Abstract
The gypsy insulator is thought to play a role in nuclear organization and the establishment of higher order chromatin domains by bringing together several individual insulator sites to form rosette-like structures in the interphase nucleus. The Su(Hw) and Mod(mdg4) proteins are components of the gypsy insulator required for its effect on enhancer–promoter interactions. Using the yeast two-hybrid system, we show that the Mod(mdg4) protein can form homodimers, which can then interact with Su(Hw). The BTB domain of Mod(mdg4) is involved in homodimerization, whereas the C-terminal region of the protein is involved in interactions with the leucine zipper and adjacent regions of the Su(Hw) protein. Analyses using immunolocalization on polytene chromosomes confirm the involvement of these domains in mediating the interactions between these proteins. Studies using diploid interphase cells further suggest the contribution of these domains to the formation of rosette-like structures in the nucleus. The results provide a biochemical basis for the aggregation of multiple insulator sites and support the role of the gypsy insulator in nuclear organization.
Keywords: chromatin/insulator/nucleus/transcription
Introduction
Insulators or boundary elements are DNA sequences defined by two functional properties: (i) the disruption of enhancer–promoter interactions when placed between them and (ii) the protection of transgenes from chromosomal position effects. Insulators may accomplish this by organizing the chromatin fiber into higher order structures and establishing independent domains of gene activity. Enhancers and promoters placed in the same domain interact with one another while those placed in different domains are unable to do so (Gerasimova and Corces, 1996; Bell and Felsenfeld, 1999). Insulator sequences may thus be important for regulating eukaryotic transcription and they have been identified in a variety of organisms, ranging from yeast to mammals (Corces and Felsenfeld, 2000; Bell et al., 2001). Well characterized insulators in Drosophila include the scs and scs′ sequences found at the boundary of the 87A heat shock locus (Kellum and Schedl, 1991; Zhao et al., 1995), the Fab-7 and Fab-8 insulators from the Abd-B region (Hagstrom et al., 1996; Zhou et al., 1996; Mihaly et al., 1998), and the insulator sequence found in the gypsy retrotransposon (Cai and Levine, 1995; Gerasimova et al., 1995; Scott and Geyer, 1995). The best-studied vertebrate insulator is the chicken β-globin insulator (Chung et al., 1993; Bell et al., 1999). Recently, boundary elements flanking the repressed HMR locus that prevent spread of silenced chromatin have been identified and characterized in yeast (Bi and Broach, 1999; Bi et al., 1999; Donze et al., 1999).
The gypsy insulator of Drosophila comprises a 350 bp sequence present in the gypsy retrotransposon and possesses properties of a classical insulator element. Insertion of this sequence between an enhancer and a promoter inhibits the activity of the enhancer (Holdridge and Dorsett, 1991; Geyer and Corces, 1992); in addition, a transgene flanked on both sides by this sequence is insulated from the repressive effect of heterochromatic sequences (Roseman et al., 1995). Genetic and molecular approaches have led to the identification and characterization of two protein components of the gypsy insulator. One of them encoded by the suppressor of Hairy wing [su(Hw)] gene is a zinc finger protein that binds to insulator DNA. In addition, Su(Hw) has two acidic domains located at the N- and C-termini of the protein, and a leucine-zipper region highly homologous to the helix 2–coiled-coil region of the bHLH-Zip proteins (Parkhurst et al., 1988; Harrison et al., 1993). The Su(Hw) protein also contains three regions termed A, B and C, which are conserved to a very high degree among several Drosophila species (Harrison et al., 1993) (Figure 1A).
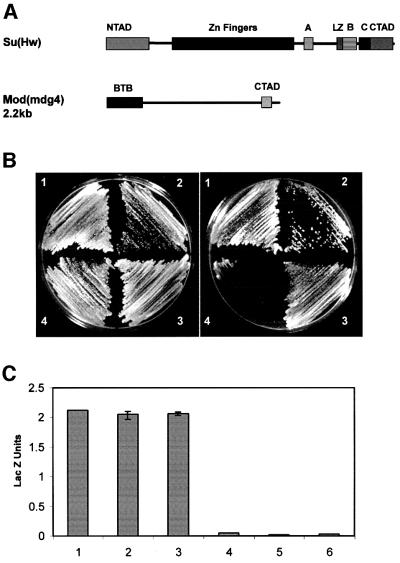
Fig. 1. Su(Hw) and mod(mdg4) interact directly with each other. (A) Schematic map of the Su(Hw) and Mod(mdg4) proteins showing the various domains described in the text. NTAD, N-terminal acidic domain; CTAD, C-terminal acidic domain; LZ, leucine zipper; A, B and C denote regions defined by homology among different Drosophila species (Harrison et al., 1993). BTB represents the BTB/POZ domain of mod(mdg4) and CTAD the C-terminal acidic domain. (B) Growth of yeast strain pJ694A expressing different Su(Hw) and/or Mod(mdg4) proteins on non-selective (left) or selective (right) media for the reporter genes used in the yeast two-hybrid assays. The numbers on the plates denote the following: 1, yeast expressing Su(Hw)-GAL4BD and Mod(mdg4)-GAL4AD; 2, yeast expressing Mod(mdg4)-GAL4BD and Su(Hw)-GAL4AD; 3, yeast expressing Mod(mdg4)-GAL4BD and Mod(mdg4)-GAL4AD; 4, yeast expressing Su(Hw)-GAL4BD and Su(Hw)-GAL4AD. (C) β-galactosidase activity, expressed as Miller units, in extracts of yeast strains carrying combinations of full-length Su(Hw) and Mod(mdg4) protein. Numbers 1–4 correspond to yeast strains described above. Numbers 5 and 6 correspond to yeast expressing Su(Hw) and Mod(mdg4) alone.
Modifier of mdg4 [Mod(mdg4)] is a second protein component of the gypsy insulator complex. Analysis of genetic interactions between mutations in mod(mdg4) and su(Hw) suggests that the proteins encoded by the two genes might interact directly or indirectly to mediate the enhancer blocking function of the gypsy insulator (Gerasimova et al., 1995; Gdula and Corces, 1997). This protein has numerous splice variants, a 2.2 kb transcript being the major form in the wild-type Canton S strain (Gerasimova et al., 1995; Buchner et al., 2000; Labrador et al., 2001). Sequence analysis of this protein reveals a 115 amino acid residue BTB motif (named for the three proteins bric-à-brac, tramtrack and broad-complex in which this motif was first found) at the N-terminal end (Figure 1A). This domain, also known as the POZ (pox virus and zinc finger) domain, is an evolutionarily conserved protein–protein interaction motif (Zollman et al., 1994; Ahmad et al., 1998). Many BTB domain-containing proteins are transcriptional regulators involved in a wide variety of developmental processes (Harrison and Travers, 1990; Xiong and Montell, 1993; Xue and Cooley, 1993; Bardwell and Treisman, 1994; Zollman et al., 1994; Philips and Herskowitz, 1998; Buchner et al., 2000). In addition to the BTB domain, the mod(mdg4) 2.2-encoded protein possesses a highly acidic domain in the C-terminus that contains 50% Asp and Glu residues (Gerasimova et al., 1995; Figure 1A). This domain is not present in most other Mod(mdg4) proteins and may impart a specific role to this particular product (Buchner et al., 2000).
