Abstract
The Ikaros family of proteins are DNA binding factors required for correct development of B and T lymphocytes. Cytogenetic studies have shown that these proteins form complexes with pericentromeric heterochromatin in B cells, and the colocalization of transcriptionally silent genes with these complexes suggests that Ikaros could silence transcription by recruiting genes to heterochromatin. Here we show that a site in the λ5 promoter that binds Ikaros and Aiolos is required for silencing of λ5 expression in activated mature B cells. Analysis of methylation and nuclease accessibility indicates that the silenced λ5 gene is not heterochromatinized in B cells, despite being associated with pericentromeric heterochromatin clusters. We also found that a promoter mutation, which affects Ikaros-mediated silencing of λ5 expression, is not rescued in a transgenic line that has the gene integrated into pericentromeric heterochromatin. Our results indicate that the Ikaros proteins initiate silencing of λ5 expression through a direct effect on the promoter with localization to pericentromeric heterochromatin likely to affect the action of Ikaros on regulatory sequences rather than causing heterochromatinization of the gene.
Keywords: heterochromatin/Ikaros family/λ5 promoter/transcription silencing
Introduction
Development of differentiated cell lineages in mammals requires a co-ordinated programme of activation and silencing of different genes. It is also necessary for gene expression programmes to be transmitted through repeated rounds of cell division in proliferating cells. The information content that allows these changes to take place resides in transcription factors and their target binding sequences. Different factors act at different levels, with some factors regulating entire programmes of gene expression leading to the differentiation of specific cell types. The proteins encoded by the Ikaros gene family have been proposed to act as master regulators of cell differentiation during the development of B and T lymphocytes (Georgopoulos et al., 1994; Wang et al., 1996).
Ikaros was initially identified as a factor that binds to the T cell receptor CD3δ gene (Georgopoulos et al., 1992) and was also shown to encode the Lyf 1 protein, which binds to the lymphocyte-specific TdT and λ5 promoters in vitro (Lo et al., 1991; Hahm et al., 1994). The Ikaros gene is essential for development of B and T cells and natural killer (NK) cells. Ikaros null mice completely lack B cells, NK cells and fetal T cells, and also show significant disruption of other T cell compartments (Wang et al., 1996). Differential splicing of the Ikaros transcript gives rise to multiple isoforms that are expressed in lymphoid cells and in haematopoietic progenitors (Hahm et al., 1994; Molnar and Georgopoulos, 1994). The largest isoform contains four zinc-finger motifs close to the N-terminus that are essential for DNA binding, and two zinc fingers at the C-terminus involved in protein–protein interactions. The C-terminal region also contains a Krüppel-like domain. Two homologous proteins, Aiolos and Helios, have been identified that show the same DNA binding specificity as Ikaros (Morgan et al., 1997; Hahm et al., 1998). Helios expression is restricted to T cells, while Aiolos is expressed in most cell types that express Ikaros, with the exception of the earliest haematopoietic progenitors. Targeting of the Aiolos gene in mouse mainly affects B-cell function, with a high incidence of B-cell lymphomas and development of autoimmune responses (Morgan et al., 1997).
There are a number of possible mechanisms by which Ikaros could affect gene expression. Binding sites for Ikaros have been found in the promoters of several lymphoid-specific genes, suggesting that it could act as a classical transcription factor by affecting the efficiency of binding of the general transcriptional machinery to promoters. Transactivation studies have shown that it has activating and repressing activity in transient expression assays of promoters that have been linked to artificial tandemly repeated Ikaros binding sites (Sun et al., 1996; Kopially et al., 1999). However, an additional striking feature of the Ikaros family of proteins is the observation that they are concentrated at centromeric heterochromatin clusters in the interphase nucleus (Brown et al., 1997, 1999; Hahm et al., 1998; Cobb et al., 2000). Several different lines of evidence have led to the suggestion that localization of transcription factors and genes in the nucleus could play a role in gene regulation. In mammalian cells, PTF and OCT1 are enriched in transcriptionally active nuclear domains and associated with specific chromosomes early in the cell cycle (Pombo et al., 1998). Proteins such as Kap-1 and polycomb, which are involved in gene silencing, have been shown to co-localize with centromeric heterochromatin in mammalian cells (Saurin et al., 1998; Ryan et al., 1999).
The large complexes that the Ikaros proteins form with heterochromatin in the nucleus co-localize with several genes that become silenced during lymphocyte differentiation (Brown et al., 1997, 1999). This finding has suggested that Ikaros might act as a repressor of transcription. The silencing mechanism could be mediated in part by recruitment of genes to a silent heterochromatic compartment in the nucleus (reviewed by Lamond and Earnshaw, 1998; Cockell and Gasser, 1999). The paradigm for this type of repression comes from yeast, where silencing at mating-type loci and telomeres has been extensively studied. Silencing of genes at telomeres and the HM loci occurs through the assembly of a silencing complex along the DNA that contains the SIR proteins (reviewed by Laurenson and Rine, 1992). In Saccharomyces cerevisiae, the telomeres are located near the nuclear periphery where the SIR proteins are concentrated (Gotta et al., 1996; Maillet et al., 1996). Defective HMR silencing is restored by artificially anchoring the locus to the nuclear membrane (Andrulis et al., 1998), suggesting that the effect of localization is due to the high level of SIR proteins at the nuclear periphery. A number of transcriptionally silent genes have also been shown to localize to the heterochromatic compartment of the nucleus in Drosophila and mammalian cells (Csink and Henikoff, 1996; Dernburg et al., 1996; Brown et al., 1997, 1999; Schubeler et al., 2000).
Although localization of genes to heterochromatin offers an attractive mechanism by which Ikaros might affect gene expression, the observation by Brown et al. (1999) that the RAG and terminal deoxytransferase (TdT) genes can undergo silencing in a transformed thymocyte cell line without being relocated to heterochromatin suggests that silencing of these genes is initiated through a more complex mechanism. It is not clear whether Ikaros is directly involved in initiating this repression and the fact that there have been no functional studies in vivo on natural target genes for the Ikaros proteins has made it difficult to determine their role in such events. The mouse λ5 and VpreB1 genes are expressed in pro-B and pre-B cells and are silenced in immature and mature B cells where they become localized to Ikaros–centromeric complexes. The genes, which are separated by only 4 kb, code for the subunits of the surrogate light chain, a component of the pre-B cell receptor (Melchers et al., 1993). Transcription of the λ5–VpreB1 locus is activated by a locus control region (LCR) that comprises a cluster of DNase I hypersensitive sites located 3′ of λ5 as well as other regulatory elements that may include the gene promoters (Sabbattini et al., 1999). The λ5 promoter contains multiple binding sites for the transcription factors early B cell factor (EBF) and the E2A proteins (E47/E12) and two binding site for Ikaros (Lo et al., 1991; Martensson and Martensson, 1997; Sigvardsson et al., 1997). One of the Ikaros binding sites overlaps with an EBF binding site that is essential for promoter activity (Martensson and Martensson, 1997; Sigvardsson et al., 1997; P.Sabbattini and N.Dillon, in preparation). In this study, we show that the λ5 gene is a target for repression by Ikaros acting on a binding site located in the λ5 promoter through a mechanism that does not require heterochromatin formation.
