Abstract
In mouse brain cDNA libraries generated from small RNA molecules we have identified a total of 201 different expressed RNA sequences potentially encoding novel small non-messenger RNA species (snmRNAs). Based on sequence and structural motifs, 113 of these RNAs can be assigned to the C/D box or H/ACA box subclass of small nucleolar RNAs (snoRNAs), known as guide RNAs for rRNA. While 30 RNAs represent mouse homologues of previously identified human C/D or H/ACA snoRNAs, 83 correspond to entirely novel snoRNAs. Among these, for the first time, we identified four C/D box snoRNAs and four H/ACA box snoRNAs predicted to direct modifications within U2, U4 or U6 small nuclear RNAs (snRNAs). Furthermore, 25 snoRNAs from either class lacked antisense elements for rRNAs or snRNAs. Therefore, additional snoRNA targets have to be considered. Surprisingly, six C/D box snoRNAs and one H/ACA box snoRNA were expressed exclusively in brain. Of the 88 RNAs not belonging to either snoRNA subclass, at least 26 are probably derived from truncated heterogeneous nuclear RNAs (hnRNAs) or mRNAs. Short interspersed repetitive elements (SINEs) are located on five RNA sequences and may represent rare examples of transcribed SINEs. The remaining RNA species could not as yet be assigned either to any snmRNA class or to a part of a larger hnRNA/mRNA. It is likely that at least some of the latter will represent novel, unclassified snmRNAs.
Keywords: cDNA library/non-messenger RNAs/RNomics/snoRNAs
Introduction
A major goal of the joint international efforts of the Human Genome Project is the sequence, identification, structure, regulation and function of all 30 000–40 000 genes and their products. To facilitate functional analysis of the encoded gene products, this endeavour has been extended to model organisms from bacteria to mouse. Furthermore, expressed sequence tags (ESTs) have been employed to catalogue all mRNAs and recent efforts to generate their full-length sequences provide essential tools to study post-transcriptional processing of transcripts including alternative splicing, identification of protein coding genes and functional analysis. In contrast, not many experimental efforts address the class of small non-messenger RNAs (snmRNAs; Kiss-Laszlo et al., 1996; Olivias et al., 1997). These molecules do not encode proteins, but have cellular functions on their own or in complex with proteins that are bound to the RNA and thus form ribonucleoprotein complexes (RNPs). Such RNPs, found in cellular compartments as diverse as the nucleolus or dendritic processes of nerve cells (Tiedge et al., 1993; Pederson, 1998), exhibit a surprisingly diverse range of functions. However, the biological role of some of them remains elusive. Moreover, most systematic genomic searches are biased against their detection, and comprehensive identification by computational analysis of the genomic sequence of any organism remains an unsolved problem (Eddy, 1999). Hundreds of genes and their RNA products may thus remain undetected. Their functions, interactions in cellular circuits and roles in disease would remain unknown and our understanding of the functioning of a cell would be incomplete. Therefore, we set out to identify directly snmRNAs and their genes in the human genome and those of various model organisms.
Here we describe our experimental approach to the discovery of novel snmRNAs in mouse. This EST-like approach has been tailored for the detection of small RNAs [starting with material that is usually discarded: small total RNA in the size range ∼50–500 nucleotides (nt)]. The resulting sequences have been termed expressed RNA sequences (ERNS). In this study, we present the first unbiased look at the small RNA population in a mammalian cell. Thus far, we have identified ∼200 candidates for novel snmRNA species via ERNS. More than half of them correspond to new members of the two expanding subclasses of small nucleolar RNAs (snoRNAs) that guide RNA ribose methylation and pseudouridylation. Interestingly, while the vast majority of previously known members of these two snoRNA classes direct the modification of rRNA, several of the novel members are able to guide modification of spliceosomal small nuclear RNAs (snRNAs). Moreover, an unexpectedly large number of them remain without identified RNA targets. Finally, some of them are not ubiquitously expressed, as expected for rRNA or snRNA modification guides, raising the possibility of tissue-specific targets, presumably mRNAs.
Results and discussion
Library construction and analysis
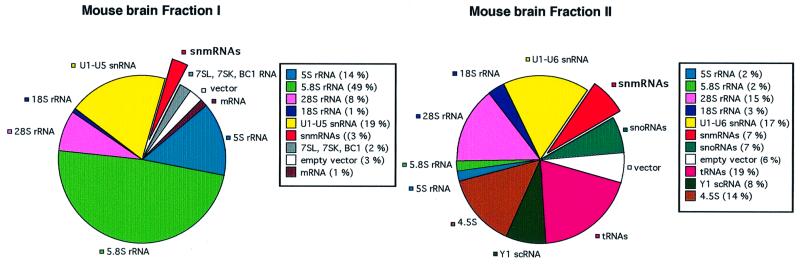
We constructed two cDNA libraries from mouse brain (see Materials and methods) based on small RNAs sized from ∼50 to ∼110 (Fraction II) and from ∼110 to ∼500 nt (Fraction I). Two separate libraries were generated to avoid potential overrepresentation of highly abundant tRNA species in Fraction II. We randomly sequenced 400 clones from each fraction and identified sequences by a BLASTN database search (Figure 1).
Fig. 1. Sequence analysis of 400 randomly chosen cDNA clones from mouse brain library Fraction I (derived from RNAs sized 500–110 nt) or Fraction II (derived from RNAs sized 110–50 nt), respectively. cDNA clones representing different RNA species or categories are shown as a percentage of total clones. The segment denoted snmRNAs identifies candidates for novel snmRNAs.
In Fraction I, many of the cDNA sequences could be assigned to genes encoding known snmRNAs (Figure 1). In addition to rRNAs or snRNAs, we identified other known small RNA species such as 7SL RNA, 7SK RNA, Y1 scRNA, RNase P or a brain-specific snmRNA, designated BC1 RNA (DeChiara and Brosius, 1987). About 1% of the sequences were derived from known mRNA fragments. Only 3% of cDNA sequences could not be assigned to known RNAs and therefore represented potentially novel snmRNAs. The Fraction II library contained, among others, sequences derived from tRNAs, 4.5S RNA and previously identified snRNAs or snoRNAs. As observed in the Fraction I cDNA libary, degradation fragments of 28S or 18S rRNA genes were also present. Compared with Fraction I, a larger number of novel, unknown cDNA clones (7%) could be identified by a BLASTN database search, thus potentially representing novel snmRNAs (Figure 1).
To enrich the fraction of novel RNA species in our analysis, cDNA clones were spotted on filters in high density arrays and hybridized to radiolabelled oligonucleotides identifying the most abundant, known snmRNAs. By this approach, we could significantly increase the amount of novel RNA species in our selection procedure from 3 to 20% in Fraction I and from 7 to 22% in Fraction II. From each library, ∼40 000 clones were screened by the hybridization procedure. Signals obtained in the filter hybridization were ranked by computer-aided analysis. Subsequently, we sequenced ∼2500 clones from each fraction exhibiting the lowest hybridization scores.
Analysis of candidates for snmRNAs
By sequence analysis, 201 novel ERNS from mouse were identified. Expression and sizes of these potential snmRNA species were confirmed by northern blot analysis. In general, they matched the sizes of the corresponding cDNAs (Tables I–VI), which were shorter by at least 5–10 bases, since the extreme 5′ ends of RNAs were not present due to the cloning strategy employed. In several cases, a complete sequence of the mouse snmRNA could be found within a mouse EST entry using a BLASTN database search. We also investigated whether novel RNAs would be expressed specifically in any of the following tissues: brain, liver, heart, kidney or testis (data not shown). The expression of a subset of ERNS could not be confirmed by northern blot analysis. This could be explained by low expression levels of the respective RNA species.
