Abstract
Transforming growth factor-β (TGF-β), a secreted factor present at high levels in bone, inhibits osteoblast differentiation in culture; yet, the mechanism of this inhibition remains unclear. We studied the effects of TGF-β and its effectors, the Smads, on the expression and function of the osteoblast transcription factor CBFA1. TGF-β inhibited the expression of the cbfa1 and osteocalcin genes, whose expression is controlled by CBFA1 in osteoblast-like cell lines. This inhibition was mediated by Smad3, which interacts physically with CBFA1 and represses its transcriptional activity at the CBFA1-binding OSE2 promoter sequence. The repression of CBFA1 function by Smad3 contrasts with previous observations that Smads function as transcription activators. This repression occurred in mesenchymal but not epithelial cells, and depended on the promoter sequence. Smad3-mediated repression of CBFA1 provides a central regulatory mechanism for the inhibition of osteoblast differentiation by TGF-β, since it inhibits both cbfa1 transcription and transcriptional activation of osteoblast differentiation genes by CBFA1. Altering Smad3 signaling influenced osteoblast differentiation in the presence or absence of TGF-β, implicating Smad3/TGF-β-mediated repression in autocrine regulation of osteoblast differentiation.
Keywords: CBFA1/osteoblast differentiation/Smad3/TGF-β
Introduction
Mesenchymal cells can differentiate into bone, cartilage, fat or muscle, depending on signals derived from the cellular environment. Extracellular signaling by members of the transforming growth factor-β (TGF-β) superfamily, which are involved in diverse cellular processes, also regulates mesenchymal differentiation. For example, TGF-β can inhibit progression of adipose and osteoblast differentiation, whereas bone morphogenetic proteins (BMPs) stimulate osteoblast differentiation (Centrella et al., 1994; Alliston and Derynck, 2000; Choy et al., 2000).
TGF-β plays a key role in osteoblast differentiation and bone development and remodeling. Osteoblasts secrete and deposit TGF-β into the bone matrix and can respond to it, thus enabling possible autocrine modes of action (Robey et al., 1987). TGF-β regulates the proliferation and differentiation of osteoblasts both in vitro and in vivo; however, the effects of TGF-β on osteoblast differentiation depend on the extracellular milieu and the differentiation stage of the cells. TGF-β stimulates proliferation and early osteoblast differentiation, while inhibiting terminal differentiation (Bonewald and Dallas, 1994; Centrella et al., 1994). Accordingly, TGF-β has been reported to inhibit expression of alkaline phosphatase and osteocalcin, among other markers of osteoblast differentiation and function (Noda, 1989; Centrella et al., 1994). While osteoblasts express cell surface receptors for TGF-β and its known effectors, Smad2 and Smad3 (Sakou et al., 1999), the signaling mechanisms that mediate the effects of TGF-β on osteoblast differentiation have not been well characterized (Alliston and Derynck, 2000). In addition, the extent to which autocrine TGF-β responsiveness regulates osteoblast differentiation remains unclear.
Progress in recent years has led to the identification and characterization of the TGF-β receptors and their intracellular signaling effectors, the Smads. TGF-β signaling is initiated by ligand binding to a heterotetrameric cell-surface receptor complex, consisting of two types of transmembrane serine/threonine kinases. Ligand-induced phosphorylation of the type I by the type II receptors results in activation of the type I receptor kinases, which in turn phosphorylate Smad2 and/or Smad3 at C-terminal serines (Heldin et al., 1997; Massagué, 1998). These Smads then dissociate from the receptor complex to form a heterotrimeric complex with Smad4, which then translocates into the nucleus. The activation and nuclear translocation of Smads are subject to regulation by other signaling pathways. In the nucleus, Smads interact with transcription factors at the promoters of TGF-β-responsive genes and regulate transcription (Derynck et al., 1998; Itoh et al., 2000; Massagué and Wotton, 2000; Miyazono, 2000; Wrana, 2000). Several types of transcription factors have been shown to interact with TGF-β-activated Smads at different promoters (Itoh et al., 2000). For example, Smad3 has been shown to associate with c-Jun (Zhang et al., 1998; Liberati et al., 1999; Qing et al., 2000), ATF-2 (Sano et al., 1999), TFE3 (Hua et al., 1998, 1999), Sp1 (Feng et al., 2000; K.Pardali et al., 2000), LEF1/TCF (Labbé et al., 2000) and the vitamin D3 receptor (Yanagisawa et al., 1999). In addition, the phosphorylated C-terminal sequence of Smad2 and Smad3 interacts with the coactivator CBP/p300 (Feng et al., 1998; Janknecht et al., 1998; Topper et al., 1998), which in turn connects the Smads to the general transcription machinery. These observations explain how TGF-β can induce or enhance transcription, i.e. through physical interactions and functional cooperation of Smads with other transcription factors.
TGF-β has been implicated in both positive and negative regulation of gene expression. However, the mechanisms of transcriptional repression in response to TGF-β have not been characterized. While Smad-mediated repression has not been reported, several corepressors have been identified that interact with Smads and decrease transcriptional activation. Evi-1 (Kurokawa et al., 1998), TGIF (Wotton et al., 1999), E1A (Nishihara et al., 1999) and Ski (Akiyoshi et al., 1999; Luo et al., 1999; Sun et al., 1999) have been shown to interact with Smad3 and to decrease or even inhibit TGF-β- and Smad3-mediated transcriptional activation. Decreased interaction of Smad3 with CBP/p300, as a result of interferon-γ signaling (Ghosh et al., 2001) or SNIP1 expression (Kim et al., 2000), likewise decreases Smad activity. In addition, activation of the JNK pathway may also inhibit Smad3-mediated transcription (Dennler et al., 2000). However, these mechanisms do not repress transcription or inhibit gene expression in response to TGF-β, but rather prevent maximal TGF-β activity.
One of the genes known to be repressed by TGF-β in osteoblast differentiation is osteocalcin, a gene specifically expressed in differentiated osteoblasts and odontoblasts (Desbois et al., 1994). The TGF-β-mediated decrease of osteocalcin has been shown to occur at the mRNA level and does not require new protein synthesis (Noda, 1989). More recently, it has been shown that transcription from the osteocalcin promoter requires binding of the transcription factor CBFA1 to a response element, named OSE2, in the osteocalcin promoter (Ducy and Karsenty, 1995; Geoffroy et al., 1995; Merriman et al., 1995; Banerjee et al., 1996). CBFA1, which is also known as RUNX2, OSF2, PEPB2αA or AML3, is a runt family transcription factor, structurally related to AML1 and AML2. Smad3 has been shown to interact and cooperate with AML1, 2 and 3 to activate transcription from the germline Ig Cα promoter in response to TGF-β (Hanai et al., 1999; Lee et al., 2000; E.Pardali et al., 2000; Zhang and Derynck, 2000; Zhang et al., 2000).
CBFA1 expression plays a key role in osteoblast differentiation and skeletal formation (Karsenty, 1999). In addition to osteocalcin, CBFA1 regulates expression of several other genes that are activated during osteoblast differentiation, including alkaline phosphatase (Harada et al., 1999), α1 and α2(I) collagen (Kern et al., 2001), osteopontin (Ducy et al., 1997) and osteoprotegerin ligand (Gao et al., 1998; Thirunavukkarasu et al., 2000). These genes also contain CBFA1-binding sites in their promoters. These observations suggest that CBFA1 is an essential transcription factor for osteoblast differentiation. This hypothesis is strongly supported by the absence of bone formation in mouse embryos in which the cbfa1 gene was inactivated (Komori et al., 1997; Otto et al., 1997). Furthermore, cleidocranial dysplasia, a human disorder in which some bones are not fully developed, has been associated with mutations in a cbfa1 allele (Lee et al., 1997; Mundlos et al., 1997). One of these mutants, which is deficient in Smad binding, can prevent cooperation of CBFA1 with BMP signaling to induce osteoblastic conversion of C2C12 myoblasts (Lee et al., 2000; Zhang et al., 2000). In addition to its role in osteoblast differentiation, CBFA1 has been implicated in the regulation of bone matrix deposition by differentiated osteoblasts (Ducy et al., 1999). The expression of cbfa1 is regulated by factors that influence osteoblast differentiation. Accordingly, BMPs can activate (Ducy et al., 1997; Gori et al., 1999; Lee et al., 2000), while Smad2 (Li et al., 1998) and glucocorticoids (Chang et al., 1998) can inhibit, cbfa1 expression. In addition, CBFA1 can bind to an OSE2 element in its own promoter, suggesting the existence of an autoregulatory feedback mechanism of transcriptional regulation during osteoblast differentiation (Ducy et al., 1999).
In this study we addressed the mechanism through which TGF-β inhibits osteoblast differentiation and function. TGF-β causes transcriptional repression of the osteocalcin and cbfa1 promoters, and this repression depends on the presence of CBFA1 and occurs through Smad3. The Smad3-mediated repression of the transcriptional function of CBFA1 stands in contrast to previously documented examples of transcriptional activation by Smads in response to TGF-β. This Smad3-mediated repression of CBFA1 function may explain the inhibitory effect of TGF-β at two stages of osteoblast differentiation, i.e. activation of cbfa1 expression and, subsequently, of CBFA1-dependent genes such as osteocalcin. Alterations in Smad3 signaling influenced the extent of osteoblast differentiation both in the presence and absence of TGF-β. Increased Smad3 signaling not only potentiated the TGF-β inhibition of osteoblast differentiation, but also suppressed differentiation in the absence of added TGF-β, thereby implicating TGF-β/Smad3 signaling in autocrine regulation of osteoblast differentiation.
Results
TGF-β represses cbfa1 transcription
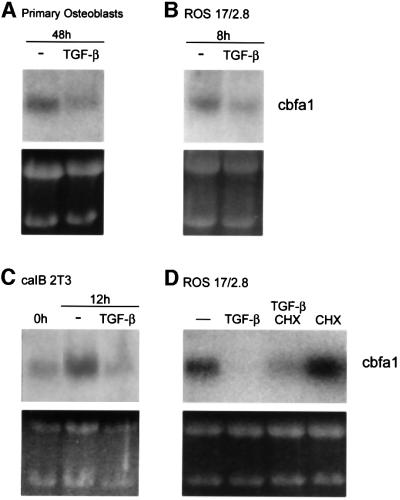
To understand the mechanism by which TGF-β inhibits osteoblast differentiation, we evaluated its effect on the expression of cbfa1 in several osteoblast differentiation systems. Primary osteoblasts, isolated from newborn mouse calvaria, were cultured under conditions that enable osteoblast differentiation, in the absence or presence of TGF-β. As shown in Figure 1A, TGF-β caused a 50% reduction of cbfa1 mRNA. ROS 17/2.8 cells, a rat osteosarcoma cell line often used for studies of osteoblast function, constitutively express cbfa1 (Ducy and Karsenty, 1995). Exposure of these cells to TGF-β similarly reduced the level of cbfa1 mRNA within 8 h (Figure 1B). Under appropriate culture conditions, caIB 2T3 cells also undergo osteoblast differentiation in a manner that resembles the pattern observed in differentiating mouse calvarial osteoblasts (Chen et al., 1998). In these cells, expression of cbfa1 mRNA was increased after 12 h in culture. Exposure to TGF-β during this time prevented the differentiation-associated increase in cbfa1 expression, as cbfa1 mRNA levels in TGF-β-treated cells were only 30% of those in untreated cells (Figure 1C). Finally, TGF-β also reduced cbfa1 mRNA expression in subclone 4 of MC3T3-E1 cells (data not shown), another cell line that undergoes osteoblast differentiation in culture (Xiao et al., 1997).
Fig. 1. TGF-β inhibits the expression of cbfa1 mRNA independently of new protein synthesis. The cbfa1 mRNA, visualized following northern blot hybridization and autoradiography, is shown. (A) Primary mouse calvarial osteoblasts cultured for 48 h in differentiation medium in the presence of TGF-β (5 ng/ml) express reduced levels of cbfa1 mRNA. (B) Treatment with TGF-β for 8 h decreases the level of cbfa1 mRNA in ROS 17/2.8 cells. (C) TGF-β treatment of caIB 2T3 cells during the 12 h period in differentiation medium represses cbfa1 mRNA expression. (D) ROS 17/2.8 cells were treated with TGF-β for 8 h as in (B) in the absence or presence of cycloheximide (CHX) (10 µg/ml) or with cycloheximide alone. Ethidium bromide-stained gels are included to show similar loading of RNA.
To determine whether the inhibition of the cbfa1 mRNA levels by TGF-β requires new protein synthesis, we assessed the effect of TGF-β in the absence or presence of cycloheximide, an inhibitor of new protein synthesis. The TGF-β-dependent decrease in cbfa1 mRNA levels in ROS 17/2.8 cells also occurred in the presence of cycloheximide, suggesting that no new protein synthesis is required for TGF-β to reduce cbfa1 transcription (Figure 1D). In ROS 17/2.8 cells, the constitutive cbfa1 mRNA level was increased in the presence of cycloheximide alone, presumably a result of stabilization of this mRNA in the presence of cycloheximide (Beelman and Parker, 1994; Alliston et al., 1997). This stabilizing effect likely explains why the TGF-β-mediated decrease in cbfa1 mRNA levels was less pronounced in the presence of cycloheximide (Figure 1D). Similar results were obtained in caIB 2T3 cells (data not shown). These results suggest that the repression of cbfa1 mRNA expression by TGF-β utilizes transcription regulators already present in the cells prior to TGF-β stimulation.
