Abstract
The evolutionarily conserved protein kinases Mec1 and Rad53 are required for checkpoint response and growth. Here we show that their role in growth is to remove the ribonucleotide reductase inhibitor Sml1 to ensure DNA replication. Sml1 protein levels fluctuate during the cell cycle, being lowest during S phase. The disappearance of Sml1 protein in S phase is due to post-transcriptional regulation and is associated with protein phosphorylation. Both phosphorylation and diminution of Sml1 require MEC1 and RAD53. More over, failure to remove Sml1 in mec1 and rad53 mutants results in incomplete DNA replication, defective mitochondrial DNA propagation, decreased dNTP levels and cell death. Interestingly, similar regulation of Sml1 also occurs after DNA damage. In this case, the regulation requires MEC1 and RAD53, as well as other checkpoint genes. Therefore, Sml1 is a new target of the DNA damage checkpoint and its removal is a conserved function of Mec1 and Rad53 during growth and after damage.
Keywords: checkpoint/Mec1/protein phosphorylation/Rad53/ribonucleotide reductase
Introduction
In the yeast Saccharomyces cerevisiae, Mec1 and Rad53 protein kinases are essential both after DNA damage and during cell growth (Zheng et al., 1993; Kato and Ogawa, 1994; Weinert et al., 1994). In response to DNA damage, they function as signal transducers in all known checkpoint pathways (reviewed in Elledge, 1996). Rad53 usually functions downstream of Mec1 and, together, they receive signals from upstream sensor proteins transmitting them to components of the cell cycle engine. Consequently, cell cycle progression is arrested or delayed, providing time for repair. In addition, Mec1 and Rad53 also increase the capacity of the cell to repair DNA lesions. One route is by the transcriptional induction of various DNA repair proteins, including ribonucleotide reductase (RNR), the enzyme that catalyzes the rate-limiting step of both deoxyribonucleotide (dNTP) and DNA synthesis (reviewed in Reichard, 1988). However, additional interfaces between the Mec1/Rad53 checkpoint pathway and DNA repair are probably required to maximize protection of genetic integrity. A better understanding of such interactions relies on the discovery of new targets of checkpoint control.
The checkpoint function of Mec1 and Rad53 is evolutionarily conserved. Their mammalian homologs, ATM/ATR and CHK2, also function as signal transducers and affect multiple components of the cell cycle and DNA repair machinery during the response to DNA damage (reviewed in Rotman and Shiloh, 1999). For example, ATM/ATR and CHK2 activate and stabilize p53, which in turn leads to the transcriptional induction of a variety of genes, including that of RNR (Tanaka et al., 2000; Zhao et al., 2000a; reviewed in Caspari, 2000).
The conservation between Mec1/Rad53 and their mammalian homologs may extend beyond their checkpoint functions. These proteins are also important for normal cell growth; in yeast, deletion of MEC1 or RAD53 is lethal (Zheng et al., 1993; Kato and Ogawa, 1994) and, in mice, deletion of ATM or ATR causes slow growth or early embryonic lethality, respectively (Xu et al., 1996; Brown and Baltimore, 2000). Nevertheless, little is known about the nature of their functions in cell growth or the relationship between their roles in growth and in DNA damage response.
A clue to their essential functions comes from studies of suppressors of mec1 and rad53 lethality in yeast. It has been reported that the essential function of Mec1 and Rad53 can be bypassed by increasing dNTP formation after overexpression of RNR genes (Desany et al., 1998), by deletion of the RNR inhibitor Sml1 (Zhao et al., 1998) or by removal of the RNR transcriptional repressor Crt1 (Huang et al., 1998). These observations raise the possibility that Mec1 and Rad53 are directly involved in dNTP regulation (Zhao et al., 1998). However, it is equally possible that they do not regulate dNTP formation but rather control a process that is sensitive to dNTP levels, such as late replication origin firing (Desany et al., 1998) or the restraint of cell cycle progression (Vallen and Cross, 1999). Until now, the genetic data have been inadequate to distinguish among these models.
