Abstract
Evidence for post-recruitment functions of yeast transcription factor (TF)IIIB in initiation of transcription was first provided by the properties of TFIIIB–RNA polymerase III–promoter complexes assembled with deletion mutants of its Brf and B” subunits that are transcriptionally inactive because they fail to open the promoter. The experiments presented here show that these defects can be repaired by unpairing short (3 or 5 bp) DNA segments spanning the transcription bubble of the open promoter complex. Analysis of this suppression phenomenon indicates that TFIIIB participates in two steps of promoter opening by RNA polymerase III that are comparable to the successive steps of promoter opening by bacterial RNA polymerase holoenzyme. B” deletions between amino acids 355 and 421 interfere with the initiating step of DNA strand separation at the upstream end of the transcription bubble. Removing an N-terminal domain of Brf interferes with downstream propagation of the transcription bubble to and beyond the transcriptional start site.
Keywords: Brf/B”/RNA polymerase III/transcription bubble/transcription factors
Introduction
Accurately initiated transcription by RNA polymerase (pol) III requires the action of three transcription factors, TFIIIA, B and C (reviewed by White, 1998). While the 10 yeast genes encoding the subunits of these three transcription factors are all essential for viability, TFIIIB is central in the sense that it alone suffices for recruiting RNA polymerase to the promoter and for securing repeated rounds of accurately initiated transcription in vitro (Kassavetis et al., 1990). The large, multi-subunit TFIIIC plays two critical accessory roles in initiation of transcription: it recruits TFIIIB to genes that lack strong TATA boxes, and it protects pol III genes against transcription-blocking encroachment by chromatin (Burnol et al., 1993; Marsolier et al., 1995; Hsieh et al., 1999; Kundu et al., 1999). TFIIIA, which binds exclusively to the promoters of 5S rRNA genes (Engelke et al., 1980), serves as the platform for binding TFIIIC to these genes (Braun et al., 1992).
Protein–protein interactions that generate these actions have been identified, but perhaps not yet fully enumerated. The 120 kDa TFIIIC subunit (Tfc4/Pcf1) interacts with the Brf and B” (Tfc5) subunits of TFIIIB. Brf engages pol III through at least three interactions: one site interacts with the pol III C17 subunit and two sites interact with the pol III C34 subunit. The latter, together with the C82 and C31 pol III subunits, forms a subassembly that is essential for accurate, transcription factor-directed initiation of transcription by pol III, but not for transcript elongation. Additional pol III interactions extend the network of contacts that allows pol III to be brought to its promoters by its core transcription factors (Khoo et al., 1994; Rameau et al., 1994; Thuillier et al., 1995; Roberts et al., 1996; Rüth et al., 1996; Chédin et al., 1998; Sethy-Coraci et al., 1998; Andrau et al., 1999; Dumay et al., 1999; Flores et al., 1999; Ferri et al., 2000). Polymerase recruitment patently is an essential function of the transcription initiation factors, but it is not their sole function.
Evidence for the existence of post-recruitment functions for TFIIIB in initiation of transcription was first provided by analyzing the properties of TFIIIB assembled with certain Brf and B” mutant subunits. These variant TFIIIB complexes retain the ability to direct accurately initiating transcription of supercoiled DNA, but are inactive or very severely defective in transcription of linear DNA. Further analysis showed that the ability to recruit and accurately place pol III over the promoter is retained, but that the promoter fails to open, so that neither abortive nor productive transcripts are made (Kassavetis et al., 1998b; Hahn and Roberts, 2000).
In the experiments reported below, we show that this defective transcription can be rescued by unpairing the DNA template so as to partially open the pol III promoter. The subsequent analysis of this suppression phenomenon provides evidence that TFIIIB intervenes in promoter opening at two steps: one of these interventions facilitates opening of an upstream segment of the promoter and is abolished by deletion of contiguous small segments of B”; the second intervention facilitates opening of a downstream segment and is abolished by removal of an N-terminal segment of Brf that includes its putative zinc ribbon.
Results
TFIIIB mutants defective in promoter opening
We have previously shown that combinations of an N-terminal truncation of Brf (NΔ164Brf), which removes the N-proximal putative zinc ribbon and the first TFIIB-related pseudo repeat element, with small (10–18 amino acid) internal deletions in B” between amino acids 272 and 310, and also between amino acids 355 and 449, destroy the ability to direct transcription of supercoiled DNA (Kassavetis et al., 1997). The defective combinations of NΔ164Brf with B” deletions between amino acids 272 and 310, and between amino acids 409 and 449, destroy the ability to form TFIIIB–DNA complexes, whereas the combinations of NΔ164Brf with B” internal deletions between amino acids 355 and 409 generate transcriptionally inactive TFIIIB–DNA complexes. TFIIIB–DNA complexes formed either with NΔ164Brf and wild-type B”, or with wild-type Brf and B” containing small internal deletions between amino acids 355 and 421, are transcriptionally competent on supercoiled DNA, but not on relaxed or linear DNA (Kassavetis et al., 1999). These defective TFIIIB–DNA complexes recruit RNA pol III and correctly position it over the start site of transcription but they do not allow localized unwinding of DNA to form an open complex.
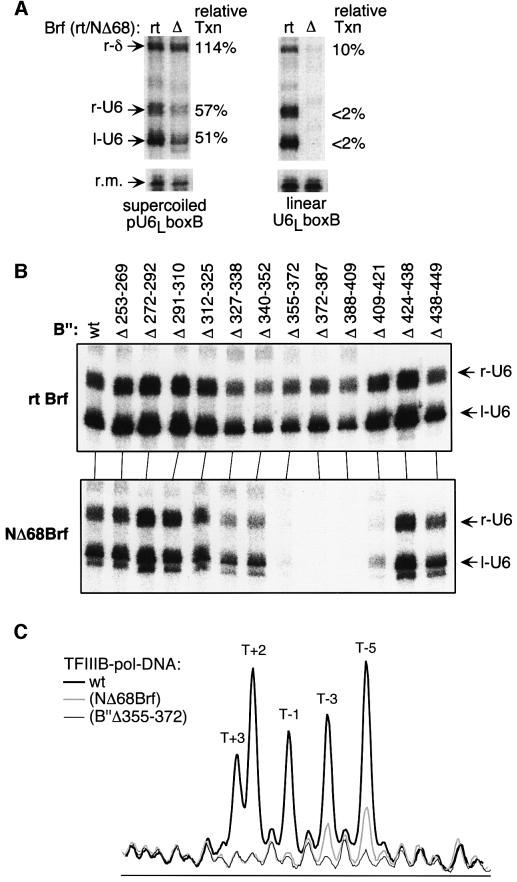
Transcriptional defects similar to those of NΔ164Brf are seen with Brf lacking only its N-terminal zinc ribbon element (amino acids 3–33) and ∼35 amino acids of adjacent sequence (NΔ68Brf) (Figure 1). Figure 1A compares the activity of NΔ68Brf and rtBrf (the reference type for NΔ68; see Materials and methods) for transcription of supercoiled and linear U6LboxB DNA. This diagnostic template for TFIIIC-independent transcription (Whitehall et al., 1995; Kassavetis et al., 1997) is derived from the U6 snRNA gene (SNR6), and contains two identical strong TATA boxes (U6 and δ) for TATA-binding protein (TBP)-mediated assembly of TFIIIB. Each TATA box generates a pair of divergent transcripts (l-U6, r-U6, l-δ and r-δ) because TBP binds to each TATA box in either orientation in the absence of TFIIIC; only the three largest transcripts are examined. TFIIIB assembled with NΔ68Brf generates 50–100% of the activity of wild-type TFIIIB on the supercoiled pU6LboxB (left panel), but is essentially inactive on the linear form (right panel), consistent with the observation that disruption of the Brf zinc ribbon domain generates a defect in promoter opening but not pol III recruitment (Hahn and Roberts, 2000). The combination of NΔ68Brf with the promoter opening-defective B” internal deletions spanning amino acids 355–421 leads to an inability to transcribe supercoiled DNA (Figure 1B, compare top and bottom panels).
