Abstract
The binding of interferons (IFNs) to their receptors leads to the phosphorylation and activation of signal transducers and activators of transcription (STATs), and their translocation from the cytoplasm to the nucleus. The mechanisms by which the STATs move to the nuclear pore are not, however, known. Here it is shown that IFN-α and -γ signalling and STAT1 translocation are independent of the actin cytoskeleton or microtubules. Using fluorescence loss in photobleaching (FLIP) and fluorescence recovery after photobleaching (FRAP) experiments, the mobility of a fusion protein of STAT1 with green fluorescent protein (STAT1–GFP) was compared with that of GFP and protein kinase C–GFP. In IFN-γ-treated and control cells, cytoplasmic STAT1–GFP shows high, energy-independent, mobility comparable to that of freely diffusible GFP. A random walk model for movement of STAT1 from the plasma membrane to the nuclear pore is, therefore, indicated. Nuclear STAT1–GFP showed similar high mobility, with exclusion from nucleoli, consistent with high rates of association and dissociation of STAT1–DNA and/or STAT1–protein complexes in the nucleoplasm of the cell.
Keywords: interferon/photobleaching/random walk/signal transduction/STAT1–GFP
Introduction
Appropriate localization of proteins is crucial for their physiological function and regulation. Proteins transmit incoming signals by interaction with other molecules with or without movement into another location in the cell. Activation of transcription factors can occur in the cytoplasm or at the cell membrane. On activation, they translocate to the nucleus through nuclear pores (Kaffman and O’Shea, 1999). Little is known, however, about the mechanisms of transport to the nuclear pores. Movement through the cytoplasm could, a priori, be active along microtubules as observed for p53 (Giannakakou et al., 2000), or passive, by diffusion, as in a random walk.
The Janus kinase (JAK)/signal transducers and activators of transcription (STAT) pathways, first identified in the interferon (IFN) systems (Darnell et al., 1994), are activated by a wide range of cytokines and growth factors. There are four known mammalian members of the JAK family of protein tyrosine kinases: JAK1, JAK2, JAK3 and Tyk2 (Wilks, 1989; Firmbach-Kraft et al., 1990; Harpur et al., 1992; Silvennoinen et al., 1993; Johnston et al., 1994; Witthuhn et al., 1994) and seven mammalian STAT genes (STATs 1–6, including STATs 5A and B; Fu, 1992; Fu et al., 1992; Schindler et al., 1992a; Akira et al., 1994; Hou et al., 1994; Wakao et al., 1994; Yamamoto et al., 1994; Zhong et al., 1994; Quelle et al., 1995). Upon ligand binding, the receptor-associated JAKs become activated by auto- and transphosphorylation, and phosphorylate the receptor. STATs are recruited to the JAK–receptor complexes, phosphorylated, released and migrate to the nucleus to activate transcription (Ihle et al., 1995; Schindler and Darnell, 1995; Duhe and Farrar, 1998; Leonard and O’Shea, 1998; Stark et al., 1998; Yeh and Pellegrini, 1999). JAK1 and Tyk2, and JAK1 and JAK2 are associated with the IFN-α/β and IFN-γ receptors, respectively (Muller et al., 1993a; Watling et al., 1993). STATs 1–5 can be activated in response to IFN-α. Here, however, we will be largely concerned with the activation of STAT1, the only STAT activated in response to IFN-γ in the human cell systems used. Retention of non-activated STAT1 in the cytoplasm does not reflect anchoring or inhibition of shuttling (McBride et al., 2000). On release from the JAK–receptor complex, activated STAT1 dimers migrate to the nuclear pore. No ‘classical’ nuclear localization sequence (NLS) has been detected in STAT1. However, interaction of activated STAT1 with importin NPI-1 initiates translocation through the nuclear pore (Sekimoto et al., 1997). The latter takes time and is energy dependent. Consequent upon these requirements, the nuclear membrane forms an effective barrier to the translocation of non-activated STAT1 (Köster and Hauser, 1999; Figure 3). Dephosphorylation of STAT1 in the nucleus (Haspel and Darnell, 1999; McBride et al., 2000) and export to the cytoplasm (McBride et al., 2000; Mowen and David, 2000) control recycling of STAT1 and maintain responsiveness of the cell.
Fig. 3. FLIP analysis. (A) Cytoplasmic and (B) nuclear fluorescence. (A) 2C4 cells stably transfected with STAT1–GFP without (row 1) and after 15 min of IFN-γ treatment (103 IU/ml; row 2), GFP (row 3) and PKC–GFP without (row 4) and after 5 min of TPA treatment (Materials and methods; row 5). Every image in a row contains the same cells. The bleach region in the cytoplasm is indicated with a white square and fluorescence intensity is shown in false colour code (vertical bar, top right). Each row shows the fluorescence prior to bleaching (0 s) and after three consecutive 90 s bleach periods (90, 180 and 270 s). In the case of PKC–GFP, after TPA the plane of focus was the membrane parallel to the coverslip (row 5). (B) 2C4 cells stably transfected with STAT1–GFP and treated with IFN-γ at 103 IU/ml for 25 min (row 1) and GFPnls (row 2). Each row shows the fluorescence prior to bleaching and after three consecutive 30 s bleach periods (0, 30, 60 and 90 s, respectively) of the nuclear area bounded by the white square.
Here we have investigated the dependence of IFN-α and -γ signalling on the cytoskeleton. Also, using a known biologically active STAT1–GFP (where GFP is green fluorescent protein) (Köster and Hauser, 1999; Figure 2), we have examined for IFN-γ how STAT1 moves from the plasma membrane JAK–receptor complex to the nuclear pore, and the nuclear mobility of STAT1. The data are consistent with random walk models for the movement of activated STAT1 in both compartments.
