Abstract
Feedback is a ubiquitous control mechanism of gene networks. Here, we have used positive feedback to construct a synthetic eukaryotic gene switch in Saccharomyces cerevisiae. Within this system, a continuous gradient of constitutively expressed transcriptional activator is translated into a cell phenotype switch when the activator is expressed autocatalytically. This finding is consistent with a mathematical model whose analysis shows that continuous input parameters are converted into a bimodal probability distribution by positive feedback, and that this resembles analog–digital conversion. The autocatalytic switch is a robust property in eukaryotic gene expression. Although the behavior of individual cells within a population is random, the proportion of the cell population displaying either low or high expression states can be regulated. These results have implications for understanding the graded and probabilistic mechanisms of enhancer action and cell differentiation.
Keywords: enhancer/genetic switch/graded response/stochastic/transcriptional activator
Introduction
Gene networks form the basis of various biological processes, such as biorhythmic oscillations, developmental pattern formation and the cell cycle. Autoregulation is a commonly used architectural element in gene networks (Freeman, 2000). Prokaryotic gene circuits mostly exploit negative feedback to ensure homeostasis (Thieffry et al., 1998; Becskei and Serrano, 2000), while eukaryotic transcriptional activators commonly regulate their own expression by both negative and positive feedback (Bateman, 1998).
Positive feedback or autocatalysis has long been recognized to underlie bistable or binary responses in chemical reaction systems. In a bistable system, transition between the two stable states can occur due to changes in the system’s input parameters. For example, transition between two maturation stages of the Xenopus oocyte is induced by progesterone. Increasing the concentration of progesterone increases the proportion of mature oocytes. Positive feedback in a mitogen-activated protein (MAP) kinase cascade of oocytes has been implicated in the conversion of a graded response to a binary cell-fate switch, although an additional mechanism may also play a role (Ferrell and Machleder, 1998).
In principle, one might expect positive feedback to generate a binary response in eukaryotic gene circuits of transcription factors. However, there are both theoretical and practical reasons to question this. In theoretical models, a binary response is not a necessary consequence of positive feedback, and positive feedback can produce a variety of dynamic behaviors other than binary responses (for example, see Walz and Caplan, 1995). Therefore, the question of whether eukaryotic transcription is capable of generating a binary response by positive feedback must be answered empirically. The verification of the mathematical model is complicated by two factors. One is that the transcription factors acting at complex promoters are varied and often still unidentified. The second is that interpretation of the experiments is difficult because the mechanism of enhancer action is still poorly understood (Bateman, 1998). Enhancers, clusters of activator binding sites, function as autonomous regulatory units to activate the usually weak eukaryotic core promoters. Hence, transcription in essentially all eukaryotic genes requires activators (Struhl, 1999). Two contrasting modes of how eukaryotic enhancers operate have been described (Fiering, 2000; Hume, 2000). In the graded response, activators bound to enhancers increase the rate of transcription in each cell in a dose-dependent manner. In the binary or probabilistic response, enhancers do not affect the transcription rate, but instead increase the probability that a reporter gene will be active, thereby increasing the proportion of expressing cells in a population. In the latter case, the cell population is divided into pools of cells with either low or high expression states. The two modes of enhancer action can be distinguished only by single-cell assays.
We have applied a novel approach to obviate the above difficulties in analysing the consequences of positive feedback. Regulatory mechanisms can be explored by the construction of gene networks using well defined components, furnished with the interactions to be studied. In this way, the properties of the regulatory mechanisms can be extracted, since a more complete control of network functioning is attained, while the interactions determining the overall behavior of the network are preserved. This approach has already been adopted to study simple prokaryotic gene circuits involving one, two or three negative interactions between repressors (Becskei and Serrano, 2000; Elowitz and Leibler, 2000; Gardner et al., 2000). In these studies, stability, toggle switch and oscillations were explored in Escherichia coli.
Here, we address the consequences of positive feedback in eukaryotic gene circuits with an integrated, experimental–theoretical approach, which takes advantage of well defined promoter elements and a well defined transcriptional activator, the tetracycline-responsive transactivator (rtTA). rtTA produces a graded response in constitutive systems, making possible the analysis of positive feedback by our mathematical model, since it provides a more complete experimental control than would enhancers responding in a probabilistic manner. We show that positive feedback is a mechanism that can convert a graded to a binary response in a eukaryotic gene circuit. The binary response itself is characterized by different degrees of separation of the two expression states (low and high), the degree of separation depending on the architecture and read-out of the autocatalytic circuit.
Results
Approach
rtTA is commonly used to regulate gene expression in eukaryotic organisms. The degree of activation by rtTA can be adjusted by changing the gene copy number, or by varying the concentration of the inducer, doxycycline. In the presence of doxycycline, rtTA takes on an active conformation and binds DNA. In this study, two series of experiments were performed, using different ways of reporting transcription activity. We used several different promoters, as well as gene circuits inserted into either plasmids or chromosomes, and reporter genes that were either regulated by the activator or directly fused to it, to eliminate artifacts possibly caused by the nature and localization of the regulatory and reporter sequences.
