Abstract
Using a genetic complementation approach we have identified disabled-2 (Dab2), a structural homolog of the Dab1 adaptor molecule, as a critical link between the transforming growth factor β (TGFβ) receptors and the Smad family of proteins. Expression of wild-type Dab2 in a TGFβ-signaling mutant restores TGFβ-mediated Smad2 phosphorylation, Smad translocation to the nucleus and Smad-dependent transcriptional responses. TGFβ stimulation triggers a transient increase in association of Dab2 with Smad2 and Smad3, which is mediated by a direct interaction between the N-terminal phosphotyrosine binding domain of Dab2 and the MH2 domain of Smad2. Dab2 associates with both the type I and type II TGFβ receptors in vivo, suggesting that Dab2 is part of a multiprotein signaling complex. Together, these data indicate that Dab2 is an essential component of the TGFβ signaling pathway, aiding in transmission of TGFβ signaling from the TGFβ receptors to the Smad family of transcriptional activators.
Keywords: Dab2/Smad/TGFβ
Introduction
The transforming growth factor β (TGFβ) superfamily consists of cytokines that modulate essential cellular functions, such as cellular proliferation, differentiation and apoptosis (Massagué, 1998). Signaling by TGFβ, the prototypic member of the TGFβ superfamily, is initiated following ligand binding to the constitutively active ser/thr kinase type II receptor (TβRII). The type I receptor (TβRI), which also possesses ser/thr kinase activity, is then recruited to TβRII, leading to the formation of an oligomeric complex (Wrana et al., 1994). The subsequent phosphorylation and activation of the type I receptor by the type II receptor leads to further propagation of TGFβ signaling by the Smad family of proteins.
The Smads can be divided into three functional groups: the receptor-activated or R-Smads, consisting of Smads 1, 2, 3, 5 and 8; the common-mediator or Co-Smad, Smad 4; and the inhibitory Smads, consisting of Smads 6 and 7 (reviewed in Heldin et al., 1997; Piek et al., 1999; Zhang and Derynck, 1999). The R-Smads are recruited to and phosphorylated by their cognate type I receptors; Smads 1, 5 and 8 transmit signaling by BMP receptors, while Smads 2 and 3 mediate signaling by TGFβ and activin receptors. Following phosphorylation, R-Smads dimerize with Smad4 and translocate to the nucleus, where the complex has been shown to activate gene transcription by binding directly to DNA (Dennler et al., 1998) as well as by interaction with other transcription factors such as FAST1, FAST2, c-Jun and c-Fos (Chen et al., 1996; Labbe et al., 1998; Zhang et al., 1998).
An accessory protein has recently been identified, SARA (Smad anchor for receptor activation), which helps recruit Smad2/3 to the activated TβRI (Tsukazaki et al., 1998). SARA binds to unphosphorylated Smad 2/3 and by virtue of its lipid-binding FYVE domain localizes Smad2/3 to the proper subcellular compartment for interaction with TβRI (Tsukazaki et al., 1998). Once phosphorylation of Smad 2/3 has occurred, the SARA– Smad complex dissociates, exposing an intrinsic nuclear import signal on Smad2/3 that results in its accumulation in the nucleus (Xu et al., 2000).
Disabled-2 (Dab2) is a signaling molecule that was first identified as DOC-2 (for differentially expressed in ovarian carcinoma) (Mok et al., 1994), and subsequently as p96, a protein whose phosphorylation was stimulated by CSF-1 (Xu et al., 1995). Dab2 contains an N-terminal phosphotyrosine binding domain (PTB) or phosphotyrosine interacting domain (PID) and a C-terminal proline-rich domain (PRD) (Xu et al., 1995), indicative of a function as an adaptor molecule (Pawson and Scott, 1997).
We demonstrate here that stable expression of Dab2 functionally complements a TGFβ-signaling mutant cell line, restoring all assayed TGFβ responses including TGFβ-induced Smad2 phosphorylation, Smad nuclear translocation and Smad-dependent transcriptional responses. Sequencing of the Dab2 message expressed by the mutant cell line demonstrates that it harbors a missense mutation in the C-terminal domain of Dab2, which causes a decrease in the stability and steady-state expression of Dab2. Dab2 associates with Smad2 and 3 in a time- and ligand-dependent manner, which is mediated by a direct interaction between the N-terminal PTB domain of Dab2 and the MH2 domain of Smad2. Mutation of a conserved phenylalanine residue in the PTB domain of Dab2 abrogates its ability to complement the mutant cell line, while expression of the PTB domain alone or Dab2 lacking the C-terminal PRD also fails to mediate complementation, demonstrating that both domains are required for TGFβ signaling. Dab2 is also found in association with both TβRI and TβRII, suggesting that it is part of a multiprotein signaling complex. Dab2 thus appears to function as an adaptor molecule, serving to bridge the TGFβ receptor complex to the Smad pathway.
Results
Dab2 functionally complements the TGFβ-signaling mutant 903 cell line
We have previously developed a genetic system consisting of a set of recessive TGFβ-mutant cell lines that could be functionally complemented to identify TGFβ signaling molecules (Hocevar and Howe, 1996). This system utilizes the human fibrosarcoma HT1080-derived parental cell line, BAHgpt, which contains the selectable marker Escherichia coli guanine phosphoribosyltransferase (gpt) linked to the TGFβ-responsive promoter 3TP (Figure 1A). Treatment of the BAHgpt cell line with TGFβ leads to expression of the gpt gene and allows for cell growth in media containing hypoxanthine, aminopterin and thymidine (HAT); conversely, in media containing 6-thioguanine (6TG), TGFβ treatment leads to cell death. Recessive mutant cell lines were generated from the parental cell line following successive rounds of chemical mutagenesis by selection in media containing 6TG and TGFβ. The mutant cell line 903, selected in this manner, has been characterized as being deficient in a variety of TGFβ-stimulated responses (Lee et al., 1997; Hocevar et al., 1999). Transfection of the 903 cell line with a cDNA expression library followed by selection in media containing HAT and TGFβ resulted in the generation of a complemented clone designated 903L2. To identify candidate genes responsible for the functional complementation, analysis of genes differentially expressed by the complemented 903L2 cell line and the mutant 903 cell line was performed using a gene filter array. One such gene identified as having differential expression corresponded to the C-terminal region of the DOC-2 gene, also known as Dab2 and p96.