Here we address this issue by examining Sml1 protein levels during the cell cycle. We found that Sml1 levels decrease at S phase concomitantly with the appearance of its phosphorylated form. Both the phosphorylation of Sml1 and the reduction of Sml1 levels require Mec1 and Rad53 function. This regulation is important since, in its absence, the cell is unable to complete DNA replication. However, Mec1 is not required for the initiation of DNA replication. Interestingly, Sml1 is regulated in a similar fashion after DNA damage. This damage response requires MEC1, RAD53 and other checkpoint genes, including RAD9, RAD17, RAD24 and MEC3, demonstrating that Sml1 is a new target of the DNA damage checkpoint.
Results
Sml1 protein levels fluctuate during the cell cycle
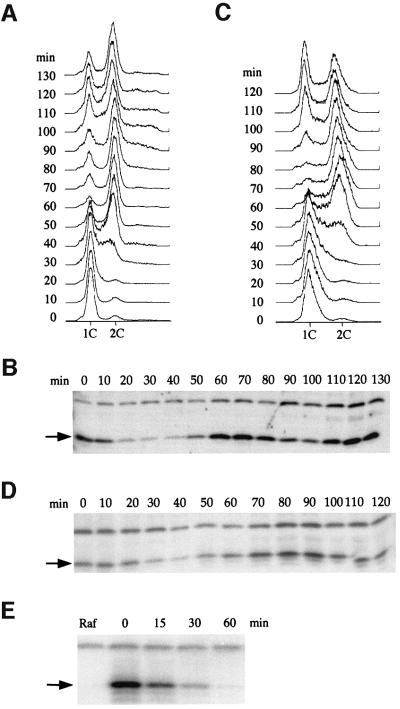
Since inhibitors of biological pathways are often subject to regulation, we examined levels of the RNR inhibitor Sml1 during the cell cycle. Wild-type cells were arrested in G1 phase and then released into the cell cycle (Figure 1A). Sml1 protein levels at various points during two cell cycles were examined and found to decrease during S phase (6-fold down compared with G1-arrested cells) and peak during G2/M phase (9-fold more than S phase cells) (Figure 1B).
Fig. 1. Sml1 protein levels fluctuate during the cell cycle. (A) The wild-type strain (W1379-3C) was arrested in G1 by α-mating factor in YPD medium and then released into the cell cycle. Samples were collected immediately after release (time zero) and every 10 min until 130 min. Cells were fixed and DNA content was measured by FACS analysis. (B) Protein extracts were made from samples collected in (A). Sml1 protein levels were examined by a protein blot using anti-Sml1 antibody. The arrow indicates the position of Sml1. The band above Sml1 cross-reacts with anti-Sml1 serum and is used as a loading control. (C) Strain W2057-11A (GAL-SML1) was arrested in G1 by α-mating factor in YPGL medium. Sml1 was induced by the addition of 2% galactose for 30 min. Cells were next released into YPGal medium and samples were collected immediately after release (time zero) and every 10 min until 120 min. Cells were fixed and DNA content was measured by FACS analysis. Note that the 10 min delay in entry into S phase compared with (A) is due to growth conditions and is not genotype specific. (D) Sml1 levels from samples collected in (C) were examined as described in (B). (E) Strain W2057-11A (GAL-SML1) was arrested in G2/M phase by nocodazole in YPRaffinose medium (Raf). Cells were next transferred to YPGal medium containing nocodazole to induce Sml1 expression and maintain their arrest. After 45 min, GAL-SML1 expression was turned off by addition of 2% glucose. Sml1 protein levels were examined by a protein blot using anti-Sml1 antibody at the time points indicated. Zero time is immediately before the addition of 2% glucose.
