Abstract
Heme controls expression of genes involved in the synthesis of globins and heme. The mammalian transcription factor Bach1 functions as a repressor of the Maf recognition element (MARE) by forming antagonizing hetero-oligomers with the small Maf family proteins. We show here that heme binds specifically to Bach1 and regulates its DNA-binding activity. Deletion studies demonstrated that a heme-binding region of Bach1 is confined within its C-terminal region that possesses four dipeptide cysteine–proline (CP) motifs. Mutations in all of the CP motifs of Bach1 abolished its interaction with heme. The DNA-binding activity of Bach1 as a MafK hetero-oligomer was markedly inhibited by heme in gel mobility shift assays. The repressor activity of Bach1 was lost upon addition of hemin in transfected cells. These results suggest that increased levels of heme inactivate the repressor Bach1, resulting in induction of a host of genes with MAREs.
Keywords: heme/Maf/NF-E2/transcription repression
Introduction
Recent progress has highlighted the role of metabolic products as regulatory molecules in gene regulation. Heme is an essential molecule that plays a central role in reactions involving molecular oxygen, as it is the prosthetic group of heme proteins. In addition, heme is known to participate in gene regulation by functioning as a ligand for transcription factors in prokaryotes (Monson et al., 1992) and yeast (Creusot et al., 1989; Pfeifer et al., 1989; Fytlovich et al., 1993; Zhang et al., 1998; Hon et al., 2000). Heme has been proposed to fulfill similar regulatory roles in higher eukaryotes as well (Sassa and Nagai, 1996). However, to date, there is no known transcription factor of higher eukaryotes that is regulated directly or indirectly by heme.
Mitochondrial transport of the mammalian erythroid-specific δ-aminolevulinic acid synthase (ALAS-E) precursor is regulated by intracellular free heme. ALAS-E possesses several heme regulatory motifs (HRMs) within its signal sequence for mitochondrial transport whose activity is inhibited by heme (Lathrop and Timko, 1993). Within the HRM, a dipeptide motif of cysteine and proline (CP motif) appeared essential for heme-mediated regulation. HRM-like CP motifs have also been found in other proteins such as yeast transcriptional activator Hap1 (Creusot et al., 1989; Pfeifer et al., 1989; Fytlovich et al., 1993; Zhang et al., 1998; Hon et al., 2000), mitochondrial heme lyases (MHLs; Steiner et al., 1996), HRI kinase (Chen et al., 1991a,b; Crosby et al., 2000; Rafie-Kolpin et al., 2000) and heme oxygenase (HO)-2 (Rotenberg and Maines, 1991; McCoubrey et al., 1997). In the case of Hap1, heme binds directly to the protein through its several CP motifs, and then activates genes that are inducible by heme and oxygen (Creusot et al., 1989; Pfeifer et al., 1989; Fytlovich et al., 1993; Zhang et al., 1998; Hon et al., 2000).
The transcription factors Bach1 and Bach2 are basic leucine zipper (bZip) proteins that form heterodimers with the Maf-related oncoprotein family (Oyake et al., 1996). Like the erythroid transcription factor NF-E2 and its related factors Nrf1, -2 and -3 (Oyake et al., 1996; Marini et al., 1997; Johnsen et al., 1998; Kobayashi et al., 1999), their heterodimers with Maf proteins bind to the Maf recognition elements (MAREs; Igarashi et al., 1994, 1995; Kataoka et al., 1995; Motohashi et al., 1997; Toki et al., 1997). MAREs are present in the regulatory region of various genes involved in heme metabolism such as oxidative stress-responsive genes (Ishii et al., 2000), globin genes (Talbot and Grosveld, 1991; Kotkow and Orkin, 1995; Igarashi et al., 1998; Yoshida et al., 1999), the HO-1 gene (Inamdar et al., 1996; Alam et al., 1999) and the ALAS-E gene (Kramer et al., 2000). Among these MARE-binding factors, Bach1 and Bach2 function primarily as repressors (Oyake et al., 1996; Igarashi et al., 1998; Muto et al., 1998; Yoshida et al., 1999; Kobayashi et al., 2000). The presence of a repressor class of molecules suggests that transcription regulation operating through MAREs is based on the balance between activation and repression. However, the physiological role of Bach1 has remained unclear. Interestingly, Bach1 possesses several CP motifs (Oyake et al., 1996). Here we show that heme inhibits the DNA-binding activity of Bach1–MafK heterodimers by directly interacting with Bach1 through the CP motifs. These results demonstrate that Bach1 is a novel mammalian heme-responsive transcription factor that regulates gene expression by altering its DNA-binding activity in response to the intracellular free heme concentration.
Results
Heme-binding activity of Bach1
We reported previously the expression of Bach1 as a fusion protein with maltose-binding protein (MBP–Bach1) and its purification using amylose resin affinity chromatography (Oyake et al., 1996). The MBP fusion protein possessed the 453 C-terminal amino acids of Bach1 (see Figure 1). During its purification, we found that the purified protein samples were a brownish color. This color appeared specific to Bach1, since other MBP fusion proteins, e.g. Nrf2, NF-E2 p45 and MafK, were not this color. The purified MBP–Bach1 protein displayed a typical hemoprotein spectrum with Soret bands by UV-visible spectroscopy (data not shown). These observations suggest the presence of heme in the purified MBP–Bach1 protein. To explore the possibility that Bach1 is a heme-binding protein, we expressed various portions of Bach1 as fusion proteins with glutathione S-transferase (GST), and carried out various binding assays.
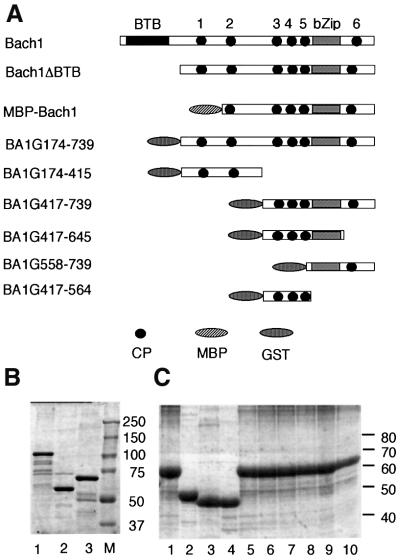
Fig. 1. Recombinant Bach1 derivatives. (A) Schematic representation of mouse Bach1, Bach1ΔBTB and fragments fused with MBP or GST. CP motifs are numbered above the first line. (B) Purified fusion proteins were analyzed by Coomassie Blue staining after SDS–PAGE. Lanes 1–3: BA1G174–739, BA1G174–415 and BA1G417–739, respectively. M, size markers. (C) Purified fusion proteins were analyzed as above. Samples were BA1G417–739 (lane 1), BA1G417–645 (lane 2), BA1G558–739 (lane 3), BA1G417–564 (lane 4), BA1G417–739CP3AP (lane 5), BA1G417–739CP4AP (lane 6), BA1G417–739CP5AP (lane 7), BA1G417–739CP6AP (lane 8), BA1G417–739CP3-5AP (lane 9) and BA1G417–739CP3-6AP (lane 10).
