Abstract
The multiprotein factor composed of XPA and replication protein A (RPA) is an essential subunit of the mammalian nucleotide excision repair system. Although XPA–RPA has been implicated in damage recognition, its activity in the DNA repair pathway remains controversial. By replacing DNA adducts with mispaired bases or non-hybridizing analogues, we found that the weak preference of XPA and RPA for damaged substrates is entirely mediated by indirect readout of DNA helix conformations. Further screening with artificially distorted substrates revealed that XPA binds most efficiently to rigidly bent duplexes but not to single-stranded DNA. Conversely, RPA recognizes single-stranded sites but not backbone bending. Thus, the association of XPA with RPA generates a double-check sensor that detects, simultaneously, backbone and base pair distortion of DNA. The affinity of XPA for sharply bent duplexes, characteristic of architectural proteins, is not compatible with a direct function during recognition of nucleotide lesions. Instead, XPA in conjunction with RPA may constitute a regulatory factor that monitors DNA bending and unwinding to verify the damage-specific localization of repair complexes or control their correct three-dimensional assembly.
Keywords: damage recognition/DNA repair/xeroderma pigmentosum/XPA–RPA
Introduction
In mammals, nucleotide excision repair (NER) is the only DNA repair pathway that eliminates bulky DNA adducts induced by UV light or other environmental carcinogens (Sancar, 1996; Wood, 1996). The NER reaction is executed by incision of damaged strands on each side of a lesion, thus releasing damaged bases as part of oligonucleotide segments that are 24–32 residues in length (Huang et al., 1992). Defects in this NER system result in failure to remove DNA adducts and cause xeroderma pigmentosum (XP) in humans, which is an inherited syndrome characterized by a >1000-fold increased risk of sunlight-induced skin cancer. Individuals affected by XP are classified into seven repair-deficient complementation groups designated XP-A to XP-G (Friedberg et al., 1995; de Boer and Hoeijmakers, 2000).
All core components that carry out the NER reaction have been identified (Aboussekhra et al., 1995; Mu et al., 1995; Araújo et al., 2000). The minimal factors necessary for removal of damaged nucleotides include XPA, replication protein A (RPA), XPC together with a human homologue of RAD23 (hHR23B), transcription factor IIH (TFIIH) and two endonucleases, i.e. XPG as well as a heterodimer composed of XPF and excision repair cross complementing 1 (ERCC1). XPC–hHR23B is a damage recognition subunit that initiates the NER pathway through binding to the adducted site (Sugasawa et al., 1998; Batty et al., 2000; Yokoi et al., 2000). Another factor with affinity for damaged DNA (UV-DDB) stimulates excision of UV lesions from non-transcribed sequences in vivo (Tang et al., 2000). XPA and RPA have also been implicated in the lesion recognition step (Jones and Wood, 1993; He et al., 1995; Burns et al., 1996), but recent studies of the role of these two core subunits produced contradictory results (Sugasawa et al., 1998; Wakasugi and Sancar, 1999). Upon recruitment of TFIIH, the damaged site is unwound by 20–25 nucleotides, thereby generating an open intermediate that precedes DNA incision (Evans et al., 1997; Mu et al., 1997). The NER pathway is completed by the synthesis of repair patches through the action of replication factor C, proliferating cell nuclear antigen, DNA polymerase δ or ε and DNA ligase I.
The function of XPA protein is intriguing because the lack of this factor in XP-A patients causes a severe deficiency in NER of both transcribed and non-transcribed sequences (Kobayashi et al., 1998). This global requirement does not extend to XPC and UV-DDB, which are dispensable for the preferential repair of template strands in transcribed genes (Venema et al., 1991; Tang et al., 2000). Additionally, XPA protein is always needed for damage excision, but NER activity can be reconstituted without UV-DDB (Kazantsev et al., 1996) and, in the presence of certain bulky lesions, even without XPC (Mu et al., 1996). Finally, XPA protein is present in the final incision complex, whereas XPC is released from this complex before DNA incision (Wakasugi and Sancar, 1998). The XPA gene product is a 32 kDa protein that associates with the 70 and 34 kDa subunits of RPA (Li et al., 1995; Mer et al., 2000). XPA and RPA not only interact with each other but also share a preference for damaged DNA (Jones and Wood, 1993; Asahina et al., 1994; He et al., 1995; Burns et al., 1996). However, it is difficult to reconcile the weak affinity of these two factors for damaged duplexes with their critical involvement in the NER pathway. In fact, XPA protein has a low DNA association constant of 105–106 M–1 and displays only a 2- to 5-fold selectivity for damaged substrates (Jones and Wood, 1993). Similarly, RPA is the major mammalian single-stranded DNA binding protein and, as a consequence, binds with low affinity to double-stranded DNA (Wold, 1997).
Previous reports suggested that the assembly of NER complexes at damaged sites is accompanied by deformation of the DNA substrate (Evans et al., 1997; Fujiwara et al., 1999). Thus, our study was instigated by the hypothesis that the critical function of XPA protein may be determined by a specific reaction intermediate that has not yet been characterized. Using a panel of synthetic DNA molecules to search for high-affinity substrates, we observed that XPA and RPA recognize distortions of the Watson–Crick helix, such that preferential binding can be induced in the absence of adducted bases or other DNA lesions. Moreover, we discovered that XPA is an architectural protein endowed with an affinity for rigidly kinked DNA double strands that is comparable to the affinity of RPA for single strands. This preferential recognition of preformed DNA kinks by a critical NER subunit implies that the mammalian NER pathway, like its prokaryotic counterpart (Shi et al., 1992; Verhoeven et al., 2001), may involve site-directed bending of the DNA substrate. In combination, XPA and RPA are able to double-check DNA bending and unwinding in the NER complex and, as a consequence, could serve as regulatory subunits that verify the damage-specific recruitment of NER factors or, alternatively, control the correct three-dimensional assembly of NER intermediates prior to endonucleolytic cleavage.
Results
XPA and RPA recognize artificially distorted DNA duplexes
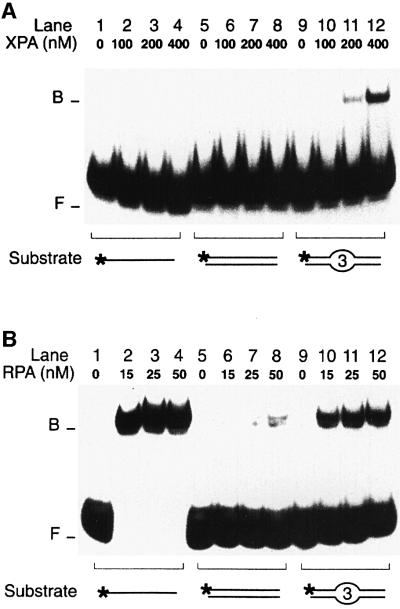
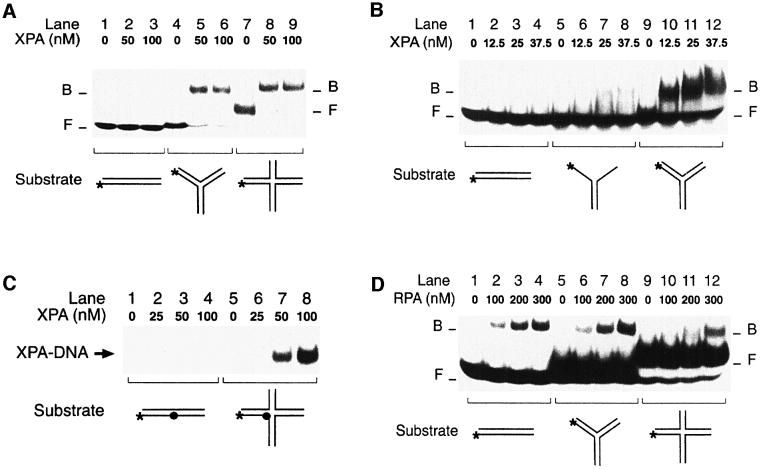
Although chemically unrelated, most NER substrates share the ability to induce conformational distortions of the DNA double helix. This common property of bulky DNA adducts prompted us to test whether XPA and RPA may recognize distorted substrates in the absence of DNA adducts or other covalent modification. For that purpose, a site-specific deformation of the Watson–Crick helix was generated by inserting three consecutive mismatches in the centre of 32P-labelled duplexes (see Materials and methods for oligonucleotide sequences). The DNA binding activity of XPA and RPA was tested in electrophoretic mobility shift assays. Figure 1 demonstrates that both XPA and RPA display an increased affinity for artificially distorted fragments over the homoduplex control. Up to a concentration of 400 nM, XPA was able to associate with DNA duplexes only in the presence of the three mismatches that simulate helical distortion (Figure 1A, compare lanes 5–8 with lanes 9–12). The same mobility shift assay also showed that XPA protein binds with higher efficiency to distorted double-stranded DNA than to single-stranded oligonucleotides of the same length (Figure 1A, compare lanes 1–4 with lanes 9–12). This finding demonstrates that the preference of XPA for conformational distortions does not result from binding to single-stranded regions of the duplex substrate.
