Abstract
The TOG/XMAP215-related proteins play a role in microtubule dynamics at its plus end. Fission yeast Alp14, a newly identified TOG/XMAP215 family protein, is essential for proper chromosome segregation in concert with a second homologue Dis1. We show that the alp14 mutant fails to progress towards normal bipolar spindle formation. Intriguingly, Alp14 itself is a component of the Mad2-dependent spindle checkpoint cascade, as upon addition of microtubule-destabilizing drugs the alp14 mutant is incapable of maintaining high H1 kinase activity, which results in securin destruction and premature chromosome separation. Live imaging of Alp14–green fluorescent protein shows that during mitosis, Alp14 is associated with the peripheral region of the kinetochores as well as with the spindle poles. This is supported by ChIP (chromatin immunoprecipitation) and overlapping localization with the kinetochore marker Mis6. An intact spindle is required for Alp14 localization to the kinetochore periphery, but not to the poles. These results indicate that the TOG/XMAP215 family may play a central role as a bridge between the kinetochores and the plus end of pole to chromosome microtubules.
Keywords: fission yeast/kinetochore/spindle checkpoint/TOG/XMAP215
Introduction
The TOG/XMAP215-related proteins constitute a conserved family. XMAP215 was purified originally as a then novel microtubule-associated protein (Gard and Kirschner, 1987). The human homologue ch-TOG was identified independently as cDNA, which was overexpressed in colonic and hepatic tumour cell lines (Charrasse et al., 1995, 1998). Mutations in the TOG/XMAP215 homologues were also identified in genetically amenable systems such as yeast, Drosophila melanogaster and Caenorhabditis elegans (Ohkura et al., 1988; Nabeshima et al., 1995; Wang and Huffaker, 1997; X.P.Chen et al., 1998; Matthews et al., 1998; Cullen et al., 1999). Biochemical and cell biological analysis in vertebrates has suggested that the TOG/XMAP215 protein plays a positive role in microtubule stability, in particular by stimulating the growth rate at the plus end (Gard and Kirschner, 1987; Vasquez et al., 1994; Tournebize et al., 2000). Genetic analysis from yeast, worm and fly has shown that this family is important for spindle formation, and localizes to the spindle poles (Nabeshima et al., 1995; Nakaseko et al., 1996; Wang and Huffaker, 1997; X.P.Chen et al., 1998; Matthews et al., 1998; Wigge et al., 1998; Cullen et al., 1999).
Pole to chromosome microtubules play a central role in proper chromosome segregation at anaphase. Whilst the minus ends of these microtubules originate from the spindle poles, the plus end specifically interacts with and is embedded in the kinetochore, the specialized structure on centromeric DNA. Pairs of the sister chromatids have to be kept together until pole to chromosome microtubules capture all the unattached kinetochores in a bivalent fashion, otherwise precocious separation of sister chromatids results in aneuploid offspring, which leads to cell death or tumorigenesis (Lengauer et al., 1998). In order to prevent such a deleterious consequence, the cell has developed a regulatory system, called the spindle assembly checkpoint. The molecular network of this pathway has been revealed by genetic approaches in budding yeast (Hoyt et al., 1991; Li and Murray, 1991; Weiss and Winey, 1996), and subsequent work shows that this surveillance mechanism is conserved ubiquitously (Li and Benezra, 1996; Taylor and McKeon, 1997; Cahill et al., 1998; Jin et al., 1998; Taylor et al., 1998).
The major, though not only, pathway of the spindle assembly checkpoint is the kinetochore-mediated network, which is composed of Mad1, 2 and 3, and Bub1 and 3 (Hoyt et al., 1991; Li and Murray, 1991). These proteins localize to the unattached kinetochores during mitosis in vertebrates (R.-H.Chen et al., 1996, 1998; Li and Benezra, 1996; Taylor and McKeon, 1997; Taylor et al., 1998; Waters et al., 1998). As the cell monitors the physical state of, or tension at, kinetochores, these checkpoint proteins bind in concert to the Slp1/Cdc20/Fizzy protein, a positive regulator of the APC/C ubiquitin ligase (anaphase-promoting complex/cyclosome) (King et al., 1995; Sudakin et al., 1995), thereby inhibiting its activation (Hwang et al., 1998; Kim et al., 1998). Two major substrates for the APC/C are mitotic cyclins and securins, the destruction of which is required for exit from mitosis and sister chromatid segregation, respectively (King et al., 1996). The central question that remains to be answered is how unattached or tensionless kinetochores are recognized and their information transduced to the Mad2-dependent checkpoint machinery. Also, the mechanism of how the plus end of spindle microtubules interacts with the kinetochores is a crucial issue that needs to be addressed.
Fission yeast alp+ genes have been identified as loci that are required for the maintenance of growth polarity control (alp1–15 loci, altered polarity) (Radcliffe et al., 1998). In line with the fact that the microtubule cytoskeleton is a crucial determinant of growth polarity in this yeast, many of the alp+ genes encode conserved proteins that are required for microtubule and spindle function (Hirata et al., 1998; Radcliffe et al., 1999; Vardy and Toda, 2000). Here we show that fission yeast Alp14, a member of the conserved TOG/XMAP215 proteins, plays an essential role as a connection between the kinetochores and pole to chromosome microtubules.
Results
Alp14 is required for bipolar spindle formation and proper chromosome segregation
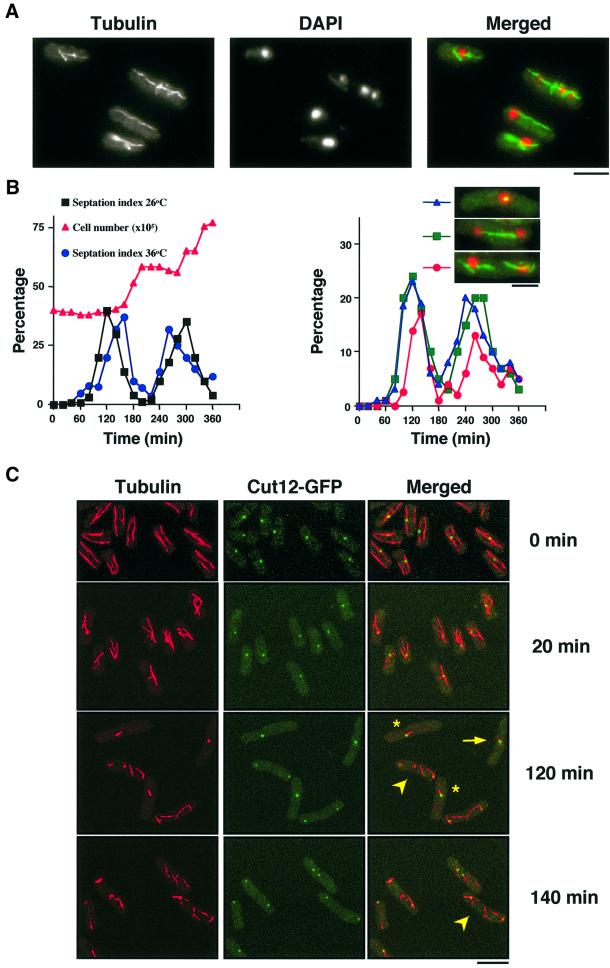
In addition to growth polarity defects, temperature- sensitive (ts) alp14-1270 mutants displayed abnormal mitotic spindles and mis-segregated chromosomes when incubated at 36°C (Figure 1A). In order to characterize the ts alp14 phenotypes in more detail, a synchronous culture was prepared by centrifugal elutriation using an alp14-1270 strain containing the integrated cut12+-GFP gene (cut12+ encodes an integral component of the spindle pole body, SPB; GFP encodes green fluorescent protein) (Bridge et al., 1998), and microtubules and the SPB were observed with confocal microscopy. Synchrony was followed in the culture incubated at 26°C (Figure 1B).
Fig. 1. Alp14 is required for bipolar spindle formation. (A) Microtubule structures and nuclear staining in alp14-1270 mutants. Exponentially growing alp14-1270 mutants at 26°C were shifted up to 36°C, incubated for 6 h and processed for immunofluorescence microscopy. (B) Synchronous culture analysis. Small early G2 cells of alp14-1270 mutants were collected by centrifugal elutriation and the cultures were divided into two parts, one part incubated at 26°C and the other shifted to 36°C. Septation index (left panel, black squares at 26°C and blue circles at 36°C) and cell number (left, at 26°C, red triangles) were plotted. Note that the peak of septation was shifted 40 min afterward at 36°C, compared with 26°C (160 min versus 120 min). The percentage of cells containing dense short spindles (right panel, blue triangles, shown in inset and arrows in C), cells with separated SPBs and continuous spindles (green squares), and cells with abnormal anaphase spindles (red circles, shown in inset and arrowheads in C) is shown on the right. (C) Abnormal microtubules and spindles in alp14 mutants. Representative images from synchronous culture analysis at 0, 20, 120 or 140 min are shown (anti-tubulin, Cut12–GFP and merged images). Cells that have dense short spindles (arrows), monopolar spindles (asterisks) and misorientated anaphase spindles (arrowheads) are marked. The bar indicates 10 µm.