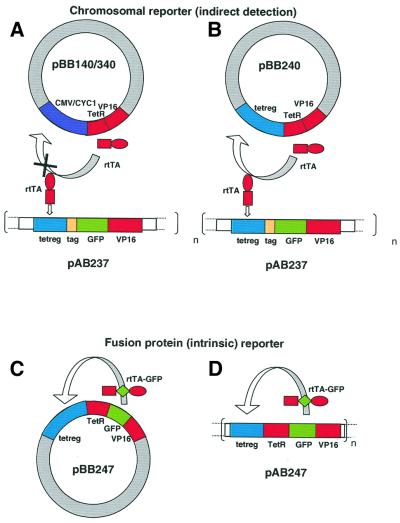
In the first series, rtTA expression was compared in a constitutive versus autocatalytic system. The protein was expressed constitutively, or by positive autoregulation, from centromeric vectors (see Materials and methods and Figure 1A and B) that are present in 1–2 copies per cell (Gunge, 1983). The activity of rtTA was assayed in reporter strains (ABY001–ABY020) carrying a chromosomally integrated green fluorescent protein (GFP) reporter construct (pAB237). Fluorescence intensity was measured in single cells by fluorescence microscopy. In the constitutive expression systems, the CMV and CYC1 core promoters were used (pBB140/340). In the autocatalytic system (pBB240), an rtTA binding sequence was linked to the CYC1 promoter to create a positive feedback by rtTA.
Fig. 1. Design of expression systems. (A) Constitutive system. (B, C and D) Autocatalytic systems. Blue colors represent promoter constructs (tetreg, CMV and CYC1), green for GFP, and red for the modules of activator. rtTA consists of a DNA binding domain, the reverse TetR (TetR) and an activator domain, VP16ad (VP16) (Gossen et al., 1995). The reporter construct trunc–GFP consists of an N-terminal tag, GFP and VP16ad. The CEN-TRP1 plasmids pBB140, pBB240, pBB340 and pBB247 contain the expression cassettes CMV-rtTA, tetreg-rtTA, CYC1-rtTA and tetreg-rtTA–GFP, respectively. The integrative LEU2 plasmids pAB237 and pAB247 are obtained by insertion of tetreg-trunc–GFP and tetreg-rtTA–GFP, respectively. Integration of pAB237 and pAB247 into the LEU2 locus of GFY259-2B resulted in yeast strains ABY001–ABY020 and ABY021–ABY040, respectively. Strains with the same number of integrated repeats (n) showed consistent behavior.
In the second series of experiments, protein expression was detected directly in two autocatalytic systems. In both cases, GFP was fused to rtTA to allow direct detection of the autocatalytically expressed rtTA (Figure 1C and D). This autocatalytic circuit was integrated either into a centromeric plasmid (pBB247) or into the chromosome (by the integrative vector pAB247).
Activation by the constitutively expressed activator
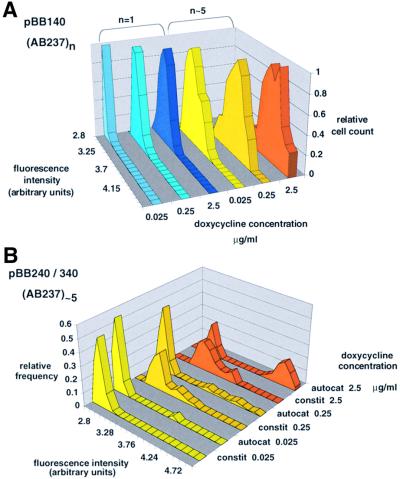
In the simple activation system, rtTA is expressed constitutively from the CMV promoter (pBB140) and activates the expression of the chromosomally integrated reporter construct in the presence of doxycycline (Figure 1A). With a single reporter construct, the average fluorescence intensity of cells is close to the detection limit at lower inducer concentrations (Figure 2A, lanes in various shades of blue). However, it was possible to amplify rtTA activity to detectable levels by the use of multiply integrated reporter constructs (Figure 2A, lanes ranging from yellow to orange). After activation of rtTA with doxycycline, every cell became fluorescent and the fluorescence level exhibited an approximately Gaussian distribution in a cell population (Figure 2A). The mean value of this unimodal distribution increased with inducer concentration (Figure 2A) and each cell displayed a different degree of fluorescence (Figure 3A and B). To determine whether the graded response to inducer concentration is peculiar to the CMV promoter, we tested another constitutive promoter, the CYC1 promoter (pBB340). In this case, the fluorescence distribution was less regular. Fluorescence still responded to inducer levels in a graded manner, but the mean values were lower than those for the CMV promoter (Figure 2B).
Fig. 2. Distribution of cell fluorescence intensities in chromosomal reporter systems. (A) The constitutive plasmid pBB140 (CMV promoter) was transformed into strains ABY001 (one copy of the reporter construct, pAB237) (blue shades) and ABY016 (more than five copies of reporter construct, pAB237) (yellow shades). Doxycycline was added to the culture to yield a final concentration of 0.025, 0.25 and 2.5 µg/ml. The distributions at 0.025 and zero doxycycline concentrations are very similar, which might be explained by the residual binding of rtTA to the enhancer in the absence of inducer. Cultures were grown in SD-Leu, Trp. Relative cell count was obtained by dividing the actual frequency by the frequency at the mode of distribution. In this way the differences in the mean intensities are visualized better. The mean value (m) and standard deviation (s) for ABY016 at 2.5 µg/ml inducer concentration are m = 4.55 and s = 0.159. The standard deviation indicates the width of distribution. (B) Comparison of the autocatalytic (pBB240) and constitutive (pBB340) systems transformed into the ABY016 strain. Both expression systems contain the CYC1 core promoter. The autocatalytic and constitutive systems are labeled with ‘autocat’ and ‘constit’ next to the indicated inducer concentration. For pBB340, m = 3.67 and s = 0.240 at 2.5 µg/ml inducer concentration.
