Abstract
Genome rearrangements can take place by a process known as replication slippage or copy-choice recombination. The slippage occurs between repeated sequences in both prokaryotes and eukaryotes, and is invoked to explain microsatellite instability, which is related to several human diseases. We analysed the molecular mechanism of slippage between short direct repeats, using in vitro replication of a single-stranded DNA template that mimics the lagging strand synthesis. We show that slippage involves DNA polymerase pausing, which must take place within the direct repeat, and that the pausing polymerase dissociates from the DNA. We also present evidence that, upon polymerase dissociation, only the terminal portion of the newly synthesized strand separates from the template and anneals to another direct repeat. Resumption of DNA replication then completes the slippage process.
Keywords: copy-choice recombination/direct repeats/DNA rearrangements/palindrome/replication slippage mechanism
Introduction
Repeated sequences are present in all genomes and are particularly abundant in eukaryotes, where runs of mono-, di- and trinucleotide repeats, known as microsatellite sequences, can represent a sizeable proportion of the genetic material. These repeats, which are present in coding and non-coding regions (Toth et al., 2000), exhibit a strong level of instability, undergoing additions or deletions of repeated units, which lead to variations in the number of copies of the repeated stretches. These variations were thus named ‘dynamic mutations’ (Richards and Sutherland, 1992). A number of studies on deletions and expansions of repeated sequences were prompted by the observation of a correlation between triplet expansion and certain human genetic diseases (for reviews see Richards and Sutherland, 1994; Hancock and Santibanez-Koref, 1998). Alteration in the number of repeats of mono-, di- or trinucleotides is linked to hereditary non-polyposis colon cancer and other human cancers (Richards and Sutherland, 1994; Djian, 1998). Neurodegenerative diseases are associated with a modification of the length polymorphism of simple repeats, where expansions of the following triplet repeats, CCG⋅CGG, CTG⋅CAG and GAA⋅TTC, account for most of the cases (for reviews see Wells, 1996; Mitas, 1997). In prokaryotes, highly repeated sequences are less common, but recombination between short repeats is also a major cause of genome instability (Ehrlich et al., 1993; van Belkum et al., 1998; Michel, 2000).
A replication slippage model, also known as copy-choice recombination, was proposed as a possible mechanism to account for formation of dynamic mutations (Tautz and Schlotterer, 1994; Wells, 1996; Pearson and Sinden, 1998a,b; Petruska et al., 1998). It was shown that replication slippage does indeed take place in vivo, in Escherichia coli, between short direct repeats (DRs) and probably also between longer tandem repeats (Lovett et al., 1993; d’Alencon et al., 1994; Bierne et al., 1997b). Direct evidence that different DNA polymerases can undergo replication slippage was also provided in vitro (Canceill and Ehrlich, 1996; Canceill et al., 1999). An experimental system that mimics the replication of the lagging DNA strand was used in these studies, involving a single-stranded DNA (ssDNA) template that carries two short DRs flanking a hairpin structure formed by annealing of two long inverted repeats (IRs). Three E.coli DNA polymerases (I, II and III) slip between the DR on this template, as do DNA polymerases from the thermophilic microorganisms Thermus aquaticus, Pyrococcus furiosus, Pyrococcus abyssi and Thermococcus littoralis, or from phages T4 (T4 pol) and T7 (T7 pol) (Canceill and Ehrlich, 1996; Canceill et al., 1999; E.Viguera, D.Canceill and S.D.Ehrlich, submitted). In contrast, the polymerases endowed with high strand displacement activity and therefore able to enter the hairpin and progress within it, such as phage Φ29 DNA polymerase and the thermostable Bacillus stearothermophilus DNA polymerase, do not slip. Furthermore, factors that stimulate the strand displacement activity of polymerases, such as the single-stranded DNA-binding protein (SSB) or specific point mutations in the polymerase exonuclease domain, interfere with slippage (Canceill et al., 1999; E.Viguera, D.Canceill and S.D.Ehrlich, submitted).
Based on these observations, we proposed the following model for the slippage process: (i) polymerase replicates a DR; (ii) it is arrested and dissociates from the newly synthesized strand; (iii) this strand separates from the template and realigns with another DR; and (iv) poly merase reloads and replication resumes. This model presents similarities with that invoked to account for so-called translesion synthesis (TLS). It is thought that during TLS, the main replicative polymerase is arrested by a bulky lesion and dissociates from the template (for a review see Baynton and Fuchs, 2000). This allows extension of the newly synthesized strand, either directly by a specialized polymerase capable of inserting a nucleotide opposite to the lesion, or, in accord with the slippage model, after the realignment of the tip of the newly synthesized strand with a homologous nucleotide positioned beyond the lesion (Napolitano et al., 2000). The latter process, which also relies upon specialized polymerases, such as E.coli DNA polymerase II, IV or V, leads to a loss of one or two nucleotides and therefore generates frameshift mutations. In contrast, two different models were proposed for slippage of mononucleotide runs by DNA polymerase III holoenzyme from E.coli (pol III HE) in vitro, which generates frameshift mutations (Seki et al., 1999): the ‘melting–misalignment model’, which involves misalignment of the tract inside the DNA polymerase and therefore no DNA polymerase dissociation; and the ‘misinsertion–misalignment model’, which involves polymerase dissociation, spontaneous realignment of the terminal nucleotide and resumption of DNA synthesis.
In this work, we addressed the two key steps of the replication slippage model: the arrest and dissociation of the polymerase, which were postulated but never directly demonstrated. We show that pausing of the polymerase within the DR is necessary for slippage and that the pausing polymerase dissociates prior to the strand realignment. These results not only strengthen the model of the replication slippage, but also lend credence to the model proposed for TLS (Napolitano et al., 2000). They lead to a better understanding of the genome rearrangements that involve short repeated sequences and to the process of mutagenesis, which involves nucleotide loss.