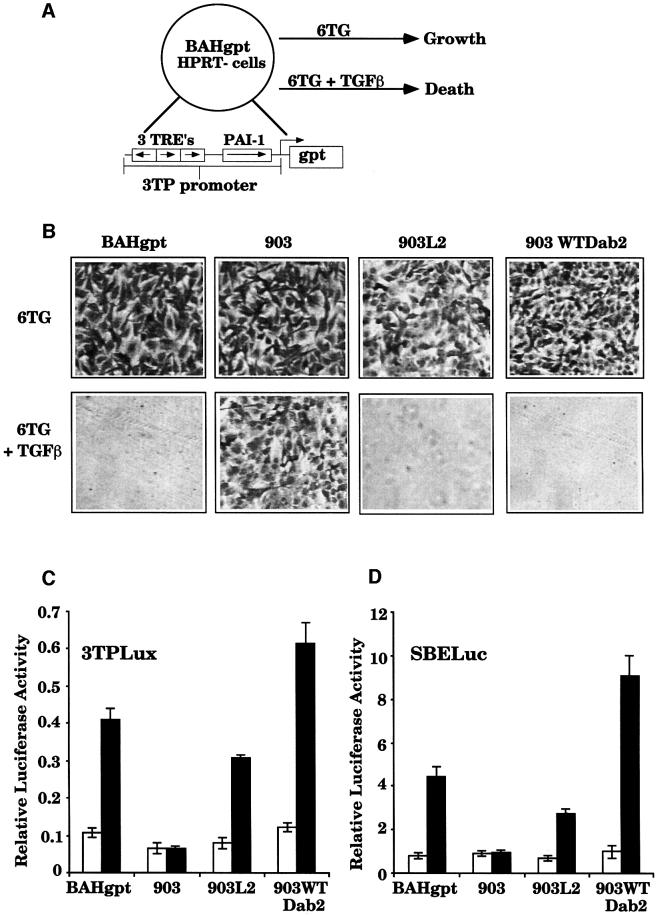
Fig. 1. Dab2 expression mediates functional complementation of the 903 TGFβ-signaling mutant cell line. (A) Diagrammatic representation of the genetic complementation approach. The parental cell line BAHgpt expresses the E.coli gpt gene driven by the 3TP promoter. In media containing 6TG, TGFβ treatment leads to cell death, indicative of TGFβ responsiveness. Mutant cell lines that do not respond to TGFβ maintain growth in media containing 6TG plus TGFβ. (B) Transfection of Dab2 into 903 cells restores TGFβ responsiveness. Parental BAHgpt, and mutant 903, 903L2 and 903 cells transfected with human wild-type Dab2 (903WTDab2) were plated into media containing 6TG (30 µM) and 6TG (30 µM) plus 5 ng/ml TGFβ. After 5 days, cells were fixed with methanol and stained with hematoxylin to visualize growth. (C and D) Dab2 expression restores TGFβ-mediated induction of the 3TPLux (C) and SBE-Luc (D) reporter constructs. BAHgpt, 903, 903L2 and 903WTDab2 cells were transiently transfected with 3TPLux or SBE-Luc and SV40-RL as a control for transfection efficiency. Cells were treated with TGFβ for 18 h, followed by lysis and determination of luciferase activity. Luciferase activity is expressed as a ratio of 3TPLux luciferase activity divided by the SV40-RL activity. Shown is the mean ± SD of duplicates from a representative experiment. Open bars represent untreated cells while black bars denote TGFβ treatment.
To assess whether expression of Dab2 was capable of functionally complementing the mutant phenotype of the 903 cell line, human Dab2 was stably introduced into the 903 cell line. Restoration of TGFβ signaling was first tested by growth of the cells in media containing 6TG and 6TG plus TGFβ (Figure 1B). In the absence of TGFβ, 6TG in the media has no effect on cell growth; however, in the presence of TGFβ, cells that are responsive to TGFβ express gpt and die in media containing 6TG (Figure 1A and B). As shown in Figure 1B, all the cell lines tested grow in media containing 6TG alone, while the parental cell line BAHgpt, the original complemented clone 903L2 and a pool of 903 cells stably expressing wild-type Dab2 (903WTDab2) die in media containing 6TG plus TGFβ. These results thus indicate that Dab2 alone can mediate restoration of the TGFβ-responsive phenotype.
To further characterize the restoration of TGFβ responsiveness mediated by Dab2 in the 903 mutant, we first performed transient transfection analysis using the luciferase reporter construct 3TPLux, which has been used to measure TGFβ responsiveness of a variety of cell lines (Wrana et al., 1992; Carcamo et al., 1994). As shown in Figure 1C, treatment of BAHgpt cells results in a 4- to 5-fold induction of the 3TPLux reporter, which is deficient in the 903 cell line. Analysis of 903L2 and 903WTDab2 cells reveals restoration of TGFβ-stimulated luciferase induction, without alteration of the basal luciferase level. These results are consistent with the ability of Dab2 to restore regulation of the gpt gene in response to TGFβ (Figure 1B).
Since the Smad family of proteins have been shown to be important intracellular mediators of the TGFβ signaling pathway, we next assayed whether Dab2 expression could influence the transcriptional activity of the Smad proteins. To assess this, we utilized a luciferase construct (p6SBE-Luc) that contains a promoter consisting of six tandem copies of the Smad-binding element (SBE) designed to monitor Smad-dependent transcriptional activity (Zawel et al., 1998). Transient transfection of the SBE-Luc reporter followed by TGFβ treatment demonstrates that Dab2 expression mediates restoration of TGFβ-stimulated SBE-Luc induction to the mutant 903 cell line (Figure 1D), suggesting that Dab2 may play a role in TGFβ-mediated Smad-dependent signaling.
The 903 cell line expresses a mutant form of Dab2 that exhibits decreased protein stability
The gene filter analysis utilized indicated that Dab2 mRNA was down-regulated, but not absent in the mutant 903 cell line. To expand this observation, we determined whether Dab2 protein levels were also decreased in the mutant cell line by western analysis. We find that the mutant 903 cells express less steady-state Dab2 protein than parental BAHgpt cells, which is restored in the 903WTDab2 cell line (Figure 2A). To assess whether this decrease in steady-state expression level is due to increased Dab2 protein turnover, we performed metabolic labeling of the BAHgpt and 903 cells to follow newly synthesized Dab2. Immunoprecipitation with a monoclonal antibody to Dab2, but not control IgG, reveals that Dab2 protein is readily detected in BAHgpt cells following a 4 h labeling period, while Dab2 is barely detectable in the 903 cells (Figure 2B).
Fig. 2. Mutant Dab2 exhibits decreased protein stability. (A) Steady-state levels of Dab2 are decreased in mutant 903 cells. Fifty micrograms of total cellular protein from BAHgpt, 903 and 903WTDab2 cells were subjected to western analysis utilizing a monoclonal antibody to Dab2 (α-p96). (B) De novo Dab2 synthesis is decreased in mutant 903 cells. BAHgpt and 903 cells were labeled with [35S]methionine for 4 h prior to lysis and immunoprecipitation with either non-immune mouse IgG or a monoclonal antibody to Dab2 (α-p96). Dab2 levels were visualized by autoradiography. (C, D and E) Mutant Dab2 is less stable than WTDab2. Pulse–chase analysis (C) was performed on COS7 cells transiently transfected with Flag-tagged WT or Mut Dab2 as described in Materials and methods. Following immunoprecipitation with anti-Flag antibody, [35S]Dab2 was visualized by autoradiography. Analysis of steady-state levels of WT or Mut Dab2 (D) was performed by western analysis utilizing anti-Flag antibody. (E) Levels of Dab2 proteins in (C) were determined by densitometric scanning and analysis with NIHImage software. Levels are expressed as a percentage of protein remaining at the various time points, with levels at time 0 designated as 100%.
To determine whether the Dab2 mRNA expressed by the 903 cell line harbors any mutations that could affect its stability, we cloned the mutant gene by RT–PCR and compared its sequence with wild-type (WT)Dab2. The Dab2 mRNA expressed by the mutant 903 cell line was found to contain a single nucleotide change of G to A, resulting in an amino acid substitution of Ser to Asn at amino acid 634 in the C-terminal domain of Dab2. To assess whether this mutation affects the stability of the Dab2 protein in vivo, Flag-tagged WT and mutant Dab2 were transiently transfected into COS7 cells and pulse– chase analysis performed. As shown in Figure 2C, WTDab2 expression is readily detected following a 30 min labeling and remains stable for 6 h. In contrast, initial lower levels of mutant Dab2 are expressed, followed by a decrease in levels by as early as 1 h post-chase. Steady-state levels of transfected Dab2 protein determined by western analysis show an accumulation of WT, but not mutant Dab2 throughout the time course (Figure 2D). Also readily apparent is the appearance of slower migrating forms of WTDab2 over time, absent from MutDab2, which may reflect phosphorylation of 634Ser (Figure 2C and D). Analysis of Dab2 levels indicates that MutDab2 exhibits a half-life of ∼10 h, while WTDab2 has a half-life of ∼20 h (Figure 2E). Taken together, these results demonstrate that the mutant 903 cells express lower de novo and steady-state levels of Dab2, which can be attributed in part to the 634Ser to Asn mutation.
