Abstract
Bacterial cell division requires accurate selection of the middle of the cell, where the bacterial tubulin homologue FtsZ polymerizes into a ring structure. In Escherichia coli, site selection is dependent on MinC, MinD and MinE. MinC acts, with MinD, to inhibit division at sites other than the midcell by directly interacting with FtsZ. Here we report the crystal structure to 2.2 Å of MinC from Thermotoga maritima. MinC consists of two domains separated by a short linker. The C-terminal domain is a right-handed β-helix and is involved in dimer formation. The crystals contain two different MinC dimers, demonstrating flexibility in the linker region. The two-domain architecture and dimerization of MinC can be rationalized with a model of cell division inhibition. MinC does not act like SulA, which affects the GTPase activity of FtsZ, and the model can explain how MinC would select for the FtsZ polymer rather than the monomer.
Keywords: cell division/crystal structure/FtsZ/MinC
Introduction
Successful bacterial cell division requires accurate spatial and temporal regulation of septum formation to ensure a complete chromosome in each daughter cell. Selection of the midcell over other sites requires MinC, MinD and MinE (de Boer et al., 1989). FtsZ, which forms a ring-like structure at the midcell, is then recruited to the midcell, followed by other components of the division machinery, including FtsA, ZipA, FtsK, FtsL, FtsI (PBP3), FtsN and FtsW. These may interact to form a hypothetical protein complex, the divisome. A successful division then requires circumferential invagination of the cytoplasmic membrane and, in bacteria with a cell wall, peptidoglycan synthesis (for recent reviews see Bramhill, 1997; Lutkenhaus and Addinall, 1997; Rothfield et al., 1999).
MinC, along with MinD and MinE, is proposed to act as the mechanism for accurate midcell selection, in favour of the potential division sites at the cell poles (Rothfield and Zhao, 1996). These are midcell sites from previous cell divisions. MinC inhibits cell division (de Boer et al., 1992; Bi and Lutkenhaus, 1993). It binds and is regulated by MinD, a 30 kDa ATPase (de Boer et al., 1991; Huang et al., 1996), which structurally resembles the nitrogenase iron proteins found in nitrogen-fixing bacteria (Cordell and Löwe, 2001). In Escherichia coli, MinC and MinD have been found to oscillate from cell pole to cell pole. This has been proposed to be a mechanism for masking potential division sites (Hu and Lutkenhaus, 1999; Raskin and de Boer, 1999a,b). In Bacillus subtilis, similar green fluorescent protein (GFP)-fusion experiments have found MinC targeted to the cell poles and retained at the midcell; as in E.coli, this requires MinD (Marston and Errington, 1999).
Relief from MinCD-induced inhibition of cell division requires MinE, which forms the earliest detectable ring structure at the midcell (Raskin and de Boer, 1997). This 10 kDa protein contains a MinD-interacting domain, which relieves MinCD inhibition, and a topological specificity domain, which is required for localization of MinE to the midcell (Zhao et al., 1995; Huang et al., 1996; King et al., 2000). It is not known how MinE finds the midcell site.
Recent analysis of E.coli MinC has found that it contains two separate domains: an N-terminal domain, which inhibits cell division and prevents FtsZ assembly in vitro, and a C-terminal domain, which interacts with MinD (Hu and Lutkenhaus, 2000). MinC was found to be an oligomer, probably a dimer, and this requires the C-terminal domain, though the N-terminal domain may enhance dimerization. MinC does not affect the GTPase activity of FtsZ, unlike another inhibitor, SulA, which is part of the SOS response (Mukherjee et al., 1998). Two different modes of MinC action have been proposed; in the first model, MinC affects polymerization of FtsZ, whereas in the second model, MinC acts later to inhibit FtsA incorporation into the FtsZ filaments. Experiments with pure MalE–MinC fusion protein and FtsZ favour the first model, as MinC affects FtsZ polymer length, leading to fewer and shorter filaments (Hu et al., 1999). An N-terminal mutant, MinC19 (E.coli Gly10Asp) had impaired ability to inhibit cell division and bind to FtsZ. The second set of experiments with cytoplasmic cell extracts did not find that MinC affects FtsZ filament length. Instead, using immunofluorescence micrographs it has been suggested that MinC acts to prevent FtsA incorporation into the FtsZ ring (Justice et al., 2000).
Here, we present the 2.2 Å crystal structure of Thermotoga maritima MinC solved by multiple anomalous dispersion (MAD) on selenomethionine-substituted crystals.
Results
Cloning, crystallization and structure determination
The minC gene from T.maritima was cloned, over expressed in C41(DE3) cells, and purified by Ni-NTA affinity and size exclusion chromatography. The protein eluted as a single peak with a retention time suggesting a dimer. The dimer is very stable, even in buffers containing 4 M NaCl (data not shown). TmMinC could be stored for several months at 4°C. SeMet-substituted protein was produced in the same, non-methionine auxotrophic C41(DE3) cells. Crystals belong to space group P212121, and the phase problem was solved using MAD with two crystals containing SeMet-substituted protein at 3 Å resolution. Crystallographic data are summarized in Table I. Four NCS related molecules were built, and the phases extended to 2.2 Å using 4-fold non-crystallographic symmetry (NCS) and two domain averaging (Figure 1).