Results
Experimental strategy
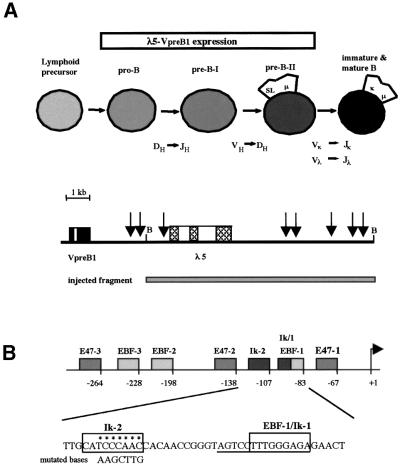
Transcription of λ5 and VpreB1 is first observed in pro-B cells prior to rearrangement of the immunoglobulin heavy chain genes, and continues through the pre-B-I and pre-B-II stages (Figure 1A) (Melchers, 1995). Silencing of expression of the genes occurs during the transition from the pre-B-II cell to the immature B cell at the time when the κ and λ light-chain loci are rearranged, and expression is not observed in mature B cells in the spleen. We set out to test whether binding of Ikaros and Aiolos to the λ5 promoter is responsible for this silencing. The λ5 gene contains two sites at positions –83 and –107 in the promoter that can bind Ikaros proteins in vitro (Lo et al., 1991) (Figure 1B). The proximal site at –83 (Ik-1) overlaps with a binding site for EBF that has been shown to be essential for efficient promoter function (Martensson and Martensson, 1997; Sigvardsson et al., 1997; P.Sabbattini and N.Dillon, in preparation). Since the binding site for Ikaros is embedded within the consensus sequence for EBF binding, it is difficult to generate mutant sites that bind EBF but not Ikaros (C.-M.Chow, unpublished data). The ability of Ikaros and Aiolos molecules to form homo- and heterodimers suggested that binding at the Ik-1 and Ik-2 sites might be co-operative. We decided therefore to test the effect of mutating the distal Ik-2 site on λ5 expression during B-cell development.
Fig. 1. Gel shift analysis of the Ik-2 site of the λ5 promoter. (A) Scheme of the stages of B-cell differentiation and temporal expression of λ5 and VpreB1 (surrogate light-chain indicated by SL) and map of the λ5-VpreB1 locus. B, BamHI. Vertical arrows indicate the position of DNase I hypersensitive sites in the locus. (B) Schematic representation of the λ5 promoter showing the position of the binding sites for EBF, the E2A proteins (E47 and E12) and Ikaros (Ik). The positions of the Ik-1 and Ik-2 sites in the sequence are indicated by boxes and the sequence of the EBF-1 site is underlined. The bases substituted in the Ik-2 mutation are indicated below the Ik-2 wild-type sequence. (C) Ikaros and Aiolos bind to the Ik-2 site of the λ5 promoter. Different antibodies were pre-incubated with WEHI-231 protein extracts (lanes 5–9). An oligonucleotide that contained a mutated Ik-2 site was added to reduce the background due to binding of proteins outside the Ik-2 site. An antibody against Elf-1 has no effect on complex B formation (lane 5). Polyclonal antibodies against the N- and C-terminal portions of Ikaros (lanes 6 and 7) and monoclonal antibodies against Ikaros (lane 8) and Aiolos (lane 9) produce a partial disruption of complex B and a weak supershift (complex C). (D) The mutated Ik-2 oligonucleotide does not compete with the wild-type oligonucleotide for the binding of complex B at the Ik-2 site. WEHI extracts were incubated with a labelled double-stranded oligonucleotide containing the wild-type Ik-2 site. Complex A is competed by high concentrations of unlabelled wild-type (wt) and mutant (mut) oligonucleotides. Complex B is competed only by the wild-type oligonucleotide and not by the mutant.
The effect of the mutation on binding of Ikaros was tested using gel retardation analysis. Nuclear extracts from the B cell line WEHI-231 were incubated with a double stranded oligonucleotide containing the wild-type Ik-2 site. A diffuse low mobility complex (complex B) was observed that was disrupted by anti-Aiolos and anti-Ikaros antibodies, but not by antibodies against the transcription factor Elf-1 (Figure 1C). Complex B is competed by an excess of unlabelled wild-type oligonucleotide but not by an oligonucleotide containing the mutated Ik-2 site (Figure 1D), demonstrating that the mutation abolishes binding of Ikaros and Aiolos.
Mutation of the Ik-2 Ikaros binding site affects λ5 silencing in mature B cells
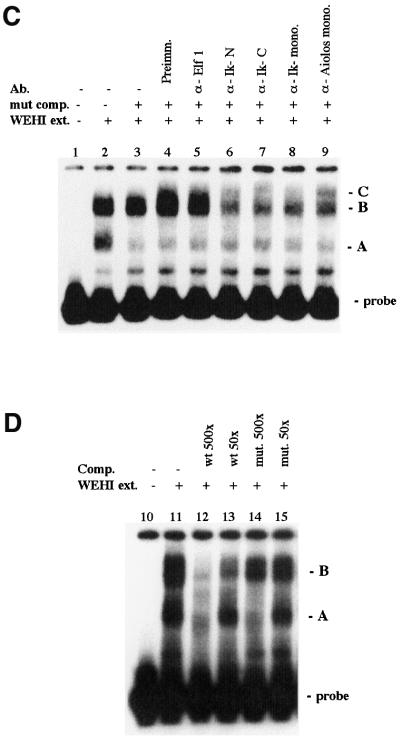
Previous studies have shown that a 12 kb region of the λ5-VpreB1 locus contains an LCR that can direct correct stage-specific expression of λ5 at levels similar to those measured for the endogenous gene (Sabbattini et al., 1999) (Figure 1A). The mutation in the distal Ik-2 site was introduced into the λ5 promoter (Figure 1B), and the 12 kb fragment containing the mutated gene was used to generate transgenic mice. A total of six lines were generated carrying the mutated gene and six control lines were also generated by injecting the wild-type gene. The level of expression of the λ5 transgene at the pre-B cell stage was determined by comparison with the expression of the endogenous λ5 gene in primary pre-B cells cultured from 16.5 day fetal liver, or in whole fetal liver (Figure 2A). To analyse expression at the mature B cell stage (Figure 2B), B cells were isolated from adult spleens by depletion with anti-CD43 antibody and activated by stimulation with anti-CD40 antibody in the presence of IL-4 for 3 days.
Fig. 2. Expression analysis of the λ5 transgene carrying the Ik-2 mutation in the promoter. (A) RNase protection analysis of λ5 expression in pre-B cells. The analysis was carried out on 25 µg of RNA from 16.5 day fetal liver (lanes 2, 4, 5, 6, 9, 10, 11 and 12) or 3 µg of RNA from primary pre-B cell cultures (lines 1, 3, 7 and 8) (NTG, non-transgenic pre-B cells). The λ5-specific probe is derived from the λ5 cDNA and contains a 36 bp oligonucleotide tag that was inserted into the λ5 transgene immediately downstream from the first ATG. The RNase protection probe is described in detail in Sabbattini et al. (1999). The protected fragments were 498 bp for the λ5 transgene (tg) and 412 bp for the λ5 endogenous gene (endog), which does not contain the tag. The transgene copy number is indicated at the bottom of each lane. Band intensities were quantified using a phosphorimager. The histograms show the expression level of the λ5 transgene, normalized to the expression of one allele of the endogenous λ5, for the wild-type (shaded bars) and mutant lines (solid bars). (B) RNase protection analysis of λ5 expression in activated mature B cells. The mature B cells were isolated from spleen and activated as described in Materials and methods. The RNase protection assay was carried out on 20 µg of RNA from activated B cell cultures using the λ5-specific probe. A β-actin-specific probe labelled to one-tenth of the specific activity of the λ5 probe was added to the reaction as a loading control (described in Sabbattini et al., 1999). The expression per copy of the mutant and wild-type transgenes is compared using an arbitrary scale in which the highest level of expression is assigned a value of 1.0.
In pre-B cells, copy-dependent expression is observed for both wild-type and mutant transgenes at levels per copy that are similar to that of the endogenous λ5 gene (Figure 2A). Expression of the λ5 transgene reaches a plateau in animals with high copy number (>10 copies) (Sabbattini et al., 1999). In mature B cells from transgenics carrying the wild-type gene, expression of the transgene is strongly repressed with very low levels of λ5 expression observed only in the high-copy transgenics (Figure 2B). In contrast, animals carrying the mutated transgene show substantially higher levels of expression in activated mature B cells. In five out of six mutant transgenic lines, expression of λ5 per transgene copy is between 5- and 10-fold higher than the expression measured in transgenics for the wild-type fragment (Figure 2B). This result provides direct in vivo evidence that the Ikaros proteins act on the promoter of the λ5 gene to mediate stage-specific silencing during B-cell development.
