Abstract
Arabidopsis Snf1-related protein kinases (SnRKs) are implicated in pleiotropic regulation of metabolic, hormonal and stress responses through their interaction with the kinase inhibitor PRL1 WD-protein. Here we show that SKP1/ASK1, a conserved SCF (Skp1-cullin-F-box) ubiquitin ligase subunit, which suppresses the skp1-4 mitotic defect in yeast, interacts with the PRL1-binding C-terminal domains of SnRKs. The same SnRK domains recruit an SKP1/ASK1-binding proteasomal protein, α4/PAD1, which enhances the formation of a trimeric SnRK complex with SKP1/ASK1 in vitro. By contrast, PRL1 reduces the interaction of SKP1/ASK1 with SnRKs. SKP1/ASK1 is co-immunoprecipitated with a cullin SCF subunit (AtCUL1) and an SnRK kinase, but not with PRL1 from Arabidopsis cell extracts. SKP1/ASK1, cullin and proteasomal α-subunits show nuclear co-localization in differentiated Arabidopsis cells, and are observed in association with mitotic spindles and phragmoplasts during cell division. Detection of SnRK in purified 26S proteasomes and co-purification of epitope- tagged SKP1/ASK1 with SnRK, cullin and proteasomal α-subunits indicate that the observed protein interactions between SnRK, SKP1/ASK1 and α4/PAD1 are involved in proteasomal binding of an SCF ubiquitin ligase in Arabidopsis.
Keywords: Arabidopsis Pleiotropic Regulatory Locus 1 (PRL1)/proteasome/SCF ubiquitin ligase/SKP1/ASK1/Snf1-related protein kinase
Introduction
AMP-activated protein kinases (AMPKs) modulated by changes in the cellular AMP/ATP ratio are important regulators of metabolic and stress responses in eukaryotes (Hardie and Carling, 1997). Members of the AMPK family recognize similar substrates and consist of homologous core subunits that show a remarkable structural and functional conservation from yeast to humans (Hardie et al., 1998; Kemp et al., 1999; Vaulon et al., 2000). A prototype of AMPKs is the Snf1 (sucrose non-fermenting) protein kinase in budding yeast. Snf1 is required for proper transcriptional control of genes that are repressed when yeast cells grow in the presence of glucose and induced in response to glucose starvation and stress (Carlson, 1998). Snf1 is also implicated in the regulation of key metabolic enzymes and essential cellular processes, including mitochondrial and peroxisome biogenesis, nuclear import, thermotolerance and meiosis (Carlson, 1999). Recently, Snf1 has been identified as a component of Srb/mediator complex of RNA polymerase II (Kutchin et al., 2000).
The conservation of the Snf1/AMPK family is illustrated by the observation that the catalytic and activator subunits of type I Snf1-related protein kinases (SnRKs) from higher plants suppress the deficiency of corresponding yeast proteins (Alderson et al., 1991; Muranaka et al., 1994; Jiang and Carlson, 1996, 1997; Kleinow et al., 2000). Despite an involvement of plant SnRKs in the regulation of some rate-limiting metabolic enzymes, their function in signalling is largely unknown (Halford and Hardie, 1998; Smeekens, 1998; Ikeda et al., 2000). Recently, Arabidopsis SnRKs AKIN10 and AKIN11 have been indirectly connected to sugar, hormone and stress signalling through the Pleiotropic Regulatory Locus 1 (PRL1). The prl1 mutation results in transcriptional derepression of many sucrose-regulated genes and causes arrested root elongation, altered leaf development and inhibition of cell elongation. In addition, the prl1 mutation results in hypersensitivity to glucose, sucrose, cold temperature, and the plant hormones cytokinin, auxin, ethylene and abscisic acid (Németh et al., 1998). PRL1 encodes an α-importin-binding nuclear WD-protein that interacts with yeast Snf1 and plant SnRKs in the two-hybrid system. Binding of PRL1 to the C-terminal regulatory domains of kinase catalytic subunits in vitro inhibits the activity of Arabidopsis SnRKs AKIN10 and AKIN11 (Bhalerao et al., 1999). In correlation with the pleiotropic phenotype, an enhanced activation of Arabidopsis SnRKs in the prl1 mutant is postulated to affect several regulators of cellular signalling.
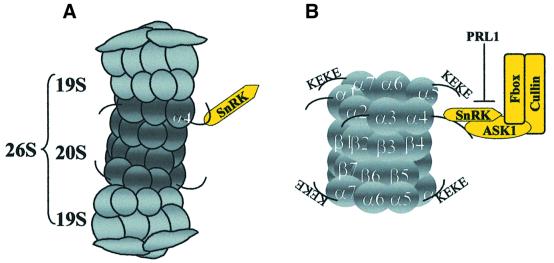
Here we show that an Arabidopsis SnRK, corresponding to either AKIN10 or AKIN11, co-purifies in the presence of ATP with the 26S proteasome, which is a large multisubunit complex controlling proteolytic degradation of ubiquitylated intracellular proteins. The 26S proteasome consists of a cylinder-shaped 20S proteolytic core and two 19S regulatory cap particles. The 20S core particle carries two copies each of seven different α and β subunits arranged in four α7β7β7α7 stacked rings (Hochstrasser, 1996; De Mot et al., 1999; Verma and Deshaies, 2000). We have found that Arabidopsis SnRKs interact with the α4/PAD1 subunit of the 20S proteasome (Fu et al., 1998; Rechsteiner, 1998; Voges et al., 1999) and the Skp1/ASK1 subunit of an SCF (Skp1-cullin-F-box) E3 ubiquitin ligase. SCF represents a conserved class of E3 enzymes that consists of core Skp1, F-box protein, Cdc53/cullin and Rbx1/Roc1 subunits, and carries an associated E2 ubiquitin conjugase in eukaryotes (Deshaies, 1999; Tyers and Jorgensen, 2000). SCF-mediated ubiquitylation and subsequent degradation of proteins occur in sequential steps. Following activation by an E1 enzyme, ubiquitin is transferred to an E2 enzyme and then to phosphorylated substrates that are specifically recognized and recruited to SCF by different F-box proteins (Hersko and Ciechanover, 1998; Patton et al., 1998). It is noteworthy that Arabidopsis SnRKs, like many E2 and E3 enzymes and ubiquitin C-terminal hydrolases, carry a ubiquitin- associated UBA domain that is postulated to confer target specificity in protein interactions (Hofmann and Bucher, 1996; Withers-Ward et al., 2000).
As in yeast and mammals, SCF components are also implicated in the regulation of essential signalling pathways in plants (del Pozo and Estelle, 1999, 2000). The Arabidopsis SCFTIR1 complex, which carries SKP1/ASK1, AtCUL1 cullin and TIR1 F-box protein subunits, plays a distinguished role in the regulation of growth responses to the plant hormone auxin (Ruegger et al., 1998; Gray et al., 1999). The ask1 mutation causes male sterility due to a chromosome segregation defect during male meiosis, whereas both tir1 and ask1 mutations result in reduction of auxin responses (Gray et al., 1999; Yang et al., 1999). SKP1/ASK1 interacts in the two-hybrid system with several Arabidopsis F-box factors that are not yet characterized in SCF complexes in vivo. These F-box factors include UFO1 (Samach et al., 1999; Zhao et al., 1999) and COI1 (Creelman, 1998; Xie et al., 1998), which control floral organ identity and jasmonate-regulated defence responses, respectively, and several other SKP1/ASK1-interacting proteins (SKIPs) described here. Association of SKP1/ASK1 with an SnRK protein kinase reinforces a key element of the SCF paradigm (Bai et al., 1996; Skowyra et al., 1997), suggesting a requirement for phosphorylation of SCF substrates. In addition, interactions of SnRKs and SKP1/ASK1 with the α4/PAD1 proteasomal subunit, and co-immunoprecipitation of 20S proteasome α-subunits with SnRK, SKP1/ASK1 and cullin, indicate that an SCF ubiquitin ligase forms a proteasomal complex in Arabidopsis.
Results
Arabidopsis SnRKs AKIN10 and AKIN11 interact with the SCF subunit SKP1/ASK1 and 20S proteasome subunit α4/PAD1
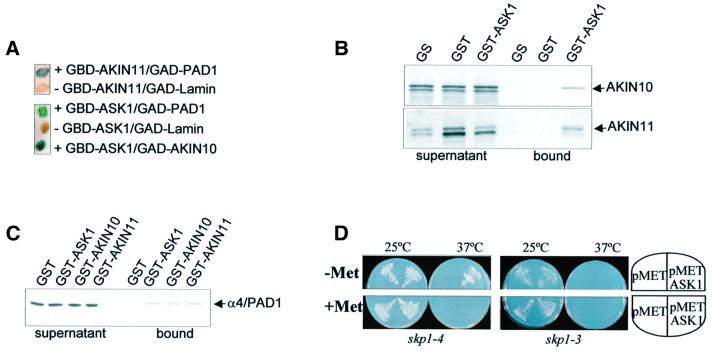
Previous studies demonstrated that the catalytic subunits of Arabidopsis SnRKs AKIN10 and AKIN11 interact through their C-terminal regulatory domains with the yeast Snf1 kinase subunits Snf4 and Sip1, and their plant orthologues in the two-hybrid system (Jiang and Carlson, 1996, 1997; Bouly et al., 1999; Kleinow et al., 2000). The regulatory domains of AKIN10 and AKIN11 were also shown to bind the Arabidopsis PRL1 WD-protein, which is an SnRK inhibitor in vitro. A role for SnRKs in signalling was suggested by genetic analysis of the prl1 mutation, which leads to enhanced activation of SnRKs, transcriptional derepression of sucrose-regulated genes, and hypersensitivity to several plant hormones, including auxin (Németh et al., 1998; Bhalerao et al., 1999). To search for novel SnRK-binding factors, AKIN10 and AKIN11 in fusion with the Gal4 DNA-binding domain (GBD) were used as baits in yeast two-hybrid screens with a pACT2 cDNA library made from cultured Arabidopsis cells (Németh et al., 1998). From 5 × 107 transformants in each screen, 59 clones with AKIN10 and 52 clones with AKIN11 were obtained that grew in the presence of 50 mM 3-aminotriazole (3-AT) HIS3-inhibitor and displayed LacZ reporter enzyme activity (Durfee et al., 1993). Following control self-activation tests and interaction assays with a range of unspecific baits (Németh et al., 1998; data not shown), sequencing of prey cDNAs identified 13 protein classes that showed two-hybrid interactions with both AKIN10 and AKIN11. One class of AKIN-interacting factors described here was represented by five cDNAs that all encoded a full-length Arabidopsis SCF ubiquitin ligase subunit (Gray et al., 1999), SKP1/ASK1 (S phase kinase-associated protein/Arabidopsis Skp1-like; DDBJ/EMBL/GenBank accession No. AF059294), in fusion with the Gal4 activator domain (GAD). Another class included a single cDNA that encoded a peptide carrying 93 C-terminal amino acids from the Arabidopsis 20S proteasome subunit α4/PAD1 (Fu et al., 1998; DDBJ/EMBL/GenBank accession No. AF043522). A previously published full-length α4/PAD1 cDNA (TAS-g64; Genschik et al., 1992) was used to construct a GAD–PAD1 prey. In control two-hybrid assays, GAD–PAD1 showed interaction with GBD– AKIN11 as expected, as well as with a GBD–ASK1 bait, indicating that α4/PAD1 could also bind to SKP1/ASK1. A reversal of bait–prey combination (GBD–ASK1 versus GAD–AKIN10) confirmed interaction of SKP1/ASK1 with AKIN10 (Figure 1A).
