likewise, a fraction of recombinant wild-type SMNrp (Figure 7C) as well as SMNrpmu1 (Figure 7D) sedimented at 17S. In contrast, SMNrpΔN could be detected exclusively in the top fractions (Figure 7E). These data suggest that SMNrp contains two domains that are essential for its function in splicing. Both domains are likely to engage in different interactions, as only the N-terminus but not the Tudor domain is required for binding to U2 snRNP.
Association of SMNrp with the [U4/U6⋅U5] tri-snRNP in vitro
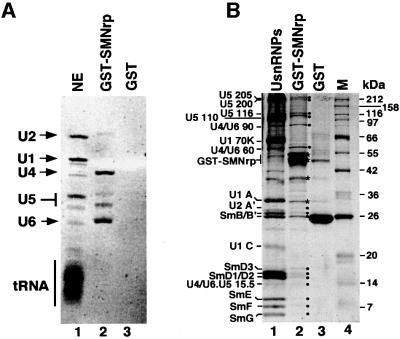
The above data strongly suggested that SMNrp may play an essential role in the conversion of complex A to the mature spliceosome. Since this step is marked by the recruitment of the [U4/U6⋅U5] tri-snRNP, we next tested whether SMNrp can also bind to this snRNP particle. Gradient centrifugation studies revealed no co-sedimentation of SMNrp with the [U4/U6⋅U5] tri-snRNP at low salt (see gradients in Figures 2B, 5B and 7), arguing against a stable association that withstands the centrifugation procedure. However, when an excess of immobilized glutathione S-transferase (GST)-tagged SMNrp was incubated with nuclear extract under splicing conditions, snRNAs U2, U4, U5 and U6 were specifically eluted with glutathione peptide (Figure 8A, lanes 2 and 3). The relatively low amount of U2 snRNA in the eluate is most likely due to a steric hindrance of the U2 snRNP binding domain by the N-terminal GST tag (see also Figure 7). Consistent with the presence of individual snRNAs, the eluate of the GST–SMNrp column contained U5-specific proteins (220, 200, 116 and 110 kDa), U4/U6-specific proteins (90 and 60 kDa), [U4/U6⋅U5] tri- snRNP-specific protein (15.5 kDa), U2-specific protein (A′) and the Sm proteins (B/B′, D1, D2, D3, E, F and G) (Figure 8B, lanes 2 and 3). Hence, these data raise the possibility that SMNrp interacts with not only the U2 snRNP, but also the [U4/U6⋅U5] tri-snRNP.
Fig. 8. Recombinant SMNrp binds to U2 snRNP and the [U4/U6⋅U5] tri-snRNP in vitro. (A) GST–SMNrp or GST tag alone was bound to glutathione–Sepharose and incubated with nuclear extract active in splicing. RNAs from the nuclear extract (lane 1), the GST–SMNrp column (lane 2) and the control column (lane 3) were separated by denaturing RNA gel electrophoresis and visualized by ethidium bromide staining. (B) Protein profile of the same fractions as in (A). Lane 2 shows proteins of U snRNPs affinity purified with H-20 monoclonal antibody.
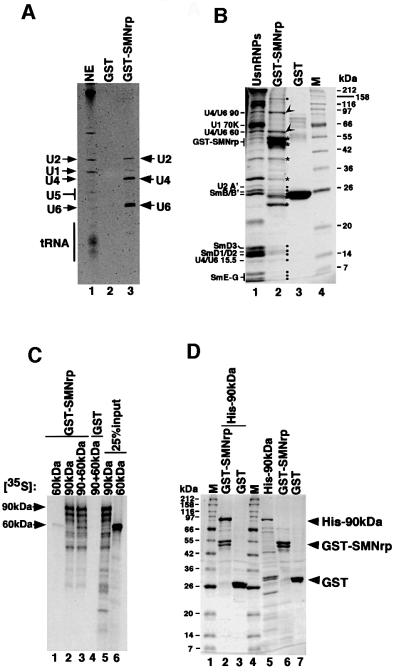
A previous study indicated that the [U4/U6⋅U5] tri-snRNP is labile upon heat treatment and can be dissociated into the snRNPs U4/U6 and U5 (Utans et al., 1992). To identify a potential SMNrp interaction partner within the [U4/U6⋅U5] tri-snRNP, we repeated the binding assay shown in Figure 8 with nuclear extract in which a substantial fraction of [U4/U6⋅U5] tri-snRNPs had been dissociated by repeated freeze–thawing. Under these conditions only U4, U6 and U2 snRNA eluted from the GST–SMNrp matrix (Figure 9A). Consistently, the Sm proteins and the U4/U6-60 kDa and 90 kDa proteins (Lauber et al., 1997) were the major U snRNP components found in the eluate (Figure 9B, compare lanes 2 and 3). This suggested that a component of the U4/U6 snRNP mediates binding to SMNrp. Using in vitro binding assays, neither Sm proteins nor the U4/U6-60 kDa protein associated to a detectable level with immobilized GST– SMNrp (Figure 9B, lane 1 and data not shown). However, SMNrp bound to in vitro translated U4/U6-90 kDa or a preformed complex composed of U4/U6-90 kDa and 60 kDa (lanes 2 and 3). Likewise, recombinant 90 kDa protein bound efficiently and in near stoichiometric amounts to immobilized GST–SMNrp (Figure 9D, lane 2), whereas no binding was detected to the GST tag alone (lane 3). These observations suggest that SMNrp interacts with the [U4/U6⋅U5] tri-snRNP by a direct interaction with one of its protein components, namely the U4/U6-90 kDa protein.
