Abstract
Fenofibrate, a peroxisome proliferator-activated receptor α (PPARα) agonist, is widely prescribed to treat hyperlipidemia and has therapeutic potential in liver and kidney diseases. However, fenofibrate is also associated with adverse effects, including elevated creatinine and liver and kidney toxicity, although the underlying mechanisms remain unclear. In addition, how fenofibrate regulates lipid metabolism differently in the liver and kidney is not well understood. Therefore, in this study, we investigated the dose-dependent effects of fenofibrate on liver and kidney metabolism in rats, with a focus on PPARα activation and potential mechanisms contributing to organ-specific toxicity. We used high-throughput transcriptomic data from 5-day rat in vivo studies, where rats were exposed to fenofibrate, and performed pathway enrichment, injury module, and detailed individual gene comparison analyses to investigate how liver and kidney metabolism were differentially altered between the two organs. Fenofibrate exposure significantly increased liver but not kidney weights and caused larger perturbations in the liver compared to the kidney transcriptome, with the majority of the changes related to PPARα regulation. Interestingly, our study revealed that the PPARα and RXRα genes are differentially regulated between the liver and kidney. In addition, we identified several differences between them in cellular and mitochondrial fatty acid transport, lipoprotein metabolism, fatty acid oxidation, branched-chain amino acid degradation, and glucose metabolism pathways. Furthermore, we identified transcriptomic inflection points at which the changes in the PPARα-mediated regulation of lipid metabolism switched from beneficial to deleterious as the fenofibrate concentration increased leading to liver injury, providing potential mechanisms of toxicity.
Keywords: Fenofibrate, Lipid metabolism, PPARα, Fatty acid uptake, Peroxisome, Branched-chain amino acids, Gluconeogenesis
Subject terms: Biochemical networks, Metabolic pathways, Chemical biology, Computational biology and bioinformatics, Systems biology, Transcriptomics
Introduction
Lipids are fundamental to living cells as they are essential structural components of biological membranes and affect multiple cellular processes that are critical for maintaining homeostasis. Lipids generate and store cellular energy (e.g., triglycerides), interact with proteins to modulate their localization and function, and play key roles in cellular signaling processes1,2. Balancing lipid synthesis, uptake, utilization, and storage controls cellular lipid homeostasis, however, an imbalance in any of these processes can lead to excessive lipid accumulation, which has been associated with cellular injury and dysfunction in diverse tissues, including the liver, kidney, heart, brain, and skeletal muscles3–5. Importantly, numerous studies have demonstrated that dysregulation of lipid homeostasis is strongly associated with the highly prevalent human metabolic syndrome nonalcoholic fatty liver disease (NAFLD) as well as with acute, chronic, and diabetic kidney diseases6–11.
Cellular lipid metabolism is regulated by peroxisome proliferator-activated receptors (PPARs), a family of nuclear receptors that play essential roles in the transcriptional regulation of a variety of biological processes, including carbohydrate metabolism, cell proliferation, and inflammation12–15. PPARs are ligand-inducible transcription factors, and three subtypes (α, β/δ, and γ) exist in mammals, each with a distinct tissue expression profile and set of functions. PPARα, which is widely expressed in tissues with high fatty-acid oxidation rates, such as the liver, kidney, heart, and skeletal muscles, functions as a major regulator of lipid hemostasis16,17. PPARα binds to DNA as a heterodimer with the retinoid X receptor (RXR) and controls the expression of numerous genes involved in a plethora of lipid metabolic pathways, including fatty acid uptake and synthesis, lipoprotein metabolism, microsomal, peroxisomal, and mitochondrial fatty acid oxidation, ketogenesis, bile acid metabolism, and gluconeogenesis, as well as various genes involved in peroxisomal processes and other metabolic pathways18–20.
A wide range of endogenous and naturally occurring biological molecules act as ligands for the activation of PPARα, including a variety of fatty acids (e.g., long-chain fatty acids), arachidonic acid metabolites, and eicosanoids21,22. In addition, PPARα can be activated by several synthetic molecules, including herbicides, surfactants, organic solvents, and fibrate drugs23–25. Importantly, fibrates are high-affinity PPARα activators that raise plasma high-density lipoprotein levels and reduce triglycerides, making them effective in the treatment of diseases like diabetes and dyslipidemia26–28. For example, fenofibrate is one of the well-known fibrate medications used to treat hypercholesterolemia in type 2 diabetes and hypertriglyceridemia and has shown positive effects on kidney function in patients with chronic kidney disease29–31. Furthermore, it is one of the drugs being evaluated for the treatment of NAFLD due to its effects on lipogenesis and catabolism of fatty acids32–34. However, fenofibrate administration has been associated with certain adverse effects on kidney function, such as elevation of plasma creatinine levels, and kidney injury has been observed in both clinical and animal studies30,35,36. Similarly, fenofibrate has also been linked to hepatotoxicity, with studies reporting adverse effects ranging from cholestasis to chronic liver injury37,38.
Although we know the beneficial and adverse effects of fenofibrate administration from clinical and animal studies, the pathogenesis of fibrate-induced liver and kidney injuries is unclear. Furthermore, while the mechanism of fenofibrate-mediated activation of PPARα in the modulation of lipid metabolism is well established in the liver, it has seldom been studied at the same level in the kidneys39–44. Therefore, in the current study, we aimed to examine the effect of fenofibrate activation on lipid metabolism in the liver and kidneys to identify potential similarities and differences in lipid regulation between the two organs. To this end, we used dose-dependent experimental data from rats exposed to fenofibrate at different concentrations and analyzed high-throughput transcriptomics data to investigate the role of PPARα in the liver and kidneys and identified key genes that were differentially regulated between the two organs as well as potential mechanisms that could lead to injury.
Materials and methods
Five-day in vivo rat exposure studies
A detailed description of the experimental protocol used for the 5-day in vivo rat studies has been published previously45. All studies were approved by the Battelle Animal Care and Use Committee, were carried out in accordance with all relevant NIH animal care and use guidelines and regulations, and adhered to the ARRIVE guidelines for the use of experimental animals. Briefly, male Sprague Dawley (Hsd: Sprague Dawley SD) rats were received at Battelle from Envigo (Haslett, MI, USA; now known as Inotiv, Inc.), were allowed to acclimate for approximately 2 weeks, and were then randomized based on body weight to ensure their initial body weights were balanced across the groups for the final experiments. After the acclimatization period, 8- to 10-week-old rats fed a normal diet received either vehicle control or fenofibrate by oral gavage. The fenofibrate concentrations (8, 16, 31.25, 62.50, 125, 250, 500, and 1000 mg/kg) were selected based on a 29-day exposure study listed in the TG-GATEs database46 and included concentrations higher and lower than those previously studied. Specifically, we selected the low concentrations of fenofibrate (8–30 mg/kg) that are equivalent to human therapeutic dose levels (145–200 mg/day). Fenofibrate was dissolved in a 0.5% aqueous methylcellulose solution (vehicle control) and administered at the selected eight different concentrations plus the control (n = 4 for each condition) once per day for 5 consecutive days starting on Day 0. The body weight of each rat was measured prior to dose administration. On Day 5, the rats were weighed and humanely terminated via exsanguination under 70% CO2:30% O2 anesthesia, and the liver and kidneys were rapidly collected. The terminal weights of the liver and kidneys were recorded, and the left lobe of the liver and the right kidney were transferred into RNAlater (Qiagen, Valencia, CA, USA) for RNA extraction.
Total RNA was extracted from the liver and kidney using the RNeasy Mini Kit (Qiagen) with a DNA digestion step, and a high-throughput transcriptomics analysis was performed using the rat S1500+ TempO-Seq platform47. Briefly, using 96-well plates, mRNA targets were hybridized with a detector oligo mix to generate a pool of amplification templates that share common polymerase chain reaction (PCR) primer-binding sites, and the plates were barcoded for proper identification following sequencing. Sample amplicons were pooled and cleaned using a PCR clean-up kit, and the libraries were then sequenced using a HiSeq 2500 Ultra-High-Throughput Sequencing System (Illumina, San Diego, CA, USA) to generate sequenced readouts in the FASTQ file format.
After quality assessment of the FASTQ files, the TempO-Seq sequences were aligned to the rat S1500+ probe sequences using Bowtie version 1.2.248. The Bowtie configuration allowed up to three mismatches and selected the single best alignment for reporting. Samples were flagged and removed from further analysis based on the following criteria: sequencing read depth < 500 K, total alignment rate < 40%, unique alignment rate < 30%, percent of probes with at least five reads < 50%, and number of aligned reads < 500 K. Furthermore, outlier samples were removed from downstream analysis based on principal component analysis, hierarchical clustering, and inter-replicate correlation analysis. Based on the attenuation factors provided in the platform documentation, unattenuated equivalent counts were calculated and normalized at the probe level by applying read counts-per-million normalization. Subsequently, a pseudo count of 1.0 was added to each normalized expression, and the values were log2 transformed to complete the normalization. Finally, the log-transformed values from the S1500 + platform were extrapolated to the whole rat transcriptome to obtain the extrapolated full transcriptome expression profiles for all the liver and kidney samples, as described in our previous study49.
Differential gene expression and KEGG pathway analysis50–52
Using the extrapolated full transcriptome expression profiles for the liver and kidney samples from the control and different fenofibrate exposure groups, we performed a one-way analysis of variance (ANOVA) to determine if there was a significant difference between the means of the control group versus the fenofibrate groups. For this analysis, we used the Anovan function together with the least significant difference test in the multcompare function in MATLAB. We then estimated the false discovery rate (FDR) to correct for multiple comparisons using the Benjamini-Hochberg method (mafdr function in MATLAB). We defined a significantly expressed gene as one with an FDR-adjusted p-value < 0.1. We used the resulting differentially expressed gene fold-change (FC) values for all our subsequent analyses. For the KEGG pathway enrichment analysis50–52, we used the aggregated fold-change (AFC) method, which indicates whether a particular KEGG pathway was up- or downregulated using the gene expression FC values and the significance values as input53,54. Briefly, given the input, the AFC method estimates the mean FC value for each gene and provides the KEGG pathway score as the total FC value of all the genes in the pathway. The sign of the pathway score represents the direction of regulation, with positive values indicating upregulation and negative values indicating downregulation in the treatment condition compared with its corresponding control.