TGF-β-mediated repression of the cbfa1 promoter depends on the presence of CBFA1
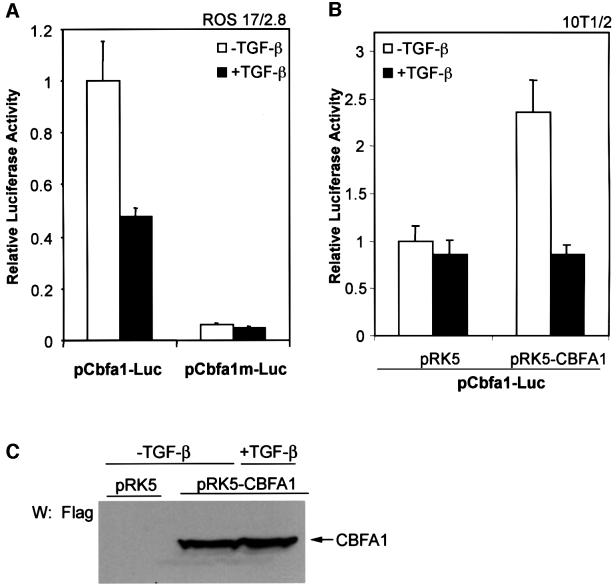
To examine the mechanism that regulates the inhibition of cbfa1 mRNA levels by TGF-β, we assessed the regulation of transcription from the cbfa1 promoter using a luciferase reporter plasmid, pCbfa1-Luc. This plasmid contains the luciferase expression unit under the control of the –89/+46 segment of the cbfa1 promoter, which contains three binding sites for CBFA1 (Ducy et al., 1999). Transient transfection of pCbfa1-Luc conferred constitutive luciferase expression in ROS 17/2.8 cells, which express CBFA1 and several other markers of osteoblast differentiation (Ducy and Karsenty, 1995), and this transcription was decreased by addition of TGF-β (Figure 2A). We also tested the transcription from the pCbfa1m-Luc reporter, in which each of the three CBFA1-binding sites is mutated to prevent CBFA1 binding (Ducy and Karsenty, 1995). As is apparent in Figure 2A, loss of CBFA1-binding ability in the mutated promoter strongly decreased luciferase expression in ROS 17/2.8 cells and abolished TGF-β-mediated repression.
Fig. 2. TGF-β inhibits transcription from the cbfa1 promoter. ROS 17/2.8 (A) or 10T1/2 (B) cells were transfected with the pCbfa1-Luc or pCbfa1m-Luc reporter plasmids and expression plasmids, as indicated. 10T1/2 cells (B) were cotransfected with the CBFA1 expression plasmid (pRK5-CBFA1) or the control empty plasmid (pRK5). Cells were cultured in the presence or absence of TGF-β (1 ng/ml) 16 h after transfection. Forty-eight hours after transfection, cells were harvested and reporter activities measured. Values normalized for transfection efficiency are shown as fold induction relative to basal promoter activity as described in Materials and methods. (C) Whole 10T1/2 cell lysates were prepared from a parallel transfection experiment for visualization of CBFA1 protein levels by western analysis using Flag antibody.
To explore this observation further, we transfected pCbfa1-Luc into 10T1/2 cells, a mesenchymal cell line that does not express CBFA1 or other osteoblast-specific genes (Ducy et al., 1997). In these cells, only a basal level of transactivation and no effect of TGF-β were observed. However, coexpression of CBFA1 enhanced transcription from the cbfa1 promoter. Under these conditions, TGF-β inhibited transcription from the cbfa1 promoter (Figure 2B), even though the level of coexpressed CBFA1 was unaffected (Figure 2C). Similar results to those in 10T1/2 cells were also observed in NIH 3T3 cells, another mesenchymal cell line (data not shown). Together, these results suggest that the inhibition of cbfa1 transcription by TGF-β requires both the presence of CBFA1 and CBFA1 binding to the cbfa1 promoter.
CBFA1-dependent repression of transcription from the osteocalcin promoter by TGF-β
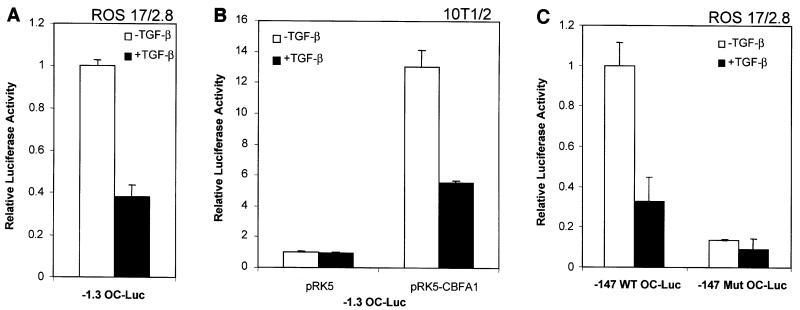
TGF-β is also known to inhibit the expression of other osteoblast-specific genes, including osteocalcin (Centrella et al., 1994). The inhibition of osteocalcin mRNA expression by TGF-β has been shown previously to occur without the requirement of new protein synthesis (Noda, 1989), suggesting that, as we have shown for cbfa1 expression (Figure 1D), the TGF-β effect on osteocalcin transcription is direct. We therefore assessed the effect of TGF-β on transcription from the osteocalcin promoter in ROS 17/2.8 and 10T1/2 cells, i.e. in the presence or absence of CBFA1. Transcriptional activation was measured by assessing luciferase expression from a 1.3 kbp promoter segment of the mouse osteocalcin II gene (OG2). This promoter segment has been shown to confer the same regulation and expression pattern in vivo as the endogenous promoter (Ducy and Karsenty, 1995). In ROS 17/2.8 cells, TGF-β inhibited the transcription from this promoter (Figure 3A). However, TGF-β did not affect the low level of basal transcription in 10T1/2 cells (Figure 3B). As in the case of the cbfa1 promoter (Figure 2B), CBFA1 coexpression in 10T1/2 cells enhanced the osteocalcin promoter activity and conferred TGF-β-dependent inhibition (Figure 3B). These results suggest that, consistent with our findings with the cbfa1 promoter, TGF-β-mediated repression of OG2 transcription also depends on CBFA1 expression.
Fig. 3. Inhibition of transcription from the osteocalcin promoter by TGF-β requires CBFA1. ROS 17/2.8 (A and C) and 10T1/2 (B) cells were transfected with the designated osteocalcin promoter/reporter plasmids followed by treatment with or without TGF-β (1 ng/ml). Luciferase expression was scored as in Figure 2. 10T1/2 cells (B) were cotransfected with the CBFA1 expression plasmid (pRK5-CBFA1) or the control empty plasmid (pRK5).
Since a CBFA1-binding site within the osteocalcin promoter (named OSE2) is particularly critical for the transcriptional inducibility of the osteocalcin promoter (Frendo et al., 1998), we assessed the role of this sequence in TGF-β-mediated repression. As shown in Figure 3C, the –147 bp proximal promoter segment contains the OSE2 site and conferred both transactivation and TGF-β-mediated repression in ROS 17/2.8 cells. Inactivation of the OSE2 site in this promoter segment strongly decreased TGF-β-mediated repression, while also decreasing the constitutive level of transcription. These data strongly suggest that the CBFA1-binding sequence confers TGF-β-dependent repression. This is consistent with our previous conclusion that TGF-β-mediated repression from the osteocalcin or cbfa1 promoters depends on the presence of CBFA1.
TGF-β decreases the transcriptional activity of CBFA1
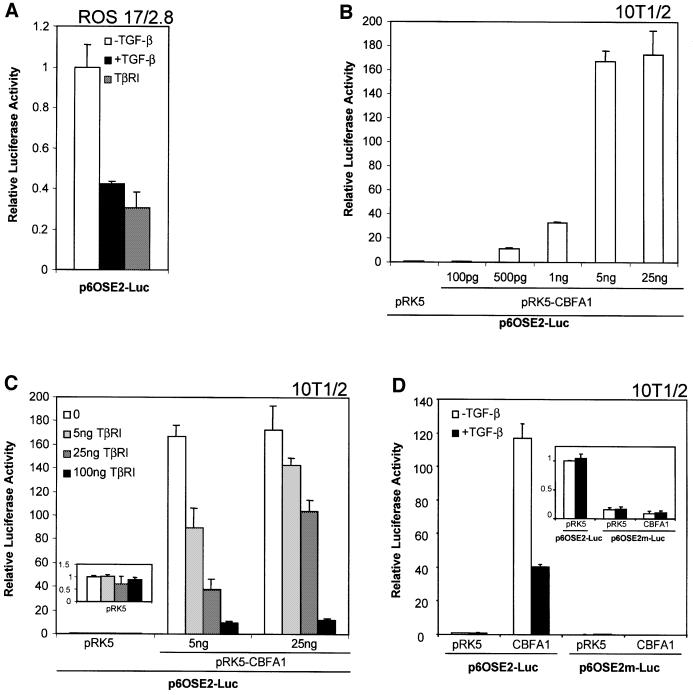
To assess better the role of CBFA1 in TGF-β-mediated repression, we utilized an artificial promoter reporter plasmid, p6OSE2-Luc, in which luciferase expression is controlled by six tandem copies of the OSE2 site, i.e. the CBFA1-binding site in the OG2 promoter (Ducy and Karsenty, 1995). In ROS 17/2.8 cells, this promoter allowed a high level of transcription, which was inhibited by TGF-β or by coexpression of a constitutively activated TGF-β type I receptor (Figure 4A). These data strongly suggest that TGF-β signaling inhibited transactivation of this reporter through a direct effect on CBFA1 function.
Fig. 4. TGF-β inhibits transcription from the OSE2 sequence by CBFA1. (A) Transiently transfected ROS 17/2.8 cells were treated with TGF-β (1 ng/ml; black bar) or cotransfected with an expression plasmid for a constitutively activated form of the TGF-β type I receptor (TβRI) (gray bar). (B) 10T1/2 cells were transfected with different quantities of pRK5-CBFA1. (C) 10T1/2 cells were transfected with 5 or 25 ng of pRK5-CBFA1, or the empty vector pRK5, in the presence of increasing quantities of TβRI expression plasmid. The inset shows the lack of effect of increased TβRI expression on basal promoter activity in 10T1/2 cells (note difference in scale). (D) A reporter plasmid containing six copies of a mutant OSE2 site, p6OSE2m-Luc, has reduced basal and CBFA1-inducible transactivation (see inset), when compared with p6OSE2-Luc. Luciferase expression was scored as in Figure 2.
As seen with the endogenous cbfa1 promoter- or osteocalcin promoter-driven reporter plasmids in 10T1/2 cells, p6OSE2-Luc conferred only a low level of transcription in 10T1/2 cells in the absence of CBFA1. Cotransfection of CBFA1, however, enhanced transcription in a dose-dependent manner (Figure 4B). While coexpression of a constitutively activated TGF-β type I receptor had no effect on basal transcription (Figure 4C, inset), TGF-β receptor signaling inhibited CBFA1-dependent transcription in a dose-dependent manner (Figure 4C). Mutation of the six copies of the OSE2 site in the p6OSE2m-Luc plasmid abolished the ability of CBFA1 to induce transcription (Figure 4D) as well as TGF-β-mediated repression (Figure 4D, inset). Together with the observations that TGF-β-mediated transcriptional repression of the cbfa1 and osteocalcin promoters does not depend on new protein synthesis (Figure 1D; Noda, 1989) and depends on the presence of CBFA1 (Figures 2 and 3) and intact CBFA1-binding sites in the promoter (Figures 2A, 3C and 4D), these data suggest that existing cellular factors interact with CBFA1 to repress its functional activity.
Smad3, but not Smad2, mediates TGF-β-dependent repression of CBFA1 function
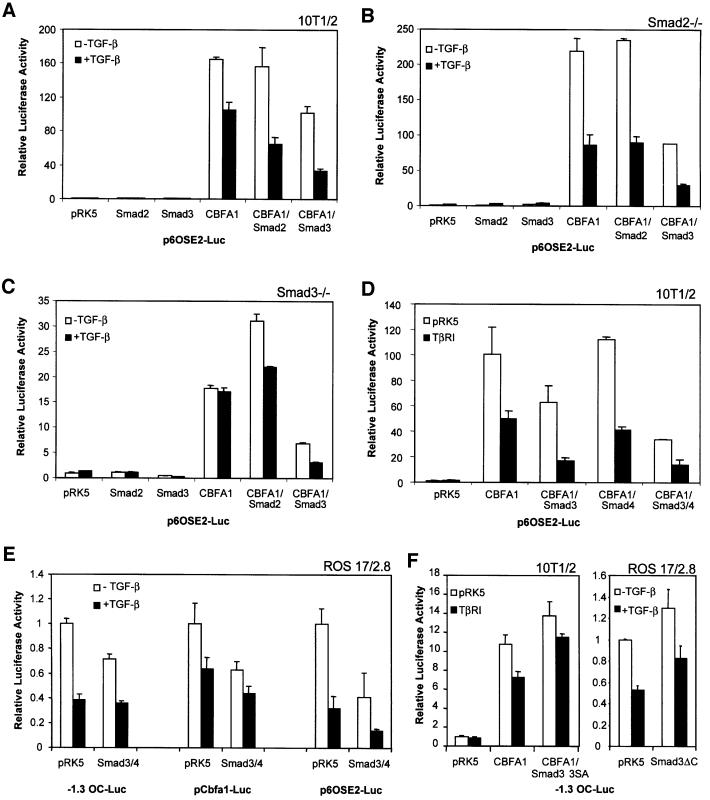
Two Smads, Smad2 and Smad3, have been implicated as downstream activators of transcription in response to TGF-β (Derynck et al., 1998; Massagué, 1998); however, Smads have not been shown to mediate transcriptional repression. We therefore tested whether Smad2 and Smad3 could act as TGF-β-induced repressors of the transcription function of CBFA1 (Figure 5A). Smad2 or Smad3 was expressed in 10T1/2 cells in the absence or presence of CBFA1 to assess their roles in the TGF-β-dependent effects on CBFA1 function. In the absence of CBFA1, Smad2 or Smad3 did not affect the basal transcription from p6OSE2-Luc, either in the presence or absence of TGF-β. However, in the presence of CBFA1, Smad3 reduced the CBFA1-mediated transactivation. This inhibitory effect on CBFA1 activity was enhanced by TGF-β, resulting in a greater TGF-β inhibition than was observed in control cells without cotransfected Smad3. Smad2 expression resulted in a milder effect, as it did not decrease CBFA1 function in the absence of TGF-β, but enhanced the TGF-β-mediated repression of CBFA1 activity to a lesser extent than Smad3 (Figure 5A).
Fig. 5. Smad3 inhibits CBFA1 function and mediates the inhibitory effect of TGF-β (1 ng/ml). Cells were transiently transfected with expression plasmids and luciferase reporter plasmids, as shown. Luciferase expression from the reporter plasmids was scored as in Figure 2. (A, D and F) 10T1/2 cells, (B) Smad2–/– mouse embryonic fibroblasts, (C) Smad3–/– mouse embryonic fibroblasts, (E and F) ROS 17/2.8 cells.