Fig. 1. SKP1/ASK1 interacts with the α4/PAD1 subunit of 20S proteasome and Snf1-related protein kinases AKIN10 and AKIN11 in the two-hybrid system and in vitro, and suppresses the yeast skp1-4 mutation. (A) LacZ filter assays show two-hybrid interactions of GBD–ASK1 with GAD–PAD1 and GAD–AKIN10, as well as GBD–AKIN11 with GAD–PAD1, but no interactions of GBD baits with a control GAD–lamin prey. (B) [35S]methionine-labelled AKIN10 and AKIN11 loaded in equal amounts (supernatant fractions) show specific binding in vitro to GST–ASK1, but not to control GS and GST matrices (bound fractions). In addition to full-size AKIN10 and AKIN11, artificial early termination of in vitro transcription–translation led to the synthesis of smaller truncated proteins (see supernatant fractions). No binding of shorter translation products to GST–ASK1 indicates that they correspond to C-terminal truncated forms of AKIN10 and AKIN11 that lack the SKP1/ASK1-binding site. (C) [35S]methionine-labelled α4/PAD1 is specifically retained on GST–ASK1, GST–AKIN10 and GST–AKIN11 resins, but not on the control GST matrix, in protein-binding assays in vitro. (D) Expression of SKP1/ASK1 by a methionine-repressible Met25 promoter (pMET-ASK1) rescues the growth defect of thermosensitive skp1-4 yeast mutant at non-permissive temperature (37°C) in methionine-free medium (–Met), but not in the presence of 1 mM methionine (+Met). By contrast, SKP1/ASK1 does not suppress the growth defect of yeast skp1-3 mutant. As controls, the skp1-3 and skp1-4 mutants were transformed with an empty p426Met25 vector (pMET; Mumberg et al., 1994).
The specificity of observed protein interactions was tested by in vitro pull-down assays using glutathione S-transferase (GST) fusion proteins. AKIN10 and AKIN11 labelled with [35S]methionine by coupled in vitro transcription–translation were incubated with GST– ASK1 and GST proteins immobilized on glutathione– Sepharose (GS), and with the GS matrix alone. AKIN10 and AKIN11 were specifically retained on GST–ASK1, but failed to bind to the control GST and GS resins (Figure 1B). The α4/PAD1 protein was similarly labelled with [35S]methionine and loaded on GS matrices carrying immobilized GST–ASK1, GST–AKIN10, GST–AKIN11 and GST proteins. α4/PAD1 showed specific binding in vitro to GST–AKIN10, GST–AKIN11 and GST–ASK1, but not to the control GST protein, supporting the results of yeast two-hybrid protein interaction assays (Figure 1B).
A two-hybrid screen performed with a GBD–ASK1 bait resulted in the identification of Arabidopsis cDNAs encoding AKIN10. In addition, seven cDNA classes of SKIPs were characterized. Six SKIP cDNAs encoded F-box proteins (Table I). These carried either leucine-rich repeats (SKIPs 1 and 2), or kelch domains (SKIPs 4 and 6; Adams et al., 2000), or no known C-terminal motifs (SKIP3 and 5). SKIP7 corresponded to fibrillarin (AtFIB1), an Arabidopsis orthologue of the nucleolar NOP1 protein (Barneche et al., 2000; Pih et al., 2000). Measurement of LacZ reporter enzyme activity in yeast cells grown under glucose limitation and on 2% glucose (Table I) indicated that all SKIP factors, except for SKIP5, interacted with SKP1/ASK1 in a glucose-regulated fashion, as was previously reported for two-hybrid interactions of yeast SCF components (Li and Johnston, 1997).
Table I. Properties of SKIPs.
| SKIP | Interaction with SKP1/ASK1 (β-GAL units) |
Structural domains | Accession No. | |
|---|---|---|---|---|
| 0.05% glucose | 2% glucose | |||
| AKIN10 | 1688.1 ± 56.8 | 266.1 ± 1.0 | SnRK | M93023 |
| AKIN11 | 113.7 ± 14.6 | 24.4 ± 0.9 | SnRK | X99279 |
| SKIP1 | 426.9 ± 33.3 | 206.4 ± 6.8 | F-box, LRR | AF263377 |
| SKIP2 | 91.4 ± 5.3 | 4.9 ± 1.4 | F-box, LRR | AF263378 |
| SKIP3 | 555.2 ± 54.3 | 58.5 ± 17.5 | F-box | AF263379 |
| SKIP4 | 1011.9 ± 56.8 | 369.1 ± 22.1 | F-box, kelch | AF263380 |
| SKIP5 | 125.8 ± 11.0 | 144.6 ± 37.7 | F-box | AF263382 |
| SKIP6 | 481.9 ± 42.5 | 20.6 ± 0.4 | F-box, kelch | AF263381 |
| SKIP7 | 636.5 ± 10.6 | 366.3 ± 17.1 | fibrillarin | AF263383 |
Glucose regulation of bait–prey interactions was assayed by measurement of β-galactosidase enzyme activities (β-GAL units).
LRR, leucine-rich repeats; kelch, kelch repeats.
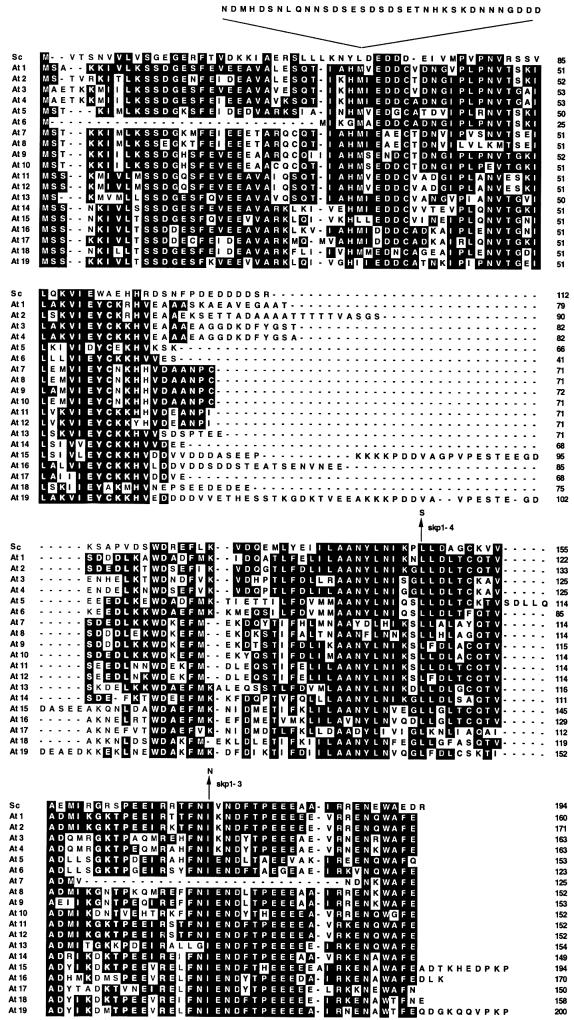
SKP1/ASK shared 35% sequence identity with budding yeast Skp1, which reflected a significant conservation of C-terminal protein domains. Amino acid positions corresponding to the yeast skp1-3 (I-172-N) and skp1-4 (L-146-S) mutations (Figure 2), causing G1–S and mitotic defects, respectively (Connelly and Hieter, 1996), were conserved within this region of SKP1/ASK1. Expression of SKP1/ASK1 by a methionine-repressible Met25 promoter (Mumberg et al., 1994) suppressed only the growth defect of the skp1-4, but not that of the skp1-3 thermosensitive yeast mutant, indicating an incomplete functional conservation between yeast Skp1 and Arabidopsis SKP1/ASK1 (Figure 1D). Another functional difference between Skp1 and SKP1/ASK1 was shown by control two-hybrid assays. These failed to reveal interactions between SKP1/ASK1 and yeast Snf1, as well as between yeast Skp1 and Snf1 (data not shown). By analogy, one of the 19 Arabidopsis SKP1/ASK homologues (Figure 2; At5), SKP1_5/ASK5, which carried a unique insertion of five amino acids (SDLLQ) in the C-terminal F-box-binding domain, displayed no interaction with AKIN10 and AKIN11, indicating that not all Arabidopsis SKP1 homologues shared the SnRK-binding property of SKP1/ASK1.
Fig. 2. Amino acid sequence comparison of yeast Skp1 (Sc) and 19 Arabidopsis SKP1/ASK1 homologues (At1–19). Conserved positions of amino acid residues corresponding to the yeast skp1-3 and skp1-4 mutations are indicated by arrows in the sequence alignment, which includes yeast Skp1 [194 amino acids (aa); AAB64763], and Arabidopsis Skp1/ASK1 sequences corresponding to the following accession Nos, BAC/P1 clones and predicted genes: 1 (160 aa, AAF26761, T4O12_17, At1g75950); 2 (171 aa, BAB08452, MJC20_30, At5g42190); 3 (163 aa, AAD31370, F3N11_15, At2g25700); 4 (163 aa, AAF79899, T20H2_8, At1g20140); 5 (153 aa, CAB75821, F24G16_290, At3g60020); 6 (123 aa, instead of annotated 85 aa, CAB86910, F8J2_230, At3g53060); 7 (125 aa, BAB00221, MSD21_15, At3g21840); 8 (152 aa, BAB00220, MSD21_14, At3g21830); 9 (153 aa, BAB00222, MSD21_16, At3g21850); 10 (152 aa, BAB00223, MSD21_17, At3g21860); 11 (152 aa, CAA17551, F28A23_30, At4g34210); 12 (152 aa, CAA18826, T4L20_50, At4g34470); 13 (154 aa, CAB75820, F24G16_280, At3g60010); 14 (149 aa, AAC34485, T18E12_16, At2g03170); 15 (194 aa, instead of annotated 177 aa, BAB00602, T5M7_7, At3g25650); 16 (170 aa, AAC34483, T18E12_14, At2g03190); 17 (150 aa, AAD24382, T2G17_4, At2g20160); 18 (158 aa, instead of annotated 183 aa, AAD32873, F14N23_11, At1g10230); and 19 (200 aa, AAC34486, T18E12_17, At2g03160). A longer SKP1-related sequence (AAC28530, F4I18_7, At2g45950, 300 aa) was not included in the alignment.
SKP1/ASK1 is recruited by the PRL1-binding regulatory domains of SnRKs
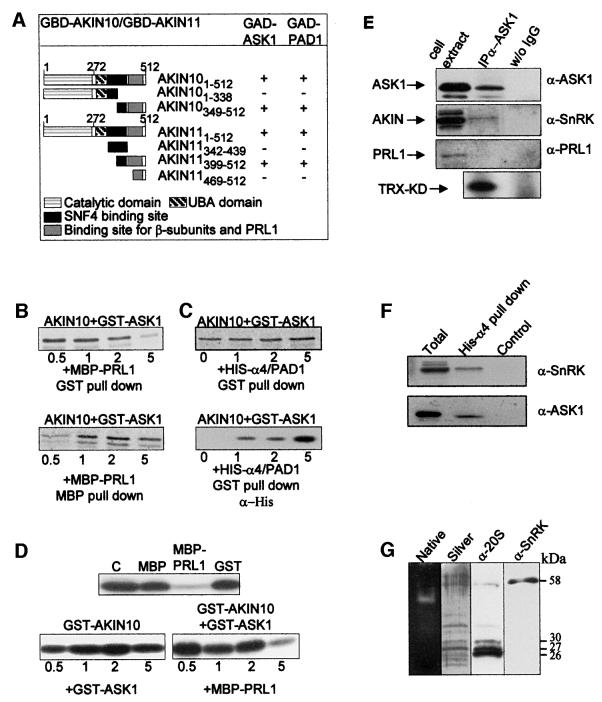
The SKP1/ASK1-binding regions of SnRKs were mapped by two-hybrid interaction assays with a GAD–ASK1 prey using a series of baits encoding different segments of AKIN10 and AKIN11 (Figure 3A). GAD–ASK1 interacted with homologous C-terminal SnRK peptides, which were located downstream of the UBA (Hofmann and Bucher, 1996) and yeast Snf4-binding regions (Bhalerao et al., 1999), between amino acid positions 349 and 512 in AKIN10, and 399 and 512 in AKIN11. These SKP1/ASK1-binding domains of AKIN10 and AKIN11 were previously observed to interact with the SnRK β and γ subunits (Kleinow et al., 2000; A.Ferrando, unpublished) and kinase inhibitor PRL1 WD-protein (Bhalerao et al., 1999). The mapping data therefore suggested that occupation of the common SnRK-binding sites by either of these factors would exclude binding of another factor to the same regions in AKIN10 and AKIN11.
