Abstract
The major route for protein export or membrane integration in bacteria occurs via the Sec-dependent transport apparatus. The core complex in the inner membrane, consisting of SecYEG, forms a protein-conducting channel, while the ATPase SecA drives translocation of substrate across the membrane. The SecYEG complex from Escherichia coli was overexpressed, purified and crystallized in two dimensions. A 9 Å projection structure was calculated using electron cryo-microscopy. The structure exhibits P121 symmetry, having two asymmetric units inverted with respect to one another in the unit cell. The map shows elements of secondary structure that appear to be transmembrane helices. The crystallized form of SecYEG is too small to comprise the translocation channel and does not contain a large pore seen in other studies. In detergent solution, the SecYEG complex displays an equilibrium between monomeric and tetrameric forms. Our results therefore indicate that, unlike other known channels, the SecYEG complex can exist as both an assembled channel and an unassembled smaller unit, suggesting that transitions between the two states occur during a functional cycle.
Keywords: electron crystallography/oligomerization/SecYEG/translocation/two-dimensional crystallization
Introduction
Most proteins are exported from bacterial cells by a ubiquitous and essential Sec-dependent protein transport pathway, the key components of which have been identified by genetic and biochemical studies in Escherichia coli (Oliver and Beckwith, 1982; Kumamoto and Beckwith, 1983; Schatz et al., 1989; for a review see Driessen et al., 1998). A core complex of the proteins SecY, SecE and SecG (SecYEG complex) is responsible for the transport of proteins to the inner membrane, the outer membrane and the periplasmic space, or for export into the medium. Homologues of SecY and SecE are found also in eukaryotes (Jungnickel et al., 1994; Rapoport et al., 1996), archaea (Pohlschroder et al., 1997) and even in chloroplast thylakoid membranes (Mant et al., 1995).
SecY and SecE are essential components of the bacterial translocase (Brundage et al., 1990; Akimaru et al., 1991). Escherichia coli and some other bacteria possess a non-essential third subunit, SecG (Nishiyama et al., 1994; Duong and Wickner, 1997a). In addition, in most bacteria, there exists a further complex of SecDFyajC (Duong and Wickner, 1997b). SecY, SecE and SecG are transmembrane proteins and their sequence hydropathy predicts that they contain 10, three and two membrane-spanning segments, respectively. Together they form the channel through which proteins are translocated (Meyer et al., 1999), presumably in the same way that the eukaryotic equivalent, the Sec61p complex, forms a protein conduit in the endoplasmic reticulum (ER) (Hanein et al., 1996; Beckmann et al., 1997; Ménetret et al., 2000). While SecYEG provides the route for protein translocation and membrane integration, the driving force is provided by SecA, a peripherally associated membrane protein, which associates with the SecYEG complex during translocation (Cabelli et al., 1988; Lill et al., 1989; Akimaru et al., 1991; Douville et al., 1995). SecA may promote translocation by transferring successive segments of a polypeptide chain through the SecYEG channel (Economou and Wickner, 1994; Economou et al., 1995; Duong et al., 1997).
At present, our structural knowledge of any transmembrane translocation apparatus is limited to low-resolution structures from single particle analysis by electron microscopy (Hanein et al., 1996; Beckmann et al., 1997; Kunkele et al., 1998; Shilton et al., 1998; Meyer et al., 1999; Manting et al., 2000; Ménetret et al., 2000). The SecYEG or SecYE complex forms circular toroid assemblies (Meyer et al., 1999; Manting et al., 2000) with the active structure possibly being a tetramer (Manting et al., 2000), although biochemical data suggest the monomer to be the active species (Yahr and Wickner, 2000). The eukaryotic homologue of the SecYEG complex, the Sec61p complex, also forms rings, probably consisting of three or four copies of the complex (Hanein et al., 1996). When bound to the ribosome, the central pore in the ring is aligned with the exit channel of the ribosome where the nascent chain emerges (Beckmann et al., 1997; Ménetret et al., 2000), suggesting that the nascent polypeptide chain traverses the ER membrane through the channel pore. Freeze-fracture electron microscopy indicates that the Sec61p channel may be assembled upon engagement with the ribosome in co-translational translocation or upon association with another membrane protein complex in post-translational translocation (Hanein et al., 1996).
The detailed architecture of the channel, the nature of the association with SecA during the translocation event and the exact conformation of substrate during transport are fundamental issues that need to be addressed by crystallographic studies. We are concentrating on the core protein-translocating complex consisting of SecY, SecE and SecG and use two-dimensional crystallization and electron cryo-microscopy for structural analysis. These methods have yielded atomic models of three membrane proteins: bacteriorhodopsin (Henderson et al., 1990; Grigorieff et al., 1996), plant light-harvesting complex (Kühlbrandt et al., 1994) and aquaporin (Murata et al., 2000). The structures of several other membrane proteins, including rhodopsin (Unger et al., 1997b), plant photosystem II reaction centre (Rhee et al., 1998), plasma membrane H+-ATPase (Auer et al., 1998), gap junction (Unger et al., 1999) and NhaA (Williams, 2000), have been determined at intermediate (6–9 Å) resolution by this technique.
Here we report on the first intermediate resolution projection structure of the SecYEG complex from E.coli. Large quantities of SecYEG were produced by homologous overexpression, yielding pure protein capable of stimulating the ATPase activity of SecA and protein translocation into vesicles. The oligomeric state of the SecYEG complex was investigated in detergent solution by analytical ultracentrifugation. Two-dimensional crystals were grown, of which images were recorded by electron cryo-microscopy. Image processing reveals a 9 Å resolution view of a protein translocation complex in projection. Our data indicate that the SecYEG complex can exist as a stable, small unit that may assemble into channels upon oligomerization.
Results
Purification of the SecYEG complex
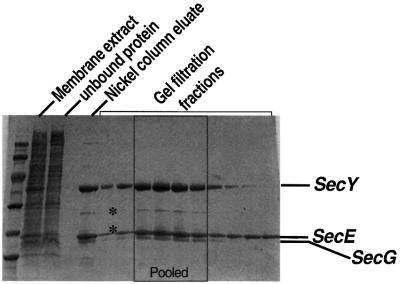
The operon coding for SecE, Y and G was modified to extend the reading frame of SecE by a His6 tag on the N-terminus. The expression was under the control of the arabinose promoter, which successfully brought about the overproduction of SecYEG. The complex was assembled appropriately into the inner membrane (W.Haase and I.Collinson, unpublished results). The overexpression of SecYEG in the membrane correlates with increased translocation activity found in isolated inner membranes (F.Duong, unpublished results). Total membranes were extracted in n-octyl-β-d-glucoside (OG), and solubilized SecYEG was bound to a nickel column, where the detergent was exchanged to nonapolyoxyethylene dodecyl ether (C12E9) by extensive washing in a new solution. Purification was completed by size exclusion and anion exchange chromatography (Figure 1), and the final protein preparation was typically 0.4 mg/ml. The preparation was pure except for two minor contaminants generated by a cleavage in SecY (Figure 1, indicated by a star). In the same way, we also purified a mutant complex, SecYprlA4EG, in which SecY carries a mutation that allows substrates with a defective signal sequence to be transported across the membrane (Bieker et al., 1990; Prinz et al., 1996; van der Wolk et al., 1998).
Fig. 1. Purification of SecYEG. SDS–PAGE of fractions during the purification of SecYEG. The detergent extract from membranes is shown on the left adjacent to the molecular weight markers (94, 67, 42, 30, 20 and 12 kDa). The unbound protein from the nickel column is next to the right and is clearly depleted in SecY and SecE. The partially pure SecYEG eluted from the same column and the eluted fractions across the main peak from the gel filtration column are also shown. The star indicates the migration position for contaminants arising from a cleavage in SecY.
