Abstract
The Balbiani ring 3 (BR3) gene contains 38 introns, and more than half of them are co-transcriptionally excised. We have determined the in situ structure of the active BR3 gene by electron tomography. Each of the 20–25 nascent transcripts on the gene is present together with splicing factors and the RNA polymerase II in a nascent transcript and splicing complex, here called the NTS complex. The results indicate that extensive changes in overall shape, substructure and molecular mass take place repeatedly within an NTS complex as it moves along the gene. The volume and calculated mass of the NTS complexes show that, maximally, one complete spliceosome is assembled on the multi-intron transcript at any given time point. The structural data show that the spliceosome is not a structurally well-defined unit in situ and that the C-terminal domain of the elongating RNA polymerase II cannot carry spliceosomal components for all introns in the BR3 transcript. Our data indicate that spliceosomal factors are continuously added to and released from the NTS complexes during transcription elongation.
Keywords: gene expression/immunoelectron microscopy/pre-mRNA processing
Introduction
Extensive biochemical and genetic analysis has shown that five small nuclear RNAs (snRNAs) and at least 50 different proteins are needed for splicing of pre-mRNA (reviewed by Will and Lührmann, 1997). In mammalian nuclear extracts, splicing takes place in a multi-component splicing complex known as the spliceosome, and this spliceosome is assembled in an ordered and stepwise pathway (reviewed by Krämer, 1996; Staley and Guthrie, 1998; Reed, 2000). The addition of specific snRNA– protein complexes (snRNPs) and proteins characterizes each step, and the assembly pathway is highly conserved.
Spliceosomes assembled in vitro have been investigated by electron microscopy in yeast (Clark et al., 1988) and in mammals (Reed et al., 1988; Sibbald et al., 1993; Furman and Glitz, 1995). Large complexes, apparently consisting of several spliceosomes, have been purified from mammalian nuclei (Sperling et al., 1997). More detailed structural information is available for purified snRNPs (Kastner, 1996; Krämer et al., 1999), in particular for individual snRNP proteins bound to small stretches of RNA (see Kambach et al., 1999a) and the snRNP protein core complex (Kambach et al., 1999b).
The assembly process and the structure of spliceosomes in vivo are less well known. A subset of pre-mRNAs may be processed in association with clusters of interchromatin granules (reviewed by Lawrence et al., 1993), but splicing seems mainly to take place at, or close to, the active genes. Spliceosomal components may therefore be recruited from storage sites to the transcribing genes (Jiménez-Garcia and Spector, 1993; Baurén et al., 1996; Misteli et al., 1997; Zeng et al., 1997). Splicing components have been located to active genes, both by light microscopy (e.g. Sass and Pederson, 1984; Wu et al., 1991; Zhang et al., 1994; Baurén et al., 1996; Neugebauer and Roth, 1997a) and by electron microscopy (EM) (Kiseleva et al., 1994; Puvion and Puvion-Dutilleul, 1996; Cmarko et al., 1999). pre-mRNA can be co-transcriptional spliced, as first demonstrated in Miller spreads (Osheim et al., 1985) and later further shown by direct analysis of isolated nascent pre-mRNA (Baurén and Wieslander, 1994; Wuarin and Schibler, 1994).
The large subunit of RNA polymerase II has a C-terminal domain (CTD), consisting of imperfect heptapeptide repeats that can be hyper- or hypophosphorylated. The hyperphosphorylated RNA polymerase II (Hirose et al., 1999), as well as the full-length CTD alone (Zeng and Berget, 2000), stimulates splicing in vitro. Several reports have further established that RNA polymerase II is involved in capping, splicing and polyadenylation (reviewed in Corden and Patturajan, 1997; Neugebauer and Roth, 1997b; Shuman, 1997; Steinmetz, 1997; Bentley, 1999). Combined, the available data show that transcription and pre-mRNA processing are closely coupled in the cell nucleus, and that the elongating RNA polymerase II should be present in a molecular complex that also contains the nascent transcript and different RNA processing factors.
In addition, rearrangements occur within the spliceosome, as has been shown by analysis of the RNA–RNA interactions, protein–protein interactions and RNA– protein interactions (Staley and Guthrie, 1998; Reed, 2000).
In the model of co-transcriptional RNA processing, processing components, including splicing factors, bind to the hyperphosphorylated CTD of the RNA polymerase II, from which they are delivered to the nascent transcript during transcription (see Neugebauer and Roth, 1997b; Bentley, 1999). However, several aspects of this model remain to be analyzed in vivo. It has, for example, not been shown how the assembly of the spliceosome on the nascent transcripts relates to the spliceosome assembly and rearrangement on a preformed transcript observed in vitro. It is also unclear where the RNA polymerase II picks up splicing factors and to what extent the elongating RNA polymerase carries splicing factors, particularly during transcription of multi-intron genes. It is not either known whether many spliceosomes do assemble on a multi-intron nascent transcript, given that the nascent transcript, spliceosomes and the elongating RNA polymerase II should be present in the same supramolecular complex.
To learn more about the structural basis of the coordinated transcription and splicing, it is important to study the in vivo structure of active genes for which we know the exon–intron structure and the splicing characteristics. The Balbiani ring 3 (BR3) gene in Chironomus tentans is 10.9 kb long and encodes a secretory protein. Thirty-eight introns are approximately evenly spaced throughout the whole gene (Paulsson et al., 1990), and more than half of these introns are co-transcriptionally excised (Wetterberg et al., 1996). In this system, it is possible to identify the actively transcribing BR3 gene in the morphologically intact cell nucleus. This has enabled us to use electron tomography to reconstruct the three-dimensional (3D) structure of the active BR3 gene. We demonstrate that splicing of the nascent transcript occurs within a defined, but dynamic, supramolecular complex. In this complex, only one complete spliceosome is assembled on a nascent BR3 transcript at a given time. Furthermore, the elongating RNA polymerase II is most unlikely to carry spliceosomal factors for all introns in the multi-intron BR3 transcript.
Results
The structure of the active BR3 gene
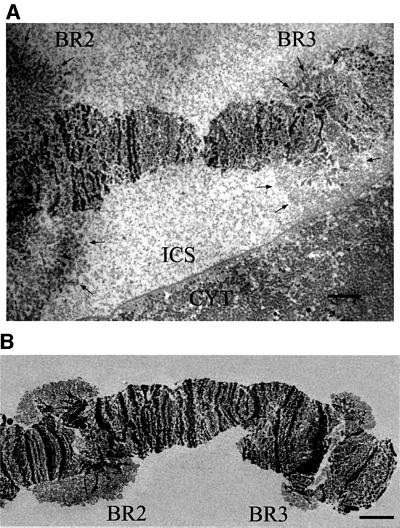
The transcriptionally active BR3 gene can be identified from the chromosome morphology, the banding pattern, and the position of the BR3 locus relative to the active Balbiani ring 1 (BR1) and Balbiani ring 2 (BR2) loci on the same chromosome. The active BR3 gene can be identified both in sections through nuclei of the intact salivary gland cells (Figure 1A) and in isolated chromosome IV (Figure 1B). There are ∼8000 copies of the BR3 gene in the polytene chromosome. The active BR3 locus (chromosomal puff) consists of a large number of decondensed transcribing chromatin fibers, radiating from and returning to central, compact chromatin (Figures 1 and 2). In agreement, it has been shown that the BR3 gene is present throughout the entire active BR3 locus (Paulsson et al., 1990). Each 50–100 nm section through the locus corresponds to 1/400–1/200 of the diameter of the whole gene locus, and consists of a large number of short segments of the active BR3 gene. Patches of compact chromatin were also seen, mainly in the center of the locus, but also farther out. The structures of the gene segments were identical in isolated chromosome IV and in chromosome IV in sections of the gland cell at the level of resolution used in our analysis (see Materials and methods; Table III). In many areas, individual gene segments were clearly seen separate from all other segments, and in this case formed granular fibers that ran in all different directions (two examples are shown in Figure 2, labeled 1 and 2). These fibers could only be followed for limited stretches on a single electron micrograph since this is a 2D projection of a sectioned 3D structure. We analyzed 43 individual fibers, located at many different positions in the locus and from several different in situ chromosome preparations. The lengths of these fibers were between 100 and 280 nm. Each fiber had a maximal diameter of ∼50 nm, but the fibers did not have a constant diameter. Instead, they consisted of similar sized granules, spaced on average 53 ± 9 nm apart. At higher magnification, the granules had an obvious substructure. They contained clusters of denser material, more or less round substructures, each with a diameter <10 nm (examples are indicated by arrowheads in Figure 2B). In the granules, there is also less dense material. The diameter of whole granules varied from 20 to 50 nm. Less dense axially placed material lay between the granules, connecting them. These observations are supported by the 3D analyses below (see Figures 3 and 5).
