Abstract
eIF5 stimulates the GTPase activity of eIF2 bound to Met-tRNAiMet, and its C-terminal domain (eIF5-CTD) bridges interaction between eIF2 and eIF3/eIF1 in a multifactor complex containing Met-tRNAiMet. The tif5-7A mutation in eIF5-CTD, which destabilizes the multifactor complex in vivo, reduced the binding of Met-tRNAiMet and mRNA to 40S subunits in vitro. Interestingly, eIF5-CTD bound simultaneously to the eIF4G subunit of the cap-binding complex and the NIP1 subunit of eIF3. These interactions may enhance association of eIF4G with eIF3 to promote mRNA binding to the ribosome. In vivo, tif5-7A eliminated eIF5 as a stable component of the pre-initiation complex and led to accumulation of 48S complexes containing eIF2; thus, conversion of 48S to 80S complexes is the rate-limiting defect in this mutant. We propose that eIF5-CTD stimulates binding of Met-tRNAiMet and mRNA to 40S subunits through interactions with eIF2, eIF3 and eIF4G; however, its most important function is to anchor eIF5 to other components of the 48S complex in a manner required to couple GTP hydrolysis to AUG recognition during the scanning phase of initiation.
Keywords: eukaryotic translation initiation factor (eIF)/GAP/Met-tRNAiMet binding/mRNA binding/scanning
Introduction
The selection of initiation codons in mRNAs during the initiation of protein synthesis is a highly regulated process in eukaryotic cells, involving the 40S ribosomal subunit and numerous eukaryotic initiation factors (eIFs). The 40S ribosome binds to the eIF2–GTP–methionyl initiator tRNA (Met-tRNAiMet) ternary complex (TC) to form the 43S pre-initiation complex. Subsequent joining of the mRNA in association with eIF4F bound to the m7G cap produces the 48S pre-initiation complex. eIF3 stimulates recruitment of Met-tRNAiMet and mRNA to the 40S ribosome (for a review see Hershey and Merrick, 2000). Formation of a 48S complex positioned at the AUG start codon is dependent on eIF1, eIF1A, eIF4A and eIF4B, in addition to eIF2, eIF3 and eIF4F (Pestova et al., 1998). In current models, eIF5 binds to a positioned 48S complex and stimulates GTP hydrolysis by eIF2, acting as a GTPase-activating protein (GAP), prior to ejection of all eIFs and joining of the 60S subunit (Chakrabarti and Maitra, 1991; Huang et al., 1997). In the yeast Saccharo myces cerevisiae, SUI (suppressor of initiation codon) mutations were isolated in eIF1, eIF5 and all three subunits of eIF2, which allow ribosomes to select UUG as the start codon at elevated frequencies (for a review see Donahue, 2000). Biochemical analysis suggests that a higher rate of GTP hydrolysis on eIF2 is responsible for the reduced accuracy of AUG selection in the Sui– mutants (Huang et al., 1997). Thus, accurate initiation requires tight coupling between the GAP activity of eIF5 and base pairing between Met-tRNAiMet and the start codon.
eIF1 and the C-terminal domain of eIF5 (eIF5-CTD) both bind to the eIF3 subunit NIP1 (Asano et al., 1998, 2000; Phan et al., 1998), suggesting that the functions of eIF1 and eIF5 in AUG recognition are coordinated by eIF3. Additionally, eIF5-CTD can bind to the β-subunit of its substrate eIF2 (Asano et al., 1999). A bipartite sequence motif of aromatic and acidic amino acids (AA-boxes) in eIF5-CTD is required for its interaction with eIF3-NIP1 and eIF2β (Asano et al., 1999; Das and Maitra, 2000), and the latter interaction is dependent on lysine-rich stretches (K-boxes) in the N-terminal half of eIF2β (Das et al., 1997; Asano et al., 1999). Alanine substitutions altering all 12 conserved residues in AA-box 1 (tif5-12A) or all seven residues in AA-box 2 (tif5-7A) of eIF5-CTD disrupted its interactions with eIF3-NIP1 and eIF2β in vitro. These mutations are lethal (tif5-12A) or confer temperature sensitivity (Ts–) and slow growth (Slg–) (tif5-7A) in yeast cells. At a semi-permissive temperature, tif5-7A disrupted interaction between eIF5 and native eIF2 and eIF3 complexes in vivo (Asano et al., 1999).
The interactions of eIF3-NIP1 with eIF1 and eIF5-CTD, and of eIF5-CTD with eIF3-NIP1 and eIF2β, occur simultaneously in vitro, suggesting that these eIFs reside in the same multifactor complexes (MFCs). We observed such complexes containing eIFs 1, 2, 3 and 5 and stoichiometric amounts of tRNAiMet in cell extracts, and showed that they were destabilized by the tif5-7A mutation in eIF5-CTD or by K-box mutations in eIF2β. As tif5-7A reduced the polyribosome content in yeast cells, it appears that the MFC is an important intermediate in translation initiation in vivo (Asano et al., 2000). The MFC could be purified free of ribosomes, suggesting that its constituent factors bind to 40S subunits as a pre-formed unit and remain associated in the 48S initiation complex during the scanning process. If so, the functions of eIFs 1, 2, 3 and 5 in recruitment of Met-tRNAiMet and mRNA to the ribosome, and in AUG recognition, might all depend on the integrity of the MFC.
In this study, we investigated which reactions in the initiation pathway are impaired when the MFC is disrupted by the tif5-7A mutation in eIF5-CTD. Consistent with the fact that eIF5-CTD bridges interaction between eIF2 and eIF3, and the known function of eIF3 in TC binding, we found that tif5-7A decreases the rate of Met-tRNAiMet binding to 40S subunits in vitro. Interestingly, mRNA binding to 40S ribosomes was also compromised by this mutation. Pursuing the latter finding, we discovered that eIF5-CTD interacts with the C-terminal half of eIF4G, the largest subunit of eIF4F (Hentze, 1997; Sachs et al., 1997), and bridges interaction between eIF3 and eIF4G both in vivo and in vitro.
In vivo, we found that tif5-7A greatly reduced the association of eIF5 with native 43–48S complexes. Moreover, 48S complexes containing all other components of the MFC accumulated in the tif5-7A mutant, indicating a block late in the pathway where eIF5 GAP function is required. Thus, while eIF5-CTD promotes Met-tRNAiMet and mRNA binding to 40S subunits in vitro, its rate-limiting function in vivo is the stable incorporation of eIF5 into pre-initiation complexes in a manner required for coupling GTP hydrolysis to AUG recognition.
Results
tif5-7A impairs binding of Met-tRNAiMet and poly(A) mRNA to 40S ribosomes in vitro
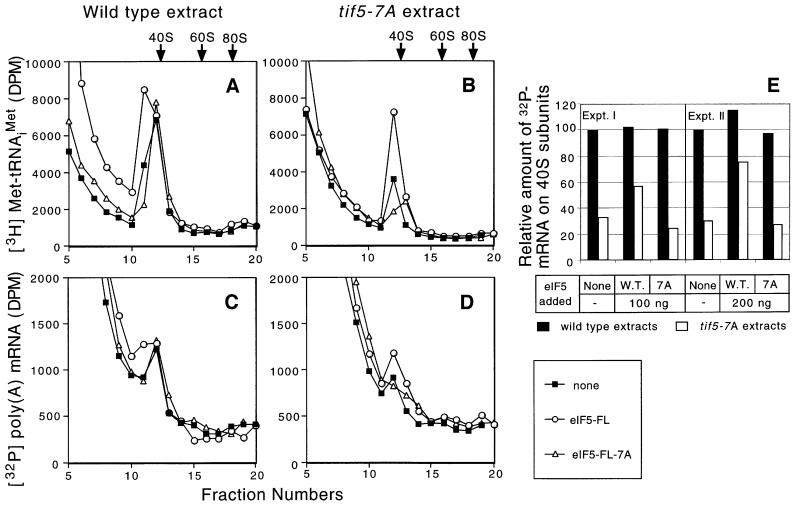
Given the function of eIF3 in stimulating TC binding to 40S ribosomes (Hinnebusch, 2000) and the role of eIF5-CTD in bridging interaction between eIF3 and eIF2 in the MFC (Asano et al., 2000), we hypothesized that eIF5-CTD would promote TC binding to 40S ribosomes. To test this idea, we prepared extracts from isogenic wild-type and tif5-7A mutants and assayed the transfer of exogenous [3H]Met-tRNAiMet to 40S ribosomes in the presence of a non-hydrolyzable GTP analog (GMPPNP). The latter was included to prevent hydrolysis of GTP in the TC and subsequent joining of 60S subunits to 48S initiation complexes (Phan et al., 1998). As shown in Figure 1A and B (filled squares), much less [3H]Met-tRNAiMet bound to 40S ribosomes in the tif5-7A extract compared with the wild-type extract. Importantly, this defect was complemented by addition of purified FLAG epitope-tagged eIF5 (eIF5-FL; open circles) but not by the tif5-7A mutant version of this protein (eIF5-FL-7A; open triangles). Thus, tif5-7A decreased the binding of Met-tRNAiMet to 40S ribosomes in cell extracts.
