Abstract
The cyclin-dependent kinase inhibitor p21WAF1/CIP1 is a key regulator of cell-cycle progression and its expression is tightly regulated at the level of transcription and by proteasome-dependent proteolysis. The turnover of p21WAF1/CIP1 by proteasomes does not always require the ubiquitylation of p21WAF1/CIP1 suggesting that there could be an alternative pathway into the proteasome. Here we show that the C8 α-subunit of the 20S proteasome interacts with the C-terminus of p21WAF1/CIP1 and mediates the degradation of p21WAF1/CIP1. A small deletion in this region that disrupts binding to C8 increased the half-life of p21WAF1/CIP1 expressed in vivo. In contrast a deletion that increased the affinity between C8 and p21WAF1/CIP1 significantly reduced the stability of the latter. These data suggest that interaction with a 20S proteasome α-subunit is a critical determinant of p21WAF1/CIP1 turn-over and show how non-ubiquitylated molecules might bypass the 19S regulator of the proteasome and become targeted directly to the 20S, core protease. Consistent with this, p21WAF1/CIP1 was degraded rapidly by purified 20S proteasomes in a manner that was dependent on the C8-interaction domain.
Keywords: C8/degradation signal/p21WAF1/CIP1/proteasome
Introduction
During the non-lysosomal turnover of many proteins in eukaryotic cells, covalent ligation of multiple ubiquitin molecules provides the signal for recognition by the 26S proteasome. This molecular machine is a multi-subunit complex in which two 19S regulatory ‘cap’ structures sit each end of a barrel-shaped chamber called the 20S proteasome. The 19S ‘caps’ mediate the recognition of polyubiquitin and also substrate unfolding through the action of a ring of ATPase molecules adjacent to the 20S chamber. The unfolded substrate is then thought to be threaded into the inner chamber of the 20S sub-complex where proteolysis takes place (reviewed in Baumeister et al., 1998; Thrower et al., 2000; Verma and Deshaies, 2000).
With the exception of ornithine decarboxylase it has been assumed that ubiquitylation and ATPase-mediated unfolding are essential pre-requisites for most proteins destined for proteasome-mediated proteolysis (Baumeister et al., 1998; Thrower et al., 2000; Verma and Deshaies, 2000). However, recently the cyclin-dependent kinase (cdk) inhibitor p21WAF1/CIP1 was shown to be degraded by the proteasome in a ubiquitin-independent manner. Specifically, p21WAF1/CIP1 in which all the potential lysine targets for ubiquitylation were mutated was shown to remain an effective substrate for proteasome-mediated degradation. In that study neither the polyubiquitin-independent targeting signal in p21WAF1/CIP1 nor the factor within the proteasome that recognizes it were identified (Sheaff et al., 2000). Although various other studies have implicated the C-terminus of p21WAF1/CIP1 in the regulation of its stability and there has been speculation that this ubiquitin-independent degradation might be mediated by an interaction with an ATPase of the 19S proteasome cap, the molecular details of p21WAF1/CIP1 turnover remain poorly defined (Cayrol and Ducommun, 1998; Rousseau et al., 1999; Verma and Deshaies, 2000). Here we show that C8, an α-subunit of the 20S proteasome, can bind to the C-terminus of p21WAF1/CIP1. In vivo, the turnover of p21WAF1/CIP1 correlates with this binding and in vitro, 20S proteasomes purified away from 19S regulatory complexes can rapidly degrade p21WAF1/CIP1 only if the C8-interaction domain is intact.
Results
The C8 subunit of 20S proteasomes binds to the C-terminus of p21WAF1/CIP1
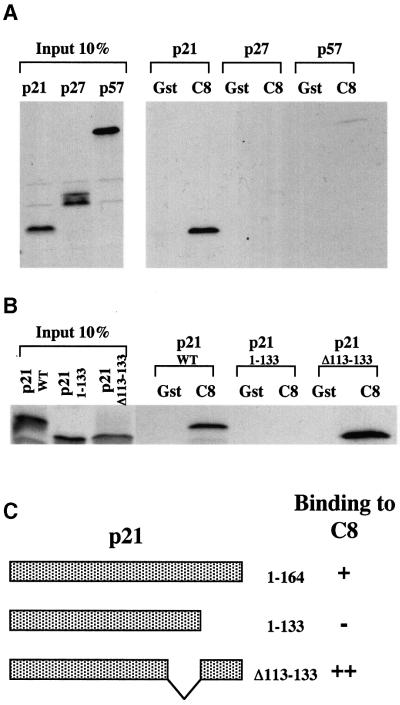
In order to gain additional insight into how the turnover of proteins that regulate the cell cycle might be controlled, interactions between a variety of regulatory factors and proteasome subunits were investigated. This revealed that in vitro-translated p21WAF1/CIP1, but not the closely related cdk inhibitors p27KIP1 and p57KIP2, could bind to a fusion between glutathione S-transferase (GST) and the C8 α-subunit of the 20S core protease (Figure 1A and data not shown). C8 (also known as HsC8, α7 and C1/PRS1) is one of the α-subunits that form the two outer seven-membered rings flanking the catalytic β-subunits in the cylindrical stack making up the 20S proteasome (Tamura et al., 1991; Akioka et al., 1995; Baumeister et al., 1998; Gerards et al., 1998). Like the other α-subunits, C8 lacks any known proteolytic activity but it may have an important role in the initiation of ring formation and 20S proteasome assembly (Gerards et al., 1998). Since it was reported that the C-terminus of p21WAF1/CIP1 is an important determinant of its stability (Cayrol and Ducommun, 1998), the ability of a truncated p21WAF1/CIP1 mutant (aa 1–133) to bind to GST–C8 was tested. This C-terminally deleted polypeptide was unable to bind to GST–C8. In contrast, another mutant of p21WAF1/CIP1 in which amino acids 113–133 are deleted showed an increased affinity for GST–C8 (Figures 1B and 3C).
Fig. 1. p21 interacts with GST–C8. (A) Bacterially expressed GST or GST–C8 fusion protein (C8) were incubated with equal amounts of [35S]methionine-labelled p21, p27 or p57. Ten per cent of the in-put protein in each binding reaction is shown. Bound proteins were resolved by SDS–PAGE (12%). (B) The last 31 aa of p21 are essential for the binding with GST–C8. Bacterially expressed GST or GST–C8 fusion protein (C8) were incubated with equal amounts of [35S]methionine-labelled p21 WT (1–164), p21 (1–133) or p21 (Δ113–133). (C) Results shown in (B) are summarized. The schematic diagram shows that the C-terminus truncated form (aa 1–133) of p21 is unable to bind to GST–C8 (–) but the deleted form of p21 (deleted from aa 113–133) is able to interact with GST–C8 (++) even better than the WT p21 (aa 1–164) (+).