Immunoprecipitation of SecYEG
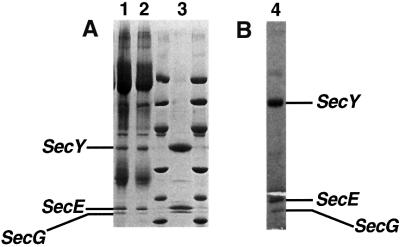
The gel profile of the SecYEG complex stained with Coomassie blue indicated that the band intensity of SecG was considerably weaker than that of SecE. To exclude that the SecG subunit had been depleted during the preparation, we performed immunoprecipitation experiments using polyclonal antibodies raised against SecY or SecG (Figure 2). With both antibodies, the ratio of the intensities of the bands appeared to be about the same (lanes 1 and 2). In addition, the ratio was similar to that in the purified complex (lane 3). Since SecG was not more abundant when SecG antibodies were used, it seems that SecG is present in every complex and was not depleted.
Fig. 2. Subunit composition of the SecYEG complex. (A) Immuno precipitation using SecY or SecG antibodies. A 5 µl aliquot (10%) of the total bound material was applied directly to the gel. Lanes 1 and 2 denote the material precipitated by SecY and SecG antibodies, respectively. Lane 3 is a sample of purified SecYEG for reference. The molecular weight markers are the same as those shown in Figure 1. The positions of SecY, E and G are indicated. (B) SDS–PAGE analysis of the crystalline SecYEG specimen (lane 4).
Activity determination of SecYEG purified in C12E9
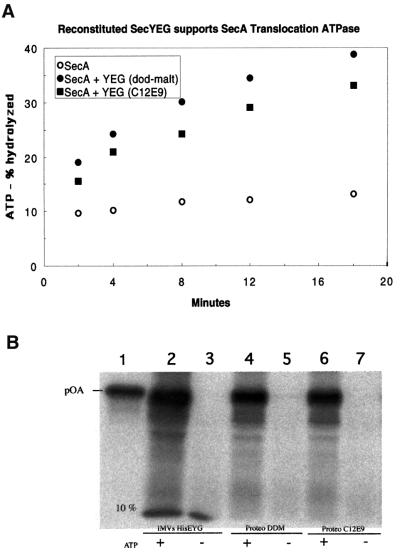
SecYEG reconstituted into phospholipid vesicles promoted stimulation of the ATPase activity of SecA in the presence of the substrate proOmpA (pro-outer membrane protein A; Figure 3A). The activity is essentially the same as that seen from material purified in dodecyl maltoside, the more commonly used detergent for the preparation of SecYEG (Figure 3A). Differences observed are probably due to variable reconstitution efficiencies of SecYEG into phospholipid vesicles. Using the data, the absolute ATPase activity can be calculated. In the absence of SecYEG, the measured ATPase activity of SecA is 4.3 mol/mol/min. In the presence of phospholipid vesicles containing SecYEG, the rate is 53 mol/mol/min, a 12-fold stimulation (only a slight stimulation is observed in the presence of empty phospholipid vesicles). These data are comparable to values previously reported (Lill et al., 1990; Bassilana and Wickner, 1993).
Fig. 3. Activity measurement of purified SecYEG. (A) ATPase assays were with either SecA alone, SecA with liposomes containing SecYEG purified in C12E9 and proOmpA, or SecA with liposomes containing SecYEG purified in dodecyl maltoside (dod-malt) and proOmpA. (B) proOmpA translocation into proteoliposomes was tested using membrane-reconstituted vesicles from dodecyl maltoside (proteo DDM; lanes 4 and 5, with and without ATP, respectively) or C12E9 (proteo C12E9, lanes 6 and 7) purified SecHisEYG. Lanes 2 and 3 represent translocation experiments using SecHisEYG-enriched inverted inner membrane vesicles (IMVs HisEYG) for comparison. Lane 1 includes proOmpA (pOA, 10% of the total reaction was applied) for reference. Reactions were incubated for 15 min at 37°C with or without ATP.
The purified complex is also active in translocation assays (Figure 3B). When SecYEG purified in C12E9 was reconstituted into phospholipid vesicles, it supported translocation of proOmpA in the presence of ATP and SecA; proOmpA translocated into the vesicles was protected from proteolysis (lane 6). In the absence of ATP, no translocation was observed (lane 7). SecYEG purified in C12E9 or dodecyl maltoside had the same translocation activity (lane 6 versus 4). For comparison, the translocation activity of inner membrane vesicles of E.coli was also tested (lane 2). In this case, partial cleavage of the signal sequence by the signal peptidase present in the membranes was observed. These results indicate that the purified SecYEG complex has a translocation activity similar to that reported previously (van der Does et al., 2000). In addition, vesicles containing reconstituted SecYEG exhibit ribosome-binding activity (Prinz et al., 2000).
In solution, SecYEG exists in an association equilibrium
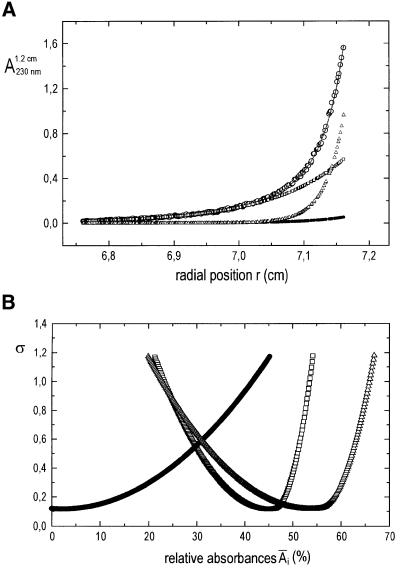
Sedimentation equilibrium experiments were carried out to determine the association state of SecYEG solubilized in the detergent C12E9 (Figure 4). D2O was added to match the density of the detergent. At a sufficiently low protein concentration, all experimental A(r) curves could be fitted perfectly according to Equation 1 (Materials and methods), applying only terms for the monomer, dimer and tetramer, with the terms for monomer and tetramer dominating. A typical example is shown in Figure 4A; the experiment described has both monomer and tetramer forms in approximately equal proportions, 52 and 48%, respectively. A model including a range of dimer of up to 5% also yielded an acceptable fit to the data. Interpretation of the experimental data based on a monomer–trimer– hexamer model of self-association led to a significant increase in the sum of the squared residuals, in most cases by >20%. Furthermore, in that case, the residuals of the fit were distributed non-statistically along the r-axis. Therefore, our data do not support this model.
