Abstract
Three serine protease zymogens, Gastrulation defective (GD), Snake (Snk) and Easter (Ea), and a nerve growth factor-like growth factor ligand precursor, Spaetzle, are required for specification of dorsal– ventral cell fate during Drosophila embryogenesis. The proteases have been proposed to function in a sequential activation cascade within the extracellular compartment called the perivitelline space. We examined biochemical interactions between these four proteins using a heterologous co-expression system. The results indicate that the three proteases do function in a sequential activation cascade, that GD becomes active and initiates the cascade and that interaction between GD and Snk is sufficient for GD to cleave itself autoproteolytically. The proteolytically active form of Ea cleaves GD at a different position, revealing biochemical feedback in the pathway. Both GD and Snk bind to heparin–Sepharose, providing a link between the pipe-defined ventral prepattern and the protease cascade. Our results suggest a model of the cascade in which initiation is by relief from inhibition, and spatial regulation of activity is due to interaction with sulfated proteoglycans.
Keywords: dorsal–ventral/embryogenesis/gastrulation defective/pattern formation/serine protease
Introduction
Proteolytic activation cascades comprise an evolutionarily conserved solution to the problem of transforming subtle molecular signals into a major reorganization of tissue architecture (Neurath, 1989). They have been implicated in numerous and diverse processes, including blood coagulation, fibrinolysis, the complement system, the phenol oxidase pathway, tumorigenesis, embryo implantation and tissue remodeling (Neurath, 1986). In many of these processes, the structures of the proteases as well as their biochemical interactions have been extensively investigated. Recently, there have been indications that proteolytic activation cascades may be involved in even more biological processes than previously appreciated. For example, genetic and molecular data from Drosophila melanogaster have suggested a central role for proteolytic cascades in elaborating the body plan during embryonic and imaginal disk morphogenesis (DeLotto and Spierer, 1986; Pino-Heiss and Schubiger, 1989). Early evidence of their involvement in embryonic development of the fruit fly came from the molecular cloning of the genes involved in dorsal–ventral patterning of the embryo (for review see Morisato and Anderson, 1995).
Specification of the dorsal–ventral axis of the Drosophila embryo involves a signal transduction pathway in which a polarized extracellular signal is generated within an extracellular compartment called the perivitelline space (PVS) (for review see Roth, 1994). Of the known components of this signaling pathway, the products of four genes function within the PVS. The genes include gastrulation defective (gd), snake (snk), easter (ea) and spaetzle (spz); epistasis indicates that their products function in this order. Molecular cloning of the genes has shown that gd, snk and ea encode the zymogen or inactive precursor forms of serine proteases (DeLotto and Spierer, 1986; Chasan and Anderson, 1989; Konrad et al., 1998). While Snk and Ea are similar in size and complexity to mammalian coagulation factor IX or X, GD is larger and resembles complement factors C2 and B (R.DeLotto, submitted). Serine protease zymogens normally require proteolytic processing to be converted to a proteolytically active form. The classical mechanism of the conversion from the inactive zymogen to the active protease has been intensively studied and is very well understood (Stroud et al., 1977).
The spz gene encodes the fourth extracellular component of the pathway and a precursor form of a nerve growth factor (NGF)-like growth factor (Morisato and Anderson, 1994). A processed form of Spz is believed to correspond to the ‘ventral specifying’ or ventralizing ligand (Schneider et al., 1994). A co-expression approach has allowed us to demonstrate that a constitutively active form of Ea can proteolytically process a Spz precursor to generate a dimeric molecule consisting of two disulfide-linked 12 kDa, 106-amino-acid C-terminal polypeptide fragments (DeLotto and DeLotto, 1998). Microinjection of the C-terminal fragment into spz-null embryos can define ventral cell fate as a function of position and polarize the dorsal–ventral axis. Additional protein isoforms of Spz have recently been described; however, their precise role in dorsal–ventral patterning is not clear (DeLotto et al., 2001).
While the hypothesis of a proteolytic cascade is certainly attractive in view of the available data, direct molecular interactions of all of the known components have not been demonstrated definitively. Reconstitution of the pathway in a heterologous expression system would be a useful way to determine whether these four proteins can interact biochemically. To address this question, we co-expressed recombinant forms of the proteins using the baculovirus system and heterologous lepidopteran cell lines, which express secreted proteins efficiently. Antibodies generated against GD, Snk, Ea and Spz were used in immunoblotting against conditioned cell culture medium after co-expressing various combinations of wild-type and altered forms of these proteins. Co-expression of all four proteins resulted in complete processing of a Spz precursor to generate the 12 kDa putative ligand form, demonstrating conclusively that they can function in a sequential proteolytic cascade. While GD when expressed alone only generated one major 72 kDa form, co-expression of GD with Snk resulted in autoproteolytic processing of GD to produce lower molecular weight species. GD processing does not require the proteolytic activity of Snk. In addition, active Ea protease feeds back upon the cascade by processing GD at an alternative position(s). Our results are incorporated into a biochemical model of the cascade and the consequences for pattern formation are discussed.
