Abstract
CD40, a member of the TNFR-1 receptor family, shares several features with LMP-1, an oncoprotein encoded by Epstein–Barr virus. CD40 and LMP-1 activate transcription by binding to TRAFs, JAK3 and/or TRADD. CD40’s association with CD40L activates signaling. However, LMP-1 signals independently of a ligand but dependently on self-association. We demonstrate that activated CD40 and LMP-1 co-localize in lipid rafts and recruit TRAF3 there, findings consistent with signals of CD40 and LMP-1 being initiated from lipid rafts. To elucidate their signaling, we compared requirements for their aggregation and subcellular localization. Targeting CD40’s monomeric C-terminal signaling domain to lipid rafts activates signaling, as does rendering it trimeric. Addition of both modifications supports signaling more efficiently. Parallel experiments with LMP-1 indicate that targeting the monomeric C-terminal signaling domain of LMP-1 to lipid rafts activates signaling, but trimerizing it does not. Fusing LMP-1’s N-terminus and membrane-spanning domains to CD40’s C-terminus supports signaling more efficiently than CD40 plus ligand or CD40’s trimerized and/or localized derivatives. An activity of LMP-1’s N-terminus and membrane-spanning domains other than trimerization must contribute to its efficient signaling.
Keywords: CD40/lipid rafts/LMP-1/receptor aggregation
Introduction
Subcellular compartmentalization and aggregation are two common modes of regulating signal transduction in eukaryotes. Signal transduction by many receptors that span the plasma membrane is initiated by aggregation of their cytosolic domains, mediated by their extracellular moieties binding multivalent ligands (Cosman, 1994; Onishi et al., 1998). Aggregation of the cytosolic domain unmasks enzymatic activities and/or recruits cytosolic adapter molecules, which induce downstream signaling events (Seed, 1995; Hibi and Hirano, 1998). Signaling is also regulated by localization to lipid rafts. Lipid rafts are specialized domains within the plasma membrane in the order of 50 nm in diameter, which contain high concentrations of glycosphingolipids and cholesterol (Harder and Simons, 1997; Simons and Toomre, 2000). Lipid rafts affect signaling by compartmentalizing a subset of cellular proteins (Brown and London, 1998). Access to this compartment fosters signaling (Simons and Toomre, 2000).
CD40 is a member of the tumor necrosis factor receptor (TNFR) family of receptors, which is involved in B-lymphocyte development (Banchereau et al., 1994; van Kooten and Banchereau, 1997). Activated CD40 and interleukin-4 (IL-4) receptors cooperate to induce proliferation of primary B lymphocytes (Banchereau et al., 1991). B lymphocytes of mice deficient in CD40 fail to switch immunoglobulin classes and produce only IgM antibodies (Kawabe et al., 1994). Consequently, the humoral immune response of these mice is severely compromised. CD40 contributes to B-cell development when it is activated by contact with its trimeric ligand, CD40L, found on the surface of activated T cells (van Kooten and Banchereau, 2000). Activation of CD40 by activated T cells or soluble CD40L trimer results in activation of NF-κB-, AP-1- and STAT-mediated transcription (Berberich et al., 1994; Francis et al., 1995; Huo and Rothstein, 1995; Karras et al., 1997). This signaling is mediated by CD40’s intracellular adapter molecules, tumor necrosis factor receptor associated factors (TRAF1, 2, 3, 5 and 6) and janus activated kinase-3 (JAK3) (Rothe et al., 1995; Ishida et al., 1996; Hanissian and Geha, 1997; Pullen et al., 1999). Structural studies indicate that both CD40L and a peptide derived from the C-terminus of CD40 bound to TRAF2 are trimers (Karpusas et al., 1995; McWhirter et al., 1999). These findings have led to the proposal that CD40L induces CD40 to trimerize, and thereby recruits TRAFs and JAK3 to activate NF-κB-, AP-1- and STAT-mediated transcription. However, like TNFR-1, CD40 may already be trimerized by its preligand association domain, PLAD, and subsequently undergo an allosteric shift upon binding its ligand that triggers signaling (Chan et al., 2000). CD40L has recently been shown to cause CD40 to co-localize with the β subunit of cholera toxin (CTxβ), and to induce TRAF2 and TRAF3 to fractionate in detergent-resistant membranes (DRMs), both of which are characteristics of proteins found in lipid rafts (Hostager et al., 2000; Vidalain et al., 2000). These observations indicate that TRAFs are recruited to lipid rafts after CD40 is activated by CD40L.
LMP-1 behaves as a ligand-independent signaling molecule, which shares multiple features with CD40 (Gires et al., 1997; Hatzivassiliou et al., 1998). LMP-1 is required for the maintenance of proliferation of B lymphocytes infected by Epstein–Barr virus (EBV) (Kilger et al., 1998). It activates NF-κB-, AP-1- and STAT-mediated transcription by binding TRAFs, tumor necrosis factor receptor associated death domain containing protein (TRADD) and JAK3 (Kaye et al., 1996; Brodeur et al., 1997; Izumi and Kieff, 1997; Kieser et al., 1997; Sandberg et al., 1997; Devergne et al., 1998; Eliopoulos and Young, 1998; Gires et al., 1999). LMP-1 can also partially restore the wild-type phenotype of mice deficient in CD40 (Uchida et al., 1999). The structure of LMP-1 differs strikingly from that of CD40 and other related receptors: LMP-1 has six membrane-spanning domains, which aggregate in the plasma membrane and support signaling via its C-terminus in the apparent absence of a ligand (Gires et al., 1997). The N-terminus and membrane-spanning domains of LMP-1 also regulate LMP-1’s signaling by inhibiting cell proliferation and gene expression in a concentration-dependent manner (Kaykas and Sugden, 2000; Sandberg et al., 2000). The extent to which LMP-1 aggregates is unknown; however, with CD40 as a model, LMP-1 would be predicted to be trimeric. LMP-1 has been shown to fractionate with DRMs and recruit TRAF3 there, as has activated CD40, which is consistent with LMP-1 being localized to lipid rafts (Clausse et al., 1997; Ardila-Osorio et al., 1999).
We have found that activated CD40 and LMP-1 co-localize with CTxβ in lipid rafts in EBV-immortalized B cells. Biochemical fractionation indicates that ∼80% of CD40 associates with DRMs when it is activated by CD40L and that the steady-state level of LMP-1 in DRMs is ∼30%. Both of these measurements have been duplicated with a fusion of Gαi to red fluorescent protein (RFP), which localizes in lipid rafts and fractionates in DRMs (Brown and London, 1998; Galbiati et al., 1999). In the presence of activated CD40 or LMP-1, TRAF3 co-localizes with GαiRFP. Similarly, at least 50% of TRAF3 translocates to DRMs in the presence of activated CD40 or LMP-1. A panel of derivatives of the C-terminal signaling domains of CD40 and LMP-1 that were trimerized by fusing them to chloramphenicol acetyltransferase (CAT) (Leslie, 1990), and derivatives of these trimers targeted to lipid rafts by fusing them with the signal for myristoylation and palmitoylation from the Src-like kinase, Yes (Koegl et al., 1994), were assessed quantitatively for their abilities to stimulate NF-κB’s activity. The C-terminal signaling domain of CD40 is activated by trimerization or localization to lipid rafts. Addition of both modifications to CD40’s C-terminal signaling domain is additive, such that it signals more efficiently than either derivative alone. This finding indicates that both trimerization and localization to lipid rafts are important for CD40’s activation of signaling. The C-terminal signaling domain of LMP-1 is not activated by trimerization and is only inefficiently activated by targeting to lipid rafts.
It is striking that although CD40 and LMP-1 aggregate, localize to lipid rafts and engage related signaling molecules, their requirements for initiating signaling differ. These differences are likely to underlie LMP-1’s ability to signal in the apparent absence of a ligand. Fusion of CD40’s C-terminal signaling domain to LMP-1’s N-terminus and membrane-spanning domains generates a constitutively active derivative of CD40 that is more active than wild-type CD40 treated with its ligand, demonstrating the efficiency of this viral mechanism.
Results
CD40, when treated with CD40L, and LMP-1 co-localize in lipid rafts
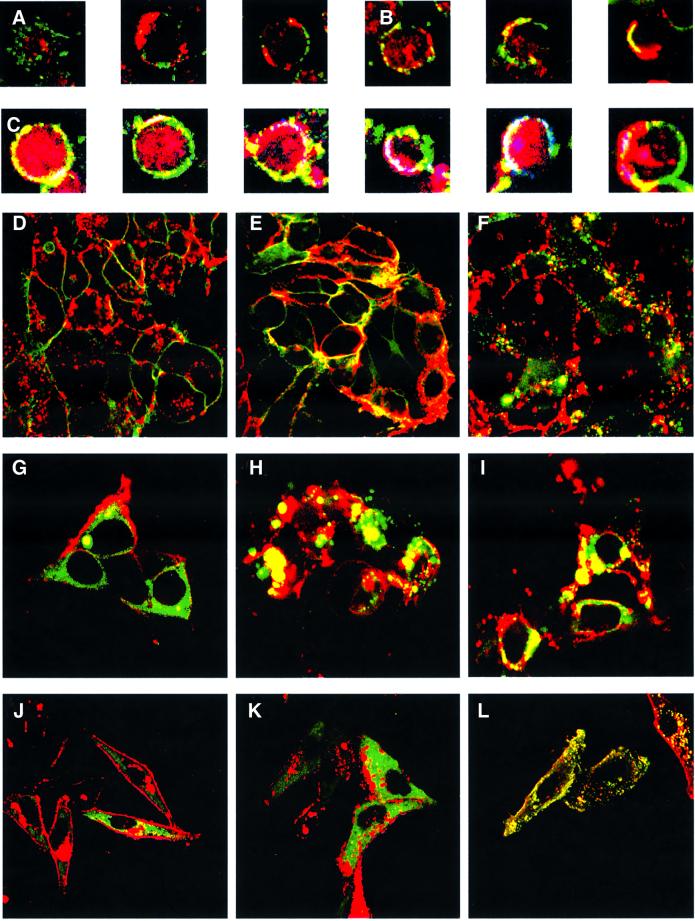
In EBV-immortalized B cells, CD40 and LMP-1 do not appear to co-localize (Figure 1A); however, when these cells are treated with CD40 ligand, an obviously detectable fraction of CD40 is induced to co-localize with the LMP-1 found at the periphery of the cell (Figure 1B). When these same cells are treated both with CD40 ligand and CTxβ, a fraction of CD40 and LMP-1 co-localize with CTxβ in lipid rafts (Figure 1C). Much of the LMP-1 at the periphery of these cells appears to co-localize with CTxβ, but much of it is not at the plasma membrane, and is internal in B-lymphoblastoid cells (Figure 1A–C). One explanation for these subcellular localizations is that CD40 (when treated with its ligand) and LMP-1 signal from lipid rafts. One observation supporting this notion is that CD40 translocates to lipid rafts shortly after the addition of ligand (Figure 1A and B). Because LMP-1 activates signaling in the apparent absence of a ligand, its movements to lipid rafts can not be readily assessed in live cells; however, some LMP-1 is found in these compartments (Figure 1C). We have characterized the subcellular localization of CD40, LMP-1 and TRAF3 to examine their possible co-localization in lipid rafts in order to test the proposition that both CD40 and LMP-1 signal from these sites.
