Abstract
T-DNA nuclear import is a central event in genetic transformation of plant cells by Agrobacterium. This event is thought to be mediated by two bacterial proteins, VirD2 and VirE2, which are associated with the transported T-DNA molecule. While VirD2 is imported into the nuclei of plant, animal and yeast cells, nuclear uptake of VirE2 occurs most efficiently in plant cells. To understand better the mechanism of VirE2 action, a cellular interactor of VirE2 was identified and its encoding gene cloned from Arabidopsis. The identified plant protein, designated VIP1, specifically bound VirE2 and allowed its nuclear import in non-plant systems. In plants, VIP1 was required for VirE2 nuclear import and Agrobacterium tumorigenicity, participating in early stages of T-DNA expression.
Keywords: Agrobacterium/nuclear import/T-DNA/tumorigenicity/VirE2
Introduction
Agrobacterium infection, the only known case of interkingdom DNA transfer (Stachel and Zambryski, 1989), elicits neoplastic growths on many plant species. This genetic transformation is achieved by transporting a single-stranded copy (T-strand) of the bacterial transferred DNA (T-DNA) from the tumor-inducing (Ti) plasmid into the plant cell nucleus followed by its integration into the host genome (reviewed by Gelvin, 2000; Tzfira et al., 2000; Zupan et al., 2000). The wild-type T-DNA carries genes involved in the synthesis of plant growth regulators and tumor-specific compounds, opines. Production of growth regulators in the transformed cell induces the formation of tumors, which then synthesize opines, a major carbon and nitrogen source for Agrobacterium. Thus, Agrobacteria are usually classified based on the type of opines specified by their T-DNA, the most common strains being nopaline- or octopine-specific. In addition to the T-DNA contents, nopaline and octopine Agrobacteria differ from each other in the composition and nucleotide sequence of the virulence (vir) region of their Ti-plasmids, which encodes the protein machinery of the T-DNA transfer (reviewed in Hooykaas and Beijersbergen, 1994).
While only the wild-type T-DNA contains tumor-inducing genes, any DNA placed between the T-DNA borders will be transported into the plant cell nucleus (reviewed by Zambryski, 1992). This lack of sequence specificity implies that a T-DNA molecule itself does not encode specific signals for nuclear import and integration. Instead, these functions are probably performed by two Agrobacterium virulence proteins, VirD2 and VirE2, which are thought to associate directly with the T-strand, forming a transport (T) complex (Zupan and Zambryski, 1997). In the T-complex, one molecule of VirD2 is covalently attached to the 5′ end of the T-strand, while VirE2, a single-stranded (ss) DNA-binding protein (SSB), is presumed to coat the rest of the ssDNA molecule cooperatively (Gietl et al., 1987; Christie et al., 1988; Citovsky et al., 1988; Das, 1988; Sen et al., 1989) and package it into a rigid coiled structure (Citovsky et al., 1997). The need for active nuclear uptake is evident from the calculated diameter of VirE2–ssDNA complexes (12.6 nm; Citovsky et al., 1997), which exceeds the diffusion limit of the nuclear pore (9 nm; reviewed by Rout and Wente, 1994). Presumably, the T-complex nuclear import is mediated by VirD2 and VirE2 proteins, which localize to the plant cell nucleus (Herrera-Estrella et al., 1990; Citovsky et al., 1992, 1994; Howard et al., 1992; Shurvinton et al., 1992; Koukolikova-Nicola et al., 1993; Rossi et al., 1993; Zupan et al., 1996). Whereas VirD2 and VirE2 accumulate in the cell nucleus even in plant species that are recalcitrant to Agrobacterium-induced tumor formation (Citovsky et al., 1994), they probably employ different pathways for nuclear import. VirD2 is imported by a mechanism conserved between animal, yeast and plant cells (Herrera-Estrella et al., 1990; Howard et al., 1992; Koukolikova-Nicola et al., 1993; Rossi et al., 1993; Citovsky et al., 1994; Guralnick et al., 1996; Ziemienowicz et al., 1999; Rhee et al., 2000), while the nuclear import of VirE2 is plant specific in living cells (Citovsky et al., 1992, 1994; Guralnick et al., 1996; Rhee et al., 2000). Consistent with this idea, VirE2 is not recognized by the Arabidopsis karyopherin α protein, AtKAPα, which has been shown to mediate nuclear import of VirD2 (Ballas and Citovsky, 1997). Interest ingly, VirE2 nuclear localizing ability is sufficient for transport of ssDNA into the plant cell nucleus even in the absence of VirD2 (Zupan et al., 1996) or for genetic transformation of plant cells by an Agrobacterium mutant strain lacking the VirD2 nuclear localization signal (NLS) (Gelvin, 1998).
To understand better the molecular mechanism by which VirE2 functions during the Agrobacterium–plant cell T-DNA transfer, it would be useful to identify and characterize plant cellular components that specifically associate with VirE2. Here, we used the yeast two-hybrid protein–protein interaction system (Fields and Song, 1989; Hollenberg et al., 1995) to identify and isolate a VirE2-interacting protein, designated VIP1, from Arabidopsis thaliana. VIP1 allowed VirE2 to be imported into the nuclei of living yeast and mammalian cells and was required for VirE2 nuclear import and Agrobacterium-induced tumor formation in tobacco plants, participating in early stages of T-DNA expression.
Results
Identification of VIP1
We used the yeast two-hybrid screen (Fields and Song, 1989; Hollenberg et al., 1995) with an Arabidopsis cDNA library and the Agrobacterium VirE2 protein as bait. Screening of ∼3 × 106 transformants resulted in identification and isolation of several independent cDNA clones producing VirE2 interactors. Two of these clones encoded the same cDNA, designated VIP1 (VirE2-interacting protein 1). The largest clone, representing the full-length cDNA of VIP1, was characterized in detail.
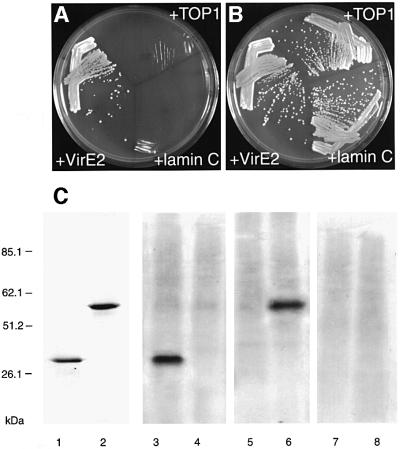
The interaction of VIP1 with VirE2 was specific because it did not occur with DNA topoisomerase I and lamin C, known as non-specific activators in the two-hybrid system best suited to eliminate false-positive interactions (Bartel et al., 1993; Park and Sternglanz, 1998). Figure 1A shows that co-expression of VIP1 and VirE2, but not of topoisomerase I or lamin C, activated the HIS3 reporter gene. Furthermore, VIP1 did not interact with VirD2 (data not shown), which is thought to function differently from VirE2 during the T-DNA nuclear import (Guralnick et al., 1996). In control experiments, under the non-selective conditions, all combinations of the tested proteins resulted in efficient cell growth (Figure 1B).
Fig. 1. Specific interaction between VIP1 and VirE2 in the two-hybrid system and in vitro. (A) Growth in the absence of histidine, tryptophan and leucine. (B) Growth in the absence of tryptophan and leucine. VIP1 was expressed from pGAD424 whereas VirE2 and negative control interactors lamin C and topoisomerase I (TOP I) were expressed from pBTM116. Growth in histidine-deficient medium represents selective conditions for protein–protein interactions. (C) VirE2 binding to immobilized VIP1 in vitro. VIP1 (lanes 1, 3, 5 and 7) and VirD2 (lanes 2, 4, 6 and 8) were electrophoresed, blotted onto a membrane, incubated with VirE2 (lanes 3 and 4), VirD2 (lanes 5 and 6) or TMV MP (lanes 7 and 8) and probed with anti-VirE2, anti-VirD2 or anti-TMV MP antibodies, respectively. Lanes 1 and 2, Coomassie blue staining of VIP1 and VirD2, respectively, after electrophoresis; lanes 3–8, autoradiographs of the binding assays. Protein molecular mass standards are indicated on the left in kDa.
In an independent approach, VIP1–VirE2 binding was examined directly using a renatured blot overlay assay for protein–protein interactions (Dorokhov et al., 1999; Chen et al., 2000). In this approach, VIP1 and a negative control protein, VirD2, are electrophoresed (Figure 1C, lanes 1 and 2), immobilized on a PVDF membrane by electroblotting, reacted with purified VirE2, and VirE2 binding is detected using anti-VirE2 antibodies. Figure 1C shows that VirE2 specifically interacted with immobilized VIP1 (lane 3) but not with VirD2 (lane 4). Furthermore, when the blot was probed with unspecific ligands, i.e. purified VirD2 (lanes 5 and 6) or cell–cell movement protein (MP) of tobacco mosaic virus (TMV) (lanes 7 and 8), no binding to Vip1 was observed. These results strengthened the notion that VirE2 specifically recognizes and binds VIP1.