Fig. 9. Binding of SMNrp to the [U4/U6⋅U5] tri-snRNP occurs via a direct interaction with the U4/U6-90 kDa protein. (A) GST–SMNrp binds specifically to U4/U6 and U2 snRNAs in nuclear extract in which the [U4/U6⋅U5] tri-snRNP had been dissociated. RNAs from the nuclear extract (lane 1), the GST–control (lane 2) and the GST–SMNrp binding reaction (lane 3) were separated and visualized as described in Figure 8. (B) Coomassie-stained protein gel of the binding experiment shown in (A). GST–SMNrp and its degradation products are marked with asterisks, U2 snRNP-specific proteins as well as Sm proteins with filled circles, and the U4/U6-specific 60 and 90 kDa proteins with arrowheads (lane 2). Lane 3 shows the protein profile of the GST–control column. (C) SMNrp binds specifically to in vitro translated U4/U6-90 kDa protein. Binding of immobilized GST–SMNrp (lanes 1–3) and GST (lane 4) to in vitro translated and 35S-labelled U4/U6-60 kDa protein (lane 1), 90 kDa protein (lane 2) or a mixture of both (lanes 3 and 4) is shown. Lanes 5 and 6 show 25% of the translated proteins used in the binding assay. (D) Direct binding of SMNrp to the U4/U6-90 kDa protein. Coomassie-stained protein gel showing binding of His-tagged recombinant U4/U6-90 kDa protein to immobilized GST–SMNrp (lane 2) or to GST tag (lane 3). Recombinant proteins used in the binding assay were separated in lanes 5–7.
Discussion
In this study we have functionally characterized SMNrp, a Tudor domain protein previously shown to share homology with the SMN protein (Talbot et al., 1998). SMNrp was recently identified in a preparation of spliceosomal complexes assembled on pre-mRNA in vitro, suggesting that it may function in pre-mRNA splicing (Neubauer et al., 1998). Here we have provided evidence that SMNrp is an essential splicing factor required for the transition from the pre-spliceosome (termed complex A) to the mature spliceosome (termed complex B). In support of this, we have shown by immuno-electron microscopy that SMNrp is associated with perichromatin fibrils, i.e. the major nuclear sites of pre-mRNA splicing (Fakan, 1994). Furthermore, upon gradient centrifugation of nuclear extract and immunoprecipitation, we were able to show that SMNrp interacts specifically with the 17S form of the U2 snRNP. During the course of a splicing reaction, SMNrp was detected in spliceosomal complexes containing the 17S U2 snRNP, namely complexes A and B. Note that due to its instability in native gels, it is unclear at present whether SMNrp interacts with complex C, the catalytically active spliceosome that also contains the 17S U2 snRNP (Krämer, 1996; Burge et al., 1999).
The most important evidence for a function of SMNrp in splicing came from our finding that splicing in nuclear extracts immunodepleted of SMNrp was inhibited at the first chemical step. Importantly, splicing could be restored by the addition of recombinant SMNrp, proving that this effect was due to depletion of SMNrp only. Furthermore, spliceosome formation in immunodepleted extract was halted at the level of complex A, and the re-addition of SMNrp allowed the progression of this complex into mature spliceosomes (complex B). Likewise, anti-SMNrp antibodies blocked the splicing reaction at the level of complex B formation (Figure 4A and B and data not shown). We conclude from these data that SMNrp is a splicing factor required for the assembly of spliceosomes.