Fig. 1. tif5-7A impairs the binding of Met-tRNAiMet and poly(A) mRNA to 40S ribosomes in vitro. Translation-competent extracts were prepared from KAY35 (TIF5) and KAY36 (tif5-7A) (Asano et al., 1999) and measured for the ability to transfer exogenous [3H]Met-tRNAiMet (A and B) and [32P]poly(A) MFA2 mRNA (C and D) to 40S ribosomes in the presence of 1.2 mM GMPPNP. Some of the reactions also contained 0.2 µg of purified native eIF5-FL or eIF5-FL-7A. The reactions were resolved by sucrose gradient–velocity sedimentation and the amounts of [3H]Met-tRNAiMet and [32P]poly(A) mRNA in each reaction were determined by scintillation counting. Arrows indicate the positions of 40S, 60S and 80S ribosomes, determined from the A254 profiles of the gradients. The data shown are typical results of several independent experiments. (E) The data from two independent experiments of the type described in (C) and (D) were quantified by summing the radioactivity in fractions 11–13 and subtracting the baseline values obtained with wild-type or mutant extracts to which [32P]poly(A) mRNA, but no eIF5, was added and the incubation at 26°C was omitted.
We also examined the effect of tif5-7A on mRNA binding to 40S ribosomes in the same extracts using exogenous [32P]MFA2 polyadenylated mRNA (Tarun and Sachs, 1995), again in the presence of GMPPNP. Interestingly, [32P]MFA2 mRNA binding was reduced significantly in the tif5-7A extract (Figure 1C and D, filled squares), and this defect was complemented by purified wild-type eIF5-FL (open circles) but not by eIF5-FL-7A (open triangles). The defect in mRNA binding could be a secondary consequence of reduced Met-tRNAiMet binding in the tif5-7A extract (Hinnebusch, 2000). Alternatively, it may signify a role for eIF5-CTD in mRNA recruitment to the 40S ribosome during 48S complex assembly.
tif5-7A reduces interaction between eIF3 and eIF4G in vivo
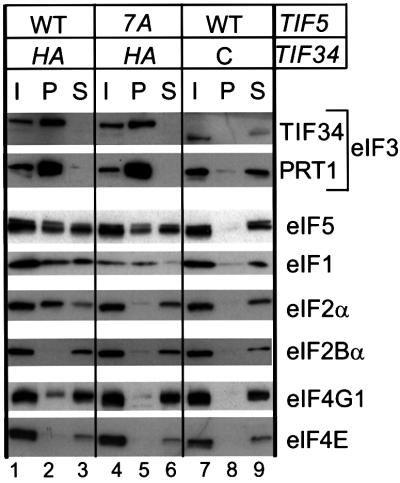
In view of the evidence that mRNA is recruited to the 40S ribosome through interaction between eIF3 and eIF4G (Sachs et al., 1997), and the fact that eIF5 is stably associated with eIF3, we considered the possibility that eIF5-CTD contributes to association between eIF4G and eIF3. If so, the mRNA binding defect conferred by tif5-7A could arise at least partly from destabilization of an eIF3–eIF5–eIF4G complex. Previously, we detected association of a small fraction of eIF4G with eIF3 in yeast cell extracts (Phan et al., 1998). To determine whether tif5-7A reduces this interaction, we immunoprecipitated eIF3 using anti-hemagglutinin (HA) antibodies directed against an HA epitope-tagged version of eIF3 subunit TIF34. As expected, the majority of eIF3 subunit PRT1 co-immunoprecipitated with HA-TIF34 from both TIF5 and tif5-7A extracts (Figure 2, lanes 1–6), whereas no PRT1 was immunoprecipitated from an extract containing untagged TIF34 (lanes 7–9). Probing the immune complexes with antibodies against eIF5, eIF1 and eIF2α confirmed that tif5-7A disrupted association between eIF2 and eIF3/eIF1 in vivo (Asano et al., 2000). Probing the same complexes for eIF4G1, one of the two isoforms of yeast eIF4G, we found that ∼3% of eIF4G1 specifically co-immunoprecipitated with HA-TIF34 (Figure 2, lanes 2 and 8), and this interaction was reduced by tif5-7A (lanes 2 and 5). Thus, it is possible that eIF5-CTD enhances interaction between eIF3 and eIF4G. None of the m7G cap-binding subunit of eIF4F, eIF4E, was co-immunoprecipitated with HA-TIF34 (Figure 2, lane 2), suggesting that yeast eIF4F complexes associated with eIF3 are relatively unstable.
Fig. 2. The eIF5-CTD mediates interaction of eIF3 with eIF2 and eIF4G in vivo. Whole-cell extracts (WCEs) were prepared from strains KAY50 (TIF34-HA TIF5-FL) (lanes 1–3), KAY51 (TIF34-HA tif5-FL-7A) (lanes 4–6) and KAY37 (TIF34 TIF5-FL) (lanes 7–9) (Asano et al., 2000) grown in YPD medium at 30°C. Aliquots of WCEs were incubated with anti-HA affinity resin and, after extensive washing, the bound proteins were analyzed by SDS–PAGE and immunoblotting with antibodies against the proteins indicated on the right. Lanes 1, 4 and 7, 20% input (I) amounts of WCE; lanes 2, 5 and 8, the entire precipitated (P) fractions; lanes 3, 6 and 9, 10% of supernatant (S) fractions. The top panel describes the presence of wild-type (WT) or tif5-7A (7A) forms of eIF5 in the extracts, and the presence (HA) or absence (C) of the HA-epitope on eIF3-TIF34.
eIF5-CTD binds directly to the C-terminal half of eIF4G2 in vitro
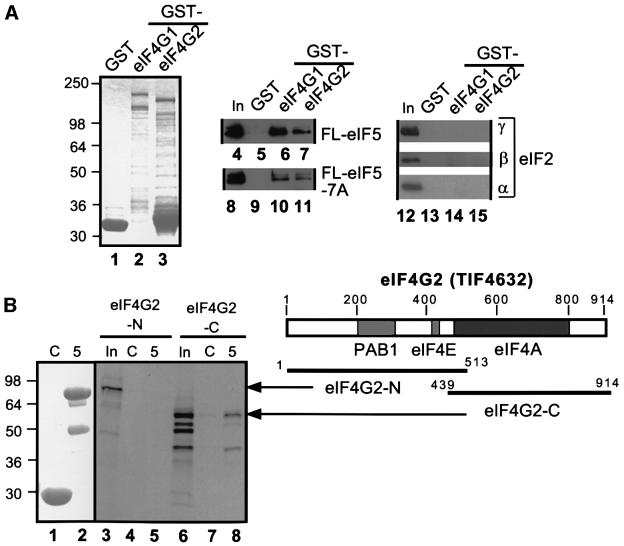
The results described above raised the possibility that eIF5-CTD binds directly to eIF4G to stabilize eIF3–eIF4G association. Accordingly, we tested purified eIF5-FL for binding to glutathione S-transferase (GST) fusions made to full-length eIF4G1 or eIF4G2. Both fusions bound specifically to wild-type eIF5-FL (Figure 3A, lanes 4–7) and this interaction was reduced to ∼30–50% by the tif5-7A mutation (lanes 8–11). In contrast, the GST–eIF4G proteins did not bind to purified eIF2 (lanes 12–15). The interactions of eIF5 with the GST–eIF4G proteins were resistant to treatment with RNases (data not shown and see Materials and methods), supporting a direct interaction between these proteins.
