Abstract
Lag1p and Lac1p are two highly homologous membrane proteins of the endoplasmic reticulum (ER). When both genes are deleted, cells cannot transport glycosylphosphatidylinositol (GPI)-anchored proteins from the ER to the Golgi at a normal rate. Here we show that microsomes or detergent extracts from lag1Δlac1Δ double mutants lack an activity transferring C26 fatty acids from C26-coenzyme A onto dihydrosphingosine or phytosphingosine. As a consequence, in intact cells, the normal ceramides and inositolphosphorylceramides are drastically reduced. lag1Δlac1Δ cells compensate for the lack of normal sphingolipids by making increased amounts of C26 fatty acids, which become incorporated into glycerophospholipids. They also contain 20- to 25-fold more free long chain bases than wild type and accumulate very large amounts of abnormally polar ceramides. They make small amounts of abnormal mild base-resistant inositolphospholipids. The lipid remodelling of GPI-anchored proteins is severely compromised in lag1Δlac1Δ double mutants since only few and mostly abnormal ceramides are incorporated into the GPI anchors. The participation of Lag1p and Lac1p in ceramide synthesis may explain their role in determining longevity.
Keywords: fumonisin/glycosylphosphatidylinositol/ lipid remodelling/longevity genes/sphingolipids
Introduction
Sphingolipids are a complex class of membrane lipids present in all eukaryotes. Essential features of the membrane architecture depend on the presence of sphingolipids, which seem to be able to aggregate laterally and form specialized areas in the membrane, which are referred to as rafts or microdomains.
The only abundant sphingolipids of Saccharomyces cerevisiae are the inositolphosphorylceramides (IPCs), mannosyl-IPCs (MIPCs) and inositolphosphoryl-MIPC [M(IP)2C)] (Figure 1). Cells that cannot make IPCs are non-viable, whereas mannosylation of IPCs is dispensable (Beeler et al., 1997; Nagiec et al., 1997). Beyond their structural role, sphingolipids of yeast have also been proposed to be signals for adaptive responses to heat and other stresses. Indeed, shifting cells from 24 to 37°C causes transient increases in the levels of dihydrosphingosine (DHS), phytosphingosine (PHS), PHS-1 phosphate (PHS-1-P), DHS-1-P and a sustained elevation of free ceramides (Dickson et al., 1997b; Jenkins et al., 1997; Skrzypek et al., 1999). The elevation of ceramide levels is inhibited by australifungin (Aus) or fumonisin B1, two well-known inhibitors of ceramide synthase, thus suggesting that heat shock induces de novo synthesis of these signalling molecules (Jenkins et al., 1997; Wells et al., 1998). Several mutants accumulating DHS-1-P and PHS-1-P survive better at elevated temperature than wild-type cells, suggesting that such compounds induce resistance to heat stress (Mandala et al., 1998; Mao et al., 1999; Skrzypek et al., 1999). It has also been proposed that the extension of the yeast replicative life span afforded by transient, sublethal heat stress and the dependence of this phenomenon on RAS1 (Shama et al., 1998) are caused by alterations in sphingolipid metabolism (Jazwinski, 1999). The rapid ubiquitylation and degradation of the surface permease Fur4p after heat stress also seem to be mediated through a heat-induced sphingolipid (Chung et al., 2000). Moreover, ceramide addition to yeast cells leads to a G1 cell cycle arrest, and this effect is mediated through a ceramide-activated protein phosphatase (Fishbein et al., 1993; Nickels and Broach, 1996).
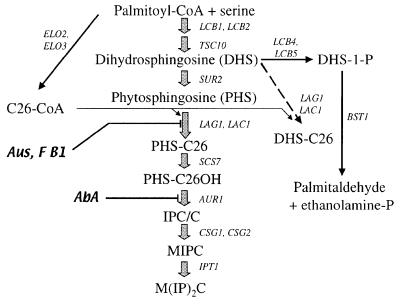
Fig. 1. A simplified scheme of sphingolipid biosynthesis in S.cerevisiae. The dashed arrow indicates the reaction observed in microsomes (see Figures 4 and 5). Aus, australifungin; AbA, aureobasidin A; F B1, fumonisin B1.
A further well-documented role of sphingolipid metabolites in yeast concerns their involvement in the vesicular transport of glycosylphosphatidylinositol (GPI)-anchored proteins out of the endoplasmic reticulum (ER) (Horvath et al., 1994; Skrzypek et al., 1997; Sütterlin et al., 1997) and in endocytosis (Zanolari et al., 2000). Transport of GPI-anchored proteins out of the ER in living cells is severely retarded if one reduces, either pharmacologically or genetically, the activity of the serine palmitoyltransferase (LCB1, LCB2), which is the key enzyme of DHS biosynthesis. It appears that the ER to Golgi transport of GPI-anchored proteins requires several dedicated components, which are dispensable for the transport of soluble proteins and ordinary membrane proteins with classical membrane-spanning domains. Dedicated components comprise sphingolipids, the cargo receptor Emp24p, components of the Golgi to ER retrotransport machinery and, as recently reported, Lag1p or Lac1p (Schimmöller et al., 1995; Skrzypek et al., 1997; Sütterlin et al., 1997; Barz and Walter, 1999; Muniz et al., 2000). Indeed, lag1Δlac1Δ cells transport GPI-anchored proteins from the ER to the Golgi at a reduced rate (Barz and Walter, 1999). Attention was drawn first to LAG1 since a gene deletion in haploid cells resulted in a pronounced increase (∼50%) in mean and maximum life span (D’mello et al., 1994). LAG1 has a close homologue in yeast, which was called LAC1 (DGT1), and orthologues of these two genes occur in a wide variety of eukaryotic cells (Jiang et al., 1998; Brandwagt et al., 2000). LAG1 and LAC1 have significant sequence similarity to TRAM, a mammalian membrane protein thought to be involved in protein translocation across the ER membrane, but no translocation defects were detected in lag1Δlac1Δ yeast cells (Barz and Walter, 1999). Furthermore, human TRAM does not complement the growth defect in a lag1Δlac1Δ double mutant, in contrast to another human gene called LAG1Hs (Jiang et al., 1998). While testing lag1Δlac1Δ cells for their capacity to remodel the lipid moiety of the GPI anchors, we realized that they have a severe remodelling defect, which is due to a general deficiency in ceramide biosynthesis.
Results
Deletion of LAG1 and LAC1 leads to drastically reduced amounts of IPCs
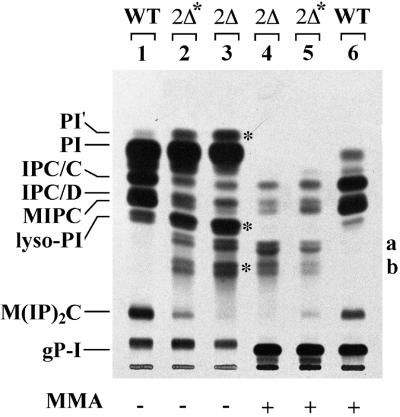
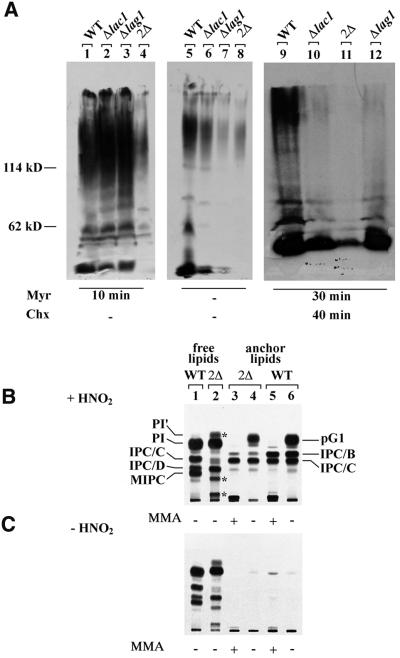
Whereas the single deletion of LAG1 or LAC1 has no phenotype, the concomitant deletion of LAG1 and LAC1 has been reported to cause a severe growth defect in the genetic background of W303 cells (Barz and Walter, 1999). The same double deletion did not yield viable cells in the background of YPK9 cells (Jiang et al., 1998). In our hands, lag1Δlac1Δ mutants generated in the W303 background grew only if the media were supplemented with osmotic stabilizers and high vitamin concentrations. However, they tended to adapt and to become overgrown by suppressors. To analyse the function of LAG1 and LAC1, we therefore used either short-term cultures of W303-1A lag1Δlac1Δ (designated 2Δ) or YPK9 lag1Δ lac1Δ pBM150:LAG1 containing a copy of LAG1 under the control of the GAL1 promoter. The latter strain has a wild-type growth phenotype when grown on galactose, but ceases to grow within 30 h when shifted to glucose, and at that stage is designated 2Δ*. Figure 2 shows one of many experiments in which the lipid extracts of [3H]inositol-labelled double mutants were compared with wild-type extracts. In all experiments, we found a very significant reduction in the classical structural sphingolipids IPC/C, IPC/D, MIPC and M(IP)2C. Quantitation of this experiment shows that the sum of these sphingolipids accounts for 26, 6 and 3% of total inositol-containing phospho lipids in wild-type, 2Δ* and 2Δ cells, respectively. Thus, structural sphingolipids are all significantly decreased, and 2Δ cells seem to be more severely affected than 2Δ* cells. Moreover, 2Δ* and 2Δ cells make several mild base-resistant [3H]inositol-labelled lipids (Figure 2, lanes 2–5), which run between MIPC and M(IP)2C and which cannot be detected in wild-type cells (Figure 2, lanes 1 and 6). In 2Δ and 2Δ* cells, the sum of these abnormal lipids (a and b) accounts for 62 and 35% of total mild base-resistant lipids, respectively. We speculate that they represent IPCs having shorter fatty acids (FAs) than the normal IPCs (see below). Another abnormality of the lipid profile of 2Δ* and 2Δ cells is the presence of a distinct, mild base-sensitive band above phosphatidylinositol (PI; band labeled PI′ in Figure 2). Moreover, 2Δ* and 2Δ cells contain a major band co-migrating with lyso-PI, which is abnormal, since it is clearly resolved from lyso-PI when other solvent systems are used (not shown).