At the restrictive temperature, the first phenotype observed was short and misorientated interphase microtubules (20 min, the second panels in Figure 1C). Note that at 0 min at 26°C, the length and structure of these microtubules were normal (top panels). Then upon entry into mitosis, three types of defects in mitotic spindles appeared sequentially. At 120 min when cells entered mitosis, dense and thick dots of microtubules were observed (24%, shown by arrows), which are never seen in wild-type cells. The SPB had not separated in these cells. Then mitotic cells with monopolar spindles (asterisk) were observed. It appeared that alp14 mutants failed to progress towards bipolar spindle formation. Despite the lack of normal bipolar spindles, anaphase and post-anaphase cells then appeared (arrowheads). It is particularly noticeable that in these cells, long anaphase spindles were rarely observed; instead, discontinuous or misorientated spindles/microtubules, some of which were associated with the SPBs, remained. This result suggested that Alp14 is required for the formation of both interphase microtubules and bipolar mitotic spindles, and plays a role in the completion of exit from anaphase at 36°C.
Fission yeast contains two TOG-related genes, alp14+ and dis1+, which play an overlapping role
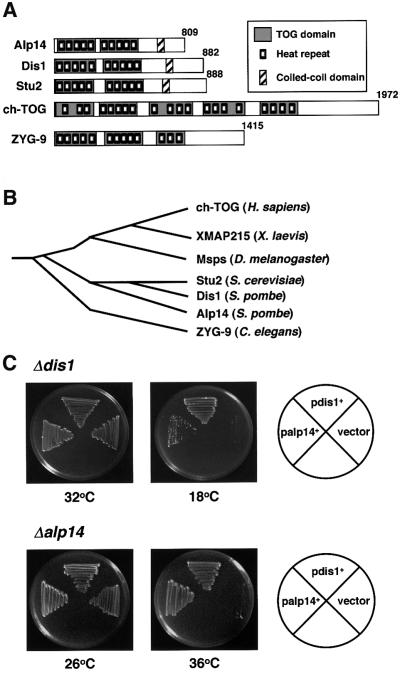
We cloned the alp14+ gene by complementation from a fission yeast genomic library. Nucleotide sequencing showed that the alp14+ gene encodes a protein that belongs to a family of conserved microtubule-binding proteins, including vertebrate ch-TOG (Charrasse et al., 1995, 1998), Xenopus XMAP215 (Gard and Kirschner, 1987), Drosophila mini spindles (Msps) (Cullen et al., 1999), C.elegans ZYG-9 (Matthews et al., 1998), budding yeast Stu2 (Wang and Huffaker, 1997) and fission yeast Dis1 (Nabeshima et al., 1995) (Figure 2A and B). The TOG/XMAP215 family contains the N-terminal repeated domains. Recently it has been shown that the conserved TOG domains comprise HEAT repeats (Andrade and Bork, 1995; Neuwald and Hirano, 2000), the hallmark of protein–protein interaction motifs (Groves et al., 1999). Proteins containing HEAT repeats include XCAP-D2 and XCAP-G (condensin), Scc2 (cohesin), COP-1 (coatomer) and PR65/A (type IIA protein phosphatase). Indeed, the TOG domains of Alp14 also consist of HEAT repeats (Figure 2A).
Fig. 2. Structural comparison between Alp14 and TOG-related proteins. (A) ‘TOG’ domains (dark boxes), HEAT-repeating units (small boxes) (Neuwald and Hirano, 2000) and coiled-coil regions (hatched) are shown in different members of the TOG/XMAP215 family from various organisms. These include Alp14, Dis1 (Schizosaccharomyces pombe), Stu2 (Saccharomyces cerevisiae), ch-TOG (Homo sapiens) and ZYG-9 (Caenorhabditis elegans). (B) Phylogenetic trees. ClustaIX dendrogram of the relationship between human, frog (XMAP215), fly (Msps), worm, budding and fission yeast proteins is shown. (C) Functional exchangeability between Alp14 and Dis1. Δdis1 (upper) or Δalp14 mutants (lower) containing an empty vector, and multicopy plasmids carrying dis1+ or alp14+ were streaked at either 18 (upper middle), 26 (lower left), 32 (upper left) or 36°C (lower middle) and incubated for 3–6 days.
Gene disruption showed that the alp14+ gene is not essential for cell viability. However, alp14-deleted cells showed ts phenotypes similar to alp14-1270 cells (Figure 2C), and defective phenotypes between Δalp14 and alp14-1270 cells were indistinguishable. Given the high degree of homology between Alp14 and Dis1, functional redundancy of these two genes was determined. It turned out that multicopy plasmids containing either gene were capable of rescuing the defects of mutations in the other gene, although suppression of Δdis1 by the alp14+ gene appeared incomplete (Figure 2C). Further more, double mutants between dis1 and alp14 were inviable at 30°C, the temperature permissive for both single mutants (Table I). Like ts stu2 mutants (Wang and Huffaker, 1997), alp14 mutants displayed strong genetic interactions with mutations in tubulin genes or alterations in tubulin gene expression (Table I). Therefore, Alp14 and Dis1 are functional, as well as structural, homologues, and Alp14 plays a major role in cells grown at high temperature (36°C), whilst Dis1 is required for growth at low temperature (18°C).
Table I. Genetic interaction between alp14 and tubulin mutants.
| Strains | Δdis1 | nda3-311a | atb2::LEU2a | nmt1-GFP- atb2+b | nmt81-GFP-atb2+b |
|---|---|---|---|---|---|
| alp14-1270 | lethal | lethal | lethal | lethalc | toxicd |
anda3+ and atb2+ encode β-tubulin and α2-tubulin, respectively.
bA thiamine-repressible strong (nmt1) or crippled promoter (nmt81) was integrated in front of the initiator methionine of the chromosomal atb2+ gene.
cStrains failed to grow at 26°C not only under derepressed conditions (without thiamine), but also under repressed conditions (in its presence).
dIn the absence of thiamine (derepressed), strains managed to form colonies, but showed slow growth (150% of doubling time) and defects in cell shape.
alp14 mutants activate the Mad2-dependent spindle assembly checkpoint transiently at the restrictive temperature
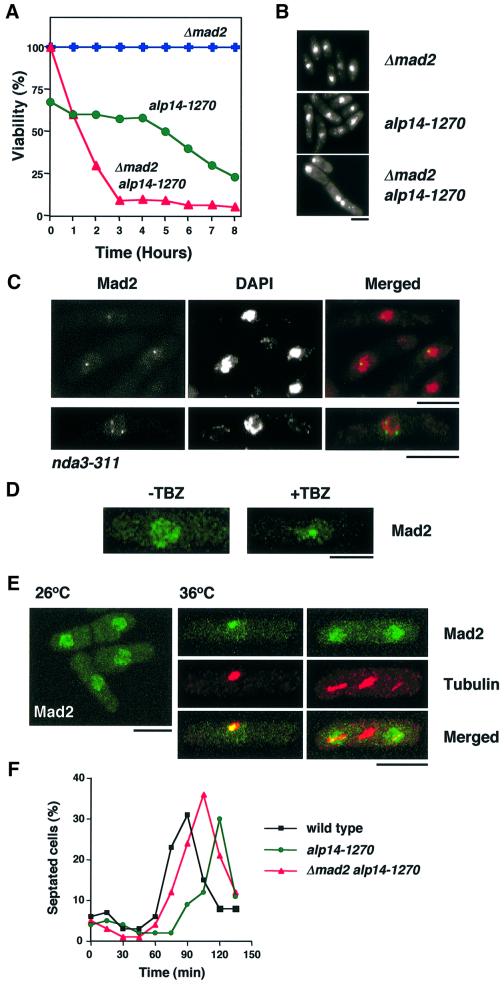
Given the appearance of abnormally segregated chromosomes and defective spindles in the alp14 mutant, whether or not the spindle assembly checkpoint pathway was activated was addressed. To this end, at first, a double mutant between the mad2 deletion and alp14-1270 was constructed, and defective phenotypes at the restrictive temperature were examined. As shown in Figure 3A, a Δmad2alp14-1270 double mutant lost viability rapidly (triangles) at 36°C, compared with the single alp14-1270 mutant (circles). Phenotypic analysis showed that the double mutant, but not the single, displayed ‘cut’ phenotypes, the combination of a chromosome attachment failure with a spindle checkpoint defect (Figure 3B). Albeit viable at 26°C, alp14-1270 mutants, as well as Δalp14, have defects in chromosome stability as, using the standard mini-chromosome assay (Niwa et al., 1989), we failed to obtain any Ade+ colonies, suggesting that mini-chromosomes are extremely unstable in this mutant.