Since 10T1/2 cells, like most cells, express endogenous Smad2 and Smad3, we performed similar CBFA1 functional assays in fibroblasts derived from mouse embryos deficient in either Smad2 or Smad3. CBFA1 expression activated transcription from the 6OSE2 promoter in both cell lines; however, TGF-β-mediated repression of CBFA1 function only occurred in the Smad2-deficient cells (Figure 5B) and not in the Smad3-deficient cells (Figure 5C). Furthermore, cotransfection of Smad2 did not affect CBFA1 activity in Smad2-deficient cells (Figure 5B) and did not restore TGF-β-mediated repression in Smad3-deficient cells (Figure 5C). In contrast, Smad3 expression in Smad2-deficient cells resulted in repression of CBFA1 function, which was enhanced by TGF-β (Figure 5B). In Smad3-deficient cells, expression of Smad3 repressed CBFA1 function and reconstituted the TGF-β-responsive repression of CBFA1 activity. These data indicate that Smad3, but not Smad2, has the ability to repress the transcriptional function of CBFA1.
Smad function has been shown to require interaction of a receptor-activated Smad with Smad4. Accordingly, Smad4 has been shown to synergize with Smad3 in activating transcription (Lagna et al., 1996; Zhang et al., 1996). While the endogenous Smad4 levels are sufficient to observe the effect of Smad3, we sought to determine the effect of increased Smad4 levels on the ability of Smad3 to repress CBFA1 function. As shown in Figure 5D, coexpression of Smad4 was unable to repress CBFA1 activity and did not enhance the TGF-β inhibition of CBFA1 function. Coexpression of Smad4 with Smad3, however, did enhance the repressive effect of Smad3 on CBFA1 transactivation of the p6OSE2-Luc reporter.
We also verified that Smad3 and Smad4 acted as transcriptional repressors of the natural cbfa1 and OG2 promoters (Figure 5E). To assess these effects in the presence of endogenous levels of CBFA1, we used ROS 17/2.8 cells. Consistent with our results shown above (Figures 2, 3 and 4), TGF-β repressed transcription from these promoters (Figure 5E). As was seen with p6OSE2-Luc in 10T1/2 cells, Smad3 and 4 repressed transcription from these promoters and this effect was further enhanced by TGF-β (Figure 5E). Finally, we assessed the effect of dominant-negative mutants of Smad3, named Smad3-3SA and Smad3ΔC, on transcriptional activation from the OG2 promoter in 10T1/2 cells and ROS 17/2.8 cells (Figure 5F). Smad3-3SA lacks the C-terminal phosphorylation sites required for TGF-β activation (Liu et al., 1997; Feng et al., 1998), while Smad3ΔC lacks the C-terminal 43 amino acids (Zhang et al., 1996). Consistent with our previous data (Figure 3), TGF-β-mediated repression of transcription was dependent on the expression of cotransfected (Figure 5F, left) or endogenous (Figure 5F, right) CBFA1. Under these conditions, expression of Smad3-3SA or Smad3ΔC decreased the repression by TGF-β (Figure 5F). Smad3-3SA and Smad3ΔC also increased somewhat the CBFA1 activity in the absence of TGF-β (Figure 5F), suggesting that CBFA1 activity is subject to auto crine downregulation by endogenous, TGF-β-activated Smad3. Thus, by repressing autocrine TGF-β signaling, Smad3-3SA may enhance CBFA1 activity.
The effect of TGF-β and Smad3 on CBFA1 function is context dependent
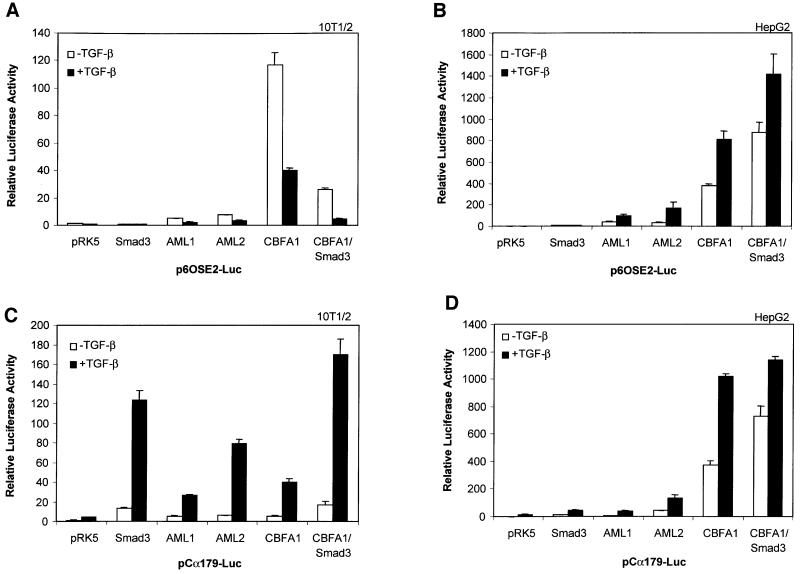
The ability of TGF-β and Smad3 to repress the transcriptional activity of CBFA1 stands in marked contrast to the reported ability of Smads to activate, but not repress, transcription. Furthermore, TGF-β and Smad3 have been reported to cooperate with two other runt family transcription factors, i.e. AML1 and AML2, in addition to AML3/CBFA1, to increase transcription (Hanai et al., 1999; E.Pardali et al., 2000; Zhang and Derynck, 2000). However, like most studies on Smad-mediated transcription, these were carried out in epithelial cells using different promoters. Since the TGF-β- and Smad3-mediated repression were observed in cells of mesenchymal origin, we compared the effects of TGF-β and Smad3 on members of the AML family in epithelial versus mesenchymal cells. As shown in Figure 6A, and consistent with our results shown above, TGF-β receptor activation decreased transcription from p6OSE2-Luc in 10T1/2 cells in the presence of CBFA1, and this decrease was enhanced when Smad3 was overexpressed. TGF-β also repressed the transactivation of p6OSE2-Luc by AML1 or AML2, although the transactivation by AML1 or AML2 was lower than with CBFA1. In contrast to mesenchymal cells, TGF-β signaling and/or Smad3 coexpression enhanced transactivation of p6OSE2-Luc by CBFA1 in HepG2 cells (Figure 6B) and Mv1Lu cells (data not shown). TGF-β also enhanced AML1- or AML2-mediated transcription from p6OSE2-Luc, although, as in 10T1/2 cells, the overall transcription levels were lower than those of CBFA1. These data suggest that mesenchymal and epithelial cells distinguish themselves from each other by the presence or absence of one or several cofac tors, which confer Smad-dependent repression versus activation.
Fig. 6. Effects of Smad3 and TGF-β on the function of CBFA1 and runt family transcription factors depend on the cell type and promoter. 10T1/2 cells (A and C) or HepG2 cells (B and D) were transiently transfected with the indicated expression plasmids and the luciferase reporter plasmids p6OSE2-Luc (A and B) or pCα179-Luc (C and D). Transactivation from these promoters was measured in the absence or presence of TGF-β (1 ng/ml). Luciferase expression from the reporter plasmids was scored as in Figure 2.
In previous studies, Smad3/4 was shown to cooperate with runt-related transcription factors at the mouse germline Ig Cα promoter (Hanai et al., 1999; Lee et al., 2000; E.Pardali et al., 2000; Zhang and Derynck, 2000; Zhang et al., 2000). The TGF-β-responsive Ig Cα promoter segment used in these assays, p179Cα-Luc, contains two putative runt-binding sites, two proposed Smad-binding sequences and a CRE sequence to which CREB can bind (Shi and Stavnezer, 1998; Xie et al., 1999; Zhang and Derynck, 2000). We therefore compared the transcriptional activation of the three AML transcription factors at the Ig Cα promoter in HepG2 and 10T1/2 cells. As shown in Figure 6C and D, TGF-β enhanced transcription from the Ig Cα promoter by AML1, AML2 or CBFA1 in both 10T1/2 and HepG2 cells. The CBFA1-mediated response was enhanced by coexpression of Smad3. Thus, whereas TGF-β/Smad3 signaling repressed the transcriptional activity of these runt transcription factors at the p6OSE2 promoter (and osteocalcin and cbfa1 promoters) in 10T1/2 cells (and ROS 17/2.8 and NIH 3T3 cells; data not shown), Smad3 and TGF-β signaling enhanced the AML-mediated transcription at the Ig Cα promoter in mesenchymal cells. In addition, this transcriptional cooperativity at the Ig Cα promoter did not depend on the cell type, consistent with previous reports (Hanai et al., 1999; Lee et al., 2000; E.Pardali et al., 2000; Zhang and Derynck, 2000; Zhang et al., 2000). These data strongly suggest that the ability of Smad3/4 to enhance or repress the transcriptional activity of CBFA1 depends not only on the cell context, but also on the promoter sequence. Specifically in the case of the Ig Cα promoter, the presence of the CRE or a Smad-binding sequence may affect the functional interactions between Smad3/4 and CBFA1.
Physical interaction of Smad3 and CBFA1
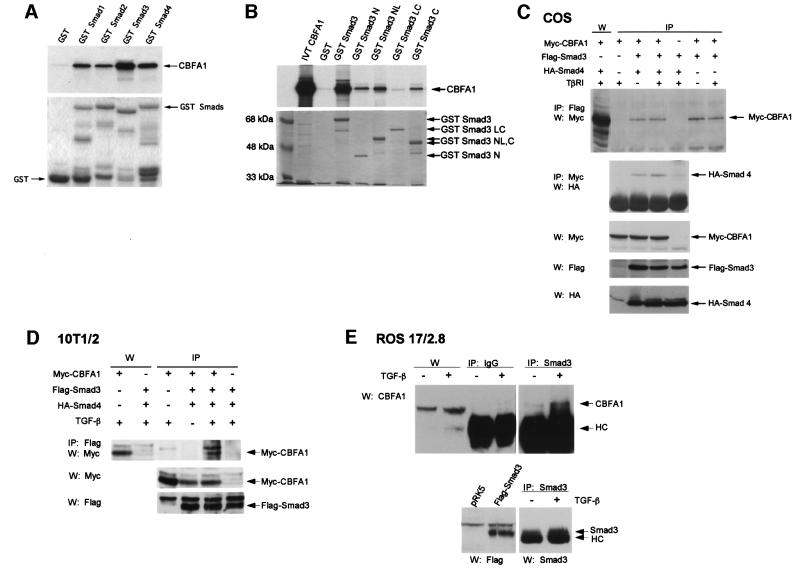
The ability of Smad3 to repress the activity of CBFA1 strongly suggested a physical interaction. We therefore assessed the ability of Smads to interact with CBFA1 in vitro using glutathione S-transferase (GST)-adsorption assays, in which in vitro translated 35S-labeled CBFA1 was incubated with Smad1, 2, 3 or 4 fused to a GST moiety. As shown in Figure 7A, CBFA1 associated with all four Smads, and this interaction was strongest with Smad3 and weakest with Smad2. Similar experiments characterized which Smad3 segments interacted with CBFA1. While other Smad-interacting transcription factors associate with either the N- or C-segment of a Smad (Itoh et al., 2000), CBFA1 interacted with both the N- and C-segments of Smad3 (Figure 7B). Together, these results indicate that CBFA1 can interact with Smad3 in vitro. However, the interaction of Smad2 with CBFA1, albeit considerably weaker than the Smad3 interaction, suggests that additional interactions may be required for Smad3-mediated repression of CBFA1.
Fig. 7. CBFA1 interacts with Smad3. (A and B) Interaction of 35S-labeled, in vitro translated CBFA1 with GST–Smad fusion proteins, as shown. Interacting 35S-labeled CBFA1 (arrow) is visualized following gel electrophoresis and autoradiography. Five percent of the input reaction volume of 35S-labeled CBFA1 (IVT CBFA1) was loaded as control in lane 1 of (B). Below the autoradiograms are photographs of Coomassie Blue-stained gels to show the integrity and equal loading of the fusion proteins. (C) Interaction of Smad3 and Smad4 with CBFA1 in vivo, as assessed using co-immunoprecipitations. Cell lysates from transfected COS-1 cells expressing the indicated tagged proteins were used for immunoprecipitations (IP), followed by western blotting (W). The top panel shows the interaction of Myc-CBFA1 with Flag-Smad3, as assessed by immunoprecipitations with anti-Flag antibody-coupled Sepharose beads, followed by western blotting using anti-Myc antibody. The first lane visualizes Myc-tagged CBFA1 by western blotting without prior immunoprecipitation. The second panel shows interaction of HA-Smad4 with Myc-CBFA1, as assessed by immunoprecipitations with anti-Myc antibody, followed by western blotting using anti-HA antibody. The three lower panels show the relative expression levels of the tagged proteins in each lysate, as assessed by western blotting. (D) Interaction of Flag-Smad3 and Myc-CBFA1 in transfected 10T1/2 cells in the presence or absence of TGF-β (5 ng/ml), as assessed in (C). The first two lanes show the expression of Myc-tagged CBFA1, visualized by western blotting without prior immunoprecipitation. The two lower panels visualize the expression of Myc-CBFA1 and Flag-Smad3 by western analysis. (E) Interaction of endogenous CBFA1 and endogenous Smad3 in ROS 17/2.8 cells. This interaction was detectable by immunoprecipitation using a Smad3 antibody, followed by anti-CBFA1 western blotting, in TGF-β-treated (5 ng/ml), but not in untreated ROS 17/2.8 cell lysates. No CBFA1 was detected when rabbit anti-mouse IgG was used instead of anti-Smad3 antibody. The IgG heavy chain band is marked HC. CBFA1 levels in the lysates were detected by western analysis (left two lanes), whereas Smad3 levels were visualized by Smad3 immunoprecipitation followed by western analysis with a Smad3 antibody (lower panel, right two lanes), using Flag-Smad3 expressed in transfected 10T1/2 cells as reference (lower panel, left two lanes).
The interactions of Smad3 and Smad4 with CBFA1 were also assessed in transfected cells using immunoprecipitations followed by western analyses. As shown in Figure 8C, Smad3 and Smad4 both interacted with CBFA1 in vivo. These interactions already occurred in the absence of TGF-β and were not enhanced in the presence of an activated TβRI, presumably because, under these experimental conditions, sufficiently high levels of the transfected Smads were already colocalized in the nucleus with CBFA1 without the need for additional receptor activation. Because TGF-β- and Smad3-mediated repression of the CBFA1 activity occurred in mesenchymal cells, we also assessed the interactions of CBFA1 with Smad3, in the presence of Smad4, in transfected 10T1/2 cells. As shown in Figure 7D, immunoprecipitation of Smad3 coprecipitated CBFA1, but only upon TGF-β treatment. The TGF-β dependence of this interaction is likely to be a result of lower Smad3 expression and higher TGF-β responsiveness of 10T1/2 cells relative to COS cells. Finally, we were also able to detect TGF-β-stimulated association of CBFA1 and Smad3 at endogenous levels in ROS 17/2.8 cells (Figure 7E), although the detection of this complex was severely limited by the quality of the antibodies and the low endogenous expression levels. Together, these data demonstrate that the TGF-β-mediated repression of CBFA1 activity by Smad3 is accompanied by a physical association of Smad3 with CBFA1.