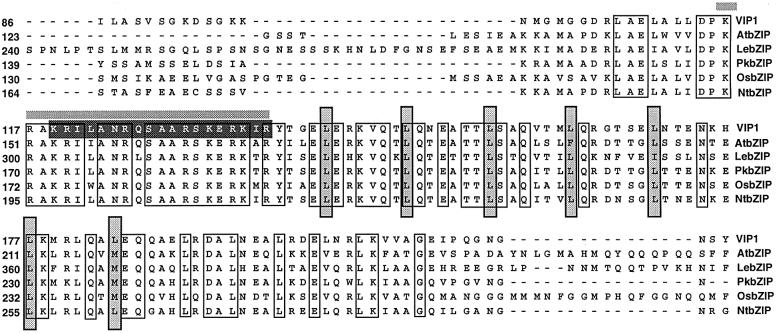
Sequence analysis of the VIP1 cDNA showed that it contained a single open reading frame (ORF) encoding a protein of 261 amino acid residues (see DDBJ/EMBL/GenBank accession Nos AF225983 for VIP1 cDNA and AC009526 for the genomic sequence containing the VIP1 gene). The deduced amino acid sequence of VIP1 contained a conserved stretch of basic amino acids (basic domain) abutting a heptad leucine repeat (leucine zipper), two structural features characteristic of the basic-zipper (bZIP) proteins (Figure 2). Because plant bZIP proteins are known to localize to the cell nucleus (van der Krol and Chua, 1991), we hypothesized that binding of VIP1 to VirE2 may function to facilitate nuclear import of VirE2. Indeed, the basic domain of the VIP1 bZIP motif contained a consensus sequence for the bipartite NLS (Dingwall and Laskey, 1991) (Figure 2). Interestingly,VIP1 exhibited a modest homology to bZIP proteins from various plantspecies, such as Arabidopsis, tomato, Paulownia kawakamii, rice and tobacco (Figure 2), whereas no animal or yeast bZIP homologs of VIP1 were found. This finding supports the notion that VIP1 may, at least partly, be responsible for the plant-specific nuclear import of VirE2. To test this hypothesis, we examined whether expression of VIP1 reconstructs nuclear import of VirE2 in non-plant systems.
Fig. 2. Alignment of the VIP1 bZIP domain with the four most homologous plant proteins identified by the BLASTA search (Altschul et al., 1990). The bZIP domain of VIP1 (DDBJ/EMBL/GenBank accession No. AF225983) was aligned using the clustal algorithm (Saitou and Nei, 1987) with similar motifs of its closest homologs from Arabidopsis thaliana (AtbZIP, accession No. AAB87576), Lycopersicon esculentum (tomato) (LebZIP, accession No. CAA52015), Paulownia kawakamii (PkbZIP, accession No. AAC04862), Oryza sativum (rice) (OsbZIP, accession No. AAC49832) and Nicotiana tabacum (tobacco) (NtbZIP, accession No. BAA97100). Regions of identity are indicated by unshaded boxes; gaps introduced for alignment are indicated by dashes. In the bZIP motif, the seven leucine repeats (leucine zipper) are indicated by shaded boxes and the basic domain is denoted by a horizontal bar above its sequence. The consensus bipartite NLS (Dingwall and Laskey, 1991) within the basic domain of the VIP1 bZIP motif is indicated by a black box.
VIP1 facilitates transport of VirE2 into the nuclei of yeast and mammalian cells
The ability of VIP1 to transport VirE2 into the yeast cell nucleus was examined using a recently developed genetic assay for functional nuclear import (Rhee et al., 2000). In this approach, a gene encoding the bacterial LexA protein was modified (mLexA), abolishing its intrinsic nuclear targeting activity, and fused to a sequence coding for the activation domain of the yeast Gal4p (Gal4AD). If a protein of interest fused to mLexA-Gal4AD (nuclear import assay hybrid, NIA) enters the yeast cell nucleus, it activates the expression of the reporter HIS3 gene, resulting in cell growth on a histidine-deficient medium (Rhee et al., 2000).
First, VirE2 and VIP1 were tested for their own nuclear import capacity. Figure 3 shows that NIA-VirE2 expressed alone did not promote cell growth, indicating the lack of nuclear uptake. As expected, NIA-VirD2, which is known to function in non-plant systems (Guralnick et al., 1996; Ziemienowicz et al., 1999), induced cell growth following its nuclear import. NIA-VirD2 nuclear import was due to the presence of the VirD2 ORF because when the latter was fused to NIA in reverse orientation, producing the NIA-Vir2D hybrid, no cell growth was observed (Figure 3). Similarly to NIA-VirD2, expression of the NIA-VIP1 fusion promoted cell growth, indicating that VIP1 is imported into the cell nucleus in yeast. In control experiments, cells harboring all NIA fusions grew in the presence of histidine, indicating that the protein hybrids did not adversely and non-specifically affect cell physiology.
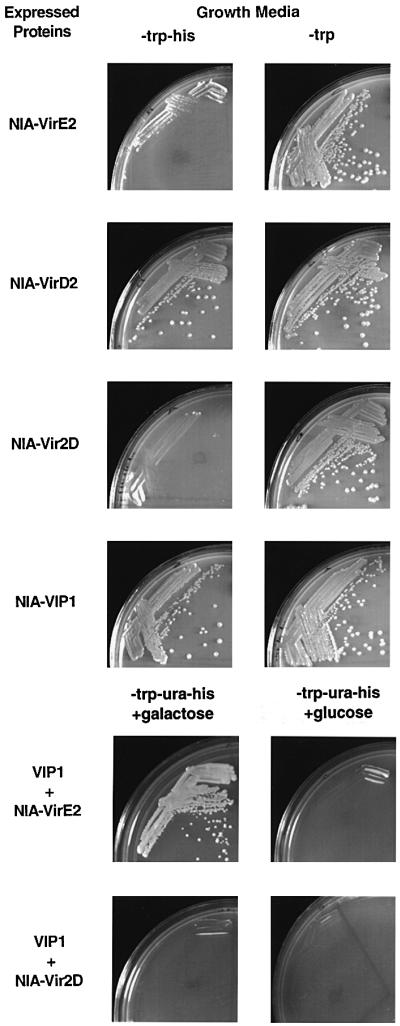
Fig. 3. A functional genetic assay for VIP1-mediated VirE2 nuclear import in yeast cells. Yeast cells expressing the indicated proteins were grown under selective (histidine and tryptophan double-dropout medium) or non-selective conditions (tryptophan single-dropout medium) for nuclear import. For co-expression with VIP1, yeast cells expressing the indicated combinations of tested proteins were grown in a histidine, tryptophan and uracil triple-dropout medium supplemented either with galactose or glucose to induce or repress the VIP1 expression, respectively.
Next, VIP1 was examined for its ability to assist nuclear import of VirE2. To this end, NIA-VirE2 was co-expressed with VIP1 driven by a galactose-inducible promoter. In the presence of galactose, cell growth was observed, indicating that NIA-VirE2 became capable of entering the cell nucleus and inducing expression of the HIS3 reporter (Figure 3). VirE2 nuclear import absolutely depended on the presence of VIP1 because, in the absence of galactose and, thus, VIP1 expression, no cell growth was observed (Figure 3). Facilitation of NIA-VirE2 nuclear import by VIP1 was due to VIP1–VirE2 interaction because co-expression of VIP1 and NIA-Vir2D did not result in nuclear import (Figure 3). These observations indicate that VIP1 facilitates nuclear import of VirE2 in yeast cells. Note that, in these experiments,VirE2 was fused to the mLexA-Gal4AD reporter while the VirE2–VIP1 binding experiments in the two-hybrid system utilized a relatively similar LexA-VirE2 fusion, providing compatibility between the protein–protein interaction and nuclear import data with respect to the VirE2 fusions used.
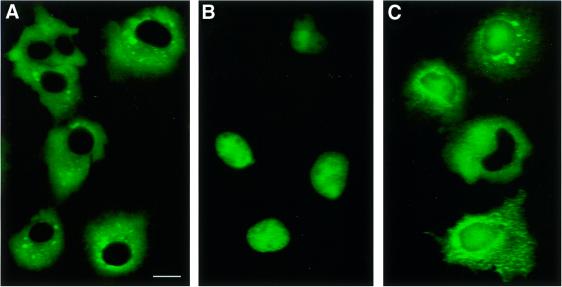
To investigate the effect of VIP1 on VirE2 nuclear import in a mammalian system, these proteins were introduced into COS-1 cells and their intracellular localization determined using confocal microscopy. First, VirE2 and VIP1 were fused to the green fluorescent protein (GFP) and expressed separately. Figure 4 shows that GFP–VirE2 expressed alone remained completely cytoplasmic (dispersed fluorescent signal surrounding fluorescence-free, black nuclei in Figure 4A), whereas GFP–VIP1 efficiently localized to the cell nucleus (fluorescent signal exclusively concentrated within cell nuclei in Figure 4B). Then, GFP–VirE2 was co-expressed with unlabeled VIP1, resulting in entry of the fluorescent signal into the cell nucleus (Figure 4C). VIP1-facilitated nuclear import of GFP–VirE2 was incomplete because the fluorescent signal was found both accumulated within the cell nucleus and dispersed in the cytoplasm areas surrounding the nucleus (Figure 4C) (see also below). It is important to note that the confocal optical sections with the plane of focus through the cell nucleus detect intranuclear accumulation of the GFP label rather than its perinuclear binding. Thus, taken together, our functional genetic and microscopic data suggest that VIP1 plays an important role in the plant-specific nuclear import of VirE2.
Fig. 4. VIP1-mediated nuclear import of VirE2 in mammalian cells. (A) COS-1 cells expressing GFP–VirE2. Dispersed fluorescence surrounding the signal-free, black cell nucleus represents the cytoplasmic localization of GFP–VirE2. (B) COS-1 cells expressing GFP–VIP1. The fluorescent signal is concentrated exclusively in the cell nucleus. (C) COS-1 cells co-expressing GFP–VirE2 and unlabeled VIP1. In most cells, part of the fluorescent signal enters the nucleus and part remains dispersed in the surrounding areas of the cytoplasm. Bar = 15 µm.
