Abstract
Human ribosomal gene repeats are distributed among five nucleolar organizer regions (NORs) on the p arms of acrocentric chromosomes. On exit from mitosis, nucleoli form around individual active NORs. As cells progress through the cycle, these mini-nucleoli fuse to form large nucleoli incorporating multiple NORs. It is generally assumed that nucleolar incorporation of individual NORs is dependent on ribosomal gene transcription. To test this assumption, we determined the nuclear location of individual human acrocentric chromosomes, and their associated NORs, in mouse> human cell hybrids. Human ribosomal genes are transcriptionally silent in this context. Combined immunofluorescence and in situ hybridization (immuno-FISH) on three-dimensional preserved nuclei showed that human acrocentric chromosomes associate with hybrid cell nucleoli. Analysis of purified nucleoli demonstrated that human and mouse NORs are equally likely to be within a hybrid cell nucleolus. This is supported further by the observation that murine upstream binding factor can associate with human NORs. Incorporation of silent NORs into mature nucleoli raises interesting issues concerning the maintenance of the activity status of individual NORs.
Keywords: human acrocentric chromosomes/nucleolar organizer regions/nucleoli/somatic cell hybrids/UBF
Introduction
The eukaryotic nucleus is functionally compartmentalized, and nowhere is this illustrated more clearly than at the nucleolus, where multiple loci from different chromosomes contribute to the formation of a functional nuclear compartment visible by light microscopy (Bridger and Bickmore, 1998; Matera, 1999). Nucleoli are the sites of rDNA transcription, rRNA processing and the assembly of ribosomes. These stages of ribosome biogenesis can be observed at the structural level (Scheer and Weisenberger, 1994; Scheer and Hock, 1999). The nucleolus can participate in many other aspects of gene expression and nuclear function (Pederson, 1998; Carmo-Fonseca et al., 2000; Olson et al., 2000).
In the human, the five chromosomal loci that encode the ∼360 copies of ribosomal genes are termed nucleolar organizer regions (NORs). NORs are located on the short arms of acrocentric chromosomes (HSA13, 14, 15, 21 and 22) (Henderson et al., 1972). Each ribosomal gene cluster is on average 3 Mb in length (80 copies of a 43 kb repeat) (Sakai et al., 1995). This represents between a quarter and a third of the DNA present on the short arms of each of the acrocentric chromosomes. The remaining DNA is largely devoid of transcribed sequences and is composed of arrays of tandem repeated satellite DNA (Waye and Willard, 1989; Choo et al., 1990; Tagarro et al., 1994; Shiels et al., 1997b). NORs can be identified as secondary constrictions on metaphase chromosomes and can be visualized by silver staining, due to the abundance of associated argyophilic proteins (Goodpasture and Bloom, 1975). However, not all NORs form secondary constrictions or can be silver stained during metaphase. The number of silver-positive NORs varies between four and 10 (Babu and Verma, 1985). In dividing HeLa cells, usually six out of 10 NORs can be silver stained (Roussel et al., 1996; G.Sullivan, data not shown). The term secondary constriction is somewhat of a misnomer since the chromatin in a silver-stained NOR is 10-fold less compact than the remainder of the metaphase chromosome and is organized in a distinct twisted loop (Heliot et al., 1997). The identity of the proteins responsible for this distinct chromatin structure is uncertain, but likely candidates include components of the RNA polymerase I (pol I) transcription machinery.
The human pol I transcription machinery is comprised of upstream binding factor (UBF), selectivity factor 1 (SL1) and pol I with its associated factors (TIF IA and TIF IC) (Grummt, 1999). SL1 is a complex of the TATA-binding protein (TBP) and three TBP-associated factors (TAFI110, TAFI63 and TAFI48) (Comai et al., 1992). SL1 binds to the core element of the ribosomal gene promoter and is the key component of the pre-initiation complex (Bell et al., 1988). There is some debate as to the role of UBF in pol I transcription. UBF was originally defined as a factor that bound to ribosomal gene promoter sequences and facilitated the interaction of SL1 (Bell et al., 1988). The abundance of UBF (up to 106 molecules per cell, see Figure 2), its relaxed DNA sequence specificity and its remarkable ability to bend and loop target DNA now lead to the view that it is performing a more generalized structural role in ribosomal gene chromatin (Reeder et al., 1995).
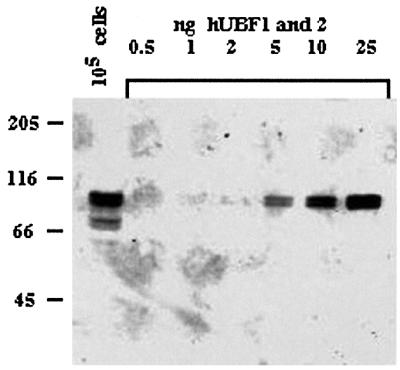
Fig. 2. Characterization of anti-UBF human autoantibodies. Human 1BR.2 primary fibroblasts were lysed directly into SDS–PAGE loading buffer at 104 cells/µl. A 10 µl aliquot of this cell lysate (equivalent to 105 cells) was electrophoresed on an 8% polyacrylamide gel alongside increasing amounts (shown above gel) of an equimolar mixture of pure recombinant hUBF1 and 2. A western blot of the gel was probed with anti-UBF human autoantibody (see Materials and methods). Note that the lower pair of bands (∼70 kDa) apparent in the first lane are proteolytic breakdown products of UBF.
As cells enter mitosis, transcription by all three classes of RNA polymerase ceases or is at least greatly decreased (reviewed in Sirri et al., 2000). In the case of pol I, it appears that mitotic repression results from changes in the phosphorylation status of UBF and SL1 (Kuhn et al., 1998; Voit et al., 1999; Sirri et al., 2000). Cdc2–cyclin B kinase is primarily responsible for this repression (Sirri et al., 2000). UBF, SL1 and at least a fraction of pol I remain associated with the metaphase NOR (Weisenberger and Scheer, 1995; Jordan et al., 1996; Roussel et al., 1996). The pol I transcription machinery is only found associated with acrocentric chromosomes that exhibit a secondary constriction and silver stain. Retention of the transcription machinery on the mitotic NOR may be responsible for its unique structure. It is generally agreed (but unproven) that acrocentric chromosomes devoid of a secondary constriction are transcriptionally silent with respect to ribosomal genes. Some human NORs contain less than the expected amount of rDNA (Shiels et al., 1997a). This raises the possibility that some NORs may be transcriptionally active despite the lack of visible silver staining.
As cells exit mitosis, nucleoli only form around transcriptionally active NORs (Ochs et al., 1985; Benavente et al., 1987; Jimenez-Garcia et al., 1994). Associated pol I transcription machinery may seed nucleolar reformation (Dousset et al., 2000). As cells progress through the cycle, the multiple small nucleoli that form around individual active NORs appear to fuse into one or a few large nucleoli, a phenomenon commonly referred to as nucleolar fusion (Anastassova-Kristeva, 1977). A consequence of this process is that multiple NORs can be found within a single nucleolus (Babu and Verma, 1985). The underlying mechanisms involved in this major dynamic nuclear reorganization involving multiple chromosome territories are not at all understood. The popular view that this phenomenon can be explained by an inherent affinity of nucleoli for each other has not been proven experimentally. A prediction of this model is that silent NORs are excluded from the nucleolus. Alternatively, one could hypothesize that human acrocentric chromosomes self-associate independently of NOR activity status. In this model, silent NORs associate with nucleoli. In support of this model, it has been shown recently in mice that large nucleoli contain apparently inactive, methylated rDNA (Akhmanova et al., 2000).