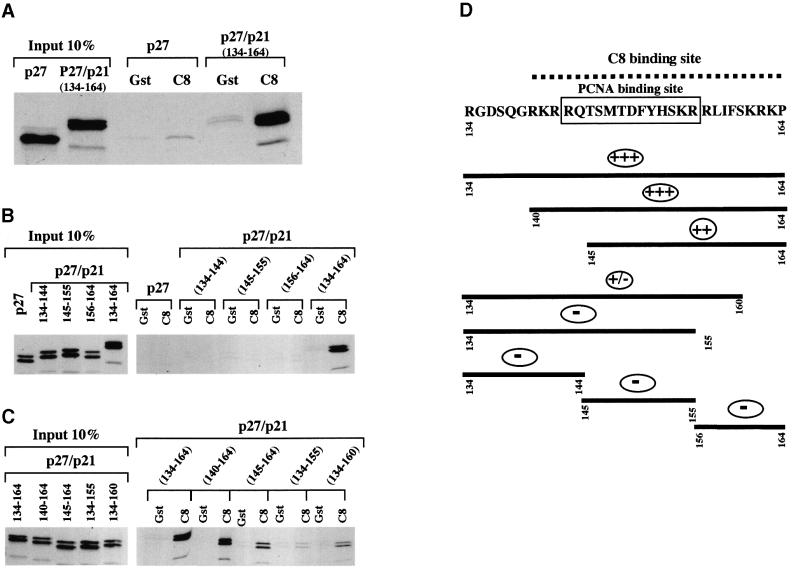
Having established that amino acids 134–164 of p21WAF1/CIP1 were necessary for the interaction with C8 (Figure 1C), a chimeric protein was produced in order to test whether these 31 residues were also sufficient for binding. A recombinant of p27KIP1 (which does not bind to C8) with amino acids 134–164 of p21WAF1/CIP1 fused at the C-terminus was found to bind GST–C8 (Figure 2A). We concluded that the C-terminal 31 amino acids of p21WAF1/CIP1 include a domain that is both necessary and sufficient for the in vitro interaction with C8.
Fig. 2. The last 31 aa of p21 are essential and sufficient for the binding with GST–C8. (A) Bacterially expressed GST or GST–C8 fusion protein (C8) were incubated with equal amounts of [35S]methionine-labelled p27 or p27/p21 (134–164) fusion protein. (B and C) Mapping of the C8 binding site in p21 shows that the region comprising aa 140–164 of p21 are essential for an efficient interaction with C8. Bacterially expressed GST or GST–C8 fusion protein (C8) were incubated with equal amounts of [35S]methionine-labelled p27, p27/p21 (134–144), p27/p21 (145–155), p27/p21 (156–164) or p27/p21 (134–164) fusion proteins (B) and equal amounts of [35S]methionine-labelled p27/p21 (134–164), p27/p21 (140–164), p27/p21 (145–164), p27/p21 (134–155) or p27/p21 (134–160) fusion proteins (C). (D) Results shown in panels B and C are summarized. The schematic diagram shows that the C8 binding site (dashed line) overlaps the PCNA binding site (boxed). (–) indicates no binding and (+) indicates binding to C8.
To define further the extent of the domain required for this interaction, shorter peptides from p21WAF1/CIP1 fused to the C-terminus of p27KIP1 were generated. The results of GST–C8 binding assays (shown in Figure 2B and C and summarized in Figure 2D) revealed that the minimal domain necessary for efficient interaction with C8 is located in the last 25 amino acids at the C-terminus of p21WAF1/CIP1. A comparison of fusion proteins including the fragments 140–164, 145–164, 134–160 and 134–155 suggested that the residues from 156–161 were critical and that the flanking residues were also necessary for efficient binding. The importance of amino acids 156–161 was confirmed when it was shown that the p21WAF1/CIP1 polypeptide Δ156–161 was highly impaired in its ability to bind GST–C8 (Figure 3A and C).
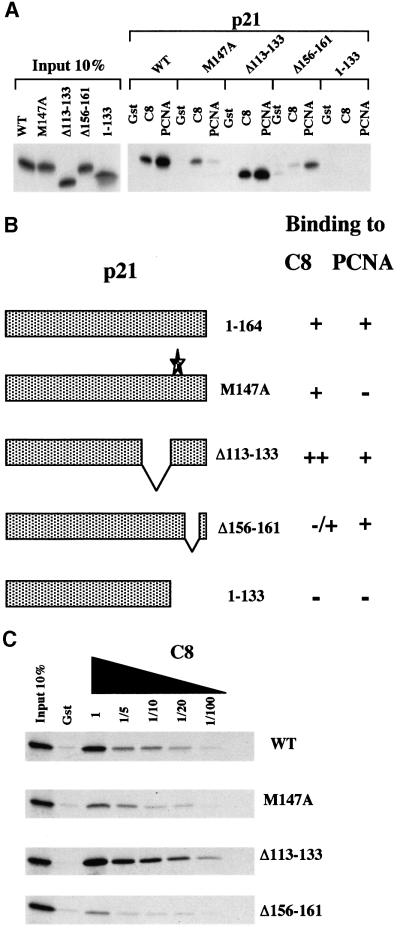
Fig. 3. (A) Comparison of p21 mutants for the interaction with GST–C8 or GST–PCNA. Bacterially expressed GST, GST–C8 (C8) and GST–PCNA (PCNA) fusion proteins were incubated with equal amounts of [35S]methionine-labelled p21 WT, p21 (M147A), p21 (Δ113–133), p21 (Δ156–161) or p21 (1–133). (B) Results shown in (A) are summarized. The schematic diagram shows that p21 (M147A), which does not interact (–) with PCNA, still interacts with C8 (+), and in contrast, p21 (Δ156–161), which does not interact with C8, still interacts with PCNA. (C) Affinity of p21 mutants for GST–C8. Bacterially expressed GST and GST–C8 (C8) were incubated with equal amounts of [35S]methionine-labelled p21 WT, p21 (M147A), p21 (Δ113–133), p21 (Δ156–161) or p21 (1–133). Various dilutions of the GST–C8 fusion protein (as indicated) were used to compare the affinity of the p21 mutants for the C8 fusion protein.
