Abstract
The double-stranded RNA (dsRNA)-activated protein kinase PKR is an interferon (IFN)-induced enzyme that controls protein synthesis through phosphorylation of eukaryotic initiation factor 2α (eIF-2α). PKR also regulates signals initiated by diverse stimuli, including dsRNA, IFN-γ, tumor necrosis factor-α, interleukin-1 and lipopolysaccharide, to different transcription factors, resulting in pro-inflammatory gene expression. Stat3 plays an essential role in promoting cell survival and proliferation by different growth factors, including platelet-derived growth factor (PDGF). Here we show that PKR physically interacts with Stat3 and is required for PDGF-induced phosphorylation of Stat3 at Tyr705 and Ser727, resulting in DNA binding and transcriptional activation. PKR-mediated phosphorylation of Stat3 on Ser727 is indirect and channeled through Erks. Although PKR is pre-associated with the PDGF β-receptor, treatment with PDGF only modestly activates PKR. However, the induction of c-fos by PDGF is defective in PKR-null cells. Taken together, these results establish PKR as an upstream regulator of activation of Stat3 and as a common mediator of both growth-promoting and growth-inhibitory signals.
Keywords: c-fos induction/ERK activation/PDGF/PKR/Stat3 phosphorylation
Introduction
The interferon (IFN)-induced double-stranded RNA (dsRNA)-dependent protein kinase PKR plays an important role in the antiviral activities of IFN (Katze, 1992; Samuel, 1993; Williams, 1995, 1997). Although first identified as a component of host antiviral defense mechanisms, PKR subsequently was found to exhibit features characteristic of a growth suppressor, inhibiting both yeast and mammalian cell proliferation (Chong et al., 1992; Dever et al., 1993). Catalytically inactive PKR mutants also act as dominant-negatives inducing malignant transformation (Koromilas et al., 1992; Meurs et al., 1993). PKR functions as a signal transducer in signaling pathways activated by different stimuli, including dsRNA, interleukin-3 (IL-3), IFN-α, IFN-γ, IL-1 and tumor necrosis factor-α (TNF-α), through a variety of effectors, including NF-κB, IRF-1, ATF-2 and eukaryotic initiation factor 2α (eIF-2α), and participates in transcription, translation and apoptosis (Kumar et al., 1994, 1997; Ito et al., 1999; Williams, 1999; Bandyopadhyay et al., 2000; Goh et al., 2000; Zamanian-Daryoush et al., 2000). PKR has also been implicated in platelet-derived growth factor (PDGF) action. 2-aminopurine, an inhibitor of PKR, can block the induction of immediate early genes by PDGF (Zinn et al., 1988; Mundschau and Faller, 1995), and a reduction in PKR levels by antisense oligonucleotides has the same effect (Mundschau and Faller, 1995). However, the mechanisms underlying PKR involvement in PDGF signaling have not been elucidated.
Stats (signal transducers and activators of transcription) mediate diverse biological functions in response to different ligands (Schindler and Darnell, 1995; Darnell, 1997). Phosphorylation on a single tyrosine residue around position 700 in all Stats is obligatory for their DNA-binding activity (Wen et al., 1995), and serine phosphorylation near this tyrosine residue towards the C-terminus is important for augmenting transcriptional activation. These complex processes of tyrosine and serine phosphorylation can be interdependent or independently regulated (Boulton et al., 1995; Chung et al., 1997; Ng and Cantrell, 1997).
In contrast to the role of Stats in cytokine signaling, less is known about their function in growth factor signal transduction. It is clear, however, that growth factors may activate both growth-stimulatory and growth-inhibitory pathways (Wagner et al., 1990; Kim et al., 1995; Chin et al., 1996; Su et al., 1997). The mechanisms leading to tyrosine and serine phosphorylation of Stats have not been clearly elucidated in this context. The tyrosine kinase activities of growth factor receptors are required for Stat activation by PDGF and epidermal growth factor (EGF), but it is not known whether these receptors directly phosphorylate Stat proteins or if this is performed indirectly in vivo by other kinases (Vignais and Gilman, 1999). JAKs are activated by growth factors but, in contrast to IFN signaling, PDGF-induced JAK phosphorylation and activation of Stat1 and Stat3 is independent of any other single JAK (Muller et al., 1993; Leaman et al., 1996; Vignais et al., 1996). Direct interaction of Stat proteins with receptor tyrosine kinases has been suggested, implying direct phosphorylation of Stats by the receptors, but in vivo evidence is lacking (Fu and Zhang, 1993; David et al., 1996; Novak et al., 1996; Chen et al., 1997). Apart from receptor tyrosine kinases and JAKs, Src family kinases also play roles in Stat phosphorylation in growth factor-treated cells (Mori et al., 1993; Cao et al., 1996). However, it has been proposed that the juxtamembrane region of the PDGF β-receptor regulates Stats and Src family kinases by independent mechanisms (Sachsenmaier et al., 1999).
PDGF- or EGF-activated Stats 1 and 3 bind to the sis-inducible element (SIE) in the c-fos promoter, conferring PDGF-dependent activation of c-fos transcription (Marais et al., 1993; Hill and Triesmann, 1995; Robertson et al., 1995). Although there are some reports to the contrary, a number of studies have shown that the SIE plays an obligatory role in the response to different stimuli including PDGF, implicating Stats as functional mediators of PDGF signaling (Robertson et al., 1995).
In spite of the participation of Stat3 in diverse physiological processes, its precise role in growth signaling remains unclear. Stat3 is activated in v-Src- or v-abl-transformed cells and, along with other Stats, in acute T-cell leukemia cells that have lost IL-2 dependency (Migone et al., 1995; Yu et al., 1995). In mycosis fungoides, a low grade cutaneous T-cell lymphoma of unknown etiology, there is constitutive activation of a slow-migrating form of Stat3 (Nielsen et al., 1997). Different studies have shown correlation of DNA-binding activities of Stat3 with degrees of malignancy (Gouilleux-Gruart et al., 1996; Garcia et al., 1997; Sartor et al., 1997; Bromberg et al., 1999). Here we show that PKR is required as an upstream regulator of activation of Stat3 by PDGF.
Results
Efficient phosphorylation of Stat3 Tyr705 and Ser727 in response to PDGF is dependent on PKR
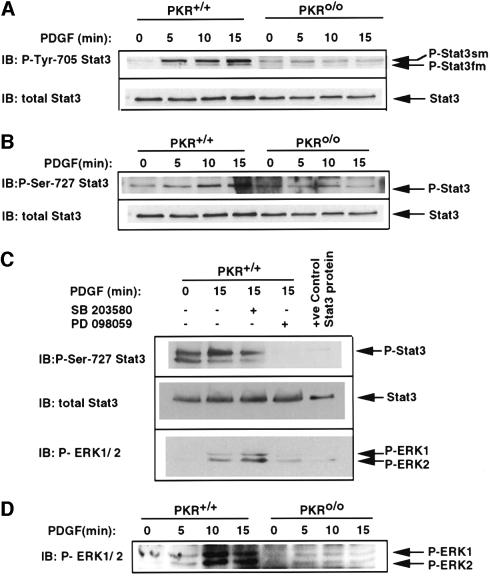
To determine whether PKR affects activation of Stat3 in response to PDGF, extracts derived from PDGF-treated PKR+/+ and PKRo/o mouse embryo fibroblasts (MEFs) were analyzed for the presence of phosphorylated Stat3 using a phosphospecific antibody that recognizes phospho-Tyr705 on the Stat3 molecule. PDGF treatment induced two Tyr705-phosphorylated forms of Stat3 in PKR+/+ cells, a faster migrating species termed Stat3fm and a slower migrating species termed Stat3sm (Figure 1A). Although the kinetics of induction of these two tyrosine-phosphorylated species were apparently similar, Stat3sm was the dominant form (Figure 1A). This slow-migrating form of tyrosine-phosphorylated Stat3 has been reported previously to be due to secondary serine/threonine phosphorylation of the faster migrating Stat3 (Stat3fm; Boulton et al., 1995). Both Stat3sm and Stat3fm have been shown to translocate to the nucleus and bind DNA in response to ligands other than PDGF (Boulton et al., 1995). Importantly, PDGF treatment did not induce either form of tyrosine-phosphorylated Stat3 in PKR-null cells (Figure 1A), implicating a requirement for PKR in both tyrosine and serine phosphorylation of Stat3. Accordingly, nuclear extracts prepared from PDGF-treated PKR- null cells were severely deficient in Stat3 containing DNA-binding complexes compared with wild-type MEFs (data not shown). Taken together, we conclude that PDGF-induced activation of Stat3 DNA binding is PKR dependent.