Dab2 restores endogenous TGFβ-mediated responses to the mutant 903 cell line
To further characterize the restoration of TGFβ responsiveness mediated by Dab2 in the 903 cell line, we next assessed the effect of Dab2 expression on TGFβ-mediated induction of the extracellular matrix proteins fibronectin and PAI-1. As shown in Figure 3A and B, the 903 cell line fails to induce PAI-1 expression following TGFβ stimulation, but while the basal level of fibronectin is decreased in the mutant cells, TGFβ can still cause an increase in fibronectin expression. Analysis of 903WTDab2 cells demonstrates that TGFβ treatment induces the expression of PAI-1 to the extent observed in BAHgpt cells, while both basal and TGFβ-stimulated levels of fibronectin are increased by Dab2 transfection. Similarly, in BAHgpt cells that stably overexpress Dab2 (BAHgptWTDab2), TGFβ-stimulated induction of fibronectin is increased; however, TGFβ-mediated PAI-1 induction does not change (Figure 3A and B). These results thus show that Dab2 can restore TGFβ-mediated induction of PAI-1 to the mutant cells and can augment TGFβ-stimulated levels of fibronectin in parental and mutant cells.
Fig. 3. Dab2 expression restores TGFβ-mediated responses to mutant 903 cells. (A and B) Dab2 expression alters TGFβ-mediated induction of the endogenous genes fibronectin (A) and PAI-1 (B). BAHgpt, BAHgpt cells stably transfected with Dab2 (BAHgptWTDab2), 903 and 903WTDab2 cells were examined for [35S]fibronectin (FN) secreted to the media (A) or [35S]PAI-1 deposited to the extracellular matrix (B) following TGFβ treatment, as described in Materials and methods. FN and PAI-1 are indicated by the arrows. (C) Dab2 expression restores TGFβ-stimulated Smad2 phosphorylation to the mutant 903 cell line. BAHgpt, BAHgptWTDab2, 903 and 903WTDab2 cells were untreated or treated with 5 ng/ml TGFβ for 1 h as indicated. Phosphorylated Smad2 was identified by western analysis using an antibody specific only for the TGFβ-stimulated phosphorylated form of Smad2 (anti-phospho Smad2 465/467; Upstate Biotechnology). (D) Equivalent expression of cellular Smad2 in BAHgpt, BAHgptWTDab2, 903 and 903WTDab2 cells was confirmed by western analysis using an anti-Smad2 antibody (Zymed Laboratories). (E and F) Dab2 restores TGFβ-stimulated Smad2 (E) and Smad3 (F) nuclear accumulation in 903 cells. Nuclear proteins were isolated from BAHgpt, 903 and 903WTDab2 cells untreated or treated with TGFβ for 1 h, as described in Materials and methods. The accumulation of Smad2 and Smad3 in the nucleus was confirmed by western analysis using anti-Smad2 (Transduction Laboratories) and anti-Smad3 antibodies (Zymed Laboratories). The positions of Smad2 and Smad3 are indicated by the arrows.
Since the transcriptional activity of both Smad2 and Smad3 is dependent on their phosphorylation by TβRI (Macías-Silva et al., 1996; Liu et al., 1997), we wished to determine whether TGFβ-mediated Smad phosphorylation is deficient in the 903 mutant cells. As shown in Figure 3C, TGFβ treatment leads to efficient phosphorylation of Smad2 in BAHgpt and BAHgptWTDab2 cells, as shown by western analysis, with an antibody that specifically recognizes TGFβ-stimulated phosphorylation sites on Smad2. While phosphorylation of Smad2 can not be detected following TGFβ treatment of 903 cells, re-introduction of Dab2 restores TGFβ-mediated Smad2 phosphorylation. This is not due to a lack of expression of Smad2 protein in 903 cells, since Smad2 levels appear to be comparable in BAHgpt and 903 cells (Figure 3D). Following phosphorylation by TβRI, Smad2 and -3 translocate to the nucleus. To assess whether the nuclear accumulation of the Smads is also deficient in mutant 903 cells, nuclear preparations derived from BAHgpt, 903 and 903WTDab2 cells stimulated with TGFβ for 1 h were analyzed for the presence of Smad2 and Smad3 (Figure 3E and F). Treatment of BAHgpt cells with TGFβ results in nuclear accumulation of Smad2 and Smad3, while in mutant 903 cells the levels of nuclear Smad2 and Smad3 do not change following TGFβ stimulation. Re-introduction of WTDab2 into 903 cells restores TGFβ-stimulated Smad2 and Smad3 nuclear accumulation (Figure 3E and F). Taken together, these results suggest that Dab2 plays a role in mediating TGFβ-stimulated Smad phosphorylation, which allows for nuclear accumulation and subsequent stimulation of Smad-dependent transcriptional responses.
The PTB and PRD domains of Dab2 are both required for TGFβ signaling
The Dab2 protein can be divided into three functional domains: the N-terminal PTB domain, a middle linker region and a C-terminal PRD. To assess the contribution of the individual domains of Dab2 in TGFβ signaling, constructs were engineered of the various domains of Dab2, including a point mutation in the PTB domain (F166VDab2) that has been shown to abrogate PTB domain function (Borg et al., 1996; Dho et al., 1998) (Figure 4A). These constructs were stably transfected into mutant 903 cells and assessed for their ability to rescue the mutant phenotype. As shown in Figure 4C and D, expression of WTDab2 restores TGFβ-stimulated 3TPLux and SBE-Luc reporter induction to 903 cells, while expression of F166VDab2, Del-PRD or the PTB domain alone fails to do so. The inability of the various constructs to restore TGFβ responsiveness was not due to differences in expression of the constructs, as verified by western analysis (Figure 4B). Expression of MutDab2 (S634N) results in restoration of ∼50% of the TGFβ-stimulated luciferase induction elicited by WTDab2, suggesting that constitutive overexpression is able to partially compensate for the decreased protein stability of MutDab2 (Figure 2). We were unable to test the ability of the PRD alone to restore TGFβ responsiveness as we could not generate a stable cell line that expressed this construct. Consistent with the reporter construct data, introduction of F166VDab2, PRD-Del and PTB constructs into 903 cells failed to restore other TGFβ-mediated responses as well, including PAI-1 induction, Smad2 phosphorylation, and Smad2 and Smad3 nuclear accumulation (data not shown). Taken together, these results indicate that both the PTB domain and PRD of Dab2 are required for restoration of TGFβ signaling in the mutant 903 cells.
Fig. 4. Restoration of TGFβ signaling requires both the PRD and PTB domain of Dab2. (A) Diagrammatic representation of various constructs of Dab2. Depicted are full-length and deletion constructs of Dab2 containing the N-terminal PTB domain and C-terminal PRD (gray shading). The asterisk designates the S634N mutation present in mutant 903 Dab2 (Mut-Dab2) and the introduced F166V mutation in full-length WTDab2 (F166V-Dab2). (B) Expression levels of various Flag-tagged Dab2 constructs from stable cell lines were determined by western analysis with α-Flag antibody (M2; Sigma). Lysate from the original mutant 903 cell line was used as a control (Cont.). (C and D) TGFβ-stimulated transcriptional activation requires both the PRD and PTB domain of Dab2. BAHgpt, 903 and 903 cells stably expressing the various forms of Dab2 described above were transiently transfected with the reporter constructs 3TPLux (C) and SBE-Luc (D), and SV40-RL as a control for transfection efficiency. After 24 h, cells were untreated (open bars) or treated (black bars) with TGFβ for 18 h. Following lysis, luciferase activity was determined and expressed as the ratio of specific luciferase activity divided by the SV40-RL activity. Shown is the mean ± SD of duplicates from a representative experiment.