Table I. Crystallographic data.
| Crystal | λ(Å) | Resolution (Å) | I/σIa | Rmb (%) | Multiplicityc | Completeness (%)d | f′/f″e |
|---|---|---|---|---|---|---|---|
| PEAK1 | 0.9790 | 3.0 | 4.9 | 0.076 | 13.8 (6.9) | 98.8 | –5/7 |
| PEAK2 | 0.9790 | 3.2 | 5.1 | 0.103 | 13.2 (6.6) | 97.8 | –5/7 |
| INFL1 | 0.9792 | 3.0 | 4.9 | 0.078 | 13.8 (6.9) | 98.8 | –9/3 |
| INFL2 | 0.9792 | 3.2 | 4.2 | 0.108 | 13.2 (6.6) | 98.4 | –9/3 |
| HREM1 | 0.9393 | 3.0 | 4.9 | 0.075 | 13.8 (6.9) | 98.8 | –3/3 |
| HREM2 | 0.9393 | 3.2 | 3.6 | 0.109 | 13.2 (6.6) | 97.6 | –3/3 |
| NATI | 0.9393 | 2.2 | 3.7 | 0.075 | 3.2 | 95.0 |
The space group is P212121, a = 51.6 Å, b = 106.1 Å, c = 162.5 Å.
aSignal to noise ratio for the highest resolution shell of intensities.
bRm = ΣhΣi|I(h,i) – I(h)|/ΣhΣi I(h,i), where I(h,i) are symmetry-related intensities and I(h) is the mean intensity of the reflection with unique index h.
cMultiplicity for unique reflections; anomalous multiplicity in parentheses.
dCompleteness for unique reflections; anomalous completeness is identical because inverse beam geometry was used.
ef′/f″ ratio, as determined from a fluorescence scan of the crystal.
Correlation coefficients of anomalous differences at different wavelengths for MAD experiment 1: PEAK1 versus INFL1, 0.54; PEAK1 versus HREM1, 0.46; INFL1 versus HREM1, 0.39.
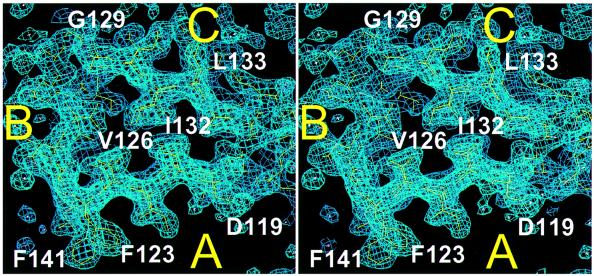
Fig. 1. Stereo drawings of the final 2Fo – Fc map, looking into the hydrophobic core of the β-helix of the C-terminal domain. Turn 2 with strands S2A, S2B and S2C is shown; turn 3 is visible underneath. Side ‘A’ of the triangular domain forms the dimer interface. Made with MAIN (Turk, 1992).
MinC monomer structure
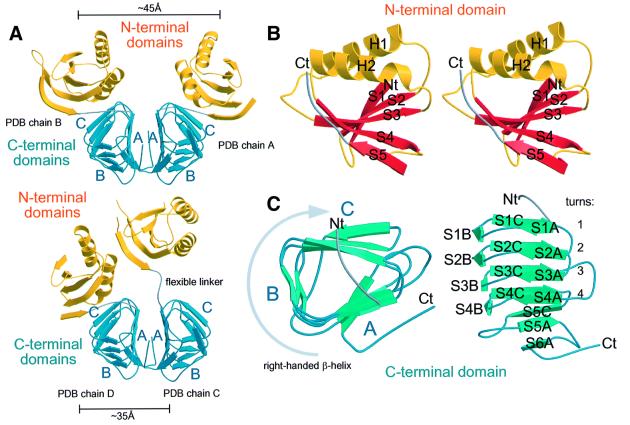
MinC consists of two domains connected by a flexible linker (Figure 2). The N-terminal domain (residues 1–95) contains two α-helices and five β-strands in the order 1a2p3p4p5a (Figure 2B). Strands S1 and S2 are followed by helix H1, strand S3, helix H2 and strands S4 and S5. Overall, the strands form a sheet with the two helices above. A three dimensional similarity search using DALI (Holm and Sander, 1995) found this region to resemble the 1A region of FtsA (PBD 1E4F; van den Ent and Lowe, 2000), a bacterial cell division protein interacting directly with FtsZ, with a Z-score of 3.7 and a root mean square deviation (r.m.s.d.) of superimposed Cα of 3.2 Å over 74 equivalent residues. The DALI search also found similarities to SpoIIAA (PDB 1AUZ; Kovacs et al., 1998), with a Z-score of 3.5 and an r.m.s.d of 3.2 Å over 74 equivalent residues. SpoIIAA is part of a four-component system that regulates σF, the key sporulation sigma factor in B.subtilis.