The C8 interaction domain of p21WAF1/CIP1 overlaps the PCNA-binding site
The C-terminus of p21WAF1/CIP1 contains the binding domain for the proliferating cell nuclear antigen (PCNA) (Nakanishi et al., 1995; Sherr and Roberts, 1995; Cayrol and Ducommun, 1998; Rousseau et al., 1999). The site which binds PCNA has been been intensively studied and is located between amino acids 143–155 (see Figure 2D) suggesting that residues required for binding to C8 overlap the PCNA interaction domain. Mutational analysis showed that although the interaction domains overlap, they are operationally distinct. Two point mutations of p21WAF1/CIP1, M147A and F150A, largely abolished the binding of p21WAF1/CIP1 to GST–PCNA, but had only a very slight effect on the interaction with GST–C8 (Figure 3A, data not shown and Cayrol and Ducommun, 1998). Conversely, the deletion mutant Δ156–161 exhibited severely reduced binding to GST–C8 but bound to GST–PCNA almost as effectively as wild-type (WT) p21WAF1/CIP1 (Figure 3A and summarized in Figure 3B). The relative affinity of these mutants was confirmed by titrating the amount of GST–C8 in the binding reactions (Figure 3C).
The stability of p21WAF1/CIP1 is determined by the C8-interaction domain but is largely independent of nuclear localization
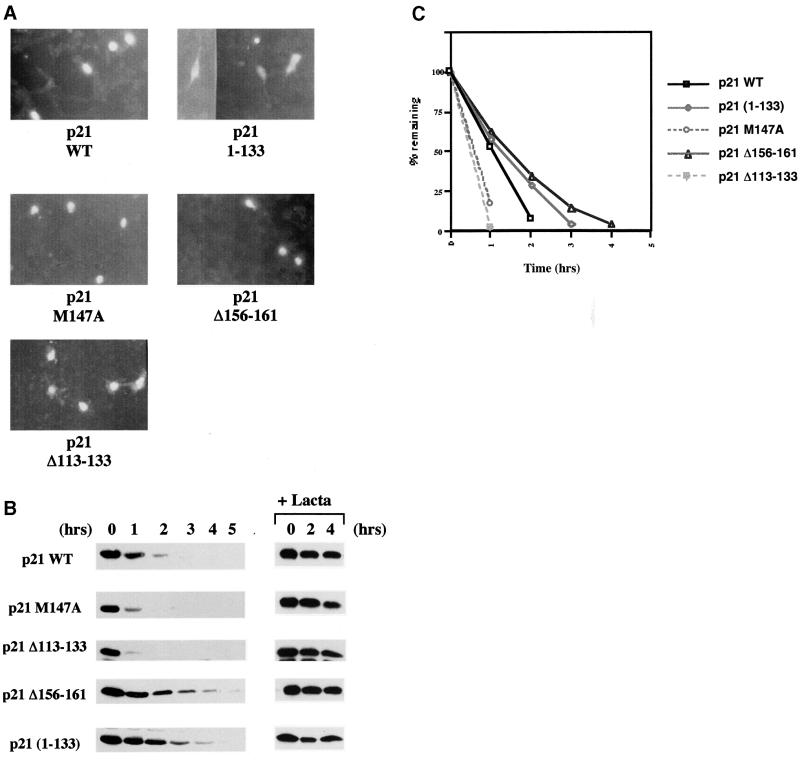
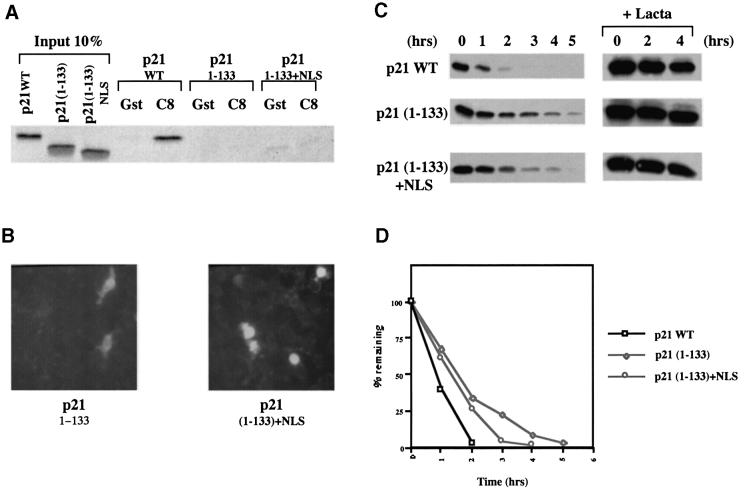
It has been suggested that the stability of p21WAF1/CIP1—so far as it is determined by proteasome-mediated proteolysis—depends on its localization to the nucleus (Sheaff et al., 2000). The cellular distribution of the various mutants analysed here was therefore established by transient transfection and immunofluorescence labelling. The nuclear localization of WT and M147A was confirmed and it was revealed that both the C8-super-binding (Δ113–133) and -weakly-binding (Δ156–161) deletion mutants were similarly expressed in the nucleus. The truncated 1–133 p21WAF1/CIP1, which lacks the putative nuclear localization signal (NLS) (Cayrol and Ducommun, 1998; Sheaff et al., 2000), was confirmed as having both a cytoplasmic and nuclear distribution (Figure 4A). The ability to bind C8 and/or PCNA per se had no direct influence on the localization of p21WAF1/CIP1 to the nucleus.
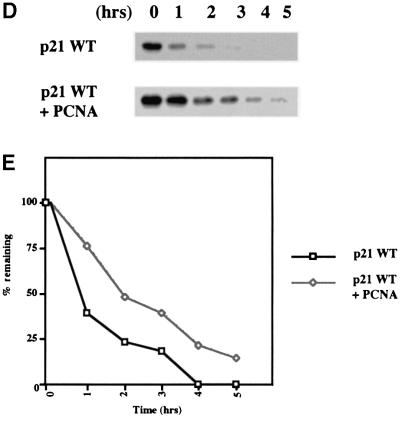
Fig. 4. (A) Immunofluorescence staining of NIH 3T3 cells transfected with 1 µg of each pSG5-p21 plasmid DNA showing the nuclear localization of p21 (M147A), p21 (Δ156–161) and p21 (Δ113–133) mutants. The truncated mutant (1–133), that lacks the predicted NLS, localized throughout the cell. (B) Effect of mutations on the half-life of p21 proteins. Twenty-four hours after transfection, DG75 cells were treated with the protein synthesis inhibitor cycloheximide. Extracts prepared from transfected cells collected at indicated times after cycloheximide treatment were analysed by western blotting with rabbit anti-p21 antibody. Endogenous p21 expression was not detected in transfected cells with pSG5 empty vector DNA (data not shown). p21 (M147A) and p21 (Δ113–133) exhibited shorter half-lives than WT p21. In contrast, p21 (Δ156–161) and p21 (1–133) exhibited extended half-lives. Treatment of cells with lactacystin (an irreversible and specific inhibitor of the proteasome activity) prior to addition of cycloheximide resulted in stabilization of proteins expressed by pSG5-p21 constructs. Protein extracts were prepared at the indicated times after treatment with cycloheximide. (C) Enhanced chemiluminescence (ECL) signals shown in example (B) were quantified using a GS 300 scanning densitometer (Hoeffer) and values were plotted as the percentage of the signal at t = 0 (given an arbitrary value of 100%) remaining at the times indicated. Similar experiments were performed at least three times and they always produced similar results. (D) Effect of PCNA expression on the half-life of p21. Twenty-four hours after cotransfection, DG75 cells were treated as described in (B) and analysed by western blotting for p21. (E) ECL signals shown in (D) were quantified as described in (C). Similar experiments were performed twice and produced comparable results.