Fig. 1. PKR is required for Erk activation and Ser727 and Tyr705 phosphorylation of Stat3 in response to PDGF. (A) PKR+/+ and PKRo/o MEFs were treated with PDGF for the indicated times, whole-cell extracts (100 µg) were separated by SDS–PAGE, and phosphorylation of Stat3 at Tyr705 was assayed by immunoblotting. The two forms of Stat3 (Stat3fm and Stat3sm) recognized by this antibody in PKR+/+ MEFs are indicated. The blot was re-probed with anti Stat3 for normalization. (B) Extracts of PDGF-treated PKR+/+ and PKRo/o MEFs were immunoblotted with antibody specific for pSer727 Stat3 (top panel), anti-Stat3 (lower panel). (C) PKR+/+ MEFs were pre-treated with 10 µM SB 203580 (p38 inhibitor) and 30 µM PD 098059 (Erk inhibitor) for 30 min before treatment with PDGF for 15 min. Extracts were separated by SDS–PAGE, transferred onto nylon membranes and immunoblotted with anti pSer727 Stat3 (top panel), anti-Stat3 (middle panel) and anti-phospho-Erk1/2 antibodies (bottom panel). (D) Extracts of PDGF-treated PKR+/+ and PKRo/o MEFs were immunoblotted with antibody specific for phosphorylated (activated) forms of Erk1/2.
PKR is required for PDGF-induced Erk activation and Ser727 phosphorylation of Stat3
The above results (Figure 1A) implied the involvement of PKR in Stat3 serine phosphorylation. Accordingly, we examined extracts of MEF cells treated with PDGF for levels of serine-phosphorylated Stat3. Whereas PKR+/+ cells exhibited a clear induction in phospho-Ser727 Stat3, PKR-null cells were defective in this process (Figure 1B). We conclude, from these results, that PKR appears to be implicated in the process of serine as well as tyrosine phosphorylation of Stat3 in response to PDGF.
There is evidence that the Erk family of mitogen-activated protein (MAP) kinases, but not JNK nor p38, phosphorylate Stat3 at Ser727 in response to growth factors (Chung et al., 1997). To determine whether PKR is a component of the Erk-mediated PDGF signaling cascade to Stat3, wild-type MEFs were treated with PDGF in the presence of PD 098059, a specific MEK (MAP kinase kinase) inhibitor. Treatment of cells for 15 min with PDGF increases Stat3 Ser727 phosphorylation but this can be inhibited by pre-treatment with PD 098059, but not with the p38 inhibitor SB 203580 (Figure 1C). Thus, Erk1/2 phosphorylation is probably responsible for PDGF-induced Ser727 phosphorylation of Stat3. Since in PKR-null MEFs there is a marked defect in Erk1 and Erk2 activation by PDGF (Figure 1D), it appears that PKR is involved in serine phosphorylation of Stat3 through the activation of Erks.
PKR physically interacts with Stat3
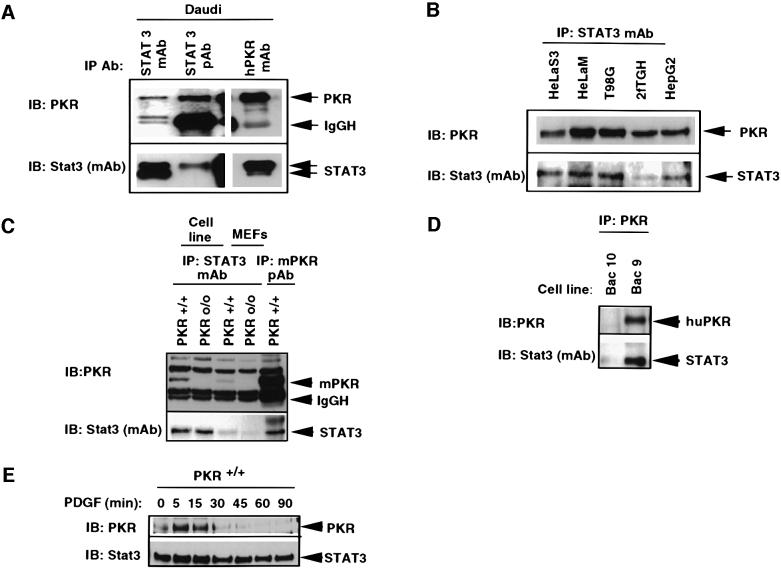
The results described thus far show that activation of Stat3 by PDGF depends on PKR. The role of PKR in activating Stat3 kinase(s) may be either direct or indirect. To determine whether PKR is present in a complex with Stat3, co-immunoprecipitation assays were performed with anti-Stat3 monoclonal or polyclonal antibodies. The results (Figure 2A) show that Stat3 and PKR can physically associate. This was confirmed in a reciprocal experiment, using anti-human PKR monoclonal antibody followed by immunoblotting with anti-Stat3 monoclonal antibody (Figure 2A). Similar co-immunoprecipitation experiments were performed with an unrelated antibody (anti-focal adhesion kinase antibody; data not shown) and this failed to immunoprecipitate either PKR or Stat3, demonstrating the specificity of PKR–Stat3 interaction. The interaction of PKR with Stat3 was ligand independent, since the physical association of PKR and Stat3 was observed in different cell lines in the absence of PDGF (Figure 2B). As expected, PKR did not immunoprecipitate with anti-Stat3 antibody in cells lacking PKR (Figure 2C). This ligand-independent constitutive association of Stat3 is unique because Stats usually homo-/heteromerize subsequent to their phosphorylation following stimulation with ligand. Although Stat3 and PKR are physically associated in untreated cells, Stat3 is not a substrate for PKR either in vitro or in vivo (data not shown). Purified PKR binds in vitro to either Stat3 full-length protein or to Stat3 without the activation domain (data not shown), indicating that, at least in vitro, the C-terminal residues from 717 to 770 are not required for binding to PKR. The interaction of PKR and Stat3 can also be restored in PKRo/o cells by stable transfection of a human PKR gene contained on a bacterial artificial chromosome (BAC; Figure 2D). Stat3 has been established as a mediator of immediate early gene induction pathways (Robertson et al., 1995; Vignais et al., 1996; Sachsenmaier et al., 1999). Since the results in Figure 2A–D implicate a potential functional link between PKR and Stat3, we investigated whether PDGF further induced PKR–Stat3 interactions. Accordingly, PKR wild-type fibroblasts were treated with PDGF for different times and cell lysates were immunoprecipitated with anti-Stat3 monoclonal antibody. Immunoblotting with anti-Stat3 as well as anti-PKR anti bodies (Figure 2E) showed an increase in interaction within 5–15 min of treatment with the growth factor, followed by a decrease to below basal level at later time points.
Fig. 2. PKR and Stat3 physically associate. (A) Whole-cell lysates (500 µg) from Daudi cells were used for immunoprecipitation with an anti-Stat3 monoclonal antibody (mAb), anti-Stat3 polyclonal antibody (pAb) and anti-PKR mAb. Immunoprecipitates were separated by SDS–PAGE and transferred to nylon membranes. The lower form of Stat3, Stat3β, is not recognized in the immunoprecipitation with anti-Stat3 pAb as it is raised against a C-terminal region of Stat3 that is missing in Stat3β. (B) Extracts from different human cell lines immunoprecipitated with anti-Stat3 antibody (mAb) and separated by SDS–PAGE, transferred to nylon membranes and immunoblotted with anti-PKR (pAb, upper panel) and anti-Stat3 (mAb, lower panel). (C) Whole-cell extracts from PKR MEFs or cell lines derived from MEFs were immunoprecipitated with anti-Stat3 antibody (mAb) followed by immunoblotting with anti-PKR (pAb, mouse, upper panel) or anti-Stat3 (mAb, lower panel). A control lane showing immuno precipitated PKR is also shown (last lane) (D) Whole-cell lysates from PKRo/o cells stably expressing human PKR from its natural promoter (Bac9) were immunoprecipitated with anti-PKR (mAb) antibody followed by immunoblotting with anti-PKR (pAb, upper panel) and anti-Stat3 (mAb, lower panel) antibodies. Bac10 cells do not express any PKR and served as control. (E) Serum-starved MEFs were treated with PDGF for different times, whole-cell lysates were immunoprecipitated with anti-Stat3 antibody (mAb), and immunoprecipitates were analyzed by immunoblotting with anti-mouse PKR (pAb, upper panel) or anti-Stat3 (mAb, lower panel) antibodies.