Here we have exploited the phenomenon of species specificity of ribosomal gene transcription to discriminate between these two alternatives. Considerable divergence between vertebrates has been observed in both the sequence of the ribosomal gene repeat and components of the pol I transcription machinery. Mouse and human ribosomal genes cannot be transcribed in vivo (Miller et al., 1976) or in vitro by the other species’ transcription machinery (reviewed in Heix and Grummt, 1995). This species specificity resides solely in the transcription factor SL1. Because of this species specificity, somatic cell hybrids provide a valuable model system to study the structure and subnuclear localization of NORs on intact human chromosomes in the certain knowledge that they are transcriptionally silent.
Using a panel of monochromosomal somatic cell hybrids, each containing an intact human acrocentric chromosome, we confirm that human ribosomal genes are transcriptionally silent within the murine cell environment. We demonstrate that mUBF can interact with human NORs to a degree similar to that observed on active murine NORs. Most surprising of all, we observe that in every case, human acrocentric chromosomes are associated with a mouse nucleolus in vivo. This is irrespective of their UBF loading status. We further demonstrate that human NORs co-purify with nucleoli isolated from hybrid cells. Thus we conclude that localization of human NORs to nucleoli can be uncoupled from ribosomal gene transcription and UBF loading. Additionally, we show that sequences from the long arms of acrocentric chromosomes co-purify with nucleoli. In the light of these findings, we favour a model in which nucleolar fusion results from the ability of NOR-containing chromosomes to associate independently of ribosomal gene transcription.
Results
Human ribosomal genes are transcriptionally silent in a mouse background
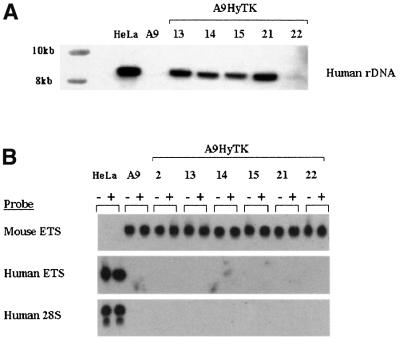
In order to study the relationship between subnuclear localization of individual human NORs and ribosomal gene transcription, we have utilized a panel of mouse> human somatic cell hybrids (Cuthbert et al., 1995). In this panel, mouse A9 cells contain individual human chromosomes 13, 14, 15, 21 and 22 (HSA13, 14, 15, 21 and 22, respectively), tagged with a selectable hygromycin resistance marker (HyTK) to maintain the stability of the hybrid. Hybrids containing the human chromosome 2 or X (HSA2 and X) served as non-acrocentric controls. Extensive characterization of these hybrids has shown that they each contain only a single intact human chromosome (Cuthbert et al., 1995). Southern blotting analysis confirms the presence of the expected amount of human rDNA in hybrids containing HSA13, 14, 15 and 21 (Figure 1A). As has been reported previously, HSA22 contains approximately one-tenth of the human rDNA observed on other human acrocentric rDNAs (Shiels et al., 1997a). Southern blots with probes derived from elsewhere in the human rDNA repeat show no detectable reorganization of the human rDNA content in hybrid cells compared with normal human cells (data not shown).
Fig. 1. Human ribosomal genes are transcriptionally silent in hybrid cell lines. (A) HindIII-digested high molecular weight DNA from hybrid, human (HeLa) and mouse (A9) cells was probed with a human rDNA-specific hybridization probe (see Materials and methods). The source of the DNA is shown above each lane and molecular weight markers are shown on the left. (B) Total RNA samples (10 µg each) from HeLa, mouse A9 and A9HyTK hybrid cells were used in S1 nuclease protection assays with probes designed to detect the 5′ ends of mouse and human 47S precursor rRNAs (Mouse ETS and Human ETS, respectively) and a probe designed specifically to detect human 28S rRNA. RNA samples from untreated and TPA-treated cells are designated – and +, respectively. The human chromosome present in each of the hybrid cell lines is shown above and the probe used is shown on the left of the appropriate panel.
Previously it has been demonstrated that in rodent> human cell hybrids, human ribosomal genes are transcriptionally silent (Miller et al., 1976). In order to demonstrate that this was the case in the hybrids used here, we determined the transcriptional status of human ribosomal genes using an S1 nuclease protection assay that detects the 5′ most 40 nucleotides of the 47S rRNA precursor. The first 3656 nucleotides of the 47S precursor specify the external transcribed spacer, which is processed and degraded rapidly in vivo. Consequently, signal generated with this probe can be considered a measure of ongoing transcription. Whereas this probe readily detected 47S precursor in HeLa cell RNA, no signal was observed with equivalent amounts of RNA prepared from A9HyTK-13, 14, 15, 21 and 22 hybrid cells or mouse A9 cells (Figure 1B, middle panel). A similar probe prepared from mouse rDNA detected mouse 47S precursor in RNA from A9, A9HyTK-13, 14, 15, 21 and 22 cells but not from HeLa cells (Figure 1B, top panel). To look at accumulated rRNA, we used a probe that can specifically detect human 28S rRNA. This oligonucleotide probe is complementary to nucleotides 3326–3369 of human 28S rRNA. This sequence represents an expansion region that is absent from murine 28S rRNA. Mature 28S rRNA is stable and provides a highly sensitive marker for accumulated human ribosomal gene transcripts. Using this probe, we readily detected human 28S rRNA in HeLa cells but not in hybrid or A9 cells (Figure 1B, bottom panel). In mixing experiments, we can detect <1 part in 1000 of human 28S rRNA (data not shown).
In more complex hybrids containing multiple human chromosomes, human ribosomal gene transcription can be reactivated by treatment with either SV40 T antigen or phorbol esters (Soprano and Baserga, 1980). The most reasonable interpretation of these experiments is that the hybrids used contain not only human acrocentric chromosomes but also human chromosomes that carry the species-specific component(s) of ribosomal gene transcription. It is envisaged that these species-specific components are able to reprogramme the mouse pol I transcription machinery to recognize and transcribe human rDNA. However, it is also formally possible that treatment of hybrids with T antigen or phorbol esters could relax the specificity of the mouse pol I transcription machinery such that it can now transcribe human rDNA. Treatment of the monochromosomal hybrids used in this study with the phorbol ester, 12-O-tetradecanoylphorbol-13-acetate (TPA), does not result in transcriptional reactivation as determined by S1 nuclease protection assays with either human probe (Figure 1B). Thus we conclude that in A9HyTK-13, 14, 15, 21 and 22 cells, human ribosomal genes are transcriptionally silent.
UBF can associate with human NORs in hybrid cells
UBF is highly conserved and functionally interchangeable between human and mouse (Grummt, 1999). In principal, mUBF is capable of recognizing a human NOR. It was of interest, therefore, to determine whether mUBF does bind to human NORs in hybrid cells. This would provide evidence that human rDNA could be recognized as an NOR despite its transcriptional silence. Association of UBF with individual chromosomes/NORs can be determined conveniently by combined immunofluorescence and in situ hybridization (immuno-FISH) performed on metaphase chromosome spreads. In order to visualize UBF, we have utilized human autoantibodies. Antibodies against UBF also known as NOR90 are common in patients with autoimmune disorders and can readily detect UBF on metaphase chromosomes (Chan et al., 1991; Roussel et al., 1993). The anti-UBF autoantibodies used here were identified using an enzyme-linked immunosorbent assay (ELISA)-based screen of human sera against recombinant hUBF (A.Gibson and B.McStay, unpublished). These autoantibodies are highly specific for UBF as demonstrated by western blotting of a human cell lysate with recombinant hUBF1 and 2 controls (Figure 2). Quantitation of this blot revealed that human primary fibroblasts contain between 5 × 105 and 1 × 106 molecules of UBF/cell. We observe similar levels of UBF in A9 and hybrid cells (data not shown).