In order to address the critical question of whether the interaction with C8 affects the turnover of p21WAF1/CIP1 in vivo, transient expression studies were performed. DG75 lymphoma-derived B cells were transfected by electroporation with plasmid DNA corresponding to WT p21WAF1/CIP1 or the various mutants described above. It should be noted that no endogenous p21WAF1/CIP1 was detected in the DG75 cells used here. After transfection, the cells were incubated for 24 h then, after the addition of cycloheximide to block further protein synthesis, the stability of the newly synthesized exogenous polypeptides was assayed by western blotting at the time points indicated. This revealed a striking inverse correlation between the ability of the p21WAF1/CIP1 polypeptides/mutants to interact with GST–C8 and their stability in the transfected cells (Figure 4B and C). A comparison of the rate of turnover (Figure 4C) shows that the p21WAF1/CIP1 mutants that are defective for C8 binding, (1–133 and Δ156–161) have an extended half-life compared with WT p21WAF1/CIP1. Although Δ156–161 has residual C8-binding activity, we suggest that it is slightly more stable than 1–133 because only the latter has lost the ability to bind PCNA and PCNA has been shown to stabilize p21WAF1/CIP1 in vivo (Cayrol and Ducommun, 1998). Another mutant polypeptide, 1–155, that fails to bind C8 is also more stable than 1–133, presumably because it also retains PCNA binding (data not shown). In contrast to the mutants that have impaired binding, the Δ113–133 mutant that has a high affinity for C8 was very unstable. The stability of the M147A mutant that no longer binds PCNA was intermediate between WT and the very unstable Δ113–133. This is also consistent with the previously published data showing PCNA can modulate the turnover of p21WAF1/CIP1 (Cayrol and Ducommun, 1998) and the results presented in Figure 4D and E. Here, cotransfection of a PCNA expression vector with pSG5-p21 in the standard half-life assay resulted in a significant stabilization of the ectopically expressed p21WAF1/CIP1. These results all support the hypothesis that the interaction with C8 directs p21WAF1/CIP1 for proteasome-mediated turnover. This C-terminal region is an important determinant of p21WAF1/CIP1 stability and behaves as a degradation signal or instability domain that can be suppressed by an interaction with PCNA. However, all these proteins were stabilized by the addition of the proteasome inhibitor lactacystin (Figure 4B) which indicates that ultimately they are all degraded by the proteasome system and that PCNA and C8 binding only modulate the rate of turnover.
As discussed above, it has been suggested that proteasomal turnover of p21WAF1/CIP1 depends largely upon its nuclear localization (Sheaff et al., 2000). A truncated polypeptide [p21WAF1/CIP1 (1–141)] that lacks both an NLS and the C8-interaction domain was shown to have an increased half-life. This was thought to be due to its exclusion from the nucleus. In the light of the analysis of the Δ156–161 mutant presented here, we suggest this interpretation is over-simplified. Since Δ156–161 has a nuclear distribution but an extended half-life, the combined data show that p21WAF1/CIP1 might require both a nuclear distribution and C8-interaction domain for efficient proteasome-mediated turnover. Alternatively, the NLS (–KKKRKV–) that was added to 1–141 by Sheaff and colleagues to redirect the protein to the nucleus may also have restored C8 binding. In order to test this possibility a recombinant molecule was created in which the same NLS was fused to p21WAF1/CIP1 1–133 (which also lacks the NLS and C8 binding site). Interaction experiments showed that this protein remained unable to bind GST–C8 (Figure 5A) and despite its nuclear localization (Figure 5B), it was much more stable than WT p21WAF1/CIP1 and only slightly less stable than p21WAF1/CIP1 1–133 (Figure 5C and D). We conclude that while targeting to the nucleus may slightly enhance the turnover of p21WAF1/CIP1, the ability to interact with C8 is undoubtedly a major determinant of stability associated with the C-terminus.
Fig. 5. (A) Addition of an SV40 NLS does not affect the ability of p21 (1–133) to interact with GST–C8. Bacterially expressed GST or GST–C8 fusion protein (C8) were incubated with equal amounts of [35S]methionine-labelled p21 WT, p21 (1–133) or p21 (1–133 + NLS). (B) Immuno fluorescence staining of NIH 3T3 cells, transfected with either 1 µg of pSG5-p21 (1–133) or 1 µg of pSG5-p21 (1–133 + NLS) plasmid DNA, showing that the addition of a SV40 NLS re-directs the protein to the nucleus. (C) The effect of an SV40 NLS on the stability of p21 (1–133) protein. Twenty-four hours after transfection, DG75 cells were treated with cycloheximide. Extracts prepared from transfected cells, collected at the indicated times after cycloheximide treatment, were analysed by western blotting with rabbit anti-p21 antibody. p21 (1–133 + NLS) exhibited a slightly shorter half-life than p21 (1–133) (that lacks the endogenous putative NLS) but still exhibited a much longer half-life than the WT p21. Treatment of cells with lactacystin prior to addition of cycloheximide resulted in stabilization of proteins expressed from the pSG5-p21 plasmids. Protein extracts were prepared at indicated times after treatment with cycloheximide. (D) ECL signals shown in example (C) were quantified using a GS 300 scanning densitometer (Hoeffer) and values were plotted as the percentage of the signal at t = 0 (given an arbitrary value of 100%) remaining at the times indicated. This experiment was performed twice and produced similar results both times.
p21WAF1/CIP1 is rapidly degraded by purified 20S proteasomes in a manner that is dependent on the C8-interaction domain
Free p21WAF1/CIP1 lacks stable secondary and tertiary structure but undergoes a transition to an ordered state when bound to a biological target such as Cdk2 (Kriwacki et al., 1996, 1997). p21WAF1/CIP1 is also unusual because it can be regulated by proteasomal activity without the need for direct ubiquitylation (Sheaff et al., 2000). These studies have fuelled speculation that free p21WAF1/CIP1 might be recognized directly by the 26S proteasome. Here we have identified C8 as a structural component of the proteasome with which p21WAF1/CIP1 can interact. The interaction domain overlaps the PCNA-binding site and this explains how C8 and PCNA might compete and therefore how PCNA could block the targeting of p21WAF1/CIP1 for degradation. Furthermore, since free p21WAF1/CIP1 is relatively unstructured and probably does not require the unfolding activity associated with 19S ATPases, the data presented here also suggest a mechanism whereby p21WAF1/CIP1 could be directed straight to the 20S protease core. This would bypass the requirements for ubiquitylation and recognition of polyubiquitin by the 19S regulatory complex.