Fig. 1. Identification of the active BR3 gene. Electron micrograph of in situ (A) and isolated (B) chromosome IV showing the location of the active BR2 and BR3 genes. Arrows in (A) indicate the border between the gene loci and the interchromatin space (ICS). The cytoplasm (CYT) is also shown in (A). Scale bar, 5 µm.
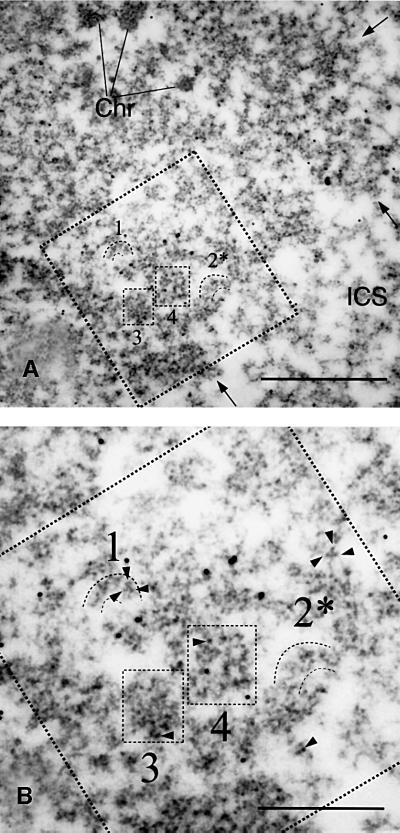
Fig. 2. 2D structure of the active BR3 gene in situ. (A) Electron micrograph of a section through the active BR3 locus. Compact chromatin (Chr) is marked. Arrows show the border between the interchromatin space (ICS) and the decondensed, actively transcribing gene locus. The large square (dashed lines) shows the digitized area. Individual gene segments are marked 1 and 2, while 3 and 4 show different levels of gene segment juxtaposition (see text). In (B), the digitized area is shown at larger magnification and the dense substructures (arrowheads) within the NTS complexes are seen. The scale bar is 1000 nm in (A) and 500 nm in (B). The 10 nm colloidal gold markers are reference points for 3D reconstruction.
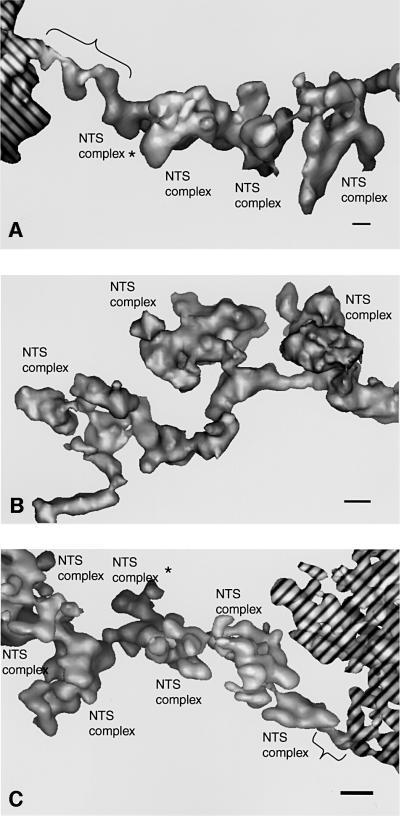
Fig. 3. 3D reconstructions of individual segments of the active BR3 gene. (A) Gene segment with a long 10 nm axis connection to compact chromatin. (B) Internal part of a gene segment (corresponding to the segment marked 2 in Figure 2 and shown in the same projection). (C) Gene segment with a short 10 nm axis connection to compact chromatin. The compact chromatin is hashed. Projections in (A) and (C) represent compromises to show both the 10 nm connection to compact chromatin (indicated by braces) and the NTS complexes. NTS complexes marked with an asterisk are partially hidden. The structures were lowpass filtered to 5 nm and shown as surface rendered objects. Scale bar, 10 nm.
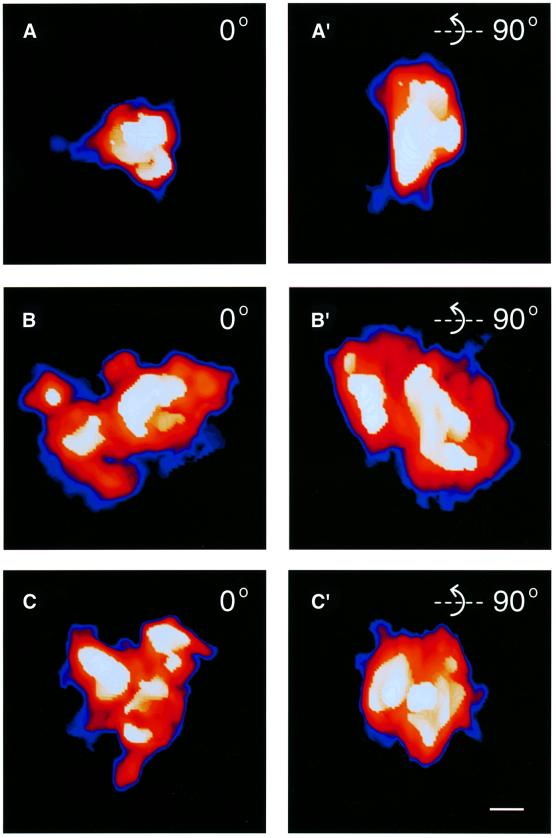
Fig. 5. Dynamic structure of the NTS complexes. The three classes of NTS complex are shown after volume rendering at 5 nm resolution and color coded for different density levels. White–yellow–orange–red corresponds to high to low electron density. The blue haloes show the gradient out to the real background. (A) Class 1. (B) Class 2. (C) Class 3. (A′), (B′) and (C′) are rotated 90° around the horizontal axis compared with (A), (B) and (C), respectively. Scale bar, 10 nm.