Results
Experimental system
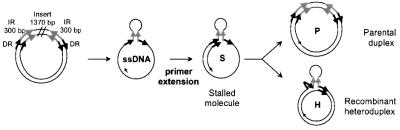
To study the mechanism of replication slippage, we used a previously developed primer extension assay (Canceill and Ehrlich, 1996; Canceill et al., 1999). Its central feature is the ssDNA template that carries a particular structure, composed of two DRs flanking two IRs of 300 bp, which are separated by a 1.4 kb insert (Figure 1). The template is replicated by a polymerase in the presence of labelled dNTPs or the labelled primer, and the reaction products are analysed by agarose gel electrophoresis followed by autoradiography. Faithful copy of the template generates a double-stranded parental molecule (P) that migrates slowly in the gel. A slippage event generates a heteroduplex molecule (H), composed of a parental and a recombinant strand lacking one of the DRs and the 2 kb region between them, which migrates ahead of parental molecules. Replication stalled at the base of the hairpin generates a molecule (S, designated stalled molecules herein) that migrates further than parental and heteroduplex molecules. Precise characterization of reaction products is done, when needed, by restriction analysis and determination of the size of different DNA fragments in sequencing gels, as described (Canceill and Ehrlich, 1996; Canceill et al., 1999).
Fig. 1. Experimental system. Schematic structure of the plasmid and of the primer extension reaction used in this work. The recombination unit of plasmids FX, FXb and FXc consists of two 27 bp DRs (black arrows), flanking a pair of 300 bp IRs (grey arrows) and a central 1370 bp region (insert).
Slippage efficiency depends on the precise position of the DR
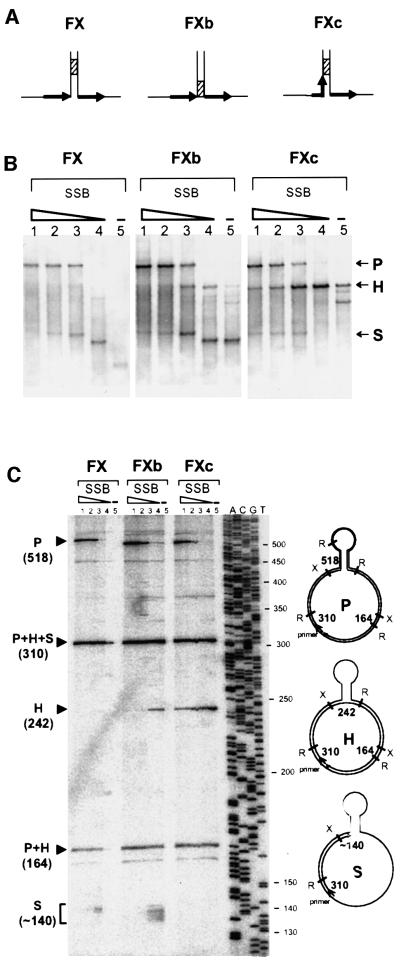
To study the effect of polymerase pausing on slippage, we used three different templates, which all carried a DR of 27 bp. The templates differed only in the base of the hairpin structure formed by annealing of the IR (Figure 2A). Templates FX and FXb had the base flanked by the DR, but contained a sequence of eight G and C residues, denoted the GC clamp, situated either 10 bp from the end or at the very end, respectively (hatched box, Figure 2A). In contrast, the base of the FXc included one DR, which ended precisely at the GC clamp, and was flanked by the other DR. We conjectured that the GC clamp would cause polymerase to pause and thus allow us to establish the relationship of pausing and slippage.
Fig. 2. Comparison of slippage efficiency of three different DNA structures. (A) Schematic representation of the base of the hairpin of each structure. DRs are represented by black arrows and the GC clamp by a hatched box. (B) Analysis of primer extension products by agarose gel electrophoresis. Assays were carried out as described in Materials and methods in the presence of 25 ng of labelled primed template, 1 U of T7 pol and decreasing amounts of SSB. Templates used were FX, FXb and FXc, as indicated at the top of the panel. Lanes 1, 750 ng of SSB (10 times more than the saturating amount); lanes 2, 375 ng of SSB (five times the saturating amount); lanes 3, 75 ng of SSB (saturating amount); lanes 4, 7.5 ng of SSB (10 times less than the saturating amount); lanes 5, no SSB. P, H and S refer to parental, heteroduplex and stalled molecules, respectively. (C) Analysis of primer extension products by denaturing polyacrylamide gel electro phoresis. Precise characterization of the products shown in (B) was done by restriction analysis and determination of the size of the different DNA fragments. Samples are in the same order as in (B). The products were cleaved with XmnI and RsaI. To the right of the figure are schematized the structures of the expected products with the relevant restriction sites (X for XmnI and R for RsaI). The numbers in bold indicate the sizes of the informative restriction segments; the size of the P segment (518) is that for FX; the two other templates, FXb and FXc, yield a segment of 508 bases. To the left of the figure are indicated the positions of migration of these segments. The ladder on the right of the gel corresponds to the sequence of M13mp18 with primer –40.
Replication was carried out by T7 pol in the presence of different SSB concentrations. SSB is required for efficient DNA synthesis by T7 pol, especially at low polymerase concentrations. It inhibits slippage when present above the saturating amount (the amount necessary to cover the ssDNA entirely), by stimulation of the strand displacement activity of T7 pol, which enables the polymerase to enter the hairpin (Canceill et al., 1999). The results are presented in Figure 2B. With FX, the parental molecules were detected only at high SSB concentrations, whereas the stalled molecules were largely predominant at low SSB concentrations (Figure 2B, left). Interestingly, almost no heteroduplex molecules were formed under any condition. A different result was obtained with FXb, where heteroduplex molecules represented a substantial proportion of the replication products, except at the highest SSB concentration at which the parental molecules were essentially the only product. Yet another result was obtained with FXc, where the heteroduplex molecules were the main product at all SSB concentrations except the highest, at which the parental molecules predominated. Precise characterization of the products shown in Figure 2B was done by restriction analysis and determination of the size of the different DNA fragments on a sequencing gel (Figure 2C). Similar results were obtained with E.coli polymerases I, II and III and with T4 pol, which all yielded a higher amount of heteroduplex molecules with FXc template compared with FXb and FX (Canceill et al., 1999 and data not shown).
Two main conclusions can be drawn from these experiments. First, the efficiency of slippage depends on the position of the DR relative to the foot of the hairpin and to the GC clamp. It is higher when the DR is part of the hairpin and right before the GC clamp, as in FXc. Secondly, irrespective of the differences, a common feature of the results obtained with the three templates is the negative correlation between the amounts of stalled and heteroduplex molecules. Such an inverse correlation would be expected if the former were precursors of the latter. An inefficient conversion would result in accumulation of the precursor and absence of the product, while an efficient conversion would lead to accumulation of the product and disappearance of the precursor.