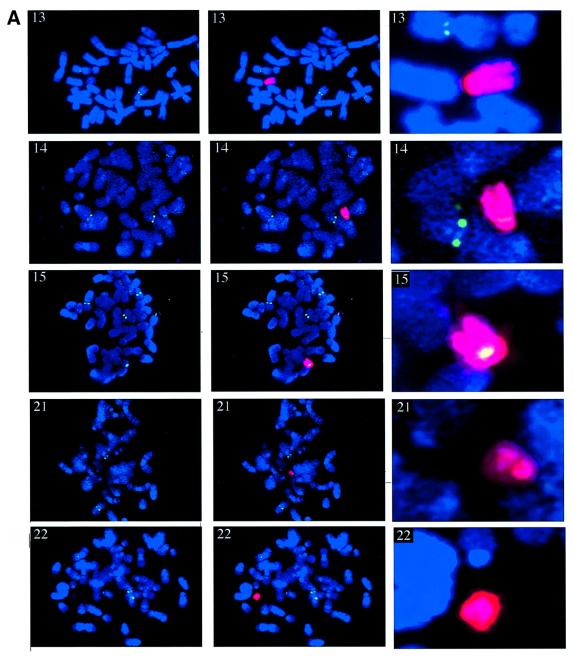
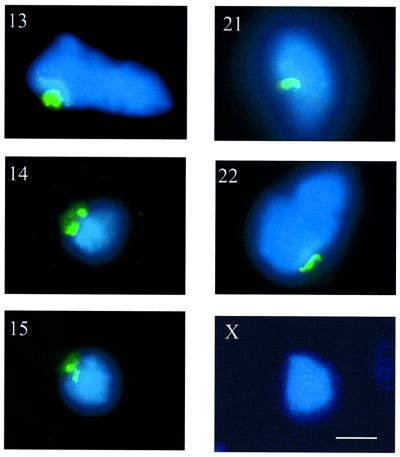
Metaphase spreads prepared for A9HyTK-13, 14, 15, 21 and 22 cells were subjected to immuno-FISH. UBF was first visualized with anti-UBF autoantibodies and fluorescein isothiocyanate (FITC)-labelled secondary antibodies (green). Following fixation of bound antibodies, human chromosomes were identified by FISH using labelled human Cot-1 (red) as a probe (Figure 3A). These experiments clearly demonstrate that HSA14 and 15 have associated UBF in a mouse background. Furthermore, the level of UBF observed on HSA15 is comparable to that observed on mouse chromosomes in the same cells (Figure 3A). No UBF appears to be associated with HSA13, 21 and 22 in A9HyTK-13, 21 and 22 cells, respectively.
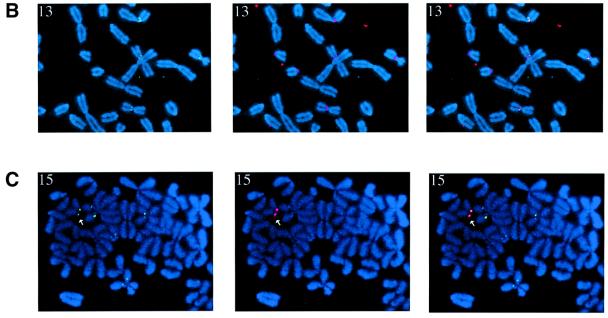
Fig. 3. UBF associates with human NORs in hybrid cells. (A) Metaphase chromosome spreads (counterstained with DAPI) from A9HyTK-13, 14, 15, 21 and 22 were subject to combined immunofluorescence using anti-UBF human autoantibody 9386 (FITC/green) and FISH with Spectrum red-labelled human Cot-1 DNA as probe. The identity of the human chromosome is indicated in the top left corner of each panel. Panels on the left show UBF and DAPI staining. In the centre panels, the signal from the human Cot-1 probe has been merged. The right hand panels show an enlarged version of the merged signal over the human chromosome. (B) Metaphase chromosome spreads (counterstained with DAPI) from A9HyTK-13 were subject to combined immunofluorescence using anti-UBF human autoantibody 9386 (FITC/green) and FISH with Spectrum red-labelled 28S rDNA probe (see Materials and methods). The left, centre and right panels show UBF staining, rDNA hybridization and the merge of both signals, respectively. (C) Metaphase chromosome spreads (counterstained with DAPI) from A9HyTK-15 were subject to combined immunofluorescence using anti-UBF human autoantibody 9386 (FITC/green) and FISH with Spectrum red-labelled human-specific rDNA probe (see Materials and methods). The left, centre and right panels show UBF staining, rDNA hybridization and the merge of both signals, respectively. Arrowheads indicate UBF and human NOR co-localization.
In order to demonstrate co-localization of UBF staining with mouse and human NORs, we performed immuno-FISH with a 28S rDNA and a human rDNA-specific probe (Figure 3B and C, respectively). The results in Figure 3B show clear co-localization of UBF (green) with mouse rDNA (red) on mouse chromosomes. The results in Figure 3C show clear co-localization of UBF (green) with human rDNA (red) on HSA15 in A9HyTK-15. For a given hybrid, all the cells in that metaphase preparation displayed similar loading of UBF on the human chromosome (data not shown). Thus the UBF loading status appears to be copied faithfully during cell division, as has been described previously in human cells (Roussel et al., 1993).
We conclude from this experiment that despite their transcriptional inactivity, human NORs can be recognized by components of the mouse pol I transcription machinery. It is interesting to speculate that the heterogeneity of UBF loading on human NORs in hybrid cells reflects that observed in the parental line (1BR.2).
Nucleolar association of human acrocentric chromosomes within the mouse nucleus
In human cells, UBF localizes exclusively to the nucleolus (Chan et al., 1991; Roussel et al., 1993). The finding that mUBF can bind to human NORs in hybrid cells indicates that these NORs may associate physically with a mouse nucleolus. To address this question, we have analysed the subnuclear localization of HSA13, 14, 15, 21, 22 and X within the three-dimensionally preserved nuclei of hybrid cells (Figure 4). The position of the nucleolus in these analyses was determined by co-immunofluorescence with an anti-fibrillarin monoclonal antibody. The probes used in these experiments were q arm-specific chromosome paints for HSA13, 14, 15, 21 and 22. A whole chromosome paint (p and q) was used for HSAX (Guan et al., 1996). Probes were verified against metaphase spreads of the appropriate hybrid (data not shown).