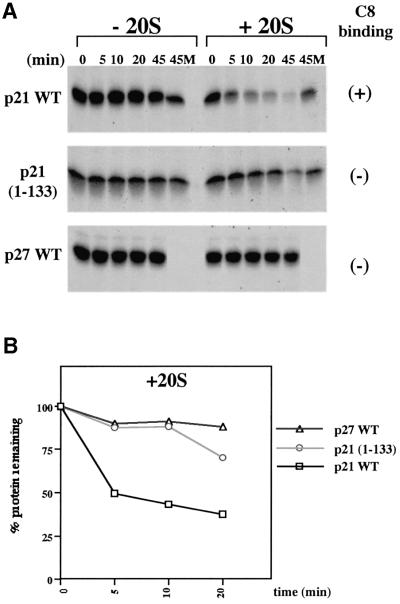
In order to test whether p21WAF1/CIP1 can be degraded by 20S proteasomes lacking the 19S complex containing ATPases and ubiquitin-recognition factors, experiments were performed with purified 20S core structures (Rivett et al., 1994). The results showed that in the absence of the 20S proteasome fraction, in vitro translated [35S]methionine-labelled p21WAF1/CIP1 was stable for >45 min at 37°C. In the sample to which 20S proteasomes were added, >50% of the p21WAF1/CIP1 was degraded in <10 min and this proteolysis was blocked by the inclusion of the proteasome-inhibitor MG-132 (Figure 6). The cdki p27KIP1 was completely stable in similar assays. This is consistent with p27KIP1 being a highly structured protein and requiring ubiquitylation and unfolding for proteasome-mediated turnover. Assays performed in parallel using a p21WAF1/CIP1 polypeptide with the C8-binding domain deleted, demonstrated the need for this domain in the interaction with, and degradation by, purified 20S proteasomes. p21(1–133) was almost as stable as p27KIP1 in the initial 10 min of the assay and only weakly degraded at later time points. These data are all consistent with p21WAF1/CIP1 binding to C8 in the assembled 20S proteasome.
Fig. 6. (A) The C-terminal end of p21 targets the protein for ubiquitin-independent proteolysis by purified 20S proteasomes. Equal volumes (5 µl/time point) of in vitro translated and [35S]methionine-labelled proteins (p21 WT, 1–133, Δ156–161 or p27 WT) were incubated at 37°C for the indicated time in the absence (–20S) or presence (+20S) of purified rat liver 20S proteasomes (1 µg/time point). Where indicated the z-LLL-H (MG-132) proteasome inhibitor was added at a final concentration of 200 µM (M). (B) [35S]signals shown in example A (+20S) were quantified using a storm 860 (Molecular Dynamics) and values were plotted as the percentage of the signal at t = 0 (given an arbitrary value of 100%) remaining at the times indicated. Multiple experiments produced similar results.
p21WAF1/CIP1 appears to bind only correctly folded C8
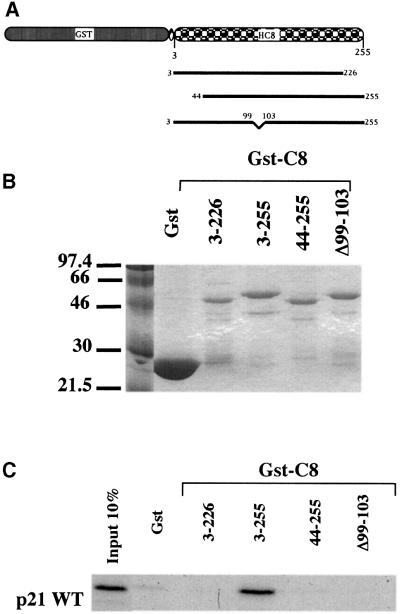
Mutants of C8 were constructed and assayed in order to determine the interaction motif in C8 that is required for p21WAF1/CIP1 binding in vitro. Several attempts to mutate C8 resulted in proteins which lost the capacity to bind to p21WAF1/CIP1. Three mutants are shown in Figure 7. Although a fusion of GST with WT C8 reproducibly binds, short deletions at the N-terminus and the C-terminus and an internal deletion (Δ99–103) all completely failed to interact with in vitro translated p21WAF1/CIP1. The most likely explanation of these data is that C8 is a small, highly conserved, essential protein (27 kDa) and any minor alteration in its primary structure destroys the ability of C8 to adopt the 3D conformation required for binding to p21WAF1/CIP1.
Fig. 7. (A) GST–C8 (aa 3–255), (aa 3–226), (aa 44–255) and (Δ99–103) fusion proteins are shown schematically. (B) Bacterially expressed and purified GST and GST–C8 (aa 3–255), (aa 3–226), (aa 44–255) or (Δ99–103) fusion proteins were separated on SDS–PAGE 12% and visualized by Coomassie blue staining. (C) Comparable amounts of GST–C8 (aa 3–255), (aa 3–226), (aa 44–255) and (Δ99–103) were incubated with an equal amount of in vitro translated [35S]methionine-labelled p21WT. Ten per cent of the total protein used in each binding reaction is shown. Bound proteins were resolved by SDS–PAGE (12%).
Discussion
p21WAF1/CIP1 belongs to the CIP/KIP family of cdk inhibitors and is a key negative regulator of cell-cycle progression, particularly at checkpoints mediated by the tumour suppressor p53. However, since p21WAF1/CIP1 also acts positively in the assembly of cyclin D/cdk complexes, its expression must be very tightly regulated both transcriptionally and post-translationally (Sherr and Roberts, 1995, 1999). Recent data suggest that the turnover of p21WAF1/CIP1 may be regulated only partly by the ubiquitin–proteasome system since it was shown that a mutant version of p21WAF1/CIP1 lacking all the target lysine residues for ubiquitylation is still degraded in a proteasome-dependent manner (Sheaff et al., 2000 and references therein). Here we show that p21WAF1/CIP1 can interact directly with a subunit of the 20S proteolytic core of the 26S proteasome and that in vitro translated p21WAF1/CIP1—but not the closely related p27KIP1—is degraded by purified 20S proteasomes. This is to our knowledge the first demonstration that p21WAF1/CIP1 can act as a substrate for the 20S proteasome. It also confirms Sheaff and colleagues’ observation that ubiquitylation is not an absolute requirement for the proteasome-mediated proteolysis of p21WAF1/CIP1. Based on all the currently available data, we propose that the degradation of p21WAF1/CIP1 is mediated by its direct association with the 20S proteasome through the interaction with C8. This interaction is probably weak or has a relatively fast off-rate, since under standard conditions of immunoprecipitation we have been unable to demonstrate complexes containing both proteins in cell extracts.