Silencing is not rescued by integration of the mutated transgene into pericentromeric heterochromatin
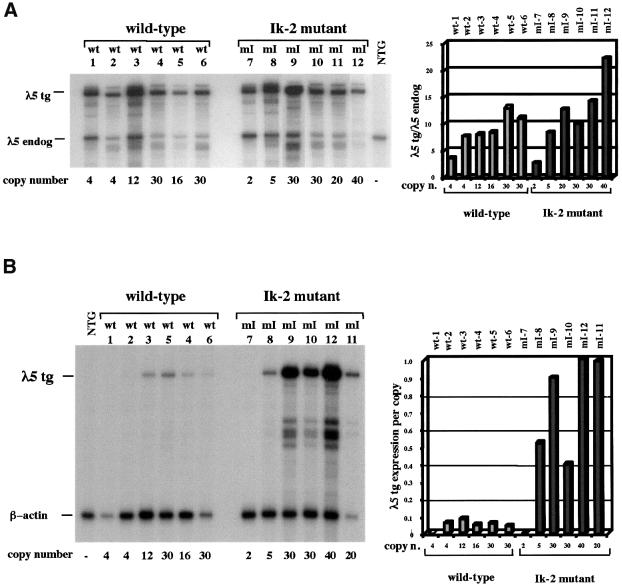
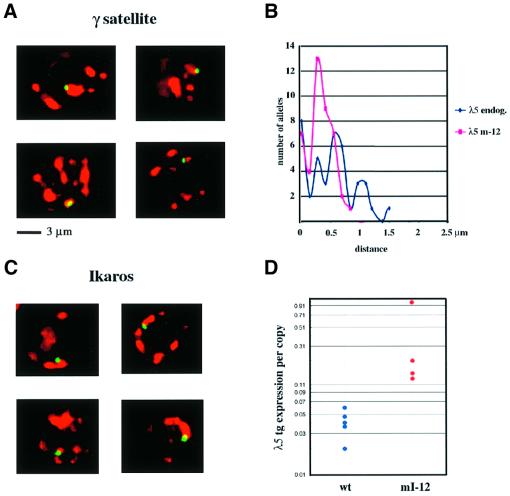
Integrations of transgenes into pericentromeric heterochromatin have been observed at varying frequencies in transgenic mice (Festenstein et al., 1996; Milot et al., 1996; Sabbattini et al., 1999). Identification of lines with this type of integration of the mutated transgene would allow direct testing of the role of centromeric localization in Ikaros-mediated silencing. The sites of integration of the transgenes were analysed in three Ikaros mutant and two wild-type lines using fluorescent in situ hybridization (FISH) of metaphase chromosomes (Figure 3). Two of the lines (mI-10 and wt-4) had the transgene integrated on the long arm of the chromosome at positions that were far away from the centromere. In two other lines (mI-9 and wt-5) the transgenes were located close to the peri-centromeric γ-satellite DNA but not in direct contact with it. This position is broadly similar to the location of the endogenous λ5 gene relative to the centromere on chromosome 16 (Sabbattini et al., 1999). In line mI-12 the transgene was integrated into pericentromeric heterochromatin. In order to confirm that the transgene in line mI-12 is derepressed in mature B cells, expression in activated B cells was analysed in a total of four transgenic animals from this line (Figure 4D). The expression levels were compared with those of five animals from the wild-type lines wt-5 and wt-6. A significant amount of variation was observed between the different animals from line mI-12, suggesting that the transgene is subject to variegation. Nevertheless, all four animals expressed the transgene at higher levels than any of the five animals analysed from the wild-type lines, and the difference between the expression for line mI-12 and the wild-type transgenics was significant (Mann–Whitney U test; P <0.02). This result shows that integration into peri-centromeric heterochromatin does not restore silencing of the Ik-2 mutant transgene at the mature B cell stage.
Fig. 3. Chromosomal localization of the transgenes. FISH of metaphase spreads from activated B cells. Spreads were hybridized with the 12 kb fragment of the λ5 locus used to generate the transgenic lines (green) and a pericentromeric γ-satellite probe (red) (Lundgren et al., 2000). The chromosomes were counterstained with DAPI (blue).
Fig. 4. Nuclear localization and expression of the pericentromeric transgene. (A) Three-dimensional FISH analysis of the location of the λ5 transgene relative to pericentromeric heterochromatin clusters in nuclei of activated B cells from line mI-12. Each panel shows a single optical section of a nucleus obtained by deconvolution microscopy. The probes used for the λ5 transgene (green) and for the γ-satellite repeats (red) were the same as in Figure 3. (B) Distance of the λ5 transgene from the nearest centromeric cluster in activated B cells from line mI-12 (red line) compared with the distance for the endogenous λ5 gene (blue line) in non-transgenic activated B cells. The distance was determined by rotating 3D images of nuclei generated by deconvolution microscopy and measuring the distance from the centre of the gene signal to the edge of the nearest cluster. The distances are shown on the x-axis, while the y-axis gives the number of alleles located at each distance. The probe used to detect the endogenous λ5 gene was the cosmid λ5 3.1 (Sabbattini et al., 1999). Forty-four alleles were analysed for the transgene and 40 for the endogenous λ5 gene. (C) Immuno-FISH analysis of the location of the λ5 transgene relative to Ikaros clusters in nuclei of activated B cells from line mI-12 (see Materials and methods). Each panel shows a single optical section of a nucleus. The λ5 transgene (green) was detected using the DNA probe described in Figure 3. The Ikaros clusters (red) were visualized using antibodies against the N- and C-terminal domains of Ikaros (Hahm et al., 1994). (D) Levels of expression of the λ5 transgene in activated B cells from four mice from the pericentromeric line mI-12 (red) compared with expression in three mice from wt-5 and two from wt-6 (blue). Expression of the λ5 transgene is normalized to the level of the endogenous β-actin transcript used as a loading control. The expression per copy of the mutant and wild-type transgenes is displayed using an arbitrary scale in which the highest level of expression was assigned a value of 1.0. The values are shown on a logarithmic scale to illustrate the variation in expression within the two groups of animals. The difference between expression in the wild-type lines and that observed for line mI-12 is significant (Mann–Whitney U test, P <0.02).
While integration into pericentromeric DNA should be effective in tethering a gene to the heterochromatin complex, it is possible that the transgene could become localized away from the heterochromatin in expressing cells through the formation of a long loop. In order to test whether this is the case, the localization of the mI-12 transgene with respect to the centromeric clusters in interphase cells was analysed using three-dimensional (3D) FISH (Brown et al., 1997) (Figure 4A). All of the cells were found to have the transgene either in direct contact with or very close to the centromeric cluster. To further confirm the location of the transgene close to heterochromatin, the distance of the transgene signal from the edge of the nearest centromeric cluster was measured and compared with the distances for the endogenous λ5 alleles (Figure 4B). The fact that the range of distances for the transgene in line mI-12 is actually smaller than for the endogenous alleles excludes the possibility that derepression of the transgene is a consequence of relocation away from the centromeric cluster. Another possible explanation for the observation that pericentromeric localization does not silence the mutated gene would be that the transgene in line mI-12 has integrated into a region that is separate from the Ikaros binding domain of the pericentromeric heterochromatin. This possibility was examined directly by carrying out immuno-FISH using an antibody against Ikaros protein and a λ5 DNA probe (Figure 4C). In 97.5% (76/78) of the cells, the transgene was directly co-localized with the Ikaros clusters, excluding the possibility that localization away from the Ikaros–heterochromatin complexes is responsible for transgene derepression.
In the other lines, which had the transgene integrated at different positions relative to the centromere (Figure 3), the transgenes showed a variable frequency of centromeric localization in interphase B cells (data not shown). The variation is likely to be due, at least in part, to the effects of sequences surrounding the sites of integration, making it difficult to directly compare localization frequencies between the mutant and wild-type transgenics.