PKR associates with the PDGF receptor
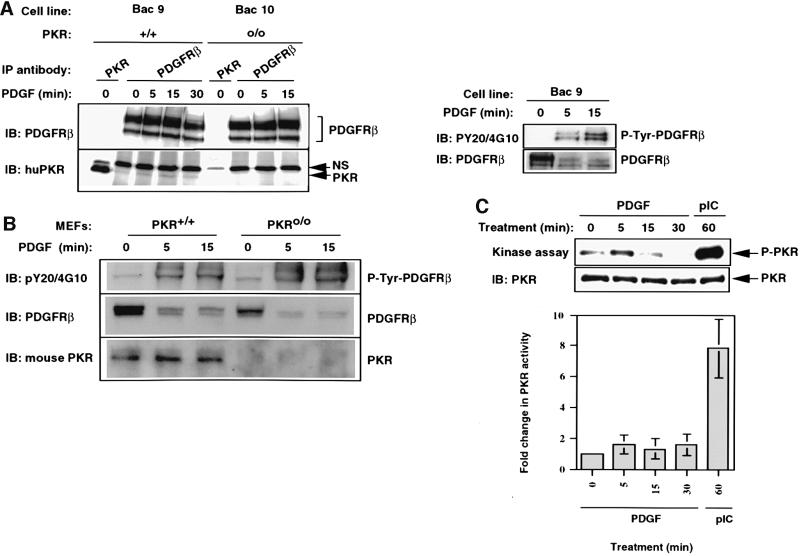
Growth factor-mediated stimulation of receptor tyrosine kinases creates binding sites for Grb2, which in turn recruits the Grb2–Sos complex to the plasma membrane (Schlessinger, 1993; Arvidsson et al., 1994; Claesson-Welsh, 1994). This recruitment of Sos activates Ras, and the activation in turn triggers the Ras–MAPK kinase–Erk pathway. Since we found that PKR is required for the activation of Erks (Figure 1D), we examined whether this signal is generated at the level of the receptor. Extracts from PDGF-treated BAC cells, derived from PKR-null MEFs and reconstituted with human PKR by stable transfection of a BAC clone containing PKR with its native promoter (Bac9), were immunoprecipitated with an antibody against the β-subunit of the PDGF receptor and immunoprobed for the presence of human PKR. PKR co-immunoprecipitated with PDGF β-receptor (Figure 3A, left panel), but this association was ligand independent since PKR co-purified with the receptor in extracts derived from untreated cells (Figure 3A, left panel). This observation was also made in PKR+/+ MEFs (Figure 3B). Since we have already shown a strong ligand-independent interaction between PKR and Stat3 (Figure 2), we examined whether Stat3 could also interact with the receptor. As for PKR, Stat3 also interacted with the PDGF β-receptor in a ligand-independent manner (data not shown; Wang et al., 2000). Despite an earlier claim (Mundschau and Faller, 1995), we were unable to detect any significant PDGF-induced activation of PKR (Figure 3C).
Fig. 3. PKR associates with the PDGF β-receptor in a constitutive manner. (A) Bac9 and Bac10 cells were serum starved overnight and then treated with PDGF (20 ng/ml) as described in Materials and methods. Whole-cell extracts (1 mg, left side; 400 µg, right side) were immunoprecipitated with anti-PDGFR β antibody (pAb). Immunoprecipitates were immunoblotted with PDGFR antibody (top panel, left side) or PKR antibody (BCI, lower panel, left side). The figure on the right shows that the receptor was tyrosine phosphorylated in response to PDGF. The membrane was first probed for phosphotyrosines (PY20 and 4G10 polyclonal) before being stripped and re-probed for total PDGFR β protein. NS refers to a non-specific cross-reacting protein band. (B) PKR+/+ and PKRo/o MEFs were serum starved for 18 h and treated with PDGF (20 ng/ml) for the indicated times. Whole-cell extracts (300 µg) were immunoprecipitated with anti-PDGFR β antibody and subjected to SDS–PAGE analysis. The top portion of the blot was probed initially for tyrosine-phosphorylated receptor (top panel) and then stripped and re-probed for total receptor (middle panel). The lower half of the blot was immunoprobed for PKR using a mouse polyclonal antibody raised against mouse PKR (lower panel). (C) Mouse PKR-null cells reconstituted for PKR by stable transfection of a BAC clone carrying human PKR gene (Bac9) were treated with PDGF (20 ng/ml) or pIC (100 µg/ml) following overnight starvation. In vitro kinase assays were performed on immunoprecipitated PKR (from 150 µg of extract) as described in Materials and methods. Following autoradiography (top panel), the membrane was immunoprobed for the presence of PKR protein in each IP (lower panel). The graph reflects the mean phosphorimager quantitation of autophosphorylated PKR from three separate experiments, each normalized for the amount of immunoprecipitated PKR.
PDGF-enhanced PKR–Stat3 interaction is required for induction of c-fos
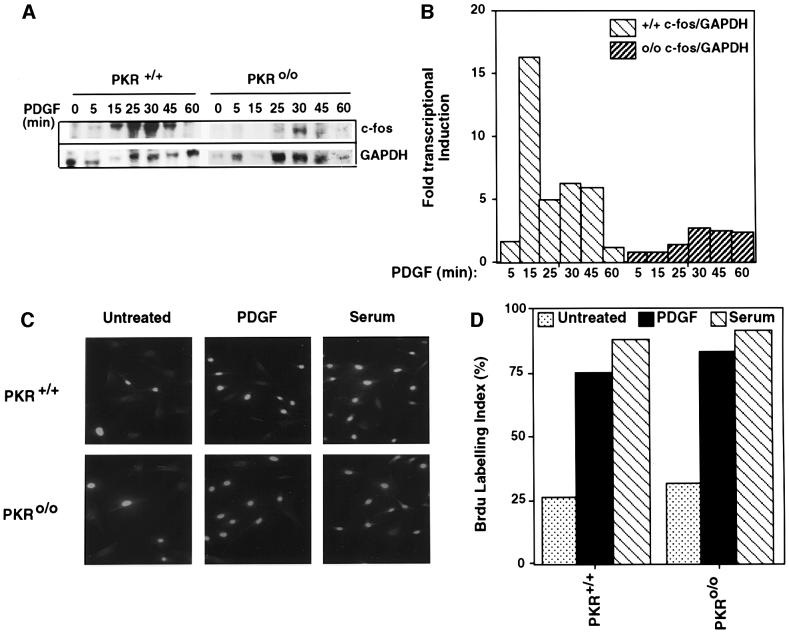
To establish whether the PDGF-enhanced interaction of PKR and Stat3 (Figure 2E) was essential for induction of immediate early gene expression, PKR+/+ and PKRo/o fibroblasts were treated with PDGF for different time periods and total RNA analyzed for c-fos induction. The results (Figure 4A and B) showed a 16-fold induction in PKR wild-type cells in contrast to a 0.8-fold induction in c-fos message in PKR-null cells at 15 min of PDGF treatment. Despite the failure to upregulate c-fos significantly, the mitogenic effects of PDGF in the PKR wild-type and null cells were indistinguishable, as observed by bromodeoxyuridine (BrdU) incorporation studies (Figure 4C and D), consistent with the normal development of PKR-null animals.