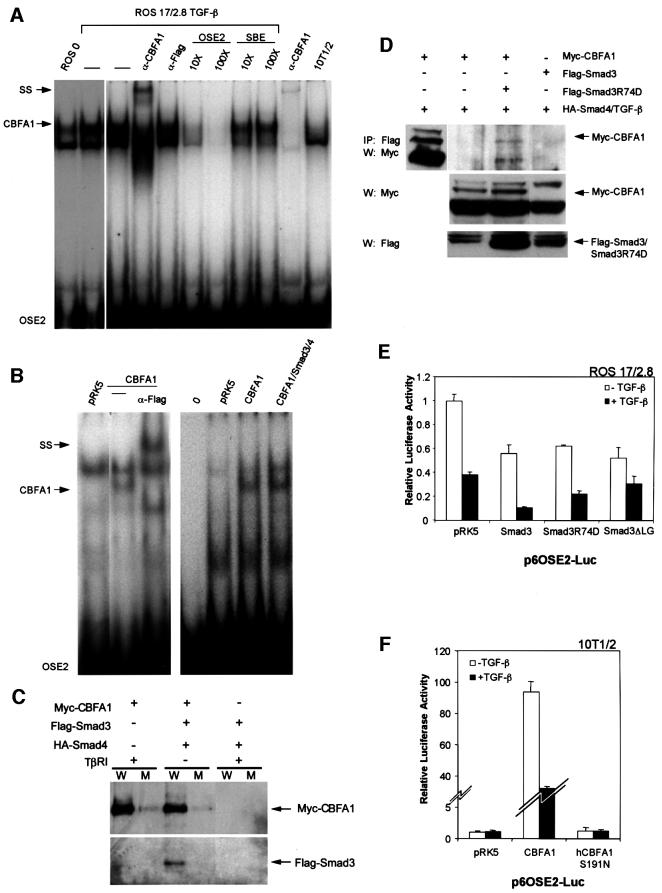
Fig. 8. Binding of CBFA1 and Smad3 to the OSE2 DNA sequence. (A) Electrophoretic mobility shift assays using untreated (lane 1) or TGF-β-treated (5 ng/ml; lane 2) ROS 17/2.8 cell extracts allowed visualization of endogenous CBFA1 binding (arrow) to a 32P-labeled OSE2 oligonucleotide. The identity of the CBFA1-containing complex was confirmed by its absence in 10T1/2 cells, which lack CBFA1 expression (lane 10), and its partial disappearance and supershift (SS) following incubation with an anti-CBFA1 antibody (α-CBFA1, lane 4), but not with an unrelated antibody (α-Flag, lane 5). Unlabeled OSE2 competitor oligonucleotide, at 10- to 100-fold molar excess, competed (lanes 6 and 7), whereas an unrelated competitor oligonucleotide did not compete (lanes 8 and 9) for the radiolabeled OSE2–CBFA1 complex. (B) Electrophoretic mobility shift assays show CBFA1 binding to an OSE2 oligonucleotide, both in the absence or presence of Smad3 and Smad4. COS-1 cells were transfected with expression plasmids for Flag-CBFA1, with or without Smad3/4 (as shown) or the pRK5 control plasmid. The radiolabeled OSE2 oligonucleotide was incubated alone (0) or in the presence of cell lysates. Expression of Flag-CBFA1 resulted in the formation of a distinct band (lanes 2 and 6), which could be supershifted (SS) using an anti-Flag antibody (lane 3). This OSE2–CBFA1 complex was also apparent in the presence of overexpressed Smad3/4 (lane 7). (C) Binding of CBFA1 to an OSE2 oligonucleotide, in the absence or presence of equimolar levels of Smad3 and Smad4. Biotinylated wild-type (W) or mutant (M) OSE2 oligonucleotides were incubated with the same lysates as in Figure 7C, and the interactions of Myc-CBFA1 and Flag-Smad3 with the oligonucleotides were assessed by western blotting. CBFA1 binds to the wild-type oligonucleotide, both in the presence or absence of Smad3/4, but not to the mutant oligonucleotide. Furthermore, Smad3 interacts with the OSE2 nucleotide only in the presence of CBFA1. (D) The DNA binding-defective Smad3R74D interacts with CBFA1 in transfected 10T1/2 cells. Immunoprecipitation assays, performed as in Figure 7C and D, revealed coprecipitation of Myc-CBFA1 (arrow) with Flag-Smad3R74D (top panel). Western analysis without prior immunoprecipitation visualized the migration of Myc-CBFA1 (top panel, left lane), and the expression levels of CBFA1 and Smad3 or Smad3 R74D (lower panels). (E) DNA binding of Smad3 is not essential for TGF-β/Smad3-mediated repression of transcription from the OSE2 sequence by CBFA1. ROS 17/2.8 cells were transfected with the p6OSE2-Luc reporter and wild-type or mutant Smad3 expression plasmids. Luciferase expression in the absence or presence of TGF-β(1 ng/ml) was scored as in Figure 2. (F) DNA binding of CBFA1 is essential for transcriptional activity and Smad3-mediated repression. 10T1/2 cells were transfected with the p6OSE2-Luc reporter and an expression plasmid for wild-type CBFA1 or the DNA binding-defective mutant hCBFA1 S191N. Luciferase expression in the absence or presence of TGF-β (1 ng/ml) was scored as in Figure 2.
Role of DNA binding in TGF-β- and Smad3-mediated repression of CBFA1 activity
The TGF-β-dependent interaction of Smad3 with CBFA1 raised the possibility that this complex formation might displace CBFA1 binding from its OSE2-binding sequence, and that this could explain the TGF-β- and Smad-mediated repression. We therefore evaluated the binding of endogenous CBFA1 in ROS 17/2.8 cells to the OSE2 DNA sequence using gel shift assays. As shown in Figure 8A, using gel shift analyses, we detected a CBFA1–DNA complex in ROS 17/2.8 cell lysates, which was not present in 10T1/2 cells that lack CBFA1 expression (last lane). This complex was supershifted using a CBFA1 antibody. TGF-β treatment did not decrease CBFA1 complex formation, but consistently increased the intensity of the CBFA1–DNA complex. This increase in intensity, which is not due to increased CBFA1 levels, may result from the ability of Smad3/4 to increase CBFA1 binding, possibly a result of a conformational change or stabilization. Similarly, the physical interaction of a Smad has been shown to increase the efficiency of Sp1 (Feng et al., 2000; K.Pardali et al., 2000) or c-Jun (J.Qing and R.Derynck, unpublished observations) binding to their respective DNA-binding sequences. Clearly, however, TGF-β did not decrease the CBFA1 binding to the OSE2 DNA sequence.
We also assessed whether Smad3 and Smad4 coexpression affected the binding of CBFA1 to the OSE2 sequence (Figure 8B). For these experiments, we used COS-1 cells because of their high transfection efficiency. Expression of CBFA1 in COS-1 cells resulted in the formation of a DNA-binding complex, which could be supershifted using an antibody specific for the tagged CBFA1. This complex was also detected when Smad3 and Smad4 were coexpressed, demonstrating that the Smad interaction with CBFA1 does not displace CBFA1 binding to the OSE2 sequence. No additional complex was observed when Smad3 and Smad4 were overexpressed, suggesting a low affinity interaction of the Smads with CBFA1 and the inability of Smad3 and 4 to bind directly to the OSE2 sequence. Nevertheless, Smad interaction with CBFA1 occurred at the DNA level, as shown in Figure 8C, in which the interaction of CBFA1 with the OSE2 sequence was assessed using biotinylated oligonucleotides. Under these conditions, using the same cell lysates as in Figure 7C, CBFA1 bound specifically to the wild-type but not the mutant OSE2 sequence, and this binding was not affected by Smad3/4 coexpression. Smad3 bound to the complex, but this interaction occurred only when CBFA1 was coexpressed and bound to the oligonucleotide. These results indicate that Smad3 and CBFA1 interact at the DNA level, in an OSE2 sequence-specific manner, again suggesting that the repression of CBFA1 activity in response to TGF-β does not result from decreased CBFA1 binding. These data also suggest that Smad3 does not interact directly with this DNA sequence, but rather that the Smad3 interaction with the OSE2 sequence depends on CBFA1 binding.
Previous results strongly suggest that direct DNA binding of a Smad complex is required for transcriptional cooperativity with an interacting transcription factor (Dennler et al., 1998; Qing et al., 2000). On the other hand, in our system of Smad-mediated repression, Smad3 interaction with the DNA is indirect and totally dependent on CBFA1 binding to the DNA. We therefore assessed whether the DNA-binding ability of Smad3 was required for its interaction with CBFA1. As shown in Figure 8D, a point-mutated Smad3 that is unable to bind DNA, i.e. Smad3R74D (Qing et al., 2000), was able to interact with CBFA1, as assessed by co-immunoprecipitation assays in transfected 10T1/2 cells. In addition, Smad3R74D was able to repress the transcriptional activity of endogenous CBFA1 from the OSE2 promoter sequence in ROS 17/2.8 cells, qualitatively similarly to wild-type Smad3 (Figure 8E). Smad3ΔLG, another mutant of Smad3 that is also defective in DNA binding (Qing et al., 2000), likewise repressed the transcriptional activity of CBFA1 (Figure 8E). Consistent with our observation that Smad3 requires CBFA1 DNA binding to interact with the OSE2 sequence (Figure 8C), these results indicate that the DNA-binding ability of Smad3 is not essential for TGF-β/Smad3-mediated repression of CBFA1 activity.
We also evaluated the requirement of the DNA binding of CBFA1 for transactivation and TGF-β-mediated repression using a CBFA1 point mutant, hCBFA1 S191N, which has been shown to lack DNA-binding ability (Lee et al., 1997). As expected, when expressed in 10T1/2 cells, hCBFA1 S191N did not activate transcription from the OSE2 promoter sequence and TGF-β treatment did not affect the basal transcription (Figure 8F). This observation is consistent with our data in Figures 2A, 3C and 4D, which show that mutation of CBFA1 DNA-binding sequences abolished both CBFA1-activated transcription and TGF-β-dependent repression.
Smad3 as repressor of osteoblast differentiation
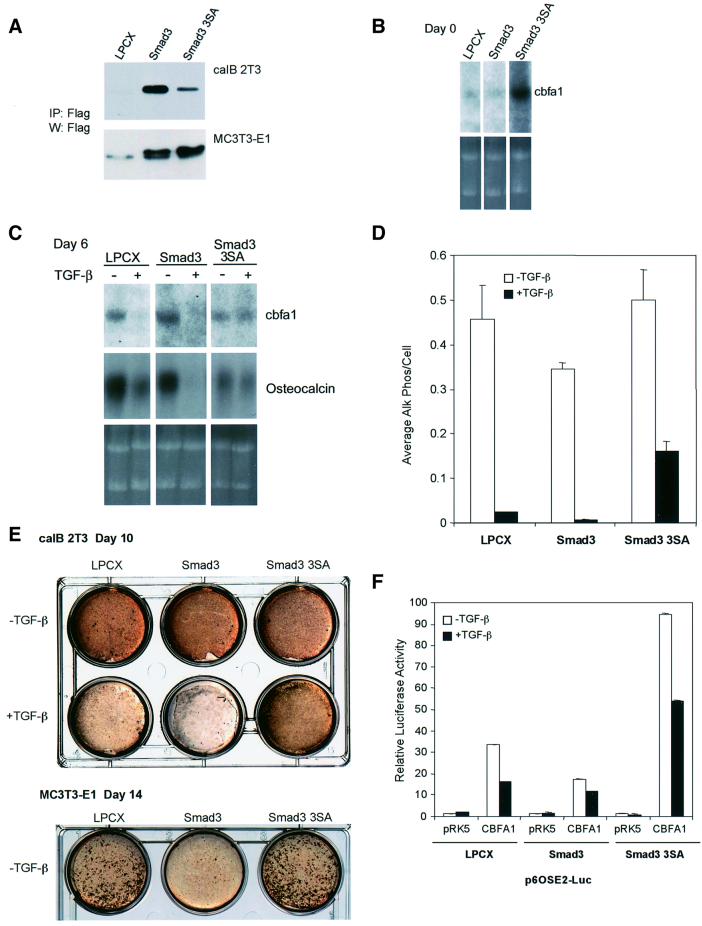
The results outlined above provide evidence that Smad3 represses the activity of CBFA1, thus downregulating cbfa1 and osteocalcin expression, and suggest that this mechanism may be at the basis of the TGF-β-mediated inhibition of osteoblast differentiation. To explore further this regulatory role of Smad3, we generated derivatives of caIB 2T3 osteoblasts, which overexpress Smad3 or a dominant-negative form of Smad3, Smad3-3SA. As mentioned, this mutant form of Smad3 has been shown to inhibit endogenous Smad3 signaling (Liu et al., 1997; Feng et al., 1998). A control cell line, infected with the empty viral expression vector, LPCX, was also generated. As shown in Figure 9A, immunoprecipitation analyses showed that Smad3 and Smad3-3SA were expressed by caIB 2T3 cells infected with the corresponding retroviral expression vector. Using these cells, we assessed the effects of increased Smad3 levels or inhibition of Smad3 function on normal or TGF-β-inhibited osteoblast differentiation.