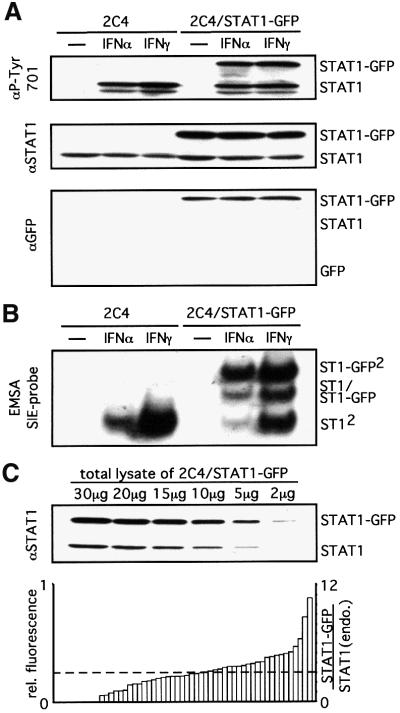
Fig. 2. STAT1–GFP is a functional transcription factor. (A and B) 2C4 (left lanes) and 2C4/STAT1–GFP cells (right lanes) were stimulated with 103 IU/ml IFN-α or -γ for 20 min. (A) Phosphorylation of STAT1–GFP. Western blot analysis of total cell lysates with an anti-P-Tyr701 specific antibody (upper panel), an anti-STAT1 antibody as loading control (middle panel) and an anti–GFP antibody to monitor for any STAT1–GFP cleavage (lower panel). No evidence for free GFP was obtained even on prolonged exposure (data not shown). (B) DNA binding of STAT1–GFP was analysed by EMSA of whole-cell extracts with an SIE probe. (C) The expression of STAT1 and STAT1–GFP in the 2C4/STAT1–GFP population was compared by analysing a series of dilutions of a total cell lysate by western blot analysis with antibody to STAT1 (upper panel). The relative fluorescence intensity of 54 single cells was analysed by confocal microscopy (left ordinate, lower panel; Materials and methods). The ratio of STAT1–GFP to endogenous STAT1 (right ordinate, lower panel) was calculated from the two data sets. The dotted line shows both the average fluorescence (left scale) and the average expression relative to STAT1 (right scale) of STAT1–GFP in the population.
Results
Interferon signalling is not dependent on an intact cytoskeleton
The inhibitors cytochalasin D, which prevents actin polymerization, and nocodazole, which disrupts the formation of microtubules, were used to determine the influence of these structural elements on IFN signalling. 2fTGH fibrosarcoma cells were pre-incubated in the absence or presence of either drug for 90 min, then stimulated with IFN-α or -γ for 15 h under continued drug treatment. The efficacy of the drugs was confirmed by changes in cell morphology and the loss of cell adherence. Transcriptional activity upon IFN stimulation was measured by RNase protection assays using protection probes for the typical IFN-stimulated genes (ISGs) p48, 6-16, IRF1 and 9-27 (Figure 1A). The data were quantified by phosphorimaging corrected for actin levels (Figure 1B). The transcriptional response to IFN-α or -γ was not affected by either drug treatment in comparison with untreated controls. In additional experiments, neither the kinetics nor the dose responses were significantly affected (data not presented). Accepting this lack of dependence on the cytoskeleton, the mobility of STAT1 was further investigated in live cells using a known functional STAT1–GFP (Köster and Hauser, 1999).
Fig. 1. Interferon signalling is not dependent on an intact cytoskeleton. (A) 2fTGH cells were incubated for 90 min with cytochalasin D or nocodazole, as indicated, then treated with 103 IU/ml IFN-α or -γ for 15 h under continued drug treatment. Expression of inducible mRNAs (p48, 6-16, IRF-1, 9-27) was monitored by RNase protection (Materials and methods). (B) Quantitation of the data in (A) by PhosphorImager analysis. Fold induction was calculated after correction for the γ-actin loading control.
STAT1–GFP is a functional transcription factor
The construction and characterization of a functional STAT1–GFP, the behaviour of which is indistinguishable from native STAT1 with respect to its activation and translocation in and out of the nucleus, have already been described (Köster and Hauser, 1999). Here, the comparable restoration of IFN responses to STAT1-negative U3A cells by STAT1 and STAT1–GFP was confirmed by RNase protection assays monitoring the induction of representative sets of IFN-γ- and IFN-α-inducible genes (Köster and Hauser, 1999 and data not shown). For the remainder of the experiments, wild-type 2C4 cells were used in preference to the mutiply mutagenized U3A cells. The function of stably transfected STAT1–GFP in 2C4 cells again appeared indistinguishable from that of the, in this case, endogenous STAT1. The STAT1– GFP is comparably tyrosine phosphorylated/activated (Figure 2A), shows comparable DNA-binding activity (Figure 2B) and is efficiently translocated to the nucleus in response to IFN-γ stimulation (Figure 5; see Köster and Hauser, 1999). Comparison of endogenous STAT1 levels with those of the stably transfected STAT1–GFP by western blotting shows an average 3-fold overexpression of STAT1–GFP in the population (Figure 2C, top). The fluorescence of single cells (Figure 2C, bottom), together with the data from the western blot analyses, indicate that the majority of single cells express STAT1–GFP to levels comparable with or up to 10-fold higher than endogenous STAT1. No significant difference was observed between high- and low-expressing cells in the single-cell-based fluorescence studies below.
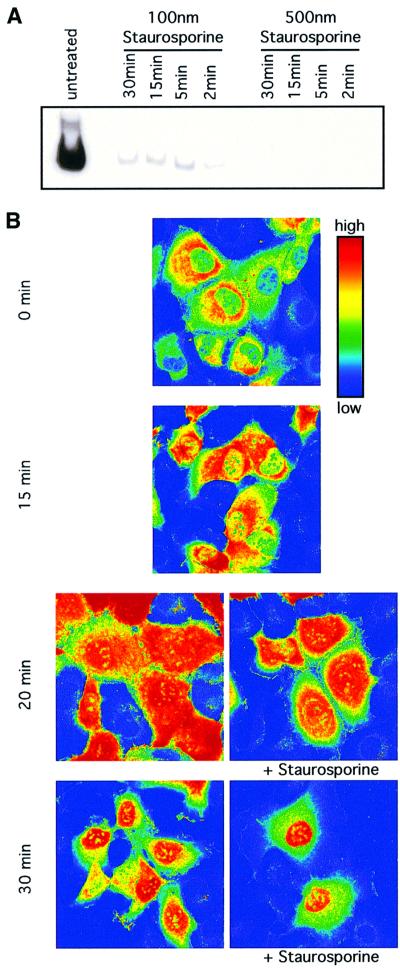
Fig. 5. Nuclear translocation of STAT1–GFP is not dependent on active JAK–receptor complexes. (A) Inhibition of JAK-dependent STAT1 activation by staurosporine. 2C4 cells were pre-treated with 100 or 500 nM staurosporine for 2, 5, 15 and 30 min, and stimulated with IFN-γ at 103 IU/ml for a further 20 min in the continued presence of the drug. Activation of STAT1 was monitored by EMSA of whole-cell extracts with an SIE probe in comparison with extracts from cells without drug treatment. [Similar results were obtained when the phosphorylation of STAT1 was monitored directly by (less sensitive) western blot analysis.] (B) Nuclear translocation of pre-activated STAT1. 2C4/STAT1–GFP cells were incubated with 103 IU/ml IFN-γ for 15 min to activate the STAT1–GFP. Staurosporine (500 nM) was added to half of the cells and both the staurosporine-treated and non-staurosporine-treated cells incubated for a further 5 and 15 min (to yield the 20 and 30 min time points, respectively). At 0, 15, 20 and 30 min, samples were fixed in paraformaldehyde and the distribution of STAT1–GFP analysed by confocal imaging. Fluorescence intensities are shown in false colour code (vertical bar, top right).