Table I. Group I: novel C/D box snoRNAs guiding a rRNA methylation; group II: novel C/D box snoRNAs guiding a snRNA methylation; and group III: novel C/D box snoRNAs with unidentified targets.
| ERNS | Copies | cDNA | RNA | Homology | Modification | Antisense element | Location/comments | Accession No. |
|---|---|---|---|---|---|---|---|---|
| Group I | ||||||||
| MBI-43 | 4 | 221 | 240 | human | Um3787 in 28S | 13 nt (5′) | very long C/D box snoRNA; intron of sortin nexin 5 gene | AF357317 |
| MBII-55 | 4 | 59 | 65 | human | Um1288 in 18S | 12 nt (5′) | AF357318 | |
| MBII-82 | 2 | 64 | 72 | – | Gm3913 in 28S | 13 nt (5′) | AF357319 | |
| MBII-95 | 1 | 63 | 75 | human EST | Gm509 in 18S | 13 nt (3′) | AF357320 | |
| MBII-99 | 4 | 66 | 95/75 | human | Gm3868 in 28S | 13 nt (5′) | functional homologue of yeast snR190 | AF357321 |
| MBII-108 | 1 | 56 | – | human | Gm683 in 18S | 12 nt (3′) | AF357322 | |
| MBII-135 | 1 | 57 | 72 | rat, bovine | Um627 in 18S | 14 nt (5′) | functional homologue of yeast snR77 | AF357323 |
| MBII-142 | 1 | 58 | 77 | human | Cm1272 in 18S | 12 nt (5′) | AF357324 | |
| MBII-180 | 2 | 89 | 80 | human EST | Cm3670 in 28S | 10 nt (5′) | functional homologue of yeast snR76 | AF357325 |
| MBII-211 | 2 | 72 | 80 | human EST | Cm3670 in 28S | 10 nt (5′) | homologue of yeast snR76, isoform of MBII-180 | AF357326 |
| MBII-202 | 1 | 48 | 70 | human, rat | Am2378 and Um428 in 18S | 14 nt (3′) and 13 nt (5′) | intron of rpL13 gene | AF357327 |
| MBII-210 | 5 | 59 | – | human EST | Gm4454 in 28S | 9 nt (3′) | AF357328 | |
| MBII-234 | 1 | 53 | 70 | – | Am512 in 18S | 12 nt (3′) | AF357329 | |
| MBII-239 | 2 | 64 | – | human | Um14 in 5.8S | 13 nt (5′) | partial and cytoplasmic 5.8S rRNA methylation | AF357330 |
| MBII-240 | 1 | 60 | 60 | human | Um4580 in 28S | 12 nt (5′) | intron of rpL37 gene | AF357331 |
| MBII-251 | 5 | 59 | 65 | – | Gm601 in 18S | 11 nt (5′) | AF357332 | |
| MBII-276 | 3 | 64 | 72 | – | Gm3713 in 28S | 11 nt (5′) | AF357333 | |
| MBII-296 | 1 | 60 | 67 | human | Gm4578 in 28S | 16 nt (5′) | AF357334 | |
| MBII-316 | 1 | 71 | 75 | – | A3836 in 28S? | 13 nt (3′) | A3836 is not a reported methylation site | AF357335 |
| MBII-324 | 1 | 94 | 80 | – | G1630 in 28S? | 10 nt (5′) | G1630 is not a reported methylation site | AF357336 |
| MBII-333 | 1 | 82 | 75 | – | Um1670 in 18S | 9 nt (3′) | AF357337 | |
| MBII-336 | 1 | 45 | 70 | human | Am576 in 18S | 13 nt (3′) | AF357338 | |
| MBII-420 | 4 | 56 | 65 | human, rat EST | Am2764 in 28S | 12 nt (3′) | AF357339 | |
| MBII-429 | 1 | 56 | 60 | rat | Gm436 in 18S | 9 nt (5′) | intron of rpS12 gene | AF357340 |
| Group II | ||||||||
| MBII-19 | 3 | 58 | 75 | – | Cm40 in U2 | 14 nt (3′) | AF357341 | |
| MBII-119 | 1 | 62 | 75 | human EST | Cm8 in U4 | 11 nt (5′) | AF357342 | |
| MBII-166 | 11 | 98 | 105 | – | Cm60 in U6 | 13 nt (3′) | AF357343 | |
| MBII-382 | 1 | 61 | 75 | human, rat EST | Cm61 and Gm11 in U2 | 8 nt (5′) and 9 nt (5′) | AF357344 | |
| Group III | ||||||||
| MBI-46 | 1 | 249 | 280 | – | n. d. | n. d. | AF357345 | |
| MBI-52 | 1 | 73 | 110 | human | n. d. | n. d. | AF357346 | |
| MBI-106 | 1 | 63 | 120 | – | n. d. | n. d. | AF357347 | |
| MBII-4 | 1 | 67 | 100 | – | n. d. | n. d. | AF357348 | |
| MBII-115 | 27 | 99 | 105 | human EST | n. d. | n. d. | AF357349 | |
| MBII-163 | 1 | 84 | 165 | human | n. d. | n. d. | AF357350 | |
| MBII-170 | 1 | 96 | – | – | n. d. | n. d. | AF357351 | |
| MBII-244 | 1 | 68 | 120 | – | n. d. | n. d. | AF357352 | |
| MBII-289 | 1 | 93 | 100 | human EST | n. d. | n. d. | AF357353 | |
| MBII-295 | 2 | 68 | 82 | human | n. d. | n. d. | AF357354 | |
| MBII-343 | 1 | 54 | 75 | – | n. d. | n. d. | AF357355 | |
| MBII-361 | 1 | 72 | – | – | n. d. | n. d. | AF357356 | |
| MBII-366 | 1 | 56 | 85 | – | n. d. | n. d. | AF357357 | |
| MBII-419 | 1 | 28 | 56 | – | n. d. | n. d. | AF357358 | |
| MBII-426 | 1 | 30 | 60 | mouse EST | n. d. | n. d. | AF357359 |
Tables I–VI: compilation of novel ERNS from Fraction I (derived from RNAs sized 500–110 nt) and Fraction II (derived from RNAs sized 110–50 nt) cDNA libraries. ERNS: expressed RNA sequences from Fraction I (MBI-) or Fraction II (MBII-); copies: number of independent cDNA clones identified from each RNA species; cDNA: length of cDNA, as assessed by sequencing; RNA: length of RNA as assessed by northern blot analysis; homology: homology of snmRNAs to genomic or EST sequences within other organisms; modification: predicted modified nucleotides within rRNAs or snRNAs (numbering according to the human RNA sequence); antisense element: for C/D box snoRNAs, the length of the antisense element is indicated in nucleotides, followed by its location in the 5′ domain (5′) or 3′ domain (3′) of the snoRNA; location/comments: genomic locus and specific features of ERNS (when applicable); Accession No.: accession number in DDBJ/EMBL/GenBank.
We determined the total number of independent cDNA clones obtained for each snmRNA. While most clones were found only once in our screen, some were present in numerous copies. This correlated well with their abundance as deduced by northern blot analysis. Homologues for more than half of the mouse ERNS could be identified in human genomic or EST sequences (sequence similarity >80%), consistent with a functional role of these RNAs. Based on structural hallmarks, expression and presumed function, the novel 201 snmRNA candidates were assigned to 13 different subgroups (see Tables I–VI).
Novel mouse snoRNAs
Based on sequence and structural features (Maxwell and Fournier, 1995; Balakin et al., 1996; Ganot et al., 1997b), we identified 72 novel snoRNA species from the C/D box and 41 from the H/ACA box type. The known function of snoRNAs is post-transcriptional processing and modification of rRNAs or snRNAs. C/D box antisense snoRNAs guide 2′-O-ribose methylation at specific sites in rRNAs or snRNAs, while H/ACA snoRNAs guide specific pseudouridylation within these RNA species (for reviews see Tollervey, 1996; Smith and Steitz, 1997; Ofengand and Fournier, 1998; Weinstein and Steitz, 1999; Bachellerie et al., 2000). Unexpectedly, a substantial number of the novel specimens of both snoRNA classes do not appear to target rRNA or a snRNA. Moreover, seven of them are not ubiquitously expressed in mouse tissues, but are specific to brain.
Novel ubiquitous C/D box snoRNAs
C/D box snoRNAs contain two short sequence motifs, box C and box D, located only a few nucleotides away from the 5′ and 3′ ends, respectively, generally as part of a typical 5′–3′ terminal stem–box structure (for a review see Bachellerie and Cavaillé, 1998). Immediately upstream from box D or from an additional box (D′) in the 5′ half, C/D snoRNAs feature sequence tracts, 10–21 nt in length, that are complementary to rRNA spanning the sites of 2′-O-ribose methylation. In the corresponding RNA duplexes, the ribose-methylated nucleotide is always at the same location, paired to the fifth snoRNA nucleotide upstream from box D or box D′ (Kiss-Laszlo et al., 1996; Nicoloso et al., 1996). In rRNA of the yeast Saccharomyces cerevisiae, cognate box C/D snoRNAs have been identified for 51 of the 55 ribose-methylated sites (Lowe and Eddy, 1999). In mammals, however, a large fraction of the 105–107 expected rRNA 2′-O-ribose methylations (Maden, 1990) remained without a known cognate guide until completion of this study. Moreover, it is now apparent that the complexity of C/D box snoRNAs might be greater than anticipated, since methylation guide snoRNAs targeting substrates other than rRNA have been identified. Thus, three 2′-O-ribose methylations of spliceosomal U6 snRNA in human are also directed by bona fide C/D box antisense snoRNAs (Tycowski et al., 1998; Ganot et al., 1999), while both a 2′-O-ribose methylation and a pseudouridylation in U5 snRNA are guided by a novel C/D–H/ACA ‘hybrid’ snoRNA (Jady and Kiss, 2001).