Fig. 9. Stable expression of wild-type or dominant-negative Smad3 in caIB 2T3 cells and MC3T3-E1 cells alters osteoblast differentiation. (A) Immunoprecipitation followed by western blot analysis demonstrates expression of Flag-tagged Smad3 or Smad3-3SA in cell lysates of stably infected caIB 2T3 and MC3T3-E1 cells. LPCX is the control empty vector. (B) Alterations in Smad3 signaling affect endogenous cbfa1 mRNA expression in caIB 2T3 cells at day 0, i.e. prior to exposure to differentiation conditions, in LPCX control cells, or cells stably expressing Smad3 or Smad3-3SA. The top panel shows cbfa1 northern hybridizations, while the lower panel shows the ethidium bromide-stained gel. (C) Alterations in Smad3 signaling affect endogenous cbfa1 and osteocalcin mRNA expression in differentiating caIB 2T3 cells at day 6 in differentiation conditions, in the absence or presence of TGF-β. The top panels show hybridizations for cbfa1 or osteocalcin mRNA using RNA from LPCX control cells or cells stably expressing Smad3 or Smad3-3SA, while the lower panel shows the ethidium bromide-stained gel. (D) Alterations in Smad3 signaling affect alkaline phosphatase activity in caIB 2T3 cells and the extent of inhibition by TGF-β (5 ng/ml). Cells were incubated in differentiation medium for 6 days in the presence or absence of TGF-β (5 ng/ml). Cell lysates were then assayed for alkaline phosphatase activity. Values are expressed per cell. (E) Alterations in Smad3 signaling affect matrix mineralization in caIB 2T3 and MC3T3-E1 cells and the extent of inhibition by TGF-β (5 ng/ml) in caIB 2T3 cells. Confluent cells were incubated for 10 (caIB 2T3 cells) or 14 days (MC3T3-E1 cells) in differentiation medium, in the presence or absence of TGF-β, and then stained for mineralization (brown) using the von Kossa method. (F) Alterations in Smad3 signaling affect the transactivation of the p6OSE2-reporter by CBFA1. Stably infected LPCX control or stable caIB 2T3 cells expressing Smad3 or Smad3-3SA were transiently transfected with the p6OSE2-Luc reporter plasmid, with or without an expression plasmid for CBFA1 (pRK5-CBFA1). Luciferase expression in the absence or presence of TGF-β (1 ng/ml) was scored as in Figure 2.
Consistent with our results in Figure 1C using parental cells, control LPCX-infected caIB 2T3 cells expressed a low level of cbfa1 mRNA at day 0 (Figure 9B). The cbfa1 mRNA level was slightly lower in Smad3-expressing cells (0.75-fold of the cbfa1 mRNA levels in LPCX cells) and was elevated in cells expressing Smad3-3SA (2.75-fold of the cbfa1 mRNA levels in LPCX cells). This is consistent with both a role for autocrine TGF-β signaling in osteoblast differentiation and the inhibitory effect of Smad3 on CBFA1 function and cbfa1 expression. After 6 days in differentiation conditions, all three stable cell populations expressed cbfa1 and osteocalcin mRNA. Treatment of the LPCX cells with TGF-β during that time inhibited cbfa1 and osteocalcin mRNA expression (Figure 9C). Increased Smad3 expression enhanced the inhibitory effect of TGF-β on the expression of these two genes, while dominant-negative interference with Smad3 signaling strongly attenuated the TGF-β response (Figure 9C). Alkaline phosphatase, another marker of the osteoblast phenotype, was similarly affected. Increased Smad3 expression decreased alkaline phosphatase activ ity in the absence of added TGF-β and enhanced its downregulation by TGF-β, while Smad3-3SA expression decreased the TGF-β response (Figure 9D). Alkaline phosphatase activity was repressed by TGF-β 20-fold in LPCX control cells, 51-fold in Smad3-overexpressing cells, but only 3-fold in Smad3-3SA-expressing cells. We also assessed the effect of Smad3 and Smad3-3SA on matrix mineralization, as visualized by von Kossa staining. While little effect on calcium deposition was detected in the absence of TGF-β, the inhibitory effect of TGF-β on matrix mineralization was accentuated by increased Smad3 expression and attenuated by Smad3-3SA expression (Figure 9E). We also used another cell system to analyze the effect of increased expression of Smad3 or the dominant-negative Smad3-3SA mutant on matrix mineralization (Figure 9A and E). In MC-3T3 cells, Smad3 overexpression reduced the level of mineralization in the absence of exogenous TGF-β, but Smad3-3SA expression exerted only a slight increase in matrix mineralization (Figure 9E).
Finally, we assessed the transcriptional activity of p6OSE2-Luc in control infected, Smad3-overexpressing or Smad3-3SA-expressing caIB 2T3 cells, in the absence or presence of CBFA1. Consistent with the data in ROS 17/2.8 and 10T1/2 cells (Figure 5), Smad3 overexpression decreased the transcriptional activity from this promoter (Figure 9F). This decrease in CBFA1 function was enhanced further by TGF-β treatment. In addition, Smad3-3SA expression enhanced the CBFA1 function in the absence of TGF-β, consistent with autocrine TGF-β signaling, while inhibiting the repressive function of TGF-β treatment. Taken together, these data illustrate the central role of Smad3 in osteoblast differentiation and in TGF-β-mediated inhibition of osteoblast differentiation.
Discussion
The characterization of mesenchymal differentiation into osteoblasts has identified several marker genes that are sequentially expressed during osteoblastogenesis and are involved in osteoblast function (Karsenty, 1999). BMPs have a striking ability to induce osteoblast differentiation in cell culture, while TGF-β is a potent inhibitor of this differentiation program (Centrella et al., 1994). TGF-β also influences multiple aspects of bone function in vivo (Bonewald and Dallas, 1994; Centrella et al., 1994; Erlebacher et al., 1998; Alliston and Derynck, 2000). While the ability of TGF-β to inhibit the expression of osteoblast genes has been well documented, the mechanism of this inhibition is as yet poorly characterized. Although BMP and activin receptors and BMP-responsive Smads 1 and 5 have been implicated as promoters of osteoblast differentiation (Fujii et al., 1999), it is not known how TGF-β inhibits osteoblast differentiation. In addition, despite the high levels of TGF-β in bone matrix, the role of autocrine TGF-β signaling in osteoblast differentiation remains unclear. In this report, we addressed the mechanism of TGF-β-mediated inhibition of osteoblast differentiation. Several important conclusions emerge from our results. We found that CBFA1, a transcription factor that is essential for osteoblast differentiation and expression of osteoblast marker genes (Karsenty, 1999), is a central target of inhibition by TGF-β. TGF-β represses the transcriptional activity of CBFA1 and this repression is mediated through regulated interaction of Smad3 with CBFA1. Because CBFA1 activates transcription from its own promoter, this mechanism also results in decreased cbfa1 expression. The ability of Smad3 to repress the transcriptional activity of CBFA1 stands in marked contrast to the well described transcriptional coactivator functions of Smads. Finally, we provide evidence that TGF-β signaling through Smad3 plays an autocrine role in osteoblast differentiation.
As a member of the runt family of transcription factors, CBFA1 has a runt DNA-binding domain and several domains that positively or negatively regulate transactivation (Thirunavukkarasu et al., 1998). During osteoblast differentiation, mesenchymal cells activate cbfa1 expression, while ectopic CBFA1 expression in mesenchymal cells induces osteoblast differentiation (Ducy et al., 1997). Targeted inactivation of the cbfa1 gene in mice results in the absence of osteoblast differentiation and bone formation (Komori et al., 1997; Otto et al., 1997). These findings illustrate the critical role of CBFA1 function in osteoblast differentiation and activation of osteoblast gene expression. CBFA1 has been shown to bind to the osteocalcin promoter at an OSE2-binding sequence and to activate osteocalcin expression (Ducy et al., 1997). Predicted CBFA1-binding sites are also present in the promoters of several other osteoblast differentiation genes, such as alkaline phosphatase (Harada et al., 1999), bone sialoprotein, α1 and α2(I) collagen (Kern et al., 2001), and osteoprotegerin ligand (Gao et al., 1998; Thirunavukkarasu et al., 2000). Accordingly, CBFA1 has been shown to activate transcription of most of these genes. Furthermore, cbfa1 expression is activated by CBFA1 itself, resulting in an autoregulatory feedback loop to induce and maintain cbfa1 expression (Ducy et al., 1999).
Based on these findings, TGF-β-mediated repression of CBFA1 function, as demonstrated by our results, has considerable consequences for osteoblast differentiation. Thus, TGF-β will inhibit not only the transcriptional activation of osteoblast differentiation genes, which are target genes of CBFA1, but also cbfa1 transcription, thus decreasing the CBFA1 mRNA and protein levels. Thus, we have shown that TGF-β decreases the expression of cbfa1 mRNA in differentiating primary calvarial osteoblasts and in several osteoblastic cell lines. Furthermore, our results show that TGF-β represses transcription from the cbfa1 promoter in a CBFA1-dependent way. These findings stand in contrast to those in C2C12 myoblasts, in which TGF-β can induce the expression of cbfa1 mRNA (M.H.Lee et al., 1999; K.S.Lee et al., 2000). However, TGF-β-induced expression of cbfa1 mRNA in myoblasts does not correlate with increased osteoblast differentiation, whereas the reduction of cbfa1 expression by TGF-β in osteoblasts is consistent with the well established inhibition of osteoblast differentiation and gene expression by TGF-β (Centrella et al., 1994; this study). In addition to cbfa1 and osteocalcin, several CBFA1-regulated genes are also inhibited by TGF-β, including alkaline phosphatase, osteopontin and osteoprotegerin ligand (Centrella et al., 1994; Takai et al., 1998), raising the possibility that this downregulation also results from TGF-β-mediated repression of CBFA1 function. This mechanism may explain the similarities in bone phenotype between transgenic mice overexpressing activated TGF-β2 (Erlebacher and Derynck, 1996) and transgenic mice with partial loss of CBFA1 expression or function (Komori et al., 1997; Otto et al., 1997), both of which exhibit characteristics of human cleidocranial dysplasia. Furthermore, because osteoprotegerin ligand regulates osteoclast determination, TGF-β inhibition of CBFA1 function may affect genes that determine osteoclast function as well as osteoblast function.
The ability of TGF-β to activate or repress expression of defined genes has been well documented, but the mechanism of transcriptional repression in response to TGF-β has not been characterized. Smads 2 and 3 were identified as effectors of transcriptional activation in response to TGF-β. So far, all gene expression functions attributed to Smads result from their ability to cooperate with other transcription factors to activate transcription (Zhang and Derynck, 1999; Massagué and Wotton, 2000; Miyazono, 2000; Wrana, 2000). Several proteins, such as Evi-1 (Kurokawa et al., 1998), TGIF (Wotton et al., 1999) and Ski (Akiyoshi et al., 1999; Luo et al., 1999; Sun et al., 1999), have been shown to interact with Smad3 to decrease or inhibit transcriptional activation by Smad3 or TGF-β. However, none of these corepressors, nor any other mechanism implicated in downregulation of TGF-β activated transcription (Dennler et al., 2000; Kim et al., 2000; Ghosh et al., 2001), has been shown to confer TGF-β- or Smad-mediated repression of gene expression. Our results now show that Smad3, but not Smad2, is able to act as a transcriptional repressor of CBFA1 activity at the OSE2-binding site.
How Smad3 represses the transcriptional function of CBFA1 is as yet unclear, although we have made several observations. Smad3 repressed the transactivation of the cbfa1 and osteocalcin promoters and the tandemly repeated 6(OSE2) promoter in mesenchymal 10T1/2 cells and ROS 17/2.8 cells, yet enhanced the activity of CBFA1 at the 6(OSE2) promoter in epithelial HepG2 cells. Consistent with these results, Smad3 repressed the transcriptional activity of the CBFA1-related AML1 and 2 at the 6(OSE2) promoter in 10T1/2 cells and enhanced their activity in HepG2 cells. These results suggest that the activator versus repressor function of Smad3 results from cell lineage-dependent differences. These differences between mesenchymal and epithelial cells could result from the presence or absence of defined proteins, which interact with the Smad3–CBFA1 transcription complex and determine whether a Smad functions as a transcriptional activator or repressor. Transcriptional repression frequently results from the recruitment of a corepressor or a deacetylase. Runt family transcription factors can interact with members of the Groucho family of corepressors (Aronson et al., 1997), and Groucho has been shown to interact with a histone deacetylase, Rpd3, to repress transcription (Chen et al., 1999). Accordingly, CBFA1 interacts with TLE2, a mammalian homolog of Drosophila Groucho, to repress CBFA1 function (Thirunavukkarasu et al., 1998; Javed et al., 2000), suggesting that a protein related to Groucho may play a role in Smad3-mediated repression of CBFA1 in mesenchymal cells. While a Smad3-interacting corepressor such as Ski could, in principle, allow for Smad-mediated repression, coexpression of Ski decreased the ability of Smad3 to repress CBFA1 function and did not repress transcription further (data not shown). This is consistent with the ability of Ski to reverse the activator function of Smad3 in other contexts (Akiyoshi et al., 1999; Luo et al., 1999; Sun et al., 1999), but argues against its role in Smad-mediated repression in our systems.
Besides the cell type, the effect of Smad3 on CBFA1 or AML1/2 activity is also dependent on the promoter context. In contrast to the OSE2-containing promoters, Smad3 cooperates with AML1, AML2 and AML3/CBFA1 to activate transcription from the mouse germline Ig Cα promoter (Hanai et al., 1999; Lee et al., 2000; E.Pardali et al., 2000; Zhang and Derynck, 2000; Zhang et al., 2000). This activation was apparent in mesenchymal 10T1/2 cells and epithelial HepG2 cells and thus did not depend on the cell type. The dependence of the transcriptional response on the promoter sequence could, in the case of the Ig Cα promoter, be due to the presence of a CRE site, which can bind CREB, or a Smad-binding sequence adjacent to the AML-binding site (Hanai et al., 1999; E.Pardali et al., 2000; Zhang and Derynck, 2000).
We also evaluated the interactions of Smad3 and CBFA1 with each other and with the promoter DNA. In the case of transcriptional activation, Smads interact physically with transcription factors in a TGF-β-dependent manner, and, accordingly, Smads have been shown to interact with AML family members (Hanai et al., 1999; E.Pardali et al., 2000). Mutation analysis suggests that physical interaction may be required for functional cooperativity at the Ig Cα promoter (Zhang et al., 2000). As in the case of transcriptional activation, we found that TGF-β induces Smad3 interaction with CBFA1 in 10T1/2 and ROS 17/2.8 cells, i.e. the cells in which we observed Smad3-mediated repression. The TGF-β-induced association of Smad3 with CBFA1 raised the possibility that this complex formation might decrease or prevent CBFA1 binding to its OSE2-binding sequence. Indeed, other stimuli have been shown to affect the DNA binding of CBFA1 (Xiao et al., 1997, 1998; Zhang et al., 1997) and it has been proposed that off-DNA interactions of c-Jun with Smad3 can decrease transcriptional activation by TGF-β and Smad3 (Verrecchia et al., 2000). In contrast, the formation of the CBFA1–Smad3 complex did not prevent CBFA1 binding to the OSE2 sequence. Instead, CBFA1 binding to the OSE2 sequence was consistently increased in response to TGF-β, possibly a consequence of a conformational change or stabilization of CBFA1 resulting from the Smad3 interaction. Similarly, a TGF-β/Smad-mediated increase in Sp1- or c-Jun-binding efficiency to their respective DNA-binding sequences has been observed in vitro (Feng et al., 2000; K.Pardali et al., 2000; J.Qing and R.Derynck, unpublished observations). We therefore conclude that TGF-β/Smad-mediated repression of transcription by CBFA1 does not result from decreased CBFA1 binding to the OSE2 sequence.
