Abstract
A hallmark of human DNA polymerase ι (polι) is the asymmetric fidelity of replication at template A and T when the enzyme extends primers annealed to a single-stranded template. Here, we report on the efficiency and accuracy of polι-dependent replication at a nick, a gap, the very end of a template and from a mispaired primer. Polι cannot initiate synthesis on a nicked DNA substrate, but fills short gaps efficiently. Surprisingly, polι’s ability to blunt-end a 1 bp recessed terminus is dependent upon the template nucleotide encountered and is highly erroneous. At template G, both C and T are inserted with roughly equal efficiency, whilst at template C, C and A are misinserted 8- and 3-fold more often than the correct base, G. Using substrates containing mispaired primer termini, we show that polι can extend all 12 mispairs, but with differing efficiencies. Polι can also extend a tandem mispair, especially when it is located within a short gap. The enzymatic properties of polι appear consistent with that of a somatic hypermutase and suggest that polι may be one of the low-fidelity DNA polymerases hypothesized to participate in the hypermutation of immunoglobulin variable genes in vivo.
Keywords: DNA polymerase η/DNA polymerase ζ/Rad30/Rad30B/somatic mutation
Introduction
In the past 18 months, the number of known eukaryotic DNA polymerases has more than doubled (Goodman and Tippen, 2000; Hubscher et al., 2000). Many of the new polymerases are related to the UmuC/DinB/Rev1/Rad30 superfamily of proteins (Gerlach et al., 1999; McDonald et al., 1999; Woodgate, 1999). These proteins share little to no sequence homology with any of the previously identified A-, B-, C-, D- or X-polymerase families (Ito and Braithwaite, 1991; Braithwaite and Ito, 1993; Cann and Ishino, 1999) and, as a consequence, it has been proposed that, collectively, the UmuC/DinB/Rev1/Rad30 superfamily be called the ‘Y-family’ of DNA polymerases (Ohmori et al., 2001). Interestingly, humans possess two RAD30 paralogs: RAD30A and RAD30B (Johnson et al., 1999a; Masutani et al., 1999b; McDonald et al., 1999). Both share roughly equal similarity to Saccharomyces cerevisiae RAD30 (McDonald et al., 1997; Roush et al., 1998) which encodes DNA polymerase η (Johnson et al., 1999b). However, biochemical characterization of the human Rad30A protein reveals that it is functionally interchangeable with its S.cerevisiae counterpart and it has therefore been designated as human polη (Johnson et al., 1999a, 2000b; Masutani et al., 1999b). As a consequence, RAD30A is also known by its approved HUGO name of POLH. The physiological role of polη appears to be the efficient and accurate bypass of many replication-blocking lesions (Johnson et al., 1999b, 2000b; Masutani et al., 1999a, 2000). In its absence, translesion replication is severely compromised, and humans with defects in polη exhibit the xeroderma pigmentosum variant (XP-V) phenotype (Johnson et al., 1999a; Masutani et al., 1999b).
RAD30B has also been shown to encode a novel DNA polymerase, polι, and it too goes under a pseudonym: POLI. The enzymatic properties of polι are very different from those reported for either S.cerevisiae or human polη (Johnson et al., 2000a; Tissier et al., 2000a,b; Zhang et al., 2000; Bebenek et al., 2001; McDonald et al., 2001). The in vivo role of polι is presently unknown. Biochemical characterization of the enzyme in vitro indicates that polι can readily misinsert bases opposite a number of replication-blocking lesions (Johnson et al., 2000a; Tissier et al., 2000a; Zhang et al., 2000, 2001; McDonald et al., 2001) and in some cases performs unassisted lesion bypass (Tissier et al., 2000a). However, a more appealing model for lesion bypass hypothesizes that it might be achieved more efficiently through the combined actions of polι together with another enzyme, such as polζ, that is better suited to elongate mispairs (Tissier et al., 2000a). Based on these observations, we suggested that one cellular function of polι (possibly in combination with polζ) is that of a backup to polη in the translesion replication of many DNA lesions (Tissier et al., 2000a). Indeed, such a role may be most evident in XP-V patients, who lack polη, and whose cells are hypermutable by UV light (Wang et al., 1993; Raha et al., 1996; McGregor et al., 1999).
Another potential clue as to polι’s cellular function comes from the recent discovery that the enzyme also possesses 5′-deoxyribose lyase (dRpase) activity and can substitute for polβ in base excision repair (BER) reactions in vitro (Bebenek et al., 2001). However, it is also clear that polι cannot fully complement all of polβ’s cellular functions in vivo as polβ-deficient mice are inviable (Gu et al., 1994; Esposito et al., 2000). Such observations imply that polι might only participate in a specialized form of BER in vivo (Bebenek et al., 2001).
In vitro replication reactions utilizing a short primer annealed to a longer single-stranded template revealed that the fidelity of polι is strictly template sequence dependent, with relatively few misinsertions occurring at template A, and most occurring at template T (Johnson et al., 2000a; Tissier et al., 2000b; Zhang et al., 2000). Overall, the average misincorporation frequency is ∼1 × 10–2. Based upon these properties, we previously hypothesized that another potential function for polι might be to generate genetic diversity during somatic hypermutation of immunoglobulin (Ig) variable (V) genes (Tissier et al., 2000b). The precise molecular mechanism of somatic hypermutation remains to be elucidated fully. However, it appears to require transcription and enhancer elements that target the process to an ∼1.5 kb region immediately downstream of the start of the rearranged IgV gene (Lebecque and Gearhart, 1990; Betz et al., 1994; Peters and Storb, 1996; Fukita et al., 1998). It was originally hypothesized that the substrate for somatic hypermutation might be a nick, a gap or a double-strand break (Brenner and Milstein, 1966) that was repaired by an error-prone DNA polymerase (Gearhart and Bogenhagen, 1983). Support for the hypothesis that a double-strand break acts as a substrate during somatic hypermutation was reported recently (Sale and Neuberger, 1998; Bross et al., 2000; Papavasiliou and Schatz, 2000). Indeed, strand breaks were shown to occur in a cell cycle-dependent manner within the IgV gene and, perhaps more importantly, most mutations were mapped to the break itself, or within 1–2 bases of it (Papavasiliou and Schatz, 2000).
Based upon its initial characterization, it appeared that polι might be a good candidate for at least one of the low-fidelity polymerase(s) hypothesized to function during somatic hypermutation of Ig genes (Goodman and Tippen, 2000; Papavasiliou and Schatz, 2000; Poltoratsky et al., 2000; Tissier et al., 2000b). We were, therefore, interested in characterizing the biochemical properties of polι further, especially its ability to replicate DNA substrates that might be expected to be generated as part of the normal somatic hypermutation process. To this end, we have analyzed the ability of polι to initiate synthesis at a nick, short gaps and at the end of a DNA template. We have also examined its ability to extend single and tandem mispairs, as well as the level of polι expression in B cells undergoing somatic hypermutation compared with resting cells.