Fig. 3. SKP1/ASK1 and α4/PAD1 interact with C-terminal domains of AKIN10 and AKIN11. In vitro binding of SnRKs to SKP1/ASK1 is competed by PRL1, but enhanced by α4/PAD1, which selectively recruits SKP1/ASK1 and SnRK from Arabidopsis cell extracts. Unlike PRL1, SnRK is co-immunoprecipitated with SKP1/ASK1 and co-purifies with 26S proteasome. (A) Mapping of SKP1/ASK1 and α4/PAD1 binding domains of AKIN10 and AKIN11 by two-hybrid interaction assays. The results of LacZ filter assays (+ or –) indicate interactions of GBD baits, expressing different segments of AKIN10 and AKIN11 (amino acid positions are indicated in subscript), with GAD–ASK1 and GAD–PAD1 preys. Boxes indicate the positions of known SnRK domains. (B) In vitro SnRK-binding assay with SKP1/ASK1 and PRL1. A preformed [35S]AKIN10–GST–ASK1 complex was challenged with increasing amounts of MBP–PRL1. Equal aliquots from each sample were bound to GS (GST pull-down) and amylose–agarose (MBP pull-down) to monitor the amount of [35S]AKIN10 present in complex with GST–ASK1 and MBP–PRL1, respectively. Recruitment of a C-terminally truncated form of AKIN10 by MBP–PRL1 (lower band in MPB pull-down assay), but not by GST–ASK1, indicates that PRL1 can also interact with AKIN10 sequences located upstream of the C-terminal SKP1/ASK1 binding site. (C) In vitro SnRK-binding assay with SKP1/ASK1 and α4/PAD1. [35S]AKIN10 was saturated with GST–ASK1, then increasing amounts of His-α4/PAD1 were added to the samples that were bound to GS. Following SDS–PAGE separation of eluted proteins, the amounts of GST–ASK1-associated [35S]AKIN10 and His-α4/PAD1 proteins were monitored by autoradiography and western blotting with an anti-His6 antibody, respectively. (D) In vitro kinase competition assay with SKP1/ASK1 and PRL1. Upper panel, phosphorylation of TRX-KD substrate by GST–AKIN10 alone (C) and in the presence of MBP, MBP–PRL1 and GST proteins. Lower panels, GST–AKIN10 was either incubated with increasing amounts of GST–ASK1 (left panel) or pre-incubated with GST–ASK1 followed by addition of increasing amounts of MBP–PRL1 (right panel) before performing the kinase assays with the TRX-KD substrate. (E) Protein extract from Arabidopsis Col-0 cells was bound to immobilized α-ASK1 IgG and protein A–Sepharose resins. Aliquots from the cell extract and proteins eluted from the IgG matrix (IP α-ASK1) and control protein A beads (w/o IgG) were immunoblotted with α-ASK1, α-SnRK and α-PRL1 antibodies and subjected to SnRK kinase assays. (F) Protein extract from Arabidopsis Col-0 cells was bound to His-α4/PAD1 immobilized on Ni-NTA–agarose and to control Ni-NTA-resin. The cell extract (Total) and protein fractions eluted from the His-α4/PAD1 (His-α4 pull-down) and Ni-NTA (Control) beads were immunoblotted with α-SnRK and α-ASK1 antibodies. (G) Purified 26S proteasome separated and stained in a non-denaturing polyacrylamide gel (Native) was eluted for separation of subunits by SDS–PAGE, which was either silver stained (Silver) or immunoblotted with α-20S proteasome and α-SnRK antibodies. Expected molecular masses for SnRK (AKIN10 or AKIN11) and proteasomal α-subunits are indicated.
To test this hypothesis, a competitive AKIN10 kinase-binding assay was performed with SKP1/ASK1 and PRL1 (Figure 3B). [35S]methionine-labelled AKIN10 was bound to GST–ASK1, then the complex was challenged with increasing amounts of PRL1 in fusion with a maltose-binding protein (MBP–PRL1). Titration of [35S]AKIN10, using a selective pull-down of GST–ASK1 with GS and MBP–PRL1 with amylose resin, showed that increasing the amount of competitor MBP–PRL1 protein resulted in concomitant loss of AKIN10 from the GST–ASK1-bound fraction and accumulation of AKIN10 in MBP–PRL1-bound form. These data indicated that PRL1 could recruit AKIN10 by disrupting its interaction with SKP1/ASK1. Protein kinase assays performed under similar conditions showed that saturation of GST–AKIN10 with increasing amounts of GST–ASK1, as well as with control MBP and GST proteins, did not alter the AKIN10 kinase activity. By contrast, MBP–PRL1 efficiently inhibited the GST– AKIN10 kinase (Figure 3D). These assays also revealed that neither SKP1/ASK1 nor PRL1 served as substrate for the AKIN10 kinase. Competition of a preformed GST– AKIN10/GST–ASK1 complex with increasing amounts of MBP–PRL1 resulted in a gradual inhibition of the kinase activity, reflecting a recruitment of AKIN10 by PRL1. As PRL1 was also found to inhibit AKIN11 (Bhalerao et al., 1999), these results suggested that the SnRK inhibitor PRL1 WD-protein and SKP1/ASK1 may not occur in common SnRK complexes.
To support this conclusion, proteins extracted from cultured Arabidopsis cells were immunoprecipitated with an antibody (α-ASK1) raised against a peptide carrying the last 21 C-terminal amino acids of SKP1/ASK1 (Figure 2). Western blotting of immunoprecipitated proteins with an α-SnRK antibody, recognizing both AKIN10 and AKIN11, showed that SKP1/ASK1 co-immunoprecipitated with a protein kinase that specifically phosphorylated the SnRK substrate peptide TRX-KD derived from sucrose-phosphate synthase (Bhalerao et al., 1999). By contrast, proteins immunoprecipitated with the α-ASK1 antibody did not cross-react with the anti-PRL1 antibody (α-PRL1; Figure 3E). These data were consistent with control experiments showing that the α-PRL1 antibody (Németh et al., 1998) immunoprecipitated an active SnRK, but not SKP1/ASK1, from the same cell extracts (data not shown).
SnRK is associated with the 26S proteasome
The α4/PAD1-binding SnRK domains were mapped as described above by assaying two-hybrid interactions of a GAD–PAD1 prey with GBD baits encoding different segments of AKIN10 and AKIN11 (Figure 3A). Similarly to SKP1/ASK1, GAD–PAD1 was found to interact with the PRL1-binding regions of SnRKs. To determine how saturation of the SnRK-binding site with SKP1/ASK1 affects subsequent binding of α4/PAD1, a kinase competition assay was performed. [35S]methionine-labelled AKIN10 was saturated by incubation with an excess of GST–ASK1. The preformed [35S]AKIN10–GST–ASK1 complex was then challenged with increasing amounts of His-α4/PAD1 protein, which carried an N-terminal His6 tag. The amount of [35S]AKIN10 in complex with GST–ASK1 was measured by pull-down assays with GS, whereas the amount of His-α4/PAD1 in the GST-bound fractions was monitored by immunoblotting with an α-His antibody (Figure 3C). A gradual increase in the amount of both [35S]AKIN10 and His-α4/PAD1 proteins in the GST–ASK1-bound fractions indicated that the α4/PAD1 protein, unlike PRL1, did not compete with SKP1/ASK1 for binding of the AKIN10 kinase. Rather, increasing the amount of α4/PAD1 proportionally increased the quantity of GST–ASK1-bound AKIN10 kinase. The His-α4/PAD1 protein could thus either recruit some residual free form of [35S]AKIN10 after saturation with GST–ASK1 or, more likely, increase the efficiency of kinase binding by forming a complex with GST–ASK1. To demonstrate the selectivity of α4/PAD1 interactions, protein extract prepared from cultured Arabidopsis cells was subjected to chromatography on Ni-NTA–agarose carrying immobilized His-α4/PAD1 protein. Whereas no specific protein binding was observed to the control empty Ni-NTA matrix, both SnRK and SKP1/ASK1 proteins were detected in the His-α4/PAD1-bound protein fraction by immunoblotting with α-SnRK and α-ASK1 antibodies (Figure 3F).
As observed for PRL1 and SKP1/ASK1, protein kinase assays with GST–AKIN10 and GST–AKIN11 indicated that α4/PAD1 was not a substrate for Arabidopsis SnRKs (data not shown). Consequently, α4/PAD1 appeared to be a proteasomal SnRK-binding protein. To test this possibility, proteasome was purified in the presence of ATP from cultured Arabidopsis cells by Q-Sepharose chromatography. The proteasomes were resolved to 26S and 20S fractions on a non-denaturing polyacrylamide gel stained by the fluorescent peptidase substrate SUC-LLVY-AMC (Umeda et al., 1997a; Glickman et al., 1998). The excised 26S proteasomal bands yielded a highly purified protein fraction in which a monoclonal anti-20S proteasome antibody (α-20S) detected the conserved core subunits α2 and α5 (both ∼26 kDa), α1, α3 and α7 (each ∼27 kDa), and α6 (30 kDa), in addition to cross-reacting with two larger subunits (Figure 3G). Western blotting with α-SnRK antibody revealed the presence of an SnRK with an expected molecular mass (58 kDa) in the 26S proteasomal fraction, clearly indicating that SnRK was not ubiquitylated as a potential proteasomal substrate. Together with the protein interaction data, this suggested that specific binding of SnRK by the α4/PAD1 proteasome subunit could mediate a recruitment of SKP1/ASK1 in potential association with other SCF ubiquitin ligase subunits.
Cellular co-localization of proteasomal and SCF subunits
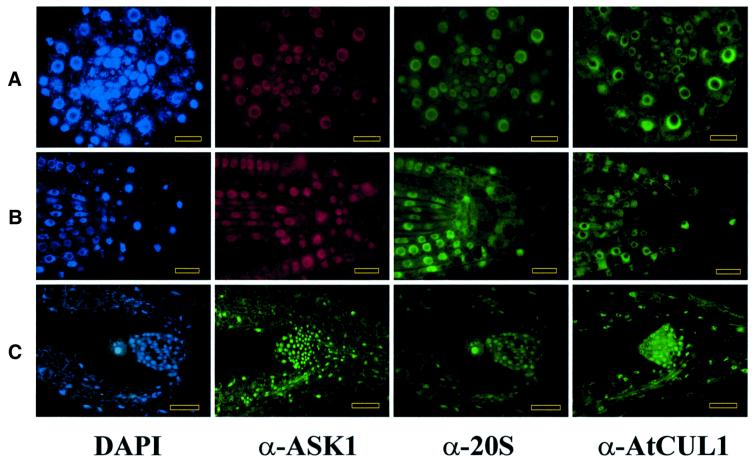
Cellular localization studies lent further support to the protein interaction data suggesting a proteasomal association of SCF ubiquitin ligase subunits. The immunofluorescence staining patterns of antibodies, which detected the proteasomal α-subunits and SCF subunits SKP1/ASK1 and cullin AtCUL1 (Gray et al., 1999), perfectly overlapped with each other and corresponded to the patterns of 4′,6-diamidino-2-phenylindole (DAPI)-stained nuclei in different Arabidopsis tissues (Figure 4). The antibodies did not stain the nucleoli, but labelled nuclear particles of ∼150 nm and a few larger speckles of ∼300 nm. Weak cytoplasmic staining was detected only in cells of meristems in which strong labelling of nuclei disappeared in late prophase of mitosis. In metaphase, the staining patterns with all three antibodies overlapped with the positions of mitotic spindles visualized by an anti-tubulin antibody, but not with the positions of DAPI-stained chromosomes (Figure 5A–C). This staining pattern was abolished by oryzalin treatment of tissues, which disrupted the mitotic spindles. In roots and shoot apices, and other vegetative and reproductive organs, the staining patterns did not overlap with peripheral and perinuclear networks of actin and tubulin (data not shown). In early anaphase, the signals were still visible on the poles of mitotic spindles, but also between the unstained moving chromosome groups. In late anaphase and telophase, the signals detected by the antibodies became equatorial and overlapped with the phragmoplasts, but not with the newly forming cytoplasmic plates (Figure 5D and E). Staining of daughter nuclei was first observed when the chromatids were decondensed and surrounded by newly formed nuclear membranes.
Fig. 4. SKP1/ASK1, AtCUL1 and proteasomal α-subunits show nuclear co-localization in Arabidopsis roots and shoots. (A) Transverse section through the meristematic zone of root (bars = 10 µm). (B) Longitudinal cross-section of root apex (bars = 10 µm). (C) Longitudinal section through the shoot apex (bars = 25 µm). DAPI, nuclei stained with the DNA dye DAPI (blue). α-ASK1, sections treated with immunoaffinity-purified polyclonal rabbit α-ASK1 antibody and stained with either fluorescein isothiocyanate (FITC)-conjugated (green) or Cy™3-conjugated (red) goat anti-rabbit IgGs. α-20 and α-AtCUL1, the sections were treated with mouse α-20S proteasome and rabbit polyclonal α-AtCUL1 antibodies followed by staining with FITC-conjugated goat anti-mouse and anti-rabbit IgGs, respectively. The first three images in (A) and (B) show identical sections double-stained with α-ASK1 and α-20S antibodies, and counterstained with DAPI. The first and third images in (C) show the same section stained with DAPI and α-20S antibody.