We only detected individual gene segments in the sections with the described structure and dimensions. In several areas of the active gene locus, more compact regions were present (see 3 and 4 in Figure 2). In 2D, all these compact areas contained granular material that had substructures of the same kind as we had seen in the granules of the individually visible gene segments. Some of these compact areas consisted of several individual, but juxtaposed gene segments (marked 3 in Figure 2) as resolved in 3D. In the even more compact areas (exemplified by 4 in Figure 2), individual gene segments could not be resolved, even in 3D. However, we found the same type of granules, including substructures, in all compact areas reconstructed. The most plausible explanation for this is that the entire active BR3 gene locus consists of granular fibers, which are BR3 genes being transcribed. These fibers are oriented more or less randomly with respect to the plane of section, and a fiber seen in one section represents only a segment of the entire BR3 gene (a section will provide samples from different genes and many different positions along the complete length of the active BR3 gene). The structural and immunological data presented below show that each granule is a growing pre-mRNA transcript with its associated transcription and splicing components. We will call each such granule a nascent transcript and splicing (NTS) complex.
We used electron tomography to provide a 3D structure of the gene segments. We also prepared serial sections and followed longer parts of individual gene segments. The 3D reconstructions showed that the segments clearly had two components: the NTS complex itself and a connecting axis (Figure 3). The axis is ∼10 nm in diameter. The NTS complexes radiate out from the axis (Figure 3), and in some segments have a distinct narrow connection to it (see the middle NTS complex in Figure 3B).
The longest gene segment was 740 nm long and had 14 NTS complexes (Table I). In our 2D analysis, the average distance between NTS complexes was 53 ± 9 nm. In 3D, it was evident that the distance between consecutive NTS complexes varied between different gene segments and even within one gene segment (between 25 and 80 nm), as measured in in situ preparations.
Table I. Characterization of segments of the active BR3 gene.
| Segment | Segment length (nm) |
No. of NTS complexes | Average distance between NTS complexes (nm) | NTS complex class order; chromatin attachment indicated (Chr) | ||
|---|---|---|---|---|---|---|
| Total length | Studded with NTS complexes | Distance from chromatin to closest NTS complex | ||||
| 1c | 740 | 650 | 90 | 14 | 46.5 | Chr-1-3-3-2-2-x-2-2-3-1-1-3-2-3 |
| 2a | 305 | 220 | 85 | 7 | 31.5 | Chr-2-3-1-3-3-1-1 |
| 3a | 190 | 110 | 80 | 3 | 36.5 | Chr-1-3-2 |
| 4c | 165 | 95 | 70 | 2 | 47.5 | Chr-1-3 |
| 5a | 315 | 300 | 15 | 9 | 33.5 | 2-2-2-2-1-2-2-2-1-3-Chr |
| 6b | 255 | 240 | 15 | 6 | 40 | 2-2-2-3-1-3-Chr |
| 7c | 215 | 200 | 15 | 5 | 40 | 3-1-2-2-1-Chr |
| 8c | 400 | 400 | – | 10 | 40 | 2-2-3-2-2-3-2-1-1-2 |
| 9c | 210 | 210 | – | 7 | 30 | 2-3-2-3-1-1-2 |
| 10c | 390 | 390 | – | 6 | 65 | 2-1-2-2-3-3 |
| 11c | 220 | 220 | – | 6 | 36.5 | 2-1-2-3-3-1 |
| 12c | 250 | 250 | – | 5 | 50 | 1-3-2-2-2 |
| 13c | 230 | 230 | – | 4 | 57.5 | 2-1-3-2 |
| 14c | 190 | 190 | – | 4 | 47.5 | 2-3-2-3 |
| 15c | 180 | 180 | – | 4 | 45 | 2-1-2-3 |
| 16c | 170 | 170 | – | 4 | 42.5 | 1-1-2-2 |
Various isolated chromosome preparations were used: anon-labeled, banti-RNA polymerase II labeled and canti-U2 snRNP B” labeled. The total segment length and the length studded with NTS complexes were measured. Some of the gene segments were connected to compact chromatin at one end (marked by Chr). The distance from the compact chromatin to the closest NTS complex on the 10 nm chromatin axis was measured. The average spacing of NTS complexes was calculated for each segment. Each NTS complex was characterized in 3D and classified as belonging to class 1, 2 or 3. The longest segment, 1c, had one NTS complex (marked x), that was split between two consecutive sections, as determined from serial sections, and this NTS complex was therefore not classified.
Several of the gene segments attached at one end to compact chromatin (Table I; Figure 3A and C). There were only two different types of structure of the gene segments closest to the compact chromatin. We have not been able to show what parts of the BR3 gene these segments contain. Based on a comparison to structurally similar connections at the 5′ and 3′ ends of the BR1 and BR2 genes (Björkroth et al., 1988; Ericsson et al., 1989), it is possible that they represent the two ends of the BR3 gene. The first type of structure consisted of a 70–90 nm long fiber, from the compact chromatin to the first identifiable NTS complex (Figure 3A). The diameter of this fiber was ∼10 nm, with small protrusions at several places. Following the first typical NTS complex, the NTS complexes were evenly spaced along the rest of the gene segment. The second type of structure also consisted of a 10 nm fiber, but the distance from the closest NTS complex to the compact chromatin was only ∼15 nm (Figure 3C). We could not resolve the continuation of the 10 nm fiber within the compact chromatin.
The expected total length of the transcribing 10.9 kb BR3 gene is ∼1000 nm. This is based on a comparison with the BR1 and BR2 genes (reviewed by Daneholt, 1992). Both BR1 and BR2 are 35–40 kb long, and are ∼4000 nm long in situ (S.Masich, unpublished data). We reconstructed one continuous gene segment extending 740 nm from one putative end of the BR3 gene, and a second segment extending 315 nm from the other putative end (Table I). We also reconstructed many gene segments of length 100–400 nm from the internal part of the active gene. The 10 nm axis could not represent the BR3 transcript. The complete, unspliced transcript, if present in a 10 nm fiber, would be maximally 400 nm in length (Lönnroth et al., 1992). Furthermore, the BR3 transcript is extensively spliced during transcription. In addition, our immunolabeling (below) shows that each NTS complex has one RNA polymerase II at its base. Thus, the length and dimensions of the reconstructed gene segments show that the 10 nm axis represents the chromatin fiber and that the growing pre-mRNA transcripts are part of the more or less regularly spaced granules. Our structural data are, therefore, representative of at least 75% and probably the entire BR3 gene. The spacing of NTS complexes suggests that there are in total 20–25 nascent transcripts on the gene, spaced 450–500 bp apart.
Immunological localization of the RNA polymerase II CTD and U2 snRNP B” protein
The locations of the CTD of RNA polymerase II and the U2 snRNP specific B” protein in the BR3 gene segments are shown in Figure 4 and Table II. More than 99% (anti-U2 snRNP B” antibody) and 97% (anti-RNA polymerase II CTD antibody) of the gold particles were associated with the active gene segments and only very few particles were found in other regions (see Figure 4A). In the 3D reconstructions of the gold particles completely located within the section, >83% (for the RNA polymerase II CTD antibody) and 97% (for the anti-U2 snRNP B” antibody) had a specific connection to NTS complexes (see below). With an unspecific antibody (data not shown), we saw very few gold particles (<1 and 3% of the number seen with the anti-U2 snRNP B” and anti-RNA polymerase II CTD antibodies, respectively) and these gold particles were randomly distributed in relation to the active BR3 gene and other, inactive parts of the chromosome.