Fig. 2. Deletion of LAG1 and LAC1 strongly reduces the amounts of IPCs. Ten OD of W303-1A (WT), YPK9 lag1Δlac1Δ harbouring pBM150:LAG1 (2Δ*), depleted of Lag1p by pre-culture on glucose for 30 h, and W303-1A lag1Δlac1Δ (2Δ) strains were radiolabelled with [3H]inositol at 30°C. The lipids were extracted, desalted, treated with monomethylamine (MMA) for saponification as indicated, and analysed on TLC using solvent system 2. Glycerolphosphorylinositol (gP-I) visible in lanes 4–6 was generated by saponification. The asterisks mark some abnormal lipids of 2Δ* and 2Δ.
lag1Δlac1Δ cells do not make PHS-C26OH
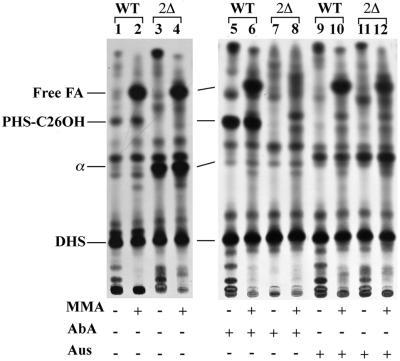
Ceramide biosynthesis was assayed by labelling cells with [3H]DHS, which is an obligatory intermediate in the biosynthesis of ceramide. Exogenously added [3H]DHS is readily incorporated into all sphingolipids (Reggiori et al., 1997). We reasoned that the lag1Δlac1Δ mutant should incorporate this physiological intermediate into sphingolipids normally if the deficiency of lag1Δlac1Δ mutants was due to a problem in the synthesis of DHS. As can be seen from Figure 3, [3H]DHS labelling of intact cells showed that 2Δ did not make any PHS-C26OH, a ceramide having a hydroxylated FA and which corresponds to the ceramide moiety of IPC/C, the main IPC in S.cerevisiae (Reggiori et al., 1997). The identity of this band was established through strong acid hydrolysis, yielding [3H]PHS, and comparison with a standard obtained through PI–phospholipase C (PLC) treatment of IPC/C (not shown). The absence of PHS-C26OH in 2Δ cells is not just due to an increased rate of utilization of this intermediate, since PHS-C26OH is also undetectable if its incorporation into IPCs is blocked by the addition of aureobasidin A (AbA), a specific inhibitor of the PI ceramide phosphorylinositol transferase AUR1 (Nagiec et al., 1997). While this drug strongly increases the relative amount of PHS-C26OH in wild-type cells, there is no appearance of this intermediate in 2Δ cells (Figure 3, compare lanes 5 and 7). On the other hand, as expected, the generation of PHS-C26OH is abrogated in the presence of Aus, a specific inhibitor of ceramide synthase (Mandala et al., 1995), which transfers FAs from acyl-CoA to DHS or PHS (Figure 3, lanes 9 and 11). In summary, the absence of IPCs in 2Δ cells can best be explained by a specific biosynthetic defect in the step transforming DHS into ceramide. The defect is not due to a general inability to utilize exogenous DHS since DHS is incorporated into glycerophospholipids efficiently through a pathway that requires phosphorylation to DHS-1-phosphate by LCB4 or LCB5 and cleavage to palmitaldehyde plus ethanolamine phosphate through the action of BST1/DPL1 (Saba et al., 1997; Nagiec et al., 1998) (Figure 1). Indeed, a prominent hydrophobic spot appearing only after mild base treatment of lipid extracts in wild-type as well as in 2Δ cells indicates the liberation of free [3H]FAs (Figure 3, lanes 1–4). Interestingly, lag1Δlac1Δ mutants make large amounts of an abnormal mild base-resistant lipid, lipid α, which may compensate for the loss of PHS-C26OH (Figure 3, lanes 3 and 4). The same lipid is also made by wild-type cells treated with Aus (Figure 3, lanes 9–12). Thus, the pharmacological inhibition of ceramide synthase not only abrogates the accumulation of PHS-C26OH, but also induces the appearance of lipid α, thus reproducing the lipid abnormalities of lag1Δlac1Δ mutants. While lipid α is resistant to mild base, it yields [3H]DHS upon strong acid hydrolysis (not shown), suggesting that it is some kind of ceramide. The fact that it is not made in the presence of AbA is presently not explained.
Fig. 3. The lag1Δlac1Δ cells cannot make PHS-C26OH. Two and a half OD of W303-1A (WT) and W303-1A lag1Δlac1Δ (2Δ) strains were pre-incubated with Aus, AbA or solvent for 10 min and were radiolabelled with 25 µCi of [3H]DHS at 30°C for 1 h. Lipid extracts were then O-deacylated with MMA or control incubated and analysed by TLC in solvent system 1.
The lag1Δlac1Δ cells up-regulate the biosynthesis of very long chain fatty acids
The absence of PHS-C26OH biosynthesis cannot be explained by deficiencies in the hydroxylation of DHS to PHS or of C26 to C26OH, since mutants defective in these hydroxylation steps have no problem in making IPCs (Haak et al., 1997). Thus, lag1Δlac1Δ mutants must either have a defect in the ceramide synthase or be deficient in making acyl-CoA. Since, in general, IPCs of S.cerevisiae contain C26 or C24 FAs or mono- or di-hydroxylated derivatives thereof, we undertook to investigate whether lag1Δlac1Δ mutants can elongate C16-CoA to C26-CoA using Elo2p, Elo3p and Tsc13p (Oh et al., 1997; Kohlwein et al., 2001). We thus analysed the content of very long chain FAs of lag1Δlac1Δ mutants by gas chromato graphy–mass spectroscopy (GC–MS). For this, the lipid extracts of cells were treated with methanolic boron fluoride (BF3) under conditions in which FAs esterified to glycerophospholipids are liberated and are methylated together with free FAs. As can be seen from Table I, in 2Δ cells the relative percentages of FAs up to C20 are not grossly altered as compared with wild type, but 2Δ cells show a discrete elevation of C20–C24 FAs and have a drastic increase in C26. After strong acid hydrolysis, which also liberates the FAs of sphingolipids, we still observed a very significant increase of C26:0 in 2Δ cells relative to wild type. Overall, it is clear that 2Δ cells are perfectly able to elongate FAs. Moreover, it seems that the lack of Lag1p and Lac1p induces the elongation pathway or inhibits the degradation pathway such as to generate an at least 4-fold higher level of C26:0 than normal. The massive excess of C26:0 is probably incorporated into glycerophospholipids, thus allowing the cells to generate C26:0-containing membrane lipids that may be able to take over some of the functions of sphingolipids, which, in normal cells, are the only lipids containing these very long chain FAs. This may explain the appearance of the [3H]inositol-labelled, mild base-sensitive lipid PI′ in Figure 2, lane 3. Indeed, preliminary experiments indicate that this lipid is a PI with abnormally long FAs (not shown).
