Abstract
The dihydroxyacetone kinase (DhaK) of Escherichia coli consists of three soluble protein subunits. DhaK (YcgT; 39.5 kDa) and DhaL (YcgS; 22.6 kDa) are similar to the N- and C-terminal halves of the ATP-dependent DhaK ubiquitous in bacteria, animals and plants. The homodimeric DhaM (YcgC; 51.6 kDa) consists of three domains. The N-terminal dimerization domain has the same fold as the IIA domain (PDB code 1PDO) of the mannose transporter of the bacterial phosphoenolpyruvate:sugar phosphotransferase system (PTS). The middle domain is similar to HPr and the C-terminus is similar to the N-terminal domain of enzyme I (EI) of the PTS. DhaM is phosphorylated three times by phosphoenolpyruvate in an EI- and HPr-dependent reaction. DhaK and DhaL are not phosphorylated. The IIA domain of DhaM, instead of ATP, is the phosphoryl donor to dihydroxyacetone (Dha). Unlike the carbohydrate-specific transporters of the PTS, DhaK, DhaL and DhaM have no transport activity.
Keywords: dihydroxyacetone kinase/phosphohistidine/protein phosphorylation/PTS
Introduction
Sugars are primed for metabolic transformation by phosphorylation. Sugar phosphates are formed by kinases at the expense of ATP, and by phosphorolytic cleavage of intracellular oligo- and polysaccharides. The equilibria of these reactions are far to the right and the intracellular concentration of free carbohydrates is kept low. In addition, in bacteria carbohydrates can be phosphorylated at the expense of phosphoenolpyruvate (PEP) (Kundig et al., 1964). Whereas phosphorolytic and ATP-dependent phosphorylation entails direct transfer of phosphate between donor and acceptor, phosphotransfer from PEP is mediated by a cascade of four proteins that are phosphorylated at histidines or cysteines (Figure 1B). The components at the end of the cascade are sugar-specific transporters that couple phosphorylation with translocation of their substrates. This transport mechanism, which results in chemical modification of the transported solute, is termed group translocation (Hays, 1978) or vectorial phosphorylation. The soluble phosphorelay proteins and the membrane-bound transporters together constitute the PEP:sugar phosphotransferase system (PTS). Several components of the phosphorylation cascade are also involved in regulation of metabolism in response to the availability of carbohydrate nutrients, hence the apparent complexity of the system (for a review see Postma et al., 1996). The first unit of the cascade, termed enzyme I (EI), is a soluble two-domain protein (LiCalsi et al., 1991). It catalyses the phosphoryl transfer from PEP to the second component: the small (9 kDa) soluble HPr (histidine-containing phosphoryl carrier protein). HPr itself then serves as phosphoryl donor to the different PTS transporters (so-called enzymes II). They consist of three to four functional units, termed IIA, IIB, IIC and IID (Saier and Reizer, 1992; Lengeler et al., 1994; Robillard and Broos, 1999), which are individual subunits of a protein complex or independently folding domains of a multidomain protein. IIA and IIB sequentially transfer a phosphoryl group from the phosphoryl carrier protein HPr to the transported substrate. IIC and IID span the membrane and mediate substrate translocation. IIC mutants with impaired translocation but normal phosphorylation activity, and mutants that catalyse facilitated diffusion uncoupled from phosphorylation, have been isolated (Ruijter et al., 1990, 1992; Buhr et al., 1992). Cytoplasmic glucose can be phosphorylated by the IICBGlc subunit of the glucose transporter without the need to exit first and then re-enter (Thompson and Chassy, 1985; Nuoffer et al., 1988). Although of interest for understanding the catalytic mechanism, these partial reactions are not physiologically relevant. Phosphorylation coupled with transport and involvement of at least one membrane-spanning IIC unit is the rule, but there is an exception.
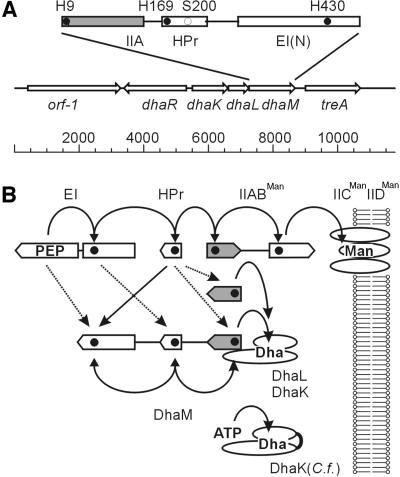
Fig. 1. The dihydroxyacetone kinase of E.coli. (A) Operon structure of the Dha PTS and domain structure of DhaM. The active site histidines are indicated with closed circles, the invariant Ser in the HPr domain with an open circle. (B) Phosphoryl flow between the general PTS proteins EI and HPr and the mannose transporter (top), and between the PTS and DhaM, respectively. Active site histidines are indicated with closed circles. Homologous domains are vertically aligned; the IIA domain is filled. Notice that IIA in DhaM is at the end of the phosphoprotein relay, whereas IIA of IIABMan is in the middle. Solid arrows indicate main phosphotransfer reactions; dotted arrows indicate cross-talk reactions. Shown at the bottom is the homologous but single-subunit DhaK of C.freundii, which utilizes ATP instead of phospho-DhaM as phosphoryl donor.