Free p21WAF1/CIP1 is a small, loosely folded protein (Kriwacki et al., 1996, 1997) therefore the C8-interaction domain in its C-terminus may be a sufficiently potent signal for degradation in the absence of ubiquitylation. Formation of a functional complex with PCNA would occlude this signal sequence and effectively stabilize p21WAF1/CIP1. Since this C-terminal region of p21WAF1/CIP1 also includes a consensus cyclin–cdk2 recognition motif (–HSKRRLIF–) it is possible that binding cyclin/cdk complexes might protect p21 in a similar manner (Adams et al., 1996). The data presented here are consistent with a model proposed recently (Verma and Deshaies, 2000) in which they speculated that all proteasomes (eukaryotic, prokaryotic and archaeal) recognize linear peptide stretches which they term class 1 degrons. The C-terminus of p21WAF1/CIP1 appears to contain such a degron and since this mode of substrate recognition is likely to be evolutionarily conserved, then a mode of recognition based on the more ancestral 20S proteasome component seems probable. However, Verma and Deshaies proposed that recognition might be via an ATPase subunit of the 19S complex. Our data show that (at least for p21WAF1/CIP1) 19S is unnecessary and are consistent with C8 being central to the recognition process. Nevertheless, because there is good evidence that (in vivo) p21WAF1/CIP1 can also be ubiquitylated (Blagosklonny et al., 1996; Maki and Howley, 1997; Cayrol and Ducommun, 1998; Rousseau et al., 1999), we cannot exclude the possibility that the interaction with C8 also favours the degradation of ubiquitylated p21WAF1/CIP1 after binding of the polyubiquitin by the 19S cap. The preliminary reports that similar mechanisms might operate in the processing of IκBα/p105 (Rousset et al., 1996; Kroll et al., 1997) suggest that substrate tethering through such an interaction with 20S may be a general feature of proteasome-dependent turnover, even when a polyubiquitin signal is also involved. Two signals operating in parallel may increase the rate of protein degradation. However, for small, relatively unfolded proteins such as p21WAF1/CIP1, a single signal may be sufficient since the signal for unfolding (polyubiquitin) may become dispensable.
Crystal structure analysis has revealed that the entrance channel at the centre of the α-ring is closed (Groll et al., 1997). In the experiments using purified 20S proteasomes (Figure 6), the isolation procedure itself activates the ‘latent’ structures—that is, the entrance site is modified so as to allow substrate molecules into the proteolytic β-ring (Rivett et al., 1994). In vivo, ‘activation’ of 20S proteasomes is achieved by association with a regulatory complex. A question that remains unanswered is, if 19S regulatory complexes are not involved, what is the alternative mechanism of ‘activation’ in vivo? Full ‘activation’ can also be achieved by an alternative regulatory complex called PA28 (or the 11S regulator). PA28 located in the cytoplasm exists as heteropolymeric structures composed of PA28α and PA28β subunits. These regulators are inducible by interferon-γ and play a central role in adaptive immunity by generating MHC class 1-binding peptides (Baumeister et al., 1998). However, a less well-characterized form of PA28 exists solely in the nucleus. This is a homopolymeric structure made up of PA28γ subunits (Murata et al., 1999; Masson et al., 2001). Nuclear PA28γ may therefore be responsible for opening the ends of nuclear 20S proteasomes in the absence of 19S regulators. With respect to p21WAF1/CIP1 turnover, it may be significant that genetic studies in both Drosophila and mice indicate that PA28γ plays an important role in cell growth and proliferation (Murata et al., 1999; Masson et al., 2001).
Finally, we note that located in the middle of the critical five amino acids (156–161) are three hydrophobic residues (leucine, isoleucine and phenylalanine) and these are flanked by predominantly basic residues. It is possible that this pattern could constitute the core of a conserved degradation signal. In this respect it may be significant that a very similar motif, a hydrophobic patch, adjacent to basic residues located in either the N- or C-terminus of a polypeptide favours degradation by Clp and Lon proteases in prokaryotes (reviewed in Wickner et al., 1999).
Materials and methods
Plasmids
p21, p27 and p57 cDNAs were cloned into BamHI-digested pGEM-2 plasmid as PCR products using primers which contain BamHI sites. They were all a gift from Dr Xin Lu, Ludwig Institute for Cancer Research, London. pGEX/PCNA was described previously (Nakanishi et al., 1995) The full-length C8 cDNA was derived from a cDNA library cloned into pACT-2 plasmid (Stratagene). The C8 restriction fragment was digested with Ecl136II and EcoRI and cloned into SmaI–EcoRI-digested pGEX-2T (Pharmacia) giving the pGEX-2T/C8 (aa 3–255). pGEX-2T/C8 (aa 3–255) was digested BamHI–XmnI and the resulting C8 restriction fragment (aa 3–226) was cloned into BamHI–SmaI pGEX-2T giving the pGEX-2T/C8 (aa 3–226). C8 (aa 44–255) was cloned into BamHI– EcoRI-digested pGEX-2T as PCR product using primers which contain either BamHI or EcoRI flanking sites giving the pGEX-2T/C8 (aa44–255). pGEX-2T/C8 (Δ99–103) was constructed using the QuickChange site-directed mutagenesis kit (Stratagene) using pGEX-2T/C8 (aa 3–255) as template.
p21 constructs, WT, (1–133), (M147A), (Δ113–133), (Δ156–161), (1–133, KB) and (1–133 + NLS) were cloned into pSG5 (for in vivo expression) and pGEM-2 (for in vitro transcription and translation). pGEM-2 was used because it contains a β-globin untranslated region which enhances in vitro translation. p21 WT and p21 (1–133) were cloned into EcoRI–BamHI-digested pSG5 plasmid as PCR products using primers which contain either EcoRI or BamHI sites. The p21 (M147A) plasmid in which Met147 of p21 was mutated to Ala was as described previously (Nakanishi et al., 1995). p21 (M147A) was then subcloned into EcoRI–BamHI-digested pSG5 plasmid as a PCR product using primers which contain either EcoRI or BamHI sites. p21 (Δ113–133) was constructed by recombinant PCR and then cloned into EcoRI–BamHI-digested pSG5 plasmid. p21(Δ156–161) was constructed using the QuickChange site-directed mutagenesis kit (Stratagene) using pSG5-p21 WT as template. p21 (1–133, KB) was cloned into EcoRI–BamHI-digested pSG5 plasmid as PCR products using primers which contain either EcoRI or KpnI–BamHI sites [KpnI was designed to facilitate cloning in-frame with p21(1–133)]. p21 (1–133 + NLS) was constructed by cloning an oligonucleotide 5′-ggtaccaaaaagaagcggaaagtgtgaggatcc-3′ encoding the SV40 NLS (–KKKRKV–) in-frame with p21(1–133). All p21 mutants were cloned into BamHI-digested pGEM-2 plasmid as PCR products using primers which contain BamHI sites and pSG5-p21 mutants as templates.