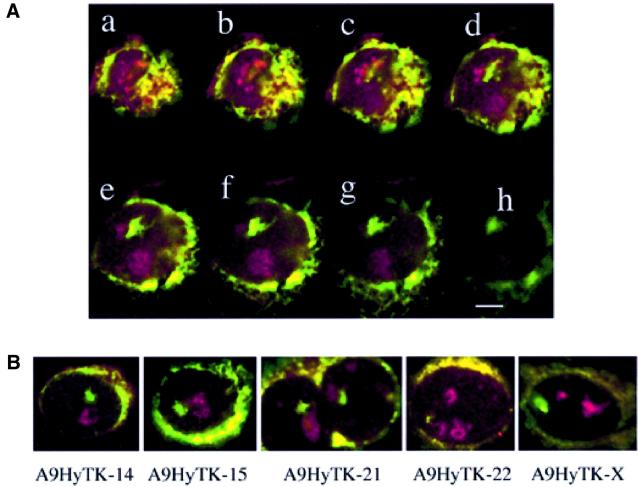
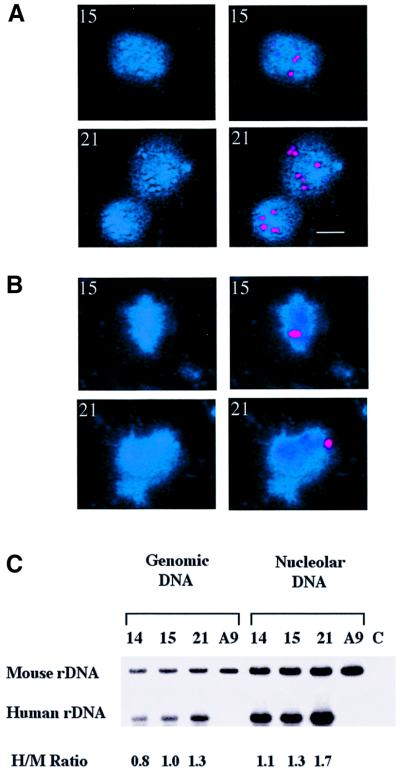
Fig. 4. Human acrocentric chromosomes associate with nucleoli in hybrid cells. (A) Optical sections (a–h) were obtained by laser-scanning confocal microscopy performed on an individual interphase A9HyTK-13 cell in which the three-dimensional structure was preserved. Nucleoli were visualized with an anti-fibrillarin mAb (red) and HSA13 with a biotinylated long arm-specific paint (green). Note that endogenous biotin present in the cytoplasm defines the nuclear–cytoplasmic boundary. The scale bar represents 5 µm. (B) Mid-sections obtained as above from A9HyTK-14, 15, 21, 22 and X. HSA14, 15, 21 and 22 were visualized with the relevant long arm-specific paint. HSAX was visualized with paint derived from both chromosome arms. Note that for A9HyTK-21, two closely adjacent cells are shown.
The three-dimensional nuclear positioning of the acrocentric and the X chromosome territories was assessed by optically sectioning nuclei using a confocal laser-scanning microscope. Galleries of the two-dimensional data (each optical section) were analysed. The gallery for A9HyTK-13 is shown in Figure 4A. Mid sections for A9HyTK-14, 15, 21, 22 and X control are shown in Figure 4B. A chromosome territory was scored as being co-localized with the nucleolar staining if the paint was found to be overlapping the nucleolar stain or immediately abutting it. In the sections d and e of HyTK-13 (Figure 4A), co-localization of HSA13 with the upper of the two nucleoli is particularly evident. Approximately 50 cells were analysed for each monochromosome hybrid, and it was found for A9HyTK-13, 14, 15, 21 and 22 that the human chromosomes were co-localized with the fibrillarin staining in 86, 84, 92, 82 and 82% of cells, respectively. In contrast, the X chromosome was only co-localized with nucleoli in 39% of cells. These numbers show that human acrocentric chromosomes co-localize with mouse nucleoli. Some of the cells that were scored negative had chromosomes extremely close to the nucleolus but, given our scoring criteria, were scored as not co-localizing. It should be noted that the p arms containing the rDNA were not painted, which may have resulted in us not visualizing all co-localizations. Both arms of the control HSAX were painted. We conclude from these experiments that each of the human acrocentrics co-localize with hybrid cell nucleoli irrespective of their UBF loading status and despite the transcriptional silence of the associated ribosomal genes.
Characterization of purified nucleoli
The above experiments demonstrate an association between human acrocentric chromosomes and nucleoli in intact cells. To demonstrate that hybrid cell nucleoli contain transcriptionally silent human NORs, we have performed analyses on purified nucleoli. Due to their mass, nucleoli can be purified conveniently by sonication and centrifugation in a high-density medium (Muramatsu et al., 1963; Maggio, 1966). Using this methodology, suspensions of essentially pure nucleoli can be prepared.
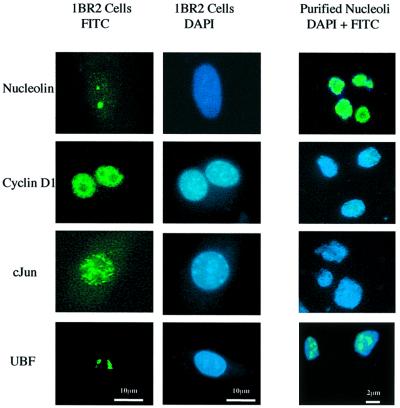
We have verified nucleolar purification in our preparations by immunofluorescence staining with antibodies directed against known nucleolar antigens (UBF and nucleolin) and nuclear antigens that are excluded from the nucleolus (c-Jun and cyclin D1). As expected, UBF and nucleolin antibodies stain purified nucleoli whereas c-Jun and cyclin D1 antibodies do not (Figure 5).
Fig. 5. Verification of nucleolar purification. 1BR.2 cells and nucleoli purified from these cells (both counterstained with DAPI) were stained with antibodies against nucleolar antigens (nucleolin and UBF) and nuclear antigens (cyclin D1 and c-Jun) that are excluded from nucleoli. Panels on the left show antibody staining of 1BR.2 cells (green). Centre panels show DAPI staining of 1BR.2 cells (blue). Right panels show combined antibody (green) and DAPI (blue) staining of nucleoli purified from 1BR.2 cells. Note that the scale bars for cell staining and nucleolar staining are different (10 and 2 µm, respectively).
Transcriptionally silent human NORs are present within purified nucleoli
To determine whether human NORs are incorporated into nucleoli in hybrid cells, FISH was performed on isolated nucleoli (Figure 6). All NORs (mouse and human) were visualized using a 28S rDNA probe and human NORs were visualized using a probe derived from the human rDNA intergenic spacer. This experiment was performed on nucleoli isolated from A9HyTK-15 and 21. Note that the NOR on HSA15 has associated UBF while that on HSA21 does not (see Figure 3). A positive signal is observed over all nucleoli with the 28S probe (Figure 6A). The presence of multiple signals in the larger nucleoli is consistent with them containing multiple mouse NORs. Given that hybrid cells each contain on average three nucleoli (data not shown) and only a single human acrocentric chromosome (Cuthbert et al., 1995), we would expect 33% of purified nucleoli to contain human rDNA sequences. Indeed, we found that 20–40% (dependent on the identity of the hybrid) of purified nucleoli did contain a single hybridization signal when utilizing the human rDNA-specific probe. Typical positive-staining nucleoli are shown in Figure 6B.
Fig. 6. Hybrid cell nucleoli contain human NORs. (A) Nucleoli isolated from HSA15 and 21 were probed with a 28S rDNA probe that detects both human and mouse NORs. Left hand panels show DAPI staining of nucleoli, right hand panels show 28S rDNA signal (red). The identity of the human chromosome present in the hybrid is shown in the top left hand corner. The scale bar represents 1 µm. (B) Nucleoli isolated from HSA15 and 21 were probed with a human-specific rDNA. Left hand panels show DAPI staining of nucleoli, right hand panels show human rDNA signal (red). (C) Quantitative PCR was performed on both genomic DNA from hybrid cells and DNA isolated from their purified nucleoli (see Materials and methods for details). PCR products generated with primer pairs that amplify mouse and human rDNA promoter sequences are labelled M and H, respectively. The source of template DNA is shown above. In control reactions (C), PCR was performed in the absence of input DNA. Signals were quantified using phosphorimaging (Bio-Rad) and the ratio of H to M signal for each template is shown below.