In Escherichia coli, dihydroxyacetone (Dha) is phosphorylated at the expense of PEP by three soluble proteins. The YcgT (SWISS-PROT P76015) and YcgS (SWISS-PROT P76014) subunits are similar to the dihydroxyacetone kinase (DhaK) of Citrobacter freundii (Daniel et al., 1995; SWISS-PROT P76015), and will henceforth be termed DhaK and DhaL. YcgC (SWISS-PROT P37349) is a PTS protein that serves as phosphoryl donor instead of ATP, and will henceforth be termed DhaM. There are multiple pathways to dihydroxyacetone phosphate (reviewed by Johnson et al., 1984): (i) formation of Dha by NAD+-dependent oxidation of glycerol and phosphorylation by an ATP-dependent kinase; (ii) flavin-dependent oxidation of glycerol-3-phosphate; and (iii) phosphorylation of Dha by a PEP-dependent PT mechanism. Whereas Klebsiella pneumoniae has all three pathways, E.coli has only (ii) and (iii). The PEP-dependent pathway was first identified by Jin and Lin (1984) in an E.coli double mutant lacking the ATP-dependent glycerol kinase and the NAD+-dependent dehydrogenase. They demonstrated that phosphorylation of Dha depends on two components: (i) EI of the PTS; and (ii) a gene at 26 min, the interruption of which abolished PEP-dependent phosphorylation of Dha. The transposon-targeted gene, termed ptsD, was predicted to encode a Dha-specific enzyme II (IIDha) of the PTS. However, no gene coding for IIDha was identified by in silico analyses of the E.coli genome, possibly because enzymes II were presumed to be multi-spanning membrane proteins. Only recently, upon re-examination of the 25 min region, were DhaK and DhaL identified as subunits of a dihydroxyacetone kinase and DhaM as a putative allosteric regulator of this kinase (Paulsen et al., 2000). We identified DhaK and DhaL on two-dimensional gels because they were both prominently upregulated in a ptsI (EI) mutant (Beutler, 2000; Beutler et al., 2001). DhaM was found because it was encoded in the same operon and could then be identified as a PTS protein by sequence comparison. We have purified the three subunits. All three are required for PEP-dependent phosphorylation of Dha, with phospho-DhaM serving as phosphoryl donor instead of ATP.
Results
Structure of the dha operon and amino acid sequence of its proteins
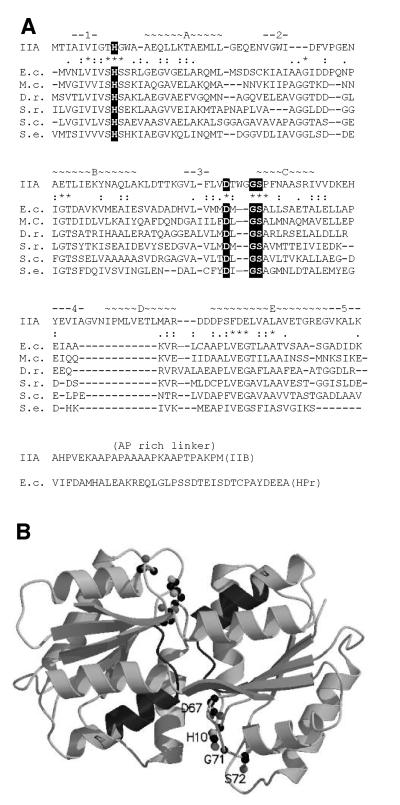
The dha operon comprises three cistrons, encoding in this order, DhaK, DhaL and DhaM (Figure 1A). The 3′ end of dhaK and the 5′ end of dhaL are separated by 10 non-coding nucleotides, dhaL and dhaM by only seven. The operon is bracketed on the 5′ side by the divergently transcribed gene dhaR, encoding a protein that is 70% identical to the Dha regulator of C.freundii (Daniel et al., 1995), and by treA on the 3′ side, which encodes a periplasmic trehalase (Gutierrez et al., 1989). DhaK (39.5 kDa) and DhaL (22.6 kDa) are 30 and 27% identical to the N- and C-terminal halves, respectively, of the DhaK (57.9 kDa) of C.freundii. DhaK shares with DhaK two histidine-containing and glycine-rich segments (SGGGSGHEPEPMH and GIHGEPG) and a third invariant sequence, KNYTGD. DhaL and the C-terminal half of DhaK have only a few short motifs in common. In view of the fact that DhaK/DhaL and DhaK have the same acceptor substrate but structurally different phosphoryl donors, the three invariant segments might be part of a common binding site for Dha. DhaM (51.6 kDa) is a multi-phosphoryl transfer protein of the PTS comparable to MTP of Rhodobacter capsulatus (Wu et al., 1990; SWISS-PROT P23388). It consists of three domains: the first (residues 1–129) is IIA like (see below); the second (residues ∼155–231) is HPr like (29% identity). It contains the GLHVRP motif of the phosphorylation site, and 31 residues further downstream the invariant serine. The third domain (residues ∼270–473) is similar to the N-terminal domain of enzymes I of the PTS (Liao et al., 1996). It displays 33% sequence identity, and contains the characteristic SHS motif of the EI phosphorylation site. Two properties of DhaM are noteworthy. First, it does not comprise a domain similar to the C-terminal domain of enzyme I that catalyses the phosphotransfer between PEP and the SHS histidine. Therefore, DhaM should be phosphorylated only in the presence of EI in trans. Secondly, the N-terminal domain of DhaM (residues ∼1–130) can be aligned (32–42% identity) with open reading frames (ORFs) of unknown function from Mycoplasma capricolum, Deinococcus radiodurans, Selenomonas ruminantium, Streptomyces coelicolor and Staphylococcus epidermidis (Figure 2A), but not with the IIA domain of the R.capsulatus MTP (<16% identity). However, using 3D-pssm, a Web-based method for protein fold recognition (http://www.bmm.icnet.uk/∼3dpssm/; Kelley et al., 2000), a striking similarity between the DhaM and the IIAMan domain of the IIABMan subunit of the mannose transporter (Erni et al., 1987, 1989; Nunn et al., 1996; PDB code 1PDO) could be detected. IIAMan has an (αβ)4αβ fold, with four parallel strands in the order 2134 and a fifth antiparallel strand donated by the opposite subunit (Figure 2B). This swap of a β-strand makes IIAMan a stable dimer (Markovic-Housley et al., 1994). His10 and Asp67 in loops β1αA and β3αC of IIAMan form a catalytic diad. These residues, invariant in all members of the IIAMan family, are also invariant in DhaM and the five similar ORFs (Figure 2A). From the similarity of design with IIABMan, DhaM is predicted to be a dimer of two interlocked subunits (Gutknecht et al., 1999).