Fig. 1. Confocal microscopy of LMP-1, CD40, their derivatives, and TRAF3. (A) RPMI-1788 cells were stained for CD40 with a fluorescein isothiocyanate (FITC)-conjugated antibody (green) and for LMP-1 with a Texas Red-conjugated antibody (red). (B) RPMI-1788 cells treated with CD40L were stained for CD40 with a FITC-conjugated antibody (green) and for LMP-1 with a Texas Red-conjugated antibody (red). The co-localization of CD40 and LMP-1 is shown in yellow. (C) RPMI-1788 cells treated with CD40L were stained for CD40 with an FITC-conjugated antibody (green), for LMP-1 with a Texas Red-conjugated antibody (red) and for CTxβ with a Cy5-conjugated antibody (cyan). The co-localization of all three proteins is shown in white. (D) Live 293 cells that express CD40GFP (green) were stained with Alexa Fluor 594-conjugated CTxβ (red) to visualize lipid rafts or (E) after treatment with CD40L for 20 min. The majority of CD40 in the absence of CD40L does not co-localize with CTxβ. However, the majority of CD40 co-localizes (yellow) with CTxβ after treatment with CD40L. (F) Live 293 cells that express LMP-1GFP (green) were stained with Alexa Fluor 594-conjugated CTxβ (red) to visualize lipid rafts. The majority of the LMP-1 in the cells does not co-localize with CTxβ. However, a portion of LMP-1 at the membrane co-localizes (yellow) with CTxβ. 293 cells were transfected with vectors encoding TRAF3GFP (green) and GαiRFP (red), and either an empty vector (G), one encoding LMP-1 (H), or one encoding CD40 and treated with CD40L (I), and visualized. The majority of TRAF3GFP in the cells transfected with the empty vector does not co-localize with GαiRFP. However, the majority of TRAF3GFP co-localizes (yellow) with GαiRFP in cells transfected with LMP-1 or with CD40 when treated with CD40L. The subcellular locations of the LMP-1 derivatives were determined by confocal microscopy of Hep2 cells that express GαiRFP (red) and CATLMP-1 (J), MYLMP-1 (K) and MYPALMP-1 (L) stained with FITC-conjugated anti-LMP-1 antiserum (green). The staining pattern of LMP-1 (CYT), CD40 (CYT) and CATCD40 is indistinguishable from CATLMP-1, as is the staining pattern of MYCATLMP-1, MYCD40 and MYCATCD40 from MYLMP-1, and MYPACATLMP-1, MYPACD40 and MYPACATCD40 from MYPALMP-1 (data not shown).
LMP-1’s localization in lipid rafts and its association with DRMs are consistent with it signaling from lipid rafts
The addition of CD40L to 293 cells expressing CD40 or a fusion of CD40 to green fluorescent protein (GFP) yields up to a 10-fold activation of NF-κB-mediated transcription (Table I). Visualization of CD40GFP in live cells treated with or without CD40L for 20 min and CTxβ demonstrates that CD40 co-localizes with CTxβ after treatment with CD40L (Figure 1D and E). After treatment with CD40L, ∼80% of CD40GFP is found in the DRM fraction of cells (Table I). This movement corresponds to a 4-fold increase in the amount of CD40GFP found in the DRM fraction of cells after treatment with CD40L. In an EBV-positive lymphoblastoid cell line, 721, which constitutively expresses LMP-1, treatment with CD40L increases the fraction of CD40 in DRMS 2-fold (Table I). These findings indicate that CD40L induces CD40 both to translocate to lipid rafts and to activate NF-κB-mediated transcription.
Table I. Biochemical fractionationa of CD40, LMP-1 and TRAF3 in LCLs and 293 cells.
| Protein detected + protein co-expressed | Treated with CD40Lf | % soluble fraction | % DRM fraction | Fold activation of NF-κBg |
|---|---|---|---|---|
| CD40b | – | 85 ± 4 | 16 ± 3 | 1 |
| CD40b | + | 21 ± 5 | 81 ± 6 | 10 ± 4 |
| CD40 LCLc | – | 72 ± 8 | 28 ± 4 | ND |
| CD40 LCLc | + | 45 ± 10 | 55 ± 15 | ND |
| LMP-1c | – | 71 ± 6 | 29 ± 3 | 95 ± 10 |
| LMP-1 LCLd | – | 70 ± 7 | 30 ± 5 | ND |
| TRAF3e | – | 90 ± 10 | 10 ± 7 | ND |
| TRAF3e + CD40 | – | 85 ± 6 | 15 ± 7 | ND |
| TRAF3e + CD40 | + | 34 ± 4 | 64 ± 8 | ND |
| TRAF3e + LMP-1 | – | 26 ± 6 | 81 ± 9 | ND |
aBiochemical fractionation was performed on cell lysates expressing various proteins shown in the first column by separation on a nycodenz step gradient into soluble and DRM fractions as described in Materials and methods. Lysates of 293 cells that express CD40GFPb and GFPTRAF3e stably were co-transfected with the expression vectors shown, separated and probed with anti-GFP antisera. Lysates of the 721 LCL were separated and probed for CD40c with anti-CD40 antisera or for LMP-1d with anti-LMP-1 antisera. fCells were treated with 10 µg/ml muCD8αCD40L for 20 min prior to harvesting. c293 cells transiently transfected with an expression vector encoding LMP-1 or LCLs that express it were lysed and separated on a nycodenz step gradient and probed with anti-LMP-1 antisera. g293 cells were transfected with an NF-κB–luciferase reporter, expression vectors encoding LMP-1 or CD40, and treated with or without CD40L as indicated, and the fold induction of luciferase was measured as described in Materials and methods. All data represent the average of at least three experiments ± SD.
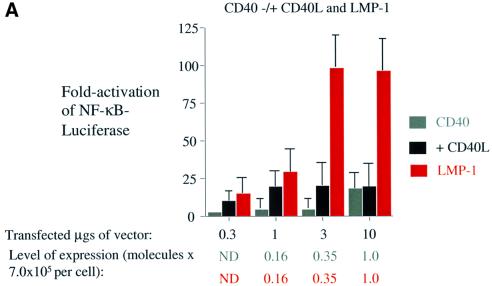
Because translocation of CD40 into lipid rafts and its association with DRMs correlate with its signaling, we tested whether LMP-1’s signaling correlates with its association with DRMs and whether it co-localizes with markers for lipid rafts. Transfection or induction of LMP-1 in 293 cells leads to up to 100-fold activation of NF-κB-mediated transcription (Figure 2A). At levels of expression of LMP-1 >2.0 × 105 molecules per cell, it inhibits gene expression (Sandberg et al., 2000). We have therefore characterized signaling by LMP-1 and its derivatives when they are expressed at levels of <2.0 × 105 molecules per cell. Visualization of LMP-1 fused to GFP in live 293 cells demonstrates that a portion of LMP-1 localizes to the plasma membrane of cells with CTxβ (Figure 1F). Surprisingly, much of LMP-1 appears to be internal and does not co-localize with CTxβ (Figure 1F). From studies with cells in which LMP-1 is expressed conditionally, LMP-1’s localization appears to be dynamic with only a fraction of the total cellular LMP-1 residing at any one time in lipid rafts (N.Lam, personal communication). The remaining fraction of LMP-1 not co-localized with CTxβ is presumably trafficking to lipid rafts or elsewhere.