Fig. 4. PKR is required for induction of c-fos but not mitogenesis in response to PDGF. (A) Total RNA was isolated from PDGF-treated PKR+/+ and PKRo/o cells and used for RNase protection assays as described in Materials and methods. Products were analyzed on 8% polyacrylamide–8 M urea gels. (B) Quantitation of normalized values for c-fos induction from phosphorimager analysis. (C and D) PKR+/+ and PKRo/o MEFs were grown in 0.3% FBS-containing medium to∼50–60% confluency before the addition of serum (10%) or PDGF (20 ng/ ml) and further incubation for 24 h in the presence of BrdU. Cells were fixed and processed for immunofluorescence as described in Materials and methods. An average of 10 fields was taken to count the percentage of cells that had incorporated BrdU. BrdU labeling index (%) is the ratio of the number of BrdU-labeled cells in a given field and the total number of cells in the same field.
Discussion
In this study, we used murine cells deficient in PKR protein to investigate whether PKR is required for the activation of Stat3 by PDGF. The results indicate that PKR plays a significant role in this signaling pathway. PKR is specifically associated with Stat3 in several cell types (Figure 2A–D), and, in response to PDGF, this interaction increases transiently (Figure 2E). PDGF-induced Stat3 DNA-binding activity is deficient in cells derived from PKR knockout mice (data not shown), and these cells are defective in PDGF-induced Tyr705 and Ser727 phosphorylation of Stat3 (Figure 1A and B). The PKR-mediated phosphorylation of Stat3 on Ser727 is indirect since Stat3 is not a direct substrate of PKR. However, Stat3 serine phosphorylation is inhibited in the presence of MEK inhibitor PD 098059, implicating Erk1 and Erk2 in this process (Figure 1C). PKR is upstream of Erk1/2 since in PKR-null cells these MAP kinases are not activated in response to PDGF (Figure 1D). Although PKR is constitutively associated with the PDGF β-receptor (Figure 3A and B), treatment with PDGF only weakly activates this kinase (Figure 3C). However, PKR-null cells exhibit a 20-fold lower expression of c-fos in response to PDGF compared with their wild-type counterparts (Figure 4A and B).
PKR was found to be associated specifically with Stat3 in several cell types in the absence of stimuli (Figure 2A–D), suggesting that it is pre-associated with monomeric Stat3. However, the Stat3–PKR interaction is increased strongly 5–15 min after PDGF exposure, and is completely lost by 30 min (Figure 2E). This suggests that PKR has increased affinity for the tyrosine-phosphorylated Stat3 homodimer. This interaction may be required to facilitate phosphorylation of Stat3 on Ser727 by Erk1/2 since, in cells lacking PKR, PDGF-induced Stat3 serine phosphorylation as well as MEK kinase activation was deficient (Figure 1B and D). PKR may act as an adaptor recruiting Stat kinases through its pre-association with Stat3, allowing for a rapid response to various stimuli. Mutations in residues both in the vicinity of Tyr705 and more distant from this site have been found to block dimerization of Stat3, suggesting that multiple interactions are involved in dimer formation (Sasse et al., 1997). However, in vitro binding assays of PKR and Stat3 show that the association is not mediated by the C-terminal region of Stat3 (amino acids 717–770) (data not shown). It is possible that the physical association of these two molecules modulates their respective activities. Homodimerization is essential for PKR activation, and heterodimerization with Stat3 could confer unique properties on either PKR or Stat3 at the level of enzyme activity, protein stabilization or subcellular localization.
Several recent publications lend support to the notion that in unstimulated cells, Stats may exist in a complex. Thus, Stat1 and Stat3 have been shown to be pre-associated in a stable complex that is primarily cytoplasmic and will only bind DNA after stimulation with ligand (Haan et al., 2000). This demonstrates that dimerization is necessary but not sufficient for DNA binding. Further more, Stat3 has been shown to homodimerize (Novak et al., 1998; Haan et al., 2000) and to exist in high molecular weight complexes of 200–400 kDa and 1–2 MDa (Ndubuisi et al., 1999) in unstimulated cells. Therefore, it is feasible that PKR exists in a higher order complex with Stat1 and Stat3 heterodimers or Stat3 homodimers. In this context, PKR has been shown to interact physically with Stat1 although, as in the case of Stat3, Stat1 is not a direct substrate for PKR (Wong et al., 1997). However, PKR has been implicated in transcriptional activation of Stat1 through phosphorylation at Ser727 in response to IFN-γ (Ramana et al., 2000) and this probably occurs through a p38MAPK-dependent pathway (Goh et al., 2000).
In MEFs, there are basal levels of phospho-Ser727 Stat3 even in the absence of PKR (Figure 1B), suggesting that constitutive phosphorylation of Stat3 is PKR independent. However, PDGF-induced serine phosphorylation of Stat3 was PKR dependent (Figure 1B). Moreover, the MEK inhibitor PD 098059 inhibits PDGF-induced Erk1/2 activation, and PKR-null cells fail to activate these MAP kinases and exhibit a significantly lower expression level in the Stat3-driven c-fos gene in response to PDGF. Accordingly, we conclude that PKR contributes in a positive manner to the PDGF-induced transcriptional potential of Stat3 through phosphorylation of Ser727 mediated by Erk1/2. These MAP kinases previously have been shown to mediate phosphorylation of Stat3 Ser727 by receptor tyrosine kinases, IL-2 and lymphocyte antigen receptors (Chung et al., 1997; Ng and Cantrell, 1997; Su et al., 1999). In mouse fibroblasts, PKR resides upstream of Erks in PDGF signaling. Recent studies have revealed that in the context of Src transformation, p38 and JNK are key serine kinases that mediate phosphorylation of Stat3 on Ser727 (Turkson et al., 1999). We recently have reported that PKR is required for p38 MAP kinase activation by dsRNA, lipopolysaccharide and pro-inflammatory cytokines (Goh et al., 2000). It remains to be determined whether PKR is upstream of p38 in Src-induced Stat3 serine phosphorylation.
Stat3 DNA-binding activity requires phosphorylation at Tyr705. In this study, we observed that the induced levels of Stat3 homodimers bound to DNA were 7-fold lower in PKR-null cells compared with wild-type cells (data not shown), implicating a role for PKR in PDGF-induced tyrosine phosphorylation of Stat3. PKR recently has been shown to be highly active in breast carcinoma cell lines compared with non-transformed control cells (Kim et al., 2000). It remains possible that the elevated levels of PKR result in Stat3 misregulation. Although PDGF-induced Tyr705 phosphorylation of Stat3 was defective in PKR-null cells, a higher constitutive level of phosphorylated Stat3 (Tyr705) was detected in extracts of these cells (Figure 1A). Since there was a deficiency in DNA binding, there must be other PKR-dependent mechanisms that control Stat3 DNA binding. The PKR-mediated signaling cascade that leads to phosphorylation of Stat3 on Tyr705 in response to PDGF remains to be identified. Several oncogenic tyrosine kinases have been reported to activate Stat3 (Bowman et al., 2000), and direct activation of Stat3 by c-Src has been suggested (Cao et al., 1996; Chaturvedi et al., 1998). Since Src is activated in response to PDGF (Gould and Hunter, 1988; Courtneidge et al., 1993), it is feasible that tyrosine phosphorylation at residue 705 is mediated by this kinase and PKR may be a required component of this event. Wang et al. (2000) have demonstrated recently that in response to PDGF, Src becomes associated with a pre-assembled PDGFR β–Stat3 complex and that ligand-dependent tyrosine phosphorylation of Stat3 on residue 705 is abolished by the chemical inhibition of Src kinase activity. We have shown that PKR associates with PDGFR β in a ligand-independent manner (Figure 3A and B) and have also observed pre-association of Stat3 with PDGFR β (data not shown). Ligand-independent interaction of Stat3 has also been observed with ErbB1 receptors (Olayioye et al., 1999). Since PDGF activation of the receptor does not result in increased interaction of Stat3 and PDGFR β (Wang et al., 2000), the interaction of Stat3 with the receptor may be phosphotyrosine independent. This is supported by results in which catalytically inactive PDGFR β is still able to bind Stat3 (our unpublished observations). Thus, stimulation with PDGF activates Src, which becomes associated with the receptor and promotes phosphorylation of Stat3. A model based on our current study of PDGF-induced c-fos gene activation and the role of PKR in this process is presented in Figure 5. PKR is pre-associated with Stat3 and PDGFR, and, by a yet to be determined mechanism, it may facilitate tyrosine phosphorylation of Stat3 by Src, perhaps by playing a scaffolding role recruiting Stat3 to Src as reported for Jak1 kinase (Zhang et al., 2000). Treatment of 3T3 cells with PDGF previously has been reported to result in activation of PKR (Mundschau and Faller, 1995). However, we were unable to observe a significant increase in PKR activity in response to PDGF in cells derived from MEFs (Figure 3C). This does not exclude an enzymatic role for PKR in PDGF-induced activation of Stat3. It is possible that the constitutive activity of PKR is sufficient to transduce the signal. Currently, there is a debate as to whether the activity of PKR is required in dsRNA signaling to NF-κB or whether it plays a structural role (Chu et al., 1999; Bonnet et al., 2000; Gil et al., 2000; Zamanian-Daryoush et al., 2000). This remains to be examined in PDGF signaling.