Quantification of GFP–VirE2 amounts on a per cell basis revealed that VIP1 redirected 40–60% of total expressed GFP–VirE2 to the cell nucleus. Because the VIP1 effect on VirE2 nuclear import is likely to be stoichiometric, it depends on the relative amounts of these proteins within the cell cytoplasm. Potentially, transient expression of GFP–VirE2 and VIP1 from separate plasmids does not generate or sustain protein concentrations necessary for the complete nuclear import of VirE2. Alternatively, in plants, VIP1 may be augmented by other cellular factors absent from the heterologous mammalian system.
VIP1 antisense plants are resistant to Agrobacterium-induced tumor formation
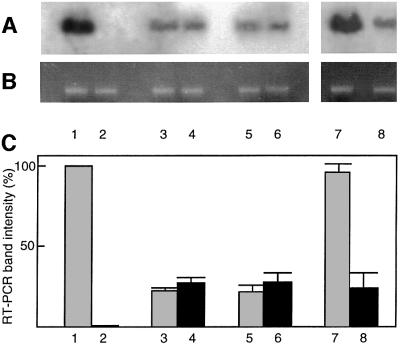
To study the biological role of VIP1 in Agrobacterium infection in planta, we generated transgenic tobacco plants expressing the VIP1 cDNA in the antisense orientation. A total of 10 independently transformed lines were produced and analyzed as described below. Five lines were not altered in their susceptibility to Agrobacterium infection, whereas the other five lines became largely resistant (data not shown). Here, we describe a detailed analysis of two of these resistant antisense lines, which were first examined for the presence of sense and antisense VIP1 RNA using quantitative RT–PCR and strand-specific oligonucleotide primers (Ni et al., 1998). Figure 5A shows that control, wild-type tobacco plants produced sense (lane 1), but not antisense (lane 2), VIP1 RNA, demonstrating the presence of VIP1 in tobacco (see also Figure 2). Two independent lines of VIP1 antisense plants, which exhibited resistance to Agrobacterium (see below), produced the antisense VIP1 RNA (Figure 5A, lanes 4 and 6) and significantly reduced amounts of the sense VIP1 RNA (Figure 5A, lanes 3 and 5). Conversely, VIP1 antisense lines that did not develop Agrobacterium, resistance retained high levels of the sense VIP1 transcript (Figure 5A, lane 7) even though the antisense transcript was also expressed (Figure 5A, lane 8).
Fig. 5. Quantitative RT–PCR analysis of wild-type and VIP1 antisense plants. (A) Detection of sense and antisense VIP1 RNA. Lanes 1 and 2, RT–PCR of sense and antisense VIP1 RNA in wild-type plants; lanes 3 and 4, RT–PCR of sense and antisense VIP1 RNA in one line of Agrobacterium-resistant VIP1 antisense plants; lanes 5 and 6, RT–PCR of sense and antisense VIP1 RNA in another line of Agrobacterium-resistant VIP1 antisense plants; lanes 7 and 8, RT–PCR of sense and antisense VIP1 RNA in a line of Agrobacterium-sensitive VIP1 antisense plants. (B) Detection of sense actin RNA-specific product in the same samples shown in (A). (C) Quantification of sense and antisense VIP1 RNA. The amount of VIP1-specific RT–PCR products is expressed as a percentage of that obtained using sense VIP1-specific primers in wild-type plants. These data represent average values of three independent experiments with the indicated standard deviations.
Quantification of these PCR products (Figure 5C) revealed that the sense VIP1 RNA, synthesized in the Agrobacterium-resistant antisense plants (bars 3 and 5), amounted to only 20% of that produced in the wild-type plants (bar 1), whereas the antisense RNA, undetectable in wild-type plants (bar 2), reached 25% of sense VIP1 RNA of the wild-type plants (bars 4 and 6). In Agrobacterium-sensitive antisense plants, the levels of the sense VIP1 RNA remained high (98% of the wild-type levels, bar 7) even in the presence of the antisense transcript (20% of sense VIP1 RNA of the wild-type plants, bar 8). In control experiments, analysis of actin-specific transcripts generated similar amounts of PCR products in all samples, indicating equal efficiencies of the RT–PCRs (Figure 5B). Thus, antisense expression of VIP1 cDNA in Agro bacterium-resistant transgenic tobacco substantially reduced transcription of the endogenous VIP1 gene and, by implication, synthesis of the VIP1 protein (data not shown). Furthermore, that Agrobacterium-sensitive antisense lines retained high levels of the sense VIP1 transcript supported the correlation between the RT–PCR data and the antisense phenotype.
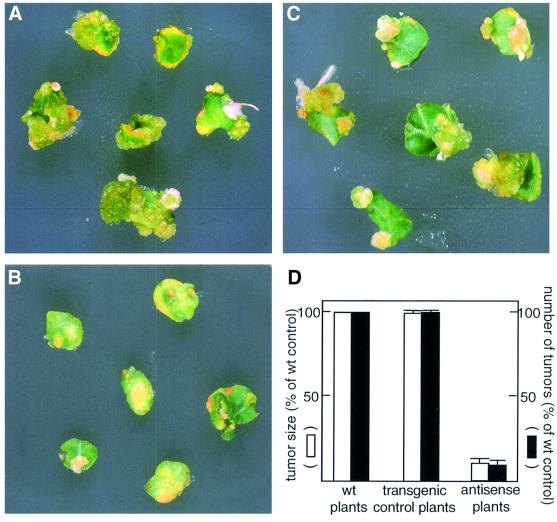
Next, we tested the ability of the VIP1 antisense plants to develop tumors following inoculation with wild-type, oncogenic Agrobacterium. Figure 6A shows that Agro bacterium elicited numerous and large tumors on leaf disks derived from the wild-type tobacco plants. Control transgenic plants transformed with an empty vector were equally susceptible to Agrobacterium-induced neoplastic growth (Figure 6C), indicating that the procedure used for generation of the transgenic plants did not render them resistant to the subsequent Agrobacterium infection. In contrast, two independent transgenic lines of VIP1 antisense plants exhibited a dramatic decrease in their susceptibility to Agrobacterium infection. Figure 6B shows that only very few and tiny tumors developed on leaf disks from one of these plants following inoculation with Agrobacterium.
Fig. 6. Reduced tumor formation in Agrobacterium-infected VIP1 antisense plants. (A) Leaf disks from the wild-type tobacco plants. (B) Leaf disks from the VIP1 antisense transgenic plants. (C) Leaf disks from the control transgenic plants. (D) Summary of the number and sizes of Agrobacterium-induced tumors developed on leaf disks from two independent lines of VIP1 antisense plants and two lines of transgenic control plants relative to the wild-type (wt) plants.
Agrobacterium infectivity was then quantified by the number and weight of tumors induced on the inoculated leaf disks. Figure 6D shows the results of these measurements averaged for both antisense lines as compared with the wild-type and transgenic control plants. Wild-type tobacco plants and transgenic control lines supported formation of multiple tumors (4–8 per leaf disk), which expanded and fused into large neoplastic growths (200–300 mg). The VIP1 antisense plants, on the other hand, developed only a very low number (1.0–1.5 per leaf disk) of small tumors (20–40 mg). Thus, the tumor-inducing activity of Agrobacterium in VIP1 antisense plants was reduced to ∼10% of that observed with the wild-type and transgenic control plants (Figure 6D). Progeny analysis demonstrated that this tumor-resistant phenotype of the VIP1 antisense plants co-segregated with the T-DNA inserts (data not shown).
All VIP1 antisense plants were indistinguishable from the wild-type plants in their morphology and seed viability (data not shown). Furthermore, shoot regeneration from uninfected VIP1 antisense leaf disks cultured on tobacco regeneration medium (Horsch et al., 1985) was identical in its rate and efficiency to that of the wild-type plants (data not shown). Thus, VIP1 antisense expression most probably did not interfere with essential plant cellular functions.
Early stages of T-DNA gene expression are blocked in VIP1 antisense plants
Expression of genes contained on the Agrobacterium T-DNA takes place in two stages, early and late. Early gene expression, which reaches its maximum 2–4 days after infection (Janssen and Gardner, 1990; Nam et al., 1999), is transient, occurring from the T-DNA molecules that have not yet integrated into the plant genome. In contrast, late gene expression, which occurs 10–14 days after infection (Janssen and Gardner, 1990), is stable, resulting from the integrated T-DNA. If VIP1 indeed participates in nuclear import of the T-DNA, VIP1 antisense plants are expected to display reduced levels of T-DNA gene expression already early in the infection process, i.e. before the T-DNA integration can take place.