Of the 72 novel mouse C/D box snoRNAs identified in this study, 66 are ubiquitously expressed, of which 24 correspond to novel C/D box snoRNAs able to guide a 2′-O-methylation within rRNA (Table I, group I) and 23 to orthologues of previously identified human snoRNAs able to guide methylation in rRNAs or in U6 snRNA (see Supplementary data available at The EMBO Journal Online; Table I, group IV). Particularly interesting is MBII-239, able to direct methylation at position U14 within ribosomal 5.8S RNA. Um14 is unique among all vertebrate rRNA ribose methylations because it is partial, takes place in the cytoplasm rather than the nucleolus, and is undermethylated in tumour tissues (Nazar et al., 1980; Munholland and Nazar, 1987). The detection of MBII-239 strongly suggests that the atypical Um14 methylation of 5.8S rRNA is catalysed by the same snoRNA-guided machinery as the remainder of rRNA ribose methylations, raising the issue of assessing the MBI-239 snoRNA intracellular site of action and its expression level in tumour tissues.
From 15 of this first subset of 24 novel mouse C/D box snoRNAs, the human orthologues can be found as genomic or EST entries in DDBJ/EMBL/GenBank, further supporting the functional relevance of the identified cDNAs, as does their location in introns (in two cases: for MBII-202 and MBII-240). Collectively, the novel C/D box snoRNAs in group I are able to direct a total of 24 rRNA methylations, since MBII-211 represents an apparent isoform of MBII-180, able to direct the same methylation in 28S rRNA, while MBII-202 can direct two methylations, corresponding to Um428 and Am2378 in human 18S rRNA.
Within group I, one particular snoRNA, MBI-43, stands out for its exceptionally large size (240 nt) for a C/D box snoRNA. So far, the only non-canonical specimen in this regard was the recently reported C/D–H/ACA ‘hybrid’ snoRNA, which exhibits a roughly similar size (Jady and Kiss, 2001). Curiously, the uridine targeted by MBI-43 is the only site that is both ribose methylated and pseudouridylated in mammalian rRNA (Maden, 1990; Ofengand and Bakin, 1997). The presumptive H/ACA snoRNA able to guide this particular pseudouridylation remains unknown so far (see below). Careful inspection of the sequence and folding potential of both MBI-43 and its human homologue in DDBJ/EMBL/GenBank did not reveal the presence of H/ACA snoRNA hallmarks in addition to C/D motifs, ruling out the possibility that the atypical snoRNA corresponds to a hybrid C/D–H/ACA snoRNA directing the two types of modification at the same nucleotide position. As a consequence of the work presented here, only 14 rRNA ribose methylations, from a total of 105–107 in mammals (Maden, 1990), remain without identified cognate guide snoRNA.
In our screen we have also discovered four novel C/D box snoRNAs (MBII-19, MBII-119, MBII-166 and MBII-382) able to direct ribose methylation within U2, U4 or U6 snRNAs [group II, Table I; see Massenet et al. (1998) for a review on snRNA nucleotide modifications). For MBII-119 and MBII-382, this assignment is supported by comparison of the complete human homologous sequences found as ESTs in DDBJ/EMBL/GenBank, which both exhibit box C and a 4 or 5 bp terminal stem in addition to conserved antisense elements. While MBII-382 could guide two distinct ribose methylations in U2 snRNA, the two cognate antisense elements are unusual, because they are both located in the 5′ half of the snoRNA and found immediately upstream of a potential D′ box carrying two deviations from the consensus.
We also discovered 15 ubiquitously expressed RNAs with structural hallmarks of C/D box snoRNAs, but devoid of complementarity to rRNAs or snRNAs at the expected position relative to the box motifs (Table I, group III). While clone MBII-426 is severely truncated (30 nt in length), it unambiguously corresponds to a bona fide C/D box snoRNA, since it matches perfectly a mouse EST sequence exhibiting box C, box C′ and a 4 bp terminal stem at the expected locations. For MBII-115, MBII-163, MBII-289 and MBII-295, human homologues are available in DDBJ/EMBL/GenBank, and in each case one of the two presumptive antisense elements is conserved between mouse and human, supporting the notion that these snoRNAs also represent bona fide methylation guides. Two ubiquitous methylation guide snoRNAs devoid of rRNA or snRNA complementarity have been reported recently (Jady and Kiss, 2000). This expanding subset of box C/D snoRNAs lacking complementarity to rRNA might be involved in rRNA processing (Tycowski et al., 1994) or other, still unknown, aspects of ribosome biogenesis or other functions. Alternatively, these snoRNAs might target cellular RNAs other than rRNAs or snRNAs, such as ubiquitous snmRNAs transiting through the nucleolus, like telomerase RNA, RNase P, SRP RNA or pre-tRNAs (for review see Pederson, 1998). However, the presence of 2′-O-ribose-methylated nucleotides has not been reported in these RNAs thus far. Furthermore, systematic searches of these snoRNA sequences did not reveal any potential antisense element of at least 8 bp that could direct 2′-O-ribose methylation within these potential targets.
Novel ubiquitous mouse H/ACA snoRNAs
The formation of pseudouridines in eukaryotic rRNA is directed by a large family of site-specific H/ACA box snoRNAs carrying an appropriate bipartite guide sequence in the internal loop of one (or both) of their two major hairpin domains (Ganot et al., 1997a; Ni et al., 1997; Ofengand and Fournier, 1998; Bortolin et al., 1999). In contrast to methylation guide snoRNAs, pseudouridylation guide snoRNAs, thus far, have been exclusively discovered by experimental approaches, as identification in genomic sequences is critically hampered by their shorter box motifs and shorter bipartite antisense elements. Of 91–93 pseudouridines of mammalian rRNAs (Maden, 1990; Ofengand and Bakin, 1997), only 15 can be guided by one of the 13 previously reported human H/ACA snoRNAs (Ganot et al., 1997a). The present study dra matically expands the repertoire of eukaryotic H/ACA snoRNAs, decreasing the number of rRNA pseudouridinylation sites without a cognate H/ACA snoRNA by 27 to 49–51.
The majority of novel H/ACA snoRNA species have no counterpart among the mammalian snoRNAs reported so far, except for seven corresponding to homologues (sequence similarity 73–90%) of human pseudouridylation guides, namely U23, E2, U64, U65, U68, U69 and U70 (see Supplementary data; Table II, group VIII). All novel mouse RNAs contain the two H and ACA box motifs at the expected locations and fold into the typical two major hairpin domains connected by a single-strand hinge region carrying the box H motif (data not shown). The vast majority also exhibit bipartite antisense elements matching known RNA pseudouridylation sites (Figure 2). This large set of novel data overwhelmingly confirms the validity of the model for the base-paired snoRNA–rRNA interaction guiding site-specific pseudouridylation (Ganot et al., 1997a).