Derepression of λ5 expression is restricted to a fraction of activated mature B cells
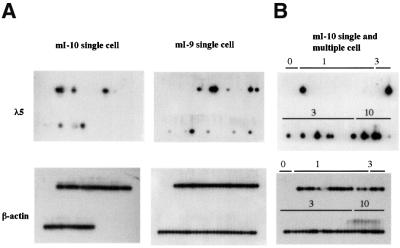
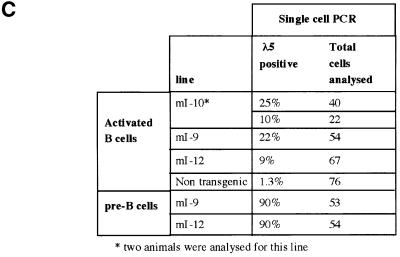
It has been hypothesized that the Ikaros proteins are involved in maintaining stable repression of silent genes, perhaps through the formation of silencing complexes. If this is the case, then mutations that destabilize silencing might be expected to give rise to variegated expression. To examine the distribution of λ5 expression resulting from disruption of Ikaros-mediated silencing, single-cell RT–PCR was used to analyse expression in individual cells. Single cells expressing the pan B cell marker B220 were isolated by fluorescence-activated cell sorter (FACS) sorting of activated B cell cultures and analysed by RT–PCR as described in Materials and methods. Two rounds of PCR were performed using nested primers specific for λ5 RNA and for β-actin RNA as a control. Analysis of pre-B cell cultures derived from line mI-9 and the pericentromeric line mI-12 showed that 90% of the cells were positive for λ5 in both lines (data not shown). In activated mature B cells from the mice carrying the transgene containing the Ik-2 mutation, λ5 expression was only detected in a fraction of the cells. The proportion of expressing cells ranged from 9 to 22% (Figure 5A and C). When three or 10 cells were deposited in each well, the number of positive wells increased but not all of the wells showed λ5 amplification, indicating that the restriction of expression to a fraction of the cells is not caused by a threshold in the sensitivity of PCR assay (Figure 5B). There is also evidence of some variation in the frequency of positive cells between animals within the same line (see line mI-10; Figure 5C). This could be due to variation at genetic modifier loci that have been shown to affect variegation in Drosophila and mammals (Weiler and Wakimoto, 1995; Festenstein and Kioussis, 2000). Variation in modifier loci might well be expected since the mice used in this study were hybrids of two inbred strains. The results of the RT–PCR analysis indicate that derepression of the mutated transgene is restricted to a fraction of activated B cells.
Fig. 5. RT–PCR analysis of λ5 expression in individual B220+ lymphocytes. Single B220+ lymphocytes from primary cultures of pre-B cells or activated B cells were sorted by FACS and single-cell RT–PCR was performed as described in Materials and methods. Reverse transcription and PCR amplification were carried out for λ5 RNA and also for β-actin RNA, which acted as a PCR control. The PCR products were blotted and hybridized to either a λ5 cDNA probe or a β-actin cDNA probe. In each panel, the first four lanes do not contain any cells and serve as negative controls. When B cells from a non-transgenic animal were analysed, one of 76 cells analysed showed a weak signal for λ5 that was not visible by ethidium bromide staining (data not shown). The single expressing cell may be due to the presence of a small number of pre-B cells in adult spleen (Rolink et al., 1993; Monroe et al., 2000). (A) λ5 and β-actin expression in single activated mature B cells from lines mI-9 and mI-10. (B) Single- and multiple-cell RT–PCR of activated mature B cells from line mI-10. The number of cells sorted into each well (0, 1, 3 or 10 cells) is indicated above the lanes. (C) Summary of RT–PCR analysis of λ5 expression in single cells.
The endogenous λ5 promoter is hypomethylated and accessible to restriction enzymes in mature B cells
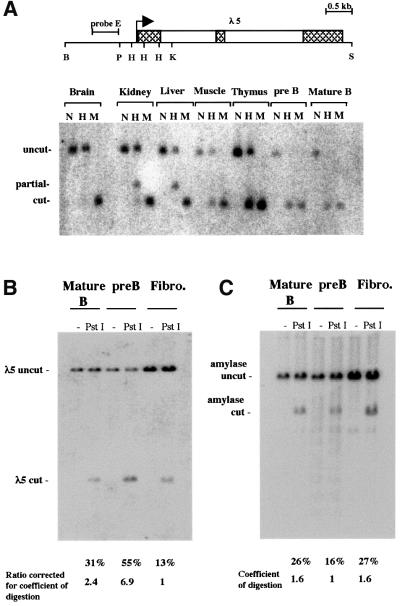
The fact that expression resulting from derepression of the λ5 transgene is restricted to a fraction of mature B cells is reminiscent of position effect variegation and raises the possibility that silencing of λ5 expression is associated with heterochromatinization of the λ5-VpreB1 locus. This hypothesis was tested by analysing the methylation status and accessibility of the λ5 gene in activated mature B cells. The analysis was carried out on the endogenous λ5 gene, which is silenced in mature B cells and is associated with centromeric heterochromatin. Methylation of CpGs has previously been shown to be a good indicator of heterochromatinization. The satellite sequences that form the bulk of centromeric heterochromatin are highly methylated and contain >40% of all methyl-CpGs in the mouse nucleus (Miller et al., 1974; Lewis et al., 1992). The formation of facultative heterochromatin during inactivation of the X chromosome in mammals is also accompanied by hypermethylation (Mohandas et al., 1981). Therefore, if the endogenous λ5 gene is heterochromatinized in mature B cells the prediction is that it should be hypermethylated. To test whether this is the case, genomic DNA from non-transgenic activated B cells was digested with the methylation-sensitive HpaII and methylation-insensitive MspI restriction enzymes and hybridized with a probe located upstream from the promoter (Figure 6A). The three HpaII sites in the λ5 promoter are methylated to different degrees in brain, kidney, liver, muscle and thymus. Complete methylation of the promoter was observed in brain. Kidney, liver and muscle showed some demethylation while the lowest level of methylation in a non-B-cell tissue was found in thymus. No methylation was detected in either pre-B cells or activated B cells. Similar results were obtained with a probe that spanned the promoter region (data not shown).
Fig. 6. Methylation and restriction accessibility analysis of the endogenous λ5 promoter. (A) Silencing of λ5 at the mature B-cell stage is not associated with hypermethylation of the promoter. The map shows the positions of the HpaII–MspI sites in the λ5 promoter. B, BamH1; P, PstI; H, HpaII–MspI; K, KpnI; S, SphI. Genomic DNA from the indicated tissues was digested with BamHI and KpnI. The DNA was further digested with the methylation-sensitive enzyme HpaII (H), the methylation-insensitive enzyme MspI (M), or left undigested (N). The hybridization was carried out with a probe located upstream of the three HpaII sites in the λ5 promoter (probe E). (B) Accessibility of the λ5 promoter to restriction enzyme digestion at the mature B-cell stage. Nuclei were isolated from primary cultures of activated mature B cells, pre-B cells and fibroblasts (Fibro.) and incubated with PstI or left untreated (–). The percentage of PstI-digested product relative to the total is indicated below the PstI lanes. The PstI digestion values for the λ5 promoter were normalized by calculating the digestion coefficient for the pancreatic amylase gene (C), which is not expressed in any of the three cell types analysed. (C) The accessibility of the pancreatic amylase gene to PstI was used as a control to measure the extent of PstI digestion of the nuclei by stripping the blot shown in (B) and reprobing it with a probe for the pancreatic amylase gene. The percentage of PstI-digested product relative to the total is indicated below the PstI lanes and was used to calculate the coefficient of digestion for each cell type.