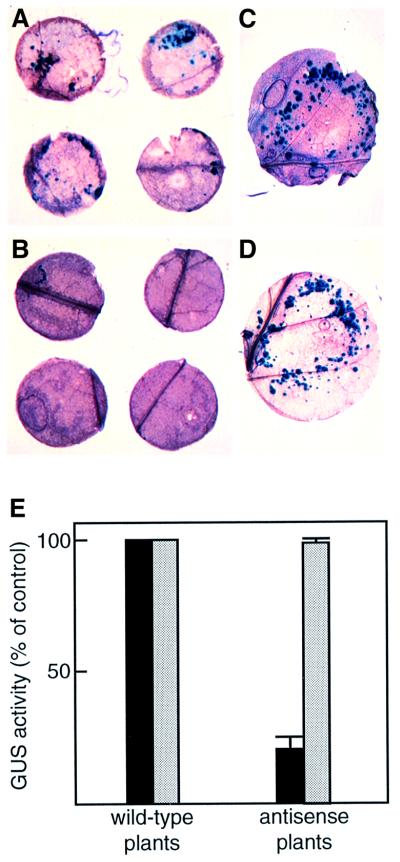
To test this idea, we determined the efficiency of transient T-DNA gene expression by inoculating leaf disks derived from the wild-type and VIP1 antisense plants with Agrobacterium carrying on its T-DNA a uidA gene encoding a reporter enzyme β-glucuronidase (GUS). Figure 7A shows multiple areas of GUS histochemical staining on leaf disks derived from the wild-type plants, indicating transient T-DNA expression within the Agrobacterium-infected cells; identical results were obtained using control transgenic plants (data not shown). VIP1 antisense plants, on the other hand, failed transiently to express GUS contained on the T-DNA (Figure 7B). Both wild-type and VIP1 antisense plants supported GUS expression resulting from biolistic delivery of the uidA gene (Figure 7C and D).
Fig. 7. Reduced transient expression of GUS activity contained within Agrobacterium T-DNA in VIP1 antisense plants. (A) Infected leaf disks from the wild-type tobacco plants. (B) Infected leaf disks from the VIP1 antisense transgenic plants. (C) Microbombarded leaf disk from the wild-type tobacco plants. (D) Microbombarded leaf disk from the VIP1 antisense transgenic plants. Note that microbombardment experiments (C and D) required larger leaf disks than that used in Agrobacterium inoculations (A and B). (E) Quantification of GUS activity. Black and white bars indicate transient GUS expression in Agrobacterium-infected and microbombarded tissues, respectively, derived from two independent lines of VIP1 antisense plants as compared with the wild-type control plants. GUS activity in control, wild-type plants was defined as 100%. All data represent average values of three independent experiments with the indicated standard deviations.
Next, early expression of GUS in two independent VIP1 antisense transgenic lines was quantified using a sensitive fluorimetric assay. Because the uidA gene contained on the T-DNA lacked regulatory sequences required for its expression in bacterial cells (Janssen and Gardner, 1990), our measurements represented the GUS activity directed by the T-DNA after its transfer to the plant rather than its potentially leaky expression in Agrobacterium. Figure 7E shows that VIP1 antisense plants inoculated with Agrobacterium displayed ∼20% transient GUS activity compared with the wild-type plants. No quantitative differences in GUS activity were detected when the uidA gene was introduced biolistically into the wild-type and VIP1 antisense plants (Figure 7E). Thus, VIP1 antisense plants specifically blocked early stages of the Agrobacterium-mediated uidA gene transfer but remained competent for efficiently expressing this gene when it was delivered by an Agrobacterium-independent technique.
Nuclear import of VirE2 is impaired in VIP1 antisense transgenic plants
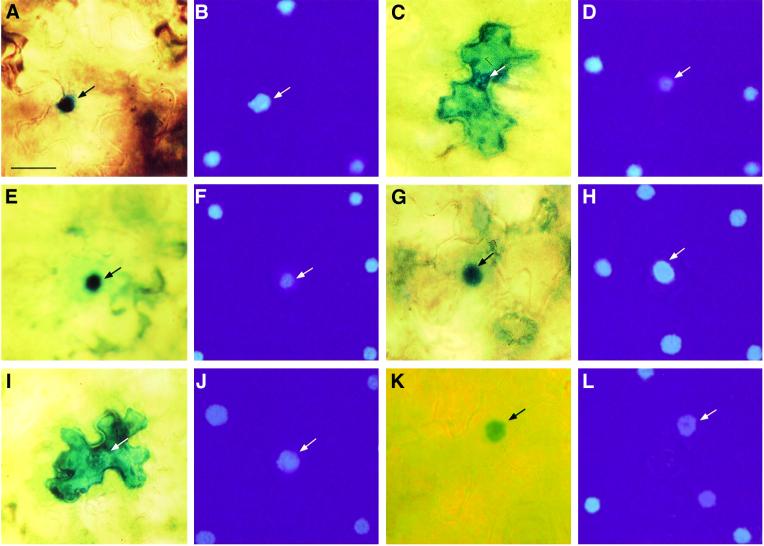
To examine directly whether the reduced susceptibility of VIP1 antisense plants to Agrobacterium-mediated gene transfer was due to a decrease in VirE2 nuclear import, we compared the accumulation of VirE2 within the cell nuclei of the wild-type and VIP1 antisense tobacco tissues. Figure 8 shows that GUS–VirE2 expressed in the mesophyll of wild-type tobacco leaves, following biolistic delivery of its encoding gene, accumulated in the cell nucleus (Figure 8A), co-localizing with the nucleus-specific stain, 4′,6-diamidino-2-phenylindole (DAPI) (Figure 8B). However, GUS–VirE2 expressed in the leaf mesophyll cells of VIP1 antisense plants remained largely cytoplasmic (Figure 8C and D), supporting the notion of VIP1 involvement in VirE2 nuclear import. Importantly, nuclear import of GUS–VirD2 expressed in wild-type plants (Figure 8E and F) was identical to that in VIP1 antisense tissues (Figure 8G and H), indicating that VirE2 and VirD2 are imported into the host cell nucleus by different mechanisms and that antisense expression of VIP1 does not interfere non-specifically with the nuclear import reactions of the cell. As expected, free GUS expressed in wild-type (data not shown but see Table I, and Citovsky et al., 1992, 1994) and VIP1 antisense plants (Figure 8I and J) remained cytoplasmic. Similarly to our observations in yeast and COS cells (see Figures 3 and 4B, respectively), VIP1 accumulated efficiently in the cell nucleus when expressed in wild-type tobacco plants (Figure 8K and L). Thus, the effect of VIP1 on nuclear transport of VirE2 most probably derives directly from the nuclear import capacity of VIP1 itself. Also, as expected, we detected no differences in nuclear import of GUS– VirE2 and GUS–VirD2 between the wild-type plants and control transgenic plants (data not shown), confirming that the procedure used for generation of the transgenic plants did not non-specifically affect their capacity for nuclear import.
Fig. 8. Nuclear import of GUS–VirE2 and GUS–VirD2 in wild-type and VIP1 antisense plants. (A and B) GUS–VirE2 expressed in wild-type plants. (C and D) GUS–VirE2 expressed in VIP1 antisense plants. (E and F) GUS–VirD2 expressed in wild-type plants. (G and H) GUS–VirD2 expressed in VIP1 antisense plants. (I and J) Free GUS expressed in VIP1 antisense plants. (K and L) GUS–VIP1 expressed in wild-type plants. (A, C, E, G, I and K) GUS staining. (B, D, F, H, J and L) DAPI staining. Bar = 25 µm.
We then analyzed the efficiency with which GUS– VirD2 and GUS–VirE2 were imported into the cell nucleus in the wild-type and VIP1 antisense plants. Table I demonstrates that, while no nuclear import of free GUS reporter was observed, all GUS–VirD2 accumulated in the nuclei of the expressing cells in both plants. In the wild-type plants, 100% of GUS–VIP1 and 97% of GUS–VirE2 was found within the nucleus, indicating efficient nuclear import of this protein. In contrast, only 30% of GUS–VirE2 was imported into the nuclei of VIP1 antisense cells (Table I); note that the expression of GUS–VIP1 in these was difficult to determine, potentially due to its suppression by the antisense transgene. These observations further support the role of VIP1 in VirE2 nuclear import and also suggest that a smaller fraction of VirE2 (30%) may enter the plant cell nucleus by another, VIP1-independent pathway. Alternatively, residual amounts of VIP1 in the antisense plants may be responsible for this low level of import.
Table I. Efficiency of nuclear localization of GUS–VirE2 and GUS–VirD2 in wild-type and VIP1 antisense plants.
| GUS activity (% of maximal) |
||
|---|---|---|
| Nucleus | Cytoplasm | |
| Wild-type plants | ||
| GUS–VirE2 | 97 (2) | 3 (2) |
| GUS–VirD2 | 100 (8) | 0 |
| GUS alone | 0 | 100 (6) |
| GUS–VIP1 | 100 (2) | 0 |
| VIP1 antisense plants | ||
| GUS–VirE2 | 30 (7) | 70 (7) |
| GUS–VirD2 | 100 (4) | 0 |
| GUS alone | 0 | 100 (5) |
The data are derived from spectral analysis of photographic images of 20 independent GUS-positive cells; average values are given, and standard error values are indicated in parentheses. All other conditions were as described in Materials and methods and in Citovsky et al. (1992) and Howard et al. (1992).
VIP1 forms a ternary complex with VirE2 and ssDNA
Our results suggest that VIP1 is involved in nuclear import of VirE2 and, by implication, the Agrobacterium T-complexes. Thus, VIP1 should be able to interact with VirE2 while the latter is bound to the ssDNA of the T-complex. We tested this assumption in vitro, using the agarose gel shift assay to monitor protein–ssDNA binding (Lohman et al., 1986; Citovsky et al., 1992). To approximate native T-complexes better, a relatively long (7.2 kb) M13mp18 ssDNA was used. Figure 9 shows that incubation of this ssDNA probe with VirE2 resulted in a gel shift due to formation of VirE2–ssDNA complexes (compare lanes 1 and 2). These results are consistent with the well-known ssDNA-binding activity of VirE2 (Gietl et al., 1987; Christie et al., 1988; Citovsky et al., 1988, 1992; Das, 1988; Sen et al., 1989). In contrast, incubation of the ssDNA probe with VIP1 produced no gel shift (Figure 9, lane 3), indicating that VIP1 does not bind ssDNA. Addition of VIP1 to the VirE2–ssDNA complexes decreased their electrophoretic mobility (Figure 9, lane 4) as compared with that observed in the absence of VIP1 (lane 2). This gel-shifted signal indeed represented protein–DNA complexes because it was not detected after treatment with proteinase K (Figure 9, lane 5). Thus, VIP1, which by itself was unable to associate with ssDNA, probably formed a ternary complex with VirE2 bound to ssDNA.