Fig. 1. Recognition of artificial DNA distortions. (A) Electrophoretic mobility shift assay demonstrating preferential binding of XPA protein to DNA duplexes containing, in the centre, three consecutive mis matches (lanes 9–12), but no binding to DNA single strands (lanes 1–4). The position of free (F) and bound (B) DNA is indicated. The asterisks denote a 32P label on the 5′ end of 19mer substrates. (B) Comparison with RPA: under identical reaction conditions, RPA retains its characteristic preference for single-stranded DNA (lanes 1–4).
Like XPA, RPA also discriminated between artificially distorted duplexes and the native control (Figure 1B, compare lanes 5–8 with lanes 9–12). However, RPA interacted most effectively with single-stranded DNA of the same length (Figure 1B, lanes 1–4), confirming that RPA binding correlates with the degree by which the distorted duplex is thermodynamically destabilized, thereby exposing single-stranded regions (Lao et al., 2000). Because short DNA fragments are prone to partial denaturation, the increased affinity of RPA for single strands also explains the more efficient binding of RPA to the oligonucleotide duplexes used in these experiments, relative to the interactions of XPA with the same DNA substrates (Figure 1, compare A, lanes 9–12 with B, lanes 9–12). Thus, even though XPA and RPA have a comparable preference for double helical deformations, their distinct response to the presence of single strands indicates that the two factors operate by different mechanisms.
XPA and RPA recognize distorted DNA by a synergic binding reaction
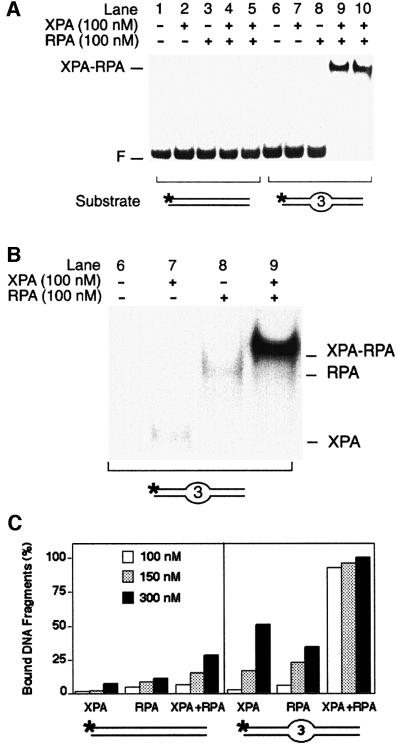
The combination of XPA and RPA has been shown to stimulate the binding of either factor alone to UV-irradiated or carcinogen-damaged DNA (He et al., 1995; Li et al., 1995). Here, we tested whether the presence of both factors may also enhance their interaction with artificially distorted substrates containing three mispaired bases. In these co-incubation experiments, a minimal substrate length of 43 bp was required to accommodate both XPA and RPA on the same DNA molecule (data not shown).
Using limiting amounts of protein relative to DNA, we found that XPA and RPA function in a cooperative way during the recognition of distorted substrates (Figure 2A). Control incubations with either factor alone (100 nM) yielded only marginal levels of nucleoprotein complexes (Figure 2A, lanes 7 and 8). However, upon combination of XPA and RPA at the same low concentration of 100 nM, >90% of the distorted fragments (lanes 9 and 10, in duplicate), but essentially no homoduplex DNA (lanes 4 and 5, in duplicate), was assembled in nucleoprotein complexes. Thus, cooperative binding by XPA and RPA can be induced in the absence of DNA adducts, just by adding low stoichiometric amounts of these two proteins to artificially distorted DNA. Figure 2B shows a longer autoradiographic exposure of lanes 6–9, illustrating the different electrophoretic mobility of the binary complexes with XPA (lane 7) or RPA (lane 8), as compared with the complexes that contain both XPA and RPA (lane 9). The modest decrease in mobility observed upon addition of XPA protein to the RPA–DNA complex is consistent with the lower molecular mass of XPA (32 kDa) relative to RPA (116 kDa).
Fig. 2. Synergic recognition of DNA distortions by XPA and RPA. (A) Electrophoretic mobility shift assay demonstrating synergic binding of XPA and RPA (100 nM each) to 43mer DNA duplexes containing three mismatches in the centre (lanes 9 and 10, in duplicate). The position of free DNA (F) and the position of nucleoprotein complexes with XPA and RPA are indicated. (B) Longer autoradiographic exposure of lanes 6–9, illustrating the different electrophoretic mobility of various complexes. The position of nucleoprotein complexes containing XPA, RPA or both XPA and RPA is indicated. (C) Quantitative evaluation of mobility shift assays (mean values of two experiments). Left panel: percentages of bound DNA obtained in the presence of the 43mer homoduplex control. Right panel: percentages of bound DNA upon incubation with 43mer DNA fragments containing three mismatches in the centre. Protein concentrations ranged from 100 to 300 nM.
When the concentration of XPA and RPA was gradually increased, we observed additive rather than cooperative binding reactions. A quantitative evaluation of these dose–response experiments is shown in Figure 2C. In all cases, XPA and RPA alone or in combination formed more nucleoprotein complexes with the duplex substrate containing three mismatches than with the homoduplex control, thereby confirming the bias of these factors for distorted DNA.
Non-hybridizing base analogues enhance recognition by XPA and RPA
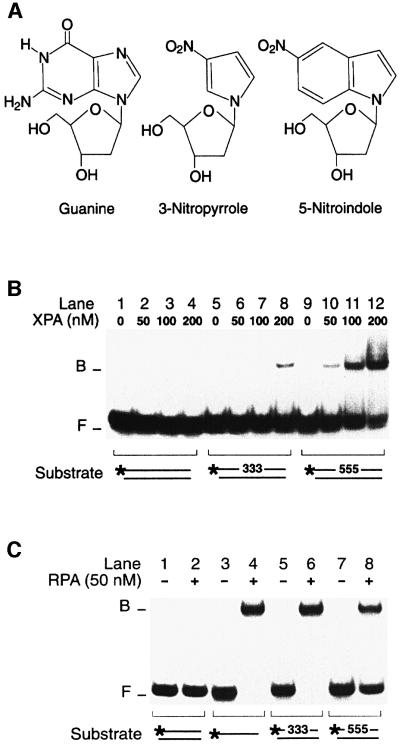
Mismatched nucleotides form aberrant hydrogen bonds with partner strands, resulting in displacement of the mispaired bases relative to the standard Watson–Crick alignment (Hunter et al., 1986). To confirm that the XPA–RPA complex recognizes such conformational defects, the mismatches used in previous binding experiments were replaced by non-hybridizing base analogues. As illustrated in Figure 3A, non-hybridizing analogues retain an aromatic ring structure similar to that of natural bases but lack hydrogen acceptor and donor groups for Watson–Crick pairing (Loakes et al., 1995). If three mismatched bases (retaining some residual hydrogen bonding) produce sufficient distortion of the double helix to recruit the XPA–RPA complex, we expected that base analogues with no residual hybridization capacity should provoke even stronger binding reactions. This expectation was confirmed by introducing three consecutive 5-nitro indoles in the centre of one 32P-labelled strand. In the presence of non-hybridizing analogues, XPA bound nearly all DNA fragments at a concentration of only 200 nM (Figure 3B, lanes 9–12), while none of the control DNA was shifted to the position of nucleoprotein complexes (lanes 1–4). Three consecutive 3-nitropyrroles were also recognized by XPA protein, but less effectively than 5-nitroindoles (Figure 3B, lanes 5–8). Previous denaturation studies demonstrated that 3-nitropyrroles reduce the melting temperature of DNA to a greater extent than 5-nitroindoles (Loakes et al., 1995). Thus, the higher affinity of XPA for 5-nitroindoles compared with 3-nitro pyrroles is consistent with the notion that destabilization of the double helix is not a predominant determinant of XPA binding. Like XPA protein, RPA also recognized the duplex substrates containing non-hybridizing base analogues but, in agreement with their stronger helix-destabilizing effect, 3-nitropyrroles resulted in more RPA binding than 5-nitroindoles (Figure 3C, compare lanes 6 and 8). In summary, the use of non-hybridizing base analogues confirmed that XPA and RPA detect distortions of the Watson–Crick double helix independently of the presence of DNA adducts.