High mobility of cytoplasmic and nuclear STAT1–GFP: FLIP and FRAP analyses
Little is known about the mobility of STAT1 in the cytoplasm or the nucleus. In order to address these issues, fluorescence loss in photobleaching (FLIP) and fluorescence recovery after photobleaching (FRAP) analyses were carried out to compare the mobilities of stably expressed STAT1–GFP, GFP and protein kinase C (PKC)–GFP in the cytoplasm and nucleus of live 2C4 cells before and after treatment with IFN-γ or phorbol ester.
FLIP analysis of cytoplasmic STAT1–GFP. In FLIP analysis, a small region of the cytoplasm (white box, Figure 3; the intensity of the fluorescence signal is indicated by a ‘false colour’ bar to the right of the images) was bleached by scanning for three consecutive periods of 90 s with maximum laser intensity and the fluoresence of the whole cell monitored. To make sure there was no generalized bleaching effect due to the imaging, every bleached cell had an unbleached neighbouring cell in the same image, which maintained high fluorescence (Figure 3). The behaviour of non-activated and IFN-γ-activated STAT1 was compared with GFP, known to diffuse freely in cells, and free and membrane-bound (more slowly diffusing) PKC–GFP. Cells expressing GFP (Figure 3A, third row), STAT1–GFP, before and after treatment with IFN-γ (Figure 3A, rows 1 and 2) and free PKC–GFP without phorbol ester treatment (Figure 3A, row 4) lost most of their cytoplasmic fluorescence after the first 90 s of bleaching. Further bleaching to totals of 180 and 270 s resulted in the entire loss of cytoplasmic fluorescence. For STAT1–GFP and PKC–GFP, the nuclear envelope imposes a barrier to free diffusion. In these short time periods, no STAT1–GFP is released from the nucleus (Figure 3A, rows 1 and 2). [There is a low level of constitutively activated STAT1–GFP in the nucleus of the control cells (Figure 3A, row 1).] Conversely, PKC–GFP does not enter the nucleus of control cells (Figure 3A, row 4). In cells treated with phorbol ester, PKC–GFP is activated and translocated to the membrane. The reduced mobility of membrane-associated PKC–GFP, visualized by focusing on the lower membrane, was obvious: even after three periods of bleaching, only the boxed area and the close surroundings showed a loss of fluorescence in these cells (Figure 3A, row 5). Depletion of ATP (see below) had no influence on the results of this type of FLIP analysis. The data indicate that most of the STAT1–GFP molecules passed through the region being bleached within 270 s, moving rapidly throughout the cytoplasm in a random ATP-independent fashion. This is in contrast to the data for the more slowly diffusing, membrane-associated, activated PKC–GFP.
FRAP analysis of cytoplasmic STAT1–GFP. In order to have a more quantitative measurement of mobility to permit the calculation of the amount of any immobile fraction of STAT1–GFP, FRAP analyses were performed on the same stably transfected cell lines as were used for the FLIP analysis A region in the cytoplasm was bleached for a period of 17.5 s and the effect on the fluorescence intensity in this area was measured for a further 60 s. In parallel, regions surrounding the bleached region were measured for the same period of time. To investigate whether the movement of the fusion proteins was an energy-dependent process, a second set of cells was depleted of ATP by pre-treatment with sodium azide and 2-deoxyglucose prior to analyses (right-hand panels, Figure 4A). This treatment is sufficient to inhibit ATP-dependent fluid-phase endocytosis of fluorescent Cy3-labelled antibodies (Wubbolts et al., 1996 and data not shown). With or without depletion of ATP, the recovery rates in the bleached regions of the cytoplasm (empty squares, Figure 4A) for GFP (Figure 4A, third panels down), STAT1–GFP before and after IFN-γ treatment (Figure 4A, top and second panels), and PKC–GFP without phorbol ester treatment (Figure 4A, fourth panels down) were similar. In contrast, the recovery of PKC–GFP after phorbol ester treatment was slower and never complete. Again, this was not influenced by the depletion of ATP (Figure 4A, bottom panels).
Fig. 4. FRAP analysis. (A) Cytoplasmic and (B) nuclear fluoresence. (A) 2C4 cells stably transfected with STAT1–GFP without (row 1) and after 15 min of IFN-γ treatment at 103 IU/ml (row 2), GFP (row 3) and PKC–GFP without (row 4) and after 5 min of TPA treatment (Materials and methods, row 5). Cells were bleached (Materials and methods) for 17.5 s in an area comparable to the bleach area in the FLIP analysis (white squares, Figure 3). The recovery of fluorescence in the bleach area (squares) and four surrounding areas (triangles) of the same size (averaged) was quantified over a period of further 60 s in 1 s intervals. The fluorescence was normalized to the remaining fluorescence in the cell at the end of the experiment. The right-hand panels show data for cells incubated for 20 min with sodium azide and 2-deoxyglucose to deplete ATP, prior to bleaching. Each graph represents the average of data from 10 single cells. (B) FRAP analysis of the nuclear fluorescence of 2C4 cells stably transfected with STAT1–GFP and treated with 103 IU/ml IFNγ for 25 min (row 1) and GFPnls (row 2). Bleaching, recovery, ATP deprivation (right hand panels) and symbols are as in (A). The apparently more extensive initial bleach level for membrane-associated PKC–GFP (row 5, squares) compared with that in all other rows reflects the slower movement of the membrane-associated PKC–GFP than the freely diffusing GFP and other GFP constructs during the switch of the laser from the bleach to the record modes.
Comparison of the bleached region (empty squares) versus the surrounding regions (filled triangles) did not show, for GFP, STAT1–GFP before and after IFN-γ treatment, and PKC–GFP without phorbol ester, a significant difference such as would have resulted from the presence of an immobile fraction, as is the case for PKC–GFP after phorbol ester (contrast rows 1–4 with row 5, Figure 4A). As little as 1% of immobile GFP would have been detected in this type of analysis. In contrast to GFP per se (Figure 4A, row 3), very small differences directly after the bleaching were detectable in the cells expressing STAT1–GFP and PKC–GFP (Figure 4A, rows 1, 2 and 4). These most likely reflect differences in size and hence in mobility of the fusion proteins versus free GFP. The comparison of the bleached region to the surrounding regions for PKC–GFP after phorbol ester treatment showed an immobile fraction of ∼5 ± 2% (n = 10) for the membrane-associated PKC– GFP (Figure 4A, bottom row).