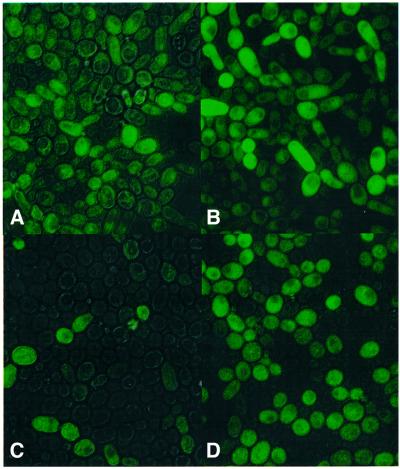
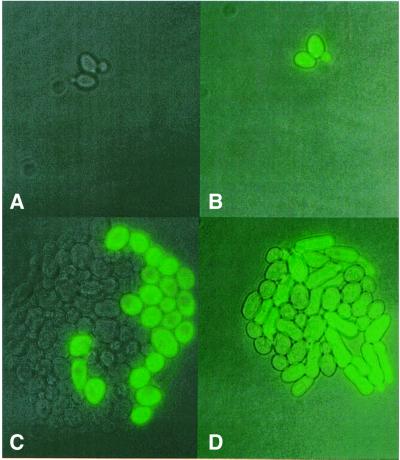
Fig. 3. Fluorescence microscope image of cells. Panels were obtained by merging phase contrast and fluorescence images of ABY016 cells. (A and B) Cells contain pBB140 and are induced with 0.25 (A) and 2.5 µg/ml (B) doxycycline, and exposed for 25 ms. (C and D) Cells contain pBB240 and are induced with 0.25 (C) and 2.5 µg/ml (D) doxycycline, and exposed for 10 ms.
Autocatalytically expressed activator detected by chromosomal reporter construct
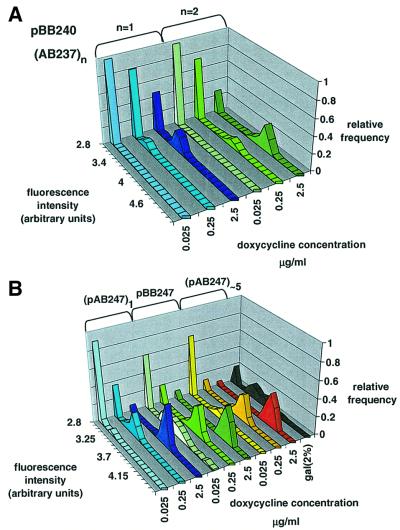
The first autocatalytic system tested is the counterpart of the simple activation system (Figure 1B) in which rtTA, after addition of doxycycline, induces its own transcription beyond the basal rate (on the plasmid pBB240) and activates the chromosomally integrated reporter construct. After the expression of rtTA was induced, the cell population consisted of two distinct pools: fluorescent cells (‘bright’ on-cells) and a proportion of cells that remained non-fluorescent (‘dark’ off-cells). The number of off-cells decreased at higher inducer concentrations, and there was a concomitant increase of on-cells. The latter was associated with an increase in mean fluorescence intensity (Figure 3C and D; Figure 4A). When a multiple-copy reporter construct was used, the distinction between dark and bright cells became more pronounced (Figure 4A), while the proportion of off-cells and on-cells did not change significantly. In other words, only the mean fluorescence intensity of on-cells increased. The shape of the fluorescence distribution of on-cells can vary slightly in different reporter strains having different copy numbers of reporter construct. For example, the distribution for ABY016 was less regular than that of ABY011 (Figure 2B; Figure 4A). Once the bimodal distribution of expression was established after induction, it stabilized; the percentage of dark and bright cells did not change during the time of examination (6–12 h). When the inducer was removed, rtTA was inactivated and GFP expression decayed to the levels in the uninduced state.
Fig. 4. Fluorescence level distributions in the autocatalytic systems. (A) Chromosomal reporter system. pBB240 was transformed into strains ABY001 (one copy of the reporter gene) and ABY011 (two copies of the reporter gene), and are represented by blue and green shades, respectively. Cells were grown in SD-Leu, Trp. (B) Intrinsic reporter system. Out of the series ABY021–040, ABY021 and ABY022 were examined with one and more than five copies of pAB247, and are represented by blue and yellow shades, respectively. Strains were grown in SD-Leu. pBB247 was transformed into GFY259-2B, grown in SD-Trp; results are represented by green shades. The black lane represents the galactose-regulated plasmid pBB407 (see Materials and methods).