Fig. 3. eIF5 interacts with eIF4G in vitro. (A) Binding of GST–eIF4G to native eIF2 or recombinant eIF5-FL. Lanes 1–3, Coomassie Blue staining following SDS–PAGE of GST, GST–eIF4G1 and GST–eIF4G2 proteins used in the assays. Aliquots containing ∼5 µg of GST or ∼1 µg of the full-length GST–eIF4G fusion proteins were incubated with 100 ng of either recombinant eIF5-FL (lanes 4–7) or eIF5-FL-7A (lanes 8–11), or 1 µg of eIF2 (lanes 12–15). Proteins bound to the GST fusions were isolated with glutathione–Sepharose beads (GST pull-down) and analyzed by immunoblotting with the appropriate polyclonal antibodies, except that anti-FLAG antibodies were used for detecting eIF5-FL and eIF5-FL-7A. Lanes 4, 8, 12 and 16, 20% of input (In) amount of the indicated proteins. (B) Binding of GST–eIF5 to segments of eIF4G2 in GST pull-down assays. Aliquots containing ∼5 µg of GST (C) or GST–eIF5 (5), shown in a Coomassie Blue-stained gel following SDS–PAGE in lanes 1 and 2, were incubated with [35S]eIF4G2-N (lanes 3–5) or eIF4G2-C (lanes 6–8), and the bound proteins were separated by SDS–PAGE, followed by autoradiography and phosphoimaging analyses. In, 50% input amount. The amino acids present in the eIF4G2 segments are indicated on the bars shown beneath the box depicting the primary structure of yeast eIF4G2. Gray boxes in the latter denote the binding sites for the indicated proteins (Tarun and Sachs, 1996; Neff and Sachs, 1999).
To localize the interaction, we synthesized the two halves of eIF4G2 in rabbit reticulocyte lysates, labeling them with [35S]methionine, and tested the labeled polypeptides for binding to GST fusions containing full-length or truncated forms of eIF5. The eIF4G2 C-terminal fragment (eIF4G2-C), but not the N-terminal fragment (eIF4G2-N), bound specifically to GST–eIF5 (Figure 3B, lanes 5 and 8). The proportion of eIF4G2-C that bound to GST–eIF5 (∼5%) was lower than the 30–50% binding of eIF3-NIP1 or eIF2β to GST–eIF5 that we observed under similar conditions (Asano et al., 1999). Thus, the eIF5–eIF4G2 interaction is weaker than the eIF5–eIF2 or eIF5–eIF3 interactions described previously.
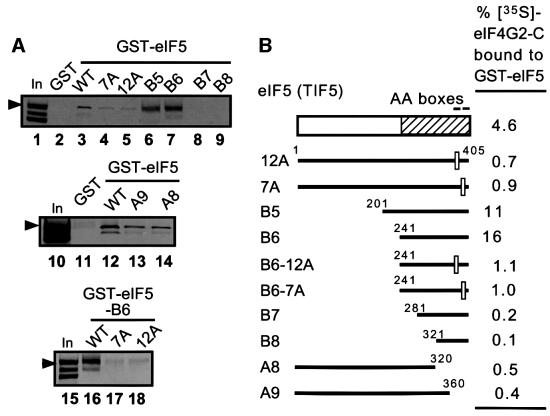
We then investigated whether eIF5-CTD mediates the interaction of eIF5 with the C-terminal half of eIF4G. Introducing the tif5-12A and tif5-7A mutations into the AA-boxes of GST–eIF5 reduced, but did not abolish, its interaction with the [35S]eIF4G2-C fragment (Figure 4A, lanes 3–5). Similar results were obtained for C-terminal deletions removing the AA-boxes from GST–eIF5 in constructs A8 and A9 (Figure 4A and B, lanes 12–14). In constrast, deletions that removed only the N-terminal half of eIF5 from the GST–eIF5 fusion (in constructs B5 and B6) enhanced the interaction with [35S]eIF4G2-C, whereas larger deletions from the N-terminus that additionally removed residues from the CTD (in constructs B7 and B8) abolished this high-level binding (Figure 4, lanes 3 and 6–9). Thus, the CTD is necessary and sufficient for interaction of GST–eIF5 with the C-terminal half of eIF4G2. Finally, introducing the tif5-12A and tif5-7A mutations into the GST–eIF5-B6 construct containing only the CTD greatly reduced its binding to [35S]eIF4G2-C (Figure 4A, lanes 16–18), confirming that the AA-boxes are important constituents of the eIF4G-binding domain in eIF5-CTD. The increased binding activity of the N-terminally truncated constructs B5 and B6 versus full-length GST–eIF5 might indicate that the N-terminus of eIF5 interferes with the ability of the CTD to interact with eIF4G2-C.
Fig. 4. AA-boxes in eIF5-CTD mediate its binding to eIF4G2. (A) Binding of GST–eIF5 derivatives to eIF4G2-C. GST, GST–eIF5 and its derivatives described in (B) were tested for binding to [35S]eIF4G2-C, as in Figure 3B. In, 50% input amount. Arrowheads indicate full-length eIF4G2-C. (B) Summary of in vitro interactions between GST–eIF5 and [35S]eIF4G2-C. The box at the top indicates the primary structure of yeast eIF5, with the minimal binding domains for eIFs 2, 3 and 4G shown by a hatched box. Bars beneath it represent the eIF5 segments used as GST fusions for in vitro binding assays in (A), with their designations shown to the left. Empty squares on the bars represent 12 and 7 alanine substitutions for AA-boxes 1 and 2 in tif5-12A and tif5-7A, respectively (Asano et al., 1999).
eIF5-CTD can bridge interactions between eIF3-NIP1 and eIF4G in vitro
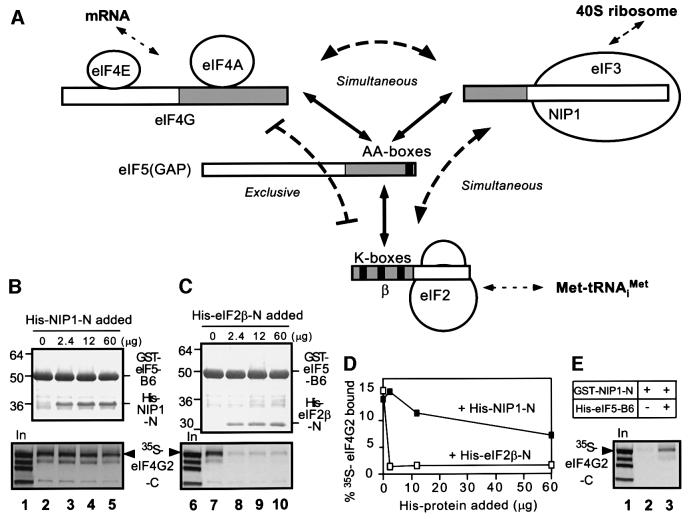
If eIF5-CTD promotes eIF4G–eIF3 interaction, we would predict that eIF5-CTD can interact simultaneously with eIF4G2-C and the N-terminus of eIF3-NIP1. In addition to testing this prediction, we wished to determine whether the eIF4G–eIF5-CTD interaction is compatible with stable association between eIF5-CTD and the K-box domain of eIF2β (Figure 5A). To address these questions, we purified polyhistidine-tagged N-terminal segments of NIP1 and eIF2β (His-NIP1N and His-eIF2β-N, respectively) containing the binding domains in these proteins for eIF5-CTD (Asano et al., 1999, 2000) and tested them for effects on the interaction between [35S]eIF4G2-C and GST–eIF5-B6 (containing the CTD). We found that GST–eIF5-B6 bound efficiently to [35S]eIF4G2-C when His-NIP1-N was present at a 25-fold molar excess over GST–eIF5-B6 (Figure 5B, bottom panel, lanes 1–5). As expected, a fraction of His-NIP1-N itself bound to GST–eIF5-B6 in this experiment (Figure 5B, top panel, lanes 3–5), confirming the ability of these two recombinant proteins to interact directly. These findings suggest that the eIF4G2–eIF5 interaction does not compete with NIP1–eIF5 association in vitro.
Fig. 5. eIF5 bridges interaction between eIF3 and eIF4G in vitro. (A) Schematic illustration of interactions involving eIF2β, eIF3-NIP1, eIF5 and eIF4G. Boxes denote primary structures of the proteins involved and the gray regions denote the minimal binding domains. Filled boxes denote the conserved motifs responsible for protein–protein interactions. Solid arrows indicate direct interactions, whereas curved dotted lines indicate whether the interactions occur simultaneously or exclusively. Straight dotted arrows indicate interactions with the other major components of the translation initiation complex. (B and C) Competition experiments. GST pull-down assays were conducted using 5 µg of GST–eIF5-B6 and [35S]eIF4G2-C in the presence of the indicated amounts of His-NIP-N or His-eIF2β-N. Top panels: Coomassie Blue staining following SDS–PAGE of GST–eIF5-B6 and bound His-tagged proteins recovered in the pull-down assays. Bottom panels: autoradiograms showing bound [35S]eIF4G2-C. Lanes 1 and 6, 50% input amounts of [35S]eIF4G2-C. (D) Graph showing the binding of [35S]eIF4G2-C plotted against the amount of each His-tagged protein added to the binding reactions. (E) Bridging experiments. GST pull-down assays using 5 µg of GST–NIP-N and [35S]eIF4G2-C in the presence (60 µg) or absence of His-eIF5-B6. Bound [35S]eIF4G2-C is shown in the autoradiogram. Lane 1, 50% input (In) amount of [35S]eIF4G2-C.