Fig. 3. Recognition of non-hybridizing base analogues. (A) Nucleoside analogues containing 3-nitropyrrole or 5-nitroindole. Non-hybridizing base analogues lack donor and acceptor groups for Watson–Crick hydrogen bonding. (B) Electrophoretic mobility shift assay demonstrating binding of XPA protein to DNA distortions generated by three consecutive 3-nitropyrroles (lanes 5–8) or three consecutive 5-nitroindoles (lanes 9–12). The asterisks denote a 32P label on the 5′ end of 19mer DNA. (C) Comparison with RPA under identical binding conditions. RPA was incubated with homoduplex DNA (lane 2), single-stranded DNA (lane 4) or duplexes containing, in the centre, either three 3-nitropyrroles (lane 6) or three 5-nitroindoles (lane 8).
XPA protein recognizes kinked backbones
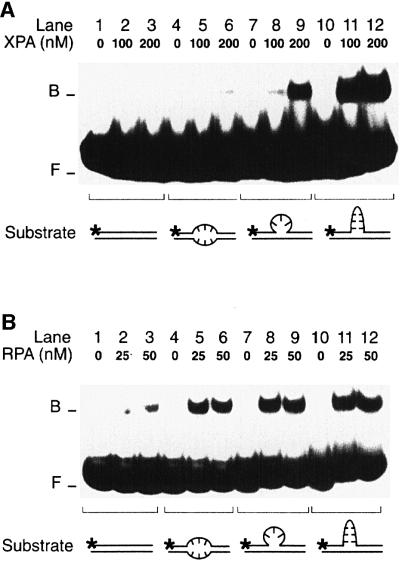
The hypothesis that XPA may recognize deformed NER intermediates, rather than the lesions themselves, prompted us to examine a broad panel of conformational distortions. First, we challenged XPA and RPA with duplex substrates containing a single-stranded loop of three nucleotides or a double-stranded loop of three GC base pairs (see Materials and methods for DNA sequences). A direct comparison revealed that XPA protein binds more efficiently to the three-nucleotide single-stranded loop (Figure 4A, lanes 7–9) than to three mismatched nucleotides (lanes 4–6). Furthermore, XPA displayed a much stronger affinity for the duplex containing a 3 bp double-stranded loop (Figure 4A, lanes 10–12). The quantitative evaluation showed that, with 200 nM XPA, only 1% of duplexes containing the mismatches, but 15% of duplexes containing the single-stranded loop and as much as 40% of duplexes containing the double-stranded loop migrated to the position of XPA–DNA complexes. A different response was obtained when the experiment was repeated with RPA. In fact, RPA showed the expected preference for substrates containing three mismatches over the homoduplex control (Figure 4B, compare lanes 1–3 with lanes 4–6). However, the affinity of RPA for distorted DNA was only marginally increased when the mismatches were replaced by a single-stranded loop (Figure 4B, lanes 7–9) or a double-stranded loop (lanes 10–12).
Fig. 4. Recognition of bulged DNA. (A) Electrophoretic mobility shift assay demonstrating the increased affinity of XPA for a single-stranded DNA loop (lanes 7–9) and, particularly, for a double-stranded loop in 19 bp DNA (lanes 10–12). The loop of lanes 7–9 results from the insertion of three unpaired nucleotides, while the loop of lanes 10–12 consists of three GC base pairs. The asterisks denote a 32P label on the 5′ end of each substrate. (B) Comparison with RPA: under identical reaction conditions, RPA binds to extra-helical loops (lanes 7–12) only slightly more efficiently than to three mismatches (lanes 4–6).
The substrate with a double-stranded loop is reminiscent of three-way DNA junctions composed of three double-helical arms radiating from a junction region. Therefore, we next examined the binding of XPA and RPA to synthetic three- and four-way DNA junctions. The three-way junction consisted of three helical stems of 20, 21 and 22 bp, while the four-way DNA junction contained four stems of 13, 20, 21 and 22 bp (see Materials and methods). On a native polyacrylamide gel, the larger four-way DNA junction was characterized by the expected lower electrophoretic mobility compared with a 43mer duplex linear control, while the three-way DNA junction migrated with intermediate mobility (data not shown). Figure 5A shows that XPA protein interacts with both three- and four-way DNA junctions more effectively than with any other DNA distortion or DNA lesion tested before. In fact, 50 nM of XPA were sufficient for nearly complete binding to three- and four-way DNA junctions (Figure 5A, lanes 4–9), while no band shift was observed with linear control DNA (lanes 1–3). Half maximal binding to three-way junctions was detected at XPA concentrations of 12.5–25 nM (Figure 5B, lanes 9–12). In contrast, we observed much less binding to Y-shaped double-stranded to single-stranded transitions consisting of a 22 bp arm with two single strand extensions of 20–21 nucleotides (Figure 5B, lanes 5–8).
Fig. 5. XPA is an architectural protein that recognizes kinked backbones. (A) Electrophoretic mobility shift assay demonstrating the extraordinary affinity of XPA protein for synthetic three- (lanes 4–6) and four-way DNA junctions (lanes 7–9). (B) Comparison between Y-shaped DNA molecules (lanes 5–8) and three-way DNA junctions (lanes 9–12). (C) XPA can be cross-linked to four-way DNA junctions. Reaction mixtures containing DNA and the indicated concentrations of XPA were pre-incubated for 10 min in the dark, followed by a 20 min exposure to UV light (366 nm). Cross-linked samples were analysed on a denaturing polyacrylamide gel. The filled circle indicates the photoreactive 4-thio-deoxythimidine. (D) Mobility shift assay showing that RPA, unlike XPA, has no increased affinity for three-way junctions (lanes 5–8) or four-way junctions (lanes 9–12) compared with linear duplex DNA (lanes 1–4).
To confirm the strong physical interaction of XPA with junction molecules, a photoreactive 4-thio-deoxythimidine residue (Green et al., 1998) was introduced in the central region of the four-way DNA substrate. As a control, the same 4-thio-deoxythimidine was inserted into the central portion of the 43mer linear homoduplex. After pre-incubation of radiolabelled DNA (either the four-way DNA junction or the linear control) with XPA protein, the reaction mixtures were UV-irradiated to cross-link XPA to DNA, and the covalent protein–DNA complexes were visualized on denaturing polyacrylamide gels. Substantial amounts of XPA–DNA cross-links could be isolated from the reactions containing four-way junction molecules (Figure 5C, lanes 5–8), but the control incubations with linear homoduplex DNA yielded only marginal levels of covalent XPA–DNA complexes (lanes 1–4). Thus, characterization of DNA binding by photo-crosslinking confirmed the extraordinary preference of XPA protein for four-way DNA junctions.