Table II. Group V: novel H/ACA box snoRNAs guiding a rRNA pseudouridylation; group VI: novel H/ACA box snoRNAs guiding a snRNA pseudouridylation; and group VII: novel H/ACA box snoRNAs with unidentified targets.
| ERNS | Copies | cDNA | RNA | Homology | Modification | Location/comments | Accession No. |
|---|---|---|---|---|---|---|---|
| Group V | |||||||
| MBI-1 | 1 | 184 | 190 | human | Ψ3727/28S | AF357383 | |
| MBI-3 | 3 | 128 | 150 | – | Ψ4391/28S; Ψ4470/28S | intron 2 of rpL23 gene | AF357384 |
| MBI-6 | 3 | 121 | 140 | human | Ψ4512/28S | functional equivalent of yeast snR42 | AF357385 |
| MBI-12 | 4 | 126 | 140 | – | Ψ3813/28S; Ψ681/18S | AF357386 | |
| MBI-13 | 5 | 122 | 130 | human | Ψ815/18S; Ψ866/18S | extremely strong expression | AF357387 |
| MBI-20 | 1 | 104 | 125 | rat | Ψ1723/28S | intron 3 of rat rpP2 gene; functional equivalent of yeast snR5 | AF357388 |
| MBI-26 | 1 | 107 | 120 | human | Ψ4633/28S; Ψ3731/28S | AF357389 | |
| MBI-28 | 5 | 120 | 130 | human | Ψ3889/28S; Ψ3928/28S | intron 3 of mouse rpL27 gene | AF357390 |
| MBI-31 | 4 | 122 | 130 | human | Ψ4588/28S | AF357391 | |
| MBI-39 | 1 | 124 | 150 | human | Ψ1003/18S | intron 4 of Tcp-1 gene; same genetic location as MBI-125 | AF357392 |
| MBI-42 | 1 | 105 | 130 | human | Ψ4956/28S | intron 4 of mouse rpS12 gene | AF357393 |
| MBI-64 | 3 | 117 | 130 | – | Ψ822/18S. | AF357394 | |
| MBI-80 | 1 | 101 | 130 | human | Ψ1237/18S; Ψ1625/18S | AF357395 | |
| MBI-89 | 2 | 118 | 130 | human | Ψ34, Ψ863/18S; Ψ4259/28S | AF357396 | |
| MBI-115 | 1 | 126 | 140 | – | Ψ1758/28S | AF357397 | |
| MBI-141 | 1 | 129 | 180/140 | human | Ψ1771/28S | intron 1 of rpL32-3A gene | AF357398 |
| MBI-142 | 1 | 94 | 120 | human | Ψ4536/28S | intron 2 of rpS16 gene | AF357399 |
| MBI-161 | 1 | 112 | – | human | Ψ218/18S; Ψ3703/28S | intron 5 of TPT1 gene for translationally controlled tumour protein (TCTP) | AF357400 |
| MBI-164 | 1 | 62 | 120 | – | U144/5.8S?; U2458/28S? | not reported pseudouridylation sites | AF357401 |
| Group VI | |||||||
| MBI-57 | 1 | 118 | – | human, Xenopus | Ψ34, Ψ44/U2 snRNA | AF357402 | |
| MBI-100 | 2 | 124 | 140 | – | Ψ40/U6 snRNA | AF357403 | |
| MBI-114 | 2 | 118 | 140 | – | Ψ40/U6 snRNA | isoform of MBI-100 | AF357404 |
| MBI-125 | 1 | 61 | 120 | – | Ψ91/U2 snRNA | intron 9 of Tcp-1 gene | AF357405 |
| Group VII | |||||||
| MBI-11 | 1 | 71 | – | – | n. d. | intron 6 of Nit1 gene, antisense direction | AF357406 |
| MBI-15 | 1 | 32 | 140 | human | n. d. | AF357407 | |
| MBI-51 | 1 | 56 | 200/120 | – | n. d. | AF357408 | |
| MBI-61 | 9 | 164 | 150 | human | n. d. | seven copies on chromosome 21, one copy on chromosome 19: retrogenes | AF357409 |
| MBI-79 | 1 | 116 | 145 | rat | n. d. | intron 10 of Cctz-1 gene for Tcp-1 protein, zeta subunit (chaperonin) | AF357410 |
| MBI-83 | 1 | 113 | 300/120 | human | n. d. | AF357411 | |
| MBI-87 | 1 | 113 | 120 | human, rat | n. d. | intron 8 of human dyskerin (DKC1) gene | AF357412 |
| MBI-137 | 1 | 112 | n.d. | human | n. d. | isoform of MBI-89, does not have the same rRNA target | AF357413 |
| MBI-147 | 1 | 77 | 200 | – | n. d. | AF357414 | |
| MBI-152 | 1 | 164 | – | – | n. d. | AF357415 |
See footnote to Table I.
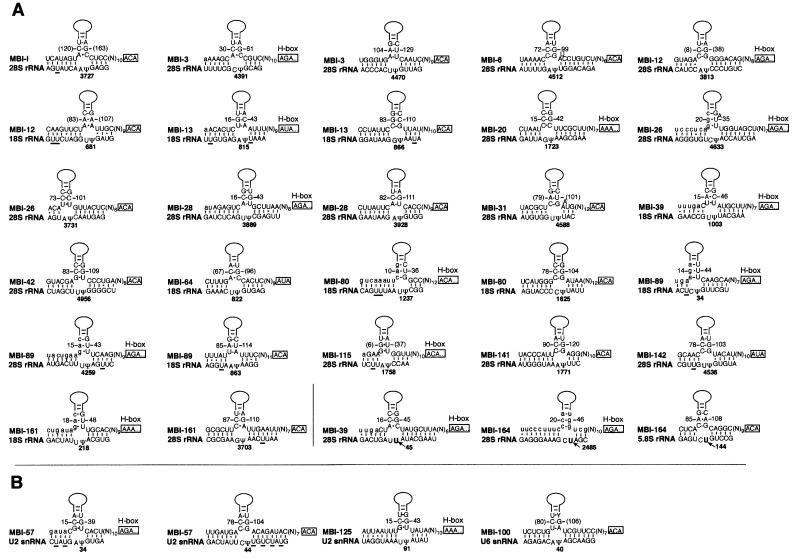
Fig. 2. Potential base-pairing interactions between novel mouse H/ACA snoRNAs and mouse rRNA (A) or snRNA (B). The snoRNA sequences in a 5′ to 3′ orientation are shown in the upper strands with the two H and ACA motifs boxed and the apical part of the snoRNA 5′ or 3′ hairpin domains schematized by a solid line. The two complementarities to the RNA target are always found within the large internal loop of one (or both) of their hairpin domains, invariably abutting its apex-proximal stem. Sequence coordinates in parentheses refer to snoRNAs for which the 5′ terminal sequence remains incomplete after database analysis. For other snoRNAs, nucleotides in the 5′ terminal sequence derived from databases are depicted as lower case letters. For rRNA or snRNA, sequence coordinates correspond to the respective human sequences to facilitate interpretation of data. Positions of pseudouridines are as reported by Maden (1990) for 18S rRNA, by Ofengand and Bakin (1997) for 28S rRNA and by Massenet et al. (1998) for mammalian snRNAs. Pseudouridines predicted to be directed by the snoRNA are denoted by Ψ, while other known pseudouridylation sites are indicated by U. In three cases, the uridine (indicated by an arrow) at the expected target position in the canonical, bipartite guide RNA duplex has not been reported to be pseudouridylated.
Nineteen entirely new specimens (Table II, group V) can collectively direct 27 of the 93–95 pseudouridylations identified in mammalian rRNAs. All but three of them display an identical location in human and mouse rRNAs (Figure 2A). Mouse homologues of seven already known human H/ACA snoRNAs show, as expected, the perfect sequence conservation of the antisense elements proposed previously (Ganot et al., 1997a), except for one. Thus, the proposal that the 3′ pseudouridylation pocket of U69 could target Ψ36 in human 18S rRNA (Ganot et al., 1997a) is not phylogenetically supported, as in comparison with its human homologue the mouse MBI-134 sequence exhibits three nucleotide differences over the presumptive bipartite 3′ antisense element of U69.
A large fraction of the novel rRNA pseudouridylation guides can each target two distinct modifications, through appropriate antisense elements in both pseudouridylation pockets. One of them, MBI-89, is even able to direct three distinct modifications: its 5′ pseudouridylation pocket contains an antisense element matching two distinct target sites, one in 18S rRNA, one in 28S rRNA. Intriguingly, in addition to the 27 rRNA pseudouridines targeted by the novel mouse snoRNAs reported previously (Ofengand and Bakin, 1997), three rRNA uridines not known to be pseudouridylated nevertheless appear as bona fide targets for two of the novel H/ACA specimens, MBI-39 (through its 5′ pseudouridylation pocket) and MBI-164 (through both pseudouridylation pockets), as shown in Figure 2A. In this regard, it is noteworthy that several rRNA 2′-O-ribose methylations had not been identified until the detection of a cognate guide snoRNA prompted further scrutiny (Qu et al., 1999).