It is unclear at present whether SMNrp is an integral component of 17S U2 snRNPs or whether this interaction is transient. SMNrp has not, to date, been identified among the 11 U2-specific proteins that are present in the U2 snRNP (Behrens et al., 1993b). However, it is possible that SMNrp may have eluded detection, since it co-migrates with the U2 snRNP-specific protein A′ in SDS gels (our unpublished observation). Nonetheless, given its low abundance in nuclear extracts, it is more likely that SMNrp interacts only transiently with the U2 snRNP.
Interactions among U2-specific proteins and their binding to the branch-point region of the pre-mRNA have been well characterized in the mammalian and the yeast splicing system (Krämer, 1996; Burge et al., 1999; Reed, 2000). The majority of these proteins are required for binding of the U2 snRNP to the commitment complex (complex E) to form the pre-spliceosome (complex A) (Hodges and Beggs, 1994; Reed, 1996; Caspary and Séraphin, 1998; Caspary et al., 1999 and references therein). In contrast to these proteins, SMNrp does not seem to be required for recruiting U2 snRNP to the pre-spliceosome. This can be inferred from our observation that U2 addition, a requirement for spliceosomal complex A formation, is not affected in mammalian extracts depleted of SMNrp (Figure 5A). Spliceosome formation was also blocked when SMNrp-depleted extract was incubated with a mutant lacking the N-terminus (SMNrpΔN), which is not incorporated into the 17S U2 snRNP (Figure 6C). It is worth noting that the mutant also displayed a dominant-negative effect on splicing in untreated nuclear extract (Figure 6D). In both cases, the electrophoretic migration of the complex formed in the presence of SMNrpΔN is identical to that of complex A in nuclear extracts containing the wild-type protein. In conclusion, these data suggest that SMNrp is not required for the addition of the U2 snRNP to the pre-spliceosome, but instead appears to function in one or more of the subsequent steps that lead to formation of the mature spliceosome.
It is attractive to speculate that binding of the [U4/U6⋅U5] tri-snRNP to the pre-spliceosome (a hallmark of complex B formation) could be mediated, at least in part, by SMNrp. Although still poorly understood, evidence has been accumulated indicating that [U4/U6⋅U5] tri-snRNP addition involves both RNA–RNA and protein–protein interactions. In metazoans, proteins of the SR class of splicing factors appear to promote the binding of this snRNP to the pre-spliceosome (Roscigno and Garcia-Blanco, 1995). In yeast, the [U4/U6⋅U5] tri-snRNP-associated protein Prp31p has been shown to engage in this step (Weidenhammer et al., 1997). At the RNA level, direct contacts between the U2 snRNP and the [U4/U6⋅U5] tri-snRNP have been identified in both experimental systems (Datta and Weiner, 1991; Wolff and Bindereif, 1992; Sun and Manley, 1995). However, it is puzzling that no protein associated with the U2 snRNP has so far been shown to be involved in this event.
A first hint that SMNrp may interact with other spliceosomal components in addition to the U2 snRNP was obtained from the analysis of mutations in its Tudor domain. As opposed to SMNrpΔN, these mutants (SMNrpmu1 and SMNrpmu2) retained their binding affinity for the 17S U2 snRNP, but severely impaired the ability of SMNrp to rescue complex B formation in SMNrp-depleted extract (Figures 6D and 7, and data not shown). Nonetheless, it was unclear whether these mutations indeed abrogated the proposed interaction with the [U4/U6⋅U5] tri-snRNP, particularly since endogenous SMNrp had not been detected so far in gradient fractions containing the [U4/U6⋅U5] tri-snRNP (Figure 2B). On the other hand, it seemed possible that an interaction between SMNrp and the [U4/U6⋅U5] tri-snRNP may not have withstood the experimental conditions previously applied. Support for the latter interpretation was obtained from pull-down experiments (Figures 8 and 9) in which nuclear extract had been incubated with immobilized GST-tagged SMNrp. Here, recombinant SMNrp was in large molar excess over the amount of [U4/U6⋅U5] tri-snRNP in nuclear extract. Under these conditions, not only the U2 snRNP, but also the snRNA and protein constituents of the [U4/U6⋅U5] tri-snRNP were significantly enriched in the eluate, whereas U1 snRNP components were absent (Figure 8). Since more of the U4 and U6 snRNAs were eluted as compared with U5 snRNA (Figure 8), particularly after repeated freeze–thawing of the extract (Figure 9), we suspected