In sharp contrast to the findings above, [35S]eIF4G2-C did not bind to GST–eIF5-B6 in the presence of His- eIF2β-N added to the reaction (Figure 5C, bottom panel, lanes 6–10). (The lowest amount of His-eIF2β-N examined in lane 8 is equimolar to the amount of GST–eIF5-B6 and should be sufficient to reduce interaction between GST–eIF5-B6 and [35S]eIF4G2-C by competition.) A portion of His-eIF2β-N was found associated with GST–eIF5-B6 (Figure 5C, top panel), confirming the ability of these two proteins to form a complex. These results suggest that the eIF4G–eIF5 interaction is mutually exclusive with the eIF2β–eIF5 interaction.
To confirm our conclusion that eIF5-CTD can bind simultaneously to eIF4G2 and eIF3-NIP1, we investigated whether the eIF5-B6 segment can bridge an interaction between [35S]eIF4G2-C and a GST fusion containing the N-terminal domain of NIP1 (GST–NIP1-N). Supporting this idea, we found that [35S]eIF4G2-C and GST–NIP1-N did not interact with one another unless the eIF5-B6 fragment was present in the reaction, being added in 25-fold molar excess to GST–NIP1-N (Figure 5E). Thus, eIF5-CTD can bridge interactions between eIF3 and either eIF2 or eIF4F through simultaneous binding to NIP1 and either eIF2β or eIF4G, respectively. In contrast, eIF5-CTD can not bridge interaction between eIF2 and eIF4F because its binding to eIF2β and eIF4G is mutually exclusive (Figure 5A). Below, we present a possible rationale for the ability of the eIF5-CTD–NIP1-N binary complex to form mutually exclusive ternary interactions with eIF4G2-C or eIF2β.
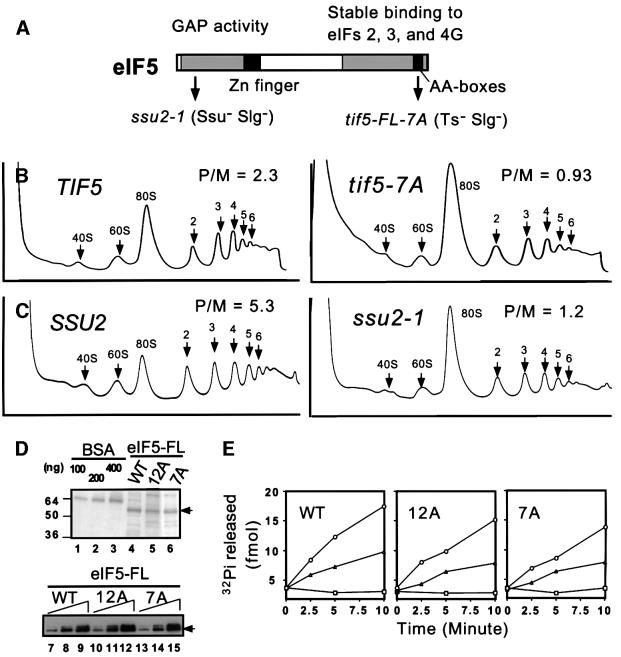
The tif5-7A mutation does not impair the GTPase-activating function of eIF5 in a model in vitro assay
eIF5 stimulates hydrolysis of GTP by the TC on base pairing between Met-tRNAiMet and the AUG start codon. Given that eIF5-CTD binds tightly to the β-subunit of eIF2, we considered that eIF5 GAP function could be dependent on this stable interaction with its substrate. Hence, we investigated whether the tif5-7A and tif5-12A mutations impair eIF5 activity in an in vitro GAP assay. In this assay, model 48S complexes containing eIF2– [γ-32P]GTP–Met-tRNAiMet ternary complexes and rAUG triplets bound to 40S ribosomes are assembled from purified components, incubated with wild-type or mutant eIF5, and assayed for hydrolysis of the [γ-32P]GTP bound to eIF2. For comparison purposes, we also analyzed the ssu2-1 allele of TIF5, which substitutes serine for Gly62 in the N-terminus of eIF5 (Figure 6A). This mutation was isolated on the basis of reverting the Ts– phenotype conferred by the sui1-17 mutation in eIF1 (T.F.Donahue, unpublished observations). ssu2-1 mutant cells exhibit Slg– phenotypes at all temperatures, and grow as slowly as do tif5-7A mutant cells at 30°C (data not shown). As shown in Figure 6B and C, the tif5-7A and ssu2-1 mutations led to similar reductions in the proportion of ribosomes present in polysomes, with a corresponding increase in the proportion of 80S monosomes.
Fig. 6. Effects of eIF5 mutations in separate domains on translation initiation in vivo and GAP activity in vitro. (A) Primary structure of yeast eIF5 (shown by box). Gray boxes denote the regions highly conserved among its eukaryotic homologs. Filled squares denote the zinc finger motif or AA-boxes, as indicated. Arrows indicate the positions of ssu2-1 (G62S) or tif5-7A mutations. The deduced roles of the N-terminal domain and CTD in GAP activity or initiation complex assembly, respectively, are indicated (see text). (B and C) Strains KAY50 (TIF5-FL), KAY51 (tif5-FL-7A) (B), or the transformants of ssu2-1 strain JRC179–4D (α ura3-53 his4– ssu2-1; T.F.Donahue, unpublished) carrying plasmid pKA235 (Table I) (SSU2) or YCplac33, the vector (ssu2-1) (C) were grown in YPD (B) or SC medium lacking uracil (C) at 30°C and cycloheximide was added just before harvesting the cells. WCEs were resolved on 5–45% sucrose gradients by centrifugation at 39 000 r.p.m. for 2.5 h, and the gradients were scanned continuously for A254. The A254 profiles of the gradients are shown from top (left) to bottom (right). The positions of 40S, 60S and 80S ribosomes and polysomes of different sizes are indicated, along with the mass ratios of polysomes to 80S ribosomes (P/M). Data for (B) were taken from Asano et al. (2000). (D) Wild-type (WT) or AA-box mutant (12A or 7A) versions of eIF5-FL were purified in one step from yeast WCEs with FLAG affinity resin (see Materials and methods). Lanes 4–6 show the Coomassie Blue staining following SDS–PAGE of the eIF5-FL preparations, along with bovine serum albumin loaded as a standard (lanes 1–3). Lanes 7–15 contain 1-, 2- and 4-fold amounts of each preparation analyzed by western blotting with anti-FLAG antibodies. (E) eIF5 GAP assay. About 100 fmol of 48S pre-initiation complexes containing the 40S ribosome, Met-tRNAiMet, rAUG, eIF2 and [γ-32P]GTP were incubated with wild-type (WT) or mutant (12A or 7A) eIF5-FL shown in (D). Aliquots were withdrawn at the indicated times and assayed for the amount of free phosphate released from the 48S complexes. Circles or triangles, 800 or 400 ng of WT or mutant eIF5-FL, respectively, were added to the reaction; squares, no eIF5 was added.
Using the in vitro GAP assay described above, we found no significant differences between the activities of purified wild-type eIF5-FL and the mutant proteins eIF5-FL-12A or eIF5-FL-7A containing multiple alanine substitutions in AA-boxes 1 and 2 of the CTD, respectively (Figure 6D and E). In contrast, the ssu2-1 mutation in the N-terminus of eIF5 led to an ∼4-fold reduction in the GAP activity of purified eIF5 (T.F.Donahue, in preparation). Although this assay is non-physiological in several respects, including the absence of eIFs 1 and 3, and with an AUG triplet replacing mRNA in the 48S complex, these results suggest that the AA-boxes in eIF5-CTD are not essential for eIF5 GAP function. It is possible, however, that stable interactions mediated by the CTD between eIF5 and other factors in native 48S complexes promote high-level GAP activity in vivo. Alternatively, the concentrations of eIF5 and 48S complexes in the GAP assay may be higher than in cells, and this could compensate for impaired substrate docking with the mutant eIF5 proteins.