Results
Active Snk proteolytically processes the Ea zymogen
To determine whether Snk can process the Ea zymogen to an active form, we co-expressed forms of these proteins and, by immunoblotting, examined whether Ea was cleaved. Lepidopteran cells were coinfected with a mixture of recombinant baculovirus expressing the Ea proform and increasing amounts of recombinant baculovirus expressing SnkΔn, a constitutively active form of Snk. As shown in Figure 1, the uncleaved Ea zymogen migrated with an Mr of 50 kDa. However, as the ratio of SnkΔn to Ea increased, a new fragment appeared at 35 kDa in an SnkΔn concentration-dependent manner. This 35 kDa polypeptide corresponds to what has been previously reported for the catalytic chain of activated Ea (Chasan et al., 1992). The 35 kDa band, as well as a lower band, also appeared when conditioned medium containing Ea proform was partially digested with trypsin in vitro. A trypsin-like activity is predicted to be required to activate Ea based upon its amino acid sequence (Chasan and Anderson, 1989). Co-expression of Ea with SnkΔn(S to A), a proteolytically inactive form of Snk, failed to generate the 35 kDa band (data not shown). We conclude that Snk can process Ea to its active form under co-expression conditions with higher specificity than trypsin has in vitro.
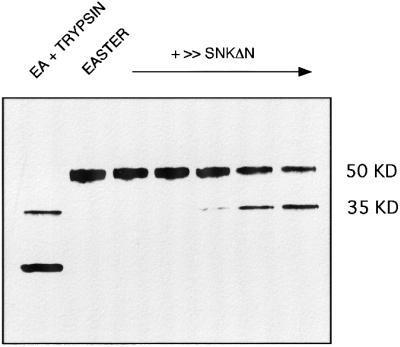
Fig. 1. Proteolytic processing of Ea by active Snk. An immunoblot, using anti-Ea specific antiserum, of conditioned cell culture medium from co-expression of Ea proenzyme (50 kDa) and increasing amounts of SnkΔn, a constitutively active form of Snk. Lanes (from left to right) correspond to: Ea partially digested in vitro with trypsin; Ea alone; and Ea with 1/100, 1/50, 1/20, one-fifth and an equal volume of SnkΔn (v/v ratio of high titer stocks). The 35 kDa polypeptide corresponds to the size previously reported for the active Ea catalytic chain.
Activated Ea proteolytically processes a Spz precursor
We have shown previously that EaΔn, a constitutively active form of Ea deleted for the proenzyme polypeptide, can process a 40 kDa form of Spz, Spz8.19, to generate a 106-amino-acid 12 kDa C-terminal fragment. To determine whether an active form of Ea retaining the covalently associated propolypeptide can also process Spz, we co-expressed Spz with various forms of Ea and Snk and immunoblotted using antisera directed against the C-terminus of Spz (Figure 2). Expression of Spz alone resulted in the appearance of a 40 kDa band. Co-expression of Spz with the Ea zymogen did not alter the size of Spz. However, co-expression with EaΔn generated the 12 kDa C-terminal fragment. Co-expression of either SnkΔn or XaSnk, two constitutively active forms of Snk, with Ea and Spz, also led to conversion of Spz to generate a 12 kDa fragment. Neither active form of Snk directly processed Spz (data not shown). This conversion appears to be more efficient than we have previously observed with EaΔn, possibly because activated Ea is more stable than EaΔn (Smith et al., 1995). We have previously shown that XaSnk, an autocatalytically activated form of Snk, is more stable than SnkΔn, a propolypeptide-deleted form. Consistent with this interpretation, co-expression of XaSnk, Ea and Spz led to a reduction in the 40 kDa polypeptide and increase in the 12 kDa fragment. Mutation of the active site serine of XaSnk to alanine eliminates conversion of Spz (Smith et al., 1995). The results demonstrate that active Snk can process Ea to generate an active form that subsequently can process Spz.
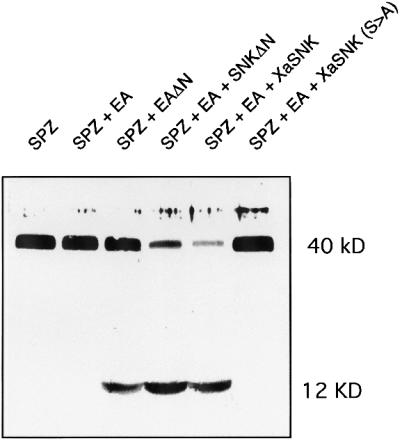
Fig. 2. Proteolytic processing of Spz 8.19 by active forms of Ea. An immunoblot, using anti-Spz C-terminal specific antiserum, of conditioned cell culture medium from co-expression of Spz 8.19 (40 kDa). While the inactive Ea zymogen had no effect upon the 40 kDa polypeptide, a constitutively active Ea catalytic chain, EaΔn, or Ea in combination with one of two different active forms of Snk, SnkΔn and XaSnk, led to generation of a 12 kDa C-terminal Spz fragment.