To understand whether the fluctuation in Sml1 levels is due to post-transcriptional regulation, the endogenous SML1 promoter was replaced by an inducible GAL1 promoter. As shown in Figure 1C and D, a similar pattern of Sml1 protein oscillation during the cell cycle is observed in this strain, demonstrating that Sml1 is post-transcriptionally regulated. Consistent with the fluctuation of Sml1 protein levels, Sml1 was shown to be a short-lived protein. It has a half-life of only 16 min in G2/M phase, the time when the highest amount of Sml1 is observed (Figure 1E).
The decrease of Sml1 levels in S phase depends on Mec1 and partially on Rad53
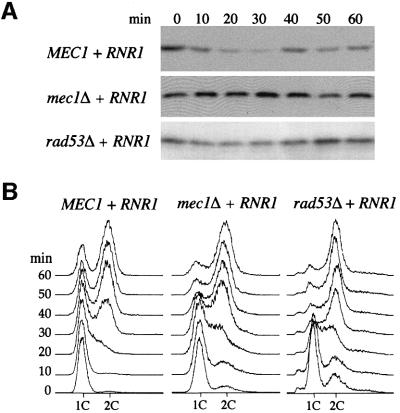
Next, we tested whether the decrease of Sml1 levels in S phase depends on Mec1 and Rad53. Since mec1Δ and rad53Δ strains are unable to grow in the presence of Sml1, their viability was maintained by the presence of a 2 µm-RNR1 plasmid (Desany et al., 1998). As shown in Figure 2A, Sml1 levels in wild-type strains decrease as the cells progress from G1 phase into S phase (3.4-fold down compared with G1-arrested cells). However, Sml1 levels are unchanged between G1 and S phase in mec1Δ strains and are reproducibly only slightly reduced at S phase in rad53Δ strains (1.2-fold down compared with G1-arrested cells). Thus, the decrease in Sml1 levels as cells enter S phase is dependent on Mec1 and partially dependent on Rad53.
Fig. 2. The decrease in Sml1 levels during S phase depends on MEC1 and partially on RAD53. (A) Strains U1476 (MEC1 + RNR1), U1195 (mec1Δ + RNR1) and U1198-10C (rad53Δ + RNR1) were arrested in G1 by α-mating factor in SC-LEU medium. Protein extracts were made immediately after release from G1 phase (time zero) and every 10 min for 60 min. Sml1 protein levels were examined by protein blots using anti-Sml1 antibody. The amount of Sml1 protein was quantified from the protein blot using a cross-reacting band (e.g. see Figure 1B) as a loading control. (B) DNA content was measured by FACS from samples collected in (A).
Sml1 protein levels decrease after DNA damage and HU treatment
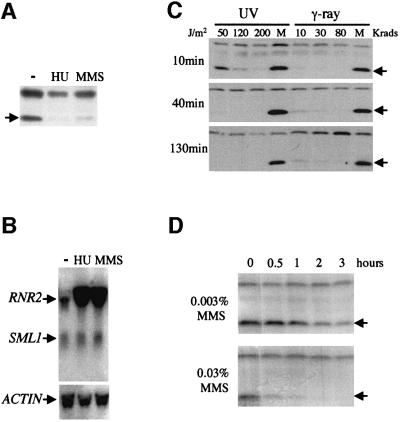
Increased RNR activity and increased dNTP pools are hypothesized to be required for the repair of DNA lesions based on the induction of the RNR genes after DNA damage (Elledge and Davis, 1989). Thus, it is possible that Mec1/Rad53 regulate Sml1 not only in S phase but also after DNA damage. Therefore, we examined Sml1 levels after methyl methanesulfonate (MMS) and hydroxyurea (HU) treatment. As shown in Figure 3A, the amount of the Sml1 protein dramatically decreases when cells are incubated with HU or MMS. In contrast, the levels of SML1 mRNA are unaffected (Figure 3B). Thus, the disappearance of Sml1 after MMS and HU treatment is due to post-transcriptional regulation.