Dab2 associates with Smad2 and Smad3
We next wished to assess whether the restoration of TGFβ responsiveness mediated by Dab2 was due to a direct interaction of Dab2 with the Smad proteins. To determine this, full-length WTDab2 was translated in vitro and incubated with bacterially expressed Smad1, Smad2, Smad3 and Smad4 glutathione S-transferase (GST) fusion proteins. Figure 5A demonstrates that while the GST moiety alone fails to precipitate Dab2, GST–Smad2 and GST–Smad3 bind to and precipitate full-length Dab2. Dab2 characteristically appears as a doublet, which represents the use of a second initiator methionine present in the sequence (Xu et al., 1995). GST–Smad1 and GST– Smad4, however, fail to interact with Dab2 (Figure 5A). To more fully characterize the interaction of Dab2 with the Smad proteins, we determined which functional domain of the Smad proteins interacts with Dab2. Incubation of in vitro translated WTDab2 with GST constructs constituting the MH1 and MH2 domains of Smad2 reveals that Dab2 preferentially binds to the MH2 domain, with negligible binding to the MH1 domain (Figure 5A). To map which domain in Dab2 mediates the binding to Smad2 and Smad3, constructs bearing these motifs (Figure 4A) were in vitro translated and tested for interaction with GST fusion proteins as above. Both constructs that contain the PTB domain, PTB and Del-PRD, are capable of interacting with Smad2 and Smad3, while the construct bearing only the PRD fails to do so. The same preference for binding to the MH2 domain over the MH1 domain of Smad2 is also maintained with the PTB-containing constructs, as observed for the full-length protein (Figure 5A), which is not due to differences in expression of the GST fusion proteins as visualized by Coomassie Blue staining (Figure 5B).
Fig. 5. Dab2 associates with Smad2 and Smad3 in vitro. (A) Dab2 interacts with the Smads in vitro. Full-length WTDab2 or the various deletion constructs depicted in Figure 4A were synthesized in vitro using [35S]methionine. Equal amounts of 35S-labeled reaction products were incubated with bacterially expressed GST constructs correspond ing to GST protein alone (GST), full-length GST fusion proteins of Smad1, Smad2, Smad3 and Smad4, or GST fusion proteins of the MH1 and MH2 domains of Smad2. Following extensive washing, bound proteins were analyzed by SDS–PAGE, subjected to fluorography and visualized by autoradiography. An aliquot (10%) of 35S-labeled reaction product input is shown in the control lane (Cont.). (B) Equal loading of the bacterially expressed GST fusion proteins utilized in (A) is demonstrated by Coomassie Blue staining of the gel following SDS–PAGE.
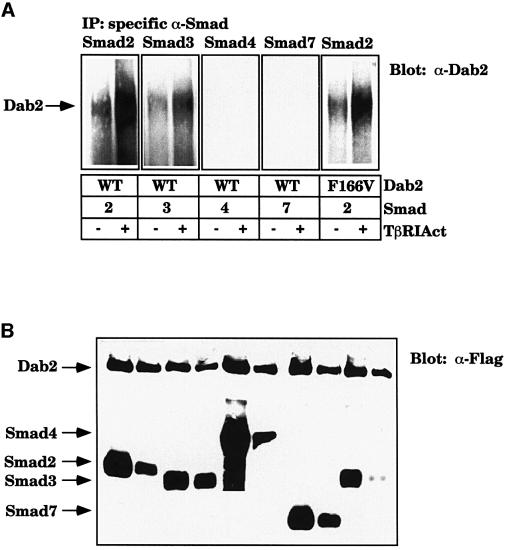
To test whether an interaction between the Smad proteins and Dab2 takes place in vivo, COS7 cells were transfected with the various Smads and Dab2 in the absence or presence of the activated form of TβRI (TβRIAct). Immunoprecipitation of the Smad proteins followed by western analysis for Dab2 reveals that although Dab2 can associate with Smad2 and Smad3 in the absence of TGFβ signaling, this association increases in the presence of TβRIAct. In contrast, Dab2 does not associate with Smad4 or Smad7 in either the absence or presence of TGFβ signaling, although the proteins are expressed at a comparable level (Figure 6B). Additionally we find that F166VDab2 is still able to associate with Smad2 and its association is stimulated in the presence of TβRIAct (Figure 6A).
Fig. 6. Dab2 associates with Smad2 and Smad3 in vivo in a ligand-dependent manner. (A and B) Smad2 and Smad3 interact with Dab2 in vivo. COS7 cells were transiently transfected with Flag-tagged WT or F166V Dab2 and various Smads with or without the activated form of TβRI (TβRIAct). After 48 h, lysates were prepared and (A) immunoprecipitated with antibodies specific to each Smad [Smad2 and Smad3 (Zymed Laboratories), Smad4 and Smad7 (Santa Cruz)]. The presence of Dab2 in the complex was confirmed by western analysis with a monoclonal antibody to Dab2 (α-p96). Expression of Dab2 and the Smads (B) was determined by western analysis with anti-Flag antibody (M2; Sigma).
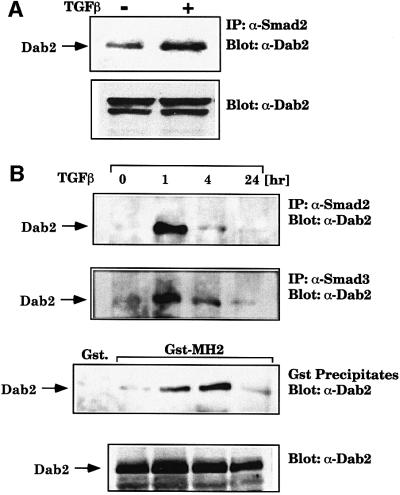
To test whether endogenous Dab2 interacts with Smad2, parental BAHgpt cells were treated with TGFβ for 1 h and lysates were immunoprecipitated with Smad2 antibody. Analysis of the immunoprecipitates for the presence of Dab2 reveals that the association of endogenous Dab2 with endogenous Smad2 is stimulated by TGFβ treatment (Figure 7A). To address the time course of association of Dab2 with Smad2 and Smad3 following TGFβ stimulation, we treated BAHgpt cells that stably overexpress WTDab2 with TGFβ for various times, followed by immunoprecipitation with Smad2 or Smad3 antibodies or incubation of lysates with the GST–MH2 domain of Smad2. Western analysis of the precipitated proteins reveals that the association of Dab2 with Smad2 and Smad3 is transiently stimulated following TGFβ treatment, reaching a peak at 1 h and returning to baseline by 24 h (Figure 7B). The association of Dab2 with the GST–MH2 domain fusion protein also follows similar kinetics, while the steady-state levels of Dab2 do not change over the course of TGFβ treatment (Figure 7B, lower panel). Together, these results indicate that the in vivo association of Dab2 with the R-Smads Smad2 and Smad3 occurs in a ligand- and time-dependent manner.
Fig. 7. Endogenous Dab2 associates with endogenous Smad2 and Smad3. (A) Dab2 interacts with Smad2 in a time- and ligand-dependent manner. BAHgpt cells were treated with TGFβ for 1 h followed by immunoprecipitation with anti-Smad2 (Transduction Laboratories) antibody. The presence of Dab2 in the immunoprecipitates was confirmed by western analysis for Dab2 (α-p96). Comparable Dab2 levels are shown by western analysis in the lower panel. (B) BAHgpt cells stably expressing WTDab2 were treated with TGFβ for various times and lysates subjected to immunoprecipitation with anti-Smad2 (Transduction Laboratories) or anti-Smad3 (Zymed Laboratories) antibodies or pull-down assay with the GST–MH2 Smad2 domain or control GST beads. The presence of Dab2 in the precipitate was confirmed by western analysis with a monoclonal antibody to Dab2 (α-p96). Equal expression of Dab2 at the various time points was verified by western analysis using the same antibody (lower panel).
Dab2 is found in association with both the TGFβ type I and type II receptors
Since we demonstrated that introduction of the Dab2 gene could restore TGFβ-specific phosphorylation of Smad2, we next wished to assess whether Dab2 could interact with the TGFβ receptors in vivo. To assess this, COS7 cells were transiently transfected with HA-tagged wild-type (WT), kinase-inactive (KR) or activated (Act.) forms of TβRI, and wild-type (WT) or kinase-inactive (KR) forms of TβRII. Immunoprecipitation of endogenous Dab2 followed by western detection for the receptors reveals that all forms of the type I and type II TGFβ receptors are found in association with Dab2 (Figure 8A). To test whether the association of Dab2 with the TGFβ receptor complex displays any dependence on ligand stimulation, we transfected the COS7 cells with both WT TβRI and TβRII, followed by stimulation with TGFβ for various times. Analysis of the association of Dab2 with the TGFβ receptor complex following ligand stimulation, however, revealed no differences in association, indicating that Dab2 is constitutively found in association with the TGFβ receptor complex (Figure 8A).