Co-expression of GD, Snk, Ea and Spz leads to efficient processing of Spz
To determine whether GD can interact with Snk, Ea and Spz, we co-expressed these proteins in various combinations and assayed for the processing of Spz to generate the 12 kDa C-terminal fragment. Figure 3 shows an immunoblot, probed with anti-Spz antiserum, of cell culture supernatants from cells expressing Spz 8.19 alone or in combination with the other proteins. Spz expressed with the zymogen forms of Ea or Snk plus Ea migrated at 40 kDa and thus is not proteolytically processed. However, when the zymogen form of GD was expressed in combination with the zymogen forms of Snk and Ea, all of the Spz 40 kDa form disappeared to yield the 12 kDa C-terminal fragment. This conversion must be dependent upon the serine protease catalytic activity of GD, since a form of GD that had been mutated in its active site serine to alanine failed to catalyze the conversion. GD does not activate Ea or Spz directly, since elimination of Snk in the co-expression resulted in no production of the Spz 12 kDa fragment. In parallel immunoblots with antisera against Snk and Ea, a band at the appropriate size for the activated form of Snk and Ea was visible when all four wild-type proteins were co-expressed (data not shown). We conclude that under the conditions of co-expression, GD serine protease activity is required for activation of Snk and the subsequent triggering of the proteolytic cascade. We do not know how the zymogen form of GD, which should be proteolytically inactive, is able to facilitate the activation of Snk and the subsequent activation of the protease cascade.
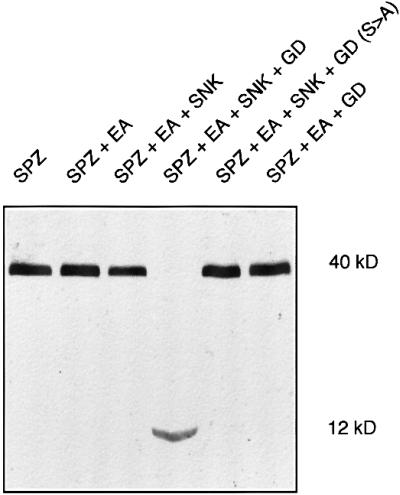
Fig. 3. Co-expression of GD, Snk, Ea and Spz leads to activation of a protease cascade. Immunoblot of Spz as in Figure 2. Expression of either Spz 8.19 alone or in combination with Ea zymogen and Snk zymogen led to no processing of Spz. However, co-expression of GD plus the other three proteins led to highly efficient processing of the Spz precursor to generate a 12 kDa fragment. Mutation of the active site serine of GD eliminated processing and GD does not bypass Snk to activate Ea.
Co-expression of GD and Snk results in autoproteolytic processing of GD
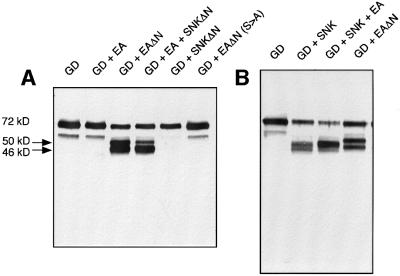
To determine what happens to GD during co-expression with Snk, various forms of GD and Snk were co-expressed and GD was examined by immunoblotting with antisera directed against GD. As shown in Figure 4, expression of GD alone resulted in a major 72 kDa polypeptide that cross-reacted with anti-GD antibodies. We attribute this to the zymogen form of GD. When GD was co-expressed with the Snk zymogen, the 72 kDa band was reduced in intensity and a doublet appeared at ∼44–48 kDa. When GD was expressed with a proteolytically inactive form of Snk, Snk(S to A), predominantly the 48 kDa band still appears. However, when a catalytically inactive form of the GD zymogen was co-expressed with either wild-type Snk or Snk(S to A), neither lower molecular weight form of GD was observed. The simplest interpretation of these results is that upon presentation with the zymogen form of Snk, GD is autoproteolytically processed into a polypeptide of 48 kDa. In the presence of an active form of Snk, GD can also process itself to generate a 44 kDa polypeptide.