Fig. 3. Sml1 protein levels, but not its mRNA levels, diminish after treatment with DNA-damaging agents. (A and B) The wild-type strain (W1588-4C) was treated with 200 mM HU and 0.05% MMS for 80 min. Sml1 protein and SML1 RNA from these samples were examined by protein blot (A) and an RNA blot (B). In (B), actin was used as loading control and RNR2 was used as a positive control for transcriptional induction by DNA damage. (C and D) The wild-type strain (W1588-4C) was irradiated with various doses of UV light and γ-rays (C) or treated with 0.003 or 0.03% MMS (D) as indicated. Samples were harvested at different time points after treatment to examine the kinetics of Sml1 protein level changes. Samples of mock treatment (M) received no irradiation. Arrows indicate the position of the Sml1 protein.
As shown in Figure 3C, Sml1 also diminishes after γ-ray and UV irradiation. Examination of Sml1 degradation kinetics reveals that: (i) Sml1 disappears rapidly after DNA damage, e.g. only 3% of Sml1 is left 10 min after 10 krads of γ-irradiation (Figure 3C); (ii) Sml1 levels decrease faster when higher doses of DNA-damaging agents are applied (Figure 3C and D); and (iii) Sml1 does not reappear even 130 min after irradiation with γ-ray or UV light (Figure 3C).
The disappearance of Sml1 after DNA damage or replication blocks is regulated by MEC1/RAD53 and other DNA damage checkpoint genes
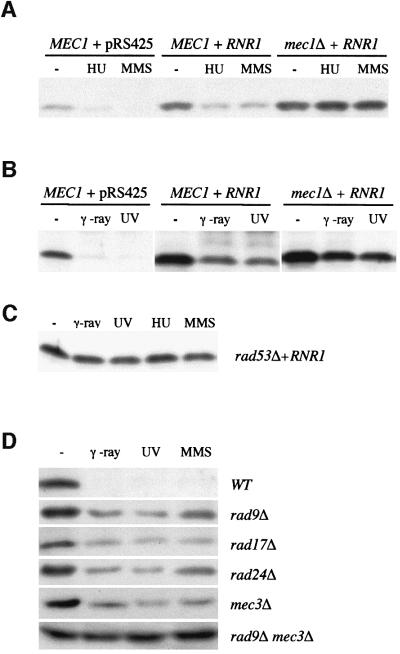
Next, Sml1 levels were examined in mec1Δ and rad53Δ strains. As mentioned before, to maintain the viability of these strains in the presence of Sml1, an RNR1 plasmid was introduced. A higher level of Sml1 protein was observed in the presence of the RNR1 plasmid and this is independent of DNA damage (compare MEC1 + pRS425 with MEC1 + RNR1 in Figure 4A and B). After DNA damage, Sml1 levels decrease in wild-type strains that contain either the vector or the RNR1 plasmid (Figure 4A and B). In contrast, the amount of Sml1 in treated mec1Δ or rad53Δ cells is similar to that seen in untreated cells (Figure 4A–C). Thus, as was observed during S phase, the diminution of Sml1 after DNA damage and replication blocks occurs in a Mec1- and Rad53-dependent manner.
Fig. 4. The diminution of Sml1 after DNA damage depends on MEC1 and RAD53, and partially on other cell cycle checkpoint genes. Sml1 protein levels from various strains were examined after cells were treated with different types of DNA-damaging agents. The strains used are: in (A) and (B), U1475 (MEC1 + pRS425), U1476 (MEC1 + RNR1) and U1195 (mec1Δ + RNR1); in (C), U1198-10C (rad53Δ + RNR1); and in (D), W1588-4A (WT), W1518-10B (rad9Δ), W1522-11B (rad17Δ), W1519-17B (rad24Δ), W1520-10B (mec3Δ) and W2617-4A (rad9Δ mec1Δ). For HU and MMS treatments, protein extracts were made after cells were incubated with 200 mM HU or 0.05% MMS for 1 h. For UV and γ-ray treatments, protein extracts were made after cells were irradiated by UV light (120 J/m2) and γ-rays (30 krads) and grown at 30°C for 30 min.