p27/p21 (134–164) was constructed by recombinant PCR and then cloned into EcoRI–BamHI-digested pSG5 plasmid. pSG5-p27/p21 (140–164) was constructed using the QuickChange site-directed mutagenesis kit (Stratagene) using pSG5-p27/p21 (134–164) as template. pSG5-p27/p21 (145–164) was constructed using the QuickChange site-directed mutagenesis kit (Stratagene) using pSG5-p27/p21 (140–164) as template. p27/p21 (134–160) and p27-p21 (134–155) were cloned into EcoRI–BamHI-digested pSG5 plasmid as PCR products using primers which contain either EcoRI or BamHI sites and pSG5-p27/p21 (134–164) as template. P27-KB was cloned into EcoRI–BamHI-digested pSG5 plasmid as PCR products using primers which contain either EcoRI or KpnI–BamHI sites (KpnI was designed to facilitate cloning in-frame with p27 full-length). pSG5-p27/p21 (134–144), (145–155) and (156–161) were all constructed by cloning oligonucleotides encoding the p21 sequences indicated flanked by KpnI or BamHI sites into KpnI–BamHI-digested pSG5-p27-KB construct. In order to create pSG5-PCNA, a cDNA corresponding to PCNA was cloned into BamH1-digested pSG5 plasmid as a PCR product produced using primers that contain BamH1 sites and pGEX-PCNA as a template (Nakanishi et al., 1995). Where appropriate, DNA constructions were validated by sequence analysis.
Cell culture and transfections
The EBV–negative human Burkitt lymphoma-derived cell line DG75 was grown at 37°C in RPMI 1640 medium (Gibco-BRL) supplemented with 10% fetal calf serum, 2 mM l-glutamine (Gibco-BRL) and 100 U/ml of penicillin and streptomycin (Gibco-BRL). The mouse fibroblast line NIH 3T3 was grown in Dulbecco’s modified Eagle medium (Gibco-BRL) supplemented as described above. Cells were cultured under a 10% CO2 humidified atmosphere.
DG75 cells were transfected by electroporation as described previously (Bain et al., 1996). In nearly all cases, 10 µg of pSG5-p21 plasmid DNA was transfected per 107 DG75 cells. For the pSG5-PCNA cotransfections, 10 µg of pSG5-p21 DNA was mixed with 10 µg of pSG5-PCNA or pSG5 empty vector DNA and 107 DG75 cells were transfected. NIH 3T3 cells were transfected by lipofectamine according to the manufacturer’s instructions (Gibco-BRL). Thirty-five mm tissue culture plates were seeded with 2 × 105 cells 24 h before transfection and in all cases 1 µg of pSG5-p21 plasmid DNA was used per plate.
Cycloheximide and lactacystin treatments
Twenty-four hours after transfection, cycloheximide [stock solution 5 mg/ml in phosphate-buffered saline (PBS)] was added at a final concentration of 50 µg/ml. Then cells were collected at indicated times. In order to inhibit proteasome activity, 24 h after transfection, lactacystin (stock solution 10mM in dimethyl sulfoxide) was added at a final concentration of 2 µM for a further 5 h. Cycloheximide was then added at a final concentration of 50 µg/ml and cells were collected at indicated times after cycloheximide treatment.
Protein extraction and western blot analysis
Protein extraction and western blot analysis was performed as described previously (Wade and Allday, 2000). In all cases 20 µg of total protein extract was loaded for SDS–PAGE (12%) using a mini-PROTEAN II cell (Bio-Rad).
Antibodies and immunofluorescence
A rabbit polyclonal anti-p21 antibody (N20, Santa Cruz) was used for western blot analysis. A mouse monoclonal anti-p21 antibody, CP68 (Cai and Dynlacht, 1998) was used for the indirect immunofluorescence. This was a gift from Dr Brian Dynlacht, Harvard University, Cambridge, MA. Immunofluorescence staining was performed as previously described (Radkov et al., 1999).
GST-fusion pull-down assays
GST pull-down experiments were performed essentially as described previously (Radkov et al., 1997, 1999). Approximately 1–2 µg of GST-fusion protein was used in each binding reaction apart from in the titration experiments when the appropriate dilutions were made.
Purification of the 20S proteasomes and in vitro degradation assays
The 20S proteasomes from rat liver were purified as described previously (Rivett et al., 1994). When analysed by SDS–PAGE, only 20S proteasome subunits of 20–30 kDa were detectable by Coomassie staining.
p21 WT, 1–133 or Δ156–161 as well as p27 WT were generated with the in vitro transcription/translation TnT-coupled Reticulocyte Lysate system (Promega). Each protein (5 µl/time point) was incubated at 37°C with or without purified 20S proteasome (1 µg/time point) for different lengths of time in PBS. When indicated the peptide aldehyde inhibitor z-LLL-H (MG-132) was added at 200 µM (final concentration) to inhibit 20S proteasome proteolytic activity.
Acknowledgments
Acknowledgements
We are grateful to Paul Farrell, Mark Wade, Gareth Inman, Roger Watson and Nico Dantuma for helpful discussions and David O’Nions for help with the purification of the 20S proteasomes. We thank Xin Lu for p21, p27 and p57 plasmids, Brian Dynlacht for antibodies and the Wellcome Trust for financial support through programme grant No. 056822 to M.J.A.