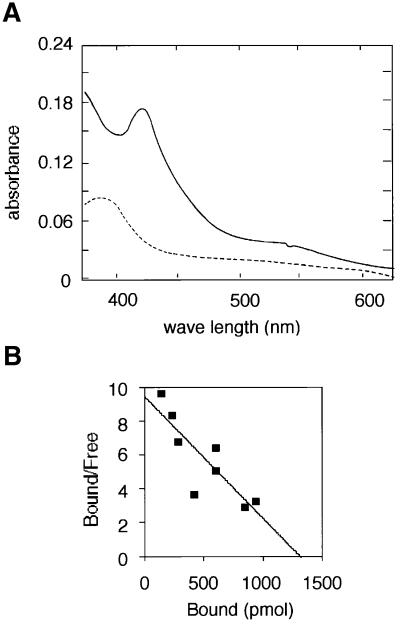
Bach1 possesses at least two functional domains, BTB/POZ and bZip domains, which are involved in protein interaction and DNA binding, respectively. To carry out a binding assay, we first expressed a portion of Bach1 (amino acids 174–739 including the putative heme-binding region but lacking the BTB/POZ domain, see Figure 1) as a fusion protein with GST. There were several other faint bands in the purified fusion protein BA1G174–739 that appeared to be degradation products of the fusion protein (Figure 1B). The BA1G174–739 preparation was found to contain an ∼0.03–0.15 molar ratio of heme, depending on the culture conditions. Upon incubating BA1G174–739 with hemin, a Soret band with a peak at 421 nm characteristic of a heme-binding protein was observed (Figure 2A). These results indicate that Bach1 is in fact a heme-binding protein. To estimate the affinity of Bach1 for heme, BA1G174–739 was immobilized on Sepharose beads and was incubated with 55Fe-labeled hemin at various concentrations. Binding of hemin to BA1G174–739 was found to occur in a concentration-dependent and saturable manner (data not shown). GST alone did not show any binding. Scatchard analysis revealed that 1.3 mol of hemin bound to 1 mol of BA1G174–739 with a Kd of 140 nM (Figure 2B).
Fig. 2. Binding of hemin to Bach1. (A) Hemin (2.7 µM) was incubated with 2.7 µM BA1G174–739 and the absorbtion spectrum was measured. The dotted line represents the absorption spectrum of hemin alone. (B) Scatchard plot analysis of heme binding to BA1G174–739 was carried out using data from several heme-binding assays. The Kd value was 140 nM using a curve-fitting program (DeltaGraph).
Identification of heme-binding motifs in Bach1
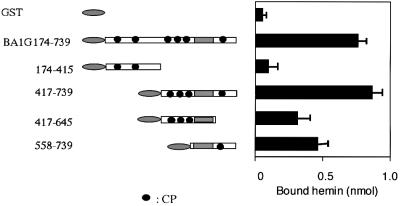
To understand the molecular basis of heme binding of Bach1, we first attempted to identify a region of Bach1 responsible for heme binding. Towards this end, we generated several GST fusions of Bach1 derivatives (Figure 3). First, two fragments (174–415 and 417–739) were tested for heme binding. The C-terminal BA1G417–739 showed a high affinity for heme that was comparable with that of BA1G174–739, whereas the N-terminal BA1G174–415 did not show any specific binding activity (Figure 3). These results indicate that a putative heme-binding domain is confined within the C-terminal region between amino acid residues 417 and 739. To define the binding region(s) more precisely, we prepared two subfragments from the heme-binding fragment (417–739). To our surprise, both fragments (417–645 and 558–739) showed comparable heme-binding activities.
Fig. 3. Binding of hemin to Bach1 subfragments. GST–Bach1 fusions (1 nmol) were immobilized onto Sepharose beads and incubated with 1 µM hemin. The amounts of bound hemin were determined using spectrofluorometry. Each data bar represents the average and SEM of several independent experiments.
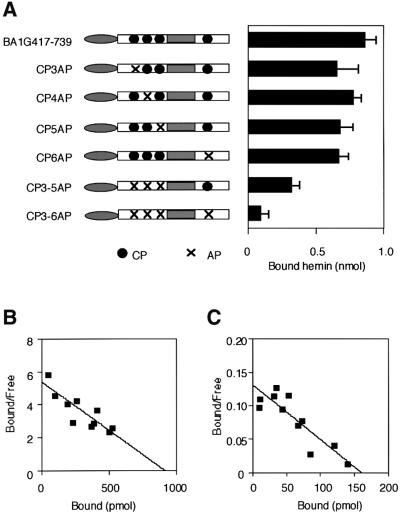
CP motifs have been found in HRMs (Creusot et al., 1989; Chen et al., 1991a; Rotenberg and Maines, 1991; Lathrop and Timko, 1993; Steiner et al., 1996; McCoubrey et al., 1997) and play an important role in heme binding in various proteins. Interestingly, Bach1 contains six CP motifs (Figure 1). There are two CP motifs downstream of the BTB domain (CP1 and CP2), three CP motifs upstream of the bZip domain (CP3, CP4 and CP5) and one CP motif downstream of the bZip domain (CP6; Figures 1 and 3). Among these CP motifs, CP3–CP6 are present within the putative heme-binding region. To test the possibility that these CP motifs are involved in heme binding, we prepared GST fusion proteins with single or multiple mutations within the CP motifs in the context of BA1G417–739 by changing a cysteine residue to alanine (Figure 4A). Single amino acid substitutions (BA1G417–739CP3AP, BA1G417–739CP4AP, BA1G417–739CP5AP and BA1G417–739CP6AP) did not influence the heme binding. In contrast, alteration of the three clustered CPs (BA1G417–739CP3–5AP) significantly decreased the heme-binding activity. Most importantly, replacement of all four CPs (CP3–CP6, BA1G417–739CP3–6AP) abolished the high affinity binding to heme. These results indicate that Bach1 binding to heme is dependent on the presence of the CP motifs, and rule out the possibility that an impurity in the preparations binds to heme. These results also indicate that there is a cooperativity among the four CP motifs of Bach1 for heme binding. Kd values were 170 and 1220 nM for BA1G417–739 and BA1G417–739CP3-6AP, respectively (Figure 4B and C).
Fig. 4. Effects of cysteine to alanine substitutions of CP motifs of Bach1 on heme binding. (A) Heme binding assays were carried out as in Figure 3 using GST fusion proteins with single or multiple mutation(s) within the CP motifs in the context of BA1G417–739. Results are the mean ± SEM of several independent experiments. (B and C) Scatchard plot analyses of BA1G417–739 (B) and BA1G417–739CP3–6AP (C). The Kd values were 170 nM (BA1G417–739) and 1220 nM (BA1G417–739CP3-6AP).