Fig. 2. Ribbon drawings of MinC. (A) An asymmetric unit contains two different MinC dimers, highlighting the flexibility of the linker region (linker, grey; N-terminal domain, yellow; C-terminal domain, blue). Face ‘A’ of the triangular C-terminal domain forms the dimer interface alone in dimer AB (top). (B) Stereo drawing of the N-terminal domain (residues 1–95) with the flexible linker (residues 96–102). (C) Top and side view of the C-terminal domain. The domain folds into a small triangular, right-handed β-helix with a hydrophobic core. The length of the sides is: A, four; B, three; and C, five residues in β-conformation. The strands in the domain have been numbered to reflect their position with respect to the turn number and the side of the β-helix. Made with MOLSCRIPT and RASTER3D (Kraulis, 1991; Merritt and Bacon, 1997).
The C-terminal domain (residues 103–206) is dominated by a right-handed β-helix. Twelve β-strands (S1A to S4C; Figure 2C) are stacked to form the parallel region of the β-helix. These form four complete turns of the helix, which are followed by three additional β-strands, S5C, S5A and S6A; these are antiparallel to the rest of the helix. The central core of the helix is exclusively hydrophobic and contains residues with small sidechains such as valine and isoleucine. The strict geometrical requirements of the helix result in a repeat that can be detected in the sequence (Figures 3 and 4). However, the repeat is less notable in the sequence compared with the very regular pattern of sidechains forming the hydrophobic core of the β-helix in three dimensions (Figure 1). A DALI search against the C-terminal domain finds proteins containing large regions of right-handed β-helix.
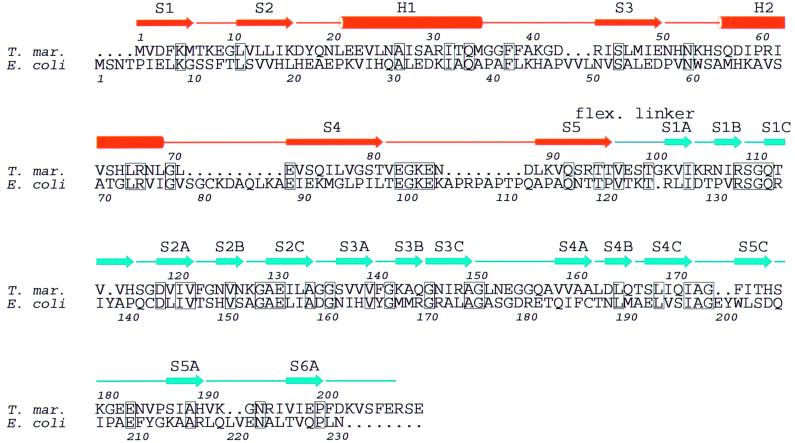
Fig. 3. Sequence alignment of T.maritima and E.coli MinC proteins. The secondary structure of the T.maritima crystal structure is indicated above. Identical residues are boxed. Made with ALSCRIPT (Barton, 1993).
Fig. 4. Sequence repeats in the C-terminal domain of T.maritima MinC giving rise to the symmetrical β-helical fold with exclusively small, hydrophobic sidechains in the core of the β-helix. Sidechain orientation can be ‘I’ with sidechains pointing into the hydrophobic core or ‘O’, pointing outwards.
MinC dimer structure
The asymmetric unit of the TmMinC crystals contains four molecules arranged in two very clear dimers, consisting of chains A and B, and C and D, respectively. These two dimers are almost exclusively formed by dimerization of the C-terminal domains. The orientation of the N-terminal domains with respect to the dimer of C-terminal domains differs between the two dimers (Figure 2A). The dimer of C-terminal domains is formed by the A surface of the β-helix (Figure 2A and C). The surface area of the C-terminal domain of molecule 1 (chain A, residues 103–206) is 5449.75 Å2, and of molecule 2 is 5416.03 Å2. The surface area of both together in the dimer is 9109.68 Å2, and at the interface the surface area is 1756.11 Å2. The surface area involved in dimerization therefore covers 32% of the surface area of one C-terminal domain. The interface holding two C-terminal domains together is of a mainly hydrophobic nature, as can be seen in Figure 4 for the residues on surface A. On all other surfaces, residues pointing outwards (O) are hydrophilic, as expected. The two A surfaces of the β-helix stack as two large, parallel β-sheets with an angle of ∼100°. This gives rise to a very regular pattern of residues pointing inwards and outwards into the dimer contact. The large extent of this surface and its hydrophobic nature make it very unlikely that the dimer can be separated in vivo. Although the two MinC dimers found in the crystal are held together by the same dimer of C-terminal domains, the N-terminal domains are found in two different arrangements (Figure 2A). The sequence region linking the two domains (residues 96–102) therefore has to be flexible by design, and this is reflected in weaker than normal electron density for these residues. The centres of mass of the two N-terminal domains are ∼45 and 35 Å apart in dimer A, B and C, D, respectively.