Table I. lag1Δlac1Δ cells strongly up-regulate the biosynthesis of very long chain fatty acidsa.
| WT | 2Δ | WT + HCl | 2Δ + HCl | |
|---|---|---|---|---|
| C14:1 | 0.235 ± 0.002 | 0.159 ± 0.061 | 0.127 | 0.046 |
| C14:0 | 1.261 ± 0.159 | 0.567 ± 0.149 | 0.831 | 0.419 |
| C16:1 | 23.59 ± 0.357 | 21.56 ± 0.387 | 25.31 | 25.07 |
| C16:0 | 18.89 ± 0.283 | 11.91 ± 0.520 | 16.2 | 9.248 |
| C18:1 | 38.73 ± 0.233 | 33.22 ± 0.548 | 44.74 | 43.65 |
| C18:0 | 14.81 ± 1.381 | 24.12 ± 2.155 | 10.22 | 14.68 |
| C20:1 | 0 ± 0.000 | 0 ± 0.000 | 0.332 | 0.544 |
| C20:0 | 0.778 ± 0.230 | 1.53 ± 0.301 | 0.291 | 0.474 |
| C22:1 | 0.479 ± 0.045 | 0.547 ± 0.086 | 0.369 | 0.966 |
| C22:0 | 0.065 ± 0.065 | 0.288 ± 0.162 | 0.143 | 0.178 |
| C24:1 | 0.438 ± 0.094 | 0.444 ± 0.107 | 0.271 | 0.691 |
| C24:0 | 0.144 ± 0.144 | 0.447 ± 0.099 | 0.209 | 0.357 |
| C26:1 | 0.038 ± 0.038 | 0.123 ± 0.123 | 0 | 0 |
| C26:0 | 0.266 ± 1.434 | 4.886 ± 0.037 | 0.892 | 3.653 |
aThe lipids of W303-1A and W303-1A lag1Δlac1Δ strains were methylated either directly (columns WT and 2Δ) or after strong acid hydrolysis (columns WT + HCl and 2Δ + HCl) and analysed by GC–MS as described in Materials and methods. C16, C18 and C26 fatty acids were used as standards. Numbers indicate the percentage of a given fatty acid species compared with the total of identified peaks in the gas chromatogram. With the BF3 methylation procedure, we were not able to identify the hydroxylated fatty acids, since we had no appropriate standards and the conditions for their quantitative methylation had not been worked out. Thus, only 80% of the signals of the GC profile could be accounted for. Standard deviations represent variations obtained in two independent experiments. The same results as shown in columns WT and 2Δ were obtained when lipid extracts were treated with MMA before being methylated with methanolic BF3, indicating that the release of fatty acids from glycerophospholipids was complete (not shown).
Microsomes of lag1Δlac1Δ mutants lack ceramide synthase activity
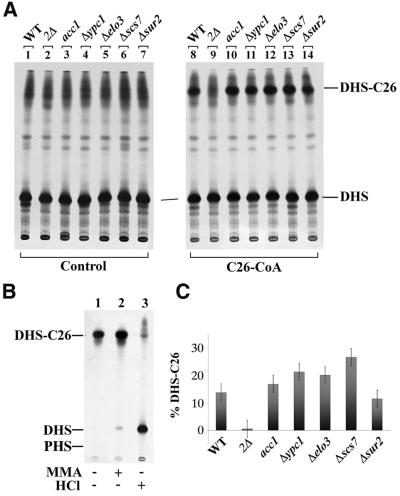
The results presented above persuaded us that the problem of lag1Δlac1Δ mutants was due to a problem with the ceramide synthase itself. Therefore, we measured ceramide synthase activity in microsomes by adding [3H]DHS and acyl-CoA with FAs of different chain length (Mandala et al., 1995). As shown in Figure 4A, lipid extracts of labelled microsomes contained lipids migrating with high mobility typical of ceramides or other unpolar compounds. In the absence of acyl-CoA, we observed a relatively diffuse smear (lanes 1–7), which was already present in the [3H]DHS that was added to microsomes (not shown), since the radiolabelled [3H]DHS is quite unstable and rapidly generates hydrophobic breakdown products after preparative purification by TLC (Lynch, 2000). However, when C26-CoA was added, a distinct band was generated on top of this material by microsomes from wild-type but not from lag1Δlac1Δ cells (Figure 4A). This band was assumed to be DHS-C26, based on the following indirect evidence: (i) the lipid is only generated when C26-CoA is added to the incubations, suggesting that it contains C26; (ii) the band resisted mild base but yielded [3H]DHS with strong acid (Figure 4B); (iii) as seen in Figure 5A, biosynthesis of this lipid in microsomes was blocked by the classical ceramide synthase inhibitors Aus and fumonisin B1 (Wu et al., 1995); and (iv) the FA was probably not hydroxylated since in scs7Δ cells, which are deficient in α-hydroxylation of FAs (Dunn et al., 1998), the band had the same mobility as in wild type (Figure 4A, lane 13; Figure 1). The band was also not generated when free C26, C16-CoA or free C16 were added in the presence of ATP and CoA (Figure 5C) or in the presence of C20-CoA (not shown). This result strongly indicates that the activity we observe requires a CoA-activated very long chain FA.
Fig. 4. Microsomes of lag1Δlac1Δ mutants lack DHS-C26 synthase activity. (A) Microsomes of W303-1A (WT), W303-1A lag1Δlac1Δ (2Δ) and the other mutants indicated at the top were radiolabelled with [3H]DHS without (Control) or in the presence of C26-CoA (0.2 mM). The thermosensitive acc1 cells were pre-incubated for 2 h at 37°C before preparing microsomes. The radiolabelled lipids were extracted, treated with MMA and analysed by TLC developed in solvent system 1. (B) The band labelled DHS-C26 in (A) was purified by two successive rounds of preparative TLC. The extract was divided into three aliquots, one of which remained untreated, one was MMA treated and the third was treated with MMA followed by strong acid hydrolysis. The three samples were then analysed by TLC developed in solvent system 1. (C) The amount of radioactivity in the spots of DHS-C26 of (A) lanes 8–14 was quantified by radioscanning and the results were expressed as the percentage of counts in [3H]DHS-C26 as compared with total radioactivity in that lane. Error bars indicate standard deviations in two entirely independent experiments.
Fig. 5. Biosynthesis of DHS-C26 is sensitive to Aus and fumonisin B1 (F B1). (A) Microsomes of W303-1A, W303-1A lag1Δlac1Δ containing a suppressor mutation (2Δsup) and acc1 were labelled with [3H]DHS in the presence of C26-CoA (0.2 mM). Microsomes were pre-incubated for 10 min with Aus, F B1 or without inhibitor (control). Lipids were extracted, MMA treated and analysed by TLC in solvent system 1. (B) The MMA-treated DHS-C26-like lipid from 2Δsup was purified by two successive rounds of preparative TLC and treated with strong HCl before being analysed by TLC with solvent system 1. (C) Microsomes of W303-1A cells were labelled with [3H]DHS under standard conditions (Materials and methods) but in the presence of C26-CoA, C26, C16-CoA, C16 and CoA as indicated.
Microsomes from acc1 and elo3Δ (deficient in FA elongation; Schneiter et al., 1996; Oh et al., 1997), from ypc1Δ (deficient in alkaline ceramidase; Mao et al., 2000) and from sur2Δ (deficient in the generation of PHS; Haak et al., 1997; Grilley et al., 1998) all contain normal or elevated ceramide synthase activity, as judged by this microsomal assay (Figures 1 and 4C). Thus, we conclude that microsomes of lag1Δlac1Δ lack a C26-CoA-dependent ceramide synthase activity in vitro. This is concordant with the absence of PHS-C26OH observed in lipid extracts from [3H]DHS-labelled cells (Figure 3) except that the microsomal in vitro system apparently does not support hydroxylation of DHS or C26:0, a feature that is not unexpected, since the microsomal system did not contain any NADPH.