RPA displayed a completely different pattern when tested with the same substrates. In fact, RPA interacted with three-way DNA junctions no more efficiently than with homoduplex linear DNA (Figure 5D, compare lanes 1–4 with lanes 5–8). Also, RPA bound to four-way DNA junctions even less efficiently than to the linear homoduplex control (Figure 5D, lanes 9–12). Moreover, RPA was unable to stimulate the intrinsic affinity of XPA for three- or four-way DNA junctions, and photo-crosslinking experiments using the 4-thio-deoxythymidine residue confirmed that RPA rejects four-way junctions as a binding substrate (data not shown). Synthetic three- or four-way DNA junctions have been shown to fold into canonical Watson–Crick helices, with the exception of a sharp backbone bend at each site of strand exchange in the junction region (Ortiz-Lombardia et al., 1999). Thus, our results indicate that the DNA binding function of XPA is determined primarily by bending of the deoxyribose–phosphate backbone in duplex DNA. RPA, on the other hand, does not share with XPA this preference for distorted backbones.
Incomplete recognition of platinum cross-links by XPA
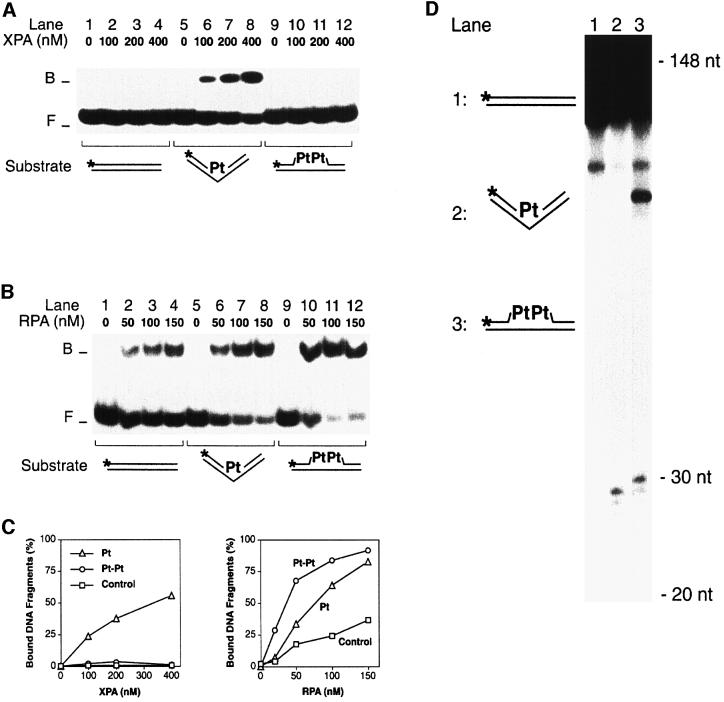
XPA is a new addition to the family of architectural proteins that bind to four-way DNA junctions, including HMG1 box proteins, winged helix proteins or the SWI/SNF complex. These unrelated proteins have the common propensity to interact with bent or kinked DNA (Zlatanova and van Holde, 1998). In subsequent experiments, we therefore exploited the rigid helical kink of 30–35° induced by a single cisplatin 1,2-d(GpG) intrastrand cross-link (Takahara et al., 1995). For comparison, we used a dinuclear analogue of the cisplatin 1,2-d(GpG) cross-link that fails to impose a rigid kink on the DNA helical axis but, instead, increases DNA flexibility in a non-directional manner (Kaspárková et al., 1996). Thermal denaturation studies showed that this flexible dinuclear 1,2-d(GpG) cross-link induces stronger destabilization of duplex DNA than the same cross-link of cisplatin (C.Hofr, N.Farrell and V.Brabec, unpublished results).
As expected (Jones and Wood, 1993), XPA was able to interact selectively with the rigid double-stranded kink generated by the cisplatin cross-link (Figure 6A, lanes 5–8), but the same protein showed no detectable binding to the flexible site resulting from the dinuclear cross-link analogue (lanes 9–12). RPA, on the other hand, recognized helical destabilization caused by the dinuclear analogue more effectively than the cisplatin-induced kink (Figure 6B, compare lanes 5–8 with lanes 9–12). A quantitative comparison emphasizing the failure of XPA to bind to the substrate containing a dinuclear platinum cross-link is shown in Figure 6C.
Fig. 6. Recognition and excision of GpG platinum cross-links. (A) Electrophoretic mobility shift assay in which XPA protein was incubated with 20mer duplexes containing a single GpG cross-link. The mononuclear cisplatin cross-link (Pt), which induces a rigid kink, is recognized (lanes 5–8), while the dinuclear analogue (Pt-Pt), in which this kink is replaced by a flexible hinge, is not recognized (lanes 9–12). (B) Comparison with RPA. Under identical reaction conditions, the mononuclear cisplatin cross-link (lanes 5–8) is recognized by RPA less efficiently than the dinuclear analogue (lanes 9–12). (C) Quantitative evaluation of two independent mobility shift assays performed with either XPA or RPA and platinated substrates. (D) Excision assay in HeLa cell extract demonstrating that both the mononuclear (lane 2) and the dinuclear GpG cross-link (lane 3) are repaired. The main excision products have a size of 29–30 nucleotides. Lane 1 shows a control reaction with undamaged DNA. The substrate preparations of lanes 1 and 3 were contaminated with a small fraction of short fragments (<148 nucleotides) resulting from incomplete oligonucleotide ligation.
The differential recognition of mono- and dinuclear platinum cross-links by XPA prompted us to test NER activity in response to these lesions. For that purpose, we constructed linear DNA fragments of 148 bp with an intrastrand platinum cross-link in the centre of one strand. The modified sequences included a 32P-labelled phosphate at the ninth phosphodiester bond on the 5′ side of the lesion. Such internally labelled substrates were mixed with a soluble HeLa cell extract containing all core NER factors. Upon addition of deoxyribonucleotides and ATP, the human NER system catalyses dual DNA incision, thereby releasing DNA damage as oligomeric segments of 24–32 nucleotides (Huang et al., 1992). The resulting excision products include the incorporated radiolabel and, as a consequence, can be visualized by gel electrophoresis and autoradiography. This repair assay in HeLa cell extract demonstrated that not only the cisplatin cross-link but also the dinuclear cross-link analogue, which is not detected by XPA, induces oligonucleotide excision (Figure 6D, lanes 2 and 3). Thus, direct recognition of the lesion by XPA is not necessary for excision activity.
XPA and RPA form unstable intermediates with NER substrates
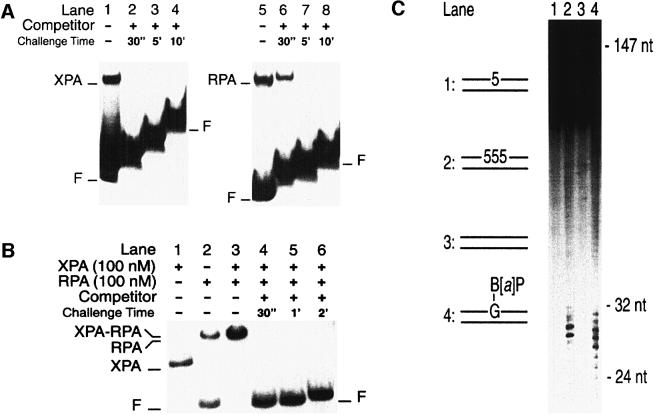
We used a competition assay to determine the stability of nucleoprotein complexes. XPA and RPA were pre-incubated with a 32P-labelled substrate to produce radiolabelled complexes. After 10 min, the preformed complexes were challenged by adding a 100-fold excess of unlabelled competitor DNA (duplex fragments containing three 5-nitroindole residues in the centre). After various time intervals following the addition of competitor DNA, the reaction mixtures were loaded on to a running gel for analysis by mobility shift. This protocol can be used to compare the stability of protein–DNA interactions because, in the presence of competitor DNA in vast excess, all protein molecules that dissociate from the radiolabelled substrate are sequestered on the unlabelled competitor (Batty et al., 2000).