that SMNrp not only binds to the fully assembled [U4/U6⋅U5] tri-snRNP, but also to its U4/U6 snRNP subunit. Our speculation that this subunit may harbour an interactor of SMNrp for binding to the [U4/U6⋅U5] tri-snRNP was indeed strengthened by the identification of an interaction between SMNrp and the U4/U6-90 kDa protein (Figure 9). SMNrp binds directly to the U4/U6-90 kDa protein, but does not interact with its binding partner U4/U6-60 kDa (Lauber et al., 1997). In fact, U4/U6-60 kDa could only be co-purified with SMNrp when U4/U6-90 kDa was present (Figure 9). Interestingly, SMNrp also did not bind to the spliceosomal Sm/LSm proteins (data not shown), which have been shown to bind to the Tudor domain of its homologue SMN (Bühler et al., 1999). These data support our view that SMNrp binds to the [U4/U6⋅U5] tri-snRNP in a weak, yet specific, manner via its interaction with the U4/U6-90 kDa protein. The essential role for SMNrp in splicing, concisely in spliceosome formation, may therefore be based on its ability to bind both the U2 snRNP and the [U4/U6⋅U5] tri-snRNP. We therefore suggest that SMNrp is one of the factors that facilitate the recruitment of the [U4/U6⋅U5] tri-snRNP to the pre-spliceosome.
Experiments are currently under way to identify the U2 snRNP protein(s) to which SMNrp binds, and to characterize the domains involved in this event. Detailed knowledge about the conditions and the temporal order of these interactions will be required to clarify whether SMNrp acts as a bridging factor between both snRNPs, or whether it actively induces a structural rearrangement required for their stable association.
Materials and methods
Localization of SMNrp by immunofluorescence and electron microscopy
Immunofluorescence studies were carried out as described previously (Liu and Dreyfuss, 1996). To control the specificity of the antibodies used, affinity-purified anti-SMNrp antibodies were saturated for 1 h with a 5-fold molar excess of recombinant SMNrp and used for immunofluorescence. For immuno-electron microscopy, HTC cells were high-pressure frozen (Hohenberg et al., 1994). The specimens were then cryo-substituted with acetone and embedded into LR White resin (von Schack and Fakan, 1993). Ultrathin sections were incubated with rabbit antiserum against SMNrp (dilution 1:50 or 1:100) following a protocol previously described (Malatesta et al., 1994), and the antigen was visualized by a goat anti-rabbit antibody conjugated with 15 nm colloidal gold particles (Aurion, The Netherlands). Control preparations were incubated in the reaction mixture that did not contain the primary antibody. The sections were contrasted with uranyl acetate and lead citrate and examined in a Philips CM 10 transmission electron microscope at 80 kV, using a 30–40 µm objective aperture.
DNA constructs and expression of recombinant proteins
For cloning of SMNrp, U4/U6-60 kDa and 90 kDa, PCR primers were designed that match the respective 5′ and 3′ ends of the full-length cDNA (Talbot et al., 1998). Using a human fetal brain cDNA library (Clontech) as a template, full-length cDNAs were amplified by PCR and subcloned into pET28a and pGEX vectors. Mutations were introduced into SMNrp cDNA by PCR-mediated mutagenesis. The peptide sequence WSEDG (amino acids 83–87 of the human SMNrp sequence) was substituted by FPDSV (SMNrpmu1) and the sequence GYGNAE (110–115) by VFVDAQ (SMNrpmu2). Expression and in vitro translation of recombinant proteins were carried out as described previously (Bühler et al., 1999).
GST pull-down assays and immunoprecipitations
GST-tagged proteins (25–100 µg) were mixed with glutathione– Sepharose (Amersham Pharmacia Biotech, Germany) and incubated for 1 h at 4°C. After washing, the beads were incubated with 4 ml of nuclear extract for 1 h at 4°C. The experiment shown in Figure 9A and B was carried out with nuclear extract that was repeatedly frozen and thawed in order to destroy the [U4/U6⋅U5] tri-snRNP. After washing with Roeder D buffer the bound proteins were eluted with 10 mM glutathione (Sigma). Fractions were phenol-extracted, and RNA and proteins were analysed by gel electrophoresis. RNAs were visualized using ethidium bromide, the proteins by Coomassie Brilliant Blue. Protein bands were identified by western blotting or by MALDI-TOF as described (Meister et al., 2000). Immunoprecipitations and 3′-labelling of the RNA were carried out as described (Bühler et al., 1999; Meister et al., 2000), except that all washes were carried out with phosphate-buffered saline pH 7.4.