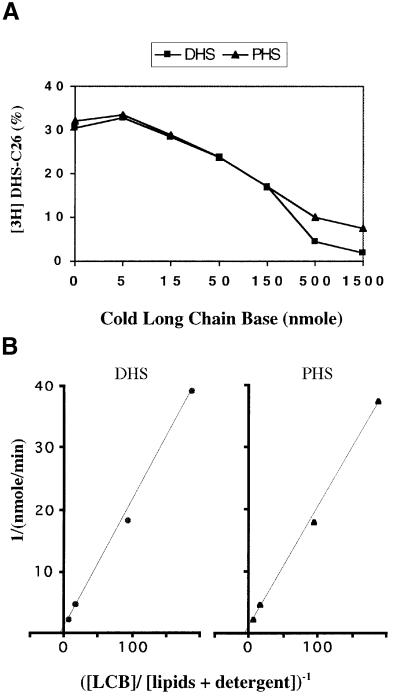
The microsomal activity generating DHS-C26 in a C26-CoA- and Lag1p/Lac1p-dependent manner could be solubilized efficiently by incubation in 1% digitonin at 4°C for 2 h with a recovery of activity of >100%. This allowed us to perform the assay in the presence of various amounts of cold DHS or PHS as shown in Figure 6A. Cold DHS and PHS both inhibit incorporation of [3H]DHS into [3H]DHS-C26. DHS, by definition, acts as a competitive inhibitor of [3H]DHS incorporation. In the physiological concentration range, PHS and DHS have overlapping inhibition curves, strongly suggesting that PHS is also a substrate which has about the same affinity for the enzyme as DHS. At long chain base/carrier lipid ratios of 0.53 and more, DHS was more inhibitory than PHS, but at these concentrations it is questionable whether the inhibition is still only competitive and whether the substrates are solubilized adequately in detergent micelles. From the data obtained using 5–155 nmol of DHS or PHS, we calculated the absolute amounts of product made per minute (Figure 6B) and thus obtained the Lineweaver–Burk plots shown in Figure 6B. They indicated that the apparent Vmax values were 1.35 nmol/min for DHS and 1.05 nmol/min for PHS. Half-maximal velocity was reached when the [long chain base]/[cellular lipid + detergent] ratio was 0.27 for DHS and 0.2 for PHS. Thus, it appears that Lag1p/Lac1p can utilize both DHS and PHS. Overexpression of Lag1p or Lac1p under the control of the GAL1 promoter did not increase the resistance of wild-type cells to Aus (Figure 7). These data are compatible with the hypothesis that a further subunit is required for ceramide synthase activity.
Fig. 6. Km and Vmax of microsomal DHS-C26 synthase activity. (A) Microsomes were lysed in 1% digitonin. A 100 µg aliquot of solubilized microsomal extract was assayed in the presence of 5 nmol of [3H]DHS, 200 nmol of C26-CoA and variable amounts (5–1500 nmol) of cold DHS or PHS in the presence of 1% digitionin. The products were extracted and analysed by TLC in solvent 1. [3H]DHS-C26 was quantitated by radioscanning and expressed as a percentage of total radioactivity in the corresponding lane. (B) The data of (A) were used to calculate enzymatic activities by calculating the total amounts of DHS-C26 made during incubations. DHS or PHS concentrations of the assays were expressed as mol/mol ratios of [long chain bases]/[total phospholipids + detergent]. The data were then transformed in double reciprocal plots according to Lineweaver–Burk. [Assays contained 120 nmol of phospholipids (estimated based on the amount of microsomal protein) and 820 nmol of detergent.]
Fig. 7. Overexpression of Lag1p does not confer resistance to Aus. YPK9 cells containing pBM150 either without an insert or harbouring LAC1 or LAG1 under the control of the GAL1 promoter were cultured on yeast nitrogen base with 2% galactose but lacking uracil, and were seeded at 5500 cells per plate containing the same medium. Then a filter paper with either solvent alone or 20 µg of Aus was placed in the centre. Plates were photographed after 3 days of incubation at 24°C.
Characterization of a suppressor growing in the absence of LAG1 and LAC1
Prolonged culture of lag1Δlac1Δ led to the appearance of spontaneous suppressors. One suppressor, labelled 2Δsup, was also assayed for microsomal ceramide synthase. As can be seen in Figure 5, lanes 2, 5 and 8, the suppressor’s microsomes had regained an activity generating lipids migrating close to DHS-C26, an activity that was insensitive to Aus and also partially resistant to fumonisin B1. These lipids are resistant to mild base and yield [3H]DHS upon strong acid hydrolysis (Figure 5B). This suggests that these lipids are DHS-containing ceramides. They are clearly different from the mild base-resistant lipid α observed in intact cells treated with Aus or in lag1Δlac1Δ (Figure 3, lanes 3, 4 and 9–12) since the lipids made by the suppressor co-migrate with DHS-C26, whereas lipid α is more polar than PHS-C26OH, a compound which itself is considerably more polar than DHS-C26. It is conceivable that the activity induced in this suppressor is mediated by Ypc1p, an alkaline ceramidase that can catalyse the reverse reaction, namely the condensation of free FAs with DHS or PHS (Mao et al., 2000). Ypc1p is not blocked by fumonisin B1 and apparently is used to make ceramide when cells are treated with ceramide synthase inhibitors. The acyl-DHS of 2Δsup can apparently functionally replace the normal ceramides as substrates for IPC biosynthesis, since when we labelled intact 2Δsup cells with [3H]DHS or [3H]inositol, they exhibited a normal profile of sphingolipids (not shown).
lag1Δlac1Δ cells accumulate free long chain bases and ceramides
It has been found previously that the treatment of yeast cells with fumonisin B1 increased the cellular concentration of DHS and PHS up to 50-fold (Wu et al., 1995). Unpolar lipids were extracted from wild type and lag1Δlac1Δ mutants, and free long chain bases were derivatized and quantitated using HPLC. As shown in Table II, while the concentrations of DHS and PHS in wild-type cells were very similar to values reported in the literature (Dickson et al., 1997b), in lag1Δlac1Δ they were 41- and 16-fold higher. Concentrations of free ceramides were measured after separating them from free long chain bases and IPCs by silica gel chromatography. As seen in Table II, while free ceramide levels in wild-type cells were in the same range as the values reported in the literature (Dickson et al., 1997b), those in lag1Δlac1Δ were drastically increased. The increase was ∼195-fold for DHS-containing ceramides and 21-fold for PHS-containing ceramides. The ceramide levels of lag1Δlac1Δ are in the range of total inositol-containing sphingolipids found in wild-type cells, i.e. ∼750 pmol/OD600 of cells (Hanson and Lester, 1980). Our procedure did not allow the identification of the FA of the ceramides, but it seems likely that these abundant free ceramides of lag1Δlac1Δ consist of lipid α (Figure 3, lanes 3 and 4), which most probably represents an abnormally polar ceramide (see above). We interpret the rise in ceramides in the sense that the lack of Lag1p and Lac1p induces the DHS levels to rise sufficientlty to permit some unknown enzyme(s) to catalyse the condensation of DHS or PHS with component X, thus generating lipid α. Whatever the exact nature of ceramides accumulating in lag1Δlac1Δ, it would appear that they are bad substrates for the IPC synthase Aur1p (Nagiec et al., 1997) or that they have difficulty in accessing the compartment in which Aur1p is active. In view of the huge accumulation of abnormal ceramides, one might consider that Lag1p and Lac1p are not catalytic subunits of the ceramide synthase, but rather are required to make it specific for C26-CoA. However, this is ruled out by the fact that microsomes from neither wild type (Figure 5C, lane 4) nor lag1Δlac1Δ (not shown) synthesize any ceramides in the presence of C16-CoA.
Table II. Free long chain base and ceramide content of lac1Δlag1Δa.
| W303-1A | W303-1A lac1Δlag1Δ | |
|---|---|---|
| Free DHS | 10 ± 3.7b | 401 ± 81 |
| Free PHS | 28 ± 7.2 | 454 ± 152 |
| DHS in Cer | 4 ± 1.7 | 974 ± 398 |
| PHS in Cer | 30 ± 7.0 | 660 ± 277 |
aAll results are given as pmol/OD600 unit of cells.
bResults are averaged from five independent experiments performed on different days and with different cell cultures.
lag1Δlac1Δ utilizes abnormal ceramides to remodel the lipid moiety of GPI anchors
The PI moiety of the GPI added to newly made GPI-anchored proteins in the ER is indistinguishable from ordinary PI and probably contains C16 or C18 FAs. This PI moiety, however, is remodelled rapidly so that more mature GPI proteins either possess a diacylglycerol with a C26 FA in the sn-2 position or contain a ceramide, namely PHS-C26 or PHS-C26OH. The latter ceramide is only found in GPI-anchored proteins that have reached the Golgi apparatus (Reggiori et al., 1997; Sipos et al., 1997). Since it has been observed that Gas1p and Yap3p, two well-characterized GPI-anchored proteins of yeast, are not transported out of the ER with normal kinetics in lag1Δlac1Δ (Barz and Walter, 1999), it was of interest to see whether they remodelled GPI anchors normally. Metabolic labelling of cells with [3H]DHS results in the labelling of a number of highly and very heterogeneously glycosylated proteins, which are all GPI-anchored proteins (Reggiori et al., 1997). In normal cells, most GPI-anchored proteins have ceramide-based anchors. A normal incorporation of [3H]DHS into proteins thus indicates that a cell makes normal amounts of GPI-anchored proteins and has no problem in introducing the ceramide moiety into GPI anchors. As can be seen in Figure 8A, the incorporation of [3H]DHS into proteins was diminished in lag1Δ, lac1Δ and lag1Δlac1Δ (lanes 5–8), but could be normalized in the single deletants by the addition of myriocin, a specific inhibitor of serine palmitoyltransferase, thus blocking the endogenous biosynthesis of DHS (lanes 1–4). Additionally, we tested remodelling reactions occurring beyond the ER by pre-incubating cells for 40 min in the presence of cycloheximide, thus allowing GPI-anchored proteins to leave the ER (Reggiori et al., 1997). This test showed that the remodelling reactions in the Golgi or at the cell surface were drastically reduced in single and double mutants (Figure 8A, lanes 9–12). Overall, the data indicated a relative deficiency in the biosynthesis and/or remodelling of GPI-anchored proteins. It should be noted that this deficiency need not be attributed to a general slowing down of metabolic processes in these slowly growing cells, since lac1Δ or lag1Δ grow normally and since the maturation kinetics of CPY and Pho8p were quite normal in lag1Δlac1Δ in spite of a strongly reduced growth rate (Barz and Walter, 1999).