The set of 34 entirely novel H/ACA snoRNAs identified in mouse also includes four outstanding specimens able to target pseudouridylation onto snRNA instead of rRNA (Table II, group VI). MBI-57, MBI-100, MBI-114 and MBI-125 have the potential to target U2 or U6 snRNAs, through the formation of guide duplexes that appear perfectly canonical as compared with those matching rRNA targets (Figure 2B). Interestingly, the sequence of a likely homologue of MBI-57, which can direct both ψ34 and ψ44 in U2 snRNA, is present in a Xenopus laevis EST (BE507485), which could provide the basis for a direct experimental analysis of the elusive function of these two U2 pseudouridylations.
Novel mouse H/ACA RNAs also include 11 species for which we could not identify any reasonable target uridine in rRNAs or snRNAs (Table II, group VII). Two of these RNAs, MBI-79 and MBI-87, are encoded in introns of ubiquitously expressed genes, like all vertebrate rRNA modification guide snoRNAs characterized so far (see below). Searches involving antisense elements of the 11 novel RNAs were also negative for potential target uridines in other stable non-coding RNAs trafficking through the nucleolus, such as telomerase RNA, RNase P or SRP RNA, the pseudouridine content of which remains unknown. We cannot rule out with certainty that novel snoRNAs might still target these RNA species; however, they could also target other cellular RNAs such as mRNA, as suggested recently in the case of a C/D box snoRNA (Cavaillé et al., 2000). Finally, recent findings that telomerase RNA in vertebrates contains a typical H/ACA domain (Mitchell et al., 1999) and that human H/ACA snoRNPs and telomerase share evolutionarily conserved proteins (Pogacic et al., 2000) expand the structural and functional diversity of H/ACA box snoRNAs, suggesting that some of the novel snoRNAs in this group might have unanticipated functions.
Tissue-specific snoRNAs
We identified six C/D box snoRNAs (MBII-13, MBII-48, MBII49, MBII-52, MBII-78 and MBII-85; Table III, group IX) and one H/ACA box snoRNA (MBI-36) that are expressed in mouse brain but not in other tissues tested so far (heart, liver, kidney, testis or muscle). Human homologues of MBI-36, MBII-13, MBII-52 and MBII-85 have been identified (Cavaillé et al., 2000; Filipowicz, 2000). In human, genes encoding MBII-52 and MBII-85 are present in multi-copy repeats on chromosome 15q11–13 and located in introns of host genes that apparently have no capacity to encode proteins (Cavaillé et al., 2000). The multi-copy repeat arrangement of these two snoRNAs is in agreement with their high abundance in cells, as deduced by northern blot analysis; in our screen, these clones could be identified 37 times (MBII-52) and 56 times (MBII-85), respectively, while most other cDNA clones encoding snoRNA genes are only found once. The human homologue of MBII-13 maps as a single-copy gene to chromosome 15q11–13 and MBI-36 maps to the large intron 2 of the serotonin 5-HT2C receptor gene, consistent with its brain-specific expression pattern with highest levels in the choroid plexus (Cavaillé et al., 2000).
Table III. Group IX: brain-specific H/ACA and C/D box snoRNAs with so far unidentified targets.
| ERNS | Copies | cDNA | RNA | Class | Homology | Modification | Location/comments | Accession No. |
|---|---|---|---|---|---|---|---|---|
| MBI-36 | 4 | 119 | 130 | H/ACA box | human | n. d. | intron 2 of serotonin receptor gene 5-HT2C | AF357423 |
| MBII-13 | 1 | 46 | 60 | C/D box | human | n. d. | chromosome 15, PWS region; PAR-5 RNA | AF357424 |
| MBII-48 | 5 | 57 | 60 | C/D box | – | n. d. | AF357425 | |
| MBII-49 | 4 | 56 | 65 | C/D box | – | n. d. | AF357426 | |
| MBII-52 | 37 | 78 | 80 | C/D box | human | n. d. | chromosome 15, PWS region, tandemly repeated genes | AF357427 |
| MBII-78 | 1 | 50 | 57 | C/D box | – | n. d. | AF357428 | |
| MBII-85 | 56 | 91 | 95 | C/D box | human | n. d. | chromosome 15, PWS region, tandemly repeated genes | AF357429 |
See footnote to Table I.
None of the brain-specific snoRNAs exhibits complementarity to ribosomal or snRNAs within their antisense element(s), in agreement with a role different from targeting these RNA species. Remarkably, the antisense element of MBII-52 snoRNA is complementary to the serotonin receptor 5-HT2C mRNA (the same gene serving as a host gene for MBI-36, see above) and is proposed to regulate editing or alternative splicing of the mRNA (Cavaillé et al., 2000). In addition, MBII-13, MBII-52 and MBII-85 C/D box snoRNAs might be involved in the aetiology of Prader–Willi syndrome, a neurodegenerative disease, thus constituting the first snoRNAs whose absence is potentially causing a human disorder (Cavaillé et al., 2000). So far, no potential targets have been identified for the remaining six brain-specific snoRNAs. Availability of the complete mouse and human genomes might reveal conserved target sites for these unusual snoRNA species.
Intronic localization of novel modification guide snoRNAs
Sequences encoding a large subset of the novel snoRNAs in mouse (or their unambiguous orthologues in another mammalian species) are located within long fragments of the mammalian genomes in sequence databases. Whenever exons and introns were annotated, the snoRNA coding region was found within an intron of a mostly ubiquitously expressed gene. This is in agreement with the observed pattern for vertebrate modification guide snoRNAs (Pelczar and Filipowicz, 1998; Smith and Steitz, 1998; Weinstein and Steitz, 1999; Bachellerie et al., 2000), which are usually processed from the debranched lariat by exonucleolytic trimming of excess intronic sequences (Kiss and Filipowicz, 1995). Characterization of our novel mouse snoRNAs largely expands the repertoire of known host genes. A majority of the novel host genes identified in this study encode ribosomal proteins, in agreement with previous observations. They include rpL3, rpL13, rpL23a, rpL37 and rpS12, as well as rpL23, rpL27a, rpL32-3A, rpP2, rpS12 and rpS16 for C/D box and H/ACA snoRNAs, respectively. In addition, detection of the mouse homologues of human C/D box U58 snoRNA and H/ACA U68 snoRNA allowed us to identify their cognate host genes, rpL17 and rpL18a, respectively. Five novel ubiquitous genes for non-ribosomal proteins have also been identified as hosts for intronic snoRNAs. One is an unidentified 5′TOP gene hosting MBII-99 C/D box snoRNA, while the other four contain intron-encoded H/ACA snoRNAs (Tables I and II). Curiously, one of them encodes dyskerin (Heiss et al., 1998), the mammalian homologue of yeast Cbf5, the pseudouridine synthase thought to catalyse the snoRNA-guided isomerization of uridine (Lafontaine et al., 1998). For a more extensive analysis of novel snoRNAs see Supplementary data.
Novel RNAs that do not exhibit snoRNA motifs
Of the 201 ERNS, 88 could not be assigned to known classes of snmRNAs and no function can be attributed to these RNA species at this point. However, hallmarks might exist at the level of secondary structure, as observed for H/ACA box RNAs. In fact, some of the RNA sequences can be folded into highly stable stem–loop structures. Since we are currently analysing cDNA libraries encoding small RNA species from organisms including Caenorhabditis elegans, Drosophila melanogaster and Arabidopsis thaliana, interspecies comparisons of the novel sequences might reveal conserved structural or sequence motifs and provide hints as to the function of these RNA species in the cell.
Novel snmRNAs located within mRNA coding regions
From 88 novel ERNS from the non-snoRNA type, 26 can be located within known or predicted mRNA or heterogeneous nuclear RNA (hnRNA) coding regions (Table IV, groups X and XI). Thereby, 16 ERNS are part of the open reading frame of mRNAs, whereas 10 are located within 5′ or 3′ untranslated regions (UTRs). At this point, the function of these RNAs remains elusive. It is noteworthy that the expression as snmRNAs of some but not all ERNS from this group can be confirmed by northern blot analysis. While ERNS derived from coding regions might correspond to more or less stable intermediates during degradation of hnRNAs or mRNAs, snmRNAs derived from the 5′ or 3′ UTRs of mRNAs could exhibit regulatory functions. Such mRNA regions have been shown previously to be involved in cis in the control of mRNA stability and intracellular localization (Schuldt et al., 1998; Saunders et al., 1999).