Results
Ability of polι to replicate at nicks, gaps and the end of a DNA template
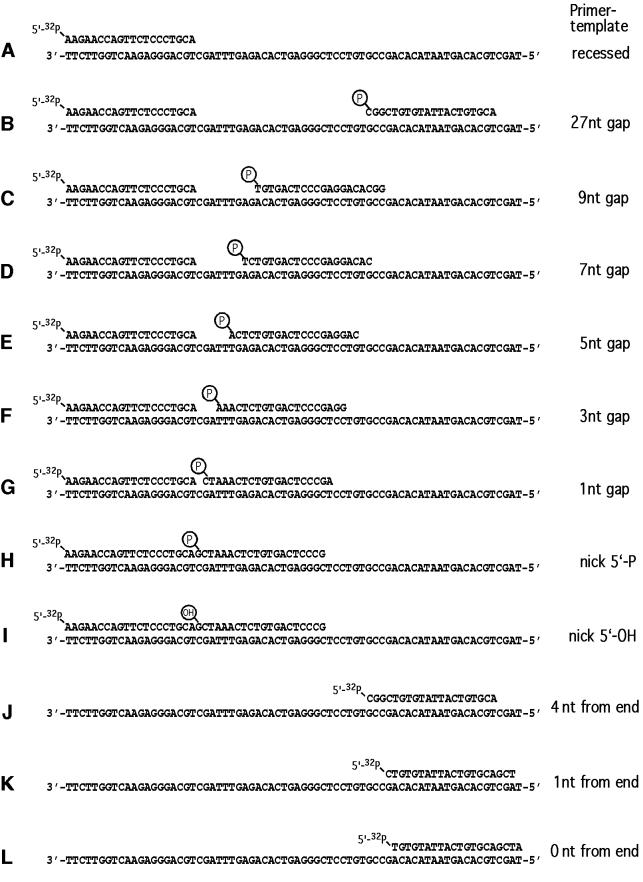
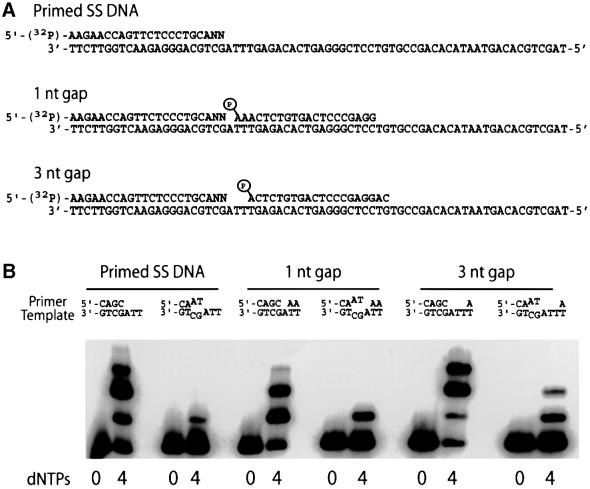
In order to analyze the ability of polι to replicate substrates that might mimic those generated during somatic hypermutation, we utilized a synthetic 70mer oligonucleotide template and a variety of short oligonucleotides, which, when annealed to the template, generated either a nicked substrate or a gapped substrate (gap sizes of 1, 3, 5, 7, 9 or 27 nucleotides; Figure 1). Furthermore, much of the sequence of the template was identical to codons 75–93 in the third framework region of the human VH6 gene (Berman et al., 1988). In particular, the three template nucleotides immediately downstream of the radiolabeled primer (3′-CGA-5′) corresponded to a known hot-spot for somatic hypermutation at codons 81–82 of the VH6 gene (Rogozin and Kolchanov, 1992; Betz et al., 1993) (Figure 1). To determine the ability of polι to replicate near the end of a template, a situation akin to that of a double-strand break, we used a single radiolabeled primer that was located 4, 1 and 0 bp from the end of the template (Figure 1).
Fig. 1. Nucleotide sequence and structure of DNA templates used in the replication assays described in Figure 2. The various primer–templates are labeled A–L on the left of the figure for ease of reference. The ‘class’ of substrate, i.e. which ones measure recessed, gapped or end-filling reactions, is given on the right of the figure.
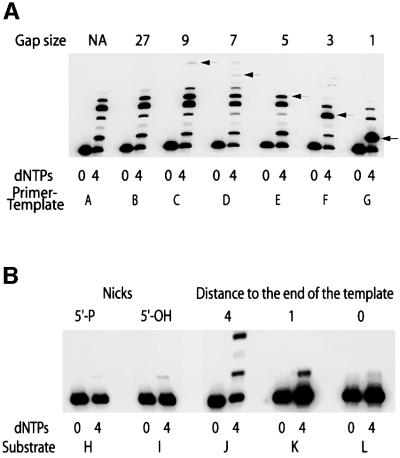
As previously reported, polι is a distributive enzyme (Tissier et al., 2000b) and only extends the singly-primed substrate by a few nucleotides (substrate A; Figure 2A). The strongest pause is 4 nucleotides from the primer, and this corresponds to template T. The pause probably reflects the efficient misincorporation of G opposite T and the reduced ability of polι to extend the G⋅T mispair relative to the correct A⋅T base pair (Tissier et al., 2000b; Zhang et al., 2000). A similar situation was observed when the template contained a second primer that generates a 27 nucleotide gap (substrate B; Figure 2A). Interestingly, faint replication products corresponding to the extension of the radiolabeled primer by 7–10 nucleotides were observed when the substrate was a short gap of 7–9 nucleotides (substrates C and D; Figure 2A), suggesting that such a context stimulates polι’s catalytic activity. Polι was also able to initiate synthesis at shorter gaps of 1, 3, and 5 nucleotides, respectively (substrates E, F and G; Figure 2A). In the case of the 7, 3 and 1 bp gaps, products longer than the expected gap size were observed, indicating that polι can also perform limited strand displacement of a downstream primer. The fidelity of the gap-filling reaction was measured at the template site immediately downstream of the radiolabeled primer (template C) and was found to be similar to that of the singly primed template, with misincorporations occurring in the range of 5–8 × 10–3 (E.G.Frank and R.Woodgate, unpublished observations).
Fig. 2. (A) Comparison of polι’s ability to replicate primed single-stranded DNA and gapped templates. For each template, polι was incubated for 30 min with the substrate in the absence (0) or presence of all four (4) deoxynucleoside triphosphates (100 µM each). The primer–template used in each reaction is denoted by a letter, A–G, under each pair of reactions, and this refers to the sequence and structure of each primer–template shown in Figure 1. Where appropriate, the gap size is indicated above each reaction. The exception are the lanes denoted ‘NA’, which represent a singly primed template in which no gap exists. In the case of the gapped substrates, the size of the expected product is indicated by an arrow at the right hand side of each pair of reactions. (B) Ability of polι to initiate synthesis at a nick or the end of a DNA template. For each template, polι was incubated for 30 min with the substrate in the absence (0) or presence of all four (4) deoxynucleoside triphosphates (100 µM each). The primer–template used in each reaction is denoted by a letter, H–L, under each pair of reactions, and this refers to the sequence and structure of each primer–template shown in Figure 1. Where appropriate, the presence of a 5′-phosphate or 5′-hydroxyl in the nicked substrate is indicated, as is the distance to the end of the template in substrates J–L, which were used to measure the efficiency of end filling.