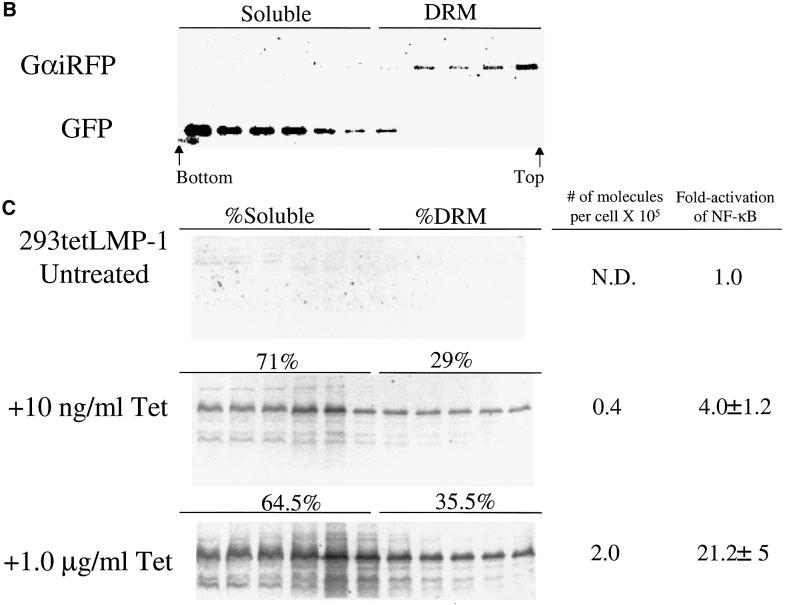
Fig. 2. LMP-1 activates NF-κB-mediated transcription more efficiently than does CD40 when treated with ligand, and a portion of LMP-1 associates with DRMs. (A) Assay for NF-κB activity in cells transfected with vectors expressing: CD40 (gray); treated with CD40L (black); and LMP-1 (red). 293 cells were transfected with the amount of expression vectors for CD40 or LMP-1 indicated, an NF-κB-responsive firefly luciferase reporter, an expression vector for Renilla luciferase or pEGFPN-1, and brought up to equal concentrations with pSG5. All transfections were normalized to Renilla luciferase levels or to the number of GFP-positive cells. The fold activation of firefly luciferase over cells transfected with pSG5 alone is shown. The relative light units (RLUs) in these experiments varied from ∼4.0 × 104 to up to ∼2.0 × 105 in cells transfected with empty vector to ∼1.0 × 107 in the presence of expression vectors for CD40 or LMP-1. The data represent the average ± SD for three separate experiments with two measurements each. The number of molecules per cell of CD40 and LMP-1 was calculated from known amounts of MYPACATCD40 or GSTLMP-1 assayed on the same blot as described in Materials and methods. ND, not detectable. (B) Biochemical fractionation of cells to separate their soluble and DRM fractions. 293 cells that express LMP-1 under the control of tetracycline were transfected with expression vectors encoding GFP and GαiRFP, extracted with Triton X-100, and separated on a nycodenz flotation gradient as described in Materials and methods. Eleven fractions were collected from the bottom of the gradient and subjected to western blot analysis. The fractions were loaded from left (which corresponds to the bottom fraction of the gradient) to right (which corresponds to the top fraction of the gradient). The DRM fraction of the cells is defined by the presence of GαiRFP and consists of the top five fractions. The soluble fraction of the cells is defined by the presence of GFP and consists of the bottom six fractions. (C) 293 cells that express LMP-1 under the control of tetracycline were left untreated, or treated with 10 ng/ml or 1 µg/ml tetracycline for 48 h, fractionated, and probed with anti-LMP-1 antiserum. The number of molecules per cell was calculated from the known amount of GSTLMP-1 assayed on the same blot as described in Materials and methods. The percentage of LMP-1 in the DRM fraction is indicated and was determined by quantifying the amount of LMP-1 by PhosphorImager analysis in the fractions that contained GαiRFP. One representative blot of three is shown. The fold activation of NF-κB in these cells was measured in parallel by introducing into them an NF-κB-responsive luciferase reporter and measuring luciferase activity as described in Materials and methods.
To extend and quantify our visual observations, we tested whether LMP-1 is associated with DRMs and whether this association correlates with its signaling. 293 cells that express LMP-1 under the control of tetracycline were induced to express LMP-1 at variable levels, extracted with Triton X-100 at 4°C, and subjected to fractionation on a nycodenz step gradient. Fractions were collected and probed for proteins that are known to reside in lipid rafts. As expected, both a fusion of Gαi to RFP and Lyn were concentrated in the top, less dense five fractions of the gradient and were used to define the DRM fraction of cells (Figure 2B and data not shown) (Cheng et al., 1999; Galbiati et al., 1999). GFP, which is found in the cytosol, was concentrated in the bottom, more dense seven fractions of the gradient, and was used to define the soluble fraction of cells (Figure 2B). The level of NF-κB in the tetracycline-inducible cell line is activated maximally to 20-fold when 1.0 µg/ml tetracycline is added, and corresponds to 2.0 × 105 molecules of LMP-1 per cell (Figure 2C). LMP-1 can be induced to an intermediate level of 4.0 × 104 molecules per cell, which corresponds to a 4-fold activation of NF-κB when 10 ng/ml tetracycline is added (Figure 2C). Transient transfection of an expression vector for LMP-1 gives 100-fold activation of NF-κB-mediated transcription (Figure 2A). The difference in the NF-κB activated in these two systems is likely to reflect the presence of low levels of LMP-1 expression in the tetracycline-inducible cells and a higher background of NF-κB activation in the absence of tetracycline. Indeed, low-level expression of LMP-1 can be detected in the absence of tetracycline (Figure 2C). However, this level of LMP-1 is below that needed for quantification. 293 cells that contain LMP-1 under the control of tetracycline were left uninduced, induced to an intermediate level or induced maximally. The activation of NF-κB-mediated transcription in these cells was measured and their DRMs isolated. The percentage of LMP-1 in the DRM fraction of cells was ∼30% irrespective of the level of LMP-1 expressed or NF-κB induced (Figure 2C). The absolute amount of LMP-1 in the DRM fraction therefore increases proportionally with its increased expression and correlates with the increased stimulation of NF-κB’s activity. This same fraction of LMP-1 is also found in DRMs in EBV-positive lymphoblastoid cells, which express LMP-1 constitutively (Table I).
CD40 treated with ligand and LMP-1 recruit their signaling molecules to rafts
CD40 and LMP-1 both signal by interacting with TRAF molecules. If lipid rafts are the site from which signaling of CD40 and LMP-1 initiates, then TRAFs should be recruited to lipid rafts in cells that express CD40 when treated with CD40L and in cells that express LMP-1. To test this proposition, we asked whether a fusion of TRAF3 with GFP, which interacts with the C-terminal signaling domains of CD40 and LMP-1, is recruited to rafts and associates with DRMs when signaling of CD40 or LMP-1 is initiated.
Visualization of TRAF3GFP and GαiRFP in 293 cells demonstrates that these two molecules do not co-localize (Figure 1G). However, if CD40 is expressed in the cells and treated with CD40L or if LMP-1 is expressed in them, TRAF3GFP co-localizes with GαiRFP (Figure 1H and I). Because GαiRFP is found in lipid rafts, where it co-localizes with CTxβ (Galbiati et al., 1999; N.Lam, personal communication), it appears that both activated CD40 and LMP-1 recruit TRAF3 to lipid rafts.
To extend and quantify these visual results, we subjected cells that express TRAF3GFP stably and were transfected with an empty vector, one encoding CD40 and treated with or without CD40 ligand, or one encoding LMP-1, to extraction with 1% Triton X-100 at 4°C to isolate DRMs. Approximately 10% of TRAF3GFP is associated with the DRM fraction of cells transfected with an expression vector for CD40 or with an empty expression vector (Table I). However, if the cells that are transfected with CD40 are treated with CD40L, then at least 30% of the TRAF3GFP becomes associated with the DRM fraction of cells (Table I). If the cells that express TRAF3GFP are transfected with an expression vector for LMP-1, then at least 40% of the TRAF3GFP becomes associated with the DRM fraction of cells (Table I). Because the transfection efficiency of the cells is in the order of 50%, as measured by uptake of GαiRFP, the actual percentage of TRAF3GFP that is relocalized to DRMs in the presence of CD40 treated with its ligand or LMP-1 is 60 and 80%, respectively. Taken together, these observations demonstrate that activated CD40 and LMP-1 recruit TRAF3 to lipid rafts, and support the proposal that signaling of CD40 and LMP-1 initiates from lipid rafts.
Localization into rafts and trimerization of the C-terminal signaling domain of CD40 activate its signaling
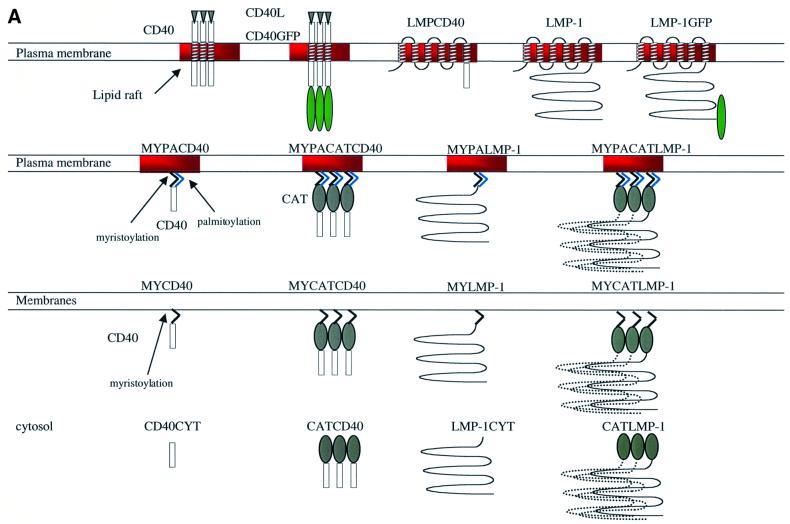
Treatment of 293 cells that express on average <5.0 × 105 molecules of CD40 per cell with CD40L induces NF-κB-mediated transcription (Figure 2A). At levels of CD40 that are >5.0 × 105 molecules per cell, CD40 becomes ligand independent and activates NF-κB in the absence of its ligand (Figure 2A). We therefore characterized the signaling of CD40 and its derivatives when they were expressed at levels <5 × 105 molecules per cell. Treatment of CD40 with CD40L induces it to translocate to lipid rafts, to become associated with DRMs and to activate NF-κB-mediated transcription. To test whether CD40’s trimerization and/or localization to lipid rafts activates its signaling, we have generated and analyzed derivatives of CD40’s C-terminal signaling domain that are trimerized and/or localized to the cytosol, to intracellular membranes and to lipid rafts (Figure 3A).