Fig. 5. SKP1/ASK1, AtCUL1 and α-subunits of 20S proteasome are co-localized with mitotic spindles and phragmoplasts during mitosis. (A) Double-staining of cotyledon cells with anti-α-tubulin (green) and α-ASK1 (red) antibodies shows co-localization of SKP1/ASK1 with the mitotic spindle in late metaphase (upper cell), and phragmoplast during telophase (lower cell). (B) Overlapping staining patterns of mitotic spindle in a cotyledon cell stained with anti-α-tubulin (green) and α-AtCUL1 (red) antibodies. (C) Root cells stained with DAPI and α-20S antibody. The lower cell in metaphase shows a dotted staining pattern with the α-20S antibody (green), which marks the position of mitotic spindle flanking the equatorially arranged DAPI-stained chromosomes that are not detected by the α-20S antibody. The upper cell in early telophase shows a dotted α-20S-staining pattern of phragmoplast between the DAPI-stained chromosomes. (D) The α-ASK1 (red) and α-20S (green) antibodies stain the phragmoplast, but not the newly forming daughter nuclei (stained by DAPI in blue) in a root cell during telophase. (E) The α-AtCUL1 antibody (green) stains the phragmoplast in a root cell during telophase. Bars = 5 µm.
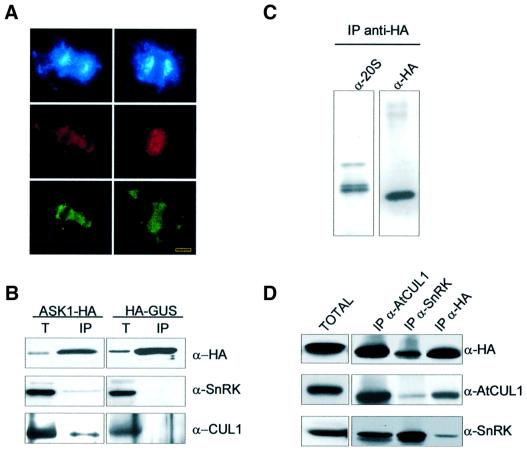
Although these data demonstrated a perfect cellular co-localization of SCF and 20S proteasomal epitopes, the co-localization of proteasome with the SCF subunit SKP1/ASK1 remained to be rigorously proven. This was necessary because the anti-SKP1/ASK1 antibody was raised against a peptide that shared at least 13 of 21 amino acids with all 19 members of the Arabidopsis SKP1/ASK family (Figure 2). For specific detection, the SKP1/ASK1 coding region was fused to an intron-tagged haemagglutinin (HA) epitope sequence. Subsequently, SKP1/ASK1 labelled with a C-terminal HA tag (ASK1-HA) was expressed under the control of a cauliflower mosaic virus 35S promoter (CaMV 35S) in cultured Arabidopsis cells by Agrobacterium-mediated transformation (Ferrando et al., 2000). Double immunostaining of transformed cells with anti-ASK1 and anti-HA antibodies resulted in identical staining patterns of nuclei, phragmoplasts and mitotic spindles (Figure 6A), confirming the previous cellular localization data obtained with the α-ASK1 antibody.
Fig. 6. SnRK is associated with SKP1/ASK1, cullin and 20S proteasome α-subunits. (A) SKP1/ASK1 is specifically detected by the polyclonal α-ASK1 antibody. Double-staining of Arabidopsis cells expressing an HA epitope-tagged form of SKP1/ASK1 protein (ASK1-HA) with polyclonal α-ASK1 (red) and monoclonal anti-HA (green) antibodies shows identical images of mitotic spindles in late anaphase (left column) and phragmoplasts in telophase (right column). Chromosomes and daughter nuclei are stained with DAPI (blue). Bars = 5 µm. (B) SKP1/ASK1 is co-immunoprecipitated with SnRK and cullin. Protein extract prepared from Arabidopsis cells expressing ASK1-HA was bound to immobilized anti-HA.11 IgG. The crude extract (T) and proteins eluted from the α-HA IgG matrix (IP) were separated by SDS–PAGE and immunoblotted with α-HA, α-SnRK and α-AtCUL1 antibodies. A control immunoprecipitation experiment was performed under identical conditions with a protein extract prepared from a cell line expressing an HA epitope-tagged β-glucuronidase enzyme (HA-GUS). (C) Purification of 20S proteasome–SCF complex. 20S proteasomal fractions co-purifying with cullin, SnRK and ASK1-HA on DEAE-Affi-Gel blue and Sephacryl S-400 were immunoaffinity purified on an anti-HA.11 IgG column. Proteins eluted with HA-peptide were separated by SDS–PAGE and immunoblotted with α-20S proteasome and α-HA antibodies. (D) Immunaffinity binding to α-HA and α-SnRK IgGs destabilizes the SCF complex. A Sephacryl S-400-purified 20S proteasome–SCF fraction (TOTAL) was immunoprecipitated using immobilized α-AtCUL1, α-SnRK and α-HA IgG antibodies, separated by SDS–PAGE and immunoblotted with α-HA, α-AtCUL1 and α-SnRK antibodies.
Proteasomal association of SKP1/ASK1, cullin and SnRK
To confirm that SKP1/ASK1 was indeed associated with cullin in an SCF complex, protein extract was prepared from the ASK1-HA-expressing cell line and loaded onto a matrix carrying immobilized anti-HA IgG. As a control, protein extract derived from a cell line expressing an HA-tagged β-glucuronidase (HA-GUS) enzyme was immunoprecipitated under identical conditions. Following specific elution of matrix-bound proteins with HA-peptide, western blotting with α-AtCUL1 and α-SnRK antibodies showed that ASK1-HA, but not the control HA-GUS protein, co-immunoprecipitated with cullin and an SnRK (Figure 6B).
To detect proteasomal association of the SCF subunits, proteasome was purified in the presence of ATP from the ASK1-HA-expressing cell line using DEAE-Affi-Gel blue Sepharose chromatography followed by Sephacryl S-400 gel filtration (see Materials and methods). Fractions cross-reacting with the α-HA, α-AtCUL1 and α-20S proteasome antibodies were subjected to immunoaffinity purification on immobilized anti-HA IgG using specific elution with HA-peptide. Western blotting of purified proteins with α-20S proteasome and α-HA antibodies showed that ASK1-HA co-purified with proteasome α-subunits (Figure 6C). In comparison, however, only a low amount of SnRK was recovered in association with ASK1-HA (as was noted earlier; Figure 6B and data not shown). To test whether SnRK was lost during immunoaffinity binding to the anti-HA IgG matrix, equal aliquots of protein fractions detected by the α-20S proteasome antibody were immunoprecipitated separately with α-AtCUL1, α-SnRK and α-HA antibodies. Western blotting of immunoprecipitated proteins showed that affinity binding to anti-HA IgG in fact resulted in a selective loss of SnRK from an immunocomplex with ASK1-HA and cullin (Figure 6D). Similarly, immunoprecipitation with the α-SnRK antibody reduced the recovery of cullin, indicating that IgG binding of certain subunits disrupted the SCF complex. By contrast, similar amounts of SnRK, cullin and SKP1/ASK1 were immunoprecipitated by the α-AtCUL1 antibody, showing that SnRK was a stoichiometric component of an SCF complex that co-purified with α-subunits of 20S proteasome and carried SKP1/ASK1 and AtCUL1 subunits.
Discussion
Phosphorylation is thought to be essential for SCF-mediated ubiquitylation of substrate proteins (Patton et al., 1998; Deshaies, 1999; Tyers and Jorgensen, 2000). Association of SCF complexes with protein kinases has been demonstrated in a few cases, including the human SCFSKP2 complex. The discovery of human S phase kinase-associated protein Skp1 and its F-box protein partner Skp2 stemmed from the observation that these proteins co-purified on a glycerol gradient with the cyclin A–CDK2 protein kinase (Zhang et al., 1995). Later, cullin1 (modified by the ubiquitin-like NEDD8 protein) and Rbx1/Roc1 were detected together with SKP1 and SKP2 in the SCFSKP2 complex, which is required in proliferating cells for ubiquitin-mediated degradation of cyclin E, E2F-1, CDK inhibitor p27Kip1 and Skp2 (Lisztwan et al., 1998; Lyapina et al., 1998; Carrano et al., 1999; Marti et al., 1999; Tsvetkov et al., 1999; Nakayama et al., 2000; Podust et al., 2000; Wirbelauer et al., 2000). It was also shown that both cullin1 and Skp2, but not Skp1, bind to cyclin A–CDK2, and that Skp2 inhibits the CDK2 kinase. In quiescent cells, cyclin A–CDK2 is associated with the CDK inhibitor p21CIP1/WAF1 and PCNA (proliferating cell nuclear antigen), indicating that p21CIP/WAF1 and SKP2 interact in a mutually exclusive manner with the cyclin A–CDK2 protein kinase (Yu et al., 1998; Yam et al., 1999).
The data presented here show that SKP1/ASK1, an Arabidopsis orthologue of human SKP1, is also associated in vivo with a protein kinase that belongs to the conserved family of eukaryotic AMPKs that are implicated in metabolic and stress signalling (Hardie et al., 1998). Originally, SKP1/ASK1 was identified in association with AtCUL1 and TIR1 F-box protein subunits of the SCFTIR1 ubiquitin ligase, which plays a distinguished role in the regulation of auxin plant hormone signalling (Gray et al., 1999). We have found that SKP1/ASK1 is co-immunoprecipitated with SnRK and cullin by α-SnRK and α-AtCUL1 antibodies. In addition, SKP1/ASK1 shows interaction with the Snf1/AMPK-related Arabidopsis protein kinases AKIN10 and AKIN11 in the two-hybrid system and in vitro. These data support the conclusion that an SnRK is an SKP1/ASK1-binding component of an SCF complex. Nonetheless, further analysis of SCF subunits (especially the substrate-binding F-box protein) is required to answer the question as to whether SnRKs contribute to the phosphorylation of SCFTIR1 substrates in the auxin signalling pathway (del Pozo and Estelle, 2000).
Mapping of the protein interaction domains indicates that SKP1/ASK1 recognizes homologous C-terminal segments of Arabidopsis SnRKs that are also implicated in binding of the SnRK inhibitor PRL1 WD-protein (Németh et al., 1998; Bhalerao et al., 1999). In analogy to exclusive interactions of human SKP2 and p21CIP1/WAF1 with the cyclin A–CDK2 kinase, we observed that SKP1/ASK1 and PRL1 show competitive kinase binding, and do not occur in common SnRK complexes in vivo. This finding is potentially important for further functional studies, because previous data show that the prl1 mutation enhances the activation of Arabidopsis SnRKs by sensitizing growth responses to auxin and other hormones (Németh et al., 1998), whereas mutations affecting the SCFTIR1 subunits SKP1/ASK1 and TIR1 reduce auxin sensitivity (Gray et al., 1999). Therefore, genetic analysis of prl1, ask1 and prl1, tir1 double mutants may be helpful in assessing whether mutually exclusive binding of PRL1 and SKP1/ASK1 to SnRKs reflects a regulatory event in hormonal signalling.
Genetic studies show that the prl1 mutation enhances the activation of SnRKs and results in transcriptional de-repression of many sucrose-regulated genes in Arabidopsis (Németh et al., 1998). It is also known that PRL1 binds to yeast Snf1, and that Arabidopsis AKIN10 and AKIN11 suppress the yeast snf1Δ10 and snf4Δ2 mutations, and interact with yeast Snf1 subunits Snf4 and Sip1 (Jiang and Carlson, 1996, 1997; Bhalerao et al., 1999; Kleinow et al., 2000). These data suggest that plant SnRKs are functionally related to yeast Snf1. Recently, Snf1 was found to interact with the CDK8–cyclin C-like Srb10/Srb11 protein kinase, which phosphorylates the C-terminal domain of RNA polymerase II and thereby inhibits the transcription of genes involved in sugar utilization, cell type specificity, thermotolerance and meiosis (Kutchin et al., 2000). Srb11 is rapidly destroyed in a proteasome-dependent manner during meiosis, suggesting a role for Snf1 in degradation of Srb11 (Cooper et al., 1997, 1999). As the Arabidopsis ask1 mutation was found to cause a meiotic chromosome segregation defect (Yang et al., 1999), we explored whether SKP1/ASK1 can complement the yeast skp1 mutations, and whether Skp1 and Snf1 interact in yeast. The results revealed significant functional differences between yeast and plant orthologues of Skp1 and Snf1. Although SKP1/ASK1 proved to be a suppressor of mitotic skp1-4 defect, it failed to rescue the G1–S block caused by the skp1-3 mutation (Connelly and Hieter, 1996) and did not interact with yeast Snf1. Similarly, no interaction between yeast Skp1 and Snf1 was detected.