Inhibition of DNA-binding activity of Bach1 by heme
The specific binding of heme to Bach1 strongly suggests that heme regulates certain functions of Bach1. Since the four CP motifs (CP3–CP6) that are involved in heme binding surround the bZip domain (see Figure 1), we examined the effects of hemin on DNA binding using electrophoretic mobility shift assays (EMSAs). In this experiment, we first used full-length recombinant Bach1 (Igarashi et al., 1998) that was overexpressed in Escherichia coli, purified from inclusion bodies and re-folded in vitro. This procedure permitted us to obtain recombinant protein that is free of heme. A MARE-containing 240 bp DNA isolated from the β-globin locus control region (LCR) was incubated with Bach1 and MafK, and the resulting DNA-binding complexes were analyzed by EMSA using agarose gels (Figure 5A). Two retarded bands were detected; the upper band contained both Bach1 and MafK whereas the lower band contained only MafK (data not shown; Igarashi et al., 1998). Addition of heme to the binding reactions resulted in a concentration-dependent inhibition of DNA binding by the Bach1–MafK hetero-oligomer. Even 0.03 µM heme caused a slight but reproducible inhibition. Heme at 1 µM almost completely inhibited the DNA-binding activity of Bach1. In contrast, there was little effect of heme on DNA binding by the MafK homodimer. To exclude the possibility that heme inhibited DNA binding by BTB domain-mediated oligomers, full-length Bach1 and Bach1ΔBTB (with amino acid residues 1–173 deleted, see Figure 1A) were compared in EMSAs using conventional polyacrylamide gels (Figure 5B). Addition of heme efficiently inhibited the DNA-binding activity of Bach1ΔBTB–MafK heterodimers. These results suggest that heme’s effect is independent of BTB domain-mediated oligomer formation (Igarashi et al., 1998). Nrf2 is distantly related to Bach1 and binds to similar DNA sequences as a heterodimer with Maf-related proteins. In contrast to Bach1, however, Nrf2 functions as a transcription activator. Therefore, we compared the role of Bach1 and Nrf2 with respect to heme. The DNA-binding activity of Nrf2–MafK heterodimers was not inhibited by heme (Figure 5C), suggesting that Bach1 is a specific regulatory target of heme among the MARE-binding proteins.
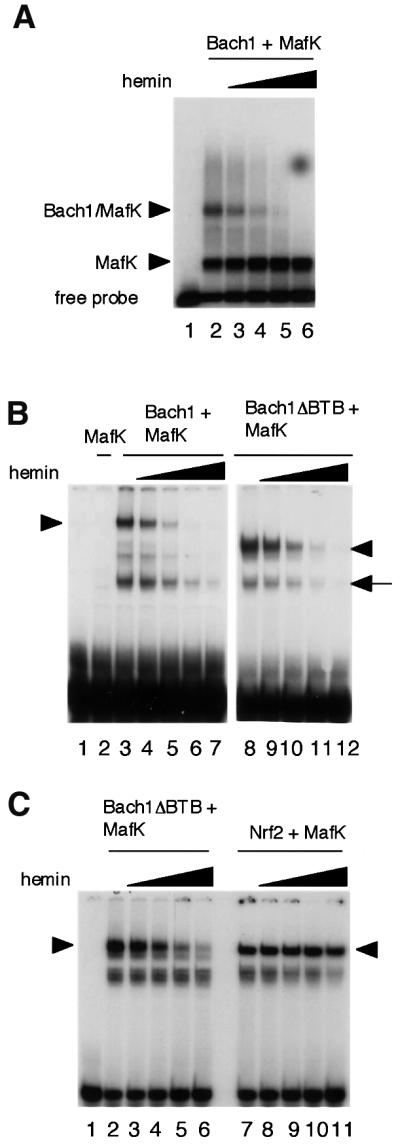
Fig. 5. Inhibition of DNA binding of Bach1 by heme. (A) EMSA was carried out using 20 ng of each recombinant Bach1 and MafK and a 240 bp enhancer fragment from the β-globin LCR containing two tandem MAREs as described previously (Igarashi et al., 1998). Complexes were separated on a 1.0% agarose gel. Heme concentrations were 0.03 µM (lane 3), 0.1 µM (lane 4), 0.3 µM (lane 5) and 1 µM (lane 6). (B) EMSA was carried out using a CβE oligonucleotide probe as described previously (Igarashi et al., 1998) using 40 ng of Bach1 or Bach1ΔBTB and 10 ng of MafK in the combinations indicated above the panels. Heme concentrations were 0.1 µM (lanes 4 and 9), 0.3 µM (lanes 5 and 10), 0.6 µM (lanes 6 and 11) and 1 µM (lanes 7 and 12). Complexes were separated through 4% polyacrylamide gels. Arrow heads indicate specific heterodimer binding complexes. The arrow indicates binding complexes that may be due to degradation products. (C) EMSA was carried out as in (B) using 40 ng of Bach1ΔBTB (lanes 2–6) or Nrf2 (lanes 7–11). Heme concentrations were 0.1 µM (lanes 3 and 8), 0.3 µM (lanes 4 and 9), 0.6 µM (lanes 5 and 10) and 1 µM (lanes 6 and 11).
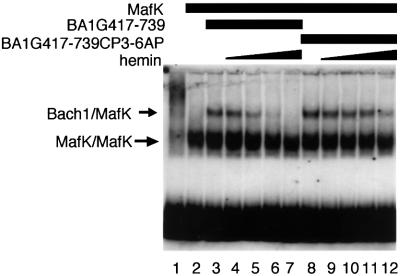
We next investigated whether the CP motifs that are essential for heme binding may also be involved in the inhibition of DNA binding by heme. We used the GST–Bach1 fusion that harbored cysteine to alanine substitutions in the four CP motifs (BA1G417–739CP3– 6AP, Figure 6). Comparison of the amounts of DNA-binding complex generated in the presence of various concentrations of heme clearly indicated that the CP3–6AP substitutions rendered Bach1 less sensitive to heme. On the other hand, in contrast to its essential lack of heme binding (see above), the CP3–6AP form still retained some sensitivity toward heme. It should be noted that Nrf2 was insensitive toward heme-mediated inhibition (see Figure 5). Taken together, these results indicate that the four CP motifs are important for both Bach1–heme interaction and heme-mediated inhibition of DNA binding. In addition, the results also suggest that other features of Bach1 may also contribute to its heme sensitivity.
Fig. 6. Involvement of the CP motifs of Bach1 in regulation of its DNA-binding activity by heme. BA1G417–739 or BA1G417–739CP3-6AP and MafK were incubated in the absence or presence of various concentrations of hemin. Binding to the CβE probe was examined by EMSA. Heme concentrations were 0.1 µM (lanes 4 and 9), 0.3 µM (lanes 5 and 10), 0.5 µM (lanes 6 and 11) and 1.0 µM (lanes 7 and 12).