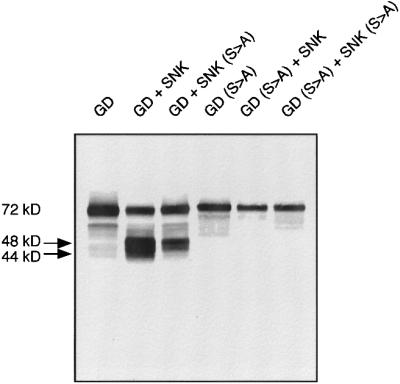
Fig. 4. Co-expression of GD and Snk results in GD autoproteolysis to generate lower molecular weight polypeptides. Western blot using anti-GD specific antisera. A GD-specific polypeptide of 72 kDa was generated when GD was expressed alone. However, when GD was expressed with either wild-type Snk zymogen or a proteolytically inactive form of Snk, two polypeptides of 44 and 48 kDa were generated. Appearance of the lower molecular weight polypeptides requires integrity of the active site serine of GD.
The correlation of the appearance of the lower molecular weight forms of GD under conditions in which GD facilitates activation of the cascade suggests that the lower molecular weight polypeptides are either active forms or a later consequence of the activation of GD. The size of either fragment is in agreement with the predicted size for the active catalytic chain of GD based upon a complement factor C2/B homology recently described (R.DeLotto, manuscript submitted). In titrations of different ratios of Snk to GD, where GD was in excess, the 48 kDa poly peptide appeared to be the predominant species generated (data not shown). However, we have not been able to show that any of the lower molecular weight forms of GD bind to any reagents with affinity for the active form of serine proteases. We therefore have no evidence at present that any of these lower molecular weight species correspond to the active form of GD.
Ea proteolytically processes GD
To determine whether activated forms of Snk and Ea can process GD, we co-expressed GD with various forms of these proteases and followed GD by immunoblotting (see Figure 5). As previously shown, co-expression of GD with the zymogen form of Snk resulted in the appearance of the 44, 48 kDa doublet while GD plus Snk(S to A) generated primarily the 48 kDa form. Co-expression of GD with SnkΔn, the constitutively active catalytic chain of Snk, had no apparent effect on the size of GD. We conclude that the activated Snk protease catalytic chain can not process GD directly.

Fig. 5. Active Snk catalytic chain does not cleave GD. The 72 kDa GD polypeptide was not cleaved by SnkΔn, a constitutively active form of Snk, or by the zymogen form of Ea.
We tested whether Ea can interact with GD (Figure 6). When GD was co-expressed with the Ea zymogen, no change in the 72 kDa polypeptide was observed. However, when GD was co-expressed in the presence of EaΔn, two new cross-reacting polypeptides appeared, of 46 and 50 kDa. The protease activity of Ea must be required, as a catalytically inactive form of Ea failed to generate the lower molecular weight forms of GD. Since the GD fragments generated by Snk-dependent autoproteolysis were similar in size to those produced by Ea, we compared the sizes of these fragments carefully. As shown in Figure 6B, the GD fragments generated by Ea did not comigrate with those generated by autoproteolysis in the presence of Snk zymogen and are therefore different in size. We concluded that the Ea protease can process GD at positions different from those cleaved after GD autoproteolysis.
Fig. 6. The active Ea protease specifically processes GD to generate 46 and 50 kDa polypeptide fragments. Western blotting with anti-GD antisera. (A) GD co-expressed with EaΔn but not that co-expressed with proteolytically inactive EaΔn(S to A) led to the generation of two lower molecular weight GD fragments. The 46 kDa band was preferentially cleaved and the 50 kDa band partially suppressed when an active form of Snk was co-expressed with Ea. (B) Ea-generated proteolytic fragments differ in size from those generated by Snk-induced GD autoproteolysis.
Co-expression of GD, Snk and Ea resulted in the appearance of a 48 kDa GD-derived polypeptide that comigrated with the upper of the Snk-induced autoproteolytic bands. Although this polypeptide is the same size as that observed upon Snk and GD co-expression, we can not determine if they are identical. Further characterization of the N- and C-terminal sequences of these processed polypeptides will be necessary to determine precisely the sites of cleavage in GD both by autoproteolysis and by Ea. These results indicate that the interaction of Snk and Ea with GD is more complex than a simple linear cascade. GD has the potential to be multiply cleaved at different sites, and the biochemical significance of each cleavage is not yet fully clear.