Figure 7A demonstrates that the interaction of XPA with a standard NER substrate, i.e. duplex DNA carrying a benzo[a]pyrene-N2-dG adduct, is very unstable. After addition of competitor DNA, the preformed radiolabelled complexes dissociated completely in <30 s (Figure 7A, lane 2), which is the minimal time interval required for mixing the reaction and loading the sample on to the gel. The complexes formed by RPA with the same damaged substrate were slightly more stable. In the case of RPA, >30 s were necessary after addition of competitor DNA for complete release of the benzo[a]pyrene-damaged duplexes (Figure 7A, lanes 5–8).
Fig. 7. XPA–RPA generates dynamic complexes with NER substrates. (A) Nucleoprotein complexes were produced by incubating XPA protein (400 nM; lanes 1–4) or RPA (50 nM; lanes 5–8) with 19mer substrates containing a benzo[a]pyrene-dG adduct. Lanes 1 and 5 show the complexes formed during the first 10 min of incubation. After this pre-incubation time, the reactions were supplemented with a 100-fold excess of cold competitor DNA (19mer duplexes with three 5-nitroindole analogues in the centre). The samples were further incubated for the time intervals indicated and loaded on to the gel for electrophoresis. The stepwise appearance of the bands reflects the delay with which the samples were applied on to the running gel. (B) Nucleoprotein complexes produced by incubating XPA and RPA (100 nM each) with 43mer substrates containing three consecutive 5-nitroindoles in the centre. The band shifts formed by either factor alone are shown in lanes 1 (XPA) and 2 (RPA). Lane 3 demonstrates that, together, XPA and RPA bind all the DNA substrate during a pre-incubation of 10 min. The samples of lanes 4–6 were loaded after the time periods indicated, following the addition of a 100-fold excess of competitor DNA. (C) Excision assay in HeLa cell extract. The human NER system excises three consecutive 5-nitroindoles (lane 2). A single 5-nitroindole (lane 1) or undamaged DNA (lane 3) is not repaired. Lane 4 shows a control reaction with substrate containing a benzo[a]pyrene-dG adduct, confirming that the excision products are within the expected size range of 24–32 nucleotides.
The same competition experiment demonstrated that XPA dissociates in <30 s from three-way DNA junctions (data not shown). Furthermore, this competition assay was used to challenge the stability of nucleoprotein complexes formed by XPA or RPA with the efficient binding substrate containing three consecutive 5-nitroindole residues, but we found again that the resulting XPA–DNA and RPA–DNA complexes dissociated in <30 s. To test whether XPA and RPA, in combination, undergo more stable interactions with DNA, we examined the dissociation rate of complexes formed when excess amounts of both subunits were incubated with duplex DNA containing three 5-nitroindoles in the centre. The resulting XPA–RPA–DNA complexes dissociated rapidly, as demonstrated by the fact that all radiolabelled DNA was released from the nucleoprotein complexes in <30 s (Figure 7B, lane 4). This striking instability raised the possibility that the site containing three consecutive 5-nitroindole residues may not represent an excision substrate. Therefore, we constructed linear DNA fragments of 147 bp that carry 5-nitroindoles in the centre of one strand. A 32P-labelled phosphate was located at a distance of eight nucleotides on the 5′ side of the base analogues. Incubation in HeLa cell extract demonstrated that a single 5-nitroindole analogue was not excised (Figure 7C, lane 1), while three consecutive 5-nitroindole residues elicited NER activity (lane 2). Control incubations showed that undamaged DNA substrate was not processed (lane 3), but the substrate containing a single benzo[a]pyrene-N2-dG adduct was excised (lane 4). We conclude that NER activity is not dependent on the formation of stable contacts between XPA or RPA and the lesion site.
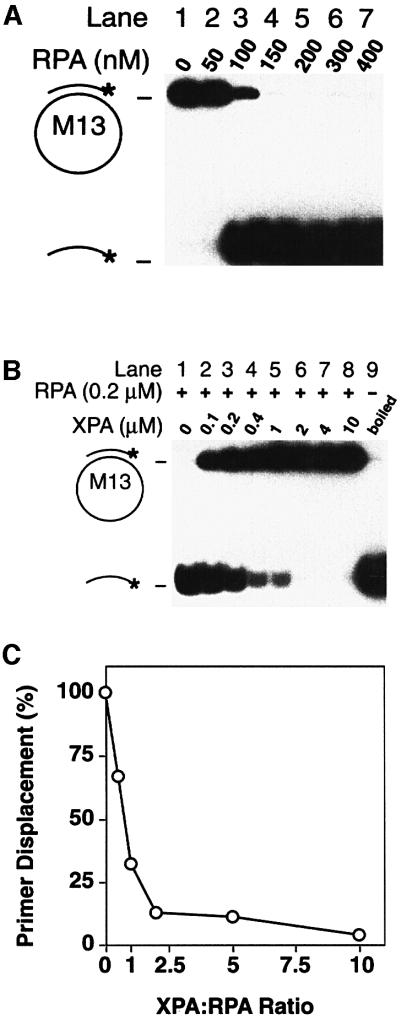
XPA inhibits the DNA unwinding activity of RPA
RPA unwinds double-stranded DNA in a reaction that is independent of ATP hydrolysis (Georgaki et al., 1992). To test whether the interaction with XPA may influence the DNA unwinding activity of RPA, partially duplex substrates were constructed by annealing a 32P-labelled primer of 24 residues to M13 single-stranded DNA. Figure 8A demonstrates that RPA (200 nM) promotes complete primer displacement (lane 5). When the partial duplex substrate was incubated with RPA (200 nM) and increasing concentrations of XPA (between 100 nM and 10 µM), XPA suppressed the DNA unwinding activity of RPA (Figure 8B, lanes 2–8). An ∼50% reduction of primer displacement was observed when XPA and RPA were co-incubated in stoichiometric amounts (Figure 8C). Control experiments under identical conditions showed that XPA does not promote hybridization of complementary primers with M13 single strands (data not shown). Thus, the observed reduction in primer release reflects inhibition of unwinding activity rather than subsequent re-annealing of displaced strands. These results indicate that XPA prevents RPA from unwinding target DNA.
Fig. 8. Inhibition of DNA unwinding. (A) ATP-independent unwinding of partial duplex DNA by RPA. The positions of circular substrate and of displaced radiolabelled primers are indicated. (B) RPA (200 nM) and partial duplex substrate were incubated with increasing concentrations of XPA protein. The sample of lane 9 was denatured by boiling for 5 min before electrophoretic analysis. (C) Quantitative evaluation of two independent unwinding assays indicating >50% inhibition at an XPA:RPA ratio of 1:1.
Discussion
In this study, we discovered that the damaged DNA binding activity of XPA and RPA reported previously is entirely attributable to indirect readout of DNA conformations. One particular helical deformation, i.e. rigid DNA bending, was identified as the specific molecular determinant that induces high-affinity interactions of XPA with DNA, but such an architectural activity is not consistent with a direct participation of XPA in damage recognition. On the other hand, the dual affinity of XPA–RPA for sharply bent and partially unwound duplexes indicates that this composite factor may exert a key regulatory function by double-checking DNA deformation during assembly of incision complexes.