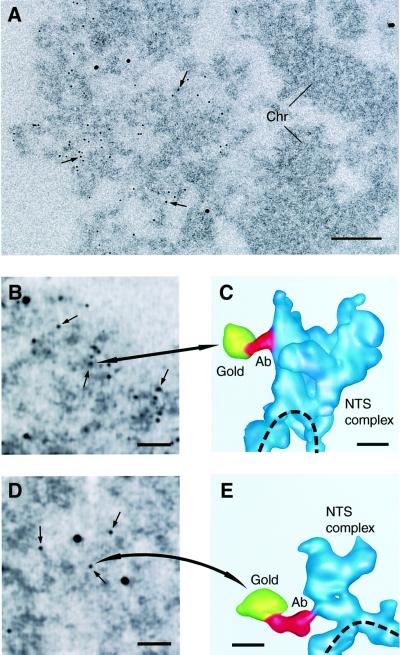
Fig. 4. Immunolocalization of the U2 snRNP B” and the RNA polymerase II CTD in NTS complexes. (A) Electron micrograph of part of a section through the active BR3 gene locus. The 6 nm gold particles (arrows) show labeling with the anti-U2 snRNP B” antibody. Only actively transcribing BR3 gene segments are labeled. No or very few gold particles are seen over the inactive, compact, chromatin (Chr) in the right half of the picture. The four large gold particles are markers used for 3D reconstruction. Scale bar, 100 nm. (B and D) Projections of a 3D reconstruction of an area containing active BR3 gene segments, labeled with the anti-U2 snRNP B” (B) or anti-RNA polymerase II CTD (D) antibodies. The reconstruction has been made from low-dose data with ∼0.5 electrons/Å2/image. Arrows indicate some of the 6 nm gold particles attached to the secondary antibody. Large alignment gold particles, used for 3D reconstruction, are also seen. Scale bars, 50 nm. (C and E) 3D reconstruction of the antibody gold particles indicated in (B) and (D) (large arrows) and the labeled NTS complexes, shown as surface rendering at 5 nm resolution. The antibody connection between the gold particle (yellow) and the NTS complex (blue) is shown in red. The dashed line indicates the chromatin axis. The 6 nm gold particles appear somewhat larger after reconstruction because they are non-transparent objects. Scale bars, 10 nm.
Table II. Immunolocalization of the RNA polymerase II CTD and the U2 snRNP B” in NTS complexes.
| Antibody | No. analyzed | NTS complex associated |
Non-specific | Non-classified | ||
|---|---|---|---|---|---|---|
| Axis side | Non-axis side | Unclear side | ||||
| Polymerase II | 33 | 13 | – | 7 | 4 | 9 |
| U2 snRNP B” | 41 | 1 | 22 | 9 | 1 | 8 |
Three kinds of antibody-bound gold particles were found. Either the gold particle was connected to an NTS complex (NTS complex associated), the gold particle was completely within the section but not connected to any structure (non-specific), or the gold particle, the connection or the labeled NTS complex was cut by the section (non-classified).
A further classification was made, based on which part of the NTS complex the gold particle was connected to (axis side, non-axis side or unclear).
The immunogold labeling was clearly specific for the transcribing BR3 gene segments (Figure 4A), but in 2D it was not possible to decide precisely which parts of the active gene segments were labeled (Figure 4B and D). The 3D reconstruction of the immunostained active BR3 gene enabled a high degree of precision in locating the antigens in the NTS complexes. In general, the combination of immunolabeling and 3D reconstruction allows epitopes to be mapped, giving significantly improved possibilities of locating individual components in supramolecular structures and in this way dissecting the composition of such structures.
We found an unambiguous, thin, electron-dense connection from most gold particles in the 3D reconstructions. This connection extended 10–20 nm from a gold particle and, with the U2 snRNP B” antibody, we observed labeling only of the NTS complexes. Most of the connections were in this case to the part of the complex facing away from the chromatin axis (Figure 4C; Table II). With the anti-RNA polymerase II CTD antibody, we also observed labeling only of the NTS complexes in the gene segments. The chromatin axis between the NTS complexes was not labeled. This supports our interpretation from the structure of the active gene segments that each NTS complex contains a nascent transcript, and therefore it follows that an RNA polymerase II is present in each NTS complex. Most connections between the gold particles attached to the anti-RNA polymerase II CTD antibodies and the NTS complex were observed at the region of the NTS complex that is closest to the chromatin axis (Figure 4E; Table II).
These immunolabeling experiments show that the CTD of the large subunit of RNA polymerase II and the U2 snRNP B” are both located in the NTS complexes along the chromatin axis. According to Table II, there is a very clear difference in the position of the two components within the NTS complexes: the RNA polymerase II CTD closest to the chromatin axis and the U2 snRNP further inside the complex. The immunolocalization data support the conclusion, based on the structural data, that the 10 nm axis is the chromatin fiber and the granules are the NTS complexes in the gene segments.
Structure and molecular mass of the NTS complexes
All NTS complexes had an overall oblate spheroid shape. Figure 5 shows a typical electron density map of the NTS complexes, pseudocolored to highlight the densities observed. Despite considerable structural variation, we could classify the NTS complexes into three classes based on overall structure and the substructures seen within the NTS complexes (Tables III and IV; Figure 5). Class 1 was small and compact, and contained a single large dense substructure. The approximate axis relationships of the class 1 NTS complex were 1:1.2:1.7 (∼20 × 28 × 33 nm). Class 2 had axis relationships of 1:1.2:1.4 (∼32 × 37 × 46 nm). It had one large dense substructure plus one or more additional small or elongated dense substructures, and appeared more loosely packed. Class 3 also had axis relationships of 1:1.2:1.4 (∼31 × 36 × 42 nm), but was even more loosely packed. Class 3 was the most structurally heterogeneous class. It had the most complex internal structure with at least two small or elongated dense substructures, but lacking any large dense substructure. The axis relationships of the NTS complexes in isolated chromosomes and in in situ sections were the same for all three classes. Our immunological labeling experiments showed that all three classes contained the U2 snRNP B” protein and the RNA polymerase II CTD. It should be pointed out that all three classes of NTS complex were present both in isolated chromosomes and in sections of the gland cells. It is, therefore, most unlikely that material is lost from the NTS complexes as a result of the preparation procedure. We have also shown by immunofluorescence microscopy that two SR proteins, two hnRNP proteins and the U1 snRNP are bound to the active BR3 gene in an RNA-dependent manner and remain bound to the BR3 gene during the preparation procedures used (data not shown). Furthermore, no loss of mass could be detected during the collection of the low-dose electron micrographs. Combined, these data show that the recorded differences in mass and structure between the three classes of NTS complexes are not the result of the preparation procedure but rather reflect biological processes.
Table III. Volumes and calculated molecular masses of NTS complexes.
| NTS complex class | Chromosome preparation | No. analyzed | Volume (nm3) | Mass (MDa) |
|---|---|---|---|---|
| 1 | in situ | 9 | 7500 ± 1400 | 3.9 ± 0.7 |
| 1 | isolated | 16 | 6800 ± 3400 | 3.6 ± 1.8 |
| 2 | in situ | 13 | 12 000 ± 3200 | 6.3 ± 1.7 |
| 2 | isolated | 20 | 12 600 ± 5800 | 6.6 ± 3.0 |
| 3 | in situ | 21 | 8700 ± 3500 | 4.6 ± 1.8 |
| 3 | isolated | 15 | 7800 ± 4600 | 4.0 ± 2.4 |
NTS complexes fell into three main classes, as described in the text. Molecular mass was estimated by using the conversion number 1.9 Å3/Da (Klug et al., 1980).
Table IV. Dimensions and calculated molecular masses of substructures within the NTS complexes.
| Substructure: size and shape | No. analyzed | Thickness (nm) | Width (nm) | Length (nm) | Volume (nm3) | Mass (MDa) |
|---|---|---|---|---|---|---|
| Small and round | 31 | 9 ± 2 | 10 ± 2 | 12 ± 3 | 500 ± 200 | 0.3 ± 0.1 |
| Elongated | 37 | 13 ± 2 | 15 ± 3 | 21 ± 4 | 1300 ± 400 | 0.7 ± 0.2 |
| Large and irregular | 22 | 18 ± 4 | 25 ± 4 | 30 ± 6 | 3400 ± 1300 | 1.8 ± 0.7 |
The diameters of the small and round substructures were all <15 nm. The elongated substructures had at least one side >15 nm and only one side >20 nm. Finally, the large and irregular substructures had at least two sides >20 nm.