Expression of the activator–reporter fusion protein with positive feedback
In order to detect the activator directly, we constructed a second class of autocatalytic circuits: the intrinsic reporter systems pBB247 and pAB247. GFP was inserted into rtTA, generating the fusion protein rtTA–GFP. Expression of the autocatalytic circuit from pBB247 (Figure 1C) resulted in a bimodal GFP distribution similar to that generated by the chromosomal reporter system (Figure 1B). However, the number of fluorescent cells was higher at all inducer concentrations, and the distance between the two peaks was smaller (Figure 4B). To see whether the switch is affected when the autocatalytic circuit is localized on the chromosome, we examined the strains ABY021–ABY040, in which pAB247 is integrated into the chromosome (Figure 1D). The results of these experiments showed that possible modifications of chromosomal chromatin do not affect the shape of bimodal distribution when compared with the extrachromosomally expressed pBB247 system. The integration of multiple repeats also permits the examination of the effect of circuit copy number on the distribution. The proportion of on-cells in this case is positively correlated with the copy number, in contrast to the situation in the chromosomal reporter system (Figure 4B versus A; see Supplementary data available at The EMBO Journal Online). This indicates that both the induction level and gene copy number contribute to the activation level. At very high activation levels, very few cells are dark. Therefore, it is difficult to distinguish a monostable system from a bistable system within this range of induction.
To compare the effects of autocatalytic expression of rtTA with indirect positive feedback, we tested the expres sion of GFP under the control of the Saccharomyces cerevisiae GAL1 promoter (see Materials and methods). In the yeast Kluyveromyces lactis, the expression of Gal4 is positively autoregulated. In S.cerevisiae there is no Gal4 binding site in the promoter of Gal4 itself. However, Gal4 enhances the expression of galactose transporters and thereby the intracellular concentration of galactose is increased, resulting in a higher Gal4 activity. In this way an indirect positive feedback is established (Lohr et al., 1995). Cell fluorescence showed a bimodal distribution also in this case (Figure 4B).
Fates of single cells during population growth
The fates of single cells were followed by monitoring growth on microscope slides. Cells were grown for 6 h in liquid culture containing 2.5 µg/ml doxycycline, and cell growth was continued on agarose-covered slides containing the same concentration of doxycycline. After 16 h of incubation, single off-cells (Figure 5A) formed colonies of 100–150 cells (Figure 5C). Few colonies consisted exclusively of off-cells, the majority being a mixture of both on- and off-cells. In the same period, single on-cells (Figure 5B) formed colonies of 20–30 cells, but all of the cells within these colonies continued to express the reporter gene (Figure 5D).
Fig. 5. Fate of single cells during growth. The division of non-fluorescent and fluorescent ABY001 cells was followed on agarose layers containing 2.5 µg/ml doxycycline. (A and B) Image of cells at the start of the experiment. (C) Colony derived from cells on (A) after 16 h of incubation. (D) Colony derived from cells on (B) after 22 h of incubation.
The proportion of on-cells on the slide in the entire cell population, estimated by counting 15–20 randomly selected colonies irrespectively of their composition, corresponded to those observed in liquid cultures.
With time, new on-cells appeared in mixed colonies, and these also divided at a slower rate than off-cells. This process gives rise to the mosaic patterning of mixed colonies.
Theoretical underpinnings for a eukaryotic switch
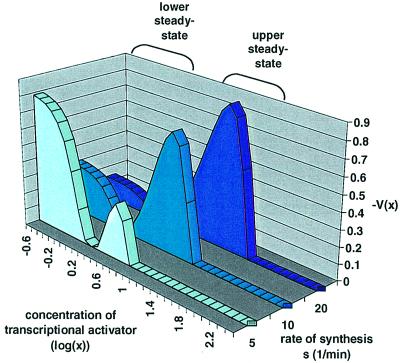
We performed a mathematical analysis, with probabilistic methods allowing for fluctuations or noise [see derivation of equation (2) in Materials and methods], to explain the conversion of graded to binary response in the positive feedback scenario tested. Biochemical processes in living organisms have a noisy character, which in part accounts for the considerable variability in the organism’s phenotype. Experimental evidence (Kringstein et al., 1998) and theoretical models show that for simple activation by a constitutively expressed activator, increasing the concentration of inducer results in a graded, sigmoid-shaped transcriptional response. The resulting expression is unimodally distributed in a cell population. Further analysis of this model shows that, in an autocatalytic system, bistability arises across a wide range of activation levels (see Materials and methods). Bistability is not an obligatory consequence of positive feedback since it depends on the parameter values characteristic to the gene circuit. Because of the high degree of cooperativity encountered in eukaryotic transcription activation, bistability arises as a robust property of eukaryotic autocatalytic gene circuits. Several mechanisms, such as nucleosomal rearrangement and protein–protein interactions, underlie the cooperativity that leads to this effect (Polach and Widom, 1996; Vashee et al., 1998; Wang et al., 1999). The lower steady-state corresponds approximately to the basal expression rate, while the upper one reflects the maximal rate. If the two stable steady-states are separated sufficiently, bistability is manifested as a bimodal distribution in a cell population. What will be the percentage of cells with low or high expression levels at a given level of activation? The probability of the lower steady-state being adopted is initially considerably higher than that for the upper one. However, as the degree of activation increases, a transition occurs and the inverse becomes true (Figure 6). The change in probability is accompanied by an upward shift in the position of the upper steady-state itself, while the lower steady-state shifts upwards only slightly at high activation levels. This theoretical model agrees with the experimental observations. Off- and on-cells correspond to the lower and upper steady-states of the model, or the left and right peaks of the bimodal fluorescence distribution, respectively. The left peak of the bimodal distribution (off-cells) overlaps with the unimodal distribution of the constitutive system and corresponds to the basal expression rate. The second peak (on-cells) is characteristic of the autocatalytic system, and corresponds to the maximally activated expression rates (Figure 2B).