Fig. 1. Defects of transcription generated by deletion of the N-terminal putative zinc ribbon domain of Brf and by B” internal deletions between amino acids 355 and 421. (A) NΔ68Brf forms TFIIIB–DNA complexes that are highly active for transcription of supercoiled DNA but inactive on linear DNA. The U6LboxB δ and SNR6 (U6 snRNA) gene TATA boxes each generate a pair of divergent transcripts (r-δ, l-δ, r-U6 and l-U6); three of these are identified at the left. The yield of each transcript in reaction mixtures containing NΔ68Brf [relative to reference type (rt) Brf; see Materials and methods] is specified to the right of each panel. The labeled DNA fragment that served as a recovery marker (r.m.) is also shown. (B) TFIIIB assembled with NΔ68Brf and B” with internal deletions between amino acids 355 and 421 is inactive for transcription of supercoiled DNA. Only the r-U6 and l-U6 transcripts are shown. The B” internal deletions are identified above each lane. Upper panel, rtBrf; lower panel, NΔ68Brf. (C) TFIIIB–pol III–DNA complexes containing B”Δ355–372 or NΔ68Brf are defective in open complex formation. The phosphorimage profiles from KMnO4 footprinting reactions of pol III–TFIIIB–DNA complexes containing wild-type TFIIIB (thick black line), B”Δ355–372 (thin black line) and NΔ68Brf (thick gray line) were aligned to the TFIIIB–DNA only profile (not shown, but indistinguishable from that of the pol III–TFIIIB–DNA complex containing B”Δ355–372). Thymines between +3 and –5 are identified (see Figure 2).
The promoter opening defect of TFIIIB–pol III–DNA complexes assembled with NΔ68Brf was explored by KMnO4 footprinting (Hayatsu and Ukita, 1967; Sasse-Dwight and Gralla, 1989) (Figure 1C). Thymines between bp +3 and –5 on the non-transcribed strand of the wild-type TFIIIB–pol III–SNR6 promoter complexes are reactive to KMnO4 (thick black line; sequence in Figure 2), whereas TFIIIB–pol–DNA complexes containing B”Δ355–372 displayed no reactivity to KMnO4 (thin black line), as reported previously (Kassavetis et al., 1998b) (above the background reactivity of DNA, which is omitted from the figure). TFIIIB–pol III–SNR6 promoter complexes assembled with NΔ68Brf generated weak KMnO4 reactivity at thymines –5 and –3, but no significant reactivity above background at thymines +3, +2 and –1 (thick gray lines).
Fig. 2. Start site-proximal DNA sequence of the heteroduplex bubble templates used in this study. The normal start site of transcription is designated +1 and the SNR6 TATA box is boxed. The transcribed strand is identical in all templates. Only the 5-nt bubbles are shown; 3-nt bubble constructs –9/–7 and –7/–5 follow the same design.
Rescuing transcription with bubble templates
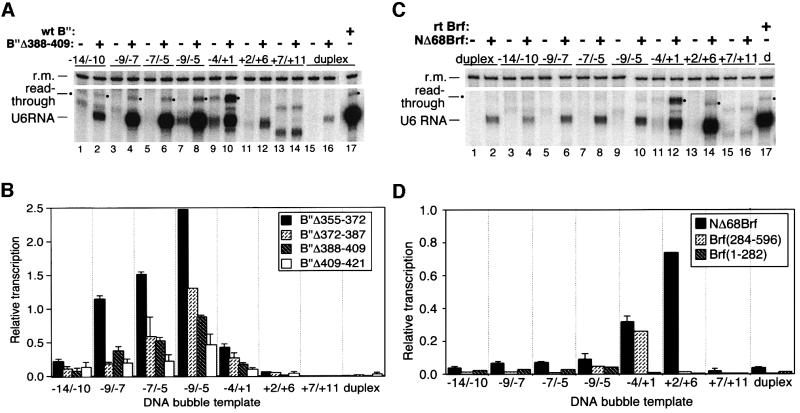
Pol III opens its promoters non-coordinately, with the upstream segment (bp –9 to –5, relative to the start site as +1) opening at a lower temperature than the downstream segment (bp –3 to +7) (Kassavetis et al., 1992). This is reminiscent of promoter opening by bacterial RNA polymerases, which are believed to nucleate strand opening near bp –11 and expand the transcription bubble to the start site of transcription (reviewed in Helmann and deHaseth, 1999). The preceding result is consistent with the possibility that pol III promoter complexes assembled with B”Δ355–372 are unable to initiate promoter opening and that promoter complexes assembled with NΔ68Brf are especially defective in fully extending promoter opening. To characterize further the contributions of the N-terminal 68 amino acids of Brf and amino acids 355–421 of B” to the formation of the open pol III promoter complex, we constructed partial heteroduplex transcription templates with 3 or 5 nt single-stranded bubbles scanning across the start site of transcription (Figure 2) and tested their ability to rescue the transcription defect of TFIIIB containing B”Δ388–409. When a preformed bubble is placed between bp –9 and –5, B” is not absolutely required for transcription under non-stringent conditions [elevated Brf and TBP concentrations and low ionic strength (40 mM NaCl)] (Kassavetis et al., 1999). This background of B”-independent transcription was reduced when a reaction buffer with 90 mM NaCl was used (Figure 3A, odd-numbered lanes 1–15).
Fig. 3. Promoter opening defects due to B” internal deletions and a Brf N-terminal deletion are rescued by 3 and 5 bp heteroduplex bubbles. (A) The –9/–7, –7/–5 and –9/–5 bubbles rescue the transcriptional defect of B”Δ388–409. DNA templates and the presence of B” are specified above each lane. The U6 transcript and recovery marker (r.m.) are designated to the left; a circle designates the read-through transcript. Only the transcript corresponding to r-U6 in Figure 1 is produced with this transcription template, which extends only to bp –60 (see Materials and methods). (B) Quantitative summary of the rescue of transcription defects generated by B” deletions through partial promoter opening. Data from nine experiments similar to (A) are presented for four internal deletions of B” covering amino acids 355–421. Transcription efficiency is normalized to a wild-type TFIIIB–fully duplex DNA transcription standard used in each experiment, after subtracting a background of B′′-independent transcription for each template. Average values and average deviations from two to three experiments are shown. (C and D) Bubbles also rescue the transcriptional defects of Brf deletions. (C) A +2/+6 bubble restores transcription factor activity of TFIIIB assembled with NΔ68Brf. The DNA template and the presence of Brf are specified above each lane. (D) Summary of the effects of bubbles on transcription with NΔ68Brf and the separate N- and C-terminal halves of Brf. Presentation of data as in (B). Data for NΔ68Brf are averages from two experiments; the rest are single experiments. The prominent read-through transcripts of the –4/+1 bubble template in (A) and (C) are probably due to RNA–DNA hybrid formation with displacement of the non-transcribed DNA strand (Campbell and Setzer, 1992).
Opening a 5-nt bubble between bp –9 and –5 effectively rescued the transcriptional defect of B”Δ388–409 (Figure 3A, compare lane 8 with lanes 16 and 17). Three-nt bubbles –9/–7 and –7/–5 also functioned well (lanes 4 and 6), while 5-nt bubbles immediately upstream (–14/–10; lane 2) or downstream (–4/+1; lane 10) were less effective in rescuing transcription. Five-nt bubbles downstream of the normal start site of transcription either barely increased transcription over the duplex DNA background (+2/+6; compare lanes 12 and 16) or yielded no increase over a no-B” background (+7/+12; lanes 13 and 14). The properties of TFIIIB assembled with other promoter opening-defective B” internal deletions were similar (Figure 3B), but with a pronounced gradient of rescue activity, B”Δ355–372 being the most effective. We have not analyzed this suppression gradient further.
The transcription defect of TFIIIB assembled with NΔ68Brf was also rescued by partially opening the promoter with 5-nt bubbles. Only the +2/+6 bubble restored transcription to nearly wild-type levels (Figure 3C, compare lane 14 with lanes 2 and 17), apparently rescuing an inability to complete promoter opening noted in Figure 1C. The –4/+1 bubble functioned half as well (lane 1 and Figure 3D), and the remaining bubbles were largely ineffective. Evidently the transcriptional defects due to these B” and Brf deletions are rescued by partially opening different segments of the transcription bubble, implying the participation of TFIIIB in at least two separable steps of promoter opening.