By comparison, polι was unable to initiate synthesis at a template containing a nick (with either a 5′-phosphate or a 5′-hydroxyl) (substrates H and I; Figure 2B). These findings are consistent with the fact that preparations of glutathione S-transferase (GST)–polι lack any measurable 5′–3′ exonuclease activity. In contrast, robust synthesis was observed with these templates when Escherichia coli polymerase I (polI) was used in control reactions (E.G.Frank and R.Woodgate, unpublished observations).
We were also interested in determining the properties of the enzyme at the end of the template. The initial experiments utilized the 70mer template with primers located 4, 1 and 0 bp from the end of the template (substrates J, K and L; Figure 2B). Under these conditions, it appeared that the primers located one and four bases from the end of the template were extended less efficiently than those annealed to the longer gapped templates used in Figure 1. Faint replication products were also detected when the primer was located at the very end of the template (substrate L; Figure 2B), which initially suggested to us that polι might possess template-independent terminal transferase activity. However, we could not exclude the possibility that the 70mer template did not contain very small amounts of a contaminating 71mer, despite the fact that the template was purified multiple times by PAGE prior to use.
Fidelity of polι replication at the end of a DNA template
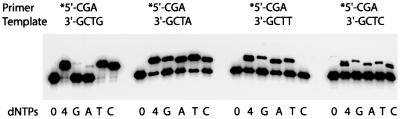
To circumvent the problems associated with the longer DNA template, we designed a shorter 20mer DNA template to control purity more accurately. In addition, the sequence of this new template was identical to that used previously in our initial fidelity studies with polι (Tissier et al., 2000b), and allowed a direct comparison between replication fidelities in the context of a long (19 nucleotides) and short (one nucleotide) DNA template. Interestingly, with the shorter template, it became apparent that the efficiency of polι-dependent replication at the end of the template is sequence specific. In the presence of all four nucleoside triphosphates (dNTPs), the template was blunt-ended most efficiently when G was replicated and least efficiently when C was the target (Figure 3). Similar results were obtained when polι extended from a correctly paired G⋅C primer–template terminus, indicating that the effects are due to the template structure rather than the sequence context (E.G.Frank and R.Woodgate, unpublished observations).
Fig. 3. Ability of polι to incorporate nucleotides at the end of a DNA template. The primer for each reaction was a radiolabeled 16mer with the sequence 5′-CTTGAAAACATAGCGA-3′. The template was a 22mer with the sequence 5′-XTCGCTATGTTTTCAAGGATTC-3′, where X was either G, A, T or C. The position of the hybridized primer on the template is underlined. The local sequence context of each primer–template is given above each panel. The extent of polι-dependent (mis)incorporation was measured at each template site in the absence of dNTPs (0), with all four dNTPs (4) or with each individual dNTP (100 µM) (G, A, T or C). Reactions were for 30 min at 37°C.
With both correctly paired primer–templates (A⋅T; Figure 3, and G⋅C; data not shown), the reaction was strictly template dependent and no replication products longer than one base were observed (Figure 3). We conclude, therefore, that the limited amount of replication products seen in Figure 2B (substrate L) represents extension of the primer annealed to a contaminating 71mer template rather than weak terminal transferase activity.
We quantitated the fidelity of the end-filling reaction using a steady-state gel assay (Boosalis et al., 1987; Creighton et al., 1995) and discovered that it is very different from that previously reported for a longer DNA template (Table I). In some cases, the fidelity increased several fold, e.g. misincorporation of G or A at template G, where the one-nucleotide end-filling reaction is ∼40-fold more accurate. However, at most sites, there was a decrease in replication fidelity. The most striking case was at template C, where C and A mispairs formed ∼8- and 3-fold more frequently than the correct base, G. In contrast, the formation of C⋅C and C⋅A mispairs on the longer template occurred at a lower frequency of ∼7 × 10–3 (Table I; Tissier et al., 2000b). Thus, when replicating C at the end of a template, polι is ∼500- to 1000-fold more likely to insert C or A than when it encounters a C in a longer template. Replication remained most accurate at template A, but overall fidelities were 5- to 13-fold lower than that observed with the longer template DNA.
Table I. Fidelity of human polι on a recessed primed DNA template and at the end of a DNA template as determined by steady-state kinetics.
| Recessed templatea |
One nucleotide from the end |
Fold difference in fidelityd | ||||
|---|---|---|---|---|---|---|
| dNTP⋅template | Vmax/Kmb (µM–1 min–1) | fincc | dNTP⋅template | Vmax/Kmb (µM–1 min–1) | fincc | |
| dGTP⋅G | 0.005 | 6.5 × 10–3 | dGTP⋅G | 0.0002 | 1.5 × 10–4 | ↑43.3 |
| dATP⋅G | 0.02 | 2.2 × 10–2 | dATP⋅G | 0.0007 | 5.4 × 10–4 | ↑40.1 |
| dTTP⋅G | 0.100 | 1.3 × 10–1 | dTTP⋅G | 0.64 | 4.9 × 10–1 | ↓3.8 |
| dCTP⋅G | 0.76 | 1 | dCTP⋅G | 1.3 | 1 | N/A |
| dGTP⋅A | 0.008 | 1.0 × 10–4 | dGTP⋅A | 0.024 | 5.1 × 10–4 | ↓5.1 |
| dATP⋅A | 0.02 | 2.0 × 10–4 | dATP⋅A | 0.11 | 2.3 × 10–3 | ↓11.5 |
| dTTP⋅A | 73.5 | 1 | dTTP⋅A | 47.2 | 1 | N/A |
| dCTP⋅A | 0.009 | 1.0 × 10–4 | dCTP⋅A | 0.06 | 1.3 × 10–3 | ↓13.0 |
| dGTP⋅T | 0.33 | 3 | dGTP⋅T | 0.006 | 3.3 × 10–1 | ↑9.1 |
| dATP⋅T | 0.11 | 1 | dATP⋅T | 0.018 | 1 | N/A |
| dTTP⋅T | 0.078 | 6.7 × 10–1 | dTTP⋅T | 0.01 | 5.6 × 10–1 | ↑0.8 |
| dCTP⋅T | nd | <1.0 × 10–5 | dCTP⋅T | nd | <1.0 × 10–5 | N/A |
| dGTP⋅C | 3.0 | 1 | dGTP⋅C | 0.0016 | 1 | N/A |
| dATP⋅C | 0.019 | 6.3 × 10–3 | dATP⋅C | 0.0055 | 3.4 | ↓540 |
| dTTP⋅C | 0.023 | 7.7 × 10–3 | dTTP⋅C | 0.0014 | 8.8 × 10–1 | ↓114 |
| dCTP⋅C | 0.022 | 7.4 × 10–3 | dCTP⋅C | 0.013 | 8.1 | ↓1095 |
N/A, not applicable.
nd, not detected.
aThe data for the recessed template were taken from Tissier et al. (2000b) and are shown for comparison.
bThe Vmax/Km ratio was the average of three or more experiments with standard deviations not exceeding 20% of the Vmax/Km value.
cThe nucleotide misincorporation ratio, finc = (Vmax/Km)incorrect/(Vmax/Km)correct..
dThe ↑ indicates that there was an increase in replication fidelity, while ↓ indicates that there was a decrease.