Fig. 6. Approximate probability distribution in an autocatalytic system. The approximation is based on the negatively signed potential V(x) function. Regions of attraction of the lower and upper steady-states are indicated. Values of V(x) <0 are shown. V(x) is obtained from ϕ(x) [equation (2), Materials and methods] by normalization by the sum of the lowest values of the two potential wells, which correspond to the lower and upper steady-states. The parameter values are d = 10, r = 0.1 min–1, k = 1 min–1. To simulate different degrees of activation, s took values of 5, 10 and 20 min–1. Concentrations and concentration-based parameters are non-dimensional. The range of bistability is 4 < s < 148 min–1, which encompasses almost two orders of magnitude of protein production rate. Outside this region, a single stable state exists as a continuation of the lower and upper steady-states at s <4 and s >148 min–1, respectively. The approximate values of the steady-states are 100.67, 100.9 and 101.3 at s = 5, 10 and 20 min–1, respectively, indicating the rise of the upper steady-state parallel with the activation level.
Thus, the degree of activation—considered to be a continuous one-dimensional parameter space—is analog information, which is converted into binary information where ‘0’ and ‘1’ correspond to lower and upper steady-states. However, the values of ‘0’ and ‘1’ are not strictly fixed.
Discussion
Graded response with various distributions in constitutive systems
rtTA produces a graded response in mammalian expression systems (Kringstein et al., 1998). In constitutive systems, rtTA drives reporter expression in a graded manner, as is also the case in S.cerevisiae. Our results show that the distribution of reporter gene expression is influenced by the nature of the constitutive promoter that drives the expression of the activator. Using the CMV promoter, the distribution is narrow and symmetric. In the case of the CYC1 promoter, the expression level in a cell population is more variable, and its distribution is skewed to high expression levels. However, the population is not separated into two sub-populations. A graded response with asymmetric distribution can be distinguished from a binary response by increasing the inducer concentration. In the graded response, the entire distribution is shifted without considerable changes in its shape (Figure 2B), while in the binary response, the area of the first peak of the bimodal distribution changes inversely in relation to the area of the second peak (Figure 2B).
Single cell assays have concentrated on the action of the enhancer rather than the mode of activator expression. Our controlled experiments show that the examination of enhancer action has to be accompanied by a careful examination of the expression properties of the activator itself.
Positive feedback converts graded into binary response in eukaryotic gene circuits
Previous studies showed that the conversion of autoregulatory gene circuits of activators into constitutive systems, by deleting the enhancers, resulted in a decrease of promoter activity. However, whether this change was accompanied by an alteration of response type was not investigated (Bateman, 1998). In single-cell assays, a large number of enhancers produces a binary response (Ko et al., 1990; Walters et al., 1995; Osheim et al., 1996; Fiering et al., 2000). Since rtTA displays a graded response in constitutive systems, it can be used to reveal cellular mechanisms involved in converting a graded transcriptional response into a binary response. When rtTA is expressed under conditions of positive feedback, the cell population is clearly divided into pools of on-cells and off-cells. Both the numbers of expressing cells and the levels of expression in these cells increased in proportion to inducer concentration. This response resembles what has been observed for the behavior of the β-globin locus control region (Bouhassira et al., 1997; Forsberg et al., 1999). In the intrinsic reporter system, the concentration of the activator was detected directly (rtTA–GFP), while in the chromosomal reporter system, the activity of autocatalytically expressed rtTA was assayed using a separate reporter construct. The separation of the two cell populations was more conspicuous in the latter system, and the number of off-cells was also higher. Two effects can account for this. First, noise sensitivity may be different in the circuit involving the rtTA–GFP fusion protein, possibly resulting in slightly altered properties compared with those of rtTA. Secondly, in the chromosomal reporter system two actions can add up: the bistability of the autocatalytic system and the sigmoidal response of the reporter system to the activator. Additivity of two sigmoidal responses was recently observed (Rossi et al., 2000), and this also resulted in a bimodal probability distribution. Although it is difficult to compare the different experimental set-ups, the combination of autocatalytic bistability with a sigmoidal response is expected to result in a better separation of the two peaks of a bimodal distribution. Positive feedback and additivity of responses might be among the mechanisms that account for the probabilistic action of the enhancers.
More efficient protein expression has been obtained in different organisms and cell strains when the activator was autocatalytically expressed (A-Mohammadi and Hawkins, 1998). However, the considerable number of non-expressing cells observed in our experiments points to the fact that a compromise must be reached between the efficiency and the homogeneity of expression.