Discussion
The MinC crystal structure has been solved by MAD and non-crystallographic averaging at 2.2 Å resolution. The structure is generally of high quality; however, the N-terminal domain for chain D is highly disordered due to a small number of crystal contacts. It is not currently possible to model this disorder, which is probably reflected in the rather high Rfree of the model (Table II). Helix H1 of the N-terminal domain of chain D is almost invisible in the electron density maps, and the whole domain seems to move around its attachment to the C-terminal domain, which is fixed by the dimer contact (Figure 2A and B). Together with the finding of two different dimers in the crystals, it is clear that residues 96–102 provide a flexible linker that allows free rotation of the N- and C-terminal domains. Unsurprisingly, the linker sequence is not highly conserved among species, as disordered regions can be created with many different sequences.I
Table II. Refinement statistics.
| Model | Four non-crystallographically related MinC molecules |
| arranged in two different dimers: A and B, and C and D | |
| A, residues 1–79, 90–206 | |
| B, residues 2–207 | |
| C, residues 1–83, 88–206 | |
| D, residues 3–78, 93–206 | |
| 661 water molecules | |
| Diffraction data | NATI, 2.2 Å, all data |
| R-factor, Rfreea | 0.2365, 0.3002 |
| Baverage, bondedb | 44.22 Åb, 4.186 Åb |
| Geometry bonds, anglesc | 0.006 Å, 1.279° |
| Ramachandrand | 83.2%/0.0% |
| Restrained NCSe | 0.55 Å |
| PDB IDf | 1HF2, R1HF2SF |
aFive percent of reflections were randomly selected for determination of the free R-factor, prior to any refinement.
bTemperature factors averaged for all atoms and r.m.s.ds of temperature factors between bonded atoms.
cR.m.s.ds from ideal geometry for bond lengths and restraint angles (Engh and Huber, 1991).
dPercentage of residues in the ‘most favoured’ region of the Ramachandran plot and percentage of outliers (PROCHECK; Laskowski et al., 1993).
eR.m.s.d. between coordinates of atoms related by non-crystallographic symmetry.
fProtein Data Bank identifiers for coordinates and structure factors, respectively.
We have used MinC from T.maritima rather than E.coli. The E.coli MinC does not behave sufficiently well for structural studies. Other experiments have used a MalE–MinC fusion of the E.coli protein to circumvent this problem (Hu et al., 1999). We are concentrating on the cell division machinery in T.maritima. This organism is a thermophilic eubacterium with an almost identical set of genes for cell division when compared with E.coli. Proteins from this organism have proved amenable to structural studies, unlike the corresponding E.coli cell division proteins.
MinC is a very tight dimer with an unusually large dimer interface of ∼32% of the C-terminal domain’s surface. This is also reflected in the stability of the dimer under conditions of extreme ionic strength. We conclude that MinC has been designed to function as a dimer in vivo. Dimerization is facilitated by the C-terminal domains, which are folded into a right-handed β-helix. Similarity searches found no convincing homologues of this domain when checked with DALI (Holm and Sander, 1995) and visually, although several right-handed β-helices have been reported in other structures. The β-helix design seems to accommodate many different sizes and shapes as well as different helical orientations, since it is only composed of the longitudinal β-sheet hydrogen bonds and some residues introducing turns in the right directions in the corners (Figure 4). For example, the spruce budworm antifreeze protein (PDB 1EWW; Graether et al., 2000) is of similar size and shape but is a left-handed β-helix. In this protein, one of the flat sides is used to bind to the hexagonal water lattice on ice crystals. In MinC, side A of the dimerization domain forms the interface that is hydrophobic, indicating that the MinC monomer cannot exist in solution. Analysis of the C-terminal domain revealed a weak sequence repeat, which may be used to search for other proteins that display a similar fold (Figure 4).
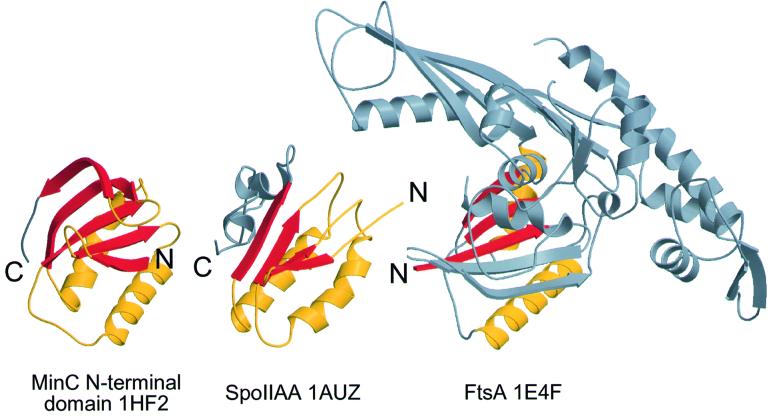
The N-terminal domain of MinC revealed a fold that is distantly related to SpoIIAA and even more distantly related to FtsA (Figure 5). SpoIIAA is the anti-anti-sigma factor of sporulation. Sporulation is a specialized form of cell division, where the cell divides asymmetrically to form a forespore and a mother cell. SpoIIAA is part of a larger domain family, called STAS after sulfate transporters and anti-sigma factor antagonists (Aravind and Koonin, 2000). MinC, however, lacks the characteristic C-terminal α-helical handle-like structure of STAS domains, though MinC and SpoIIAA share a S1-S2-H1-S3-H2 arrangement, forming a central β-sheet region with two α-helices above it. Finding two similar structures for proteins involved in sporulation and cell division is intriguing, but further biochemical analysis will be required to find out whether there are functional similarities between SpoIIAA and MinC.