The volume was measured and the molecular mass calculated of the individual NTS complexes (Table III; Materials and methods). The calculated average molecular masses of the three classes of NTS complex were 3.9, 6.3 and 4.6 MDa, respectively.
The dense substructures within each NTS complex varied in size and fell into three classes: small and approximately round; elongated; or large and irregular (Table IV; Figure 5). The dense substructures in the 3D reconstructions corresponded to the clustered substructures building up the NTS complexes seen in the original micrographs (see above and Figure 2B). A close inspection of the micrographs showed that all NTS complexes contained dense substructures. The volumes were measured and the calculated molecular masses in the in situ preparations were 300 kDa, 700 kDa and 1.8 MDa for the small, elongated and large substructures, respectively (Table IV).
The order of the three classes of NTS complexes was random along the active gene (Table I). Also, the putative ends of the gene had one of the three classes in the observed data. At the resolution attained in this study, we could not detect any significant gradual size difference between the NTS complexes along the active gene (Table I). Owing to the transcription process, the NTS complexes must contain an increasing mass of nascent transcript along the gene. The precision of our volume measurements, however, allows an increasing nascent transcript to be present in the NTS complexes. Neighboring NTS complexes should differ maximally by ∼500 bases of RNA, corresponding to ∼0.17 MDa, and a completely spliced BR3 transcript of 5.5 kb has an approximate mass of 1.9 MDa.
Discussion
The exon–intron structure of the BR3 gene (Paulsson et al., 1990), the co-transcriptional splicing characteristics of the nascent BR3 gene transcripts (Wetterberg et al., 1996) and the rate of transcription of the BR3 gene (E.Egyhàzi, personal communication) have been determined previously. Here, we characterize the in situ structure of the NTS complexes on the actively transcribing BR3 gene, and show that they contain both a splicing factor and the hyperphosphorylated RNA polymerase II CTD. Altogether, these data allow us to interpret the structure of the NTS complexes in functional terms. We argue that the structure and dynamics of the NTS complexes mainly reflect the co-transcriptional splicing of the nascent transcripts (see below). Our structural data have implications for understanding co-transcriptional splicing of the multi-intron BR3 pre-mRNA.
Only one spliceosome is assembled on a multi-intron nascent BR3 transcript at a given time
The mass of a nascent pre-mRNP must increase during transcription, as clearly shown for the BR1 and BR2 pre-mRNP, where continuous addition of exon sequences results in a gradual increase in size of these complexes (Skoglund et al., 1983). In the BR3 gene, we did not observe a statistically significant increase in the mass of the NTS complex along the active gene. Instead, large amounts of material, one to several megadaltons (Tables I and III), are repeatedly gained and lost, while the addition of pre-mRNA during the same period amounts to only 150–200 kDa (see below). The growth of the nascent transcript, continuously being spliced (Wetterberg et al., 1996), therefore cannot account for changes seen in the NTS complexes and is likely to be largely hidden in the NTS complex. The change in molecular mass, the internal structural reorganization and the change in overall shape between consecutive NTS complexes therefore suggest that spliceosomes repeatedly assemble within, carry out splicing and at least partly leave one NTS complex during transcription of the BR3 gene.
Our structural data suggest that more than one complete spliceosome is not formed on a given BR3 pre-mRNA at each moment. The data also suggest that there is not a full set of spliceosomal components in many of the NTS complexes. We base these interpretations on a comparison of the mass and structural characteristics of the NTS complexes, and the available knowledge on the mass and dimensions of in vitro assembled spliceosomes and isolated snRNPs.
First, there is not enough mass (3.9–6.3 MDa) in the NTS complexes to accommodate more than one complete spliceosome. Individual snRNPs have been extensively characterized in mammals (Kastner, 1996; Will and Lührmann, 1997) and are largely conserved in Drosophila (Paterson et al., 1991). Based on the sum of the individual components, a complete spliceosome can be estimated to be ∼4 MDa. In addition, each kilobase of the pre-mRNA corresponds to 340 kDa of RNA, and this RNA is associated with different hnRNP proteins, which are known to bind to the RNA co-transcriptionally (Matunis et al., 1993; Visa et al., 1996). RNA polymerase II with its associated elongation factors adds a mass of at least 0.5 MDa (Young, 1991) and may also carry polyadenylation factors (Bentley, 1999).
Secondly, the dimensions of spliceosomes and snRNPs make it unlikely that more than one complete spliceosome is present in an NTS complex. Whole spliceosomes or assembly intermediates have been analyzed by electron microscopy in Drosophila (Osheim et al., 1985; Beyer and Osheim, 1988), yeast (Clark et al., 1988) and human cells (Reed et al., 1988; Sibbald et al., 1993; Furman and Glitz, 1995). The dimensions of the ellipsoid or ovoid discs varied between 18–25 nm × 24–60 nm (compare to dimensions of NTS complexes above). Also, the structure of individual snRNPs argues against the possibility that more than one set of spliceosomal snRNPs could be present in an NTS complex (Achsel et al., 1999; Kambach et al., 1999b). The total maximum length is in the range from 8 to 25 nm (reviewed by Kastner, 1996). The dimensions, calculated mass and the dynamic reorganizations of the substructures seen within the BR3 NTS complexes (see Figure 5) suggest that these more highly stained substructures could be snRNPs.
There could be several reasons why we detect maximally one complete spliceosome at a given time on the multi-intron BR3 pre-mRNA. First, the splicing process could be so fast in vivo that more than one intron is never present on the nascent transcript. A variant of this possibility is that some introns are excised much faster than others and are rapidly removed from the NTS complexes. We regard both these possibilities as being unlikely. Introns do accumulate to some extent in the BR3 pre-mRNA during transcription (Wetterberg et al., 1996). We should also most likely have caught at least some NTS complexes with structures consistent with the presence of multiple spliceosomes.
Secondly, even though two spliceosomes can form simultaneously on a double intron mRNA in vitro (Christofori et al., 1987), steric interference between spliceosomes on neighboring introns in the compact NTS complex could possibly prevent the formation of more than one spliceosome.
A third reason could be that parts of the spliceosome are reused and never leave the NTS complex. The initial recognition of the splice sites is rapid in vitro and involves the U1 snRNP and a number of proteins in the E complex (reviewed by Reed, 2000), as well as the U2 snRNP (Das et al., 2000). It is possible that all the exon–intron borders are recognized very rapidly in a multi-intron pre-mRNA and marked, while other parts of the spliceosome, e.g. the U4/U6.U5 tri-snRNP, could be repeatedly used for several introns. The observed cooperation between introns in vivo may reflect such a mechanism (Neel et al., 1993). The U1 and U2 snRNPs, especially the U1 snRNP, are the most abundant of the spliceosomal snRNPs (Yu et al., 1999). It should be pointed out that if the distance between introns is long, snRNPs that contain the Sm epitope appear to leave the pre-mRNA (Kiseleva et al., 1994).