Autocatalytic gene circuits produce a random switch
Single off-cells can switch to the on-state in a stochastic way. While the theoretical model does not preclude the switch in the other direction, we did not observe the conversion of on- to off-cells within the period tested. Since basal expression levels are heterogeneous in the constitutive expression system pBB340 (Figure 2B), one might anticipate that in the autocatalytic system, only ‘predifferentiated’ cells with a high basal expression level will switch to the on-state after induction. This possibility can be excluded because the overwhelming majority of single off-cells eventually grew into mixed colonies, even if they initially exhibited lower expression levels (Figure 5). The continual switching from the off- to the on-state over the entire range of inducer concentration might classify the autocatalytic switch as a noise-based switch to differentiate it from the toggle switch (Hasty et al., 2000). In the toggle switch, when one of two mutually inhibiting repressors is transiently inactivated by inducer, the other repressor is expressed in the entire cell population, even after the removal of the inducer (Gardner et al., 2000). Although the basic properties of bistability for the toggle and autocatalytic switch are similar in theoretical models, they are different in that the toggle switch remains stable and random transition does not occur.
Autocatalytic expression underlies cell differentiation
The cell-doubling time of off-cells is shorter than the average time required to switch to the on-state. This difference between cell division and switching rates maintains a relatively large pool of off-cells, even at high inducer concentration. The relatively slow switching rate might be attributable to the robust bistability of eukaryotic autocatalytic gene expression (Figure 6). The proportion of off-cells is also influenced by the lower growth rate of on-cells compared with off-cells. Interest ingly, in the autocatalytic system of the lac operon of E.coli, on-cells expressing the endogenous β-galacto sidase also divided more slowly (Novick and Weiner, 1957). Theoretically, toxic effects of high reporter concentration might have an impact, if they lead to the presence of off-cells that somehow fail to ‘express’. However, this possibility can be excluded, since cells with constitutive rtTA expression from the CMV promoter have a high concentration of reporter gene, yet no off-cells were observed (Figure 2A).
The switching time itself is random. In growing colonies starting from single off-cells, on-cells can appear as early as when a colony reaches a total cell number of 6–10, or much later at a 50–60-cells stage. However, statistically the overall percentage of on- and off-cells in a cell population is controlled by the inducer concentration. Off-cells can be considered to serve as a pool of precursor cells, since this pool is self-maintained and also produces or ‘differentiates’ on-cells. Similar processes might play a role in blood stem cell differentiation. In this process, a stochastic decision leads to the initiation of genetic programs that give rise to distinct differentiated blood cell lineages, while a pool of undifferentiated stem cells is maintained. At the same time, the proportion of each cell type within the population is regulated.
Materials and methods
Construction of gene circuits
The modules of the expression systems were obtained from vectors pCM172, pCM240, pCM243 and yEGFP by restriction digestion or PCR (Cormack et al., 1997; Gari et al., 1997; Belli et al., 1998). Fidelity of PCR was checked by sequencing. The tetreg promoter construct consists of ADH1 transcriptional terminator, two copies of tetO box and CYC1 TATA region. The terminator serves for prevention of transcriptional readthrough from upstream sequences. The CMV promoter is the human cytomegalovirus promoter IE. rtTA–GFP is obtained by inserting in-frame the translated region of yEGFP into the KpnI site of rtTA; thus, the resulting fusion protein consists of the reverse Tet repressor, spacer region from the λ bacteriophage cI gene, yEGFP, spacer region (with amino acid sequence GENLYFQSGG) and the VP16 activator domain. The reporter construct (trunc–GFP) is the truncated version of rtTA–GFP, in which only the first eight amino acids of Tet repressor and the VP16 domain are retained. Thereby the DNA binding was abolished. However, transcriptional initiation and termination occurred in the same sequence context as in the intrinsic reporter systems. Since the initiation and termination processes affect the properties of the entire transcription, this reporter construct enabled the comparison of chromosomal reporter with the intrinsic reporter system. The autocatalytic steps are essentially the same in both reporter systems, while in the chromosomal reporter system the autocatalytic step is followed by an additional transcriptional step to express the gene reporter. However, this additional step has the same sequence context in both the initiation and termination process. So, the effects of the two steps add up in a controlled way.
KasI–PstI fragments of pCM172 and pCM243 were used as CEN-TRP1 and integrative LEU2 vectors, respectively, for the insertion of expression cassettes. pBB240 is derived from pCM172 by replacement of tTA by rtTA under the control of the constitutive CMV promoter, and retained the lacZ reporter. yEGFP was inserted into CEN-HIS pYX123 vector to obtain pBB407, expressed under the control of GAL1 promoter.
Nucleic acid manipulation
Cloning steps were performed in E.coli DH5α. Plasmids were transformed into S.cerevisiae strain GFY259-2B (MATa, his3, leu2, lys2, trp1, ura3) or its derivatives. Yeast genomic DNA was isolated in small scale; Southern analysis was performed with radioactive probes prepared by random priming. The number of repeats integrated into the chromosome of each of the yeast strains was determined by two series of Southern blots of digested genomic DNA. Restriction digestion by BamHI, which cuts only outside the integrated DNA, results in a single band of differing lengths, while in the case of KasI, which cuts inside the integrated DNA at a single site, two or three bands of constant lengths appear, with differing relative intensities. However, the number of integrated copies can not be determined accurately above 5, but in the strains used here was between 5–10.