Fig. 3. alp14 mutants transiently activate the Mad2-dependent spindle assembly checkpoint at the restrictive temperature. (A) Dependence of viability on Mad2. alp14-1270 (circles), mad2-deleted (crosses) or mad2alp14-1270 mutants (triangles) were grown at 26°C and shifted up to 36°C. Cell cultures were spread on rich plates after appropriate dilution and incubated at 26°C. At the same time, cell number was counted, and viability (number of viable cells per total number of cells) was calculated. (B) Nuclear staining. Cells were stained with 4′,6-diamidino-2-phenylindole (DAPI) after 6 h at 36°C. Note that chromosomes were condensed in alp14-1270 cells (middle), whilst in mad2alp14-1270 double mutants (bottom), decondensed chromosomes are evident. (C) Localization of Mad2 in tubulin mutants. nda3-311 mutants containing chromosomally tagged mad2+-GFP were incubated for 10 h at 20°C. (D) Localization of Mad2 upon depolymerization of microtubules. Wild-type cells containing chromosomally tagged mad2+-GFP were treated with TBZ (50 µg/ml) and incubated for 90 min. Localization of Mad2–GFP was examined before (–TBZ) and after incubation (+TBZ). (E) Mad2 localization in alp14 mutants at 36°C. alp14 mutant cells containing chromosomally tagged mad2+-GFP were synchronized with HU at 26°C, washed and released to HU-free medium at 36°C. Signals from Mad2–GFP were observed after 60 and 90 min. Anti-tubulin staining was also performed. The bar indicates 10 µm. (F) Mad2-dependent division delay in alp14 mutants. Wild-type (black squares), alp14 (green circles) and alp14Δmad2 mutant cells (red triangles) were synchronized as in (E). The percentage of septated cells was counted every 15 min.
In vertebrates, when the spindle checkpoint is activated, Mad2 localizes to unattached kinetochores (R.-H.Chen et al., 1996, 1998; Li and Benezra, 1996; Taylor and McKeon, 1997; Taylor et al., 1998; Waters et al., 1998). As Mad2 localization has not been reported in fission yeast, its similarities to mammalian cells were examined. In cold-sensitive nda3-311 mutants, which are defective in β-tubulin (Hiraoka et al., 1984) and activate the checkpoint, the chromosomal mad2+ gene was tagged with green fluorescent protein (GFP) at the C-terminus. It was concentrated on a single dot, or sometimes three dots, on condensed chromosomes (Figure 3C). It was also found that Mad2 was concentrated to a nuclear dot when the spindle checkpoint was activated in wild-type cells by adding the microtubule-destabilizing drug thibendazole (TBZ; Figure 3D, right), whilst in interphase cells (exponentially growing wild-type cells or alp14 mutants at the permissive temperature), it localizes mostly to the entire nucleus in a discontinuous patched pattern (see Figure 3D, left, and E, left). This localization pattern suggested that, like higher eukaryotes, fission yeast Mad2 localizes to the unattached kinetochores when the spindle checkpoint is activated.
Having established Mad2 localization upon checkpoint activation, its localization in alp14 mutants at 36°C was examined. When alp14 mutant cells were arrested with hydroxyurea (HU) at early S phase and released in the absence of HU at 36°C synchronously, Mad2–GFP was found to localize specifically to a spot in abnormal mitotic cells which showed dense and thick dots of microtubules (Figure 3E, middle panels). In normally dividing cells at 26°C, Mad2 localized to the nucleus (left). Despite this specific localization, consistent with cell cycle progression at 36°C shown before (Figure 1B and C), Mad2 was subsequently delocalized and dispersed in the divided nuclei at anaphase (Figure 3E, right panels). Although the cell cycle proceeded, checkpoint activation did result in a delay in cell division, which was evident in the percentage of septated cells in the elutriated culture (see left panel in Figure 1B). Under these conditions, the frequency of septated cells peaked 40 min later than those at the permissive temperature (120 min at 26°C versus 160 min at 36°C).
In order to examine the requirement for Mad2 for the division delay in alp14 mutants, a similar synchronous culture experiment was repeated in wild-type, alp14 and alp14Δmad2 mutants. As shown in Figure 3F, septation took place earlier in alp14Δmad2 double mutants (peaked at 105 min) than in a single alp14 mutant (120 min), indicating that the mitotic delay is attributed to activation of the Mad2 checkpoint. However it should be noted that the timing of septation was not completely reverted in this double mutant, compared with wild-type cells (90 min). This suggested that there is an additional Mad2-independent mechanism, which leads to the division delay in alp14 mutants. Taken together, these results indicated that the absence of Alp14 results in spindle damage, which activates the Mad2 checkpoint pathway, leading to division delay; however, this activation was only transient and cell cycle progression continued.
Alp14 is a novel component of the spindle assembly checkpoint and functions in a linear manner upstream of Mad2
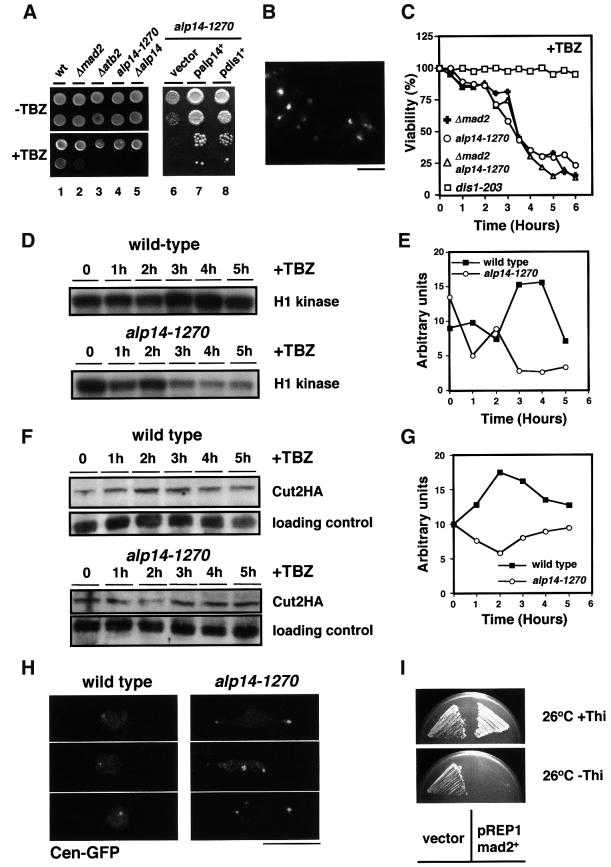
Mutants defective in microtubule and/or spindle function often show hypersensitivity to microtubule-destabilizing drugs. We found that the alp14 mutant was hypersensitive to TBZ even at the permissive temperature (Figure 4A, lanes 4 and 5). This hypersensitivity was rescued by the introduction of plasmids containing either the alp14+ or the dis1+ gene (lanes 6–8). Cell morphology showed that, surprisingly, alp14 mutants in the presence of TBZ displayed a cut phenotype (20% after 6 h at 26°C, Figure 4B), rather than a mitotic arrest. In line with this, unlike wild-type cells, viability fell sharply upon TBZ addition (Figure 4C). This suggested that alp14 mutant cells are defective in the spindle checkpoint.
Fig. 4. Alp14 is a component of the Mad2-dependent spindle checkpoint pathway. (A) Hypersensitivity to TBZ. Wild-type (lane 1), mad2-deleted (lane 2), atb2-deleted (lane 3), alp14-1270 (lane 4) or alp14-deleted cells (lane 5) were spotted (105 and 104 cells) on rich plates in the absence (upper) or presence of 30 µg/ml TBZ for 5 days and incubated at 26°C. Also, alp14-1270 cells containing an empty vector (pREP1, lane 6), and multicopy plasmids carrying the alp14+ (lane 7) or dis1+ gene (lane 8) were spotted after serial dilution on rich plates containing 10 µg/ml TBZ and incubated at 26°C for 3 days. (B) Lethal ‘cut’ phenotypes in the presence of TBZ. DAPI staining of alp14-1270 cells incubated for 6 h in the presence of 50 µg/ml TBZ at 26°C is shown. The bar indicates 10 µm. (C) Viability loss. The indicated strains were grown in rich medium at 26°C and TBZ (50 µg/ml) was added. After appropriate dilution, cells were plated on rich YES plates to examine colony forming ability. Viability was calculated by dividing the number of colonies by the number of cells per culture. While dis1-203 cells (squares) did not lose viability, alp14-1270 (circles), mad2-deleted (crosses) or mad2alp14-1270 cells (triangles) did sharply, but with similar kinetics. (D) Decline of H1 kinase activity in alp14-1270 in the presence of TBZ. (E) Quantification of H1 kinase activity from the data in (D) (wild-type, squares; alp14-1270, circles). (F) Securin destruction. Immunoblotting was performed in wild-type cells and alp14 mutants (containing integrated cut2+-HA) upon addition of TBZ. (G) Quantification of securin levels from the data in (F). (H) Separation of sister chromatids. Centromere-marked GFP was observed after 3 h upon TBZ addition in wild-type (left) and alp14 mutants (right), which contain cen-LacO and GFP-LacI-NLS. (I) Toxicity of overproduced Mad2 in alp14-1270 cells. alp14-1270 mutants were transformed with a vector or plasmid in which mad2+ is under the control of the thiamine-repressible nmt1 promoter. Transformants were streaked on minimal plates in the presence (upper) or absence (lower) of thiamine and incubated for 4 days at 26°C.