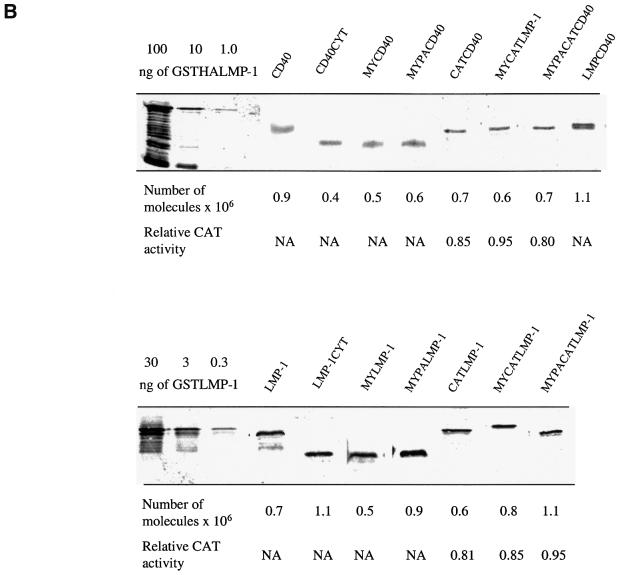
Fig. 3. The proposed structure, proposed location of signaling, and expression level of wild-type CD40, wild-type LMP-1 and their derivatives. (A) Wild-type CD40 with its ligand, CD40L, is depicted as a trimer and localizes in lipid rafts (red) after binding CD40L. CD40GFP is a fusion between full-length CD40 (amino acids 1–277), a 10-amino-acid linker and GFP (amino acids 1–240) (green ellipse). Wild-type LMP-1 (amino acids 1–386) is depicted in lipid rafts as a monomer because the extent to which LMP-1 aggregates is unknown. LMPGFP is a fusion of wild-type LMP-1, a 10-amino-acid linker and GFP (amino acids 1–240). LMPCD40 is a fusion of LMP-1’s N-terminus and transmembrane-spanning domains (amino acids 1–190) fused to the C-terminus of CD40 (amino acids 215–277). LMP-1CYT and CD40CYT consist of the C-terminal signaling domains of LMP-1 (amino acids 182–386) and CD40 (amino acids 215–277), respectively, and are depicted as monomers located in the cytosol. CATCD40 and CATLMP-1 consist of CAT (amino acids 1–220) (gray ellipse) fused to CD40CYT and LMP-1CYT, and are depicted as trimers located in the cytosol. MYPACD40 and MYPALMP-1 consist of the 10-amino-acid N-terminal myristoylation (MY) sequence (black membrane anchor) and palmitoylation (PA) sequence (light blue membrane anchor) derived from Yes fused to CD40CYT and LMP-1CYT. MYPACATCD40 and MYPACATLMP-1 consist of the 10-amino-acid myristoylation and palmitoylation sequence derived from Yes fused to CATCD40 and CATLMP-1, and are depicted as trimers located in lipid rafts. MYCD40 and MYLMP-1 consist of the 10-amino-acid N-terminal myristoylation sequence derived from Src fused to CD40CYT and LMP-1CYT, and are depicted as monomers located in all cellular membranes. MYCATCD40 and MYCATLMP-1 consist of the N-terminal 10-amino-acid myristoylation sequence derived from Src protein to CATCD40 and CATLMP-1, and are depicted as trimers located in all cellular membranes. All derivatives except wild-type CD40, LMP-1, CATLMP-1 and CATCD40 contain HA epitope tags. (B) Quantitative western blots and CAT assays were performed to measure the expression and CAT activity of CD40, LMP-1 and their derivatives after 10 µg of the expression vectors encoding them were introduced into 293 cells as described in Materials and methods. The number of molecules per cell of CD40 and LMPCD40 was calculated from known amounts of MYPACATCD40 assayed on the same blot. The number of molecules per cell of MYPACATCD40 and the other fusions, which contain an HA epitope tag, was calculated from known amounts of GSTHALMP-1 assayed on the same blot. The number of molecules per cell of LMP-1 and its derivatives, which contain LMP-1-epitopes, was calculated from known amounts of GSTLMP-1 assayed on the same blot. CAT activity was measured by quantifying the percentage of [14C]chloramphenicol acetylated in extracts containing equal numbers of molecules of CAT or its derivatives as described in Materials and methods. The levels shown are relative to wild-type CAT, which is set at 1.0. NA, not applicable.
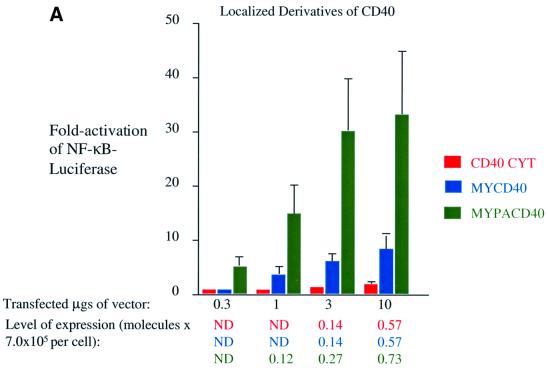
The localization of the C-terminal signaling domain of CD40, CD40CYT, is consistent with it being in the cytosol (data not shown). CD40CYT at most gives a 3-fold activation of NF-κB, even when it is expressed at levels at which wild-type CD40 is ligand independent (Figure 4A). Targeting the C-terminal signaling domain of CD40 to intracellular membranes by addition of sequences derived from Src for myristoylation (MY) increases its activation of NF-κB-mediated transcription. MYCD40’s localization is consistent with it being in intracellular membranes (data not shown). It induces a 5-fold activation of NF-κB-mediated transcription, which is ∼20% the activity of wild-type CD40 treated with CD40 ligand, at concentrations at which CD40’s activation of NF-κB is ligand dependent (Figure 4A). Targeting the C-terminal signaling domain of CD40 to lipid rafts by addition of sequences derived from Yes for myristoylation and palmitoylation (MYPA) increases the C-terminal signaling domain’s activation of NF-κB-mediated transcription. MYPACD40’s localization is consistent with it being in lipid rafts (data not shown). It activates NF-κB-mediated transcription to similar levels to wild-type CD40 in the presence of ligand at concentrations at which its activation of NF-κB is ligand dependent (Figure 4A). These data indicate that localization of CD40 to lipid rafts contributes to its signaling.
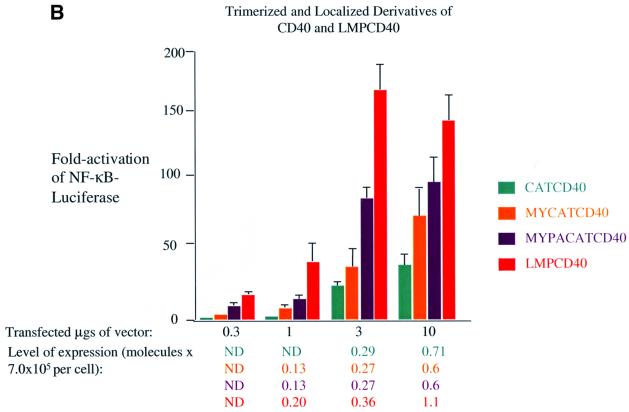
Fig. 4. Localization to lipid rafts and trimerization of CD40’s C-terminal signaling domains activate NF-κB-mediated transcription. Induction of NF-κB activity was measured as described in the legend to Figure 2 and Materials and methods in 293 cells transfected with vectors expressing: (A) CD40CYT (red), MYCD40 (blue) and MYPACD40 (green); (B) CATCD40 (teal), MYCATCD40 (orange), MYPACATCD40 (purple) and LMPCD40 (orange–red). The RLUs in these experiments varied from ∼4.0 × 104 up to ∼2.0 × 105 in cells transfected with empty vector to ∼4.0 × 107 in the presence of expression vectors for derivatives of CD40. The level of expression of each protein was determined as described in the legend to Figure 3 and Materials and methods.
The PLAD of CD40 favors its aggregation in the plasma membrane (Chan et al., 2000). Co-crystallization of TRAF2 with a peptide derived from its binding domain on CD40 yields a trimeric complex (McWhirter et al., 1999). The crystal structure of CD40L is also a trimer (Karpusas et al., 1995). These structural data indicate that trimerization of CD40 and an allosteric alteration mediated by CD40L induce its signaling. To test whether trimerization of CD40 is sufficient to activate its signaling, we fused CD40’s C-terminal signaling domain to CAT. CAT is a trimeric enzyme and its fusion to TNFR-1 has been demonstrated to activate TNFR-1’s induction of apoptosis and NF-κB-mediated transcription (Vandevoorde et al., 1997). Because localization of CD40 to lipid rafts is important for its signaling, we also localized CATCD40 to intracellular membranes in general and to lipid rafts in particular. Trimerization of CD40’s C-terminal signaling domain in the cytosol activates NF-κB-mediated transcription. The location of CATCD40 is in the cytosol, as predicted (data not shown). CATCD40 induces ∼20-fold activation of NF-κB-mediated transcription, which is slightly less than the levels of NF-κB activated by wild-type CD40 treated with CD40L, under conditions where the expression of CD40’s activation of NF-κB is ligand dependent (Figure 4B). Targeting of CATCD40 to intracellular membranes further increases its ability to activate NF-κB-mediated transcription. MYCATCD40 activates the levels of NF-κB ∼25-fold, which is similar to wild-type CD40’s activation of NF-κB-mediated transcription when it is present at levels where it is dependent on ligand for signaling (Figure 4B). Targeting of CATCD40 to lipid rafts further increases its activation of NF-κB-mediated transcription. MYPACATCD40 activated NF-κB-mediated transcription ∼75-fold, which is a higher level than that activated by wild-type CD40 when its activation is ligand dependent (Figure 4B). Interestingly, a fusion of LMP-1’s N-terminus and transmembrane-spanning domains to CD40’s C-terminus activates signaling more efficiently than CD40 when treated with ligand or its derivatives. LMPCD40 activated NF-κB-mediated transcription ∼150-fold, which is more than any other derivative of CD40 (Figure 4B). This fusion protein inhibits gene expression when expressed at levels >2.0 × 105 molecules per cell (Figure 4B), which is characteristic of molecules containing LMP-1’s N-terminus and membrane-spanning domains (Sandberg et al., 2000). These data indicate that both trimerization and localization of CD40’s C-terminal signaling domain to lipid rafts activate CD40’s signaling. The data also demonstrate that the N-terminus and transmembrane-spanning domains of LMP-1, when fused to the C-terminus of CD40, activate CD40’s signaling more efficiently than does CD40L.