Effect of heme on Bach1–MafK interaction
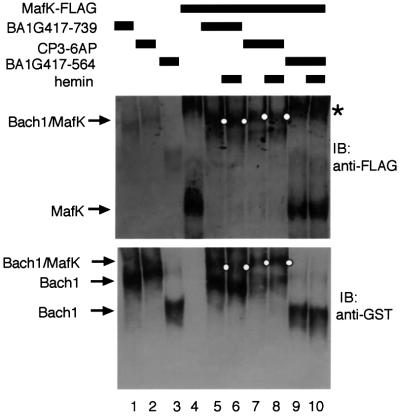
Heme may inhibit DNA binding of Bach1 by influencing heterodimer formation with MafK through the leucine zippers since Bach1 cannot bind to DNA by itself (Oyake et al., 1996; Igarashi et al., 1998). To test this possibility, we investigated the effect of heme on the leucine zipper-mediated Bach1–MafK interaction. FLAG-tagged MafK was incubated with or without GST-fused Bach1 (BA1G417–739) and separated through non-denaturing polyacrylamide gels. Proteins were transferred onto membranes and were detected using anti-FLAG antibody (Figure 7, upper panel). In the absence of BA1G417–739, a single high mobility band was observed that appeared to be a monomer or homodimer of MafK (lane 4). Another slower mobility band, shown with an asterisk, may be due to conformational differences in native gels or formation of a MafK homo-oligomer. In the presence of BA1G417–739, both of the complexes disappeared with the concomitant appearance of the third distinct band (lane 5, indicated with a dot). This band was judged to reflect leucine zipper-mediated formation of a Bach1–MafK complex since addition of BA1G417–564 lacking the bZip domain did not change the mobility of MafK complexes (compare lanes 4, 5 and 9). On the other hand, similar changes in mobility of MafK complexes were also observed by adding BA1G417–739CP3–6AP (lane 7). To confirm further the formation of a Bach1–MafK complex, the same blot was reacted with anti-GST antibody to detect Bach1 proteins (lower panel). Single bands were observed when Bach1 proteins were electrophoresed alone (lanes 1–3). In the presence of MafK, slower mobility bands were detected with BA1G417–739 or BA1G417–739CP3–6AP (lanes 5 and 7, indicated with dots). In contrast, the mobility of BA1G417–564 did not change by adding MafK (compare lanes 3 and 9). Using this experimental system, we examined the effects of heme. The amounts of slower mobility complexes in the presence of BA1G417–739 or BA1G417–739CP3–6AP did not change in either the absence or presence of 1 µM heme (compare lanes 5–8). We thus concluded that heme did not inhibit Bach1–MafK interaction through the leucine zippers, but that rather it inhibited the DNA-binding activity of the resulting heterodimer.
Fig. 7. Effects of heme on formation of Bach1–MafK heterodimers. FLAG-tagged MafK and GST–Bach1 fusion proteins (BA1G417–739, BA1G417–739CP3–6AP or BA1G417–564) were incubated in the presence or absence of 1 µM heme. The resulting complexes were resolved in native polyacrylamide gels, transferred onto membranes and detected with anti-FLAG (upper panel) and anti-GST (lower panel) antibodies. Bach1–MafK complexes are indicated with small dots along the lanes. Asterisks indicate the slower mobility complex of MafK.
Heme abrogates transcription repression by Bach1 in vivo
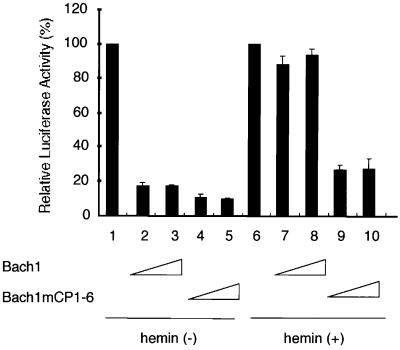
To probe the functional consequences of the interaction between heme and Bach1 within a cell, we compared the regulatory activities of Bach1 and a Bach1 derivative with multiple CP mutations. Bach1 mCP1–6 harbors replacement of a cysteine by an alanine in all six CP motifs, i.e. CP1–CP6. Each of the expression plasmids was transfected into 293 cells along with a synthetic MARE-dependent reporter plasmid, and their effects on reporter gene expression in the presence or absence of 10 µM hemin in the medium were compared (Figure 8). The amount of hemin is similar to that utilized in experiments examining heme-mediated gene induction in cultured cells (Alam et al., 1999). Without addition of hemin, both Bach1 and Bach1 mCP1–6 repressed expression of the reporter gene. Bach1 mCP1–6 appeared to show more efficient repression than the wild-type (compare columns 2–5). When hemin was added to the medium, repression by Bach1 was markedly abrogated (columns 6–8). In contrast, Bach1 mCP1–6 remained effective in transcription repression even in the presence of hemin (columns 9 and 10). Taken together with the fact that the CP motifs play an important role in Bach1–heme interaction, these results indicated that the repressor activity of Bach1 within a cell is negatively regulated by heme.
Fig. 8. Effects of heme on the transcriptional activity of Bach1. Reporter gene assays were carried out using 293 cells to compare repressor activity of the wild-type Bach1 (lanes 2, 3, 7 and 8) or Bach1 with mutations in the CP motifs (Bach1 mCP1–6, lanes 4, 5, 9 and 10) in the absence (lanes 1–5) or presence (lanes 6–10) of 10 µM hemin. Either 300 or 900 ng of the Bach1 expression plasmids were used. Reporter gene expression in the absence of any effector plasmid was set to 100% for each experimental group.
Discussion
Heme plays various critical roles within cells because of its ability to bind molecular oxygen and to mediate electron transfer. Besides these functions, heme is known to induce expression of ALAS-E (Fujita et al., 1991), globin (Fukuda et al., 1994; Harigae et al., 1998) and HO-1 genes (Yoshida et al., 1988; Shibahara et al., 1993) while it decreases the mRNA of non-specific ALAS (ALAS-N) (Yamamoto et al., 1988; Fujita et al., 1991). These proteins are involved in metabolism of heme or hemoglobin synthesis, and proper control of their expression is important for survival under various conditions. These observations led to the conjecture that heme plays a role in gene regulation (Sassa and Nagai, 1996). While this hypothesis predicts the presence of a heme-regulated transcription factor, the yeast transcription factor Hap1 has been the only known example in eukaryotes to date (Creusot et al., 1989; Pfeifer et al., 1989; Fytlovich et al., 1993; Zhang et al., 1998; Hon et al., 2000), with no other factors with such function known in higher eukaryotes. We report here the biochemical characterization of the heme–Bach1 interaction. Most importantly, DNA-binding and repressor activities of Bach1 are negatively regulated by heme. These results suggest that heme may regulate gene expression through shifting the balance between activators and repressors that bind to MAREs.