Fig. 8. lag1Δlac1Δ utilizes abnormal ceramides to remodel the lipid moiety of GPI anchors. (A) Two and a half OD of W303-1A (WT), W303-1A lac1Δ, W303-1A lag1Δ and W303-1A lag1Δlac1Δ were radiolabelled at 30°C with 25 µCi of [3H]DHS, and proteins were analysed by SDS–PAGE/fluorography. Before addition of the label, cells were pre-incubated for the indicated times with the inhibitors myriocin (myr) or cycloheximide (Chx). (B and C) Ten OD600 of W303-1A (WT) and W303-1A lag1Δlac1Δ (2Δ) were radiolabelled with [3H]inositol at 30°C and GPI-anchored peptides free of contaminating lipids were prepared. Anchor peptides were treated with HNO2 (B) or control incubated (C). The liberated lipids were desalted by butanol extraction and incubated with our without MMA. Anchor lipids (lanes 3–6) together with the free lipids obtained by the delipidation procedure (lanes 1 and 2) were analysed on TLC using solvent system 3. pG1 indicates the typical phosphatidylinositol found on GPI anchors. The asterisks marks the several abnormal lipids of 2Δ already pointed out in Figure 2. The absence of anchor lipids in lanes 3–6 of (C) indicates that the delipidation of GPI-anchored proteins was complete.
For further analysis, we labelled cells with [3H]inositol, a metabolic labelling procedure that specifically labels GPI-anchored proteins, irrespective of the type of anchor lipid. The incorporation of [3H]inositol into GPI anchors in lag1Δlac1Δ was only 28% of that observed in the wild type, while the incorporation into PI was unaffected. After extensive delipidation, the labelled proteins were digested with pronase, thus generating anchor peptides, and their [3H]inositol-phosphoryl-lipid moieties were liberated by treatment with nitrous acid (Guillas et al., 2000). The released anchor lipids were analysed by TLC and compared with the cell’s free lipids as shown in Figure 8B. Anchors of normal cells contain the previously characterized phosphoinositides pG1, IPC/B and IPC/C (Sipos et al., 1997) (Figure 8B, lane 6). pG1 is a PI with a C26 FA in sn-2 of its glycerol moiety and therefore exhibits a higher mobility than the bulk of the cell’s free PI. Thus, although the absolute amount of incorporation of [3H]inositol into proteins is significantly reduced in the lag1Δlac1Δ mutants, the relative amount of diacylglycerol-based anchors (pG1) is normal (55 instead of 52%). A major difference appears in the mild base-resistant ceramide-based anchors. GPI anchors of wild-type cells contain mainly IPC/B and IPC/C, their ceramide moieties being PHS-C26:0 and PHS-C26OH, respectively (Reggiori et al., 1997). In wild-type cells, IPC/B and IPC/C accounted for 68 and 31%, respectively, of base-resistant anchors (quantitation of Figure 8B, lane 5). Anchors from lag1Δlac1Δ contained three base-resistant lipids, two of which co-migrated with IPC/B and IPC/C and accounted for 9.5 and 86% of base-resistant anchors, respectively (quantitation of Figure 8B, lane 3). It is thus apparent that the main base-resistant lipid moiety of GPI anchors of lag1Δlac1Δ is not IPC/B as in wild-type cells, but rather a less hydrophobic lipid co-migrating with IPC/C. In normal cells, IPC/C is only found in GPI-anchored proteins that have reached the Golgi apparatus. Since lag1Δlac1Δ specifically delays the transport of GPI-anchored proteins to the Golgi, the pulse labelling protocol used in Figure 8 should, in principle, lead to a reduced relative amount of IPC/C in the anchor population. However, we observe the opposite. We therefore suspect that, in spite of their co-migration with IPC/B and IPC/C, the base-resistant lipids used for remodelling in lag1Δlac1Δ consist of abnormal ceramides having C16 FAs or some other mild-base-resistant lipid, but further studies will have to confirm this hypothesis.
Discussion
The data presented here indicate that Lag1p and Lac1p are either coding for a catalytic subunit of ceramide synthase or are obligatory activators for the ceramide synthase activity in yeast. Indeed, they fulfil many of the criteria that we would want to see met by any candidate gene coding for ceramide synthase itself. (i) Homologues have been found in mammals, insects, worms, higher fungi and plants (Jiang et al., 1998; Brandwagt et al., 2000). (ii) They contain ER retention signals (KKXX or KXKXX), and tagged versions of Lag1p and Lac1p have been localized in yeast to the ER by immunofluorescence (Barz and Walter, 1999; P.A.Kirchman and S.M.Jazwinski, unpublished). This is the expected location of ceramide synthase, since mammalian ceramide synthase has been localized to the ER (Mandon et al., 1992) and since other sphingolipid biosynthesis enzymes, such as keto-DSH reductase (Tsc10p) or DSH-hydroxylase (Sur2p), have ER retention motifs (Haak et al., 1997; Beeler et al., 1998) (Figure 1). ER retention signals are also present in LAG1 homologues from Schizosaccharomyces pombe, man, mice, Drosophila melanogaster and Caenorhabditis elegans. (iii) The human homologue LAG1Hs, in spite of very limited homology to yeast LAG1 and LAC1, can restore viability and a normal life span to yeast lag1Δlac1Δ mutants. Functional conservation is seen more often with enzymes than with regulatory subunits (Jiang et al., 1998). (iv) As frequently found for housekeeping genes, the human LAG1Hs lacks a TATA box in the promoter (Jiang et al., 1998). (v) The hydrophobic transmembrane domains of LAG1 and its homologues can be predicted not only to have membrane-anchoring function, but also to carry out some specific function, since a large part of the conserved residues including the so-called Lag1p motif are found within the transmembrane domains (Jiang et al., 1998; Barz and Walter, 1999).
Cells remaining viable in spite of the concomitant deletion of LAG1 and LAC1 show interesting adaptive phenomena. lag1Δlac1Δ mutants make novel mild base-resistant, inositol-containing lipids in vivo (Figure 2, lipids a and b), and they also generate huge amounts of an abnormal, [3H]DHS-labelled, base-resistant lipid, probably some kind of ceramide with a shorter chain FA (Figure 3, lipid α). Further structural studies will have to be undertaken in order to determine whether these lipids are related, i.e. whether lipids a and b contain lipid α. It is conceivable that the latter is a ceramide made by Ypc1p, a newly discovered alkaline ceramidase that can also catalyse the reverse reaction, namely the condensation of a FA with DHS or PHS (Mao et al., 2000). Mao et al. (2000) show that microsomes from cells that overexpress Ypc1p make significantly increased amounts of labelled ceramide from [3H]palmitic acid and PHS, and they provide evidence suggesting that Ypc1p synthesizes ceramide in intact cells if they are treated with fumonisin B1. It is noteworthy that YBR183w (YPC1) was isolated as a multicopy suppressor of a lag1Δlac1Δ (P.A.Kirchman and S.M.Jazwinski, in preparation). In our microsomal assay, the synthesis of ceramide in the presence of C16 was, at best, very minor in comparison with the LAG1/LAC1-dependent synthesis of DHS-C26 (Figure 5C, lanes 1, 2 and 5). However, the conditions in our assays were quite different from those of Mao et al. (2000) since here Ypc1p was not overexpressed, incubations lasted only for 1 h, contained no detergent and no Ca2+ but rather contained EGTA and ATP, and since the label was supplied as [3H]DHS. A compensatory mechanism also seems to provide some ceramide in the 2Δsup strain, although for the moment we cannot tell what enzyme is synthesizing this lipid. In summary, our study lends support to the notion that yeast cells have alternative ways of making ceramides, so that functional studies using inhibitors such as fumonisins or Aus have to be interpreted with great care.