Yeast two-hybrid screens for SKIPs identified AKIN10, in addition to several novel F-box proteins and fibrillarin AtFIB1/NOP1. Recent structural analysis of the human SKP1–SKP2 complex suggests that the recognition of F-box motifs by SKP1 may be selective (Schulman et al., 2000). Nonetheless, it remains to be determined whether the identified SKIP F-box proteins occur in SKP1/ASK1-containing SCF complexes in vivo, and whether their interaction with SKP1/ASK1 is glucose regulated in Arabidopsis, as in yeast. Although SKP1_5/ASK5, which carries divergent C-terminal sequences (Figure 2), showed no SnRK binding in the two-hybrid system, we also need to answer the questions whether other members of the Arabidopsis SKP1/ASK1 family can interact with SnRKs or other protein kinases, and whether these interactions are conserved in other eukaryotic systems.
Proteasomal association of an SCF complex in Arabidopsis is indicated by selective two-hybrid and in vitro interactions of SKP1/ASK1 and SnRKs with the α4/PAD1 proteasomal subunit, association of an SnRK with purified 26S proteasome, and co-immunoprecipitation of SKP1/ASK1, SnRK and cullin with 20S proteasomal α-subunits. These data show that proteasomal binding of SnRK and SKP1/ASK1 is mediated by recognition of the C-terminus of the α4/PAD1 proteasomal subunit (Figure 7). The 20S proteasomal subunits show structural and functional conservation between Arabidopsis and other eukaryotes (Parmentier et al., 1997; Fu et al., 1998). Studies of subunit topography and crystal structure demonstrate an identical arrangement of α-subunits in the outer rings of eukaryotic 20S core particles (Groll et al., 1997; Dahlmann et al., 1999). The ends of the 20S particle are sealed by the N-termini of α-subunits, whereas the C-terminal ends of the α1, α3, α4, α5 and α7 proteins, carrying lysine (K)- and glutamine (E)-rich KEKE motifs, stick out from the particle surface (Rechsteiner, 1998; Voges et al., 1999). The KEKE motifs, also present in the Arabidopsis α4/PAD1 protein, are postulated to mediate interactions with 11S activators and 19S regulatory cap particles that open the gated channel (alpha-annulus) of the 20S particle formed by the α-subunits (Realini et al., 1994; Groll et al., 2000; Whitby et al., 2000). The α2 (C3), α3 (C9), α4, α5 (ζ), α6 (C2) and α7 (C8) subunits are known to be phosphorylated in several organisms (for review see Bose et al., 1999). Casein kinase II (CKII) was found to co-purify with the 26S proteasome and phosphorylate the rat α3, rice α6 and yeast α7 proteasomal subunits (Ludemann et al., 1993; Umeda et al., 1997b; Pardo et al., 1998). As CKII-mediated phosphorylation of rat α7 subunit had no effect on the 26S proteasome activity, it was suggested that phosphorylation modulates the interaction of proteasomal α-subunits with regulatory proteins (Castaño et al., 1996). As the sole example so far, this model is supported by the observation that the C-terminus of human α4 (XAPC7) subunit interacts with and regulates transcriptional activation by the HBX hepatitis virus B protein (Huang et al., 1996). In further support of this model, we have shown that the α4/PAD1 subunit interacts in vivo and in vitro with SKP1/ASK1 and an SnRK protein kinase. α4/PAD1 is not phosphorylated by the Arabidopsis SnRKs in vitro. However, in our experiments, proteasome was isolated in the presence of ATP. Thus, further analysis is required to test whether SnRK binding to α4/PAD1 facilitates the phosphorylation of other proteasomal subunits, such as α3 or α5, and whether SnRK-mediated phosphorylation of any α-subunit leads to opening of the gated channel of the 20S proteasomal cylinder.
Fig. 7. Schematic model showing protein interactions in the proteasomal–SCF complex. (A) The α4/PAD1 proteasome subunit interacts with Snf1-related Arabidopsis protein kinases AKIN10 and AKIN11 in the yeast two-hybrid system and in vitro. SnRK protein kinase is detected in association with purified 26S proteasomes. (B) Epitope-tagged SKP1/ASK1 co-purifies and co-immunoprecipitates with SnRK, cullin and 20S proteasome α-subunits. SnRKs interact with the C-terminus of the α4/PAD1 proteasome subunit, which carries characteristic KEKE motifs present in several other α-subunits. The α4/PAD1 and SKP1/ASK1 proteins interact with each other and bind to the C-terminal regulatory domains of SnRKs. The activity and interaction of SnRKs with SKP1/ASK1 are inhibited by the PRL1 WD-protein.
Until recently, it has been an intriguing question as to how the ubiquitylation system delivers the substrates for degradation to the proteasome. A model suggesting recognition of ubiquitylated substrates by the ubiquitin-binding 19S cap protein Rnp10 now has less support because Rnp10 proved to be non-essential for proteolysis in budding yeast (van Nocker et al., 1996). Our data suggest that SCF ubiquitin ligase may directly target substrate proteins to the proteasome. This conclusion is supported by the data of Tongaonkar et al. (2000), who demonstrated that E2 ubiquitin conjugases Ubc1, Ubc2, Ubc4 and Ubc5 can directly interact with the 26S proteasome. Recently, Xie and Varshavsky (2000), reported that none-SCF-type E3 ubiquitin ligases, Ubr1 (N-recognin) and Ufd4 (required for the ubiquitin fusion degradation pathway) can also interact with the proteasome. Ufd4 binds to Rpt6, whereas Ubr1 recognizes the Rpn2, Rpt1 and Rpt6 proteins in the 19S cap particle. By mass spectrometry analysis of epitope-tagged yeast proteasomal particles, Verma et al. (2000) showed that components of SCF-ubiquitin ligases co-purify and assemble with the 19S cap. Our data provide evidence that the C-terminus of α4/PAD1 subunit in the 20S core particle can bind an SnRK protein kinase in association with the SKP1/ASK1 subunit of an SCF complex in Arabidopsis. Since SnRK showed an expected molecular mass in the 26S proteasome and 20S proteasome–SCF complexes, it is likely that SnRK is not ubiquitylated as a substrate protein when associated with active proteasome and SCF.
The observation that SKP1/ASK1, cullin and α-subunits of the 20S proteasome are co-localized with tubulin during mitosis suggests that components of the mitotic spindle may serve as substrates for the SCF–proteasomal complex. The fact that the ask1 mutation causes a chromosome segregation defect in male meiosis supports this model, but also predicts that either SKP1/ASK1 or one of its interacting partners has a unique meiosis-specific function. Therefore, it is noteworthy that the α4 proteasome subunit is encoded by duplicated genes, PAD1 and PAD2, in Arabidopsis (Fu et al., 1999), as well as in Drosophila melanogaster, where both α4-subunit genes (PROS28.1A and B) are expressed specifically during male spermatogenesis (Yuan et al., 1996). By contrast, Drosophila virilis has two separate genes for specific expression in somatic and sperm cells (Belote et al., 1998). In fertilized eggs of Xenopus and goldfish, α4 homologues display G2-specific phosphorylation and dephosphorylation in meiotic metaphase (Tokumoto et al., 1999, 2000). Further structural and functional dissection of the proteasomal–SCF complex is expected to clarify whether the α4 proteasome subunits show a similar meiotic regulation in Arabidopsis.
Materials and methods
Protein interaction assays in the yeast two-hybrid system
Construction of GBD–AKIN10 and GBD–AKIN11 baits in pAS2, preparation of a pACT2 cDNA library using mRNA from Arabidopsis cell suspension, and detection of GBD fusion protein in the yeast two-hybrid host Y190 by western blotting with anti-Gal4p-DB (Clontech) and α-SnRK (α-NPK5; Muranaka et al., 1994) antibodies were described previously (Németh et al., 1998; Bhalerao et al., 1999). Following selection of transformants on SD medium (lacking leucine, histidine and tryptophan, but containing 50 mM 3-AT), yeast colonies were grown on nitrocellulose filters on SD plates with 25 mM 3-AT to perform LacZ filter lift assays as described (Durfee et al., 1993; Ausubel et al., 1999). The pACT2 clones, encoding putative AKIN-interacting factors, were isolated and transformed into yeast strains Y187 and Y190 (Durfee et al., 1993) that either carried no bait, or harboured GBD–AKIN10, GBD–AKIN11, pAS2-lamin (Matchmaker System; Clontech), and other control baits described previously (Németh et al., 1998). The cDNAs were sequenced using an automatic ABI377 sequencer and pAS2-specific oligonucleotide primers. To map the SKP1/ASK1 and α4/PAD1 binding domains of SnRKs, GBD baits carrying PCR-amplified segments of AKIN10 and AKIN11 cDNAs were constructed. Amino acid positions of AKIN10 and AKIN11 sequences expressed by these baits are shown in Figure 3A. Interactions of AKIN10 and AKIN11 baits with a GAD–ASK1 prey were tested by LacZ filter assays. Similar assays were performed with a GAD–PAD1 prey, which carried a full-length cDNA from pTAS-g64 (Genschik et al., 1992).
The SKP1/ASK1 cDNA was cloned as a BamHI–BglII fragment from pACT2 in pAS2 to construct a GBD–ASK1 bait, which was used in a two-hybrid screen to identify SKIPs. The pACT2-SKIP cDNA clones were transformed into yeast strains to test their interaction with GBD–ASK1, GBD–AKIN10, GBD–AKIN11 and GBD–lamin baits. The longest sequence from each class of SKIP cDNAs is displayed by the DDBJ/EMBL/GenBank accession Nos: SKIP1, AF263377; SKIP2, AF263378; SKIP3, AF263379; SKIP4, AF263380; SKIP5, AF263382; SKIP6, AF263381; and SKIP7, AF263383. Glucose regulation of protein interactions between GBD–ASK1 and each GAD–SKIP prey, as well as between GBD–AKIN10 and GBD–AKIN11 baits and GAD–ASK1, was assayed by measuring β-galactosidase enzyme levels in six independent transformants grown in SC medium with either 2% glucose, or 0.05% glucose and 2% each of galactose, glycerol and ethanol, as described (Jiang and Carlson, 1996, 1997; Table I).
In vitro SnRK binding and kinase competition assays with SKP1/ASK1 and PRL1
The SKP1/ASK1 cDNA was cloned as a BamHI–BglII fragment in pGEX-3X-2 (Pharmacia) to purify a GST–ASK1 fusion protein (Ausubel et al., 1999). DNA templates for in vitro transcription–translation (Promega TNT) were generated by PCR amplification of AKIN10 and AKIN11 coding domains using a 5′ primer with a T7 promoter sequence (5′-AATACGACTCACTATACGGAGACCACATGGGAGGCCCGGGATCCGAAT-3′), and a 3′ primer (5′-ATGCACAGTTGAAGTGAACTT-3′). GST–ASK1 and control GST proteins (10 µg of each) were immobilized on GS beads (Pharmacia) in binding buffer R [20 mM Tris–HCl pH 7.5, 150 mM NaCl, 2 mM EDTA, 0.1% (v/v) IGEPAL] for 30 min at 4°C. Equal aliquots of GST matrices and GS beads washed with binding buffer were incubated with [35S]methionine-labelled AKIN10 and AKIN11 for 60 min at 4°C. Following the removal of unbound proteins and washing the beads, the matrix-bound proteins were eluted with SDS sample buffer, separated together with the unbound protein fractions on SDS–polyacrylamide gels, and detected by autoradiography.