Nuclear translocation of pre-activated STAT1 does not depend on continued activity of JAK–receptor complexes
At any given instant in time, a small percentage (below the 1% that would be detected by FRAP) of the STAT1–GFP might be associated with a ‘hard-wired’ directional transport mechanism linking active JAK–receptor complexes to nuclear pores. In the presence of the kinase inhibitor staurosporine (Haspel and Darnell, 1999) and hence the absence of continued JAK–receptor activity, pre-activated STAT1–GFP is efficiently translocated into the nucleus. In initial electrophoretic mobility shift assay (EMSA) analyses in 2C4 cells, adding staurosporine before stimulation with IFN-γ showed that the drug is effective in inhibiting JAK activation of STAT1 in <2 min under the conditions to be used (Figure 5A). In subsequent experiments, 2C4/STAT1–GFP cells were stimulated with IFN-γ and, after 15 min, were incubated with or without staurosporine for a further 5–15 min (to yield the 20 and 30 min time points). The cells were fixed and imaged (Figure 5B). Up to 15 min, only a small amount of STAT1–GFP is translocated. Between 15 and 20 min, fluorescence intensities in the cytoplasm and nucleus are similar, and after 30 min most of the STAT1–GFP is translocated to the nucleus (Figure 5B). Translocation was comparable in the drug-treated and control cells. Accordingly, inhibition of the activation of STAT1 at the membrane JAK–receptor complex by staurosporine was without effect on the translocation of randomly distributed, pre-activated, cytoplasmic STAT1. It seems that a substantial portion of STAT1 molecules are activated within 15 min of ligand stimulation and remain randomly distributed in the cytoplasm until translocated through the nuclear pore. These data also confirm that at least 50% of the cytoplasmic STAT1–GFP molecules in the FRAP and FLIP experiments in IFN-γ-treated 2C4/STAT1–GFP cells were indeed pre-activated at the time of analysis.
FLIP and FRAP analyses of nuclear STAT1–GFP
The mobility of STAT1–GFP in the nucleus of 2C4 cells after IFN-γ treatment was studied in comparison to 2C4 cells expressing GFP tagged with a nuclear localization signal (GFPnls; Materials and methods) rather than wild-type GFP. In the FLIP experiments (Figure 3B), the boxed area was bleached for shorter periods of 30 s, in accordance with the smaller volume of the nucleus. There was no detectable difference between STAT1–GFP and GFPnls, with most of the fluorescence in the nucleus being bleached after 30 s. A further two consecutive 30 s bleach periods were required for a complete loss of fluorescence. No loss of fluorescence was observed in the cytoplasm of the 2C4/STAT1–GFP cells, reflecting the barrier presented to STAT1–GFP by the nuclear envelope (Figure 3B, top row). FRAP analysis of STAT1–GFP and GFPnls in the nucleus showed comparable recovery rates for the two molecules with or without depletion of ATP (Figure 4B). Furthermore, no immobile fraction was detectable in either case. Movement of STAT1–GFP in the nucleoplasm appears, therefore, to be rapid and random.
Dynamic interactions exclude STAT1 from nucleoli
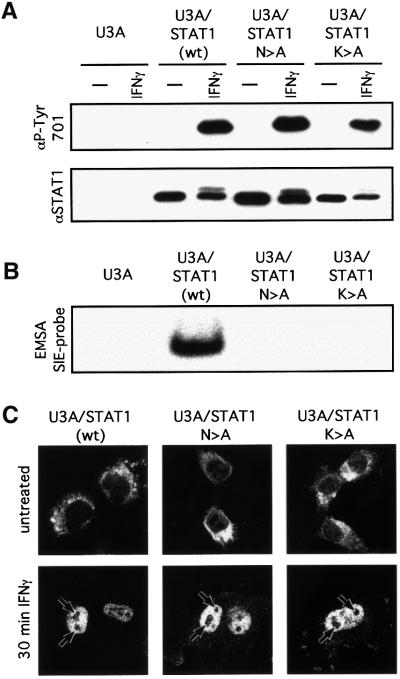
During the FLIP analyses, it was noted that in contrast to free GFP or GFPnls, STAT1–GFP was excluded from nucleoli (Figure 3). Rapid association and dissociation of STAT1 with the DNA could account for both the retention and the high mobility of nucleoplasmic STAT1. The behaviour of two DNA-binding mutants was therefore investigated. When expressed in STAT1-negative U3A cells, the mutant STAT1s are phosphorylated/activated, but do not bind DNA or support transcriptional activity in response to IFN-γ (Figure 6A and B and data not presented). The mutant STATs were translocated to the nucleus comparably to wild-type STAT1 (Figure 6C). Both the mutant and the wild-type STATs were, however, excluded from the nucleoli (Figure 6C). Nucleoplasmic GAS-element-specific DNA binding alone cannot, therefore, account for the nucleolar exclusion of STAT1.
Fig. 6. Exclusion of STAT1 and STAT1 DNA-binding mutants from nucleoli. STAT1-negative U3A cells stably transfected with STAT1, STAT1-N460A or STAT1-K336A were stimulated for 20 (A and B) or 30 (C) min with 103 IU/ml IFN-γ. (A) STAT phosphorylation/activation. Proteins were immunoprecipitated with anti-STAT1 antibody and western blots performed with anti-STAT1-P-Tyr701-specific antibody (upper panel) and anti-STAT1 antibody (lower panel). (B) STAT DNA binding. EMSAs of whole-cell extracts with an SIE probe. (C) Nuclear translocation. Cells were fixed and stained with antibody to STAT1 (Materials and methods). Arrows indicate the nucleoli.
Discussion
It remains unclear to what extent signal transduction pathways are ‘soft’ versus ‘hard’ wired involving random diffusion versus directional movement on a scaffold or cytoskeletal element. Here, we investigated how STAT1 moves from the JAK–receptor complex at the cell membrane to the nuclear pore, and from the nuclear pore to the DNA.
The induction of representative sets of IFN-α/β- and IFN-γ-inducible mRNAs was not affected by disruption of the actin cytoskeleton or microtubules (Figure 1). Accordingly, for the cells used here, JAK/STAT signalling and hence the movement of STAT1 from the cell membrane to the nuclear pore in response to the IFNs is independent of the cytoskeleton. In contrast, translocation of p53 is dependent on interaction with microtubules and dynein, a minus-ended microtubule motor, which would move p53 along microtubules towards the nucleus in response to stress (Giannakakou et al., 2000).
In light of the absence of dependence on the cytoskeleton, STAT1–GFP was used to monitor the movement of STAT1 in live cells. STAT1–GFP behaves indistinguishably from endogenous STAT1: it is activated, translocates and initiates gene expression with comparable efficiency to STAT1 (Köster and Hauser, 1999; Figure 2). It is not detectably cleaved to yield free GFP (Figure 2). There was considerable cell-to-cell variation in the levels of expression of the stably transfected STAT1–GFP (Figure 2). Throughout the photobleaching experiments, similar results were obtained with high (>5-fold wild type) and low (≈ wild type) expressing cells. The results do not, therefore, reflect overexpression artefacts.