The C-terminal half of Brf contributes most of the affinity of Brf for TBP and B”. The N-terminal, TFIIB-related half, Brf(1–282), generates unstable TFIIIB–DNA complexes that are transcriptionally nearly fully active on the supercoiled SNR6 gene, but are inactive on linear DNA (Kassavetis et al., 1998a,b). Partial promoter opening did not rescue this transcriptional defect of Brf(1–282) (Figure 3D and data not shown). Brf(284–596) retains only a trace of transcriptional activity, even on the supercoiled SNR6 gene [∼1% (l-U6 and r-U6) to 10% (r-δ) (Kassavetis et al., 1998a)]. Nevertheless, opening the promoter between bp –4 and +1 enhanced the transcriptional activity of this Brf deletion significantly; other bubble templates had little or no effect (Figure 3D and data not shown).
We have noted previously that bubble-containing DNA functions less well than fully duplex DNA for transcription with wild-type TFIIIB and pol III (Kassavetis et al., 1999). This is also true of some of the bubble constructs used in the current analysis, which differs from previous work in the use of higher salt concentrations (Figure 4); in particular, the –4/+1 and +7/+11 constructs functioned quite poorly. The bases of these lower activities are of potential interest but have not been fully elucidated. The +7/+11 template displays an unusually high level of factor-independent pol III binding (higher than pol III– TFIIIB–DNA complexes; data not shown), and may sequester polymerase or form transcriptionally inactive TFIIIB–pol III–DNA complexes. The lower activity of –4/+1 template is partly accounted for by a failure of RNA strand displacement, leading to the formation of an RNA–DNA hybrid duplex that continues elongation through the terminator (Kassavetis et al., 1999). Pol III that transcribes to the end of a linear DNA duplex recycles for another round of transcription at a lower rate (Dieci and Sentenac, 1996).
Fig. 4. Transcription of bubble-containing templates directed by wild-type TFIIIB. A sample experiment is shown, and averages of two experiments (quantified and presented as in Figure 3) are shown below each lane.
The effect of B” and Brf deletions on selection of the start site of transcription in the context of bubble templates was also examined by primer extension analysis with reverse transcriptase (data not shown). As noted previously (Kassavetis et al., 1999), wild-type TFIIIB directed initiation at bp +1 on all bubble templates, with approximately equal levels of initiation also occurring at bp +5 on construct +2/+6 and at bp +7 on construct +7/+11. Start site selection directed by wild-type TFIIIB and TFIIIB containing B”Δ372–387, B”Δ388–409 or B”Δ355–372 did not differ significantly. (Levels of initiation with duplex DNA and with templates –14/–10, –4/+1 and +2/+6 were greatly reduced, consistent with Figure 3A; template +7/+11 yielded too little RNA to analyze.) TFIIIB assembled with NΔ68Brf differed markedly from wild-type TFIIIB in start site selection with the +2/+6 bubble template, specifying initiation almost exclusively at bp +5. In fact, primer extension analysis of transcription of supercoiled DNA templates with TFIIIB–DNA complexes containing NΔ68Brf also demonstrated significant levels of initiation at bp +5 (Figure 1B and data not shown). Alterations of start site selection for pol II have also been noted for zinc ribbon domain-proximal mutations in the related transcription factor TFIIB (Pinto et al., 1994).
Two post-recruitment steps to initiation of transcription
No specific region of B” or Brf is absolutely essential for transcription of supercoiled DNA, yet both factors are required for transcription of duplex DNA (Kumar et al., 1997; Kassavetis et al., 1998a, 1999). The simplest explanation for this apparent contradiction calls for multiple sites of interaction between pol III and TFIIIB. Indeed, Brf has been shown or inferred to interact with both the C34 and C17 subunits of pol III (Werner et al., 1993; Khoo et al., 1994; Andrau et al., 1999; Ferri et al., 2000), and a weak but specific interaction between B” and pol III has been noted (Kassavetis et al., 1995). Although the N-terminal half of Brf retains nearly full transcriptional activity (Kassavetis et al., 1998a), the predominant interaction with C34 appears to be through the C-terminal half of Brf (Andrau et al., 1999). The experiments summarized in Figure 3 imply the existence of separate post-recruitment steps at which TFIIIB intervenes in promoter opening—one compromised by the B” deletions, and another compromised by the NΔ68 deletion in Brf. The KMnO4 footprint analysis in Figure 1C suggests that the TFIIIB–pol III–promoter complex assembled on duplex DNA with B”Δ355–372 is transcriptionally inactive because it does not open the promoter, while the complex assembled with NΔ68Brf is defective because it does not propagate promoter opening downstream. That conclusion is confirmed by KMnO4 footprinting of TFIIIB–pol III complexes on –9/–5 bubble DNA (analyzed in the non-transcribed strand and shown in Figure 5). The reactivity of T–8 and –7 in the bubble was very great in DNA alone (data not shown), as expected, and in the wtTFIIIB–DNA complex. The wtTFIIIB– pol III–DNA complex opened the promoter at T–3, –1, +2 and +3; the complex assembled with NΔ68Brf also opened the promoter at T–3, but did so poorly at T–1 and not at all at T+2 and T+3. The TFIIIB–pol III–DNA complex assembled with B”Δ355–372 opened the promoter fully, consistent with its rescue of transcription with this bubble template (data not shown).
Fig. 5. The TFIIIB–pol III complex assembled with NΔ68Brf is defective in propagating promoter opening to its downstream end. Phosphorimage profiles from KMnO4 footprints (non-transcribed strand) of TFIIIB–pol III complexes assembled on –9/–5 DNA (Figure 2) are shown. Complexes were assembled with wild-type TFIIIB (thick gray line) and with NΔ68Brf (thick black line), and footprints were aligned to the wild-type TFIIIB(only)–DNA complex. Thymines between T–12 and +3 are identified. The reactivity of T–8 and T–7 in the –9/–5 bubble is elevated in DNA and in all of these complexes.
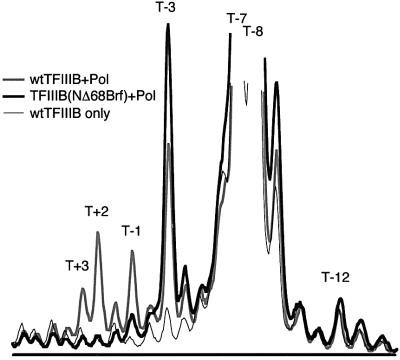
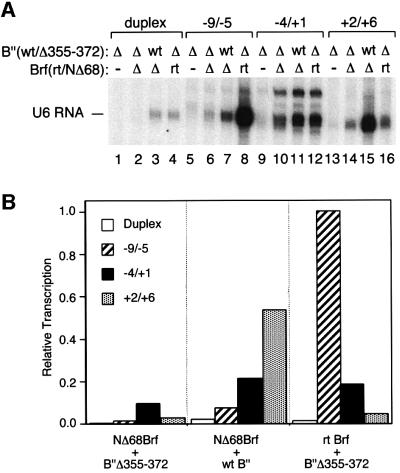
The next experiments examined the residual capacity for pol III recruitment and transcription when TFIIIB is assembled with the defective NΔ68Brf and the defective B”Δ355–372 (Figure 6). As anticipated, TFIIIB assembled with NΔ68Brf and B”Δ355–372 was transcriptionally inert on duplex DNA (Figure 6A, lane 2 and 6B). For transcription of the –9/–5 template (optimal for B”Δ355– 372; lane 8) and the +2/+6 template (optimal for NΔ68Brf; lane 15), only very low levels of TFIIIB-dependent transcription remained (lanes 6 and 14, respectively). Replacing B”Δ355–372 with B”Δ372–387 or B”Δ388–409 in the preceding experiment did not change the outcome (data not shown). Residual transcription was retained for the –4/+1 construct, at 45–50% of activity of TFIIIB assembled with NΔ68Brf or B”Δ355–372, respectively (data not shown). Quantitative interpretation is made problematic by the low activity of the –4/+1 construct in conjunction with wild-type TFIIIB (Figure 4), but the result does indicate some residuum of competence for transcription that is specific to the bubble placed at this position.