Results
Fenofibrate exposure resulted in higher absolute and relative liver weights compared to kidney weights
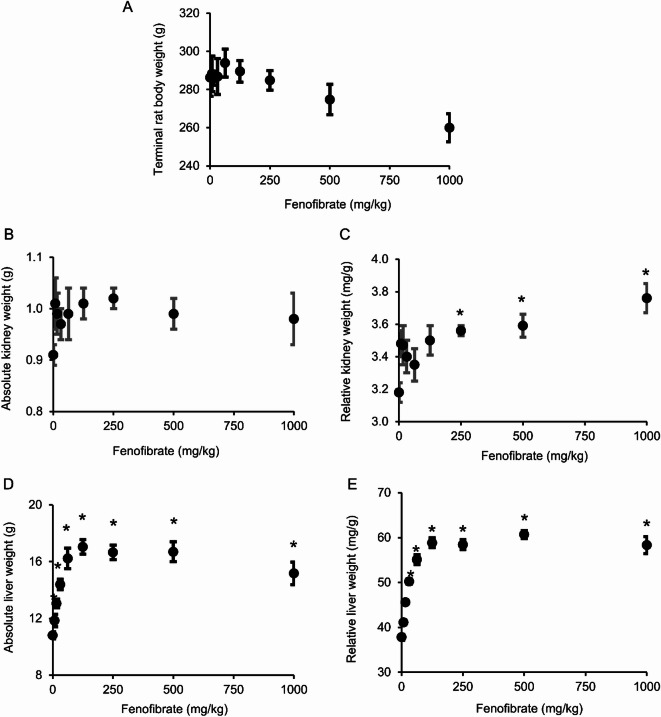
Figure 1 shows the alterations in terminal body weights as well as absolute and relative rat liver and kidney weights at the end of the 5-day fenofibrate exposure for the different concentrations. During the 5-day treatment, we did not observe a significant change in rat body weights, but we did observe a decreasing trend as the concentration of fenofibrate increased (Fig. 1A). Similarly, we did not see any change in the absolute kidney weight (Fig. 1B), although the relative kidney weight, which is the ratio of the absolute kidney weight to the terminal body weight, did increase significantly for the high-dose fenofibrate exposures (Fig. 1C) due to the decreasing trend observed in the terminal body weights (Fig. 1A). In contrast, compared to the control group, fenofibrate exposure resulted in significant increases in absolute (Fig. 1D) and relative liver weights (Fig. 1E), starting with a low dose (16 mg/kg), and the response was dose dependent, with increasing fenofibrate concentrations leading to greater increases in absolute and relative liver weights until saturation was reached at the higher dose levels. These results suggest that fenofibrate exposure leads to significant enlargement of liver cells, consistent with previous observations of hepatomegaly observed with its use in other studies55,56.
Fig. 1.
Alterations in rat terminal body weight and liver and kidney weights with increasing concentrations of fenofibrate at the end of a 5-day exposure study. (A) Terminal rat body weight. (B) Absolute kidney weight. (C) Relative kidney weight, which is the ratio of the absolute kidney weight to the terminal body weight. (D) Absolute liver weight. (E) Relative liver weight, which is the ratio of the absolute liver weight to the terminal body weight. *Statistical significance between the fenofibrate group and the vehicle control (p < 0.01).
Fenofibrate exposure induced significant dose-dependent alterations in the liver and kidney transcriptome
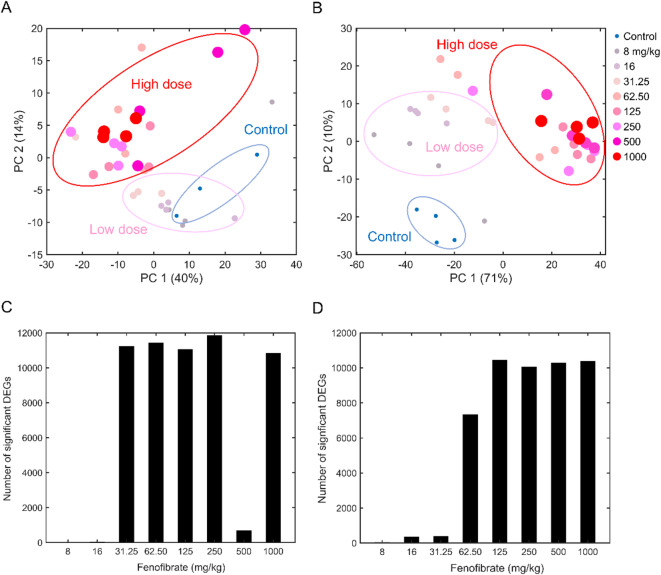
To obtain a global view of the effects of fenofibrate on liver and kidney metabolism, we used the targeted RNA-sequencing platform TempO-seq combined with extrapolating the targeted gene expression changes to the rat whole transcriptome. Each control and treatment group contained four biological replicates per dose. First, we performed a principal component analysis (PCA) to assess the global transcriptomic changes in kidney (Fig. 2A) and liver metabolism (Fig. 2B) at the mRNA abundance levels. As expected, PCA analysis revealed clear separation of the control groups from the fenofibrate treatment groups. Furthermore, our observations from the PCA analysis indicated that the low-dose fenofibrate groups segregated from the high-dose groups and that the alterations in liver metabolism were much stronger, with 71% of the total variance in the data explained by the first component in the liver compared to 40% in the kidney. Next, we performed a one-way ANOVA to assess the magnitude of the effect of fenofibrate treatment on the liver and kidney transcriptome. Exposure to fenofibrate induced only minor changes in kidney metabolism at low concentrations (up to 16 mg/kg), however, the changes exponentially increased beginning at 31.25 mg/kg, with more than 10,000 genes altered significantly for the rest of the high-dose groups when we applied an FDR threshold of less than 0.1 (Fig. 2C). We observed a similar magnitude of gene expression changes in the liver, however, the changes exponentially increased beginning at 62.50 mg/kg fenofibrate compared to the kidney (Fig. 2D).
Fig. 2.
(A, B) Identification of global gene perturbations in rat liver and kidney metabolism with fenofibrate treatment. We separated the control and fenofibrate-treated groups (N = 4 each) for the (A) kidney and (B) liver using principal component (PC) analysis. (C, D) Identification of the number of significant differentially expressed genes (DEGs) (false discovery rate < 0.1) in the (C) kidney and (D) liver using analysis of variance (ANOVA). Here, we performed a one-way ANOVA to identify differentially expressed genes and corrected for multiple comparisons using the Benjamini-Hochberg method to calculate the false discovery rate.
Use of an FDR threshold alone resulted in a large number of genes being significantly altered with increasing fenofibrate dose, however, the magnitude of the change for a majority of these genes was much smaller. Therefore, to identify genes that showed biologically meaningful changes, we also implemented an FC threshold, with an absolute log2(FC) of greater than 0.5. With this criterion, we observed a drastic reduction in the number of significantly altered genes in both the kidney and liver across the different dose levels. Figure 3A shows the total number of significantly altered genes that satisfy both the FDR and FC criteria at the highest fenofibrate dose and the genes that are common between the kidney and liver. A hierarchical clustering analysis of the common altered genes between the kidney and liver indicated a significant overlap, with many genes perturbed similarly with increasing fenofibrate concentration (Fig. 3B; Clusters 1, 3, and 4). However, we observed a set of genes that were significantly downregulated in the kidney but upregulated in the liver upon fenofibrate exposure, indicating potential differences in metabolism (Fig. 3B; Cluster 2). Taken together, the gene expression data suggest that high-dose fenofibrate exposure significantly affected liver more than kidney metabolism, with the total number of significantly altered genes approximately 4 times higher in the liver than the kidney.
Fig. 3.

Fenofibrate induced unique and common gene and pathway alterations in rat liver and kidney metabolism. (A) Venn diagram of altered genes in the kidney and liver based on logarithmic fold-change (FC) value [|log2(FC)| > 0.5] and statistical significance criteria [false discovery rate (FDR) < 0.1] at the highest dose. (B) Hierarchical clustering of significantly altered logarithmic FC values for genes common between the kidney and liver for increasing doses of fenofibrate. (C) Significantly altered biological pathways in the kidney (orange) and liver (blue) at the highest dose. AFC, aggregated fold change.
Hepatic lipid metabolism was strongly affected by fenofibrate exposure
To obtain more insight on the functional impact of fenofibrate exposure on biological pathways, we performed a KEGG enrichment analysis using the kidney and liver genes that were significantly altered at the highest dose. Figure 3C shows a summary of the significantly upregulated cellular pathways, as indicated by their positive AFC z-score values, for the liver (blue) and the kidney (orange). As expected, we observed several common upregulated cellular pathways for lipid metabolism, indicating that it is a common target for fenofibrate in the liver and kidney. Interestingly, our pathway analysis indicated that the PPARα signaling pathway was significantly upregulated in both the liver and kidney upon fenofibrate exposure, suggesting its role in the activation of PPARα. In addition, we observed enrichment of several biological pathways unique to each organ. For example, histidine metabolism and the AMP-activated protein kinase signaling pathway were significantly upregulated only in the liver, whereas the terpenoid backbone biosynthesis and propanoate metabolism pathways were upregulated only in the kidney. These results suggest that fenofibrate exposure significantly perturbed biological pathways related to lipid metabolism in the liver and the kidney, with the magnitude of the perturbations much greater in the liver.
