Abstract
Protein degradation by proteasomes is the source of most antigenic peptides presented on MHC class I molecules. To determine whether proteasomes generate these peptides directly or longer precursors, we developed new methods to measure the efficiency with which 26S and 20S particles, during degradation of a protein, generate the presented epitope or potential precursors. Breakdown of ovalbumin by the 26S and 20S proteasomes yielded the immunodominant peptide SIINFEKL, but produced primarily variants containing 1–7 additional N-terminal residues. Only 6–8% of the times that ovalbumin molecules were digested was a SIINFEKL or an N-extended version produced. Surprisingly, immunoproteasomes which contain the interferon-γ-induced β-subunits and are more efficient in antigen presentation, produced no more SIINFEKL than proteasomes. However, the immunoproteasomes released 2–4 times more of certain N-extended versions. These observations show that the changes in cleavage specificity of immunoproteasomes influence not only the C-terminus, but also the N-terminus of potential antigenic peptides, and suggest that most MHC-presented peptides result from N-terminal trimming of larger proteasome products by aminopeptidases (e.g. the interferon-γ-induced enzyme leucine aminopeptidase).
Keywords: antigen processing/immunoproteasomes/MHC class I/proteasomes/protein degradation
Introduction
An important function of protein breakdown in mammalian cells is to generate the 8- to 10-residue peptides that are presented to the immune system on MHC class I molecules (Rock and Goldberg, 1999). Once generated in the cytosol, antigenic peptides are transported by the TAP complex into the endoplasmic reticulum (ER), where they bind to class I molecules and are delivered to the cell surface. The clearest evidence that most antigenic peptides are generated by proteasomes was the finding that inhibitors of the proteasome reduce or prevent antigen presentation (Rock et al., 1994). While it is clear that proteasomes play a key role in generating many antigenic peptides, it is uncertain whether these particles produce the class I-presented peptide directly (Dick et al., 1998; Lucchiari-Hartz et al., 2000), or release larger precursors, which are then trimmed by other peptidases to the presented epitopes.
In model experiments, isolated 20S proteasomes are capable of making the cleavages needed to release some antigenic peptides from larger oligopeptides (Niedermann et al., 1995, 1999; Lucchiari-Hartz et al., 2000). However, these experiments used activated 20S proteasomes, which generate a different pattern of products than 26S protea somes (Kisselev et al., 1999; Emmerich et al., 2000), the complexes that catalyze protein degradation in vivo. Moreover, these studies used as precursors 20- to 44-residue peptides, whose uptake and cleavage may well differ from what occurs during the processive, ATP-dependent breakdown of full-length proteins.
A variety of recent observations indicate that if N-extended versions of the antigenic peptide are released by proteasomes, these peptides can be efficiently trimmed in the cytosol or ER to the mature epitope by aminopeptidases (Falk et al., 1990; Craiu et al., 1997; Stoltze et al., 1998; Mo et al., 1999). Furthermore, interferon-γ (IFN-γ), which stimulates antigen presentation, induces leucine aminopeptidase, the enzyme that in cell extracts is most active in removing additional N-terminal residues from precursors of antigenic peptides (Beninga et al., 1998). These findings raise the possibility that many epitopes arise by N-terminal trimming of proteasome products. However, it has never been shown that such N-extended forms are in fact produced by proteasomes at significant rates, and appreciable controversy has developed on this issue. The present studies were undertaken to determine (i) whether during protein breakdown 26S particles do in fact generate the presented epitope or N-extended precursors and (ii) the efficiency with which such antigenic peptides or longer versions are produced. These questions can not be addressed in vivo, where nearly all the proteasome’s products are rapidly digested to amino acids, and precursors of antigenic peptides are processed rapidly.
Resolution of these questions is also important if we are to understand the greater efficiency of antigen presentation in immune tissues and in infected states, where this process is stimulated by IFN-γ. This cytokine promotes class I presentation through multiple actions, including the induction of three novel proteasome β-subunits (Rock and Goldberg, 1999). Their incorporation in place of homologous subunits enhances the proteasome’s capacity to generate some antigenic peptides (Van Kaer et al., 1994; Groettrup et al., 1995; Sewell et al., 1999; Schwarz et al., 2000; Sijts et al., 2000). Consequently, these alternative forms are often called ‘immunoproteasomes’. Experi ments with small fluorogenic substrates showed that immunoproteasomes have a greater capacity to cleave after hydrophobic and basic residues, and a lower capacity to cleave after acidic residues (Gaczynska et al., 1993). Consequently, peptides generated by immunoproteasomes should have a higher percentage of hydrophobic and basic C-termini, both of which favor uptake by TAP transporters and are essential for tight binding to MHC class I molecules (Rock and Goldberg, 1999). However, direct evidence that immunoproteasomes actually generate more antigenic peptides from proteins is lacking.
To address these questions, we have established conditions in which pure 26S proteasomes and immunoproteasomes degrade a full-length protein in a processive, linear manner. Denatured ovalbumin (Ova) was studied because it can be processed in vivo by proteasomes to antigenic peptides without ubiquitylation (Michalek et al., 1996), and we have found treatments that make it a good substrate for 26S proteasomes without ubiquitylation. To compare the products of proteasomes and immunoproteasomes, we isolated these particles from muscle and spleen, respectively, because these tissues contain these forms exclusively (Van Kaer et al., 1994; Eleuteri et al., 1997). In addition, we have developed new analytical procedures to measure what fraction of the Ova degradation yields the MHC ligand SIINFEKL directly, and versions that contain additional N- or C-terminal residues. Subsequent studies tested whether our findings with 26S complexes are due to inherent properties of the 20S core proteasome.
Results
Protein degradation by proteasomes and immunoproteasomes
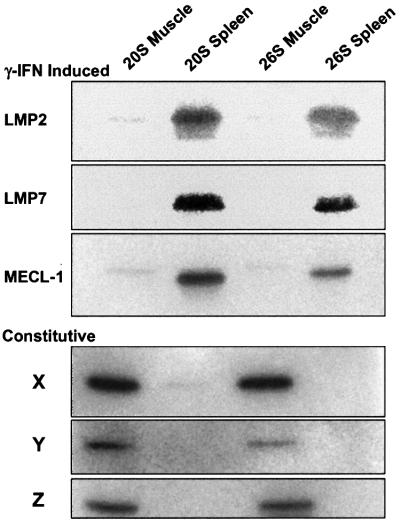
In order to compare their abilities to degrade proteins and to generate MHC class I-presented peptides, we purified 26S and 20S proteasomes from rabbit muscle and immunoproteasomes from rabbit spleen. These preparations appeared to be nearly homogeneous by native PAGE, and upon SDS–PAGE showed the characteristic subunits of the 20S and 26S particles. Western blot analysis showed that proteasomes from muscle contained only the constitutive β-subunits (X, Y and Z) and that the particles from spleen contained only the IFN-γ-induced subunits (LMP2, LMP7 and MECL-1) (Figure 1), as was found previously for proteasomes from bovine (Eleuteri et al., 1997) and mouse (Van Kaer et al., 1994) spleen. Presumably, immunoproteasomes are expressed constitutively in lymphoid organs to ensure efficient class I presentation. In accord with prior findings (Rock and Goldberg, 1999), the immunoproteasomes cleaved the hydrophobic substrate Z-GGL-amc 50% faster, the basic substrate Boc-LRR-amc 100% faster and the acidic substate Ac-YVAD-amc at 20% the rate of proteasomes. To study proteasomal production of specific peptides, it is essential that there be no contamination by peptidases that might modify the products generated. Our final preparations did not show any tripeptidyl peptidase II, leucine aminopeptidase or thimet oligopeptidase activities (as assayed with fluorogenic substrates), all of which contaminated cruder preparations. The purified particles also did not contain the proteasome activator PA28, as assayed with antibodies against PA28α and PA28β (not shown).