In kinase competition assays (Figure 3B and D), samples of [35S]AKIN10 were bound to GST–ASK1 (5 µg) in binding buffer for 30 min and incubated with increasing amounts of MBP–PRL1 protein (0.5, 1, 2 or 5 µg) for 30 min at 24°C. Half of each sample was bound to GS, whereas the other half was bound to amylose resin (New England Biolabs) for 60 min at 4°C. After washing with binding buffer, the matrix-bound proteins were eluted, separated and detected as described above. In protein kinase assays, GST–AKIN10 (5 µg) was incubated either alone or with 5 µg of either GST, MBP or MBP–PRL1 in kinase buffer [50 mM Tris–HCl pH 8.0, 15 mM MgCl2, 5 mM EGTA, 1 mM dithiothreitol (DTT)] for 30 min at 25°C before initiating the kinase reaction by addition of 5 µCi of [γ-32P]ATP and 1 µg of SnRK substrate TRX-KD (Bhalerao et al., 1999). To test the effect of SKP1/ASK1 binding on the activity of SnRKs, increasing amounts of GST–ASK1 (0.5, 1, 2 and 5 µg) were incubated with 1 µg of GST–AKIN10. In competition assays, 1 µg of GST–AKIN10 was saturated with 5 µg of GST–ASK1, then incubated with 0.5, 1, 2 or 5 µg of MBP–PRL1 for 30 min at 25°C before performing the kinase reactions for 1 h at 25°C. The phosphorylated TRX-KD peptide was bound to Ni-NTA–agarose (Qiagen), washed, eluted, separated by SDS–PAGE, stained, and detected by autoradiography as described (Bhalerao et al., 1999).
In vitro binding assays with α4/PAD1
For in vitro transcription–translation, the α4/PAD1 coding region was PCR amplified using a T7 promoter primer (see above). GST–ASK1, GST–AKIN10, GST–AKIN11 and control GST proteins (10 µg of each) were bound to GS for 30 min at 4°C in binding buffer T (25 mM Tris–HCl pH 7.5, 1 g/l milk powder) and incubated with [35S]α4/PAD1 for 60 min at 4°C. The supernatant and eluted matrix-bound [35S]α4/PAD1 fractions were separated by SDS–PAGE and detected as described above.
To purify α4/PAD1 in fusion with an N-terminal His6 tag, the coding domain was PCR amplified and cloned in pRSETb (Invitrogen) to obtain pHis-α4/PAD1 in Escherichia coli BL21(DE3) LysS. Cells grown at 28°C to an OD660 of 0.6 were induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 2 h, harvested, resuspended in lysis buffer (50 mM Tris–HCl pH 7.8, 2 mM EDTA, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, 10 µg/ml aprotinin) and passed through a French press to prepare a crude lysate. A cleared lysate obtained by centrifugation (12 000 g for 45 min at 4°C) was purified on a DEAE Fast Flow Sepharose column XK/16X40 using a linear gradient of 0–1 M NaCl in lysis buffer. The His-α4/PAD1 fractions were pooled, dialysed in buffer (50 mM Tris–HCl pH 7.8, 250 mM NaCl, 10 mM imidazole) and loaded on a Ni-NTA–agarose (Bio-Rad) column. After washing with binding buffer and elution with buffer L (50 mM Tris–HCl pH 7.8, 250 mM NaCl, 250 mM imidazole), His-α4/PAD1 was obtained in apparently homogeneous form.
In protein binding assays (Figure 3C), 2 µg of GST–ASK1 were first incubated in binding buffer R (see above) with [35S]AKIN10 for 30 min at 20°C, then with increasing amounts of His-α4/PAD1 (1, 2 or 5 µg) for 30 min, and bound to GS matrix for 60 min at 4°C. Proteins were eluted from the washed GST beads with SDS sample buffer and separated on two identical SDS–polyacrylamide gels. The first gel was subjected to autoradiography to detect the GST–ASK1-bound [35S]AKIN10 protein, whereas the second gel was immunoblotted with a monoclonal anti-RGSHis6 antibody (Boehringer-Roche) to detect His-α4/PAD1 in association with GST–ASK1.
To determine whether α4/PAD1 can recruit SKP1/ASK1 and SnRK from a plant protein extract, 5 µg of His-α4/PAD1 were immobilized on Ni-NTA–agarose for 1 h at 4°C in extraction buffer [50 mM Tris–HCl pH 7.6, 10% (v/v) glycerol, 20 mM β-mercaptoethanol, 20 µl/ml Sigma plant protease inhibitor mix]. Protein extract (6 mg) from cultured Arabidopsis Col-0 cells was prepared in extraction buffer, cleared by centrifugation (20 000 g for 30 min and 50 000 g for 45 min), supplemented with 10 mM imidazole and 150 mM NaCl, precleared with Ni-NTA–agarose for 2 h, and incubated with His-α4/PAD1-Ni-NTA resin for 4 h at 4°C. The resin was washed with extraction buffer containing 10 mM imidazole and 250 mM NaCl, then the matrix-bound proteins were eluted with SDS sample buffer and subjected to western blotting with α-ASK1 and α-SnRK antibodies (Figure 3F).
Suppressor assays with budding yeast skp1 mutants
The SKP1/ASK1 cDNA was cloned as a BamHI–BglII fragment in a yeast gene expression cassette that carried a methionine-repressible Met25 promoter in plasmid p426Met25 (Mumberg et al., 1994). Budding yeast strains (Connelly and Hieter, 1996) YPH1161 [Mata, ura3-52, lys2-801, ade2-101, trp1Δ63, his3-Δ200, leu2-Δ1, skp1Δ1::TRP1, skp1-4:: LEU2 CFIII(CEN3.LYPH983), HIS3, SUP1] and YPH1172 (as YPH1161, but skp1-3::LEU2) were transformed with p426Met25 carrying the SKP1/ASK1 cDNA and assayed on ura– SC medium (Bhalerao et al., 1999) for suppression of the thermosensitive skp1-3 and skp1-4 mutations at 25 and 37°C in the absence or presence of 1 mM methionine inhibiting the Met25 promoter.
Epitope tagging of SKP1/ASK1 and immunoprecipitation of ASK1-HA protein complexes
The SKP1/ASK1 cDNA was cloned in pPILY and expressed by a cauliflower mosaic virus 35S promoter in Agrobacterium-transformed Arabidopsis cells as described (Ferrando et al., 2000). A polyclonal rabbit antibody (α-ASK1) raised against a C-terminal SKP1/ASK1-peptide, CKNDFTPEEEEEVRRE, was affinity purified using Affi-Prep10 (Harlow and Lane, 1988). For immunoprecipitation, 30 mg of protein extract from Arabidopsis Col-0 cells were precleared with protein A– Sepharose (Bhalerao et al., 1999) and bound to 350 µg of α-ASK1 IgG immobilized on protein A–Sepharose with dimethylpimelimidate (Harlow and Lane, 1988). The IgG-bound proteins were eluted with 1 ml of ASK1 peptide [0.5 mg/ml in TBSG buffer: 20 mM Tris–HCl pH 7.5, 150 mM NaCl and 10% (v/v) glycerol], concentrated with AMICON YM10, separated by SDS–PAGE, and subjected to either immunoblotting with α-ASK1, α-PRL1 (Németh et al., 1998) and α-SnRK antibodies, or used in SnRK protein kinase assays. For immunoprecipitation of ASK1-HA protein complexes, 30 mg of protein extract from an ASK1-HA-expressing cell line were precleared with protein G–Sepharose (Sigma), and loaded in repeated cycles on a column containing anti-HA.11 IgG–beads (Babco) at 4°C for 12 h using a peristaltic pump. The column was washed with buffer A [TBS buffer containing 0.1% (v/v) IGEPAL], buffer B [20 mM Tris–HCl pH 7.5, 300 mM NaCl, 0.1% (v/v) IGEPAL] and buffer A again. Proteins were eluted from the anti-HA.11 IgG with HA-peptide (Genosys; 0.5 mg/ml, in TBSG buffer); resolved by SDS–PAGE, immunoblotted separately with α-SnRK, α-AtCUL1 (Gray et al., 1999), α-20S proteasome [mAB(MCP231); Affinity Research], and peroxidase-conjugated anti-HA (Boehringer) antibodies, followed by enhanced chemiluminescence (ECL; Amersham-Pharmacia) detection.
Plant growth conditions and indirect immunofluorescence microscopy
Seed sterilization, in vitro culture and growth conditions for Arabidopsis thaliana seedlings and plants, as well as establishment, maintenance and transformation of cell suspensions, were described previously (Mathur et al., 1998; Németh et al., 1998; Ferrando et al., 2000). For sampling of roots, seeds were sown on a nylon mesh placed on MS agar medium (Gibco-BRL) containing 0.5% sucrose, and germinated in horizontal position. Samples from cultured cells, 6-day-old seedlings, and different organs of 5-week-old soil-grown plants were fixed, embedded, sectioned and treated with antibodies as described previously (Vitha et al., 1997; Ferrando et al., 2000). The following antibodies were used: affinity-purified rabbit polyclonal α-ASK1 (Ferrando et al., 2000; dilution 1:300); rabbit polyclonal α-AtCUL1 (Gray et al., 1999; 1:1000); mouse monoclonal anti-human 20S proteasome [Affinity Research; mAB (MCP231); 1:600]; and mouse monoclonal anti-HA IgG (Babco; 1:500). After incubation with primary antibodies, the sections were washed with phosphate-buffered saline (PBS) containing 0.1% Tween-20, and treated for 1 h at 20°C with either fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit IgG (Sigma; 1:150), to visualize the SKP1/ASK1 and AtCUL1 epitopes, or FITC-conjugated goat anti-mouse IgG (Sigma; 1:150), to detect the 20S proteasome α-subunits. In double-staining experiments, the rabbit α-ASK1 antibody was used in combination with either mouse α-20S proteasome, or mouse anti-actin (clone C4; ICN Biomedicals; 1:150), or mouse anti-α-tubulin (clone DM1A; Sigma; 1:150) antibodies. The sections were stained with FITC-conjugated goat anti-mouse IgG in combination with a Cy™3-conjugated anti-rabbit IgG (H+L; Jackson ImmunoResearch; 1:500). Arabidopsis cells expressing ASK1-HA were similarly treated with rabbit α-ASK1 and mouse α-HA IgGs, then with FITC-conjugated goat anti-mouse and Cy™3-conjugated anti-rabbit IgGs. In control experiments, the primary antibodies were either omitted or replaced by rabbit serum. In competition experiments, the α-ASK1 and α-HA antibodies were blocked with purified GST–ASK1 protein and HA peptide, respectively. All secondary antibodies were tested for cross-reaction. For microscopy, the sections were washed with PBS containing 0.1% Tween-20 and mounted with p-phenylenediamine-glycerol antifade solution. Fluores cence images were examined using Leica Aristoplan and DMRB microscopes with FITC, rhodamine (Cy™3), DAPI and combined BGR filters, and recorded with a Hitachi HV-20 camera.
Proteasome purification from wild-type- and ASK1-HA-expressing Arabidopsis cells
Proteasome was purified from Arabidopsis Col-0 cell suspension using a modification of the procedure described by Umeda et al. (1997b). A crude lysate was prepared by homogenization of cells with liquid nitrogen in buffer H (25 mM Tris–HCl pH 7.5, 2 mM ATP, 5 mM MgCl2, 1 mM DTT, 10% glycerol, 10 mM creatinine phosphate, 10 µg/ml creatine phosphokinase). A cleared lysate (50 mg/ml protein) obtained by differential centrifugation (20 000 g for 30 min and 50 000 g for 45 min at 4°C) was fractionated on a Q-Sepharose column using a linear gradient of 0–1 M NaCl. The column fractions were subjected to electrophoresis on a non-denaturing polyacrylamide gel, which was stained for 15 min using an overlay of 0.1 mM Suc-LLVY-AMIC (Bachem) fluorescent peptidase substrate in buffer H to detect the 26S and 20S proteasomal bands by UV (360 nM) illumination (Glickman et al., 1998). Gel sections containing 26S proteasomes were excised, resuspended in buffer I [50 mM Tris–HCl pH 7.5, 5 mM MgCl2, 2 mM DTT, 10% (v/v) glycerol], and homogenized to obtain a cleared protein sample, which was resolved by 12% SDS–PAGE for silver staining (Roth) and western blotting with α-20S proteasome and α-SnRK antibodies (Figure 3G).