A further interesting compensatory mechanism seems to be the up-regulation of the biosynthesis of long chain FAs and their incorporation into glycerophospholipids (Table I). It has been reported previously that lcb1slc1-1 cells, being deficient in serine palmitoyltransferase, start to incorporate C26 FAs into PI when long chain bases are removed from their culture medium (Lester et al., 1993; Nagiec et al., 1993). Thus, these cells made a lipid that may be identical to PI′ observed in [3H]inositol-labelled lag1Δlac1Δ cells (Figure 2, lanes 1–3). This previous study also showed that PI with a C26 FA in the sn-2 position of the glycerol moiety could serve as a substrate for the mannosyltransferase (MIPC synthase CSG1, CSG2; Zhao et al., 1994; Beeler et al., 1997) and M(IP)2C synthase (IPT1; Figure 1; Dickson et al., 1997a), thus generating a class of lipids that resembles the natural sphingolipids with regard to the head groups as well as the presence of C26. Further studies into the exact nature of PI′ and other abnormal lipids of lag1Δlac1Δ are under way. It is quite conceivable that the potential to induce compensatory mechanisms depends on the genetic background, thus explaining why the double deletion of LAG1 and LAC1 is lethal in some but not all genetic backgrounds.
The vesicular transport of GPI-anchored proteins out of the ER is singularly dependent on ongoing sphingolipid biosynthesis (Horvath et al., 1994; Skrzypek et al., 1997; Sütterlin et al., 1997). The delay in transport of GPI-anchored proteins from ER to Golgi, which was observed in lag1Δlac1Δ cells (Barz and Walter, 1999), is likely to be secondary to the defect in ceramide biosynthesis in this mutant. This delay may result from inhibition or delay of GPI anchor attachment, a possibility raised by the 4-fold lower [3H]inositol incorporation into GPI-anchored proteins (Figure 8). Alternatively, and more probably, this low incorporation may be a reflection of a corresponding decrease in GPI-anchored protein biosynthesis in these slowly growing cells. Thus, it is not yet clear whether the apparent GPI protein transport deficiency of lag1Δlac1Δ cells is due to a problem with the anchoring or with the actual ER to Golgi transport of GPI-anchored proteins. Only if it is established that it is transport that is at fault can we interpret the data to mean that ceramide and not simply DHS, PHS, DHS-1-P or PHS-1-P is required for ER to Golgi transport of GPI-anchored proteins.
The identification of the role of LAG1 and LAC1 in sphingolipid metabolism provides a rationale for their impact on life span.
Materials and methods
Strains, growth conditions and materials
Strains used in this study are listed in Table III; the plasmids were described previously (Jiang et al., 1998). Yeast cells were grown in YPD, SD or SDC medium (Benghezal et al., 1996), supplemented as necessary, with the exception of W303-1A lag1Δlac1Δ, which was grown on SDCUA supplemented with 1 M sorbitol and 10 times the normal concentration of vitamins. Photometric determination of cell density was performed as described (Benghezal et al., 1996), 1 OD600 of cells corresponding to 1–2 × 107 cells. [3H]DHS was synthesized by NEN (Du Pont De Nemours, Les Ulis, France) and purified as described (Reggiori et al., 1997). AbA was from Takara Shuzo Co. (Shiga, Japan) and fumonisin B1 from Alexis Biochemicals (Läufelfingen, Switzerland). Aus was a generous gift of Suzanne Mandala (Merck, Rahway, NJ) and myriocin was a kind gift of Dr N.Rao Movva (Novartis, Basel, Switzerland).
Table III. Strains of S.cerevisiae used in this study.
| Strain | Genotype | Source |
|---|---|---|
| W303-1A | MATa can1-100 ade2-1 his3-11,15 leu2-3 112 trp1-1 ura3-1 | S.M.Jazwinski |
| YPK9 | MATa ade2-101ochre his3-Δ200 leu2-Δ1 lys2-801amber trp1-Δ63 ura3-52 | S.M.Jazwinski |
| W3030-1A lac1Δ | same as W303-1A, but lac1Δ::LEU2 | S.M.Jazwinski |
| W303-1A lag1Δ | same as W303-1A, but lag1Δ::TRP1 | S.M.Jazwinski |
| W303-1A lac1Δlag1Δ | same as W303-1A, but lac1Δ::LEU2 lag1Δ::TRP1 | S.M.Jazwinski |
| YPK9 lac1Δ | same as YPK9, but lac1Δ::LEU2 | S.M. Jazwinski |
| YPK9 lag1Δ | same as YPK9, but lag1Δ::TRP1 | S.M.Jazwinski |
| YPK9 lac1Δlag1Δ pBM150:LAG1 | same as YPK9, but lac1Δ::LEU2 lag1Δ::TRP1 containing pBM150:LAG1 | S.M.Jazwinski |
| YPK9 lac1Δlag1Δ pBM150:LAC1 | same as YPK9, but lac1Δ::LEU2 lag1Δ::TRP1 containing pBM150:LAC1 | S.M.Jazwinski |
| YPK9 pBM150:LAG1 | same as YPK9, but containing pBM150:LAG1 | S.M.Jazwinski |
| YPK9 pBM150:LAC1 | same as YPK9, but containing pBM150:LAC1 | S.M.Jazwinski |
| YPK9 pBM150 | same as YPK9, but containing empty pBM150 | S.M.Jazwinski |
| ypc1Δ | MATα ura3-52 HIS3 leu2-3,112 TRP1 YBR183w(4948)::kanMX4 | EUROSCARF |
| acc1 ts | MATa acc1-2150 | R.Schneiter |
| elo3Δ | MATa ura3 trp1 leu2 elo3::URA3 | G.Loison and R.Schneiter |
| scs7Δ | MATa trp1 leu2 his4 scs7Δ::URA3 | T.Dunn |
| sur2Δ | MATa ura3-52 leu2 his4 sur2Δ::TRP1 | T.Dunn |
Cell labelling, lipid extraction, treatment and thin-layer chromatography
Aus, AbA, fumonisin B1, myriocin and cycloheximide were added at concentrations of 2, 1, 10, 40 and 200 µg/ml, respectively. Unless stated otherwise, labelling, cell disruption, lipid extraction and analysis were carried out as described in Reggiori et al. (1997). Where indicated, lipids were subjected to mild base treatment using monomethylamine (MMA). For this, lipids were dissolved in MMA:water (10:3) and incubated for 1 h at 53°C. The samples were allowed to cool before being evaporated under vacuum in a Speed Vac evaporator. Strong HCl hydrolysis was carried out at 80°C in 2 M HCl for 16 h. Solvents 1, 2 or 3 were used for ascending TLC, namely 1 = chloroform:methanol:NH4OH (40:10:1), 2 = chloroform:methanol:0.25%KCl (55:45:10) and 3 = chloroform: methanol:0.25%KCl (55:45:5). Radioactivity was detected and quantitated by one-dimensional radioscanning and fluorography. Lipids to be analysed by GC–MS were always dried under a nitrogen stream instead of being vacuum evaporated, and were kept under nitrogen until injection. Labelling of cells with [3H]inositol for purification and analysis of GPI-anchored peptides was carried out as described (Guillas et al., 2000).
GC–MS analysis of fatty acids
Dry lipids obtained from 100 OD of exponentially growing cells were incubated, either directly or after strong acid hydrolysis (see above), with 1 ml of 14% BF3 in methanol for 2 min at 95°C. After methylation, 1 ml of water was added, and the methanol–water phase was extracted three times with 3 ml of petroleum ether. Pooled petroleum ether phases were washed with 2 ml of water before being dried under nitrogen. For injection, samples were resuspended in hexane:anhydrous ethyl ether (1:1). GC–MS analysis was performed as described (Oh et al., 1997), using a 30 m × 0.25 mm Optima 05ms column with a film thickness of 0.25 µm. Gas chromatography data and mass spectra were analysed using the Xcalibur™ Finnigan corporation software.
Quantitation of long chain bases, free ceramides and total sphingolipids
Lipids were extracted as described in Nagiec et al. (1997) with 5 ml of chloroform:methanol (1:1) at 50°C. Lipids from 100 OD600 were dried under a stream of nitrogen and were derivatized directly with the o-phthalaldehyde (OPA) reagent (Merrill et al., 1988) to quantify free long chain bases. Alternatively, to isolate free ceramides, dried lipids were resuspended in 0.5 ml of chloroform and applied to a 1 ml silica gel 100–200 mesh (Sigma, S4133) column equilibrated in chloroform. After washing with 3 ml of chloroform, the free ceramides were eluted with 3 ml of chloroform:methanol (9:1) (Nagiec et al., 1997). The eluted ceramides were dried and subjected to strong acid hydrolysis before OPA derivatization. Derivatized PHS and DHS were separated and quantified by HPLC on a 25 × 0.46 Nucleosil 100-5 C18 column, eluted with methanol–5 mM K2HPO4 pH 7.0 (90:10) at a flow rate of 0.8 ml/min and monitored at 340 nm.