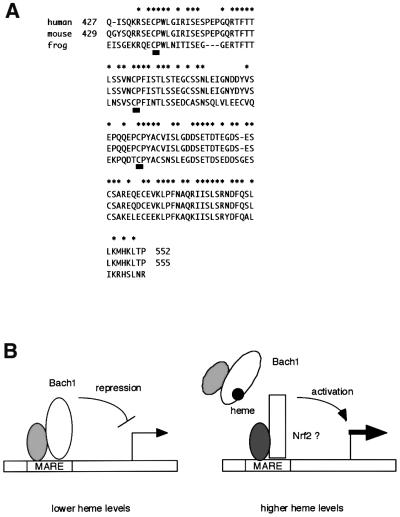
Despite the fact that Bach1 is not related to Hap1, there is a resemblance between Hap1 and Bach1 in that they interact with heme through their CP motifs, i.e. both proteins bind to heme with high affinity via their CP motifs. However, no single CP motif of Bach1 is essential for heme binding, indicating their functional redundancy. Consistent with this notion, a single CP motif on Hap1 is known to bind to heme when tested using a synthetic peptide (Zhang and Guarente, 1995). Thus, it is reasonable to assume that each CP motif on Bach1 possesses the potential to interact with heme. This may explain the heme-binding activity of the C-terminal fragment containing one CP motif (BA1G417–739CP3–5AP and BA1G558–739). Paradoxically, the results of Scatchard plot analyses indicated that Bach1 possessed roughly one heme-binding site. While each of the CP motifs may be capable of binding to heme, binding of one molecule of heme to Bach1 is likely to preclude further heme binding in the context of Bach1. Evolutionary comparison also underscores the importance of the Bach1 CP motifs. The amino acid sequences around CP3–CP5 are almost identical between mouse (Oyake et al., 1996) and human (Blouin et al., 1998; Ohira et al., 1998). Furthermore, this domain is also well conserved in Xenopus Bach1 (DDBJ/EMBL/GenBank accession No. AW147501, see Figure 9A). The clustering of the CP motifs are also conserved: there are 24 and 29 animo acid spacers between CP3, CP4 and CP5, respectively. Together with CP6, they form a CP cluster surrounding the bZip domain. Binding of hemin may induce structural changes within or around the bZip domain that inhibit DNA binding but not the leucine zipper interaction. Clustered CP motifs are also found in mammalian ALAS-E precursor (Lathrop and Timko, 1993). Thus, the clustering of CP motifs may be important for high-affinity heme binding by increasing the local concentration of binding sites.
Fig. 9. Transcription regulation by heme through Bach1. (A) Sequence alignment of human, mouse and Xenopus Bach1. Amino acid sequences surrounding the three clustered CP motifs (CP3–CP5, underlined) of human, mouse and Xenopus (deduced from AW147501) Bach1 are compared. Asterisks indicate residues that are conserved among the three species. (B) A model based on our observations. In the presence of lower concentrations of heme within a cell, Bach1 represses genes with MAREs together with small Maf proteins. In the presence of higher concentrations of heme, Bach1 is inactivated by binding to heme, leaving MAREs available for the activator class of factors such as Nrf2 or other MARE-binding proteins. In this way, heme directly regulates the balance of repression and activation of gene expression.
There is still a remaining question of whether CP3–6 and their clustering are the sole determinant for heme binding. This question stems from the observation that hemin exerts a regulatory effect on Bach1 in vitro at relatively lower concentrations as compared with other mammalian heme-regulated proteins. For example, hemin is known to inhibit mitochondrial transport of ALAS-E precursor at 5–25 µM (Lathrop and Timko, 1993). It should also be noted that there are two other CP motifs, CP1 and CP2, on Bach1. Even though BA1G417–739 and BA1G174–739 showed similar heme-binding activity and BA1G174–415 showed no detectable heme binding, the possibility that the N-terminal region of Bach1 also contributes to heme–Bach1 interaction cannot be excluded. Another observation that cannot be explained fully at present is that BA1G417–739CP3-6AP, with the four CP–AP substitutions, still showed reduced but reproducible sensitivity to heme in the DNA-binding assays. Because Nrf2 was insensitive to heme, it is likely that there may be an additional mechanism for heme–Bach1 interaction that eluded our analyses. In this respect, it should be noted that the CP motifs of heme lyase are suggested to be involved in increasing the local concentration of heme, providing there is a sufficient substrate concentration at the catalytic center (Steiner et al., 1996). Thus, it remains possible that Bach1 possesses another heme-binding site(s) that mediates a weak and/or transient protein–heme interaction, and we are currently testing this possibility.
Comparison of various heme-related molecules revealed that hemin and Fe mesoporphyrin IX (FeMePIX) bound to Bach1 and inhibited its DNA binding, whereas porphyrin derivatives without Fe at the center, including protoporphyrin IX (PPIX) and MePIX, did not (data not shown). Furthermore, other derivatives such as octaethylporphine did not show Bach1 binding or inhibitory effects on DNA binding (data not shown). These results suggest that only heme and porphyrin derivatives with chelated iron can bind to Bach1 specifically. Since FeMPIX, which does not have vinyl groups, bound to Bach1, the binding is likely to be non-covalent, like the iron–imidazole interaction at the histidyl site, unlike the covalent binding of cytochrome c to heme via two cysteine residues.
Based on our observation and previous reports, we propose a hypothetical mechanism by which heme functions as an inducer of gene expression by inhibiting the repressor molecule Bach1 (Figure 9B). In this model, Bach1 represses genes with MAREs in their regulatory regions in the presence of lower levels of heme by competing with activators such as Nrf2 (Alam et al., 1999; Ishii et al., 2000) and NF-E2 p45 (Igarashi et al., 1998) for binding to MAREs. Higher levels of heme cause inhibition of the DNA-binding activity of Bach1, resulting in dissociation of Bach1 from the enhancers and derepression of the target genes. The next step will be identification of physiological target genes for Bach1. Bach1 targets will be found among genes that possess MAREs in their regulatory regions and that are induced by heme. It may be worth noting that the LCR of the β-globin genes (Talbot and Grosveld, 1991; Kotkow and Orkin, 1995; Igarashi et al., 1998; Yoshida et al., 1999) and the heme-responsive enhancers of HO-1 (Inamdar et al., 1996; Alam et al., 1999) contain multiple MAREs. Since Bach1 forms mutlivalent DNA-binding complexes through its BTB domain that can bind cooperatively to reiterated MARE motifs (Yoshida et al., 1999), β-globin and HO-1 enhancers may allow effective binding by Bach1. Together with the fact that these genes are directly or indirectly regulated by heme (Shibahara et al., 1993; Fukuda et al., 1994; Harigae et al., 1998; Yoshida et al., 1988), β-globin and HO-1 genes are prime candidates. Further studies obviously are necessary to test this hypothesis. Nevertheless, our observations provide, for the first time, a molecular mechanism for the long-standing contention that intracellular free heme regulates gene expression in mammalian cells.