The Mod(mdg4) 2.2 protein is present in ∼500 sites on polytene chromosomes. About 200 of these sites also contain the Su(Hw) protein. These sites of co-localization do not contain copies of the gypsy retrotransposon and are presumed to be endogenous insulator sites similar to the one found in gypsy. The sites where Mod(mdg4) and Su(Hw) do not co-localize may be binding sites for proteins that bind to Mod(mdg4) other than Su(Hw). Studies in diploid Drosophila nuclei using antibodies against Su(Hw) and Mod(mdg4), however, do not show a diffuse staining pattern for these proteins, as would be expected from the numerous binding sites present on polytene chromosome. Instead, 20–25 sites of localization are observed per diploid nucleus (Gerasimova and Corces, 1998; Gerasimova et al., 2000). This result has been interpreted to suggest that several individual binding sites could come together in a single nuclear location, forming a rosette-like structure and causing the punctate nuclear staining observed in diploid nuclei (Gerasimova et al., 2000). This implies that several insulator sequences located on different parts of the chromosome are brought together, presumably through interactions between their protein components. The rosette structures are formed by multiple loops, each representing a higher order domain of chromatin organization. To further our understanding of the nature of the interactions involved in the establishment of these domains, we performed a thorough analysis of the functional regions of both Su(Hw) and Mod(mdg4) proteins mediating intermolecular interactions. Here, we present evidence that the Su(Hw) and Mod(mdg4) proteins interact directly under in vivo conditions. We also demonstrate that the Mod(mdg4) protein is able to interact with itself. Finally, immunofluorescence studies done in imaginal disc cells suggest that interactions between these proteins might be responsible for organizing the chromatin fiber into higher order domains.
Results
Su(Hw) and mod(mdg4) interact directly with each other
We used the yeast two-hybrid system to demonstrate that the gypsy insulator Su(Hw) and Mod(mdg4) proteins interact with one another directly under in vivo conditions. To test this idea we made plasmids containing the full-length Su(Hw) coding region fused in-frame to the yeast Gal4 activation domain (GAL4AD) and the coding region of Mod(mdg4) 2.2 fused in-frame with the yeast Gal4 DNA binding domain (GAL4BD). Both plasmids were then co-transfected into yeast cells. In this and all experiments described below, western blot analyses were carried out with yeast whole-cell extracts to ensure that the expected proteins were expressed at similar levels. The results from all experiments are summarized in Table I. Interaction between the Su(Hw) and Mod(mdg4) proteins results in activation of the two nutritional reporter genes used in the two-hybrid assay, histidine and adenine, and hence growth on selective plates lacking these amino acids (Figure 1B). To confirm this result, we carried out the reciprocal two-hybrid test where Su(Hw) was fused to GAL4AD and Mod(mdg4) was fused to GAL4BD. Growth on selective plates indicates that the two proteins also interact strongly with each other in this case (Figure 1B). No growth occurred after transformation with single plasmids, indicating that interactions between the proteins are required for expression of the reporter genes (data not shown).
Table I. Summary of interactions between full-length and modified Su(Hw) and Mod(mdg4) 2.2 proteins.
| Interacting proteins | Strength of interaction |
|---|---|
| Su(Hw)–Mod(mdg4) | +++ |
| Su(Hw)–Su(Hw) | – |
| Mod(mdg4)–Mod(mdg4) | +++ |
| Su(Hw)ΔNTAD–Mod(mdg4) | +++ |
| Su(Hw)ΔCTAD–Mod(mdg4) | +++ |
| Su(Hw)ΔNoAD2–Mod(mdg4) | +++ |
| Su(Hw)ΔLZ–Mod(mdg4) | – |
| Su(Hw)ΔA–Mod(mdg4) | +++ |
| Su(Hw)ΔB–Mod(mdg4) | – |
| Su(Hw)ΔC–Mod(mdg4) | – |
| Mod(mdg4)–Mod(mdg4)ΔBTB | – |
| Mod(mdg4)–Mod(mdg4)ΔCTAD | +++ |
| Su(Hw)–Mod(mdg4)ΔBTB | + |
| Su(Hw)–Mod(mdg4)ΔCTAD | – |
| LZ + B + C + CTAD–Mod(mdg4) | +++ |
| LZ + B + C–Mod(mdg4) | +++ |
| LZ + B–Mod(mdg4) | – |
| LZ–Mod(mdg4) | – |
| Su(Hw)–BTB | – |
| LZ + B + C–BTB | – |
| Su(Hw)–Mod(mdg4) CTAD | ++ |
| BTB–Mod(mdg4) | +++ |
| BTB–Mod(mdg4)ΔBTB | – |
| BTB–BTB | ++ |
Yeast colonies transformed with Su(Hw)–GAL4BD/Mod(mdg4)–GAL4AD or Su(Hw)–GAL4AD/Mod(mdg4)– GAL4BD also tested positive when analyzed for β-galactosidase activity, which is the third reporter gene of the system. The strength of the interaction was quantitated by liquid culture assays of β-galactosidase enzymatic activity. Yeast co-transformed with Su(Hw) and Mod(mdg4) in either orientation produced ∼75-fold higher β-galactosidase units than when expressing either protein alone, which serves as the negative control (Figure 1C). In conclusion, Su(Hw) and Mod(mdg4) exhibit strong direct interaction under the in vivo conditions of the yeast two-hybrid system (Table I).
We next sought to test whether either the Su(Hw) or the Mod(mdg4) proteins interact with themselves. The Su(Hw) protein was a likely candidate since it possesses a region homologous to the helix 2–leucine-zipper region of bHLH proteins, which has been shown to mediate the formation of homodimers or heterodimers (Brownlie et al., 1997). Constructs containing the full-length Su(Hw) protein coding region were fused to yeast GAL4AD and GAL4BD. After co-transformation of these two plasmids into yeast, we failed to observe colony growth on selective plates, indicating that the Su(Hw) protein is not able to interact with itself (Figure 1B). This result was also confirmed by the β-galactosidase assays in liquid culture (Figure 1C). This lack of interaction was not due to toxicity or lack of protein production, as western blot analysis of yeast extracts showed that the fusion proteins were properly expressed (Figure 2C and data not shown).
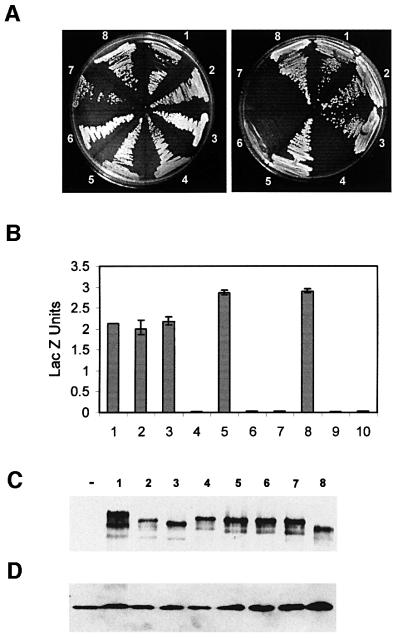
Fig. 2. Interactions between Su(Hw) and Mod(mdg4) are mediated by the leucine zipper and regions B and C of Su(Hw). (A) Growth of yeast strains carrying full-length Mod(mdg4) fused to GAL4BD and different deletion constructs of Su(Hw) fused to GAL4AD on non-selective (left) or selective (right) media. The numbers on the plates denote yeast expressing Mod(mdg4)–GAL4AD and either full-length or deletions of Su(Hw) fused to GAL4BD. 1, full-length Su(Hw); 2, Su(Hw)ΔNTAD; 3, Su(Hw)ΔCTAD; 4, Su(Hw)ΔLZ; 5, Su(Hw)ΔA; 6, Su(Hw)ΔB; 7, Su(Hw)ΔC; 8, Su(Hw)ΔNoAD2. (B) β-galactosidase activities, expressed as Miller units, corresponding to strains carrying combinations of full-length Mod(mdg4) and different deletion constructs of Su(Hw). Numbers denote the same strains described above. Numbers 9 and 10 denote yeast expressing Su(Hw) and Mod(mdg4) alone. (C) Western analyses of yeast extracts carrying different deletions of Su(Hw). The panel shows the expression of Su(Hw) detected with polyclonal anti-Su(Hw) antibody. Numbers are as in the previous panels. The ‘–’ symbol represents extracts from yeast cells not expressing Su(Hw) or Mod(mdg4). (D) Same western as in (C), after the filter was stripped and re-probed with a porin monoclonal antibody. The presence of mitochondrial porin was used as a loading control.