Fenofibrate induced distinct effects on kidney and liver PPARα expression, lipoprotein metabolism, and fatty acid transport
As described in Fig. 3C, fenofibrate exposure activated several common lipid metabolism-related pathways in both the kidney and liver, however, the magnitude of change for each pathway in each tissue indicated subtle differences between them. Therefore, we explored dose-dependent alterations in several genes involved in lipid metabolism for the fenofibrate groups compared to their controls. Figure 4 shows the effect of fenofibrate exposure on the expression of the PPARα and RXRα genes as well as the genes for lipoprotein metabolism and fatty acid transport. Our analysis revealed that fenofibrate consistently downregulated the PPARα and RXRα genes in the kidney and that the changes were significant (FDR < 0.1, bars in green) starting with a low dose (16 mg/kg) (Fig. 4A, top panel). Similarly, although fenofibrate exposure downregulated these genes in the liver at low dose levels, they were not statistically significant (Fig. 4A, bottom panel). However, in contrast, expression of these genes was significantly upregulated (FDR < 0.1; bars in red) in the liver as the fenofibrate concentration increased to higher dose levels (125 mg/kg), suggesting differential modes of regulation for these transcription factor genes between the liver and kidney upon fenofibrate activation.
Fig. 4.
Effect of fenofibrate on PPARα and RXRα, lipoprotein metabolism, and fatty acid transport in rat kidney and liver. Fenofibrate induced dose-dependent alterations in the logarithmic fold-change (FC) values of the following genes: (A) Ppara and Rxra; (B) lipoprotein lipase (Lpl) and phospholipid transport protein (Pltp); (C) apolipoprotein A1 (Apoa1), apolipoprotein A2 (Apoa2), and apolipoprotein C3 (Apoc3); (D) solute carrier family 27 member 1 and member 2 (Slc27a1 and Slc27a2); (E) fatty acid translocase (Cd36) and fatty acid binding proteins (Fabp1 and Fabp7); and (F) mitochondrial carnitine palmitoyltransferases (Cpt1b and Cpt2). For each gene, individual bars represent fenofibrate dose groups ranging from a low concentration (left) to the highest concentration (right). Green and red indicate significant gene downregulation and upregulation (false discovery rate < 0.1), respectively, and gray bars indicate no change.
Upon activation by fenofibrate, PPARα modulates lipoprotein metabolism to lower plasma triglycerides (TGs), and clearance of TG-rich lipoproteins is mediated by the enzyme product of the genes lipoprotein lipase (Lpl) and apolipoprotein c3 (Apoc3). In our 5-day rat fenofibrate exposure study, we observed a significant dose-dependent upregulation of Lpl gene expression in the liver starting with the 16 mg/kg dose (Fig. 4B, bottom panel), whereas it was consistently unchanged at any dose in the kidney, indicating major differences in lipoprotein metabolism between the liver and kidney. We observed a similar behavior for the Apoc3 gene in the liver and kidney (Fig. 4C), except Apoc3 gene expression was significantly downregulated in the liver while unchanged in the kidney. In addition, fenofibrate-induced alterations in genes involved in high-density lipoprotein (HDL) metabolism showed a similar trend (Fig. 4C). We observed consistent downregulation of the genes apolipoprotein a1 and a2 (Apoa1 and Apoa2) in the rat liver compared to the kidney upon fenofibrate exposure. However, in contrast, we observed that the phospholipid transport protein gene (Pltp), which is involved in lipoprotein metabolism, was significantly downregulated in the kidney at the highest fenofibrate dose but was unchanged in the liver, indicating contrasting roles of these genes between the organs.
Induction of the Lpl gene leads to conversion of TG-rich lipoproteins into free fatty acids, which need to be transferred across the cell membrane before they can be metabolized in the liver or kidney, and there are several proteins involved in fatty acid transport across the plasma membrane with both transporter and acyl-CoA synthetase activity. Our studies showed that the genes for fatty acid transport proteins belonging to the solute carrier family (Slc27a1 and Slc27a2) were significantly upregulated in both the liver and kidney, with stronger perturbations in the liver compared to the kidney (Fig. 4D). However, in contrast, fenofibrate exposure significantly upregulated expression of the fatty acid translocase gene (Cd36) in the liver whereas it was unchanged in the kidney (Fig. 4E). We observed a similar behavior for a group of high-affinity fatty acid binding proteins (Fabp1 and Fabp7) that play a significant role in partitioning fatty acids to specific lipid metabolic pathways for further processing. In addition, we observed that a rate-limiting gene involved in fatty acid uptake into the mitochondria (Cpt1b) was significantly upregulated in the liver yet unchanged in the kidney upon fenofibrate exposure (Fig. 4F), indicating major differences in fatty acid uptake between the liver and kidney. However, the gene responsible for the subsequent step in fatty acid uptake into the mitochondria (Cpt2) was upregulated in both the liver and kidney, with higher perturbations in the liver than the kidney, indicating a systematic subdued regulation of fatty acid uptake in rat kidney metabolism.
PPARα differentially regulated lipid synthesis and fatty acid oxidation in the kidney and liver with fenofibrate exposure
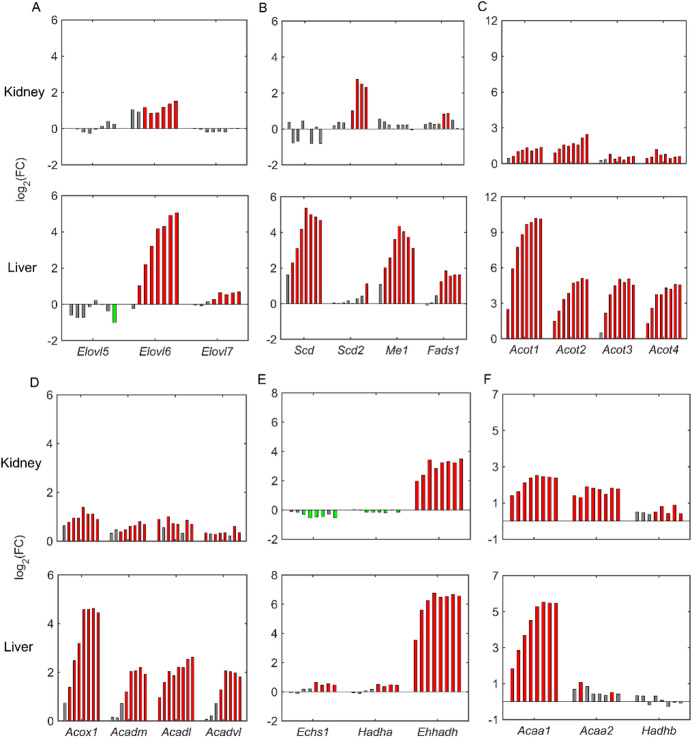
Pharmacological activation of PPARα with fenofibrate leads to a significant upregulation of numerous genes involved in fatty acid elongation (Elovl family) and fatty acid desaturation (Scd). Figure 5 compares the logarithmic FC values of the genes involved in lipogenesis and fatty acid oxidation between the liver and kidney. We observed a dose-dependent upregulation of the fatty acid elongation genes Elovl6 and Elovl7 in the liver, indicating a major increase in fatty acid synthesis in liver. In contrast, only the Elvol6 gene was upregulated in the kidney, and the magnitude of change was three times lower than that in the liver at the highest fenofibrate concentration (Fig. 5A). Similarly, we observed a strong upregulation of the fatty acid desaturation (Scd and Fads1) and malic enzyme (Me1) genes in the liver, whereas none of them was significantly altered in the kidney (Fig. 5B). However, one of the isoforms of the Scd gene (i.e., Scd2) was significantly upregulated in the kidney due to fenofibrate exposure, indicating notable differences in lipogenesis between the kidney and liver. Furthermore, we observed that numerous peroxisomal acyl-CoA thioesterase genes (Acot1-4), which convert acyl-CoAs into fatty acids, were significantly upregulated both in the liver and the kidney, but with a higher magnitude of change in the liver compared to the kidney (Fig. 5C).
Fig. 5.
Effect of fenofibrate on lipid synthesis and fatty acid oxidation in rat kidney and liver. Fenofibrate induced dose-dependent alterations in the logarithmic fold-change (FC) values of the following genes for lipid synthesis: (A) Elovl5, Elovl6, and Elovl7; (B) Scd, Scd2, Me1, and Fads1; (C) Acot1–4; and fatty acid oxidation: (D) Acox1, Acadm, Acadl, and Acadvl; (E) Echs1, Hadha, and Ehhadh; and (F) Acaa1, Acaa2, and Hadhb. For each gene, individual bars represent fenofibrate dose groups ranging from a low concentration (left) to the highest concentration (right). Green and red indicate significant gene downregulation and upregulation (false discovery rate < 0.1), respectively, and gray bars indicate no change.