Localization of the C-terminal signaling domain of LMP-1 to lipid rafts, but not its trimerization, activates its signaling
LMP-1 binds TRAFs and TRADD, and efficiently activates NF-κB-mediated transcription. At 1–2 × 105 molecules per cell, which is near its physiologically expressed levels, LMP-1 activates a 100-fold increase in NF-κB-mediated transcription in 293 cells (Figure 2A). We generated derivatives of LMP-1’s C-terminal signaling domain, similar to those of CD40, which are localized to the cytosol, intracellular membranes and lipid rafts and/or trimerized (Figure 3A).
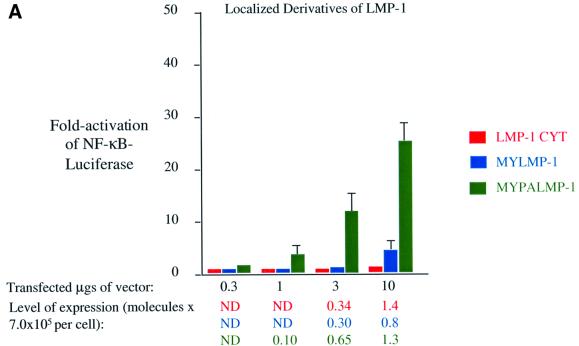
When expressed in cells, the C-terminal signaling domain of LMP-1 is localized to the cytoplasm, as predicted (data not shown). LMP-1CYT, similarly to CD40CYT, gives only minimal activation of NF-κB even when it is expressed at high levels in cells (Figure 5A). Targeting the C-terminal signaling domain of LMP-1 to intracellular membranes by adding the sequences derived from Src for myristoylation increases its ability to activate NF-κB-mediated transcription slightly. MYLMP-1 is localized to intracellular membranes in cells, as predicted (Figure 1K). It activates an ∼2-fold increase in NF-κB-mediated transcription when it is expressed at physiological levels, which is 2% the activity of wild-type LMP-1 (Figure 5A). Targeting the C-terminal signaling domain of LMP-1 to lipid rafts by addition of sequences derived from Yes increases its activation of NF-κB-mediated transcription. MYPALMP-1 is localized to lipid rafts, as predicted (Figure 1L). It activates an ∼15-fold increase in NF-κB-mediated transcription when expressed at physiological levels, which is 15% the activity of wild-type LMP-1 (Figure 5A). These data indicate that localization of LMP-1 to lipid rafts contributes to its signaling. Neither the structure nor the extent to which LMP-1 aggregates is known. On one hand, it seems likely that LMP-1 signals as a trimer because of its similarities to CD40. On the other hand, both its ligand independence and multiple membrane-spanning domains render its structure in the plasma membrane enigmatic. We tested whether fusion of LMP-1’s C-terminal signaling domain to CAT activates NF-κB-mediated transcription, and whether targeting of this CATLMP-1 fusion to intracellular membranes and lipid rafts further activates its signaling.
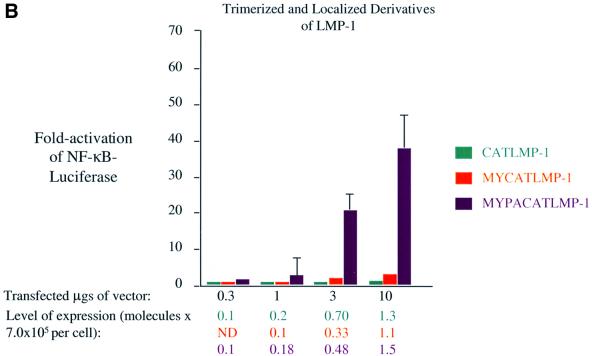
Fig. 5. Localization to lipid rafts but not trimerization of LMP-1’s C-terminal signaling domains activates NF-κB-mediated transcription. Induction of NF-κB activity was measured as described in the legend to Figure 2 and Materials and methods in 293 cells transfected with vectors expressing: (A) LMP-1CYT (red), MYLMP-1 (blue) and MYPALMP-1 (green); (B) CATLMP-1 (teal), MYCATLMP-1 (orange) and MYPACATLMP-1 (purple). The RLUs in these experiments varied from ∼4.0 × 104 to ∼2.0 × 105 in cells transfected with empty vector to up to ∼2.25 × 107 in the presence of expression vectors for derivatives of LMP-1. The level of expression of each protein was determined as described in the legend to Figure 3 and Materials and methods.
Unexpectedly, CATLMP-1, which localizes to the cytosol as predicted (Figure 1J), does not efficiently activate NF-κB-mediated transcription. Even when CATLMP-1 is expressed at higher than physiological levels, it only activates NF-κB-mediated transcription 4-fold, which is 4% that of wild-type LMP-1 (Figure 5B). Targeting CATLMP-1 to intracellular membranes by addition of the sequence for myristoylation from Src to CATLMP-1 slightly increases its ability to activate NF-κB-mediated transcription, but only to the level of MYLMP-1 (Figure 5B). Targeting CATLMP-1 to lipid rafts by addition of the sequences for myristoylation and palmitoylation from Yes increases its activation of NF-κB-mediated transcription, but again only to the level of MYPALMP-1 (Figure 5B). These data indicate that trimerization of the C-terminal signaling domain of LMP-1 does not contribute to its signaling, so long as the CAT fusions are, in fact, trimeric. We therefore tested each of the CATCD40 and CATLMP-1 derivatives for CAT activity. The CAT fusion proteins acetylate chloramphenicol to the same level as wild-type CAT (Figure 3B). Because CAT is only enzymatically active as a trimer and all of the fusion proteins are expressed at similar levels in cells (Figure 3B), these data demonstrate that each of the fusion proteins forms trimers in cells. The measurements of activation of NF-κB by these derivatives of LMP-1 (Figure 5) indicate that one C-terminus of LMP-1 signals as efficiently when fused to CAT and homed to lipid rafts as when monomeric and homed to lipid rafts. Fusion to CAT, therefore, neither activates nor inhibits LMP-1’s signaling. Derivatives of LMP-1 competent to signal, therefore, do not share the trimeric structure formed by CD40 and its derivatives. LMP-1’s structure not only allows it to home to lipid rafts in the absence of a ligand, but also to bind TRAFs, TRADD and JAK3 in a particularly efficient signaling complex.
Discussion
CD40 and LMP-1 activate similar signal transduction pathways and share many characteristics, including their subcellular localization. Our study demonstrates that lipid rafts are the site where signaling of CD40 and LMP-1 is initiated, as identified by their localizing TRAF3 to that site. Consistent with this conclusion, both activated CD40 and LMP-1 co-localize with two markers known to reside in lipid rafts, and also induce TRAF3 to co-localize with these markers. Approximately 80% of activated CD40 and ∼30% of LMP-1 are associated with DRMs, and >60% of TRAF3 becomes associated with DRMs in the presence of activated CD40 and LMP-1 in 293 cells. Localization of signaling domains of CD40 and LMP-1 to lipid rafts induces them to signal, albeit to different levels. The structural requirements for C-terminal signaling domains of CD40 and LMP-1 to signal differ. The trimeric structure that CAT imposes when fused to CD40’s C-terminus induces CD40’s signaling. CATCD40 can signal from the cytosol, but signals more efficiently when homed to lipid rafts. However, a parallel construction, CATLMP-1, does not signal. The N-terminus and membrane-spanning domains of LMP-1 provide a structure that both targets the C-terminus of LMP-1 to lipid rafts and supports its signaling in the absence of a ligand. These observations lead to a model underscoring the importance in lipid rafts for CD40 and LMP-1 to signal.
CD40 and LMP-1 signal by relocalization of TRAFs: the ‘shuttling’ model
In the absence of its ligand, CD40 is found in the plasma membrane as a trimer as a result of its PLAD. Trimeric CD40L induces a conformational change in the CD40 complex that increases its affinity for TRAFs and lipid rafts. The entire receptor–TRAF complex shuttles to lipid rafts. How TRAFs’ binding to CD40’s C-terminus might induce downstream signaling is unclear. One tantalizing possibility is that the environment in lipid rafts fosters signaling. For example, inhibitors of TRAFs, such as MIP-3T (Ling and Goeddel, 2000), could be excluded from lipid rafts, or, on the other hand, molecules required for TRAF-mediated signaling, such as NF-κB inducing kinase (NIK) (Song et al., 1997), could be concentrated in lipid rafts. This model proposes that it is the relocalization of TRAFs to lipid rafts that induces signaling by juxtaposing TRAFs with their targets. Consistent with this model, relocalization of TRAF3 to intracellular membranes activates AP-1-mediated transcription (Dadgostar and Cheng, 2000). TRAF3 has not been targeted to lipid rafts, but given our observation it is likely that targeting of TRAF3 to lipid rafts would signal more efficiently than the derivative targeted to intracellular membranes.
LMP-1 activates signaling in the absence of ligand and therefore its movement to lipid rafts can not be readily detected in live cells. However, LMP-1’s localization is consistent with observing its entire life cycle from synthesis, movement to lipid rafts and degradation. One-third of LMP-1 appears to signal from lipid rafts at any one time in cells. It is possible that the remaining two-thirds of LMP-1 is trafficking to or from these signaling compartments in lipid vesicles. This model proposes that after LMP-1’s synthesis, it translocates to the plasma membrane where it acquires a ‘modification’ that increases its affinity for TRAFs, TRADD and lipid rafts. This ‘modification’ is intrinsically encoded by LMP-1’s N-terminus and membrane-spanning domains. We hypothesize that this ‘modification’ results from LMP-1’s peculiar structure combining the self-association of the PLAD with the allosteric changes mediated by CD40L. This structure promotes efficient binding to TRAFs at the plasma membrane and movement to lipid rafts. LMP-1 aggregates spontaneously and moves spontaneously to lipid rafts to localize the TRAFs it binds there. It is fascinating that while CATCD40 fusions signal and CATLMP-1 fusions do not, a fusion of LMP-1’s N-terminus and membrane-spanning domains fused to CD40’s C-terminus signals more efficiently than CD40 plus its ligand, i.e. LMP-1’s modification also activates CD40 efficiently. Elucidating this modification is likely to illuminate the intricacies of signaling by members of the TNFR family.