Fig. 1. Subunit composition of the preparations of proteasomes and immunoproteasomes. 26S and 20S were analyzed by SDS–PAGE and western blot analysis with antibodies against different β-subunits.
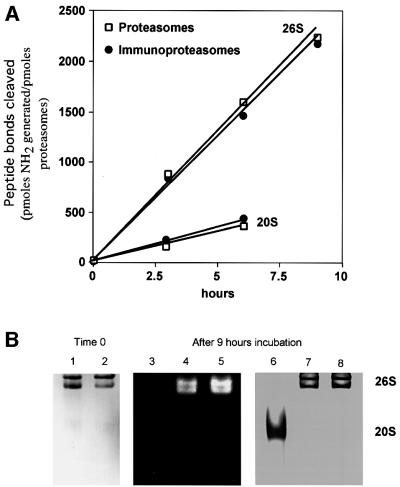
A major problem that has prevented studies of the generation of antigenic peptides from full-length proteins has been their very low rates of degradation by proteasomes in vitro. We showed previously that although ubiquitylation of microinjected Ova is important for efficient presentation of SIINFEKL on MHC class I molecules (Michalek et al., 1993), if this protein is denatured before microinjection, it can be degraded by proteasomes in a ubiquitin-independent process and serve in antigen presentation (Michalek et al., 1996). Therefore, we oxidized Ova with performic acid (Hirs, 1967) and further denaturated it with guanidine–HCl to promote its degradation in vitro. Although native Ova was not degraded at significant rates by 26S proteasomes, the hydrolysis of denatured substrate was easily measurable and linear for up to 9 h (Figure 2A).
Fig. 2. Ova degradation by proteasomes and immunoproteasomes. (A) Ova was incubated with SDS-activated 20S particles or with 26S particles in the presence of ATP. Aliquots were analyzed using fluorescamine. (B) 26S proteasomes or immunoproteasomes were incubated at 37°C for 9 h in the presence of Ova. The samples were then subjected to non-denaturing PAGE at 4°C, and the gel analyzed by overlay of fluorogenic substrate (100 µM Suc-LLVY-amc). The same gel was then Commassie Blue stained. Lanes 1 and 2, 26S proteasomes and immunoproteasomes. The two bands correspond to the double and single capped forms of the 26S particle. Lanes 4 and 7, 26S immunoproteasomes after 9 h of incubation at 37°C; lanes 5 and 8, 26S proteasomes after 9 h of incubation at 37°C; lanes 3 and 6, 20S as control.
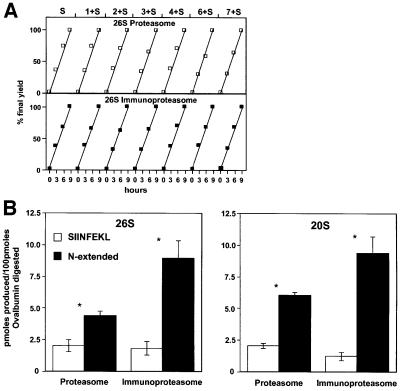
This degradation was by the 26S particles, since even after incubation for 9 h no 20S proteasome could be detected (Figure 2B), and without SDS present the 20S particles are completely unable to degrade Ova (not shown). After activation with 0.01% SDS, the 20S particles also digested the denatured Ova, but only at 20% the rate of the 26S particles. In these incubations, Ova was present in large excess over proteasomes to ensure that the peptides released by the proteasomes were not digested further by these particles. Accordingly, the immunodominant peptide SIINFEKL and various N-extended versions all accumulated at linear rates (Figure 6A). Although the 26S particle is the active form in vivo, we also studied the production of different variants of SIINFEKL by 20S proteasomes in order to see whether findings made on 26S proteasomes were due to inherent properties of the core 20S particle.
Fig. 6. Immunoproteasomes generate similar amounts of SIINFEKL but greater amounts of N-extended versions than proteasomes. (A) All peptides analyzed increased at linear rates, as Ova was digested. At 3, 6 and 9 h, aliquots were analyzed for the content of SIINFEKL and N-extended versions. To measure the amounts of each peptide, the areas under individual peaks were compared with those obtained upon analyzing known amounts of each. (B) Yields of SIINFEKL and all N-extended versions combined. The data shown are the averages of three independent experiments for the 26S proteasomes and of two for the 20S particles. *SIINFEKL versus N-extended versions, p = 0.0495, Mann–Whitney U-test.
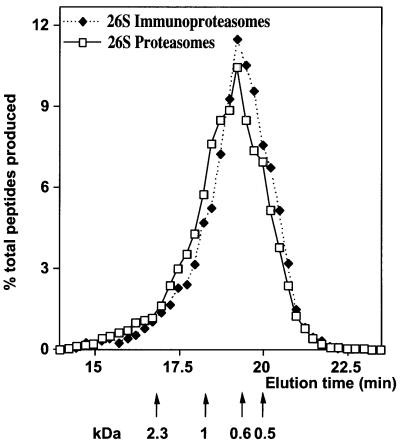
The 26S proteasomes and immunoproteasomes hydrolyzed Ova at very similar rates (Figure 2A), as indicated by similar rates of appearance of new NH2 groups. The much slower rates of degradation of Ova by the SDS-activated 20S proteasomes and immunoproteasomes were also indistinguishable (Figure 2A). Thus, the different β-subunits, while altering the sites of cleavage in proteins (see below), do not affect the rates of protein digestion and the number of cleavages per protein. Accordingly, the sizes of peptides produced from Ova by the 26S proteasomes and immunoproteasomes were quite similar, as measured by size-exclusion HPLC (Figure 3). In both cases, the products ranged between 3 and 22 residues, and fit a log–normal distribution with a mean size of 7–8, in accord with our prior findings (Kisselev et al., 1999). The 20S particles also yielded products ranging between 3 and 22 residues (not shown), although their mean size of 8–9 residues was slightly longer than that of the 26S particles, as we found previously (Kisselev et al., 1999).
Fig. 3. Sizes of peptides generated from Ova by 26S proteasomes and immunoproteasomes. After 9 h incubation, the peptide products were separated from the undegraded substrate on a C18 column and then run on a polyhydroxyethyl aspartamide column. The molar amounts of peptides in each fraction were determined using fluorescamine. Each curve is an average of two independent incubations and peptide analyses.
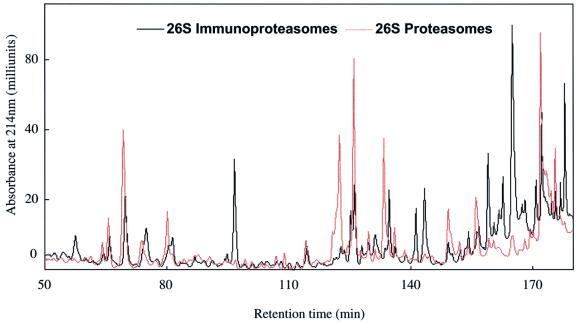
Despite these similar properties, the specific pattern of peptides generated during the breakdown of proteins was clearly different with 26S particles from muscle and spleen. With both, the number of peptides produced from Ova was too large to allow resolution of individual products by HPLC. However, when the small polypeptide insulin-like growth factor-1 (IGF-1) (8000 Da) was used as the substrate, the number of peptides produced was much smaller, and individual peaks could be resolved by reverse-phase (RP)-HPLC. The patterns of products generated from IGF-1 by 26S proteasomes and immunoproteasomes differed dramatically (Figure 4). Also, the peptides produced by the 20S proteasomes and immunoproteasomes differed from one another (not shown). Interestingly, with both 26S and 20S immunoproteasomes, the elution patterns showed more peptides eluting at higher concentrations of acetonitrile, which probably reflects the generation of more peptides with hydrophobic C-termini (see Discussion; Table I).