Smad3 interaction with the OSE2 promoter was dependent on CBFA1 binding to DNA, and presumably resulted from the physical association of Smad3 with CBFA1. Consistent with this result, mutations in Smad3 that abolish DNA binding did not prevent TGF-β/Smad3-mediated repression. This may be in contrast to transcriptional activation by TGF-β and Smad3, which appears to require the DNA-binding ability of Smad3 (Dennler et al., 1998; Qing et al., 2000), although this remains to be further evaluated for the cooperation of Smad3 with AML transcription factors. While DNA binding of Smad3 was not essential for repression, DNA binding of CBFA1 to the OSE2 sequence was absolutely required for TGF-β/Smad3-mediated repression, as assessed using mutations in the OSE2 sequence and a point-mutated CBFA1. Further studies will be needed to define how Smad3 mediates transcriptional repression.
Our findings also suggest that autocrine TGF-β signaling regulates osteoblast differentiation in culture. Osteoblasts express TGF-β receptors and Smad2 and Smad3, which mediate the TGF-β responses. Alterations in the levels of Smad3 signaling by overexpressing wild-type Smad3 or a dominant-negative Smad3 mutant resulted in changes in the osteoblast differentiation program. In these cells, the changes in basal cbfa1 expression, CBFA1 function at the OSE2 promoter and matrix calcification are consistent with an autocrine inhibitory effect of endogenous TGF-β signaling on osteoblast differentiation. In addition, changes in cbfa1 and osteocalcin expression, alkaline phosphatase activity and matrix calcification revealed that increased Smad3 levels conferred increased sensitivity to exogenous TGF-β, while decreased signaling impaired the differentiation responses to exogenous TGF-β. Conceptually similar conclusions have been reached in the case of mesenchymal differentiation into adipocytes (Choy et al., 2000). In this differentiation program, decreased levels of TGF-β receptor or Smad3 signaling enhance adipocyte differentiation, while increased Smad3 signaling inhibits adipogenic differentiation (Choy et al., 2000). Thus, autocrine TGF-β signaling regulates both types of mesenchymal differentiation in culture, and in both cases exerts a negative role. Additional studies will be required to dissect the roles of the TGF-β signaling pathway components in osteoblast differentiation and to evaluate the autocrine role of TGF-β signaling in mesenchymal differentiation in vivo.
Materials and methods
Expression plasmids
The mammalian expression plasmids encoding TβRI-T202D, a partially activated type I TGF-β receptor (Feng and Derynck, 1996; Weis-Garcia and Massagué, 1996), N-terminally Flag-tagged Smad2 or Smad3 (Zhang et al., 1998), hemagglutinin (HA)-tagged Smad4 (Feng et al., 1998), Flag-tagged Smad3ΔC (Zhang et al., 1996), Smad3-3SA (Feng et al., 1998), Smad3R74D and Smad3ΔLG (Qing et al., 2000) have been described previously. The expression plasmid for hCBFA1 S191N, a mutant human CBFA1 that lacks DNA-binding ability, has also been described (Lee et al., 1997). Retroviral LPCX vectors encoding Smad3 or Smad3-3SA were described by Choy et al. (2000). Bacterial expression plasmids for GST-fused Smads or Smad fragments have also been described (Zhang et al., 1996, 1998). Plasmid 6Myc-PEPB2αA, an expression plasmid for Myc-tagged CBFA1 (Hanai et al., 1999), was provided by Y.Ito (University of Kyoto, Japan) and K.Miyazono (Cancer Institute, Tokyo, Japan). pCMV5-AML1b, an expression vector for AML1 (Meyers et al., 1995), was obtained from S.Hiebert (Vanderbilt University Medical School, Nashville, TN), whereas pRK5-HA-AML2, which expresses AML2, was described previously (Zhang and Derynck, 2000). Finally, using pCMV-OSF2 as template (Ducy et al., 1997), we generated pRK5-CBFA1 using a PCR-based approach to express C-terminally Flag-tagged CBFA1 from the CMV promoter in pRK5 (Graycar et al., 1989). Details of plasmid construction will be provided upon request.
Reporter plasmids
The pCbfa1-Luc plasmid contains a 135 bp fragment of the mouse cbfa1 promoter from –89 to +46, driving the expression of luciferase (Ducy et al., 1999). The OSE2 sites within this cbfa1 promoter segment are mutated in the pCbfa1m-Luc reporter as previously described (Ducy et al., 1999). The –1.3 OC-Luc plasmid expresses luciferase under the control of the proximal 1.3 kbp mouse osteocalcin gene 2 promoter segment (Ducy and Karsenty, 1995). –147 WT-OC-Luc and –147 Mut-OC-Luc direct luciferase expression under the control of the –147 to +13 osteocalcin promoter segment, but in the latter case, the OSE2 site has been inactivated by mutation (Ducy and Karsenty, 1995). The p6OSE2-Luc and p6OSE2m-Luc plasmids contain six copies of the wild-type or mutant OSE2 sequence of the osteocalcin promoter, respectively, followed by a minimal promoter, which directs the expression of luciferase (Ducy and Karsenty, 1995). pCα179-Luc directs luciferase expression from the –179 to +46 segment of the mouse germline Ig Cα promoter (Lin and Stavnezer, 1992) and was kindly provided by J.Stavnezer (University of Massachusetts Medical School, Worcester, MA). pRK5-βgal expresses β-galactosidase under the control of a human CMV promoter (Feng et al., 1995).
Cell culture
Primary osteoblasts were isolated from calvaria of 2-day-old mice as described previously (Ducy and Karsenty, 1995). Two days after isolation, cells were passaged for differentiation in α-MEM, 10% fetal bovine serum (FBS), 1× penicillin/streptomycin (P/S), 100 µg/ml ascorbic acid, 5 mM β-glycerophosphate in the presence or absence of 5 ng/ml TGF-β.
Several cell lines were used in our experiments, all of which were maintained at 37°C in the presence of 5% CO2. ROS 17/2.8 (rat osteosarcoma) cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM)/F12 medium with 10% FBS, 1× P/S. C3H-10T1/2 cells (mouse embryonic fibroblasts) were maintained in subconfluent conditions in basal MEM, 10% heat-inactivated FBS, 1× P/S. COS-1 cells, Phoenix E cells (obtained from G.Nolan, Stanford University, CA) and HepG2 cells were grown in DMEM, 10% FBS, 1× P/S. caIB 2T3 cells (obtained from S.E.Harris, University of Texas, San Antonio, TX) and subclone 4 of MC3T3-E1 cells (obtained from R.Franceschi, University of Michigan, Ann Arbor, MI) were grown in α-MEM, 10% FBS, 1× P/S. caIB 2T3 cells were derived from transgenic mice expressing SV40 large T antigen under the control of the osteocalcin promoter (Ghosh-Choudhury et al., 1996) and were engineered further to express a constitutively active BMP-RIB receptor. caIB 2T3 cells can undergo osteoblast differentiation in culture without the need for exogenous BMP addition (Chen et al., 1998). MC3T3-E1 cells (subclone 4) have been shown to express high levels of osteoblast markers under differentiation conditions (Xiao et al., 1997). Smad2–/– fibroblasts (obtained from E.Böttinger, Albert Einstein College of Medicine, NY) and Smad3–/– fibroblasts (Datto et al., 1999; obtained from X.-F.Wang, Duke University Medical School, NC), were cultured in DMEM, 20% FBS, 1× P/S.
Stably infected caIB 2T3 cells and MC3T3 cells were maintained in α-MEM, 10% FBS, 1× P/S, supplemented with 2 µg/ml puromycin (Calbiochem). For differentiation studies, these cells were switched to differentiation medium upon reaching confluence. This medium consisted of α-MEM, 2% FBS, 100 µg/ml ascorbic acid, 5 mM β-glycerophosphate, 1× P/S, 2 µg/ml puromycin, with or without 5 ng/ml TGF-β.
Transfections and reporter assays
Cells were transfected using commercial reagents according to the manufacturers’ recommendations. COS-1 cells and 10T1/2 cells used to generate whole-cell lysates for co-immunoprecipitation or DNA binding analyses were transfected using Lipofectamine (Gibco-BRL). 10T1/2 cells used in transcriptional activity assays and all other cell lines were transfected using Effectene (Qiagen). For each transfection with subsequent reporter assay, the total quantity of transfected plasmid DNA was kept constant by the addition, when required, of pRK5 plasmid DNA. Forty-eight hours after transfection, cells were harvested and lysed in Reporter Lysis Buffer (Promega) prior to analysis of reporter activity using a luciferase assay system (Pharmingen) or β-galactosidase detection kit (Tropix). All luciferase values, measured using a luminometer, were normalized for transfection efficiency against the β-galactosidase activities from the cotransfected pRK5-βgal plasmid. Each assay was performed in duplicate or triplicate and is representative of at least three experiments. All values, means and standard deviations were expressed relative to the basal promoter activity as fold induction.
Generation of stably infected cell lines
Cells stably infected with an LPCX-based retroviral vector were generated as described (Choy et al., 2000). Specifically, caIB 2T3 cells and MC3T3-E1 cells were plated at a density of 50 000 or 42 500 cells, respectively, per 35 mm diameter dish 1 day prior to infection. Cells were then overlaid with undiluted viral supernatant and infected using a spinfection procedure. Selection with puromycin (2 µg/ml), starting 48 h after infection with LPCX, LPCX-Smad3 or LPCX-Smad3-3SA, yielded stable caIB 2T3 or MC3T3-E1 cell populations expressing N-terminally Flag-tagged Smad3 or Smad3-3SA or control-infected cells. The expression levels of Smad3 and Smad3-3SA were verified using Flag antibody western analyses. All differentiation experiments utilized cells within four passages of the generation of stable cell lines.
Northern blot analyses
Total RNA was isolated from cell cultures at the indicated times by scraping cells in RNA extraction buffer (140 mM NaCl, 5 mM KCl, 3 mM MgCl2, 25 mM Tris–HCl pH 7.5, 1% NP-40) at 4°C. Nuclei were removed by centrifugation and RNA was prepared from the resulting supernatant in the presence of 1% SDS. Following two extractions with phenol/chloroform (1:1) and chloroform, the RNA was ethanol precipitated and redissolved. RNA (10–20 µg) was denatured and separated on a 1% formaldehyde–agarose gel, and then transferred to a 0.2 µm nylon membrane (Biotrans). Northern analyses using cbfa1 or osteocalcin cDNA as hybridization probe were performed as previously described (Ausubel et al., 1994). Probes were labeled using [α-32P]dCTP (6000 C/mmol; NEN) using the random primer method. Results were visualized by autoradiography and quantified using a PhosphorImager (Molecular Dynamics).
GST interaction assays
GST fusion proteins, in which full-size Smads or defined segments of Smads were fused to GST, were expressed in Escherichia coli as described (Ausubel et al., 1994). These fusion proteins were then purified by adsorption to glutathione–Sepharose (Pharmacia). 35S-labeled CBFA1 was obtained by in vitro translation in the presence of [35S]methionine using the TNT kit (Promega). The translation mixture, containing in vitro translated [35S]CBFA1, was precleared by incubation with glutathione–Sepharose prior to incubation with glutathione–Sepharose, to which the GST–Smad protein of interest was adsorbed. After a 1 h incubation at 4°C in 50 mM Tris–HCl pH 7.5, 120 mM NaCl, 2 mM EDTA, 0.1% NP-40, the beads were extensively and repeatedly washed in the same solution. The adsorbed protein complexes were then analyzed by SDS–PAGE. Staining of the gels with Coomassie Blue allowed verification of the amounts and integrity of the fusion proteins, whereas subsequent incubation with Amplify (Amersham) and autoradiography of the dried gel revealed the adsorption of 35S-labeled CBFA1.
Immunoprecipitations
Transfected COS-1 or 10T1/2 cells were harvested 48 h after transfection and lysed in 20 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.5% Triton X-100, containing a cocktail of protease inhibitors (Minicomplete, BMB). ROS 17/2.8 cells were harvested in the same manner 48 h after treatment with or without TGF-β (5 ng/ml). After clarification by centrifugation, lysates were precleared for 1 h at 4°C with protein A–Sepharose. Anti-Flag immunoprecipitations were performed using anti-Flag antibody coupled to Sepharose (Sigma). For anti-myc and anti-HA immunoprecipitations, lysates were incubated with 2 µg/ml 9E10 antibody (Covance) or 12CA5 antibody (Sigma), respectively. For Smad3 or IgG immunoprecipitations, ROS 17/2.8 lysates were incubated with 1 µg/ml rabbit anti-Smad3 antibody (Zymed) or rabbit anti-mouse IgG antibody (Pierce). In each case, lysates were incubated for 1 h at 4°C prior to incubation with protein A–Sepharose. After extensive washing of the Sepharose-adsorbed protein complexes in the lysis buffer mentioned above, immunoprecipitates were analyzed by SDS–PAGE and western blot analyses. The expression levels of the cotransfected proteins of interest were verified by western analyses of the cell lysates using epitope tag-specific antibodies. Endogenous Smad3 and CBFA1 were detected using rabbit anti- Smad3 (Zymed) or rabbit anti-CBFA1 (Thirunavukkarasu et al., 1998) antibodies.
Electrophoretic mobility shift assays and biotinylated oligonucleotide interactions
Previously characterized wild-type and mutant OSE2 oligonucleotides corresponding to the –156/–112 segment of the osteocalcin promoter (Ducy and Karsenty, 1995) were obtained from Genosys. These oligonucleotides had the following sequences: WT OSE2, 5′-CCCA GGCAGCTGCAATCACCAACCACAGCATCCTTTGGGTTTGAC-3′; Mut OSE2, 5′-CCCAGGCAGCTGCAATCACCAAGAACAGCATCC TTTGGGTTTGAC-3′. For biotinylated oligonucleotide interaction experiments, the oligonucleotides were modified by the addition of biotin to the 5′ end. All oligonucleotides were annealed with non-biotinylated complementary oligonucleotides.