Synthesis of C26-CoA
The protocol was adapted from Hajra and Bishop (1986) for non-radioactive FAs. Briefly, 30 µmol of C26 were dried thoroughly three times with 1 ml of dry benzene under a gentle stream of nitrogen. A 1.6 ml aliquot of dry benzene and 0.8 ml of oxalyl chloride were added to the FA residue and incubated for 90 min under nitrogen at room temperature. Liquids were evaporated with dry nitrogen and the oxalyl chloride residue was dried three times with dry benzene under dry nitrogen. A 35 µmol concentration of CoA lithium salt dissolved in 2.6 ml of tetrahydrofuran– aqueous 150 mM NaHCO3 (2.2:1.0) (adjusted to pH 8.8 with NaOH) was added to the acyl chloride and incubated for 2 h at 37°C under nitrogen with vigorous stirring. The reaction was stopped by adding 40 µl of 10% HClO4. The tetrahydrofuran was evaporated under dry nitrogen. The residue was then washed by repeated resuspension and centrifugation (18 000 g, 15 min at 4°C): twice with 5 ml of 1.3% HClO4, once with 8 ml of acetone, twice with 8 ml of anhydrous ether and finally with 8 ml of tetrahydrofuran. The residue was dried under nitrogen and resuspended in 0.5 mM Zwittergent 3-16. The concentration of C26-CoA was then evaluated by its absorbance at 260 nm (ε = 16 800).
Labelling of microsomes with [3H]DHS
Microsomes from 2.5 OD of cells were prepared as described (Canivenc-Gansel et al., 1998). They were labelled with 50 µCi of [3H]DHS for 1 h at 37°C in labelling buffer containing 100 mM Tris–HCl pH 7.5, 1 mM EGTA, 3 mM MgCl2, 0.5 mM MnCl2, 1 mM ATP, 30 mM creatine phosphate and 1 mg/ml creatine kinase. CoA, C26-CoA, C16-CoA, C26 and C16 were added to achieve final concentrations of 1.0, 0.1, 0.14, 0.1 and 0.1 mM, respectively, only where indicated.
Acknowledgments
Acknowledgements
We would like to thank Drs Suzanne Mandala and Rao Movva and their respective companies for inhibitors, and Dr Philippe Renaud for help with the synthesis of C26-CoA. We would also like to thank Patrick Fraering and Isabelle Flury for preparing [3H]DHS, and Carole Roubaty for technical help. This work was supported by grant No. 3100-032515 from the Swiss National foundation to A.C. and by grant AG06168 from the National Institute on Aging of the National Institutes of Health (USPHS) to S.M.J.
References
- Barz W.P. and Walter,P. (1999) Two endoplasmic reticulum (ER) membrane proteins that facilitate ER-to-Golgi transport of glycosylphosphatidylinositol-anchored proteins. Mol. Biol. Cell, 10, 1043–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeler T.J., Fu,D., Rivera,J., Monaghan,E., Gable,K. and Dunn, T.M. (1997) SUR1 (CSG1/BCL21), a gene necessary for growth of Saccharomyces cerevisiae in the presence of high Ca2+ concentrations at 37°C, is required for mannosylation of inositolphosphorylceramide. Mol. Gen. Genet., 255, 570–579. [DOI] [PubMed] [Google Scholar]
- Beeler T., Bacikova,D., Gable,K., Hopkins,L., Johnson,C., Slife,H. and Dunn,T. (1998) The Saccharomyces cerevisiae TSC10/YBR265w gene encoding 3-ketosphinganine reductase is identified in a screen for temperature-sensitive suppressors of the Ca2+-sensitive csg2Δ mutant. J. Biol. Chem., 273, 30688–30694. [DOI] [PubMed] [Google Scholar]
- Benghezal M., Benachour,A., Rusconi,S., Aebi,M. and Conzelmann,A. (1996) Yeast Gpi8p is essential for GPI anchor attachment onto proteins. EMBO J., 15, 6575–6583. [PMC free article] [PubMed] [Google Scholar]
- Brandwagt B.F., Mesbah,L.A., Takken,F.L., Laurent,P.L., Kneppers,T.J., Hille,J. and Nijkamp,H.J. (2000) A longevity assurance gene homolog of tomato mediates resistance to Alternaria alternata f. sp. lycopersici toxins and fumonisin B1. Proc. Natl Acad. Sci. USA, 97, 4961–4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canivenc-Gansel E., Imhof,I., Reggiori,F., Burda,P., Conzelmann,A. and Benachour,A. (1998) GPI anchor biosynthesis in yeast: phosphoethanolamine is attached to the α1,4-linked mannose of the complete precursor glycophospholipid. Glycobiology, 8, 761–770. [DOI] [PubMed] [Google Scholar]
- Chung N., Jenkins,G., Hannun,Y.A., Heitman,J. and Obeid,L.M. (2000) Sphingolipids signal heat stress-induced ubiquitin-dependent proteolysis. J. Biol. Chem., 275, 17229–17232. [DOI] [PubMed] [Google Scholar]
- Dickson R.C., Nagiec,E.E., Wells,G.B., Nagiec,M.M. and Lester,R.L. (1997a) Synthesis of mannose-(inositol-P)2-ceramide, the major sphingolipid in Saccharomyces cerevisiae, requires the IPT1 (YDR072c) gene. J. Biol. Chem., 272, 29620–29625. [DOI] [PubMed] [Google Scholar]
- Dickson R.C., Nagiec,E.E., Skrzypek,M., Tillman,P., Wells,G.B. and Lester,R.L. (1997b) Sphingolipids are potential heat stress signals in Saccharomyces. J. Biol. Chem., 272, 30196–30200. [DOI] [PubMed] [Google Scholar]
- D’mello N.P., Childress,A.M., Franklin,D.S., Kale,S.P., Pinswasdi,C. and Jazwinski,S.M. (1994) Cloning and characterization of LAG1, a longevity-assurance gene in yeast [published erratum appears in J. Biol. Chem., 269, 28522]. J. Biol. Chem., 269, 15451–15459. [PubMed] [Google Scholar]
- Dunn T.M., Haak,D., Monaghan,E. and Beeler,T.J. (1998) Synthesis of monohydroxylated inositolphosphorylceramide (IPC-C) in Saccharomyces cerevisiae requires Scs7p, a protein with both a cytochrome b5-like domain and a hydroxylase/desaturase domain. Yeast, 14, 311–321. [DOI] [PubMed] [Google Scholar]
- Fishbein J.D., Dobrowsky,R.T., Bielawska,A., Garrett,S. and Hannun,Y.A. (1993) Ceramide-mediated growth inhibition and CAPP are conserved in Saccharomyces cerevisiae. J. Biol. Chem., 268, 9255–9261. [PubMed] [Google Scholar]
- Grilley M.M., Stock,S.D., Dickson,R.C., Lester,R.L. and Takemoto,J.Y. (1998) Syringomycin action gene SYR2 is essential for sphingolipid 4-hydroxylation in Saccharomyces cerevisiae. J. Biol. Chem., 273, 11062–11068. [DOI] [PubMed] [Google Scholar]
- Guillas I., Pfefferli,M. and Conzelmann,A. (2000) Analysis of ceramides present in glycosylphosphatidylinositol anchored proteins of Saccharomyces cerevisiae. Methods Enzymol., 312, 506–515. [DOI] [PubMed] [Google Scholar]
- Haak D., Gable,K., Beeler,T. and Dunn,T. (1997) Hydroxylation of Saccharomyces cerevisiae ceramides requires Sur2p and Scs7p. J. Biol. Chem., 272, 29704–29710. [DOI] [PubMed] [Google Scholar]
- Hajra A.K. and Bishop,J.E. (1986) Preparation of radioactive acyl coenzyme A. Methods Enzymol., 122, 50–53. [DOI] [PubMed] [Google Scholar]
- Hanson B.A. and Lester,R.L. (1980) Effects of inositol starvation on phospholipid and glycan syntheses in Saccharomyces cerevisiae. J. Bacteriol., 142, 79–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath A., Sütterlin,C., Manning-Krieg,U., Movva,N.R. and Riezman,H. (1994) Ceramide synthesis enhances transport of GPI-anchored proteins to the Golgi apparatus in yeast. EMBO J., 13, 3687–3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazwinski S.M. (1999) Molecular mechanisms of yeast longevity. Trends Microbiol., 7, 247–252. [DOI] [PubMed] [Google Scholar]
- Jenkins G.M., Richards,A., Wahl,T., Mao,C., Obeid,L. and Hannun,Y. (1997) Involvement of yeast sphingolipids in the heat stress response of Saccharomyces cerevisiae. J. Biol. Chem., 272, 32566–32572. [DOI] [PubMed] [Google Scholar]
- Jiang J.C., Kirchman,P.A., Zagulski,M., Hunt,J. and Jazwinski,S.M. (1998) Homologs of the yeast longevity gene LAG1 in Caenorhabditis elegans and human. Genome Res., 8, 1259–1272. [DOI] [PubMed] [Google Scholar]