To test whether Mod(mdg4) interacts with itself, we made similar constructs in which the coding region of mod(mdg4) 2.2 was fused in-frame to either GAL4AD or GAL4BD. Upon co-transfection, colony growth on selective plates suggests that Mod(mdg4) 2.2 interacts strongly with itself (Figure 1B). Quantitation of β-galactosidase activity suggests that this interaction is as strong as that found between Su(Hw) and Mod(mdg4) (Figure 1C).
Interactions between Su(Hw) and Mod(mdg4) are mediated by the leucine zipper and regions B and C of Su(Hw)
The Su(Hw) protein has several conserved structural motifs that are good candidates to mediate interactions with other proteins. These domains include two acidic- rich regions, a region homologous to the helix 2–coiled-coil region of bHLH leucine-zipper proteins (LZ) and three regions highly conserved among several Drosophila species (Figure 1A). These regions are termed conserved regions A, B, and C, and are located in the C-terminal part of the protein adjacent to the leucine-zipper domain; these conserved regions do not show homology to any functional domain found in the comprehensive DNA and protein databases. However, the high degree of conservation suggests that these domains could have an important functional role.
We constructed plasmids expressing Su(Hw) proteins, bearing precise deletions of each of these different regions, fused to yeast GAL4BD. We then tested their ability to interact with full-length Mod(mdg4) 2.2 protein in the yeast two-hybrid system in order to determine the contribution of each of these domains to the intermolecular interactions between the two gypsy insulator proteins. Su(Hw) protein lacking either the 48 residue N-terminal acidic domain (amino acids 154–202) [Su(Hw)ΔNTAD] or the 82 residue C-terminal acidic domain (amino acids 862–944) [Su(Hw)ΔCTAD] are able to interact with Mod(mdg4), indicating that these two domains are dispensable for the interaction between the two proteins (Figure 2A). It is possible that the two acidic domains could have overlapping functions and one region could compensate for the absence of the other. Hence we constructed a Su(Hw) protein that lacks both the N- as well as the C-terminal acidic domains and fused it in-frame with GAL4BD. This construct, called Su(Hw)ΔNoAD2, is still able to interact with mod(mdg4) 2.2, indicating that both acidic domains of the Su(Hw) protein are dispensable for its interaction with Mod(mdg4) 2.2 (Figure 2A). Next we tested a Su(Hw) protein carrying a 19 amino acid residue deletion spanning the region homologous to the coiled-coil leucine-zipper region (amino acids 760–778) [Su(Hw)ΔLZ]. This protein failed to interact with Mod(mdg4) 2.2 in the yeast two-hybrid system (Figure 2A). Finally, to test the contribution of each of the three conserved regions to the interaction between Su(Hw) and Mod(mdg4), we made three different Su(Hw) constructs, termed Su(Hw)ΔA, Su(Hw)ΔB and Su(Hw)ΔC, lacking each of these domains. Su(Hw) protein lacking region A was able to interact with Mod(mdg4) (Figure 2A). However, proteins lacking regions B and C failed to interact with full-length Mod(mdg4) (Figure 2A). These results suggest that the leucine zipper and conserved regions B and C, all located at the C-terminal end of the protein and also adjacent to one another (amino acids 760–860), are required to mediate interactions with Mod(mdg4) 2.2 (Table I). The ability of Su(Hw) proteins carrying different deletions to interact with Mod(mdg4) was confirmed by measuring β-galactosidase (Figure 2B). The failure to observe growth of yeast cells carrying deletions in Su(Hw) is not due to lack of expression of the deleted proteins. Western blot analysis of extracts obtained from yeast strains carrying plasmids containing the various deletions described above shows that they produce similar levels of Su(Hw) protein (Figure 2C). In addition, negative controls with yeast cells expressing only Su(Hw) deletion constructs do not show any trans-activation of the reporter genes (data not shown).
The BTB domain of Mod(mdg4) is necessary for homodimerization, whereas the C-terminal acidic domain mediates interactions with Su(Hw)
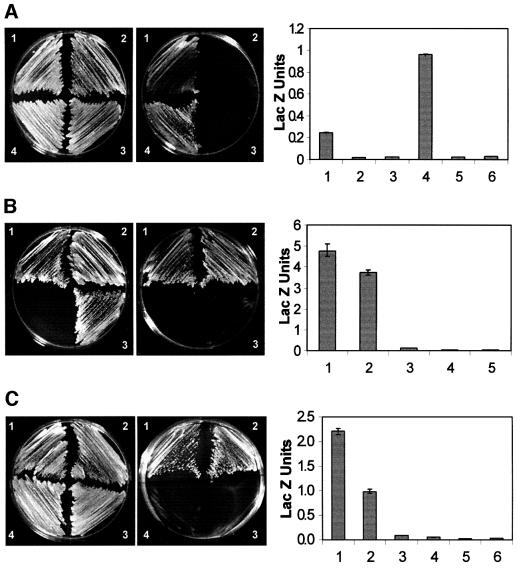
We next performed a deletion analysis of the Mod(mdg4) 2.2 protein to identify domains responsible for homo- and heterodimeric interactions. The Mod(mdg4) 2.2 protein has two domains that might possibly be involved in interactions with itself or other proteins: a BTB domain at the N-terminus and a highly acidic region at the C-terminus. We made constructs of Mod(mdg4) 2.2 lacking either the BTB domain [Mod(modg4)ΔBTB] or the C-terminal acidic domain [Mod(mdg4)ΔCTAD] and tested their interactions with either full-length Mod(mdg4) 2.2 or full-length Su(Hw). The Mod(mdg4) 2.2 protein lacking the BTB domain failed to interact with full-length Mod(mdg4) 2.2 (Figure 3A). However, Mod(mdg4) 2.2 protein lacking the C-terminal acidic domain was able to interact with intact Mod(mdg4) 2.2 (Figure 3A). The strength of this interaction was quantitated by measuring the enzymatic activity of β-galactosidase. Yeast cells co-transformed with Mod(mdg4)ΔBTB and full-length Mod(mdg4) 2.2 produce levels of β-galactosidase units comparable to those of the negative control, whereas cells co-transformed with Mod(mdg4)ΔCTAD and the full- length protein have much higher levels of β-galactosidase activity, suggesting a stronger interaction (Figure 3A).