Ability of polι to extend single and tandem base mispairs
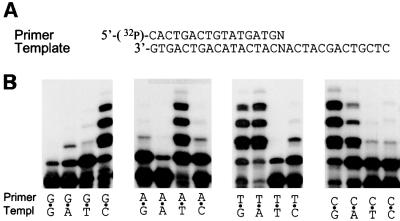
To determine whether polι can extend past mispairs, primer–templates were designed so as to generate all 12 mispairs in addition to the four correct base pairs (Figure 4A). We then analyzed the ability of polι to extend the paired/mispaired primers in the presence of all four dNTPs. In these experiments, the first base encountered by the polymerase is template A. This template base was chosen as polι prefers to insert T opposite A with catalytic efficiencies greater than any other nucleotide (Johnson et al., 2000a; Tissier et al., 2000b; Zhang et al., 2000; Bebenek et al., 2001), and we hypothesized that this would most probably represent the best sequence context for mismatch extension. As can be seen in Figure 4B, all mispairs are extended, but some mispairs are clearly favored over others. The T⋅G mispair appears to be the most efficiently extended, and in this qualitative assay appears similar to that of the correct A⋅T base pair. Many of the mispairs were extended by one nucleotide, but no further, suggesting that the mispair still represents a kinetic block to elongation even when it is no longer in the active site of the enzyme. The G⋅G, A⋅A and T⋅T mispairs were extended with the lowest efficiency. A more detailed analysis of polι-dependent extension from a mispair and the effects of nearest-neighbor bases will be presented elsewhere (A.Vaisman, A.Tissier, E.G.Frank, M.F. Goodman and R.Woodgate, submitted).
Fig. 4. (A) Nucleotide sequence of the primer and templates utilized to measure polι-dependent primer extension. In both cases, N is either G, A, T or C. (B) Comparison of polι’s ability to extend a correctly paired or mispaired primer terminus. The sequence of the pair/mispair is given below each track. These reactions were performed for 15 min in the presence of all four dNTPs.
Our studies with mice deficient in the PMS2 mismatch repair protein suggested that tandem mutations occur in the IgV gene and may be repaired by a PMS2-dependent pathway (Winter et al., 1998). Polι clearly has the ability to insert an erroneous base as well as extend it (albeit with varying efficiencies). We were, therefore, interested in determining whether polι might be able to extend a tandem mispair. The template for these studies was the 70mer used in Figure 1, and we followed the ability of polι to extend correctly paired and tandem mismatched primers on a single-stranded template as well as within the context of a short gap (Figure 5A). The A⋅C and T⋅G mispairs were chosen as they represent the most common mutational events recovered at the somatic hypermutation hot-spot found at codons 81–82 within the VH6 gene (Rosner et al., 2001). Given the structural distortions imposed by the tandem mispair, it was not surprising that the mispaired primer was poorly extended when in the context of single-stranded DNA (Figure 5B). Interestingly, however, extension from the tandem mispair was stimulated greatly when it was located within the context of a 1 or 3 bp gap (Figure 5B).
Fig. 5. (A) Nucleotide sequence and structure of DNA templates used in replication assays to measure polι-dependent extension from a correctly paired 22mer and a 22mer with two terminal mismatches. (B) Ability of polι to extend a tandem mispair. For each template, polι was incubated for 30 min with the substrate in the absence (0) or presence of all four (4) deoxynucleoside triphosphates (100 µM each).
Tissue-specific expression of polι
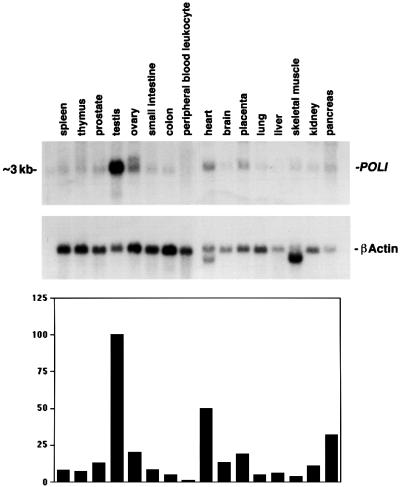
The expression pattern of human POLI (encoding polι) in various adult tissues was examined initially by northern blot analysis and was detected in most tissues as an ∼3 kb transcript, except in ovary, where the transcript appeared slightly longer (Figure 6). As previously noted, expression of human POLI is highest in testis, and in this respect is similar to the related mouse Poli gene, which is highly expressed in post-meiotic round spermatids (McDonald et al., 1999). The human POLI messenger is also abundant in heart and pancreas, and is present at low levels in other tissues. In addition, nearly 50 human expressed sequence tag (EST) sequences have been identified in the EST database that correspond to POLI. These ESTs come from various human tissue sources including fetal heart (DDBJ/EMBL/GenBank accession No. AI14880), uterus (AA156602), placenta (AI277458), neuroendocrine lung carcinoid (AA878207), testis (AI1223233), parathyroid tumor cells (AI167389), umbilical vein endothelial cells (AA181126) and germinal center B cells (AA856713) and, as a consequence, we conclude that the expression of POLI is ubiquitous but elevated in certain tissues.
Fig. 6. Northern blot analysis of POLI mRNA in adult tissues. Upper panel: human POLI cDNA probes were hybridized with 2 µg of poly(A)+ RNA from each tissue as indicated. A β-actin cDNA was used as a control. Lower panel: autoradiograms were quantitated and relative expression was normalized to the level of β-actin in each tissue. The extent of POLI expression in the various tissue sources is indicated as a percentage of that observed in the testis (100%).
The fact that human POLI was identified in germinal center B cells is of particular importance as that is where hypermutation of IgV genes takes place (Jacob et al., 1991, 1993). We therefore sought to quantitate its in vivo expression more accurately. To do so, the level of murine Poli transcript was measured in resting, non-germinal center B cells (B220+PNA–) and activated, germinal center B cells (B220+PNA+) by relative quantitative RT–PCR, which compares the levels of transcripts with an internal 18S rRNA standard. As shown in Table II, polι is expressed 1.6-fold more in the germinal center cells compared with the non-germinal center cells. In contrast, the catalytic subunit of polα is not expressed at detectable levels in the resting cells, but is up-regulated in the activated cells, and control transcripts of polβ and β-actin are expressed equally in both groups of cells.