It is known that mutants defective in the spindle checkpoint pathway fail to maintain H1 kinase activity to high levels under conditions of checkpoint activation (Li and Murray, 1991; He et al., 1997). We next sought to examine whether Alp14 function is required for the maintenance of high H1 kinase in the presence of TBZ. Wild-type and alp14 mutant cells were synchronized with HU, released in the presence of TBZ upon washout of HU, and H1 kinase activity was measured. The H1 kinase activity increased (3 h) and remained at high levels in wild-type cells, whereas alp14 mutants failed to maintain high activity and declined to low levels (Figure 4D and E).
Another phenotype of spindle checkpoint mutants is premature separation of sister chromatids, which is attributed to the untimely degradation of securin (Cut2 in fission yeast) (King et al., 1996). In order to examine securin levels in the alp14 mutant, immunoblotting using strains containing C-terminally tagged Cut2 (Cut2-HA) (Funabiki et al., 1996) was performed. As shown in Figure 4F and G, unlike wild-type cells, Cut2 levels were reduced in alp14 mutants in the presence of TBZ. Consistent with this result, sister chromatids segregated abnormally in alp14 mutants, which was visualized using centromere marking GFP–LacI (Cen–GFP) (Straight et al., 1996; Nabeshima et al., 1998) (Figure 4H), and Mad2 failed to accumulate at the kinetochores (data not shown). These results demonstrate that Alp14 is required for the activation of the spindle checkpoint pathway in the presence of TBZ and strongly suggest that Alp14 is a component of this pathway.
In order to examine a functional relationship between Alp14 and Mad2, cell viability of the mad2alp14-1270 mutant in the presence of TBZ was compared with each single mutant. It was found that the kinetics of viability loss were indistinguishable between each single and double mutant (Figure 4C). This result showed that Alp14 functions in a linear manner with Mad2. Ectopic overexpression of mad2+ activates the checkpoint without spindle damage and arrests the cells in metaphase (He et al., 1997; Kim et al., 1998). We next asked whether Mad2 overproduction is still capable of activating the checkpoint in alp14 mutants. As shown in Figure 4I, overexpression of mad2+ was still toxic and arrested alp14 cells in mitosis (data not shown). Overexpression of alp14+, on the other hand, cannot be used for this type of analysis, as it disrupts mitotic spindles and activates the checkpoint (data not shown). Taken together, these results show that Alp14 functions either upstream of or in concert with the Mad2 checkpoint protein.
Alp14 localizes to the spindle poles and to dots along mitotic spindles
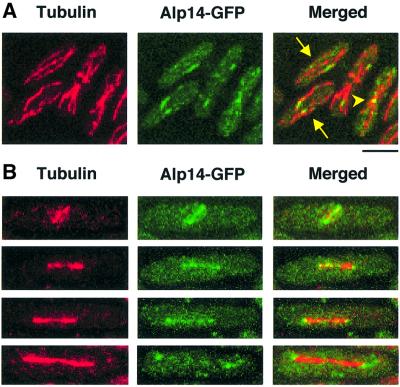
In order to examine the cellular localization of Alp14, the GFP gene was fused to the C-terminus of the chromosomal alp14+ gene. GFP tagging did not interfere with protein function as strains containing Alp14–GFP grew as well as wild-type strains at 36°C. GFP signals were examined in fixed samples from exponentially growing wild-type cells. Results are shown in Figure 5, and summarized below. During interphase, Alp14 localized along cytoplasmic microtubules (Figure 5A, arrows) as discontinuous patches, rather than as filamentous structures. Upon entry into mitosis, mitotic spindles were stained (Figure 5B). In addition, the two distal dots, likely to be the SPB, were stained. Upon exit from mitosis, Alp14–GFP appeared to dissociate from long anaphase spindles, and instead co-localized with astral microtubules (Figure 5B, bottom panels, see below). During post-anaphase, Alp14 relocalized to microtubules at the central ring (Figure 5A, arrowhead).
Fig. 5. Cellular localization of Alp14. Alp14 localization during the cell cycle. An integrated alp14+-GFP strain was grown at 30°C, fixed with methanol (A) or formaldehyde (B) and processed for immuno fluorescence microscopy. Anti-α-tubulin antibody (TAT-1) was used to visualize microtubules. A confocal microscope was used to observe each image. (A) Localization of Alp14 in interphase cells. Dotted staining along interphase microtubules (arrows) and localization at the equatorial ring in a post-anaphase cell (arrowhead) are shown. (B) Localization of Alp14 in mitotic cells. Phase 2 (top) or elongating anaphase cells (phase 3, lower three panels) are shown. The bar indicates 10 µm.
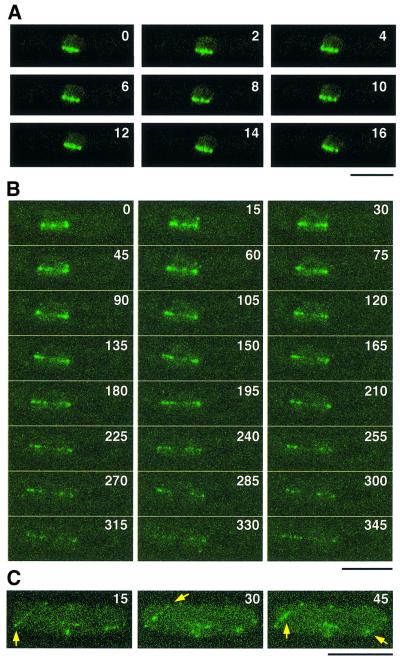
To delineate further the cellular localization of Alp14 during the cell cycle in detail, particularly focusing on mitotic behaviour, time-lapse live imaging of Alp14–GFP was performed using a conventional and confocal laser scanning microscope. As shown in Figure 6A, during phase 2, in which spindle length remains constant (Nabeshima et al., 1998), Alp14–GFP localized to four or five intranuclear dots along spindles (2 s intervals). Upon elongation of the spindle (Figure 6B, 75 s and later, phase 3, also see Supplementary data available at The EMBO Journal Online), these dots appeared to be pulled polewards and could hardly be localized to the equator of the anaphase spindle. This dynamic movement is very similar to that of centromeric DNAs reported previously (Nabeshima et al., 1998). In late anaphase, Alp14 localizes to astral microtubules (arrows in Figure 6C). This analysis establishes the dynamic nature of Alp14 localization along spindles during mitotic phase 2 and phase 3, and suggests that Alp14 localizes to the kinetochores.
Fig. 6. Time-lapse imaging of Alp14–GFP during mitosis. (A–C) Live image analysis of Alp14–GFP. Time-lapse imaging of Alp14–GFP was performed using a conventional fluorescence microscope with 2 s intervals (A) or using confocal z-stack series, comprising 13 sections with a 0.36 µm step size with 15 s intervals (B and C). Mitotic cells of phase 2 (Nabeshima et al., 1998) corresponding to prometaphase and metaphase are shown in (A). Spindles started to elongate in phase 3 of mitosis (at 75 s, in B). Post-anaphase cells are shown in (C). Alp14 co-localizing with astral microtubules is shown by arrows. See Supplementary data for live images. The bar indicates 10 µm.
Alp14 localizes to the kinetochore periphery during metaphase in a microtubule-dependent manner
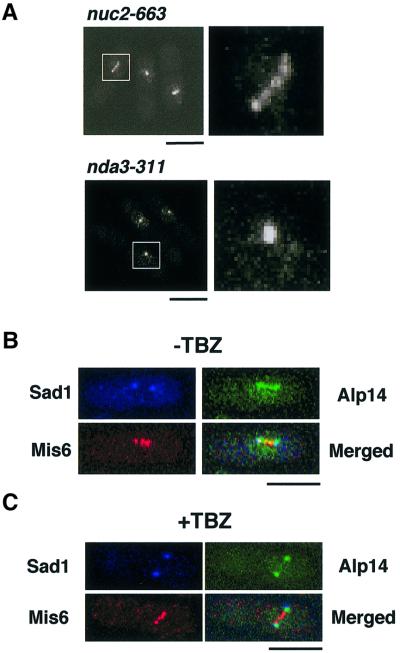
Given the involvement of Alp14 in the Mad2-mediated checkpoint pathway and the localization of Alp14 to the intranuclear dots along mitotic spindles, we were interested in whether Alp14 localizes to the kinetochores. To address this question, the following approaches were undertaken. First, Alp14–GFP localization was examined in two mitotically arresting mutants. A nuc2-663 strain defective in the homologue of Cdc27/Apc3 displays a uniform metaphase arrest at 36°C (Hirano et al., 1988), whilst the nda3-311 mutant arrests at prophase at 20°C (Hiraoka et al., 1984). In nuc2-arrested cells, Alp14–GFP showed, in addition to the two distal SPBs, three intranuclear dots (Figure 7A, upper) similar to those seen in live images (see Figure 6A), and these dots may represent the kinetochores. In contrast, in nda3 mutants, only a single dot instead of multiple dots was detected (Figure 7A, lower), which probably corresponded to the unseparated SPB (Ding et al., 1997). This suggested that Alp14–GFP localization to the kinetochore periphery requires the mitotic spindle.