Promoter accessibility of the endogenous λ5 gene was also measured by digesting isolated nuclei from activated B cells with the restriction enzyme PstI (Figure 6B). Nuclei from pre-B cells and primary embryonic fibroblasts were also subjected to the same treatment and the relative accessibility in the three cell types was compared. The blot was reprobed with a probe for the pancreatic amylase gene as a control for the extent of PstI digestion and the values were used to normalize the values obtained for λ5 so that they could be directly compared. The relative level of digestion of λ5 in pre-B cells was 6.9 times higher than in fibroblasts, while digestion in mature B cells was 2.4 times greater than in fibroblasts (Figure 6B). The high level of accessibility in pre-B cells is expected as the promoter is active in these cells and contains a DNase I hypersensitive site. However, it is significant that the silent promoter in mature B cells is still relatively accessible to the restriction enzyme compared with fibroblasts. In a separate study, a λ5 transgene integrated into centromeric heterochromatin was shown to be four times less accessible than the endogenous λ5 gene in fibroblasts (Lundgren et al., 2000). Taken together, the methylation and accessibility results provide strong evidence that the endogenous λ5 gene is not heterochromatinized in activated mature B cells.
Discussion
The Ikaros family of proteins have been the focus of attention because of their importance for correct B- and T-cell development, and also because they form large nuclear complexes with pericentromeric heterochromatin in actively dividing cells of haematopoietic lineages. The finding that inactive genes co-localize with the Ikaros–heterochromatin complexes has led to the suggestion that the Ikaros proteins could be involved in maintaining genes in a repressed state by promoting the formation of heterochromatin at the repressed gene (reviewed by Lamond and Earnshaw, 1998). However, the fact that no target genes for Ikaros had been identified in functional studies has made it difficult to determine the mechanism by which the Ikaros proteins affected gene expression.
The results described here provide direct in vivo evidence of a role for Ikaros/Aiolos in the silencing of a specific target gene. Mutation of a binding site for the proteins in the λ5 promoter was found to affect silencing of the promoter in activated B cells of transgenic mice. The same mutation had no discernible effect on the activation of λ5 transcription in pre-B cells. The identification of an in vivo target for Ikaros-mediated repression indicates that silencing of specific genes forms at least one part of the role of Ikaros proteins in controlling lymphocyte differentiation. What is the mechanism by which Ikaros silences transcription? The finding that mutation of a single binding site in the λ5 promoter causes a loss of repression points to a direct effect of Ikaros binding on promoter function rather than a mechanism involving heterochromatin formation. This conclusion is further supported by the finding that the repressed endogenous λ5 promoter is hypomethylated and relatively accessible to restriction enzyme digestion in activated mature B cells, indicating that it is not heterochromatinized.
The overlap of an Ikaros binding site in the λ5 promoter with the EBF-1 binding site at position –83 (Ik-1/EBF-1), which has been shown to be essential for the efficient activation of the λ5 promoter (Martensson and Martensson, 1997; Sigvardsson et al., 1997; P.Sabbattini, in preparation), suggests that initiation of silencing could be due to competition for occupancy of this site. As interactions between the Ikaros proteins have been shown to increase their affinity for DNA (Sun et al., 1996), we propose that co-operativity between the Ik-1 site and the neighbouring Ik-2 site at –107 enhances the ability of Ikaros to bind to both sites, thereby helping it to compete EBF binding at the Ik-1/EBF-1 site. As a result, mutation of the Ik-2 binding site would also impair Ikaros binding to the Ik-1 site, allowing binding of EBF and derepression. Interestingly, the overlap of Ikaros binding sites with the sites for other transcription factors has also been observed in the TdT promoter, where an Ikaros site overlaps with an Ets site (Ernst et al., 1996) and in the DNase I hypersensitive sites that form part of the λ5 LCR, with several E2A binding sites overlapping with sites that bind Ikaros (C.-M.Chow, S.Minaee and P.Sabbattini, unpublished data). The dosage of EBF and E2A has been shown previously to affect λ5 expression (O’Riordan and Grosschedl, 1999), and overexpression of these factors induces transcription of λ5 (Sigvardsson et al., 1997; Kee and Murre et al., 1998). Competition could therefore be an important component of the mechanism by which Ikaros represses transcription.
Such a mechanism has parallels in a number of other systems. For example, competition between factors for overlapping binding sites is well established as a mechanism of repression in Drosophila. The even-skipped stripe 2 enhancer contains six activator binding sites that bind bicoid and hunchback, and four of these sites overlap with giant or Krüppel repressor binding sites. Krüppel in particular has a consensus binding site that shares seven out of 10 bases with the binding site for bicoid in a situation that has similarities to the overlap of the Ikaros and EBF binding sites (Small et al., 1991). However, there is also evidence that Krüppel can repress transcription through a short-range quenching mechanism that does not depend on competition but does require that the Krüppel binding site is located within 100 bp of the repressed activator binding site (Gray and Levine, 1996). It is possible that the repressive effects of the Ikaros proteins on the λ5 promoter could be the result of both competition and quenching. In addition, Ikaros and Aiolos have both been shown to interact with the mSin3A and mSin3B histone deacetylases and with the Mi-2 chromatin remodelling complex (Kopially et al., 1999). Silencing could involve recruitment of these proteins to the promoter.
What role does pericentromeric localization play in Ikaros-mediated silencing? As we have already discussed, the hypomethylation and relative accessibility of the silent endogenous λ5 promoter in activated mature B cells argue against a model where localization promotes heterochromatinization. In addition, the finding that integration of the mutated transgene into pericentromeric heterochromatin does not rescue silencing is further evidence that silencing is not a direct result of localization to the pericentromeric compartment. Although the observation that silencing is not rescued by pericentromeric localization is based on a single integration, we were able to demonstrate that the expressing pericentromeric transgene remained closely associated with the pericentromeric Ikaros complexes in interphase B cells. Additional evidence for our conclusions comes from a recent study by Lundgren et al. (2000) showing that a pericentromeric λ5 transgene remains closely associated with the heterochromatin complex in pre-B cells where the transgene is expressed. Recent studies have shown that the chromatin structure and expression of pericentromeric transgenes are sensitive to transcription factor dosage (Lundgren et al., 2000; McMorrow et al., 2000) and to the level of the heterochromatin binding protein HP1 (Festenstein et al., 1999). These results suggest that it is the equilibrium between positive and negative factors that controls the transcriptional status of the gene. Changes in this equilibrium could be influenced by movement to a different compartment, but they can also occur within the heterochromatic compartment, either as a result of changing levels of factors, or changes in binding sites. Therefore, while localization of a gene to heterochromatin might influence the transcriptional state of the gene, it is unlikely to act as the sole, or even the major determinant of repression. Instead, the critical parameter that could be influenced by heterochromatin localization of the λ5 gene is efficient occupancy of Ikaros binding sites in the promoter, caused by the increased local concentration of Ikaros. Mutation of the Ik-2 site would remove an essential target of Ikaros, eliminating the effect of heterochromatin localization.
The mutation in the promoter results in expression of λ5 in a proportion of cells while leaving the gene silenced in the remainder. Together with the observation that the silenced λ5 gene co-localizes with heterochromatin clusters in mature B cells (Brown et al., 1997, 1999), this finding suggests that Ikaros is involved in stable repression of λ5. However, the results of this study also indicate that silencing of the endogenous λ5 gene in mature B cells does not involve heterochromatinization since the promoter remains demethylated and relatively accessible to digestion. While stable repression is often equated with the formation of heterochromatin, evidence exists from other systems that stable silencing complexes can form in the absence of heterochromatinization. For example, the complexes that give rise to stable repression by the Polycomb group proteins have several features that differentiate them from pericentromeric heterochromatin. These include the absence from Polycomb silencing complexes of histone H1, an important constituent of heterochromatin (Franke et al., 1992). Polycomb is also unevenly distributed on target genes (Orlando and Paro, 1993) and possesses a significant degree of promoter and enhancer specificity, unlike the non-specific spreading effect of heterochromatin (reviewed by Bienz and Muller, 1995). If sequence-specific binding of Ikaros to the λ5 gene does result in the formation of stable silencing complexes, then localization to heterochromatin would be expected to promote the formation of these complexes by increasing the local concentration of Ikaros.