Materials and methods
Recombinant proteins
FLAG-tagged MafK, His-tagged Bach1, and His-tagged Bach1ΔBTB, a Bach1 variant lacking the N-terminal 173 amino acids including the BTB domain, were prepared as described previously (Igarashi et al., 1998). To construct GST fusion protein expression plasmids, portions of mouse Bach1 cDNA were amplified by PCR and the resulting fragments were ligated with pGEX-2T vector (Amersham Pharmacia Biotech AB, Uppsala, Sweden). The plasmids thus constructed were pGEX2/Ba1G693–2393, pGEX2/Ba1G693–1418, pGEX2/Ba1G1422–1865, pGEX2/Ba1G1422–2108, pGEX2/Ba1G1422–2393 and pGEX2/Ba1G1845–2393 (the numbers indicate the base positions of mouse bach1 cDNA; Oyake et al., 1996). These plasmids encoded Bach1 subfragments fused with GST (BA1G174–739, BA1G174–415, BA1G417–564, BA1G417–645, BA1G417–739 and BA1G558–739, respectively; the numbers indicate the amino acid positions of mouse Bach1 protein). Cysteine to alanine substitutions in the CP motifs were introduced into BA1G417–739 cDNA using the Transformer™ Site-Directed Mutagenesis Kit (Clontech, Palo Alto, CA) to express BA1G417–739CP3AP, BA1G417–739CP4AP, BA1G417–739CP5AP and BA1G417–739CP6AP, bearing mutations at amino acids 438, 464, 495 and 649, respectively. These proteins were expressed in E.coli (strains SG12036 or BL21) either at 37°C for 2 h or at 25°C for 12 h, followed by purification with glutathione–Sepharose 4B (Amersham Pharmacia Biotech AB, Uppsala, Sweden) according to the manufacturer’s protocols. Purified GST fusion proteins were eluted from glutathione–Sepharose beads with 10 mM glutathione, 50 mM Tris–HCl pH 8.0 and dialyzed with Slide-A-Lyzer 10K Dialysis Cassettes (Pierce) in 10 mM Tris–HCl pH 8.0 containing 0.1 mM EDTA, 0.1 mM dithiothiothreitol (DTT) and 4% glycerol. The purity and integrity of the prepared proteins were checked by SDS–PAGE.
Spectral analysis
Hemin (2.7 µM) was incubated with or without 2.7 µM BA1G174–739 in 10 mM Tris–HCl pH 8.0, 150 mM NaCl (Tris-buffered saline, TBS). The resulting mixtures were scanned using a UV-1200 UV-VIS spectrophotometer (Shimidazu, Kyoto, Japan).
Hemin-binding assays
[55Fe]Hemin (5.5 × 104 d.p.m./nmol) was synthesized from PPIX and 55Fe (>3 Ci/g, NEN Life Science Products) by the non-enzymatic method (Taketani and Tokunaga, 1984). GST fusion proteins (1 nmol) immobilized on glutathione–Sepharose beads were suspended in 1 ml of TBS–0.1% Tween-20 (TBS-T). [55Fe]hemin dissolved in dimethysulfoxide (DMSO) was added to the mixture and incubated at room temperature for 30 min. After washing four times with 500 µl of TBS-T, the radioactivity of the beads was counted. Alternatively, non-labeled porphyrin derivatives including hemin were used as ligands in the same binding conditions described above. In this case, after washing, beads were suspended in 2 ml of 2 M oxalic acid and heated at 110°C for 10 min to remove the metal from metalloporphyrins. The amounts of bound porphyrins were determined from the peak values of fluorescence emission scanning 550–700 nm with excitation at 400 nm.
Electrophoretic mobility shift assay (EMSA)
Chicken β-globin enhancer oligonucleotide (CβE; Igarashi et al., 1994) or 240 bp DNA from the β-globin LCR containing the tandem MARE (HS-2; Igarashi et al., 1998) was labeled with 32P using polynucleotidyl kinase. Alternatively, DNA was labeled with digoxigenin (DIG) using the DIG Oligonucleotide 3′-End Labeling Kit (Boehringer Mannheim). A 40 ng aliquot of GST fusion or His-tagged Bach1 proteins (BA1G417–739, BA1G417–739CP3-6AP, Bach1 or Bach1ΔBTB) and 10 ng of MafK-FLAG were incubated on ice for 20 min in 10 or 20 µl of gel shift buffer containing 20 mM HEPES pH 7.9, 0.05 µg/µl dIdC, 0.05 µg/µl dAdT, 0.1 µg/µl bovine serum albumin (BSA), 5 mM DTT, 5 mM MgCl2, 1 mM EDTA, 20 mM KCl and 4% glycerol. After addition of hemin or porphyrin derivatives, the reaction mixtures were left on ice for 10 min. Labeled CβE or HS-2 was then added to the mixture and incubated at 37°C for 10 min. The resulting solutions were separated through 4% polyacrylamide gels or 1% agarose gels. When 32P-labeled probe was used, gels were dried and exposed to X-ray films. When DIG-labeled probe was used, protein–DNA complexes were transferred onto nylon membranes (Boehringer Mannheim) and were detected using the DIG Luminescent Detection Kit (Boehringer Mannheim).
Native gel western blotting
BA1G417–739 (90 ng), BA1G417–739CP3-6AP (90 ng) or BA1G417–563 (60 ng) were incubated with MafK-FLAG (15 ng) in 20 µl of the gel shift buffer containing 0.2 µg/µl BSA and left at room temperature for 30 min. After adding hemin to 1 µM, the reaction mixtures were incubated further at room temperature for 10 min, and were then separated through 6% polyacrylamide gels. Proteins were transferred onto Immobilon-P membrane (Millipore). Membranes were reacted with anti-FLAG M2 monoclonal antibody (Sigma) and anti-mouse IgG–horseradish peroxidase (HRP) conjugate (Promega) to detect MafK-FLAG or with anti-GST antibody B-14 (Santa Cruz Biotechnology) to detect GST–Bach1 fusion proteins. Immune complexes were detected using ECL Western Blotting Detection Reagents (Amersham Pharmacia Biotech).
Transfection reporter assays
The synthetic reporter plasmid pRBGP2 was reported previously (Igarashi et al., 1994) and contained three copies of MARE in front of a minimal TATA box promoter and the firefly luciferase gene. Bach1 mCP1–6 cDNA, containing multiple mutations that replace all of the CP motifs (CP1–CP6) by AP, was constructed by two-step site-directed mutagenesis using the Transformer™ Site-Directed Mutagenesis Kit (Clontech). First, CP3–CP6 were mutagenized simultaneously. After verifying the mutations, CP1 and CP2 were then converted simultaneously to AP. To express wild-type mouse Bach1 cDNA or Bach1 mCP1–6, each cDNA was cloned into pcDNA3.1/Myc-His A vector (Invitrogen). 293 cells were seeded in 6-well dishes 24 h before transfection. The cells were transfected with 500 ng of the reporter and 300 or 900 ng of the effector plasmids using FuGENE 6 transfection reagent (Roche Diagnostic, Corp). After incubating cells with the plasmids for 18 h, hemin was added to the medium to 10 µM by supplying 2 µl of 10 mM hemin dissolved in DMSO. As a control, 2 µl of DMSO were added to the wells that did not receive hemin. After further incubation for 4 h, cell lysates were prepared using the Luciferase Assay System (Promega) following the supplier’s protocol. Luciferase activities were measured with a Biolumat Luminometer (Berthold). Firefly luciferase activity was normalized for transfection efficiency as determined by a control sea pansy luciferase activity (Muto et al., 1998). Three independent experiments, each carried out in duplicate, were performed, and the results were averaged and diagrammed with the standard errors.