Fig. 6. Transcription with TFIIIB–DNA complexes containing both NΔ68Brf and B”Δ355–372. (A) TFIIIB–DNA complexes were formed with wild-type B” (wt) or B”Δ355–372 (Δ) and rt Brf (rt) or NΔ68Brf (Δ) as designated for each template above the figure. (B) U6 RNA synthesis is quantified and presented as in Figure 3.
Open and closed pol III–TFIIIB–DNA complexes assembled with wild-type TFIIIB are stable to electrophoretic mobility shift assay (EMSA) separation (Kassavetis et al., 1992, 1998b). Substantial reductions of the ability to form an EMSA-stable pol III–TFIIIB–DNA complex reflect weaker interaction between pol III and TFIIIB. Combining EMSA with DNA footprinting additionally specifies the site of pol III placement on DNA, and may provide indications of altered structure. A substantial loss of transcriptional activity that is not reflected strictly in reduced binding points to a defective step subsequent to polymerase binding. The next experiments investigate the capacity of TFIIIB assembled with NΔ68Brf and B”Δ355–372 to recruit pol III to the promoter.
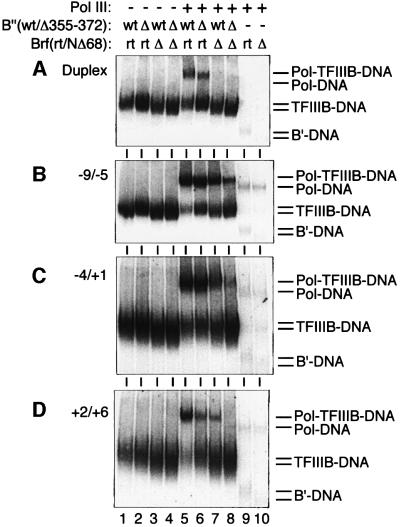
Entry of pol III into wild-type and variant TFIIIB–DNA complexes was examined by EMSAs, which are summarized in Figure 7. TFIIIB assembled with rtBrf, NΔ68Brf, wild-type B” and B”Δ355–372, in all combinations, bound stably to duplex and bubble DNA (Figure 7A–D, lanes 1–4). Formation of stable pol III complexes on duplex or bubble DNA required TFIIIB (Figure 7A–D, lanes 9 and 10; a very faint background of pol III–DNA complexes was barely detected in the original images, and is not seen in the reproduced figure). TFIIIB assembled with B”Δ355–372 recruited pol III to duplex DNA (Figure 7A, lane 6), but this promoter complex was transcriptionally inactive and in a closed state (Figures 3A and 1C; Kassavetis et al., 1998b). The NΔ68 deletion in Brf eliminated stable pol III binding to the corresponding TFIIIB complex with duplex DNA (Figure 7A, lanes 7 and 8), but partial promoter opening restored TFIIIB-dependent pol III recruitment, more effectively with the –9/–5 construct and less effectively with the –4/+1 and +2/+6 constructs (Figure 7B–D, lane 7). In contrast, the pol III complex with +2/+6 DNA was the most active transcriptionally and the –9/–5 construct was inactive (Figure 3D). Thus, fully opening the bp –9/–5 segment of the promoter ameliorated pol III recruitment by TFIIIB assembled with NΔ68Brf only to reveal an additional, post-recruitment defect that was manifested in the absence of downstream bubble propagation (Figure 5). On the +2/+6 construct, NΔ68Brf-containing TFIIIB and B”Δ355–372-containing TFIIIB generated comparable levels of pol III recruitment (Figure 7D, lanes 6 and 7), but only the promoter complex assembled with NΔ68Brf was transcriptionally active (Figure 3).
Fig. 7. Pol III recruitment. EMSA with (A) duplex, (B) –9/–5 bubble, (C) –4/+1 bubble and (D) +2/+6 bubble DNA probes was performed with pol III, and TFIIIB assembled with wild-type B” (wt) or B”Δ355–372 (Δ) and rt Brf (rt) or NΔ68Brf (Δ), as designated above (A). The mobilities of B′(Brf+TBPm3)–, TFIIIB–, pol– and pol–TFIIIB–DNA complexes are specified at the right of each panel. Free DNA is not shown.
TFIIIB assembled with NΔ68Brf and B”Δ355–372 recruited pol III to the –9/–5 and –4/+1 constructs relatively poorly (Figure 7B and C, lane 8), and pol III complexes with the +2/+6 construct were not detected. This loss of complex formation greatly exceeded the additive effects of the individual NΔ68Brf and B”Δ355–372 deletions (compare lane 8 with lanes 6 and 7, and data not shown). Experiments like Figure 7 were also carried out with the other B” deletion proteins, with essentially identical results. EMSA-stable TFIIIB–DNA and TFIIIB–pol III–DNA complexes formed with B”Δ372–387, wild-type B”, NΔ68Brf and rtBrf, in all combinations, were selected for examination by methid iumpropyl EDTA-Fe(II) [MPE-Fe(II)] footprinting. All TFIIIB–DNA footprints on duplex and bubble DNA probes were identical. Pol III assembled by wild-type TFIIIB or TFIIIB containing either B”Δ372–387 or NΔ68Brf also generated identical footprints (data not shown).
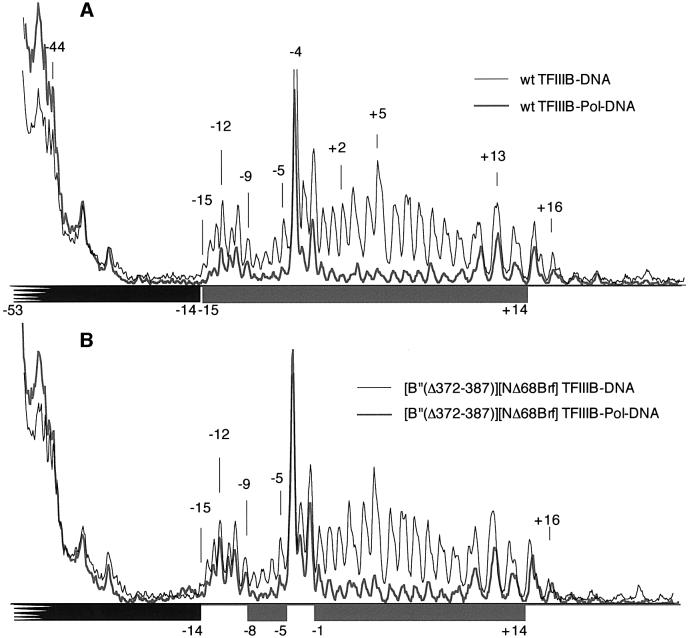
MPE-Fe(II) footprinting of the –9/–5 bubble probe did expose differences of DNA protection between pol III complexes assembled with wild-type and doubly mutant TFIIIB, with loss of protection between bp –14 and –9 at the upstream TFIIIB-interacting end, and also between bp –4 and –2 for the complex with doubly mutant TFIIIB (Figure 8, compare A and B). Despite partial promoter opening and pol III placement over the start site, this promoter complex was transcriptionally inactive. A parallel MPE-Fe(II) footprinting analysis of the –4/+1 bubble probe yielded similar results (data not shown): the footprints of pol III promoter complexes assembled with wild-type TFIIIB and with TFIIIB containing the individual Brf or B” deletion proteins were indistinguishable. TFIIIB assembled with both NΔ68Brf and B”Δ355–372 was able to direct accurate placement of pol III, but the footprints showed a loss of DNA protection (relative to the pol III complex assembled with wild-type TFIIIB) principally between bp –12 and –10. Thus, the pol III footprints of the promoter complexes on –9/–5 and –4/+1 bubble DNA were essentially indistinguishable, but only the complex assembled on the –4/+1 bubble template exhibited residual transcriptional activity, as stated above.