Fig. 4. 26S proteasomes and immunoproteasomes generate different patterns of peptides from IGF-1. Denatured IGF-1 was incubated with 26S particles. Peptides generated were separated on a C8 Vydac column equilibrated with 0.06% trifluoroacetic acid. Peptides were eluted at a flow rate of 0.15 ml/min with a gradient of acetonitrile from 0 to 8% in 20 min, to 28% in the next 100 min, to 36% in the subsequent 20 min, and to 44% in the last 10 min.
Table I. Sequence of ovalbumin surrounding the immunodominant epitope, SIINFEKL, and peptides produced by proteasomes.
| Ovalbumin sequence | S | G | L | E | Q | L | E | SIINFEKL | T | E | W |
| Peptides studied | 7+S | 6+S | 5+S | 4+S | 3+S | 2+S | 1+S | S | S+1 | S+2 | |
| Cleavages with immunoproteasome | ↑ | ↑ | ↑ | ||||||||
| predicted to increase | yes | yes | yes | ||||||||
| found to increase | yes | yes | yes | ||||||||
| ↑ | ↑ | ↑ | |||||||||
| predicted to decrease | yes | yes | yes | ||||||||
| found to decrease | yes | yes | yes |
Generation of SIINFEKL from Ova by proteasomes
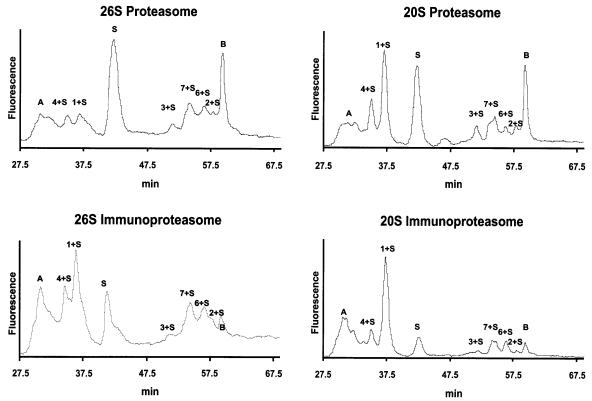
A major problem in evaluating rates of generation of antigenic peptides has been the inability to measure specifically the amounts of the MHC class I-presented epitopes and their precursors in the midst of the very large number of other peptides generated from proteins. Our prior studies indicated that, in vivo, proteasomes make the critical cleavages that determine the C-termini of the presented peptides, but that their N-termini can be generated by aminopeptidases that trim longer peptides released by proteasomes (Craiu et al., 1997; Beninga et al., 1998; Mo et al., 1999). Therefore, it was important to quantitate all N-extended versions of the immunodominant epitope, SIINFEKL. We developed a sensitive new approach to isolate and quantify accurately all the peptides generated by proteasomes from Ova that contain the sequence SIINFEKL at their C-termini. All such peptides were isolated from the hundreds of proteasome products using an immunoaffinity column containing a monoclonal antibody against this antigenic peptide. This specific antibody was chosen because it bound equally strongly SIINFEKL and various peptides containing up to 12 additional residues at its N-terminus, but interacted only weakly with versions of SIINFEKL containing additional residues at its C-terminus (Hilton et al., 2001). Such C-terminal extensions do not bind to the column and are not of immunological importance, since in vivo they can not serve as precursors of the MHC-presented epitopes unless they are further cleaved by proteasomes (Craiu et al., 1997). This possibility is unlikely in vivo because these short peptides are rapidly hydrolyzed by cytosolic peptidases and are only slowly cleaved by proteasomes (Dolenc et al., 1998). Never theless, since the yield of these C-extended versions is also important for understanding the mechanism of proteolysis and its regulation by IFN-γ, we also developed a distinctive method to quantify them (see below).
After their isolation with the antibody column, the peptides terminating with SIINFEKL were chemically modified by reaction with fluorescamine, so that their amounts could be precisely quantitated by fluorescence. The derivatized peptides were then fractionated by RP-HPLC, and their elution times were compared with those of synthetic standards corresponding to SIINFEKL or longer peptides containing 1–7 additional N-terminal residues, as found in Ova (Table I). After reaction with fluorescamine, the amount of each peptide was assayed by comparison of the area under each peak with that of the fluorescamine-derivatized standard peptide. Various control experiments indicated that the recoveries of these different N-extended peptides from the column and their subsequent assays were quantitative (see Materials and methods).
To validate this approach, the main products of Ova degradation were also identified by sequencing using mass spectrometry. This analysis showed the presence in each peak of only the expected peptide, and that the ε-amino group of the lysine in SIINFEKL and in the N-extended versions does not react with fluorescamine. In the column eluate, two other fluorescent peaks (A and B in Figure 5) were detected that did not correspond to any of the standards. By mass spectrometry, peak A was identified as the peptide TNGIIRN (OVA154–160). Presumably, this peptide cross-reacts with the anti-SIINFEKL antibody. The nature of peak B is unclear, since mass spectrometry failed to detect the presence of any peptide. This peak does not represent a product of Ova digestion, since its area did not increase during prolonged incubations with proteasomes (not shown). Presumably, peak B is due to a non-peptide contaminant of the proteasome preparations that binds non-specifically to the affinity column.
Fig. 5. 26S and 20S proteasomes and immunoproteasomes generate from Ova SIINFEKL and different amounts of N-extended variants. An aliquot of peptides generated by the proteasomes was loaded onto the antibody column, which binds peptides with the C-terminal sequence SIINFEKL. These eluted peptides were analyzed by HPLC.
During the 9 h incubation with 26S proteasomes, the generation of SIINFEKL and of the various N-extended versions all occurred at identical linear rates (Figure 6A), which were proportional to the rate of Ova hydrolysis (not shown). During this process, both 26S and 20S particles generated the MHC-presented epitope SIINFEKL (Figure 5). We previously found that in degrading an Ova molecule, the 26S proteasome makes 51 cuts and the 20S proteasome 40 cuts (Kisselev et al., 1999). These findings were confirmed here, both for proteasomes and immunoproteasomes (not shown). By measuring the total amount of peptides produced from Ova, we were able to calculate the frequency with which an SIINFEKL molecule was generated for every molecule of Ova degraded. About 2% of the time when the 26S proteasomes hydrolyzed Ova, a molecule of SIINFEKL was produced. With the activated 20S particles also, ∼2% of the Ova molecules degraded yielded an SIINFEKL molecule. These rates were very similar in three independent incubations with the 26S proteasome and in two incubations with the 20S particle using two different proteasome preparations. However, both forms also generated peptides that contained from one to seven additional N-terminal residues (Figure 5), and the combined production of the N-extended peptides exceeded the amount of SIINFEKL production. Thus, ∼6% of the Ova molecules degraded by the 26S proteasome yielded either an SIINFEKL or an SIINFEKL-containing peptide. With the 20S particle, ∼8% of the Ova molecules degraded yielded either SIINFEKL or an N-extended version. It is noteworthy that although the 8mer was the single most abundant form, the total yield of the N-extended versions was consistently 2- to 3-fold higher than that of SIINFEKL with both 26S and 20S proteasomes (Tables II and III; Figure 6B). There fore, these properties must reflect the proteolytic mechanism of the core particle.