Table II. Expression of polι message in B cells from murine germinal centers and non-germinal centers.
| Germinal center:non-germinal center ratioa | |
|---|---|
| Polι | 1.61 (0.04) |
| Polα | >7.40 (0.38)b |
| Polβ | 1.02 (0.02) |
| β-actin | 1.01 (0.02) |
aQuantity of RT–PCR product for genes compared with 18s rRNA for each cell type. The standard deviation for two experiments is shown in parentheses.
bPolα was strongly expressed in germinal center cells and was not detectable in non-germinal center cells.
Discussion
The role of polι within the cell is presently unknown, and the focus of the current study is to characterize the enzyme better at the biochemical level, so as to provide clues to possible cellular functions. The experiments were designed initially to investigate a potential role for polι in somatic hypermutation, but it should be mentioned that they also serendipitously provide insights into a possible role of polι in BER. Support for such a role comes from the recent discovery that polι possesses an intrinsic dRpase activity, which allows it to substitute for polβ during in vitro BER reactions (Bebenek et al., 2001). In the experiments described here, the catalytic activity of polι was stimulated by a gapped substrate and was particularly robust with a one-nucleotide gap (Figure 2). Indeed, under these conditions, polι, like polβ (Dianov et al., 1999; Prasad et al., 2000), is able to perform limited strand displacement of a downstream primer, indicating that the enzyme can participate in both ‘short patch’ and ‘long patch’ gap-filling/BER reactions (Figure 2). In vitro, polβ-dependent long patch repair is stimulated greatly by additional cofactors, such as the flap-endonuclease FEN1 (Dianov et al., 1999; Prasad et al., 2000), and it will be interesting to determine whether FEN1 also stimulates the polι-dependent long patch BER reaction.
To date, the hallmark of polι-dependent replication is the asymmetry of its fidelity when extending from a primed single-stranded template. Under these conditions, most errors occur at template T, where misinsertion of G is favored over the correct base, A (Johnson et al., 2000a; Tissier et al., 2000b; Zhang et al., 2000). In contrast, polι is relatively accurate at template A, with misinsertion fidelities in the range of normal replicative polymerases (Kunkel and Bebenek, 2000). This asymmetry is reminiscent of the strand bias found in IgV genes (Smith et al., 1996; Foster et al., 1999), where there are more mutations from A (due to misincorporations opposite template T) and few mutations from T (i.e. T is incorporated accurately opposite template A), and led us to hypothesize that polι might participate in somatic hypermutation (Tissier et al., 2000b; McDonald et al., 2001). To test such a hypothesis, we generated synthetic substrates that we hypothesized might mimic those that arise naturally during the mutagenic processing of IgV genes. In particular, it is now evident that many mutations are generated at, or very close to, a double-stranded break (Bross et al., 2000; Papavasiliou and Schatz, 2000). Intriguingly, under conditions where polι replicates at the end of a template, we observed a dramatic change in its fidelity (Table I). In particular, there was a 100- to 1000-fold decrease in fidelity at template C, where the enzyme generally was unable to determine which base to insert. We also observed changes in fidelity at template G, where T was misinserted with almost the same frequency as the correct base C, and misincorporations of A and G decreased significantly (Table I).
Analysis of the spectra of mutations generated during somatic hypermutation reveals that transitions are favored 2:1 over transversions, and it has been suggested that the spectra might be generated through the actions of separate G/C and A/T mutator polymerases (Rada et al., 1998; Spencer et al., 1999). It is now known that cells contain many low-fidelity DNA polymerases (Goodman and Tippen, 2000), so it is plausible that one or more DNA polymerase are indeed involved in the hypermutagenic process. However, it may be more than a passing coincidence that our in vitro studies indicate that polι, acting on a primer terminus with a long template overhang, will give rise to few mutations from T, while generating a high incidence of A to G transitions. Similarly, at the end of a template, we expect a preponderance of C to T transitions, while both transition and transversions from G are predicted to occur with roughly similar frequency. Thus, polι may fulfill the role of an A/T mutator in the middle of a template and a G/C mutator at the end of a template.
In addition to exhibiting extremely low template-specific fidelity, which results in frequent misinsertions, polι is able to extend both single and tandem mispairs, with the latter occurring more efficiently within the context of a short gap, where the catalytic activity of the enzymes is enhanced. The ability of the enzyme to extend a tandem mispair indicates that the active site of the enzyme is sufficiently flexible to accommodate the distorted primer terminus imposed by the tandem mispair, and is consistent with the ability of the enzyme to also incorporate nucleotides opposite a variety of bulky adducts in vitro (Tissier et al., 2000a; Johnson et al., 2000a; Zhang et al., 2000, 2001; McDonald et al., 2001). Our experiments with PMS2 mismatch repair-deficient mice revealed that tandem mutations are observed during somatic hypermutation, but may be repaired by mismatch repair (Winter et al., 1998). Thus, a prerequisite for the somatic hypermutase is that it be able to generate and elongate both single and tandem mispairs, which, as demonstrated in Figures 4 and 5, is a property that can be attributed to polι.
Finally, while human POLI mRNA was found to be overexpressed in testis, heart and pancreas, it was also expressed ubiquitously in other tissues. RT–PCR of the mouse Poli gene revealed that it is modestly elevated in proliferating germinal center B cells compared with the non-germinal center cells. The expression of polι in the quiescent non-germinal center cells correlates with the ubiquitous expression of the human POLI gene in many non-dividing tissues and suggests that basal expression of the enzyme is required for its putative role in translesion replication (Tissier et al., 2000a) and/or BER (Bebenek et al., 2001). Its elevated expression in activated germinal center cells may be due to an association of polι with the replication fork so as to facilitate lesion bypass (Tissier et al., 2000a) and/or the BER of deoxyuracil incorporated during normal replication (Nilsen et al., 2000; Bebenek et al., 2001). Another formal possibility is that polι is elevated in germinal center cells because it participates in somatic hypermutation.
To date, the characterization of polι has been entirely at the biochemical level. The data obtained from these experiments suggest that polι may participate in at least three discrete processes: translesion replication of replication-blocking lesions in DNA; a specific subset of BER reactions; and somatic hypermutation. Of course, the possibility exists that the true function of polι has yet to be identified. Whatever the ultimate biological role(s) of this enzyme, it presumably must provide some benefit to higher eukaryotes as it is not found in lower eukaryotes, and the distantly related Drosophila melanogaster polι enzyme exhibits biochemical properties quite distinct from those found with the human enzyme (Ishikawa et al., 2001). Clearly, the best way to demonstrate a biological role for polι in any of the aforementioned processes is to demonstrate an absence of function in polι-deficient cell lines or knockout mice. Indeed, such experiments are currently in progress in a number of laboratories, and the results from such experiments are eagerly awaited.