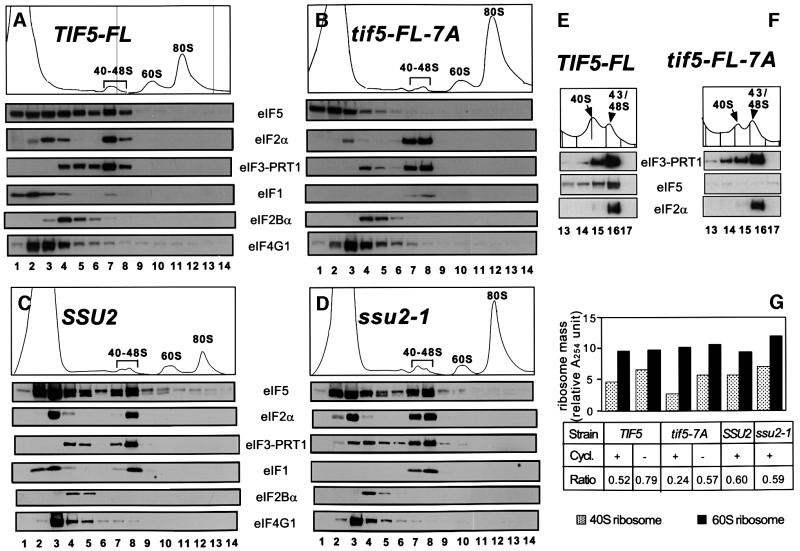
tif5-7A impairs association of eIF5 with 43–48S complexes and impedes a step late in the initiation pathway in vivo
To investigate the rate-limiting defect in translation initiation produced by the tif5-7A mutation in vivo, we compared the amounts of different initiation factors present on free 40S ribosomes in extracts prepared from isogenic TIF5 and tif5-FL-7A strains. If the Met-tRNAiMet binding defect observed in vitro (Figure 1) is rate limiting in vivo, we would expect to find reduced amounts of eIF2 bound to free 40S ribosomes in 43–48S initiation complexes. If, however, a defect in eIF5 GAP function is rate limiting, we should observe accumulation of 48S complexes containing eIFs 1, 2 and 3 that are blocked at the step of AUG recognition prior to joining of 60S subunits. The TIF5-FL and SSU2 wild-type extracts contained sizeable proportions of eIFs 2, 3 and 5 that co-sedimented with free 40S subunits (Figure 7A and C), and also non-ribosomal pools of these factors that were present in fractions located closer to the top of the gradient. At most, only a small proportion of eIF4G co-sedimented with 40S subunits in both wild-type extracts. These results provide the first evidence that eIF5 is a stable component of 43S or 48S pre-initiation complexes in vivo.
Fig. 7. The tif5-7A mutation impairs stable association of eIF5 with 43–48S initiation complexes and leads to accumulation of 48S complexes. (A–D) The TIF5-FL (A), tif5-FL-7A (B), SSU2 (C) and ssu2-1 (D) strains described in Figure 6 were grown in YPD (A and B) or SC medium lacking uracil (C and D) at 30°C, cycloheximide was added just prior to harvesting the cells and extracts were prepared in the presence of cycloheximide. Twenty A260 units of WCEs were fractionated on 15–40% sucrose gradients by centrifugation at 39 000 r.p.m. for 4.5 h. Top panels depict the A254 absorbance profiles of the gradients, and the panels below show the results of immunoblot analyses of the gradient fractions using antibodies against the factors listed next to the panels. Similar results were obtained when the strains shown in (A) and (B) were grown in SC versus YPD medium. (E and F) The same extracts analyzed in (A) and (B) were fractionated on 7.5–30% sucrose gradients by centrifugation at 41 000 r.p.m. for 5 h, and fractions 13–17 from the top of the gradient were analyzed by immunoblotting. Top panels show the A254 absorbance profiles for the 40–48S region of the gradient. (G) Histogram showing the free 40–48S and 60S subunit masses, quantitated by the area under the A254 profiles shown in (A)–(D) and from several independent experiments using 20 A260 units of extracts prepared in the presence or absence of cycloheximide. The bottom panel indicates the strains used (designated as in A–D), the presence (+) or absence (–) of cycloheximide in preparing the extracts, and the calculated 40–48S/60S ribosome mass ratio. In all these experiments, WCEs were prepared in the presence of heparin, an essential component to stabilize the 43–48S complex during sucrose gradient fractionation. The 43–48S complexes are unstable once isolated from yeast, and dissociate into the MFC and 40S subunits in the absence of heparin, as shown previously (Asano et al., 2000).
It is striking that in the tif5-7A extract, no eIF5 co-sedimented with 40S subunits (Figure 7B), indicating that this mutation impairs stable association of eIF5 with pre-initiation complexes. Thus, eIF5-CTD is required for anchoring eIF5 to the initiation complex. In contrast, we observed increased proportions of eIF2, eIF3 and eIF1 in pre-initiation complexes in the tif5-7A extract versus the wild-type extract (Figure 7A and B). This was also true for eIF1 even though the total amount of eIF1 was lower in the mutant extract (Figure 7B). These results suggest that the weakened association of eIF5 with 43–48S pre-initiation complexes in tif5-7A cells impaired a step following recruitment of eIF3, eIF1 and TC to 40S ribosomes as the rate-limiting defect in initiation.
As opposed to the results obtained for tif5-7A, the ssu2-1 mutation in the N-terminus of eIF5 did not reduce the amount of eIF5 that co-sedimented with the 43–48S complexes. Nor did we detect any accumulation of eIFs 1, 2 or 3 in the 40–48S region (Figure 7C and D). Because the N-terminal two-thirds of eIF5 are dispensable for tight interactions with eIFs 2 and 3 in vitro (Asano et al., 2000), these findings are consistent with the idea that only the C-terminal domain of eIF5 is required for its stable incorporation into pre-initiation complexes. [The non-ribosomal form of eIF1 was absent in the ssu2-1 extract, as observed in the tif5-7A extract (Figure 7B and D). Subsequent experiments revealed that degradation of the non-ribosomal eIF1 occurred during centrifugation, as wild-type levels of the protein were present in the unfractionated mutant extracts prepared from cycloheximide-treated cells (data not shown). Presumably, eIF1 dissociated from the MFC during centrifugation of the mutant extracts and was degraded by proteases.]
Our final experiments were based on the observation that the mass of free 40S ribosomes present in the 40–48S complexes relative to the amount of free 60S ribosomes was consistently lower in the tif5-7A mutant versus the wild-type (Figure 7A, B, E and F, top panels). Quantitation of the data from several independent experiments showed that the ratio of 40S subunits in the 40–48S fractions to free 60S subunits was 0.24 ± 0.019 in the tif5-7A mutant compared with 0.52 ± 0.056 in the wild type (Figure 7G). Because the proportions of non-ribosomal eIF2 and eIF3 present near the top of the gradient were also reduced in the tif5-7A mutant (Figure 7A and B, lanes 2–5), it seemed likely that 40S ribosomes were sequestered in the polysomes in the form of 48S complexes containing the TC. If this interpretation is correct, then 40S ribosomes lacking bound eIF2 should be depleted from the 40–48S region in the tif5-7A extract. To test this prediction, we used centrifugation conditions that produced a greater separation of ribosomal species in the 40–48S region and probed the fractions for eIFs 2, 3 and 5. As shown in Figure 7E, there were two peaks evident in the 40–48S region of the wild-type extract, of which the smaller lacked eIF2 and had relatively low levels of eIFs 3 and 5. This 40S peak lacking eIF2 was specifically depleted in the tif5-7A mutant, consistent with a depletion of free 40S subunits lacking the TC (Figure 7F). As expected, eIF5 was absent from the 43–48S ribosomes in the mutant (Figure 7F).
To test our interpretation that the free 40S subunits absent in the tif5-7A mutant are sequestered in polysomes as 48S complexes, we asked whether disruption of the polysomes would restore the amount of free 40S ribosomes in the tif5-7A extract to the wild-type level. When the extracts were prepared in the absence of cycloheximide, most of the polysomes were lost, with concomitant accumulation of vacant 80S couples, as a consequence of polysome run-off (data not shown) (Foiani et al., 1991). The amount of free 60S subunits was not altered by the polysome run-off (Figure 7G), suggesting that most of the 80S ribosomes were released from polysomes without being dissociated into free subunits. In accordance with our prediction, the amount of 40S ribosomes in the 40–48S fractions increased significantly in the mutant following polysome run-off, approaching the value observed in the wild-type strain under the same conditions (Figure 7G). Furthermore, the non-ribosomal pool of eIF2, which is depleted in the mutant (Figure 7B), increased to the wild-type level in the absence of cycloheximide (data not shown). These findings support the idea that a greater proportion of 40S ribosomes are distributed throughout the polysome fractions, most likely as 48S complexes, in the tif5-7A mutant versus the wild-type strain.