To understand whether this regulation is a unique function of Mec1/Rad53 or requires other DNA damage checkpoint genes, we examined Sml1 levels in several other checkpoint mutants, including rad9Δ, rad17Δ, rad24Δ and mec3Δ. These mutant strains all contain significant amounts of Sml1 after DNA damage, indicating that Rad9, Rad17, Rad24 and Mec3 are required for the regulation of Sml1 (Figure 4D). The fact that some reduction in Sml1 levels is observed in these mutants suggests that these genes are functionally redundant. It has been shown previously that there are two additive branches of the DNA damage checkpoint pathway: one defined by RAD9 and the other by RAD17, RAD24 and MEC3 (de la Torre-Ruiz et al., 1998). We therefore examined Sml1 levels in three double mutant strains, each containing rad9 and one of the three other checkpoint mutations. In these strains, Sml1 levels remain unchanged after DNA damage (Figure 5D and data not shown). Taken together, these results show that Sml1 is a new target of the DNA damage checkpoint response and that its regulation requires the bifurcated Rad9 and the Rad17/Rad24/Mec3 pathways.
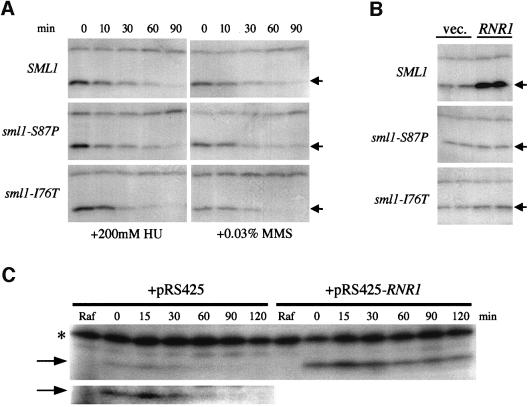
Fig. 5. Wild-type and mutant Sml1 protein levels after DNA damage and in the presence of RNR1 overexpression. (A) Sml1 protein levels were examined in strains W1588-4C (SML1), W2097-63B (sml1-S87P) and W2099-63D (sml1-I76T) at various time points after incubation with HU and MMS at the concentrations indicated. Arrows show the position of wild-type or mutant Sml1 proteins. (B) Sml1 protein levels were examined in strains W1588-4C (SML1), W2097-63B (sml1-S87P) and W2099-63D (sml1-I76T) in the presence of either an empty vector pRS425 (vec.) or a 2 µm-RNR1 plasmid (RNR1). (C) W2057-11A (GAL-SML1) strains that contain either an empty vector (pRS425) or a 2 µm-RNR1 plasmid (pRS425-RNR1) were first grown in raffinose-Leu medium to early-log phase (Raf). Expression of Sml1 proteins was then induced by addition of 2% galactose for 30 min. The induction was inhibited by the addition of 2% glucose and the Sml1 proteins levels were examined at various time points. The zero time point is the time of addtion of glucose. The lower panel in (C) is an overexposure of the Sml1 portion of the protein blot from the strain containing pRS425, which illustrates the absence of Sml1 protein at later time points. The increased signal of the cross-reacting band (*) in (C) compared with (A) and (B) is due to different batches of antibody.