The role of lipid rafts and TRAFs in ubiquitylation and internalization of CD40 and LMP-1
Shortly after binding to their ligand, some receptors that span the plasma membrane are ubiquitylated and internalized from the plasma membrane. In the case of epidermal growth factor receptor (EGFR), ubiquitylation is required for its signaling (Stang et al., 2000). Alternatively, ubiquitylation may trigger internalization and degradation, and play a role in down-regulating receptor signaling (Hicke, 1997, 1999). Lipid rafts may not only coordinate signaling of CD40 and LMP-1, but also their ubiquitylation, internalization and subsequent down-regulation. TRAFs contain ring finger motifs, which can act as ubiquitin ligases, and TRAF6 has been shown to be a ubiquitin ligase (Deng et al., 2000). Concentrating TRAFs in lipid rafts with CD40 and LMP-1 may induce their ubiquitylation, internalization and degradation. For instance, ubiquitin-activating and -conjugating enzymes could reside in lipid rafts; CD40 and LMP-1 could relocalize TRAFs there to induce ubiquitylation. In support of this notion, the same regions of LMP-1, its N-terminus and transmembrane-spanning domains, are required for its targeting to lipid rafts, ubiquitylation, rapid internalization from the plasma membrane and short half-life (Martin and Sugden, 1991a,b; Aviel et al., 2000). Although CD40’s ubiquitylation, internalization and degradation have not been studied, it seems likely that homing to lipid rafts and binding TRAFs contribute to these processes for CD40 too. Coupling the initiation of signaling by a receptor with its degradation in a single compartment would allow temporal definition of ligand-dependent signaling. Whatever the intricacies of CD40’s signaling are, LMP-1 appears to have appropriated some facets of them and evolved to use them particularly efficiently.
Materials and methods
Cell culture
293, a human embryonic kidney cell line, was obtained from the American Type Culture Collection (ATCC) (CRL 1573) and grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Cells that express LMPGFP and CD40GFP stably were generated by selecting clones of 293 cells transfected with a vector expressing either of the proteins and resistance to puromycin. Cells that express TRAF3GFP stably were generated by selecting clones of 293 cells that were transfected with a vector that expresses TRAF3GFP and resistance to neomycin. All cells were transfected via calcium phosphate precipitation (Graham et al., 1977). The puromycin- and neomycin-resistant clones were selected and maintained in DMEM, 10% FBS and 1 µg/ml puromycin or 750 µg/ml neomycin. The derivatives of 293 cells that express LMP-1 under the regulation of tetracycline were described previously (Kaykas and Sugden, 2000). Hep2 is a human epithelial cell line obtained from the ATCC (CCL 23) and grown in DMEM supplemented with 10% calf serum. RPMI-1788, 721 and GM-2783 are EBV-positive lymphoblastoid cell lines, which express EBV’s latent genes, including LMP-1, and were grown in RPMI 1640 supplemented with 10% FBS. All cell culture media were supplemented with 200 U/ml penicillin and 200 µg/ml streptomycin, and all cells were grown at 37°C in a humidified 5% CO2 atmosphere.
Recombinant DNA constructs
p1958 is an SV40 expression vector derived from pSG5 (Stratagene) modified to have an expanded multiple cloning site, and was used as an empty vector to normalize all transfections to the same concentration of DNA. The wild-type LMP-1 expression vector (p1990) was generated by inserting the LMP-1 cDNA derived from the B958 strain of EBV into p1958. All of the following expression vectors were generated from p1990: LMP-1CYT (p2525), MYLMP-1 (p2524), MYPALMP-1 (p2484), CATLMP-1 (p2411), MYCATLMP-1 (p2409) and MYPACATLMP-1 (p2391). They were generated by replacing the N-terminus and membrane-spanning domains of LMP-1 (amino acids 1–180) with combinations of commercially synthesized DNAs and PCR products encoding the following inserts: a hemagglutinin (HA) epitope tag (YPYDVPDYA), the sequence for myristoylation (MY) from Src (MGSSKSKPKD), the sequence for myristoylation and palmitoylation (MYPA) from Yes (MGCIKSKENK) and/or the sequence for CAT from pMXCAT provided by W.Fiers (Vandevoorde et al., 1997). The expression vector encoding LMPCD40 (p2060) encodes LMP-1 (amino acids 1–190) fused to CD40 (amino acids 215–277) under the control of the SV40 early promoter and was provided by W.Hammerschmidt (Gires et al., 1997). The expression vector for wild-type CD40 (p2534) was generated by cloning the human cDNA for CD40 provided by W.Hammerschmidt (Gires et al., 1997) into p1958. All of the following expression vectors were generated from p2534: CD40CYT (p2526), MYCD40 (p2523), MYPACD40 (p2519), CATCD40 (p2449), MYCATCD40 (p2435) and MYPACATCD40 (p2434). They were generated by replacing the N-terminus and membrane-spanning domain of CD40 (amino acids 1–214) with commercially synthesized DNAs and PCR products as described above for LMP-1. The vector encoding wild-type CAT was generated by cloning CAT from pMXCAT into p1958. The vectors encoding LMP-1GFP (p2527) and CD40GFP (p2594) were generated by fusing EGFP from pEGFPN-1 (Clontech) plus a linker (GQSGPGGA) generated by PCR to full-length LMP-1 (p1990) and CD40 (p2534), respectively. The vector encoding GαiRFP was generated by fusing commercially synthesized DNAs encoding the first 32 residues of Gαi (MGCTLSAEDKAAVERSKMIDRNLREDGEKAAE) (Galbiati et al., 1999) to pDsREDN-1 (Clontech). The vector encoding TRAF3GFP (p2635) was generated by fusing the EGFP from pEGFPC-1 (Clontech) in-frame to a TRAF3 expression vector (p1572) (Sandberg et al., 1997).
SDS–PAGE and quantitative western blot analysis
GSTLMP-1 and GSTHALMP-1 fusion proteins (where GST is glutathione S-transferase), used to quantify the number of molecules of LMP-1- or HA epitope-containing proteins, were described previously (Kaykas and Sugden, 2000; Sandberg et al., 2000). GSTLMP-1, GSTHALMP-1, LMP-1, CD40 and their derivatives were resolved by electrophoresis through a 10% polyacrylamide gel, except for CD40CYT, MYCD40 and MYPACD40, which were separated through an 8–16% TrisTrycine polyacrylamide gel. Proteins resolved by SDS–PAGE were transferred to nitrocellulose and blocked with Blotto (1% non-fat dry milk and 0.05% Tween-20 in phosphate-buffered saline) for 20 min. Blots were probed with affinity-purified polyclonal anti-LMP-1 antiserum that recognizes epitopes in the C-terminus of LMP-1 at a 1:200 dilution, with a RAT monoclonal anti-HA antibody (Boehringer-Ingelheim) that recognizes the sequence (YPYDVPDYA) at a 1:500 dilution, with rabbit polyclonal anti-CD40 antiserum that recognizes the C-terminus of CD40 at a 1:500 dilution, or with rabbit polyclonal anti-GFP or anti-RFP (Clontech) antiserum at a 1:250 dilution. The blots were probed with the corresponding anti-species antibodies (Kirkegaard Perry) conjugated to biotin at a 1:2000 dilution and 35S-labeled streptavidin (Amersham) at a 1:1000 dilution (0.5 µCi per blot). The blots were probed for 45 min with each antibody and with streptavidin at room temperature. The blots were then washed once with Blotto for 10 min at room temperature and exposed to a PhosphorImager screen (Molecular Dynamics). The level of protein expression was quantified using Imagequant software (Molecular Dynamics).
Assays for NF-κB and CAT activity
The assay for NF-κB activity was described previously (Mitchell and Sugden, 1995) with a few modifications. In short, 50–80% confluent 10 cm or 6-well dishes of 293 cells were transfected via calcium phosphate precipitation. The precipitate was made as a 1 ml slurry and all of it was used for a 10 cm dish or 250 µl for one well of a 6-well dish. One milliliter of precipitate contains 50 ng of an NF-κB-luciferase reporter, which contained four copies of an NF-κB-responsive element upstream of luciferase (p1242), 20 ng of an expression vector for Renilla luciferase (pLR) (Promega) or 1 µg of an expression vector for EGFP or RFP. The DNA was brought up to a concentration of 30 µg/ml with p1958 and expression vectors encoding LMP-1, CD40 or their derivatives. Four to 12 h after transfection, fresh medium was added to the cells. Forty-eight hours after transfection, the cells were harvested. The 10 cm dishes were split and half the cells were used for SDS–PAGE/western analysis. A total of ∼1.0 × 105 cells from the 10 cm dish or one well of a 6-well dish were lysed in passive lysis buffer (Promega) and counted on a monolight 3010 luminometer. All transfection efficiencies are normalized to Renilla, GFP or RFP levels, and the fold induction refers to that over p1958 alone.