Stalled molecules are intermediates in replication slippage
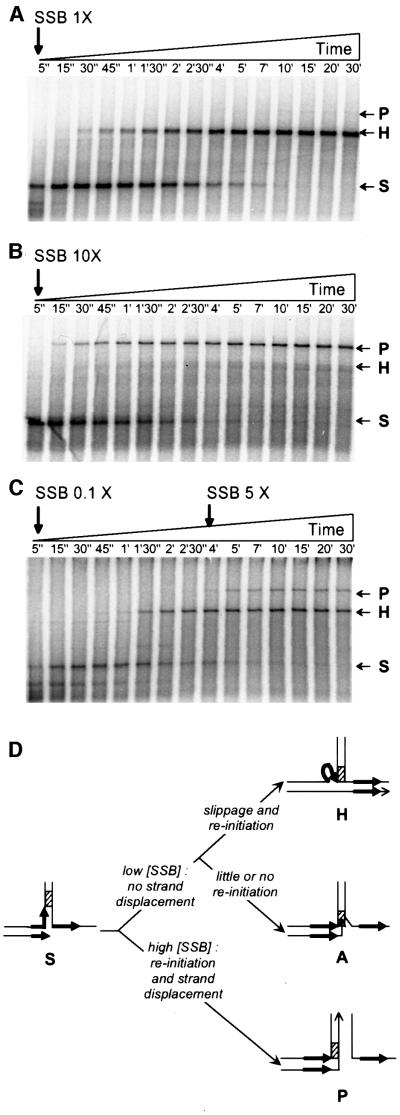
The experiments presented above suggest a precursor– product relationship between the stalled and heteroduplex molecules. To confirm this hypothesis and establish that stalled molecules are intermediates in replication slippage, we replicated the FXc template with T7 pol at low SSB concentration, where the final products are only the heteroduplex molecules (see above; Figure 2B, right). A kinetics experiment revealed that stalled molecules were formed first and disappeared thereafter, concomitant with the appearance of heteroduplex molecules (Figure 3A). As expected, no parental molecules were detected. This shows that stalled molecules are precursors of heteroduplex molecules and are thus intermediates in replication slippage (see scheme in Figure 3D, top).
Fig. 3. Stalled molecules can be converted into either heteroduplex or parental molecules. Primer extension was carried out as described in Materials and methods in the presence of 7.5 U of T7 pol, 375 ng of labelled primed template FXc and different amounts of SSB. Aliquots were withdrawn at the times indicated at the top of each panel and loaded on agarose gels. (A) A 1.12 µg aliquot of SSB (saturating amount). (B) An 11.2 µg aliquot of SSB (10 times more than the saturating amount). (C) A 112.5 ng aliquot of SSB at the beginning of the reaction followed by the addition of 2.6 µg after 4 min. (D) Schematic representation of the base of the hairpin showing the possible evolution of stalled molecules. DRs are represented by black arrows and the GC clamp by a hatched box. As above, P, H and S refer to parental, heteroduplex and stalled molecules, respectively, whereas A stands for abortive products (see text for details).
A similar experiment was carried out at high SSB concentration, which favours synthesis of parental molecules. Stalled molecules were again formed first and disappeared when the parental molecules appeared (Figure 3B). This shows that stalled molecules can be converted into parental molecules under appropriate conditions (see scheme in Figure 3D, bottom). Finally, to confirm that stalled molecules can be converted into either heteroduplex or parental molecules, replication was initiated at low SSB concentration to allow formation of only heteroduplex molecules, and an excess of SSB was added later (Figure 3C). As expected, stalled molecules were formed first and were partially converted to heteroduplex molecules. Addition of SSB arrested further synthesis of heteroduplex molecules and led to the appearance of the parental molecules in parallel with the disappearance of the remaining stalled molecules. Occasionally, a limited amount of stalled molecules was not converted into either parental or heteroduplex molecules (abortive products, see scheme in Figure 3D, middle). These molecules result from inefficient progression of the polymerase within the hairpin, leading to their accumulation at the end of the reaction (see below).
We conclude that formation of heteroduplex molecules by replication slippage and of parental molecules can occur upon pausing of the DNA polymerase at the foot of the hairpin structure.
Multiple pausing sites within a repeated region favour slippage
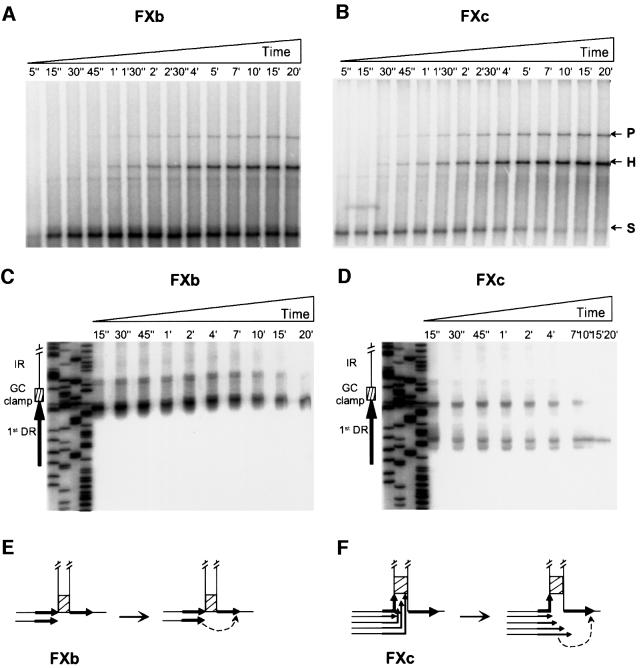
We observed that slippage efficiency depends on the position of the DR relative to the foot of the hairpin and the GC clamp (Figure 2). This observation may be explained by the occurrence of polymerase stalling at the GC clamp. The stalling would take place outside the DR on FX and at its end on FXb and FXc. The tip of the newly synthe sized strand would not be complementary to the second DR on FX and would therefore be unable to anneal to it, leading to the formation of abortive products (see scheme in Figure 3D, middle). In contrast, on FXb and FXc, annealing of the tip of the newly synthesized strand to the second DR would be possible and would lead to the appearance of heteroduplex molecules at the expense of the stalled molecules. To test the above hypothesis and to clarify the difference between the slippage efficiency on FXb and FXc, we followed the kinetics of replication of these two templates by T7 pol in the presence of SSB amounts that allow formation of both the parental and heteroduplex molecules. With both FXb and FXc, stalled molecules appeared very early (Figure 4A and B). However, with FXb, they persisted throughout the reaction, diminishing only slightly in amount with the appearance of the heteroduplex and parental molecules (Figure 4A), while with FXc they disappeared almost totally with the appearance of the heteroduplex and parental molecules (Figure 4B). Similar results were obtained with E.coli pol III HE and T4 pol (data not shown).