Materials and methods
Human DNA polι
Wild-type GST-tagged polι was purified by glutathione–agarose affinity chromatography and hydroxylapatite ion-exchange chromatography as previously described (Tissier et al., 2000b).
DNA templates
The synthetic oligonucleotides used as primers or templates in the in vitro assays were synthesized by Loftstrand Laboratories (Gaithersburg, MD) using standard techniques and were gel purified prior to use. The sequence of each oligonucleotide is given either in the figure or the legend of the respective experiment in which it was used. Where indicated, the primer was 5′ labeled with [γ-32P]ATP (5000 Ci/mmol; 1 Ci = 37 GBq) (Amersham Pharmacia Biotech, Piscataway, NJ) using T4 polynucleotide kinase (Life Technologies, Gaithersburg, MD).
Replication reactions
Radiolabeled primer–template DNAs were prepared by annealing the 5′-32P-labeled primer to the unlabeled template DNA at a molar ratio of 1:1.5
Where indicated, a second unlabeled oligonucleotide was also annealed to the same template in a ratio of 2:1, so as to generate a gapped substrate. Standard replication reactions (10 µl) contained 40 mM Tris–HCl pH 8.0, 5 mM MgCl2, 100 µM of each ultrapure dNTP (Amersham Pharmacia Biotech, NJ), 10 mM dithiothreitol (DTT), 250 µg/ml bovine serum albumin (BSA), 60 mM KCl, 2.5% glycerol, 10 nM 5′-[32P]primer–template DNA and 3 nM GST–polι. Reactions were incubated at 37°C for 30 min (unless noted otherwise), and were terminated by the addition of 10 µl of 95% formamide/10 mM EDTA containing 0.1% xylene cyanol and 0.1% bromophenol blue. The samples were heated to 100°C for 5 min and 5 µl of the reaction mixture subjected to 20% polyacrylamide–7 M urea gel electrophoresis. Replication products were subsequently visualized by autoradiography or PhosphorImager analysis.
Kinetic analysis of replication products
Reactions were performed essentially as described previously (Boosalis et al., 1987; Creighton et al., 1995; Tissier et al., 2000b). Briefly, they were first optimized so as to ensure that the reaction remained within a linear range and, in subsequent reactions, the concentration of each nucleotide in the reaction mixture was varied from 0.1 to 100 µM. Reaction products were separated in a 20% polyacrylamide gel containing 7 M urea, and gels were dried prior to quantitative PhosphorImager analysis using ImageQuant software (Molecular Dynamics, CA). Nucleotide misincorporation frequencies were calculated as described previously and the data presented are the average of 2–3 separate experiments (Boosalis et al., 1987; Creighton et al., 1995; Tissier et al., 2000b).
Northern analysis of the human POLI transcripts
Human multiple-tissue northern blots (Clontech, Palo Alto, CA), containing 2 µg of poly(A)+ RNA per track, were hybridized with random primed probes generated from the coding region of the POLI cDNA (McDonald et al., 1999). After overnight hybridization at 42°C in Hybrisol I buffer (Oncor, Gaithersburg, MD), membranes were washed twice in 2× SSC at room temperature for 15 min, and subsequently twice in 2× SSC containing 2% SDS at 65°C for 45 min. More stringent conditions were obtained by washing the blots in 0.1× SSC at 65°C for 15–30 min.
Analysis of murine Poli expression in resting and proliferating B cells
In two separate experiments using different BALB/cJ mice (Jackson Laboratories, Bar Harbor, ME), splenic lymphocytes were prepared 11 days after immunization with phenyl-oxazolone–chicken serum albumin, as previously described (Winter et al., 2000). Briefly, the cells were stained with phycoerythrin-labeled antibody to B220 (PharMingen, San Diego, CA) and fluorescein-labeled peanut agglutinin (PNA) (E-Y Laboratories, San Mateo, CA). B220+PNA+ germinal center B cells and B220+PNA– non-germinal center B cells were isolated by flow cytometry; RNA was prepared from ∼10 000 cells using RNA STAT-60 (Tel-Test B, Friendswood, TX), and cDNA was made (RETROscript; Ambion, Austin, TX). The expression of transcripts for polι, polα and β-actin in the two cell populations was analyzed using relative quantitative RT–PCR (QuantumRNA; Ambion, Austin, TX), where the quantity of transcripts is standardized by co-amplification of 18S rRNA as an internal standard. For the PCR analysis, cDNA from ∼1000 cells was amplified in the linear range using a first set of primers for 40 cycles (94°C for 30 s, 62°C for 30 s and 72°C for 30 s) and a second set of nested primers for 25 cycles. The primers for Poli spanned several introns (J.P.McDonald and R.Woodgate, in preparation), and were located at nucleotide positions 1088–1797 within the cDNA clone (McDonald et al., 1999): first set, forward 5′-GTATGCCAGGATGGAAGGAAGCC, reverse 5′-GATAC CCGCTTGCTAGGGAGCG; second set, forward 5′-GGTACTCTGAC AAACACTGTAATCG, reverse 5′-CCTCTAGAGGCATGCAGCG GAC. The primers for the polα catalytic subunit, polβ and β-actin have been described (Winter et al., 2000). The PCR products were separated by electrophoresis on 1.2% agarose gels in the presence of ethidium bromide, visualized on a Fluorimager SI (Molecular Dynamics, Sunnyvale, CA) and quantified using ImageQuant software (Molecular Dynamics, Sunnyvale, CA). The background around each band was subtracted using the local median technique.
Acknowledgments
Acknowledgements
We would like to thank Rick Wood for initially suggesting that we investigate the link between polι and somatic hypermutation by assaying the properties of polι on gaps, nicks and at the end of a DNA template; we thank Samuel Wilson for polβ that was used as a control in many of the replication reactions; and Alexandra Vaisman, Antonio Rodriguez, Mary McLenigan and François Boudsocq for their helpful comments and suggestions during the course of this work. This work was supported by the NIH Intramural research program.