The phenotype of Ikaros and Aiolos null mutations indicates that the Ikaros proteins play a key role in regulating B- and T-cell development. Our findings show that at least one of the functions of the Ikaros proteins is to act as repressors of transcription by binding to promoters of target genes. In developing B cells, this silencing effect would be antagonistic to the effects of other determinants of B cell fate such as EBF and the E2A proteins that are known to activate transcription of B-cell-specific genes. The results of this study suggest that specification of the B- and T cell lineages occurs through a fine balance between activation and repression of specific genes. They also suggest that while localization to heterochromatin could influence this balance, this is likely to be a secondary effect, which is subordinate to direct interference with transcriptional activation caused by binding of Ikaros to the promoter.
Materials and methods
Electrophoretic mobility shift assay
The oligonucleotide CCCGGTAGTCGGTTGGGATGCAAGCCTA containing the Ik-2 site was 32P-labelled using T4 kinase and annealed to its complementary strand. Nuclear extracts from WEHI 231 cells were prepared as described previously (Dignam et al., 1983). The binding assay was performed in 10 mM Tris–HCl pH 7.5, 50 mM NaCl, 1 mM EDTA and 1 mM dithiothreitol (DTT). Probe (0.1 pmol) was incubated with 2.5 µg of nuclear extract in the presence of 0.1 µg of double-stranded poly dI–dC. Competition was carried out using the double-stranded oligonucleotide CCCGGTTGTGCAAGCTTTGCAAGCCTA containing the mutated Ik-2 site (underlined). Competitors and antibodies were pre-incubated with extract on ice for 15 and 25 min, respectively. Samples were separated on 4% native PAGE in 0.5× TBE buffer.
DNA fragments used for generation of transgenic mice
The Ik-2 mutation was introduced into the promoter of the λ5 gene on a 12 kb BamHI fragment from the λ5-VpreB1 locus using the PCR overlap extension method (Ho et al., 1989). Fragments containing the mutated or the wild-type λ5 promoter were purified and injected into pronuclei of C5BL6/CBA F1 mouse eggs as previously described (Dillon and Grosveld, 1993). Transgenic lines were established by breeding founders with non-transgenic animals and individual transgenic mice were identified by Southern blotting tail DNA.
Primary pre-B cell culture system
Livers from 16.5 day fetuses were disaggregated and fetal liver cells were grown in 1× RPMI, 20% fetal calf serum (FCS), 50 µM 2-mercaptoethanol, 50 µg gentamicin/ml on ST-2 stromal cell feeder layers as described in Sabbattini et al. (1999).
Selection and activation of mature B cells
Spleens from 6- to 10-week-old mice were disaggregated and the single-cell suspension was subjected to centrifugation on a Ficoll cushion to remove erythrocytes. Cells from the interface were collected and incubated with streptavidin–anti-CD43 antibodies (Dynal), washed and incubated with magnetic beads (Dynal) conjugated to biotin. CD43-positive cells were removed by magnetic separation to enrich the cell suspension for resting B cells. B-cell activation was induced by culturing the cells in 1× RPMI, 15% FCS, 50 µM 2-mercaptoethanol, 50 µg/ml gentamicin, 4 ng/ml IL-4, 10 µg/ml anti-CD40 (monoclonal antibody FGK45) for 3 days.
RNase protection assay
RNA was obtained from 16.5 day fetal livers, pre-B cell primary cultures or activated splenic mature B cells by lithium chloride extraction (Auffray and Rougeon, 1980; Dillon and Grosveld, 1993) and subjected to RNase protection analysis as described in detail in Sabbattini et al. (1999). The relative intensities of bands obtained by RNase protection were quantitated using a phosphorimager.
Fluorescent in situ hybridization (FISH) analysis
DNA FISH of metaphase chromosomes was carried out as previously described (Sabbattini et al., 1999). Splenic B cells were activated for 2 days in 1× RPMI, 15% FCS, 50 µM 2-mercaptoethanol, 50 µg/ml gentamicin, 10 µg/ml lipopolysaccharide. The cells were hypotonically swollen in 0.056 M KCl, fixed in 3:1 methanol–acetic acid and distributed on slides. The λ5-specific probe (12 kb λ5 BamHI fragment used to generate transgenic mice cloned in pUC19) was labelled with digoxigenin and the γ-satellite probe (eight copies of the 234 bp γ-satellite repeat cloned in pBluescript) was labelled with fluoroRED (Amersham-Pharmacia) by standard nick translation.
Three-dimensional (3D) FISH was performed on activated B cells or pre-B cells according to Brown et al. (1997) with minor modifications (Lundgren et al., 2000). Immuno-FISH, which allowed simultaneous detection of Ikaros protein and the λ5 transgene, was carried out as described by Brown et al. (1997). Z-series images were collected with a 100× plan-Apochromat objective fitted on an Axiovert 100 microscope (Zeiss). Images were captured using a MicroMax 5 MHz camera (Princeton Instruments Inc.). The image stacks were deblurred by blind deconvolution with AutoDeblur software (AutoQuant Imaging, Inc.) and 3D reconstructions were made with MetaMorph software (Universal Imaging Corp.).
Analysis of single cells by RT–PCR
Activated B cells or pre-B cells were incubated with anti-B220 antibodies and single B220+ cells were sorted twice by FACS (Cell Quest software and cytoclone system and software) and deposited in 96-well PCR plates. The plates contained 4 µl of lysis buffer [0.4% NP-40, 60 µM dNTPs, 25 µM DTT, 0.5 U/µl RNasin (Promega)] and the cells were lysed for 15 min on ice. The single-cell RT–PCR was carried out following the method of Hu et al. (1997) with the modifications described by Delassus et al. (1999). Mature B-cell lysates were reverse transcribed in the presence of the λ5-specific primers TCTCCTCCTGCTGCTGCTGTTGG and CTTGGGCTGACCTAGGATTGTGAG and the β-actin primers GGCACCACACCTTCTACAATGAGC and CCCGGCCAGCCAGGT CCAG using 48 U of MMLV-RT per well in buffer provided by the supplier (Gibco-BRL). The first round of PCR was performed by the addition of 40 µl of PCR buffer and 1.25 U of AmpliTaq polymerase (Perkin-Elmer). The PCR cycles were as follows: 1 min at 95°C, 1 min at 54°C, 2 min at 72°C for 35 cycles. One microlitre of the first PCR was further amplified independently for λ5 and β-actin using either the λ5-specific nested primers CTGTTGGGTCTAGTGGATGGTGTC and CAAAACTGGGGCTTAGATGGA or the nested primers specific for β-actin (TGAACCCTAAGGCCAACCGTGAAA and GCAGGATGG CGTGAGGGAGAGC) and 0.625 U of AmpliTaq per reaction. The PCR conditions were: 1 min at 95°C, 1 min at 53°C, 1 min at 72°C for 35 cycles for the λ5 amplification, and 1 min at 95°C, 1 min at 58°C, 1 min at 72°C for 35 cycles of the β-actin amplification. Aliquots of the second-round PCRs were subjected to gel electrophoresis, blotted and hybridized to a 0.7 kb λ5 cDNA probe or to a 1.3 kb β-actin cDNA probe. When lysates from pre-B cells were amplified, the reverse transcription and the first PCR round were carried out using a λ5 5′ oligonucleotide (AGCTGATTTCTGAGGAGGATCTGG) complementary to the transgene tag to avoid amplification of the endogenous transcript. The second PCR round for these cells was performed as described above. The efficiency of amplification of the two λ5 5′ primers was tested in mature B cells and the standard primer was found to amplify at least as efficiently as the tag primer (data not shown).