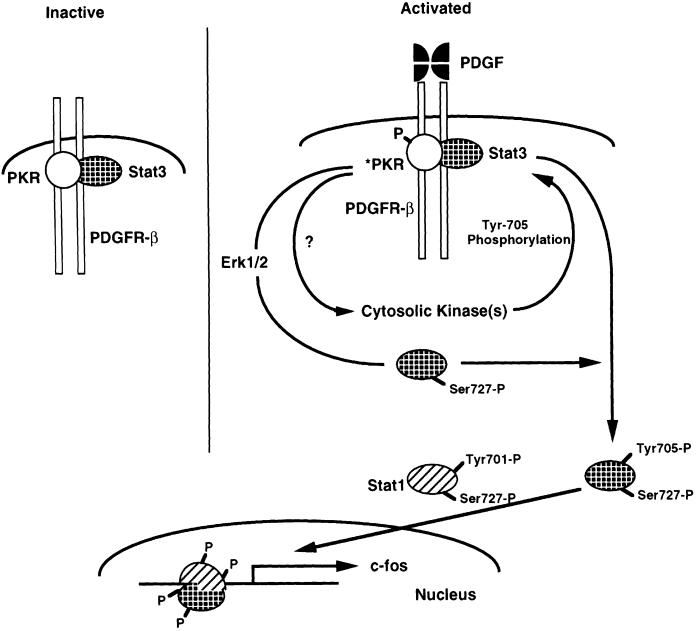
Fig. 5. Schematic representation of the role of PKR in PDGF signaling. PKR and Stat3 are constitutively associated with the PDGF β-receptor. PKR is required for both serine and tyrosine phosphorylation of Stat3 in response to PDGF. The interaction of PKR and Stat3 on the PDGF β-receptor may be a part of a large complex involving additional factors not shown in the model. The open, hatched and striped molecules represent PKR, Stat3 and Stat1 proteins, respectively.
Materials and methods
Cell culture
Cell lines and MEFs derived from isogenic PKR+/+ and PKRo/o littermates were grown as previously described (Yang et al., 1995; Kumar et al., 1997). The mouse cell line Bac9 was derived from PKR-null MEFs and stably transfected with a BAC clone harboring the gene for human PKR and its native promoter using zeocin selection. The cell line Bac10 was selected for similarly but does not express any PKR. Bac cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS) with zeocin (100 µg/ml; InVitrogen) (Z.Xu and B.R.G.Williams, unpublished). Human cell lines except Daudi were grown in DMEM supplemented with 10% heat-inactivated FBS, 2 mM glutamine, 100 U/ml penicillin and 100 µg/ml streptomycin (Gibco-BRL). Daudi cells were maintained at 2.5–10 × 105 cells/ml in RPMI 1640 containing 10% FBS. Unless otherwise specified, cells were grown in 10 cm dishes to ∼60% confluence before placing in 0.3% serum-containing medium for 14–16 h. Serum-deprived cells were treated with PDGF (20 ng/ml) as specified. For assaying Tyr705 and Ser727 phosphorylation of Stat3, MEFs were grown in large plates (150 mm) to 40–60% confluence and treated with PDGF with pre-serum starvation of 16 h.
Reagents
PDGF BB (20 ng/ml; Upstate Biotechnology) was added after serum deprivation for 14–16 h. Cells were pre-treated with 10 µM SB 203580 (p38 inhibitor; Calbiochem) or 30 µM PD 098059 (Erk1/2 inhibitor; Calbiochem) for 30 min prior to stimulation with PDGF BB (20 ng/ml) for 15 min as indicated. Antibodies were obtained from Upstate Biotechnology (monoclonal 4G10), Santa Cruz Biotechnology (polyclonal β PDGFR antibody), Transduction Labs (monoclonal Stat3 and monoclonal PY20), New England Biolabs (phospho-Erk 1/2, pSer727-Stat3, pTyr705-Stat3 and polyclonal Stat3) and Dr Ara Hovanessian (human PKR monoclonal antibody). The rabbit polyclonal antibody against human PKR (BCI) was generated using the dsRNA-binding domain as the immunogen (our laboratory). The polyclonal antibody against mouse PKR was generated in PKR-null mice (our laboratory).
Immunoprecipitation
Cell lysates (500 µg) in 500 µl of ice-cold lysis buffer [50 mM HEPES pH 7.5, 0.5% NP-40, 0.25% deoxycholate, 150 mM NaCl, 2 mM EDTA, 2 µM phenylmethylsulfonyl fluoride (PMSF), 20 µM leupeptin, 50 µM NaF, 2 µM Na3VO4] and extracts were passed through a 25 gauge needle, centrifuged at 10 000 g for 20 min, and the supernatant pre-cleared with pre-immune serum and used for immunoprecipitation with 2–5 µg of Stat3 monoclonal antibody overnight on a rotatory wheel at 4°C. The immune complex was pulled down by 30 µl of protein G–Sepharose (1:1 in lysis buffer, protein G–Sepharose 4 fast flow; Gibco), incubated for 3–4 h at 4°C, centrifuged, and beads were washed three times with 1× lysis buffer, boiled in 1× SDS sample buffer with β-mercaptoethanol and electrophoresed on a 10% SDS–polyacrylamide gel. The proteins were electrotransferred in Immobilon P (Millipore) and visualized by enhanced chemiluminescence (ECL, Amersham Life Science).
Electrophoresis and immunoblotting of protein samples
Protein samples were separated by SDS–PAGE, transferred to polyvinylidene difluoride membranes (Immobilon-P, Millipore), blocked with buffer [1× TBS; 0.1% Triton X-100 containing 5% bovine serum albumin (BSA)] for 1–2 h and then probed with the indicated primary antibody for 3–5 h. The membranes were washed with three changes of wash buffer and proteins detected using the enhanced chemiluminescence reagent (Amersham). Immunoblotting with pSer727-Stat3 and pTyr705-Stat3 antibodies was carried out according to the manufacturer’s instructions (New England Biolabs).