The FLIP and FRAP experiments provide insight into the nature of STAT1 movement. STAT1–GFP mobility, with or without ligand stimulation, is comparable to that of freely diffusible GFP (Figures 3 and 4). It is rapid, with all of the STAT1 molecules moving through all locations in the cytoplasm or nucleus within minutes, and independent of ATP. An immobile fraction of STAT1–GFP was not detected and can be excluded down to a level of ∼1% of the total STAT1–GFP (Figure 4). A priori, at any given instant in time, a small percentage of the STAT1–GFP might be associated with a ‘hard-wired’ directional transport mechanism linking the JAK–receptor complex to the nuclear pore. However, in the presence of staurosporine and hence the absence of an enzymatically active JAK–receptor complex, a substantial fraction (∼50%) of total STAT1–GFP is translocated into the nucleus (Figure 5). Thus, pre-activated, randomly distributed STAT1–GFP is translocated to the nucleus as efficiently as in non-staurosporine-treated cells (Figure 5). One still cannot exclude the possibility that, at any given time, a small percentage of STAT1–GFP (too low to be detected by FLIP or FRAP) is transported directionally to the nuclear pore. Access to any such putative transport system would, however, have to be available to randomly distributed pre-activated STAT1–GFP. In this model, the putative translocation system would conceptually be an extension of the nuclear pore–importin complex, with random access through free diffusion after release from the receptor. In an alternative approach, STAT1 generated from a completely foreign receptor can sustain an IFN-γ-like response. This favours modular signalling and ‘soft’ rather than ‘hard’ wiring of the IFN-γ-induced cytoplasmic signal transduction pathway(s) (V.Arulampalam, B.Strobl, H.Is’harc and I.M.Kerr, in preparation). Overall, the data are consistent with free diffusion of the STAT1 from the membrane JAK–receptor complex to the nuclear pore. Activated STAT1 forms a dimer, dimerization being required for recognition by importin NPI-1 and translocation through the nuclear pore (Sekimoto et al., 1997). Larger STAT1 complexes have been reported (Lackmann et al., 1998; Ndubuisi et al., 1999), and highly dynamic interactions with randomly distributed cytoplasmic complexes remain perfectly possible. Such interactions would not, however, confer directionality to the movement of STAT1.
The behaviour of STAT1–GFP in the nucleus is also of interest. Nucleoplasmic movement is again rapid, random, independent of ATP and comparable to that of freely diffusible GFP (Figures 3B and 4B). STAT1–GFP, in contrast to free GFP, is, however, excluded from nucleoli (Figures 3 and 6), presumably through interaction with nucleoplasmic protein–DNA complexes. Such interactions would have to be highly dynamic, with high rates of association and dissociation, to sustain both the high mobility and localization of the STAT1–GFP. Interactions of this type with similar effects on localization have recently been described for a number of nuclear proteins (Phair and Misteli, 2000). More particularly, for the glucocorticoid receptor, association with transcriptional complexes is similarly highly dynamic, in accord with a ‘hit and run’ model rather than the formation of stable initiation complexes (McNally et al., 2000). As the loss of specific GAS site DNA-binding function does not influence nucleoplasmic localization, direct interaction with GAS elements in the DNA is not solely responsible for exclusion from the nucleoli (Figure 6). The retention of STAT1–GFP in the nucleoplasm may, therefore, reflect a ‘scanning’ process for specific sites in addition to the formation of a small percentage of dynamic or relatively more stable specific transcrptional initiation complexes. (Accepting the limit of sensitivity of the FRAP analysis, relatively stable complexes presenting <1% of the nucleoplasmic STAT1–GFP clearly cannot be excluded.)
Although peripheral to the main argument, the contrast in the results obtained here with the K336A and N460A STAT1 DNA-binding mutants with those of others for the EE428/429AS DNA-binding mutant is also of potential interest. Nuclear localization of the latter mutant in response to IFN-γ was only observable in the presence of leptomycin B, an inhibitor of CRM1 interaction and nuclear export (McBride et al., 2000). Both K336A and N460A lie outside the STAT1 sequence, AA392–413, known to be required for CRM1 binding (McBride et al., 2000). Taken together, these data, like those for the retention of the STAT1 mutants in the nucleoplasm, suggest that factors besides GAS-element-specific DNA binding and interaction with CRM1 may contribute to retention of STAT1 in the nucleus. A much more detailed analysis of the characteristics of the mutants, with respect, for example, to the kinetics of dephosphorylation and interaction with transcriptional co-activators, will be required to establish whether or not this is the case and, if so, reveal the nature of the factors involved.
Currently, there is increasing evidence for both ‘hard’ and ‘soft’ wiring of cytoplasmic signal transduction pathways (Teruel and Meyer, 2000; Vousden and Woude, 2000). Moreover, there is no a priori reason why both types of element should not be involved in a given pathway. Indeed, arguably, the nuclear pore represents a ‘hard-wired’ section to what appears to be an otherwise ‘soft-wired’ JAK/STAT1 pathway. The data presented here, taken together with available data on nuclear import and export, lead to a model in which for STAT1 there are a series of random walks interrupted by dynamic interactions with membrane JAK–receptor complexes, nuclear pores and nucleoplasmic protein–DNA complexes. More specifically, randomly diffusing cytoplasmic STAT1 is activated through dynamic interaction with activated JAK–receptor complexes at the cell membrane. The activated STAT1 dimerizes and, with or without interaction with additional proteins, diffuses to the nuclear pore where interaction with importin NPI-1 initiates translocation into the nucleus. On release from the nuclear pore, the STAT1 diffuses freely through the nucleoplasm, to which it is localized by dynamic interaction with protein/DNA. These interactions probably involve both the scanning of chromatin for potential transcriptional initiation sites and the formation of specific initiation complexes. Dephosphorylation of bound or unbound STAT1 leaves non-phosphorylated unbound STAT1 to diffuse freely to nuclear pores, where interaction with CRM1 mediates export to the cytoplasm, rendering inactive STAT1 available for further rounds of activation. Superimposed upon this basic cycle will be additional controls. Although at first sight arguably unappealing, a model involving random walks interspersed with dynamic interactions has the advantage of potential flexibility, rapid modulation and fine tuning, which might be necessary for the maintenance of an appropriate balance in response to multiple stimuli.
Materials and methods
Cell culture
Human fibrosarcoma cell lines 2C4, 2fTGH and U3A (McKendry et al., 1991; Watling et al., 1993) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% heat-inactivated fetal calf serum and 5 µM l-glutamine. Hygromycin-resistant cells were maintained in medium containing 250 µg/ml hygromycin and G418-resistant cells in 400 µg/ml G418. IFN-α was a highly purified mixture of human subspecies [Wellferon 1.5 × 108 IU/mg protein (Allen et al., 1982), provided by Wellcome Research Laboratories, Beckenham, Kent, UK]. Recombinant IFN-γ (4 × 107 IU/mg protein) was generously provided by Dr G.Adolf, Ernst Boehringer Institute für Arzneimittelforschung, Vienna, Austria. Staurosporine, nocodazole, cytochalasin D and 12-O-tetradecanoylphorbol-13-acetate (TPA) (all Sigma) were dissolved in DMSO and used at final concentrations of 500 nM, 5 µg/ml, 4 µg/ml and 400 nM, respectively [the final DMSO concentration (<0.1%) was without effect on parallel controls]. Where indicated, cells were cultured in sodium azide (0.05%) and 2-deoxyglucose (50 µM) for 20 min prior to experiments to deplete ATP.