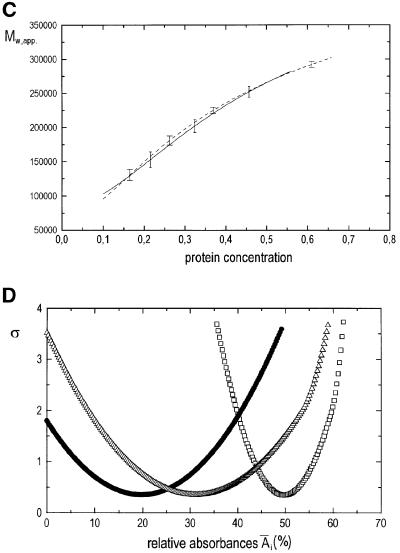
Fig. 4. Sedimentation equilibrium analysis of SecYEG. (A) Experimental absorbance versus radius data (open circles), best fit to the data assuming a monomer–dimer–tetramer model of self-association (—), and calculated local contributions of SecYEG monomers (open squares), dimers (filled circles) and tetramers (open triangles) to A(r). Initial protein concentration: 4.8 µM (A1.2 cm230 nm = 0.36). Rotor speed: 13 000 r.p.m. Rotor temperature: 4°C. (B) Statistical accuracy of the calculated relative absorbance contributions Âi, of the different SecYEG oligomers [obtained from the Ai(ro) values by integrating over the sample volume]: changes in the sum of the squared residuals, σ, of fits to the data from (A), which result from one non-optimal absorbance parameter (Schuck, 1994; Schuck et al., 1995). The minima of the curves correspond to the best fit figures for Âi. The nomenclature is the same as in (A). (C) Dependency of the local weight average molar mass, Mw,app., of SecYEG on the local protein concentration for two samples with loading concentrations of 0.14 mg/ml (- – -) and 0.28 mg/ml (—). Rotor speed, 11 000 r.p.m.; rotor temperature, 4°C. (D) Oligomer distribution of the mutant protein SecYprlA4EG. The graph corresponds to that shown in (B) for the wild-type protein. Initial protein concentration: 4.8 µM. Rotor speed, 15 000 r.p.m.; rotor temperature, 4°C.
At ∼10-fold higher protein concentrations, the occurrence of larger oligomers (around octamers) was observed. However, in these cases, the number of parameters required to fit the data was too large to allow an unambiguous identification of the oligomers present.
The dependency of the local weight average molar mass in the centrifugation cell on local protein concentration, Mw,app.(c) is shown in Figure 4C for two different loading concentrations. The two curves are identical within the limits of error. This shows that the different oligomers of SecYEG present are linked by an association equilibrium.
Similar experiments were performed on the mutant SecYprlA4EG complex that is able to transport substrates with defective or missing signal sequences (Figure 4D). The SecYprlA4EG had a different oligomeric distribution in detergent solution, namely 50% monomers, 20% dimers and 30% tetramers.
Two-dimensional crystallization of SecYEG and projection structure
Two-dimensional crystals of SecYEG were obtained after detergent removal by dialysis, at lipid to protein ratios of 0.05–0.8 (w/w). They could be recognized by their ability to distort the shape of vesicles and were typically between 0.5 and 2 µm in length (Figure 5). The presence of a lattice was usually indicated by straight-edged, tubular or triangular shapes. These crystals flatten on the electron microscope grid to form two planar lattices, which can be processed independently. Coherent crystalline patches were usually ∼0.4 µm in diameter. Analysis of the crystals by SDS–PAGE indicated that they contained all three subunits in the same proportion as the purified SecYEG and the immunoprecipitated complexes (Figure 2B).
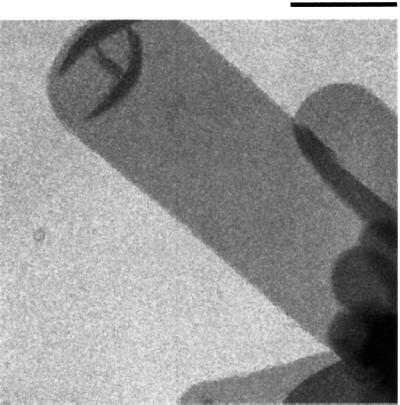
Fig. 5. Electron micrograph of a negatively stained two-dimensional crystal of SecYEG. Crystals stained by uranyl acetate were visualized using a Philips CM12 electron microscope. The tubular vesicle shown is flattened to the carbon surface and contains a two-dimensional crystal on each face. Bar, 0.5 µm.
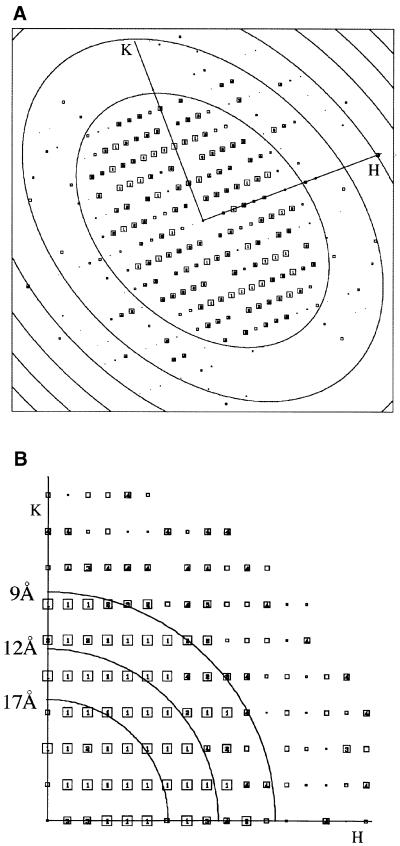
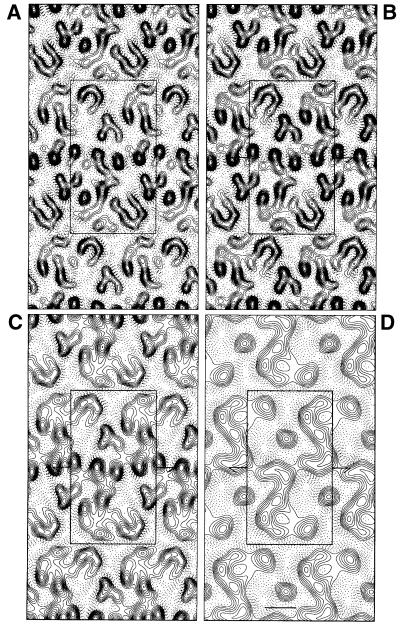
A computed Fourier transform from an image of a crystal after correction of the lattice distortions is shown in Figure 6. Significant reflections appear up to 9 Å resolution. The phase errors associated with each Fourier component after merging 12 images, and the overall phase residuals (Table I) show that the data are complete and isotropic up to 9 Å resolution. The overall phase residual to 8.9 Å was 15.6°. As suggested by the weak or absent IQ (intensity quality) values of the [0, k (odd)] reflections in the Fourier transform indicate that the phases obeyed the P121–b symmetry (revealed by ALLSPACE). Thus, the unit cell (a = 102 Å, b = 56 Å, γ = 90°) contains two asymmetric units, related to each other by a 2-fold screw axis along the b-direction. In the two-dimensional crystals, the SecYEG assemblies form rows, which alternate with respect to one another. Figure 7A shows the projection map of SecYEG drawn in P1 (without imposed symmetry) to 9 Å calculated from 12 independent lattices; Figure 7B and D shows projections maps drawn at 9 and 15 Å, respectively, with the P121–b symmetry imposed.
Fig. 6. Crystallographic data of SecYEG embedded in trehalose. (A) Computer-generated Fourier transform of an image of a SecYEG crystal. Each symbol represents a reflection, and the size of the box and the number within it (IQ values) relate to the quality of the data: a large box and a low number reflect a high signal to noise reflection (Henderson et al., 1986, 1990). The program MMBOXA was used to generate the data (Crowther et al., 1996). The rings show the position where the CTF is zero, and the axes refer to the reciprocal lattice vectors. The edge of the plot corresponds to a resolution of 6 Å. (B) Phase error of unique Fourier components plotted as a function of resolution. The size of the box indicates the error associated with each measurement (1 <8°, 2 <14°, 3 <20°, 4 <30°, 5 <40°, 6 <50°, 7 <70°, 8 <90°, where 90° is random). Values from 1 to 4 are shown as numbers, and decreasing box size indicates higher values. In this representation, measurements for reflections that are forbidden for P121 symmetry ([0, k (odd)]) were not removed, but were omitted upon imposing the P121 symmetry.