Fig. 3. Domains of Su(Hw) and Mod(mdg4) necessary and sufficient for interactions. The photographs on the left show growth of yeast strains carrying deletions of Mod(mdg4) and full-length Su(Hw) or full-length Mod(mdg4) on media containing (left panel) or lacking (right panel) histidine and adenine. The graphs on the right show β-galactosidase activity, expressed as Miller units, in extracts from strains carrying the same combinations of Mod(mdg4) and Su(Hw). For each graph, numbers 1–4 are as on the photographs to the left, and numbers 5 and 6 correspond to yeast expressing Su(Hw) and Mod(mdg4) alone. (A) The numbers on the plates denote yeast expressing the following constructs: 1, yeast expressing Su(Hw)–GAL4BD and Mod(modg4)ΔBTB–GAL4AD; 2, Mod(mdg4)–GAL4BD and Mod(mdg4)ΔBTB–GAL4AD; 3, Su(Hw)–GAL4BD and Mod(mdg4)ΔCTAD–GAL4AD; 4, Mod(mdg4)–GAL4BD and Mod(mdg4)ΔCTAD–GAL4AD. (B) The numbers on the plates denote the following: 1, yeast transformed with Mod(mdg4)–GAL4BD and LZ + B + C + CTAD–GAL4AD; 2, Mod(mdg4)–GAL4BD and LZ + B + C–GAL4AD; 3, BTB–GAL4BD and LZ + B + C–GAL4AD. Numbers 4 and 5 in the graph correspond to yeast expressing Su(Hw) and Mod(mdg4) alone. (C) The numbers on the plates correspond to yeast transformed with: 1, BTB–GAL4BD and Mod(mdg4)–GAL4AD; 2, BTB–GAL4BD and BTB–GAL4AD; 3, BTB–GAL4BD and Mod(mdg4)ΔBTB–GAL4AD; 4, BTB–GAL4AD and Su(Hw)–GAL4AD.
The ability of the deleted Mod(mdg4) 2.2 proteins to interact with Su(Hw) was tested using the same assays. Fewer colonies appeared on selective plates upon co-transfection of Mod(mdg4)ΔBTB and full-length Su(Hw) compared with growth on non-selective plates, indicative of a weak interaction (Figure 3A). Yeast cells expressing full-length Su(Hw) and Mod(mdg4)ΔBTB produce levels of β-galactosidase activity greater than that of the negative control but 8-fold less than those produced by yeast cells expressing the intact proteins (Figure 3A). These results suggest that the BTB domain of Mod(mdg4) 2.2 is partially required for its interaction with Su(Hw). In addition, Mod(mdg4)ΔCTAD failed to interact with full-length Su(Hw) (Figure 3A). In conclusion, the Mod(mdg4) 2.2 protein appears to form homodimers through the BTB domain, whereas interactions with Su(Hw) are mostly mediated by its C-terminal acidic region with some participation of the BTB domain (Table I). One possible interpretation of this dual requirement of two separate domains of Mod(mdg4) for its interaction with Su(Hw) is that dimerization of Mod(mdg4) through the BTB domain is required to form an interface at the C-terminal region through which interaction with Su(Hw) takes place.
Domains of Su(Hw) and Mod(mdg4) sufficient for interactions
After identification of the domains necessary for the interaction between Su(Hw) and Mod(mdg4) 2.2, we wanted to test whether these domains were also sufficient for the interactions under in vivo conditions. We first tested the domains of Su(Hw) responsible for interactions with Mod(mdg4) by making a construct carrying the C-terminal end of the protein including the leucine zipper, regions B and C, and the C-terminal acidic domain fused to the GAL4AD domain. This region was able to interact with full-length Mod(mdg4) 2.2 in the yeast two-hybrid assay, and the strength of the interaction is similar to that of the intact Su(Hw) protein (Figure 3B). A construct containing only the leucine zipper and regions B and C, but not the C-terminal acidic region is also able to interact with full-length mod(mdg4) 2.2 in the two-hybrid assay (Figure 3B). Elimination of either region C or regions B and C results in a protein unable to interact with Mod(mdg4) (data not shown; summarized in Table I). From these results and the deletion analysis described above we conclude that the region of Su(Hw) containing the leucine zipper and regions B and C (amino acids 760–860) is both necessary and sufficient for its interaction with Mod(mdg4) 2.2 (Table I).
Deletion of the BTB domain in Mod(mdg4)ΔBTB results in a weak interaction with full-length Su(Hw). Hence, we decided to test whether this domain by itself is sufficient to mediate this interaction. The BTB domain fails to interact with a protein containing the LZ and regions B and C of Su(Hw) (Figure 3B) or the intact Su(Hw) protein (Figure 3C). This result, and the failure of Mod(mdg4)ΔCTAD to interact with full-length Su(Hw), suggests that the BTB domain might be involved in homodimeric interactions required for subsequent binding to Su(Hw) through the C-terminal acidic domain of Mod(mdg4) 2.2. This conclusion is supported by the fact that a construct expressing only the C-terminal acidic domain of Mod(mdg4) 2.2 is able to interact with the full-length Su(Hw) protein in the yeast two-hybrid assay (Table I). The role of the BTB domain of Mod(mdg4) 2.2 in homodimeric interactions was confirmed by the observation that this domain in isolation is able to interact with itself (Figure 3C). In addition, the BTB domain failed to interact with Mod(mdg4)ΔBTB (Figure 3C). This further confirms that the BTB domain is both necessary and sufficient for mediating interactions between Mod(mdg4) 2.2 molecules, but it is not responsible for interaction with Su(Hw). The strength of the interaction of the BTB domain with full-length Mod(mdg4) 2.2 is comparable to that seen with the full-length proteins (Figure 3C), whereas interactions between the BTB domains are significantly higher than those of the negative controls but lower than those of the full-length proteins (Figure 3C). This is not unexpected, since residues outside of the BTB domain might render stability to the protein and thus produce a more stable interacting interface. The levels of β-galactosidase activity in yeast cells expressing the BTB domain and Mod(mdg4)ΔBTB or full-length Su(Hw) are comparable to those of the negative controls (Figure 3C) (see Table I for a summary of these results).