Table II. Comparison of efficiency of production of the immunodominant epitope SIINFEKL and N-extended variants by 26S proteasomes and immunoproteasomes.
| Peptide | Proteasomes | Immunoproteasomes |
|---|---|---|
| (pmol produced/100 pmol ovalbumin digested) | ||
| S | 2.0 ± 0.5 | 1.8 ± 0.5 |
| 1+S | 1.4 ± 0 | 4.1 ± 0.7 |
| 2+S | 0.4 ± 0.1 | 0.3 ± 0.1 |
| 3+S | 0.6 ± 0.1 | 0.7 ± 0.2 |
| 4+S | 0.6 ± 0 | 1.5 ± 0.2 |
| 6+S | 0.4 ± 0.2 | 0.8 ± 0.2 |
| 7+S | 1.0 ± 0.1 | 1.6 ± 0.3 |
| Total S-containing peptides | 6.4 ± 0.6 | 10.8 ± 1.8 |
| Total N-extended versions | 4.4 ± 0.3 | 9.0 ± 1.4 |
| % extended | 69% | 83% |
Values are mean ± SE.
Analysis of proteasome products by the hybridoma assay
We obtained quite similar results in independent experiments in which we analyzed the production of SIINFEKL and extended versions using HPLC to fractionate the proteasome products (without first isolating the peptides with the immunoaffinity column), and with this approach we also could quantify C-extended variants of SIINFEKL. The fractions corresponding to elution times of the different SIINFEKL-containing standards were then assayed using a T-cell hybridoma based upon measurement of interleukin-2 (IL-2) production by T cells. Under these conditions, aminopeptidases and carboxypeptidases associated with the cells or medium converted longer peptides to the MHC-bound ligand. Standard curves were established with increasing amounts of each extended peptide. By this approach, 26S proteasomes were found to produce SIINFEKL directly ∼0.8% of the time an Ova molecule was degraded, but they generated N-extended versions (especially 1+S and 4+S) ∼3.5 times more often. Similarly, with the 20S particles, the yield of SIINFEKL per Ova degraded was 1.3%, but the yield of peptides containing 1–4 additional N-terminal residues was again 3.5-fold greater, due largely to production of 1+S and 4+S (Table IV).
Table IV. Comparison by T-T hybridoma assay of the efficiency of production of the immunodominant epitope SIINFEKL and N- and C-extended variants by 26S and 20S proteasomes and immunoproteasomes.
| Peptide | 26S proteasomes | 26S immunoproteasomes |
|---|---|---|
| (pmol produced/100 pmol ovalbumin digested) | ||
| S | 0.8 ± 0.04 | 1.2 ± 0.21 |
| 1+S | 1.2 ± 0.14 | 2.6 ± 0.10 |
| 2+S | 0.2 ± 0.09 | 0.4 ± 0.16 |
| 3+S | 0.3 ± 0.16 | 0.5 ± 0.08 |
| 4+S | 1.1 ± 0.14 | 3.8 ± 1.16 |
| S+1 | 0.3 ± 0.08 | 0.2 ± 0.05 |
| S+2 |
0.9 ± 0.15 |
0.4 ± 0.03 |
| Peptide |
20S proteasomes |
20S immunoproteasomes |
| S | 1.3 ± 0.02 | 1.3 ± 0.18 |
| 1+S | 2.5 ± 0.31 | 4.1 ± 0.47 |
| 2+S | 0.0 ± 0.00 | 0.0 ± 0.00 |
| 3+S | 0.2 ± 0.06 | 0.0 ± 0.00 |
| 4+S | 2.2 ± 0.27 | 3.7 ± 0.62 |
| S+1 | 1.4 ± 0.16 | 0.3 ± 0.02 |
| S+2 | 8.1 ± 2.06 | 2.6 ± 0.46 |
Values are mean ± SE.
This T-cell hybridoma assay also allowed us to measure the rates of production of peptides containing one or two additional residues on the C-terminus of SIINFEKL. Total production of these peptides by the 26S proteasomes occurred 1% of the time an Ova molecule was degraded. Surprisingly, with the 20S particles, both S+1 and S+2 were produced at much higher rates, and these peptides together were generated 10% of the time an Ova was digested (Table IV).
Production of antigenic peptides by immunoproteasomes
When similar incubations were carried out with 26S and 20S particles from spleen, SIINFEKL was generated, and the total amount of N-extended versions was higher than SIINFEKL itself (Figures 5 and 6B; Tables II and III), as was found with muscle proteasomes. However, the total amount of SIINFEKL-containing peptides generated per Ova degraded was consistently greater with 26S immunoproteasomes than with the corresponding particles from muscle. The 20S particles from spleen also generated more total SIINFEKL-containing peptides than those from muscle, but the differences appeared smaller than with 26S forms. These findings are the first direct evidence that the replacement of the proteasome’s normal β-subunits by LMP2, LMP7 and MECL-1 does in fact enhance the efficiency of production of potential antigenic peptides from a full-length protein.
Table III. Comparison of efficiency of production of the immunodominant epitope SIINFEKL and N-extended variants by 20S proteasomes and immunoproteasomes.
| Peptide | Proteasomes | Immunoproteasomes |
|---|---|---|
| (pmol produced/100 pmol ovalbumin digested) | ||
| S | 2.0 ± 0.2 | 1.2 ± 0.3 |
| 1+S | 2.8 ± 0.6 | 5.8 ± 1.5 |
| 2+S | 0.3 ± 0.1 | 0.1 ± 0 |
| 3+S | 0.6 ± 0 | 0.5 ± 0 |
| 4+S | 1.1 ± 0.2 | 1.3 ± 0.2 |
| 6+S | 0.3 ± 0 | 0.5 ± 0 |
| 7+S | 0.9 ± 0 | 1.2 ± 0.1 |
| Total S-containing peptides | 8.0 ± 0 | 10.6 ± 0.9 |
| Total N-extended versions | 6.0 ± 0.2 | 9.4 ± 1.2 |
| % extended | 75% | 89% |
Values are mean ± SE.
It is noteworthy that with the immunoproteasomes this increase in total antigenic peptides was not due to greater production of SIINFEKL, but was due to increased production of N-extended versions. With the immunoproteasomes, generation of these N-extended variants was 100% greater by the 26S particles (p <0.05) and 60% greater by the 20S particles. By contrast, production of SIINFEKL did not change (or seemed to fall in the experiments with 20S). As a result, with the immunoproteasomes, the MHC-presented epitope was not the most abundant single SIINFEKL-containing species generated. Instead, the 1+S peptide was produced by the 20S particles in 3-fold greater amounts, and 4+S and 7+S in similar amounts to SIINFEKL itself. In other words, although an SIINFEKL was generated only 2% of the times that an Ova molecule was degraded by 26S immunoproteasomes, the yield of all peptides containing SIINFEKL at their C-termini corresponded to ∼11% of the Ova molecules degraded (compared with 6% with muscle proteasomes). Similarly, with activated 20S immunoproteasomes, SIINFEKL was generated from ∼1% of the Ova molecules digested, but the yield of all peptides containing SIINFEKL at their C-termini was actually 11%.
Increased production of N-extended versions containing 1–4 additional residues with little or no change in production of SIINFEKL was also observed when the peptides were fractionated by HPLC and assayed using the T-cell hybridoma and measurement of IL-2 production (Table IV). This assay showed even larger increases in the production of N-extended versions by 26S immunoproteasomes over rates with proteasomes. The hybridoma approach also allowed us to assay the production of peptides containing an additional one or two residues on the C-terminus of SIINFEKL. Interestingly, their production was consistently lower in the immunoproteasomes than in the corresponding muscle proteasomes.