Plasmids and DNA transfections
STAT1–GFP/pMBC has been described previously by Köster and Hauser (1999). PKCα–GFP/pcDNA3 was from P.J.Parker [Imperial Cancer Research Fund (ICRF), London, UK; Ng et al., 1999] and STAT1-N460A/pRcCMV and STAT1-K336A/pRcCMV were from J.E.Darnell, Jr (The Rockefeller University, New York, NY). GFP and GFPnls were purchased from Clontech. Stable cell lines were generated by transfection of the appropriate construct with SuperFect™ (Qiagen) and drug selection and/or fluorescence-activated cell sorting (FACS).
Antibodies
A monoclonal antibody against STAT1 (Novacastra) was used for immunoprecipitation (8 µl/1 ml lysate), immunohistochemestry (1:100 dilution) and western blotting (1:2000 dilution). Phosphorylation of STAT1 was detected with an antibody against the phosphorylated form of tyrosine 701 (New England Biolabs). The monoclonal antibody against GFP was produced at the ICRF, UK.
RNase protection assay
Total cellular RNA was prepared from monolayer cells by NP-40 lysis and phenol–chloroform extraction (Porter et al., 1988). RNase protection assays (Melton et al., 1984) were performed using probes (Muller et al., 1993b) synthesized from SP6/T7 transcription vectors. Probes were labelled with [32P]UTP to a specific activity of 2–5 × 108 c.p.m. per microgram of input DNA. Aliquots equivalent to ∼1–3 × 105 c.p.m. of each probe and 10 µg of cytoplasmic RNA were used per assay. The intensities of radioactive bands in dried gels were quantified using a PhosphorImager (Molecular Dynamics). The bands of interest were quantitated, corrected for background and normalized to the control band (γ-actin). The data (Figure 1B) are expressed as fold induction compared with the uninduced samples.
EMSA
Whole-cell extracts were prepared on ice by lysis of cells in 0.5% NP-40, 10% glycerol, 50 mM Tris–HCl pH 8.0, 150 mM NaCl, 0.1 mM EDTA, 1 mM sodium orthovanadate, 0.5 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM dithiothreitol (DTT), 3 µg/ml aprotinin and 1 µg/ml leupeptin (Schindler et al., 1992b), and the sequence of the oligonucleotide probe used corresponded to the high affinity SIE (hSIE) of the c-fos gene (SIEM67) (Sadowski et al., 1993). Probes were end labelled with [γ-32P]ATP and aliquots equivalent to ∼30 000 c.p.m. used per reaction. Binding reactions were performed in a total volume of 20 µl, in 10 mM HEPES pH 7.9, 1.5 mM MgCl2, 0.1 mM EGTA, 5% glycerol, 1 mg/ml bovine serum albumin (BSA), 0.125 mg/ml pd(N5), 0.25 mg/ml tRNA, 2% Ficoll. Extracts were pre-incubated for 10 min at room temperature with 150 µg/ml poly(dI–dC⋅dI–dC) prior to incubation with probe for an additional 20 min at room temperature. Complexes were separated on 6% non-denaturing acrylamide gels and detected by autoradiography.
Immunoprecipitation, SDS–PAGE and western blotting
Cells were lysed as above and nuclei removed by centrifugation. Lysates were either used directly for SDS–PAGE (Laemmli, 1970) or for immunoprecipitation. For the latter, the approriate antibody and protein A–Sepharose were added to the lystes and incubated for 18 h at 4°C. The ‘precipitates’ were washed in ice-cold lysis buffer. Proteins eluted in loading buffer were separated on 6.5 or 12% SDS–PAGE gels and transferred electrophoretically to Immobilon™ PVDF (Millipore) membranes. The membranes were blocked by incubation with 5% BSA (fraction V, Sigma) in TBST (10 mM Tris–HCl pH 7.4, 75 mM NaCl, 1 mM EDTA, 0.1% Tween-20) with 1 mM sodium orthovanadate for 18 h at 4°C, incubated with the relevant primary antibody in TBST for 30 min, washed in TBST and incubated for 30 min with peroxidase-conjugated secondary antibody (Amersham Life Science), washed in TBST and exposed to enhanced chemiluminescence (Amersham Life Science) followed by fluorography. Membranes were stripped in 2 M glycine pH 2.5, 1% SDS, 1 mM DTT and 0.05% azide for 18 h at room temperature, washed with TBST, and re-analysed as above, with alternative antibody where indicated.
Confocal analysis and bleaching
Cells were plated on glass coverslips and fixed with 4% paraformaldehyde in phosphate-buffered saline pH 7.4 (PBS) for 20 min at room temperature. Cells were permeabilized with 0.2% Triton X-100 in PBS for 5 min, washed in PBS and blocked with 0.2% fish skin gelatine in PBS for 1 h. The slides were incubated for 20 min with the primary antibody, washed three times with PBS, incubated with Cy3-labelled secondary antibody (P.Bastiaens, ICRF, London, UK), washed three times with PBS and mounted on glass slides with Mowiol (Calbiochem). Living cells were analysed in glass-bottom tissue culture dishes (MatTek Corporation) at 37°C in DMEM with low bicarbonate (2.2 g NaHCO3/l), no fluorescent agents and 25 mM HEPES pH 7.4. An Axiovert 100 M confocal microscope equipped with argon and helium–neon lasers (Zeiss, Germany) was used for analysis. Green fluorescence was detected at λ >505 nm after excitation at λ = 488 nm. Cy3-labelled antibodies were detected at λ >560 nm after excitation at λ = 543 nm. Endosomes were labelled by internalization of Cy3-labelled mouse IgG (50 µg/ml) in living cells. To bleach GFP-tagged proteins in living cells, a small region of the cytoplasm or the nucleus was scanned with maximum laser power for the times indicated.