Fig. 7. Localization of Alp14 at the kinetochore periphery in metaphase. (A) Alp14 localization in nuc2- or nda3-arrested cells. nuc2-663 (upper) or nda3-311 (lower) cells containing integrated alp14+-GFP are arrested at 36°C for 3 h or at 18°C for 8 h, respectively, fixed with formaldehyde and observed by confocal microscopy. Three individual cells are shown in each left panel and enlarged GFP images (6-fold) are shown on the right. The bar indicates 10 µm. (B) Localization of Alp14 and Mis6 in metaphase-arrested cells. nuc2-663 cells containing integrated alp14+-GFP and mis6+-HA were arrested at 36°C for 3 h. Then cells were fixed, and signals from Alp14–GFP, anti-HA and anti-Sad1 antibodies were observed. (C) Dependence of kinetochore localization of Alp14 upon spindles. nuc2-663 arrested cells in (B) were treated with TBZ (50 µg/ml) and, after 15 min, cells were processed for immunofluorescence microscopy. The bar indicates 10 µm.
Next, co-localization between Alp14 and Mis6, a component of the fission yeast centromeres (Saitoh et al., 1997), was addressed. To this end, ts nuc2-663 cells containing doubly integrated alp14+-GFP and mis6+-HA were used. The dots of Alp14–GFP were visible along spindles, in addition to the two distal SPBs (visualized by anti-Sad1 antibodies; Hagan and Yanagida, 1995) (Figure 7B). Importantly, these inner dots overlapped, if not completely, with Mis6-HA. It is of note that the intranuclear dotted structures of Alp14–GFP appeared less defined in this image compared with the fixed samples shown before (Figure 7A) or non-fixed live images (see Figure 6A), which might be attributable to the procedures used for sample preparations for immunofluorescence microscopy.
The addition of TBZ to these nuc2-arrested cells resulted in the disappearance of Alp14–GFP dots (Figure 7C), providing further support that kinetochore localization of Alp14–GFP is dependent upon spindle integrity. On the other hand, SPB localization of Alp14–GFP and centromere localization of Mis6-HA were not disrupted by this treatment. Taken together, these results strongly suggest that Alp14 localizes to the kinetochore periphery at metaphase in a spindle-dependent fashion.
Alp14 is associated with the centromere DNA
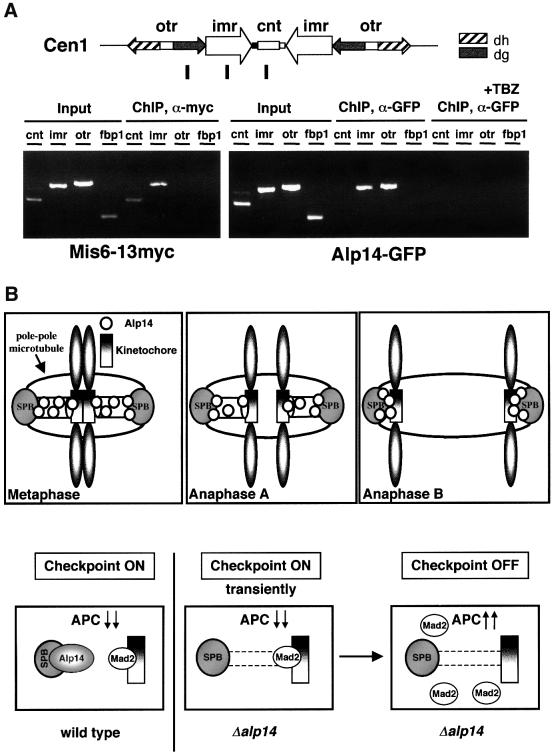
Finally, to confirm the kinetochore localization of Alp14, the ChIP method (chromatin immunoprecipitation) (Hecht et al., 1996; Ekwall et al., 1997; Meluh and Koshland, 1997; Saitoh et al., 1997; Ekwall and Partridge, 1999) was applied to nuc2-663 mutants containing Alp14–GFP. Upon incubation at 36°C for 3 h, cells were fixed with formaldehyde and immunoprecipitation was performed with an anti-GFP antibody. A PCR then followed. The primers used were those encompassing the central region of the centromeres of chromosome I (cnt1) (Takahashi et al., 1992; Saitoh et al., 1997; Partridge et al., 2000) and the flanking repeated region (imr1 and otr1). As a control, a Mis6-Myc strain and primers for the non-centromeric region (fbp1) were also used. ChIP showed that Mis6 precipitated, as reported previously, with cnt1 and imr1, but not with otr1 nor fbp1 (Saitoh et al., 1997; Partridge et al., 2000). In nuc2-arrested cells, Alp14 precipitated with either imr1 or otr1 centromeric DNA (Figure 8A). We have failed so far to show any binding between Alp14 and cnt1 DNA, the reasons for which might be technical in nature. Alternatively, Alp14 is indeed associated with the kinetochore periphery.
Fig. 8. Association of Alp14 with the centromere DNA and model for Alp14 function at kinetochores. (A) Association of Alp14 with the centromeric DNA. ChIP was performed in nuc2-663 arrested cells containing integrated alp14+-GFP. Cells were incubated at 36°C for 3 h and fixed with 1% formaldehyde. A schematic diagram showing the centromeric region of chromosome I is also shown. (B) A role for Alp14 in the kinetochores and the spindle checkpoint. Alp14 localizes to mitotic spindles, the SPBs and the kinetochore periphery during mitosis (upper). In the absence of Alp14, the spindle checkpoint failed to maintain the activation status, although Mad2 is capable of localizing to the kinetochores transiently. When microtubules and spindles are damaged or depolymerized, Alp14 is required for the activation of the Mad2-mediated spindle checkpoint.
Next, ChIP was performed in the presence of TBZ in nuc2-663 mutants (the same condition as in Figure 7C). Consistent with the dissociation of Alp14 from spindles and kinetochores, the binding of Alp14 to imr1 or otr1 was diminished (Figure 8A, four rightmost lanes). These results established that Alp14 is a mitosis-specific kinetochore-associated protein, the localization of which requires intact spindles.
Discussion
We have shown that the conserved Alp14 protein localizes to the kinetochore periphery and the poles during the mitotic stage and is a component of the Mad2-mediated spindle checkpoint (Figure 8B). We have also shown that the absence of Alp14 results in transient activation of this checkpoint pathway and this activation leads to mitotic delay in a Mad2-dependent fashion. Therefore, Alp14 apparently plays dual roles in this kinetochore-mediated pathway. One involves a regulatory function, in which Alp14 acts in a linear fashion with Mad2, probably upstream of it. The other role is structural, in which Alp14 is required for the interaction between the kinetochores and pole to chromosome microtubules. The dense short spindles seen at early mitosis in alp14 mutants could be a result of either the inherent instability of pole to chromosome microtubules or the failure to capture the kinetochore, or in fact a combination of both. Mad2 localizes to these dots, which is consistent with the notion that these represent unattached kinetochores. In addition to activation of the Mad2-dependent spindle checkpoint, alp14 mutants show Mad2-independent division delay. Although the molecular basis of this second block is currently elusive, it might be related to a post-anaphase delay; it is possible that the aberrant anaphase spindle assembled in the alp14 mutant slows down the anaphase process and, as a result, leads to cell division delay.
Activation of the Mad2 checkpoint and loss of checkpoint activation in alp14 mutants might seem to be a contradiction. However, it should be noted that the involvement of Alp14 in the Mad2-dependent checkpoint pathway becomes most evident under conditions when microtubules are depolymerized at the permissive temperature in alp14 mutants, as the Mad2-dependent checkpoint pathway is dispensable for normally dividing cells in yeast. In contrast, the activation of the checkpoint in the alp14 mutant is observed at high temperature, in which Alp14 is essential for cell division and required for the formation of functional pole to kinetochore spindles. These two phenotypes, therefore, would not be conflicting, but rather represent distinct aspects of Alp14 function. It is also of note that activation of the checkpoint in alp14 mutants is transient (Figure 8B). In other words, in the absence of Alp14 function, the Mad2 checkpoint could sense the spindle damage, but the unattached kinetochores fail to maintain Mad2 localization and mitotic arrest. Similar, if not identical, checkpoint-deficient phenotypes have been reported recently in other conserved components of the kinetochores (e.g. Spc24) and, interestingly, these proteins were identified originally as the components of the SPB (Wigge et al., 1998; Janke et al., 2001; Wigge and Kilmartin, 2001).