The rate and dynamics of spliceosome function in vivo
The transcription time for the BR3 gene at 18°C is 5–6 min, based on the size relationship between the BR3 and the BR1 and BR2 genes. Clearance of nascent transcripts from the BR3 gene after DRB treatment indicates that the transcription time may, in fact, be somewhat longer (E.Egyhàzi, personal communication). The number of introns excised, the transcription time and the fact that only one spliceosome is present in an NTS complex at a given time allow us to conclude that the average excision time for an intron in the BR3 gene is 15–25 s. This is also the approximate time for polymerase II to travel 450–550 bp on the BR3 gene, which is the distance between two NTS complexes. Thus, on average, at least one intron has been excised when we go from one NTS complex to the next in our reconstructions of the active gene. At the same time, on average, two new introns have been added to the pre-mRNA, according to the lengths of exons and introns (Paulsson et al., 1990). These structural observations, combined with earlier biochemical data (Wetterberg et al., 1996), show that a full cycle of spliceosome formation and splicing takes place, apparently independently in each transcript, in <25 s. The extensive structural changes in the BR3 NTS complexes show that the active spliceosome is highly dynamic in vivo. A single, well-defined spliceosome structure does not seem to exist unless the splicing reaction is frozen at a specific stage. The spliceosome should, therefore, not be regarded as a defined structural unit, but as a functional entity.
The elongating RNA polymerase II does not carry spliceosomal components for all introns in the multi-intron BR3 nascent transcript
The rate of splicing of the multi-intron BR3 pre-mRNA is much faster than the rate of splicing in vitro (seconds compared with minutes). It is also somewhat faster than the rates observed for splicing of individual introns in Drosophila (Osheim et al., 1985; Beyer and Osheim, 1988) and in the BR1 pre-mRNA in C.tentans in vivo (Baurén and Wieslander, 1994). Several results suggest that the CTD of the RNA polymerase II large subunit coordinates pre-mRNA splicing (reviewed in Corden and Patturajan, 1997; Neugebauer and Roth, 1997b; Shuman, 1997; Steinmetz, 1997; Bentley, 1999). The CTD may only recruit factors essential for splicing, or it may have a more active role in splicing, e.g. in the assembly of the spliceosome. It is not known whether splicing factors are already associated with the CTD close to the promoter, or whether the CTD coordinates splicing factor recruitment during transcription elongation. Our structural characterization of the NTS complexes is incompatible with a model in which the CTD carries a full set of splicing factors for many of the 38 introns in the BR3 gene. Our data do not rule out the possibility that a subset of splicing factors may be bound to the elongating RNA polymerase II CTD, but our molecular mass estimations put an upper limit. For example, it is not compatible with our data that more than one or a few U1 snRNPs could be carried by the CTD.
The extensive exchange of material that we observe as the NTS complex moves along the BR3 gene indicates that there is instead a very rapid and dynamic exchange of splicing factors between the NTS complex and the surroundings, possibly controlled by the CTD of the RNA polymerase II. We could not detect any consistent structures connecting the NTS complexes and the surroundings of the gene. The exchange of splicing factors, therefore, could not be associated with any defined structures (or any such structures are transient). It has been suggested that splicing factors are recruited to the active genes from clusters of interchromatin granules, which may be found close to the genes (e.g. Misteli and Spector, 1998). Interchromatin granule clusters have not been observed in the polytene nuclei of C.tentans (Vazquéz-Nin et al., 1993).
Materials and methods
Isolation of salivary glands
Salivary glands from fourth instar C.tentans larvae were fixed in 2% glutaraldehyde in 0.1 M sodium cacodylate buffer containing 0.05 M sucrose pH 7.2 for 2 h at 4°C and then washed four times for 15 min in the same buffer, without glutaraldehyde.
Isolation of chromosomes and antibody labeling
Salivary glands were fixed for 2 min in 2% paraformaldehyde in TKM (10 mM triethanolamine–HCl pH 7.0, 100 mM KCl, 1 mM MgCl2) at 4°C, before isolation of chromosome IV by pipetting up and down with small glass pipettes (Björkroth et al., 1988; Kiseleva et al., 1994). Chromosome IV was then transferred to a siliconized objective glass and fixed for 30 min in TKM containing 4% paraformaldehyde at room temperature before the immunoreaction was carried out as described. The chromosomes were post-fixed in 2% glutaraldehyde in TKM for 1 h. The non-immunolabeled chromosomes were directly fixed in glutaraldehyde in TKM after isolation.
Goat anti-Drosophila RNA polymerase IIo antibody (i.e. the hyperphosphorylated CTD of the large subunit of RNA polymerase II), gAPα-PCTD, was a generous gift from Dr Arno L.Greenleaf (Duke University Medical Center, USA). Mouse anti-U2 snRNP B” antibody was purchased from ICN Pharmaceuticals, Inc. Both antibodies are specific in western blots. Secondary IgG antibodies conjugated with 6 nm gold particles were purchased from Jackson ImmunoResearch Labs., Inc.
Embedding, staining and sectioning
Following glutaraldehyde fixation, all specimens (intact glands and isolated chromosomes) were dehydrated and embedded in the same way (Björkroth et al., 1988). The isolated chromosomes were embedded on top of a siliconized glass slide. The plastic with the isolated chromosomes was removed from the glass with a razor after cooling in liquid nitrogen. The area of interest was cut out, mounted and sectioned into either 60 or 100 nm sections. The sections were put onto EM grids (serial sections were ordered) and stained with 2% uranylacetate in ethanol for 5 min. Colloidal 10 nm gold markers (Auroprobe EM GAD lgG G19; Amersham), used as markers in the alignment, were added on top of the sections. The gold markers were diluted 1:3 and incubated with the grids for 8–10 min. The grids were finally washed three times in water and air dried.
Tilt series and digitization
The tilt series of the chromosomes in sectioned nuclei were recorded manually on a Zeiss EM902 transmission electron microscope at 80 kV acceleration voltage using the inelastic scattering filter, after pre-irradiation of the preparations, resulting in an initial 20% shrinkage in the beam direction and prevention of any further shrinkage during the tilt series. A magnification of 32 000× was used and every 5° from +60° to –60° was recorded, giving a total of 25 projections. Two extra 0° tilts were recorded, before and after the tilt series, to control for specimen radiation damage. Only approved tilt series were processed further, and the negatives were scanned with an Optronics P-1000 drum scanner with a 25 µm raster size.
The isolated chromosome tilt series, including immunolabeled chromosomes, were recorded on a Philips CEM200 FEG transmission electron microscope automatically, using the TVIPS (TVIPS GmbH, Gauting, Germany) detection system and the EMMENU software. This software controls the tilting, focusing, tracking and data collection. The acceleration voltage was 200 kV, the magnification 26 700× and the pictures recorded directly on a 2048 × 2048 pixel CCD chip with a raster size of 14 µm. Sixty-one pictures were recorded, every 2° between +60° and –60°. The total dose for a full tilt series was <30 electrons/Å2. No shrinkage can be detected during these conditions (Miralles et al., 2000).
The tilt series of chromosomes in situ were digitized at a pixel size of 7.8 Å and the tilt series of isolated chromosomes were digitized at a pixel size of 5.2 Å. The total size of the digitized area for the tilt series of chromosomes in sectioned nuclei (see Figure 2) was 1.25 × 1.25 µm2 and that for the isolated chromosomes 1.07 × 1.07 µm2.
3D reconstruction
The data were processed (aligned, reconstructed and refined) using the ET software package version 3.4 (Skoglund et al., 1996b). All image processing was performed on a Compaq workstation (500a) and a GS60 server.
Colloidal gold markers (10 nm) were used for the alignment of the different projections. The average alignment error was <5 Å. The entire digitized in situ area was reconstructed using the filtered back-projection principle. The individual reconstructions were further refined using COMET (Skoglund et al., 1996a) to give a better fit to the experimental data, improve the signal/noise ratio and to keep only significant densities. Finally, the reconstructed densities were lowpass filtered to a homogeneous resolution.