References
- Adams P.D., Sellers,W.R., Sharma,S.K., Wu,A.D., Nalin,C.M. and Kaelin,W. (1996) Identification of a cyclin–cdk2 recognition motif present in substrates and p21-like cyclin-dependent kinase inhibitors. Mol. Cell Biol., 16, 6623–6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akioka H., Forseberg,N.E., Ishida,N., Okumura,K., Nogami,M., Taguchi,H., Noda,C. and Tanaka,K. (1995) Isolation and characterization of the HC8 subunit gene of the human proteasome. Biochem. Biophys. Res. Commun., 207, 318–323. [DOI] [PubMed] [Google Scholar]
- Bain M., Watson,R.J., Farrell,P.J. and Allday,M.J. (1996) Epstein–Barr virus nuclear antigen 3C is a powerful repressor of transcription when tethered to DNA. J. Virol., 70, 2481–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumeister W., Walz,J., Zuhl,F. and Seemuller,E. (1998) The proteasome: paradigm of a self-compartmentalizing protease. Cell, 92, 367–380. [DOI] [PubMed] [Google Scholar]
- Blagosklonny M.V., Wu,G.S., Omura,S. and El-Deiry,W.S. (1996) Proteasome-dependent regulation of p21WAF1/CIP1 expression. Biochem. Biophys. Res. Commun., 227, 564–569. [DOI] [PubMed] [Google Scholar]
- Cai K. and Dynlacht,B.D. (1998) Activity and nature of p21WAF1 complexes during the cell cycle. Proc. Natl Acad. Sci. USA, 95, 12254–12259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayrol C. and Ducommun,B. (1998) Interaction with cyclin-dependent kinases and PCNA modulates proteasome-dependent degradation of p21. Oncogene, 17, 2437–2444. [DOI] [PubMed] [Google Scholar]
- Gerards W.L.H., de Jong,W.W., Bloemendal,H. and Boelens,W. (1998) The human proteasomal subunit HsC8 induces ring formation of other A-type subunits. J. Mol. Biol., 275, 113–121. [DOI] [PubMed] [Google Scholar]
- Groll M., Ditzel,L., Lowe,J., Stock,D., Bochtler,M., Bartunick,H.D. and Huber,R. (1997) Structure of 20S proteasome from yeast at 2.4 Å resolution. Nature, 386, 463–471. [DOI] [PubMed] [Google Scholar]
- Kriwacki R.W., Hengst,L., Tennant,L., Reed,S.I. and Wright,P.E. (1996) Structural studies of p21Waf1/Cip1/Sdi1 in the free and Cdk2-bound state: conformational disorder mediates binding diversity. Proc. Natl Acad. Sci. USA, 93, 11504–11509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriwacki R.W., Wu,J., Tennant,L., Wright,P.E. and Siuzdack,G. (1997) Proteolysis, matrix assisted laser desorption/ionization mass spectrometry, high performance liquid chromatography and size exclusion chromatography of p21Waf1/Cip1/Sdi1. J. Chromatogr. A, 777, 23–30. [DOI] [PubMed] [Google Scholar]
- Kroll M. et al. (1997) The carboxy-terminus of IκBα determines susceptibility to degradation by the catalytic core of the proteasome. Oncogene, 15, 1841–1850. [DOI] [PubMed] [Google Scholar]
- Maki C.G. and Howley,P.M. (1997) Ubiquitination of p53 and p21 is differentially affected by ionizing and UV radiation. Mol. Cell. Biol., 17, 355–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson P., Andersson,O., Petersen,U.-M. and Young,P. (2001) Identification and characterisation of a Drosophila nuclear proteasome regulator. J. Biol. Chem., 276, 1383–1390. [DOI] [PubMed] [Google Scholar]
- Murata S., Kawahara,H., Tohma,S., Yamamoto,K., Kasahara,M., Nabeshima,Y., Tanaka,K. and Chibas,T., (1999) Growth retardation in mice lacking the proteasome activator PA28γ. J. Biol. Chem., 274, 38211–38215. [DOI] [PubMed] [Google Scholar]
- Nakanishi M., Robetorye,R.S., Pereira-Smith,O.M. and Smith,J.R. (1995) The C-terminal region of p21SDI1/WAF1/CIP1 is involved in proliferating cell nuclear antigen binding but does not appear to be required for growth inhibition. J. Biol. Chem., 270, 17060–17063. [DOI] [PubMed] [Google Scholar]
- Radkov S.A., Bain,M., Farrell,P.J., West,M., Rowe,M. and Allday,M.J. (1997) Epstein–Barr virus EBNA 3C represses Cp, the major promoter for EBNA expression, but has no effect on the promoter of the cell gene CD21. J. Virol., 71, 8552–8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radkov S.A., Touitou,R., Brehm,A., Rowe,M., West,M., Kouzarides,T. and Allday,M.J. (1999) Epstein–Barr virus nuclear antigen 3C interacts with histone deacetylase to repress transcription. J. Virol., 73, 5688–5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivett A.J., Savory,P.J. and Djaballah,N. (1994) Multicatalytic endopeptidase complex (proteasome). Methods Enzymol., 244, 331–350. [DOI] [PubMed] [Google Scholar]
- Rousseau D., Cannnella,D., Boulaire,J., Fitzgerald,P., Fotedar,A. and Fotedar,R. (1999) Growth inhibition by CDK–cyclin and PCNA binding domains of p21 occurs by distinct mechanisms and is regulated by ubiquitin–proteasome pathway. Oncogene, 18, 4313–4325. [DOI] [PubMed] [Google Scholar]
- Rousset R., Desbois,C., Bantignies,F. and Jalinot,P. (1996) Effects on NFκB1/p105 processing of the interaction between the HTLV1 transactivator Tax and the proteasome. Nature, 381, 328–331. [DOI] [PubMed] [Google Scholar]
- Sheaff R.J., Singer,J.D., Swanger,J., Smitherman,M., Roberts,J.M. and Clurman,B. (2000) Proteasomal turnover of p21Cip1 does not require p21Cip1 ubiquitination. Mol. Cell, 5, 403–410. [DOI] [PubMed] [Google Scholar]
- Sherr C.J. and Roberts,J.M. (1995) Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev., 9, 1149–1163. [DOI] [PubMed] [Google Scholar]
- Sherr C.J. and Roberts,J.M. (1999) CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev., 13, 1501–1512. [DOI] [PubMed] [Google Scholar]
- Tamura T., Lee,D.H., Osaka,F., Fujiwara,T., Shin,S., Chung,C.H., Tanaka,K. and Ichihara,A. (1991) Molecular cloning and sequence analysis of cDNAs for five major subunits of human proteasomes (multi-catalytic proteinase complexes). Biochim. Biophys. Acta, 1089, 95–102. [DOI] [PubMed] [Google Scholar]
- Thrower J.S., Hoffman,L., Rechsteiner,M. and Pickart,C.M. (2000) Recognition of the polyubiquitin proteolytic signal. EMBO J., 19, 94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R. and Deshaies,R.J. (2000) A proteasome howdunit: the case of the missing signal. Cell, 101, 341–344. [DOI] [PubMed] [Google Scholar]
- Wade M. and Allday,M.J. (2000) Epstein–Barr virus suppresses a G2/M checkpoint activated by genotoxins. Mol. Cell. Biol., 20, 1344–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner S., Maurizi,M.R. and Gottesman,S. (1999) Post-translational quality control: folding, refolding and degrading proteins. Science, 286, 1888–1893. [DOI] [PubMed] [Google Scholar]