For electrophoretic mobility shift assays, whole-cell extracts were generated as described previously from ROS 17/2.8 cells, 10T1/2 cells or transfected COS-1 cells (Alliston et al., 1997). Probes were end-labeled with [α-32P]dCTP (6000 Ci/mmol; NEN). Approximately 1 ng of labeled oligonucleotide probe was incubated with 5 µg of whole-cell extract for 30 min at room temperature (RT) in the presence of 1× gel shift binding buffer containing 15 mM Tris pH 7.5, 1 mM MgCl2, 0.5 mM EDTA, 0.5 mM dithiothreitol (DTT), 50 mM NaCl, 10% glycerol and 1 µg of poly(dI–dC)⋅poly(dI–dC). Competition was performed with either a 10 or 100× molar excess of unlabeled OSE2 or unrelated SBE oligonucleotides (Qing et al., 2000). Antibodies against CBFA1 (1 µl) or Flag (1 µg) were pre-incubated with whole-cell extracts and binding buffer for 30 min at 4°C prior to the addition of probe. Reactions were separated on 5% polyacrylamide, 2.5% glycerol, 0.5× TBE gels.
For each biotinylated oligonucleotide binding reaction, 200 ng of annealed oligonucleotide were coupled to 50 µl of streptavidin magnetic beads (Promega), washed in 10 mM Tris pH 7.5, 1 mM EDTA, 2 M NaCl. After coupling the oligonucleotides for 10 min at RT, the oligonucleotide-coupled beads were washed repeatedly in the same solution using a magnetic stand to precipitate the beads between each wash. Washed beads were added to 200 µg of cell lysates (see Immunoprecipitations) and 5 µg of poly(dI–dC)⋅poly(dI–dC) with a final concentration of 4 mM Tris pH 7.5, 20 mM HEPES pH 7.5, 5% glycerol, 170 mM NaCl, 0.5 mM EDTA, 0.5 mM DTT, 1 mM MgCl2 and 0.1% Triton X-100. After incubating for 1 h at 4°C, beads were washed repeatedly in 16 mM HEPES pH 7.6, 100 mM NaCl, 0.4 mM EDTA, 1 mM MgCl2 and 1% glycerol. Precipitated proteins were analyzed by SDS–PAGE and western blotting.
Alkaline phosphatase assays
To assay alkaline phosphatase activity, cells were lysed in phosphate-buffered saline (PBS), 0.01% SDS. Following clarification by centrifugation, equal volumes of cell lysate were assayed for alkaline phosphatase activity using an Alkaline Phosphatase Assay Kit (Sigma Diagnostics). The colorimetric reactions, executed in the linear range, were stopped by addition of 0.05 M NaOH, and the absorbance was measured at 420 nm. Background absorbance (assessed spectrophotometrically after addition of HCl) was subtracted from the total value. Phosphatase activities were expressed per cell, after counting the number of cells trypsinized on a parallel plate.
von Kossa staining
Cells cultured for 10 (caIB 2T3 cells) or 14 days (MC3T3-E1 cells) in differentiation conditions were assayed for mineralization using the histochemical von Kossa staining method (Bhargava et al., 1988). Briefly, cells were washed in Ca/Mg-free PBS and fixed for 30 min in 4% formalin/PBS. Fixed cells were incubated with 5% silver nitrate solution for 30 min in the dark, followed by exposure to UV light. Unincorporated silver nitrate was removed using 5% sodium thiosulfate followed by several washes with water.
Acknowledgments
Acknowledgements
We thank S.E.Harris (University of Texas, San Antonio, TX) for the caIB 2T3 cells, E.Böttinger (Albert Einstein College of Medicine, NY) for Smad2–/– fibroblasts, X.-F.Wang (Duke University Medical School, NC) for Smad3–/– fibroblasts, R.Franceschi (University of Michigan, Ann Arbor, MI) for subclone 4 MC3T3-E1 cells, and Y.Ito (University of Kyoto, Japan) and K.Miyazono (Cancer Institute, Tokyo, Japan) for the 6Myc PEPB2αA expression plasmid.This research was supported by a grant from the Arthritis Foundation to R.D., NIH grants RO1-CA63101 and P60-DE13058 (Project III) to R.D., and AR-45548 to G.K. T.A. and L.C. were supported by postdoctoral fellowships from the Arthritis Foundation and American Heart Association, respectively.
References
- Akiyoshi S., Inoue,H., Hanai,J., Kusanagi,K., Nemoto,N., Miyazono,K. and Kawabata,M. (1999) c-Ski acts as a transcriptional co-repressor in transforming growth factor-β signaling through interaction with Smads. J. Biol. Chem., 274, 35269–35277. [DOI] [PubMed] [Google Scholar]
- Alliston T.N. and Derynck,R. (2000) Transforming growth factor-β in skeletal development and maintenance. In Canalis,E. (ed.), Skeletal Growth Factors. Lippincott Williams & Wilkins, Philadelphia, PA, pp. 233–249.
- Alliston T.N., Maiyar,A.C., Buse,P., Firestone,G.L. and Richards,J.S. (1997) Follicle stimulating hormone-regulated expression of serum/glucocorticoid-inducible kinase in rat ovarian granulosa cells: a functional role for the Sp1 family in promoter activity. Mol. Endocrinol., 11, 1934–1949. [DOI] [PubMed] [Google Scholar]
- Aronson B.D., Fisher,A.L., Blechman,K., Caudy,M. and Gergen,J.P. (1997) Groucho-dependent and -independent repression activities of Runt domain proteins. Mol. Cell. Biol., 17, 5581–5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (1994) Current Protocols in Molecular Biology. John Wiley & Sons, New York, NY.
- Banerjee C., Hiebert,S.W., Stein,J.L., Lian,J.B. and Stein,G.S. (1996) An AML-1 consensus sequence binds an osteoblast-specific complex and transcriptionally activates the osteocalcin gene. Proc. Natl Acad. Sci. USA, 93, 4968–4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beelman C.A. and Parker,R. (1994) Differential effects of translational inhibition in cis and in trans on the decay of the unstable yeast MFA2 mRNA. J. Biol. Chem., 269, 9687–9692. [PubMed] [Google Scholar]
- Bhargava U., Bar-Lev,M., Bellows,C.G. and Aubin,J.E. (1988) Ultrastructural analysis of bone nodules formed in vitro by isolated fetal rat calvaria cells. Bone, 9, 155–163. [DOI] [PubMed] [Google Scholar]
- Bonewald L.F. and Dallas,S.L. (1994) Role of active and latent transforming growth factor-β in bone formation. J. Cell. Biochem., 55, 350–357. [DOI] [PubMed] [Google Scholar]
- Centrella M., Horowitz,M.C., Wozney,J.M. and McCarthy,T.L. (1994) Transforming growth factor-β gene family members and bone. Endocr. Rev., 15, 27–39. [DOI] [PubMed] [Google Scholar]
- Chang D.J., Ji,C., Kim,K.K., Casinghino,S., McCarthy,T.L. and Centrella,M. (1998) Reduction in transforming growth factor β receptor I expression and transcription factor CBFA1 on bone cells by glucocorticoid. J. Biol. Chem., 273, 4892–4896. [DOI] [PubMed] [Google Scholar]
- Chen D., Ji,X., Harris,M.A., Feng,J.Q., Karsenty,G., Celeste,A.J., Rosen,V., Mundy,G.R. and Harris,S.E. (1998) Differential roles for bone morphogenetic protein (BMP) receptor type IB and IA in differentiation and specification of mesenchymal precursor cells to osteoblast and adipocyte lineages. J. Cell Biol., 142, 295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Fernandez,J., Mische,S. and Courey,A.J. (1999) A functional interaction between the histone deacetylase Rpd3 and the corepressor groucho in Drosophila development. Genes Dev., 13, 2218–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy L., Skillington,J. and Derynck,R. (2000) Roles of autocrine TGF-β receptor and Smad signaling in adipocyte differentiation. J. Cell Biol., 149, 667–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datto M.B., Frederick,J.P., Pan,L., Borton,A.J., Zhuang,Y. and Wang,X.F. (1999) Targeted disruption of Smad3 reveals an essential role in transforming growth factor β-mediated signal transduction. Mol. Cell. Biol., 19, 2495–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennler S., Itoh,S., Vivien,D., ten Dijke,P., Huet,S. and Gauthier,J.M. (1998) Direct binding of Smad3 and Smad4 to critical TGF β-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J., 17, 3091–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennler S., Prunier,C., Ferrand,N., Gauthier,J.M. and Atfi,A. (2000) c-Jun inhibits transforming growth factor-β-mediated transcription by repressing Smad3 transcriptional activity. J. Biol. Chem., 275, 28858–28865. [DOI] [PubMed] [Google Scholar]
- Derynck R., Zhang,Y. and Feng,X.H. (1998) Smads: transcriptional activators of TGF-β responses. Cell, 95, 737–740. [DOI] [PubMed] [Google Scholar]
- Desbois C., Hogue,D.A. and Karsenty,G. (1994) The mouse osteocalcin gene cluster contains three genes with two separate spatial and temporal patterns of expression. J. Biol. Chem., 269, 1183–1190. [PubMed] [Google Scholar]
- Ducy P. and Karsenty,G. (1995) Two distinct osteoblast-specific cis-acting elements control expression of a mouse osteocalcin gene. Mol. Cell. Biol., 15, 1858–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducy P., Zhang,R., Geoffroy,V., Ridall,A.L. and Karsenty,G. (1997) Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell, 89, 747–754. [DOI] [PubMed] [Google Scholar]
- Ducy P., Starbuck,M., Priemel,M., Shen,J., Pinero,G., Geoffroy,V., Amling,M. and Karsenty,G. (1999) A Cbfa1-dependent genetic pathway controls bone formation beyond embryonic development. Genes Dev., 13, 1025–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlebacher A. and Derynck,R. (1996) Increased expression of TGF-β2 in osteoblasts results in an osteoporosis-like phenotype. J. Cell Biol., 132, 195–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlebacher A., Filvaroff,E.H., Ye,J.Q. and Derynck,R. (1998) Osteoblastic responses to TGF-β during bone remodeling. Mol. Biol. Cell, 9, 1903–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X.H. and Derynck,R. (1996) Ligand-independent activation of transforming growth factor (TGF) β signaling pathways by heteromeric cytoplasmic domains of TGF-β receptors. J. Biol. Chem., 271, 13123–13129. [DOI] [PubMed] [Google Scholar]
- Feng X.H., Filvaroff,E.H. and Derynck,R. (1995) Transforming growth factor-β (TGF-β)-induced down-regulation of cyclin A expression requires a functional TGF-β receptor complex. Characterization of chimeric and truncated type I and type II receptors. J. Biol. Chem., 270, 24237–24245. [DOI] [PubMed] [Google Scholar]
- Feng X.H., Zhang,Y., Wu,R.Y. and Derynck,R. (1998) The tumor suppressor Smad4/DPC4 and transcriptional adaptor CBP/p300 are coactivators for Smad3 in TGF-β-induced transcriptional activation. Genes Dev., 12, 2153–2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X.H., Lin,X. and Derynck,R. (2000) Smad2, Smad3 and Smad4 cooperate with Sp1 to induce p15Ink4B transcription in response to TGF-β. EMBO J., 19, 5178–5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frendo J.L., Xiao,G., Fuchs,S., Franceschi,R.T., Karsenty,G. and Ducy,P. (1998) Functional hierarchy between two OSE2 elements in the control of osteocalcin gene expression in vivo. J. Biol. Chem., 273, 30509–30516. [DOI] [PubMed] [Google Scholar]
- Fujii M. et al. (1999) Roles of bone morphogenetic protein type I receptors and Smad proteins in osteoblast and chondroblast differentiation. Mol. Biol. Cell, 10, 3801–3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y.H., Shinki,T., Yuasa,T., Kataoka-Enomoto,H., Komori,T., Suda,T. and Yamaguchi,A. (1998) Potential role of cbfa1, an essential transcriptional factor for osteoblast differentiation, in osteoclastogenesis: regulation of mRNA expression of osteoclast differentiation factor (ODF). Biochem. Biophys. Res. Commun., 252, 697–702. [DOI] [PubMed] [Google Scholar]
- Geoffroy V., Ducy,P. and Karsenty,G. (1995) A PEBP2α/AML-1-related factor increases osteocalcin promoter activity through its binding to an osteoblast-specific cis-acting element. J. Biol. Chem., 270, 30973–30979. [DOI] [PubMed] [Google Scholar]
- Ghosh A.K., Yuan,W., Mori,Y., Chen,S. and Varga,J. (2001) Antagonistic regulation of type I collagen gene expression by interferon-γ and transforming growth factor-β: integration at the level of p300/CBP transcriptional coactivators. J. Biol. Chem., 276, 11041–11048. [DOI] [PubMed] [Google Scholar]
- Ghosh-Choudhury N., Windle,J.J., Koop,B.A., Harris,M.A., Guerrero,D.L., Wozney,J.M., Mundy,G.R. and Harris,S.E. (1996) Immortalized murine osteoblasts derived from BMP 2-T-antigen expressing transgenic mice. Endocrinology, 137, 331–339. [DOI] [PubMed] [Google Scholar]
- Gori F., Thomas,T., Hicok,K.C., Spelsberg,T.C. and Riggs,B.L. (1999) Differentiation of human marrow stromal precursor cells: bone morphogenetic protein-2 increases OSF2/CBFA1, enhances osteoblast commitment and inhibits late adipocyte maturation. J. Bone Miner. Res., 14, 1522–1535. [DOI] [PubMed] [Google Scholar]
- Graycar J.L., Miller,D.A., Arrick,B.A., Lyons,R.M., Moses,H.L. and Derynck,R. (1989) Human transforming growth factor-β3: recombinant expression, purification and biological activities in comparison with transforming growth factors-β1 and -β2. Mol. Endocrinol., 3, 1977–1986. [DOI] [PubMed] [Google Scholar]
- Hanai J. et al. (1999) Interaction and functional cooperation of PEBP2/CBF with Smads. Synergistic induction of the immunoglobulin germline Cα promoter. J. Biol. Chem., 274, 31577–31582. [DOI] [PubMed] [Google Scholar]
- Harada H., Tagashira,S., Fujiwara,M., Ogawa,S., Katsumata,T., Yamaguchi,A., Komori,T. and Nakatsuka,M. (1999) Cbfa1 isoforms exert functional differences in osteoblast differentiation. J. Biol. Chem., 274, 6972–6978. [DOI] [PubMed] [Google Scholar]