References
- Bebenek K., Tissier,A., Frank,E.G., McDonald,J.P., Prasad,R., Wilson,S.H., Woodgate,R. and Kunkel,T.A. (2001) 5′-deoxyribose phosphate lyase activity of human DNA polymerase ι in vitro. Science, 291, 2156–2159. [DOI] [PubMed] [Google Scholar]
- Berman J.E. et al. (1988) Content and organization of the human Ig VH locus: definition of three new VH families and linkage to the Ig CH locus. EMBO J., 7, 727–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz A.G., Rada,C., Pannell,R., Milstein,C. and Neuberger,M.S. (1993) Passenger transgenes reveal intrinsic specificity of the antibody hypermutation mechanism: clustering, polarity and specific hot spots. Proc. Natl Acad. Sci. USA, 90, 2385–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz A.G., Milstein,C., Gonzalez-Fernandez,A., Pannell,R., Larson,T. and Neuberger,M.S. (1994) Elements regulating somatic hypermutation of an immunoglobulin κ gene: critical role for the intron enhancer/matrix attachment region. Cell, 77, 239–248. [DOI] [PubMed] [Google Scholar]
- Boosalis M.S., Petruska,J. and Goodman,M.F. (1987) DNA polymerase insertion fidelity. Gel assay for site-specific kinetics. J. Biol. Chem., 262, 14689–14696. [PubMed] [Google Scholar]
- Braithwaite D.K. and Ito,J. (1993) Compilation, alignment and phylogenetic relationships of DNA polymerases. Nucleic Acids Res., 21, 787–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. and Milstein,C. (1966) Origin of antibody variation. Nature, 211, 242–243. [PubMed] [Google Scholar]
- Bross L., Fukita,Y., McBlane,F., Demolliere,C., Rajewsky,K. and Jacobs,H. (2000) DNA double-strand breaks in immunoglobulin genes undergoing somatic hypermutation. Immunity, 13, 589–597. [DOI] [PubMed] [Google Scholar]
- Cann I.K. and Ishino,Y. (1999) Archaeal DNA replication: identifying the pieces to solve a puzzle. Genetics, 152, 1249–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton S., Bloom,L.B. and Goodman,M.F. (1995) Gel fidelity assay measuring nucleotide misinsertion, exonucleolytic proofreading and lesion bypass efficiencies. Methods Enzymol., 262, 232–256. [DOI] [PubMed] [Google Scholar]
- Dianov G.L., Prasad,R., Wilson,S.H. and Bohr,V.A. (1999) Role of DNA polymerase β in the excision step of long patch mammalian base excision repair. J. Biol. Chem., 274, 13741–13743. [DOI] [PubMed] [Google Scholar]
- Esposito G., Texido,G., Betz,U.A., Gu,H., Muller,W., Klein,U. and Rajewsky,K. (2000) Mice reconstituted with DNA polymerase β-deficient fetal liver cells are able to mount a T cell-dependent immune response and mutate their Ig genes normally. Proc. Natl Acad. Sci. USA, 97, 1166–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster S.J., Dorner,T. and Lipsky,P.E. (1999) Targeting and subsequent selection of somatic hypermutations in the human Vκ repertoire. Eur. J. Immunol., 29, 3122–3132. [DOI] [PubMed] [Google Scholar]
- Fukita Y., Jacobs,H. and Rajewsky,K. (1998) Somatic hypermutation in the heavy chain locus correlates with transcription. Immunity, 9, 105–114. [DOI] [PubMed] [Google Scholar]
- Gearhart P.J. and Bogenhagen,D.F. (1983) Clusters of point mutations are found exclusively around rearranged antibody variable genes. Proc. Natl Acad. Sci. USA, 80, 3439–3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach V.L., Aravind,L., Gotway,G., Schultz,R.A., Koonin,E.V. and Friedberg,E.C. (1999) Human and mouse homologs of Escherichia coli DinB (DNA polymerase IV), members of the UmuC/DinB superfamily. Proc. Natl Acad. Sci. USA, 96, 11922–11927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman M.F. and Tippen,B. (2000) The expanding polymerase universe. Nature Rev. Mol. Cell. Biol., 1, 101–109. [DOI] [PubMed] [Google Scholar]
- Gu H., Marth,J.D., Orban,P.C., Mossmann,H. and Rajewsky,K. (1994) Deletion of a DNA polymerase β gene segment in T cells using cell type-specific gene targeting. Science, 265, 103–106. [DOI] [PubMed] [Google Scholar]
- Hubscher U., Nasheuer,H.-P. and Syvaoja,J.E. (2000) Eukaryotic DNA polymerases, a growing family. Trends Biochem. Sci., 25, 143–147. [DOI] [PubMed] [Google Scholar]
- Ishikawa T., Uematsu,N., Mizukoshi,T., Iwai,S., Masutani,C., Hanaoka,F., Ueda,R., Ohmori,H. and Todo,T. (2001) Mutagenic and non-mutagenic bypass of DNA lesions by Drosophila DNA polymerases dpolη and dpolι. J. Biol. Chem., 276, 15155–15163. [DOI] [PubMed] [Google Scholar]
- Ito J. and Braithwaite,D.K. (1991) Compilation and alignment of DNA polymerase sequences. Nucleic Acids Res., 19, 4045–4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob J., Kelsoe,G., Rajewsky,K. and Weiss,U. (1991) Intraclonal generation of antibody mutants in germinal centres. Nature, 354, 389–392. [DOI] [PubMed] [Google Scholar]
- Jacob J., Przylepa,J., Miller,C. and Kelsoe,G. (1993) In situ studies of the primary immune response to (4-hydroxy-3-nitrophenyl)acetyl. III. The kinetics of V region mutation and selection in germinal center B cells. J. Exp. Med., 178, 1293–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R.E., Kondratick,C.M., Prakash,S. and Prakash,L. (1999a) hRAD30 mutations in the variant form of xeroderma pigmentosum. Science, 285, 263–265. [DOI] [PubMed] [Google Scholar]
- Johnson R.E., Prakash,S. and Prakash,L. (1999b) Efficient bypass of a thymine–thymine dimer by yeast DNA polymerase, polη. Science, 283, 1001–1004. [DOI] [PubMed] [Google Scholar]
- Johnson R.E., Washington,M.T., Haracska,L., Prakash,S. and Prakash,L. (2000a) Eukaryotic polymerases ι and ζ act sequentially to bypass DNA lesions. Nature, 406, 1015–1019. [DOI] [PubMed] [Google Scholar]
- Johnson R.E., Washington,M.T., Prakash,S. and Prakash,L. (2000b) Fidelity of human DNA polymerase η. J. Biol. Chem., 275, 7447–7450. [DOI] [PubMed] [Google Scholar]
- Kunkel T.A. and Bebenek,K. (2000) DNA replication fidelity. Annu. Rev. Biochem., 69, 497–529. [DOI] [PubMed] [Google Scholar]
- Lebecque S.G. and Gearhart,P.J. (1990) Boundaries of somatic mutation in rearranged immunoglobulin genes: 5′ boundary is near the promoter and 3′ boundary is approximately 1 kb from V(D)J gene. J. Exp. Med., 172, 1717–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutani C., Araki,M., Yamada,A., Kusumoto,R., Nogimori,T., Maekawa,T., Iwai,S. and Hanaoka,F. (1999a) Xeroderma pigmentosum variant (XP-V) correcting protein from HeLa cells has a thymine dimer bypass DNA polymerase activity. EMBO J., 18, 3491–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutani C. et al. (1999b) The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature, 399, 700–704. [DOI] [PubMed] [Google Scholar]