Fig. 8. Dab2 associates with the TGFβ receptor complex in vivo. (A and B) Dab2 interacts with the TGFβ receptor complex. COS7 cells were transiently transfected with HA-tagged wild-type (WT), kinase-deficient (KR) or activated (Act.) type I TGFβ receptors or HA-tagged wild-type (WT) or kinase-deficient (KR) type II TGFβ receptors as indicated. Cells were untreated or treated with TGFβ for the various times, as indicated, and lysates were subjected to immunoprecipitation with control IgG or antibodies to endogenous Dab2 (p96). The presence of co-precipitating TβRI or TβRII was confirmed by western analysis (α-HA 12CA5). Expression levels of the type I and type II TGFβ receptors in (A) were confirmed by western analysis of cell lysates using anti-HA antibodies (B). The positions of TβRI and TβRII are indicated by arrows. (C) Dab2 interacts with the TGFβ receptor complex. Mv1Lu, BAHgpt and 903 cells were incubated with [125I]TGFβ1. Following crosslinking, cells were lysed and an aliquot analyzed for receptor expression and crosslinking efficiency, left panel. The remainder of the lysate was immunoprecipitated with control mouse IgG or a monoclonal antibody to Dab2 (α-p96). Receptor complexes were resolved by SDS–PAGE and visualized by autoradiography. The positions of TβRI and TβRII are indicated by arrows.
We next wished to determine whether endogenous Dab2 is found in association with the TGFβ receptors in vivo. To investigate this, BAHgpt, mutant 903 and TGFβ-responsive mink lung epithelial (Mv1Lu) cells were incubated with 125I-labeled TGFβ, followed by crosslinking and immunoprecipitation with an antibody to Dab2. As a control for the efficiency of [125I]TGFβ crosslinking to the TGFβ receptors, an aliquot of the lysate prior to immunopreciptation was analyzed, demonstrating the presence of type I, II and III TGFβ receptors in all of these cell lines (Figure 8C). Immunoprecipitation with Dab2 antibody following crosslinking revealed that Dab2 interacts with the TGFβ receptor complex in Mv1Lu and the parental BAHgpt cells; however, Dab2 does not associate with the TGFβ type I or type II receptors in the mutant 903 cells (Figure 8C). Taken together, these results indicate that Dab2 is part of a multiprotein signaling complex that contains the TGFβ receptors.
Discussion
Using a genetic complementation approach to identify molecules that function in the TGFβ signaling pathway, we have demonstrated that Dab2, a member of the disabled gene family, can restore TGFβ responsiveness to a signaling-deficient cell line. The ability of Dab2 to restore TGFβ-stimulated Smad2 phosphorylation as well as other Smad-dependent responses thus places Dab2 downstream of the TGFβ receptors but upstream of the Smad proteins in the TGFβ signaling pathway. Mutant Dab2 contains a single point mutation in the C-terminal PRD that causes a decrease in protein stability. Consistent with this observation, overexpression of MutDab2 is able to partially overcome this decrease in protein stability to mediate complementation of the 903 cell line. Mutant Dab2 fails to associate with the TGFβ receptor complex, leading to a loss of TGFβ-stimulated R-Smad phosphorylation, Smad translocation to the nucleus and activation of Smad-dependent transcription. Dab2 re-introduction into the mutant cell line is able to restore these responses and also endogenous TGFβ-mediated gene regulation, demonstrating the essential role of Dab2 in TGFβ signaling.
The role of PTB domain-containing proteins in signal transduction
Receptors, which are usually expressed at low levels on the cell surface, commonly use adaptor and docking proteins to amplify signaling initiated following receptor activation (Pawson and Scott, 1997). Evolutionarily conserved PTB domains are commonly found in adaptor proteins, including: Dab1, which has been shown to serve as an adaptor for the Reelin/VLDL receptor (D’Arcangelo et al., 1999); FRS2, which binds to both the FGF and NGF receptors (Ong et al., 2000); Shc, which can bind to a variety of growth factor receptors; and IRS-1, which binds to the insulin receptor (reviewed in van der Geer and Pawson, 1995). Structure analysis of both the PTB domain of Shc (Zhou et al., 1995) and the PTB domain of Numb, in complex with a peptide (Li et al., 1998), demonstrated that a critical phenylalanine residue of the PTB domain makes direct contact with the target binding sequence. Mutation of this conserved phenylalanine to valine has been shown to abrogate the function of PTB-containing proteins (Borg et al., 1996; Yajnik et al., 1996; Dho et al., 1998; Howell et al., 1999). We also find that mutation of this conserved phenylalanine residue in Dab2 (F166) renders Dab2 unable to restore TGFβ responsiveness to the mutant cell line; however, it does not prevent the association of Dab2 with the Smad proteins. While the PTB domain was first characterized in Shc and IRS-1 as a module that binds to phosphorylated tyrosine residues (Kavanaugh et al., 1995), the majority of other proteins that contain this motif bind to sequences that do not contain a phosphorylated residue (Borg et al., 1996; Dho et al., 1998; Howell et al., 1999). Although the actual target binding sequences for the PTB domains differ, they commonly contain one or more aromatic residues that promote β-turn structures in proteins (Li et al., 1998). We show here that one binding partner of the Dab2 PTB domain is the MH2 domain of Smad2 and Smad3. The exact residues that mediate the Dab2 interaction have yet to be determined; however, several sequences that fit the requirement for a binding interaction are present. Due to the fact that PTB-containing proteins appear to have multiple binding partners (Dho et al., 1998; Howell et al., 1999), we also expect Dab2 to bind to other proteins that may also regulate TGFβ signaling.
The PTB domain of Dab1 has recently been shown to bind to PtdIns4P or PtdIns4,5P2-containing membranes, localizing it to the plasma membrane while simultaneously binding to the amyloid precursor protein (APP), members of the low-density lipoprotein receptor family and Ship1 (Howell et al., 1999). Since the PTB domain of Dab1 and Dab2 exhibits 66% overall identity, we postulate that the PTB domain of Dab2 may also mediate its membrane association. Recently, the PTB domain of Dab2 was found to interact with the cytoplasmic tail of the gp600/megalin endocytic receptor (Oleinikov et al., 2000). In addition, Dab2 was found to co-localize with LDLR in clathrin-coated pits, primarily due to its binding to the clathrin adaptor molecule AP-2 (Morris and Cooper, 2001). It may be that this association with the endocytic machinery helps to localize Dab2 to the same subcellular compartment as the TGFβ receptors, which have also been shown to reside in endosomes (Doré et al., 1998). Our data support this hypothesis, in that Dab2 is found in association with both the type I and type II TGFβ receptors, which would not be expected if Dab2 formed a direct contact with either receptor.