Fig. 4. Analysis of stalled molecules on two different DNA structures. Primer extension was carried out as described in Materials and methods in the presence of 375 ng of labelled primed template FXb (A and C) or FXc (B and D), 1.12 µg of SSB (saturating amount) and 7.5 U of T7 pol. Aliquots were withdrawn at the times indicated at the top of each panel and loaded on either agarose gels (A and B) or sequencing gels (C and D). An interpretation of the results is schematized in (E) and (F) for each structure (only the bases of the hairpins are represented). DRs are represented by black arrows, the GC clamp by a hatched box and the annealing event by a dotted arrow. In (E), only one replication intermediate is represented because only one main stop position (at the end of the DR) was found. In (F), several replication intermediates are represented because multiple stops occurred inside the DR. See text for details.
The above data indicate that T7 pol stalls with similar efficiency on FXb and FXc, since the amount of stalled molecules at the early times of replication is similar. However, stalling on FXc apparently generates molecules that can be converted to a terminal replication product (heteroduplex and parental molecules), whereas stalling on FXb generates molecules that are refractory to such conversion (Figure 3D, middle). To determine whether these differences are due to a difference in polymerase pause sites on the two templates, the samples from Figure 4A and B were analysed on a sequencing gel. As shown in Figure 4C, at the beginning of the reaction, polymerase paused on FXb mostly at the very end of the first DR, just in front of the GC clamp. However, as the reaction progressed, pausing took place one or two nucleotides further along, within the GC clamp. In contrast, a large proportion of pause sites on FXc were within the first DR, in a region included in the hairpin, or at its end, right in front of the GC clamp (Figure 4D). The difference in the pause sites suggests that the polymerase is arrested not only at the GC clamp, but also upon encounter with the duplex DNA formed by annealing of the IR. We propose that the molecules formed by polymerase stalling on the FXc template can be converted to heteroduplex molecules because most of the arrests are within the first DR (Figure 4F). The tip of the newly synthesized strand can therefore anneal to the second DR and be extended. Consequently, replication slippage on FXc is efficient (Figure 4F). In contrast, with FXb, annealing and extension can occur on a fraction of stalled molecules only, in which polymerase arrest occurred at the first DR and not beyond it, and the slippage is therefore less efficient overall (Figure 4E).
Replication slippage requires DNA polymerase dissociation
After pausing at the base of the hairpin, the polymerase could either remain bound to the DNA template or dissociate from the DNA. To distinguish between the two possibilities, we replicated FXc template with T7 pol in the presence of heparin, a trapping agent able to bind free DNA polymerase (Reddy et al., 1992). Heparin addition prevents the polymerase from reassociating with the labelled primed template once it has dissociated and, thus, any given primed template is extended by only one polymerase molecule. A limiting amount of SSB was used to allow the formation of heteroduplex molecules only. A labelled primer was annealed to FXc and the replication was initiated in the absence of dTTP. Only six nucleotides should be added to the primer under these conditions and the polymerase should remain bound to the primed template thereafter. Heparin was then added simultaneously with dTTP, which allows the synthesis to proceed; aliquots were withdrawn at different times and analysed on an agarose gel (Figure 5A). Before heparin addition, a single fast migrating band was observed, corresponding to the primed template elongated in the presence of three nucleotides. After addition of heparin and dTTP, a different band was found, corresponding to the stalled molecules, formed by replication of the 1.2 kb region that separates the primer from the hairpin (Figure 5B). The intensity of this band remained constant, both at the initial SSB concentrations and upon addition of excess SSB, which should lead to the synthesis of parental molecules. We conclude that the polymerase arrested by the hairpin structure dissociates from DNA and is thus trapped by the heparin. This shows that replication slippage involves dissociation of the polymerase upon arrest.
Fig. 5. Stalled molecules accumulate in the presence of a DNA polymerase trapping agent. (A) Primer extension was carried out as described in Materials and methods in the presence of 5 U of T7 pol, 250 ng of labelled primed template FXc, 75 ng of SSB (10 times less than the saturating amount) and three deoxynucleotides (dATP, dCTP and dGTP). The reaction was started by the simultaneous addition of the fourth deoxynucleotide (dTTP) and 5–10 µg of heparin. Aliquots were withdrawn at the times indicated at the top of each well and loaded on an agarose gel. After 4 min, 2.25 µg of SSB were added (five times more than the saturating amount), and 10 µl aliquots were withdrawn until 25 min. The last lane is a control sample incubated for 25 min in the presence of 0.5 U of T7 pol, 25 ng of primed template FXc, 375 ng of SSB and the four dNTPs, but in the absence of heparin. p/t refers to the primer–template. (B) Schematic representation of the base of the hairpin showing the interpretation of the results. In the presence of heparin, the polymerase replicates 1.2 kb until encounter ing the hairpin, where it dissociates. DRs are represented by black arrows, the GC clamp by a hatched box, the labelled primer by an asterisk and the DNA polymerase by a sphere. The cross on the sphere indicates the trapping of the dissociated polymerase by heparin.
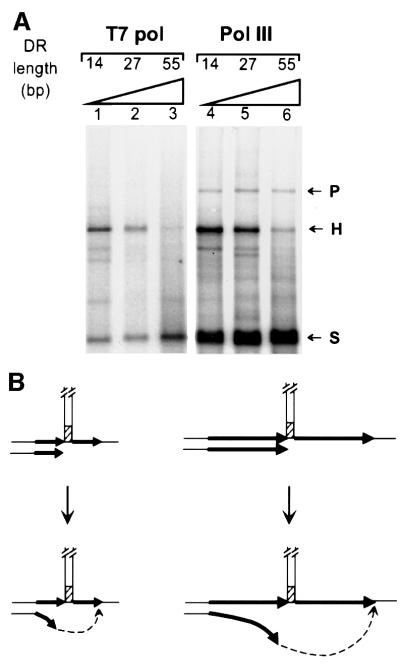
Slippage efficiency is inversely related to the length of the DR
After pausing and dissociation of the polymerase, replication slippage requires remodelling of the replication intermediate, which involves separation of the newly synthesized DR from the template and its pairing to the complementary sequence present at the other side of the hairpin. We analysed this process by studying the effect of the length of the DR on the slippage efficiency of T7 pol. The templates were FXb, which has a DR of 27 bp, and two derivatives, which had DRs of 14 or 55 bp (FXb-14 or FXb-55, respectively). Heteroduplex molecules were detected with the three templates (Figure 6A, lanes 1–3), but their amounts decreased with increasing DR length. Similar results were obtained with E.coli pol III HE (Figure 6A, lanes 4–6). We propose that the decrease in the slippage efficiency is due to the lower efficiency of the pairing process when the distance between the end of the newly synthesized DR and the complementary part of the second DR is greater (Figure 6B). This result suggests that only the tip of the new strand, when pausing occurs within the first DR, is implicated in the remodelling of the replication intermediate required for replication slippage.