- Kohlwein S.D., Eder,S., Oh,C.S., Martin,C.E., Gable,K., Bacikova,D. and Dunn,T. (2001) Tsc13p is required for fatty acid elongation and localizes to a novel structure at the nuclear–vacuolar interface in Saccharomyces cerevisiae. Mol. Cell. Biol., 21, 109–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester R.L., Wells,G.B., Oxford,G. and Dickson,R.C. (1993) Mutant strains of Saccharomyces cerevisiae lacking sphingolipids synthesize novel inositol glycerophospholipids that mimic sphingolipid structures. J. Biol. Chem., 268, 845–856. [PubMed] [Google Scholar]
- Lynch D.V. (2000) Enzymes of sphingolipid metabolism in plants. Methods Enzymol., 311, 130–149. [DOI] [PubMed] [Google Scholar]
- Mandala S.M. et al. (1995) The discovery of australifungin, a novel inhibitor of sphinganine N-acyltransferase from Sporormiella australis. Producing organism, fermentation, isolation, and biological activity. J. Antibiot., 48, 349–356. [DOI] [PubMed] [Google Scholar]
- Mandala S.M., Thornton,R., Tu,Z., Kurtz,M.B., Nickels,J., Broach,J., Menzeleev,R. and Spiegel,S. (1998) Sphingoid base 1-phosphate phosphatase: a key regulator of sphingolipid metabolism and stress response. Proc. Natl Acad. Sci. USA, 95, 150–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandon E.C., Ehses,I., Rother,J., van Echten,G. and Sandhoff,K. (1992) Subcellular localization and membrane topology of serine palmitoyl transferase, 3-dehydrosphinganine reductase, and sphinganine N-acyltransferase in mouse liver. J. Biol. Chem., 267, 11144–11148. [PubMed] [Google Scholar]
- Mao C., Saba,J.D. and Obeid,L.M. (1999) The dihydrosphingosine-1-phosphate phosphatases of Saccharomyces cerevisiae are important regulators of cell proliferation and heat stress responses. Biochem. J., 342, 667–675. [PMC free article] [PubMed] [Google Scholar]
- Mao C., Xu,R., Bielawska,A. and Obeid,L.M. (2000) Cloning of an alkaline ceramidase from Saccharomyces cerevisiae. An enzyme with reverse (CoA-independent) ceramide synthase activity. J. Biol. Chem., 275, 6876–6884. [DOI] [PubMed] [Google Scholar]
- Merrill A.H., Wang,E., Mullins,R.E., Jamison,W.C., Nimkar,S. and Liotta,D.C. (1988) Quantitation of free sphingosine in liver by high-performance liquid chromatography. Anal. Biochem., 171, 373–381. [DOI] [PubMed] [Google Scholar]
- Muniz M., Nuoffer,C., Hauri,H.P. and Riezman,H. (2000) The Emp24 complex recruits a specific cargo molecule into endoplasmic reticulum-derived vesicles. J. Cell Biol., 148, 925–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagiec M.M., Wells,G.B., Lester,R.L. and Dickson,R.C. (1993) A suppressor gene that enables Saccharomyces cerevisiae to grow without making sphingolipids encodes a protein that resembles an Escherichia coli fatty acyltransferase. J. Biol. Chem., 268, 22156–22163. [PubMed] [Google Scholar]
- Nagiec M.M., Nagiec,E.E., Baltisberger,J.A., Wells,G.B., Lester,R.L. and Dickson,R.C. (1997) Sphingolipid synthesis as a target for antifungal drugs. Complementation of the inositol phosphoryl ceramide synthase defect in a mutant strain of Saccharomyces cerevisiae by the AUR1 gene. J. Biol. Chem., 272, 9809–9817. [DOI] [PubMed] [Google Scholar]
- Nagiec M.M., Skrzypek,M., Nagiec,E.E., Lester,R.L. and Dickson,R.C. (1998) The LCB4 (YOR171c) and LCB5 (YLR260w) genes of Saccharomyces encode sphingoid long chain base kinases. J. Biol. Chem., 273, 19437–19442. [DOI] [PubMed] [Google Scholar]
- Nickels J.T. and Broach,J.R. (1996) A ceramide-activated protein phosphatase mediates ceramide-induced G1 arrest of Saccharomyces cerevisiae. Genes Dev., 10, 382–394. [DOI] [PubMed] [Google Scholar]
- Oh C.S., Toke,D.A., Mandala,S. and Martin,C.E. (1997) ELO2 and ELO3, homologues of the Saccharomyces cerevisiae ELO1 gene, function in fatty acid elongation and are required for sphingolipid formation. J. Biol. Chem., 272, 17376–17384. [DOI] [PubMed] [Google Scholar]
- Reggiori F., Canivenc-Gansel,E. and Conzelmann,A. (1997) Lipid remodeling leads to the introduction and exchange of defined ceramides on GPI proteins in the ER and Golgi of Saccharomyces cerevisiae. EMBO J., 16, 3506–3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saba J.D., Nara,F., Bielawska,A., Garrett,S. and Hannun,Y.A. (1997) The BST1 gene of Saccharomyces cerevisiae is the sphingosine-1-phosphate lyase. J. Biol. Chem., 272, 26087–26090. [DOI] [PubMed] [Google Scholar]
- Schimmöller F., Singer-Kruger,B., Schroder,S., Kruger,U., Barlowe,C. and Riezman,H. (1995) The absence of Emp24p, a component of ER-derived COPII-coated vesicles, causes a defect in transport of selected proteins to the Golgi. EMBO J., 14, 1329–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiter R., Hitomi,M., Ivessa,A.S., Fasch,E.V., Kohlwein,S.D. and Tartakoff,A.M. (1996) A yeast acetyl coenzyme A carboxylase mutant links very-long-chain fatty acid synthesis to the structure and function of the nuclear membrane–pore complex. Mol. Cell. Biol., 16, 7161–7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shama S., Lai,C.-Y., Antoniazzi,J.M., Jiang,J.C. and Jazwinski,S.M. (1998) Heat stress-induced life span extension in yeast. Exp. Cell Res., 245, 379–388. [DOI] [PubMed] [Google Scholar]
- Sipos G., Reggiori,F., Vionnet,C. and Conzelmann,A. (1997) Altern ative lipid remodelling pathways for glycosylphosphatidylinositol membrane anchors in Saccharomyces cerevisiae. EMBO J., 16, 3494–3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrzypek M., Lester,R.L. and Dickson,R.C. (1997) Suppressor gene analysis reveals an essential role for sphingolipids in transport of glycosylphosphatidylinositol-anchored proteins in Saccharomyces cerevisiae. J. Bacteriol., 179, 1513–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrzypek M.S., Nagiec,M.M., Lester,R.L. and Dickson,R.C. (1999) Analysis of phosphorylated sphingolipid long-chain bases reveals potential roles in heat stress and growth control in Saccharomyces. J. Bacteriol., 181, 1134–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sütterlin C., Doering,T.L., Schimmöller,F., Schroder,S. and Riezman,H. (1997) Specific requirements for the ER to Golgi transport of GPI-anchored proteins in yeast. J. Cell Sci., 110, 2703–2714. [DOI] [PubMed] [Google Scholar]
- Wells G.B., Dickson,R.C. and Lester,R.L. (1998) Heat-induced elevation of ceramide in Saccharomyces cerevisiae via de novo synthesis. J. Biol. Chem., 273, 7235–7243. [DOI] [PubMed] [Google Scholar]
- Wu W.I., McDonough,V.M., Nickels,J.T.J., Ko,J., Fischl,A.S., Vales,T.R., Merrill,A.H.J. and Carman,G.M. (1995) Regulation of lipid biosynthesis in Saccharomyces cerevisiae by fumonisin B1. J. Biol. Chem., 270, 13171–13178. [DOI] [PubMed] [Google Scholar]
- Zanolari B., Friant,S., Funato,K., Sütterlin,C., Stevenson,B.J. and Riezman,H. (2000) Sphingoid base synthesis requirement for endocytosis in Saccharomyces cerevisiae. EMBO J., 19, 2824–2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Beeler,T. and Dunn,T. (1994) Suppressors of the Ca(2+)-sensitive yeast mutant (csg2) identify genes involved in sphingolipid biosynthesis. Cloning and characterization of SCS1, a gene required for serine palmitoyltransferase activity. J. Biol. Chem., 269, 21480–21488. [PubMed] [Google Scholar]