A well-known function of PPARα upon physiological or pharmacological activation is to regulate fatty acid oxidation in peroxisomes, mitochondria, and microsomes. Our results show that the acyl-CoA oxidase gene (Acox1), which encodes the first enzyme in peroxisomal long-chain fatty acid oxidation, was significantly dose-dependently upregulated in the liver upon fenofibrate exposure (Fig. 5D, bottom panel). We observed a similar behavior with the acyl-CoA dehydrogenase genes (Acadm, Acadl, and Acadvl) that are involved in the first step of mitochondrial fatty acid oxidation. In contrast, although we observed upregulation of these peroxisomal and mitochondrial genes in the kidney, the magnitude of the change was much lower compared to the liver (Fig. 5D, top panel). In addition, we did not see a clear dose-dependent behavior in the kidney as expression was saturated at low fenofibrate concentrations. However, we observed comparable responses between the kidney and liver for the subsequent steps downstream of Acox1 in the peroxisomal fatty acid oxidation pathway, i.e., for the bifunctional 3-hydroxyacyl-CoA dehydrogenase gene (Ehhadh) and the peroxisomal 3-ketoacyl-CoA thiolase activity gene (Acaa1) (Fig. 5E, F). We did not see a similar response for the downstream genes involved in mitochondrial fatty acid oxidation. For example, the genes Echs1 and Hadha were marginally upregulated in the liver but downregulated in the kidney (Fig. 5E). Furthermore, the genes Acaa2 and Hadhb were significantly upregulated in the kidney even at a low fenofibrate concentration but were unchanged in the liver (Fig. 5F), suggesting crucial differences in fatty acid oxidation between the two organs.
Fenofibrate significantly upregulated branched-chain amino acid degradation in the liver and ketogenesis in both the liver and kidney
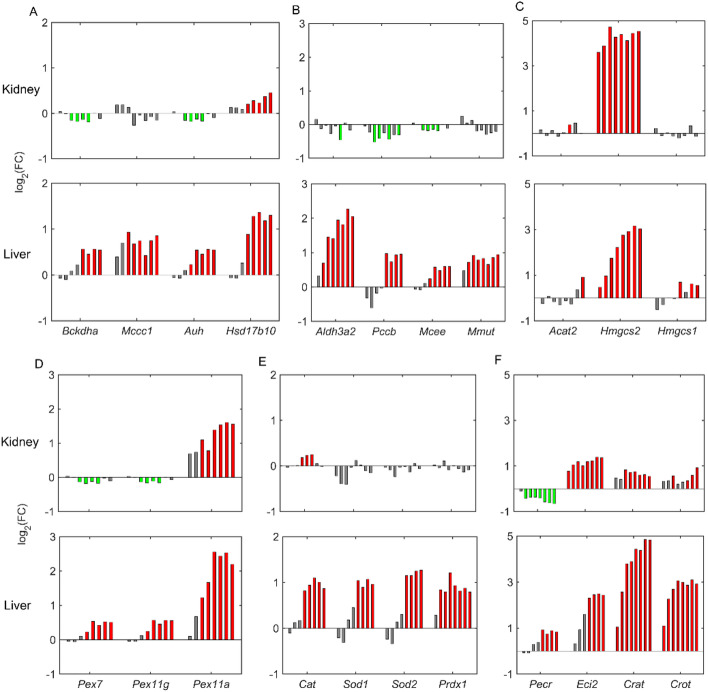
Upregulation of peroxisomal and mitochondrial fatty acid oxidation can result in excess production of acetyl-CoA, which feeds into ketogenesis pathways. Figure 6 shows the expression of several genes involved in ketogenesis and branched-chain amino acid (BCAA) degradation pathways that lead to production of ketone bodies. Our results show that fenofibrate exposure resulted in dose-dependent upregulation of several essential genes in BCAA catabolism in the liver, including the rate-limiting gene branched-chain α-ketoacid dehydrogenase complex (Bckdha) and several intermediate genes, such as Mccc1, Auh, Hsd17b10, Aldh3a2, Pccb, Mcee, and Mmut, that are part of BCAA degradation (Fig. 6A, B). Interestingly, except for Hsd17b10, none of the BCAA degradation genes was upregulated in the kidney, indicating that BCAA catabolism is not directly controlled by PPARα in kidney tissue. However, we observed a predominant upregulation of the HMG-CoA synthase gene (Hmgcs2) in both tissues, with a marginally higher FC value in the kidney, indicating similarity between the liver and kidney with respect to PPARα regulation of the ketogenesis pathway (Fig. 6C). These results indicate that the kidney plays an important role in the overall production of ketone bodies upon fenofibrate treatment.
Fig. 6.
Effect of fenofibrate on branched-chain amino acid degradation, ketogenesis, and peroxisomes in rat liver and kidney. Fenofibrate induced dose-dependent alterations in logarithmic fold-change (FC) values of the following genes for branched-chain amino acid degradation: (A) Bckdha, Mccc1, Auh, and Hsd17b10; (B) Aldh3a2, Pccb, Mcee, and Mmut; ketogenesis: (C) Acat2, Hmgcs2, and Hmgcs1; peroxisomal division: (D) Pex7, Pex11g, and Pex11a; antioxidant systems: (E) Cat, Sod1, Sod2, and Prdx1; and fatty acid oxidation: (F) Pecr, Eci2, Crat, and Crot. For each gene, individual bars represent fenofibrate dose groups ranging from a low concentration (left) to the highest concentration (right). Green and red indicate significant gene downregulation and upregulation (false discovery rate < 0.1), respectively, and gray bars indicate no change.
Fenofibrate induced stronger peroxisomal perturbations in the liver compared to the kidney
Pharmacological activation of PPARα is known to cause massive peroxisome proliferation in rodents via induction of several genes involved in fatty acid oxidation and peroxisomal biogenesis. Our 5-day rat exposure study results show that several genes involved in import of peroxisomal proteins and peroxisomal division (Pex family) were consistently upregulated in the liver with fenofibrate exposure (Fig. 6D, bottom panel). However, we observed downregulation of genes Pex7 and Pex11g in the kidney, indicating differences between the liver and kidney. We observed a similar behavior with respect to alterations in the antioxidant system in the liver, where the catalase (Cat), superoxide dismutase (Sod1 and Sod2), and peroxiredoxin (Prdx1) genes were significantly dose-dependently upregulated in the liver but were unchanged in the kidney with fenofibrate exposure (Fig. 6E). Furthermore, our results also indicated upregulation of several other genes involved in peroxisomal fatty acid oxidation, such as Eci2, Crat, and Crot, both in the liver and kidney, with the exception of Pecr, which was downregulated in the kidney compared to the liver (Fig. 6F). These results show that fenofibrate exposure resulted in much stronger perturbations in liver peroxisomes compared to the kidney.
Effect of fenofibrate exposure on energy metabolism, gluconeogenesis, and histidine metabolism
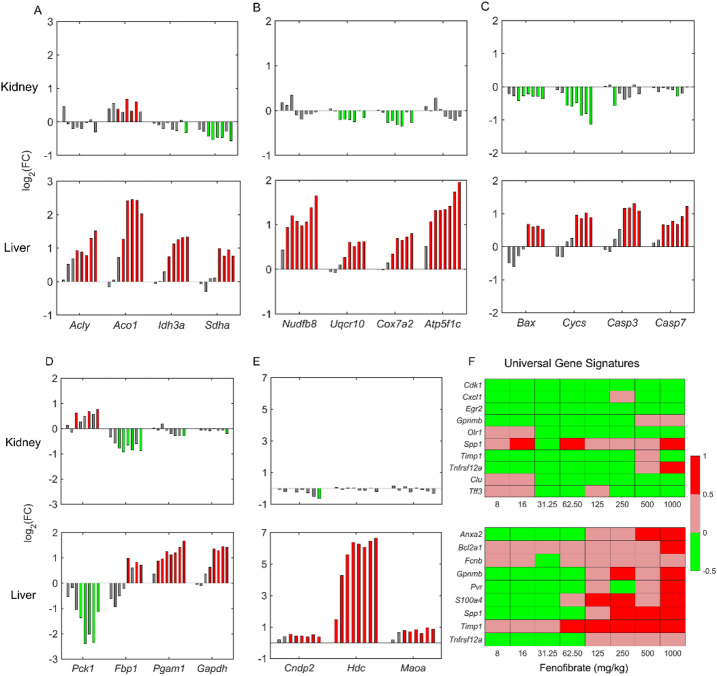
Our observation of upregulated fatty acid oxidation in peroxisomes and mitochondria leads to excess production of acetyl-CoA, which can be utilized in the mitochondrial cellular ATP generation process. To test whether fenofibrate had any effect on mitochondrial energy metabolism, we explored alterations in several genes in the tricarboxylic acid (TCA) cycle, oxidative phosphorylation, and other mitochondrial processes related to cell death (Fig. 7A-C). Our results show a dose-dependent increase in several genes in the TCA cycle in the liver but not in the kidney (Fig. 7A), indicating a strong upregulation of the TCA cycle in the liver due to fenofibrate exposure. The TCA cycle generates electron carriers, such as NADH and FADH2, which are transferred to oxidative phosphorylation in the mitochondria to generate ATP via the electron transport chain (ETC). Similar to the genes in the TCA cycle, we observed a dose-dependent increase in a majority of the genes that participate in the ETC in the liver, but they were either not changed or downregulated in the kidney (Fig. 7B). Furthermore, we observed a dose-dependent upregulation of several genes in the mitochondrial processes related to cell death at high fenofibrate concentrations in the liver but not in the kidney (Fig. 7C), indicating perturbed redox balance in the liver mitochondria that may result in oxidative stress.
Fig. 7.
Effect of fenofibrate on energy metabolism, gluconeogenesis, histidine metabolism, and the overall expression pattern of universal gene signatures for necrosis in rat kidney and liver. Fenofibrate induced dose-dependent alterations in logarithmic fold-change (FC) values of the following genes for the tricarboxylic acid cycle: (A) Acly, Aco1, Idh3a, and Sdha; oxidative phosphorylation: (B) Nudfb8, Uqcr10, Cox7a2, and Atp5f1c; mitochondrial cell death: (C) Bax, Cycs, Casp3, and Casp7; gluconeogenesis: (D) Pck1, Fbp1, Pgam1, and Gapdh; and histidine metabolism: (E) Cndp2, Hdc, and Maoa. (F) Universal gene signatures for necrosis in the kidney and liver. For each gene, individual bars represent fenofibrate dose groups ranging from a low concentration (left) to the highest concentration (right). Green and red indicate significant gene downregulation and upregulation (false discovery rate < 0.1), respectively, and gray bars indicate no change.