Proteasomes were similarly prepared from the ASK1-HA-expressing Arabidopsis cell line, with the exception that the cleared lysate (2 g of protein) was fractionated on a DEAE-Affi-Gel blue Sepharose column (Bio-Rad) using a linear gradient of 0–500 mM NaCl in buffer E [25 mM Tris–HCl pH 7.5, 2 mM ATP, 5 mM MgCl2, 1 mM DTT, 10% (v/v) glycerol]. Fractions containing ASK1-HA were detected by western blotting with anti-HA IgG, combined, simultaneously desalted and concentrated in a Vivaspin 20 concentrator (10 kDa cut-off; Sartorius), and further fractionated on a Sephacryl S-400 column (Pharmacia) equilibrated with buffer E containing 100 mM NaCl. Fractions detected by α-20S proteasome, α-HA, α-AtCUL1 and α-SnRK antibodies were further purified on anti-HA.11 IgG. A 20S proteasome–SCF fraction (15 mg of protein) was precleared with protein G–Sepharose for 4 h, then incubated with anti-HA.11 IgG matrix for 12 h at 4°C. The resin was placed in a column, washed with buffer E containing 150 mM NaCl, and then the ASK1-HA-associated proteins were eluted from the α-HA.11 IgG matrix with HA-peptide (0.5 mg/ml in TBSG buffer) and subjected to western blotting with α-20S proteasome and α-HA antibodies (Figure 6C). Alternatively, the Sephacryl S-400-purified 20S proteasome–SCF fraction (5 mg of protein) was precleared and subjected to immunoprecipitation separately with anti-HA.11, α-AtCUL1, and α-SnRK antibodies for 4 h at 4°C. The immunocomplex obtained with anti-HA.11 IgG was bound to protein G–Sepharose, whereas immunocomplexes formed with the α-AtCUL1 and α-SnRK antibodies were bound to protein A–Sepharose for 2 h at 4°C. The matrices were washed with buffer E containing 150 mM NaCl. The matrix-bound proteins were eluted by SDS sample buffer and divided into three equal aliquots, which were resolved by SDS–PAGE and subjected to western blotting separately with α-HA, α-SnRK and α-AtCUL1 antibodies followed by ECL detection (Figure 6D).
Acknowledgments
Acknowledgements
We thank M.Estelle and T.Muranaka for providing antibodies; C.Connelly and P.Hieter for the yeast skp1 mutants; P.Genschik for the α4/PAD1 plasmid p64; R.P.Bhalerao for the AKIN10 and AKIN11 clones; L.Bakó for help in proteasome purification; and F.Salamini, S.Schwartz-Sommer, R.Thompson, L.Szabados, G.Coupland and P.Dudley for their critical comments on the manuscript. This work was supported by grants from the Deutsche Forschungsgemeinschaft (KO1483/3-1) and Deutsches Zentrum für Luft und Raumfahrt (UNG-027-97), and a stipend for J.J. by the Alexander-von-Humboldt Stifftung.
References
- Adams J., Kelso,R. and Cooley,L (2000) The kelch repeat superfamily of proteins: propellers of cell function. Trends Cell Biol., 10, 17–24. [DOI] [PubMed] [Google Scholar]
- Alderson A., Sabelli,P.A., Dickinson,R.J., Cole,D., Richardson,M., Kreis,M., Shewry,P.R. and Halford,N.G. (1991) Complementation of snf1, a mutation affecting global carbon metabolism in yeast, by a plant protein kinase cDNA. Proc. Natl Acad. Sci. USA, 88, 8602–8605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (1999) Current Protocols in Molecular Biology. John Wiley & Sons, New York, NY.
- Bai C., Sen,P., Hofmann,K., Ma,L., Goebl,M., Harper,J.W. and Elledge,S.J. (1996) SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell, 86, 263–274. [DOI] [PubMed] [Google Scholar]
- Barneche F., Steinmetz,F. and Echeverria,M. (2000) Fibrillarin genes encode both a conserved nucleolar protein and a novel small nucleolar RNA involved in ribosomal RNA methylation in Arabidopsis thaliana. J. Biol. Chem., 275, 27212–27220. [DOI] [PubMed] [Google Scholar]
- Belote J.M., Miller,M. and Smyth,K.A. (1998) Evolutionary conservation of a testes-specific proteasome subunit gene in Drosophila. Gene, 215, 93–100. [DOI] [PubMed] [Google Scholar]
- Bhalerao R.P., Salchert,K., Bakó,L., Ökrész,L., Szabados,L., Muranaka,T., Machida,Y., Schell,J. and Koncz,C. (1999) Regulatory interaction of PRL1 WD protein with Arabidopsis SNF1-like protein kinases. Proc. Natl Acad. Sci. USA, 96, 5322–5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose S., Mason,G.G.F. and Rivett,J.A. (1999) Phosphorylation of proteasomes in mammalian cells. Mol. Biol. Rep., 26, 11–14. [DOI] [PubMed] [Google Scholar]
- Bouly J.-P., Gissot,L., Lessard,P., Kreis,M. and Thomas,M. (1999) Arabidopsis thaliana proteins related to the yeast SIP and SNF4 interact with AKINα1, an SNF1-like protein kinase. Plant J., 18, 541–550. [DOI] [PubMed] [Google Scholar]
- Carlson M. (1998) Regulation of glucose utilization in yeast. Curr. Opin. Genet. Dev., 8, 560–564. [DOI] [PubMed] [Google Scholar]
- Carlson M. (1999) Glucose repression in yeast. Curr. Opin. Microbiol., 2, 202–207. [DOI] [PubMed] [Google Scholar]
- Carrano A.C., Eytan,E., Hersko,A. and Pagano,M. (1999) SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nature Cell Biol., 1, 193–199. [DOI] [PubMed] [Google Scholar]
- Castaño J.G., Mahillo,E., Arizti,P. and Arribas,J. (1996) Phosphorylation of C8 and C9 subunits of the multicatalytic proteinase by casein kinase II and identification of the C8 phosphorylation sites by direct mutagenesis. Biochemistry, 35, 3782–3789. [DOI] [PubMed] [Google Scholar]
- Connelly C. and Hieter,P. (1996) Budding yeast SKP1 encodes an evolutionarily conserved kinetochore protein required for cell cycle progression. Cell, 86, 275–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper K.F., Mallory,M.J., Smith,J.B. and Strich,R. (1997) Stress and developmental regulation of the yeast C-type cyclin Ume3p (Srb11/Ssn8p). EMBO J., 16, 4665–4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper K.F., Mallory,M.J. and Strich,R. (1999) Oxidative stress-induced destruction of the yeast C-type cyclin Ume3p requires phosphatidyl inositol-specific phospholipase C and the 26S proteasome. Mol. Cell. Biol., 19, 3338–3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creelman R.A. (1998) Jasmonate perception: characterization of COI1 mutants provides the first clues. Trends Plant Sci., 3, 367–368. [Google Scholar]
- Dahlmann B., Kopp,F., Kristensen,P. and Hendil,K.B. (1999) Identical subunit topographies of human and yeast 20S proteasomes. Arch. Biochem. Biophys., 363, 296–300. [DOI] [PubMed] [Google Scholar]
- del Pozo C. and Estelle,M. (1999) Function of the ubiquitin–proteosome pathway in auxin response. Trends Plant Sci., 4, 107–112. [DOI] [PubMed] [Google Scholar]
- del Pozo C.J. and Estelle,M. (2000) F-box proteins and protein degradation: an emerging theme in cellular regulation. Plant Mol. Biol., 44, 123–128. [DOI] [PubMed] [Google Scholar]
- De Mot R., Nagy,I., Walz,J. and Baumeister,W. (1999) Proteasomes and other self-compartmentalizing proteases in prokaryotes. Trends Microbiol., 7, 88–92. [DOI] [PubMed] [Google Scholar]
- Deshaies R.J. (1999) SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol., 15, 435–467. [DOI] [PubMed] [Google Scholar]
- Durfee R., Becherer,K., Chen,P.L., Yeh,S.H., Yang,Y., Kilburn,A., Lee,W.H. and Elledge,S.J. (1993) The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev., 7, 555–569. [DOI] [PubMed] [Google Scholar]
- Ferrando A., Farrás,R., Jásik,J., Schell,J. and Koncz,C. (2000) Intron-tagged epitope: a tool for facile detection and purification of proteins expressed in Agrobacterium-transformed plant cells. Plant J., 22, 553–560. [DOI] [PubMed] [Google Scholar]
- Fu H., Doelling,J.H., Arendt,C.S., Hochstrasser,M. and Vierstra,R.D. (1998) Molecular organization of the 20S proteasome gene family from Arabidopsis thaliana. Genetics, 149, 677–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H., Girod,P.-A., Doelling,J.H., van Nocker,S., Hochstrasser,M., Finley,D. and Vierstra,R.D. (1999) Structure and functional analyses of the 26S proteasome subunits from plants. Mol. Biol. Rep., 26, 137–146. [DOI] [PubMed] [Google Scholar]
- Genschik P., Philipps,G., Gigot,C. and Fleck,J. (1992) Cloning and sequence analysis of a cDNA from Arabidopsis thaliana homologous to a proteasome α subunit from Drosophila. FEBS Lett., 309, 311–315. [DOI] [PubMed] [Google Scholar]
- Glickman M.H., Rubin,D.M., Fried,V.A. and Finley,D. (1998) The regulatory particle of the Saccharomyces cerevisiae proteasome. Mol. Cell. Biol., 18, 3149–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray W.M. et al. (1999) Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev., 13, 1678–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groll M., Ditzel,L., Lowe,J., Stock,D., Bochtler,M., Bartunik,H.D. and Huber,R. (1997) Structure of 20S proteasome from yeast at 2.4 Å resolution. Nature, 386, 463–471. [DOI] [PubMed] [Google Scholar]
- Groll M., Bajorek,M., Köhler,A., Moroder,L., Rubin,D.M., Huber,R., Glickman,M.H. and Finley,D. (2000) A gated channel into the proteasome core particle. Nature Struct. Biol., 7, 1062–1067. [DOI] [PubMed] [Google Scholar]
- Halford N.G. and Hardie,D.G. (1998) SNF1-related protein kinases: global regulators of carbon metabolism in plants? Plant Mol. Biol., 37, 735–748. [DOI] [PubMed] [Google Scholar]
- Hardie D.G. and Carling,D. (1997) The AMP-activated protein kinase. Fuel gauge of the mammalian cell? Eur. J. Biochem., 246, 259–273. [DOI] [PubMed] [Google Scholar]
- Hardie D.G., Carling,D. and Carlson,M. (1998) The AMP-activated/SNF1 protein kinase subfamily: metabolic sensors of the eukaryotic cell? Annu. Rev. Biochem., 67, 821–855. [DOI] [PubMed] [Google Scholar]
- Harlow E. and Lane,D. (1988) Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- Hersko A. and Ciechanover,A. (1998) The ubiquitin system. Annu. Rev. Biochem., 67, 425–479. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. (1996) Ubiquitin-dependent protein degradation. Annu. Rev. Genet., 30, 405–439. [DOI] [PubMed] [Google Scholar]
- Hofmann K. and Bucher,P. (1996) The UBA domain: a sequence motif present in multiple enzyme classes of the ubiquitination pathway. Trends Biochem. Sci., 21, 172–173. [PubMed] [Google Scholar]
- Huang J., Kwong,J., Sun,E.C.-Y. and Liang,T.J. (1996) Proteasome complex as a potential cellular target of hepatitis B virus X protein. J. Virol., 70, 5582–5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda Y., Koizumi,N., Kusano,T. and Sano,H. (2000) Specific binding of a 14-3-3 protein to autophosphorylated WPK4, an SNF1-related wheat protein kinase and to WPK4-phosphorylated nitrate reductase. J. Biol. Chem., 275, 31695–31700. [DOI] [PubMed] [Google Scholar]
- Jiang R. and Carlson,M. (1996) Glucose regulates protein interactions within the yeast SNF1 protein kinase complex. Genes Dev., 10, 3105–3115. [DOI] [PubMed] [Google Scholar]
- Jiang R. and Carlson,M. (1997) The Snf1 protein kinase and its activating subunit Snf4 interact with distinct domains of the Sip1/Sip2/Gal83 component in the kinase complex. Mol. Cell. Biol., 17, 2099–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp B.E., Mitchelhill,K.I., Stapleton,D., Michell,B.J., Chen,Z.-P. and Witters,L.A. (1999) Dealing with energy demand: the AMP-activated protein kinase. Trends Biochem. Sci., 24, 22–25. [DOI] [PubMed] [Google Scholar]