Fig. 8. MPE-Fe(II) footprinting of pol III–TFIIIB complexes bound to –9/–5 bubble DNA. Pol III was assembled onto TFIIIB–DNA complexes containing: (A) wild-type B”+rtBrf and (B) B”Δ372–387+NΔ68Brf. The transcribed DNA strand was analyzed. Aligned phosphorimage profiles of TFIIIB–DNA complexes (thin black lines) and pol III–TFIIIB–DNA complexes (thick gray lines) are shown. The upstream border of protection of the labeled transcribed strand by TFIIIB is not defined due to the proximity of uncleaved DNA(bp –56) on denaturing gels, but the downstream border for all TFIIIB–DNA complexes is at bp –14, as indicated by the black bar below each panel. All TFIIIB–DNA complexes enhanced DNA cleavage by MPE-Fe(II) at bp –4, –2 and –1. (The baseline cleavage pattern of free DNA is omitted for the sake of clarity of presentation.) The extent of DNA protection by pol III is indicated by the gray bar below each panel.
Discussion
TFIIIB participates in promoter opening (Kassavetis et al., 1998b). Normal promoter opening by pol III (at the SUP4 tRNA gene) displays a characteristic temperature dependence in which the upstream portion of the transcription bubble (extending to ∼bp –9) melts at lower temperature (Kassavetis et al., 1992). This has suggested that promoter opening by pol III may nucleate at the upstream end of the transcription bubble. Promoter opening by bacterial RNA polymerase (Chen and Helmann, 1997) and by pol I (Kahl et al., 2000) shows the same non-coordinacy of promoter melting. Insight into the process of open complex formation by bacterial RNA polymerases (for reviews see Helmann and deHaseth, 1999; Fiedler and Timmers, 2000) provides a model framework for pol III. Open complex formation by Escherichia coli RNA polymerase-σ70 holoenzyme is proposed to be initiated by flipping out the non-transcribed strand –11 adenine of the –10 promoter element (TATAAT) for interaction with aromatic amino acid side chains in region 2.3 of the initiation factor subunit σ70 (Fenton et al., 2000; Matlock and Heyduk, 2000; Panaghie et al., 2000). The temperature dependence of open complex formation has long been thought to reflect the need to destabilize base pairing, but now appears to involve other conformational changes (Guo et al., 2000). σ54 also plays a comparable role in initiating promoter opening (Cannon et al., 2000; Guo et al., 2000).
The experiments that are reported here pursue the question of how TFIIIB participates in this process. The implications of evidence that a group of deletions introduced into Brf and B” leads to failures of promoter opening (Kassavetis et al., 1998b) have been explored by examining whether artificially opening small segments of the transcription bubble would restore transcription. Five-bp segments of the promoter have been unpaired, while keeping the sequence of the transcribed DNA strand unchanged; 3-bp unpairing has also been examined. The total span of these bubble constructs, covering bp –14 to +11 (Figure 2), exceeds the span of the (SUP4 gene) transcription bubble. The analysis focuses on one Brf deletion and on a cluster of small B” deletions covering amino acids 355–421 (Figure 1) that impair the ability of the corresponding TFIIIB to direct transcription of linear DNA (Figures 1A and 3B), and in combination also eliminate the transcription of supercoiled DNA (Figure 1B).
Brf
We have previously observed that TFIIIB–DNA complexes containing NΔ164Brf do not assemble pol III stably as measured by EMSA, which requires stability during electrophoretic separation, while pol III assembly is readily detected by photochemical cross-linking, which monitors site occupancy at unperturbed equilibrium (Kassavetis et al., 1998b). Likewise, TFIIIB–DNA complexes containing NΔ68Brf do not stably assemble pol III on fully duplex DNA (Figure 7A). This is somewhat surprising, since the known pol III interaction sites in Brf do not lie N-terminal to the TFIIB-related imperfect repeats (Khoo et al., 1994; Andrau et al., 1999; Ferri et al., 2000). Moreover, disruption of the Brf zinc ribbon domain by a small N-terminal deletion (NΔ12Brf) does not eliminate the ability of TFIIIB–DNA complexes to stably assemble pol III (Hahn and Roberts, 2000).
Nevertheless, a role for the Brf N-terminus in assembly of pol III initiation complexes is consistent with known properties of the related TFB and pol II transcription factor TFIIB: TFB of the archaeon Sulfolobus acidocaldarius with an N-terminal truncation similar in extent to NΔ68Brf retains the ability to form stable TFB–TBP–DNA complexes that are indistinguishable from the wild-type TFB–TBP–DNA complex (in terms of their DNase I footprint), but can not assemble the cognate RNA polymerase (Bell and Jackson, 2000). Mutations in another segment of the yeast TFIIB zinc ribbon or within a C-terminally adjacent sequence also elicit a pol II binding defect (Bushnell et al., 1996; Pardee et al., 1998).
Additional effects of TFB and TFIIB mutations in the zinc ribbon domain and C-terminally adjacent sequence also extend to Brf. Mutations in a segment C-terminal to the zinc ribbon that is conserved between human TFIIB, yeast TFIIB and archaeal TFBs [(P/S)EWR(A/T)F…SRVG] alter start site selection by the cognate polymerase and/or cause the loss of transcriptional activity at a step subsequent to polymerase recruitment (Pinto et al., 1994; Pardee et al., 1998; Cho and Buratowski, 1999; Hawkes and Roberts, 1999; Ranish et al., 1999; Bell and Jackson, 2000). NΔ68Brf likewise generates altered start site selection (Figure 1B and 5′ end determinations by primer extension, not shown). The TFIIB/TFB conserved sequence just referred to is not found in Brf, but a similarly positioned motif that is conserved among Brfs [IVSEV(T/Q)F(G/V)E] might play a related role. Certainly, the similarity of effect of mutations in the zinc ribbon domain and adjacent sequence of Brf, TFB and TFIIB reinforces the supposition that this region serves a similar function in all three transcription systems. If this region serves as a site of direct interaction with the cognate polymerase, the target has yet to be identified. It is also conceivable that the effects of mutations in the zinc ribbon domains of Brf, TFIIB and TFB on polymerase recruitment are indirect.
Partial promoter opening compensates for the defect in opening the downstream segment of the transcription bubble caused by the NΔ68 deletion in Brf (Figures 1C and 3). Opening the downstream end of the promoter (bp +2 to +6) is most effective, and opening bp –4 to +1 also generates substantial suppression, while unpairing bp –9 to –5 has little effect on either transcription (Figure 3) or downstream propagation of the transcription bubble (Figure 5). Since the –4/+1 bubble construct considerably reduces transcription with wild-type TFIIIB (Figure 4), it is difficult to specify exactly where the optimal suppression of the NΔ68Brf defect lies, but it clearly resides in the downstream side of the transcription bubble.
The effects of preformed bubbles on transcription with the N- and C-terminal halves of Brf (Figure 3D) are also instructive. Brf(1–282) is transcriptionally active on supercoiled DNA, but inert on linear DNA (Kassavetis et al., 1998a,b). The defect is not rescued by either the –4/+1 start site-proximal bubble or any other bubble, because it involves a step preceding promoter opening: Brf(1–282) assembles TFIIIB aberrantly on linear DNA (Figure 4 of Kassavetis et al., 1998a), and there is no expectation that partial promoter opening would repair this defect. On the other hand, Brf(284–596) is barely active transcriptionally with supercoiled DNA and inert with linear DNA, but the –4/+1 bubble restores significant transcription (Figure 3D). In fact, wild-type TFIIIB and TFIIIB assembled with Brf(284–596), NΔ68Brf or B” with internal deletions all generate comparable transcription of the –4/+1 template (Figures 3B and D, and 4). This centrally located bubble may destabilize both its upstream and downstream neighboring segments in the fully opened transcription bubble. The level of transcription obtained with Brf(284–596) also provides clear evidence for the existence of significant polymerase-binding epitopes in the C-terminal half of Brf and/or B”.