Interactions between Su(Hw) and Mod(mdg4) in Drosophila polytene chromosomes
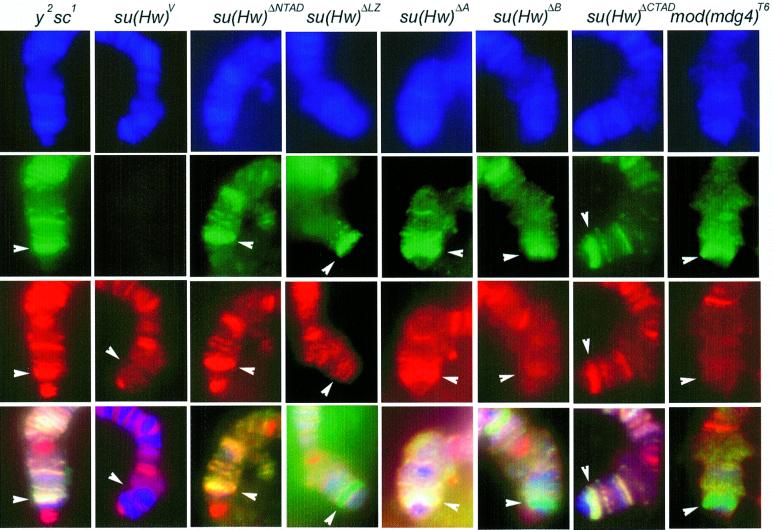
Upon identification of domains of the Su(Hw) and Mod(mdg4) 2.2 protein involved in homo- and heterodimeric interactions using the two-hybrid assay we wished to confirm the results in Drosophila under in vivo conditions. To this end, we used immunolocalization of these two proteins on polytene chromosomes as an assay system to measure interactions. Independent fly strains were generated carrying deletions in each of the functional domains of Su(Hw) (Harrison et al., 1993; Gerasimova et al., 1995; Gdula and Corces, 1997). We have previously shown that only mutations in the Zn fingers affect the ability of Su(Hw) to bind to polytene chromosomes, and deletion of other protein domains do not affect DNA binding or chromosome localization (Harrison et al., 1993; Gdula and Corces, 1997). To test the ability of Mod(mdg4) 2.2 to interact with Su(Hw) proteins lacking various functional domains, we then examined the distribution of Mod(mdg4) 2.2 protein on polytene chromosomes of flies carrying deletions in these different domains of Su(Hw). In wild-type flies, Su(Hw) protein is present at ∼200 sites on polytene chromosomes and Mod(mdg4) 2.2 is present at all of these sites; the Mod(mdg4) 2.2 protein is present in ∼300 additional sites without Su(Hw) (Harrison et al., 1993; Gerasimova and Corces, 1998). As expected, flies wild type for the su(Hw) gene show co-localization of Su(Hw) and Mod(mdg4) proteins (Figure 4). To facilitate the analysis, strains used in these studies carried the gypsy-induced y2 mutation in the yellow gene; since this gene is located at the tip of the X chromosome, the presence of this mutation allowed us to easily identify a site in the chromosome where the two proteins normally overlap (Figure 4). In the background of a null mutation of su(Hw), su(Hw)V, the Mod(mdg4) 2.2 protein is unable to bind to the chromosomes at sites where the two proteins normally overlap, including the yellow locus (Figure 4). However, Mod(mdg4) binds to other sites on polytene chromosomes, suggesting it might have other interacting partners. Flies carrying a deletion of the N- or C-terminal acidic domains, su(Hw)ΔNTAD and su(Hw)ΔCTAD, show the presence of normal levels of the truncated Su(Hw) protein at the yellow locus, confirming the observation that deletion of these domains does not interfere with DNA binding. In addition, the Mod(mdg4) protein is also present at the yellow locus, suggesting that the interaction of this protein with Su(Hw) is not affected by the absence of these two acidic domains (Figure 4 and data not shown). The same results were obtained with flies expressing Su(Hw)ΔA, which lacks region A located immediately upstream from the leucine-zipper domain. This protein also binds to polytene chromosomes at the yellow locus and is able to interact with Mod(mdg4) (Figure 4).
Fig. 4. Interactions between Su(Hw) and Mod(mdg4) in Drosophila polytene chromosomes. Localization of Su(Hw) (green) and Mod(mdg4) (red) on polytene chromosomes; 4′,6-diamidine-2-phenylindole (DAPI) stains DNA and is indicated in blue. The lower panels represent the overlap of the three individual images. All strains carry a gypsy-induced mutation in the yellow gene and they are either wild type for su(Hw) and mod(mdg4) (y2) or carry different mutations in these genes as indicated at the top of the various panels. Su(Hw) and Mod(mdg4) proteins were detected using FITC- and Texas red-conjugated secondary antibodies, respectively. Sites where Su(Hw) is present alone should be labeled in green, whereas sites for Mod(mdg4) should be marked in red; sites where both proteins are present appear yellow. The location of the yellow locus is indicated by a white arrowhead.
We also tested the ability of Mod(mdg4) to interact with a Su(Hw) protein lacking the leucine-zipper domain. Flies expressing the Su(Hw)ΔLZ protein show normal localization of this protein on polytene chromosomes, but, in this case, the Mod(mdg4) protein fails to co-localize with Su(Hw)ΔLZ at the yellow locus (Figure 4). Examination of flies expressing the Su(Hw)ΔB protein also shows that Mod(mdg4) is unable to interact with a Su(Hw) protein lacking region B (Figure 4).
A similar approach was used to determine whether the C-terminal acidic domain of Mod(mdg4) 2.2 is required to interact with Su(Hw) on polytene chromosomes. To this end, we examined the co-localization of the two proteins in flies carrying the mod(mdg4)T6 allele. This allele results from a point mutation at amino acid residue 588 of the Mod(mdg4) 2.2 coding region, producing a truncated protein that lacks the last 32 residues at the C-terminal acidic domain. Polytene chromosome staining shows that su(Hw) protein binds to the chromosomes as expected in flies carrying this mutation, but Mod(mdg4) fails to co-localize with Su(Hw) (Figure 4).
These results confirm the observations made using the yeast two-hybrid system. Results obtained using immunofluorescence microscopy only give qualitative evidence of the ability or failure of the two proteins to interact, but this information is a better indication of the actual in vivo interactions that take place in Drosophila nuclei. In general, the results of the immunofluorescence experiments follow the same pattern as those obtained in the yeast two-hybrid assay, i.e. the leucine zipper and the conserved region B are responsible for mediating interactions between Su(Hw) and Mod(mdg4), whereas the C-terminal acidic domain of Mod(mdg4) 2.2 is responsible for interacting with Su(Hw). Nevertheless, slight differences between the two methods are worth noting. The immunofluorescence data appear to suggest that the Su(Hw) LZ and Mod(mdg4) CTAD domains are more critical for the recruitment of Mod(mdg4) protein to polytene chromosomes than the Su(Hw) region B. Similarly, although none of these domains are critical for the interactions, the Su(Hw) CTAD domain appears to be more important than the NTAD and A regions. It is unclear at this time whether these differences are real or merely a consequence of the techniques employed.
Distribution of Su(Hw) and Mod(mdg4) in diploid cells
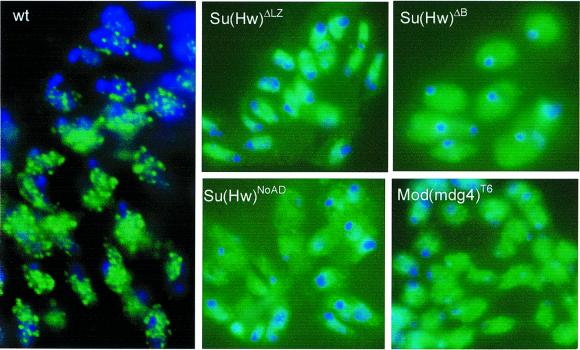
Previous studies in Drosophila imaginal disc cells have shown that the Su(Hw) and Mod(mdg4) proteins are not diffusely localized throughout the nucleus; instead, these two proteins are present in ∼20–25 specific sites located mostly around the nuclear periphery. The punctate pattern observed in interphase diploid nuclei is created by several individual insulator sites coming together at specific nuclear locations (Gerasimova and Corces, 1998; Gerasimova et al., 2000). These interactions between individual insulator sites should be mediated by the protein components of the insulator, including Su(Hw) and Mod(mdg4). The picture that emerges from the results discussed above is that Su(Hw) binds to the DNA and interacts with the C-terminal region of Mod(mdg4) through the leucine zipper and regions B and C. Two individual insulator sites could come together in the nucleus through interactions mediated by the BTB domain of Mod(mdg4) 2.2. Disruption of domains of the Su(Hw) protein involved in interactions with Mod(mdg4) should then affect the ability of individual sites to come together and cause alterations in the punctate pattern of Su(Hw) nuclear localization. To test this possibility, we examined whether deletion of the LZ domain of Su(Hw) affects its punctuated distribution in diploid nuclei of interphase cells. We have shown above that this altered Su(Hw)ΔLZ protein is able to bind DNA and shows a normal distribution on polytene chromosomes. As predicted by the hypothesis, since deletion of this region affects interactions between Su(Hw) and Mod(mdg4), imaginal disc cells from flies expressing the Su(Hw)ΔLZ protein show a diffuse nuclear localization of this protein instead of the standard punctuated pattern observed in wild-type cells (Figure 5). The same result was obtained with Su(Hw)ΔB and Su(Hw)ΔNoAD, which bear precise deletions of the leucine zipper and regions B, C and CTAD, respectively (Figure 5 and data not shown).