Areas of ∼300 × 300 × 100 nm were first analyzed at 7.5 nm resolution. Selected structures were further reconstructed and analyzed at 5 nm.
Bob (Ken Chin-Purcell, Minnesota Supercomputing Center Inc., freeware) was used for volume rendering, and XTV (Skoglund et al., 1996b) and AVS/Express version 4.0 (Advanced Visual Systems, Inc.) were both used for surface rendering.
The volumes of individual NTS complexes were measured by adding the voxels within a tailor-made envelope (only the density of interest was cut out using the XTV software). Only the voxels above a threshold density value, usually 2 SD above average density, were added. The linear extension of gene segments from embedded isolated chromosomes compared with in situ sections showed an ∼20% shrinkage in the isolated chromosomes, most probably due to the different treatments of the specimens. The linear shrinkage corresponds to an isotropic shrinkage in volume of ∼50% compared with the in situ sections. The volume measurements for the isolated chromosomes were therefore corrected by a factor of two. To estimate mass from volume, we used the conversion number 1.9 Å3/Da. This value has been used for nucleosomes (Klug et al., 1980) and for pre-mRNP in the related BR1 and BR2 genes in pre-irradiated in situ preparations (Lönnroth et al., 1992), both of which are nucleic acid–protein complexes.
Acknowledgments
Acknowledgements
We thank Birgitta Ivarsson, Birgitta Björkroth and Jan Strandh for excellent technical assistance. We also thank Neus Visa and Lars-Göran Öfverstedt for valuable discussions, and Bertil Daneholt for advice and technical support. The Swedish Natural Science Research Council and the Swedish Research Council for Engineering Sciences supported this work.
References
- Achsel T., Brahms,H., Kastner,B., Bachi,A., Wilm,M. and Lührmann,R. (1999) A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J., 18, 5789–5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baurén G. and Wieslander,L. (1994) Splicing of Balbiani Ring 1 gene pre-mRNA occurs simultaneously with transcription. Cell, 76, 183–192. [DOI] [PubMed] [Google Scholar]
- Baurén G., Jiang,W.-Q., Bernholm,K., Gu,F. and Wieslander,L. (1996) Demonstration of a dynamic, transcription dependent organization of pre-mRNA splicing factors in polytene nuclei. J. Cell Biol., 133, 929–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley D. (1999) Coupling RNA polymerase II transcription with pre-mRNA processing. Curr. Opin. Cell Biol., 11, 347–351. [DOI] [PubMed] [Google Scholar]
- Beyer A.L. and Osheim,Y.N. (1988) Splice site selection, rate of splicing and alternative splicing on nascent transcripts. Genes Dev., 2, 754–765. [DOI] [PubMed] [Google Scholar]
- Björkroth B., Ericsson,C., Lamb,M.M. and Daneholt,B. (1988) Structure of the chromatin axis during transcription. Chromosoma, 96, 333–340. [Google Scholar]
- Christofori G., Frendewey,D. and Keller,W. (1987) Two spliceosomes can form simultaneously and independently on synthetic double-intron messenger RNA precursors. EMBO J., 6, 1747–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark M.W., Goelz,S. and Abelson,J. (1988) Electron microscopic identification of the yeast spliceosome. EMBO J., 7, 3829–3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cmarko D., Verschure,P.J., Martin,T.E., Dahmus,M.E., Krause,S., Fu,X.-D., van Driel,R. and Fakan,S. (1999) Ultrastructural analysis of transcription and splicing in the cell nucleus after bromo-UTP microinjection. Mol. Biol. Cell, 10, 211–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corden J.L. and Patturajan,M. (1997) A CTD function linking transcription to splicing. Trends Biochem. Sci., 22, 413–416. [DOI] [PubMed] [Google Scholar]
- Daneholt B. (1992) The transcribed template and the transcription loop in Balbiani rings. Cell Biol. Int. Rep., 16, 709–715. [DOI] [PubMed] [Google Scholar]
- Das R., Zhou,Z. and Reed,R. (2000) Functional association of U2 snRNP with the ATP-independent spliceosomal complex E. Mol. Cell, 5, 779–787. [DOI] [PubMed] [Google Scholar]
- Ericsson C., Mehlin,H., Björkroth,B., Lamb,M.M. and Daneholt,B. (1989) The ultrastructure of upstream and downstream regions of an active Balbiani ring gene. Cell, 56, 631–639. [DOI] [PubMed] [Google Scholar]
- Furman E. and Glitz,D.G. (1995) Purification of the spliceosome A-complex and its visualization by electron microscopy. J. Biol. Chem., 270, 15515–15522. [DOI] [PubMed] [Google Scholar]
- Hirose Y., Tacke,R. and Manley,J.L. (1999) Phosphorylated RNA polymerase II stimulates pre-mRNA splicing. Genes Dev., 13, 1234–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-Garcia L.F. and Spector,D.L. (1993) In vivo evidence that transcription and splicing are coordinated by a recruiting mechanism. Cell, 73, 47–59. [DOI] [PubMed] [Google Scholar]
- Kambach C., Walke,S. and Nagai,K. (1999a) Structure and assembly of the spliceosomal small nuclear ribonucleoprotein particles. Curr. Opin. Struct. Biol., 9, 222–230. [DOI] [PubMed] [Google Scholar]
- Kambach C., Walke,S., Young,R., Avis,J.M., de la Fortelle,E., Raker,V.A., Lührmann,R., Li,J. and Nagai,K. (1999b) Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell, 96, 375–387. [DOI] [PubMed] [Google Scholar]
- Kastner B. (1996) Purification and electron microscopy of spliceosomal snRNPs. In Schenkel,J. (ed.), RNP Particles, Splicing and Immunodiseases. Springer-Verlag, Heidelberg, Germany, pp. 95–140.