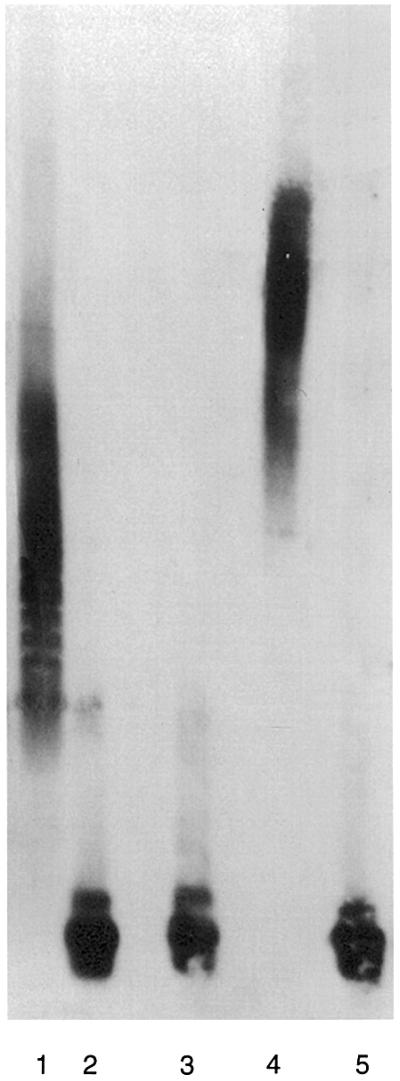
Fig. 9. In vitro formation of ternary complexes between VIP1, VirE2 and ssDNA. Gel shift assays were performed as described in Materials and methods. Lane 1, ssDNA incubated with VirE2; lane 2, ssDNA incubated alone; lane 3, ssDNA incubated with VIP1; lane 4, ssDNA incubated with VirE2 and VIP1; lane 5, ssDNA incubated with VirE2 and VIP1 and treated for 30 min at 37°C with 1 mg/ml of proteinase K.
Discussion
T-DNA transport into the host cell nucleus is the central event in genetic transformation of plants by Agro bacterium. The bacterial protein VirE2 is one of the major players in this process. VirE2 cooperatively binds the transported DNA molecule, shapes it into a transferable form and, together with VirD2, mediates its nuclear uptake (reviewed by Tzfira and Citovsky, 2000; Tzfira et al., 2000). Furthermore, VirE2 alone was shown actively to import ssDNA into the plant cell nucleus (Zupan et al., 1996). Nuclear localization of the nopaline-type VirE2 is especially intriguing because, unlike nuclear import of VirD2, it occurs in plant but not in animal or yeast cells (Citovsky et al., 1992, 1994; Guralnick et al., 1996; Rhee et al., 2000). On the other hand, octopine VirE2 has been shown to enter animal cell nuclei in vitro (Ziemienowicz et al., 1999), suggesting that nopaline and octopine VirE2 proteins may differ in their ability to function in non-plant systems. Alternatively, nuclear import of VirE2 in a cell-free system may differ from that within living cells. Interestingly, however, octopine VirE2 function in animal cells may still be impaired because it was unable to mediate nuclear import of ssDNA in this system (Ziemienowicz et al., 1999).
The lack of nuclear import of VirE2 in living animal and yeast cells suggests involvement of plant-specific host cellular factors that interact with VirE2. Using the yeast two-hybrid system to probe protein–protein interactions (Fields and Song, 1989), we have isolated an Arabidopsis cDNA that encodes a VirE2-interacting protein. The deduced amino acid sequence of this protein, designated VIP1, contained a bZIP motif composed of a long basic domain followed by a leucine zipper, seven leucine repeats evenly separated from each other by six amino acid residues. Although the bZIP sequence is found in many transcription factors (van der Krol and Chua, 1991), the protein database search did not identify any yeast or animal homologs of VIP1. In contrast, bZIP proteins from several diverse plant species showed homology to VIP1. That VIP1 was more related to bZIP proteins of plant rather than animal or yeast origin and that plant bZIP proteins are known to localize to the cell nucleus (van der Krol and Chua, 1991) suggested (i) a role for VIP1 in plant-specific nuclear import of VirE2 and (ii) that bZIP motifs per se also found in non-plant proteins are not sufficient to mediate the VirE2 nuclear import.
Genetic and confocal microscopy experiments indeed demonstrated that VIP1 localized efficiently to the nucleus in yeast and mammalian cells. Co-expression of VIP1 with VirE2 in these non-plant systems allowed VirE2 to be imported into the cell nucleus, suggesting that VIP1 may represent a cellular factor involved in the plant-specific nuclear uptake of VirE2. However, because the VIP1-mediated VirE2 nuclear import in mammalian cells was, although significant, still incomplete, other proteins may be involved in the VirE2 import pathway in planta, augmenting the VIP1 function. Facilitation of VirE2 nuclear import by VIP1 in yeast and mammalian cells also indicates that these two proteins specifically recognize and bind each other in vivo, within the cell cytoplasm.
If VIP1 is required for nuclear import of VirE2, which, in turn, is required for Agrobacterium tumorigenicity (Citovsky et al., 1992; Gelvin, 1998), inactivation of VIP1 should lead to a decrease in Agrobacterium-induced tumor formation. This notion was tested using transgenic tobacco plants in which the expression of their VIP1 gene was repressed by antisense expression of the Arabidopsis VIP1 cDNA. The rationale for this heterologous antisense approach is 2-fold. First, it has been successful both in plant and animal systems; for example, antisense expression of an alfalfa gene suppressed its tobacco homolog (Schiene et al., 2000) and a human gene was repressed by antisense expression of its mouse homolog (Oku et al., 1998). This is because antisense expression of a transgene is well known to silence not only the same gene but also its closely related homologs (reviewed by Vaucheret et al., 1998). Secondly, tobacco plants are much more suitable for standard assays of Agrobacterium–host interaction, e.g. tumorigenicity and transient and stable T-DNA expression, than Arabidopsis plants. Our results demonstrated that one-half of all produced VIP1 antisense tobacco plants (five out of 10 lines) exhibited a considerably reduced production of the VIP1 RNA. That not all transgenic lines develop antisense suppression is common and has been observed before with other transgenes (see, for example, Stam et al., 2000). Importantly, all antisense plants in which VIP1 expression was reduced developed a remarkable resistance to infection by Agrobacterium. Agrobacterium resistance of the VIP1 antisense plants was due to a blockage in early stages of T-DNA gene expression, which precede integration. Because these plants also failed to support efficient nuclear import of VirE2, we suggest that VIP1 activity during nuclear import of the invading T-complexes is a prerequisite for both transient T-DNA gene expression and tumor formation.
Specifically, we propose that VIP1 represents a cellular interactor for Agrobacterium VirE2. During Agro bacterium infection, VIP1 possibly recognizes VirE2 within the host cell cytoplasm. Because VIP1 itself is a nuclear protein, which is probably imported into the nucleus through the basic domain of its bZIP motif, binding of VIP1 to VirE2 may result in a ‘piggyback’ transport of VirE2 into the host cell nucleus. VirE2, in turn, is presumed to be bound cooperatively to the T-strand (Citovsky et al., 1989; Sen et al., 1989); thus, VIP1, most probably in concert with other cellular factors, may participate in the nuclear import of the entire Agro bacterium T-complex. This idea is consistent with our observations that VIP1 can bind to the VirE2–ssDNA complexes in vitro.
Interestingly, another protein component of the T-complex, VirD2, is transported into the plant cell nucleus by AtKAPα, a member of the Arabidopsis karyopherin α family (Ballas and Citovsky, 1997). That AtKAPα does not interact with VirE2 (Ballas and Citovsky, 1997) whereas VirD2 is not recognized by VIP1 suggests involvement of two distinct pathways in the T-complex nuclear import. Indeed, nuclear import of VirE2 but not of VirD2 was compromised in the VIP1 antisense plants. This dual pathway transport mechanism makes biological sense. It minimizes competition between a single molecule of VirD2 at the 5′ end of the T-strand and the more abundant VirE2 that coats the rest of the ssDNA molecule. Thus, VirD2 would have a better chance to lead the T-complex into the nucleus and specify its import polarity, which is thought to be important for the subsequent integration event (Zambryski, 1992; Tinland and Hohn, 1995; Sheng and Citovsky, 1996; De Neve et al., 1997).
In uninfected cells, similarly to many other plant bZIP proteins (van der Krol and Chua, 1991), VIP1 may be involved in transcription, associating with the chromosomal DNA either directly or through other components of transcription complexes. Thus, it is tempting to speculate that VIP1 and VirE2 may function in a multiprotein complex, which performs a dual function: it first facilitates nuclear targeting of VirE2 and then mediates intranuclear transport of VirE2 and its cognate T-strand to the site of integration. A similar dual role in nuclear and intranuclear transport has been suggested for the yeast Kap114p protein that functions to import the TATA-binding protein (TBP) into the cell nucleus and target it to the promoters of genes to be transcribed (Pemberton et al., 1999). This model for VIP1-mediated intranuclear transport addresses the long-standing question of how the invading T-complex finds its way to the host genome. Furthermore, because VIP1 probably interacts with the host cell chromatin during transcription, it may bring the T-complex to chromosomal regions where the host DNA is more exposed and, thus, more suitable for T-DNA integration.