Role of Dab2 in Smad-dependent and Smad-independent signaling
The ability of Dab2 to rescue TGFβ-induced Smad2 phosphorylation and Smad-dependent transcriptional responses in the mutant 903 cell line points to a critical role of Dab2 in mediating Smad-dependent responses. Additionally, we show that the PTB domain of Dab2 binds directly to the MH2 domain of Smad2 in vitro, and that the association of Dab2 with Smad2 and Smad3 occurs in a ligand- and time-dependent manner. Other receptor systems, namely EGF and HGF, have recently been shown to activate Smad2-dependent transcription (de Caestecker et al., 1998), while transfection of MEKK1, the upstream activator of the c-Jun N-terminal kinase (JNK) pathway, has been shown to activate Smad2, resulting in its increased association with Smad4 and subsequent nuclear accumulation (Brown et al., 1999). These results thus suggest functional crosstalk between signaling pathways, which is also supported by the ability of the Smads to bind to and augment the activity of transcription factors, such as c-Jun and c-Fos, which are known targets of MAPK kinase signaling pathways (reviewed in Zhang and Derynck, 1999). The C-terminal region of Dab2 contains proline-rich PXXP sequences that have been shown to bind to SH3-containing signaling proteins (Yu et al., 1994). Recently, Dab2 has been shown to interact with Grb2 through this region (Xu et al., 1998). Dab2 may also play a role in activation of other signaling pathways through recruitment of SH3-containing signaling molecules to its C-terminal PRD, which may explain why Dab2 can modulate basal as well as TGFβ-mediated fibronectin levels in the 903 mutant cell line, a response that we have recently shown to be dependent on the JNK pathway and independent of Smad4 expression (Hocevar et al., 1999). The requirement for the C-terminal domain of Dab2 for efficient TGFβ signaling is demonstrated by the inability of constructs that lack this domain to complement the mutant cells. It may be that Dab2 serves as a bridge to link the Smad and the JNK signaling pathways, a relationship that has recently been postulated to be required for TGFβ-mediated transcriptional responses (Engel et al., 1999). Identification of the binding partners for the C-terminal region of Dab2 may thus help to clarify the role of other signaling pathways in TGFβ signaling.
Role of Dab2 as a tumor suppressor
Loss of responsiveness to the growth-inhibitory effects of TGFβ is commonly observed in human carcinomas, indicating that inactivation of the TGFβ signaling pathway is a common target that may permit cancer initiation and progression. This may occur as a result of mutations in either the type I or type II TGFβ receptors or a mutation in a component of the signaling pathway. Dab2 was initially identified as a transcript that was down-regulated in ovarian carcinoma, but was present in normal ovarian epithelial cells (Mok et al., 1994, 1998; Fazili et al., 1999). Subsequently, Dab2 expression has been demonstrated to be down-regulated in breast (Schwahn and Medina, 1998) and prostate carcinoma as well (Tseng et al., 1998). Loss of expression, which is seen early in tumor progression (Fazili et al., 1999), is not due to loss or gross chromosomal rearrangements of the gene (Mok et al., 1998). Re-introduction of Dab2 in ovarian, prostate and choriocarcinoma cell lines resulted in a decreased growth rate (Fulop et al., 1998; Mok et al., 1998; Tseng et al., 1998), while Dab2-transfected SKOV3 ovarian carcinoma cells formed tumors 50% smaller compared with parental cells when injected into nude mice, demonstrating that Dab2 acts as a tumor suppressor gene (Mok et al., 1998). Whether the re-introduction of Dab2 in these cell lines mediates restoration of TGFβ signaling is unknown at this time. Dab2 inactivation by mutation or down-regulation, an early event in cancer progression, may thus represent a new mechanism by which cancer cells acquire resistance to the growth-inhibitory effects of TGFβ. Further investigation of the role of Dab2 in TGFβ signaling may therefore provide important new insight as to how loss of TGFβ signaling leads to cancer initiation and progression.
Materials and methods
Cell culture, TGFβ treatment and DNA constructs
The generation of BAHgpt and 903 cell lines has been described previously (Hocevar and Howe, 1996; Lee et al., 1997). Cells were cultured in DME/F12 media supplemented with 10% newborn calf serum and 100 µg/ml hygromycin B (Gibco-BRL). 903L2 and stable BAHgpt and 903 cells transfected with Dab2 were grown in DME/F12 media supplemented with 10% newborn calf serum, 100 µg/ml hygromycin B (Gibco-BRL) and 500 µg/ml G418 (Gibco-BRL). COS7 and Mv1Lu cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% newborn calf serum. For TGFβ treatment of cells, recombinant human TGFβ2 generously provided by Genzyme (Cambridge, MA) was utilized at a concentration of 5 ng/ml. This TGFβ2 was found to be equivalent to TGFβ1 in all responses tested. The 3TPLux (Wrana et al., 1992) and p6SBE-Luc (Le Dai et al., 1998) luciferase reporter constructs have been described previously. GST fusion constructs of Smad1, Smad4, MH1 and MH2 domains of Smad2, and HA-tagged versions of TβRI and TβRII have been described previously (Wrana et al., 1992; Carcamo et al., 1994; Wieser et al., 1995; Zhang et al., 1996; Tsukazaki et al., 1998).
Generation of DNA constructs
Human WT and mutant Dab2 cDNA were obtained by RT–PCR and inserted into a Flag-tag expression vector pRK5 (Zhang et al., 1996). The F166V mutation in WTDab2 was introduced using the QuikChange site-directed mutagenesis kit (Stratagene) as directed. The various domain constructs of Dab2 were generated by standard PCR methods using murine Dab2 cDNA in pBluescript KS (Xu et al., 1995) as a template and inserted into the pcDNA3 vector that contained a Flag epitope tag. The sequences for PCR amplification are available upon request. GST–Smad2 and GST–Smad3 were constructed in the pGEX4T-1 vector following PCR amplification using pRK5-Smad2 and pRK5-Smad3 as templates (Zhang et al., 1996). Each construct was verified by sequencing analysis.
Generation of stable complemented cell lines
903 cells were stably transfected with a human liver cDNA expression library in the pcDNA3.1 vector (Invitrogen) by polybrene as previously described (Hocevar and Howe, 1996). Following selection in media containing 500 µg/ml G418, pools of cells were plated in media containing a HAT supplement (Sigma) and 5 ng/ml TGFβ. Clones able to grow in HAT plus TGFβ were expanded and tested for death in media containing 30 µM 6TG (Sigma) and 5 ng/ml TGFβ. One clone, designated 903L2, met the criteria for restoration of the parental TGFβ-responsive phenotype with regards to proper growth/death in the various selection media. Differential display analysis was performed with a cDNA probe prepared from mRNA samples of 903 and 903L2 cells treated with 5 ng/ml TGFβ for 4 h using the Gene Discovery Array (Human I, Genome Systems, Inc.) as per the manufacturer’s protocols. Hybridization results were imaged by PhosphorImager and results sent to Genome Systems for analysis, which identified the 3′ region of the Dab2 gene as a differentially expressed gene. To generate stable 903 and BAHgpt cell lines that overexpress Flag-tagged mouse and human Dab2, the constructs were stably introduced into the cell lines by polybrene transfection. Cells were selected in media containing 500 µg/ml G418 and maintained as pools. Expression of the constructs was verified by western analysis using anti-Flag antibody (M2; Sigma) and anti-Dab2 antibody (p96; Transduction Laboratories).
Transient transfections and reporter gene measurements
For transient transfection and luciferase assays, cells were transfected with Fugene6 (Roche Diagnostics) following manufacturer’s protocols. For luciferase assays, 2 × 105 cells/well were transfected with 0.5 µg/well of the specific luciferase construct (3TPLux or p6SBE-Luc) along with 50 ng/well SV40-RL (Promega) as an internal control for transfection efficiency. After 24 h, the medium was changed and TGFβ (5 ng/ml) was added for an additional 18 h. Cells were lysed and luciferase activity determined using the Promega Dual Luciferase Reporter Assay as per the manufacturer’s instructions in a Dynex model ML-2250 luminometer.