In contrast, the ssu2-1 mutation did not lead to a low level of free 40S subunits (Figure 7C and D), and the 40–48S:60S free subunit ratios for the isogenic ssu2-1 and SSU2 strains were virtually identical (Figure 7G). These results are in accordance with the fact that eIF2 and eIF3 did not accumulate in the 40–48S fractions in the ssu2-1 mutant (Figure 7C and D). Hence, it appears that a catalytic defect in eIF5 GAP activity produced by ssu2-1 does not lead to accumulation of 48S complexes stalled at the AUG start codon. Presumably, these complexes decay quickly into free eIFs and 40S subunits, rather than accumulating in the polysome fractions. The fact that 48S complexes lacking eIF5 accumulated in the tif5-7A mutant may have important implications concerning the steps in AUG recognition and GTP hydrolysis that are impaired when eIF5 is not stably anchored to the pre-initiation complex.
Discussion
Role of eIF5-CTD in stabilizing 43S and 48S complexes
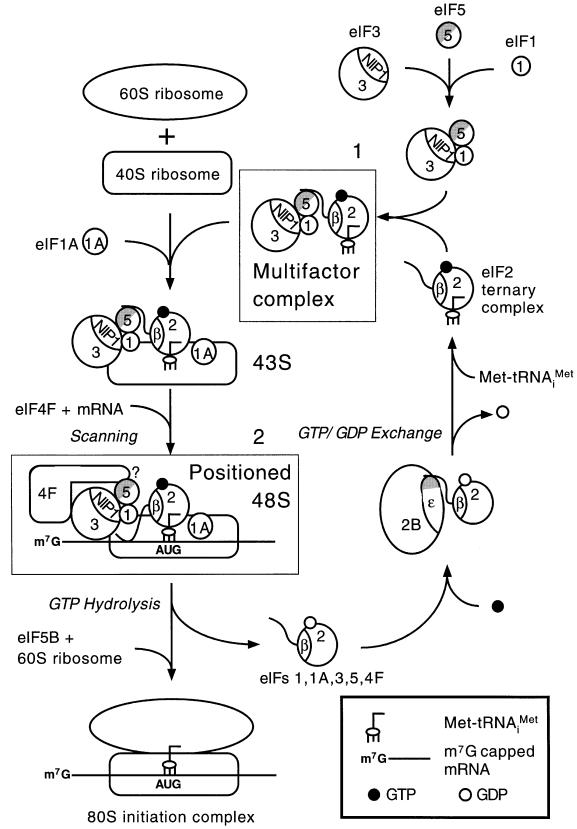
eIF3 can bind to 40S ribosomes and is required for recruitment of the TC in cell extracts (Trachsel et al., 1977; Benne and Hershey, 1978; Phan et al., 1998), but the molecular interactions involved in this activity are poorly understood. The tif5-7A mutation altering eIF5-CTD is known to destabilize formation of an MFC containing eIFs 1, 2, 3 and 5, and tRNAiMet (Asano et al., 2000) (box 1 in Figure 8). We found that binding of exogenous Met-tRNAiMet to the 40S ribosome was diminished in an extract from the tif5-7A mutant, and this defect was complemented by addition of purified eIF5 (Figure 1A and B). These results suggest that, by linking eIF2 and eIF3 in the MFC, eIF5-CTD stimulates binding of TC to the 40S subunit to form the 43S pre-initiation complex.
Fig. 8. Hypothetical model for the role of eIF5-CTD in assembling the translation initiation complex in S.cerevisiae. The CTD (gray half circle) of eIF5 (5), containing the conserved AA-boxes, bridges interaction between eIF3-NIP1 (3) and eIF2β (2) and mediates formation of the MFC, also containing Met-tRNAiMet and eIF1 (box 1) (Asano et al., 2000). The wavy line on eIF2β represents the K-box domain, the binding site for eIF5-CTD. As the MFC occurs free of the ribosomes, it could carry out the recruitment of TC, eIFs 1, 3 and 5 in a single step, to form the 43S complex. The eIF1A (1A) may bind directly to the 40S ribosome (Hershey and Merrick, 2000). Capped poly(A) mRNA bound to eIF4F is recruited to the 43S complex by interactions between eIF3 and the eIF4G subunit of eIF4F. The eIF2β–eIF5 interaction in the MFC may be replaced by eIF4G–eIF5 interaction in the 48S complex, possibly when eIF2β interacts with mRNA. The eIF4G–eIF5 and eIF2β–mRNA interactions help stabilize the 48S complex. The 48S complex scans to the AUG start codon. Base pairing between AUG and Met-tRNAiMet triggers hydrolysis of GTP bound to eIF2, dependent on the eIF5 N-terminal domain (white half circle), followed by ejection of eIF2-GDP and other eIFs. Joining of the 60S subunit is stimulated by eIF5B. The GDP bound to eIF2 is subsequently replaced with GTP by the guanine nucleotide exchange factor eIF2B (2B). This last interaction is mediated at least partly by a second AA-box-containing motif (shown as gray shading) in the catalytic (ε) subunit of eIF2B (Asano et al., 1999). Our results indicate that formation of the MFC is required for efficient binding of Met-tRNAiMet and mRNA to 40S ribosomes. It is also required for stable incorporation of eIF5 into 48S complexes and conversion of 48S to 80S complexes. The latter appears to be the rate-limiting defect in initiation produced by the tif5-7A mutation in eIF5-CTD, which destabilizes the MFC.
Binding of exogenous mRNA to the 40S ribosome was also defective in the tif5-7A extract, and could be rescued with purified eIF5 (Figure 1C–E). While this mRNA binding defect may result indirectly from reduced TC binding, it led us to investigate whether eIF5-CTD interacts with eIF4G, an adaptor subunit of the eIF4F complex that promotes mRNA binding to 40S ribosomes. Using purified proteins, we showed that eIF4G binds directly to eIF5-CTD, in a manner stimulated by the AA-boxes (Figures 3 and 4), and this interaction can occur simultaneously with association between eIF5-CTD and eIF3-NIP1 (Figure 5B and E). It is thought that direct interaction between eIF3 and eIF4G promotes mRNA binding to ribosomes in mammalian cells (Hentze, 1997; Sachs et al., 1997). The ability of eIF5-CTD to interact simultaneously with eIF3-NIP1 and eIF4G may help to stabilize the eIF3–eIF4G interaction and promote mRNA binding. Supporting this idea, co-immunoprecipitation of native eIF4G and eIF3 was reduced by the tif5-7A mutation in the eIF5-CTD (Figure 2).
The eIF5–eIF4G interaction was of lower affinity, and mutually exclusive with the eIF5–eIF2β interaction (Figure 5C and D), suggesting that the two interactions occur at different steps in the initiation pathway. The eIF2β–eIF5-CTD association occurs in the MFC free of the ribosome (Asano et al., 2000), and helps to recruit eIF5 to the 43S complex in proper juxtaposition with TC, eIF1 and eIF3. This interaction may give way to eIF4G– eIF5-CTD interaction in the 48S complex to promote mRNA binding or facilitate scanning (Figure 8). Because the K-boxes in eIF2 mediate mRNA binding by eIF2 (Laurino et al., 1999), the mRNA–eIF2β interaction in the 48S complex may displace eIF5 from the K-boxes, allowing the lower affinity eIF5–eIF4G interaction to proceed (box 2 in Figure 8).
Distinct requirements for the C- and N-terminal domains of eIF5 in initiation complex formation and GAP activity
The tif5-7A mutation reduced Met-tRNAiMet binding to 40S ribosomes in vitro (Figure 1), suggesting that eIF5-CTD enhances the rate of TC binding to 40S subunits, or the stability of the 43S complex in cell extracts. Ostensibly at odds with this finding, we observed accumulation of eIF2 on free 40S subunits and a reduction in the non-ribosomal pool of this factor in tif5-7A cells (Figure 7A and B). Moreover, we deduced that polysome-associated 48S complexes accumulated in tif5-7A cells, accounting for the observed depletion of free 40S subunits lacking bound eIF2 (Figure 7B, F and G). Thus, it appears that conversion of 48S to 80S initiation complexes is the rate-limiting defect responsible for the reduced rate of translation initiation in this mutant. We presume that the rate of 43S complex formation is also reduced in tif5-7A cells, as seen in vitro (Figure 1), but, because the defect in 48S to 80S conversion is more severe, we observed accumulation of 48S complexes. Importantly, eIF5 was lost from 43–48S complexes in tif5-7A cells, showing that eIF5-CTD is crucial for stable incorporation of eIF5 into the initiation complex (Figure 7A and B). Hence, the absence of eIF5 from 48S complexes impedes their conversion to 80S complexes. In principle, this could occur by a reduction in the rate of scanning from the 5′ cap, reduced activation of GTP hydrolysis on AUG recognition or impaired 60S subunit joining following GTP hydrolysis.