Production of anti-SMNrp antiserum and western blotting
A polyclonal antiserum against SMNrp was generated by repeated immunization of rabbits using an emulsion of complete Freund’s adjuvant and purified His-SMNrp (500 µg) or a purified N-terminal fragment of SMNrp (amino acids 1–72). Three different animals were injected with a recombinant fragment (amino acids 72–140 of the human SMNrp sequence) to produce antibodies directed against the Tudor domain. Serum was harvested from final bleeds and used for affinity purification of SMNrp-specific antibodies (Bühler et al., 1999).
Splicing assays
HeLa nuclear extracts were prepared from fresh cells as described by Dignam et al. (1983). To deplete SMNrp from nuclear extract, 200 µg of affinity-purified anti-SMNrp antibodies were bound to protein G– Sepharose beads in the presence of saturation buffer (0.1 mg/ml BSA, 0.1 µg/ml tRNA and 0.1 µg/ml glycogen). Control beads were incubated with saturation buffer only. Five hundred microlitres of high-salt nuclear HeLa extract (500 mM KCl, 20% glycerol, 20 mM HEPES–KOH pH 7.9, 1.5 mM MgCl2, 0.5 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride) were incubated with the protein G–Sepharose beads for 1 h at 4°C. The beads were subsequently pelleted by centrifugation. The supernatant was dialysed against Roeder D60 buffer and used for splicing assays.
For in vitro splicing we used the 32P-labelled pre-mRNA substrate (Fischer et al., 1995). In vitro splicing reactions were essentially performed as described (Ségault et al., 1995). Reactions contained 30% (v/v) nuclear extract, 2 × 104 c.p.m. of 32P-labelled pre-mRNA, 3 mM MgCl2, 40 mM KCl, 2 mM ATP and 10 mM creatine phosphate in a total volume of 12.5 µl. Pre-incubation with affinity-purified antibodies was carried out for 30 min at 30°C. Analysis of SMNrpΔN and SMNrpmu1 in splicing was performed by adding 5 µg of recombinant His-tagged protein to the splicing reaction. In reconstitution assays, the splice reaction was carried out with 40–50% nuclear extract and complemented with 3 or 5 µg of recombinant His-tagged SMNrp. Reactions were analysed on 5% polyacrylamide–8 M urea gels. For the analysis of spliceosomal complexes, 2 µl of glycerol [86% (w/v)] and 2 µl of heparin (5 mg/ml) were added to the splicing reactions and loaded on to native composite gels containing 0.5% agarose, 3.5% acrylamide (80:1), 10% glycerol and 0.3× TBE (Nelson and Green, 1986). For reconstitution assays, 3 µg of recombinant His-tagged SMNrp, SMNrpmu1 or SMNrpΔN were added to the splicing reactions. Splicing in oocytes was analysed as described in Fischer et al. (1995).
Sedimentation experiments
Five hundred microlitres of nuclear extract were layered on a 15–45% linear sucrose gradient containing 150 mM NaCl, 50 mM Tris–HCl pH 7.4 and 5 mM MgCl2. After centrifugation at 40 000 r.p.m. for 18 h in a Beckman SW41 rotor, fractions were collected and analysed by western blotting using anti-SMNrp antibodies. The RNA of each gradient fraction was phenol-extracted, ethanol-precipitated and visualized by silver staining (Merril et al., 1983). For reconstitution of 17S U2 snRNPs, 500 µl of SMNrp depleted nuclear extract were incubated with 0.5 µg of recombinant His-SMNrp, His-SMNrpmu1 or His-SMNrpΔN for 20 min at 30°C. The sample was layered on to a gradient under the same conditions as described above. Fractions were analysed by western blotting using an affinity-purified antibody directed against SMNrp.
Supplementary data
Supplementary data for this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We are grateful to D.Bühler, C.Will and R.Lührmann for helpful discussions, G.Sowa, J.Köhler, J.Fakan, V.Mamin, D.Studer and F.Voinesco for technical support and R.Reed for antibodies. This work was supported by the MPI, the DFG (DFG Fi 573/2-1 to U.F.) and the Swiss National Science foundation (31-53944.98 to S.F.). B.L. receives an FCI Liebig-Scholarship.