B”
The defect generated by the B” deletions is also compensated by partial promoter opening (Figure 3). Suppression is most effectively provided by the –9/–5 bubble; opening just 3 bp of DNA substantially restores transcription, whose accuracy of initiation (at +1) is retained (Figure 3A and primer extension analysis not shown). Opening the downstream end of the transcription bubble (+2/+6) fails to restore transcription, and KMnO4 footprinting analysis indicated that the +2/+6 bubble provides no help in opening the upstream segment of the transcription bubble (data not shown). Evidently, these B” deletions generate a defect in opening the upstream segment of the transcription bubble.
TFIIIB participates in two steps of promoter opening
The rescue, by heteroduplex bubbles, of promoter opening defects that result from B” and Brf deletions recapitulates the bacterial RNA polymerase pathway to open complex formation (Figure 9). The engagement of DNA with the RNA polymerase channels that accommodate the separated strands (Korzheva et al., 2000; Naryshkin et al., 2000) proceeds unidirectionally from the upstream end of the transcription bubble. B” deletions between amino acids 355 and 421 interfere with either the nucleation of strand opening or subsequent melting of the upstream portion of the transcription bubble; N-terminal Brf deletions or mutations prevent the downstream propagation of the transcription bubble (Figure 1C) that completes the engagement of the template strand with the catalytic site (Figure 9A). Preforming the –9/–5 bubble bypasses a role of B” in the formation of the initial upstream bubble, and also bypasses the absolute requirement for B” in transcription (Kassavetis et al., 1999), but the N-terminal domain of Brf is still required for downstream propagation of the bubble (line B). The –9/–5 bubble most effectively alleviates the polymerase recruitment defect of NΔ68Brf (Figure 7), probably due to the additional stabilizing interactions generated by entry of the upstream portion of the bubble into the single-stranded DNA-accommodating channels of pol III.
Fig. 9. Points of intervention of TFIIIB in the reaction sequence leading to initiation of transcription.
Preforming the +2/+6 bubble bypasses the role of the N-terminal domain of Brf in the downstream propagation of the bubble, but B” is still required to open the upstream portion of the bubble (Figure 9C), perhaps reflecting the need to open the upstream end of the bubble in order to initiate the engagement of the non-transcribed and transcribed DNA strands with their appropriate channels in pol III. Thus, the +2/+6 bubble of transcriptionally inactive pol III–TFIIIB–promoter complex containing B”Δ372–387 may be held at a location characteristic of the closed complex, or it may be misaligned relative to the catalytic site. At the SUP4 tRNA gene promoter, the downstream part of the transcription bubble is maintained in the presence of the initiating ribonucleotide upon temperature downshift to 0°C, but the upstream segment collapses (Kassavetis et al., 1992). That the initiating nucleotide allows maintenance of the downstream bubble segment indicates that the latter retains its proper orientation relative to the catalytic site, supporting the notion that the upstream bubble segment is required for initiating engagement with the pol III channels that accommodate melted DNA.
Whether the participation of TFIIIB in these two steps of promoter opening is direct or indirect remains to be determined. Opening the –9/–5 bubble ameliorates pol III recruitment by TFIIIB assembled with NΔ68Brf (Figure 7), indicating the existence of a protein interaction driving DNA strand separation at the upstream end of the transcription bubble. The potential sites of that interaction are the two large core pol III subunits, Brf and B”, as well as the C34 and C82 pol III subunits, all of which sit in the vicinity of the DNA segment that opens up to form the transcription bubble (Bartholomew et al., 1993, 1994).
Materials and methods
DNA templates and probes
Plasmid pU6LboxB (Whitehall et al., 1995; Yieh et al., 2000) and its 366 bp linear fragment (bp –211 to +155) have been described (Kassavetis et al., 1998b). The construction of 198 bp (bp –60 to +138) pU6RboxB-derived transcription templates containing 3–5 nt heteroduplex bubbles has also been described (Kassavetis et al., 1999). Fully duplex and bubble-containing 86 bp (bp –56 to +30) probes for MPE-Fe(II) footprinting and EMSA differed in sequence from the corresponding transcription templates at three positions: an upstream (–29)A→G substitution to generate a TGTA mutant TATA box for TFIIIB binding in a single orientation with the TBP variant TBPm3 (Whitehall et al., 1995), as well as downstream (+8) and (+9)A→T substitutions to allow the formation of an elongation complex halting at bp +11 in the absence of ATP. (Changes are specified for the non-transcribed strand.) Probes were prepared by annealing separated strands (Kassavetis et al., 1999) with the transcribed strand 5′-32P-labeled with T4 polynucleotide kinase.
Proteins
The purification and quantification of the following proteins have been described: monoQ-purified pol III (Kassavetis et al., 1990); recombinant proteins TBP (Joazeiro et al., 1994), TBPm3 (Whitehall et al., 1995), full-length Brf (N- and C-terminally His6 tagged), Brf(1–282) and Brf(284–596) (N-His6 tagged) (Kassavetis et al., 1998a), NΔ164Brf (C-His6 tagged) (Kassavetis et al., 1997), BrfΔ366–408 and NΔ68BrfΔ366–408 (N-His7 tagged) (Kassavetis et al., 1999), full-length B” (C-His6 tagged, purified under denaturing conditions) (Kassavetis et al., 1995), full-length B” (C-His6 tagged, purified under native conditions) (Kassavetis et al., 1997) and B” containing small internal deletions (C-His6 tagged, purified under denaturing conditions) (Kumar et al., 1997). B” internal deletion proteins, purified under native conditions as described for B”(138–594) in Kumar et al. (1997) were also used, as specified below. Quantities of pol III are specified as fmol of enzyme active for specific transcription (Kassavetis et al., 1989); quantities of the other proteins are specified as fmol of protein (Kassavetis et al., 1998b). For the experiments shown in Figures 3 and 4, full-length and internal deletion B” were purified under denaturing conditions; B” were purified under native conditions for the remaining experiments. Experiments involving NΔ68BrfΔ366–408 used BrfΔ366–408 as reference type (rt) in place of full-length Brf (wt) (Figures 1, 3 and 6–8).
Nomenclature
BrfΔ366–408 removes a segment that is not present in fungal (Candida albicans and Kluyveromyces lactis) Brf homologs (Khoo et al., 1994), and is referred to as the reference type (rt) to distinguish it from the wild-type full-length protein. Full-length Brf and rtBrf are indistinguishable in every aspect except that rtBrf retains 1.7-fold more active molecules following purification (Kassavetis et al., 1999). For these experiments, the NΔ68 deletion has been introduced into rtBrf only; the corresponding protein is referred to simply as NΔ68Brf. The distinction between rtBrf and wild-type Brf is dropped when referring to TFIIIB (as in Figure 7).
Assays
TFIIIB–DNA complexes for transcription and EMSA were formed for 40–60 min at 20°C in 18 µl of reaction buffer [40 mM Tris–HCl pH 8, 80–100 mM NaCl, 7 mM MgCl2, 3 mM dithiothreitol, 100 µg/ml bovine serum albumin, 5 µg/ml poly(dG–dC)⋅poly(dG–dC), 6–8% (v/v) glycerol] containing 50 fmol of DNA (for transcription) or 6 fmol of DNA (for EMSA), 200 fmol of TBP (for transcription) or 800 fmol of TBPm3 (for EMSA), 200 fmol of Brf, and 100–150 fmol of B” (except as specified in the figure legends, or below). Pol III (2 µl; 10 fmol) was added for an additional 10–20 min. Multiple round transcription during 30 min (Kassavetis et al., 1999) was initiated by adding ATP, GTP, CTP and [α-32P]UTP to 200, 100, 100 and 25 µM, respectively. Samples were processed, and transcripts were analyzed as described (Kassavetis et al., 1989) and quantified by phosphorimage plate analysis. For the less active full-length Brf, 350 fmol (64 fmol of active protein) were used in experiments shown in Figures 3A and B, 4 and 5. Primer extension analysis of unlabeled transcription reactions with reverse transcriptase followed Kassavetis et al. (1999). For EMSA, samples assembled and incubated as specified above were subjected to challenge with 20 µg/ml poly(dA–dT)⋅poly(dA–dT) for 5 min before electrophoresis, as described (Kassavetis et al., 1998b). For KMnO4 footprinting, TFIIIB–pol III–DNA complexes were formed essentially as described for transcription, but with 15 fmol of pol III and 12 fmol of the 198 bp duplex or +2/+6 bubble transcription templates, 5′-32P-labeled in the non-transcribed strand. KMnO4 treatment for 30 s and processing of samples followed Kassavetis et al. (1992). For MPE-Fe(II) footprinting, TFIIIB–pol III–DNA complexes were formed, as for EMSA, in 30 µl of reaction buffer containing 12 fmol of DNA probe, 1.6 pmol of TBPm3, 400 fmol of Brf, 450 fmol of B” and 15 fmol of pol III. Following post-assembly challenge with poly(dA–dT)⋅poly(dA–dT), complexes were incubated with 1 mM sodium ascorbate and 2 µM MPE-Fe(II) for 2 min and then directly loaded onto a native gel. Elution and processing for subsequent denaturing gel electrophoresis followed Kassavetis et al. (1998a). A particular feature of this two-step footprinting procedure is the under-representation in the footprint of DNA cleavage events close to the 32P-labeled end of the footprinted DNA strand. This is due to partial dissociation of short (<12 nt) DNA strands during gel-isolation of protein–DNA complexes. All footprinting was done under single-hit conditions.