Fig. 2. (A) Secondary structure assignment for the IIA domain of DhaM and sequence alignment. The secondary structure elements (α-helices A–E and β-strands 1–5) and the sequence of IIAMan are given at the top. Active site residues invariant between IIAMan and the six similar sequences are boxed. Sequence alignments between the IIA domain of DhaM (E.c.) and five similar ORFs of unknown function are given below. Residues that are invariant (*) and similar (:.) between these six sequences are indicated. The linker regions connecting IIA with IIB (Ala Pro rich) and the IIA-like with the HPr-like domains of DhaM, respectively, are shown at the bottom. E.c., E.coli; M.c., M.capricolum; D.r., D.radiodurans; S.r., S.ruminantium; S.c., S.coelicolor; S.e. S.epidermidis. (B) Model of the IIAMan dimer (PDB code 1PDO). Shown in ball and stick representation are the active site residues. Shown in dark grey is the part of helix D that is presumed to be shortened in DhaM. The shortening of helix D might widen the cleft between the two subunits and thereby facilitate the access to the phosphoryl donor His for Dha bound to the DhaK–DhaL complex.
Production and purification of DhaK, DhaL and DhaM
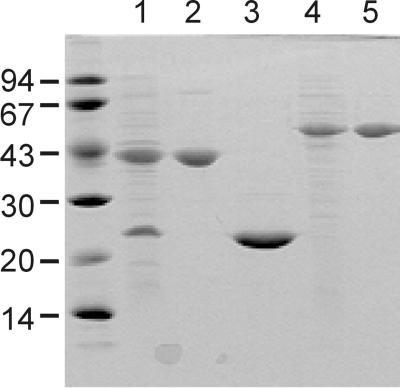
Full-length proteins were produced without affinity tags. DhaK and DhaL were synthesized together because it was initially not clear whether the two subunits would be stable independently of each other. The two subunits are overproduced in comparable amounts and in completely soluble form (Figure 3, lane 1). DhaK binds quantitatively to the anion exchange resin, DhaL only partly, although DhaK and DhaL have comparable pIs (4.7 and 5.1). DhaL and DhaK might form a weak complex, which is dissociated during purification. DhaK and DhaL can be completely separated by gel filtration in 0.15 M NaCl (Figure 3, lanes 2 and 3). DhaK elutes after the marker protein IIABMan (2 × 35 kDa), DhaL shortly before IIAGlc (18 kDa), suggesting a monomeric state for both proteins (results not shown). Production and purification of DhaM are straightforward (Figure 3, lanes 4 and 5). As expected of a 2 × 52 kDa homodimer, DhaM elutes between the dimeric EI (2 × 63 kDa) and IIABMan (2 × 35 kDa) on gel filtration (results not shown).
Fig. 3. Purification of DhaK, DhaL and DhaM. Whole-cell lysates containing DhaK and DhaL (lane 1) and DhaM (lane 4). Separated DhaK (lane 2), DhaL (lane 3) and DhaM (lane 5) after gel filtration purification. The 15% polyacrylamide gel is stained with Coomassie Blue.
Dha phosphotransferase (PT) activity
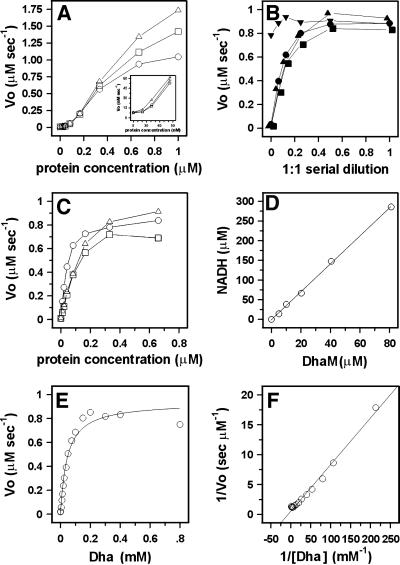
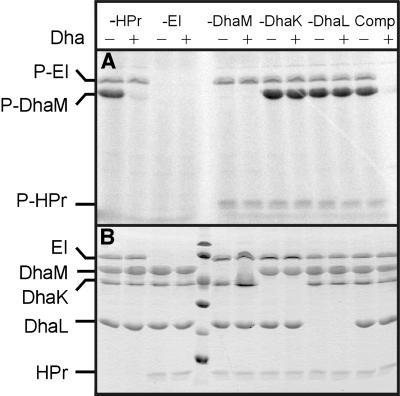
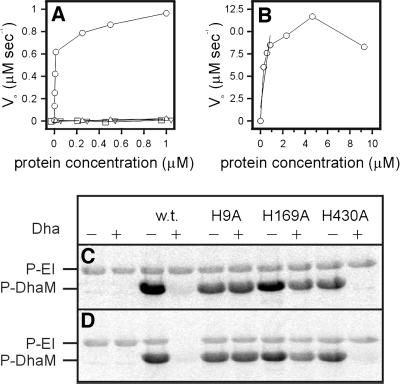
All three subunits, DhaM, DhaK and DhaL, are necessary for PEP-dependent phosphorylation of Dha. PT activity increases non-linearly at subunit concentrations <0.1 µM (Figure 4A, insert), as expected if the catalytic complex is formed of two or more randomly colliding subunits. The reaction rate starts to level off when the reduction of Dha phosphate by glycerol-3-phosphate dehydrogenase becomes rate limiting. PT activity further depends on PEP, EI and HPr, and the single omission of either EI or HPr completely abolishes the PT activity (Figure 4B). This indicates that the HPr-like domain of DhaM cannot efficiently substitute for the general phosphoryl carrier protein HPr (Figure 1B). ATP cannot serve as phosphoryl donor to DhaK and DhaL (Figure 4B). All three subunits of the DhaK complex must be present in ∼1:1:1 molar ratio. PT activity decreases as soon as the concentration of one of the three is reduced (Figure 4C). The KM of the purified DhaK–DhaL–DhaM complex for Dha is 45 µM and kcat is 2.8 s–1 (Figure 4E and F).