References
- Achsel T., Brahms,H., Kastner,B., Bachi,A., Wilm,M. and Lührmann,R. (1999) A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J., 18, 5789–5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens S.E. and Lührmann,R. (1991) Immunoaffinity purification of a U4/U6⋅U5 tri-snRNP from human cells. Genes Dev., 5, 1439–1452. [DOI] [PubMed] [Google Scholar]
- Behrens S.E., Gallison,F., Legrain,P. and Lührmann,R. (1993a) Evidence that the 60-kDa protein of 17S U2 small nuclear ribonucleoprotein is immunologically and functionally related to the yeast PRP9 splicing factor and is required for the efficient formation of prespliceosome. Proc. Natl Acad. Sci. USA, 90, 8229–8233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens S.E., Tyc,K., Kastner,B., Reichelt,J. and Lührmann,R. (1993b) Small nuclear ribonucleoprotein (RNP) U2 contains numerous additional proteins and has a bipartite RNP structure under splicing conditions. Mol. Cell. Biol., 13, 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringmann P., Rinke,J., Appel,B., Reuter,R. and Lührmann,R. (1983) Purification of snRNPs U1, U2, U4, U5 and U6 with 2,2,7-trimethylguanosine-specific antibody and definition of their constituent proteins reacting with anti-Sm and anti-(U1)RNP antisera. EMBO J., 2, 1129–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bühler D., Raker,V., Lührmann,R. and Fischer,U. (1999) Essential role for the Tudor domain of SMN in spliceosomal U snRNP assembly: implications for spinal muscular atrophy. Hum. Mol. Genet., 8, 2351–2357. [DOI] [PubMed] [Google Scholar]
- Burge C.B., Tuschl,T. and Sharp,P.A. (1999) Splicing of precursors to mRNAs by the spliceosome. In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 525–560.
- Caspary F. and Séraphin,B. (1998) The yeast U2A′/U2B′′; complex is required for pre-spliceosomal formation. EMBO J., 17, 6348–6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspary F., Shevchenko,A., Wilm,M. and Séraphin,B. (1999) Partial purification of the yeast U2 snRNP reveals a novel yeast pre-mRNA splicing factor required for pre-spliceosomal assembly. EMBO J., 18, 3463–3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R., Zhou,Z. and Reed,R. (2000) Functional association of U2 snRNP with the ATP-independent spliceosomal complex E. Mol. Cell, 5, 779–787. [DOI] [PubMed] [Google Scholar]
- Datta B. and Weiner,A.M. (1991) Genetic evidence for base pairing between U2 and U6 snRNA in mammalian mRNA splicing. Nature, 352, 821–824. [DOI] [PubMed] [Google Scholar]
- Dignam J.D., Lebivitz,R.M. and Roeder,G. (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res., 11, 1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakan S. (1994) Perichromatin fibrils are in situ forms of nascent transcripts. Trends Cell Biol., 4, 86–90. [DOI] [PubMed] [Google Scholar]
- Fischer U., Huber,J., Boelens,W.C., Mattaj,I.W. and Lührmann,R. (1995) The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell, 82, 475–483. [DOI] [PubMed] [Google Scholar]
- Fischer U., Liu,Q. and Dreyfuss,G. (1997) The SMN–SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell, 90, 1023–1029. [DOI] [PubMed] [Google Scholar]
- Fu X.D. and Maniatis,T. (1990) Factor required for mammalian spliceosome assembly is localised to distinct regions in the nucleus. Nature, 343, 437–441. [DOI] [PubMed] [Google Scholar]
- Hermann H., Fabrizio,P., Raker,V.A., Foulaki,K., Hornig,H., Brahms,H. and Lührmann,R. (1995) snRNP Sm proteins share two evolutionarily conserved sequence motifs which are involved in Sm protein–protein interactions. EMBO J., 14, 2076–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges P.E. and Beggs,J.D. (1994) RNA splicing. U2 fulfils a commitment. Curr. Biol., 4, 264–267. [DOI] [PubMed] [Google Scholar]