The unbound form of Sml1 is targeted for degradation after DNA damage and replication blocks
To investigate further the regulation of Sml1 after DNA damage, we asked whether the Rnr1-bound or unbound form of Sml1 is the target of degradation. Using two-hybrid and in vitro binding analysis, we isolated sml1 mutations that abolish the interaction with Rnr1 (Zhao et al., 2000b). In these mutant strains, Sml1 probably exists only in the unbound state and, therefore, changes in Sml1 protein levels will reflect only that of this unbound form. Two such mutant strains were examined after MMS and HU treatment. We found that the mutant proteins show similar degradation kinetics to those of the wild-type protein (Figure 5A). Thus, the decrease in Sml1 levels after DNA damage does not depend on the Sml1–Rnr1 interaction. As noted above, the steady-state levels of wild-type Sml1 increase ∼4-fold when Rnr1 is overexpressed (Figures 4A and B, and 5B). This effect is eliminated by the sml1 mutations that abolish Sml1–Rnr1 interaction (Figure 5B), suggesting that Sml1 protein is stabilized by binding to Rnr1. This conclusion is strengthened further by the fact that Sml1 protein levels are stabilized when RNR1 is overexpressed (Figure 5C).
Sml1 is phosphorylated after DNA damage and at S phase in a Mec1/Rad53-dependent manner
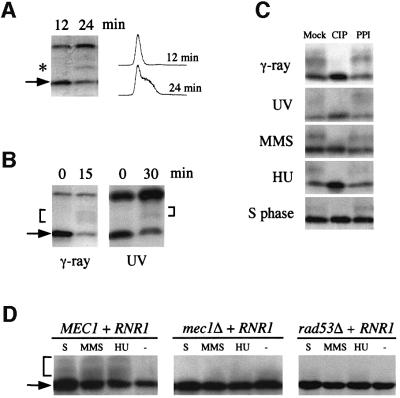
Regulation of target proteins by the Mec1/Rad53 kinase cascade usually involves protein phosphorylation (Zhou and Elledge, 1993; Sidorova and Breeden, 1997; Sanchez et al., 1999). In many cases, phosphoproteins exhibit decreased mobility during electrophoresis. Interestingly, a slower migrating Sml1 band was observed from cells entering S phase when phosphatase inhibitors were added during extraction (Figure 6A). Slower migrating bands of Sml1 were also observed after DNA damage, and these bands are more evident from strains that contain a higher amount of Sml1 due to either the presence of an RNR1 plasmid or GAL-SML1 on the chromosome (Figure 6B and data not shown). It is noteworthy that additional slower migrating Sml1 bands are observed after γ-ray and UV irradiation compared with S phase. Similar bands were also detected from cells that were treated with MMS and HU (Figure 6C). In all cases, the slower migrating band(s) of Sml1 represent phosphorylated forms, as they can be converted to the Sml1-sized band by phosphatase treatment and this conversion is blocked by the addition of a phosphatase inhibitor (Figure 6C). The phosphorylated forms of Sml1 were not observed in mec1Δ and rad53Δ strains after γ-ray and UV irradiation (Figure 4B and C) or after MMS or HU treatment and in S phase (Figure 6D), indicating that Sml1 phosphorylation is dependent on Mec1 and Rad53.
Fig. 6. Sml1 is phosphorylated in S phase and after DNA damage. (A) Wild-type strain W1588-4C was arrested in G1 phase by α-mating factor and then released into the cell cycle. Samples were taken 12 and 24 min after release. Protein extracts were made with phosphatase inhibitors included in the boiling buffer. Sml1 protein levels were examined by protein blots using anti-Sml1 antibody, and the DNA content was measured by FACS analysis. The arrow indicates the position of Sml1 protein and the star marks the position of a slower migrating Sml1 band. (B) Strain U1476 (wild-type strain containing a 2 µm-RNR1 plasmid) was irradiated by γ-rays (30 krads) or UV light (120 J/m2). Protein extracts were made from samples before (0 min) and after irradiation (times as indicated) as described in (A). Sml1 was detected by a protein blot using anti-Sml1 antibody. The arrow indicates the position of the Sml1 protein and the brackets mark the position of slower migrating Sml1 bands. (C) Cell samples from (A) and (B) or from HU- (200 mM, 1 h) and MMS- (0.05%, 1 h) treated U1476 cells were collected. Protein extracts were made using the TCA method and were incubated with calf intestinal phosphatase (CIP) at 37°C for 15 min. The phosphatase inhibitor β-glycerophosphate was added together with CIP (PPI). Mock reactions did not have either CIP or β-glycerophosphate. (D) Strains U1476 (MEC1 + RNR1), U1195 (mec1Δ + RNR1) and U1198-10C (rad53Δ + RNR1) were treated with 200 mM HU or 0.05% MMS for 1 h. Protein extracts were made using the TCA method and analyzed by protein blots using anti-Sml1 antibody. For the S phase extract (S), cells were first arrested in G1 and then released into the cell cycle. Samples were collected 25 min after release when the majority of the cells were in S phase.