Acknowledgments
Acknowledgements
We thank Makoto Kobayashi and Tetsuro Ishii (University of Tsukuba) for continuing and stimulating discussions and critical comments on the manuscript. This work was supported by Grants-in-aid from the Ministry of Education, Science, Sport and Culture, the Ministry of Health and Welfare, Japan and an RFTF grant from the Japanese Society for the Promotion of Science, as well as grants from the Uehara Memorial Foundation, the Inamori Foundation, Yamanouchi Foundation for Research on Metabolic Disorders, Japan Foundation of Cardiovascular Research and a USPHS grant (DK-32890).
References
- Alam J., Stewart,D., Touchard,C., Boinapally,S., Choi,A.M. and Cook,J.L. (1999) Nrf2, a Cap’n’collar transcription factor, regulates induction of the heme oxygenase-1 gene. J. Biol. Chem., 274, 26071–26078. [DOI] [PubMed] [Google Scholar]
- Blouin J.L., Duriaux Sail,G., Guipponi,M., Rossier,C., Pappasavas,M.P. and Antonarakis,S.E. (1998) Isolation of the human BACH1 transcription regulator gene, which maps to chromosome 21q22.1. Hum. Genet., 102, 282–288. [DOI] [PubMed] [Google Scholar]
- Chen J.J., Pal,J.K., Petryshyn,R., Kuo,I., Yang,J.M., Throop,M.S., Gehrke,L. and London,I.M. (1991a) Amino acid microsequencing of internal tryptic peptides of heme-regulated eukaryotic initiation factor 2α subunit kinase: homology to protein kinases. Proc. Natl Acad. Sci. USA, 88, 315–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.J., Throop,M.S., Gehrke,L., Kuo,I., Pal,J.K., Brodsky,M. and London,I.M. (1991b) Cloning of the cDNA of the heme-regulated eukaryotic initiation factor 2α (eIF-2α) kinase of rabbit reticulocytes: homology to yeast GCN2 protein kinase and human double-stranded-RNA-dependent eIF-2α kinase. Proc. Natl Acad. Sci. USA, 88, 7729–7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creusot F., Verdiere,J., Gaisne,M. and Slonimski,P.P. (1989) CYP1 (HAP1) regulator of oxygen-dependent gene expression in yeast. I. Overall organization of the protein sequence displays several novel structural domains. J. Mol. Biol., 204, 263–276. [DOI] [PubMed] [Google Scholar]
- Crosby J.S., Chefalo,P.J., Yeh,I., Ying,S., London,I.M., Leboulch,P. and Chen,J.J. (2000) Regulation of hemoglobin synthesis and proliferation of differentiating erythroid cells by heme-regulated eIF-2α kinase. Blood, 96, 3241–3248. [PubMed] [Google Scholar]
- Fujita H., Yamamoto,M., Yamagami,T., Hayashi,N. and Sassa,S. (1991) Erythroleukemia differentiation. Distinctive responses of the erythroid-specific and the nonspecific δ-aminolevulinate synthase mRNA. J. Biol. Chem., 266, 17494–17502. [PubMed] [Google Scholar]
- Fukuda Y., Fujita,H., Garbaczewski,L. and Sassa,S. (1994) Regulation of β-globin mRNA accumulation by heme in dimethyl sulfoxide (DMSO)-sensitive and DMSO-resistant murine erythroleukemia cells. Blood, 83, 1662–1667. [PubMed] [Google Scholar]
- Fytlovich S., Gervais,M., Agrimonti,C. and Guiard,B. (1993) Evidence for an interaction between the CYP1(HAP1) activator and a cellular factor during heme-dependent transcriptional regulation in the yeast Saccharomyces cerevisiae. EMBO J., 12, 1209–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harigae H., Suwabe,N., Weinstock,P.H., Nagai,M., Fujita,H., Yamamoto,M. and Sassa,S. (1998) Deficient heme and globin synthesis in embryonic stem cells lacking the erythroid-specific δ-aminolevulinate synthase gene. Blood, 91, 798–805. [PubMed] [Google Scholar]
- Hon T., Hach,A., Lee,H.C., Cheng,T. and Zhang,L. (2000) Functional analysis of heme regulatory elements of the transcriptional activator Hap1. Biochem. Biophys. Res. Commun., 273, 584–591. [DOI] [PubMed] [Google Scholar]
- Igarashi K., Kataoka,K., Itoh,K., Hayashi,N., Nishizawa,M. and Yamamoto,M. (1994) Regulation of transcription by dimerization of erythroid factor NF-E2 p45 with small Maf proteins. Nature, 367, 568–572. [DOI] [PubMed] [Google Scholar]
- Igarashi K., Itoh,K., Motohashi,H., Hayashi,N., Matuzaki,Y., Nakauchi,H., Nishizawa,M. and Yamamoto,M. (1995) Activity and expression of murine small Maf family protein MafK. J. Biol. Chem., 270, 7615–7624. [DOI] [PubMed] [Google Scholar]
- Igarashi K., Hoshino,H., Muto,A., Suwabe,N., Nishikawa,S., Nakauchi,H. and Yamamoto,M. (1998) Multivalent DNA binding complex generated by small Maf and Bach1 as a possible biochemical basis for β-globin locus control region complex. J. Biol. Chem., 273, 11783–11790. [DOI] [PubMed] [Google Scholar]
- Inamdar N.M., Ahn,Y.I. and Alam,J. (1996) The heme-responsive element of the mouse heme oxygenase-1 gene is an extended AP-1 binding site that resembles the recognition sequences for MAF and NF-E2 transcription factors. Biochem. Biophys. Res. Commun., 221, 570–576. [DOI] [PubMed] [Google Scholar]
- Ishii T., Itoh,K., Takahashi,S., Sato,H., Yanagawa,T., Katoh,Y., Bannai,S. and Yamamoto,M. (2000) Transcription factor Nrf2 coordinately regulates a group of oxidative stress-inducible genes in macrophages. J. Biol. Chem., 275, 16023–16029. [DOI] [PubMed] [Google Scholar]
- Johnsen O., Murphy,P., Prydz,H. and Kolsto,A.B. (1998) Interaction of the CNC-bZIP factor TCF11/LCR-F1/Nrf1 with MafG: binding-site selection and regulation of transcription. Nucleic Acids Res., 26, 512–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka K., Igarashi,K., Itoh,K., Fujiwara,K.T., Noda,M., Yamamoto,M. and Nishizawa,M. (1995) Small Maf proteins heterodimerize with Fos and may act as competitive repressors of the NF-E2 transcription factor. Mol. Cell. Biol., 15, 2180–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi A., Ito,E., Toki,T., Kogame,K., Takahashi,S., Igarashi,K., Hayashi,N. and Yamamoto,M. (1999) Molecular cloning and functional characterization of a new cap’n’collar family transcription factor Nrf3. J. Biol. Chem., 274, 6443–6452. [DOI] [PubMed] [Google Scholar]