Fig. 6. Effect of the length of the DR on replication slippage. (A) A primer extension assay was carried out as described in Materials and methods in the presence of 25 ng of labelled primed template, 75 ng of SSB (saturating amount) and either 0.5 U of T7 pol (lanes 1–3) or 10 U of pol III HE (lanes 4–6). DNA templates were FXb-14 (lanes 1 and 4), FXb-27 (lanes 2 and 5) and FXb-55 (lanes 3 and 6), having DRs of 14, 27 and 55 bp, respectively. (B) Schematic representation of the base of the hairpin indicating the interpretation of the results. Left, short DR (black arrows); right, longer DR (longer black arrows). Dotted arrows represent why increasing the length of the DR increases the distance for the tip of the newly synthesized strand to pair to the second DR after unpairing from the first DR.
Discussion
In this work, we show that replication slippage involves three steps: polymerase pausing within a DR, polymerase dissociation and DNA re-annealing.
Replication slippage requires DNA polymerase pausing within a DR
Our results show that replication slippage requires polymerase pausing within a DR. First, heteroduplex molecules are produced at the expense of the molecules that are formed by polymerase stalling. We have shown in kinetic experiments that molecules resulting from stalling accumulate first and then disappear while the heteroduplex molecules are formed. Secondly, slippage frequency is correlated with the frequency of pausing within the DR, as determined by comparing templates FXc and FXb. We have shown that there are multiple pausing sites within the DR of the former, whereas there is only a single pausing site within the DR of the latter. In parallel, we have observed a higher rate of replication slippage on the former than on the latter template.
In our experiments, polymerase stalling occurs at the foot of the hairpin structure and at a GC-rich sequence close to the base of the hairpin. Previous in vitro and in vivo experiments have shown that secondary structures act as preferential pausing sites of DNA polymerases (Weaver and DePamphilis, 1982; LaDuca et al., 1983; Weaver and DePamphilis, 1984; Bedinger et al., 1989). Secondary structures that cause polymerase pausing include hairpin structures, triplexes and tetraplexes (Ohshima and Wells, 1998). DNA secondary structures are known to be recombination hot spots (Sinden et al., 1991; Leach, 1994; Pinder et al., 1998) and replication fork pause sites are deletion hot spots (Bierne et al., 1991, 1997a; Bebenek et al., 1993; Bierne and Michel, 1994). Secondary structures are formed frequently in microsatellite sequences, which are known to be highly unstable tracts (Mitas, 1997). Recently, pausing of replication forks has been shown to occur at trinucleotide repeats in vivo (Samadashwily et al., 1997) and in vitro (Kang et al., 1995; Ohshima and Wells, 1997). Several triplet repeats have been shown in vitro to generate hairpin structures (Mitas et al., 1995; Hartenstine et al., 2000), and evidence that they are able to adopt such secondary structure is also found in vivo (Darlow and Leach, 1998; Moore et al., 1999; Spiro et al., 1999), and reviewed in Sinden et al. (1999). Our demonstration that pausing is the first step of replication slippage accounts for the reported correlation between replication pausing and frequent rearrangements in repeated sequences.
After pausing, DNA polymerase dissociates
We report here that the presence of a trapping agent in primer extension experiments prevents DNA synthesis from proceeding further than the pause site. This shows that, upon stalling, the polymerase dissociates from the primer–template complex. Previous studies have already suggested polymerase dissociation upon stalling: T4 pol HE dissociates rapidly when it encounters a hairpin (Hacker and Alberts, 1994) and pol III HE dissociates when a challenger template with the accessory protein is present (O’Donnell, 1987; O’Donnell and Studwell, 1990). The β-subunit of pol III HE (the sliding clamp), which encircles the DNA, is unable to traverse secondary structures larger than a few nucleotides (Yao et al., 2000). Consequently, the observation that pol III HE is able to slip on molecules that carry hairpins (Canceill and Ehrlich, 1996) implies that at least the core polymerase dissociates from β and thus eventually from DNA. The same is likely to hold true for T4 pol HE and its gp45 sliding clamp, as well as for T7 pol, which encircles the DNA with the help of the thioredoxin subunit.
Polymerase dissociation upon arrest is also central to the TLS, since the main replicative polymerase arrested by a lesion must be replaced temporarily by a specialized polymerase that is capable of lesion bypass, either directly or after realigning the tip of the newly synthesized strand (Napolitano et al., 2000). The latter process leads to a loss of one or two nucleotides and therefore generates frameshift mutations.
Pairing to the second DR involves only the end of the newly synthesized repeat
In our experimental system, increasing the length of the DR from 14 to 55 bp decreases the slippage efficiency of T7 pol and pol III HE. The observation that pairing of the newly synthesized DR to its complementary sequence is less efficient when DRs are longer, hence further apart, suggests that only the end of the new strand is involved in the pairing step. We suggest that the tip of the new strand melts spontaneously (‘breathes’) after polymerase dissociation, which allows its annealing to the complementary sequence. Efficiency of the annealing might be the limiting step of the slippage process, since only upon annealing can a polymerase re-attach to the tip of the newly synthesized strand and resume replication.