In summary, proteasomes do not cleave out the immunodominant epitope from Ova stochiometrically, but instead release at least seven different N-extended forms. The production of peptides terminating in SIINFEKL is a rare event, and the great majority of the time SIINFEKL is cleaved internally. As expected, the immunoproteasomes are more efficient in antigen generation, but surprisingly, this effect is due to their greater rates of production of the N-extended versions.
Discussion
A full understanding of the pathway for class I antigen presentation will require quantitative information on how often, during the degradation of a protein, the 26S proteasome actually generates the MHC-presented peptide or larger precursors. Using a variety of new methods, we found that 26S proteasomes generate either an SIINFEKL molecule or an N-extended version from only ∼6% of the Ova molecules degraded. The efficiency of proteasomes in producing the MHC-presented peptide or potential precursors thus appears quite low, and even with 26S immunoproteasomes the efficiency is still low (11%). Thus, the proteasomal cleavages that can yield antigenic peptides are rare events, and antigen generation is not a stoichiometric process that occurs most of the times a protein is digested. This conclusion is consistent with those of Pamer and Cresswell (1998), who estimated that between 3 and 30 class-I presented peptides, depending on the epitope, were generated in vivo from 100 molecules of Listeria proteins degraded. The low rate of generation of antigenic peptides by proteasomes emphasizes that their primary function is to rapidly degrade proteins to amino acids, and only an occasional peptide product escapes this fate and serves in antigen presentation. It remains to be established whether the present findings about SIINFEKL also apply to the production of immunodominant epitopes from other proteins (Mo et al., 2000), and of subdominant or cryptic epitopes (Yewdell and Bennink, 1999), whose inability to elicit an effective T-cell response could be due to a low efficiency in their generation by proteasomes.
The present finding that proteasomes yield primarily N-extended versions of SIINFEKL makes it very likely that many, perhaps most, of the MHC-presented peptides are derived by trimming longer precursors. Accordingly, Paz et al. (1999), who studied antigen generation from a 99 residue polypeptide containing the synthetic epitope SIINFEHL, found only N-extended versions in the cytosol (mainly 2+S and 3+S) and only SIINFEHL in the ER. Our prior studies (Craiu et al., 1997; Mo et al., 1999) and those of others (Stoltze et al., 1998) indicated that when N-extended versions of SIINFEKL or of four viral epitopes were introduced into cells, they were trimmed and presented on MHC molecules with high efficiency. As shown here, production of these N-extended versions can potentially increase the amount of immunogenic peptides 2- to 4-fold. Therefore, it would appear highly advantageous for cells to utilize aminopeptidases to trim these potential epitopes to the forms capable of tight binding to class I molecules. The present observations fit nicely with the discovery that the major cytosolic activity capable of trimming these precursors, leucine aminopeptidase, is induced by IFN-γ (Beninga et al., 1998), although trimming may also involve other aminopeptidases in the cytosol (Stoltze et al., 2000) or ER (Snyder et al., 1994; Craiu et al., 1997).
It has generally been assumed that proteasomes generate mature epitopes directly, because in model experiments activated 20S proteasomes are capable of making the appropriate cleavages in 20- to 44-residue-long oligopeptide precursors or even in a protein (Dick et al., 1994) to produce some of the final epitope. However, these particles generate from proteins a different spectrum of peptides than is generated during the processive, ATP-dependent degradation by 26S (Kisselev et al., 1999; Emmerich et al., 2000). The present study is the first to systematically quantitate the yields during protein breakdown by 26S proteasomes of various peptides that may serve in antigen presentation. Under the conditions used here, proteasomes degraded Ova and generated various peptide products at linear rates for many hours, and these products were not metabolized further.
It has often been assumed that studies like the present one would be impossible, based on the incorrect impression that ubiquitylation of protein is absolutely essential for degradation by 26S proteasomes, and because it has not been possible to enzymatically ubiquitylate substrates in sufficient amounts to allow chemical analysis of peptide products. Previously, we had shown that if native Ova is microinjected into cells, it has to be ubiquitylated for SIINFEKL to be presented rapidly on surface MHC molecules (Michalek et al., 1993). However, if Ova was denatured prior to microinjection, it could also be degraded by proteasomes without ubiquitylation, and SIINFEKL could be presented efficiently (Michalek et al., 1996). A key development in this study was our finding of in vitro conditions that denatured Ova so as to make it a good substrate for pure proteasomes. Presumably, this denaturation facilitates proteasome recognition and mimics the unfolding that is catalyzed by the ATPases of the 19S complex (Benaroudj and Goldberg, 2000).
In any case, it seems very likely that the present conclusions will also hold if an Ova molecule initially binds to the 26S particle in a ubiquitylated form, because the main new properties discussed here appear to result from inherent proteolytic behavior of the 20S particle. Even though the 26S proteasomes degraded proteins faster and generated different amounts of each peptide, both forms produce primarily N-extended versions of SIINFEKL and with comparable efficiencies. Moreover, both 20S and 26S immunoproteasomes are more efficient than their normal counterparts, specifically in the production of the N-extended versions. Thus, these important properties reflect the core particle’s mode of degradation of a polypeptide and should also hold for ubiquitylated substrates.
For studies of antigen generation, the use of denatured proteins offers clear advantages over the insertion of SIINFEKL into an unrelated protein whose breakdown is independent of ubiquitin (Ben-Shahar et al., 1999), since in such an artificial construct the sequences flanking the epitope are completely unnatural. It is noteworthy that denatured proteins are likely to be important substrates for intracellular proteolysis since a significant fraction of ribosomal products are degraded before successful folding (Reits et al., 2000; Schubert et al., 2000). Furthermore, there are a growing number of other examples where native proteins can be degraded by 26S proteasomes in the absence of ubiquitylation (Tarcsa et al., 2000; Verma and Deshaies, 2000). Therefore, it seems likely that a significant fraction of proteasome-mediated proteolysis occurs in a ubiquitin-idependent mode in vivo.
Effect of IFN-γ on the generation of antigenic peptides
IFN-γ stimulates class I presentation through multiple actions: it increases the expression of MHC class I molecules, TAP transporter, the PA28 complex, leucine aminopeptidase and the three novel catalytic β-subunits of the proteasome. These subunits alter peptidase specificities and thus are believed to increase the production of peptides with C-termini capable of binding to MHC class I molecules. Although these IFN-γ-induced subunits alter the rates of cleavage of fluorogenic peptides and of model peptide precursors to epitopes (Groettrup et al., 1995; Sijts et al., 2000), there have been no prior studies of their capacity to generate in vitro antigenic peptides from proteins. As anticipated, the total yield of potential antigenic peptides was greater with the immunoproteasomes, even though they digested polypeptides at the same rates as normal particles. However, to our surprise, SIINFEKL was not produced in higher amounts by the immunoproteasomes, which instead only generated greater amounts of N-extended versions (especially 1+S and 4+S), which were produced ∼3 times more rapidly than by muscle proteasomes. These findings provide further strong evidence that the N-extended peptides are likely to be important intermediates in antigen presentation in vivo.