Fig. 5. Structural alignment of the N-terminal domain of MinC from T.maritima, SpoIIAA from B.subtilis (PDB 1AUZ; Kovacs et al., 1998) and FtsA from T.maritima (PDB 1E4F; van den Ent and Löwe, 2000). FtsA shows the highest DALI score of 3.7, r.m.s.d. 3.2 Å over 74 residues. SpoIIAA has a DALI score against the N-terminal domain of MinC of 3.5, r.m.s.d. 3.6 Å over 72 almost consecutive residues. Aligned stretches are coloured, all other residues are shown in grey. Made with MOLSCRIPT and RASTER3D (Kraulis, 1991; Merritt and Bacon, 1997).
A DALI (Holm and Sander, 1995) search for similar structures found FtsA to be the closest, but a weak homologue of the N-terminal domain of MinC. The region of similarity is a small part of the FtsA structure, and even in the ribbon plot (Figure 5), the similarity is not striking. Analysis of the two structures indicates that the surface residues are conserved and that the underlying fold is less so. Although the similarity is not very strong, we believe this could be significant since evidence suggests that both FtsA (Wang et al., 1997; Yan et al., 2000) and MinC (Hu et al., 1999) bind FtsZ, and that MinC prevents FtsA from binding to FtsZ (Justice et al., 2000). A tempting speculation would then be to assume that the structural similarity reflects a common function: binding of FtsA and MinC to the same site on the FtsZ polymer, though there is no direct evidence for this. A gel filtration experiment indicates that MinC does not bind to monomeric FtsZ (data not shown). Together with data showing MinC binding to FtsZ polymers in pelleting assays (Hu and Lutkenhaus, 2000), this suggests that MinC preferentially binds FtsZ polymer.
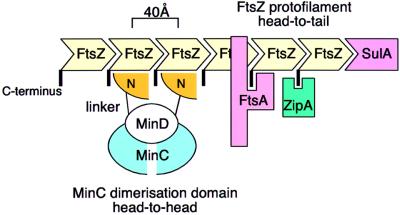
We propose a model of how MinC might function in the cell by binding to the FtsZ polymer (Figure 6); FtsZ forms a polymer in the cell of which only the protofilament interaction is known (Erickson et al., 1996; Löwe and Amos, 1999). In a protofilament, FtsZ molecules form a linear polymer in a head-to-tail arrangement. This protofilament formation is shared with eukaryotic tubulin (Nogales et al., 1998). MinC, however, is a head-to-head dimer. The flexible linker between the N- and C-terminal domains would allow the N-terminal domains to be rotated by up to 180° to each other. The N-terminal domains, which have been shown to bind to FtsZ on their own (Hu and Lutkenhaus, 2000), could then both bind to the FtsZ protofilament. The distances between the N-terminal domains found in the two different dimers (45 and 35 Å) are compatible with the 40 Å repeat of the FtsZ protofilament. Having two FtsZ-binding domains would enable MinC to select for the FtsZ polymer instead of the monomer. The sequence conservation is rather low in the N-terminal domain, but a single lysine residue (Lys5) is 100% conserved in all known MinC sequences (data not shown). Lys5 is next to Met6, which is Gly10 in E.coli and has been found to be involved in the FtsZ interaction (Hu and Lutkenhaus, 2000). This suggests, that the FtsZ-binding surface is opposite to the flexible linker and the dimerization domain on the MinC N-terminal domain (Figure 2B, bottom). Sequence conservation is higher in the C-terminal domain, but this may only reflect more stringent requirements of sequence conservation for a β-helical structure. The C-terminal part of that domain has the lowest sequence conservation (strands S5C–S6A), making interactions with other molecules in that region unlikely. We have no evidence for this, but an intuitive way of interpreting biochemical data would be to place the MinD binding site in MinC somewhere in the upper part of the C-terminal domain, maybe reaching up to the N-terminal domains. This would allow for different orientations of the N-terminal domains to be selected by different nucleotide states of MinD. Although we also have a MinD structure (Cordell and Löwe, 2001), there are no apparent clues as to how MinC and MinD interact.
Fig. 6. Schematic overview of the model of MinC function. FtsZ forms linear head-to-tail polymers (protofilaments) with a repeat of 40 Å (Löwe and Amos, 1999). MinC forms tight head-to-head dimers via the C-terminal domain (blue). To enable binding of MinC to FtsZ protofilaments, the N–terminal domains of MinC (orange) have to be rotated, which is facilitated by a flexible linker region. By forming a dimer, MinC will bind much more tightly to FtsZ protofilaments than FtsZ monomers. Although the binding site of MinD on MinC is not known, we speculate that it may pre-orient the N-terminal domains of MinC, thereby greatly enhancing binding affinity to FtsZ. MinD’s ATPase activity could be required for changing conformation in MinC to switch between different affinities or FtsZ polymers. MinC’s binding site on FtsZ is thought to overlap with FtsA’s binding site (Justice et al., 2000), which includes the conserved C-terminal peptide in FtsZ. ZipA can bind to FtsZ in the presence of MinCD, whereas FtsA cannot (Justice et al., 2000). SulA, an SOS response factor, interferes with FtsZ polymerization directly (Mukherjee et al., 1998). By inhibiting FtsA binding, MinC can block septum formation, since the recruitment of all other septum proteins is dependent on localization of FtsZ and FtsA.