Growth conditions and microscopy
Cultures were grown in synthetic dropout (SD) media at 30°C. Overnight saturated cultures were diluted 1:20 to 1:50 and doxycycline was added. Six to twelve hours later, cells were harvested and examined by fluorescence microscopy. Cells containing the galactose-responsive pBB407 plasmid were grown in SD-His overnight, diluted 1:20 into synthetic medium-His supplemented with 2% galactose and examined after 6 h. For microscopic examination, 10 µl of 1% agar were dropped on the microscope slide to form a thin layer. For the single cell follow-up SD medium was mixed with 1% agarose and doxycycline was added to yield the appropriate final concentration. Immediately after drying of the drop, 1 µl of cell suspension was pipetted onto it and covered with a coverslip. Microscopy was carried out using a Leica DMIRB/E microscope equipped with a highly light-sensitive Hamamatsu C4742–95 digital CCD camera and an automatic light shutter. Image acquisition and the control of the light shutter were controlled by Openlab imaging software 2.0.6. A Chroma GFP 513852 photocube transmitting a wavelength of 485–495 nm was used to stimulate GFP fluorescence. Fluorescent images of single fields of cells were obtained using a phase contrast 63×/1.32 oil-immersion lens. Different exposure times were used to ensure the linearity of the detection and absolute values of fluorescence intensity were normalized as if exposure of 200 ms was applied in all cases. Logarithms of the normalized values are used in this work. At exposure of 200 ms there is visible autofluorescence of control cells, which corresponds to a fluorescence intensity of 2.8–2.9. The autofluorescence limits the detection of GFP below these levels. For statistical analysis fluorescence levels of 150–200 cells were measured in each experiment.
Analysis of bistability
The properties of the autocatalytic switch are explained by the following model:
Where x is the concentration of activator, s is the maximal rate and r is the basal rate of synthesis of activator, d is the dissociation constant of activator from its DNA binding site, n is the cooperativity of activation, and k is the degradation rate of the activator. The model is based on the kinetic equations of autocatalytic gene expression (Keller, 1995; Smolen et al., 1998). s is a lumped parameter that includes the copy number of gene circuit, the inducer concentration and the proportionality constant between transcription and translation. Experimentally it was tuned by doxycycline and gene dosage to attain different degrees of activation. Eukaryotic transcriptional activation is characterized by a high degree of cooperativity. Cooperativity accounts for the sigmoidal transcriptional response in a simple activation. The autocatalytic system displays multistability for a wide range of s: two stable steady-states at low and high concentrations and an unstable steady-state at intermediate values. Any degree of positive cooperativity (n >1) is sufficient for conversion of graded to binary response in positive feedback. Higher order of cooperativity widens the range of bistability. In order to compare how the two stable states are attracting at different parameter values, the potential (ϕ) of equation (1) was calculated by taking dϕ/dx = –f(x), a = d1/3, n = 3, and the integration constant was set to zero.
The potential considered as an energy landscape is negatively correlated with the probability distribution of the concentration of transcriptional activator. The distribution is also influenced by the nature and intensity of noise (Hasty et al., 2000). The unstable steady-state is not expected to be observed in vivo owing to the noisy nature of gene expression.
Supplementary data
Supplementary data for this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank E.Herrero, E.Bertrand and G.Faye for the plasmids and the yeast strain, and J.Mendes, A.Westernholm, M.Petukhov and S.Fisinger for reading the manuscript. A.B. is supported by the Louis-Jeantet Foundation.
References
- A-Mohammadi S. and Hawkins,R.E. (1998) Efficient transgene regulation from a single tetracycline-controlled positive feedback regulatory system. Gene Ther., 5, 76–84. [DOI] [PubMed] [Google Scholar]
- Bateman E. (1998) Autoregulation of eukaryotic transcription factors. Prog. Nucleic Acid Res. Mol. Biol., 60, 133–168. [DOI] [PubMed] [Google Scholar]
- Becskei A. and Serrano,L. (2000) Engineering stability in gene networks by autoregulation. Nature, 405, 590–593. [DOI] [PubMed] [Google Scholar]
- Belli G., Gari,E., Piedrafita,L., Aldea,M. and Herrero,E. (1998) An activator/repressor dual system allows tight tetracycline-regulated gene expression in budding yeast [published erratum appears in Nucleic Acids Res., 1998, 26, 1855]. Nucleic Acids Res., 26, 942–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouhassira E.E., Westerman,K. and Leboulch,P. (1997) Transcriptional behavior of LCR enhancer elements integrated at the same chromosomal locus by recombinase-mediated cassette exchange. Blood, 90, 3332–3344. [PubMed] [Google Scholar]