In spite of the evolutionary conservation of overall protein structure, kinetochore localization has not been reported before in other members of the TOG/XMAP215 proteins; instead, what is common is their localization to microtubules and the spindle poles (centrosomes) (Popov et al., 2001). This could simply be due to species-specific diversity between related proteins. Alternatively, the localization to the kinetochores may be too dynamic to be detected with conventional fluorescence microscopy of fixed samples. As shown in this study, only time-lapse live images using chromosomally integrated Alp14–GFP and also ChIP performed in metaphase-arrested cells have provided convincing data of kinetochore localization. Support for a role of Alp14 in kinetochore function comes from a number of previous reports (Gard and Kirschner, 1987; Vasquez et al., 1994; Tournebize et al., 2000), which pointed out the close involvement of XMAP215 in plus end-dependent microtubule dynamics, and, importantly, the plus end is where the kinetochore binds the mitotic spindle. We show that kinetochore localization of Alp14 is dependent upon the presence of intact spindles, which suggests that Alp14 passes through microtubules to attach to the kinetochores and its attachment would be transient. Whether or not all the TOG/XMAP215 family members attach to the kinetochores awaits careful localization studies in other organisms.
Although most of Alp14 dissociates from the kinetochores in the absence of intact spindles, it must execute a crucial role in activation of the checkpoint at the kinetochores. It should be pointed out that the SPB and the centromeres locate in close proximity under conditions of checkpoint activation (Funabiki et al., 1993). It is possible that Alp14 interacts directly or indirectly with checkpoint proteins and/or other regulators of kinetochore function. In dividing cells, Mad2 localization at the kinetochores appears to utilize microtubules and it travels between the kinetochores and the poles (Howell et al., 2000). Vertebrate CENP-E kinesin and yeast Spc24 play a role similar to Alp14 (Abrieu et al., 2000; Yao et al., 2000; Janke et al., 2001; Wigge and Kilmartin, 2001). It would be of interest to see whether TOG/XMAP215 interacts with these proteins. Although there are no obvious homologues to CENP-E in fission yeast, the dynamic behaviour of Alp14 on microtubules and spindles suggests the existence of motor molecules, which directly or indirectly interact with Alp14. Identification of such motor molecule(s) would be the next goal towards understanding the molecular anatomy of the kinetochore.
The kinetochore-mediated mitotic role of Alp14 is not its sole function. As alp14 mutants show growth polarity defects (Radcliffe et al., 1998) and shorter interphase microtubules, we think that like XMAP215 (Gard and Kirschner, 1987; Vasquez et al., 1994), Alp14 also regulates the dynamics of cytoplasmic microtubules. The TOG/XMAP215-related proteins could be general plus end regulators of both interphase microtubules and mitotic spindles.
Materials and methods
Strains, media and genetic methods
Strains used in this study are listed in Table II. The standard methods were followed as described (Moreno et al., 1991).
Table II. Strain list.
| Strains | Genotypes | Derivations |
|---|---|---|
| HM123 | h–leu1 | our stock |
| DH1270 | h–leu1alp14-1270 | our stock |
| Δatb2 | h–leu1atb2::LEU2 | our stock |
| NC102 | h–leu1nuc2-663 | our stock |
| Δdis1 | h–leu1ura4dis1::ura4+ | Nabeshima et al. (1995) |
| AE148 | h–leu1ura4mad2::ura4+ | Kim et al. (1998) |
| HF178 | h+leu1cut2+-3HA-LEU2 | Funabiki et al. (1996) |
| GFP-nmt1-atb2 | h–leu1kanr-nmt1-GFP-atb2+ | this study |
| GFP-nmt81-atb2 | h–leu1kanr-nmt81-GFP-atb2+ | this study |
| HM248 | h–his2ade6-M210 containing Ch16 (mini-chromosome) | Niwa et al. (1989) |
| NK04 | h–leu1ura4 alp14::kanr | this study |
| MKY7A-4 | h+leu1ura4lys1his7LacO-lys1+GFP-LacI-NLS-his7+ | Nabeshima et al. (1995) |
| MAG000 | h–leu1ura4 alp14-1270mad2::ura4+ | this study |
| MAG004 | h–leu1ura4his7alp14+-GFP-kanr | this study |
| MAG031 | h–leu1nuc2-663alp14+-GFP-kanr | this study |
| MAG037 | h+leu1nda3-KM311alp14+-GFP-kanr | this study |
| MAG040 | h+leu1ura4his2alp14-1270 cut12+-GFPura4+ | this study |
| MAG050 | h–leu1alp14-1270LacO-lys1+GFP-LacI-NLS-his7+ | this study |
| MAG068 | h+leu1alp14-1270cut2+-3HA-LEU2 | this study |
| MAG071 | h–leu1ura4mad2+-GFP-kanr | this study |
| MAG074 | h–leu1ura4mis6+-13myc-kanr | this study |
| MAG090 | h–leu1ura4alp14-1270mad2+-GFP-kanr | this study |
| MAG092 | h–leu1nuc2-663alp14+-GFP-kanrmis6+-3HA-kanr | this study |
| MAG117 | h–leu1nda3-311mad2+-GFP-kanr | this study |
Synchronous culture analysis
Small early G2 cells were collected from exponentially growing cultures at 26°C by centrifugal elutriation, and were divided into two halves. One half continued to be incubated at 26°C to monitor the degree of synchrony, whilst the other half was shifted up to 36°C to observe defective phenotypes.
Cloning of the alp14+ gene
A Schizosaccharomyces pombe genomic library in a LEU2-based multicopy vector pAL-KS (obtained from Taro Nakamura) was used for the isolation of the alp14+ gene. Seven transformants of an alp14-1270 strain, which were Ts+ in a plasmid-dependent manner, were obtained from 10 000 Leu+ transformants. Plasmid DNAs were recovered from each transformant, and restriction mapping showed that all the plasmids contained an overlapping insert. Identification of the cloned gene as alp14+ was confirmed by genetic crosses between tagged strains (alp14+-GFP-kanr) and the original ts alp14-1270 mutant.
Preparation and manipulation of nucleic acids
Enzymes were used as recommended by the suppliers (New England Biolabs). Nucleotide sequence data reported in this paper are in the DDBJ/EMBL/GenBank databases under the accession No. AB032409 (alp14+).
C-terminal epitope tagging
C-terminal tagging of Alp14 with GFP, Mis6 with 13Myc or 3HA, or Mad2 with GFP was performed by a PCR-based gene targeting method (Bähler et al., 1998).
Histone H1 kinase assay
H1 kinase assay was performed using p13suc1 agarose beads (Oncogene) in the presence of [γ-32P]ATP. Kinase activities were quantified by a Molecular Dynamics PhosphorImager.
Immunochemical assays
Affinity-purified rabbit polyclonal anti-Sad1 antibody was obtained from Dr Mizuki Shimanuki. Mouse monoclonal anti-α-tubulin antibody (TAT-1) was provided by Dr Keith Gull. Mouse monoclonal anti-Myc (9E10; BAbCO), anti-HA (16B12; BAbCO) and anti-GFP (Clones 7.1 and 13.1; Boehringer Mannheim) antibodies were also used.
Indirect immunofluorescence microscopy
Cells were fixed with methanol or formaldehyde, and primary antibodies (TAT-1 1:50, anti-Sad1 antibody 1:10 000 or anti-HA antibody 1:1000) were applied, followed by Cy3-conjugated goat anti-rabbit or anti-mouse IgG (Sigma), fluorescein-linked sheep anti-mouse IgG (Amersham) or Cy5-conjugated donkey anti-mouse IgG (Jackson Laboratories). Immunofluorescence images were viewed with a chilled video-rated CCD camera (model C5985; Hamamatsu) connected to a computer (Apple Power Macintosh G3/400) or with a laser scanning confocal microscope LSM510 (Zeiss Co.) and processed by use of Adobe® Photoshop (version 5.5).
Time-lapse live imaging
Rich medium containing 2% agar was solidified on a glass slide and one drop of the culture of a strain containing the chromosomal alp14+-GFP was placed on top of the agar and sandwiched with a coverslip. Live images were taken at room temperature with a Zeiss Axioplan equipped with a chilled video CCD camera (C4742-95) and the PC computer containing kinetic image AQM software (Kinetic Imaging Ltd) or with a Zeiss LSM510 laser scanning confocal microscope under conditions as described previously (Brunner and Nurse, 2000). Z-stacks (200–400 nm steps) were taken every 15 s for maximally 12 min for a single preparation.