Receptor association studies
Bac9 and Bac10 cells were seeded at 2.2 × 106 on 150 mm tissue culture dishes and grown in 10% FBS-containing DMEM with zeocin (100 µg/ml) to ∼60% confluence (overnight growth). Cells were starved for 18–20 h in 0.3% FBS-containing DMEM in the absence of zeocin before treatment with PDGF (20 ng/ml) in starvation medium for the indicated times. Cells were washed (twice) in cold phosphate-buffered saline (PBS) containing 0.2 mM sodium orthovanadate and scraped in 500 µl of the appropriate lysis buffer. For receptor association, the lysis buffer contained 50 mM HEPES pH 7.5, 0.5% NP-40, 0.25% sodium deoxycholate, 10% glycerol, 150 mM NaCl, 2 mM EDTA, 1 mM PMSF, 1 µg/ml leupeptin, 50 mM NaF and 2 mM sodium orthovanadate. To assess the tyrosine phosphorylation status of the PDGF receptor, cells were lysed in a buffer containing 50 mM Tris–HCl pH 7.4, 1% NP-40, 0.25% sodium deoxycholate, 150 mM NaCl, 1 mM EGTA, 1 mM sodium orthovanadate, 1 mM NaF, 10% glycerol, 1 mM PMSF and 1 µg/ml each of aprotinin, leupeptin and pepstatin. Extracts were stored on ice for 20 min before clarification by centrifugation (18 000 g, 20 min, 4°C). Immunoprecipitations (IPs) were set up on fresh extracts using an anti-PDGFR β polyclonal antibody (sc-432) on 1 mg (for receptor association) or 400 µg of extract (for tyrosine phosphorylation) in a 300 µl volume. Control IPs for PKR were set up on 100 µg of extract using monoclonal antibody against human PKR. The mixture was incubated on ice for 1 h before adding protein G–Sepharose and rotating on a rotatory shaker overnight. IPs were centrifuged at 8000 g for 2–3 min and washed three times with lysis buffer before being subjected to SDS–PAGE analysis (7.5% acrylamide gel). Following transfer to an Immobilon membrane, the top portion was probed for total PDGFR β (association) using polyclonal PDGFR β antibody, or for tyrosine-phosphorylated PDGFR β using a 1:1 mixture of PY20 and 4G10 monoclonal antibodies. The lower portion of the membrane for association studies was immunoprobed for human PKR using the BCI polyclonal antibody. The phosphotyrosine blot was stripped and reprobed for total receptor protein. A similar experiment was performed in PKR wild-type and PKR-null MEFs using 300 µg of whole-cell extracts. PKR was detected with a mouse polyclonal antibody raised against mouse PKR.
Determination of PKR activity by in vitro kinase assay
Bac9 cells seeded on 100 mm tissue culture dishes were grown to 60% confluence before being starved overnight in 0.3% FBS-containing DMEM in the absence of zeocin. Following treatment (20 ng/ml PDGF up to 30 min; 100 µg/ml pIC for 60 min), cells were washed twice in PBS, scraped and lysed in 300 µl of IP lysis buffer [50 mM Tris–HCl pH 7.6, 150 mM NaCl, 10% glycerol, 1% NP-40, 5 mM EDTA, 1 mM dithiothreitol (DTT), 100 mM NaF, 2 mM sodium pyrophosphate, 2 mM sodium orthovanadate, 1 mM PMSF, 10 µg each of aprotinin and leupeptin per ml] and stored on ice for 20 min before clarification by centrifugation (18 000 g, 20 min, 4°C). PKR was immunoprecipitated from 150 µg of total cell extract using a PKR monoclonal antibody. After incubation on ice for 30 min, two volumes of IP lysis buffer and 30 µl of protein G–Sepharose (1:1 in the lysis buffer; Life Technologies) were added. The mixture was rotated on a rotatory shaker overnight, centrifuged at 8000 g for 2–3 min and washed twice with lysis buffer. The beads were washed further (twice) in DBGA buffer (10 mM Tris–HCl pH 7.6, 50 mM KCl, 2 mM MgOAc, 20% glycerol and 7 mM β-mercaptoethanol). A 30 µl aliquot of DBGA was added to the beads with 20 µl of DBGB (2.5 µl of 1 M MnCl2 in 1 ml of DBGA) followed by addition of 5 µl of ATP mix {200 µl of DBGA, 2.5 µl of [γ-32P]ATP (6000 Ci/mmol and 150 µCi/µl), 2 µl of 1 mM ATP}. The mixture was incubated for 30 min at 30°C. After centrifuging at 1000 g for 5 min, the supernatant was discarded, an equal volume of SDS–PAGE buffer was added and phosphoproteins were resolved on 10% SDS–polyacrylamide gels and transferred onto an Immobilon membrane. Following autoradiography and exposure to a phosphorimager screen, the membrane was subjected to immunodetection for the presence of PKR using a polyclonal antibody (BCI; Zamanian-Daryoush et al., 2000). Phosphoproteins were quantitated on a PhosphorImager (Molecular Dynamics).
RNase protection assay
Total RNA was isolated from cells using the Trizol™ reagent as described by the manufacturer (Gibco-BRL). A 5 µg aliquot of total RNA was used for protection assays (Ambion). Riboprobe 1 (299 bases) for c-fos (pTRI-c-fos/exon 4-mouse probe template, Ambion) protected a 250-nucleotide fragment specific for mouse c-fos, and riboprobe 2 (383 bases) for GAPDH (pTRI-GAPDH-mouse antisense control template; Ambion) protected a 316-nucleotide fragment specific for mouse GAPDH. Products were analyzed on 8% polyacrylamide–8 M urea gels. The signal intensity was quantitated by PhosphorImager (Molecular Dynamics).
BrdU labeling and immunofluorescence
PKR+/+ and PKRo/o fibroblasts were grown on coverslips. Cells were seeded at 0.5 × 105 in medium containing 0.3% FBS to ∼50–60% confluence before being serum (10% FBS)/PDGF (20 ng/ml) stimulated for 24 h along with BrdU (100 µM; Amersham Inc.). Cells were washed twice with PBS and fixed for 10 min in cold methanol at –20°C. To analyze for DNA synthesis, cells were incubated for 10 min in 1.5 M HCl, washed three times in PBS and the coverslips were blocked with 3% BSA/0.1% Tween-20 in PBS (PBST). Cells were stained with monoclonal anti-BrdU antibody (Boehringer) and finally with Texas red-conjugated anti-mouse antibody (Vector Labs). Coverslips were mounted on glass slides using Vectashield (Vector Labs) and were examined under a Nikon fluorescence microscope (Model Nikon Microphot FXA).
Acknowledgments
Acknowledgements
This work is supported by grants P01-CA62220 and RO1-AI34039 from the National Institutes of Health.
References
- Arvidsson A.K. et al. (1994) Tyr-716 in the platelet-derived growth factor β-receptor kinase insert is involved in GRB2 binding and Ras activation. Mol. Cell. Biol., 14, 6715–6726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay S.K., de La Motte,C.A. and Williams,B.R. (2000) Induction of E-selectin expression by double stranded RNA and TNF-α is attenuated in murine aortic endothelial cells derived from protein kinase PKR-null mice. J. Immunol., 164, 2077–2083. [DOI] [PubMed] [Google Scholar]
- Bonnet M.C., Weil,R., Dam,E., Hovanessian,A.G. and Meurs,E.F. (2000) PKR stimulates NF-κB irrespective of its kinase function by interacting with IκB kinase complex. Mol. Cell. Biol., 20, 4532–4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton T.G., Zhong,Z., Wen,Z., Darnell,J.E.,Jr, Stahl,N. and Yancopoulos,G.D. (1995) Stat3 activation by cytokines utilizing gp130 and related transducers involves a secondary modification requiring an H7 sensitive kinase. Proc. Natl Acad. Sci. USA, 92, 6915–6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman T., Garcia,R., Turkson,J. and Jove,R. (2000) STATs in oncogenesis. Oncogene, 19, 2474–2488. [DOI] [PubMed] [Google Scholar]
- Bromberg J.F., Wrzeszczynska,M.H., Devgan,G., Zhao,Y., Pestell,R.G., Albanese,C. and Darnell,J.E.,Jr (1999) Stat3 as an oncogene. Cell, 98, 295–303. [DOI] [PubMed] [Google Scholar]