Deletions produced by replication slippage in vivo
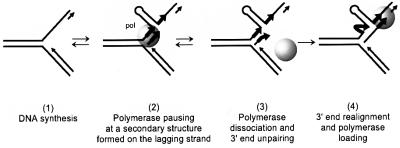
We have observed that slippage occurs during replication of an ssDNA template that carries a hairpin structure flanked by repeated sequences. We speculate that similar slippage takes place in vivo, during synthesis of the lagging DNA strand (Figure 7). Current models of DNA replication in E.coli or phage T7 imply a coordinated leading and lagging strand DNA synthesis carried out by a dimeric polymerase (McHenry, 1991; Debyser et al., 1994). Hairpin structures may form in the lagging strand template rendered single stranded by the progression of the leading strand polymerase (Cromie et al., 2000), and impede the progression of the lagging strand polymerase (Figure 7, step 2). If the polymerase has a low strand displacement activity, it will be unable to enter the hairpin and progress within it, and would dissociate from the DNA (step 3). The 3′ end of the newly synthesized strand would then be able to realign beyond the hairpin, on a complementary sequence, and the polymerase could load again and resume the synthesis (step 4).
Fig. 7. Schematic representation of the slippage process at a replication fork. During DNA synthesis of a repeated sequence (step 1), the polymerase reaches a barrier on the lagging strand (a hairpin structure in our experimental system) and pauses (step 2). Polymerase dissociation occurs (step 3). If the polymerase is not able to disrupt the barrier (with its strand displacement activity), then the tip of the newly synthesized strand can unpair from its template and anneal to the second repeat beyond the barrier, allowing polymerase re-loading and resumption of the synthesis (step 4).
In conclusion, our report provides a deeper insight into molecular events that take place during the process of replication slippage, which underlies many instances of genome rearrangements, in both prokaryotes and eukaryotes. Furthermore, it points to the resemblance of this process to mutagenesis that involves the newly recognized class of polymerases, specialized in bypass of lesions (Friedberg et al., 2000; Napolitano et al., 2000). It thus provides a unifying view of a possible fate of arrested replication polymerases, by either a secondary structure or a chemical lesion, where dissociation of the polymerase allows remodelling of the substrate and thus resumption of DNA synthesis, albeit at the expense of the integrity of the genetic material.
Materials and methods
Proteins
T7 pol and T4 pol were purchased from New England Biolabs. Pol III HE was purified as described (Canceill and Ehrlich, 1996). DNA sequencing was carried out with T7 pol exo– (Sequenase™ Version2, APBiotech) according to the furnished protocol. Escherichia coli SSB was purchased from United States Biochemical (APBiotech). Proteinase K was from Roche Molecular Biochemicals. For all polymerases except pol III, 1 U of enzyme catalyses the incorporation of 10 nmol of total nucleotide into acid-insoluble material in 30 min at 37°C. For pol III HE, 1 U of enzyme catalyses the incorporation of 1 pmol of total nucleotide into acid-insoluble material in 1 min at 37°C.
Chemicals
[α-32P]dATP (3000 Ci/mmol), [α-32P]dCTP (3000 Ci/mmol) and [γ-32P]ATP (6000 Ci/mmol) were purchased from APBiotech or Dupont-NEN. Unlabelled nucleotides were from Pharmacia. Heparin was from Sigma.
ssDNA templates
Plasmids pHP727FX, pHP727FXb and pHP727FXc are represented in Figure 1. Their construction and the preparation of the ssDNA templates have been described previously (Canceill and Ehrlich, 1996).
Primer extension reactions
The primer extension assay has been described previously (Canceill and Ehrlich, 1996; Canceill et al., 1999). Briefly, a primer designated #1233 (24mer) was annealed 1235 bases from the palindrome. All primer extension reactions contained in 10 µl: 25 ng of primed ssDNA, 250 µM dNTP (each) if 32P-labelled primer was used, or 250 µM dGTP and dTTP (each) and 50 µM (2.5 µCi) [α-32P] dATP and [α-32P] dCTP if unlabelled primer was used. SSB and DNA polymerase were added to the reaction mixture as indicated in the figure legends. Reactions were pre-incubated for 5 min at 37°C in the presence or absence of SSB, with all the other components, before DNA polymerase addition. The reaction buffers contained, in addition to 30 mM NaCl brought by the primed ssDNA, the following ingredients. For T7 pol: 20 mM Tris–HCl pH 7.5, 10 mM MgCl2, 1 mM dithiothreitol (DTT), 50 µg/ml bovine serum albumin (BSA), 10% glycerol. For pol III HE: 20 mM Tris–HCl pH 7.5, 10 mM MgCl2, 2 mM DTT, 2 mM ATP, 100 µg/ml BSA, 5–30 mM NaCl, 10% glycerol. After 15 min at 37°C, the synthesis was arrested by the addition of 25 mM EDTA and 500 µg/ml proteinase K and the mixture was incubated further for 15 min at 55°C. Reaction products were then analysed by electrophoresis through 0.8% agarose gels (Seakem GTG or Ultrapure BRL), run in TAE buffer (40 mM Tris acetate, 1 mM EDTA pH 8.3) at 2 V/cm for 16 h. For analysis of the reaction products on a sequencing gel, proteinase K was inactivated by addition of 2 mM 4-(2-aminoethyl)-benzenesulfonyl fluoride hydrochloride (AEBSF; ICN Biomedicals), incubated for 10 min at room temperature and the mixture dialysed on a micromembrane (Millipore VS; 0.025 µM) for 20 min. The products were then cleaved with restriction enzymes (XmnI and RsaI) and analysed by electrophoresis through 6% acrylamide–urea sequencing gels (National Diagnostics) run in TBE buffer (90 mM Tris borate, 2 mM EDTA pH 8.3) at 60 W/65 mA for 2–3 h. DNA was visualized by direct exposure of the dried gels to Storage Phosphor Screens and analysed on a STORM (Molecular Dynamics).
Primer extension was performed as described above, except that the reaction volume and the amounts of all the components were scaled up 10–15 times (final volume of 100–150 µl) and 10–15 µl aliquots were withdrawn at the times indicated in the figures.
DNA polymerase dissociation
To analyse DNA polymerase dissociation during slippage, primer extensions were performed using labelled primed ssDNA, a limiting amount of DNA polymerase and heparin as a trapping agent. In a volume of 50 µl, 250 ng of primed labelled ssDNA, 75 ng–1.5 µg of SSB and 200 µM dATP, dCTP and dGTP were pre-incubated for 2 min at 37°C in T7 pol buffer, before addition of 5 U of T7 pol and further incubation for 5 min. A second 50 µl reaction mixture, containing 200 µM dATP, dCTP and dGTP, 400 µM dTTP and 50–100 µg/ml heparin in T7 pol buffer, was pre-incubated for 5 min at 37°C and then added to the first mixture. Incubation was continued at 37°C and 10 µl aliquots were withdrawn at the times indicated in the figure legends. For each aliquot, the reactions were stopped and analysed on an agarose gel as described above.