Immunoprecipitation, western blot analysis, metabolic labeling and affinity labeling
For immunoprecipitation and western blot analysis, cells were lysed in buffer D and immunoprecipitation carried out as previously described (Hocevar et al., 1999). Nuclear extracts were prepared as previously described (Patil et al., 2000). To detect the de novo synthesis of Dab2, BAHgpt and 903 cells were labeled with 100 µCi/ml of [35S]methionine for 4 h, lysed, and immunoprecipitation performed with a monoclonal antibody to Dab2 (anti-p96; Transduction Laboratories). Immuno complexes were resolved by SDS–PAGE on 10% acrylamide gels, subjected to fluorography and visualized by autoradiography. For pulse–chase analysis, COS7 cells were transfected with WT or MutDab2 using Fugene6 as per the manufacturer’s protocols. After 48 h, cells were labeled with 100 µCi/ml of [35S]methionine for 30 min and either lysed immediately or chased in regular media for various times. Transfected Dab2 was immunoprecipitated with an anti-Flag antibody (M2; Sigma) and resolved as above. Densitometric scanning of the autoradiograph and analysis by NIHImage software were used to quantitate the levels of immunoprecipitated Dab2 protein. TGFβ-stimulated fibronectin and PAI-1 induction were determined as previously described (Hocevar and Howe, 1996). Affinity labeling of confluent cell monolayers with [125I]TGFβ1 (ICN) followed by crosslinking to the receptors with disuccinimidyl suberate (DSS; Pierce) was performed as described (Howe et al., 1990). Detergent-soluble affinity-crosslinked complexes were either analyzed directly or immunoprecipitated with anti-Dab2 antibody (α-p96) prior to separation on 6% SDS–PAGE gels and visualization by PhosphorImager analysis.
GST fusion protein interactions
In vitro 35S-labeled proteins were generated using the T7-Quick coupled TNT system (Promega) as per the manufacturer’s instructions. Equal amounts of labeled proteins were incubated with bacterially expressed GST fusion constructs in binding buffer (50 mM Tris–HCl pH 8.0, 60 mM octylglycoside, 1 mM EDTA, 150 mM NaCl, 1% Triton X-100 and EDTA-free protease inhibitor cocktail Complete; Roche Diagnostics). Following incubation at 4°C for 3 h, complexes were washed six times with binding buffer minus octylglycoside, resolved by SDS–PAGE, subjected to fluorography and visualized by autoradiography.
Acknowledgments
Acknowledgements
We thank Dr Joan Massagué and Dr Scott Kern for the 3TPLux and SBELuc constructs, respectively. We also thank Dr Jeff Wrana for the receptor constructs Smad1, Smad2 MH1 and MH2 GST constructs, and Dr Rik Derynck for GST–Smad4, pRK5 Smad2 and Smad3 plasmids. We thank Drs Bruce Pratt and Steve Ledbetter at Genzyme Inc. for generous provision of TGFβ2. We acknowledge Drs Ed Leof and Jeff Wrana for helpful discussions. This work was supported by grant CA55536 from the National Cancer Institute to P.H.H. and a Scientist Development Grant from the American Heart Association to B.A.H.
References
- Borg J.-P., Ooi,J., Levy,E. and Margolis,B. (1996) The phosphotyrosine interaction domains of X11 and FE65 bind to distinct sites on the YENPTY motif of amyloid precursor protein. Mol. Cell. Biol., 16, 6229–6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J.D., DiChiara,M.R., Anderson,K.R., Gimbrone,M.A.,Jr and Topper,J.N. (1999) MEKK-1, a component of the stress (stress-activated protein kinase/c-Jun N-terminal kinase) pathway, can selectively activate Smad2-mediated transcriptional activation in endothelial cells. J. Biol. Chem., 274, 8797–8805. [DOI] [PubMed] [Google Scholar]
- Carcamo J., Weis,F.M., Ventura,F., Wieser,R., Wrana,J.L., Attisano,L. and Massagué,J. (1994) Type I receptors specify growth-inhibitory and transcriptional responses to transforming growth factor β and activin. Mol. Cell. Biol., 14, 3810–3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Rubock,M.J. and Whitman,M. (1996) A transcriptional partner for MAD proteins in TGF-β signalling. Nature, 383, 691–696. [DOI] [PubMed] [Google Scholar]
- D’Arcangelo G., Homayouni,R., Keshvara,L., Rice,D.S., Sheldon,M. and Curran,T. (1999) Reelin is a ligand for lipoprotein receptors. Neuron, 24, 471–479. [DOI] [PubMed] [Google Scholar]
- de Caestecker M.P., Parks,W.T., Frank,C.J., Castagnino,P., Bottaro,D.P., Roberts,A.B. and Lechleider,R.J. (1998) Smad2 transduces common signals from receptor serine-threonine and tyrosine kinases. Genes Dev., 12, 1587–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennler S., Itoh,S., Vivien,D., ten Dijke,P., Huet,S. and Gauthier,J.-M. (1998) Direct binding of Smad3 and Smad4 to critical TGFβ-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J., 17, 3091–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dho S.E., Jacob,S., Wolting,C.D., French,M.B., Rohrschneider,L.R. and McGlade,C.J. (1998) The mammalian numb phosphotyrosine-binding domain. Characterization of binding specificity and identification of a novel PDZ domain-containing numb binding protein, LNX. J. Biol. Chem., 273, 9179–9187. [DOI] [PubMed] [Google Scholar]
- Doré J.J.E. Jr, Edens,M., Garamszegi,N. and Leof,E.B. (1998) Heteromeric and homomeric transforming growth factor-β receptors show distinct signaling and endocytic responses in epithelial cells. J. Biol. Chem., 273, 31770–31777. [DOI] [PubMed] [Google Scholar]
- Engel M.E., McDonnell,M.A., Law,B.K. and Moses,H.L. (1999) Interdependent SMAD and JNK signaling in transforming growth factor-β-mediated transcription. J. Biol. Chem., 274, 37413–37420. [DOI] [PubMed] [Google Scholar]
- Fazili Z., Sun,W., Mittelstaedt,S., Cohen,C. and Xu,X.-X. (1999) Disabled-2 inactivation is an early step in ovarian tumorigenicity. Oncogene, 18, 3104–3113. [DOI] [PubMed] [Google Scholar]
- Fulop V., Colitti,C.V., Genest,D., Berkowitz,R.S., Yiu,G.K., Ng,S.-W., Szepesi,J. and Mok,S.C. (1998) DOC-2/hDAB2, a candidate tumor suppressor gene involved in the development of gestational trophoblastic diseases. Oncogene, 17, 419–424. [DOI] [PubMed] [Google Scholar]
- Heldin C.H., Miyazono,K. and ten Dijke,P. (1997) TGF-β signalling from cell membrane to nucleus through SMAD proteins. Nature, 390, 465–471. [DOI] [PubMed] [Google Scholar]
- Hocevar B.A. and Howe,P.H. (1996) Isolation and characterization of mutant cell lines defective in transforming growth factor β signaling. Proc. Natl Acad. Sci. USA, 93, 7655–7660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocevar B.A., Brown,T.L. and Howe,P.H. (1999) TGF-β induces fibronectin synthesis through a c-Jun N-terminal kinase-dependent, Smad4-independent pathway. EMBO J., 18, 1345–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe P.H., Cunningham,M.R. and Leof,E.B. (1990) Inhibition of mink lung epithelial cell proliferation by transforming growth factor-β is coupled through a pertussis-toxin-sensitive substrate. Biochem. J., 266, 537–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell B.W., Lanier,L.M., Frank,R., Gertler,F.B. and Cooper,J.A. (1999) The disabled 1 phosphotyrosine-binding domain binds to the internalization signals of transmembrane glycoproteins and to phospholipids. Mol. Cell. Biol., 19, 5179–5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanaugh W.M., Turck,C.W. and Williams,L.T. (1995) PTB domain binding to signaling proteins through a sequence motif containing phosphotyrosine. Science, 268, 1177–1179. [DOI] [PubMed] [Google Scholar]