GD and Snk bind to heparin
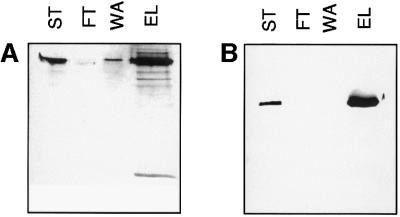
To determine whether any of these four proteins were able to interact with sulfated glycosaminoglycans, we tested the ability of recombinant forms of GD, Snk, Ea and Spz to bind to heparin–Sepharose in the presence of 150 mM NaCl, conditions that should reduce non-specific binding. The results for GD and Snk are illustrated in Figure 7. The 72 kDa form of GD was present in the dialysate that was loaded on the column (ST), but not in the flowthrough (FT) (Figure 7A). Elution with 2 M NaCl removed GD from the column. Similarly a 52 kDa form of the Snk zymogen bound to heparin–Sepharose and eluted at 2 M NaCl (Figure 7B). Since the elution volumes were 16-fold smaller than the starting volume, the signal in the ‘elution’ lanes is more intense than in the ‘starting material’ lane. The binding is specific, since both GD and Snk could also be eluted by 5 mg/ml heparin under isotonic salt conditions (data not shown). Under similar binding conditions neither Ea nor Spz 8.19 appeared to interact with heparin–Sepharose (data not shown). These results suggest a strong potential of both GD and Snk to interact with sulfated proteoglycan.
Fig. 7. Binding of GD and Snk to heparin–Sepharose. Western blotting with anti-GD antiserum (A) or monoclonal anti-Snk antibody (mAb53) (B) of column chromatography on a 0.5 ml bed volume heparin–Sepharose column. (A) GD binding and elution. (B) Snk binding. Lanes in both panels are as follows: ST, starting material; FT, flowthrough; WA, wash; and EL, eluate. For details see Materials and methods.
Discussion
The results presented here provide the first biochemical demonstration of direct interaction among the three known dorsal–ventral serine proteases and presumptive ligand precursor Spz. When expressed in a heterologous system, GD, Snk and Ea function together in a sequential proteolytic activation cascade, which results in the proteolytic processing of a precursor form of Spz to generate a molecule that has previously been demonstrated to have the properties of the ventralizing ligand (DeLotto and DeLotto, 1998). These four proteins together are clearly sufficient for processing of Spz to occur. Consequently, we can rule out an absolute requirement for any additional proteases in the cascade. However, our results do not exclude the possibility that, in vivo, additional components regulate the activation of the protease cascade. Nor do our results indicate whether binding to membrane surfaces or receptors is a prerequisite for efficient interaction, since intact cell membranes are present under the conditions of co-expression. In spite of these limitations, the co-expression system described here is clearly a powerful and convenient tool for analyzing direct interactions among any of the identified components of the dorsal–ventral pathway that can be expressed using recombinant baculovirus.
A heterologous expression system was specifically chosen for these studies to avoid potential complications that might arise due to endogenous background expression of any of these proteins by the host cells. Schneider S2 cells, one of the most common Drosophila tissue culture cell lines, can translocate Dorsal from cytoplasm to nucleus in response to Toll (Kubota and Gay, 1995). They also constitutively express several components of the dorsal–ventral signaling pathway (R.DeLotto and B.Voldborg, unpublished results). By choosing a heterologous system, we sought to minimize the contribution of endogenous protein expression to the analysis. The results presented here were generated in Mammestra brassica IZD-MB0503 host cells (Hink et al., 1991). We were able to reproduce the interactions described here using High-5 cells derived from Trichoplusia ni (data not shown). Consequently, we argue that the interactions defined here represent the potential of the expressed proteins to interact directly and are not an artifact of the cell line used.
nudel, a somatically required dorsal group gene, encodes a 320 kDa mosaic protein with a centrally located serine protease catalytic domain (Hong and Hashimoto, 1995). It has been suggested that nudel might activate a dorsal–ventral proteolytic cascade (LeMosy et al., 1998). We investigated this possibility using our assay system by co-expressing either the full-length form of Nudel, or a constitutively active Nudel serine protease catalytic chain (ndlΔn) in different combinations with the four proteins described here. In all experiments conducted thus far, we failed to observe any reproducible effect of Nudel or nudΔn upon any of the other proteins nor an effect of any of the other proteins upon Nudel-specific polypeptides (data not shown). Consequently we have no evidence for direct biochemical interaction of Nudel with GD, Snk, Ea or Spz and therefore can not assign a direct role for Nudel within the protease cascade. We favor the idea that Nudel does not directly activate the cascade but rather is required earlier for proper establishment or maintenance of the ventral prepattern (for review see LeMosy et al., 1999).
GD appears to play a critical role in the proteolytic cascade since it can initiate the cascade yet does not appear to require classical zymogen activation in order to do so. Our data suggest that exposure to the zymogen form of Snk is sufficient for GD to become active and activate Snk, triggering the cascade, and for GD to generate lower molecular weight polypeptides. With respect to a mechanism of activation, it is interesting to note that GD bears some similarity to mammalian complement factors C2 and B (R.DeLotto, submitted). These proteases have novel activation mechanisms requiring complex formation and a conformational change as a prerequisite to activa tion (Arlaud et al., 1998). An alternative explanation for how GD functions is that it has some intrinsic activity as a zymogen. Upon binding to and activating Snk, it then proteolytically processes itself to generate lower molecular weight inactive forms.