- Kiseleva E., Wurtz,T., Visa,N. and Daneholt,B. (1994) Assembly and disassembly of spliceosomes along a specific pre-messenger RNP fiber. EMBO J., 13, 6052–6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug A., Rhodes,D., Smith,J., Finch,J. and Thomas,J.O. (1980) A low resolution structure for the histone core of the nucleosome. Nature, 287, 509–516. [DOI] [PubMed] [Google Scholar]
- Krämer A. (1996) The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu. Rev. Biochem., 65, 367–409. [DOI] [PubMed] [Google Scholar]
- Krämer A., Gruter,P., Groning,K. and Kastner,B. (1999) Combined biochemical and electron microscopic analyses reveal the architecture of the mammalian U2 snRNP. J. Cell Biol., 145, 1355–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence J.B., Carter,K.C. and Xing,X. (1993) Probing functional organization within the nucleus: is genome structure integrated with RNA metabolism? Cold Spring Harb. Symp. Quant. Biol., 58, 807–818. [DOI] [PubMed] [Google Scholar]
- Lönnroth A., Alexciev,K., Mehlin,H., Wurtz,T., Skoglund,U. and Daneholt,B. (1992) Demonstration of a 7 nm RNP fiber as the basic structural element in a premessenger RNP particle. Exp. Cell Res., 199, 292–296. [DOI] [PubMed] [Google Scholar]
- Matunis E.L., Matunis,M.J. and Dreyfuss,G. (1993) Association of individual hnRNP proteins and snRNPs with nascent transcripts. J. Cell Biol., 121, 219–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miralles F., Öfverstedt,L.-Ö., Sabri,N., Aissouni,Y., Hellman,U., Skoglund,U. and Visa,N. (2000) Electron tomography reveals posttranscriptional binding of pre-mRNPs to specific fibers in the nucleoplasm. J. Cell Biol., 148, 271–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. and Spector,D.L. (1998) The cellular organization of gene expression. Curr. Opin. Cell Biol., 10, 323–331. [DOI] [PubMed] [Google Scholar]
- Misteli T., Caceres,J.F. and Spector,D.L. (1997) The dynamics of a pre-mRNA splicing factor in living cells. Nature, 387, 523–527. [DOI] [PubMed] [Google Scholar]
- Neel H., Weil,D., Giansante,C. and Dautry,F. (1993) In vivo cooperation between introns during pre-mRNA processing. Genes Dev., 7, 2194–2205. [DOI] [PubMed] [Google Scholar]
- Neugebauer K.M. and Roth,M.B. (1997a) Distribution of pre-mRNA splicing factors at sites of RNA polymerase II transcription. Genes Dev., 11, 1148–1159. [DOI] [PubMed] [Google Scholar]
- Neugebauer K.M. and Roth,M.B. (1997b) Transcription units as RNA processing units. Genes Dev., 11, 3279–3285. [DOI] [PubMed] [Google Scholar]
- Osheim Y.N., Miller,O.L. and Beyer,A.L. (1985) RNP particles at splice junction sequences on Drosophila chorion transcripts. Cell, 43, 143–151. [DOI] [PubMed] [Google Scholar]
- Paterson T., Beggs,J.D., Finnegan,D.J. and Lührmann,R. (1991) Polypeptide components of Drosophila small nuclear ribonucleoprotein particles. Nucleic Acids Res., 19, 5877–5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsson G., Lendahl,U., Galli,J., Ericsson,C. and Wieslander,L. (1990) The Balbiani ring 3 gene in Chironomus tentans has a diverged repetitive structure split by many introns. J. Mol. Biol., 211, 331–349. [DOI] [PubMed] [Google Scholar]
- Puvion E. and Puvion-Dutilleul,F. (1996) Ultrastructure of the nucleus in relation to transcription and splicing: role of perichromatin fibrils and interchromatin granules. Exp. Cell Res., 229, 217–225. [DOI] [PubMed] [Google Scholar]
- Reed R. (2000) Mechanisms of fidelity in pre-mRNA splicing. Curr. Opin. Cell Biol., 12, 340–345. [DOI] [PubMed] [Google Scholar]
- Reed R., Griffith,J. and Maniatis,T. (1988) Purification and visualization of native spliceosomes. Cell, 53, 949–961. [DOI] [PubMed] [Google Scholar]
- Sass H. and Pedersen,T. (1984) Transcription-dependent localization of U1 and U2 small nuclear ribonucleoproteins at major sites of gene activity in polytene chromosomes. J. Mol. Biol., 180, 911–926. [DOI] [PubMed] [Google Scholar]
- Shuman S. (1997) Origins of mRNA identity: capping enzymes bind to the phosphorylated C-terminal domain of RNA polymerase II. Proc. Natl Acad. Sci. USA, 94, 12758–12760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibbald M.J., Carlemalm,E.C., Beer,M. and Sproat,B.S. (1993) Imaging of RNA–protein interactions in splicing complexes with dark-field STEM. J. Struct. Biol., 110, 111–121. [DOI] [PubMed] [Google Scholar]
- Skoglund U., Andersson,K., Björkroth,B., Lamb,M.M. and Daneholt,B. (1983) Visualization of the formation and transport of a specific hnRNP particle. Cell, 34, 847–855. [DOI] [PubMed] [Google Scholar]
- Skoglund U., Öfverstedt,L.-G., Burnett,R. and Bricogne,G. (1996a) Maximum-entropy three-dimensional reconstruction with deconvolution of the contrast transfer function, a test application with adenovirus. J. Struct. Biol., 117, 173–188. [DOI] [PubMed] [Google Scholar]
- Skoglund U., Öfverstedt,L.-G. and Daneholt,B. (1996b) Procedures for three-dimensional reconstruction from thin slices with electron tomography. In Schenkel,J. (ed.), RNP Particles, Splicing and Immunodiseases. Springer-Verlag, Heidelberg, Germany, pp. 72–94.
- Sperling R., Koster,A.J., Malamed-Bessudo,C., Rubinstein,A., Angenitzki,M., Berkovitch-Yellin,Z. and Sperling,J. (1997) Three dimensional image reconstruction of large nuclear RNP (lnRNP) particles by automated electron tomography. J. Mol. Biol., 267, 570–583. [DOI] [PubMed] [Google Scholar]
- Staley J.P. and Guthrie,C. (1998) Mechanical devices of the spliceosome, motors, clocks, springs and things. Cell, 92, 315–326. [DOI] [PubMed] [Google Scholar]
- Steinmetz E.J. (1997) pre-mRNA processing and the CTD of RNA polymerase II: The tail that wags the dog. Cell, 89, 491–494. [DOI] [PubMed] [Google Scholar]
- Vazquéz-Nin G.H., Echeverria,O.M., Rouelle-Rossier,V.B. and Fakan,S. (1993) A new type of ribonucleoprotein constituent of the polytene nucleus of the salivary glands of Chironomus thummi and Chironomus tentans. Chromosoma, 102, 693–699. [DOI] [PubMed] [Google Scholar]
- Visa N., Alzhanova-Ericsson,A.T., Sun,X., Kiseleva,E., Björkroth,B., Wurtz,T. and Daneholt,B. (1996) A pre-mRNA-binding protein accompanies the RNA from the gene through the nuclear pores and into polysomes. Cell, 84, 253–264. [DOI] [PubMed] [Google Scholar]
- Wetterberg I., Baurén,G. and Wieslander,L. (1996) The intranuclear site of excision of each intron in the Balbiani Ring 3 pre-mRNA is influenced by the time remaining to transcription termination and different excision efficiencies for the various introns. RNA, 2, 641–651. [PMC free article] [PubMed] [Google Scholar]
- Will C.L. and Lührmann,R. (1997) Protein functions in pre-mRNA splicing. Curr. Opin. Cell Biol., 9, 320–328. [DOI] [PubMed] [Google Scholar]
- Wu Z., Murphy,C., Callan,H.G. and Gall,J.G. (1991) Small nuclear ribonuceloproteins and heterogeneous nuclear ribonucleoproteins in the amphibian germinal vesicle: loops, spheres and snurposomes. J. Cell Biol., 113, 465–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuarin J. and Schibler,U. (1994) Physical isolation of nascent RNA chains transcribed by RNA polymerase II: evidence for cotranscriptional splicing. Mol. Cell. Biol., 14, 7219–7225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R.A. (1991) RNA polymerase II. Annu. Rev. Biochem., 60, 689–715. [DOI] [PubMed] [Google Scholar]
- Yu X.-T., Schari,E.C., Smith,C.M. and Steitz,J.A. (1999) The growing world of nuclear ribonucleoproteins. In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 487–524.
- Zeng C., Kim,E., Warren,S.L. and Berget,S.M. (1997) Dynamic relocation of transcription and splicing factors dependent upon transcriptional activity. EMBO J., 16, 1401–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C. and Berget,S.M. (2000) Participation of the C-terminal domain of RNA polymerase II in exon definition during pre-mRNA splicing. Mol. Cell. Biol., 20, 8290–8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Taneja,K.L., Singer,R.H. and Green,M.R. (1994) Localization of pre-mRNA splicing in mammalian nuclei. Nature, 372, 809–812. [DOI] [PubMed] [Google Scholar]