Formaldehyde cross-linked ChIP
The method previously described (Ekwall and Partridge, 1999) was followed with the following modifications: immunoprecipitates were washed sequentially with 1 ml of lysis buffer, 1 ml of lysis buffer containing 0.5 M NaCl and 1 ml of TE buffer. nuc2-663 containing the chromosomal alp14+-GFP or wild-type containing the chromosomal mis6+-13myc was grown at 36°C for 3 h, fixed with 1% formaldehyde and soluble chromatin was prepared. Immunoprecipitations were performed with anti-GFP or anti-Myc antibody. Recovered DNA was assayed by PCR (35 cycles). Primer pairs used are as follows: 5′-AACAATAAACACGAATGCCTC-3′ plus 5′-ATAGTACCATGCGATTGTCTG-3′ for cnt1, 5′-GATGGAGAGTTACGAGGTAAAATG-3′ plus 5′-AATTGAATGAGAATCTGTGTACAG-3′ for imr1, 5′-CAACATTCGAAAATTCAAGGGAAA-3′ and 5’-CCGCAGCATATTGGTCGTCGTTTTTAA-3′ for otr1, and 5′-AATGACAATTCCCCACTAGCC-3′ plus 5′-ACTTCAGCTAGGATTCACCTGG-3′ for fbp1.
Supplementary data
Live images of Alp14–GFP can be seen as Supplementary data available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Drs Iain M.Hagan, Tomohiro Matsumoto, Taro Nakamura, Yukinobu Nakaseko, Osami Niwa, Paul Nurse, Shelley Sazer, Mizuki Shimanuki, Chikashi Shimoda and Mitsuhiro Yanagida for providing materials used in this study, Satoko Yamaguchi and Hiroyuki Yamano for showing us the H1 kinase assay, Heidi Browning, Damian Brunner and Teresa Niccoli for instructions in live imaging, and Peter Jordan for help with confocal microscopy. Special thanks are given to Drs Robin C.Allshire and Janet F.Partridge for instruction in the ChIP method. We are grateful to Drs Anabelle Decottignies, Tatsuya Hirano, Tomohiro Matsumoto, Yukinobu Nakaseko and Hiroyuki Ohkura for informing us of unpublished results prior to publication, Kanji Furuya and Mitsuhiro Yanagida for information on Dis1 and Mad2 localization, Satoru Uzawa for stimulating discussion, Drs Jacqueline Hayles and Frank Uhlman for critical reading of the manuscript and useful suggestions, and the two anonymous referees, who helped improve this paper considerably. M.A.G. was supported by an EMBO long-term fellowship. This work is supported by the ICRF and the HFSP research grant.
Note added in proof
While this paper was under review, two papers appeared related to this work. One paper [Nakaseko,Y., Goshima,G., Morishita,J. and Yanagida,M. (2001) M phase-specific kinetochore proteins in fission yeast: microtubule-associated Dis1 and Mtc1 display rapid separation and segregation during anaphase. Curr. Biol., 11, 537–549] shows kinetochore localization of Dis1, whilst the other paper [Severin,F., Habermann,B., Huffaker,T. and Hyman,T. (2001) Stu2 promotes mitotic spindle elongation in anaphase. J. Cell Biol., 153, 435–442] shows activation of the spindle checkpoint in budding yeast ts stu2 mutants.
References
- Abrieu A., Kahana,J.A., Wood,K.W. and Cleveland,D.W. (2000) CENP-E as an essential component of the mitotic checkpoint in vitro. Cell, 102, 817–826. [DOI] [PubMed] [Google Scholar]
- Andrade M.A. and Bork,P. (1995) HEAT repeats in the Huntington’s disease protein. Nature Genet., 11, 115–116. [DOI] [PubMed] [Google Scholar]
- Bähler J., Wu,J., Longtine,M.S., Shah,N.G., McKenzie,A.,III, Steever,A.B., Wach,A., Philippsen,P. and Pringle,J.R. (1998) Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast, 14, 943–951. [DOI] [PubMed] [Google Scholar]
- Bridge A.J., Morphew,M., Bartlett,R. and Hagan,I.M. (1998) The fission yeast SPB component Cut12 links bipolar spindle formation to mitotic control. Genes Dev., 12, 927–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner D. and Nurse,P. (2000) CLIP170-like tip1p spatially organizes microtubular dynamics in fission yeast. Cell, 102, 695–704. [DOI] [PubMed] [Google Scholar]
- Cahill D.P., Lengauer,C., Yu,J., Riggins,G.J., Willson,J.K.V., Markowitz,S.D., Kinzler,K.W. and Vogelstein,B. (1998) Mutations of mitotic checkpoint genes in human cancers. Nature, 392, 300–303. [DOI] [PubMed] [Google Scholar]
- Charrasse S., Mazel,M., Taviaux,S., Berta,P., Chow,T. and Larroque,C. (1995) Characterization of the cDNA and pattern of expression of a new gene over-expressed in human hepatomas and colonic tumors. Eur. J. Biochem., 234, 406–413. [DOI] [PubMed] [Google Scholar]
- Charrasse S., Schroeder,M., Gauthier-Rouviere,C., Ango,F., Cassimeris,L., Gard,D.L. and Larroque,C. (1998) The TOGp protein is a new human microtubule-associated protein homologous to the Xenopus XMAP215. J. Cell Sci., 111, 1371–1383. [DOI] [PubMed] [Google Scholar]
- Chen R.-H., Waters,J.C., Salmon,E.D. and Murray,A.W. (1996) Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science, 274, 242–246. [DOI] [PubMed] [Google Scholar]
- Chen R.-H., Shevchenko,A., Mann,M. and Murray,A.W. (1998) Spindle checkpoint protein Xmad1 recruits Xmad2 to unattached kinetochores. J. Cell Biol., 143, 283–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.P., Yin,H. and Huffaker,T.C. (1998) The yeast spindle pole body component Spc72p interacts with Stu2p and is required for proper microtubule assembly. J. Cell Biol., 141, 1169–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen C.F., Deák,P., Glover,D.M. and Ohkura,H. (1999) mini spindles: a gene encoding a conserved microtubule-associated protein required for the integrity of the mitotic spindle in Drosophila. J. Cell Biol., 146, 1005–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding R., West,R.R., Morphew,M., Oakley,B.R. and McIntosh,J.R. (1997) The spindle pole body of Schizosaccharomyces pombe enters and leaves the nuclear envelope as the cell cycle proceeds. Mol. Biol. Cell, 8, 1461–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekwall K. and Partridge,J.F. (1999) Fission yeast chromosome analysis: fluorescence in situ hybridisation (FISH) and chromatin immunoprecipitation (CHIP). In Bickmore,W.A. (ed.), Chromosome Structural Analysis: A Practical Approach. Oxford University Press, Oxford, UK, pp. 39–57.
- Ekwall K., Olsson,T., Turner,B.M., Cranston,G. and Allshire,R.C. (1997) Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres. Cell, 91, 1021–1032. [DOI] [PubMed] [Google Scholar]
- Funabiki H., Hagan,I.M., Uzawa,S. and Yanagida,M. (1993) Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J. Cell Biol., 121, 961–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H., Yamano,H., Kumada,K., Nagao,K., Hunt,T. and Yanagida,M. (1996) Cut2 proteolysis required for sister-chromatid separation in fission yeast. Nature, 381, 438–441. [DOI] [PubMed] [Google Scholar]
- Gard D.L. and Kirschner,M.W. (1987) A microtubule-associated protein from Xenopus eggs that specifically promotes assembly at the plus-end. J. Cell Biol., 105, 2203–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves M.R., Hanlon,N., Turowski,P., Hemmings,B.A. and Barford,D. (1999) The structure of the protein phosphatase 2A PR65/A subunit reveals the conformation of its 15 tandemly repeated HEAT motifs. Cell, 96, 99–110. [DOI] [PubMed] [Google Scholar]
- Hagan I. and Yanagida,M. (1995) The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J. Cell Biol., 129, 1033–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Patterson,T.E. and Sazer,S. (1997) The Schizosaccharomyces pombe spindle checkpoint protein mad2p blocks anaphase and genetically interacts with the anaphase-promoting complex. Proc. Natl Acad. Sci. USA, 94, 7965–7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht A., Strahl-Bolsinger,S. and Grunstein,M. (1996) Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature, 383, 92–96. [DOI] [PubMed] [Google Scholar]
- Hirano T., Hiraoka,Y. and Yanagida,M. (1988) A temperature-sensitive mutation of the S.pombe gene nuc2+ that encodes a nuclear scaffold-like protein blocks spindle elongation in mitotic anaphase. J. Cell Biol., 106, 1171–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka Y., Toda,T. and Yanagida,M. (1984) The NDA3 gene of fission yeast encodes β-tubulin: a cold-sensitive nda3 mutation reversibly blocks spindle formation and chromosome movement in mitosis. Cell, 39, 349–358. [DOI] [PubMed] [Google Scholar]
- Hirata D., Masuda,H., Eddison,M. and Toda,T. (1998) Essential role of tubulin-folding cofactor D in microtubule assembly and its association with microtubules in fission yeast. EMBO J., 17, 658–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell B.J., Hoffman,D.B., Fang,G., Murray,A.W. and Salmon,E.D. (2000) Visualization of Mad2 dynamics at kinetochores, along spindle fibers and at spindle poles in living cells. J. Cell Biol., 150, 1233–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt M.A., Totis,L. and Roberts,B.T. (1991) S.cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell, 66, 507–117. [DOI] [PubMed] [Google Scholar]