Indirect readout by the XPA–RPA complex
Previous reports indicated that XPA and RPA, alone or in combination, bind with weak selectivity to damaged DNA and, as a consequence, these two factors have been implicated in the primary lesion recognition step of the NER pathway (Jones and Wood, 1993; Asahina et al., 1994; He et al., 1995; Burns et al., 1996; Wakasugi and Sancar, 1999). Alternatively, it has been proposed that the affinity of XPA for damaged substrates may be used in a secondary recognition step to verify the presence of DNA lesions (Sugasawa et al., 1998; Yokoi et al., 2000). Here, we observed preferential binding of XPA and RPA in response to artificial DNA distortions produced by mismatched nucleotides, non-hybridizing base analogues or other unusual conformations, indicating that both XPA and RPA operate by indirect readout of the DNA double helix. Even the cooperative binding of XPA and RPA, observed previously with UV or carcinogen lesions (He et al., 1995), can be induced in the presence of artificial DNA distortions with substrates that are completely devoid of base adducts or other chemical modifications. Thus, the damage recognition function of XPA and RPA reflects an affinity for conformational defects of the Watson–Crick double helix rather than direct recognition of DNA lesions. Many different base adducts are able to stimulate DNA binding by XPA and RPA because bulky lesions distort the double helix at least to some extent, thereby generating a signal for indirect readout by these two factors.
Double-check probing of DNA helix conformation by XPA–RPA
Our results indicate that XPA and RPA use distinct mechanisms for recognition of distorted DNA. In fact, XPA protein is unable to interact with single-stranded oligomers, whereas RPA exerts a strong single-stranded DNA binding activity. The finding that XPA rejects single-stranded substrates is consistent with the NMR model of XPA revealing a central DNA binding cleft, which has the correct dimensions to accommodate duplex DNA (Ikegami et al., 1998). Instead, RPA uses its single-stranded DNA binding domain to promote interactions with regions of duplex destabilization (Lao et al., 2000). Also, XPA binds preferentially to double-stranded DNA containing the thermodynamically more stable 5-nitroindole analogues, while RPA prefers the more destabilizing 3-nitropyrrole residues. This comparison between different non-hybridizing base analogues confirmed that single-stranded regions are the main determinant for recognition by RPA but not for recognition by XPA. Moreover, the use of three- and four-way DNA junctions revealed an extraordinary affinity of XPA protein for sharply bent backbones. From the 50% maximal binding point in the mobility shift assays, we estimate that XPA protein interacts with synthetic three- and four-way junctions two orders of magnitude more efficiently than with linear controls. In contrast, RPA binds to four-way DNA junctions even less efficiently than to linear duplex DNA. Finally, XPA protein recognizes the stable double-stranded kink (Takahara et al., 1995) induced by a single GpG cross-link of cisplatin, while RPA binds preferentially to the flexible site (Kaspárkova et al., 1996) introduced by a dinuclear cross-link analogue. XPA is dependent on a preformed kink for its high-affinity interaction, because no XPA binding could be detected when the platinum-induced kink was replaced by a flexible hinge.
In summary, the XPA–RPA complex combines two DNA binding modules for indirect readout of DNA conformations. Each subunit binds to a distinct structural component of the distorted substrate: XPA is specialized for the recognition of backbone bends but has no affinity for single-stranded sites. Conversely, RPA recognizes single-stranded sites but has no special preference for kinked backbones. In combination, XPA and RPA form a composite sensor that is able to monitor, simultaneously, the degree of DNA bending and unwinding.
Molecular function of XPA protein
Several findings argue against a function of XPA protein mediated by direct interactions with base lesions. First, XPA binds to DNA distortions (for example three consecutive mismatches) in the complete absence of covalent DNA modifications. Secondly, XPA is an architectural protein with a stronger affinity for kinked DNA duplexes than for other types of structural DNA anomaly. Thirdly, the challenge with platinum cross-links showed that XPA exerts its function in the absence of direct contacts with the target lesion. In fact, XPA binds to the rigid kink generated by a cisplatin cross-link but fails to recognize the flexible hinge induced by a dinuclear cross-link analogue, yet both platinum lesions are processed by the human NER system. Finally, XPA on its own forms very unstable nucleoprotein complexes, even when the target lesion is successfully excised. The striking instability of this primary interaction with DNA indicates that the recruitment of XPA to damaged sites depends not only on the formation of a kinked DNA intermediate but also on additional protein–protein contacts with other components of the NER complex (Sancar, 1996; Wood, 1996).
If the activity of XPA is not mediated by direct recognition of DNA lesions, what molecular function could account for the essential role of this factor in excision repair? Because XPA binds to backbone kinks and RPA binds to single-stranded DNA, the two different modules of XPA–RPA are potentially able to monitor, simultaneously, the degree of DNA bending and unwinding during assembly of the NER complex. One possibility is that XPA in conjunction with RPA carries out a damage verification step by sensing damage-induced distortion of the DNA helix. In this model, the two DNA binding modules of XPA–RPA may complement each other to enhance the range of detectable DNA distortions. An alternative licensing hypothesis is suggested by the observation that the affinity of many architectural proteins for kinked DNA reflects the structural similarity with a physiological DNA target (Zlatanova and van Holde, 1998). Hence, our finding that a critical NER subunit binds to sharply bent backbones leads to the prediction that the mammalian NER pathway involves site-specific kinking of the DNA molecule, as has been shown for the prokaryotic NER system (Shi et al., 1992; Verhoeven et al., 2001). There is also evidence for DNA bending in a human NER intermediate (Fujiwara et al., 1999) and local DNA unwinding in the ultimate incision complex has been demonstrated (Evans et al., 1997; Mu et al., 1997). Therefore, probing of the DNA helix conformation by XPA–RPA could provide a checkpoint mechanism to control the three-dimensional organization of NER complexes prior to endonucleolytic cleavage. A regulatory function in controlling the endonuclease activity of XPF-ERCC1 and XPG has already been proposed for RPA (de Laat et al., 1998). A similar regulatory function of XPA could explain the critical requirement of this architectural subunit for NER proficiency.
In conclusion, the architectural activity of XPA protein may be necessary for a damage verification step that aborts DNA incision when NER complexes have been erroneously assembled at undamaged sites. The other possibility is that XPA is needed for a licensing step that controls DNA architecture in the NER complex and prevents endonucleolytic processing of NER intermediates in which the individual DNA strands are not correctly wrapped around the recognition proteins. Finally, we identified another function of XPA, i.e. the inhibition of DNA unwinding by RPA. This inhibitory activity may be important to avoid excessive opening of the DNA substrate around the lesion site and, hence, limit the size of NER patches to a length of 24–32 nucleotides.
Materials and methods
Binding substrates
Unmodified 19mer homo- or heteroduplex DNA was prepared by annealing ON1 (5′-ACCACCCTTCGAACCACAC-3′) with ON2 (5′-GTGTGGTTCGAAGGGTGGT-3′) or ON3 (5′-GTGTGGTTCTTT GGGTGGT-3′). The melting temperature of 19mer duplexes under the conditions used in this report is 66.5°C and, when these duplexes contain three mismatches in the centre, their melting temperature is 51.1°C (Hess et al., 1997a). Unmodified 43mer homoduplex or heteroduplex DNA was obtained by annealing ON4 (5′-CGACTGCAGACGTCGAGCCAT CGCTACCGTGGAATTCTAGAGC-3′) with either ON5 (5′-GCTCTA GAATTCCACGGTAGCGATGGCTCGACGTCTGCAGTCG-3′) or ON6 (5′-GCTCTAGAATTCCACGGTAGTTTTGGCTCGACGTCTGCAG TCG-3′). To obtain substrates containing three non-hybridizing base analogues, the three thymines in the centre of ON3 and ON6 were replaced by 5-nitroindoles or 3-nitropyrroles (Loakes et al., 1995). The 19mer oligonucleotide (5′-CGAGCCATCGCTACCGGTG-3′) containing a site-directed (+)-cis-benzo[a]pyrene-dG adduct was constructed as described (Hess et al., 1997b). A three-nucleotide single-stranded loop was obtained by annealing ON1 with ON7 (5′-GTGTGGTTCGTT CAAGGGTGGT-3′). The 3 bp double-stranded loop was generated by hybridizing ON1 and ON8 (5′-GTGTGGTTCGCGCGCGAAGGG TGGT-3′). Three-way DNA junctions resulted from the annealing of ON9 (5′-CGACTGCAGACGTACCACCCTTCGAACCACACGAAT TCTAGAG-3′) with ON10 (5′-CGATACGTCCCCAATATCCCAAGG GAGGTACGTCTCCAGTCG-3′) and ON11 (5′-CTCTAGAATTCG TGTGGTTCGGGGATATTGGGGACGTATCG-3′). For construc tions of Y-shaped molecules, ON9 was annealed with ON10. The linear control DNA of Figure 5 consisted of ON9 and ON12 (5′-CTCTAGAAT TCGTGTGGTTCGAAGGGAGGTAC GTCTCCAGTCG-3′). Four-way DNA junctions resulted from the annealing of ON9, ON10, ON13 (5′-CTCTAGAATTCGTGTGGTTCGTATCACGACTAGC-3′) and ON14 (5′-GCTAGTCGTGATAGGGATATTGGGGACGTATCG-3′). Alternatively, four-way DNA junctions contained a single 4-thio-deoxythymidine in the centre of ON9. The 20mer oligonucleotide 5′-CCTCTCTCTGGTCTTCTTCT-3′ was used to generate site-specific d(GpG) platinum cross-links (Kaspárková et al., 1996). Oligonucleotides were 32P-labelled at the 5′ end and, before each mobility shift assay, annealed with unlabelled complementary oligonucleotides. All hybridizations were performed in 50 mM Tris–HCl pH 7.4, 10 mM MgCl2 and 1 mM dithiothreitol (DTT) by heating to 80°C for 10 min and then incubating for another 3 h at 25°C.