Fig. 4. Dha phosphorylation and DhaM active site titration. (A) Rate of Dha phosphorylation by DhaM–DhaK–DhaL at a constant molar ratio of 1:1:1 and three different concentrations of glycerol-3-phosphate dehydrogenase (open circle, 1.0 µg/ml; open square, 2.0 µg/ml and open triangle, 4.0 µg/ml) The insert shows the non-linearity at low concentration. (B) Dependence of Dha phosphorylation upon 1:1 serial dilution of ATP (upside-down triangle, ≤1.0 mM), PEP (closed triangle, ≤1.0 mM), EI (closed circle, ≤10 nM) and HPr (closed square, ≤180 nM). The numbers in parentheses indicate the maximum concentrations used at dilution 1. (C) Rate of Dha phosphorylation by DhaK, DhaM and DhaL. The concentration of two subunits was kept constant at 0.33 µM, while the third (open circle, DhaM; open square, DhaK; open triangle, DhaL) was varied from 0 to 0.66 µM. (D) Active site titration of DhaM. DhaM was incubated with PEP and catalytic concentrations of EI and HPr. The amount of PEP consumed was determined from the NADH consumed during the reduction of pyruvate with lactate dehydrogenase (Mukhija and Erni, 1997). The ratio NADH/DhaM obtained from the linear regression is 3.5. (E) Michaelis–Menten plot. The rate-limiting concentrations of DhaM, DhaK and DhaL were 0.33 µM each. KM = 45 ± 6 µM and kcat = 2.8 ± 0.1 s–1 were calculated by non-linear least squares fit to a hyperbola. (F) Double-reciprocal plot of the Michaelis–Menten plot from (E).
Protein phosphorylation and active site titration
Equimolar amounts of EI, HPr, DhaM, DhaK and DhaL were incubated with [33P]PEP, and analysed by gel electrophoresis and autoradiography. DhaM was strongly phosphorylated, whereas no traces of phosphorylated DhaK or DhaL could be detected (Figure 5A). DhaM is six times more strongly phosphorylated than EI and HPr, indicative of multiple phosphorylation of DhaM. Active site titration of DhaM with PEP in the presence of catalytic amounts of EI and HPr revealed that ∼3.5 mol PEP are consumed per mole of DhaM (Figure 4D). This number is consistent with three phosphorylation sites, which are presumed to be the invariant His9, His169 and His430 (Figure 1B). Neither DhaK nor DhaL caused a burst of pyruvate formation in this assay, indicating that the absence of an autoradiographic signal is not due to phosphoprotein hydrolysis during gel electrophoresis (results not shown) and confirming that these proteins are indeed not phosphorylated. Phosphorylation of DhaM depends strictly upon EI but not upon HPr (Figure 5, first and second lanes). Contamination by HPr of the purified EI, DhaK, DhaL or DhaM preparations can be excluded, because all proteins were produced in ptsH (HPr deletion) strains. We conclude that slow transfer of phosphoryl groups also occurs from EI to DhaM. The transfer rate suffices to radiolabel 0.1 nmol of protein, but not several micromoles of Dha.
Fig. 5. Phosphorylation of DhaM. Equimolar amounts of purified DhaM, DhaK, DhaL, EI and HPr were incubated with [33P]PEP for 10 min at 37°C either together (Comp) or with one component at a time omitted. To one aliquot was added a molar excess of Dha (+) to dephosphorylate the PTS proteins. One polyacrylamide gel was autoradiographed (A). Note that HPr is dispensible for slow phosphotransfer from PEP to Dha, but that both DhaK and DhaL are essential. The second gel was stained with Coomassie Blue (B).
Functional characterization of active site mutants and the isolated IIA-like domain of DhaM
The general phosphoryl carrier protein HPr is obviously necessary for efficient phosphorylation of Dha (Figure 4B). There are two likely pathways (Figure 1B) for the fast phosphotransfer between HPr and DhaM: (i) HPr(His15) → DhaM(His430 → His169 → His9) → Dha; (ii) HPr(His15) → DhaM(His9) → Dha (with a possibility of backward phosphorylation of His169 and His430). For the slow phosphorylation of DhaM observed in the absence of HPr (Figure 5A, lanes 1 and 2), there are also two possible pathways: (i) PEP → DhaM(His430) mediated by the C-terminal domain of EI; and (ii) PEP → EI(His189) → DhaM(His169). The third possibility, direct phosphotransfer between HPr and the HPr-like domain or between the N-terminal domain of EI and the EI-like domain of DhaM, is less plausible because of the non-complementarity of the interacting sites (Schauder et al., 1998).
To characterize different pathways of phosphotransfer, the active site histidines of DhaM were mutated to alanine, one at a time, to afford the three mutants DhaM(H9A), DhaM(H169A) and DhaM(H430A). In addition, the IIA-like domain of DhaM (residues 1–129) was produced as protein subunit DhaM*.
The three His→Ala mutants are completely inactive (Figure 6A). This outcome is expected of DhaM(H9A) if phospho-His9 is the immediate phosphoryl donor to Dha. That DhaM(H169A) and DhaM(H430A) are also inactive indicates that HPr(His15) → DhaM(His430 → His169 → His9) → Dha is the only route of fast phosphotransfer (Figure 1B, solid arrows), and that EI(His189) → DhaM(His169) and HPr(His15) → DhaM(His9) are not alternative pathways. DhaM*, the separate IIA-like domain, has <10 nmol/µmol/s PT activity, which is <0.3% of DhaM-specific activity (Figure 6B). This indicates that direct phosphotransfer between His15 of HPr and His9 of DhaM is possible, but slow.