Activated Ea can proteolytically process GD, suggesting that a second form of feedback occurs within the cascade. Ea cleaves GD at a novel position to generate a GD polypeptide that is slightly larger than the predominant band generated by GD itself. The significance of processing by Ea is not altogether clear from our data. However, it is reasonable to assume that cleavage by Ea is a way to modify GD’s biochemical properties. We propose that Ea may feed back negatively on the precursor form and/or the active form of GD. This would provide a means of down-regulating the protease cascade to prevent amplification from ‘running away’, resulting in overproduction of the ventralizing signal.
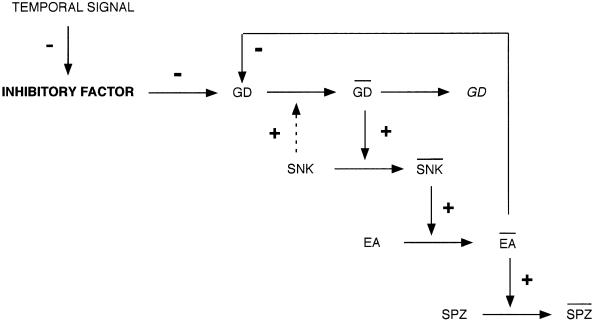
Since GD can autoactivate the cascade, we need not postulate a requirement for an upstream protease to activate the cascade. Rather, our data might argue that a mechanism exists to prevent GD from becoming activated too early in embryogenesis before the Toll receptor is completely expressed on the plasma membrane. This idea has been incorporated into a biochemical model of the cascade, illustrated in Figure 8. We propose the existence of an inhibitory factor that prevents GD from activating Snk. The inhibitory factor must itself be inactivated in a spatially or temporally regulated way for proper activation of the cascade. However, the net result must be to permit the activation of GD within the ventral PVS at the proper time for accurate elaboration of the ventralizing signal. This region of the PVS may correspond to the ventral stripe prepattern recently described from gd mRNA injection experiments (R.DeLotto, submitted).
Fig. 8. Biochemical model of the dorsal–ventral protease cascade. Based upon our co-expression results, we propose a model in which GD, Snk, Ea and Spz sequentially activate each other. GD, SNK and EA indicate active forms of the proteases. SPZ indicates the ligand form of SPZ. GD, which can become active in the absence of a specific activator, must be kept inactivated via an inhibitory factor. Activation of the cascade would therefore require relief from inhibition by inactivation of this inhibitory factor. Upon complexing with and activating Snk, GD undergoes auto proteolysis to generate cleaved and proteolytically inactive forms represented as GD. An active form of Ea feeds back upon either the proform or the active form of GD to process it and down-regulate the signal.
The ability of both GD and Snk to bind to heparin– Sepharose suggests that their activities may be regulated in vivo by sulfated proteoglycans. Since pipe expression in somatic follicle cells is ventrally restricted and the gene encodes a heparan sulfate 2-O-sulfotransferase, sulfate modification of an as yet unknown proteoglycan may provide the ventral cue in the egg (Sen et al., 1998). GD and Snk may interact directly with this sulfated proteoglycan and this interaction may provide the ventral restriction to activation of the cascade.
It has been suggested that the protease cascade may enable an initial asymmetry in the form of a ventral stripe prepattern to be converted into a graded distribution of processed Spz ligand (R.DeLotto, manuscript submitted). The potential for both (i) amplification with subsequent steps and (ii) feedback after activation could enable the cascade to self-regulate the shape of the Dorsal protein gradient. This property could provide plasticity in the patterning process and a means of compensating for minor variation in the size and shape of individual embryos. Such a mechanism would also be sufficiently adaptable that it could be conserved evolutionarily. Our data suggest some remarkable similarities between the dorsal–ventral protease cascade and the classical complement and blood coagulation pathways.