Proteins
Human RPA was expressed in Escherichia coli strain BL21 harbouring plasmid p11d-tRPA, which co-expresses the three RPA subunits, and purified by Affi-Gel blue and Mono-Q chromatography as described (Henricksen et al., 1994). Human XPA protein was overexpressed with an N-terminal polyhistidine tag and purified to homogeneity through a Ni2+ affinity column followed by hydroxylapatite chromatography (Jones and Wood, 1993).
DNA binding
Electrophoretic mobility shift assays were performed by incubating, at 20°C, 32P-labelled DNA substrate (20–40 fmol), unlabelled homoduplex competitor DNA (19mer, 1.4 pmol) and the indicated amounts of XPA or RPA in reactions of 20 µl containing 25 mM HEPES–KOH pH 8.3, 30 mM KCl, 4 mM MgCl2, 1 mM EDTA, 0.9 mM DTT, 45 µg/ml bovine serum albumin and 10% (v/v) glycerol. To assess binding at equilibrium, reactions were stopped after 30 min by cooling the samples to 0°C. Following addition of gel loading buffer (4 µl) containing 100 mM Tris–HCl pH 8.3, 10% (v/v) glycerol and 0.05% (w/v) Orange G, the extent of binding was determined on 5% native polyacrylamide gels. Electrophoresis was performed at 1.5 mA/cm for 50 min at 4°C, using 45 mM Tris–HCl pH 8.3, 45 mM boric acid, 1 mM EDTA as the running buffer. Gels were dried and subjected to autoradiography on X-ray films. The relative levels of binding were determined by densitometric analysis of X-ray films, and the linearity of each quantification was confirmed by counting Cerenkov radiations of the corresponding gel slices. The photo-crosslinking reactions were performed in the absence of DTT. After exposure to UV light (366 nm) for 20 min, the cross-linking was stopped by the addition of denaturing loading buffer. After boiling for 5 min, the cross-linked products were separated by 12% denaturing PAGE and visualized by autoradiography (Green et al., 1998).
DNA unwinding and excision repair
Internally 32P-labelled DNA duplexes of 147 or 148 bp were constructed as described by Matsunaga et al. (1995). Repair reactions were performed in extracts prepared from HeLa cells, and excision products were separated on 10% denaturing polyacrylamide gels and visualized by autoradiography (Kazantsev et al., 1996; Hess et al., 1997a). Partial duplex DNA substrates were obtained by hybridizing M13mp8 single-stranded DNA with a complementary radiolabelled 24mer oligonucleotide. Unwinding reactions and subsequent analysis on native 5% polyacrylamide gels were performed as described (Georgaki et al., 1992).
Acknowledgments
Acknowledgements
We thank J.T.Reardon and A.Sancar for HeLa cell extracts (used in the experiment of Figure 6D), R.D.Wood for plasmid pET15b/XPA and N.E.Geacintov for the benzo[a]pyrene-modified oligonucleotide. This research was supported by the Swiss National Science Foundation grant 31-61494.00, by the Krebsliga of the Kanton Zürich and the Stiftung für Zeitgemässe Ernährung.
References
- Aboussekhra A. et al. (1995) Mammalian DNA nucleotide excision repair reconstituted with purified components. Cell, 80, 859–868. [DOI] [PubMed] [Google Scholar]
- Araújo S.J., Tirode,F., Coin,F., Pospiech,H., Syväoja,J.E., Stucki,M., Hübscher,U., Egly,J.-M. and Wood,R.D. (2000) Nucleotide excision repair of DNA with recombinant human proteins: definition of the minimal set of factors, active forms of TFIIH, and modulation by CAK. Genes Dev., 14, 349–359. [PMC free article] [PubMed] [Google Scholar]
- Asahina H., Kuraoka,I., Shirakawa,M., Morita,E.H., Miura,N., Miyamoto,I., Ohtsuka,E., Okada,Y. and Tanaka,K. (1994) The XPA protein is a zinc metalloprotein with an ability to recognize various kinds of DNA damage. Mutat. Res., 315, 229–237. [DOI] [PubMed] [Google Scholar]
- Batty D., Rapic’-Otrin,V., Levine,A.S. and Wood,R.D. (2000) Stable binding of human XPC complex to irradiated DNA confers strong discrimination for damaged sites. J. Mol. Biol., 300, 275–290. [DOI] [PubMed] [Google Scholar]
- Burns J.L., Guzder,S.N., Sung,P., Prakash,S. and Prakash,L. (1996) An affinity of human replication protein A for ultraviolet-damaged DNA. J. Biol. Chem., 271, 11607–11610. [DOI] [PubMed] [Google Scholar]
- De Boer J. and Hoeijmakers,J.H. (2000) Nucleotide excision repair and human syndromes. Carcinogenesis, 21, 453–460. [DOI] [PubMed] [Google Scholar]
- De Laat W.L, Appeldoorn,E., Sugasawa,K., Weterings,E., Jaspers,N.G. and Hoeijmakers,J.H. (1998) DNA-binding polarity of human replication protein A positions nucleases in nucleotide excision repair. Genes Dev., 12, 2598–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans E., Moggs,J.G., Hwang,J.R., Egly,J.-M. and Wood,R.D. (1997) Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. EMBO J., 16, 6559–6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg E.C., Walker,G.C. and Siede,W. (1995) DNA Repair and Mutagenesis. ASM Press, Washington DC.