Table I. Electron crystallographic image statistics.
| Plane group symmetry | P121–b |
| Unit cell dimensions | a = 101.9 ± 1.8 Å |
| b = 56.4 ± 0.6 Å | |
| γ = 90.2 ± 0.8° | |
| No. of lattices | 12 |
| No. of images | 11 |
| Range of defocus | 6.800–2.700 Å |
| Magnification | 70 000–53 000× |
| Overall phase residual to 8.9 Åa | 15.6° |
| Resolution range (Å) | No. of unique reflectionsa | Phase residual (90° is random)a |
|---|---|---|
| 200.0–20.0 |
14 |
12.0° |
| 20.0–14.0 | 16 | 13.8° |
| 14.0–11.5 | 12 | 13.3° |
| 11.5–9.9 | 12 | 11.8° |
| 9.9–8.9 | 14 | 26.0° |
| 8.9–8.1 | 11 | 35.0° |
| 200.0–8.9 | 68 | 15.6° |
aIncluding reflections with IQ ≤7.
Fig. 7. Projection structure of SecYEG. Projection maps calculated from merged amplitudes and phases from 12 independent lattices of crystals embedded in trehalose. (A) Map drawn to 9 Å in P1 (no symmetry imposed). (B) With P121 symmetry imposed. An isotropic temperature factor of –600 Å2 was applied in (A) and (B) to compensate partially for the resolution-dependent degradation of image-derived amplitudes (Unger et al., 1997a). (C) Map at 9 Å resolution with imposed P121 symmetry without temperature factor adjustments. (D) Map at 15 Å resolution with P121 symmetry imposed. In all cases, four unit cells are shown, and one unit cell (containing two symmetry-related asymmetric units) is boxed. The solid lines indicate densities above the mean. Scale bar, 20 Å.
Discussion
We have overexpressed and purified large quantities of SecYEG complex and have used this material to grow two-dimensional crystals. The crystals have yielded the first detailed view of a bacterial core translocase. The purified complex is active according to different criteria. Upon reconstitution into phospholipid vesicles, the complex promotes the translocation-associated ATPase activity of SecA, and supports translocation of substrate across the bilayer. Both activities are comparable to those in similar experiments with SecYEG purified in the more usually employed detergent dodecyl maltoside. Although the purified SecYEG complex shows only a weakly stainable SecG band, this is not likely to be due to depletion of SecG during the preparation; this has been demonstrated by immunoprecipitation experiments. SecG probably binds Coomassie blue only poorly, like other membrane proteins which, on account of their hydrophobic nature, sometimes do not even stain at all (Collinson et al., 1994). Alternatively, the complex may have an unusual subunit stoichiometry. We see the same staining pattern in preparations purified from at least five alternative detergents, including dodecyl maltoside, and also with the crystalline preparations. Furthermore, the complex appears to constitute a homogeneous population since it elutes as a symmetrical peak from a gel filtration column. Moreover, a population of complexes with different subunit compositions would not be conducive to the growth of crystals. Taken together, it seems that the purified protein and the crystals have the same full complement of all three subunits.
The oligomeric state of the protein in detergent solution has been studied by analytical ultracentrifugation. The complex exhibits an association equilibrium between monomers and tetramers, with few or no dimeric intermediates present. In addition, higher oligomers were evident at elevated concentrations. Interestingly, the mutant SecYprlA4EG, which can translocate substrates with a defective or missing signal sequence, has a significantly increased dimer component in solution. Since the mutant complex is believed to be pre-primed for translocation (Duong and Wickner, 1999), it is possible that the dimer constitutes an intermediate during the assembly of an active channel. A recent study using electron microscopy and single particle analysis of negatively stained complexes (Manting et al., 2000) also highlights the presence of monomers, dimers and tetramers. However, the authors report that SecA is required for tetramer formation and they propose that the active translocation channel consists of four copies of SecYEG and two of SecA. Another study reported that the active complex consists of one SecYEG (Yahr and Wickner, 2000). Our experiments were done in the absence of SecA and in a detergent different from that used the former studies. Therefore, it is difficult to make a direct comparison.
If the asymmetric unit contained only one SecYEG complex with a total of 15 membrane-spanning α-helices, the calculated surface area per α-helix (199 Å2/α-helix) would be only slightly above that of the most tightly packed two-dimensional crystals of membrane proteins [bacteriorhodopsin 186 Å2/α-helix (Henderson et al., 1990; Grigorieff et al., 1996), NhaA 179 Å2/α-helix (Williams, 2000) and rhodopsin 184 Å2/α-helix (Krebs et al., 1998)]. In the unit cell, the asymmetric elements have an inverted orientation with respect to each another, with the contacts between them being crystallographic and not related to physiological associations.
The dimensions of the peaks and bands of density observed in the projection map and their spacings of ∼10 Å are consistent with a predominantly α-helical architecture. At 9 Å resolution, some densities are resolved that are probably transmembrane α-helices running perpendicular to the membrane. Other densities are likely to represent more tilted helices. Clearly it would be premature at this point to comment on the number of transmembrane helices or to assign any density peak to any particular helix. Tilted helices can project in variable ways, as has been observed for many other membrane proteins, e.g. bacteriorhodopsin (Henderson et al., 1986), aquaporin (Murata et al., 2000), NhaA (Williams, 2000) and gap junctions (Unger et al., 1999). The hydrophilic loops connecting the transmembrane helices of SecYEG are not expected to contribute much to a projection map at this resolution, as they are quite short and tend not to be α-helical. Further interpretation will require a three-dimensional map, which will be obtained from images of tilted crystals (work in progress).
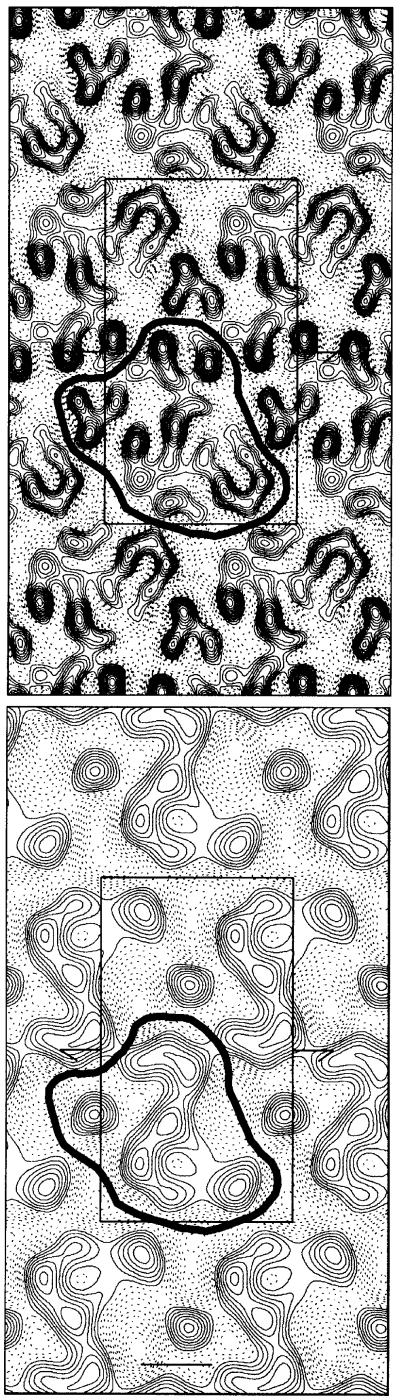
Although the molecular boundaries in the crystal cannot be drawn with any certainty at this stage, one possibility is shown in Figure 8. The outline was sketched using the 15 Å projection as a guide (Figure 7D), as often the structure presented at a lower resolution can be used to define the molecular boundary more clearly. Of course, any other possible outline would cover roughly the same area in projection.