Table IV. Group X: novel RNAs located within coding regions of known mRNAs; and group XI: novel RNAs located within 5′ or 3′ UTRs of mRNAs.
| ERNS | Copies | cDNA | RNA | Homology | Location/comments | Accession No. |
|---|---|---|---|---|---|---|
| Group X | ||||||
| MBI-45 | 1 | 90 | – | – | mitochondrial cyt c mRNA | AF357430 |
| MBI-50 | 1 | 201 | – | human | RPA16 mRNA | AF357431 |
| MBI-69 | 1 | 210 | 240 | human, rat | M-PFK mRNA | AF357432 |
| MBI-85 | 1 | 98 | 110 | human, rat | ATP1B2 mRNA | AF357433 |
| MBI-93 | 1 | 34 | – | human, rat | GAD 65 mRNA | AF357434 |
| MBI-112 | 1 | 112 | 120 | human, rat | S27 mRNA | AF357435 |
| MBI-122 | 1 | 311 | – | human | C1/C2 mRNA | AF357436 |
| MBII-8 | 1 | 56 | 56 | – | GARP45 mRNA | AF357437 |
| MBII-26 | 1 | 96 | – | human | Ilf3 mRNA | AF357438 |
| MBII-51 | 1 | 53 | – | rat | PTP-NP mRNA | AF357439 |
| MBII-193 | 1 | 56 | 2000 | human | snap 25 mRNA but homology only from pos. 4–38 to snap | AF357440 |
| MBII-198 | 1 | 39 | 2000 | – | SGP-1 mRNA | AF357441 |
| MBII-208 | 1 | 17 | n. d. | human | glutamate receptor channel mRNA | AF357442 |
| MBII-228 | 1 | 74 | – | human | Pam mRNA (protein associated with Myc) | AF357443 |
| MBII-267 | 1 | 86 | 60 | human | Eph receptor A4 (Epha4) mRNA, snmRNA corresponds to signal peptide sequence | AF357444 |
| MBII-339 | 1 | 90 | – | human, rat | thrombomodulin mRNA | AF357445 |
| Group XI | ||||||
| MBI-129 | 2 | 156 | – | human | mRNA DKFZp566B183 3′ UTR | AF357446 |
| MBI-145 | 1 | 130 | – | human | scg mRNA 3′ UTR | AF357447 |
| MBI-151 | 1 | 167 | – | human | cdk mRNA 3′ UTR | AF357448 |
| MBI-154 | 1 | 109 | – | human, rat | cd24 mRNA 3′ UTR | AF357449 |
| MBI-156 | 1 | 98 | – | human | HSPC218 mRNA 5′ UTR | AF357450 |
| MBI-163 | 1 | 25 | – | human, rat | cam III mRNA 5′ UTR (calmodulin) | AF357451 |
| MBII-84 | 1 | 65 | – | human | UDP-glucuronosyltransferase mRNA 3′ UTR | AF357452 |
| MBII-283 | 1 | 88 | – | rat | Podxl mRNA 3′ UTR | AF357453 |
| MBII-395 | 1 | 83 | 135 | rat | RC3 mRNA 3′ UTR (calmodulin binding protein) | AF357454 |
| MBII-396 | 1 | 68 | 265/115 | rat | add2 mRNA 5′ UTR | AF357455 |
See footnote to Table I.
Novel snmRNAs resembling repetitive elements
Clones MBI-2, MBI-56, MBI-160, MBII-133 and MBII-373 contain sequences derived from short interspersed repetitive elements (SINEs; Table V, group XII). A common denominator between sequences from this group of clones is sequence similarity to nuclear 4.5SH RNA, 4.5SI RNA or the related B1, B2 or B4 retronuons. These sequences, in turn, are related to an ancestral SRP RNA or tRNA (Jurka, 2000). The small RNAs that served as templates for the aforementioned cDNAs may be related to 4.5SH RNA, 4.5SI RNA or directly transcribed B-type SINEs. Alternatively they may reflect degradation or processing products from larger hnRNAs or mRNAs that harbour such sequences. B1 and B2 SINEs, for example, can often be found in 3′ UTRs of mature mRNAs in both orientations (Brosius, 1999). Further work is necessary to establish whether the clones from this category reflect novel snmRNAs related to 4.5SH RNA, 4.5SI RNA, B1, B2 or B4 RNAs.
Table V. Group XII: novel RNAs resembling repetitive elements.
| ERNS | Copies | cDNA | RNA | Homology | Location/comments | Accession No. |
|---|---|---|---|---|---|---|
| MBI-2 | 1 | 306 | – | human | SINE B4 | AF357456 |
| MBI-56 | 1 | 84 | 300/140 | – | SINE B4 | AF357457 |
| MBI-160 | 1 | 122 | 80 | human | SINE B1; homology to MBI-2 | AF357458 |
| MBII-133 | 1 | 65 | n. d. | human | SINE B2 | AF357459 |
| MBII-373 | 1 | 91 | n. d. | human | SINE B2; homology to MBII-133 | AF357460 |
See footnote to Table I.
Novel ERNS without known sequence or structural motifs
Of the 88 novel snmRNAs identified without snoRNA motifs, 57 did not exhibit any sequence or structural motifs that would have made it possible to assign a genomic location within the mouse genome or a specific function to these RNAs. A notable exception is clone MBI-44, which maps to the mitochondrial pro/D loop (Table VI, group XIII). Twenty snmRNAs from the group of 57 were expressed, as assessed by northern blot analysis, while the expression of the remaining 37 snmRNAs could not be confirmed. From three randomly chosen clones of that group, however, we could amplify cDNA fragments of the expected size by RT–PCR, demonstrating their expression. Again, at this point, we cannot exclude the possibility that some of the cDNA sequences of this class represent degradation products of unknown hnRNAs or mRNAs rather than snmRNAs. If this turns out to be the case, these sequences are still useful in providing ESTs for novel mRNAs in mouse. Further analysis of the human and mouse genomes should provide a better insight as to whether these sequences represent novel snmRNAs, as at least some of them will.
Table VI. Group XIII: novel snmRNAs without known sequence or structural motifs.