- Kobayashi A., Yamagiwa,H., Hoshino,H., Muto,A., Sato,K., Morita,M., Hayashi,N., Yamamoto,M. and Igarashi,K. (2000) A combinatorial code for gene expression generated by transcription factor Bach2 and MAZR (MAZ-related factor) through the BTB/POZ domain. Mol. Cell. Biol., 20, 1733–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotkow K.J. and Orkin,S.H. (1995) Dependence of globin gene expression in mouse erythroleukemia cells on the NF-E2 heterodimer. Mol. Cell. Biol., 15, 4640–4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer M.F., Gunaratne,P. and Ferreira,G.C. (2000) Transcriptional regulation of the murine erythroid-specific 5-aminolevulinate synthase gene. Gene, 247, 153–166. [DOI] [PubMed] [Google Scholar]
- Lathrop J.T. and Timko,M.P. (1993) Regulation by heme of mitochondrial protein transport through a conserved amino acid motif. Science, 259, 522–525. [DOI] [PubMed] [Google Scholar]
- Marini M.G., Chan,K., Casula,L., Kan,Y.W., Cao,A. and Moi,P. (1997) hMAF, a small human transcription factor that heterodimerizes specifically with Nrf1 and Nrf2. J. Biol. Chem., 272, 16490–16497. [DOI] [PubMed] [Google Scholar]
- McCoubrey W.K. Jr, Huang,T.J. and Maines,M.D. (1997) Heme oxygenase-2 is a hemoprotein and binds heme through heme regulatory motifs that are not involved in heme catalysis. J. Biol. Chem., 272, 12568–12574. [DOI] [PubMed] [Google Scholar]
- Monson E.K., Weinstein,M., Ditta,G.S. and Helinski,D.R. (1992) The FixL protein of Rhizobium meliloti can be separated into a heme-binding oxygen-sensing domain and a functional C-terminal kinase domain. Proc. Natl Acad. Sci. USA, 89, 4280–4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi H., Shavit,J.A., Igarashi,K., Yamamoto,M. and Engel,J.D. (1997) The world according to Maf. Nucleic Acids Res., 25, 2953–2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto A. et al. (1998) Identification of Bach2 as a B-cell-specific partner for small maf proteins that negatively regulate the immunoglobulin heavy chain gene 3′ enhancer. EMBO J., 17, 5734–5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohira M., Seki,N., Nagase,T., Ishikawa,K., Nomura,N. and Ohara,O. (1998) Characterization of a human homolog (BACH1) of the mouse Bach1 gene encoding a BTB-basic leucine zipper transcription factor and its mapping to chromosome 21q22.1. Genomics, 47, 300–306. [DOI] [PubMed] [Google Scholar]
- Oyake T., Itoh,K., Motohashi,H., Hayashi,N., Hoshino,H., Nishizawa,M., Yamamoto,M. and Igarashi,K. (1996) Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Mol. Cell. Biol., 16, 6083–6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer K., Kim,K.S., Kogan,S. and Guarente,L. (1989) Functional dissection and sequence of yeast HAP1 activator. Cell, 56, 291–301. [DOI] [PubMed] [Google Scholar]
- Rafie-Kolpin M., Chefalo,P.J., Hussain,Z., Hahn,J., Uma,S., Matts,R.L. and Chen,J.J. (2000) Two heme-binding domains of heme-regulated eukaryotic initiation factor-2α kinase. N terminus and kinase insertion. J. Biol. Chem., 275, 5171–5178. [DOI] [PubMed] [Google Scholar]
- Rotenberg M.O. and Maines,M.D. (1991) Characterization of a cDNA-encoding rabbit brain heme oxygenase-2 and identification of a conserved domain among mammalian heme oxygenase isozymes: possible heme-binding site? Arch. Biochem. Biophys., 290, 336–344. [DOI] [PubMed] [Google Scholar]
- Sassa S. and Nagai,T. (1996) The role of heme in gene expression. Int. J. Hematol., 63, 167–178. [DOI] [PubMed] [Google Scholar]
- Shibahara S., Yoshizawa,M., Suzuki,H., Takeda,K., Meguro,K. and Endo,K. (1993) Functional analysis of cDNAs for two types of human heme oxygenase and evidence for their separate regulation. J. Biochem., 113, 214–218. [DOI] [PubMed] [Google Scholar]
- Steiner H., Kispal,G., Zollner,A., Haid,A., Neupert,W. and Lill,R. (1996) Heme binding to a conserved Cys–Pro–Val motif is crucial for the catalytic function of mitochondrial heme lyases. J. Biol. Chem., 271, 32605–32611. [DOI] [PubMed] [Google Scholar]
- Taketani S. and Tokunaga,R. (1984) Non-enzymatic heme formation in the presence of fatty acids and thiol reductants. Biochim. Biophys. Acta, 798, 226–230. [DOI] [PubMed] [Google Scholar]
- Talbot D. and Grosveld,F. (1991) The 5′HS2 of the globin locus control region enhances transcription through the interaction of a multimeric complex binding at two functionally distinct NF-E2 binding sites. EMBO J., 10, 1391–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toki T. et al. (1997) Human small Maf proteins form heterodimers with CNC family transcription factors and recognize the NF-E2 motif. Oncogene, 14, 1901–1910. [DOI] [PubMed] [Google Scholar]
- Yamamoto M., Kure,S., Engel,J.D. and Hiraga,K. (1988) Structure, turnover and heme-mediated suppression of the level of mRNA encoding rat liver δ-aminolevulinate synthase. J. Biol. Chem., 263, 15973–15979. [PubMed] [Google Scholar]
- Yoshida C. et al. (1999) Long range interaction of cis-DNA elements mediated by architectural transcription factor Bach1. Genes to Cells, 4, 643–655. [DOI] [PubMed] [Google Scholar]
- Yoshida T., Biro,P., Cohen,T., Muller,R.M. and Shibahara,S. (1988) Human heme oxygenase cDNA and induction of its mRNA by hemin. Eur. J. Biochem., 171, 457–461. [DOI] [PubMed] [Google Scholar]
- Zhang L. and Guarente,L. (1995) Heme binds to a short sequence that serves a regulatory function in diverse proteins. EMBO J., 14, 313–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Hach,A. and Wang,C. (1998) Molecular mechanism governing heme signaling in yeast: a higher-order complex mediates heme regulation of the transcriptional activator HAP1. Mol. Cell. Biol., 18, 3819–3828. [DOI] [PMC free article] [PubMed] [Google Scholar]