A number of genes involved in liver gluconeogenesis are known to be targets of PPARα regulation. Figure 7D compares the effect of fenofibrate exposure on the genes involved in gluconeogenesis between the kidney and liver. Interestingly, one of the crucial genes involved in gluconeogenesis, phosphoenolpyruvate carboxykinase (Pck1), was consistently downregulated in the liver but upregulated in the kidney. However, in contrast, several genes downstream of Pck1 showed the opposite behavior, being dose-dependently upregulated in the liver (Fig. 7D, bottom panel) but either downregulated or unchanged in the kidney (Fig. 7D, top panel), indicating differential regulation of gluconeogenesis between the two organs. We observed a similar behavior for the histidine metabolism genes upon fenofibrate treatment (Fig. 7E). Specifically, the histidine decarboxylase gene (Hdc) was significantly upregulated (FC change > 6) only in the liver, indicating a stronger PPARα regulation in the liver than the kidney.
Fenofibrate exposure caused cellular degeneration or necrosis at higher doses in the liver
Glaab et al.57 previously identified a set of tissue-specific universal genes and their upregulated expression patterns that serve as an indicator of cellular degeneration or necrosis. To examine the effect of fenofibrate treatment on liver and kidney injury, we performed a universal toxicity gene signature analysis57 using the gene expression alterations from the 5-day rat studies. Figure 7F shows the dose-dependent expression patterns of the genes that are part of the kidney and liver universal gene signatures. Our results show that the kidney universal toxicity genes were consistently downregulated across the fenofibrate concentrations, with low FC values (Fig. 7F, top panel). However, in contrast, the liver universal toxicity genes were downregulated (indicated in green) at low concentrations of fenofibrate (Fig. 7F, bottom panel) but significantly upregulated (indicated in red) as the fenofibrate concentration increased above 125 mg/kg. These results indicate that fenofibrate exposure did not cause kidney cell necrosis at any concentration but led to liver cell degeneration at higher dose levels, indicating a potential injury initiation process that can result in liver injury.
Discussion
Fibrates, such as fenofibrate, have been known as activators of peroxisome proliferation in rodents for many years21,25,58. They have been approved for the treatment of dyslipidemia and act via PPARα, which controls lipid metabolism and glucose homeostasis59. The PPARα receptor is highly expressed in the liver, kidney, and heart, and fibrates decrease plasma lipid levels and induce hepatomegaly and hepatic peroxisome proliferation in a PPARα-dependent manner, as proven using a PPARα knockout mouse60. In addition, several differences in PPARα function between murine species and humans have been identified in PPARα expression, ligand activation, and biological responses, particularly its role in peroxisome proliferation in rats and mice but not humans61. Although fenofibrate-induced alterations are well elucidated in the mouse liver and to some extent in the rat liver, there are few studies that examined fenofibrate exposure in the rat kidney. Furthermore, the majority of these studies were conducted in mice to explore the protective role of fibrates in various liver and kidney diseases62–65. Therefore, in this study, using healthy rats under normal feeding conditions, we aimed to explore how fenofibrate regulates lipid metabolism and its potential to cause toxicity in the rat kidney compared to the liver by performing a high-throughput transcriptomics analysis. To this end, we explored gene expression data from rats that received either vehicle control or fenofibrate for 5 days, with fenofibrate concentrations per group (n = 4) ranging from low to high doses. We determined how fenofibrate exposure altered the rats’ body and organ weights, explored dose-dependent alterations in significant genes and pathways, and identified potential commonalities and differences in gene expression changes related to PPARα regulation across all dose levels.
The 5-day rat exposure data used in this study were collected for eight different concentrations of fenofibrate45, which allowed us to determine its effect across low and high dose levels. The selected low dose levels allowed us to examine fenofibrate-induced alterations that are relevant for human therapeutic doses, while the high dose levels can be useful to probe fenofibrate’s effect on the liver and kidney under overdose conditions. Consistent with previous studies, exposure to fenofibrate resulted in a dose-dependent enlargement of liver cells, leading to hepatocellular hypertrophy60, which we did not see in the kidney. Furthermore, we observed larger gene perturbations in the kidney at a lower fenofibrate concentration (31.25 mg/kg) than in the liver (62.50 mg/kg), indicating differential sensitivity to fenofibrate exposure (Fig. 2). However, based on the magnitude of the change in gene expression, most of these changes at low concentrations in the kidney were not biologically meaningful, and fenofibrate induced more gene perturbations in the liver than the kidney at higher dose levels, based on FC value thresholds. Furthermore, we observed several common biological pathways in lipid metabolism that were significantly upregulated upon high-dose fenofibrate exposure, indicating similarities between the liver and kidney (Fig. 3C). For example, we observed upregulation of the pathways for biosynthesis of unsaturated fatty acids, fatty acid degradation, PPAR signaling, fatty acid elongation, and peroxisome proliferation, which are known to be upregulated by fenofibrate-induced PPARα activation. However, differences in the magnitude of the perturbations for these pathways between the liver and kidney suggest potential subtle differences in PPARα regulation.
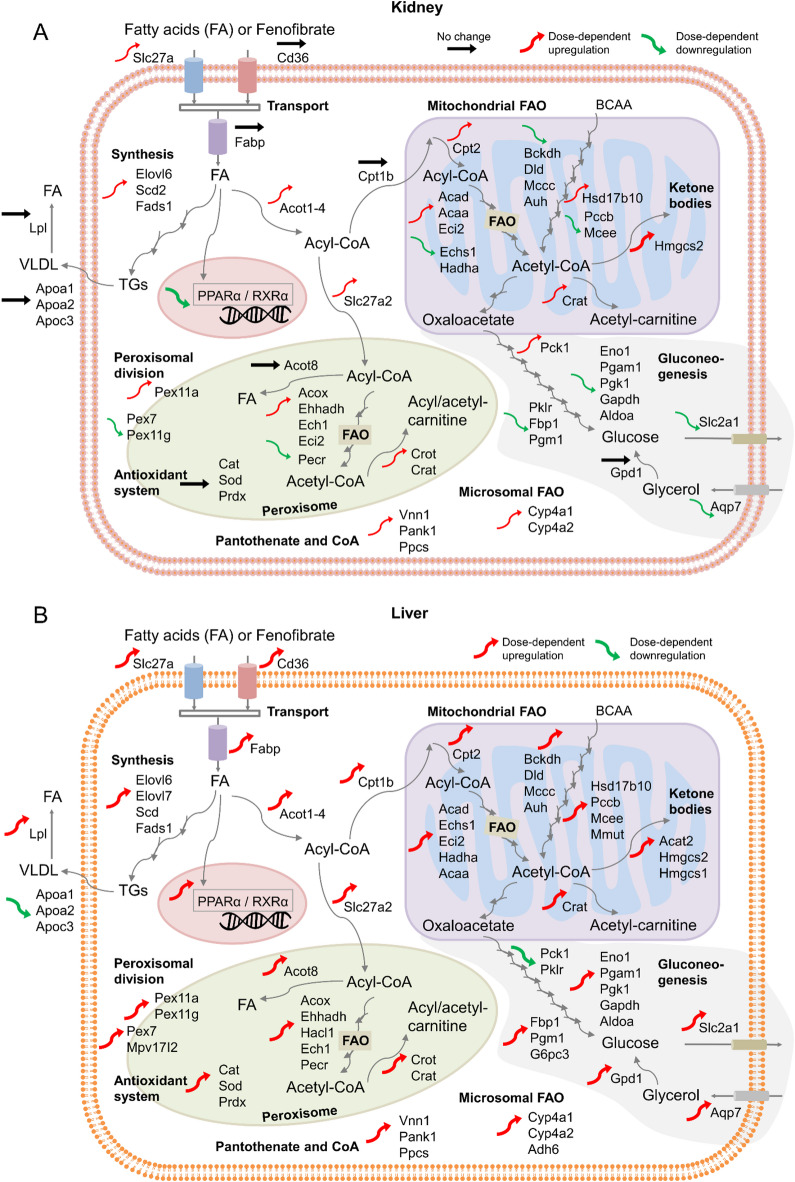
We compared the dose-dependent fenofibrate exposure data to identify potential similarities and differences in the magnitude of perturbations in PPARα target genes in the liver and kidney. Figure 8 summarizes the overall directionality of the changes in the genes regulated by PPARα for different cellular metabolic processes in the kidney and liver, including changes in PPARα and RXRα genes, lipoprotein metabolism, lipid transport, lipid synthesis, fatty acid oxidation, peroxisome proliferation, and gluconeogenesis. Since the tissue expression levels of PPARα are tightly connected to its function, we first compared how fenofibrate altered the expression of the PPARα and RXRα genes. Interestingly, we found the expression of these genes to be regulated in opposite directions, with consistent downregulation in the kidney and upregulation in the liver (Fig. 4A), indicating tissue-specific regulation of PPARα between the two organs in healthy rats under normal feeding conditions. Indeed, studies using primary cultures of human hepatocytes reported a marked upregulation of PPARα mRNA levels upon fenofibrate exposure, suggesting that PPARα positively autoregulates its own expression in the liver compared to the kidney66. We hypothesize that, in addition to tissue-specific regulation, the downregulated PPARα expression in the kidney can be associated with the subdued responses of its target genes to fenofibrate exposure.
Fig. 8.