Materials and methods
Expression of recombinant proteins
Proteins were produced using recombinant baculovirus generated by standard methods using the pBacPak8 vector and BstEII-linearized Bac6 viral DNA (Invitrogen). A reverse transcription–polymerase chain reaction (RT–PCR) product, gdcD7, produced from GD cDNA based on the sequence data of Konrad et al. (1998) and R.DeLotto (submitted), was subcloned into pGEM4 to generate pGEM4gd. The insert of pGEM4gd was digested with XbaI and KpnI and subcloned into XbaI- and KpnI-digested pBacPaK8, to generate pBacPaK8gd. Recombination between pBacPaK8gd and Bac6 was used to generate recombinant baculovirus, which was plaque purified and amplified after assay with GD-specific rat antisera using standard methods (Summers and Smith, 1987). The production of recombinant Snk, Snk(S to A) XaSnk, snkΔn and snkΔn(S to A) have been previously described (Smith and DeLotto, 1994; Smith et al., 1994, 1995). An Ea cDNA was generated by RT–PCR using 5′-GAATTCAGATCTATGCTAAAGCCATCGATTATCTGCCTCTTTTTGGGC-3′ and 5′-CTCAAGCTTGGTACCTCAGGACTCAATAGTGTTTTGTATCCAATC-3′ under reaction conditions previously described (DeLotto and DeLotto, 1998). The Ea fragment was digested with BglII and KpnI and subcloned into pBacPak8 digested with BamHI and KpnI. Recombination and characterization were conducted as for GD and analysis was done by western blotting with rat anti-Ea sera. Recombinant Spz 8.19 and 11.7 proteins have been described previously (DeLotto and DeLotto, 1998).
Antibodies
Antisera to Snk and Spz have been described previously (Smith et al., 1995; DeLotto and DeLotto, 1998). Rat polyclonal antisera were generated by immunization using insoluble aggregate preparations of bacterial fusion proteins expressed in pATH10 and pATH11. A PstI– BamHI fragment of gdcD7 was subcloned into PstI- and BamHI-digested pATH11. For Ea, a SacI–HindIII fragment of the Ea cDNA (Chasan and Anderson, 1989) was subcloned into SacI- and HindIII-digested pATH10. Insoluble aggregate proteins were purified by sodium dodecyl sulfate (SDS) gel electrophoresis; bands were excised from the gel and injected for immunization. Sera from immunized animals (Pocono Rabbit Farms) were tested against the bacterial fusion protein and against supernatants from baculovirus-expressed proteins by western blotting.
Co-expression and western blotting
IZD-HQ cells were grown in Hyclone CCM-3 serum-free medium at 27°C. Cells grown to 75% confluence were infected with either recombinant baculovirus or mixtures of recombinant baculovirus and incubated for 72 h. Infected cells were removed by centrifugation at 900 r.p.m. for 5 min in an swinging bucket rotor in a Dupont RT6000 centrifuge, clear supernatants were loaded on a 12.5 or 15% SDS– polyacrylamide gel and western blotting was performed as described previously (Driever and Nuesslein-Volhard, 1988). Alternative host cells for expression were High-5 (Invitrogen) grown in Ex-cell 401 serum-free medium (JRH Biosciences).
Heparin–Sepharose chromatography
GD or Snk was expressed as described above and 16 ml of conditioned cell culture medium were dialyzed into 50 mM MOPS pH 6.8, 150 mM NaCl, 2 mM EDTA and loaded on to a 0.5 ml bed volume of heparin–Sepharose (Pharmacia) in a Bio-Rad disposable column. The column was washed with 1 ml of dialysis buffer and eluted with 1 ml of 50 mM MOPS pH 6.8, 2 M NaCl. Ten microlitres of each sample were electrophoresed on a Hoefer minigel apparatus and western blotted using either rat anti-GD polyclonal antiserum or Snk mAb53 monoclonal antibodies as described above.
Acknowledgments
Acknowledgements
The authors would like to thank Stanley Brown for helpful discussions and comments on the manuscript. R.D. would like to thank Yvonne DeLotto for superb technical assistance. This work was supported by the Danish Natural Science Research Council, the Danish Cancer Fund, the Vera and Carl Johan Michaelsens Legacy and the US National Science Foundation.