Acknowledgments
Acknowledgements
We are deeply grateful to Bénédicte Michel for invaluable help during preparation of this manuscript. We acknowledge Marie-Agnès Petit for encouragement during this work. E.V. was supported by a postdoctoral fellowship from the Spanish Ministerio de Educación y Cultura.
References
- Baynton K. and Fuchs,R.P. (2000) Lesions in DNA: hurdles for polymerases. Trends Biochem. Sci., 25, 74–79. [DOI] [PubMed] [Google Scholar]
- Bebenek K., Abbotts,J., Wilson,S.H. and Kunkel,T.A. (1993) Error-prone polymerization by HIV-1 reverse transcriptase. Contribution of template–primer misalignment, miscoding and termination probability to mutational hot spots. J. Biol. Chem., 268, 10324–10334. [PubMed] [Google Scholar]
- Bedinger P., Munn,M. and Alberts,B.M. (1989) Sequence-specific pausing during in vitro DNA replication on double-stranded DNA templates. J. Biol. Chem., 264, 16880–16886. [PubMed] [Google Scholar]
- Bierne H. and Michel,B. (1994) When replication forks stop. Mol. Microbiol., 13, 17–23. [DOI] [PubMed] [Google Scholar]
- Bierne H., Ehrlich,S.D. and Michel,B. (1991) The replication termination signal terB of the Escherichia coli chromosome is a deletion hot spot. EMBO J., 10, 2699–2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne H., Ehrlich,S.D. and Michel,B. (1997a) Deletions at stalled replication forks occur by two different pathways. EMBO J., 16, 3332–3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne H., Vilette,D., Ehrlich,S.D. and Michel,B. (1997b) Isolation of a dnaE mutation which enhances RecA-independent homologous recombination in the Escherichia coli chromosome. Mol. Microbiol., 24, 1225–1234. [DOI] [PubMed] [Google Scholar]
- Canceill D. and Ehrlich,S.D. (1996) Copy-choice recombination mediated by DNA polymerase III holoenzyme from Escherichia coli. Proc. Natl Acad. Sci. USA, 93, 6647–6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canceill D., Viguera,E. and Ehrlich,S.D. (1999) Replication slippage of different DNA polymerases is inversely related to their strand displacement efficiency. J. Biol. Chem., 274, 27481–27490. [DOI] [PubMed] [Google Scholar]
- Cromie G.A., Millar,C.B., Schmidt,K.H. and Leach,D.R. (2000) Palindromes as substrates for multiple pathways of recombination in Escherichia coli. Genetics, 154, 513–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Alencon E., Petranovic,M., Michel,B., Noirot,P., Aucouturier,A., Uzest,M. and Ehrlich,S.D. (1994) Copy-choice illegitimate DNA recombination revisited. EMBO J., 13, 2725–2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlow J.M. and Leach,D.R. (1998) Evidence for two preferred hairpin folding patterns in d(CGG).d(CCG) repeat tracts in vivo. J. Mol. Biol., 275, 17–23. [DOI] [PubMed] [Google Scholar]
- Debyser Z., Tabor,S. and Richardson,C.C. (1994) Coordination of leading and lagging strand DNA synthesis at the replication fork of bacteriophage T7. Cell, 77, 157–166. [DOI] [PubMed] [Google Scholar]
- Djian P. (1998) Evolution of simple repeats in DNA and their relation to human disease. Cell, 94, 155–160. [DOI] [PubMed] [Google Scholar]
- Ehrlich S.D., Bierne,H., d’Alencon,E., Vilette,D., Petranovic,M., Noirot,P. and Michel,B. (1993) Mechanisms of illegitimate recombination. Gene, 135, 161–166. [DOI] [PubMed] [Google Scholar]
- Friedberg E.C., Feaver,W.J. and Gerlach,V.L. (2000) The many faces of DNA polymerases: strategies for mutagenesis and for mutational avoidance. Proc. Natl Acad. Sci. USA, 97, 5681–5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker K.J. and Alberts,B.M. (1994) The rapid dissociation of the T4 DNA polymerase holoenzyme when stopped by a DNA hairpin helix. A model for polymerase release following the termination of each Okazaki fragment. J. Biol. Chem., 269, 24221–24228. [PubMed] [Google Scholar]
- Hancock J.M. and Santibanez-Koref,M.F. (1998) Trinucleotide expansion diseases in the context of micro- and minisatellite evolution. EMBO J., 17, 5521–5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartenstine M.J., Goodman,M.F. and Petruska,J. (2000) Base stacking and even/odd behavior of hairpin loops in DNA triplet repeat slippage and expansion with DNA polymerase. J. Biol. Chem., 275, 18382–18390. [DOI] [PubMed] [Google Scholar]
- Kang S., Ohshima,K., Shimizu,M., Amirhaeri,S. and Wells,R.D. (1995) Pausing of DNA synthesis in vitro at specific loci in CTG and CGG triplet repeats from human hereditary disease genes. J. Biol. Chem., 270, 27014–27021. [DOI] [PubMed] [Google Scholar]
- LaDuca R.J., Fay,P.J., Chuang,C., McHenry,C.S. and Bambara,R.A. (1983) Site-specific pausing of deoxyribonucleic acid synthesis catalyzed by four forms of Escherichia coli DNA polymerase III. Biochemistry, 22, 5177–5188. [DOI] [PubMed] [Google Scholar]
- Leach D.R. (1994) Long DNA palindromes, cruciform structures, genetic instability and secondary structure repair. BioEssays, 16, 893–900. [DOI] [PubMed] [Google Scholar]
- Lovett S.T., Drapkin,P.T., Sutera,V.A.,Jr and Gluckman-Peskind,T.J. (1993) A sister-strand exchange mechanism for recA-independent deletion of repeated DNA sequences in Escherichia coli. Genetics, 135, 631–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHenry C.S. (1991) DNA polymerase III holoenzyme. Components, structure and mechanism of a true replicative complex. J. Biol. Chem., 266, 19127–19130. [PubMed] [Google Scholar]