Summary of fenofibrate-induced alterations of lipid metabolism in the rat kidney and liver. Black, red, and green arrows indicate genes that showed no change, dose-dependent upregulation, and dose-dependent downregulation, respectively. Thick arrows indicate a stronger response with a higher fold-change value. BCAA, branched-chain amino acid; FAO, fatty acid oxidation; PPAR, peroxisome proliferator-activated receptor; RXR, retinoid X receptor; TGs, triglycerides; VLDL, very-low-density lipoprotein.
In our study, we also observed some fundamental differences in the activation of PPARα by fenofibrate between the liver and kidney with respect to its therapeutic effect on lipoprotein metabolism. The hypotriglyceridemic action of fenofibrate involves a combined effect on Lpl and Apoc3 gene expression, resulting in increased lipolysis28,67. Consistent with literature studies, we observed increased Lpl gene expression and reduced expression of Apoc3, a natural inhibitor of Lpl activity, in the rat liver. However, we did not see any change in these genes in the kidney with fenofibrate (Fig. 8A), suggesting no effect of fenofibrate on kidney lipoprotein metabolism. Similarly, we also observed differences in HDL metabolism: the major HDL apolipoproteins, Apoa1 and Apoa2, were downregulated in the liver but unchanged in the kidney with fenofibrate. In contrast, in humans, fenofibrate increased the expression of these two genes, contributing to increased plasma HDL levels40, suggesting differences in fenofibrate activation between the species.
Lipid metabolism in the liver and kidney is comprised of four primary processes: uptake, synthesis, fatty acid oxidation, and ketogenesis68,69. Interestingly, we identified distinct effects of fenofibrate exposure on these processes between the rat liver and kidney and other species. For example, fatty acid uptake in the rat kidney is governed by the Fatp genes (Slc27a1 and Slc27a2) upon fenofibrate exposure (Fig. 8A), whereas it is controlled by multiple transport genes (Fatp, Cd36, and Fabp) in the rat liver, as reported in mouse and human liver studies70,71. In addition, indicating differences in the rate-limiting gene for fatty acid import into the mitochondria, we found that fenofibrate exposure upregulated the expression of the Cpt1b gene in the rat liver but upregulated the Cpt1a gene in mouse and human studies72. Furthermore, neither of these genes was affected in the rat kidney, indicating major differences in the fatty acid uptake mechanisms between the organs and species. However, we did observe similarities in several PPARα-governed fatty acid catabolism genes that can be rate limiting, e.g., the β-oxidation of peroxisomes (Acox, Ehhadh, and Acot), mitochondria (Acad and Acaa), and microsomes [cytochrome P450 family 4 subfamily A (Cyp4A)]. In addition, we also found similarities across the species and organs in the expression of ketogenic enzymes required to convert acetyl-CoA to ketone bodies (Hmgcs2) and in genes involved in de novo lipogenesis (Elovl6, Scd, and Fads1), including the gene sterol regulatory element binding transcription factor 1 (Srebf1), which regulates lipogenesis. We also observed similarities in upregulation of the gene Vanin-1 (Vnn1) that catalyzes the hydrolysis of pantetheine into pantothenic acid, which is a precursor in the synthesis of CoA. All of these genes were significantly upregulated in both the liver and kidney; however, they were expressed at a higher level in the rat liver, indicating a stronger response in the liver compared to the kidney.
Beyond the transcriptional regulation of genes involved in lipid metabolism, the PPARα agonist fenofibrate affects glucose and amino acid metabolism73–75. Several studies have shown that the expression of numerous genes involved in amino acid metabolism and urea synthesis was markedly decreased in the liver upon treatment with fibrates75. In our study, although some of the genes involved in these pathways were downregulated, the magnitude of the change was below the imposed FC threshold in both the liver and kidney. However, we observed that several genes in the BCAA degradation pathway were significantly upregulated in the liver but not in the kidney, indicating tissue-specific effects of fenofibrate exposure. Similarly, our study showed that the gluconeogenesis pathways were differentially regulated between the liver and kidney (Fig. 8), with genes involved in the production of glucose upregulated in the liver, including the conversion of glycerol to glucose, and either downregulated or unchanged in the kidney. In addition, we observed marked differences in histidine metabolism only in the liver. Particularly, we observed a more than six-fold change in Hdc, an essential gene in liver histidine metabolism that converts histidine to histamine. Previous studies demonstrated that Hdc expression increased with repeated fibrate administration and hypothesized that it could be involved in hepatic cell proliferation and hypertrophy induced by PPARα agonists76.
In addition to controlling peroxisomal fatty acid oxidation, fibrate-induced PPARα activation has been shown to cause massive peroxisome proliferation in rodents77. In our study, we observed substantial PPARα-mediated upregulation of peroxisomal division (Pex11 genes) and the antioxidant system in the rat liver compared to the kidney. Furthermore, our observations of a dose-dependent increase in absolute and relative liver weights at the end of the 5-day exposure confirm that fenofibrate exposure leads to increases in hepatocyte size due to peroxisome proliferation in the liver. In contrast, although fenofibrate induced the gene Pex11 in humans78, multiple studies reported no increase in hepatocyte size in humans, indicating species-specific functionalities associated with fenofibrate exposure42,61,79.
Finally, despite fenofibrate’s use in hyperlipidemia management and its therapeutic potential for various liver and kidney diseases80,81, its effects on hepatomegaly and liver regeneration as well as its mechanisms of action in liver and kidney injuries remain unclear44,82,83. In our study, we investigated fenofibrate’s potential to cause cellular degeneration or necrosis based on universal gene signatures identified for liver and kidney injuries57. Our results revealed that fenofibrate exposure clearly leads to liver necrosis and potential injury at high doses but may not induce cellular necrosis in the kidney in rats under normal feeding conditions. Indeed, our previous study showed that cellular degeneration was a highly activated injury phenotype for fenofibrate exposure, confirming its potential for hepatotoxicity in rats49. We hypothesize that excessive upregulation of fatty acid uptake and fatty acid oxidation in the liver leads to upregulated mitochondrial ATP generation via an upregulated TCA cycle and oxidative phosphorylation (Fig. 7A, B), which can result in excess mitochondrial superoxide and hydrogen peroxide production. This may result in cellular redox imbalance, leading to oxidative stress and cellular damage. In addition, we also observed a substantial increase in peroxisomal proliferation in the liver, which can also lead to excess hydrogen peroxide production that can contribute to lipid peroxidation. Our results indeed show that the genes responsible for programmed cell death, such as Bax, Cycs, Casp3, Casp7, and Casp8, were significantly upregulated at fenofibrate concentrations above 125 mg/kg (Fig. 7C), a potential transcriptomic inflection point, where the universal gene signatures indicated initiation of cellular necrosis in the liver (Fig. 7F). These results suggest that mitochondrial damage due to oxidative stress may be one of the mechanisms through which fenofibrate exposure leads to liver injury at high concentrations, which is likely a secondary manifestation of the pharmacological effects of fenofibrate that have been shown to be specific to rodents.
In conclusion, our results show dose-dependent alterations in lipid metabolism in the rat liver and kidney with fenofibrate exposure. The transcriptional changes identified in our study are largely consistent with fenofibrate’s effect on the activation of PPARα in rat liver and kidney metabolism. However, using our 5-day exposure study data, we identified some novel differences in fenofibrate’s action in the modulation of lipid metabolism between the liver and kidney. Particularly, our study revealed that fenofibrate differentially modulates the expression of PPARα and RXRα genes between the liver and kidney, which could explain the observed relatively lower magnitude of change in several lipid metabolism-related genes in the kidney compared to the liver. Furthermore, our observations of differences in cellular and mitochondrial fatty acid uptake, peroxisomal division and antioxidant systems, BCAA metabolism, histidine metabolism, and gluconeogenesis between the liver and kidney may play an important role in deciphering the beneficial effects of fenofibrate at low doses from its potential to cause liver injury at high dose levels. To the best of our knowledge, this is the first study to compare dose-dependent fenofibrate exposure data between the liver and kidney, provide a detailed summary of differences in PPARα-mediated regulation of liver metabolism, and identify potential mechanisms through which fenofibrate may lead to liver injury at high concentrations. Therefore, our study provides an improved understanding of PPARα activation by its agonists and may help in designing future therapeutic developments targeted at modulating lipid metabolism in the liver and kidney.
Acknowledgements
The opinions and assertions contained herein are the private views of the authors and are not to be construed as official or as reflecting the views of the Defense Health Agency, the U.S. Department of Defense, or The Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc. This paper was approved for public release with unlimited distribution.
Author contributions
Conceptualization, V.R.P. and A.W.; methodology and analysis, V.R.P.; data curation, processing, and quality control, M.R.B.-M., D.M., D.P.P., E.H.S., R.R.S., W.C., and S.S.A.; writing—original draft preparation, V.R.P.; writing—review and editing, V.R.P., M.R.B.-M., D.M., D.P.P., E.H.S., R.R.S., W.C., S.S.A., and A.W.; supervision, A.W.; funding acquisition, A.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the NIH, National Institute of Environmental Health Sciences, through Intramural Research Project ZIAES103385 and Interagency Agreement No. AES22010-001-00001 between NIEHS and BHSAI. The Henry M. Jackson Foundation was supported by the U.S. Army Medical Research and Development Command under Contract Nos. W81XWH20C0031 and HT942524F0189.
Data availability
The datasets presented in this study are derived from the original study, which is openly available in the NCBI’s GEO database gene repository for rats under accession number GSM4415261. The derived datasets supporting the conclusions of this article will be made available by the authors on request.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Venkat R. Pannala, Email: vpannala@bhsai.org
Anders Wallqvist, Email: sven.a.wallqvist.civ@health.mil.