Fluorescence recovery after photobleaching
A boxed region in the cytoplasm or the nucleus was bleached and the effect on the fluorescence in this box versus four adjoined boxed regions was quantitated. The later regions were averaged to compensate for local variations. These data were plotted in a graph to compare recovery of fluorescence in the different regions (through diffusion of GFP or GFP fusion proteins into the boxed areas). Any immobile fraction of the GFP-tagged proteins can be determined using the formula: % immobile = [(FBafter/FBbefore) – (FAafter/FAbefore)] × 100. FA is the fluorescence in the bleach box and FB the average fluorescence in the boxes outside this region. The fluorescence is determined before and after bleaching, and whereas both FA and FB will decrease due to equal bleaching of the mobile fraction, any immobile fraction will be bleached only for FA (Edidin et al., 1976; Cole et al., 1996). For all data, the average of 10 measurements is presented.
Ratio of STAT1–GFP to STAT1 in 2C4/STAT1–GFP cells
To quantify the total fluorescence in single cells expressing GFP-tagged STAT1 (Figure 2C), 54 cells from the FACS sorted population were sectioned in Z-stacks of 0.5 µm height. The fluorescence intensity of each stack was corrected for the background fluorescence and the area of the stack. The total fluorescence of a cell was the sum of the fluorescence of all stacks for the cell. The average fluorescence of the 54 measured cells was set equal to the ratio of STAT1–GFP to STAT1, which was determined by western blotting (Figure 2C, top). From these data, the ratio of STAT1–GFP to STAT1 in each of the 54 cells was calculated (Figure 2C, bottom).
Acknowledgments
Acknowledgements
We thank Peter Jordan and Daniel Zicha, Light Microscopy Laboratory, ICRF, for invaluable advice and technical assistance, Peter Bastiaens for the anti-Cy3 antibody, Gunter Adolf for the recombinant IFN-γ, and Hansjoerg Hauser, Peter Parker and James Darnell, Jr for the STAT1–GFP, PKC–GFP and STAT1 DNA-binding mutant constructs, respectively. B.F.L. was supported in part by a fellowship from Boehringer Ingelheim Fonds.
References
- Akira S. et al. (1994) Molecular cloning of APRF, a novel IFN-stimulated gene factor 3 p91-related transcription factor involved in the gp130-mediated signaling pathway. Cell, 77, 63–71. [DOI] [PubMed] [Google Scholar]
- Allen G., Fantes,K.H., Burke,D.C. and Morser,J. (1982) Analysis and purification of human lymphoblastoid (Namalwa) interferon using a monoclonal antibody. J. Gen. Virol., 63, 207–212. [DOI] [PubMed] [Google Scholar]
- Cole N.B., Smith,C.L., Sciaky,N., Terasaki,M., Edidin,M. and Lippincott-Schwartz,J. (1996) Diffusional mobility of Golgi proteins in membranes of living cells. Science, 273, 797–801. [DOI] [PubMed] [Google Scholar]
- Darnell J.E.,Jr, Kerr,I.M. and Stark,G.R. (1994) Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science, 264, 1415–1421. [DOI] [PubMed] [Google Scholar]
- Duhe R.J. and Farrar,W.L. (1998) Structural and mechanistic aspects of Janus kinases: how the two-faced god wields a double-edged sword. J. Interferon Cytokine Res., 18, 1–15. [DOI] [PubMed] [Google Scholar]
- Edidin M., Zagyansky,Y. and Lardner,T.J. (1976) Measurement of membrane protein lateral diffusion in single cells. Science, 191, 466–468. [DOI] [PubMed] [Google Scholar]
- Firmbach-Kraft I., Byers,M., Shows,T., Dalla-Favera,R. and Krolewski,J.J. (1990) tyk2, prototype of a novel class of non-receptor tyrosine kinase genes. Oncogene, 5, 1329–1336. [PubMed] [Google Scholar]
- Fu X.Y. (1992) A transcription factor with SH2 and SH3 domains is directly activated by an interferon α-induced cytoplasmic protein tyrosine kinase(s). Cell, 70, 323–335. [DOI] [PubMed] [Google Scholar]
- Fu X.Y., Schindler,C., Improta,T., Aebersold,R. and Darnell,J.E.,Jr (1992) The proteins of ISGF-3, the interferon α-induced transcriptional activator, define a gene family involved in signal transduction. Proc. Natl Acad. Sci. USA, 89, 7840–7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannakakou P., Sackett,D.L., Ward,Y., Webster,K.R., Blagosklonny,M.V. and Fojo,T. (2000) p53 is associated with cellular microtubules and is transported to the nucleus by dynein. Nature Cell Biol., 2, 709–717. [DOI] [PubMed] [Google Scholar]
- Harpur A.G., Andres,A.C., Ziemiecki,A., Aston,R.R. and Wilks,A.F. (1992) JAK2, a third member of the JAK family of protein tyrosine kinases. Oncogene, 7, 1347–1353. [PubMed] [Google Scholar]
- Haspel R.L. and Darnell,J.E.,Jr (1999) A nuclear protein tyrosine phosphatase is required for the inactivation of Stat1. Proc. Natl Acad. Sci. USA, 96, 10188–10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J., Schindler,U., Henzel,W.J., Ho,T.C., Brasseur,M. and McKnight,S.L. (1994) An interleukin-4-induced transcription factor: IL-4 Stat. Science, 265, 1701–1706. [DOI] [PubMed] [Google Scholar]
- Ihle J.N., Witthuhn,B.A., Quelle,F.W., Yamamoto,K. and Silvennoinen,O. (1995) Signaling through the hematopoietic cytokine receptors. Annu. Rev. Immunol., 13, 369–398. [DOI] [PubMed] [Google Scholar]
- Johnston J.A., Kawamura,M., Kirken,R.A., Chen,Y.Q., Blake,T.B., Shibuya,K., Ortaldo,J.R., McVicar,D.W. and O’Shea,J.J. (1994) Phosphorylation and activation of the Jak-3 Janus kinase in response to interleukin-2. Nature, 370, 151–153. [DOI] [PubMed] [Google Scholar]
- Kaffman A. and O’Shea,E.K. (1999) Regulation of nuclear localization: a key to a door. Annu. Rev. Cell Dev. Biol., 15, 291–339. [DOI] [PubMed] [Google Scholar]
- Köster M. and Hauser,H. (1999) Dynamic redistribution of STAT1 protein in IFN signaling visualized by GFP fusion proteins. Eur. J. Biochem., 260, 137–144. [DOI] [PubMed] [Google Scholar]
- Lackmann M. et al. (1998) Biomolecular interaction analysis of IFN γ-induced signaling events in whole-cell lysates: prevalence of latent STAT1 in high-molecular weight complexes. Growth Factors, 16, 39–51. [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Leonard W.J. and O’Shea,J.J. (1998) Jaks and STATs: biological implications. Annu. Rev. Immunol., 16, 293–322. [DOI] [PubMed] [Google Scholar]