Fig. 6. Activities of DhaM mutants. (A) Rate of Dha phosphorylation by DhaM wild type (open circle), H9A (open square), H169A (open triangle) and H430A (upside-down triangle). The concentrations of DhaK and DhaL were kept constant at 0.33 µM. (B) Rate of Dha phosphorylation by DhaM*, the recombinant IIA-like domain of DhaM. The concentration of DhaK and DhaL was 0.33 µM. (C and D) Phosphorylation of DhaM mutants. DhaM wild type and mutants were phosphorylated with [33P]PEP in the absence (C) and presence (D) of HPr. To one of two identical aliquots was added a molar excess of Dha (+) to allow for dephosphorylation of the proteins. Shown are the EI- and DhaM-containing sectors from the autoradiogram. The complete autoradiograms and Coomassie Blue-stained gels had the same appearance as in Figure 5. For experimental details see legend to Figure 5.
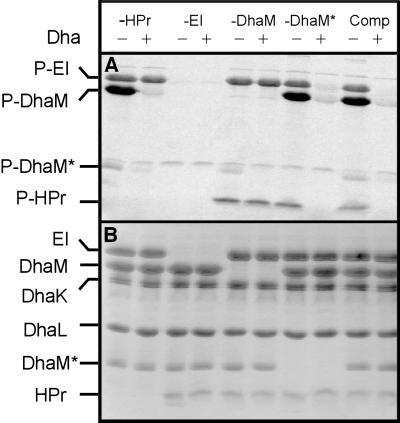
To identify additional pathways of slow transfer, the H9A, H169A and H430A mutants of DhaM and DhaM* were phosphorylated in the absence and presence of HPr (Figures 6C, D and 7). DhaM(H430A) is phosphorylated without and with HPr (Figure 6C and D, lanes 9 and 10), indicating that phosphotransfer from His189 of EI to His169 on the HPr-like domain of DhaM, and from His15 of HPr to His9 of DhaM, occur at measurable rates. DhaM(H430A) is dephosphorylated by Dha (Figure 6C and D, lane 10), as expected of DhaM with a functional IIA-like domain. The H169A and H9A mutants of DhaM are also phosphorylated both in the absence and presence of HPr (Figure 6C and D, lanes 5–8). In the absence of HPr, DhaM must be phosphorylated by direct phosphotransfer from PEP to His430 catalysed by the C-terminal domain of EI. In the presence of HPr, His9 of DhaM in addition is phosphorylated by HPr (see below and Figure 7). Unlike the H430A mutant, the H9A and H169A mutants cannot be dephosphorylated by Dha, as expected if His9 is the phosphoryl donor to Dha and if phosphotransfer between His430 and His9 depends on His169. The isolated IIA-like domain, DhaM*, is weakly phosphorylated by the general phosphoryl carrier protein HPr (Figure 7A, lane 5), and in the absence of HPr also by full-length DhaM (Figure 7A, lane 1), indicating intermolecular phosphotransfer between His169 of a DhaM subunit and His9 of truncated DhaM* as an additional pathway of phosphotransfer.
Fig. 7. Phosphorylation of the IIA-like domain of DhaM. Stoichiometric amounts of purified IIA-like domain (DhaM*), DhaM, DhaK, DhaL, EI and HPr were incubated with [33P]PEP for 10 min at 37°C, either together (Comp) or with the indicated proteins omitted. To one aliquot was added a molar excess of Dha (+) to dephosphorylate the PTS proteins. Note that the IIA-like domain (DhaM*) is phosphorylated by HPr (in the absence of DhaM) as well as by DhaM (in the absence of HPr), and is completely dephosphorylated by Dha. (A) Autoradiography; (B) gel stained with Coomassie Blue.
Discussion
Dha is phosphorylated by PEP in a reaction catalysed by components of the bacterial PEP:sugar PTS. The Dha binding moiety is a soluble heterodimeric complex homologous to the ATP-dependent dihydroxyacetone kinase, and not a membrane-spanning enzyme II of the PTS as originally suspected (Jin and Lin, 1984). ATP-dependent dihydroxyacetone kinases are ubiquitous in bacteria (Daniel et al., 1995), yeast, animals and plants. It therefore appears that in the course of evolution the enzyme now found in E.coli has switched from ATP as a source of high-energy phosphate to a phosphohistidine protein of the PTS. Why a phosphoprotein instead of ATP? ATP is a highly connected metabolite (Jeong et al., 2000) and therefore not suitable as a sensor to monitor the availability of a particular metabolite. In principle, a phosphoprotein such as DhaM, dedicated to phosphorylating a single or a few similar substrates, can assume this function, as demonstrated for other PTS proteins (for reviews see Amster-Choder et al., 1989; Schnetz and Rak, 1990; Stülke et al., 1998; Görke and Rak, 1999). The multiple phosphorylation and oligomeric structure of DhaM might permit non-linear and time-delayed responses to intracellular metabolic cues. For example, dephosphorylated DhaM might act directly as a co-inducer, perhaps together with the nearby DhaR regulator.
The general phosphoryl carrier protein HPr and all three active site histidines of DhaM are required for efficient transfer, suggesting the sequence of transfer reactions depicted by the solid arrows in Figure 1B. However, cross-talk between EI/HPr on one side and the phosphorylation sites of DhaM on the other can be detected if protein phosphorylation is measured (Figure 6C and D). Slow phosphotransfer occurs by all routes between complementary domains on EI and DhaM. Whether protein phosphorylation and dephosphorylation in the presence of Dha are detectable depends strongly on the relationship between PEP concentration and incubation time (results not shown). Because all three active site mutants can be phosphorylated, but at the same time do not have catalytic activity, the phosphotransfer reactions detected by protein phosphorylation are most likely not relevant physiologically.
DhaM is composed of three functional modules with similarities to the IIABMan subunit of the mannose transporter and the general PTS proteins EI and HPr. Whereas the EI- and HPr-like domains share 30% sequence identity with other PTS proteins, the similarity of fold and sequence between the IIA domain and IIAMan could only be detected by 3D template matching (Figure 2A). Sequence alignment of DhaM with IIAMan shows a deletion of 12 residues extending from loop β4αD into helix D of IIAMan (Figure 2B). These modifications could be an adaptation of the structure for better binding of the Dha–DhaK–DhaL ternary complex. A docking model of the IIAMan–IIBMan complex showed that the imidazole residue of the acceptor His on IIB is on a convex surface that can reach deep into the narrow cleft between the subunits of the IIA dimer (Schauder et al., 1998). Dha, in contrast to the acceptor His of IIB, is not covalently bound. Non-covalent binding requires several weak interactions and the Dha binding site is, therefore, likely to be extended if not a shallow cleft. Truncation of helix D of the IIA domain of DhaM could widen the cleft and create space for docking with the flat (less convex binding) surface of the Dha–DhaK–DhaL ternary complex.