To measure CAT activity, 50–80% confluent 10 cm dishes of 293 cells were transfected with 10 µg of expression vectors for CAT or its fusion proteins and 1 µg of pEGFPN-1, to measure transfection efficiency, via calcium phosphate precipitation. Forty-eight hours later, the cells were counted and 1.0 × 105 GFP-positive cells were used for SDS–PAGE/western analysis with a mouse anti-CAT antibody (Sigma) and goat anti-mouse secondary antibody conjugated to alkaline phosphatase to confirm the level of expression. A total of 2.5 × 106 GFP-positive cells were harvested in 1 ml of 0.25 M Tris–HCl pH 8.0 and lysed by freeze–thawing four times on dry ice/ethanol. Extracts containing 1.0 × 103 cell equivalents were diluted in 50 µl of 0.25 M Tris–HCl pH 8.0, incubated at 65°C for 10 min to inactivate endogenous deacetylase activity, and added to a solution containing 10 µl of [14C]chloramphenicol (0.05 mCi/ml), 20 µl of acetyl coenzyme A (3.5 mg/ml), and incubated for 30 min at 37°C. Ethyl acetate (900 µl) was added to the solution and the organic phase was separated, evaporated in a speedvac, and resuspended in 25 µl of ethyl acetate. The products were separated on a silica gel thin-layer plate in a chromatographic chamber containing chloroform:methanol in a 19:1 ratio. The products were detected and quantified by PhosphorImager analysis.
Microscopy
All microscopy was performed on a Bio-Rad MRC 1024 laser scanning confocal microscope equipped with a mixed gas (argon/kryton) laser operated by 24-bit Lasersharp software, allowing simultaneous display of red, green and blue signals. Where more than one fluorophore was used, each one was displayed separately, and simultaneously merged to minimize non-specific excitation of overlapping fluorophores. 293 cells or their derivatives or Hep2 cells were plated on 18 × 18 mm coverslips and imaged live or fixed, as indicated. Cells were fixed with 4.0% neutral basic formalin for 20 min at room temperature or methanol:acetone at –20°C, permeabilized with 0.1% Triton X-100 for 10 min, and mounted on slides with one eyedrop full of vectashield (Vector Labs). Cells were imaged for GFP and/or RFP and/or stained as indicated with the following reagents: biotinylated CTxβ (Sigma) at a concentration of 10 µg/ml was detected with either CY5 or Alexa Fluor 594-conjugated streptavidin (Molecular Probes) at a 1:10 dilution; mouse anti-CD40 antibody (Pharmagin) at a 1:100 dilution was detected with a rabbit anti-mouse IgG conjugated to Alexa Fluor 488; rabbit anti-LMP-1 antiserum at a 1:100 dilution was detected with a mouse anti-rabbit IgG conjugated to Alexa Fluor 594. Some cells were treated for 20 min at room temperature with a fusion of muCD8α to the extracellular domain of CD40L (Ancell) at a concentration of 10 µg/ml prior to staining.
Biochemical fractionation
293 cells (3.0 × 107) or lymphoid cells (1.0 × 108) were grown on 150 mm dishes. The 293tetLMP-1-inducible clones were grown in tetracycline for 24 h prior to harvesting. The 293 cells that express CD40GFP stably or lymphoid cells were treated with 10 µg/ml muCD8αCD40L for 20 min prior to harvesting. The cells were treated with l ml of ice-cold buffer containing 150 mM NaCl, 25mM Tris–HCl pH 7.5, 5 mM EDTA, 1% Triton X-100 on ice for 30 min. The samples were diluted 1:1 with 70% nycodenz (Sigma) in TNE (25 mM Tris–HCl pH 7.5, 150 mM NaCl, 5 mM EDTA). The sample was then loaded at the bottom of a 6 ml ultracentrifuge tube (Beckman) and overlayed with 500 µl of 25, 22.5, 20, 18, 15, 12, 8% nycodenz. The samples were spun at 200 000 g at 4°C in a Beckman SWi50 rotor for 4 h and 11 fractions of 500 µl were collected from the bottom of the gradient. Ten microliters of each fraction were subjected to SDS–PAGE and western analysis. The 293 cells that express TRAF3GFP stably were fractionated by extraction with Triton X-100. In short, the cells were transfected with a vector expressing GαiRFP. Forty-eight hours after transfection, the cells were lysed in 1× RIPA buffer (Kaykas and Sugden, 2000) or treated with a buffer containing 150 mM NaCl, 25 mM Tris–HCl pH 7.5, 5 mM EDTA, 1% Triton X-100 on ice for 30 min. The Triton X-100-treated samples were separated by centrifugation into soluble (S) and insoluble (P) fractions by centrifugation at 14 000 r.p.m. in a microfuge for 10 min. To determine the DRM fraction, western blots were performed with anti-RFP antibodies on 1.0 × 105 cell equivalents from the S, P and RIPA-treated samples. At least 95% of the total Gαi in the RIPA lysed sample is found in the P fraction of the Triton X-100-treated sample and therefore shows that this method accurately separated the DRM and soluble fractions.
Acknowledgments
Acknowledgements
We would like to thank John Young for his critical review of this manuscript. We would also like to thank N.Lam and M.Sandberg for discussion of the experiments and data presented in this manuscript. This work was supported by Public Health Service grants CA-22443, CA-07175, T32-CA-09135 and CA-70723. B.S. is a Research Professor of the American Cancer Society.
References
- Ardila-Osorio H., Clausse,B., Mishal,Z., Wiels,J., Tursz,T. and Busson,P. (1999) Evidence of LMP1–TRAF3 interactions in glyco sphingolipid-rich complexes of lymphoblastoid and nasopharyngeal carcinoma cells. Int. J. Cancer, 81, 645–649. [DOI] [PubMed] [Google Scholar]
- Aviel S., Winberg,G., Massucci,M. and Ciechanover,A. (2000) Degradation of the Epstein–Barr virus latent membrane protein 1 (LMP1) by the ubiquitin–proteasome pathway. Targeting via ubiquitination of the N-terminal residue. J. Biol. Chem., 275, 23491–23499. [DOI] [PubMed] [Google Scholar]
- Banchereau J., de Paoli,P., Valle,A., Garcia,E. and Rousset,F. (1991) Long-term human B cell lines dependent on interleukin-4 and antibody to CD40. Science, 251, 70–72. [DOI] [PubMed] [Google Scholar]
- Banchereau J., Bazan,F., Blanchard,D., Briere,F., Galizzi,J.P., van Kooten,C., Liu,Y.J., Rousset,F. and Saeland,S. (1994) The CD40 antigen and its ligand. Annu. Rev. Immunol., 12, 881–922. [DOI] [PubMed] [Google Scholar]
- Berberich I., Shu,G.L. and Clark,E.A. (1994) Cross-linking CD40 on B cells rapidly activates nuclear factor-κB. J. Immunol., 153, 4357–4366. [PubMed] [Google Scholar]
- Brodeur S.R., Cheng,G., Baltimore,D. and Thorley-Lawson,D. (1997) Localization of the major NF-κB-activating site and the sole TRAF3 binding site of LMP-1 defines two distinct signaling motifs. J. Biol. Chem., 272, 19777–19784. [DOI] [PubMed] [Google Scholar]
- Brown D.A. and London,E. (1998) Functions of lipid rafts in biological membranes. Annu. Rev. Cell Dev. Biol., 14, 111–136. [DOI] [PubMed] [Google Scholar]
- Chan F.K., Chun,H.J., Zheng,L., Siegel,R.M., Bui,K.L. and Lenardo,M.J. (2000) A domain in TNF receptors that mediates ligand-independent receptor assembly and signaling. Science, 288, 2351–2354. [DOI] [PubMed] [Google Scholar]
- Cheng P.C., Dykstra,M.L., Mitchell,R.N. and Pierce,S.K. (1999) A role for lipid rafts in B cell antigen receptor signaling and antigen targeting. J. Exp. Med., 190, 1549–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausse B., Fizazi,K., Walczak,V., Tetaud,C., Wiels,J., Tursz,T. and Busson,P. (1997) High concentration of the EBV latent membrane protein 1 in glycosphingolipid-rich complexes from both epithelial and lymphoid cells. Virology, 228, 285–293. [DOI] [PubMed] [Google Scholar]
- Cosman D. (1994) A family of ligands for the TNF receptor superfamily. Stem Cells, 12, 440–455. [DOI] [PubMed] [Google Scholar]
- Dadgostar H. and Cheng,G. (2000) Membrane localization of TRAF3 enables JNK activation. J. Biol. Chem., 275, 2539–2544. [DOI] [PubMed] [Google Scholar]
- Deng L., Wang,C., Spencer,E., Yang,L., Braun,A., You,J., Slaughter,C., Pickart,C. and Chen,Z.J. (2000) Activation of the IκB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell, 103, 351–361. [DOI] [PubMed] [Google Scholar]
- Devergne O., McFarland,E.C., Mosialos,G., Izumi,K.M., Ware,C.F. and Kieff,E. (1998) Role of the TRAF binding site and NF-κB activation in Epstein–Barr virus latent membrane protein 1-induced cell gene expression. J. Virol., 72, 7900–7908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliopoulos A.G. and Young,L.S. (1998) Activation of the cJun N-terminal kinase (JNK) pathway by the Epstein–Barr virus-encoded latent membrane protein 1 (LMP1). Oncogene, 16, 1731–1742. [DOI] [PubMed] [Google Scholar]
- Francis D.A., Karras,J.G., Ke,X.Y., Sen,R. and Rothstein,T.L. (1995) Induction of the transcription factors NF-κB, AP-1 and NF-AT during B cell stimulation through the CD40 receptor. Int. Immunol., 7, 151–161. [DOI] [PubMed] [Google Scholar]
- Galbiati F., Volonte,D., Meani,D., Milligan,G., Lublin,D.M., Lisanti,M.P. and Parenti,M. (1999) The dually acylated NH2-terminal domain of gi1α is sufficient to target a green fluorescent protein reporter to caveolin-enriched plasma membrane domains. Palmitoylation of caveolin-1 is required for the recognition of dually acylated g-protein α subunits in vivo. J. Biol. Chem., 274, 5843–5850. [DOI] [PubMed] [Google Scholar]