For a more quantitative comparison, DNAs extracted from A9HyTK-14, 15 and 21 nucleoli were used as templates in quantitative PCR (Figure 6C). A9 nucleoli provided a control. Primer pairs that specifically recognize either human or mouse ribosomal gene promoter sequences were utilized. Primers were labelled isotopically in order to facilitate quantitation of PCR products by phosphorimaging. As A9HyTK-14, 15 and 21 cells contain only a single human NOR (i.e. one-tenth of the rDNA that is found in a normal human cell), the human primer pairs used in PCR were labelled to 10 times the specific activity of the mouse primer pairs. Using cloned templates, we have demonstrated that the PCR conditions used here were in the linear range and that the amount of product was proportional to the amount of target sequence present (data not shown). To establish a ratio of human to mouse rDNA present in each hybrid cell, quantitative PCR was performed using total genomic DNA as template. As expected, we observe a human to mouse signal ratio of ∼1 for each of the hybrids (see Figure 6C). No product was observed with human primers using control A9 genomic DNA as template. The ratio of signal obtained from human and mouse primer pairs using nucleolar DNA prepared from A9HyTK-14, 15 and 21 was equivalent to that obtained with genomic DNA. Thus we can conclude that human NORs are within hybrid cell nucleoli, and that this occurs with a probability comparable to that of a mouse NOR.
Hybrid cell nucleoli contain sequences derived from the long arm of human acrocentric chromosomes
Figure 4A and B shows that the q arms of HSA13, 14, 15, 21 and 22 co-localize with nucleoli in the nuclei of the relevant hybrid cells. To determine whether this reflects a genuine physical association, we used the same probes in FISH experiments on isolated nucleoli (Figure 7). As above, we found that 20–40% (dependent on the identity of the hybrid) of purified nucleoli did contain hybridization signals from the appropriate q arm human acrocentric chromosome paint. Typical positively hybridizing nucleoli are shown in Figure 7. In contrast, a chromosome paint derived from both the p and q arms of HSAX did not hybridize significantly to nucleoli purified from A9HyTK-X (<5%) (Figure 7).
Fig. 7. Sequences from the q arms of human acrocentric chromosomes co-purify with hybrid cell nucleoli. Nucleoli purified from A9HyTK-13, 14, 15, 21, 22 and X were probed with paints derived from the q arms of HSA13, 14, 15, 21 and 22, and the p and q arms of HSAX, respectively (green). Nucleoli were stained with DAPI (blue). The identity of each nucleolus is shown in the top left corner of each panel. The scale bar represents 1 µm.
Discussion
Here we have demonstrated that the NOR present on each of the human acrocentric chromosomes can associate with a nucleolus irrespective of its ribosomal gene transcription status. This conclusion is supported by the following observations. Human NORs are transcriptionally silent in a mouse cell background (Figure 1). Nevertheless, three-dimensional immuno-FISH showed clearly that each of the human acrocentrics associated with hybrid cell nucleoli in vivo (Figure 4A and B). We visualized human NORs within isolated hybrid cell nucleoli (Figure 6B). Quantitative PCR illustrated that human and mouse NORs are equally likely to be present within nucleoli (Figure 6C). FISH clearly demonstrated that sequences from the long arm of human acrocentric chromosomes are physically associated with hybrid cell nucleoli (Figure 7). Finally, the observation that HSA14 and 15 have the nucleolar pol I transcription factor UBF associated with their NORs underscores their nucleolar address (Figure 3).
Our investigation warrants reconsideration of current views on nucleolar formation. In mitotic cells, nucleolar proteins such as nucleolin, fibrillarin and small nucleolar RNAs are located within a specific domain termed the perichromosomal layer (Scheer and Weisenberger, 1994; Olson et al., 2000). This is thought to facilitate their equal partitioning to daughter cells during mitosis. At the end of mitosis, these nucleolar proteins coalesce in pre-nucleolar bodies (PNBs), which are distributed through the telophase nucleus (Scheer and Weisenberger, 1994; Olson et al., 2000). PNBs can be considered as pre-packaged nucleolar complexes whose primary function is base modification and processing of rRNA. As cells enter G1, rRNA gene transcription resumes and PNBs fuse to NORs (Ochs et al., 1985; Jimenez-Garcia et al., 1994). This results in the formation of multiple small nucleoli. This fusion appears to be dependent on ribosomal gene transcription. On inhibition of transcription by injection of antibodies recognizing pol I into metaphase cells or pharmacologically with low concentrations of actinomycin D (act D), nucleoli are not reformed in the G1 nuclei of daughter cells. Instead, nuclei are filled with numerous PNBs (Ochs et al., 1985; Benavente et al., 1987). It has been proposed that PNB targeting is dependent on rDNA transcription and, consequently, that nucleoli only form on acrocentric chromosomes with an active NOR (Scheer and Hock, 1999). Similarly, it has been demonstrated that nascent snRNA transcripts mediate association of snRNA genes with coiled bodies (Matera, 1999). Interestingly, it has been demonstrated recently that in act D-treated cells, UBF-associated NORs may recruit processing factors directly, together with pre-rRNA that was synthesized in the previous G2 (Dousset et al., 2000). This process does not involve PNBs and results in formation of mini-nucleolar structures on these NORs, even when ongoing ribosomal gene transcription is inhibited. It should be stressed, however, that formation of nucleolar structures on inactive NORs (i.e. those that have no associated transcription machinery) was not addressed (Dousset et al., 2000).
At first glance, the results we have presented here appear to contradict the above models of nucleolar formation. Our data are best interpreted by a consideration of subsequent events in the nucleolar cycle. As cells progress through the cycle, the multiple small nucleoli that form around individual active NORs appear to fuse into one or a few large nucleoli, a phenomenon commonly referred to as nucleolar fusion (Anastassova-Kristeva, 1977). A consequence of this process is that multiple NORs can be found within a single nucleolus. The mechanism underlying nucleolar fusion is unclear. If it results from an inherent affinity between nucleoli that form on individual acrocentric chromosomes, inactive NORs should remain excluded from nucleoli. We believe that our data provide support for an alternative model for the nucleolar cycle. In this model, nucleoli initially form only around transcriptionally active (or UBF-loaded) NORs in early G1. Subsequently, there is a dynamic reorganization of acrocentric chromosomes within the interphase nucleus that is independent of the activity status of linked NORs. A consequence of this chromosomal reorganization is nucleolar fusion. The question then arises, what are the chromosomal elements that mediate these associations? It is unlikely that DNA sequences themselves facilitate this chromosomal reorganization. It is more likely that it is the associated chromatin or higher order structure that is recognized. As NORs display a heterogeneous chromatin structure, manifest by the presence or absence of a secondary constriction, we think it unlikely that they directly facilitate nucleolar association of acrocentric chromosomes.
We consider it more likely that chromosomal elements other than rDNA mediate acrocentric chromosome clustering. It has been known for some time that a fraction of human centromeres cluster around nucleoli in vivo (Park and De Boni, 1992; Leger et al., 1994; Bridger et al., 1998). Furthermore, we (data not shown) and others have demonstrated that centromeres are integral components of purified nucleoli (Ochs and Press, 1992). This is not surprising given the proximity of rDNA and centromeres on human acrocentric chromosomes. However, centromeres from some non-acrocentric chromosomes also appear to associate with nucleoli. Pericentromeric heterochromatic regions of HSA1, 9 and Y have been shown to associate with nucleoli in vivo (Manuelidis and Borden, 1988; Leger et al., 1994; Bridger et al., 1998).