References
- 1.Chandel, N. S. Lipid metabolism. Cold Spring Harb Perspect. Biol.13, a040576. 10.1101/cshperspect.a040576 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Santos, A. L. & Preta, G. Lipids in the cell: organisation regulates function. Cell. Mol. Life Sci.75, 1909–1927. 10.1007/s00018-018-2765-4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schaffer, J. E. Lipotoxicity: when tissues overeat. Curr. Opin. Lipidol.14, 281–287. 10.1097/00041433-200306000-00008 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Goldberg, I. J., Trent, C. M. & Schulze, P. C. Lipid metabolism and toxicity in the heart. Cell. Metab.15, 805–812. 10.1016/j.cmet.2012.04.006 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weinberg, J. M. & Lipotoxicity Kidney Int.70, 1560–1566, doi:10.1038/sj.ki.5001834 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Monteillet, L. et al. Intracellular lipids are an independent cause of liver injury and chronic kidney disease in non alcoholic fatty liver disease-like context. Mol. Metab.16, 100–115. 10.1016/j.molmet.2018.07.006 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ipsen, D. H., Lykkesfeldt, J. & Tveden-Nyborg, P. Molecular mechanisms of hepatic lipid accumulation in non-alcoholic fatty liver disease. Cell. Mol. Life Sci.75, 3313–3327. 10.1007/s00018-018-2860-6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geng, Y., Faber, K. N., de Meijer, V. E., Blokzijl, H. & Moshage, H. How does hepatic lipid accumulation lead to lipotoxicity in non-alcoholic fatty liver disease? Hepatol. Int.15, 21–35. 10.1007/s12072-020-10121-2 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giardini, E., Moore, D., Sadlier, D., Godson, C. & Brennan, E. The dual role of lipids in chronic kidney disease: pathogenic culprits and therapeutic allies. Atherosclerosis398, 118615. 10.1016/j.atherosclerosis.2024.118615 (2024). [DOI] [PubMed] [Google Scholar]
- 10.Gai, Z. et al. Lipid accumulation and chronic kidney disease. Nutrients11, 722. 10.3390/nu11040722 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Agrawal, S., Zaritsky, J. J., Fornoni, A. & Smoyer, W. E. Dyslipidaemia in nephrotic syndrome: mechanisms and treatment. Nat. Rev. Nephrol.14, 70. 10.1038/nrneph.2017.175 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Walczak, R. & Tontonoz, P. PPARadigms and PPARadoxes: expanding roles for PPARgamma in the control of lipid metabolism. J. Lipid Res.43, 177–186 (2002). [PubMed] [Google Scholar]
- 13.Varga, T., Czimmerer, Z. & Nagy, L. PPARs are a unique set of fatty acid regulated transcription factors controlling both lipid metabolism and inflammation. Biochim. Biophys. Acta. 1812, 1007–1022. 10.1016/j.bbadis.2011.02.014 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harmon, G. S., Lam, M. T. & Glass, C. K. PPARs and lipid ligands in inflammation and metabolism. Chem. Rev.111, 6321–6340. 10.1021/cr2001355 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chinetti, G., Fruchart, J. C. & Staels, B. Peroxisome proliferator-activated receptors and inflammation: from basic science to clinical applications. Int. J. Obes. Relat. Metab. Disord. 27 (Suppl 3), S41–S45. 10.1038/sj.ijo.0802499 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Bookout, A. L. et al. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell126, 789–799. 10.1016/j.cell.2006.06.049 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feige, J. N., Gelman, L., Michalik, L., Desvergne, B. & Wahli, W. From molecular action to physiological outputs: peroxisome proliferator-activated receptors are nuclear receptors at the crossroads of key cellular functions. Prog Lipid Res.45, 120–159. 10.1016/j.plipres.2005.12.002 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Kersten, S. & Stienstra, R. The role and regulation of the peroxisome proliferator activated receptor alpha in human liver. Biochimie136, 75–84. 10.1016/j.biochi.2016.12.019 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Bougarne, N. et al. Molecular actions of PPARalpha in lipid metabolism and inflammation. Endocr. Rev.39, 760–802. 10.1210/er.2018-00064 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Bocher, V., Pineda-Torra, I., Fruchart, J. C. & Staels, B. PPARs: transcription factors controlling lipid and lipoprotein metabolism. Ann. Acad. Sci.967, 7–18. 10.1111/j.1749-6632.2002.tb04258.x (2002). [DOI] [PubMed] [Google Scholar]
- 21.Schoonjans, K., Staels, B. & Auwerx, J. Role of the peroxisome proliferator-activated receptor (PPAR) in mediating the effects of fibrates and fatty acids on gene expression. J. Lipid Res.37, 907–925 (1996). [PubMed] [Google Scholar]
- 22.Hostetler, H. A., Petrescu, A. D., Kier, A. B. & Schroeder, F. Peroxisome proliferator-activated receptor alpha interacts with high affinity and is conformationally responsive to endogenous ligands. J. Biol. Chem.280, 18667–18682. 10.1074/jbc.M412062200 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Forman, B. M., Chen, J. & Evans, R. M. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors alpha and delta. Proc. Natl. Acad. Sci. USA. 94, 4312–4317. 10.1073/pnas.94.9.4312 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Willson, T. M., Brown, P. J., Sternbach, D. D. & Henke, B. R. The PPARs: from orphan receptors to drug discovery. J. Med. Chem.43, 527–550. 10.1021/jm990554g (2000). [DOI] [PubMed] [Google Scholar]
- 25.Lalloyer, F. & Staels, B. Fibrates, glitazones, and peroxisome proliferator-activated receptors. Arterioscler. Thromb. Vasc Biol.30, 894–899. 10.1161/ATVBAHA.108.179689 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taniguchi, A. et al. Effects of bezafibrate on insulin sensitivity and insulin secretion in non-obese Japanese type 2 diabetic patients. Metabolism50, 477–480. 10.1053/meta.2001.21028 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Martin, A. et al. Management of dyslipidemia in patients with non-alcoholic fatty liver disease. Curr. Atheroscler Rep.24, 533–546. 10.1007/s11883-022-01028-4 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Staels, B. et al. Mechanism of action of fibrates on lipid and lipoprotein metabolism. Circulation98, 2088–2093. 10.1161/01.cir.98.19.2088 (1998). [DOI] [PubMed] [Google Scholar]
- 29.Davis, T. M. et al. Effects of fenofibrate on renal function in patients with type 2 diabetes mellitus: the Fenofibrate Intervention and Event Lowering in Diabetes (FIELD) study. Diabetologia54, 280–290. 10.1007/s00125-010-1951-1 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Kostapanos, M. S., Florentin, M. & Elisaf, M. S. Fenofibrate and the kidney: an overview. Eur. J. Clin. Invest.43, 522–531. 10.1111/eci.12068 (2013). [DOI] [PubMed] [Google Scholar]
- 31.McKeage, K. & Keating, G. M. Fenofibrate: a review of its use in dyslipidaemia. Drugs71, 1917–1946. 10.2165/11208090-000000000-00000 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Sumida, Y. & Yoneda, M. Current and future pharmacological therapies for NAFLD/NASH. J. Gastroenterol.53, 362–376. 10.1007/s00535-017-1415-1 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mahmoudi, A., Jamialahmadi, T., Johnston, T. P. & Sahebkar, A. Impact of fenofibrate on NAFLD/NASH: a genetic perspective. Drug Discov Today. 27, 2363–2372. 10.1016/j.drudis.2022.05.007 (2022). [DOI] [PubMed] [Google Scholar]
- 34.Rosenson, R. S. Fenofibrate: treatment of hyperlipidemia and beyond. Expert Rev. Cardiovasc. Ther.6, 1319–1330. 10.1586/14779072.6.10.1319 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Kim, S., Ko, K., Park, S., Lee, D. R. & Lee, J. Effect of fenofibrate medication on renal function. Korean J. Fam Med.38, 192–198. 10.4082/kjfm.2017.38.4.192 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hernandez-Arroyo, C. F., Kanduri, S. R., Justiniano, R., Martinez-Pitre, P. J. & Velez, J. C. Q. Improvement in kidney function after discontinuation of fenofibrate in outpatient nephrology consultation for chronic kidney disease. Kidney Blood Press. Res.47, 586–591. 10.1159/000522081 (2022). [DOI] [PubMed] [Google Scholar]
- 37.Ganne-Carrie, N. et al. Autoimmune hepatitis induced by fibrates. Gastroenterol. Clin. Biol.22, 525–529 (1998). [PubMed] [Google Scholar]
- 38.Ho, C. Y. et al. Fenofibrate-induced acute cholestatic hepatitis. J. Chin. Med. Assoc.67, 245–247 (2004). [PubMed] [Google Scholar]
- 39.Martin, G., Schoonjans, K., Lefebvre, A. M., Staels, B. & Auwerx, J. Coordinate regulation of the expression of the fatty acid transport protein and acyl-CoA synthetase genes by PPARalpha and PPARgamma activators. J. Biol. Chem.272, 28210–28217. 10.1074/jbc.272.45.28210 (1997). [DOI] [PubMed] [Google Scholar]
- 40.Vu-Dac, N. et al. Fibrates increase human apolipoprotein A-II expression through activation of the peroxisome proliferator-activated receptor. J. Clin. Invest.96, 741–750. 10.1172/JCI118118 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rakhshandehroo, M. et al. Comprehensive analysis of PPARalpha-dependent regulation of hepatic lipid metabolism by expression profiling. PPAR Res. 2007, 26839. 10.1155/2007/26839 (2007). [DOI] [PMC free article] [PubMed]