Fig. 8. Proposed molecular boundary of SecYEG. Projection map of SecYEG (Figure 7B and D) with the molecular boundaries outlined. The upper panel corresponds to the 9 Å projection structure (Figure 7B) and the lower to 15 Å (Figure 7D).
We have compared our structure cautiously with the limited resolution information that is available from single particle analysis for protein translocation channels of SecYE (Meyer et al., 1999), SecYEG (Manting et al., 2000) and Sec61p (Beckmann et al., 1997; Ménetret et al., 2000). Clearly, the structure we have crystallized is not the same ring-like assembly seen before. Not only is our structure significantly smaller, but it also does not contain an obvious large pore. Therefore, the actual channel is likely to be formed by the assembly of more than one copy of the structure that we have crystallized. However, it would be premature to state how many SecYEG complexes constitute the functional protein channel. Conformational changes that seem likely upon oligomerization and channel formation may complicate the picture further. Nevertheless, the oligomerization of the SecYEG complex from a monomeric to a tetrameric species is supported by our analytical ultracentrifugation experiments, and this finding is consistent with experiments performed elsewhere (Manting et al., 2000). The monomeric species we identify in solution also has to be too small to form the observed translocation channel. The assembly of the translocation channel from smaller units is also supported by freeze-fracture electron microscopy employing the Sec61p complex (Hanein et al., 1996). The addition of ribosomes to proteoliposomes containing the Sec61p complex or the co-reconstitution with another membrane protein complex induced the appearance of ring structures in the plane of the membrane. Therefore, it is possible that the actual channel is assembled from smaller units when protein translocation is initiated and disassembled upon termination. Assembly and disassembly of the channel may be triggered by the binding and release of its partners, such as the ribosome or SecA, and/or by the occupancy with a translocation substrate.
The reversible formation of a channel from smaller units appears to be unique to the Sec complex. Other known channels that form by oligomerization of monomers assemble after biosynthesis and remain assembled during their lifetime. The different behaviour of the protein-conducting channel may be related to its particularly large pore size. Disassembly of the channel upon termination of translocation may allow maintenance of the membrane barrier for small molecules. The large pore may not present a problem when the channel is in the process of translocating polypeptides because the latter may at least partially plug the hole. The reversible assembly of the translocation channel may thus be a novel mechanism of channel gating.
Knowledge of the mechanism of protein translocation will, of course, require a structural understanding to augment results from biochemical and genetic studies. Our medium-resolution projection map of SecYEG represents the first steps towards the elucidation of the structural details, which are needed to really understand in molecular detail how proteins are translocated through membranes. The next step towards this goal will be to determine a three-dimensional map of the complex.
Materials and methods
C12E9 was purchased from Sigma and OG from Anatrace. Chelating Sepharose Fast Flow, a pre-packed Superdex 200 preparation grade gel filtration column, Q-Sepharose High Performance media and protein-A/G-coupled Sepharose media were obtained from Pharmacia Biotech. Lipids were from Avanti Polar Lipids Inc. BioBeads were purchased from BioRad. All other chemicals were obtained from Sigma.
Analytical methods
SDS–PAGE was performed using 15 × 8 × 0.075 cm gels with 15% acrylamide (Laemmli, 1970) and proteins were visualized by staining with Coomassie blue R-250.
Overexpression and purification
To allow overproduction and purification of SecYEG, six histidines were attached to the N-terminus of SecE. Briefly, the HA-SecE gene carried by pHA-EYG [pBAD22, overexpressing clone containing HASecE, SecY and SecG in that order driven from the same (ara) promoter; Douville et al., 1995] was replaced by a 430 bp EcoRI–SalI PCR fragment coding for His6-SecE. Plasmid pHis-EYG expresses His6-tagged SecE, SecY and SecG under the control of the araBAD promoter of pBAD22. Plasmid pHis-EY4G is a derivative of pHis-EYG carrying the prlA4 mutations (F286Y and I408N) in SecY.
A colony of freshly transformed E.coli strain c43 (DE3) (Miroux and Walker, 1996) with pHis-EYG or pHisEY4G was inoculated into 50 ml of LB growth medium containing 100 µg/ml ampicillin and grown with shaking overnight at 37°C as a pre-culture. Cultures (12 × 1 l) were then grown at 30°C overnight in 2× YT broth also containing 100 µg/ml ampicillin, 1% (w/v) l-(+)-arabinose and 4 ml of pre-culture. Cells were harvested by centrifugation and used without freezing by resuspending in 300 ml of buffer TSG [20 mM Tris–HCl pH 8, 0.3 M NaCl, 10% (v/v) glycerol]. The cells were lysed by one passage through a 40 ml pre-chilled French pressure cell. The lysate was kept on ice and phenylmethylsulfonyl fluoride (PMSF) added to a final concentration of 0.01% (w/v). Total membranes were crudely prepared by centrifugation (142 000 g, 90 min, 4°C) and washed by resuspension in 200 ml of TSG buffer before re-centrifugation. Washed membranes were extracted by resuspension in the same buffer (100 ml) plus 1.25% (w/v) OG and stirred in the cold for 1 h at 4°C. Insoluble material was removed by centrifugation (142 000 g, 60 min, 4°C). Purification proceeded from the extract making use of the hexa-histidine tag. An Ni2+-chelated Sepharose column was employed to bind the SecYEG complex. After binding, the column was washed with TSG buffer containing 30 mM imidazole and 0.2% (v/v) C12E9. This procedure served both to remove the majority of contaminants and to exchange the detergent bound to SecYEG, so all subsequent steps were performed in C12E9. Bound protein was eluted in the same buffer with an elevated concentration of imidazole (330 mM). Further purification was achieved by gel filtration (Superdex 200pg XK 26/60) and anion exchange (Q-Sepharose HP XK 26/20) chromatography. The final buffer consisted of 20 mM Tris–HCl pH 8, 0.2% (v/v) C12E9 and 0.12 M NaCl.
Immunoprecipitation of SecYEG
Polyclonal antibodies were raised in rabbits against peptides corresponding to the N-terminal regions of SecY and SecG. After several boosts, the sera were assayed by western blotting until an acceptable titre was attained. To 100 µl of SecYEG [0.4 mg/ml in 20 mM Tris–HCl pH 8, 0.2% (v/v) C12E9 and 0.12 M NaCl], 100 µl of undiluted antiserum was added and the sample was mixed gently overnight at 4°C (temperature maintained throughout). A mixture of protein A and protein G coupled to Sepharose (25 µl each, in the same buffer) was then added and mixed for 4 h. The beads were also washed extensively with buffer and the bound material was released with SDS sample buffer (50 µl) at 40°C for 10 min. Fractions of the bound material were examined by SDS–PAGE.
Activity determination of SecYEG
ATPase activation of SecA. SecYEG was reconstituted into phospholipid vesicles as follows. BioBeads SM2 (200 µl) washed in water and reconstitution buffer [50 mM K-HEPES pH 7.5, 200 mM K-acetate, 1 mM dithiothreitol (DTT), 12.5% glycerol] were added to the reaction mixture, which included 17 µg of purified SecYEG (purified in C12E9 or dodecyl maltoside) and 33 µl of E.coli phospholipids (20 mg/ml, in 6% deoxyBig-CHAP, 50 mM K-HEPES pH 7.5, 1 mM DTT). The volume of the reaction mixture was topped up to 200 µl with reconstitution buffer. After incubation overnight at 4°C, the beads were removed by low-speed centrifugation and washed with 1 ml of 50 mM K-HEPES pH 7.5, 1 mM DTT. The proteoliposomes were harvested from the combined supernatants by high-speed centrifugation (Beckmann TLA 100.3 rotor, 70 000 r.p.m., 30 min, 4°C). The pellet was resuspended in 50 µl of 50 mM K-HEPES pH 7.5, 1 mM DTT, 10% glycerol.