Methylation analysis of the λ5 promoter
Genomic DNA from mouse tissues or cells was digested with BamHI and KpnI. Aliquots of the digested DNAs were further digested with the methylation-sensitive HpaII or the methylation-insensitive MspI restriction enzymes or left untreated. The DNA was subjected to gel electrophoresis, blotted and hybridized to probe E (Figure 6A).
Restriction endonuclease sensitivity analysis
Nuclei from pre-B cells, activated B cells and fibroblasts were isolated as described previously (Forrester et al., 1990). Restriction enzyme accessibility was performed on the isolated nuclei using the method described by Boyes and Felsenfeld (1996). Nuclei (2 × 107) were digested with 300 U of PstI for 30 min. Genomic DNA was extracted and digested with SphI and BamHI and resolved by Southern blotting. The restriction fragments were probed with probe E (Figure 6). The probe was stripped from the filter, which was then rehybridized to a probe specific for the pancreatic amylase gene (Schubeler et al., 2000).
Acknowledgments
Acknowledgements
We are grateful to Amanda Fisher and Matthias Merkenschlager for helpful advice and discussions and critical comments on the manuscript. We thank Matthias Merkenschlager for drawing our attention to the possibility that Ikaros might repress transcription through a competitive mechanism similar to that of Krüppel. We also thank Karen Brown for advice on the 3D FISH procedure, Steven Smale for providing antibodies to Ikaros and Aiolos, Tariq Enver and Sylvie Delassus for advice on single-cell PCR, Mauro Santibanez-Koref for advice on statistical analysis and Richard Festenstein for comments on the manuscript. This work was supported by the Leukaemia Research Fund and the Medical Research Council, UK.
References
- Andrulis E., Neiman,A., Zappulla,D. and Sternglatz,R. (1998) Perinuclear localisation of chromatin facilitates transcriptional silencing. Nature, 394, 592–595. [DOI] [PubMed] [Google Scholar]
- Auffray C. and Rougeon,F. (1980) Purification of mouse immunoglobulin heavy-chain messenger RNAs from total myeloma tumour RNA. Eur. J. Biochem., 107, 303–314. [DOI] [PubMed] [Google Scholar]
- Bienz M. and Muller,J. (1995) Transcriptional silencing of homeotic genes in Drosophila. BioEssays, 17, 775–784. [DOI] [PubMed] [Google Scholar]
- Boyes J. and Felsenfeld,G. (1996) Tissue-specific factors additively increase the probability of the all-or-none formation of a hypersensitive site. EMBO J., 15, 2496–2507. [PMC free article] [PubMed] [Google Scholar]
- Brown K.E., Guest,S.S., Smale,S.T., Hahm,K., Merkenschlager,M. and Fisher,A.G. (1997) Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell, 91, 845–854. [DOI] [PubMed] [Google Scholar]
- Brown K.E., Baxter,J., Graf,D., Merkenschlager,M. and Fisher,A.G. (1999) Dynamic repositioning of genes in the nucleus of genes preparing for cell division. Mol. Cell, 3, 207–217. [DOI] [PubMed] [Google Scholar]
- Cobb B.S., Morales-Alcelay,S., Kleiger,G., Brown,K.E., Fisher,A.G. and Smale,S.T. (2000) Targeting of Ikaros to pericentromeric heterochromatin by direct DNA binding. Genes Dev., 14, 2146–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockell M. and Gasser,S. (1999) Nuclear compartments and gene regulation. Curr. Opin. Genet. Dev., 9, 199–205. [DOI] [PubMed] [Google Scholar]
- Csink A.K. and Henikoff,S. (1996) Genetic modification of heterochromatic association and nuclear organization in Drosophila. Nature, 381, 529–531. [DOI] [PubMed] [Google Scholar]
- Delassus S., Titley,I. and Enver,T. (1999) Functional and molecular analysis of hematopoietic progenitors derived from the aorta–gonad–mesonephros region of the mouse embryo. Blood, 94, 1495–1503. [PubMed] [Google Scholar]
- Dernburg A.F., Broman,K.W., Fung,J.C., Marshall,W.F., Philips,J., Agard,D.A. and Sedat,J.W. (1996) Perturbation of nuclear architecture by long-distance chromosome interactions. Cell, 85, 745–759. [DOI] [PubMed] [Google Scholar]
- Dignam J.D., Lebovitz,R.M. and Roeder,R.G. (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian cells. Nucleic Acids Res., 11, 1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon N. and Grosveld,F. (1993) Transcriptional analysis using transgenic animals. In Hames,B.D. and Higgins,S.J. (eds), Gene Transcription: A Practical Approach. IRL Press, Oxford, UK, pp. 153–188.
- Ernst P., Hahm,K., Trinh,L., Davis,J., Roussel,M., Turck,C. and Smale,S. (1996) A potential role for Elf-1 in terminal transferase gene regulation. Mol. Cell. Biol., 16, 6121–6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festenstein R. and Kioussis,D. (2000) Locus control regions and epigenetic modifiers. Curr. Opin. Genet. Dev., 10, 199–203. [DOI] [PubMed] [Google Scholar]
- Festenstein R., Tolaini,M., Corbella,P., Mamalaki,C., Parrington,J., Fox,M., Miliou,A., Jones,M. and Kioussis,D. (1996) Locus control region function and heterochromatin-induced position effect variegation. Science, 271, 1123–1125. [DOI] [PubMed] [Google Scholar]
- Festenstein R., Sharghi-Namini,S., Fox,M., Roderick,K., Tolaini,M., Norton,T., Saveliev,A., Kioussis,D. and Singh,P. (1999) Heterochromatin protein 1 modifies mammalian PEV in a dose and chromosomal context dependent manner. Nature Genet., 23, 457–461. [DOI] [PubMed] [Google Scholar]
- Forrester W.C., Epner,E., Driscoll,M.C., Enver,T., Brice,M., Papayannopoulou,T. and Groudine,M. (1990) A deletion of the human β-globin locus activation region causes a major alteration in chromatin structure and replication across the entire β-globin locus. Genes Dev., 4, 1637–1649. [DOI] [PubMed] [Google Scholar]
- Franke A., DeCamillis,M., Zink,B., Cheng,N., Brock,H.W. and Paro,R. (1992) Polycomb and polyhomeotic are constituents of a multimeric protein complex in chromatin of Drosophila melanogaster. EMBO J., 11, 2941–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgopoulos K., Moore,D. and Derfler,B. (1992) Ikaros, an early lymphoid-specific transcription factor and a putative mediator for T cell commitment. Science, 258, 808–812. [DOI] [PubMed] [Google Scholar]
- Georgopoulos K., Bigby,M., Wang,J., Molnar,A., Wu,P., Winandy,S. and Sharpe,A. (1994) The Ikaros gene is required for the development of all lymphoid lineages. Cell, 79, 143–156. [DOI] [PubMed] [Google Scholar]
- Gotta M., Laroche,T., Formenton,A., Maillet,L., Scherthan,H. and Gasser,S. (1996) The clustering of telomeres and colocalisation with Rap1, Sir3 and Sir4 proteins in wild-type Saccharomyces cerevisiae. J. Cell Biol., 134, 1349–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray S. and Levine,M. (1996) Short-range transcriptional repressors mediate both quenching and direct repression within complex loci in Drosophila. Genes Dev., 10, 700–710. [DOI] [PubMed] [Google Scholar]
- Hahm K., Ernst,P., Lo,K., Kim,G.S., Turck,C. and Smale,S.T. (1994) The lymphoid transcription factor LyF-1 is encoded by specific, alternatively spliced mRNAs derived from the Ikaros gene. Mol. Cell. Biol., 14, 7111–7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahm K. et al. (1998) Helios, a T cell restricted Ikaros family member that quantitatively associates with Ikaros at centromeric heterochromatin. Genes Dev., 12, 782–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich B. and Bird,A. (1998) Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol. Cell. Biol., 18, 6538–6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho S.N., Hunt,H.D., Horton,R.M., Pullen,J. and Pease,L.R. (1989) Site directed mutation by overlap extension using the polymerase chain reaction. Gene, 77, 51–59. [DOI] [PubMed] [Google Scholar]