- Heldin C.H., Miyazono,K. and ten Dijke,P. (1997) TGF-β signalling from cell membrane to nucleus through SMAD proteins. Nature, 390, 465–471. [DOI] [PubMed] [Google Scholar]
- Hua X., Liu,X., Ansari,D.O. and Lodish,H.F. (1998) Synergistic cooperation of TFE3 and smad proteins in TGF-β-induced transcription of the plasminogen activator inhibitor-1 gene. Genes Dev., 12, 3084–3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X., Miller,Z.A., Wu,G., Shi,Y. and Lodish,H.F. (1999) Specificity in transforming growth factor β-induced transcription of the plasminogen activator inhibitor-1 gene: interactions of promoter DNA, transcription factor muE3 and Smad proteins. Proc. Natl Acad. Sci. USA, 96, 13130–13135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh S., Itoh,F., Goumans,M.J. and ten Dijke,P. (2000) Signaling of transforming growth factor-β family members through Smad proteins. Eur. J. Biochem., 267, 6954–6967. [DOI] [PubMed] [Google Scholar]
- Janknecht R., Wells,N.J. and Hunter,T. (1998) TGF-β-stimulated cooperation of Smad proteins with the coactivators CBP/p300. Genes Dev., 12, 2114–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javed A. et al. (2000) Groucho/TLE/R-esp proteins associate with the nuclear matrix and repress RUNX (CBFα/AML/PEBP2α) dependent activation of tissue-specific gene transcription. J. Cell Sci., 113, 2221–2231. [DOI] [PubMed] [Google Scholar]
- Karsenty G. (1999) The genetic transformation of bone biology. Genes Dev., 13, 3037–3051. [DOI] [PubMed] [Google Scholar]
- Kern B., Shen,J., Starbuck,M. and Karsenty,G. (2001) Cbfa1 contributes to the osteoblast-specific expression of Type I collagen genes. J. Biol. Chem., 276, 7101–7107. [DOI] [PubMed] [Google Scholar]
- Kim R.H. et al. (2000) A novel smad nuclear interacting protein, SNIP1, suppresses p300-dependent TGF-β signal transduction. Genes Dev., 14, 1605–1616. [PMC free article] [PubMed] [Google Scholar]
- Komori T. et al. (1997) Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell, 89, 755–764. [DOI] [PubMed] [Google Scholar]
- Kurokawa M., Mitani,K., Irie,K., Matsuyama,T., Takahashi,T., Chiba,S., Yazaki,Y., Matsumoto,K. and Hirai,H. (1998) The oncoprotein Evi-1 represses TGF-β signalling by inhibiting Smad3. Nature, 394, 92–96. [DOI] [PubMed] [Google Scholar]
- Labbé E., Letamendia,A. and Attisano,L. (2000) Association of Smads with lymphoid enhancer binding factor 1/T cell-specific factor mediates cooperative signaling by the transforming growth factor-β and Wnt pathways. Proc. Natl Acad. Sci. USA, 97, 8358–8363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagna G., Hata,A., Hemmati-Brivanlou,A. and Massagué,J. (1996) Partnership between DPC4 and SMAD proteins in TGF-β signalling pathways. Nature, 383, 832–836. [DOI] [PubMed] [Google Scholar]
- Lee B., Thirunavukkarasu,K., Zhou,L., Pastore,L., Baldini,A., Hecht,J., Geoffroy,V., Ducy,P. and Karsenty,G. (1997) Missense mutations abolishing DNA binding of the osteoblast-specific transcription factor OSF2/CBFA1 in cleidocranial dysplasia. Nature Genet., 16, 307–310. [DOI] [PubMed] [Google Scholar]
- Lee K.S. et al. (2000) Runx2 is a common target of transforming growth factor β1 and bone morphogenetic protein 2 and cooperation between runx2 and smad5 induces osteoblast-specific gene expression in the pluripotent mesenchymal precursor cell line C2C12. Mol. Cell. Biol., 20, 8783–8792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.H. et al. (1999) Transient upregulation of CBFA1 in response to bone morphogenetic protein-2 and transforming growth factor β-1 in C2C12 myogenic cells coincides with suppression of the myogenic phenotype but is not sufficient for osteoblast differentiation. J. Cell. Biochem., 73, 114–125. [PubMed] [Google Scholar]
- Li J., Tsuji,K., Komori,T., Miyazono,K., Wrana,J.L., Ito,Y., Nifuji,A. and Noda,M. (1998) Smad2 overexpression enhances Smad4 gene expression and suppresses CBFA1 gene expression in osteoblastic osteosarcoma ROS17/2.8 cells and primary rat calvaria cells. J. Biol. Chem., 273, 31009–31015. [DOI] [PubMed] [Google Scholar]
- Liberati N.T., Datto,M.B., Frederick,J.P., Shen,X., Wong,C., Rougier-Chapman,E.M. and Wang,X.F. (1999) Smads bind directly to the Jun family of AP-1 transcription factors. Proc. Natl Acad. Sci. USA, 96, 4844–4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.C. and Stavnezer,J. (1992) Regulation of transcription of the germ-line Ig α constant region gene by an ATF element and by novel transforming growth factor-β1-responsive elements. J. Immunol., 149, 2914–2925. [PubMed] [Google Scholar]
- Liu X., Sun,Y., Constantinescu,S.N., Karam,E., Weinberg,R.A. and Lodish,H.F. (1997) Transforming growth factor β-induced phosphorylation of Smad3 is required for growth inhibition and transcriptional induction in epithelial cells. Proc. Natl Acad. Sci. USA, 94, 10669–10674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo K., Stroschein,S.L., Wang,W., Chen,D., Martens,E., Zhou,S. and Zhou,Q. (1999) The Ski oncoprotein interacts with the Smad proteins to repress TGF-β signaling. Genes Dev., 13, 2196–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J. (1998) TGF-β signal transduction. Annu. Rev. Biochem., 67, 753–791. [DOI] [PubMed] [Google Scholar]
- Massagué J. and Wotton,D. (2000) Transcriptional control by the TGF-β/Smad signaling system. EMBO J., 19, 1745–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merriman H.L., van Wijnen,A.J., Hiebert,S., Bidwell,J.P., Fey,E., Lian,J., Stein,J. and Stein,G.S. (1995) The tissue-specific nuclear matrix protein, NMP-2, is a member of the AML/CBF/PEBP2/runt domain transcription factor family: interactions with the osteocalcin gene promoter. Biochemistry, 34, 13125–13132. [DOI] [PubMed] [Google Scholar]
- Meyers S., Lenny,N. and Hiebert,S.W. (1995) The t(8;21) fusion protein interferes with AML-1B-dependent transcriptional activation. Mol. Cell. Biol., 15, 1974–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazono K. (2000) Positive and negative regulation of TGF-β signaling. J. Cell Sci., 113, 1101–1109. [DOI] [PubMed] [Google Scholar]
- Mundlos S. et al. (1997) Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell, 89, 773–779. [DOI] [PubMed] [Google Scholar]
- Nishihara A., Hanai,J., Imamura,T., Miyazono,K. and Kawabata,M. (1999) E1A inhibits transforming growth factor-β signaling through binding to Smad proteins. J. Biol. Chem., 274, 28716–28723. [DOI] [PubMed] [Google Scholar]
- Noda M. (1989) Transcriptional regulation of osteocalcin production by transforming growth factor-β in rat osteoblast-like cells. Endocrinology, 124, 612–617. [DOI] [PubMed] [Google Scholar]
- Otto F. et al. (1997) Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell, 89, 765–771. [DOI] [PubMed] [Google Scholar]
- Pardali E., Xie,X.Q., Tsapogas,P., Itoh,S., Arvanitidis,K., Heldin,C.H., ten Dijke,P., Grundstrom,T. and Sideras,P. (2000) Smad and AML proteins synergistically confer transforming growth factor β1 responsiveness to human germ-line IgA genes. J. Biol. Chem., 275, 3552–3560. [DOI] [PubMed] [Google Scholar]
- Pardali K., Kurisaki,A., Moren,A., ten Dijke,P., Kardassis,D. and Moustakas,A. (2000) Role of smad proteins and transcription factor Sp1 in p21Waf1/Cip1 regulation by transforming growth factor-β. J. Biol. Chem., 275, 29244–29256. [DOI] [PubMed] [Google Scholar]
- Qing J., Zhang,Y. and Derynck,R. (2000) Structural and functional characterization of the transforming growth factor-β-induced Smad3/c-Jun transcriptional cooperativity. J. Biol. Chem., 275, 38802–38812. [DOI] [PubMed] [Google Scholar]
- Robey P.G., Young,M.F., Flanders,K.C., Roche,N.S., Kondaiah,P., Reddi,A.H., Termine,J.D., Sporn,M.B. and Roberts,A.B. (1987) Osteoblasts synthesize and respond to transforming growth factor-type β (TGF-β) in vitro. J. Cell Biol., 105, 457–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakou T., Onishi,T., Yamamoto,T., Nagamine,T., Sampath,T. and ten Dijke,P. (1999) Localization of Smads, the TGF-β family intracellular signaling components during endochondral ossification. J. Bone Miner. Res., 14, 1145–1152. [DOI] [PubMed] [Google Scholar]
- Sano Y., Harada,J., Tashiro,S., Gotoh-Mandeville,R., Maekawa,T. and Ishii,S. (1999) ATF-2 is a common nuclear target of Smad and TAK1 pathways in transforming growth factor-β signaling. J. Biol. Chem., 274, 8949–8957. [DOI] [PubMed] [Google Scholar]
- Shi M.J. and Stavnezer,J. (1998) CBF α3 (AML2) is induced by TGF-β1 to bind and activate the mouse germline Igα promoter. J. Immunol., 161, 6751–6760. [PubMed] [Google Scholar]
- Sun Y., Liu,X., Eaton,E.N., Lane,W.S., Lodish,H.F. and Weinberg,R.A. (1999) Interaction of the Ski oncoprotein with Smad3 regulates TGF-β signaling. Mol. Cell, 4, 499–509. [DOI] [PubMed] [Google Scholar]
- Takai H., Kanematsu,M., Yano,K., Tsuda,E., Higashio,K., Ikeda,K., Watanabe,K. and Yamada,Y. (1998) Transforming growth factor-β stimulates the production of osteoprotegerin/osteoclastogenesis inhibitory factor by bone marrow stromal cells. J. Biol. Chem., 273, 27091–27096. [DOI] [PubMed] [Google Scholar]
- Thirunavukkarasu K., Mahajan,M., McLarren,K.W., Stifani,S. and Karsenty,G. (1998) Two domains unique to osteoblast-specific transcription factor Osf2/Cbfa1 contribute to its transactivation function and its inability to heterodimerize with Cbfβ. Mol. Cell. Biol., 18, 4197–4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirunavukkarasu K., Halladay,D.L., Miles,R.R., Yang,X., Galvin,R.J., Chandrasekhar,S., Martin,T.J. and Onyia,J.E. (2000) The osteoblast-specific transcription factor Cbfa1 contributes to the expression of osteoprotegerin, a potent inhibitor of osteoclast differentiation and function. J. Biol. Chem., 275, 25163–25172. [DOI] [PubMed] [Google Scholar]
- Topper J.N., DiChiara,M.R., Brown,J.D., Williams,A.J., Falb,D., Collins,T. and Gimbrone,M.A.,Jr (1998) CREB binding protein is a required coactivator for Smad-dependent, transforming growth factor β transcriptional responses in endothelial cells. Proc. Natl Acad. Sci. USA, 95, 9506–9511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrecchia F., Pessah,M., Atfi,A. and Mauviel,A. (2000) Tumor necrosis factor-α inhibits transforming growth factor-β/Smad signaling in human dermal fibroblasts via AP-1 activation. J. Biol. Chem., 275, 30226–30231. [DOI] [PubMed] [Google Scholar]
- Weis-Garcia F. and Massagué,J. (1996) Complementation between kinase-defective and activation-defective TGF-β receptors reveals a novel form of receptor cooperativity essential for signaling. EMBO J., 15, 276–289. [PMC free article] [PubMed] [Google Scholar]
- Wotton D., Lo,R.S., Lee,S. and Massagué,J. (1999) A Smad transcriptional corepressor. Cell, 97, 29–39. [DOI] [PubMed] [Google Scholar]
- Wrana J.L. (2000) Regulation of Smad activity. Cell, 100, 189–192. [DOI] [PubMed] [Google Scholar]
- Xiao G., Cui,Y., Ducy,P., Karsenty,G. and Franceschi,R.T. (1997) Ascorbic acid-dependent activation of the osteocalcin promoter in MC3T3-E1 preosteoblasts: requirement for collagen matrix synthesis and the presence of an intact OSE2 sequence. Mol. Endocrinol., 11, 1103–1113. [DOI] [PubMed] [Google Scholar]
- Xiao G., Wang,D., Benson,M.D., Karsenty,G. and Franceschi,R.T. (1998) Role of the α2-integrin in osteoblast-specific gene expression and activation of the Osf2 transcription factor. J. Biol. Chem., 273, 32988–32994. [DOI] [PubMed] [Google Scholar]
- Xie X.Q., Pardali,E., Holm,M., Sideras,P. and Grundstrom,T. (1999) AML and Ets proteins regulate the Iα1 germ-line promoter. Eur. J. Immunol., 29, 488–498. [DOI] [PubMed] [Google Scholar]
- Yanagisawa J. et al. (1999) Convergence of transforming growth factor-β and vitamin D signaling pathways on SMAD transcriptional coactivators. Science, 283, 1317–1321. [DOI] [PubMed] [Google Scholar]
- Zhang R., Ducy,P. and Karsenty,G. (1997) 1,25-dihydroxyvitamin D3 inhibits osteocalcin expression in mouse through an indirect mechanism. J. Biol. Chem., 272, 110–116. [DOI] [PubMed] [Google Scholar]
- Zhang Y. and Derynck,R. (1999) Regulation of Smad signalling by protein associations and signalling crosstalk. Trends Cell Biol., 9, 274–279. [DOI] [PubMed] [Google Scholar]
- Zhang Y. and Derynck,R. (2000) Transcriptional regulation of the transforming growth factor-β-inducible mouse germ line Ig α constant region gene by functional cooperation of smad, CREB and AML family members. J. Biol. Chem., 275, 16979–16985. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Feng,X., Wu,R. and Derynck,R. (1996) Receptor-associated Mad homologues synergize as effectors of the TGF-β response. Nature, 383, 168–172. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Feng,X.H. and Derynck,R. (1998) Smad3 and Smad4 cooperate with c-Jun/c-Fos to mediate TGF-β-induced transcription. Nature, 394, 909–913. [DOI] [PubMed] [Google Scholar]
- Zhang Y.W. et al. (2000) A RUNX2/PEBP2αA/CBFA1 mutation displaying impaired transactivation and Smad interaction in cleidocranial dysplasia. Proc. Natl Acad. Sci. USA, 97, 10549–10554. [DOI] [PMC free article] [PubMed] [Google Scholar]