- Kleinow T., Bhalerao,R., Breuer,F., Umeda,M., Salchert,K. and Koncz,C. (2000) Functional identification of an Arabidopsis Snf4 ortholog by screening for heterologous multicopy suppressors of snf4 deficiency in yeast. Plant J., 23, 115–122. [DOI] [PubMed] [Google Scholar]
- Kutchin S., Treich,I. and Carlson,M. (2000) A regulatory shortcut between the Snf1 protein kinase and RNA polymerase II holoenzyme. Proc. Natl Acad. Sci. USA, 97, 7916–7920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F.N. and Johnston,M. (1997) Grr1 of Saccharomyces cerevisiae is connected to the ubiquitin proteolysis machinery through Skp1: coupling glucose sensing to gene expression and the cell cycle. EMBO J., 16, 5629–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisztwan J., Marti,A., Sutterlüty,H., Gstaiger,M., Wirbelauer,C. and Krek,W. (1998) Association of human CUL1 and ubiquitin-conjugating enzyme CDC34 with the F-box protein p45SKP2: evidence for evolutionary conservation in the subunit composition of the CDC34-SCF pathway. EMBO J., 17, 368–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludemann R., Lerea,K.M. and Etlinger,J.D. (1993) Copurification of casein kinase II with 20S proteasomes and phosphorylation of a 30 kDa proteasome subunit. J. Biol. Chem., 268, 17413–17417. [PubMed] [Google Scholar]
- Lyapina S.A., Correl,C.C., Kipreos,E.T. and Deshaies,R.J. (1998) Human CUL1 forms an evolutionarily conserved ubiquitin ligase complex (SCF) with SKP1 and an F-box protein. Proc. Natl Acad. Sci. USA, 95, 7451–7456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti A., Wirbelauer,C., Scheffner,M. and Krek,W. (1999) Interaction between ubiquitin-ligase SCFSKP2 and E2F-1 underlies the regulation of E2F-1 degradation. Nature Cell Biol., 1, 14–19. [DOI] [PubMed] [Google Scholar]
- Mathur J., Szabados,L., Schaefer,S., Grunenberg,B., Lossow,A., Jonas-Straube,E., Schell,J., Koncz,C. and Koncz-Kálmán,Z. (1998) Gene identification with sequenced T-DNA tags generated by transformation of Arabidopsis cell suspensions. Plant J., 13, 707–716. [DOI] [PubMed] [Google Scholar]
- Mumberg D., Müller,R. and Funk,M. (1994) Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res., 22, 5767–5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muranaka T., Banno,H. and Machida,Y. (1994) Characterization of tobacco protein kinase NPK5, a homolog of Saccharomyces cerevisiae SNF1 that constitutively activates expression of the glucose-repressible SUC2 gene for a secreted invertase of S. cerevisiae. Mol. Cell. Biol., 14, 2958–2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama K. et al. (2000) Targeted disruption of SKP2 results in accumulation of cyclin E and p27Kip1, polyploidy and centrosome overduplication. EMBO J., 19, 2069–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Németh K. et al. (1998) Pleiotropic control of glucose and hormone responses by PRL1, a nuclear WD protein, in Arabidopsis. Genes Dev., 12, 3059–3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo P.S., Murray,P.F., Walz,K., Franco,L. and Passerson,S. (1998) In vivo and in vitro phosphorylation of the α7/PSR1 subunit of Saccharomyces cerevisiae 20S proteasome: in vitro phosphorylation by protein kinase CK2 is absolutely dependent on polylysine. Arch. Biochem. Biophys., 349, 397–401. [DOI] [PubMed] [Google Scholar]
- Parmentier Y., Bouchez,D., Fleck,J. and Genschik,P. (1997) The 20S proteasome gene family in Arabidopsis thaliana. FEBS Lett., 416, 281–285. [DOI] [PubMed] [Google Scholar]
- Patton E.E., Willems,A.R. and Tyers,M. (1998) Combinatorial control in ubiquitin-dependent proteolysis: don’t Skp the F-box hypothesis. Trends Genet., 14, 236–243. [DOI] [PubMed] [Google Scholar]
- Pih K.T., Yi,M.J., Liang,Y.S., Shin,B.J., Cho,M.J., Hwang,I. and Son,D. (2000) Molecular cloning and targeting of a fibrillarin homolog from Arabidopsis. Plant Physiol., 123, 51–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podust V.N., Brownell,J.E., Gladysheva,T.B., Luo,R.-S., Wang,C., Coggins,M.B., Pierce,J.W., Lightcap,E.S. and Chau,V. (2000) A Nedd8 conjugating pathway is essential for proteolytic targeting of p27Kip1 by ubiquitination. Proc. Natl Acad. Sci. USA, 97, 4579–4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Realini C., Rogers,S.W. and Rechsteiner,M. (1994) Proposed roles in protein–protein association and presentation of peptides by MHC class I receptors. FEBS Lett., 348, 109–113. [DOI] [PubMed] [Google Scholar]
- Rechsteiner M. (1998) The 26S proteasome. In Peters,J.-M., Harris,R.J. and Finley,D. (eds), Ubiquitin and the Biology of the Cell. Plenum Press, New York, NY, pp. 147–189.
- Ruegger M., Dewey,E., Gray,W.M., Hobbie,L., Turner,J. and Estelle,M. (1998) The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast Grr1p. Genes Dev., 12, 198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach A., Klenz,J.E., Kohalmi,S.E., Risseeuw,E., Haughn,G.W. and Crosby,W.L. (1999) The UNUSUAL FLORAL ORGANS gene of Arabidopsis thaliana is an F-box protein required for normal patterning and growth in the floral meristem. Plant J., 20, 433–445. [DOI] [PubMed] [Google Scholar]
- Schulman B.A. et al. (2000) Insights into SCF ubiquitin ligases from the structure of the Skp1–Skp2 complex. Nature, 408, 381–386. [DOI] [PubMed] [Google Scholar]
- Skowyra D., Craig,K.L., Tyers,M., Elledge,S.J. and Harper,J.W. (1997) F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell, 91, 209–219. [DOI] [PubMed] [Google Scholar]
- Smeekens S. (1998) Sugar regulation of gene expression in plants. Curr. Opin. Plant Biol., 1, 230–234. [DOI] [PubMed] [Google Scholar]
- Tokumoto M., Horiguchi,R., Nagahama,Y. and Tokumoto,T. (1999) Identification of the Xenopus 20S proteasome α4 subunit which is modified in the meiotic cell cycle. Gene, 239, 301–308. [DOI] [PubMed] [Google Scholar]
- Tokumoto M., Horiguchi,R., Nagahama,Y., Ishikawa,K. and Takumoto,T. (2000) Two proteins, a goldfish 20S proteasome subunit and the protein interacting with 26S proteasome, change in the meiotic cell cycle. Eur. J. Biochem., 267, 97–103. [DOI] [PubMed] [Google Scholar]
- Tongaonkar P., Chen,L., Lamberston,D., Ko,B. and Madure,K. (2000) Evidence for an interaction between ubiquitin-conjugating enzymes and the 26S proteasome. Mol. Cell. Biol., 20, 4691–4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsvetkov L.M., Yeh,K.H., Lee,S.J., Sun,H. and Zhang,H. (1999) p27KIP1 ubiquitination and degradation is regulated by the SCFSkp2 complex through phosphorylated Thr187 in p27. Curr. Biol., 9, 661–664. [DOI] [PubMed] [Google Scholar]
- Tyers M. and Jorgensen,P. (2000) Proteolysis and the cell cycle: with this RING I do thee destroy. Curr. Opin. Genet. Dev., 10, 54–64. [DOI] [PubMed] [Google Scholar]
- Umeda M., Manabe,Y. and Uchimiya,H. (1997a) Phosphorylation of the C2 subunit of the proteasome in rice (Oryza sativa L.). FEBS Lett., 403, 313–317. [DOI] [PubMed] [Google Scholar]
- Umeda M., Fujii,N., Manabe,Y. and Uchimiya,H. (1997b) Molecular and biochemical characterization of a proteasome subunit from rice and carrot cells. Mol. Gen. Genet., 255, 19–27. [DOI] [PubMed] [Google Scholar]
- van Nocker S., Sadis,S., Rubin,D.M., Glickman,M., Fu,H., Coux,O., Wefes,I., Finley,D. and Vierstra,R.D. (1996) The multiubiquitin-chain-binding protein Mcb1 is a component of the 26S proteasome in Saccharomyces cerevisiae and plays a nonessential, substrate-specific role in protein turnover. Mol. Cell. Biol., 16, 6020–6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaulon S., Vasseur-Cognet,M. and Kahn,A. (2000) Glucose regulation of gene transcription. J. Biol. Chem., 275, 31555–31558. [DOI] [PubMed] [Google Scholar]
- Verma R. and Deshaies,R.J. (2000) A proteasome howdunit; the case of the missing signal. Cell, 101, 341–344. [DOI] [PubMed] [Google Scholar]
- Verma R., Chen,S., Feldman,R., Schieltz,D., Yates,J., Dohmen,J. and Deshaies,R.J. (2000) Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol. Biol. Cell, 11, 3425–3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitha S., Baluska,F., Mews,M. and Volkmann,D. (1997) Immuno fluorescence detection of F-actin on low melting point wax sections from plant tissues. J. Histochem. Cytochem., 45, 89–95. [DOI] [PubMed] [Google Scholar]
- Voges D., Zwickl,P. and Baumeister,W. (1999) The 26S proteasome, a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem., 68, 1015–1068. [DOI] [PubMed] [Google Scholar]
- Wirbelauer C., Sutterlüty,H., Blondel,M., Gsteiger,M., Peter,M., Reymond,F. and Krek,W. (2000) The F-box protein Skp2 is a ubiquitylation target of a Cul1-based core ubiquitin ligase complex: evidence for a role of Cul1 in the suppression of Skp2 expression in quiescent fibroblasts. EMBO J., 19, 5362–5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitby F.G., Masters,E.I., Kramer,L., Knowlton,J.R., Yao,Y., Wang,C.C. and Hill,C.P. (2000) Structural basis for the activation of 20S proteasomes by 11S regulators. Nature, 408, 115–120. [DOI] [PubMed] [Google Scholar]
- Withers-Ward E.S., Mueller,T.D., Chen,I.S. and Feigon,J. (2000) Biochemical and structural analysis of the interaction between the UBA(2) domain of the DNA repair protein HHR23A and HIV-1 Vpr. Biochemistry, 39, 14103–14112. [DOI] [PubMed] [Google Scholar]
- Xie D.-X., Feys,B.F., James,S., Nieto-Rostro,M. and Turner,J.G. (1998) COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science, 280, 1091–1094. [DOI] [PubMed] [Google Scholar]
- Xie Y. and Varshavsky,A. (2000) Physical association of ubiquitin ligases and the 26S proteasome. Proc. Natl Acad. Sci. USA, 97, 2497–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yam C.H., Ng,R.W.M., Siu,W.Y., Lau,A.W.S. and Poon,R.Y.C. (1999) Regulation of cyclin A–CDK2 by SCF component Skp1 and F-box protein Skp2. Mol. Cell. Biol., 19, 635–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Hu,Y., Lodhi,M., McCombie,R. and Ma,H. (1999) The Arabidopsis SKP1-LIKE1 gene is essential for male meiosis and may control homologue separation. Proc. Natl Acad. Sci. USA, 96, 11416–11421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z.-K., Gervais,J.L.M. and Zhang,H. (1998) Human CUL1 associates with the SKP1/SKP2 complex and regulates p21CIP1/WAF1 and cyclin D proteins. Proc. Natl Acad. Sci. USA, 95, 11324–11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X., Miller,M. and Belote,J.M. (1996) Duplicated proteasome subunit genes in Drosophila melanogaster encoding testes-specific isoforms. Genetics, 144, 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Kobayashi,R., Galaktionov,K. and Beach,D. (1995) p19Skp1 and p45Skp2 are essential elements of the cyclin A–CDK2 S phase kinase. Cell, 82, 915–925. [DOI] [PubMed] [Google Scholar]
- Zhao D., Yang,M., Solava,J. and Ma,H. (1999) The ASK1 gene regulates development and interacts with the UFO gene to control floral organ identity in Arabidopsis. Dev. Genet., 25, 209–223. [DOI] [PubMed] [Google Scholar]