- Hwang L.H., Lau,L.F., Smith,D.L., Mistrot,C.A., Hardwick,K.G., Hwang,E.S., Amon,A. and Murray,A.W. (1998) Budding yeast Cdc20: a target of the spindle checkpoint. Science, 279, 1041–1044. [DOI] [PubMed] [Google Scholar]
- Janke C., Ortiz,J., Lechner,J., Shevchenko,A., Shevchenko,A., Magiera,M.M., Schramm,C. and Schiebel,E. (2001) The budding yeast proteins Spc24p and Spc25p interact with Ndc80p and Nuf2p at the kinetochore and are important for kinetochore clustering and checkpoint control. EMBO J., 20, 777–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin D.-Y., Spencer,F. and Jeang,K.-T. (1998) Human T cell leukemia virus type 1 oncoprotein tax targets the human mitotic checkpoint protein MAD1. Cell, 93, 81–91. [DOI] [PubMed] [Google Scholar]
- Kim S.H., Lin,D.P., Matsumoto,S., Kitazono,A. and Matsumoto,T. (1998) Fission yeast Slp1: an effector of the Mad2-dependent spindle checkpoint. Science, 279, 1045–1047. [DOI] [PubMed] [Google Scholar]
- King R.W., Peters,J.-M., Tugendreich,S.T., Rolfe,M., Hieter,P. and Kirschner,M.W. (1995) A 20S complex containing CDC27 and CDC16 catalyzes the mitotic-specific conjugation of ubiquitin to cyclin B. Cell, 81, 279–288. [DOI] [PubMed] [Google Scholar]
- King R.W., Deshaies,R.J., Peters,J.-M. and Kirschner,M.W. (1996) How proteolysis drives the cell cycle. Science, 274, 1652–1659. [DOI] [PubMed] [Google Scholar]
- Lengauer C., Kinzler,K.W. and Vogelstein,B. (1998) Genetic instabilities in human cancers. Nature, 396, 643–649. [DOI] [PubMed] [Google Scholar]
- Li R. and Murray,A.W. (1991) Feedback control of mitosis in budding yeast. Cell, 66, 519–531. [DOI] [PubMed] [Google Scholar]
- Li Y. and Benezra,R. (1996) Identification of a human mitotic checkpoint gene: hsMAD2. Science, 274, 246–248. [DOI] [PubMed] [Google Scholar]
- Matthews L.R., Carter,P., Thierry-Mieg,D. and Kemphues,K. (1998) ZYG-9, a Caenorhabditis elegans protein required for microtubule organization and function, is a component of meiotic and mitotic spindle poles. J. Cell Biol., 141, 1159–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh P.B. and Koshland,D. (1997) Budding yeast centromere composition and assembly as revealed by in vivo cross-linking. Genes Dev., 11, 3401–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S., Klar,A. and Nurse,P. (1991) Molecular genetic analyses of fission yeast Schizosaccharomyces pombe. Methods Enzymol., 194, 795–823. [DOI] [PubMed] [Google Scholar]
- Nabeshima K., Kurooka,H., Takeuchi,M., Kinoshita,K., Nakaseko,Y. and Yanagida,M. (1995) p93dis1, which is required for sister chromatid separation, is a novel microtubule and spindle pole body-associating protein phosphorylated at the Cdc2 target sites. Genes Dev., 9, 1572–1585. [DOI] [PubMed] [Google Scholar]
- Nabeshima K., Nakagawa,T., Straight,A.F., Murray,A., Chikashige,Y., Yamashita,Y.M., Hiraoka,Y. and Yanagida,M. (1998) Dynamics of centromeres during metaphase–anaphase transition in fission yeast: Dis1 is implicated in force balance in metaphase bipolar spindle. Mol. Biol. Cell, 9, 3211–3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaseko Y., Nabeshima,K., Kinoshita,K. and Yanagida,M. (1996) Dissection of fission yeast microtubule associating protein p93dis1: regions implicated in regulated localization and microtubule interaction. Genes Cells, 1, 633–644. [DOI] [PubMed] [Google Scholar]
- Neuwald A.F. and Hirano,T. (2000) HEAT repeats associated with condensins, cohesins and other complexes involved in chromosome-related functions. Genome Res., 10, 1445–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa O., Matsumoto,T., Chikashige,Y. and Yanagida,M. (1989) Characterization of Schizosaccharomyces pombe minichromosome deletion derivatives and a functional allocation of their centromere. EMBO J., 8, 3045–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkura H., Adachi,Y., Kinoshita,N., Niwa,O., Toda,T. and Yanagida,M. (1988) Cold-sensitive and caffeine supersensitive mutants of the Schizosaccharomyces pombe dis genes implicated in sister chromatid separation during mitosis. EMBO J., 7, 1465–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge J.F., Borgstrøm,B. and Allshire,R.C. (2000) Distinct protein interaction domains and protein spreading in a complex centromere. Genes Dev., 14, 783–791. [PMC free article] [PubMed] [Google Scholar]
- Popov A.V., Pozniakovsky,A., Arnal,I., Antony,C., Ashford,A., Kinoshita,K., Tournebize,R., Hyman,A.A. and Karsenti,E. (2001) XMAP215 regulates microtubule dynamics through two distinct domains. EMBO J., 20, 397–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radcliffe P., Hirata,D., Childs,D., Vardy,L. and Toda,T. (1998) Identification of novel temperature-sensitive lethal alleles in essential β-tubulin and nonessential α2-tubulin genes as fission yeast polarity mutants. Mol. Biol. Cell, 9, 1757–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radcliffe P.A., Hirata,D., Vardy,L. and Toda,T. (1999) Functional dissection and hierarchy of tubulin-folding cofactor homologues in fission yeast. Mol. Biol. Cell, 10, 2987–3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh S., Takahashi,K. and Yanagida,M. (1997) Mis6, a fission yeast inner centromere protein, acts during G1/S and forms specialized chromatin required for equal segregation. Cell, 90, 131–143. [DOI] [PubMed] [Google Scholar]
- Straight A.F., Belmont,A.S., Robinett,C.C. and Murray,A.W. (1996) GFP tagging of budding yeast chromosomes reveals that protein–protein interactions can mediate sister chromatid cohesion. Curr. Biol., 6, 1599–1608. [DOI] [PubMed] [Google Scholar]
- Sudakin V., Ganoth,D., Dahan,A., Heller,H., Hershko,J., Luca,F.C., Ruderman,J.V. and Hershko,A. (1995) The cyclosome, a large complex containing cyclin-selective ubiquitin ligase activity, targets cyclins for destruction at the end of mitosis. Mol. Biol. Cell, 6, 185–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Murakami,S., Chikashige,Y., Funabiki,H., Niwa,O. and Yanagida,M. (1992) A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol. Biol. Cell, 3, 819–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S.S. and McKeon,F. (1997) Kinetochore localization of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage. Cell, 89, 727–735. [DOI] [PubMed] [Google Scholar]
- Taylor S.S., Ha,E. and McKeon,F. (1998) The human homologue of Bub3 is required for kinetochore localization of Bub1 and a Mad3/Bub1-related protein kinase. J. Cell Biol., 142, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournebize R. et al. (2000) Control of microtubule dynamics by the antagonistic activities of XMAP215 and XKCM1 in Xenopus egg extracts. Nature Cell Biol., 2, 13–19. [DOI] [PubMed] [Google Scholar]
- Vardy L. and Toda,T. (2000) The fission yeast γ-tubulin complex is required in G1 phase and is a component of the spindle-assembly checkpoint. EMBO J., 19, 6098–6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasquez R.J., Gard,D.L. and Cassimeris,L. (1994) XMAP from Xenopus eggs promotes rapid plus end assembly of microtubules and rapid microtubule polymer turnover. J. Cell Biol., 127, 985–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P.J. and Huffaker,T.C. (1997) Stu2p: a microtubule-binding protein that is an essential component of the yeast spindle pole body. J. Cell Biol., 139, 1271–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters J.C., Chen,R.-H., Murray,A.W. and Salmon,E.D. (1998) Localization of Mad2 to kinetochores depends on microtubule attachment, not tension. J. Cell Biol., 141, 1181–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss E. and Winey,M. (1996) The Saccharomyces cerevisiae spindle pole body duplication gene MPS1 is part of a mitotic checkpoint. J. Cell Biol., 132, 111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge P.A. and Kilmartin,J.V. (2001) The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J. Cell Biol., 152, 349–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge P.A., Jensen,O.N., Holmes,S., Souès,S., Mann,M. and Kilmartin,J.V. (1998) Analysis of the Saccharomyces spindle pole by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. J. Cell Biol., 141, 967–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X., Abrieu,A., Zheng,Y., Sullivan,K.F. and Cleveland,D.W. (2000) CENP-E forms a link between attachment of spindle microtubules to kinetochores and the mitotic checkpoint. Nature Cell Biol., 2, 484–491. [DOI] [PubMed] [Google Scholar]