Fig. 5. Subnuclear localization of Su(Hw) in nuclei. Diploid cells in interphase were obtained from imaginal discs of Drosophila wild type (wt) or strains carrying different mutations in Su(Hw) and Mod(mdg4) as indicated in the panels. Distribution of Su(Hw) detected with FITC-conjugated secondary antibody (green). DNA was stained with DAPI and is represented in blue.
These results suggest that an interaction between Su(Hw) and Mod(mdg4) is required for the punctate nuclear distribution of Su(Hw) protein, which is a consequence of the aggregation of individual insulator sites into rosette-like structures (Gerasimova et al., 2000). To confirm further this conclusion, we examined the effect of deletion of the C-terminal domain of Mod(mdg4) 2.2 in flies carrying the mod(mdg4)T6 mutation. Flies expressing this altered Mod(mdg4) 2.2 protein also show a diffuse pattern of Su(Hw) localization, confirming the need for interactions between Su(Hw) and Mod(mdg4) for proper arrangement of insulator sites in the nucleus (Figure 5).
Discussion
Boundary or insulator elements disrupt enhancer–promoter interactions in a polar fashion when placed between them. Insulators also buffer transgenes from chromosomal position effects. Current models to explain insulator function are based on the assumption that the normal role of insulators in the nucleus is defined by the two properties described above. The promoter decoy model assumes that insulators are regulatory sequences of the same class as enhancers and promoters, and their function is to regulate promoter–enhancer communication. Insulators might accomplish this by assembling a protein complex similar to the transcription complex, thus fooling the enhancer into interacting with the insulator instead of the promoter (Geyer, 1997). The second model suggests that the normal role of insulators is to organize the chromatin into distinct domains that establish independent regions of gene activity, such that regulatory regions present in one domain are unable to interact with promoters located in a different one. Support for the latter model comes from the analysis of the gypsy insulator, and the observation that mutations in the mod(mdg4) gene act both as enhancers of position-effect variegation and trithorax-group genes, suggesting an involvement of the Mod(mdg4) protein in chromatin-related phenomena. In addition, analyses of the distribution of Su(Hw) and Mod(mdg4) show that these proteins are localized in 20–25 specific sites in diploid nuclei, suggesting that several individual insulator sites come together at single nuclear locations, forming loop-like structures that might represent the higher order chromatin domains hypothesized to define the role of insulators (Gerasimova and Corces, 1998; Gerasimova et al., 2000). The existence of these loops is also supported by functional studies showing that paired insulator sites can not interfere with enhancer– promoter interactions; these results can best be explained by the formation of loops between individual insulators (Cai and Shen, 2001; Muravyova et al., 2001).
The formation of loops or higher order domains of chromatin structure requires the individual insulator sites from different chromosomal locations to come together in the nucleus. This organization must be mediated by interactions among protein components of the insulator. Here we show that these interactions are indeed possible and take place in vivo in the case of the gypsy insulator of Drosophila. Mapping the domains of the Su(Hw) and Mod(mdg4) proteins involved in this interaction might shed light on how insulators could be involved in the establishment of higher order chromatin organization. Disruption of the leucine zipper and regions B and C of Su(Hw) renders the gypsy insulator unable to interfere with enhancer–promoter interactions (Gerasimova et al., 1995; Gdula and Corces, 1997). Results presented here indicate that disruption of this region of Su(Hw) also abolishes its interaction with Mod(mdg4) and eliminates the punctate nuclear staining pattern, suggesting that interaction between the two proteins is required for establishing domains in the nucleus, and that the establishment of these domains correlates with the functionality of the insulator.
The second characterized component of the gypsy insulator Mod(mdg4) has at least 21 different isoforms generated by alternative splicing (Buchner et al., 2000). All the proteins contain a common N-terminus of 402 amino acids that includes a BTB/POZ domain, whereas the C-terminus of the protein is variable. Most of these Mod(mdg4) proteins are present in a few sites on polytene chromosomes and only the Mod(mdg4) 2.2 protein appears to be a general component of the gypsy insulator (Gerasimova et al., 1995; Buchner et al., 2000). Our experiments show that the Su(Hw) protein interacts with Mod(mdg4) 2.2 through the C-terminal domain of the Mod(mdg4) 2.2 protein. Since this domain is specific to this form of the protein and it is not present in any of the other variants, this result supports the idea that Mod(mdg4) 2.2 is the component of the gypsy insulator, whereas other mod(mdg4)-encoded proteins might have more specific roles in the cell. Deletion of the BTB domain eliminates homodimeric interactions between Mod(mdg4) 2.2 and results in weakened interactions between Su(Hw) and Mod(mdg4) 2.2. This result could be interpreted as suggesting that Su(Hw) and Mod(mdg4) 2.2 interact through the BTB domain. However, this domain by itself is not able to interact with the full-length Su(Hw) protein or with the LZ-B-C region; this is not due to incorrect folding of the protein, since the BTB domain by itself is able to fold properly and mediate interaction with full-length Mod(mdg4) or another BTB domain. We interpret these results to suggest that the BTB domain mediates the formation of Mod(mdg4) 2.2 dimers, which in turn are required to mediate the interaction with Su(Hw).
BTB domain-containing proteins frequently have zinc fingers involved in DNA binding. The Mod(mdg4) 2.2 protein is unusual in the sense that it does not possess any such DNA-binding domain at the C-terminus. However, the presence of a domain that mediates interactions with Su(Hw), which binds DNA through its zinc fingers, might serve the purpose of recruiting this protein to chromatin. The BTB domain is responsible for self-oligomerization of proteins such as GAGA, promyelocytic leukemia zinc finger protein (PLZF) and ZID in vitro (Bardwell and Treisman, 1994; Zollman et al., 1994; Espinas et al., 1999; Katsani et al., 1999). Interestingly, although the BTB domain-containing promyelocytic leukemia zinc finger protein appears to form only dimers in solution, a short four-stranded antiparallel β-sheet between two symmetry-related dimers can be observed in the crystal (Ahmad et al., 1998). This interaction involves four different peptide chains and, therefore, can give rise to the formation of tetramers and oligomers of higher stoichiometry, suggesting that BTB-containing proteins can form large multimers. This observation is especially significant in the context of proposed models for insulator function, which require multiple insulator sites to come together in one large aggregate. It might be possible for Mod(mdg4) 2.2 to interact with several Mod(mdg4) 2.2 molecules, thus helping to bring together several Mod(mdg4) binding sites to form insulator aggregates as observed in interphase diploid cells. Alternatively, Mod(mdg4) 2.2 might interact with other BTB domain-containing proteins, which might be an integral part of the gypsy insulator complex. The BTB domain forms an extensive dimer interface that is a possible binding site for other proteins. Since the presence of the BTB domain is only partially required for binding of Su(Hw), there might possibly be other as yet unidentified partners of Mod(mdg4) that interact with the BTB domain. Alternatively, the BTB dimer interface might stabilize the interaction of Su(Hw) with the C-terminal region of Mod(mdg4).