| ERNS | Copies | cDNA | RNA | Abundance | Homology | Location/comments | Accession No. |
|---|---|---|---|---|---|---|---|
| MBI-44 | 1 | 97 | 100/60 | +++ | – | mitochondrial pro/D loop | AF357461 |
| MBI-68 | 1 | 117 | 130 | +++ | – | AF357462 | |
| MBI-82 | 2 | 145 | 160 | +++ | human | AF357463 | |
| MBII-147 | 1 | 75 | 50 | +++ | – | AF357464 | |
| MBII-156 | 1 | 103 | 500/78 | +++ | human | AF357465 | |
| MBII-367 | 1 | 70 | 140 | +++ | – | AF357466 | |
| MBII-386 | 1 | 81 | 75 | +++ | – | AF357467 | |
| MBII-433 | 1 | 73 | 65 | +++ | – | AF357468 | |
| MBI-7 | 1 | 153 | 160 | + | – | AF357469 | |
| MBI-16 | 1 | 112 | 110 | + | – | AF357470 | |
| MBI-22 | 2 | 164 | 160 | + | – | AF357471 | |
| MBI-40 | 1 | 67 | 200/150/60 | + | – | AF357472 | |
| MBI-58 | 1 | 278 | 300 | + | – | AF357473 | |
| MBI-88 | 1 | 60 | 160/70 | + | – | AF357474 | |
| MBI-149 | 1 | 70 | 220 | + | – | AF357475 | |
| MBII-104 | 1 | 68 | 75 | + | human | AF357476 | |
| MBII-106 | 1 | 58 | 93 | + | – | AF357477 | |
| MBII-157 | 1 | 59 | 95 | + | – | AF357478 | |
| MBII-201 | 1 | 66 | 150 | + | human | AF357479 | |
| MBII-302 | 1 | 65 | 65 | + | – | AF357480 | |
| MBI-4 | 1 | 204 | – | – | – | AF357481 | |
| MBI-10 | 1 | 29 | – | – | – | AF357482 | |
| MBI-14 | 1 | 30 | – | – | – | AF357483 | |
| MBI-21 | 1 | 107 | – | – | – | AF357484 | |
| MBI-25 | 1 | 82 | – | – | – | AF357485 | |
| MBI-27 | 1 | 151 | – | – | – | AF357486 | |
| MBI-32 | 1 | 181 | – | – | human | AF357487 | |
| MBI-54 | 1 | 105 | – | – | – | detectable by RT–PCR | AF357488 |
| MBI-66 | 1 | 212 | – | – | – | AF357489 | |
| MBI-67 | 1 | 228 | – | – | human | AF357490 | |
| MBI-72 | 1 | 48 | – | – | – | AF357491 | |
| MBI-73 | 1 | 163 | – | – | – | AF357492 | |
| MBI-75 | 1 | 48 | – | – | – | AF357493 | |
| MBI-76 | 1 | 119 | – | – | – | AF357494 | |
| MBI-86 | 1 | 76 | – | – | – | AF357495 | |
| MBI-102 | 1 | 85 | – | – | – | AF357496 | |
| MBI-105 | 1 | 123 | – | – | – | AF357497 | |
| MBI-107 | 2 | 56 | – | – | – | AF357498 | |
| MBI-109 | 1 | 256 | – | – | – | AF357499 | |
| MBII-5 | 1 | 50 | – | – | – | AF357500 | |
| MBII-11 | 1 | 51 | – | – | – | AF357501 | |
| MBII-37 | 1 | 82 | – | – | – | detectable by RT–PCR | AF357502 |
| MBII-60 | 1 | 58 | – | – | – | AF357503 | |
| MBII-65 | 1 | 62 | – | – | – | RNA exhibits pseudoknot structure | AF357504 |
| MBII-97 | 1 | 65 | – | – | – | AF357505 | |
| MBII-109 | 1 | 80 | – | – | human | detectable by RT–PCR | AF357506 |
| MBII-122 | 1 | 64 | – | – | – | AF357507 | |
| MBII-123 | 1 | 94 | – | – | – | AF357508 | |
| MBII-148 | 1 | 55 | – | – | – | AF357509 | |
| MBII-158 | 1 | 82 | – | – | – | AF357510 | |
| MBII-161 | 1 | 55 | – | – | – | AF357511 | |
| MBII-223 | 1 | 73 | – | – | – | AF357512 | |
| MBII-247 | 1 | 66 | – | – | – | AF357513 | |
| MBII-279 | 1 | 55 | – | – | – | AF357514 | |
| MBII-280 | 1 | 93 | – | – | – | AF357515 | |
| MBII-293 | 2 | 18 | – | – | – | AF357516 | |
| MBII-298 | 1 | 69 | – | – | – | AF357517 |
See footnote to Table I.
Conclusion
This study represents a first unbiased look at the population of snmRNA species in a mammalian cell, providing the basis for a comprehensive understanding of genomic, cellular and organismal function. By our experimental approach, we could identify a large set of novel snoRNAs of the C/D or H/ACA box type guiding ribose methylation or pseudouridylation not only in rRNA, as expected, but also in snRNAs. For the first time, we report the detection of guide snoRNAs directing ribose methylations in U2 and U4 snRNAs, as well as snoRNA guides for pseudouridylations in U2 and U6 snRNAs. In addition, we identified a surprisingly large number of snoRNA species from both classes without the potential to target rRNAs or snRNAs, as deduced from their lack of appropriate complementarity. Especially intriguing was the identification of several brain-specific snmRNAs, all of which belong to the snoRNA type. This might lead to further studies to identify snoRNAs expressed tissue specifically in tissues other than brain. One of the brain-specific snoRNAs (MBII-52) has been suggested to target serotonin receptor 5-HT2C hnRNA or mRNA, which in turn is expressed specifically in brain (Cavaillé et al., 2000). This may be indicative of a novel function of snoRNAs, namely the regulation of gene expression by binding to and/or modifying mRNAs or their hnRNA precursors via their antisense elements. At this stage, it is difficult to speculate about the function of potential snmRNAs of the non-snoRNA type. As demonstrated, some of these novel species are derived from hnRNAs or mRNAs and might therefore correspond to degradation products of larger transcripts. Alternatively, they could regulate the expression of mRNAs by as yet unknown mechanisms, especially when located within their 5′ or 3′ UTRs. Their detection sets the stage for direct experimental testing of these hypotheses.
Materials and methods
Identification of novel RNA species
We prepared total RNA from mouse brain by the TRIzol method (Gibco-BRL). Total RNA was subsequently fractionated on a denaturing 8% polyacrylamide gel (7 M urea, 1× TBE buffer). RNAs in the size range ∼50 to ∼110 (Fraction II) or ∼110 to ∼500 nt (Fraction I) were excised from the gel, passively eluted and ethanol precipitated. Subsequently, 5 µg of RNA were tailed with CTP using poly(A) polymerase, as described by DeChiara and Brosius (1987). RNAs were reverse transcribed into cDNAs using primer GIBCO1 (see Supplementary data) and cloned into pSPORT 1 vector employing the GIBCO Superscript™ system (Gibco-BRL). cDNAs were amplified by PCR using primers FSP and RSP (see Supplementary data). PCR products were spotted by robots in high density arrays onto filters by the method of Schmitt et al. (1999), performed at the Resource Center of the German Human Genome Project (Berlin, Germany).
Filter hybridization and isolation of clones
For exclusion of the most abundant, known, small RNA species, we end-labelled oligonucleotides (see Supplementary data) derived from these sequences with [33P]ATP and T4 polynucleotide kinase, and hybridized oligonucleotides to DNA arrays spotted on filters (see above). We performed hybridization in 0.5 M sodium phosphate pH 7.2, 7% SDS, 1 mM EDTA at 53°C for 12 h. We washed filters twice at room temperature for 15 min in 40 mM sodium phosphate buffer pH 7.2, 0.1% SDS, exposed filters to a phosphoimaging screen and analysed filters by computer-aided determination of hybridization signals (Maier et al., 1994).
Accession numbers of sequences
The sequence data have been submitted to the DDBJ/EMBL/GenBank databases under the accession numbers AF357317–AF357517.
Supplementary data
For additional methods see Supplementary data available at The EMBO Journal Online. These data will also be available and periodically updated at our web page: http://exppc01.uni-muenster.de/expath/ snmrnas.htm
Acknowledgments
Acknowledgements
We would like to thank Dr Stefan Hennig for his support with computer analysis of data and Christine Mersmann for technical assistance during the initial phase of the project. This work was supported by the German Human Genome Project through the BMBF (#01KW9616 and #01KW9966) to J.B. and A.H., by an IZKF grant (Teilprojekt F3, Münster) to A.H. and a grant from the Association pour la Recherche sur le Cancer and laboratory funds from the Centre National de la Recherche Scientifique and Université Paul-Sabatier, Toulouse, to J.-P.B.
References
- Bachellerie J.P. and Cavaillé,J. (1998) Small nucleolar RNAs guide the ribose methylations of eukaryotic rRNAs. In Grosjean,H. and Benne,R. (eds), Modification and Editing of RNA: The Alteration of RNA Structure and Function. ASM Press, Washington, DC, pp. 255–272.
- Bachellerie J.P., Cavaillé,J. and Qu,L.H. (2000) Nucleotide modifications of eukaryotic rRNAs: the world of small nucleolar RNAs revisited. In Garrett,R., Douthwaite,S., Liljas,A., Matheson,A., Moore,P.B. and Noller,H. (eds), Ribosome: Structure, Function, Antibiotics and Cellular Interactions. ASM Press, Washington, DC, pp. 191–203.