Studies with fluorogenic substrates had indicated that immunoproteasomes have a greater maximal ability to cleave after large hydrophobic and basic residues, and a lesser ability to cleave after acidic residues. These tendencies should lead to enhanced production of peptides capable of binding to MHC molecules, but the relevance of such data on fluorogenic peptides to protein breakdown has been questioned. However, our findings on the products of 26S and 20S immunoproteasomes are in excellent agreement with the changes in peptidase specificity that we and others found with fluorogenic substrates (Rock and Goldberg, 1999) and with recent findings on proteolysis by 20S immunoproteasomes (Cardozo and Kohanski, 1998). The greater capacity of immunoproteasomes to cleave after leucine residues would predict a greater total yield of peptides terminating with SIINFEKL, as we observed (Table I), and even larger increases in the yields of 1+S and 4+S, whose production requires two post-leucyl cleavages, as we found. In addition, the reduced tendency of immunoproteasomes to cleave after acidic residues should lead to decreased yields of S+2, which we observed. Finally, these changes can account for the surprising finding that immunoproteasomes did not generate more SIINFEKL. Its production probably does not change because the tendency of immunoproteasomes to cleave more after the C-terminal leucine is balanced by their decreased capacity to cleave after a glutamate, as is essential to generate the N-terminus of SIINFEKL. These findings clearly confirm an important influence of the P1 residue on the cuts made by proteasomes. Therefore, during breakdown of a protein, whether a cleavage occurs after a particular residue has a certain probability that is determined largely by the P1, and that clearly differs after incorporation of IFN-γ-induced subunits.
Prior studies of the effects of the IFN-γ-induced subunits have focused on their abilities to influence the C-termini of antigenic peptides. However, as shown here, these subunits clearly also determine the N-termini of the proteasomal products. The increased generation of N-extended versions of SIINFEKL by immunoproteasomes fits nicely with our prior finding that IFN-γ also induces the major trimming enzyme, leucine aminopeptidase (Beninga et al., 1998). Although the increase in the total amount of SIINFEKL-containing products by the 26S immunoproteasome was <2-fold, the production of 1+S and 4+S increased 3- to 4-fold. These various N-extended versions may differ in their rates of processing by leucine aminopeptidase and transport by TAP. Because the efficiency of these steps is unknown, it is unclear how the greater yield of N-extended versions affects the amount of SIINFEKL reaching the cell surface. The TAP complex transports rapidly or even preferentially N-extended versions of antigenic peptides (Momburg et al., 1994; Lauvau et al., 1999). If cytosolic aminopeptidases also attack SIINFEKL, they would remove residues essential for its binding to MHC molecules. Therefore, an N-extended epitope may even have a greater chance of reaching TAP and serving in antigen presentation than a mature epitope released by the proteasome. If so, the increased production of N-extended variants by immunoproteasomes would have a major influence in promoting class I responses.
Materials and methods
Peptide standards
Peptides with the same sequences as are found in chicken Ova, SIINFEKL (S), ESIINFEKL (1+S), LESIINFEKL (2+S), QLESIINFEKL (3+S), EQLESIINFEKL (4+S), LEQLESIINFEKL (5+S), SIINFEKLT (S+1) and SIINFEKLTE (S+2), were synthesized by Macromolecular Resources (Colorado State University, CO). The peptides GLEQLESIINFEKL (6+S) and SGLEQLESIINFEKL (7+S) were prepared by Res. Gen. Inc. (Huntsville, AL). The peptides were dissolved at 10 mg/ml in DMSO and stored at –80°C. The concentration of each peptide was confirmed by amino acid analysis.
Proteasome purification
26S and 20S proteasomes were purified from 500 g of frozen rabbit muscle (Kisselev et al., 1999). The immunoproteasomes were purified in a similar manner from 250 g of frozen rabbit spleen (purchased from Pel Freez Biologicals). The amount of Affi-Gel Blue (Bio-Rad) necessary to bind all the proteasomes from spleen and muscle was 200 and 60 ml, respectively. The immunoproteasomes were eluted from the UNO-Q column (Bio-Rad) at a slightly lower ionic strength than the corresponding muscle particles.
Electrophoretic and immunoblot analyses
The amounts of constitutive (X, Y, Z) and IFN-γ-induced (LMP2, LMP7, MECL-1) β-subunits were measured by immunoblot analysis. Three micrograms of 26S and 1 µg of 20S were separated on a 12% SDS–PAGE gel, and the proteins were transferred onto an Immobilon P membrane. The filters were blocked for 1 h with 5% bovine serum albumin in phosphate-buffered saline, and incubated with rabbit antisera against X, Y, LMP2, LMP7 and with monoclonal antibodies against Z and MECL-1, kindly provided by K.Tanaka (Tokyo Metropolitan Research Institute). Bound antibodies were detected with anti-rabbit IgG antibodies conjugated with horseradish peroxidase (Promega, WI) and visualized using the ECL method (Amersham Pharmacia).
Protein digestion and peptide analysis
Ova (Sigma) was oxidized with performic acid (Kisselev et al., 1999), lyophilized and immediately before use resuspended in 6 M guanidine– HCl, 100 mM dithiothreitol (DTT). Guanidine was removed by gel filtration using a column of Sepharose G-25 equilibrated with 50 mM BTP (bis-Tris–propane) pH 7.5, 1 mM DTT, 0.1 mM EDTA. With the 20S particles (100 nM), the digestion of Ova (7 µM) was performed typically for 4 h in 50 mM BTP pH 7.5, 50 mM KCl, 5 mM EGTA and 0.01% SDS to activate the 20S proteasomes. For the 26S particles (20 nM), the buffer contained 50 mM BTP pH 7.5, 5 mM MgCl2, 1 mM DTT and 2 mM ATP, and the incubation was for 9 h. To assay peptides generated during Ova degradation, we measured the appearance of new amino groups using fluorescamine (Akopian et al., 1997). Recombinant human IGF-1 (kindly provided by Dr W.Prouty, Eli Lilly) was denatured by reduction of disulfide bonds and carboxymethylation of the cysteines (Akopian et al., 1997). Denatured IGF (560 µM) was incubated with 26S proteasomes and immunoproteasomes (20 nM) at 37°C as described above for 5 h. Peptides generated were separated on a C8 column. Size-exclusion chromatography of peptides generated during the degradation of reductively methylated Ova was performed as described (Kisselev et al., 1999).
Analysis of peptide products with immunoaffinity column validation
Monoclonal antibodies (mAb) against the antigenic peptide H-2Kb-restricted SIINFEKL (chicken OVA257–264) were raised by immunization of mice with keyhole limpet hemocyanin conjugated with Biotin-SIINFEKL (Hilton et al., 2001). The mAb (isotype IgG2) were purified from B-cell hybridoma culture supernatant and bound to protein G–Sepharose with dimethylpimelimidate (Pierce). A total of 200 µl of settled antibody–protein G–Sepharose complex were used for each affinity column. Between 15 and 40 nmol of peptides generated by the proteasomes were loaded onto the immunoaffinity column. After washing the column with 10 mM phosphate buffer pH 6.8, the bound peptides were eluted with 10 mM ammonium acetate pH 2.8 and lyophilized. To identify and measure the amount of peptides generated, the samples were resuspended in 30 µl of 200 mM phosphate buffer pH 6.8, and 15 µl of fluorescamine (0.3 mg/ml acetone) added. The reaction was terminated after 1 min with 185 µl of H2O. The sample was resolved by HPLC using a C18 column (2.1 × 250 mm, 5 µm; Vydac, CA) equilibrated with 14% acetonitrile in 10 mM phosphate buffer pH 6.8 and analyzed by detection of fluorescence. Peptides were eluted by an isocratic step of 20 min, followed by a gradient of acetonitrile from 14 to 21.7% in 1 min, to 22.05% within the next 20 min, to 45.5% within the subsequent 20 min, and to 70% in the last 10 min at a flow rate of 0.2 ml/min.