- Gires O., Zimber,S.U., Gonnella,R., Ueffing,M., Marschall,G., Zeidler,R., Pich,D. and Hammerschmidt,W. (1997) Latent membrane protein 1 of Epstein–Barr virus mimics a constitutively active receptor molecule. EMBO J., 16, 6131–6140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gires O. et al. (1999) Latent membrane protein 1 of Epstein–Barr virus interacts with JAK3 and activates STAT proteins. EMBO J., 18, 3064–3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F.L., Smiley,J., Russell,W.C. and Nairn,R. (1977) Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol., 36, 59–74. [DOI] [PubMed] [Google Scholar]
- Hanissian S.H. and Geha,R.S. (1997) Jak3 is associated with CD40 and is critical for CD40 induction of gene expression in B cells. Immunity, 6, 379–387. [DOI] [PubMed] [Google Scholar]
- Harder T. and Simons,K. (1997) Caveolae, DIGs and the dynamics of sphingolipid-cholesterol microdomains. Curr. Opin. Cell Biol., 9, 534–542. [DOI] [PubMed] [Google Scholar]
- Hatzivassiliou E., Miller,W.E., Raab-Traub,N., Kieff,E. and Mosialos,G. (1998) A fusion of the EBV latent membrane protein-1 (LMP1) transmembrane domains to the CD40 cytoplasmic domain is similar to LMP1 in constitutive activation of epidermal growth factor receptor expression, nuclear factor-κB and stress-activated protein kinase. J. Immunol., 160, 1116–1121. [PubMed] [Google Scholar]
- Hibi M. and Hirano,T. (1998) Signal transduction through cytokine receptors. Int. Rev. Immunol., 17, 75–102. [DOI] [PubMed] [Google Scholar]
- Hicke L. (1997) Ubiquitin-dependent internalization and down-regulation of plasma membrane proteins. FASEB J., 11, 1215–1226. [DOI] [PubMed] [Google Scholar]
- Hicke L. (1999) Gettin’ down with ubiquitin: turning off cell-surface receptors, transporters and channels. Trends Cell Biol., 9, 107–112. [DOI] [PubMed] [Google Scholar]
- Hostager B.S., Catlett,I.M. and Bishop,G.A. (2000) Recruitment of CD40 and tumor necrosis factor receptor-associated factors 2 and 3 to membrane microdomains during CD40 signaling. J. Biol. Chem., 275, 15392–15398. [DOI] [PubMed] [Google Scholar]
- Huo L. and Rothstein,T.L. (1995) Receptor-specific induction of individual AP-1 components in B lymphocytes. J. Immunol., 154, 3300–3309. [PubMed] [Google Scholar]
- Ishida T.K., Tojo,T., Aoki,T., Kobayashi,N., Ohishi,T., Watanabe,T., Yamamoto,T. and Inoue,J. (1996) TRAF5, a novel tumor necrosis factor receptor-associated factor family protein, mediates CD40 signaling. Proc. Natl Acad. Sci. USA, 93, 9437–9442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi K.M. and Kieff,E.D. (1997) The Epstein–Barr virus oncogene product latent membrane protein 1 engages the tumor necrosis factor receptor-associated death domain protein to mediate B lymphocyte growth transformation and activate NF-κB. Proc. Natl Acad. Sci. USA, 94, 12592–12597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpusas M., Hsu,Y.M., Wang,J.H., Thompson,J., Lederman,S., Chess,L. and Thomas,D. (1995) 2 Å crystal structure of an extracellular fragment of human CD40 ligand. Structure, 3, 1426. [PubMed] [Google Scholar]
- Karras J.G., Wang,Z., Huo,L., Frank,D.A. and Rothstein,T.L. (1997) Induction of STAT protein signaling through the CD40 receptor in B lymphocytes: distinct STAT activation following surface Ig and CD40 receptor engagement. J. Immunol., 159, 4350–4355. [PubMed] [Google Scholar]
- Kawabe T., Naka,T., Yoshida,K., Tanaka,T., Fujiwara,H., Suematsu,S., Yoshida,N., Kishimoto,T. and Kikutani,H. (1994) The immune responses in CD40-deficient mice: impaired immunoglobulin class switching and germinal center formation. Immunity, 1, 167–178. [DOI] [PubMed] [Google Scholar]
- Kaye K.M., Devergne,O., Harada,J.N., Izumi,K.M., Yalamanchili,R., Kieff,E. and Mosialos,G. (1996) Tumor necrosis factor receptor associated factor 2 is a mediator of NF-κB activation by latent infection membrane protein 1, the Epstein–Barr virus transforming protein. Proc. Natl Acad. Sci. USA, 93, 11085–11090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaykas A. and Sugden,B. (2000) The N-terminus and membrane-spanning domains of LMP-1 inhibit cell proliferation. Oncogene, 19, 1400–1410. [DOI] [PubMed] [Google Scholar]
- Kieser A., Kilger,E., Gires,O., Ueffing,M., Kolch,W. and Hammerschmidt,W. (1997) Epstein–Barr virus latent membrane protein-1 triggers AP-1 activity via the c-Jun N-terminal kinase cascade. EMBO J., 16, 6478–6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilger E., Kieser,A., Baumann,M. and Hammerschmidt,W. (1998) Epstein–Barr virus-mediated B-cell proliferation is dependent upon latent membrane protein 1, which simulates an activated CD40 receptor. EMBO J., 17, 1700–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koegl M., Zlatkine,P., Ley,S.C., Courtneidge,S.A. and Magee,A.I. (1994) Palmitoylation of multiple Src-family kinases at a homologous N-terminal motif. Biochem. J., 303, 749–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie A.G. (1990) Refined crystal structure of type III chloramphenicol acetyltransferase at 1.75 Å resolution. J. Mol. Biol., 213, 167–186. [DOI] [PubMed] [Google Scholar]
- Ling L. and Goeddel,D.V. (2000) MIP-T3, a novel protein linking tumor necrosis factor receptor-associated factor 3 to the microtubule network. J. Biol. Chem., 275, 23852–23860. [DOI] [PubMed] [Google Scholar]
- Martin J. and Sugden,B. (1991a) The latent membrane protein oncoprotein resembles growth factor receptors in the properties of its turnover. Cell Growth Differ., 2, 653–660. [PubMed] [Google Scholar]
- Martin J. and Sugden,B. (1991b) Transformation by the oncogenic latent membrane protein correlates with its rapid turnover, membrane localization and cytoskeletal association. J. Virol., 65, 3246–3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWhirter S.M., Pullen,S.S., Holton,J.M., Crute,J.J., Kehry,M.R. and Alber,T. (1999) Crystallographic analysis of CD40 recognition and signaling by human TRAF2. Proc. Natl Acad. Sci. USA, 96, 8408–8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell T. and Sugden,B. (1995) Stimulation of NF-κB-mediated transcription by mutant derivatives of the latent membrane protein of Epstein–Barr virus. J. Virol., 69, 2968–2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi M., Nosaka,T. and Kitamura,T. (1998) Cytokine receptors: structures and signal transduction. Int. Rev. Immunol., 16, 617–634. [DOI] [PubMed] [Google Scholar]
- Pullen S.S., Dang,T.T., Crute,J.J. and Kehry,M.R. (1999) CD40 signaling through tumor necrosis factor receptor-associated factors (TRAFs). Binding site specificity and activation of downstream pathways by distinct TRAFs. J. Biol. Chem., 274, 14246–14254. [DOI] [PubMed] [Google Scholar]
- Rothe M., Sarma,V., Dixit,V.M. and Goeddel,D.V. (1995) TRAF2-mediated activation of NF-κB by TNF receptor 2 and CD40. Science, 269, 1424–1427. [DOI] [PubMed] [Google Scholar]
- Sandberg M., Hammerschmidt,W. and Sugden,B. (1997) Characterization of LMP-1’s association with TRAF1, TRAF2 and TRAF3. J. Virol., 71, 4649–4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg M.L., Kaykas,A. and Sugden,B. (2000) Latent membrane protein 1 of Epstein–Barr virus inhibits as well as stimulates gene expression. J. Virol., 74, 9755–9761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seed B. (1995) Initiation of signal transduction by receptor aggregation: role of nonreceptor tyrosine kinases. Semin. Immunol., 7, 3–11. [DOI] [PubMed] [Google Scholar]
- Simons K. and Toomre,D. (2000) Lipid rafts and signal transduction. Nature Rev., 1, 31–39. [DOI] [PubMed] [Google Scholar]
- Song H.Y., Regnier,C.H., Kirschning,C.J., Goeddel,D.V. and Rothe,M. (1997) Tumor necrosis factor (TNF)-mediated kinase cascades: bifurcation of nuclear factor-κB and c-jun N-terminal kinase (JNK/SAPK) pathways at TNF receptor-associated factor 2. Proc. Natl Acad. Sci. USA, 94, 9792–9796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stang E., Johannessen,L.E., Knardal,S.L. and Madshus,I.H. (2000) Polyubiquitination of the epidermal growth factor receptor occurs at the plasma membrane upon ligand-induced activation. J. Biol. Chem., 275, 13940–13947. [DOI] [PubMed] [Google Scholar]
- Uchida J., Yasui,T., Takaoka-Shichijo,Y., Muraoka,M., Kulwichit,W., Raab-Traub,N. and Kikutani,H. (1999) Mimicry of CD40 signals by Epstein–Barr virus LMP1 in B lymphocyte responses. Science, 286, 300–303. [DOI] [PubMed] [Google Scholar]
- Vandevoorde V., Haegeman,G. and Fiers,W. (1997) Induced expression of trimerized intracellular domains of the human tumor necrosis factor (TNF) p55 receptor elicits TNF effects. J. Cell Biol., 137, 1627–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kooten C. and Banchereau,J. (1997) Functional role of CD40 and its ligand. Int. Arch. Allergy Immunol., 113, 393–399. [DOI] [PubMed] [Google Scholar]
- van Kooten C. and Banchereau,J. (2000) CD40–CD40 ligand. J. Leukoc. Biol., 67, 2–17. [DOI] [PubMed] [Google Scholar]
- Vidalain P.O., Azocar,O., Servet-Delprat,C., Rabourdin-Combe,C., Gerlier,D. and Manie,S. (2000) CD40 signaling in human dendritic cells is initiated within membrane rafts. EMBO J., 19, 3304–3313. [DOI] [PMC free article] [PubMed] [Google Scholar]