The ATPase assay was performed in the following way. SecA (7.5 pmol, with respect to dimer) in a final volume of 20 µl [50 mM K-HEPES pH 7.5, 50 mM KCl, 1.5 mM MgCl2, 1 mM DTT, 0.5 mg/ml bovine serum albumin (BSA)] was mixed on ice with 9 pmol (2 µl) of SecYEG reconstituted into phospholipid vesicles and then with 0.7 µl of proOmpA (3 mg/ml, 8 M urea, 50 mM Tris–HCl pH 7.5, 2 mM DTT). Assuming that we have 50% of SecYEG in the correct orientation and that all the SecYEG was reconstituted, leaves 4.5 pmol to activate SecA. The reaction was initiated by adding [γ-32P]ATP to 1.5 mM followed by transfer to 37°C. Samples (3.5 µl) were withdrawn at different time points and adsorbed onto polyethyleneimine-cellulose TLC sheets, which were processed and analysed as described elsewhere (Misselwitz et al., 1999).
In vitro translocation of proOmpA by SecYEG. For reconstitution of SecYEG into liposomes for translocation experiments, 50 µl of purified SecHisEYG proteins (∼0.3 mg/ml in dodecyl maltoside or C12E9) were diluted into 1 ml of reconstitution buffer (50 mM Tris–HCl pH 7.9, 50 mM KCl, 1 mM DTT) containing 0.2% dodecyl maltoside and 2 mg/ml of E.coli phospholipids. Proteoliposome formation was then allowed by adsorption of the detergents onto SM2 Bio-Beads (100 mg of wet Bio-Beads/ml) during overnight incubation at 4°C. Proteoliposomes were collected by centrifugation (120 000 g; 30 min, 4°C) and resuspended in 200 µl of reconstitution buffer. The final protein concentration was ∼35 µg/ml.
Translocation assays were performed in 100 µl of translocation buffer (50 mM Tris–HCl pH 7.9, 50 mM KCl, 5 mM MgCl2, 1 mM DTT) containing 2 mM ATP, 10 mM creatine phosphate, 0.5 µg of creatine kinase, 1.0 µg of SecB, 2.0 µg of SecA, 25 µg of BSA and ∼30 000 c.p.m. of 125I-labelled proOmpA (∼105 c.p.m./µg). The reaction was initiated by addition of 10 µl of SecHisEYG proteoliposomes (∼0.35 µg) or 10 µl of SecHisEYG-enriched inverted inner membrane vesicles (∼10 µg) and incubated for 15 min at 37°C. Samples were treated with proteinase K (0.1 mg/ml) and precipitated with trichloroacetic acid. The translocated [125I]proOmpA molecules were analysed by 15% SDS–PAGE and fluorography.
Analytical ultracentrifugation
Sedimentation equilibrium experiments were performed in a Beckman Optima XL-A analytical ultracentrifuge using Epon 6-channel or double Sector cells with a pathlength of 1.2 cm in combination with an An-50Ti rotor. The rotor speed was between 11 000 and 15 000 r.p.m., and the temperature 4°C. The absorbance versus radius distributions A(r) were recorded at 230 or 280 nm. Purified SecYEG (or SecYprlA4EG) was applied in 140 or 250 µl volumes and initial protein concentrations were between 0.03 and 0.3 mg/ml in 8.6 mM Tris–HCl pH 8, 50 mM NaCl, 0.1% (w/v) C12E9 containing 57% (v/v) D2O to match the detergent density (Schubert and Schuck, 1991).
The experimental sedimentation equilibrium profiles were analysed according to the following equation (Schubert and Schuck, 1991):
with i, number of protomers per oligomer; Ai, absorbance of corresponding species; M1, molar mass of the protomer, v¯, partial specific volume of the protein (assumed to be independent of the oligomeric state); ρo, solvent density; ω, angular velocity of the rotor; r, radial position; ro, fixed radial position. The evaluations were performed according to Schuck (1994). They included tests for the statistical accuracy of the best fit parameters, considering the minimal increase in the sum of the squared residuals of the fit, A(r), that results from one non-optimal parameter Ai(ro); all other Aj(ro) were varied to compensate for the imposed constraint. The sharp minima of peaks obtained in the plots (Figure 2B and D) indicate very accurate values for each of the corresponding parameters. The partial specific volume of SecYEG at 0.75 ml/g was calculated according to Durchschlag, using the data set of Cohn and Edsall, as tabulated in Durchschlag (1986).
Two-dimensional crystallization
Purified protein was crystallized in two dimensions by the dialysis method (Kühlbrandt, 1992). Dilute complex (0.4 mg/ml in 0.2% C12E9) was added to detergent-solubilized lipid (phosphatidylethanolamine purified from E.coli, also solubilized in C12E9) at varying lipid:protein ratios (0.05–0.8, w/w). The mixtures, with 0.01% (w/v) PMSF, were dialysed against 200 ml of 20 mM Tris–HCl pH 8, 0.12 M NaCl, 1 mM EDTA, 1 mM NaN3, without detergent at 23°C, and the outside buffer was changed daily. The dialysis serves to remove the detergent, which in turn results in the formation of phospholipid bilayers (in our case, vesicles), wherein two-dimensional crystals form. Crystal trials were negatively stained and screened using a Philips CM12 electron microscope. Samples (2–4 µl) were allowed to sit for 2 min on a carbon-coated copper grid. Excess solution was blotted off, and the grid was stained with 1% uranyl acetate. Crystalline samples were seen over many defined lipid:protein ratios, with 0.2 being the best, and they took several weeks to grow. Promising crystalline samples were subjected to electron cryo-microscopy.
Electron cryo-microscopy and image processing
Frozen samples were prepared on grids using the back injection method, i.e. 2.5 µl of sample was pipetted into the same volume of a 4% (w/v) trehalose solution on a carbon-coated copper grid, blotted for 15 s face on the filter paper, before rapidly freezing in liquid nitrogen.
Grids were mounted in a Leica KF80 apparatus for transfer into a JEOL 3000 SFF electron microscope operated at 300 kV, equipped with a top-entry cryo-stage cooled to 4°K with liquid helium. Images were then taken using a spot-scan procedure with a total electron dose of 20–30 e/Å2, at magnifications of 60 000 and 53 000×. Kodak SO-163 films were used to record images, and developed for 12 min with full-strength Kodak D19.
Optical diffraction was used to screen the micrographs, and areas giving rise to symmetrical distribution of sharp spots were digitized using a Zeiss SCAI scanner with a 7-µm step size over an area of 6000 × 6000 pixels. Images were then corrected for distortion of the crystal lattice and the effects of the contrast transfer function (CTF) using the MRC image-processing programs (Crowther et al., 1996). Projection maps were calculated, using the standard crystallographic computer programs in the CCP4 package (CCP4, 1994). The symmetry was revealed by the program ALLSPACE (Valpuesta et al., 1994) and the P121-b space group was imposed on the merged data. Projection maps were scaled to a maximum density of 250, and contoured in steps of 0.25 × r.m.s. density. A negative temperature factor of –600 was applied in Figures 7A, B and D, and 8 to compensate partially for the resolution-dependent degradation of image-derived amplitudes (Unger et al., 1997a).