This approach was validated in several different ways. (i) Five, 10 and 50 pmol of synthetic peptides S, 1+S, 4+S and S+1 were loaded separately and also all together on the immunoaffinity column. After washing the column, the amounts of S, 1+S and 4+S recovered were almost identical to the amount applied. By contrast, S+1 did not bind to the column, and was recovered in the flow-through. (ii) In addition, a mixture of 10 pmol of each of these peptides (S, 1+S, 2+S, 3+S, 4+S, 5+S, 6+S, 7+S) was loaded onto the immunoaffinity column. All these peptides bound to the column and were recovered quantitatively, and none were detected in the flow-through. (iii) Finally, S, 1+S and 4+S were added to a mixture of peptides produced from casein by 26S proteasomes, and the samples were then loaded onto the immunoaffinity column. No peptide from casein bound to the column. Only the peaks of S, 1+S and 4+S were detected after elution, while all the peptides from casein were in the flow-through. To obtain standard curves for each peptide, we tagged S, 1+S, 2+S, 3+S, 4+S, 5+S, 6+S and 7+S with fluorescamine and introduced increasing amounts of each into the HPLC. The areas under the peaks corresponding to each peptide were proportional to the amounts added.
To measure the amounts of peptide products that contained the C-terminal SIINFEKL, for each analysis we loaded a mixture containing 10 pmol of each synthetic peptide on the immunoaffinity column and treated the mixture exactly as the proteasome digest. The areas under the peaks obtained with the standard mixture were used to assay the amounts generated by the proteasomes. This internal control also allowed us to rule out any possible artifacts due to technical variation in specific experiment (e.g. deterioration of the antibody or decreased solubility of certain lyophilized peptides).
Analysis of immunogenic peptides by hybridoma assay
A peptide sample generated by the proteasomes (between 15 and 40 nmol) was fractionated by HPLC on a C18 column (4.6 × 250mm, 5 µm;Vydac, CA), and the bound peptides were eluted using a gradient from 0 to 60% acetonitrile in 20 mM CH3COONH4 in 60 min at a flow rate of 0.750 ml/min. The fractions where SIINFEKL and the various N-extended variants should be present were collected, and tested for the presence of SIINFEKL-containing peptides by the T-T hybridoma assay. Individual RP-HPLC fractions were mixed with Kb-expressing antigen-presenting cells and tested for antigen presentation with the appropriate T-T hybridoma (Rock et al., 1990). This assay can detect 2-fold differences in the amounts of different antigenic peptides. It was used to identify fractions that contained such peptides and for their quantitation by comparison to standards.
Analysis by mass spectrometry
The main peaks detected by HPLC after derivatization with fluorescamine were collected individually, lyophilized and subjected to sequencing by tandem mass spectrometry on a Finningan LCQ Quadrupole Ion Trap Mass Spectrometer.
Acknowledgments
Acknowledgements
We thank Dr K.Tanaka for antibodies, Dr T.Jagoe for statistical advice, T.Jagoe, S.Lecker, O.Kandror, N.Benaroudj for critical reading of this manuscript, and are grateful to Joshua Burton for assistance in preparation of this manuscript. This research was supported by grants from the NIGMS to A.L.G. and the NIH to K.L.R.
References
- Akopian T.N., Kisselev,A.F. and Goldberg,A.L. (1997) Processive degradation of proteins and other catalytic properties of the proteasome from Thermoplasma acidophilum. J. Biol. Chem., 272, 1791–1798. [DOI] [PubMed] [Google Scholar]
- Benaroudj N. and Goldberg,A.L. (2000) PAN, the proteasome-activating nucleotidase from archaebacteria, is a protein-unfolding molecular chaperone. Nature Cell Biol., 2, 833–839. [DOI] [PubMed] [Google Scholar]
- Beninga J., Rock,K.L. and Goldberg,A.L. (1998) Interferon-γ can stimulate post-proteasomal trimming of the N terminus of an antigenic peptide by inducing leucine aminopeptidase. J. Biol. Chem., 273, 18734–18742. [DOI] [PubMed] [Google Scholar]
- Ben-Shahar S., Komlosh,A., Nadav,E., Shaked,I., Ziv,T., Admon,A., DeMartino,G.N. and Reiss,Y. (1999) 26 S proteasome-mediated production of an authentic major histocompatibility class I-restricted epitope from an intact protein substrate. J. Biol. Chem., 274, 21963–21972. [DOI] [PubMed] [Google Scholar]
- Cardozo C. and Kohanski,R.A. (1998) Altered properties of the branched chain amino acid-preferring activity contribute to increased cleavages after branched chain residues by the ‘immunoproteasome’. J. Biol. Chem., 273, 16764–16770. [DOI] [PubMed] [Google Scholar]
- Craiu A., Akopian,T., Goldberg,A. and Rock,K.L. (1997) Two distinct proteolytic processes in the generation of a major histocompatibility complex class I-presented peptide. Proc. Natl Acad. Sci. USA, 94, 10850–10855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick L.R. et al. (1994) Proteolytic processing of ovalbumin and β-galactosidase by the proteasome to yield antigenic peptides. J. Immunol., 152, 3884–3894. [PMC free article] [PubMed] [Google Scholar]
- Dick T.P., Stevanovic,S., Keilholz,W., Ruppert,T., Koszinowski,U., Schild,H. and Rammensee,H.G. (1998) The making of the dominant MHC class I ligand SYFPEITHI. Eur. J. Immunol., 28, 2478–2486. [DOI] [PubMed] [Google Scholar]
- Dolenc I., Seemuller,E. and Baumeister,W. (1998) Decelerated degradation of short peptides by the 20S proteasome. FEBS Lett., 434, 357–361. [DOI] [PubMed] [Google Scholar]
- Eleuteri A.M., Kohanski,R.A., Cardozo,C. and Orlowski,M. (1997) Bovine spleen multicatalytic proteinase complex (proteasome)—replacement of X, Y and Z subunits by Lmp7, Lmp2 and Mecl1 and changes in properties and specificity. J. Biol. Chem., 272, 11824–11831. [DOI] [PubMed] [Google Scholar]
- Emmerich N.P., Nussbaum,A.K., Stevanovic,S., Priemer,M., Toes,R.E., Rammensee,H.G. and Schild,H. (2000) The human 26 S and 20 S proteasomes generate overlapping but different sets of peptide fragments from a model protein substrate. J. Biol. Chem., 275, 21140–21148. [DOI] [PubMed] [Google Scholar]
- Falk K., Rotzschke,O. and Rammensee,H.G. (1990) Cellular peptide composition governed by major histocompatibility complex class I molecules. Nature, 348, 248–251. [DOI] [PubMed] [Google Scholar]
- Gaczynska M., Rock,K.L. and Goldberg,A.L. (1993) γ-interferon and expression of MHC genes regulate peptide hydrolysis by proteasomes. Nature, 365, 264–267. [DOI] [PubMed] [Google Scholar]
- Groettrup M., Ruppert,T., Kuehn,L., Seeger,M., Standera,S., Koszinowski,U. and Kloetzel,P.M. (1995) The interferon-γ-inducible 11 S regulator (Pa28) and the Lmp2/Lmp7 subunits govern the peptide production by the 20 S proteasome in vitro. J. Biol. Chem., 270, 23808–23815. [DOI] [PubMed] [Google Scholar]