Materials and methods
Plasmid construction
The overlapping coding regions of dhaK and dhaL were PCR amplified in one piece, and dhaM separately with genomic DNA as template and primers encoding NdeI and HindIII restriction sites. The PCR fragments were digested with NdeI and HindIII, and ligated with the expression vector pMSEH2 (Beutler et al., 2000). The plasmid pUBIIA encoding the IIA-like domain of DhaM (residues 1–129) with an N-terminal His tag was prepared from a PCR fragment. The sequences of the N- and C-termini are NH2mgsshhhhhhssglvprgshM1VNL/HALE129-COOH. The active site mutations H9A, H169A and H430A of DhaM were introduced by primer extension mutagenesis (Ho et al., 1989) using primers encoding the His to Ala mutation and a diagnostic NheI restriction site. Successful mutagenesis was verified by DNA sequencing. Standard procedures were used for plasmid purification, restriction analysis, ligation and transformation.
Production and purification of DhaL, DhaK and DhaM
Escherichia coli K-12 WA2127ΔHIC (ΔmanXYZ ΔptsHIcrr) (Mao et al., 1995) was used to overproduce the protein subunits. Cells were grown in 1 l of Luria–Bertani (LB) medium in an Erlenmeyer flask on a rotary shaker at 37°C. When the cells had reached A550 = 1.0, gene expression was induced with 0.05 mM isopropyl-β-d-thiogalactopyranoside and incubation was continued for 5 h.
Purification of DhaM. Cells were harvested by centrifugation (7500 g for 20 min at 4°C), resuspended in 2.5 ml/g wet weight of buffer A [20 mM Tris–HCl pH 7.6, 1 mM EDTA, 0.5 mM dithiothreitol (DTT), 0.2 mM phenylmethylsulfonyl fluoride (PMSF)] and lysed by two passages through a French pressure cell (1000 p.s.i.). Cell debris was removed by low-speed centrifugation (12 000 g for 10 min at 4°C), membranes by high-speed centrifugation (360 000 g for 1 h at 4°C), and the supernatants containing the soluble proteins were applied to ion-exchange columns as follows. Extracts containing DhaM from 1 l of culture were applied to a HighQ anion exchange column (10 ml Macro Prep, HighQ Support; Bio-Rad) equilibrated with buffer A. The column was developed with a gradient of 0–800 mM NaCl in buffer A (flow rate 1.5 ml/min, 4.5 ml/fraction). DhaM eluted at 0.5 M NaCl. The trailing half of the peak was pooled, concentrated in a Centriprep-30 concentrator to 5 ml and 2.5 ml were applied to a gel filtration column (Superdex 200 16/60; Pharmacia) equilibrated with buffer B (20 mM NaPi pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.5 mM DTT) and eluted at a flow rate of 1 ml/min. The peak fractions were pooled. The yield was ∼50 mg of pure DhaM per litre of cell culture.
Purification of DhaK and DhaL. Cells were lysed in buffer A pH 8.6 as described for DhaM. Extracts from 1 l were applied to 15 ml DEAE–cellulose (C545, Fluka) equilibrated with buffer A pH 8.6. More than 50% of DhaL did not bind and eluted in the void volume. The DhaL-containing fractions were pooled and concentrated in a Centriprep-10 concentrator. DhaK was eluted together with the remaining DhaL in a gradient of 0–700 mM NaCl at a flow rate of 1 ml/min. DhaK-containing fractions were pooled and concentrated in a Centriprep-30 concentrator. DhaL from the flow-through and DhaK were further purified by gel filtration (Superdex 75 16/60, equilibrated with buffer B, flow rate 1 ml/min). DhaK and DhaL, which eluted well separated, were rechromatographed to remove minor contaminants. The yield per litre was 60 mg of DhaK and 3 mg of DhaL.
Purification of DhaM*, the IIA-like domain of DhaM. Escherichia coli K-12 BL21 transformed with plasmid pUBIIA were grown in 300 ml of LB in an Erlenmeyer flask on a rotary shaker at 28°C. When the cells had reached A550 = 1.0, gene expression was induced with 0.3 mM IPTG and incubation was continued at 22°C (to minimize the formation of inclusion bodies) for 4 h. Cells were lysed by one passage through a French pressure cell (8 ml buffer G, 50 mM NaPi pH 8.0, 0.5 M NaCl, 5 mM imidazole, 10 mM β-mercaptoethanol) and the high-speed supernatant mixed with 4 ml of Ni2+-NTA–agarose equilibrated in buffer G. After 30 min at 22°C, the slurry was cooled to 4°C and poured into a column. The column was washed with 15 ml each of buffers G, H (50 mM NaPi pH 5.5, 0.5 M NaCl, 10 mM β-mercaptoethanol) and I30 (30 mM imidazole, 50 mM NaPi pH 7.4, 0.5 M NaCl, 10 mM β-mercapto ethanol). DhaM* was eluted with buffer I150 (150 mM imidazole in buffer I). The yield was between 5 and 10 mg of 80% pure protein.
Assay for PEP: Dha PT activity
Reactions were performed in 96-well microtitre plates. DhaP was reduced with NADH by glycerol-3-phosphate dehydrogenase. The disappearance of NADH was monitored in a Spectramax-250 plate reader, continuously during 10 min at 30°C. Initial rates were calculated from the maximal slope of the absorption at 340 nm. One millilitre of reaction mixture contained 9 µg of EI, 25 µg of HPr (purified as described by Mao et al., 1995), DhaK, DhaL and DhaM as specified in the figure legends, 1 µg of rabbit glycerol-3-phosphate dehydrogenase (Fluka), 0.8 mM Dha, 1.0 mM PEP, 0.5 mM NADH, 50 mM KPi pH 7.5, 2.5 mM DTT and 2.55 mM MgCl2. The reaction volume was 150 µl/well and the reactions were started by the addition of PEP.