- Cao X., Tay,A., Guy,G.R. and Tan,Y.H. (1996) Activation and association of Stat3 with Src in v-Src-transformed cell lines. Mol. Cell. Biol., 16, 1595–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi P., Reddy,M.V. and Reddy,E.P. (1998) Src kinases and not JAKs activate STATs during IL-3 induced myeloid cell proliferation. Oncogene, 16, 1749–1758. [DOI] [PubMed] [Google Scholar]
- Chen J., Sadowski,H.B., Kohanski,R.A. and Wang,L.H. (1997) Stat5 is a physiological substrate of the insulin receptor. Proc. Natl Acad. Sci. USA, 94, 2295–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin Y.E., Kitagawa,M., Su,W.C., You,Z.H., Iwamoto,Y. and Fu,X.Y. (1996) Cell growth arrest and induction of cyclin-dependent kinase inhibitor p21 WAF1/CIP1 mediated by Stat1. Science, 272, 719–722. [DOI] [PubMed] [Google Scholar]
- Chong K.L., Feng,L., Schappert,K., Meurs,E., Donahue,T.F., Friesen,J.D., Hovanessian,A.G. and Williams,B.R. (1992) Human p68 kinase exhibits growth suppression in yeast and homology to the translational regulator GCN2. EMBO J., 11, 1553–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu W.M., Ostertag,D., Li,Z.W., Chang,L., Chen,Y., Hu,Y., Williams,B., Perrault,J. and Karin,M. (1999) JNK2 and IKKβ are required for activating the innate response to viral infection. Immunity, 11, 721–731. [DOI] [PubMed] [Google Scholar]
- Chung J., Uchida,E., Grammer,T.C. and Blenis,J. (1997) Stat3 serine phosphorylation by Erk-dependent and -independent pathways negatively modulates its tyrosine phosphorylation. Mol. Cell. Biol., 17, 6508–6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesson-Welsh L. (1994) Platelet-derived growth factor receptor signals. J. Biol. Chem., 269, 32023–32026. [PubMed] [Google Scholar]
- Courtneidge S.A., Fumagalli,S., Koegl,M., Superti-Furga,G. and Twamley-Stein,G.M. (1993) The Src family of protein tyrosine kinases: regulation and functions. Dev. Suppl., 57–64. [PubMed] [Google Scholar]
- Darnell J.E.,Jr (1997) STATs and gene regulation. Science, 277, 1630–1635. [DOI] [PubMed] [Google Scholar]
- David M., Wong,L., Flavell,R., Thompson,S.A., Wells,A., Larner,A.C. and Johnson,G.R. (1996) Stat activation by epidermal growth factor (EGF) and amphiregulin. Requirement for the EGF receptor kinase but not for tyrosine phosphorylation sites of JAK1. J. Biol. Chem., 271, 9185–9188. [DOI] [PubMed] [Google Scholar]
- Dever T.E., Chen,J.J., Barber,G.N., Cigan,A.M., Feng,L., Donahue,T.F., London,I.M., Katze,M.G. and Hinnebusch,A.G. (1993) Mammalian eucaryotic initiation factor 2α kinases functionally substitute for GCN2 protein kinase in the GCN4 translational control mechanism of yeast. Proc. Natl Acad. Sci. USA, 90, 4616–4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X.Y. and Zhang,J.J. (1993) Transcription factor p91 interacts with epidermal growth factor receptor and mediates activation of c-fos gene promoter. Cell, 74, 1135–1145. [DOI] [PubMed] [Google Scholar]
- Garcia R., Yu,C.L., Hudnall,A., Catlett,R., Nelson,K.L., Smithgall,T., Fujita,D.J., Ethier,S.P. and Jove,R. (1997) Constitutive activation of Stat3 in fibroblasts transformed by diverse oncoproteins and in breast carcinoma cells. Cell Growth Differ., 8, 1267–1276. [PubMed] [Google Scholar]
- Gil J., Alcami,J. and Esteban,M. (2000) Activation of NF-κB by the dsRNA-dependent protein kinase, PKR, involves the IκB kinase complex. Oncogene, 19, 1369–1378. [DOI] [PubMed] [Google Scholar]
- Goh K.C., deVeer,M.J. and Williams,B.R. (2000) The protein kinase PKR is required for p38 MAPK activation and the innate immune response to bacterial endotoxin. EMBO J., 19, 4292–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouilleux-Gruart V. et al. (1996) Stat-related transcription factors are constitutively activated in peripheral blood cells from acute leukemia patients. Blood, 87, 1692–1697. [PubMed] [Google Scholar]
- Gould K.L. and Hunter,T. (1988) Platelet-derived growth factor induces multisite phosphorylation of pp60c-src and increases its protein-tyrosine kinase activity. Mol. Cell. Biol., 8, 3345–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haan S., Kortylewski,M., Behrmann,I., Muller-Esterl,W., Heinrich,P.C. and Schaper,F. (2000) Cytoplasmic STAT proteins associate prior to activation. Biochem. J., 345, 417–421. [PMC free article] [PubMed] [Google Scholar]
- Hill C.S. and Triesmann,R. (1995) Differential activation of the c-fos promoter elements by serum, lyophosphatidic acid, G proteins and polypeptide growth factors. EMBO J., 14, 5037–5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Yang,M. and May,W.S. (1999) RAX, a cellular activator for double-stranded RNA-dependent protein kinase during stress signaling. J. Biol. Chem., 274, 15427–15432. [DOI] [PubMed] [Google Scholar]
- Katze M.G. (1992) The war against the interferon-induced dsRNA-activated protein kinase: can viruses win? J. Interferon Res., 12, 241–248. [DOI] [PubMed] [Google Scholar]
- Kim H.R., Upadhyay,S., Li,G., Palmer,K.C. and Deuel,T.F. (1995) Platelet-derived growth factor induces apoptosis in growth-arrested murine fibroblasts. Proc. Natl Acad. Sci. USA, 92, 9500–9504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.S., Forman,A.P., Mathews,M.B. and Gunnery,S. (2000) Human breast cancer cells contain elevated levels and activity of the protein kinase, PKR. Oncogene, 19, 3086–3094. [DOI] [PubMed] [Google Scholar]
- Koromilas A.E., Roy,S., Barber,G.N., Katze,M.G. and Sonenberg,N. (1992) Malignant transformation by a mutant of the IFN-inducible dsRNA-dependent protein kinase. Science, 257, 1685–1689. [DOI] [PubMed] [Google Scholar]
- Kumar A., Haque,J., Lacoste,J., Hiscott,J. and Williams,B.R. (1994) Double-stranded RNA-dependent protein kinase activates transcription factor NF-κB by phosphorylating IκB. Proc. Natl Acad. Sci. USA, 91, 6288–6292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A. et al. (1997) Deficient cytokine signaling in mouse embryo fibroblasts with a targeted deletion in the PKR gene: role for IRF-1 and NF-κB. EMBO J., 16, 406–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaman D.W., Pisharody,S., Flickinger,T.W., Commane,M.A., Schlessinger,J., Kerr,I.M., Levy,D.E. and Stark,G.R. (1996) Roles of JAKs in activation of Stats and stimulation of c-fos gene expression by epidermal growth factor. Mol. Cell. Biol., 16, 369–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marais R., Wyne,J. and Triesmann,R. (1993) The SRF accessory protein Elk-1 contains a growth factor-regulated transcriptional activation domain. Cell, 73, 381–393. [DOI] [PubMed] [Google Scholar]
- Meurs E.F., Galabru,J., Barber,G.N., Katze,M.G. and Hovanessian,A.G. (1993) Tumor suppressor function of the interferon-induced double stranded RNA-activated protein kinase. Proc. Natl Acad. Sci. USA, 90, 232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migone T.S., Lin,J.X., Cereseto,A., Mulloy,J.C., O’Shea,J.J., Franchini,G. and Leonard,W.J. (1995) Constitutively activated Jak–Stat pathway in T-cells transformed with HTLV-1. Science, 269, 79–81. [DOI] [PubMed] [Google Scholar]
- Mori S., Ronnstrand,L., Yokote,K., Engstrom,A., Courtneidge,S.A., Claesson-Welsh,L. and Heldin,C.H. (1993) Identification of two juxtamembrane autophosphorylation sites in the PDGF β-receptor; involvement in the interaction with Src family of tyrosine kinases. EMBO J., 12, 2257–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M. et al. (1993) The protein tyrosine kinase JAK1 complements defects in interferon-α/β and -γ signal transduction. Nature, 366, 129–135. [DOI] [PubMed] [Google Scholar]