- Michel B. (2000) Replication fork arrest and DNA recombination. Trends Biochem. Sci., 25, 173–178. [DOI] [PubMed] [Google Scholar]
- Mitas M. (1997) Trinucleotide repeats associated with human disease. Nucleic Acids Res., 25, 2245–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitas M., Yu,A., Dill,J. and Haworth,I.S. (1995) The trinucleotide repeat sequence d(CGG)15 forms a heat-stable hairpin containing Gsyn⋅Ganti base pairs. Biochemistry, 34, 12803–12811. [DOI] [PubMed] [Google Scholar]
- Moore H., Greenwell,P.W., Liu,C.P., Arnheim,N. and Petes,T.D. (1999) Triplet repeats form secondary structures that escape DNA repair in yeast. Proc. Natl Acad. Sci. USA, 96, 1504–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napolitano R., Janel-Bintz,R., Wagner,J. and Fuchs,R.P. (2000) All three SOS-inducible DNA polymerases (pol II, pol IV and pol V) are involved in induced mutagenesis. EMBO J., 19, 6259–6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell M.E. (1987) Accessory proteins bind a primed template and mediate rapid cycling of DNA polymerase III holoenzyme from Escherichia coli. J. Biol. Chem., 262, 16558–16565. [PubMed] [Google Scholar]
- O’Donnell M. and Studwell,P.S. (1990) Total reconstitution of DNA polymerase III holoenzyme reveals dual accessory protein clamps. J. Biol. Chem., 265, 1179–1187. [PubMed] [Google Scholar]
- Ohshima K. and Wells,R.D. (1997) Hairpin formation during DNA synthesis primer realignment in vitro in triplet repeat sequences from human hereditary disease genes. J. Biol. Chem., 272, 16798–16806. [DOI] [PubMed] [Google Scholar]
- Ohshima K. and Wells,R. (1998) In vitro DNA synthesis of triplet repeat sequences. In Wells,R.D. and Warren,S.T. (eds), Genetic Instabilities and Hereditary Neurological Diseases. Academic Press, San Diego, CA, pp. 717–735.
- Pearson C.E. and Sinden,R.R. (1998a) Slipped strand DNA, dynamic mutations and human disease. In Wells,R.D. and Warren,S.T. (eds), Genetic Instabilities and Hereditary Neurological Diseases. Academic Press, San Diego, CA, pp. 585–621.
- Pearson C.E. and Sinden,R.R. (1998b) Trinucleotide repeat DNA structures: dynamic mutations from dynamic DNA. Curr. Opin. Struct. Biol., 8, 321–330. [DOI] [PubMed] [Google Scholar]
- Petruska J., Hartenstine,M.J. and Goodman,M.F. (1998) Analysis of strand slippage in DNA polymerase expansions of CAG/CTG triplet repeats associated with neurodegenerative disease. J. Biol. Chem., 273, 5204–5210. [DOI] [PubMed] [Google Scholar]
- Pinder D.J., Blake,C.E., Lindsey,J.C. and Leach,D.R. (1998) Replication strand preference for deletions associated with DNA palindromes. Mol. Microbiol., 28, 719–727. [DOI] [PubMed] [Google Scholar]
- Reddy M.K., Weitzel,S.E. and von Hippel,P.H. (1992) Processive proofreading is intrinsic to T4 DNA polymerase. J. Biol. Chem., 267, 14157–14166. [PubMed] [Google Scholar]
- Richards R.I. and Sutherland,G.R. (1992) Dynamic mutations: a new class of mutations causing human disease. Cell, 70, 709–712. [DOI] [PubMed] [Google Scholar]
- Richards R.I. and Sutherland,G.R. (1994) Simple repeat DNA is not replicated simply. Nature Genet., 6, 114–116. [DOI] [PubMed] [Google Scholar]
- Samadashwily G.M., Raca,G. and Mirkin,S.M. (1997) Trinucleotide repeats affect DNA replication in vivo. Nature Genet., 17, 298–304. [DOI] [PubMed] [Google Scholar]
- Seki M., Akiyama,M., Sugaya,Y., Ohtsubo,E. and Maki,H. (1999) Strand asymmetry of +1 frameshift mutagenesis at a homopolymeric run by DNA polymerase III holoenzyme of Escherichia coli. J. Biol. Chem., 274, 33313–33319. [DOI] [PubMed] [Google Scholar]
- Sinden R.R., Zheng,G.X., Brankamp,R.G. and Allen,K.N. (1991) On the deletion of inverted repeated DNA in Escherichia coli: effects of length, thermal stability and cruciform formation in vivo. Genetics, 129, 991–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinden R.R., Hashem,V.I. and Rosche,W.A. (1999) DNA-directed mutations. Leading and lagging strand specificity. Ann. N. Y. Acad. Sci., 870, 173–189. [DOI] [PubMed] [Google Scholar]
- Spiro C. et al. (1999) Inhibition of FEN-1 processing by DNA secondary structure at trinucleotide repeats. Mol. Cell, 4, 1079–1085. [DOI] [PubMed] [Google Scholar]
- Tautz D. and Schlotterer,C. (1994) Simple sequences. Curr. Opin. Genet. Dev., 4, 832–837. [DOI] [PubMed] [Google Scholar]
- Toth G., Gaspari,Z. and Jurka,J. (2000) Microsatellites in different eukaryotic genomes: survey and analysis. Genome Res., 10, 967–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Belkum A., Scherer,S., van Alphen,L. and Verbrugh,H. (1998) Short-sequence DNA repeats in prokaryotic genomes. Microbiol. Mol. Biol. Rev., 62, 275–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver D.T. and DePamphilis,M.L. (1982) Specific sequences in native DNA that arrest synthesis by DNA polymerase α. J. Biol. Chem., 257, 2075–2086. [PubMed] [Google Scholar]
- Weaver D.T. and DePamphilis,M.L. (1984) The role of palindromic and non-palindromic sequences in arresting DNA synthesis in vitro and in vivo. J. Mol. Biol., 180, 961–986. [DOI] [PubMed] [Google Scholar]
- Wells R.D. (1996) Molecular basis of genetic instability of triplet repeats. J. Biol. Chem., 271, 2875–2878. [DOI] [PubMed] [Google Scholar]
- Yao N., Hurwitz,J. and O’Donnell,M. (2000) Dynamics of β and proliferating cell nuclear antigen sliding clamps in traversing DNA secondary structure. J. Biol. Chem., 275, 1421–1432. [DOI] [PubMed] [Google Scholar]