Materials and methods
Plasmid construction
To use as bait in the yeast two-hybrid system, nopaline VirE2 ORF (Citovsky et al., 1992) was amplified by PCR as a BamHI fragment and fused with LexA by cloning into pBTM116 (TRP1+, Hollenberg et al., 1995), producing pBTM116-VirE2.
For studies of nuclear import in yeast cells, VIP1, VirE2 (Citovsky et al., 1992) and VirD2 ORFs (Ballas and Citovsky, 1997) were amplified by PCR and subcloned as BamHI–SalI, BamHI–XhoI or BamHI fragments, respectively, into the NIA plasmid pNIA (TRP1+; Rhee et al., 2000), producing pNIA-VIP1, pNIA-VirE2 and pNIA-VirD2 constructs. VirD2 ORF was also inserted in the reverse orientation, resulting in the pNIA-Vir2D plasmid. For co-expression with VirE2, VIP1 ORF was subcloned as a PCR-amplified SalI–BamHI fragment into a galactose-inducible expression vector pSJ101 (UrRA3+; Kironmai et al., 1998), producing pJS101-VIP1.
For expression in mammalian cells, VIP1 and VirE2 ORFs were PCR amplified and fused as SalI–BamHI or XhoI–BamHI fragments, respectively, to the GFP reporter gene in pEGFP-C1 (Clontech), resulting in pEGFP-C1-VIP1 and pEGFP-C1-VirE2 constructs. For co-expression with GFP–VirE2, the VIP1 ORF was cloned as a SalI–BamHI fragment into a mammalian expression vector pCB6 that contains a cytomegalovirus promoter (obtained from Dr D.Brown, SUNY, Stony Brook), producing pCB6-VIP1.
Free GUS, GUS–VirE2 and GUS–VirD2 were expressed in plant tissues from pRTL2-GUS (Restrepo et al., 1990), pRTL-GUSE2 (Citovsky et al., 1992) and pGD plasmids (Howard et al., 1992), respectively. Finally, for generation of transgenic VIP1 antisense plants, the VIP1 ORF was first inserted in reverse orientation as a PCR-amplified SalI fragment into a plant expression vector, pCd, containing the 35S promoter of cauliflower mosaic virus, tobacco mosaic virus translational enhancer (Gallie et al., 1989) and the nopaline synthase poly(A) signal. Then, the entire antisense expression cassette was subcloned as a BamHI–XbaI fragment into the binary vector pBIN19, carrying a kanamycin selection marker, to produce pBIN19/VIP1-AS. All PCRs were performed using a high fidelity Pfu DNA polymerase (Promega) and their products were verified by dideoxynucleotide sequencing (Kraft et al., 1988).
Yeast two-hybrid assay
Saccharomyces cerevisiae strain L40 [MATa his3Δ200 trp1-901 leu2-3 112 ade2 lys2-801am URA3::(lexAop)8-lacZ LYS2::(lexAop)4-HIS3; Hollenberg et al., 1995] was grown in yeast extract/peptone/dextrose or the appropriate selective minimal medium using standard conditions (Kaiser et al., 1994). Plasmids were introduced into yeast cells using a standard lithium acetate protocol (Kaiser et al., 1994).
The A.thaliana (ecotype Colombia) cDNA library in pGAD424 (LEU3+, Clontech) (Ballas and Citovsky, 1997) was screened with pBTM116-VirE2 as bait as described (Hollenberg et al., 1995; Ballas and Citovsky, 1997) and positive clones were selected on a histidine-deficient selective medium and confirmed by the β-galactosidase assay as described (Durfee et al., 1993). False-positives were eliminated using human lamin C and topoisomerase I, known to function as non-specific activators in the two-hybrid system (Bartel et al., 1993; Hollenberg et al., 1995), as baits.
VIP1–VirE2 binding in vitro
Renatured blot overlay assay (Dorokhov et al., 1999; Chen et al., 2000) was used to examine interaction between VIP1 and VirE2 in vitro. Briefly, VirE2, VIP1, VirD2 and TMV MP were purified to near homogeneity (95–98% pure as determined by silver-stained SDS–polyacrylamide gels) following expression in Escherichia coli (Citovsky et al., 1988, 1989, 1990; Chen et al., 2000). VIP1 and VirD2 were then electrophoresed through a 12.5% SDS–polyacrylamide gel, electroblotted onto a PVDF membrane, depleted of SDS by guanidine hydrochloride extraction, renatured, blocked with 1% bovine serum albumin (BSA) and incubated with 10 µg/ml of VirE2 as described (Chen et al., 2000). Following incubation, the membrane was washed and protein binding was detected using anti-VirE2 antibody and the ECL western blotting kit (Amersham). In control experiments, blotted VIP1 and VirD2 were incubated with purified VirD2 or TMV MP, and protein binding was detected using anti-VirD2 or anti-TMV MP antibodies, respectively.
Nuclear import in yeast and mammalian cells
For yeast nuclear import, L40 cells were transformed with pNIA-VIP1, pNIA-VirE2, pNIA-VirD2 or pNIA-Vir2D and grown in the presence or absence of histidine; histidine prototrophy indicated functional nuclear import (Rhee et al., 2000). To examine the effect of VIP1 on nuclear import proteins expressed from pNIA constructs, they were co-transformed with pJS101-VIP1 into L40 cells and VIP1 expression was induced by growing cells in the presence of 10 mg/ml galactose.
For nuclear import in mammalian cells, pEGFP-C1-VIP1, pEGFP-C1-VirE2 or a mixture of pEGFP-C1-VirE2 and pCB6-VIP1 were introduced into 24-h-old COS-1 cells (2.5 × 104 cells/cm2) grown on 20 mm coverslips in 6-well tissue culture plates. Prior to transformation, cells were grown for 4 h at 37°C in 3 ml of fresh Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% bovine calf serum (BCS). The cells were then transformed with the expression constructs using FuGENE 6 (Roche) according to the manufacturer’s instructions, and cultured for 24 h at 37°C to allow expression of the transfected genes. To visualize GFP expression, cells were washed briefly in phosphate-buffered saline (PBS), mounted on a fresh coverslip and observed under a Zeiss LSM 410 laser scanning confocal microscope equipped with a 485 nm excitation argon laser and a 527 nm GFP emission filter. For quantification of nuclear import, the signal intensity of intranuclear and cytoplasmic fluorescence was determined using the recorded images.
Generation of VIP1 antisense tobacco plants and assays for Agrobacterium tumorigenicity and transient T-DNA gene expression
The binary vector pBIN19/VIP1-AS was introduced into the disarmed Agrobacterium strain EHA105, which was then used to transform tobacco plants (Nicotiana tabacum cv. Turk) as described (Horsch et al., 1985). Transgenic tobacco plants expressing VIP1 in the antisense orientation were selected on a kanamycin-containing medium and maintained for 1 month in sterile conditions on an MS basal medium (Murashige and Skoog, 1962) with no exogenous growth regulators. For transgenic controls, the plants were transformed with an empty pBIN19 binary vector and maintained under the same conditions as the antisense plants. Plants were then transferred to soil in a greenhouse, allowed to set seed, and the transgenic progeny were selected by germinating the seeds on MS agar in the presence of kanamycin.
For tumor assays, 9 mm disks were excised from mature leaves of 1-month-old wild-type plants and transgenic control and VIP1 antisense plants, submerged in an overnight culture of Agrobacterium strain C58 (OD600 = 0.5), and incubated for 30 min at 25°C. The disks were then transferred to a hormone-free MS medium (Murashige and Skoog, 1962). After cultivation for 48 h at 25°C, the disks were washed three times in sterile distilled water, blotted dry and cultured on a hormone-free MS medium supplemented with 300 mg/l carbenicillin, to eliminate the remaining Agrobacterium. Four weeks later, the developed tumors were counted and their weight was determined.
For transient T-DNA gene expression, the leaf disks were co-cultivated for 48 h at 25°C on a hormone-free MS medium with Agrobacterium strain EHA105 (OD600 = 0.5) harboring a GUS-expressing binary vector pKIWI105 (Janssen and Gardner, 1990). For histochemical detection of GUS activity, the leaf disks were stained with the chromatogenic substrate X-Gluc as described (Nam et al., 1999). For quantitative analysis, 20 leaf disks per experimental condition were combined, ground and assayed for GUS activity using the fluorescent substrate 4-methyl umbelliferyl β-d-galactoside (MUG) as described (Nam et al., 1999). In control experiments, GUS activity was determined in leaf disks microbombarded with pRTL2-GUS (Carrington et al., 1991) followed by incubation for 24 h at 25°C to express the transfected DNA. All experiments were performed in triplicate and the resulting data represent average values with the indicated standard deviations.