- Hu M., Krause,D., Greaves,M., Sharkis,S., Dexter,M., Heyworth,C. and Enver,T. (1997) Multilineage gene expression precedes commitment in the hemopoietic system. Genes Dev., 11, 774–785. [DOI] [PubMed] [Google Scholar]
- Kee B.L. and Murre,C. (1998) Induction of early B cell factor (EBF) and multiple B lineage genes by the basic helix–loop–helix transcription factor E12. J. Exp. Med., 188, 699–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopially J., Renold,A., Kim,J. and Georgopoulos,K. (1999) Repression by Ikaros and Aiolos is mediated through histone deacetylation complexes. EMBO J., 18, 3090–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond A. and Earnshaw,W. (1998) Structure and function in the nucleus. Science, 280, 547–553. [DOI] [PubMed] [Google Scholar]
- Laurenson P. and Rine,J. (1992) Silencers, silencing and heritable transcriptional states. Microbiol. Rev., 56, 543–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J., Meehan,R., Henzel,W., Maurer-Fogy,I., Jeppesen,P., Klein,F. and Bird,A. (1992) Purification, sequence and cellular localisation of a novel chromosomal protein that binds to methylated DNA. Cell, 69, 905–914. [DOI] [PubMed] [Google Scholar]
- Lo K., Landau,N.R. and Smale,S.T. (1991) LyF-1, a transcriptional regulator that interacts with a novel class of promoters for lymphocyte-specific genes. Mol. Cell. Biol., 11, 5229–5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundgren M., Chow,CM., Sabbattini,P., Georgiou,A., Minaee,S. and Dillon,N. (2000) Transcription factor dosage affects sequential changes in higher order chromatin structure associated with activation of a heterochromatic gene. Cell, 103, 733–743. [DOI] [PubMed] [Google Scholar]
- Maillet L.B.C., Gotta,M., Marcand,S., Gilson,E. and Gasser,S.M. (1996) Evidence for silencing compartments within the yeast nucleus: a role for telomere proximity and Sir protein concentration in silencer-mediated repression. Genes Dev., 10, 1796–1811. [DOI] [PubMed] [Google Scholar]
- Martensson A. and Martensson,I.L. (1997) Early B cell factor binds to a site critical for λ5 core enhancer activity. Eur. J. Immunol., 27, 315–320. [DOI] [PubMed] [Google Scholar]
- McMorrow T. et al. (2000) Activation of the β globin locus by transcription factors and chromatin modifiers. EMBO J., 19, 4986–4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchers F. (1995) The role of B cell and pre-B cell receptors in development and growth control of the B-lymphocyte cell lineage. In Honjo,T. and Alt,F.W. (eds), Immunoglobulin Genes. Academic Press, London, UK, pp. 33–56.
- Melchers F., Karasuyama,H., Haasner,D., Bauer,S., Kudo,A., Sakaguchi,N., Jameson,B. and Rolink,A. (1993) The surrogate light chain in B-cell development. Immunol. Today, 14, 60–68. [DOI] [PubMed] [Google Scholar]
- Miller O., Schnedl,W., Allen,J. and Erlanger,B. (1974) 5-methylcytosine localised in mammalian constitutive heterochromatin. Nature, 251, 636–637. [DOI] [PubMed] [Google Scholar]
- Milot E. et al. (1996) Heterochromatin effects on the frequency and duration of LCR-mediated gene transcription. Cell, 87, 105–114. [DOI] [PubMed] [Google Scholar]
- Mohandas T., Sparkes,R. and Shapiro,L. (1981) Reactivation of an inactive human X chromosome: evidence for X inactivation by DNA methylation. Science, 211, 393–396. [DOI] [PubMed] [Google Scholar]
- Molnar A. and Georgopoulos,K. (1994) The Ikaros gene encodes a family of functionally diverse zinc finger DNA-binding proteins. Mol. Cell. Biol., 14, 8292–8303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe R. et al. (2000) RAG2:GFP knockin mice reveal novel aspects of RAG2 expression in primary and peripheral lymphoid tissues. Immunity, 11, 201–212. [DOI] [PubMed] [Google Scholar]
- Morgan B., Sun,L., Avitahl,N., Andrikopoulos,K., Gonzales,E., Nichogiannopoulou,A., Wu,P., Neben,S. and Georgopoulos,K. (1997) Aiolos, a lymphoid-restricted transcription factor that interacts with Ikaros to regulate lymphocyte differentiation. EMBO J., 16, 2004–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando V. and Paro,R. (1993) Mapping Polycomb-repressed domains in the bithorax complex using in vivo formaldehyde cross-linked chromatin. Cell, 75, 1187–1198. [DOI] [PubMed] [Google Scholar]
- O’Riordan M. and Grosschedl,R. (1999) Coordinate regulation of B cell differentiation by the transcription factors EBF and E2A. Immunity, 11, 21–31. [DOI] [PubMed] [Google Scholar]
- Pombo A., Cuello,P., Schul,W., Yoon,J.B., Roeder,R.G., Cook,P.R. and Murphy,S. (1998) Regional and temporal specialization in the nucleus: a transcriptionally-active nuclear domain rich in PTF, Oct1 and PIKA antigens associates with specific chromosomes early in the cell cycle. EMBO J., 17, 1768–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolink A., Haasner,D., Nishikawa,S. and Melchers,F. (1993) Changes in frequencies of clonable pre-B cells during life in different lymphoid organs of mice. Blood, 81, 2290–2300. [PubMed] [Google Scholar]
- Ryan R., Singh,P., Schultz,D., Freidman,J. and Rauscher,F. (1999) KAP-1 core-repressor protein interacts and co-localises with heterochromatic and euchromatic HP1 proteins: a potential role for Krüppel-associated box-zinc finger proteins in heterochromatin-mediated silencing. Mol. Cell. Biol., 19, 4366–4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbattini P., Georgiou,A., Sinclair,C. and Dillon,N. (1999) Analysis of mice with single copies and multiple copies of transgenes reveals a novel arrangement for the λ5-VpreB1 locus control region. Mol. Cell. Biol., 19, 671–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurin A., Shiels,C., Williamson,J., Satijn,D., Otte,A., Sheer,D. and Freemont,P. (1998) The human polycomb group complex associates with pericentromeric heterochromatin to form a novel nuclear domain. J. Cell Biol., 142, 887–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubeler D., Francastel,C., Cimbora,D., Reik,A., Martin,D. and Groudine,M. (2000) Nuclear localisation and histone acetylation: a pathway for chromatin opening and transcriptional activation of the human β-globin locus. Genes Dev., 14, 940–950. [PMC free article] [PubMed] [Google Scholar]
- Sigvardsson M., O’Riordan,M. and Grosschedl,R. (1997) EBF and E47 collaborate to induce expression of the endogenous immunoglobulin surrogate light-chain genes. Immunity, 7, 25–36. [DOI] [PubMed] [Google Scholar]
- Small S., Kraut,T., Hoey,R., Warrior,R. and Levine,M. (1991) Transcriptional regulation of a pair-rule stripe in Drosophila. Genes Dev., 5, 827–839. [DOI] [PubMed] [Google Scholar]
- Sun L., Liu,A. and Georgopoulos,K. (1996) Zinc finger proteins modulate Ikaros activity, a molecular control of lymphocyte development. EMBO J., 15, 5358–5369. [PMC free article] [PubMed] [Google Scholar]
- Wang D.M., Taylor,S. and Levy-Wilson,B. (1996) Evaluation of the function of the human apolipoprotein B gene nuclear matrix association regions in transgenic mice. J. Lipid Res., 37, 2117–2124. [PubMed] [Google Scholar]
- Weiler K. and Wakimoto,B. (1995) Heterochromatin and gene expression in Drosophila. Annu. Rev. Genet., 29, 577–605. [DOI] [PubMed] [Google Scholar]