Long arm sequences are unlikely candidates for mediating chromosome associations as they are not conserved between the different acrocentrics. It is difficult to rule centromeres in or out since no defining characteristic has yet been ascribed to those of acrocentric chromosomes. It is tempting to speculate that heterochromatic sequences both proximal and distal to the NOR on the p arms of human acrocentric chromosomes are responsible for their association. These heterochromatic sequences comprise arrays of tandemly repeated satellite DNA, including β-satellite and satellites 1, 2 and 3 (Waye and Willard, 1989; Choo et al., 1990; Tagarro et al., 1994; Sakai et al., 1995; Shiels et al., 1997b). Interestingly, some of these satellites are also present at the large blocks of pericentric heterochromatin present on HSA1, 9 and Y (Waye and Willard, 1989) that have been demonstrated to associate with nucleoli (Manuelidis and Borden, 1988). This provides further suggestive evidence for a model in which acrocentric chromosome associations are facilitated by acrocentric p arm heterochromatin. Recent work in yeast has uncovered a role for rDNA-associated heterochromatin in the maintenance of nucleolar structure (reviewed in Carmo-Fonseca et al., 2000). Silent rDNA repeats are interspersed with active repeats in yeast (Dammann et al., 1995). Given the proposed role of rDNA heterochromatin in nucleolar structure, this interspersion may be critical to nucleolar function. The observations that in certain cell types NORs can be found outside the nucleolus (Weipoltshammer et al., 1996) or that multiple nucleoli fail to associate (Krystosek, 1998) raise the possibility that association of acrocentric chromosomes may be a regulated event.
The generally accepted view that inactive NORs are excluded from the nucleolus provides a simplistic mechanism for the maintenance of their silent status. They are never exposed to the pol I transcription machinery, which is exclusively nucleolar in location. The implication of the work presented here is that in human cells, all NORs can be localized to a nucleolar environment regardless of their transcriptional status. This raises an interesting question. What is the mechanism by which certain NORs can associate with the pol I transcription factor UBF while others cannot? This is all the more puzzling given the abundance of UBF (up to 106 molecules per cell). An epigenetic mechanism provides the most likely explanation. In the future, these monochromosomal hybrids will provide a valuable model system for studying epigenetic differences between active and inactive NORs, since we can study human NORs of a particular class (e.g. with or without associated UBF).
Materials and methods
Cell culture
Mouse A9 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum (FCS, Gibco). Monochromosome hybrids A9HyTK-13, A9HyTK-14, A9HyTK-15, A9HyTK-21, A9HyTK-22 and A9HyTK-X (A9 cells containing human chromosomes 13, 14, 15, 21, 22 and X, respectively) were cultured in DMEM supplemented with 10% FCS and 400 U/ml hygromycin B (Calbiochem). The normal adult male fibroblast strain 1BR.2 was maintained in DMEM supplemented with 15% FCS.
DNA analysis
A 1.2 kb fragment present ∼8 kb upstream of the human ribosomal gene promoter was used as a probe in the Southern blotting experiement shown in Figure 1A. This fragment encompasses the cdc25 pseudogene present at this location and is devoid of both Alu and other repetitive elements found in the human rDNA intergenic spacer.
RNA analysis
The 5′ ends of human 47S rRNA were detected with the antisense strand of a subcloned human rDNA fragment extending from position –224 to +51, end-labelled with T4 polynucleotide kinase and [γ-32P]ATP (3000 Ci/mM Amersham). The 5′ ends of mouse 47S rRNA were detected with the antisense strand of a labelled PCR product generated from mouse rDNA with the primer pair described below. To detect human 28S rRNA specifically, the oligonucleotide 5′-GACCGCTCC CGCCCCCAGCGGACGCGCGCGCGACCGAGACGTG-3′ (complementary to nucleotides 3326–3369 of human 28S rRNA) was end-labelled as above. Total RNA was isolated from dividing cells using Trizol (Gibco), and S1 nuclease protection assays were performed as described previously (Labhart and Reeder, 1986).
Isolation of nucleoli
Cells (2 × 150 mm dishes) were grown to ∼70% confluence and fixed by addition of formaldehyde to the medium to a final concentration of 0.2%. After incubation at room temperature for 10 min, the medium was removed and the cells were washed in phosphate-buffered saline (PBS) and harvested by scraping into a total volume of 40 ml of PBS. After centrifugation at 200 g for 5 min, the cell pellet was resuspended into 1.0 ml of high magnesium buffer [HM: 10 mM HEPES pH 7.5, 0.88 M sucrose, 12 mM MgCl2, 1 mM dithiothreitol (DTT) plus protease inhibitors]. Nucleoli were released by sonication on ice (2–3 bursts of 10 s each at full power) using a Soniprep 150 (MSE) with a fine probe. Release of nucleoli was monitored microscopically. Following sonication, nucleoli were pelleted by centrifugation in a microfuge (15 000 g for 20 s). The nucleolar pellet was resuspended in 0.5 ml of low magnesium buffer (LM: 10 mM HEPES pH 7.5, 0.88 M sucrose, 1 mM MgCl2, 1 mM DTT plus protease inhibitors). Nucleoli resuspended in LM were subjected to a further sonication (10 s full power) and pelleted as before. Nucleoli to be used immediately for immunofluorescent staining, FISH or DNA extraction were resuspended in 20/2 TE (20 mM Tris pH 7.5, 2 mM EDTA). For long-term storage, nucleoli were resuspended in 20/2 TE plus 50% glycerol and stored at –20°C.
Quantitative PCR
The primers used to amplify human ribosomal gene sequences were 5′-TTTCGCTCCGAGTCGGCA-3′ and 5′-TCTCCAGCGACAGGTC GCC-3′. These primers yield a 77 bp product encoding nucleotides –40 to +37 from the human ribosomal gene promoter. The primers used to amplify mouse ribosomal gene sequences were 5′-GTTGTCAGGTCG ACCAGTTGT-3′ and 5′-GTGTCCTTTAGTGTTAATAGG-3′. These primers yield a 216 bp product encoding nucleotides –176 to + 40 of the mouse ribosomal gene promoter. PCRs (50 µl) contained 10 pM of each primer and 2.5 U of Taq DNA polymerase, and were performed in the following buffer: 50 mM KCl, 10 mM Tris pH 9.0, 0.1% Triton X-100, 2.5 mM MgCl2, 0.2 mM dNTPs, 5% dimethylsulfoxide (DMSO). Reactions also contained 0.5 or 0.05 pM of the human or mouse primer pairs, respectively, end-labelled with T4 polynucleotide kinase and [γ-32P]ATP (3000 Ci/mM; Amersham). Nucleolar DNA equivalent to that isolated from ∼105 cells or 50 ng of the relevant total genomic DNA was used as template. Reactions were either 20 or 23 cycles of 0.5 min at 95°C followed by 0.5 min at 45°C then 1.0 min at 72°C. Aliquots of PCRs were subjected to electrophoresis on 8% polyacrylamide gels in 1× TBE. Signals were quantified using a phosphorimager (Bio-Rad).
Antibodies
The human anti-UBF antibody (NOR90/9386) used here was identified from a panel of autoimmune sera on the basis of reactivity towards recombinant hUBF. This antibody was used at a 1:100 dilution for immunofluorescent staining and 1:500 for western blotting. The anti-fibrillarin mouse monoclonal antibody D77 (gift of John Aris, University of California) was used at a dilution of 1:1000. Anti-nucleolin rabbit polyclonal antibodies have been described previously (Cairns and McStay, 1995) and were used at a 1:100 dilution for immunofluorescence staining. Anti c-Jun (SC822) mouse monoclonal and anti-cyclin D1 (SC718) rabbit polyclonal antibodies were purchased from Santa Cruz Biotechnology Inc. (Santa Cruz, CA) and were used at 1:100 and 1:50 dilutions, respectively. Goat anti-rabbit IgG–FITC, goat anti-mouse IgG–FITC, goat anti-human IgG–FITC and goat anti-human IgG–horseradish peroxidase (HRP) conjugate were purchased from Sigma Chemical Co. All secondary antibodies were used at the recommended dilution.