A recurring theme in bacterial cell division is regulation at the level of the FtsZ polymer, including the proposed mechanism of MinC (Figure 6). Other proteins that interact with FtsZ are SulA, ZipA and FtsA. FtsZ polymer formation can be inhibited by SulA (Justice et al., 2000). It is part of the SOS response and its function is to switch off cell division after damage to the DNA has occurred (Gottesman et al., 1981). ZipA, a modulator of FtsZ filaments, binds to the FtsZ polymer in the presence of MinCD; therefore, its binding site is likely to be different from MinCD and FtsA (Justice et al., 2000). FtsA is required for incorporation of the other components of the septal machinery. Although FtsA and ZipA share binding to the C-terminal conserved peptide of FtsZ (Mosyak et al., 2000; Yim et al., 2000), additional regions on ZipA have been implicated in FtsZ binding (Mosyak et al., 2000; Yim et al., 2000). MinC has been found to bind to the FtsZ polymer (Hu and Lutkenhaus, 2000) and interferes with FtsA binding to FtsZ (Justice et al., 2000). Based on the finding of some structural similarity between MinC and FtsA, we propose that the proteins share a common binding site on the FtsZ protofilament, enabling inhibition of septum formation at all sites other than the midcell.
Materials and methods
Protein expression and purification
Genomic DNA from T.maritima (DSMZ No. 3109, freeze dried) was extracted using a commercially available kit (Promega Wizard). The minC gene (WWW.TIGR.ORG: TM1047, SWALL:Q9X0D7) was amplified by PCR using primers that introduce unique cleavage sites for NdeI and BamHI into the product. After digestion, the fragment was cloned into pHis17. This enables the protein to be expressed under the control of the T7 promoter and adds 8 residues to the C-terminus (GSHHHHHH). C41(DE3) cells (Miroux and Walker, 1996) were transformed, grown at 37°C and induced in log phase (OD600 = 0.6) with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Three hours after induction the cells were harvested and frozen in liquid nitrogen. Cells were thawed and lysed by the addition of lysozyme and by sonication, in a buffer of 50 mM Tris pH 7.4. DNase I and RNase A were added, and after centrifugation the lysate was applied to a Ni-NTA column (Qiagen). Buffer A was 300 mM NaCl and 50 mM Tris pH 7.0, buffer B was 1 M imidazole and 50 mM Tris pH 7.0. After washing with 5% buffer B, the protein was eluted with 30% buffer B. Peak fractions were pooled and concentrated before loading onto a Sephacryl S200 column (Amersham Pharmacia) equilibrated in 20 mM Tris, 1 mM EDTA and 1 mM NaN3 pH 7.0.
SeMet-substituted MinC
The SeMet-substituted protein was grown as for MukB (van den Ent et al., 1999), which was based on conditions described previously (Van Duyne et al., 1993). The protein was more susceptible to proteolysis than the native protein and so was purified in a different manner. The frozen cell pellet was ground under liquid nitrogen and poured directly into 100 ml of boiling 50 mM Tris pH 8.0 and 300 mM NaCl, and stirred for 90 s. Ice was added to 400 ml along with DNase I, RNase A and 5 mM β-mercaptoethanol. After centrifugation, the protein was purified as for the native but with the addition of 5 mM β-mercapto ethanol for the Ni-NTA column buffers and 5 mM dithiothreitol for the Sephacryl 200 column buffer. Electrospray mass spectroscopy was used to confirm SeMet incorporation. (Native MinC: observed 23 694.03 Da, calculated 23 694.00 Da; SeMet: observed 23 881.29 Da, calculated 23 881.58 Da.)
Crystallization and data collection
Native crystals were grown using the sitting drop vapour diffusion technique using 21% PEG 2000, 0.1 M sodium citrate pH 5.0 and 5.8% ethanol as the crystallization solution. Drops composed of 1 µl of protein at 10 mg/ml and 1 µl of crystallization solution were equilibrated for a minimum of 3 days at 19°C. SeMet-substituted crystals were grown in the same manner as for the native protein but with 24% PEG 2000, 0.1 M sodium citrate pH 4.8, 3.6% ethanol for the crystallization solution, and additionally 200 mM NaI in the drop.
Crystals were frozen in the mother liquor with 20% glycerol. Crystals belong to space group P212121 and have cell dimensions a = 51.6 Å, b = 106.1 Å, c = 162.5 Å. The two MAD datasets and the native dataset were collected at ID14-4 ESRF, Grenoble. Crystals were indexed and integrated using the MOSFLM package (Leslie, 1991), and data were further processed using the CCP4 package (Collaborative Computational Project No. 4, 1994).