Acknowledgments
Acknowledgements
We thank A.Grove and M.Ouhammouch for helpful discussions, and M.Bartlett, M.Ouhammouch and O.Schröder for advice on the manuscript. Support of this research by a grant from the NIGMS is gratefully acknowledged.
References
- Andrau J.-C., Sentenac,A. and Werner,M. (1999) Mutagenesis of yeast TFIIIB70 reveals C-terminal residues critical for interaction with TBP and C34. J. Mol. Biol., 288, 511–520. [DOI] [PubMed] [Google Scholar]
- Bartholomew B., Durkovich,D., Kassavetis,G.A. and Geiduschek,E.P. (1993) Orientation and topography of RNA polymerase III in transcription complexes. Mol. Cell. Biol., 13, 942–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomew B., Braun,B.R., Kassavetis,G.A. and Geiduschek,E.P. (1994) Probing close DNA contacts of RNA polymerase III transcription complexes with the photoactive nucleoside 4-thiodeoxythymidine. J. Biol. Chem., 269, 18090–18095. [PubMed] [Google Scholar]
- Bell S.D. and Jackson,S.P. (2000) The role of transcription factor B in transcription initiation and promoter clearance in the archaeon Sulfolobus acidocaldarius. J. Biol. Chem., 275, 12934–12940. [DOI] [PubMed] [Google Scholar]
- Braun B.R., Bartholomew,B., Kassavetis,G.A. and Geiduschek,E.P. (1992) Topography of transcription factor complexes on the Saccharomyces cerevisiae 5S RNA gene. J. Mol. Biol., 228, 1063–1077. [DOI] [PubMed] [Google Scholar]
- Burnol A.F., Margottin,F., Huet,J., Almouzni,G., Prioleau,M.N., Méchali,M. and Sentenac,A. (1993) TFIIIC relieves repression of U6 snRNA transcription by chromatin. Nature, 362, 475–477. [DOI] [PubMed] [Google Scholar]
- Bushnell D.A., Bamdad,C. and Kornberg,R.D. (1996) A minimal set of RNA polymerase II transcription protein interactions. J. Biol. Chem., 271, 20170–20174. [DOI] [PubMed] [Google Scholar]
- Campbell F.E. Jr and Setzer,D.R. (1992) Transcription termination by RNA polymerase III: uncoupling of polymerase release from termination signal recognition. Mol. Cell. Biol., 12, 2260–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon W.V., Gallegos,M.T. and Buck,M. (2000) Isomerization of a binary σ-promoter DNA complex by transcription activators. Nature Struct. Biol., 7, 594–601. [DOI] [PubMed] [Google Scholar]
- Chédin S., Ferri,M.L., Peyroche,G., Andrau,J.C., Jourdain,S., Lefebvre,O., Werner,M., Carles,C. and Sentenac,A. (1998) The yeast RNA polymerase III transcription machinery: a paradigm for eukaryotic gene activation. Cold Spring Harb. Symp. Quant. Biol., 63, 381–389. [DOI] [PubMed] [Google Scholar]
- Chen Y.F. and Helmann,J.D. (1997) DNA-melting at the Bacillus subtilis flagellin promoter nucleates near –10 and expands unidirectionally. J. Mol. Biol., 267, 47–59. [DOI] [PubMed] [Google Scholar]
- Cho E.J. and Buratowski,S. (1999) Evidence that transcription factor IIB is required for a post-assembly step in transcription initiation. J. Biol. Chem., 274, 25807–25813. [DOI] [PubMed] [Google Scholar]
- Dieci G. and Sentenac,A. (1996) Facilitated recycling pathway for RNA polymerase III. Cell, 84, 245–252. [DOI] [PubMed] [Google Scholar]
- Dumay H., Rubbi,L., Sentenac,A. and Marck,C. (1999) Interaction between yeast RNA polymerase III and transcription factor TFIIIC via ABC10α and τ131 subunits. J. Biol. Chem., 274, 33462–33468. [DOI] [PubMed] [Google Scholar]
- Engelke D.R., Ng,S.Y., Shastry,B.S. and Roeder,R.G. (1980) Specific interaction of a purified transcription factor with an internal control region of 5S RNA genes. Cell, 19, 717–728. [DOI] [PubMed] [Google Scholar]
- Fenton M.S., Lee,S.J. and Gralla,J.D. (2000) Escherichia coli promoter opening and –10 recognition: mutational analysis of σ70. EMBO J., 19, 1130–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferri M.L., Peyroche,G., Siaut,M., Lefèbvre,O., Carles,C., Conesa,C. and Sentenac,A. (2000) A novel subunit of yeast RNA polymerase III interacts with the TFIIB-related domain of TFIIIB70. Mol. Cell. Biol., 20, 488–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler U. and Timmers,H.T.M. (2000) Peeling by binding or twisting by cranking: models for promoter opening and transcription initiation by RNA polymerase II. BioEssays, 22, 316–326. [DOI] [PubMed] [Google Scholar]
- Flores A. et al. (1999) A protein–protein interaction map of yeast RNA polymerase III. Proc. Natl Acad. Sci. USA, 96, 7815–7820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y.L., Lew,C.M. and Gralla,J.D. (2000) Promoter opening by σ54 and σ70 RNA polymerases: σ factor-directed alterations in the mechanism and tightness of control. Genes Dev., 14, 2242–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn S. and Roberts,S. (2000) The zinc ribbon domains of the general transcription factors TFIIB and Brf: conserved functional surfaces but different roles in transcription initiation. Genes Dev., 14, 719–730. [PMC free article] [PubMed] [Google Scholar]
- Hawkes N.A. and Roberts,S.G.E. (1999) The role of human TFIIB in transcription start site selection in vitro and in vivo. J. Biol. Chem., 274, 14337–14343. [DOI] [PubMed] [Google Scholar]
- Hayatsu H. and Ukita,T. (1967) The selective degradation of pyrimidines in nucleic acids by permanganate oxidation. Biochem. Biophys. Res. Commun., 29, 556–561. [DOI] [PubMed] [Google Scholar]
- Helmann J.D. and deHaseth,P.L. (1999) Protein–nucleic acid interactions during open complex formation investigated by systematic alteration of the protein and DNA binding partners. Biochemistry, 38, 5959–5967. [DOI] [PubMed] [Google Scholar]
- Hsieh Y.J., Kundu,T.K., Wang,Z.X., Kovelman,R. and Roeder,R.G. (1999) The TFIIIC90 subunit of TFIIIC interacts with multiple components of the RNA polymerase III machinery and contains a histone-specific acetyltransferase activity. Mol. Cell. Biol., 19, 7697–7704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joazeiro C.A., Kassavetis,G.A. and Geiduschek,E.P. (1994) Identical components of yeast transcription factor IIIB are required and sufficient for transcription of TATA box-containing and TATA-less genes. Mol. Cell. Biol., 14, 2798–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahl B.F., Li,H. and Paule,M.R. (2000) DNA melting and promoter clearance by eukaryotic RNA polymerase I. J. Mol. Biol., 299, 75–89. [DOI] [PubMed] [Google Scholar]
- Kassavetis G.A., Riggs,D.L., Negri,R., Nguyen,L.H. and Geiduschek,E.P. (1989) Transcription factor IIIB generates extended DNA interactions in RNA polymerase III transcription complexes on tRNA genes. Mol. Cell. Biol., 9, 2551–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis G.A., Braun,B.R., Nguyen,L.H. and Geiduschek,E.P. (1990) S. cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell, 60, 235–245. [DOI] [PubMed] [Google Scholar]