The finding of specific domains of the Su(Hw) and Mod(mdg4) proteins that mediate intermolecular interactions provides a strong biochemical foundation for the involvement of these proteins in the establishment of chromosomal loops. These loops are the basis for the proposed role of insulators in the formation of higher order chromatin domains and nuclear organization of the chromosomes during interphase. These studies also provide support for the involvement of other proteins in insulator function. The identification of these proteins will provide additional evidence to understand the mechanisms by which these important sequences control eukaryotic gene expression.
Materials and methods
Drosophila strains and plasmid constructions
Fly stocks were maintained at 22.5°C and 65% relative humidity. Transgenic fly lines expressing different Su(Hw) mutant proteins were maintained as homozygous stocks in a y2; su(Hw)V background.
Bait constructs were made by cloning PCR products into pAS2 to express fusion proteins containing the GAL4BD. Target constructs were cloned into pACT2 to express fusion proteins containing the yeast GAL4AD. Full-length Su(Hw), and truncated forms containing the LZ + B + C + CTAD or LZ + B + C domains of Su(Hw) were obtained by PCR using primers located at positions 1–2835, 2064–2835 and 2064–2667 of the Su(Hw) cDNA, respectively, and subsequently cloned into either the pACT2 or pAS2 vectors. Deletion constructs of Su(Hw) were made by PCR of previously made Su(Hw) deletions cloned in pCaSpER and subsequently cloned into pACT2. PCR products for Su(Hw)ΔCTAD, Su(Hw)ΔNoAD2, Su(Hw)ΔA, Su(Hw)ΔB and Su(Hw)ΔC were obtained from deletion constructs described in Gdula and Corces (1997). PCR products for Su(Hw)ΔLZ and Su(Hw)ΔNTAD were obtained by amplification of Su(Hw)Δ283 and Su(Hw)Δ100, respectively, described in Harrison et al. (1993). Full-length Mod(mdg4) 2.2, the BTB domain of Mod(mdg4) 2.2, Mod(mdg4)ΔBTB and Mod(mdg4)ΔCTAD constructs were made by PCR of the Mod(mdg4) 2.2 kb cDNA using primers at nucleotide positions 1–1824, 1–480, 361–1824 and 1–1653, respectively (Gerasimova et al., 1995); the PCR fragments were then cloned in the yeast vectors described above.
Yeast two-hybrid assays
Two-hybrid assays were performed using yeast strain pJ694A and plasmids and protocols obtained from Clontech (Palo Alto, CA). For growth assays, plasmids were transformed into yeast strain pJ694A by the lithium acetate method described in the yeast protocols handbook (Clontech). Bait and target fusion proteins were produced constitutively under the control of the ADH1 promoter. Co-transformants were plated on media lacking tryptophan and leucine (non-selective plates for the reporter gene) or on plates lacking tryptophan, leucine, histidine and adenine (selective plates for the reporter gene). After incubation at 30°C for 3–4 days, growth on both plates was compared. Appearance of transformants on the selective plates indicates a positive interaction. Single colonies were subsequently streaked out on selective plates to obtain the plates shown in the figures.
Liquid culture assays were performed according to protocols described in the yeast protocols handbook (Clontech). Briefly, single yeast colonies were grown in media lacking appropriate amino acids. Cells were harvested in mid-log phase, pelleted and broken by the freeze–thaw method by placing the tubes in liquid nitrogen. After breaking the cells open, buffer 1 (39 mM Na2HPO4, 61 mM NaH2PO4, 10 mM KCl and 1 mM MgSO4) was added and the exact time of addition was recorded. The reaction was stopped by addition of 1 M Na2CO3 when the color of the reaction turned yellow, and the stop time was recorded. Tubes were centrifuged to obtain cell pellets and supernatants were transferred to fresh tubes. OD578 were measured for each supernatant. All assays were done in triplicate and β-galactosidase units were calculated as Miller units (1 unit is defined as the amount that hydrolyzes 1 µmol of ONPG to O-nitrophenol and d-galactose per minute per cell).
Western and immunofluorescence analyses
Yeast whole-cell extracts were prepared according to the yeast protocols handbook (Clontech). Yeast strains were grown in appropriate selection media and harvested in mid-log phase. Appropriate amounts of pre-warmed cracking buffer (8 M urea, 5% SDS, 40 mM Tris–HCl pH 6.8, 0.1 mM EDTA and 0.4 mg/ml bromophenol blue) and glass beads were added to cell pellets. Pellets were vortexed vigorously for 10 min and centrifuged at 14 000 r.p.m. in a microcentrifuge. Supernatants were either frozen in liquid nitrogen and stored at –70°C or loaded on a 10% SDS–polyacrylamide gel for western blot analysis. Proteins were electrophoresed and electroblotted onto a nitrocellulose membrane using standard procedures. Membranes were blocked and incubated with polyclonal antibodies generated either against Mod(mdg4) or Su(Hw) (Gerasimova et al., 1995). Filters were developed by standard procedures using an ECL kit (Amersham Pharmacia Biotech, Piscataway, NJ) and visualized with Kodak X-ray film. Membranes were stripped of the primary antibody and incubated with yeast mitochondrial porin antibody (obtained from Dr Beverly Wendland) and re-developed.
Antibodies against Su(Hw) and Mod(mdg4) were generated in rabbits and rats as described previously (Gerasimova et al., 1995). Immuno localization of proteins in polytene chromosomes and imaginal disc cells was carried out as previously described (Gerasimova and Corces, 1998; Gerasimova et al., 2000). Proteins were visualized using FITC- or Texas red-conjugated secondary antibodies; tissues and/or chromosomes were examined using a Zeiss Axiophot microscope.
Acknowledgments
Acknowledgements
We would like to thank Dr Kyle Cunningham for a gift of strain PJ69-4A, Dr Beverly Wendland for help with the yeast two-hybrid assays and porin antibodies, and Kelly Baxter for help with western blot analysis. Work reported here was supported by US Public Health Service Award GM35463 from the National Institutes of Health.
References
- Ahmad K.F., Engel,C.K. and Prive,G.G. (1998) Crystal structure of the BTB domain from PLZF. Proc. Natl Acad. Sci. USA, 95, 12123–12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell V.J. and Treisman,R. (1994) The POZ domain: a conserved protein–protein interaction motif. Genes Dev., 8, 1664–1677. [DOI] [PubMed] [Google Scholar]
- Bell A.C. and Felsenfeld,G. (1999) Stopped at the border: boundaries and insulators. Curr. Opin. Genet. Dev., 9, 191–198. [DOI] [PubMed] [Google Scholar]
- Bell A.C., West,A.G. and Felsenfeld,G. (1999) The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell, 98, 387–396. [DOI] [PubMed] [Google Scholar]
- Bell A.C., West,A.G. and Felsenfeld,G. (2001) Insulators and boundaries: Versatile regulatory elements in the eukaryotic genome. Science, 291, 447–450. [DOI] [PubMed] [Google Scholar]
- Bi X. and Broach,J.R. (1999) UASrpg can function as a heterochromatin boundary element in yeast. Genes Dev., 13, 1089–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X., Braunstein,M., Shei,G.J. and Broach,J.R. (1999) The yeast HML I silencer defines a heterochromatin domain boundary by directional establishment of silencing. Proc. Natl Acad. Sci. USA, 96, 11934–11939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownlie P., Ceska,T., Lamers,M., Romier,C., Stier,G., Teo,H. and Suck,D. (1997) The crystal structure of an intact human Max–DNA complex: new insights into mechanisms of transcriptional control. Structure, 5, 509–520. [DOI] [PubMed] [Google Scholar]
- Buchner K., Roth,P., Schotta,G., Krauss,V., Saumweber,H., Reuter,G. and Dorn,R. (2000) Genetic and molecular complexity of the position effect variegation modifier mod(mdg4) in Drosophila. Genetics, 155, 141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H. and Levine,M. (1995) Modulation of enhancer–promoter interactions by insulators in the Drosophila embryo. Nature, 376, 533–536. [DOI] [PubMed] [Google Scholar]
- Cai H.N. and Shen,P. (2001) Effects of cis arrangement of chromatin insulators on ennhancer-blocking activity. Science, 291, 493–495. [DOI] [PubMed] [Google Scholar]
- Chung J.H., Whiteley,M. and Felsenfeld,G. (1993) A 5′ element of the chicken β-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell, 74, 505–514. [DOI] [PubMed] [Google Scholar]
- Corces V.G. and Felsenfeld,G. (2000) Chromatin boundaries. In Elgin,S.C.R. and Workman,J.L. (eds), Chromatin Structure and Gene Expression. Oxford University Press, Oxford, UK, pp. 278–299.