Immunofluorescence staining of 1BR.2 cells and purified nucleoli
1BR.2 cells grown on coverslips were washed three times in PBS and fixed in methanol (–20°C) for 4 min. Following fixation, cells were washed as above. Cells were incubated with 30 µl of primary antibody (diluted in PBS/1% FCS as indicated above) for 30 min at 37°C in a humidified chamber. The slides were then washed three times in PBS/1% FCS (2 min/wash). A 30 µl aliquot of secondary antibody was then applied (diluted as indicated above) and incubated for 30 min at 37°C in a humidified chamber. The slides were then washed three times in PBS/ 1% FCS, once with PBS and once with water. Coverslips were then mounted in Vectashield (Vector Labs, Burlingame, CA) containing 4′,6′-diamidino-2-phenylindole (DAPI).
Purified nucleoli were dropped onto slides, air dried, washed in PBS/1% FCS and incubated with primary and secondary antibodies as above.
Combined immunofluorescence and FISH on metaphase spreads
UBF was visualized using NOR90/9386. Human chromosomes were identified using labelled Cot-1 DNA. NORs (mouse and human) were detected with a 28S rDNA probe and human NORs were identified with a human-specific rDNA probe. (For details of probes, metaphase chromosomes preparation and FISH see Supplementary data, available at The EMBO Journal Online.)
FISH-isolated nucleoli
Purified nucleoli were dropped onto slides, air dried and incubated in 2× SSC containing 100 µg/ml RNase A at 37°C for 2 h. Slides were then processed for hybridization, and FISH was performed as described above except that formamide denaturation was performed for only 2 min at 70°C.
Combined immunofluorescence and FISH on three-dimensionally preserved nuclei
Immuno-FISH on three-dimensionally preserved nuclei was performed essentially as described by Bridger and Lichter (1999). Human acrocentric chromosomes were visualized with q arm-specific paints and human X was visualized with a whole chromosome paint (kind gift of M.Bittner; see Guan et al., 1996) (see Supplementary data).
Supplementary data
Supplementary data for this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
The authors thank Mike Kerr for human autoantisera, Mark Sallis and Norman Pratt (Ninewells Hospital, Dundee) for help with image acquisition, Beth Sullivan (Salk Institute) for the CREST sera, Mike Bittner and Jeff Trent (NIH, Bethesda, MD) for human chromosome paints, John Aris (University of California, San Francisco CA) for D77 anti-fibrillarin monoclonal antibody, Deborah Trott (Brunel) for hybrid cell lines and Paul Perry for personalized scripts. W.A.B. is a Centennial fellow in human genetics of the James S.McDonnell foundation. B.M. is funded by a Program Grant from the MRC.
References
- Akhmanova A., Verkerk,T., Langeveld,A., Grosveld,F. and Galjart,N. (2000) Characterisation of transcriptionally active and inactive chromatin domains in neurons. J. Cell Sci., 113, 4463–4474. [DOI] [PubMed] [Google Scholar]
- Anastassova-Kristeva M. (1977) The nucleolar cycle in man. J. Cell Sci., 25, 103–110. [DOI] [PubMed] [Google Scholar]
- Babu K.A. and Verma,R.S. (1985) Structural and functional aspects of nucleolar organizer regions (NORs) of human chromosomes. Int. Rev. Cytol., 94, 151–176. [DOI] [PubMed] [Google Scholar]
- Bell S.P., Learned,R.M., Jantzen,H.M. and Tjian,R. (1988) Functional cooperativity between transcription factors UBF1 and SL1 mediates human ribosomal RNA synthesis. Science, 241, 1192–1197. [DOI] [PubMed] [Google Scholar]
- Benavente R., Rose,K.M., Reimer,G., Hugle-Dorr,B. and Scheer,U. (1987) Inhibition of nucleolar reformation after microinjection of antibodies to RNA polymerase I into mitotic cells. J. Cell Biol., 105, 1483–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridger J.M. and Bickmore,W.A. (1998) Putting the genome on the map. Trends Genet., 14, 403–409. [DOI] [PubMed] [Google Scholar]
- Bridger J.M. and Lichter,P. (1999) Analysis of mammalian interphase chromosomes by FISH and immunofluorescence. In Bickmore,W.A. (ed.), Chromosome Structural Analysis. Oxford University Press, Oxford, UK, pp. 103–121.
- Bridger J.M., Kill,I.R. and Lichter,P. (1998) Association of pKi-67 with satellite DNA of the human genome in early G1 cells. Chromosome Res., 6, 13–24. [DOI] [PubMed] [Google Scholar]
- Cairns C. and McStay,B. (1995) Identification and cDNA cloning of a Xenopus nucleolar phosphoprotein, xNopp180, that is the homolog of the rat nucleolar protein Nopp140. J. Cell Sci., 108, 3339–3347. [DOI] [PubMed] [Google Scholar]
- Carmo-Fonseca M., Mendes-Soares,L. and Campos,I. (2000) To be or not to be in the nucleolus. Nature Cell Biol., 2, E107–E112. [DOI] [PubMed] [Google Scholar]
- Chan E.K., Imai,H., Hamel,J.C. and Tan,E.M. (1991) Human autoantibody to RNA polymerase I transcription factor hUBF. Molecular identity of nucleolus organizer region autoantigen NOR-90 and ribosomal RNA transcription upstream binding factor. J. Exp. Med., 174, 1239–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo K.H., Earle,E. and McQuillan,C. (1990) A homologous subfamily of satellite III DNA on human chromosomes 14 and 22. Nucleic Acids Res., 18, 5641–5648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L., Tanese,N. and Tjian,R. (1992) The TATA-binding protein and associated factors are integral components of the RNA polymerase I transcription factor, SL1. Cell, 68, 965–976. [DOI] [PubMed] [Google Scholar]
- Cuthbert A.P., Trott,D.A., Ekong,R.M., Jezzard,S., England,N.L., Themis,M., Todd,C.M. and Newbold,R.F. (1995) Construction and characterization of a highly stable human:rodent monochromosomal hybrid panel for genetic complementation and genome mapping studies. Cytogenet. Cell Genet., 71, 68–76. [DOI] [PubMed] [Google Scholar]
- Dammann R., Lucchini,R., Koller,T. and Sogo,J.M. (1995) Transcription in the yeast rRNA gene locus: distribution of the active gene copies and chromatin structure of their flanking regulatory sequences. Mol. Cell. Biol., 15, 5294–5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dousset T., Wang,C., Verheggen,C., Chen,D., Hernandez-Verdun,D. and Huang,S. (2000) Initiation of nucleolar assembly is independent of RNA polymerase I transcription. Mol. Biol. Cell, 11, 2705–2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodpasture C. and Bloom,S.E. (1975) Visualization of nucleolar organizer regions in mammalian chromosomes using silver staining. Chromosoma, 53, 37–50. [DOI] [PubMed] [Google Scholar]
- Grummt I. (1999) Regulation of mammalian ribosomal gene transcription by RNA polymerase I. Prog. Nucleic Acid Res. Mol. Biol., 62, 109–154. [DOI] [PubMed] [Google Scholar]
- Guan X.Y., Zhang,H., Bittner,M., Jiang,Y., Meltzer,P. and Trent,J. (1996) Chromosome arm painting probes. Nature Genet., 12, 10–11. [DOI] [PubMed] [Google Scholar]
- Heix J. and Grummt,I. (1995) Species specificity of transcription by RNA polymerase I. Curr. Opin. Genet. Dev., 5, 652–656. [DOI] [PubMed] [Google Scholar]