- Balakin A.G., Smith,L. and Fournier,M.J. (1996) The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell, 86, 823–834. [DOI] [PubMed] [Google Scholar]
- Bortolin M.L., Ganot,P. and Kiss,T. (1999) Elements essential for accumulation and function of small nucleolar RNAs directing site-specific pseudouridylation of ribosomal RNA. EMBO J., 18, 457–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J. (1999) Genomes were forged by massive bombardments with retroelements and retrosequences. Genetica, 107, 209–238. [PubMed] [Google Scholar]
- Cavaillé J., Buiting,K., Kiefmann,M., Lalande,M., Brannan,C.I., Horsthemke,B., Bachellerie,J.-P., Brosius,J. and Hüttenhofer,A. (2000) Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl Acad. Sci. USA, 97, 14311–14316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeChiara T.M. and Brosius,J. (1987) Neural BC1 RNA: cDNA clones reveal nonrepetitive sequence content. Proc. Natl Acad. Sci. USA, 84, 2624–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy S.R. (1999) Noncoding RNA genes. Curr. Opin. Genet. Dev., 9, 695–699. [DOI] [PubMed] [Google Scholar]
- Filipowicz W. (2000) Imprinted expression of small nucleolar RNAs in brain: time for RNomics. Proc. Natl Acad. Sci. USA, 97, 14035–14037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganot P., Bortolin,M.-L. and Kiss,T. (1997a) Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell, 89, 799–809. [DOI] [PubMed] [Google Scholar]
- Ganot P., Caizergues-Ferrer,M. and Kiss,T. (1997b) The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev., 11, 941–956. [DOI] [PubMed] [Google Scholar]
- Ganot P., Jady,B.E., Bortolin,M.L., Darzacq,X. and Kiss,T. (1999) Nucleolar factors direct the 2′-O-ribose methylation and pseudo uridylation of U6 spliceosomal RNA. Mol. Cell. Biol., 19, 6906–6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiss N.S., Knight,S.W., Vulliamy,T.J., Klauck,S.M., Wiemann,S., Mason,P.J., Poustka,A. and Dokal,I. (1998) X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nature Genet., 19, 32–38. [DOI] [PubMed] [Google Scholar]
- Jady B.E. and Kiss,T. (2000) Characterisation of the U83 and U84 small nucleolar RNAs: two novel 2′-O-ribose methylation guide RNAs that lack complementarities to ribosomal RNAs. Nucleic Acids Res., 28, 1348–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jady B.E. and Kiss,T. (2001) A small nucleolar guide RNA functions both in 2′-O-ribose methylation and pseudouridylation of the U5 spliceosomal RNA. EMBO J., 20, 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J. (2000) Repbase Update; a database and an electronic journal of repetitive elements. Trends Genet., 16, 418–420. [DOI] [PubMed] [Google Scholar]
- Kiss T. and Filipowicz,W. (1995) Exonucleolytic processing of small nucleolar RNAs from pre-mRNA introns. Genes Dev., 9, 1411–1424. [DOI] [PubMed] [Google Scholar]
- Kiss-Laszlo Z., Henry,Y., Bachellerie,J.P., Caizergues-Ferrer,M. and Kiss,T. (1996) Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell, 85, 1077–1088. [DOI] [PubMed] [Google Scholar]
- Lafontaine D.L., Bousquet-Antonelli,C., Henry,Y., Caizergues-Ferrer,M. and Tollervey,D. (1998) The box H + ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev., 12, 527–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe T.M. and Eddy,S.R. (1999) A computational screen for methylation guide snoRNAs in yeast. Science, 283, 1168–1171. [DOI] [PubMed] [Google Scholar]
- Maden B.E. (1990) The numerous modified nucleotides in eukaryotic ribosomal RNA. Prog. Nucleic Acid Res. Mol. Biol., 39, 241–301. [DOI] [PubMed] [Google Scholar]
- Maier E., Meier-Ewert,S., Ahmadi,A., Curtis,J. and Lehrach,H.J. (1994) Application of robotic technology to automated sequence fingerprint analysis by oligonucleotide hybridisation. J. Biotechnol., 35, 191–203. [DOI] [PubMed] [Google Scholar]
- Massenet S., Mougin,A. and Branlant,C. (1998) Posttranscriptional modifications in the U snRNAs. In Grosjean,H. and Benne,R. (eds), Modification and Editing of RNA: The Alteration of RNA Structure and Function. ASM Press, Washington, DC, pp. 201–228.
- Maxwell E.S. and Fournier,M.J. (1995) The small nucleolar RNAs. Annu. Rev. Biochem., 64, 897–934. [DOI] [PubMed] [Google Scholar]
- Mitchell J.R., Cheng,J. and Collins,K. (1999) A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol. Cell. Biol., 19, 567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munholland J.M. and Nazar,R.N. (1987) Methylation of ribosomal RNA as a possible factor in cell differentiation. Cancer Res., 47, 169–172. [PubMed] [Google Scholar]
- Nazar R.N., Sitz,T.O. and Somers,K.D. (1980) Cytoplasmic methylation of mature 5.8S ribosomal RNA. J. Mol. Biol., 142, 117–121. [DOI] [PubMed] [Google Scholar]
- Ni J., Tien,A.L. and Fournier,M.J. (1997) Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell, 89, 565–573. [DOI] [PubMed] [Google Scholar]
- Nicoloso M., Qu,L.H., Michot,B. and Bachellerie,J.P. (1996) Intron-encoded, antisense small nucleolar RNAs: the characterization of nine novel species points to their direct role as guides for the 2′-O-ribose methylation of rRNAs. J. Mol. Biol., 260, 178–195. [DOI] [PubMed] [Google Scholar]
- Ofengand J. and Bakin,A. (1997) Mapping to nucleotide resolution of pseudouridine residues in large subunit ribosomal RNAs from representative eukaryotes, prokaryotes, archaebacteria, mitochondria and chloroplasts. J. Mol. Biol., 266, 246–268. [DOI] [PubMed] [Google Scholar]
- Ofengand J. and Fournier,M.J. (1998) The pseudouridine residues of rRNA: number, location, biosynthesis, and function. In Grosjean,H. and Benne,R. (eds), Modification and Editing of RNA: The Alteration of RNA Structure and Function. ASM Press, Washington, DC, pp. 229–253.
- Olivias W.M., Muhlrad,D. and Parker,P. (1997) Analysis of the yeast genome: identification of new non-coding and small ORF-containing RNAs. Nucleic Acids Res., 22, 4619–4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T. (1998) The plurifunctional nucleolus. Nucleic Acids Res., 26, 3871–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelczar P. and Filipowicz,W. (1998) The host gene for intronic U17 small nucleolar RNAs in mammals has no protein-coding potential and is a member of the 5′-terminal oligopyrimidine gene family. Mol. Cell. Biol., 18, 4509–4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogacic V., Dragon,F. and Filipowicz,W. (2000) Human H/ACA small nucleolar RNPs and telomerase share evolutionarily conserved proteins NHP2 and NOP10. Mol. Cell. Biol., 20, 9028–9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu L.H. et al. (1999) Seven novel methylation guide small nucleolar RNAs are processed from a common polycistronic transcript by Rat1p and RNase III in yeast. Mol. Cell. Biol., 19, 1144–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders C. and Cohen,R.S. (1999) The role of oocyte transcription, the 5′UTR, and translation repression and derepression in Drosophila gurken mRNA and protein localization. Mol. Cell, 3, 43–54. [DOI] [PubMed] [Google Scholar]
- Schmitt A.O., Herwig,R., Meier-Ewert,S. and Lehrach,H. (1999) High density cDNA grids for hybridization fingerprinting experiments. In Innis,M.A., Gelfand,D.H. and Sninsky,J.J. (eds), PCR Applications: Protocols for Functional Genomics. Academic Press, San Diego, CA, pp. 457–472.
- Schuldt A.J., Adams,J.H., Davidson,C.M., Micklem,D.R., Haseloff,J., St Johnston,D. and Brand,A.H. (1998) Miranda mediates asymmetric protein and RNA localization in the developing nervous system. Genes Dev., 12, 1847–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C.M. and Steitz,J.A. (1997) Sno storm in the nucleolus: new roles for myriad small RNPs. Cell, 89, 669–672. [DOI] [PubMed] [Google Scholar]
- Smith C.M. and Steitz,J.A. (1998) Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5′-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol. Cell. Biol., 18, 6897–6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiedge H., Chen,W. and Brosius,J. (1993) Primary structure, neural-specific expression, and dendritic location of human BC200 RNA. J. Neurosci., 13, 2382–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D. (1996) Trans-acting factors in ribosome synthesis. Exp. Cell Res., 229, 226–232. [DOI] [PubMed] [Google Scholar]
- Tycowski K.T., Shu,M.D. and Steitz,J.A. (1994) Requirement for intron-encoded U22 small nucleolar RNA in 18S ribosomal RNA maturation. Science, 266, 1558–1561. [DOI] [PubMed] [Google Scholar]
- Tycowski K.T., You,Z.H., Graham,P.J. and Steitz,J.A. (1998) Modification of U6 spliceosomal RNA is guided by other small RNAs. Mol. Cell, 2, 629–638. [DOI] [PubMed] [Google Scholar]
- Weinstein L.B. and Steitz,J.A. (1999) Guided tours: from precursor snoRNA to functional snoRNP. Curr. Opin. Cell Biol., 11, 378–384. [DOI] [PubMed] [Google Scholar]