- Hohenberg H., Mannweiler,K. and Müller,M. (1994) High-pressure freezing of cell suspensions in cellulose capillary tubes. J. Microsc., 175, 34–43. [DOI] [PubMed] [Google Scholar]
- Krämer A. (1996) The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu. Rev. Biochem., 65, 367–409. [DOI] [PubMed] [Google Scholar]
- Lamm G.M., Blencowe,B.J., Sproat,B.S., Iribarren,A.M., Ryder,U. and Lammond,A.I. (1991) Antisense probes containing 2-aminoadenosine allow efficient depletion of U5 snRNP from HeLa splicing extracts. Nucleic Acids Res., 19, 3193–3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber J., Plessel,G., Prehn,S., Will,C.L., Fabrizio,P., Groning,K., Lane,W.S. and Lührmann,R. (1997) The human U4/U6 snRNP contains 60 and 90 kDa proteins that are structurally homologous to the yeast splicing factors Prp4p and Prp3p. RNA, 3, 926–941. [PMC free article] [PubMed] [Google Scholar]
- Lefebvre S. et al. (1995) Identification and characterization of a spinal muscular atrophy-determining gene. Cell, 80, 155–165. [DOI] [PubMed] [Google Scholar]
- Lefebvre S., Bürglen,L., Frezal,J., Munnich,A. and Melki,J. (1998) The role of the SMN gene in proximal spinal muscular atrophy. Hum. Mol. Genet., 7, 1531–1536. [DOI] [PubMed] [Google Scholar]
- Legrain P., Séraphin,B. and Rosbash,M. (1988) Commitment of yeast pre-mRNA to the spliceosome pathway does not require U2 snRNP. Mol. Cell. Biol., 8, 3755–3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q. and Dreyfuss,G. (1996) A novel nuclear structure containing the survival of motor neurons protein. EMBO J., 15, 3555–3565. [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Fischer,U., Wang,F. and Dreyfuss,G. (1997) The spinal muscular atrophy disease gene product, SMN and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell, 90, 1013–1021. [DOI] [PubMed] [Google Scholar]
- Malatesta M., Zancanaro,C., Martin,T.E., Chan,E.K.L., Amalric,F., Lührmann,R., Vogel,P. and Fakan,S. (1994) Cytochemical and immunocytochemical characterization of nuclear bodies during hibernation. Eur. J. Cell Biol., 65, 82–93. [PubMed] [Google Scholar]
- Maroney P.A., Romfo,C.M. and Nilsen,T.W. (2000) Functional recognition of the 5′ splice site by U4/U6⋅U5 tri-snRNP defines a novel ATP dependent step in early spliceosome assembly. Mol. Cell, 6, 317–328. [DOI] [PubMed] [Google Scholar]
- Meister G., Bühler,D., Laggerbauer,B., Zobawa,M., Lottspeich,F. and Fischer,U. (2000) Characterisation of a nuclear 20S complex containing the survival of motor neurons (SMN) protein and a specific subset of spliceosomal Sm proteins. Hum. Mol. Genet., 9, 1977–1986. [DOI] [PubMed] [Google Scholar]
- Merril C.R., Goldman,D. and Van Keuren,M.L. (1983) Silver staining methods for polyacrylamide gel electrophoresis. Methods Enzymol., 96, 230–239. [DOI] [PubMed] [Google Scholar]
- Michaud S. and Reed,R. (1991) An ATP-independent complex commits pre-mRNA to the mammalian spliceosome assembly pathway. Genes Dev., 5, 2534–2546. [DOI] [PubMed] [Google Scholar]
- Nelson K. and Green,M. (1988) Splice site selection and ribonucleo protein complex assembly during in vitro pre-mRNA splicing. Genes Dev., 2, 319–329. [DOI] [PubMed] [Google Scholar]
- Neubauer G., King,A., Rappsilber,J., Calvio,C., Watson,M., Ajuh,P., Sleeman,J., Lamond,A. and Mann,M. (1998) Mass spectrometry and EST-database searching allows characterization of the multi-protein spliceosome complex. Nature Genet., 20, 46–50. [DOI] [PubMed] [Google Scholar]
- Pellizzoni L., Kataoka,N., Charroux,B. and Dreyfuss,G. (1998) A novel function of SMN, the spinal muscular atrophy disease gene product, in pre-mRNA splicing. Cell, 95, 615–624. [DOI] [PubMed] [Google Scholar]
- Ponting C.P. (1997) Tudor domains in proteins that interact with RNA. Trends Biochem. Sci., 22, 51–52. [DOI] [PubMed] [Google Scholar]