- Cormack B.P., Bertram,G., Egerton,M., Gow,N.A., Falkow,S. and Brown,A.J. (1997) Yeast-enhanced green fluorescent protein (yEGFP)a reporter of gene expression in Candida albicans. Microbiology, 143, 303–311. [DOI] [PubMed] [Google Scholar]
- Elowitz M.B. and Leibler,S. (2000) A synthetic oscillatory network of transcriptional regulators. Nature, 403, 335–338. [DOI] [PubMed] [Google Scholar]
- Ferrell J.E.,Jr and Machleder,E.M. (1998) The biochemical basis of an all-or-none cell fate switch in Xenopus oocytes. Science, 280, 895–898. [DOI] [PubMed] [Google Scholar]
- Fiering S., Whitelaw,E. and Martin,D.I. (2000) To be or not to be active: the stochastic nature of enhancer action. BioEssays, 22, 381–387. [DOI] [PubMed] [Google Scholar]
- Forsberg E.C., Zaboikina,T.N., Versaw,W.K., Ahn,N.G. and Bresnick,E.H. (1999) Enhancement of β-globin locus control region-mediated transactivation by mitogen-activated protein kinases through stochastic and graded mechanisms. Mol. Cell. Biol., 19, 5565–5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman M. (2000) Feedback control of intercellular signalling in development. Nature, 408, 313–319. [DOI] [PubMed] [Google Scholar]
- Gardner T.S., Cantor,C.R. and Collins,J.J. (2000) Construction of a genetic toggle switch in Escherichia coli. Nature, 403, 339–342. [DOI] [PubMed] [Google Scholar]
- Gari E., Piedrafita,L., Aldea,M. and Herrero,E. (1997) A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast, 13, 837–848. [DOI] [PubMed] [Google Scholar]
- Gossen M., Freundlieb,S., Bender,G., Muller,G., Hillen,W. and Bujard,H. (1995) Transcriptional activation by tetracyclines in mammalian cells. Science, 268, 1766–1769. [DOI] [PubMed] [Google Scholar]
- Gunge N. (1983) Yeast DNA plasmids. Annu. Rev. Microbiol., 37, 253–276. [DOI] [PubMed] [Google Scholar]
- Hasty J., Pradines,J., Dolnik,M. and Collins,J.J. (2000) Noise-based switches and amplifiers for gene expression. Proc. Natl Acad. Sci. USA, 97, 2075–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hume D.A. (2000) Probability in transcriptional regulation and its implications for leukocyte differentiation and inducible gene expression. Blood, 96, 2323–2328. [PubMed] [Google Scholar]
- Keller A.D. (1995) Model genetic circuits encoding autoregulatory transcription factors. J. Theor. Biol., 172, 169–185. [DOI] [PubMed] [Google Scholar]
- Ko M.S., Nakauchi,H. and Takahashi,N. (1990) The dose dependence of glucocorticoid-inducible gene expression results from changes in the number of transcriptionally active templates. EMBO J., 9, 2835–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kringstein A.M., Rossi,F.M., Hofmann,A. and Blau,H.M. (1998) Graded transcriptional response to different concentrations of a single transactivator. Proc. Natl Acad. Sci. USA, 95, 13670–13675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D., Venkov,P. and Zlatanova,J. (1995) Transcriptional regulation in the yeast GAL gene family: a complex genetic network. FASEB J., 9, 777–787. [DOI] [PubMed] [Google Scholar]
- Novick A. and Weiner,M. (1957) Enzyme induction as an all-or-none phenomenon. Proc. Natl Acad. Sci. USA, 43, 553–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osheim Y.N., Mougey,E.B., Windle,J., Anderson,M., O’Reilly,M., Miller,O.L.,Jr, Beyer,A. and Sollner-Webb,B. (1996) Metazoan rDNA enhancer acts by making more genes transcriptionally active. J. Cell Biol., 133, 943–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polach K.J. and Widom,J. (1996) A model for the cooperative binding of eukaryotic regulatory proteins to nucleosomal target sites. J. Mol. Biol., 258, 800–812. [DOI] [PubMed] [Google Scholar]
- Rossi F.M., Kringstein,A.M., Spicher,A., Guicherit,O.M. and Blau,H.M. (2000) Transcriptional control: rheostat converted to on/off switch. Mol. Cell, 6, 723–728. [DOI] [PubMed] [Google Scholar]
- Smolen P., Baxter,D.A. and Byrne,J.H. (1998) Frequency selectivity, multistability and oscillations emerge from models of genetic regulatory systems. Am. J. Physiol., 274, C531–C542. [DOI] [PubMed] [Google Scholar]
- Struhl K. (1999) Fundamentally different logic of gene regulation in eukaryotes and prokaryotes. Cell, 98, 1–4. [DOI] [PubMed] [Google Scholar]
- Thieffry D., Huerta,A.M., Perez-Rueda,E. and Collado-Vides,J. (1998) From specific gene regulation to genomic networks: a global analysis of transcriptional regulation in Escherichia coli. BioEssays, 20, 433–440. [DOI] [PubMed] [Google Scholar]
- Vashee S., Melcher,K., Ding,W.V., Johnston,S.A. and Kodadek,T. (1998) Evidence for two modes of cooperative DNA binding in vivo that do not involve direct protein–protein interactions. Curr. Biol., 8, 452–458. [DOI] [PubMed] [Google Scholar]
- Walters M.C., Fiering,S., Eidemiller,J., Magis,W., Groudine,M. and Martin,D.I. (1995) Enhancers increase the probability but not the level of gene expression. Proc. Natl Acad. Sci. USA, 92, 7125–7129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walz D. and Caplan,S.R. (1995) Chemical oscillations arise solely from kinetic nonlinearity and hence can occur near equilibrium. Biophys. J., 69, 1698–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Ellwood,K., Lehman,A., Carey,M.F. and She,Z.S. (1999) A mathematical model for synergistic eukaryotic gene activation. J. Mol. Biol., 286, 315–325. [DOI] [PubMed] [Google Scholar]