Acknowledgments
Acknowledgements
This work was initiated at the Max Planck Institut für Biophysik, Frankfurt am Main where help from Dr Janet Vonck and especially from Deryck Mills was greatly appreciated during cryo-EM data collection. We are grateful to Dr Christopher Akey for the use of his CM12 electron microscope. I.C. appreciates Adam Cliffe for enthusiastic technical support in Frankfurt, and Pavan Bendapudi in Boston. The E.coli strain c43 was a kind gift of Drs J.E.Walker and B.Miroux. I.C. was the recipient of a Long-Term EMBO Fellowship. In Boston, I.C. was supported from a Human Frontiers Science Program Long-Term Fellowship. C.B. and E.O. were also recipients of an EMBO Long-Term Fellowship. T.R. is a Howard Hughes Medical Institute Investigator.
Note added in proof
Recently, a 3D structure has been calculated at 8 Å resolution (C.Breyton, W.Kühlbrandt and I.Collinson, unpublished results). Surprisingly, the crystal is constituted by a double membrane. The unit cell spans both membranes and is twice the volume we originally thought; thus, the asymmetric units must contain more than a single SecYEG copy. In fact, we see a well defined SecYEG dimer, exhibiting a 2-fold non-crystallographic symmetry. Within the unit cell, the two dimers, one in each membrane, are related to each other by the screw axis symmetry. Nevertheless, the SecYEG dimer is too small to constitute the translocating channel, and the conclusions concerning the possible reversible assembly as a means of channel gating remain unaffected.
References
- Akimaru J., Matsuyama,S., Tokuda,H. and Mizushima,S. (1991) Reconstitution of a protein translocation system containing purified SecY, SecE and SecA from Escherichia coli.Proc. Natl Acad. Sci. USA, 88, 6545–6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auer M., Scarborough,G. and Kühlbrandt,W. (1998) Three-dimensional map of the plasma membrane H+-ATPase in the open conformation. Nature, 392, 840–843. [DOI] [PubMed] [Google Scholar]
- Bassilana M. and Wickner,W. (1993) Purified Escherichia coli preprotein translocase catalyzes multiple cycles of precursor protein translocation. Biochemistry, 32, 2626–2630. [DOI] [PubMed] [Google Scholar]
- Beckmann R., Bubeck,D., Grassucci,R., Penczek,P., Verschoor,A., Blobel,G. and Frank,J. (1997) Alignment of conduits for the nascent polypeptide chain in the ribosome–Sec61 complex. Science, 278, 2123–2126. [DOI] [PubMed] [Google Scholar]
- Bieker K.L., Phillips,G.J. and Silhavy,T.J. (1990) The sec and prl genes of Escherichia coli.J. Bioenerg. Biomembr., 22, 291–310. [DOI] [PubMed] [Google Scholar]
- Brundage L., Hendrick,J., Schiebel,E., Driessen,A. and Wickner,W. (1990) The purified E.coli integral membrane protein SecY/E is sufficient for reconstitution of SecA-dependent precursor protein translocation. Cell, 62, 649–657. [DOI] [PubMed] [Google Scholar]
- Cabelli R.J., Chen,L., Tai,P.C. and Oliver,D.B. (1988) SecA protein is required for secretory protein translocation into E.coli membrane vesicles. Cell, 55, 683–692. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project No. 4. (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Collinson I., Runswick,M., Buchanan,S., Fearnley,I., Skehel,J., van,R.M., Griffiths,D. and Walker,J. (1994) Fo membrane domain of ATP synthase from bovine heart mitochondria: purification, subunit composition and reconstitution with F1-ATPase. Biochemistry, 33, 7971–7978. [DOI] [PubMed] [Google Scholar]
- Crowther R., Henderson,R. and Smith,J. (1996) MRC image processing programs. J. Struct. Biol., 116, 9–16. [DOI] [PubMed] [Google Scholar]
- Douville K., Price,A., Eichler,J., Economou,A. and Wickner,W. (1995) SecYEG and SecA are the stoichiometric components of preprotein translocase. J. Biol. Chem., 270, 20106–20111. [DOI] [PubMed] [Google Scholar]
- Driessen A., Fekkes,P. and van der Wolk,J. (1998) The Sec system. Curr. Opin. Microbiol., 1, 216–222. [DOI] [PubMed] [Google Scholar]
- Duong F. and Wickner,W. (1997a) Distinct catalytic roles of the SecYE, SecG and SecDFyajC subunits of preprotein translocase holoenzyme. EMBO J., 16, 2756–2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong F. and Wickner,W. (1997b) The SecDFyajC domain of preprotein translocase controls preprotein movement by regulating SecA membrane cycling. EMBO J., 16, 4871–4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong F. and Wickner,W. (1999) The PrlA and PrlG phenotypes are caused by a loosened association among the translocase SecYEG subunits. EMBO J., 18, 3263–3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong F., Eichler,J., Price,A., Leonard,M. and Wickner,W. (1997) Biogenesis of the gram-negative bacterial envelope. Cell, 91, 567–573. [DOI] [PubMed] [Google Scholar]
- Durchschlag H. (1986) Specific volumes of biological macromolecules and some other molecules of biological interest. In Hinz,H.-J. (ed), Thermodynamic Data for Biochemistry and Biotechnology. Springer-Verlag, Berlin, pp. 45–128.
- Economou A. and Wickner,W. (1994) SecA promotes preprotein translocation by undergoing ATP-driven cycles of membrane insertion and deinsertion. Cell, 78, 835–843. [DOI] [PubMed] [Google Scholar]
- Economou A., Pogliano,J., Beckwith,J., Oliver,D. and Wickner,W. (1995) SecA membrane cycling at SecYEG is driven by distinct ATP binding and hydrolysis events and is regulated by SecD and SecF. Cell, 83, 1171–1181. [DOI] [PubMed] [Google Scholar]
- Grigorieff N., Ceska,T., Downing,K., Baldwin,J. and Henderson,R. (1996) Electron-crystallographic refinement of the structure of bacteriorhodopsin. J. Mol. Biol., 259, 393–421. [DOI] [PubMed] [Google Scholar]
- Hanein D., Matlack,K., Jungnickel,B., Plath,K., Kalies,K., Miller,K., Rapoport,T. and Akey,C. (1996) Oligomeric rings of the Sec61p complex induced by ligands required for protein translocation. Cell, 87, 721–732. [DOI] [PubMed] [Google Scholar]
- Henderson R., Baldwin,J., Downing,K. and Zemlin,F. (1986) Structure of purple membrane from Halobacterium halobium. Recording, measurement and evaluation of electron micrographs at 3.5 Å resolu tion. Ultramicroscopy, 19, 147–178. [Google Scholar]
- Henderson R., Baldwin,J., Ceska,T., Zemlin,F., Beckmann,E. and Downing,K. (1990) Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J. Mol. Biol., 213, 899–929. [DOI] [PubMed] [Google Scholar]
- Jungnickel B., Rapoport,T. and Hartmann,E. (1994) Protein translocation: common themes from bacteria to man. FEBS Lett., 346, 73–77. [DOI] [PubMed] [Google Scholar]
- Krebs A., Villa,C., Edwards,P.C. and Schertler,G.F. (1998) Characterisation of an improved two-dimensional p22121 crystal from bovine rhodopsin. J. Mol. Biol., 282, 991–1003. [DOI] [PubMed] [Google Scholar]
- Kühlbrandt W. (1992) Two-dimensional crystallization of membrane proteins. Q. Rev. Biophys., 25, 1–49. [DOI] [PubMed] [Google Scholar]