- Reed R. (1996) Initial splice-site recognition and pairing during pre-mRNA splicing. Curr. Opin. Genet. Dev., 6, 215–220. [DOI] [PubMed] [Google Scholar]
- Reed R. (2000) Mechanisms of fidelity in pre-mRNA splicing. Curr. Opin. Cell Biol., 12, 340–345. [DOI] [PubMed] [Google Scholar]
- Roscigno R.F. and Garcia-Blanco,M.A. (1995) SR proteins escort the U4/U6⋅U5 tri-snRNP to the spliceosome. RNA, 1, 692–706. [PMC free article] [PubMed] [Google Scholar]
- Salgado-Garrido J., Bragado-Nilsson,E., Kandel-Lewis,S. and Séraphin,B. (1999) Sm and Sm-like proteins assemble in two related complexes of deep evolutionary origin. EMBO J., 18, 3451–3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ségault V., Will,C.L., Sproat,B.S. and Lührmann,R.L. (1995) In vitro reconstitution of mammalian U2 and U5 snRNPs active in splicing: Sm proteins are functionally interchangeable and are essential for the formation of functional U2 and U5 snRNPs. EMBO J., 14, 4010–4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selenko P., Sprangers,R., Stier,G., Bühler,D., Fischer,U. and Sattler,M. (2001) Structure and Sm protein interaction of the SMN Tudor domain. Nature Struct. Biol., 8, 27–31. [DOI] [PubMed] [Google Scholar]
- Séraphin B. (1995) Sm and Sm-like proteins belong to a large family: identification of proteins of the U6 as well as the U1, U2, U4 and U5 snRNPs. EMBO J., 14, 2089–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séraphin B. and Rosbash,M. (1989) Identification of functional U1 snRNP–pre-mRNA complexes committed to spliceosome assembly and splicing. Cell, 59, 349–358. [DOI] [PubMed] [Google Scholar]
- Smith C.W.J., Porro,E.B., Patton,J.G. and Nadal-Ginard,B. (1989) Scanning from an independently specified branch point defines the 3′ splice site of mammalian introns. Nature, 342, 243–247. [DOI] [PubMed] [Google Scholar]
- Staley J.P. and Guthrie,C. (1998) Mechanical devices of the spliceosome: motors, clocks, springs and things. Cell, 92, 315–326. [DOI] [PubMed] [Google Scholar]
- Sun J.S. and Manley,J.L. (1995) A novel U2-U6 snRNA structure is necessary for mammalian mRNA splicing. Genes Dev., 9, 843–854. [DOI] [PubMed] [Google Scholar]
- Talbot K., Miguel-Aliaga,I., Mohaghegh,P., Ponting,C.P. and Davies,K.E. (1998) Characterization of a gene encoding survival motor neuron (SMN)-related protein, a constituent of the spliceosome complex. Hum. Mol. Genet., 7, 2149–2156. [DOI] [PubMed] [Google Scholar]
- Utans U., Behrens,S.E., Lührmann,R., Kole,R. and Krämer,A. (1992) A splicing factor that is inactivated during in vivo heat shock is functionally equivalent to the [U4/U6⋅U5] triple snRNP-specific proteins. Genes Dev., 6, 631–641. [DOI] [PubMed] [Google Scholar]
- Von Schack M.L. and Fakan,S. (1993) The study of the cell nucleus using cryofixation and cryosubstitution. Micron, 24, 507–519. [Google Scholar]
- Weidenhammer E.M., Ruiz-Noriega,M. and Woolford,J.L. (1997) Prp31p promotes the association of the U4/U6⋅U5 tri-snRNP with prespliceosome to form spliceosomes in Saccharomyces cerevisiae. Mol. Cell. Biol., 17, 3580–3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will C.L. and Lührmann,R. (1997) Protein functions in pre-mRNA splicing. Curr. Opin. Cell Biol., 9, 320–328. [DOI] [PubMed] [Google Scholar]
- Will C.L., Behrens,S.E. and Lührmann,R. (1993) Protein composition of mammalian spliceosomal snRNPs. Mol. Biol. Rep., 18, 121–126. [DOI] [PubMed] [Google Scholar]
- Wolff T. and Bindereif,A. (1992) Reconstituted mammalian U4/U6 snRNP complements splicing: a mutational analysis. EMBO J., 11, 345–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore P.D., Patton,J.D. and Green,M.R. (1992) Cloning and domain structure of the mammalian splicing factor U2AF. Nature, 355, 609–614. [DOI] [PubMed] [Google Scholar]