Quantitative RT–PCR
Total RNA was extracted from 2.0 g of leaf tissue, treated with RQ1 RNase-free DNase and reverse-transcribed with M-MLV reverse transcriptase using strand-specific primers (i.e. forward or reverse to detect antisense or sense transcripts, respectively) derived from VIP1 and tobacco actin (DDBJ/EMBL/GenBank accession No. X63603; Thangavelu et al., 1993) gene sequences. The resulting cDNAs were PCR amplified as described (Kang et al., 1995; Ni et al., 1998), using a mixture of both forward and reverse primers. VIP1-specific RT–PCR products were then detected by Southern blot analysis using DIG-labeled full-length VIP1 cDNA as probe followed by autoradiography. The intensity of hybridized signals was quantified by scanning densitometry of the autoradiograms. Actin-specific RT–PCR products in the same reaction mixtures were detected by ethidium bromide staining of agarose gels. VIP1 forward and reverse primers, 5′-CGAACGGTGTTGTTCCTCCTAATTCTCTT-3′ and 5′-GCTCAGCAAGTCTATCACC-3′, respectively, generated a 290 bp product, while actin forward and reverse primers, 5′-TCACTGAAGCACCTCTTAACC-3′ and 5′-CAGCTTCCATTCCAATCATTG-3′, respectively, generated a 500 bp product. Note that the forward VIP1 primer was designed to span one of the introns predicted based on the genomic VIP1 sequence on Arabidopsis chromosome I (DDBJ/EMBL/GenBank accession No. AC009526).
Microbombardment
For expression of GUS–VirE2 and GUS–VirD2 fusion proteins, and GUS alone in plant tissues, 2 µg of DNA was adsorbed onto 50 µl of 1 µm gold particles according to the instructions of the manufacturer (Bio-Rad, CA) and microbombarded into the leaf mesophyll of greenhouse-grown wild-type and VIP1 antisense tobacco plants. Microbombardment was performed at a pressure of 150 p.s.i. using a portable Helios gene gun system (Model PDS-1000/He; Bio-Rad, CA). After incubation for 16 h at 25°C to allow expression of the transfected DNA, the leaf disks were stained for GUS activity for 3 h (Nam et al., 1999) and observed under a Zeiss Axiophot microscope. DAPI staining was used to verify the location of the cell nucleus. Each experiment was repeated at least six times. The intensity of indigo dye formed during the GUS assay in the cell cytoplasm and nucleus was quantified by photodensitometry of the images recorded on a 35 mm film exactly as described (Citovsky et al., 1992; Howard et al., 1992). Nuclear localization of GUS–VirD2 was defined as 100% activity, and GUS alone was defined as 0% activity. Average values and standard deviations were calculated based on 20 independent measurements.
Gel shift assay for protein–ssDNA binding
Purified VirE2 (3 µg) and/or VIP1 (10 µg) proteins were incubated for 30 min at 4°C in 30 µl of buffer L [10 mM Tris–HCl pH 8.0, 200 mM NaCl, 1 mM EDTA, 10% glycerol, 1 mM dithiothreitol and 1 mM phenylmethylsulfonyl fluoride (PMSF)] with 0.25 µg of M13mp18 ssDNA (New England Biolabs). For formation of VIP1–VirE2–ssDNA complexes, VirE2 was first allowed to bind to ssDNA for 10 min at 4°C and then VIP1 was added, and the incubation continued for another 20 min. After incubation, samples were electrophoresed on a 0.3% agarose gel as described (Lohman et al., 1986; Citovsky et al., 1992) and analyzed by Southern blotting (Ausubel et al., 1987), using an M13mp18 ssDNA labeled with DIG according to the manufacturer’s instructions (Amersham).
DDBJ/EMBL/GenBank accession number
The accession number for the sequence reported in this paper is AF225983.
Acknowledgments
Acknowledgements
We thank Dr Nancy Hollingsworth for help and guidance with the yeast growth experiments. Esaak Mullaev and Miron Kristos are also acknowledged for their participation in this research. This work was supported by grants from National Institutes of Health, National Science Foundation Functional Genomic Initiative, US Department of Agriculture, US–Israel Binational Research and Development Fund (BARD) and US–Israel Binational Science Foundation (BSF) to V.C., by a postdoctoral fellowship from the US–Israel Binational Research and Development Fund (BARD) to T.T. and, in part, by a Research Fellow Award from BARD to V.C.
Note added in proof
After submission of this manuscript, Tzfira and Citovsky (2001) showed that nopaline- and octopine-specific VirE2 proteins exhibit similar features of nuclear import in living cells (Mol. Plant Pathol., 2, in press).
References
- Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Smith,J.A., Seidman,J.G. and Struhl,K. (1987) Current Protocols in Molecular Biology. Greene Publishing–Wiley Interscience, New York, NY.
- Ballas N. and Citovsky,V. (1997) Nuclear localization signal binding protein from Arabidopsis mediates nuclear import of Agrobacterium VirD2 protein. Proc. Natl Acad. Sci. USA, 94, 10723–10728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel P., Chien,C., Sternglanz,R. and Fields,S. (1993) Elimination of false positives that arise in using the two-hybrid system. Biotechniques, 14, 920–924. [PubMed] [Google Scholar]
- Carrington J.C., Freed,D.D. and Leinicke,A.J. (1991) Bipartite signal sequence mediates nuclear translocation of the plant potyviral NIa protein. Plant Cell, 3, 953–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M.-H., Sheng,J., Hind,G., Handa,A. and Citovsky,V. (2000) Interaction between the tobacco mosaic virus movement protein and host cell pectin methylesterases is required for viral cell-to-cell movement. EMBO J., 19, 913–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie P.J., Ward,J.E., Winans,S.C. and Nester,E.W. (1988) The Agrobacterium tumefaciens virE2 gene product is a single-stranded-DNA-binding protein that associates with T-DNA. J. Bacteriol., 170, 2659–2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citovsky V., De Vos,G. and Zambryski,P. (1988) Single-stranded DNA binding protein encoded by the virE locus of Agrobacterium tumefaciens. Science, 240, 501–504. [DOI] [PubMed] [Google Scholar]
- Citovsky V., Wong,M.L. and Zambryski,P. (1989) Cooperative interaction of Agrobacterium VirE2 protein with single stranded DNA: implications for the T-DNA transfer process. Proc. Natl Acad. Sci. USA, 86, 1193–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citovsky V., Knorr,D., Schuster,G. and Zambryski,P. (1990) The P30 movement protein of tobacco mosaic virus is a single-strand nucleic acid binding protein. Cell, 60, 637–647. [DOI] [PubMed] [Google Scholar]
- Citovsky V., Zupan,J., Warnick,D. and Zambryski,P. (1992) Nuclear localization of Agrobacterium VirE2 protein in plant cells. Science, 256, 1802–1805. [DOI] [PubMed] [Google Scholar]
- Citovsky V., Warnick,D. and Zambryski,P. (1994) Nuclear import of Agrobacterium VirD2 and VirE2 proteins in maize and tobacco. Proc. Natl Acad. Sci. USA, 91, 3210–3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citovsky V., Guralnick,B., Simon,M.N. and Wall,J.S. (1997) The molecular structure of Agrobacterium VirE2–single stranded DNA complexes involved in nuclear import. J. Mol. Biol., 271, 718–727. [DOI] [PubMed] [Google Scholar]
- Das A. (1988) Agrobacterium tumefaciens virE operon encodes a single-stranded DNA-binding protein. Proc. Natl Acad. Sci. USA, 85, 2909–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Neve M., De Buck,S., Jacobs,A., Van Montagu,M. and Depicker,A. (1997) T-DNA integration patterns in co-transformed plant cells suggest that T-DNA repeats originate from co-integration of separate T-DNAs. Plant J., 11, 15–29. [DOI] [PubMed] [Google Scholar]
- Dingwall C. and Laskey,R.A. (1991) Nuclear targeting sequences—a consensus? Trends Biochem. Sci., 16, 478–481. [DOI] [PubMed] [Google Scholar]
- Dorokhov Y.L., Makinen,K., Frolova,O.Y., Merits,A., Saarinen,J., Kalkkinen,N., Atabekov,J.G. and Saarma,M. (1999) A novel function for a ubiquitous plant enzyme pectin methylesterase: the host-cell receptor for the tobacco mosaic virus movement protein. FEBS Lett., 461, 223–228. [DOI] [PubMed] [Google Scholar]
- Durfee T., Becherer,K., Chen,P.-L., Yeh,S.-H., Yang,Y., Kilburn,A.E., Lee,W.-H. and Elledge,S.J. (1993) The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev., 7, 555–569. [DOI] [PubMed] [Google Scholar]
- Fields S. and Song,O.-K. (1989) A novel genetic system to detect protein–protein interactions. Nature, 340, 245–246. [DOI] [PubMed] [Google Scholar]
- Gallie D.R., Lucas,W.J. and Walbot,V. (1989) Visualizing mRNA expression in plant protoplasts: factors influencing efficient mRNA uptake and translation. Plant Cell, 1, 303–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelvin S.B. (1998) Agrobacterium VirE2 proteins can form a complex with T strands in the plant cytoplasm. J. Bacteriol., 180, 4300–4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelvin S.B. (2000) Agrobacterium and plant genes involved in T-DNA transfer and integration. Annu. Rev. Plant Physiol. Plant Mol. Biol., 51, 223–256. [DOI] [PubMed] [Google Scholar]
- Gietl C., Koukolikova-Nicola,Z. and Hohn,B. (1987) Mobilization of T-DNA from Agrobacterium to plant cells involves a protein that binds single-stranded DNA. Proc. Natl Acad. Sci. USA, 84, 9006–9010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guralnick B., Thomsen,G. and Citovsky,V. (1996) Transport of DNA into the nuclei of Xenopus oocytes by a modified VirE2 protein of Agrobacterium. Plant Cell, 8, 363–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera-Estrella A., Van Montagu,M. and Wang,K. (1990) A bacterial peptide acting as a plant nuclear targeting signal: the amino-terminal portion of Agrobacterium virD2 protein directs a β-galactosidase fusion protein into tobacco nuclei. Proc. Natl Acad. Sci. USA, 87, 9534–9537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenberg S.M., Sternglanz,R., Cheng,P.F. and Weintraub,H. (1995) Identification of a new family of tissue-specific basic helix–loop–helix proteins with a two-hybrid system. Mol. Cell. Biol., 15, 3813–3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooykaas P.J.J. and Beijersbergen,A.G.M. (1994) The virulence system of Agrobacterium tumefaciens. Annu. Rev. Phytopathol., 32, 157–179. [Google Scholar]
- Horsch R.B., Fry,J.E., Hoffman,N.L., Eichholtz,D., Rogers,S.G. and Fraley,R.T. (1985) A simple and general method for transferring genes into plants. Science, 227, 1229–1231. [DOI] [PubMed] [Google Scholar]
- Howard E., Zupan,J., Citovsky,V. and Zambryski,P. (1992) The VirD2 protein of A.tumefaciens contains a C-terminal bipartite nuclear localization signal: implications for nuclear uptake of DNA in plant cells. Cell, 68, 109–118. [DOI] [PubMed] [Google Scholar]
- Janssen B.J. and Gardner,R.C. (1990) Localized transient expression of GUS in leaf discs following cocultivation with Agrobacterium. Plant Mol. Biol., 14, 61–72. [DOI] [PubMed] [Google Scholar]
- Kaiser C., Michaelis,S. and Mitchell,A. (1994) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Kang J., Kuhn,J.E., Schafer,P., Immelmann,A. and Henco,K. (1995) Quantification of DNA and RNA by PCR. In McPherson,M.J., Hames,B.D. and Taylor,G.R. (eds), PCR 2: A Practical Approach. IRL Press at Oxford University Press, Oxford, UK.