- Kassavetis G.A., Blanco,J.A., Johnson,T.E. and Geiduschek,E.P. (1992) Formation of open and elongating transcription complexes by RNA polymerase III. J. Mol. Biol., 226, 47–58. [DOI] [PubMed] [Google Scholar]
- Kassavetis G.A., Nguyen,S.T., Kobayashi,R., Kumar,A., Geiduschek,E.P. and Pisano,M. (1995) Cloning, expression, and function of TFC5, the gene encoding the B” component of the Saccharomyces cerevisiae RNA polymerase III transcription factor TFIIIB. Proc. Natl Acad. Sci. USA, 92, 9786–9790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis G.A., Bardeleben,C., Kumar,A., Ramirez,E. and Geiduschek,E.P. (1997) Domains of the Brf component of RNA polymerase III transcription factor IIIB (TFIIIB): functions in assembly of TFIIIB–DNA complexes and recruitment of RNA polymerase to the promoter. Mol. Cell. Biol., 17, 5299–5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis G.A., Kumar,A., Ramirez,E. and Geiduschek,E.P. (1998a) The functional and structural organization of Brf, the TFIIB-related component of the RNA polymerase III transcription initiation complex. Mol. Cell. Biol., 18, 5587–5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis G.A., Kumar,A., Letts,G.A. and Geiduschek,E.P. (1998b) A post-recruitment function for the RNA polymerase III transcription initiation factor TFIIIB. Proc. Natl Acad. Sci. USA, 95, 9196–9201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis G.A., Letts,G.A. and Geiduschek,E.P. (1999) A minimal RNA polymerase III transcription system. EMBO J., 18, 5042–5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoo B., Brophy,B. and Jackson,S.P. (1994) Conserved functional domains of the RNA polymerase III general transcription factor BRF. Genes Dev., 8, 2879–2890. [DOI] [PubMed] [Google Scholar]
- Korzheva N., Mustaev,A., Kozlov,M., Malhotra,A., Nikiforov,V., Goldfarb,A. and Darst,S.A. (2000) A structural model of transcription elongation. Science, 289, 619–625. [DOI] [PubMed] [Google Scholar]
- Kumar A., Kassavetis,G.A., Geiduschek,E.P., Hambalko,M. and Brent,C.J. (1997) Functional dissection of the B” component of RNA polymerase III transcription factor (TF)IIIB: a scaffolding protein with multiple roles in assembly and initiation of transcription. Mol. Cell. Biol., 17, 1868–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu T.K., Wang,Z.X. and Roeder,R.G. (1999) Human TFIIIC relieves chromatin-mediated repression of RNA polymerase III transcription and contains an intrinsic histone acetyltransferase activity. Mol. Cell. Biol., 19, 1605–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsolier M.C., Tanaka,S., Livingstone-Zatchej,M., Grunstein,M., Thoma,F. and Sentenac,A. (1995) Reciprocal interferences between nucleosomal organization and transcriptional activity of the yeast SNR6 gene. Genes Dev., 9, 410–422. [DOI] [PubMed] [Google Scholar]
- Matlock D.L. and Heyduk,T. (2000) Sequence determinants for the recognition of the fork junction DNA containing the –10 region of promoter DNA by E. coli RNA polymerase. Biochemistry, 39, 12274–12283. [DOI] [PubMed] [Google Scholar]
- Naryshkin N., Revyakin,A., Kim,Y.G., Mekler,V. and Ebright,R.H. (2000) Structural organization of the RNA polymerase–promoter open complex. Cell, 101, 601–611. [DOI] [PubMed] [Google Scholar]
- Panaghie G., Aiyar,S.E., Bobb,K.L., Hayward,R.S. and deHaseth,P.L. (2000) Aromatic amino acids in region 2.3 of Escherichia coli σ 70 participate collectively in the formation of an RNA polymerase–promoter open complex. J. Mol. Biol., 299, 1217–1230. [DOI] [PubMed] [Google Scholar]
- Pardee T.S., Bangur,C.S. and Ponticelli,A.S. (1998) The N-terminal region of yeast TFIIB contains two adjacent functional domains involved in stable RNA polymerase II binding and transcription start site selection. J. Biol. Chem., 273, 17859–17864. [DOI] [PubMed] [Google Scholar]
- Pinto I., Wu,W.H., Na,J.G. and Hampsey,M. (1994) Characterization of sua7 mutations defines a domain of TFIIB involved in transcription start site selection in yeast. J. Biol. Chem., 269, 30569–30573. [PubMed] [Google Scholar]
- Rameau G., Puglia,K., Crowe,A., Sethy,I. and Willis,I. (1994) A mutation in the second largest subunit of TFIIIC increases a rate-limiting step in transcription by RNA polymerase III. Mol. Cell. Biol., 14, 822–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranish J.A., Yudkovsky,N. and Hahn,S. (1999) Intermediates in formation and activity of the RNA polymerase II preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes Dev., 13, 49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts S., Miller,S.J., Lane,W.S., Lee,S. and Hahn,S. (1996) Cloning and functional characterization of the gene encoding the TFIIIB90 subunit of RNA polymerase III transcription factor TFIIIB. J. Biol. Chem., 271, 14903–14909. [DOI] [PubMed] [Google Scholar]
- Rüth J., Conesa,C., Dieci,G., Lefèbvre,O., Düsterhoft,A., Ottonello,S. and Sentenac,A. (1996) A suppressor of mutations in the class III transcription system encodes a component of yeast TFIIIB. EMBO J., 15, 1941–1949. [PMC free article] [PubMed] [Google Scholar]
- Sasse-Dwight S. and Gralla,J.D. (1989) KMnO4 as a probe for lac promoter DNA melting and mechanism in vivo. J. Biol. Chem., 264, 8074–8081. [PubMed] [Google Scholar]
- Sethy-Coraci I., Moir,R.D., López-de-León,A. and Willis,I.M. (1998) A differential response of wild type and mutant promoters to TFIIIB70 overexpression in vivo and in vitro. Nucleic Acids Res., 26, 2344–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuillier V., Stettler,S., Sentenac,A., Thuriaux,P. and Werner,M. (1995) A mutation in the C31 subunit of Saccharomyces cerevisiae RNA polymerase III affects transcription initiation. EMBO J., 14, 351–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner M., Chaussivert,N., Willis,I.M. and Sentenac,A. (1993) Interaction between a complex of RNA polymerase III subunits and the 70-kDa component of transcription factor IIIB. J. Biol. Chem., 268, 20721–20724. [PubMed] [Google Scholar]
- White R.J. (1998) RNA Polymerase III Transcription. Springer-Verlag & Landes Bioscience, New York & Georgetown.
- Whitehall S.K., Kassavetis,G.A. and Geiduschek,E.P. (1995) The symmetry of the yeast U6 RNA gene’s TATA box and the orientation of the TATA-binding protein in yeast TFIIIB. Genes Dev., 9, 2974–2985. [DOI] [PubMed] [Google Scholar]
- Yieh L., Kassavetis,G.A., Geiduschek,E.P. and Sandmeyer,S.B. (2000) The Brf and TATA-binding protein subunits of the RNA polymerase III transcription factor IIIB mediate position-specific integration of the gypsy-like element, Ty3. J. Biol. Chem., 275, 29800–29807. [DOI] [PubMed] [Google Scholar]