- Hilton C.J., Dahl,A.M. and Rock,K.L. (2001) Anti-peptide antibody blocks peptide binding to MHC class I molecules in the endoplasmic reticulum. J. Immunol., 166, 3952–3956. [DOI] [PubMed] [Google Scholar]
- Hirs C.H.W. (1967) Performic acid oxidation. Methods Enzymol., 11, 197–198. [Google Scholar]
- Kisselev A.F., Akopian,T.N., Woo,K.M. and Goldberg,A.L. (1999) The sizes of peptides generated from protein by mammalian 26 and 20 S proteasomes. Implications for understanding the degradative mechanism and antigen presentation. J. Biol. Chem., 274, 3363–3371. [DOI] [PubMed] [Google Scholar]
- Lauvau G., Kakimi,K., Niedermann,G., Ostankovitch,M., Yotnda,P., Firat,H., Chisari,F.V. and van Endert,P.M. (1999) Human transporters associated with antigen processing (TAPs) select epitope precursor peptides for processing in the endoplasmic reticulum and presentation to T cells. J. Exp. Med., 190, 1227–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchiari-Hartz M., van Endert,P.M., Lauvau,G., Maier,R., Meyerhans,A., Mann,D., Eichmann,K. and Niedermann,G. (2000) Cytotoxic T lymphocyte epitopes of HIV-1 Nef: generation of multiple definitive major histocompatibility complex class I ligands by proteasomes. J. Exp. Med., 191, 239–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalek M.T., Grant,E.P., Gramm,C., Goldberg,A.L. and Rock,K.L. (1993) A role for the ubiquitin-dependent proteolytic pathway in MHC class I-restricted antigen presentation. Nature, 363, 552–554. [DOI] [PubMed] [Google Scholar]
- Michalek M.T., Grant,E.P. and Rock,K.L. (1996) Chemical denaturation and modification of ovalbumin alters its dependence on ubiquitin conjugation for class I antigen presentation. J. Immunol., 157, 617–624. [PubMed] [Google Scholar]
- Mo X.Y., Cascio,P., Lemerise,K., Goldberg,A.L. and Rock,K. (1999) Distinct proteolytic processes generate the C and N termini of MHC class I-binding peptides. J. Immunol., 163, 5851–5859. [PubMed] [Google Scholar]
- Mo A.X., van Lelyveld S.F., Craiu,A. and Rock,K.L. (2000) Sequences that flank subdominant and cryptic epitopes influence the proteolytic generation of MHC class I-presented peptides. J. Immunol., 164, 4003–4010. [DOI] [PubMed] [Google Scholar]
- Momburg F., Roelse,J., Hammerling,G.J. and Neefjes,J.J. (1994) Peptide size selection by the major istocompatibility complex-encoded peptide transporter. J. Exp. Med., 179, 1613–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedermann G., Butz,S., Ihlenfeldt,H.G., Grimm,R., Lucchiari,M., Hoschutzky,H., Jung,G., Maier,B. and Eichmann,K. (1995) Contribution of proteasome-mediated proteolysis to the hierarchy of epitopes presented by major histocompatibility complex class I molecules. Immunity, 2, 289–299. [DOI] [PubMed] [Google Scholar]
- Niedermann G., Geier,E., Lucchiari-Hartz,M., Hitziger,N., Ramsperger,A. and Eichmann,K. (1999) The specificity of proteasomes: impact on MHC class I processing and presentation of antigens. Immunol. Rev., 172, 29–48. [DOI] [PubMed] [Google Scholar]
- Pamer E. and Cresswell,P. (1998) Mechanisms of MHC class I-restricted antigen processing. Annu. Rev. Immunol., 16, 323–358. [DOI] [PubMed] [Google Scholar]
- Paz P., Brouwenstijn,N., Perry,R. and Shastri,N. (1999) Discrete proteolytic intermediates in the MHC class I antigen processing pathway and MHC I-dependent peptide trimming in the ER. Immunity, 11, 241–251. [DOI] [PubMed] [Google Scholar]
- Reits A.J., Vos,J.C., Gromme,M. and Neefjes J. (2000) The major substrates for TAP in vivo are derived from newly synthesized proteins. Nature, 404, 774–778. [DOI] [PubMed] [Google Scholar]
- Rock K.L. and Goldberg,A.L. (1999) Degradation of cell proteins and the generation of MHC class I-presented peptides. Annu. Rev. Immunol., 17, 739–779. [DOI] [PubMed] [Google Scholar]
- Rock K.L., Rothstein,L. and Gamble,S. (1990) Generation of class I MHC-restricted T-T hybridomas. J. Immunol., 145, 804–811. [PubMed] [Google Scholar]
- Rock K.L., Gramm,C., Rothstein,L., Clark,K., Stein,R., Dick,L., Hwang,D. and Goldberg,A.L. (1994) Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell, 78, 761–771. [DOI] [PubMed] [Google Scholar]
- Schubert U., Anton,L., Gibbs,J., Norbury,C., Yewdell,J.W. and Bennink,J.R. (2000) Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature, 404, 770–774. [DOI] [PubMed] [Google Scholar]
- Schwarz K., van Den Broek,M., Kostka,S., Kraft,R., Soza,A., Schmidtke,G., Kloetzel,P.M. and Groettrup,M. (2000) Over expression of the proteasome subunits LMP2, LMP7 and MECL-1, but not PA28 α/β, enhances the presentation of an immunodominant lymphocytic choriomeningitis virus T cell epitope. J. Immunol., 165, 768–778. [DOI] [PubMed] [Google Scholar]
- Sewell A.K., Price,D.A., Teisserenc,H., Booth,B.L.,Jr, Gileadi,U., Flavin,F.M., Trowsdale,J., Phillips,R.E. and Cerundolo,V. (1999) IFN-γ exposes a cryptic cytotoxic T lymphocyte epitope in HIV-1 reverse transcriptase. J. Immunol., 162, 7075–7079. [PubMed] [Google Scholar]
- Sijts A.J., Ruppert,T., Rehermann,B., Schmidt,M., Koszinowski,U. and Kloetzel,P.M. (2000) Efficient generation of a hepatitis B virus cytotoxic T lymphocyte epitope requires the structural features of immunoproteasomes. J. Exp. Med., 191, 503–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder H.L., Yewdell,J.W. and Bennink,J.R. (1994) Trimming of antigenic peptides in an early secretory compartment. J. Exp. Med., 180, 2389–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltze L., Dick,T.P., Deeg,M., Pommerl,B., Rammensee,H.G. and Schild,H. (1998) Generation of the vesicular stomatitis virus nucleoprotein cytotoxic T lymphocyte epitope requires proteasome-dependent and -independent proteolytic activities. Eur. J. Immunol., 28, 4029–4036. [DOI] [PubMed] [Google Scholar]
- Stoltze L. et al. (2000) Two new proteases in the MHC class I processing pathway. Nature Immunol., 1, 413–418. [DOI] [PubMed] [Google Scholar]
- Tarcsa E., Szymanska,G., Lecker,S.H., O’Connor,C.M. and Goldberg,A.L. (2000) Ca2+-free calmodulin and calmodulin damaged by in vitro aging are selectively degraded by 26S proteasomes without ubiquitination. J. Biol. Chem., 275, 20295–20301. [DOI] [PubMed] [Google Scholar]
- Van Kaer L., Ashtonrickardt,P.G., Eichelberger,M., Gaczynska,M., Nagashima,K., Rock,K.L., Goldberg,A.L., Doherty,P.C. and Tonegawa,S. (1994) Altered peptidase and viral-specific T-cell response in LMP2 mutant mice. Immunity, 1, 533–541. [DOI] [PubMed] [Google Scholar]
- Verma R. and Deshaies,R.J. (2000) A proteasome howdunit: the case of the missing signal. Cell, 101, 341–344. [DOI] [PubMed] [Google Scholar]
- Yewdell J.T. and Bennink,J.R. (1999) Immunodominance in major histocompatibility complex class I-restricted T lymphocyte responses. Annu. Rev. Immunol., 17, 51–88. [DOI] [PubMed] [Google Scholar]