- Fujiwara Y., Masutani,C., Mizukoshi,T., Kondo,J., Hanaoka,F. and Iwai,S. (1999) Characterization of DNA recognition by the human UV-damaged DNA-binding protein. J. Biol. Chem., 274, 20027–20033. [DOI] [PubMed] [Google Scholar]
- Georgaki A., Strack,B., Podust,V. and Hübscher,U. (1992) DNA unwinding activity of replication protein A. FEBS Lett., 308, 240–244. [DOI] [PubMed] [Google Scholar]
- Green R., Switzer,C. and Noller,H.F. (1998) Ribosome-catalyzed peptide-bond formation with an A-site substrate covalently linked to 23S ribosomal RNA. Science, 280, 286–289. [DOI] [PubMed] [Google Scholar]
- He Z., Henricksen,L.A., Wold,M.S. and Ingles,C.J. (1995) RPA involvement in the damage-recognition and incision steps of nucleotide excision repair. Nature, 374, 566–569. [DOI] [PubMed] [Google Scholar]
- Henricksen L.A., Umbricht,C.B. and Wold,M.S. (1994) Recombinant replication protein A: expression, purification, and functional characterization. J. Biol. Chem., 269, 11121–11132. [PubMed] [Google Scholar]
- Hess M.T., Schwitter,U., Petretta,M., Giese,B. and Naegeli,H. (1997a) Bipartite substrate discrimination by human nucleotide excision repair. Proc. Natl Acad. Sci. USA, 94, 6664–6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess M.T., Gunz,D., Luneva,N., Geacintov,N.E. and Naegeli,H. (1997b) Base pair conformation-dependent excision of benzo[a]pyrene diol epoxide–guanine adducts by human nucleotide excision repair enzymes. Mol. Cell. Biol., 17, 7069–7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J.-C., Svoboda,D., Reardon,J.T. and Sancar,A. (1992) Human nucleotide excision nuclease removes thymine dimers from DNA by incising the 22nd phosphodiester bond 5′ and the 6th phosphodiester bond 3′ to the photodimer. Proc. Natl Acad. Sci. USA, 89, 3664–3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter W.N., Brown,T., Anand,N.N. and Kennard,O. (1986) Structure of an adenine.cytosine base pair in DNA and its implications for mismatch repair. Nature, 320, 552–555. [DOI] [PubMed] [Google Scholar]
- Ikegami T., Kuraoka,I., Saijo,M., Kodo,N., Kyogoku,Y., Morikawa,K., Tanaka,K. and Shirakawa,M. (1998) Solution structure of the DNA- and RPA-binding domain of the human repair factor XPA. Nature Struct. Biol., 5, 701–706. [DOI] [PubMed] [Google Scholar]
- Jones C.J. and Wood,R.D. (1993) Preferential binding of the xeroderma pigmentosum group A complementing protein to damaged DNA. Biochemistry, 32, 12096–12104. [DOI] [PubMed] [Google Scholar]
- Kaspárková J., Mellish,K.J., Brabec,V. and Farrell,N. (1996) Site-specific d(GpG) intrastrand crosslinks formed by dinuclear platinum complexes. Bending and NMR studies. Biochemistry, 35, 16705–16713. [DOI] [PubMed] [Google Scholar]
- Kazantsev A., Mu,D., Nichols,A.F., Zhao,X., Linn,S. and Sancar,A. (1996) Functional complementation of xeroderma pigmentosum group E by replication protein A in an in vitro system. Proc. Natl Acad. Sci. USA, 93, 5014–5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Takeuchi,S., Saijo,M., Nakatsu,Y., Morioka,H., Otsuka,E., Wakasugi,M., Nikaido,O. and Tanaka,K. (1998) Mutational analysis of a function of xeroderma pigmentosum group A (XPA) protein in strand-specific repair. Nucleic Acids Res., 26, 4662–4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao Y., Gomes,X.V., Ren,Y., Taylor,J.-S. and Wold,M.S. (2000) Replication protein A interactions with DNA. III. Molecular basis of recognition of damaged DNA. Biochemistry, 39, 850–859. [DOI] [PubMed] [Google Scholar]
- Li L., Lu,X., Peterson,C.A. and Legerski,R.J. (1995) An interaction between the DNA repair factor XPA and replication protein A appears essential for nucleotide excision repair. Mol. Cell. Biol., 15, 5396–5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loakes D., Brown,D.M., Linde,S. and Hill,F. (1995) 3-nitropyrrole and 5-nitroindole as universal bases in primers for DNA sequencing and PCR. Nucleic Acids Res., 23, 2361–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga T., Mu,D., Park,C.-H., Reardon,J.T. and Sancar,A. (1995) Human DNA repair excision nuclease. J. Biol. Chem., 270, 20862–20869. [DOI] [PubMed] [Google Scholar]
- Mer G., Bochkarev,A., Gupta,R., Bochkareva,E., Frappier,L., Ingles,C.J., Edwards,A.M. and Chazin,W.J. (2000). Structural basis for the recognition of DNA repair proteins UNG2, XPA, and RAD52 by replication factor RPA. Cell, 103, 449–456. [DOI] [PubMed] [Google Scholar]
- Mu D., Park,C.H., Matsunaga,T., Hsu,D.S., Reardon,J.T. and Sancar,A. (1995) Reconstitution of human DNA repair excision nuclease in a highly defined system. J. Biol. Chem., 270, 2415–2418. [DOI] [PubMed] [Google Scholar]
- Mu D., Hsu,D.S. and Sancar,A. (1996) Reaction mechanism of human DNA repair excision nuclease. J. Biol. Chem., 271, 8285–8294. [DOI] [PubMed] [Google Scholar]
- Mu D., Wakasugi,M., Hsu,D.S. and Sancar,A. (1997) Characterization of reaction intermediates of human excision repair nuclease. J. Biol. Chem., 272, 28971–28979. [DOI] [PubMed] [Google Scholar]
- Ortiz-Lombardia M., González,A., Eritja,R., Aymami,J., Azorin,F. and Coll,M. (1999) Crystal structure of a DNA Holliday junction. Nature Struct. Biol., 6, 913–917. [DOI] [PubMed] [Google Scholar]
- Sancar A. (1996) DNA excision repair. Annu. Rev. Biochem., 65, 43–81. [DOI] [PubMed] [Google Scholar]
- Shi Q., Thresher,R., Sancar,A. and Griffith,J. (1992) An electron microscopic study of (A)BC excinuclease. J. Mol. Biol., 226, 425–432. [DOI] [PubMed] [Google Scholar]
- Sugasawa K., Ng,J.M.Y., Masutani,C., Iwai,S., van der Spek,P.J., Eker,A.P.M., Hanaoka,F., Bootsma,D. and Hoeijmakers,J.H. (1998) Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol. Cell, 2, 223–232. [DOI] [PubMed] [Google Scholar]
- Takahara P.M., Rosenzweig,A.C., Frederick,C.A. and Lippard,S.J. (1995) Crystal structure of double-stranded DNA containing the major adduct of the anticancer drug cisplatin. Nature, 377, 649–652. [DOI] [PubMed] [Google Scholar]
- Tang J.Y., Hwang,B.J., Ford,J.M., Hanawalt P.C. and Chu,J. (2000) Xeroderma pigmentosum p48 gene enhances genomic repair and suppresses UV-induced mutagenesis. Mol. Cell, 5, 737–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema J., van Hoffen,A., Karcagi,V., Natarajan,A.T., van Zeeland,A.A. and Mullenders,L.H. (1991) Xeroderma pigmentosum complementation group C cells remove pyrimidine dimers selectively from the transcribed strand of active genes. Mol. Cell. Biol., 11, 4128–4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoeven E.E., Wyman,C., Moolenaar,G., Hoeijmakers,J.H., and Gooseen,N. (2001) Architecture of nucleotide excision repair complexes: DNA is wrapped by UvrB before and after damage recognition. EMBO J., 20, 601–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakasugi M. and Sancar,A. (1998) Assembly, subunit composition, and footprint of human DNA repair excision nuclease. Proc. Natl Acad. Sci. USA, 95, 6669–6674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakasugi M. and Sancar,A. (1999) Order of assembly of human DNA repair excision nuclease. J. Biol. Chem., 274, 18759–18768. [DOI] [PubMed] [Google Scholar]
- Wold M.S. (1997) Replication protein A. Annu. Rev. Biochem., 66, 61–92. [DOI] [PubMed] [Google Scholar]
- Wood R.D. (1996) DNA repair in eukaryotes. Annu. Rev. Biochem., 65, 135–167. [DOI] [PubMed] [Google Scholar]
- Yokoi M., Masutani,C., Maekawa,T., Sugasawa,K., Ohkuma,Y. and Hanaoka,F. (2000) The xeroderma pigmentosum group C protein complex XPC–HR23B plays an important role in the recruitment of transcription factor IIH to damaged DNA. J. Biol. Chem., 275, 9870–9875. [DOI] [PubMed] [Google Scholar]
- Zlatanova J. and van Holde,K. (1998) Binding to four-way junction DNA: a common property of architectural proteins? FASEB J., 12, 421–431. [PubMed] [Google Scholar]