References
- Arlaud G.J., Volanakis,J.E., Thielens,N.M., Narayana,S.V., Rossi,V. and Xu,Y. (1998) The atypical serine proteases of the complement system. Adv. Immunol., 69, 249–307. [PubMed] [Google Scholar]
- Chasan R. and Anderson,K.V. (1989) The role of easter, an apparent serine protease, in organizing the dorsal–ventral axis of the Drosophila embryo. Cell, 56, 391–400. [DOI] [PubMed] [Google Scholar]
- Chasan R., Jin,Y. and Anderson,K.V. (1992) Activation of the easter zymogen is regulated by five other genes to define dorsal–ventral polarity in the Drosophila embryo. Development, 115, 607–616. [DOI] [PubMed] [Google Scholar]
- DeLotto R. and Spierer,P. (1986) A gene required for the specification of dorsal–ventral pattern in Drosophila appears to encode a serine protease. Nature, 323, 688–692. [DOI] [PubMed] [Google Scholar]
- DeLotto Y. and DeLotto,R. (1998) Proteolytic processsing of the Drosophila Spaetzle protein by Easter generates a dimeric NGF-like molecule with ventralizing activity. Mech. Dev., 72, 141–148. [DOI] [PubMed] [Google Scholar]
- DeLotto Y., Smith,C. and DeLotto,R. (2001) Multiple isoforms of the NGF-like growth factor ligand are encoded as alternatively spliced mRNAs in precellular blastoderm embryos. Mol. Gen. Genet., 264, 643–652. [DOI] [PubMed] [Google Scholar]
- Driever W. and Nuesslein-Volhard,C. (1988) A gradient of bicoid protein in Drosophila embryos. Cell, 54, 83–93. [DOI] [PubMed] [Google Scholar]
- Hink W.F., Thomsen,D.R., Davidson,D.J., Meyer,A.L. and Castellino,F.J. (1991) Expression of three recombinant proteins using baculovirus vectors in 23 insect cell lines. Biotechnol. Prog., 7, 9–14. [DOI] [PubMed] [Google Scholar]
- Hong C. and Hashimoto,C. (1995) An unusual mosaic protein with a protease domain encoded by the nudel gene is involved in defining dorsoventral polarity in Drosophila. Cell, 82, 785–794. [DOI] [PubMed] [Google Scholar]
- Konrad K.D., Goralski,T.J., Mahowald,A.P. and Marsh,J.L. (1998) The gastrulation defective gene of Drosophila melanogaster is a member of the serine protease superfamily. Proc. Natl Acad. Sci. USA, 95, 6819–6824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota K. and Gay,N.J. (1995) Calcium destabilises Drosophila cactus protein and dephosphorylates the dorsal transcription factor. Biochem. Biophys. Res. Commun., 214, 1191–1196. [DOI] [PubMed] [Google Scholar]
- LeMosy E.K., Kemler,D. and Hashimoto,C. (1998) Role of Nudel protease activation in triggering dorsoventral polarization of the Drosophila embryo. Development, 125, 4045–4053. [DOI] [PubMed] [Google Scholar]
- LeMosy E.K., Hong,C.C. and Hashimoto,C. (1999) Signal transduction by a protease cascade. Trends Cell Biol., 9, 102–107. [DOI] [PubMed] [Google Scholar]
- Morisato D. and Anderson,K. (1994) The spaetzle gene encodes a component of the extracellular signalling pathway establishing the dorsal–ventral pattern of the Drosophila embryo. Cell, 76, 677–688. [DOI] [PubMed] [Google Scholar]
- Morisato D. and Anderson,K.V. (1995) Signaling pathways that establish the dorsal–ventral pattern of the Drosophila embryo. Annu. Rev. Genet., 29, 371–399. [DOI] [PubMed] [Google Scholar]
- Neurath H. (1986) The versatility of proteolytic enzymes. J. Cell. Biochem., 32, 35–49. [DOI] [PubMed] [Google Scholar]
- Neurath H. (1989) Proteolytic processing and physiological regulation. Trends Biochem. Sci., 14, 268–271. [DOI] [PubMed] [Google Scholar]
- Pino-Heiss S. and Schubiger,G. (1989) Extracellular protease production by Drosophila imaginal discs. Dev. Biol., 132, 282–291. [DOI] [PubMed] [Google Scholar]
- Roth S. (1994) Axis determination. Proteolytic generation of a morphogen. Curr. Biol., 4, 755–757. [DOI] [PubMed] [Google Scholar]
- Schneider D., Jin,Y., Morisato,D. and Anderson,K. (1994) A processed form of the Spaetzle protein defines dorsal–ventral polarity in the Drosophila embryo. Development, 120, 1243–1250. [DOI] [PubMed] [Google Scholar]
- Sen J., Goltz,J.S., Stevens,L. and Stein,D. (1998) Spatially restricted expression of pipe in the Drosophila egg chamber defines embryonic dorsal–ventral polarity. Cell, 95, 471–481. [DOI] [PubMed] [Google Scholar]
- Smith C. and DeLotto,R. (1994) Ventralizing signal determined by protease activation in Drosophila embryogenesis. Nature, 368, 548–551. [DOI] [PubMed] [Google Scholar]
- Smith C., Giordano,H. and DeLotto,R. (1994) Mutational analysis of the Drosophila Snake protease: an essential role for domains within the proenzyme polypeptide chain. Genetics, 136, 1355–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C., Giordano,H., Schwartz,M. and DeLotto,R. (1995) Spatial regulation of Drosophila Snake protease activity in the generation of dorsal–ventral polarity. Development, 121, 4127–4135. [DOI] [PubMed] [Google Scholar]
- Stroud R., Kossiakoff,A. and Chambers,J. (1977) Mechanisms of zymogen activation. Annu. Rev. Biophys. Bioeng., 6, 177–193. [DOI] [PubMed] [Google Scholar]
- Summers M. and Smith,G. (1987) A Manual of Methods for Baculovirus Vectors and Insect Cell Culture Procedures. Texas Agricultural Station Bulletin, No. 1555, Houston, TX.