- McBride K.M., McDonald,C. and Reich,N.C. (2000) Nuclear export signal located within theDNA-binding domain of the STAT1 transcription factor. EMBO J., 19, 6196–6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKendry R., John,J., Flavell,D., Muller,M., Kerr,I.M. and Stark,G.R. (1991) High-frequency mutagenesis of human cells and characterization of a mutant unresponsive to both α and γ interferons. Proc. Natl Acad. Sci. USA, 88, 11455–11459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally J.G., Muller,W.G., Walker,D., Wolford,R. and Hager,G.L. (2000) The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science, 287, 1262–1265. [DOI] [PubMed] [Google Scholar]
- Melton D.A., Krieg,P.A., Rebagliati,M.R., Maniatis,T., Zinn,K. and Green,M.R. (1984) Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res., 12, 7035–7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowen K. and David,M. (2000) Regulation of STAT1 nuclear export by Jak1. Mol. Cell. Biol., 20, 7273–7281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M. et al. (1993a) The protein tyrosine kinase JAK1 complements defects in interferon-α/β and -γ signal transduction. Nature, 366, 129–135. [DOI] [PubMed] [Google Scholar]
- Muller M., Laxton,C., Briscoe,J., Schindler,C., Improta,T., Darnell,J.E.,Jr, Stark,G.R. and Kerr,I.M. (1993b) Complementation of a mutant cell line: central role of the 91 kDa polypeptide of ISGF3 in the interferon-α and -γ signal transduction pathways. EMBO J., 12, 4221–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndubuisi M.I., Guo,G.G., Fried,V.A., Etlinger,J.D. and Sehgal,P.B. (1999) Cellular physiology of STAT3: Where’s the cytoplasmic monomer? J. Biol. Chem., 274, 25499–25509. [DOI] [PubMed] [Google Scholar]
- Ng T. et al. (1999) Imaging protein kinase Cα activation in cells. Science, 283, 2085–2089. [DOI] [PubMed] [Google Scholar]
- Phair R.D. and Misteli,T. (2000) High mobility of proteins in the mammalian cell nucleus. Nature, 404, 604–609. [DOI] [PubMed] [Google Scholar]
- Porter A.C., Chernajovsky,Y., Dale,T.C., Gilbert,C.S., Stark,G.R. and Kerr,I.M. (1988) Interferon response element of the human gene 6-16. EMBO J., 7, 85–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quelle F.W. et al. (1995) Cloning of murine Stat6 and human Stat6, Stat proteins that are tyrosine phosphorylated in responses to IL-4 and IL-3 but are not required for mitogenesis. Mol. Cell. Biol., 15, 3336–3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski H.B., Shuai,K., Darnell,J.E.,Jr and Gilman,M.Z. (1993) A common nuclear signal transduction pathway activated by growth factor and cytokine receptors. Science, 261, 1739–1744. [DOI] [PubMed] [Google Scholar]
- Schindler C. and Darnell,J.E.,Jr (1995) Transcriptional responses to polypeptide ligands: the JAK–STAT pathway. Annu. Rev. Biochem., 64, 621–651. [DOI] [PubMed] [Google Scholar]
- Schindler C., Fu,X.Y., Improta,T., Aebersold,R. and Darnell,J.E.,Jr (1992a) Proteins of transcription factor ISGF-3: one gene encodes the 91-and 84-kDa ISGF-3 proteins that are activated by interferon α. Proc. Natl Acad. Sci. USA, 89, 7836–7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler C., Shuai,K., Prezioso,V.R. and Darnell,J.E.,Jr (1992b) Interferon-dependent tyrosine phosphorylation of a latent cytoplasmic transcription factor. Science, 257, 809–813. [DOI] [PubMed] [Google Scholar]
- Sekimoto T., Imamoto,N., Nakajima,K., Hirano,T. and Yoneda,Y. (1997) Extracellular signal-dependent nuclear import of Stat1 is mediated by nuclear pore-targeting complex formation with NPI-1, but not Rch1. EMBO J., 16, 7067–7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvennoinen O., Witthuhn,B.A., Quelle,F.W., Cleveland,J.L., Yi,T. and Ihle,J.N. (1993) Structure of the murine Jak2 protein-tyrosine kinase and its role in interleukin 3 signal transduction. Proc. Natl Acad. Sci. USA, 90, 8429–8433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark G.R., Kerr,I.M., Williams,B.R., Silverman,R.H. and Schreiber,R.D. (1998) How cells respond to interferons. Annu. Rev. Biochem., 67, 227–264. [DOI] [PubMed] [Google Scholar]
- Teruel M.N. and Meyer,T. (2000) Translocation and reversible localization of signaling proteins: a dynamic future for signal transduction. Cell, 103, 181–184. [DOI] [PubMed] [Google Scholar]
- Vousden K.H. and Woude,G.F. (2000) The ins and outs of p53. Nature Cell Biol., 2, E178–180. [DOI] [PubMed] [Google Scholar]
- Wakao H., Gouilleux,F. and Groner,B. (1994) Mammary gland factor (MGF) is a novel member of the cytokine regulated transcription factor gene family and confers the prolactin response [published erratum appears in EMBO J. (1995), 14, 854–855]. EMBO J., 13, 2182–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watling D. et al. (1993) Complementation by the protein tyrosine kinase JAK2 of a mutant cell line defective in the interferon-γ signal transduction pathway. Nature, 366, 166–170. [DOI] [PubMed] [Google Scholar]
- Wilks A.F. (1989) Two putative protein-tyrosine kinases identified by application of the polymerase chain reaction. Proc. Natl Acad. Sci. USA, 86, 1603–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witthuhn B.A., Silvennoinen,O., Miura,O., Lai,K.S., Cwik,C., Liu,E.T. and Ihle,J.N. (1994) Involvement of the Jak-3 Janus kinase in signalling by interleukins 2 and 4 in lymphoid and myeloid cells. Nature, 370, 153–157. [DOI] [PubMed] [Google Scholar]
- Wubbolts R., Fernandez-Borja,M., Oomen,L., Verwoerd,D., Janssen,H., Calafat,J., Tulp,A., Dusseljee,S. and Neefjes,J. (1996) Direct vesicular transport of MHC class II molecules from lysosomal structures to the cell surface. J. Cell Biol., 135, 611–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K., Quelle,F.W., Thierfelder,W.E., Kreider,B.L., Gilbert,D.J., Jenkins,N.A., Copeland,N.G., Silvennoinen,O. and Ihle,J.N. (1994) Stat4, a novel γ interferon activation site-binding protein expressed in early myeloid differentiation. Mol. Cell. Biol., 14, 4342–4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh T.C. and Pellegrini,S. (1999) The Janus kinase family of protein tyrosine kinases and their role in signaling. Cell Mol. Life Sci., 55, 1523–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z., Wen,Z. and Darnell,J.E.,Jr (1994) Stat3 and Stat4: members of the family of signal transducers and activators of transcription. Proc. Natl Acad. Sci. USA, 91, 4806–4810. [DOI] [PMC free article] [PubMed] [Google Scholar]