- Masutani C., Kusumoto,R., Iwai,S. and Hanaoka,F. (2000) Mechanisms of accurate translesion synthesis by human DNA polymerase η. EMBO J., 19, 3100–3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald J.P., Levine,A.S. and Woodgate,R. (1997) The Saccharomyces cerevisiae RAD30 gene, a homologue of Escherichia coli dinB and umuC, is DNA damage inducible and functions in a novel error-free postreplication repair mechanism. Genetics, 147, 1557–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald J.P., Rapic-Otrin,V., Epstein,J.A., Broughton,B.C., Wang,X., Lehmann,A.R., Wolgemuth,D.J. and Woodgate,R. (1999) Novel human and mouse homologs of Saccharomyces cerevisiae DNA polymerase η. Genomics, 60, 20–30. [DOI] [PubMed] [Google Scholar]
- McDonald J.P., Tissier,A., Frank,E.G., Iwai,S., Hanaoka,F. and Woodgate,R. (2001) DNA polymerase ι and related Rad30-like enzymes. Philos. Trans. R. Soc. Lond. B Biol. Sci., 356, 53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGregor W.G., Wei,D., Maher,V.M. and McCormick,J.J. (1999) Abnormal, error-prone bypass of photoproducts by xeroderma pigmentosum variant cell extracts results in extreme strand bias for the kinds of mutations induced by UV light. Mol. Cell. Biol., 19, 147–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen H. et al. (2000) Uracil-DNA glycosylase (UNG)-deficient mice reveal a primary role of the enzyme during DNA replication. Mol. Cell, 5, 1059–1065. [DOI] [PubMed] [Google Scholar]
- Ohmori H. et al. (2001) The Y-family of DNA polymerases. Mol. Cell, in press. [DOI] [PubMed] [Google Scholar]
- Papavasiliou F.N. and Schatz,D.G. (2000) Cell-cycle-regulated DNA double-stranded breaks in somatic hypermutation of immunoglobulin genes. Nature, 408, 216–221. [DOI] [PubMed] [Google Scholar]
- Peters A. and Storb,U. (1996) Somatic hypermutation of immunoglobulin genes is linked to transcription initiation. Immunity, 4, 57–65. [DOI] [PubMed] [Google Scholar]
- Poltoratsky V., Goodman,M.F. and Scharff,M.D. (2000) Error-prone candidates vie for somatic mutation. J. Exp. Med., 192, F27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad R., Dianov,G.L., Bohr,V.A. and Wilson,S.H. (2000) FEN1 stimulation of DNA polymerase β mediates an excision step in mammalian long patch base excision repair. J. Biol. Chem., 275, 4460–4466. [DOI] [PubMed] [Google Scholar]
- Rada C., Ehrenstein,M.R., Neuberger,M.S. and Milstein,C. (1998) Hot spot focusing of somatic hypermutation in MSH2-deficient mice suggests two stages of mutational targeting. Immunity, 9, 135–141. [DOI] [PubMed] [Google Scholar]
- Raha M., Wang,G., Seidman,M.M. and Glazer,P.M. (1996) Mutagenesis by third-strand-directed psoralen adducts in repair-deficient human cells: high frequency and altered spectrum in a xeroderma pigmentosum variant. Proc. Natl Acad. Sci. USA, 93, 2941–2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogozin I.B. and Kolchanov,N.A. (1992) Somatic hypermutagenesis in immunoglobulin genes. II. Influence of neighbouring base sequences on mutagenesis. Biochim. Biophys. Acta, 1171, 11–18. [DOI] [PubMed] [Google Scholar]
- Rosner K., Winter,D.B., Kasmer,C., Skovgaard,G.L., Tarone,R.E., Bohr,V.A. and Gearhart,P.J. (2001) Impact of age on hypermutation of immunoglobulin variable genes in humans. J. Clin. Immunol., 21, 102–115. [DOI] [PubMed] [Google Scholar]
- Roush A.A., Suarez,M., Friedberg,E.C., Radman,M. and Siede,W. (1998) Deletion of the Saccharomyces cerevisiae gene RAD30 encoding an Escherichia coli DinB homolog confers UV radiation sensitivity and altered mutability. Mol. Gen. Genet., 257, 686–692. [DOI] [PubMed] [Google Scholar]
- Sale J.E. and Neuberger,M.S. (1998) TdT-accessible breaks are scattered over the immunoglobulin V domain in a constitutively hypermutating B cell line. Immunity, 9, 859–869. [DOI] [PubMed] [Google Scholar]
- Smith D.S., Creadon,G., Jena,P.K., Portanova,J.P., Kotzin,B.L. and Wysocki,L.J. (1996) Di- and trinucleotide target preferences of somatic mutagenesis in normal and autoreactive B cells. J. Immunol., 156, 2642–2652. [PubMed] [Google Scholar]
- Spencer J., Dunn,M. and Dunn-Walters,D.K. (1999) Characteristics of sequences around individual nucleotide substitutions in IgVH genes suggest different GC and AT mutators. J. Immunol., 162, 6596–6601. [PubMed] [Google Scholar]
- Tissier A., Frank,E.G., McDonald,J.P., Iwai,S., Hanaoka,F. and Woodgate,R. (2000a) Misinsertion and bypass of thymine–thymine dimers by human DNA polymerase ι. EMBO J., 19, 5259–5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissier A., McDonald,J.P., Frank,E.G. and Woodgate,R. (2000b) Polι, a remarkably error-prone human DNA polymerase. Genes Dev., 14, 1642–1650. [PMC free article] [PubMed] [Google Scholar]
- Wang Y.C., Maher,V.M., Mitchell,D.L. and McCormick,J.J. (1993) Evidence from mutation spectra that the UV hypermutability of xeroderma pigmentosum variant cells reflects abnormal, error-prone replication on a template containing photoproducts. Mol. Cell. Biol., 13, 4276–4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D.B. et al. (1998) Altered spectra of hypermutation in antibodies from mice deficient for the DNA mismatch repair protein PMS2. Proc. Natl Acad. Sci. USA, 95, 6953–6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D.B., Phung,Q.H., Wood,R.D. and Gearhart,P.J. (2000) Differential expression of DNA polymerase ε in resting and activated B lymphocytes is consistent with an in vivo role in replication and not repair. Mol. Immunol., 37, 125–131. [DOI] [PubMed] [Google Scholar]
- Woodgate R. (1999) A plethora of lesion-replicating DNA polymerases. Genes Dev., 13, 2191–2195. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Yuan,F., Wu,X. and Wang,Z. (2000) Preferential incorporation of G opposite template T by the low-fidelity human DNA polymerase ι. Mol. Cell. Biol., 20, 7099–7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Yuan,F., Wu,X., Taylor,J.-S. and Wang,Z. (2001) Response of human DNA polymerase ι to DNA lesions. Nucleic Acids Res., 29, 928–935. [DOI] [PMC free article] [PubMed] [Google Scholar]