- Labbe E., Silvestri,C., Hoodless,P.A., Wrana,J.L. and Attisano,L. (1998) Smad2 and Smad3 positively and negatively regulate TGFβ-dependent transcription through the forkhead DNA-binding protein FAST2. Mol. Cell, 2, 109–120. [DOI] [PubMed] [Google Scholar]
- Le Dai J., Turnacioglu,K.K., Schutte,M., Sugar,A.Y. and Kern,S.E. (1998) Dpc4 transcriptional activation and dysfunction in cancer cells. Cancer Res., 58, 4592–4597. [PubMed] [Google Scholar]
- Lee Y.J. et al. (1997) TGF-β suppresses IFN-γ induction of class II MHC gene expression by inhibiting class II transactivator messenger RNA expression. J. Immunol., 158, 2065–2075. [PubMed] [Google Scholar]
- Li S.-C., Zwahlen,C., Vincent,S.J.F., McGlade,C.J., Kay,L.E., Pawson,T. and Forman-Kay,J.D. (1998) Structure of a Numb PTB domain–peptide complex suggests a basis for diverse binding specificity. Nature Struct. Biol., 5, 1075–1083. [DOI] [PubMed] [Google Scholar]
- Liu X., Sun,Y., Constantinescu,S.N., Karam,E., Weinberg,R.A. and Lodish,H.F. (1997) Transforming growth factor β-induced phosphorylation of Smad3 is required for growth inhibition and transcriptional induction in epithelial cells. Proc. Natl Acad. Sci. USA, 94, 10669–10674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macías-Silva M., Abdollah,S., Hoodless,P.A., Pirone,R., Attisano,L. and Wrana,J.L. (1996) MADR2 is a substrate of the TGFβ receptor and its phosphorylation is required for nuclear accumulation and signaling. Cell, 87, 1215–1224. [DOI] [PubMed] [Google Scholar]
- Massagué J. (1998) TGF-β signal transduction. Annu. Rev. Biochem., 67, 753–791. [DOI] [PubMed] [Google Scholar]
- Mok S.C., Wong,K.-K., Chan,R.K.W., Lau,C.C., Tsao,S.-W., Knapp,R.C. and Berkowitz,R.S. (1994) Molecular cloning of differentially expressed genes in human epithelial ovarian cancer. Gynecol. Oncol., 52, 247–252. [DOI] [PubMed] [Google Scholar]
- Mok S.C. et al. (1998) DOC-2, a candidate tumor suppressor gene in human epithelial ovarian cancer. Oncogene, 16, 2381–2387. [DOI] [PubMed] [Google Scholar]
- Morris S.M. and Cooper,J.A. (2001) Disabled-2 colocalizes with the LDLR in clathrin-coated pits and interacts with AP-2. Traffic, 2, 111–123. [DOI] [PubMed] [Google Scholar]
- Oleinikov A.V., Zhao,J. and Makker,S.P. (2000) Cytosolic adaptor protein Dab2 is an intracellular ligand of endocytic receptor gp600/megalin. Biochem. J., 347, 613–621. [PMC free article] [PubMed] [Google Scholar]
- Ong S.H., Guy,G.R., Hadari,Y.R., Laks,S., Gotoh,N., Schlessinger,J. and Lax,I. (2000) FRS2 proteins recruit intracellular signaling pathways by binding to diverse targets on fibroblast growth factor and nerve growth factor receptors. Mol. Cell. Biol., 20, 979–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil S., Wildey,G.M., Brown,T.L., Choy,L., Derynck,R. and Howe,P.H. (2000) Smad7 is induced by CD40 and protects WEHI 231 B-lymphocytes from TGFβ-induced growth inhibition and apoptosis. J. Biol. Chem., 275, 38363–38370. [DOI] [PubMed] [Google Scholar]
- Pawson T. and Scott,J.D. (1997) Signaling through scaffold, anchoring, and adaptor proteins. Science, 278, 2075–2080. [DOI] [PubMed] [Google Scholar]
- Piek E., Heldin,C.-H. and Dijke,P.T. (1999) Specificity, diversity, and regulation in TGF-β superfamily signaling. FASEB J., 13, 2105–2124. [PubMed] [Google Scholar]
- Schwahn D.J. and Medina,D. (1998) p96, a MAPK-related protein, is consistently downregulated during mouse mammary carcinogenesis. Oncogene, 17, 1173–1178. [DOI] [PubMed] [Google Scholar]
- Tseng C.-P., Ely,B.D., Li,Y., Pong,R.-C. and Hsieh,H.-T. (1998) Regulation of rat DOC-2 gene during castration-induced rat ventral prostate degeneration and its growth inhibitory function in human prostatic carcinoma cells. Endocrinology, 139, 3542–3553. [DOI] [PubMed] [Google Scholar]
- Tsukazaki T., Chiang,T.A., Davison,A.F., Attisano,L. and Wrana,J.L. (1998) SARA, a FYVE domain protein that recruits Smad2 to the TGFβ receptor. Cell, 95, 779–791. [DOI] [PubMed] [Google Scholar]
- van der Geer P. and Pawson,T. (1995) The PTB domain: a new protein module implicated in signal transduction. Trends Biochem. Sci., 20, 277–280. [DOI] [PubMed] [Google Scholar]
- Wieser R., Wrana,J.L. and Massagué,J. (1995) GS domain mutations that constitutively activate TβR-I, the downstream signaling component in the TGF-β receptor complex. EMBO J., 14, 2199–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrana J.L., Attisano,L., Carcamo,J., Zentella,A., Doody,J., Laiho,M., Wang,X.F. and Massagué,J. (1992) TGFβ signals through a heteromeric protein kinase receptor complex. Cell, 71, 1003–1014. [DOI] [PubMed] [Google Scholar]
- Wrana J.L., Attisano,L., Wieser,R., Ventura,F. and Massagué,J. (1994) Mechanism of activation of the TGF-β receptor. Nature, 370, 341–347. [DOI] [PubMed] [Google Scholar]
- Xu L., Chen,Y.-G. and Massagué,J. (2000) The nuclear import function of Smad2 is masked by SARA and unmasked by TGFβ-dependent phosphorylation. Nature Cell Biol., 2, 559–562. [DOI] [PubMed] [Google Scholar]
- Xu X.-X., Yang,W., Jackowski,S. and Rock,C.O. (1995) Cloning of a novel phosphoprotein regulated by colony-stimulating factor 1 shares a domain with the Drosophila disabled gene product. J. Biol. Chem., 270, 14184–14191. [DOI] [PubMed] [Google Scholar]
- Xu X.-X., Yi,T., Tang,B. and Lambeth,J.D. (1998) Disabled-2 (Dab2) is an SH3 domain-binding partner of Grb2. Oncogene, 16, 1561–1569. [DOI] [PubMed] [Google Scholar]
- Yajnik V., Blaikie,P., Bork,P. and Margolis,B. (1996) Identification of residues within the SHC phosphotyrosine binding/phosphotyrosine interaction domain crucial for phosphopeptide interaction. J. Biol. Chem., 271, 1813–1816. [DOI] [PubMed] [Google Scholar]
- Yu H., Chen,J.K., Feng,S., Dalgarno,D.C., Brauer,A.W. and Schreiber,S.L. (1994) Structural basis for the binding of proline-rich peptides to SH3 domains. Cell, 76, 933–945. [DOI] [PubMed] [Google Scholar]
- Zawel L., Le Dai,J., Buckhaults,P., Zhou,S., Kinzler,K.W., Vogelstein, B. and Kern,S.E. (1998) Human Smad3 and Smad4 are sequence-specific transcription activators. Mol. Cell, 1, 611–617. [DOI] [PubMed] [Google Scholar]
- Zhang Y. and Derynck,R. (1999) Regulation of Smad signalling by protein associations and signalling crosstalk. Trends Cell Biol., 9, 274–279. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Feng,X., We,R. and Derynck,R. (1996) Receptor-associated Mad homologues synergize as effectors of the TGF-β response. Nature, 383, 168–172. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Feng,X.-H. and Derynck,R. (1998) Smad3 and Smad4 cooperate with c-Jun/c-Fos to mediate TGF-β-induced transcription. Nature, 394, 909–913. [DOI] [PubMed] [Google Scholar]
- Zhou M.-M. et al. (1995) Structure and ligand recognition of the phosphotyrosine binding domain of Shc. Nature, 378, 584–592. [DOI] [PubMed] [Google Scholar]