- Donze D., Adams,C.R., Rine,J. and Kamakaka,R.T. (1999) The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes Dev., 13, 698–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinas M.L., Jimenez-Garcia,E., Vaquero,A., Canudas,S., Bernues,J. and Azorin,F. (1999) The N-terminal POZ domain of GAGA mediates the formation of oligomers that bind DNA with high affinity and specificity. J. Biol. Chem., 274, 16461–16469. [DOI] [PubMed] [Google Scholar]
- Gdula D.A. and Corces,V.G. (1997) Characterization of functional domains of the su(Hw) protein that mediate the silencing effect of mod(mdg4) mutations. Genetics, 145, 153–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimova T.I. and Corces,V.G. (1996) Boundary and insulator elements in chromosomes. Curr. Opin. Genet. Dev., 6, 185–192. [DOI] [PubMed] [Google Scholar]
- Gerasimova T.I. and Corces,V.G. (1998) Polycomb and trithorax group proteins mediate the function of a chromatin insulator. Cell, 92, 511–521. [DOI] [PubMed] [Google Scholar]
- Gerasimova T.I., Gdula,D.A., Gerasimov,D.V., Simonova,O. and Corces,V.G. (1995) A Drosophila protein that imparts directionality on a chromatin insulator is an enhancer of position-effect variegation. Cell, 82, 587–597. [DOI] [PubMed] [Google Scholar]
- Gerasimova T.I., Byrd,K. and Corces,V.G. (2000) A chromatin insulator determines the nuclear localization of DNA. Mol. Cell, 6, 1025–1035. [DOI] [PubMed] [Google Scholar]
- Geyer P.K. (1997) The role of insulator elements in defining domains of gene expression. Curr. Opin. Genet. Dev., 7, 242–248. [DOI] [PubMed] [Google Scholar]
- Geyer P.K. and Corces,V.G. (1992) DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev., 6, 1865–1873. [DOI] [PubMed] [Google Scholar]
- Hagstrom K., Muller,M. and Schedl,P. (1996) Fab-7 functions as a chromatin domain boundary to ensure proper segment specification by the Drosophila bithorax complex. Genes Dev., 10, 3202–3215. [DOI] [PubMed] [Google Scholar]
- Harrison D.A., Gdula,D.A., Coyne,R.S. and Corces,V.G. (1993) A leucine zipper domain of the suppressor of Hairy-wing protein mediates its repressive effect on enhancer function. Genes Dev., 7, 1966–1978. [DOI] [PubMed] [Google Scholar]
- Harrison S.D. and Travers,A.A. (1990) The tramtrack gene encodes a Drosophila finger protein that interacts with the ftz transcriptional regulatory region and shows a novel embryonic expression pattern. EMBO J., 9, 207–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdridge C. and Dorsett,D. (1991) Repression of hsp70 heat shock gene transcription by the suppressor of hairy-wing protein of Drosophila melanogaster. Mol. Cell. Biol., 11, 1894–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsani K.R., Hajibagheri,M.A. and Verrijzer,C.P. (1999) Co-operative DNA binding by GAGA transcription factor requires the conserved BTB/POZ domain and reorganizes promoter topology. EMBO J., 18, 698–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellum R. and Schedl,P. (1991) A position-effect assay for boundaries of higher order chromosomal domains. Cell, 64, 941–950. [DOI] [PubMed] [Google Scholar]
- Labrador M., Mongelard,F., Baxter,E.M., Plata-Rengifo,P., Corces,V.G. and Gerasimova,T.I. (2001) Protein encoding by both DNA strands. Nature, 409, 1000. [DOI] [PubMed] [Google Scholar]
- Mihaly J. et al. (1998) Chromatin domain boundaries in the Bithorax complex. Cell. Mol. Life Sci., 54, 60–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muravyova E., Golovnin,A., Gracheva,E., Parshikov,A., Belenkaya,T., Pirrotta,V. and Georgiev,P. (2001) Loss of insulator activity by paired Su(Hw) chromatin insulators. Science, 291, 495–498. [DOI] [PubMed] [Google Scholar]
- Parkhurst S.M., Harrison,D.A., Remington,M.P., Spana,C., Kelley,R.L., Coyne,R.S. and Corces,V.G. (1988) The Drosophila su(Hw) gene, which controls the phenotypic effect of the gypsy transposable element, encodes a putative DNA-binding protein. Genes Dev., 2, 1205–1215. [DOI] [PubMed] [Google Scholar]
- Philips J. and Herskowitz,I. (1998) Identification of Kel1p, a kelch domain-containing protein involved in cell fusion and morphology in Saccharomyces cerevisiae. J. Cell Biol., 143, 375–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseman R.R., Swan,J.M. and Geyer,P.K. (1995) A Drosophila insulator protein facilitates dosage compensation of the X chromosome min-white gene located at autosomal insertion sites. Development, 121, 3573–3582. [DOI] [PubMed] [Google Scholar]
- Scott K.S. and Geyer,P.K. (1995) Effects of the su(Hw) insulator protein on the expression of the divergently transcribed Drosophila yolk protein genes. EMBO J., 14, 6258–6267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong W.C. and Montell,C. (1993) tramtrack is a transcriptional repressor required for cell fate determination in the Drosophila eye. Genes Dev., 7, 1085–1096. [DOI] [PubMed] [Google Scholar]
- Xue F. and Cooley,L. (1993) kelch encodes a component of intercellular bridges in Drosophila egg chambers. Cell, 72, 681–693. [DOI] [PubMed] [Google Scholar]
- Zhao K., Hart,C.M. and Laemmli,U.K. (1995) Visualization of chromosomal domains with boundary element-associated factor BEAF-32. Cell, 81, 879–889. [DOI] [PubMed] [Google Scholar]
- Zhou J., Barolo,S., Szymanski,P. and Levine,M. (1996) The Fab-7 element of the bithorax complex attenuates enhancer–promoter interactions in the Drosophila embryo. Genes Dev., 10, 3195–3201. [DOI] [PubMed] [Google Scholar]
- Zollman S., Godt,D., Prive,G.G., Couderc,J.L. and Laski,F.A. (1994) The BTB domain, found primarily in zinc finger proteins, defines an evolutionarily conserved family that includes several developmentally regulated genes in Drosophila. Proc. Natl Acad. Sci. USA, 91, 10717–10721. [DOI] [PMC free article] [PubMed] [Google Scholar]