- Mundschau L.J. and Faller,D.V. (1995) Platelet derived growth factor signal transduction through the interferon inducible kinase PKR. J. Biol. Chem., 270, 3100–3106. [DOI] [PubMed] [Google Scholar]
- Ndubuisi M.I., Guo,G.G., Fried,V.A., Etlinger,J.D. and Sehgal,P.B. (1999) Cellular physiology of STAT3: where’s the cytoplasmic monomer? J. Biol. Chem., 274, 25499–25509. [DOI] [PubMed] [Google Scholar]
- Ng J. and Cantrell,D. (1997) Stat3 is a serine kinase target in T lymphocytes. Interleukin 2 and T cell antigen receptor signals converge upon serine 727. J. Biol. Chem., 272, 24542–24549. [DOI] [PubMed] [Google Scholar]
- Nielsen M., Kaltoft,K., Nordahl,M., Ropke,C., Geisler,C., Mustelin,T., Dobson,P., Svejgaard,A. and Odum,N. (1997) Constitutive activation of a slowly migrating form of Stat3 in mycosis fungoides: tyrphostin AG490 inhibits Stat3 activation and growth of mycosis fungoides tumor cell lines. Proc. Natl Acad. Sci. USA, 94, 6764–6769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak U., Nice,E., Hamilton,J.A. and Paradiso,L. (1996) Requirement for Y706 of the murine (or Y708 of the human) CSF-1 receptor for Stat1 activation in response to CSF-1. Oncogene, 13, 2607–2613. [PubMed] [Google Scholar]
- Novak U., Ji,H., Kanagasundaram,V., Simpson,R. and Paradiso,L. (1998) STAT3 forms stable homodimers in the presence of divalent cations prior to activation. Biochem. Biophys. Res. Commun., 247, 558–563. [DOI] [PubMed] [Google Scholar]
- Olayioye M.A., Beuvink,I., Horsch,K., Daly,J.M. and Hynes,N.E. (1999) ErbB receptor-induced activation of stat transcription factors is mediated by Src tyrosine kinases. J. Biol. Chem., 274, 17209–17218. [DOI] [PubMed] [Google Scholar]
- Ramana C.V., Grammatikakis,N., Chernov,M., Nguyen,H., Goh,K.C., Williams,B.R. and Stark,G.R. (2000) Regulation of c-myc expression by IFN-γ through Stat1-dependent and -independent pathways. EMBO J., 19, 263–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson L.M., Kerppola,T.K., Vendrell,M., Luk,D., Smeyne,R.J., Bocchiaro,C., Morgan,J.I. and Curran,T. (1995) Regulation of c-fos expression in transgenic mice requires multiple interdependent transcription control elements. Neuron, 14, 241–252. [DOI] [PubMed] [Google Scholar]
- Sachsenmaier C., Sadowski,H.B. and Cooper,J.A. (1999) STAT activation by the PDGF receptor requires juxtamembrane phosphorylation sites but not Src kinase activation. Oncogene, 18, 3583–3592. [DOI] [PubMed] [Google Scholar]
- Samuel C.E. (1993) The eIF-2α protein kinase, regulators of translation in eucaryotes from yeasts to humans. J. Biol. Chem., 268, 7603–7606. [PubMed] [Google Scholar]
- Sartor C.I., Dziubinski,M.L., Yu,C.L., Jove,R. and Ethier,S.P. (1997) Role of epidermal growth factor receptor and Stat3 activation in autonomous proliferation of SUM-102PT human breast cancer cells. Cancer Res., 57, 978–987. [PubMed] [Google Scholar]
- Sasse J., Hemmann,U., Schwartz,C., Schniertshauer,U., Heesel,B., Landgraf,C., Schneider-Mergener,J., Heinrich,P.C. and Horn,F. (1997) Mutational analysis of acute-phase response factor/Stat3 activation and dimerization. Mol. Cell. Biol., 17, 4677–4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler C. and Darnell,J.E.,Jr (1995) Transcriptional responses to polypeptide ligands: the JAK–STAT pathway. Annu. Rev. Biochem., 64, 621–651. [DOI] [PubMed] [Google Scholar]
- Schlessinger J. (1993) How receptor tyrosine kinases activate Ras. Trends Biochem. Sci., 18, 273–275. [DOI] [PubMed] [Google Scholar]
- Su L., Rickert,R.C. and David,M. (1999) Rapid STAT phosphorylation via the B cell receptor. Modulatory role of CD19. J. Biol. Chem., 274, 31770–31774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W.C., Kitagawa,M., Xue,N., Xie,B., Garofalo,S., Cho,J., Deng,C., Horton,W.A. and Fu,X.Y. (1997) Activation of Stat1 by mutant fibroblast growth-factor receptor in thanatophoric dysplasia type II dwarfism. Nature, 386, 288–292. [DOI] [PubMed] [Google Scholar]
- Turkson J. et al. (1999) Requirement for Ras/Rac1-mediated p38 and c-Jun N-terminal kinase signaling in Stat3 transcriptional activity induced by the Src oncoprotein. Mol. Cell. Biol., 19, 7519–7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignais M.L. and Gilman,M. (1999) Distinct mechanisms of activation of Stat1 and Stat3 by platelet-derived growth factor receptor in a cell free system. Mol. Cell. Biol., 19, 3727–3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignais M.L., Sadowski,H.B., Watling,D., Rogers,N.C. and Gilman,M. (1996) Platelet-derived growth factor induces phosphorylation of multiple JAK family of kinases and Stat proteins. Mol. Cell. Biol., 16, 1759–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner B.J., Hayes,T.E., Hoban,C.J. and Cochran,B.H. (1990) The SIF binding element confers sis/PDGF inducibility onto c-fos promoter. EMBO J., 9, 4477–4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.-Z., Wharton,W., Garcia,R., Kraker,A., Jove,R. and Pledger,W.J. (2000) Activation of Stat3 preassembled with platelet-derived growth factor β receptors requires Src kinase activity. Oncogene, 19, 2075–2085. [DOI] [PubMed] [Google Scholar]
- Wen Z., Zhong,Z. and Darnell,J.E.,Jr (1995) Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell, 82, 241–250. [DOI] [PubMed] [Google Scholar]
- Williams B.R.G. (1995) The role of the dsRNA-activated kinase, PKR, in signal transduction. Semin. Virol., 6, 191–202. [Google Scholar]
- Williams B.R.G. (1997) Role of the double-stranded RNA-activated protein kinase (PKR) in cell regulation. Biochem. Soc. Trans, 25, 509–513. [DOI] [PubMed] [Google Scholar]
- Williams B.R.G. (1999) PKR; a sentinal kinase for cellular stress. Oncogene, 18, 6112–6120. [DOI] [PubMed] [Google Scholar]
- Wong A.H., Tam,N.W., Yang,Y.L., Cuddihy,A.R., Li,S., Kirchhoff,S., Hauser,H., Decker,T. and Koromilas,A.E. (1997) Physical association between Stat1 and the interferon-inducible protein kinase PKR and implications for interferon and double stranded RNA signaling pathways. EMBO J., 16, 1291–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.L., Reis,L.F., Pavlovic,J., Aguzzi,A., Schafer,R., Kumar,A., Williams,B.R., Aguet,M. and Weissmann,C. (1995) Deficient signaling in mice devoid of double stranded RNA-dependent protein kinase. EMBO J., 14, 6095–6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.L., Meyer,D.J., Campbell,G.S., Larner,A.C., Carter-Su,C., Schwartz,J. and Jove,R. (1995) Enhanced DNA binding activity of a Stat3-related protein in cells transformed by the Src oncoprotein. Science, 269, 81–83. [DOI] [PubMed] [Google Scholar]
- Zamanian-Daryoush M., Mogensen,T.H., DiDonato,J.A. and Williams,B.R. (2000) NF-κB activation by double-stranded-RNA-activated protein kinase (PKR) is mediated through NF-κB-inducing kinase and IκB kinase. Mol. Cell. Biol., 20, 1278–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Turkson,J., Carter-Su,C., Smithgall,T., Levitzki,A., Kraker,A., Krolewski,J.J., Medveczky,P. and Jove,R. (2000) Activation of Stat3 in v-Src-transformed fibroblasts requires cooperation of Jak1 kinase activity. J. Biol. Chem., 275, 24935–24944. [DOI] [PubMed] [Google Scholar]
- Zinn K., Keller,A., Whittemore,L.A. and Maniatis,T. (1988) 2-Aminopurine selectively inhibits the induction of β-interferon, c-fos and c-myc gene expression. Science, 240, 210–213. [DOI] [PubMed] [Google Scholar]