Structural determination and refinement
An initial 3 Å MAD density map was generated by locating 11 selenium sites in the data sets PEAK1, INFL1, HREM1, and from the second crystal, PEAK2, INFL2, HREM2, using the program SOLVE (Terwilliger and Berendzen, 1999), which was also used to calculate phases. RESOLVE (Terwilliger, 2000) was used for solvent flattening, assuming a 48% solvent content. A polyalanine model was built using MAIN (Turk, 1992) for four NCS related molecules. DMMULTI was used to extend the phases to 2.2 Å using 4–fold non-crystallographic symmetry and two-domain averaging, as different NCS operators were required for the N- and C-terminal domains. The resulting electron density map was of superb quality and shows most carbonyls. The two MinC dimers were built using MAIN and refined using CNS (Brünger et al., 1998) with Engh and Huber stereochemical parameters (Engh and Huber, 1991). Details of the final model are summarized in Table II.
Acknowledgments
Acknowledgements
We thank Gordon Leonard at beamline ID14-4, ESRF (Grenoble) for assistance with MAD data collection, and Sew Peak-Chew (MRC-LMB, Cambridge) and Ian Fearnley (MRC Dunn Nutrition Unit, Cambridge) for performing mass spectrometry.
References
- Aravind L. and Koonin,E.V. (2000) The STAS domain—a link between anion transporters and antisigma-factor antagonists. Curr. Biol., 10, R53–R55. [DOI] [PubMed] [Google Scholar]
- Barton G.J. (1993) ALSCRIPT: a tool to format multiple sequence alignments. Protein Eng., 6, 37–40. [DOI] [PubMed] [Google Scholar]
- Bi E. and Lutkenhaus,J. (1993) Cell division inhibitors SulA and MinCD prevent formation of the FtsZ ring. J. Bacteriol., 175, 1118–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramhill D. (1997) Bacterial cell division. Annu. Rev. Cell. Dev. Biol., 13, 395–424. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project No. 4 (1994) The CCP4 Suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Cordell S.C. and Löwe,J. (2001) Crystal structure of the bacterial cell division regulator MinD. FEBS Lett., 492, 160–165. [DOI] [PubMed] [Google Scholar]
- de Boer P.A.J., Crossley,R.E. and Rothfield,L.I. (1989) A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in Escherichia coli. Cell, 56, 641–649. [DOI] [PubMed] [Google Scholar]
- de Boer P.A.J., Crossley,R.E., Hand,A.R. and Rothfield,L.I. (1991) The MinD protein is a membrane ATPase required for the correct placement of the Escherichia coli division site. EMBO J., 10, 4371–4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer P.A.J., Crossley,R.E. and Rothfield,L.I. (1992) Roles of MinC and MinD in the site-specific septation block mediated by the MinCDE system of Escherichia coli. J. Bacteriol., 174, 63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engh R.A. and Huber,R. (1991) Accurate bond and angle parameters for X-ray protein-structure refinement. Acta Crystallogr. A, 47, 392–400. [Google Scholar]
- Erickson H.P., Taylor,D.W., Taylor,K.A. and Bramhill,D. (1996) Bacterial cell division protein FtsZ assembles into protofilament sheets and minirings, structural homologs of tubulin polymers. Proc. Natl Acad. Sci. USA, 93, 519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S., Halpern,E. and Trisler,P. (1981) Role of SulA and SulB in filamentation By Lon mutants of Escherichia coli K-12. J. Bacteriol., 148, 265–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graether S.P., Kuiper,M.J., Gagne,S.M., Walker,V.K., Jia,Z.C., Sykes,B.D. and Davies,P.L. (2000) β-helix structure and ice-binding properties of a hyperactive antifreeze protein from an insect. Nature, 406, 325–328. [DOI] [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1995) Dali—a network tool for protein-structure comparison. Trends Biochem. Sci., 20, 478–480. [DOI] [PubMed] [Google Scholar]
- Hu Z.L. and Lutkenhaus,J. (1999) Topological regulation of cell division in Escherichia coli involves rapid pole-to-pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Mol. Microbiol., 34, 82–90. [DOI] [PubMed] [Google Scholar]
- Hu Z.L. and Lutkenhaus,J. (2000) Analysis of MinC reveals two independent domains involved in interaction with MinD and FtsZ. J. Bacteriol., 182, 3965–3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z.L., Mukherjee,A., Pichoff,S. and Lutkenhaus,J. (1999) The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc. Natl Acad. Sci. USA, 96, 14819–14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Cao,C. and Lutkenhaus,J. (1996) Interaction between FtsZ and inhibitors of cell division. J. Bacteriol., 178, 5080–5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice S.S., Garcia-Lara,J. and Rothfield,L.I. (2000) Cell division inhibitors SulA and MinC/MinD block septum formation at different steps in the assembly of the Escherichia coli division machinery. Mol. Microbiol., 37, 410–423. [DOI] [PubMed] [Google Scholar]
- King G.F., Shih,Y.L., Maciejewski,M.W., Bains,N.P.S., Pan,B.L., Rowland,S.L., Mullen,G.P. and Rothfield,L.I. (2000) Structural basis for the topological specificity function of MinE. Nature Struct. Biol., 7, 1013–1017. [DOI] [PubMed] [Google Scholar]
- Kovacs H., Comfort,D., Lord,M., Campbell,I.D. and Yudkin,M.D. (1998) Solution structure of SpoIIAA, a phosphorylatable component of the system that regulates transcription factor σF of Bacillus subtilis. Proc. Natl Acad. Sci. USA, 95, 5067–5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraulis P.J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Laskowski R.A., Macarthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK—a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Leslie A.G.W. (1991) Recent changes to the MOSFLM package for processing film and image plate data. SERC Laboratory, Daresbury, Warrington, UK.