Assay for protein phosphorylation
[33P]PEP was a gift of Dr Seema Mukhija (Arpida AG, Münchenstein). Assay mixtures (20 µl) contained approximately equimolar amounts (0.5–5 µg) of EI, HPr, DhaK, DhaL and DhaM, 10 mM NaPi pH 7.0, 5 mM MgCl2, 0.65 mM [33P]PEP and 1.3 mM Dha where indicated. Incubation was for 10 min at 37°C. The reaction was stopped by dilution with gel electrophoresis sample buffer. Duplicate gels were run, one of which was stained with Coomassie Blue and the other dried and analysed on a phosphoimager.
Active site titration of DhaM
The amount of PEP consumed upon phosphorylation of a known amount of DhaM was determined in a coupled assay with pyruvate dehydrogenase. NADH consumption was determined colorimetrically at 340 nm in a Spectramax-250 plate reader. One millilitre of reaction mixture contained catalytic amounts of EI, HPr, 1.0 mM PEP, 0.5 mM NADH, 1.5 µg of lactate dehydrogenase (Boehringer Mannheim), 50 mM KPi pH 7.5, 2.5 mM DTT, 2.5 mM NaF, 2.5 mM MgCl2, and between 0 and 80 µM purified DhaM. The reaction was started by the addition of PEP. In order to exclude the remote possibility that DhaK is phosphorylated but that the phosphoprotein is too labile for detection by autoradiography, DhaK was also tested by active site titration in the presence of catalytic amounts of EI, HPr and DhaM. No PEP was consumed (results not shown).
Acknowledgments
Acknowledgements
We thank Dr S.Mukhija (ARPIDA AG, Münchenstein) for the gift of [33P]PEP. This work was supported by grants 31-45838.95 from the Swiss National Science Foundation and a grant from the Secretaria de Estado de Educacion y Universidades to L.F.G.-A.
References
- Amster-Choder O., Houman,F. and Wright,A. (1989) Protein phosphorylation regulates transcription of the β-glucoside utilization operon in E. coli. Cell, 58, 847–855. [DOI] [PubMed] [Google Scholar]
- Beutler R. (2000) Bacterial phosphotransferase system (PTS): topological variants of a multispanning membrane protein and a proteomic approach to investigate PTS mediated signal transduction. PhD thesis, University of Bern, Bern, Switzerland.
- Beutler R., Ruggiero,F. and Erni,B. (2000) Folding and activity of circularly permuted forms of a polytopic membrane protein. Proc. Natl Acad. Sci. USA, 97, 1477–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutler R., Kämpfer,U., Schaller,J. and Erni,B. (2001) Heterodimeric dihydroxyacetone kinase from a ptsI mutant of Escherichia coli. Microbiology, 147, 249–250. [DOI] [PubMed] [Google Scholar]
- Buhr A., Daniels,G.A. and Erni,B. (1992) The glucose transporter of Escherichia coli. Mutants with impaired translocation activity that retain phosphorylation activity. J. Biol. Chem., 267, 3847–3851. [PubMed] [Google Scholar]
- Daniel R., Stuertz,K. and Gottschalk,G. (1995) Biochemical and molecular characterization of the oxidative branch of glycerol utilization by Citrobacter freundii. J. Bacteriol., 177, 4392–4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erni B., Zanolari,B. and Kocher,H.P. (1987) The mannose permease of Escherichia coli consists of three different proteins: amino-acid sequence and function in sugar transport, sugar phosphorylation, and penetration of phage λ DNA. J. Biol. Chem., 262, 5238–5247. [PubMed] [Google Scholar]
- Erni B., Zanolari,B., Graff,P. and Kocher,H.P. (1989) Mannose permease of Escherichia coli. Domain structure and function of the phosphorylating subunit. J. Biol. Chem., 264, 18733–18741. [PubMed] [Google Scholar]
- Görke B. and Rak,B. (1999) Catabolite control of Escherichia coli regulatory protein BglG activity by antagonistically acting phosphorylations. EMBO J., 18, 3370–3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez C., Ardourel,M., Bremer,E., Middendorf,A., Boos,W. and Ehmann,U. (1989) Analysis and DNA sequence of the osmoregulated treA gene encoding the periplasmic trehalase of Escherichia coli K12. Mol. Gen. Genet., 217, 347–354. [DOI] [PubMed] [Google Scholar]
- Gutknecht R., Flükiger,K., Lanz,R. and Erni,B. (1999) Mechanism of phosphoryl transfer in the dimeric IIABMan subunit of the Escherichia coli mannose transporter. J. Biol. Chem., 274, 6091–6096. [DOI] [PubMed] [Google Scholar]
- Hays J.B. (1978) Group translocation transport systems. In Rosen,B. (ed.), Bacterial Transport. Marcel Dekker, New York, NY, pp. 43–102.