- 42.Kersten, S. Integrated physiology and systems biology of PPARalpha. Mol. Metab.3, 354–371. 10.1016/j.molmet.2014.02.002 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharma, R. K. et al. Differential induction of peroxisomal and microsomal fatty-acid-oxidising enzymes by peroxisome proliferators in rat liver and kidney. Characterisation of a renal cytochrome P-450 and implications for peroxisome proliferation. Eur. J. Biochem.184, 69–78. 10.1111/j.1432-1033.1989.tb14991.x (1989). [DOI] [PubMed] [Google Scholar]
- 44.Wronska, A., Kiezun, J. & Kmiec, Z. High-dose fenofibrate stimulates multiple cellular stress pathways in the kidney of old rats. Int. J. Mol. Sci.25, 3038. 10.3390/ijms25053038 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gwinn, W. M. et al. Evaluation of 5-day in vivo rat liver and kidney with high-throughput transcriptomics for estimating benchmark doses of apical outcomes. Toxicol. Sci.176, 343–354. 10.1093/toxsci/kfaa081 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Igarashi, Y. et al. Open TG-GATEs: a large-scale toxicogenomics database. Nucleic Acids Res.43, D921–D927. 10.1093/nar/gku955 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mav, D. et al. A hybrid gene selection approach to create the S1500 + targeted gene sets for use in high-throughput transcriptomics. PLoS One. 13, e0191105. 10.1371/journal.pone.0191105 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol.10, R25. 10.1186/gb-2009-10-3-r25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pannala, V. R. et al. Correction: Pannala et al. High-throughput transcriptomics differentiates toxic versus non-toxic chemical exposures using a rat liver model. Int. J. Mol. Sci.25, 7108. https://doi.org/10.3390/ijms25137108 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci.28, 1947–1951. 10.1002/pro.3715 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kanehisa, M., Furumichi, M., Sato, Y., Matsuura, Y. & Ishiguro-Watanabe, M. KEGG: biological systems database as a model of the real world. Nucleic Acids Res.53, D672–D677. 10.1093/nar/gkae909 (2025). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res.28, 27–30. 10.1093/nar/28.1.27 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu, C. et al. A strategy for evaluating pathway analysis methods. BMC Bioinform.18, 453. 10.1186/s12859-017-1866-7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schyman, P., Xu, Z., Desai, V. & Wallqvist, A. TOXPANEL: a gene-set analysis tool to assess liver and kidney injuries. Front. Pharmacol.12, 601511. 10.3389/fphar.2021.601511 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fan, S. et al. Fenofibrate-promoted hepatomegaly and liver regeneration are PPARalpha-dependent and partially related to the YAP pathway. Acta Pharm. Sin B. 14, 2992–3008. 10.1016/j.apsb.2024.03.030 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamamoto, K., Fukuda, N., Zhang, L. & Sakai, T. Altered hepatic metabolism of fatty acids in rats fed a hypolipidaemic drug, fenofibrate. Pharmacol. Res.33, 337–342. 10.1006/phrs.1996.0046 (1996). [DOI] [PubMed] [Google Scholar]
- 57.Glaab, W. E. et al. Universal toxicity gene signatures for early identification of drug-induced tissue injuries in rats. Toxicol. Sci.181, 148–159. 10.1093/toxsci/kfab038 (2021). [DOI] [PubMed] [Google Scholar]
- 58.Issemann, I. & Green, S. Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature347, 645–650. 10.1038/347645a0 (1990). [DOI] [PubMed] [Google Scholar]
- 59.Katsiki, N. et al. The role of fibrate treatment in dyslipidemia: an overview. Curr. Pharm. Des.19, 3124–3131. 10.2174/1381612811319170020 (2013). [DOI] [PubMed] [Google Scholar]
- 60.Lee, S. S. et al. Targeted disruption of the alpha isoform of the peroxisome proliferator-activated receptor gene in mice results in abolishment of the pleiotropic effects of peroxisome proliferators. Mol. Cell. Biol.15, 3012–3022. 10.1128/MCB.15.6.3012 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cheung, C. et al. Diminished hepatocellular proliferation in mice humanized for the nuclear receptor peroxisome proliferator-activated receptor alpha. Cancer Res.64, 3849–3854. 10.1158/0008-5472.CAN-04-0322 (2004). [DOI] [PubMed] [Google Scholar]
- 62.de la Rosa Rodriguez, M. A. et al. The whole transcriptome effects of the PPARalpha agonist fenofibrate on livers of hepatocyte humanized mice. BMC Genom.19, 443. 10.1186/s12864-018-4834-3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sohn, M. et al. Delayed treatment with fenofibrate protects against high-fat diet-induced kidney injury in mice: the possible role of AMPK autophagy. Am. J. Physiol. Ren. Physiol.312, F323–F334. 10.1152/ajprenal.00596.2015 (2017). [DOI] [PubMed] [Google Scholar]
- 64.Lakhia, R. et al. PPARalpha agonist fenofibrate enhances fatty acid beta-oxidation and attenuates polycystic kidney and liver disease in mice. Am. J. Physiol. Ren. Physiol.314, F122–F131. 10.1152/ajprenal.00352.2017 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tanaka, Y. et al. Fenofibrate, a PPARalpha agonist, has renoprotective effects in mice by enhancing renal lipolysis. Kidney Int.79, 871–882. 10.1038/ki.2010.530 (2011). [DOI] [PubMed] [Google Scholar]
- 66.Pineda Torra, I., Jamshidi, Y., Flavell, D. M., Fruchart, J. C. & Staels, B. Characterization of the human PPARalpha promoter: identification of a functional nuclear receptor response element. Mol. Endocrinol.16, 1013–1028. 10.1210/mend.16.5.0833 (2002). [DOI] [PubMed] [Google Scholar]
- 67.Staels, B. et al. Fibrates downregulate apolipoprotein C-III expression independent of induction of peroxisomal acyl coenzyme A oxidase. A potential mechanism for the hypolipidemic action of fibrates. J. Clin. Invest.95, 705–712. 10.1172/JCI117717 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zou, P. & Wang, L. Dietary pattern and hepatic lipid metabolism. Liver Res.7, 275–284. 10.1016/j.livres.2023.11.006 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee, L. E., Doke, T., Mukhi, D. & Susztak, K. The key role of altered tubule cell lipid metabolism in kidney disease development. Kidney Int.106, 24–34. 10.1016/j.kint.2024.02.025 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang, X. et al. CD36 in chronic kidney disease: novel insights and therapeutic opportunities. Nat. Rev. Nephrol.13, 769–781. 10.1038/nrneph.2017.126 (2017). [DOI] [PubMed] [Google Scholar]
- 71.Su, X. & Abumrad, N. A. Cellular fatty acid uptake: a pathway under construction. Trends Endocrinol. Metab.20, 72–77. 10.1016/j.tem.2008.11.001 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Louet, J. F. et al. Long-chain fatty acids regulate liver carnitine palmitoyltransferase I gene (L-CPT I) expression through a peroxisome-proliferator-activated receptor alpha (PPARalpha)-independent pathway. Biochem. J.354, 189–197. 10.1042/0264-6021:3540189 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xu, J. et al. Peroxisome proliferator-activated receptor alpha (PPARalpha) influences substrate utilization for hepatic glucose production. J. Biol. Chem.277, 50237–50244. 10.1074/jbc.M201208200 (2002). [DOI] [PubMed] [Google Scholar]
- 74.Patsouris, D. et al. PPARalpha governs glycerol metabolism. J. Clin. Invest.114, 94–103. 10.1172/JCI20468 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kersten, S. et al. The peroxisome proliferator-activated receptor alpha regulates amino acid metabolism. FASEB J.15, 1971–1978. 10.1096/fj.01-0147com (2001). [DOI] [PubMed] [Google Scholar]
- 76.Amagase, Y., Mizukawa, Y. & Urushidani, T. Peroxisome proliferator-activated receptor alpha agonist-induced histidine decarboxylase gene expression in the rat and mouse liver. J. Toxicol. Sci.45, 475–492. 10.2131/jts.45.475 (2020). [DOI] [PubMed] [Google Scholar]
- 77.Rakhshandehroo, M., Hooiveld, G., Muller, M. & Kersten, S. Comparative analysis of gene regulation by the transcription factor PPARalpha between mouse and human. PLoS One. 4, e6796. 10.1371/journal.pone.0006796 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Palmer, C. N., Hsu, M. H., Griffin, K. J., Raucy, J. L. & Johnson, E. F. Peroxisome proliferator activated receptor-alpha expression in human liver. Mol. Pharmacol.53, 14–22 (1998). [PubMed] [Google Scholar]
- 79.Gervois, P. et al. A truncated human peroxisome proliferator-activated receptor alpha splice variant with dominant negative activity. Mol. Endocrinol.13, 1535–1549. 10.1210/mend.13.9.0341 (1999). [DOI] [PubMed] [Google Scholar]
- 80.Fernandez-Miranda, C. et al. A pilot trial of Fenofibrate for the treatment of non-alcoholic fatty liver disease. Dig. Liver Dis.40, 200–205. 10.1016/j.dld.2007.10.002 (2008). [DOI] [PubMed] [Google Scholar]
- 81.Nseir, W., Mograbi, J. & Ghali, M. Lipid-lowering agents in nonalcoholic fatty liver disease and steatohepatitis: human studies. Dig. Dis. Sci.57, 1773–1781. 10.1007/s10620-012-2118-3 (2012). [DOI] [PubMed] [Google Scholar]
- 82.Ahmad, J. et al. Identification and characterization of fenofibrate-induced liver injury. Dig. Dis. Sci.62, 3596–3604. 10.1007/s10620-017-4812-7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Emami, F., Hariri, A., Matinfar, M. & Nematbakhsh, M. Fenofibrate-induced renal dysfunction, yes or no? J. Res. Med. Sci.25, 39. 10.4103/jrms.JRMS_772_19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets presented in this study are derived from the original study, which is openly available in the NCBI’s GEO database gene repository for rats under accession number GSM4415261. The derived datasets supporting the conclusions of this article will be made available by the authors on request.