- Heliot L. et al. (1997) Electron tomography of metaphase nucleolar organizer regions: evidence for a twisted-loop organization. Mol. Biol. Cell, 8, 2199–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson A.S., Warburton,D. and Atwood,K.C. (1972) Location of ribosomal DNA in the human chromosome complement. Proc. Natl Acad. Sci. USA, 69, 3394–3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Garcia L.F., Segura-Valdez,M.L., Ochs,R.L., Rothblum,L.I., Hannan,R. and Spector,D.L. (1994) Nucleologenesis: U3 snRNA-containing prenucleolar bodies move to sites of active pre-rRNA transcription after mitosis. Mol. Biol. Cell, 5, 955–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan P., Mannervik,M., Tora,L. and Carmo-Fonseca,M. (1996) In vivo evidence that TATA-binding protein/SL1 colocalizes with UBF and RNA polymerase I when rRNA synthesis is either active or inactive. J. Cell Biol., 133, 225–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krystosek A. (1998) Repositioning of human interphase chromosomes by nucleolar dynamics in the reverse transformation of HT1080 fibrosarcoma cells. Exp. Cell Res., 241, 202–209. [DOI] [PubMed] [Google Scholar]
- Kuhn A., Vente,A., Doree,M. and Grummt,I. (1998) Mitotic phosphorylation of the TBP-containing factor SL1 represses ribosomal gene transcription. J. Mol. Biol., 284, 1–5. [DOI] [PubMed] [Google Scholar]
- Labhart P. and Reeder,R.H. (1986) Characterization of three sites of RNA 3′ end formation in the Xenopus ribosomal gene spacer. Cell, 45, 431–443. [DOI] [PubMed] [Google Scholar]
- Leger I., Guillaud,M., Krief,B. and Brugal,G. (1994) Interactive computer-assisted analysis of chromosome 1 colocalization with nucleoli. Cytometry, 16, 313–323. [DOI] [PubMed] [Google Scholar]
- Maggio R. (1966) Some properties of isolated nucleoli from guinea-pig liver. Biochim. Biophys. Acta, 119, 641–644. [DOI] [PubMed] [Google Scholar]
- Manuelidis L. and Borden,J. (1988) Reproducible compartmentalization of individual chromosome domains in human CNS cells revealed by in situ hybridization and three-dimensional reconstruction. Chromosoma, 96, 397–410. [DOI] [PubMed] [Google Scholar]
- Matera A.G. (1999) Nuclear bodies: multifaceted subdomains of the interchromatin space. Trends Cell Biol., 9, 302–309. [DOI] [PubMed] [Google Scholar]
- Miller O.J., Miller,D.A., Dev,V.G., Tantravahi,R. and Croce,C.M. (1976) Expression of human and suppression of mouse nucleolus organizer activity in mouse–human somatic cell hybrids. Proc. Natl Acad. Sci. USA, 73, 4531–4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu M., Smetana,K. and Busch,H. (1963) Quantitative aspects of isolation of nucleoli of the Walker carcinosarcoma and liver of the rat. Cancer Res., 23, 510–522. [PubMed] [Google Scholar]
- Ochs R.L. and Press,R.I. (1992) Centromere autoantigens are associated with the nucleolus. Exp. Cell Res., 200, 339–350. [DOI] [PubMed] [Google Scholar]
- Ochs R.L., Lischwe,M.A., Shen,E., Carroll,R.E. and Busch,H. (1985) Nucleologenesis: composition and fate of prenucleolar bodies. Chromosoma, 92, 330–336. [DOI] [PubMed] [Google Scholar]
- Olson M.O., Dundr,M. and Szebeni,A. (2000) The nucleolus: an old factory with unexpected capabilities. Trends Cell Biol., 10, 189–196. [DOI] [PubMed] [Google Scholar]
- Park P.C. and De Boni,U. (1992) Spatial rearrangement and enhanced clustering of kinetochores in interphase nuclei of dorsal root ganglion neurons in vitro: association with nucleolar fusion. Exp. Cell Res., 203, 222–229. [DOI] [PubMed] [Google Scholar]
- Pederson T. (1998) The plurifunctional nucleolus. Nucleic Acids Res., 26, 3871–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder R.H., Pikaard,C.S. and McStay,B. (1995) UBF, an architectural element for RNA polymerase I promoters. In Eckstein,F. and Lilley,D.M.J. (eds), Nucleic Acids and Molecular Biology. Vol. 9. Springer-Verlag, Berlin, Heidelberg, Germany, pp. 251–263.
- Roussel P., Andre,C., Masson,C., Geraud,G. and Hernandez,V.D. (1993) Localization of the RNA polymerase I transcription factor hUBF during the cell cycle. J. Cell Sci., 104, 327–337. [DOI] [PubMed] [Google Scholar]
- Roussel P., Andre,C., Comai,L. and Hernandez-Verdun,D. (1996) The rDNA transcription machinery is assembled during mitosis in active NORs and absent in inactive NORs. J. Cell Biol., 133, 235–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai K., Ohta,T., Minoshima,S., Kudoh,J., Wang,Y., de Jong,P.J. and Shimizu,N. (1995) Human ribosomal RNA gene cluster: identification of the proximal end containing a novel tandem repeat sequence. Genomics, 26, 521–526. [DOI] [PubMed] [Google Scholar]
- Scheer U. and Hock,R. (1999) Structure and function of the nucleolus. Curr. Opin. Cell Biol., 11, 385–390. [DOI] [PubMed] [Google Scholar]
- Scheer U. and Weisenberger,D. (1994) The nucleolus. Curr. Opin. Cell Biol., 6, 354–359. [DOI] [PubMed] [Google Scholar]
- Shiels C., Coutelle,C. and Huxley,C. (1997a) Analysis of ribosomal and alphoid repetitive DNA by fiber-FISH. Cytogenet. Cell Genet., 76, 20–22. [DOI] [PubMed] [Google Scholar]
- Shiels C., Coutelle,C. and Huxley,C. (1997b) Contiguous arrays of satellites 1, 3 and β form a 1.5-Mb domain on chromosome 22p. Genomics, 44, 35–44. [DOI] [PubMed] [Google Scholar]
- Sirri V., Roussel,P. and Hernandez-Verdun,D. (2000) In vivo release of mitotic silencing of ribosomal gene transcription does not give rise to precursor ribosomal RNA processing. J. Cell Biol., 148, 259–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soprano K.J. and Baserga,R. (1980) Reactivation of ribosomal RNA genes in human–mouse hybrid cells by 12-O-tetradecanoylphorbol 13-acetate. Proc. Natl Acad. Sci. USA, 77, 1566–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagarro I., Wiegant,J., Raap,A.K., Gonzalez-Aguilera,J.J. and Fernandez-Peralta,A.M. (1994) Assignment of human satellite 1 DNA as revealed by fluorescent in situ hybridization with oligonucleotides. Hum. Genet., 93, 125–128. [DOI] [PubMed] [Google Scholar]
- Voit R., Hoffmann,M. and Grummt,I. (1999) Phosphorylation by G1-specific cdk–cyclin complexes activates the nucleolar transcription factor UBF. EMBO J., 18, 1891–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waye J.S. and Willard,H.F. (1989) Concerted evolution of α satellite DNA: evidence for species specificity and a general lack of sequence conservation among alphoid sequences of higher primates. Chromosoma, 98, 273–279. [DOI] [PubMed] [Google Scholar]
- Weipoltshammer K., Schofer,C., Almeder,M., Sylvester,J. and Wachtler,F. (1996) Spatial distribution of sex chromosomes and ribosomal genes: a study on human lymphocytes and testicular cells. Cytogenet. Cell Genet., 73, 108–113. [DOI] [PubMed] [Google Scholar]
- Weisenberger D. and Scheer,U. (1995) A possible mechanism for the inhibition of ribosomal RNA gene transcription during mitosis. J. Cell Biol., 129, 561–575. [DOI] [PMC free article] [PubMed] [Google Scholar]