- Löwe J. and Amos,L.A. (1999) Tubulin-like protofilaments in Ca2+-induced FtsZ sheets. EMBO J., 18, 2364–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhaus J. and Addinall,S.G. (1997) Bacterial cell division and the Z ring. Annu. Rev. Biochem., 66, 93–116. [DOI] [PubMed] [Google Scholar]
- Marston A.L. and Errington,J. (1999) Selection of the midcell division site in Bacillus subtilis through MinD-dependent polar localization and activation of MinC. Mol. Microbiol., 33, 84–96. [DOI] [PubMed] [Google Scholar]
- Merritt E.A. and Bacon,D.J. (1997) Raster3D: Photorealistic molecular graphics. Methods Enzymol., 277, 505–524. [DOI] [PubMed] [Google Scholar]
- Miroux B. and Walker,J.E. (1996) Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol., 260, 289–298. [DOI] [PubMed] [Google Scholar]
- Mosyak L., Zhang,Y., Glasfeld,E., Haney,S., Stahl,M., Seehra,J. and Somers,W.S. (2000) The bacterial cell-division protein ZipA and its interaction with an FtsZ fragment revealed by X-ray crystallography. EMBO J., 19, 3179–3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A., Cao,C.N. and Lutkenhaus,J. (1998) Inhibition of FtsZ polymerization by SulA, an inhibitor of septation in Escherichia coli. Proc. Natl Acad. Sci. USA, 95, 2885–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales E., Downing,K.H., Amos,L.A. and Löwe,J. (1998) Tubulin and FtsZ form a distinct family of GTPases. Nature Struct. Biol., 5, 451–458. [DOI] [PubMed] [Google Scholar]
- Raskin D.M. and de Boer,P.A.J. (1997) The MinE ring: an FtsZ-independent cell structure required for selection of the correct division site in E.coli. Cell, 91, 685–694. [DOI] [PubMed] [Google Scholar]
- Raskin D.M. and de Boer,P.A.J. (1999a) MinDE-dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. J. Bacteriol., 181, 6419–6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin D.M. and de Boer,P.A.J. (1999b) Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc. Natl Acad. Sci. USA, 96, 4971–4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothfield L.I. and Zhao,C.R. (1996) How do bacteria decide where to divide? Cell, 84, 183–186. [DOI] [PubMed] [Google Scholar]
- Rothfield L., Justice,S. and Garcia-Lara,J. (1999) Bacterial cell division. Annu. Rev. Genet., 33, 423–448. [DOI] [PubMed] [Google Scholar]
- Terwilliger T.C. (2000) Maximum-likelihood density modification. Acta Crystallogr. D, 56, 965–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T.C. and Berendzen,J. (1999) Automated MAD and MIR structure solution. Acta Crystallogr. D, 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk D. (1992) Weiterentwicklung eines Programms für Molekülgrafik und Elektrondichte-Manipulation und seine Anwendung auf verschiedene Protein-Strukturaufklärungen. PhD Thesis, Technische Universität München.
- van den Ent F. and Löwe,J. (2000) Crystal structure of the cell division protein FtsA from Thermotoga maritima. EMBO J., 19, 5300–5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Ent F., Lockhart,A., Kendrick-Jones,J. and Löwe,J. (1999) Crystal structure of the N-terminal domain of MukB: a protein involved in chromosome partitioning. Structure Fold. Des., 7, 1181–1187. [DOI] [PubMed] [Google Scholar]
- Van Duyne G.D., Standaert,R.F., Karplus,P.A., Schreiber,S.L. and Clardy,J. (1993) Atomic structures of the human immunophilin Fkbp-12 complexes with Fk506 and rapamycin. J. Mol. Biol., 229, 105–124. [DOI] [PubMed] [Google Scholar]
- Wang X.D., Huang,J.A., Mukherjee,A., Cao,C. and Lutkenhaus,J. (1997) Analysis of the interaction of FtsZ with itself, GTP, and FtsA. J. Bacteriol., 179, 5551–5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan K., Pearce,K.H. and Payne,D.J. (2000) A conserved residue at the extreme C-terminus of FtsZ is critical for the FtsA–FtsZ interaction in Staphylococcus aureus. Biochem. Biophys. Res. Commun., 270, 387–392. [DOI] [PubMed] [Google Scholar]
- Yim L., Vandenbussche,G., Mingorance,J., Rueda,S., Casanova,M., Ruysschaert,J.M. and Vicente,M. (2000) Role of the carboxy terminus of Escherichia coli FtsA in self-interaction and cell division. J. Bacteriol., 182, 6366–6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C.R., de Boer,P.A.J. and Rothfield,L.I. (1995) Proper placement of the Escherichia coli division site requires two functions that are associated with different domains of the MinE protein. Proc. Natl Acad. Sci. USA, 92, 4313–4317. [DOI] [PMC free article] [PubMed] [Google Scholar]