- Kühlbrandt W., Wang,D. and Fujiyoshi,Y. (1994) Atomic model of plant light-harvesting complex by electron crystallography. Nature, 367, 614–621. [DOI] [PubMed] [Google Scholar]
- Kumamoto C. and Beckwith,J. (1983) Mutations in a new gene, secB, cause defective protein localization in Escherichia coli.J. Bacteriol., 154, 253–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkele K. et al. (1998) The preprotein translocation channel of the outer membrane of mitochondria. Cell, 93, 1009–1019. [DOI] [PubMed] [Google Scholar]
- Laemmli U. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Lill R., Cunningham,K., Brundage,L., Ito,K., Oliver,D. and Wickner,W. (1989) SecA protein hydrolyzes ATP and is an essential component of the protein translocation ATPase of Escherichia coli.EMBO J., 8, 961–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill R., Dowhan,W. and Wickner,W. (1990) The ATPase activity of SecA is regulated by acidic phospholipids, SecY and the leader and mature domains of precursor proteins. Cell, 60, 271–280. [DOI] [PubMed] [Google Scholar]
- Mant A., Schmidt,I., Herrmann,R., Robinson,C. and Klosgen,R. (1995) Sec-dependent thylakoid protein translocation. ΔpH requirement is dictated by passenger protein and ATP concentration. J. Biol. Chem., 270, 23275–23281. [DOI] [PubMed] [Google Scholar]
- Manting E., van,D.D.C., Remigy,H., Engel,A. and Driessen,A. (2000) SecYEG assembles into a tetramer to form the active protein translocation channel. EMBO J., 19, 852–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ménetret J., Neuhof,A., Morgan,D., Plath,K., Radermacher,M., Rapoport,T. and Akey,C. (2000) The structure of ribosome-channel complexes engaged in protein translocation. Mol. Cell, 6, 1219–1232. [DOI] [PubMed] [Google Scholar]
- Meyer T., Ménetret,J., Breitling,R., Miller,K., Akey,C. and Rapoport,T. (1999) The bacterial SecY/E translocation complex forms channel-like structures similar to those of the eukaryotic Sec61p complex. J. Mol. Biol., 285, 1789–1800. [DOI] [PubMed] [Google Scholar]
- Miroux B. and Walker,J. (1996) Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol., 260, 289–298. [DOI] [PubMed] [Google Scholar]
- Misselwitz B., Staeck,O., Matlack,K. and Rapoport,T. (1999) Interaction of BiP with the J-domain of the Sec63p component of the endoplasmic reticulum protein translocation complex. J. Biol. Chem., 274, 20110–20115. [DOI] [PubMed] [Google Scholar]
- Murata K., Mitsuoka,K., Hirai,T., Walz,T., Agre,P., Heymann,J.B., Engel,A. and Fujiyoshi,Y. (2000) Structural determinants of water permeation through aquaporin-1. Nature, 407, 599–605. [DOI] [PubMed] [Google Scholar]
- Nishiyama K., Hanada,M. and Tokuda,H. (1994) Disruption of the gene encoding p12 (SecG) reveals the direct involvement and important function of SecG in the protein translocation of Escherichia coli at low temperature. EMBO J., 13, 3272–3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver D. and Beckwith,J. (1982) Identification of a new gene (secA) and gene product involved in the secretion of envelope proteins in Escherichia coli.J. Bacteriol., 150, 686–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlschroder M., Prinz,W., Hartmann,E. and Beckwith,J. (1997) Protein translocation in the three domains of life: variations on a theme. Cell, 91, 563–566. [DOI] [PubMed] [Google Scholar]
- Prinz A., Behrens,C., Rapoport,T., Hartmann,E. and Kalies,K. (2000) Evolutionarily conserved binding of ribosomes to the translocation channel via the large ribosomal RNA. EMBO J., 19, 1900–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz W., Spiess,C., Ehrmann,M., Schierle,C. and Beckwith,J. (1996) Targeting of signal sequenceless proteins for export in Escherichia coli with altered protein translocase. EMBO J., 15, 5209–5217. [PMC free article] [PubMed] [Google Scholar]
- Rapoport T., Jungnickel,B. and Kutay,U. (1996) Protein transport across the eukaryotic endoplasmic reticulum and bacterial inner membranes. Annu. Rev. Biochem., 65, 271–303. [DOI] [PubMed] [Google Scholar]
- Rhee K., Morris,E., Barber,J. and Kuhlbrandt,W. (1998) Three-dimensional structure of the plant photosystem II reaction centre at 8 Å resolution. Nature, 396, 283–286. [DOI] [PubMed] [Google Scholar]
- Schatz P., Riggs,P., Jacq,A., Fath,M. and Beckwith,J. (1989) The secE gene encodes an integral membrane protein required for protein export in Escherichia coli.Genes Dev., 3, 1035–1044. [DOI] [PubMed] [Google Scholar]
- Schubert D. and Schuck,P. (1991) Analytical ultracentrifugation as a tool for studying membrane proteins. Prog. Colloid Polym. Sci., 86, 12–22. [Google Scholar]
- Schuck P. (1994) Simultaneous radial and wavelength analysis with the Optima XL-A analytical ultracentrifuge. Prog. Colloid Polym. Sci., 94, 1–13. [Google Scholar]
- Schuck P., Legrum,B., Passow,H. and Schubert,D. (1995) The influence of two anion-transport inhibitors, 4,4′-diisothiocyanatodihydro stilbene-2,2′-disulfonate and 4,4′-dibenzoylstilbene-2,2′-disulfonate, on the self-association of erythrocyte band 3 protein. Eur. J. Biochem., 230, 806–812. [DOI] [PubMed] [Google Scholar]
- Shilton B., Svergun,D., Volkov,V., Koch,M., Cusack,S. and Economou,A. (1998) Escherichia coli SecA shape and dimensions. FEBS Lett., 436, 277–282. [DOI] [PubMed] [Google Scholar]
- Unger V., Kumar,N., Gilula,N. and Yeager,M. (1997a) Projection structure of a gap junction membrane channel at 7 Å resolution. Nature Struct. Biol., 4, 39–43. [DOI] [PubMed] [Google Scholar]
- Unger V.M., Hargrave, PA., Baldwin,J.M. and Schertler,G.F. (1997b) Arrangement of rhodopsin transmembrane α-helices. Nature, 389, 203–206. [DOI] [PubMed] [Google Scholar]
- Unger V., Kumar,N., Gilula,N. and Yeager,M. (1999) Three-dimensional structure of a recombinant gap junction membrane channel. Science, 283, 1176–1180. [DOI] [PubMed] [Google Scholar]
- Valpuesta J., Carrascosa,J. and Henderson,R. (1994) Analysis of electron microscope images and electron diffraction patterns of thin crystals of M-X29 connectors in ice. J. Mol. Biol., 240, 281–287. [DOI] [PubMed] [Google Scholar]
- van der Does, C., Swaving,J., van,K.W. and Driessen,A. (2000) Non-bilayer lipids stimulate the activity of the reconstituted bacterial protein translocase. J. Biol. Chem., 275, 2472–2478. [DOI] [PubMed] [Google Scholar]
- van der Wolk J., Fekkes,P., Boorsma,A., Huie,J., Silhavy,T. and Driessen,A. (1998) PrlA4 prevents the rejection of signal sequence defective preproteins by stabilizing the SecA–SecY interaction during the initiation of translocation. EMBO J., 17, 3631–3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K. (2000) Three-dimensional structure of the ion-coupled transport protein NhaA. Nature, 403, 112–115. [DOI] [PubMed] [Google Scholar]
- Yahr T. and Wickner,W. (2000) Evaluating the oligomeric state of SecYEG in preprotein translocase. EMBO J., 19, 4393–4401. [DOI] [PMC free article] [PubMed] [Google Scholar]