We observed little or no effect of the tif5-7A or tif5-12A mutations on the ability of purified eIF5 to stimulate GTP hydrolysis by model 48S complexes containing the TC and AUG triplets bound to 40S subunits (Figure 6D and E). Although these assay conditions are non-physiological in several respects, our data suggest that the AA-boxes in eIF5-CTD are not required for the catalytic activity of eIF5. In the same assay, the ssu2-1 mutation in the N-terminus of eIF5 (G62S) leads to a substantial reduction in GAP activity. Consistently, ssu2-1 reduced the rate of translation initiation (Figure 6C) but did not diminish the amount of eIF5 associated with the 43–48S complexes in vivo (Figure 7D). Accordingly, we propose that ssu2-1 impairs the GAP activity of eIF5, lodged in the N-terminus of the protein, but not its interactions with other eIFs in 43–48S initiation complexes. In contrast, tif5-7A would leave the catalytic activity of eIF5 largely intact while eliminating this factor as an integral component of 48S complexes. This model is consistent with the previous finding that the Sui– mutation in eIF5 altering Gly31 to arginine increased GAP activity in vitro by ∼2-fold (Huang et al., 1997).
Even if the CTD is not required for eIF5 catalytic function, the loss of eIF5 from 48S pre-initiation complexes in the tif5-7A mutant might reduce the rate of GTP hydrolysis in vivo by decreasing the concentration of eIF5 in the vicinity of 48S complexes paired with start codons. In principle, a defect in this ‘substrate docking’ function of the CTD should have been detected in our GAP assays, and indeed Das and Maitra (2000) reported that alteration of acidic residues in the AA-boxes of rat eIF5 reduced GAP activity in vitro with concomitant decreases in binding to recombinant eIF2β. These workers used a model 48S complex composed of mammalian components, whereas ours contained the corresponding yeast components. Perhaps the catalytic step, rather than substrate binding, is rate limiting in GAP assays using model 48S complexes from yeast.
Another way to explain the defect in 48S to 80S conversion in tif5-7A mutants is to propose that eIF5 must be positioned precisely in native 48S complexes in order to trigger GTP hydrolysis with maximum efficiency. It is known that eIF5 can not stimulate GTP hydrolysis by the TC unless both factors are bound to the 40S subunit and the Met-tRNAiMet is base paired with AUG (Chakrabarti and Maitra, 1991). Hence, GTP hydrolysis may require conformational changes on recognition of the start codon that depend on proper juxtaposition of eIF5 with other components of the MFC or with eIF4G. This hypothetical function of eIF5-CTD could be bypassed in model 48S complexes containing only the TC, or at the non-physiological Mg2+ concentrations used for in vitro GAP assays.
The fact that 48S complexes accumulated in tif5-7A but not in ssu2-1 cells (Figure 7B, D and G) suggests that different steps in the maturation of 48S complexes are disrupted by these mutations. Perhaps impaired interaction of eIF5 with eIF4G in the tif5-7A mutant impedes the rate of scanning, so that base pairing between the Met-tRNAiMet and AUG codon is delayed. Considering that eIF1 also interacts with eIF5-CTD (Asano et al., 2000) and has been implicated in AUG recognition (Pestova et al., 1998; Donahue, 2000), tif5-7A may impair the function of eIF1 in AUG recognition during scanning. The reduced rate of scanning would account for the accumulation of 48S complexes observed in tif5-7A cells. If ssu2-1 impairs only the GAP activity of eIF5, then 48S complexes should be stalled at the AUG start codon in this mutant. The fact that 48S complexes did not accumulate in ssu2-1 cells suggests that 48S complexes positioned at an AUG will decay to free 40S subunits if GTP hydrolysis does not occur in a prescribed period of time.
Materials and methods
Materials
Plasmids employed in this study are listed in Table I. Details of their construction are available upon request. His-eIF2β-N, His-NIP1-N and His-eIF5-B6 were expressed in Escherichia coli strain BL21(DE3) from the appropriate pET15b derivatives (Table I), purified with Ni2+ affinity resin (Novagen) as recommended by the manufacturer and dialyzed against the GST pull-down buffer (Asano et al., 1998) without milk. To purify eIF5-FL proteins from yeast, 10 mg of WCEs were prepared in buffer A (Asano et al., 1999) from KAY39, KAY40 and KAY58, encoding wild-type, tif5-7A and tif5-12A forms of eIF5 in high copy (Asano et al., 1999), and incubated with 100 µl of anti-FLAG affinity resin (Sigma) at 4°C for 2 h. After washing with buffer A, beads were eluted with 400 ng/µl FLAG peptide (Sigma) in 200 µl of buffer A. eIF5-FL or its tif5-7A derivative were purified similarly from BL21(DE3) transformants carrying pT7-TIF5 or pT7-TIF5-7A grown in the presence of isopropyl-β-d-thiogalactopyranoside (IPTG). Yeast eIF2 (Pavitt et al., 1998), [3H]Met-tRNAiMet (Phan et al., 1998), uncapped [32P]poly(A) MFA2 mRNA (Tarun and Sachs, 1995) and 40S ribosomes (Huang et al., 1997) were prepared as described previously.
Table I. Plasmids employed in this study.
| Plasmid | Descriptiona | Product | Source |
|---|---|---|---|
| pGEX vectors | expression vectors for GST fusions | GST | Pharmacia |
| pAS466 | full-length TIF4631 ORF in pGEX | GST–eIF4G1 | Tarun and Sachs (1996) |
| pAS467 | full-length TIF4632 ORF in pGEX | GST–eIF4G2 | Tarun and Sachs (1996) |
| pGEX-TIF5 | full-length TIF5 ORF in pGEX | GST–eIF5 | Phan et al. (1998) |
| pGEX-B6 | TIF5 ORF (241–405) in pGEX | GST–eIF5-B6 | Asano et al. (1999) |
| pGEX-B6-12A | pGEX-B6 carrying tif5-12A | GST–eIF5-B6-12A | this study |
| pGEX-B6-7A | pGEX-B6 carrying tif5-7A | GST–eIF5-B6-7A | this study |
| pGEX-NIP1-N | NIP1 ORF (1–156) in pGEX | GST–NIP1-N | Asano et al. (2000) |
| pT7-7 | expression vector with T7 promoter | Tabor and Richardson (1987) | |
| pT7-4G2ΔN | TIF4632 ORF (1–513) in pT7-7 | eIF4G2-N | this study |
| pT7-4G2ΔS | TIF4632 ORF (439–914) in pT7-7 | eIF4G2-C | this study |
| pT7-TIF5 | full-length TIF5-FL in pT7-7 | eIF5-FL | Asano et al. (2000) |
| pT7-TIF5-7A | pT7-TIF5 carrying tif5-7A | eIF5-FL-7A | Asano et al. (2000) |
| pET15b | expression vector for polyhistidine-tagged proteins | Novagen | |
| pHis-NIP1-N | NIP1 ORF (1–156) in pET15b | His-NIP1-N | Asano et al. (2000) |
| pHis-TIF5-B6 | TIF5 ORF (241–405) in pET15b | His-eIF5-B6 | Asano et al. (2000) |
| pHis-SUI3ΔS | SUI3 ORF (1–140) in pET15b | His-eIF2β-N | this study |
| pKA235 | CEN plasmid carrying TIF5 URA3 | Asano et al. (1999) |
aNumbers in parentheses indicate amino acid positions at the termini of the relevant protein segment.
Biochemical assays
Immunoprecipitations with antibodies against the HA epitope were conducted as described (Asano et al., 1998, 1999). Rabbit polyclonal antibodies used for immunoblot analysis are listed in Asano et al. (1999), except those against eIF1 (Yoon and Donahue, 1992), eIF5 (Huang et al., 1997), eIF2Bα (Cigan et al., 1991), eIF4G1 (Wells et al., 1998) and eIF4E (Lang et al., 1994). GST pull-down assays were conducted as described previously (Asano et al., 1999, 2000). For RNase treatment in pull-down assays, GST–eIF4G–eIF5-FL complexes attached to glutathione–Sepharose were incubated with 20 µg of RNase A (Sigma) at 26°C for 5 min or 120 U of micrococcal nuclease (USB) at 4°C for 30 min and washed extensively, prior to separation by SDS–PAGE (Tarun and Sachs, 1996; Winstall et al., 2000).