- Kironmai K.M., Muniyappa,K., Friedman,D.B., Hollingsworth,N.M. and Byers,B. (1998) DNA-binding activities of Hop1 protein, a synaptonemal complex component from Saccharomyces cerevisiae. Mol. Cell. Biol., 18, 1424–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koukolikova-Nicola Z., Raineri,D., Stephens,K., Ramos,C., Tinland,B., Nester,E.W. and Hohn,B. (1993) Genetic analysis of the virD operon of Agrobacterium tumefaciens: a search for functions involved in transport of T-DNA into the plant cell nucleus and in T-DNA integration. J. Bacteriol., 175, 723–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft R., Tardiff,J., Kranter,K.S. and Leinwand,L.A. (1988) Using miniprep plasmid DNA for sequencing double stranded templates with Sequenase. Biotechniques, 6, 544–547. [PubMed] [Google Scholar]
- Lohman T.M., Overman,L.B. and Datta,S. (1986) Salt-dependent changes in the DNA binding cooperativity of Escherichia coli single strand binding protein. J. Mol. Biol., 187, 603–615. [DOI] [PubMed] [Google Scholar]
- Murashige T. and Skoog,F. (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant., 15, 473–497. [Google Scholar]
- Nam J., Mysore,K.S., Zheng,C., Knue,M.K., Matthysse,A.G. and Gelvin,S.B. (1999) Identification of T-DNA tagged Arabidopsis mutants that are resistant to transformation by Agrobacterium. Mol. Gen. Genet., 261, 429–438. [DOI] [PubMed] [Google Scholar]
- Ni M., Tepperman,J.M. and Quail,P.H. (1998) PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix–loop–helix protein. Cell, 95, 657–667. [DOI] [PubMed] [Google Scholar]
- Oku T., Tjuvajev,J.G., Miyagawa,T., Sasajima,T., Joshi,A., Joshi,R., Finn,R., Claffey,K.P. and Blasberg,R.G. (1998) Tumor growth modulation by sense and antisense vascular endothelial growth factor gene expression: effects on angiogenesis, vascular permeability, blood volume, blood flow, fluorodeoxyglucose uptake and proliferation of human melanoma intracerebral xenografts. Cancer Res., 58, 4185–4192. [PubMed] [Google Scholar]
- Park H. and Sternglanz,R. (1998) Two separate conserved domains of eukaryotic DNA topoisomerase I bind to each other and reconstitute enzymatic activity. Chromosoma, 107, 211–215. [DOI] [PubMed] [Google Scholar]
- Pemberton L.F., Rosenblum,J.S. and Blobel,G. (1999) Nuclear import of the TATA-binding protein: mediation by the karyopherin Kap114p and a possible mechanism for intranuclear targeting. J. Cell Biol., 145, 1407–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo M.A., Freed,D.D. and Carrington,J.C. (1990) Nuclear transport of plant potyviral proteins. Plant Cell, 2, 987–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee Y., Gurel,F., Gafni,Y., Dingwall,C. and Citovsky,V. (2000) A genetic system for detection of protein nuclear import and export. Nature Biotechnol., 18, 433–437. [DOI] [PubMed] [Google Scholar]
- Rossi L., Hohn,B. and Tinland,B. (1993) The VirD2 protein of Agrobacterium tumefaciens carries nuclear localization signals important for transfer of T-DNA to plant. Mol. Gen. Genet., 239, 345–353. [DOI] [PubMed] [Google Scholar]
- Rout M.P. and Wente,S.R. (1994) Pores for thought: nuclear pore complex proteins. Trends Cell Biol., 4, 357–365. [DOI] [PubMed] [Google Scholar]
- Saitou N. and Nei,M. (1987) The neighbor joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol., 4, 406–425. [DOI] [PubMed] [Google Scholar]
- Schiene K., Puhler,A. and Niehaus,K. (2000) Transgenic tobacco plants that express an antisense construct derived from a Medicago sativa cDNA encoding a Rac-related small GTP-binding protein fail to develop necrotic lesions upon elicitor infiltration. Mol. Gen. Genet., 263, 761–770. [DOI] [PubMed] [Google Scholar]
- Sen P., Pazour,G.J., Anderson,D. and Das,A. (1989) Cooperative binding of Agrobacterium tumefaciens VirE2 protein to single-stranded DNA. J. Bacteriol., 171, 2573–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng J. and Citovsky,V. (1996) Agrobacterium–plant cell interaction: have virulence proteins—will travel. Plant Cell, 8, 1699–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shurvinton C.E., Hodges,L. and Ream,W. (1992) A nuclear localization signal and the C-terminal omega sequence in the Agrobacterium tumefaciens VirD2 endonuclease are important for tumor formation. Proc. Natl Acad. Sci. USA, 89, 11837–11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachel S.E. and Zambryski,P. (1989) Bacteria–yeast conjugation. Generic trans-kingdom sex. Nature, 340, 190–191. [DOI] [PubMed] [Google Scholar]
- Stam M., de Bruin,R., van Blokland,R., van der Hoorn,R.A., Mol,J.N. and Kooter,J.M. (2000) Distinct features of post-transcriptional gene silencing by antisense transgenes in single copy and inverted T-DNA repeat loci. Plant J., 21, 27–42. [DOI] [PubMed] [Google Scholar]
- Thangavelu M., Belostotsky,D., Bevan,M.W., Flavell,R.B., Rogers,H.J. and Lonsdale,D.M. (1993) Partial characterization of the Nicotiana tabacum actin gene family: evidence for pollen-specific expression of one of the gene family members. Mol. Gen. Genet., 240, 290–295. [DOI] [PubMed] [Google Scholar]
- Tinland B. and Hohn,B. (1995) Recombination between prokaryotic and eukaryotic DNA: integration of Agrobacterium tumefaciens T-DNA into the plant genome. Genet. Eng., 17, 209–229. [PubMed] [Google Scholar]
- Tzfira T. and Citovsky,V. (2000) From host recognition to T-DNA integration: the function of bacterial and plant genes in the Agrobacterium–plant cell interaction. Mol. Plant Pathol., 1, 201–212. [DOI] [PubMed] [Google Scholar]
- Tzfira T., Rhee,Y., Chen,M.-H. and Citovsky,V. (2000) Nucleic acid transport in plant–microbe interactions: the molecules that walk through the walls. Annu. Rev. Microbiol., 54, 187–219. [DOI] [PubMed] [Google Scholar]
- van der Krol A.R. and Chua,N.-H. (1991) The basic domain of plant B-ZIP proteins facilitates import of a reporter protein into plant nuclei. Plant Cell, 3, 667–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H., Beclin,C., Elmayan,T., Feuerbach,F., Godon,C., Morel,J.B., Mourrain,P., Palauqui,J.C. and Vernhettes,S. (1998) Transgene-induced gene silencing in plants. Plant J., 16, 651–659. [DOI] [PubMed] [Google Scholar]
- Zambryski P. (1992) Chronicles from the Agrobacterium–plant cell DNA transfer story. Annu. Rev.Plant Physiol. Plant Mol. Biol., 43, 465–490. [Google Scholar]
- Ziemienowicz A., Gorlich,D., Lanka,E., Hohn,B. and Rossi,L. (1999) Import of DNA into mammalian nuclei by proteins originating from a plant pathogenic bacterium. Proc. Natl Acad. Sci. USA, 96, 3729–3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zupan J. and Zambryski,P. (1997) The Agrobacterium DNA transfer complex. Crit. Rev. Plant Sci., 16, 279–295. [Google Scholar]
- Zupan J., Citovsky,V. and Zambryski,P. (1996) Agrobacterium VirE2 protein mediates nuclear uptake of ssDNA in plant cells. Proc. Natl Acad. Sci. USA, 93, 2392–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zupan J., Muth,T.R., Draper,O. and Zambryski,P. (2000) The transfer of DNA from Agrobacterium tumefaciens into plants: a feast of fundamental insights. Plant J., 23, 11–28. [DOI] [PubMed] [Google Scholar]