- Ho S.N., Hunt,H.D., Horton,R.M., Pullen,J.K. and Pease,L.R. (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene, 77, 51–59. [DOI] [PubMed] [Google Scholar]
- Jeong H., Tombor,B., Albert,R., Oltvai,Z.N. and Barabasi,A.L. (2000) The large-scale organization of metabolic networks. Nature, 407, 651–654. [DOI] [PubMed] [Google Scholar]
- Jin R.Z. and Lin,E.C.C. (1984) An inducible phosphoenolpyruvate: dihydroxyacetone phosphotransferase system in Escherichia coli. J. Gen. Microbiol., 130, 83–88. [DOI] [PubMed] [Google Scholar]
- Johnson E.A., Burke,S.K., Forage,R.G. and Lin,E.C. (1984) Purification and properties of dihydroxyacetone kinase from Klebsiella pneumoniae. J. Bacteriol., 160, 55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley L.A., MacCallum,R.M. and Sternberg,M.J. (2000) Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol., 299, 499–520. [DOI] [PubMed] [Google Scholar]
- Kundig W., Gosh,S. and Roseman,S. (1964) Phosphate bound to histidine in protein as an intermediate in a novel phosphotransferase system. Proc. Natl Acad. Sci. USA, 52, 1067–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengeler J.W., Jahreis,K. and Wehmeier,U.F. (1994) Enzymes II of the phosphoenolpyruvate-dependent phosphotransferase systems: their structure and function in carbohydrate transport. Biochim. Biophys. Acta, 1188, 1–28. [DOI] [PubMed] [Google Scholar]
- Liao D.I., Silverton,E., Seok,Y.J., Lee,B.R., Peterkofsky,A. and Davies,D.R. (1996) The first step in sugar transport: crystal structure of the amino terminal domain of enzyme I of the E.coli PEP:sugar phosphotransferase system and a model of the phosphotransfer complex with HPr. Structure, 4, 861–872. [DOI] [PubMed] [Google Scholar]
- LiCalsi C., Crocenzi,T.S., Freire,E. and Roseman,S. (1991) Sugar transport by the bacterial phosphotransferase system. Structural and thermodynamic domains of enzyme I of Salmonella typhimurium. J. Biol. Chem., 266, 19519–19527. [PubMed] [Google Scholar]
- Mao Q., Schunk,T., Gerber,B. and Erni,B. (1995) A string of enzymes, purification and characterization of a fusion protein comprising the four subunits of the glucose phosphotransferase system of Escherichia coli. J. Biol. Chem., 270, 18295–18300. [DOI] [PubMed] [Google Scholar]
- Markovic-Housley Z., Cooper,A., Lustig,A., Flükiger,K., Stolz,B. and Erni,B. (1994) Independent folding of the domains in the hydrophilic subunit IIAB(man) of the mannose transporter of Escherichia coli. Biochemistry, 33, 10977–10984. [DOI] [PubMed] [Google Scholar]
- Mukhija S. and Erni,B. (1997) Phage display selection of peptides against enzyme I of the phosphoenolpyruvate sugar phospho transferase system (PTS). Mol. Microbiol., 25, 1159–1166. [DOI] [PubMed] [Google Scholar]
- Nunn R.S., Markovic-Housley,Z., Génovésio-Taverne,J.C., Flükiger,K., Rizkallah,P.J., Jansonius,J.N., Schirmer,T. and Erni,B. (1996) Structure of the IIA domain of the mannose transporter from Escherichia coli at 1.7 Å resolution. J. Mol. Biol., 259, 502–511. [DOI] [PubMed] [Google Scholar]
- Nuoffer C., Zanolari,B. and Erni,B. (1988) Glucose permease of Escherichia coli. The effect of cysteine to serine mutations on the function, stability, and regulation of transport and phosphorylation. J. Biol. Chem., 263, 6647–6655. [PubMed] [Google Scholar]
- Paulsen I.T., Reizer,J., Jin,R.Z., Lin,E.C. and Saier,M.H. (2000) Functional genomic studies of dihydroxyacetone utilization in Escherichia coli. Microbiology, 146, 2343–2344. [DOI] [PubMed] [Google Scholar]
- Postma P.W., Lengeler,J.W. and Jacobson,G.R. (1996) Phospho enolpyruvate: carbohydrate phosphotransferase systems. In Neidhardt,F.C. et al. (eds), Escherichia coli and Salmonella: Cellular and Molecular Biology. ASM Press, Washington, DC, pp. 1149–1174.
- Robillard G.T. and Broos,J. (1999) Structure/function studies on the bacterial carbohydrate transporters, enzymes II, of the phospho enolpyruvate-dependent phosphotransferase system. Biochim. Biophys. Acta, 1422, 73–104. [DOI] [PubMed] [Google Scholar]
- Ruijter G.J.G., Postma,P.W. and van Dam,K. (1990) Adaptation of Salmonella typhimurium mutants containing uncoupled enzyme IIGlc to glucose-limited conditions. J. Bacteriol., 172, 4783–4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruijter G.J.G., van Meurs,G., Verwey,M.A., Postma,P.W. and van Dam, K. (1992) Analysis of mutations that uncouple transport from phos phorylation in enzyme-IIGlc of the Escherichia coli phospho enolpyruvate-dependent phosphotransferase system. J. Bacteriol., 174, 2843–2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier M.H. and Reizer,J. (1992) Proposed uniform nomenclature for the proteins and protein domains of the bacterial phosphoenol pyruvate:sugar phosphotransferase system. J. Bacteriol., 174, 1433–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauder S., Nunn,R.S., Lanz,R., Erni,B. and Schirmer,T. (1998) Crystal structure of the IIB subunit of a fructose permease (IIBLev) from Bacillus subtilis. J. Mol. Biol., 276, 591–602. [DOI] [PubMed] [Google Scholar]
- Schnetz K. and Rak,B. (1990) β-glucoside permease represses the bgl operon of Escherichia coli by phosphorylation of the antiterminator protein and also interacts with glucose-specific enzyme III, the key element in catabolite control. Proc. Natl Acad. Sci. USA, 87, 5074–5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stülke J., Arnaud,M., Rapoport,G. and Martin-Verstraete,I. (1998) PRD—a protein domain involved in PTS-dependent induction and carbon catabolite repression of catabolic operons in bacteria. Mol. Microbiol., 28, 865–874. [DOI] [PubMed] [Google Scholar]
- Thompson J. and Chassy,B.M. (1985) Intracellular phosphorylation of glucose analogs via the phosphoenolpyruvate:mannose phospho transferase system in Streptococcus lactis. J. Bacteriol., 162, 224–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L.F., Tomich,J.M. and Saier,M.H. (1990) Structure and evolution of a multidomain multiphosphoryl transfer protein. Nucleotide sequence of the fruB(HI) gene in Rhodobacter capsulatus and comparisons with homologous genes from other organisms. J. Mol. Biol., 213, 687–703. [DOI] [PubMed] [Google Scholar]