Binding of [3H]Met-tRNAiMet and [32P]poly(A) MFA2 mRNA to 40S ribosomes was assayed in cell extracts as described previously (Phan et al., 1998). A 20 µl aliquot of the extracts was pre-incubated with 6 µl of buffer A (Asano et al., 1999) containing the indicated amounts of eIF5 on ice for 5 min, and then incubated with 26 µl of 2× buffer (Phan et al., 1998) containing 2.4 mM GMPPNP, [3H]Met-tRNAiMet (0.77 µCi, 84 Ci/mmol) and [32P]poly(A) MFA2 mRNA (2.0 µCi, 5.6 Ci/mmol) for 20 min at 26°C. After adding 6 µl of 3% formaldehyde, the sample was loaded on a 7.5–30% sucrose gradient prepared in buffer (20 mM Tris–HCl pH 7.5, 100 mM KCl, 1 mM MgCl2) and centrifuged at 41 000 r.p.m. for 5 h at 4°C in a Beckman SW41 rotor. The gradient was separated into 20 fractions of 0.6 ml with an ISCO gradient fractionater while scanning continuously at 254 nm, and 0.2 ml of each fraction was diluted in 1 ml of water and mixed with 10 ml of Ecolite™ cocktail for liquid scintillation counting of 3H and 32P radioactivity. A model 48S complex containing the 40S ribosome, rAUG and eIF2–[γ-32P]GTP–Met-tRNAiMet ternary complex was prepared and employed as substrate in GAP assays of purified eIF5 as described previously (Huang et al., 1997).
Polysome analysis was conducted as previously described (Asano et al., 2000).
Acknowledgments
Acknowledgements
We are indebted to Alan Sachs for his gifts of materials and to Han Huang for advice on the GAP assay. We thank Thanuja Krishnamoorthy for purified yeast eIF2, Salvador Tarun for advice on eIF4G binding experiments, Gota Kawai for discussion, Tom Dever for comments on the manuscript, and members of the Hinnebusch and Dever laboratories for advice. A.S. was supported by an Endocrine Fellowship from the Diabetes Branch, NIDDK, NIH.
References
- Asano K., Phan,L., Anderson,J. and Hinnebusch,A.G. (1998) Complex formation by all five homologues of mammalian translation initiation factor 3 subunits from yeast Saccharomyces cerevisiae. J. Biol. Chem., 273, 18573–18585. [DOI] [PubMed] [Google Scholar]
- Asano K., Krishnamoorthy,T., Phan,L., Pavitt,G.D. and Hinnebusch,A.G. (1999) Conserved bipartite motifs in yeast eIF5 and eIF2Bε, GTPase-activating and GDP–GTP exchange factors in translation initiation, mediate binding to their common substrate eIF2. EMBO J., 18, 1673–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K., Clayton,J., Shalev,A. and Hinnebusch,A.G. (2000) A multifactor complex of eukaryotic initiation factors eIF1, eIF2, eIF3, eIF5 and initiator tRNAMet is an important translation initiation intermediate in vivo. Genes Dev., 14, 2534–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benne R. and Hershey,J.W.B. (1978) The mechanism of action of protein synthesis initiation factors from rabbit reticulocytes. J. Biol. Chem., 253, 3078–3087. [PubMed] [Google Scholar]
- Chakrabarti A. and Maitra,U. (1991) Function of eukaryotic initiation factor 5 in the formation of an 80S ribosomal polypeptide chain initiation complex. J. Biol. Chem., 266, 14039–14045. [PubMed] [Google Scholar]
- Cigan A.M., Foiani,M., Hannig,E.M. and Hinnebusch,A.G. (1991) Complex formation by positive and negative translational regulators of GCN4. Mol. Cell. Biol., 11, 3217–3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S. and Maitra,U. (2000) Mutational analysis of mammalian translation initiation factor 5 (eIF5): role of interaction between the β subunit of eIF2 and eIF5 in eIF5 function in vitro and in vivo. Mol. Cell. Biol., 20, 3942–3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S., Maiti,T., Das,K. and Maitra,U. (1997) Specific interaction of eukaryotic translation initiation factor 5 (eIF5) with the β-subunit of eIF2. J. Biol. Chem., 272, 31712–31718. [DOI] [PubMed] [Google Scholar]
- Donahue T. (2000) Genetic approaches to translation initiation in Saccharomyces cerevisiae. In Sonenberg,N., Hershey,J.W.B. and Mathews,M.B. (eds), Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 487–502.
- Foiani M., Cigan,A.M., Paddon,C.J., Harashima,S. and Hinnebusch,A.G. (1991) GCD2, a translational repressor of the GCN4 gene, has a general function in the initiation of protein synthesis in Saccharomyces cerevisiae. Mol. Cell. Biol., 11, 3203–3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze M.W. (1997) eIF4G: a multipurpose ribosome adapter. Science, 275, 500–501. [DOI] [PubMed] [Google Scholar]
- Hershey J.W.B. and Merrick,W.C. (2000) Pathway and mechanism of initiation of protein synthesis. In Sonenberg,N., Hershey,J.W.B. and Mathews,M.B. (eds), Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 33–88.
- Hinnebusch A.G. (2000) Mechanism and regulation of initiator methionyl-tRNA binding to ribosomes. In Sonenberg,N., Hershey,J.W.B. and Mathews,M.B. (eds), Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 185–243.
- Huang H., Yoon,H., Hannig,E.M. and Donahue,T.F. (1997) GTP hydrolysis controls stringent selection of the AUG start codon during translation initiation in Saccharomyces cerevisiae. Genes Dev., 11, 2396–2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang V., Zanchin,N.I., Lunsdorf,H., Tuite,M. and McCarthy,J.E. (1994) Initiation factor eIF-4E of Saccharomyces cerevisiae. Distribution within the cell, binding to mRNA and consequences of its overproduction. J. Biol. Chem., 269, 6117–6123. [PubMed] [Google Scholar]
- Laurino J.P., Thompson,G.M., Pacheco,E. and Castilho,B.A. (1999) The β subunit of eukaryotic translation initiation factor 2 binds mRNA through the lysine repeats and a region comprising the C2–C2 motif. Mol. Cell. Biol., 19, 173–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff C.L. and Sachs,A.B. (1999) Eukaryotic translation initiation factors eIF4G and eIF4A from Saccharomyces cerevisiae physically and functionally interact. Mol. Cell. Biol., 19, 5557–5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavitt G.D., Ramaiah,K.V.A., Kimball,S.R. and Hinnebusch,A.G. (1998) eIF2 independently binds two distinct eIF2B subcomplexes that catalyze and regulate guanine-nucleotide exchange. Genes Dev., 12, 514–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova T.V., Borukhov,S.I. and Hellen,C.U.T. (1998) Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature, 394, 854–859. [DOI] [PubMed] [Google Scholar]
- Phan L., Zhang,X., Asano,K., Anderson,J., Vornlocher,H.P., Greenberg,J.R., Qin,J. and Hinnebusch,A.G. (1998) Identification of a translation initiation factor 3 (eIF3) core complex, conserved in yeast and mammals, that interacts with eIF5. Mol. Cell. Biol., 18, 4935–4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs A.B., Sarnow,P. and Hentze,M.W. (1997) Starting at the beginning, middle and end: translation initiation in eukaryotes. Cell, 89, 831–838. [DOI] [PubMed] [Google Scholar]
- Tabor S. and Richardson,C.C. (1987) DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc. Natl Acad. Sci. USA, 84, 4767–4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarun S.Z. and Sachs,A.B. (1995) A common function for mRNA 5′ and 3′ ends in translation initiation in yeast. Genes Dev., 9, 2997–3007. [DOI] [PubMed] [Google Scholar]
- Tarun S.Z. and Sachs,A.B. (1996) Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J., 15, 7168–7177. [PMC free article] [PubMed] [Google Scholar]
- Trachsel H., Erni,B., Schreier,M.H. and Staehelin,T. (1977) Initiation of mammalian protein synthesis: the assembly of the initiation complex with purified initiation factors. J. Mol. Biol., 116, 755–767. [DOI] [PubMed] [Google Scholar]
- Wells S.E., Hillner,P.E., Vale,R.D. and Sachs,A.B. (1998) Circulariz ation of mRNA by eukaryotic translation initiation factors. Mol. Cell, 2, 135–140. [DOI] [PubMed] [Google Scholar]
- Winstall E., Sadowski,M., Kuhn,U., Wahle,E. and Sachs,A.B. (2000) The Saccharomyces cerevisiae RNA-binding protein Rbp29 functions in cytoplasmic mRNA metabolism. J. Biol. Chem., 275, 21817–21826. [DOI] [PubMed] [Google Scholar]
- Yoon H.J. and Donahue,T.F. (1992) The sui1 suppressor locus in Saccharomyces cerevisiae encodes a translation factor that functions during tRNAiMet recognition of the start codon. Mol. Cell. Biol., 12, 248–260. [DOI] [PMC free article] [PubMed] [Google Scholar]