Mec1 and Rad53 are required to complete DNA replication but not initiate S phase
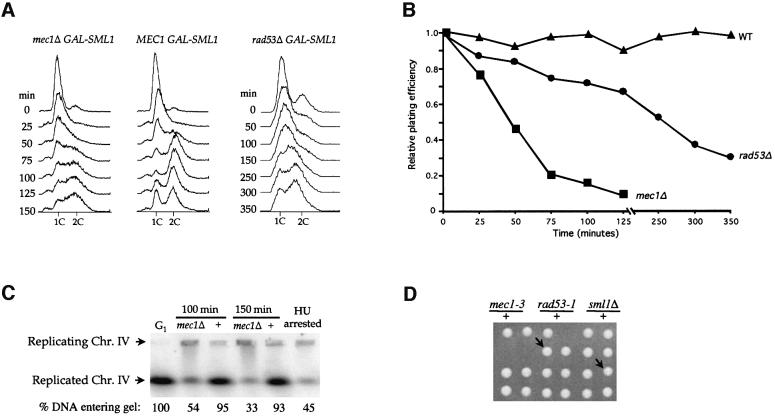
To understand the biological significance of the regulation of Sml1 by Mec1 and Rad53 at S phase, we examined the cellular consequences of removal of this regulatory circuit. We utilized a regulatable Sml1 construct (GAL-SML1 strains). GAL-SML1 mec1Δ strains grow normally in non-inducible medium but not after galactose induction. After arresting cells in G1, Sml1 expression was induced to near endogenous levels (data not shown). Next, the G1 block was removed and cells were released into the cell cycle. DNA content and cell viability were tested at various time points after the release (Figure 7A and B). Interestingly, mec1 cells enter S phase at the same time as wild-type cells. However, they do not complete DNA replication even after 150 min, while wild-type cells finish DNA replication in 50 min (Figure 7A). The prolonged S phase in mec1 cells is associated with cell death since cell viability decreases even after turning off Sml1 expression (Figure 7B). Thus, Mec1 is not required to enter S phase but is needed to finish S phase. This requirement is completely bypassed in the absence of Sml1 as mec1Δ sml1Δ strains progress through the cell cycle normally (data not shown).
Fig. 7. mec1 mutations cause incomplete DNA replication and exhibit synthetic lethality with rad53-1. (A–C) Strains W2057-2B (mec1Δ GAL-SML1), W2057-11A (MEC1 GAL-SML1) and W2079-11B (rad53 GAL-SML1) were arrested in G1 phase by α-mating factor in YPGL medium. Sml1 was induced by addition of 2% galactose for 30 min. Cells were then released into YPGal medium and samples taken at different time points. In (A), cells were fixed in ethanol and analyzed by FACS. Note that due to the slower growth rate of the rad53 GAL-SML1 strain, longer time point intervals were used. (B) Cells were washed in YPD medium, plating units were determined by microscopic examination and appropriate dilutions were spread onto YPD plates. After incubation at 30°C for 3 days, colonies were counted and the plating efficiency was calculated using the number of colonies divided by the number of plating units. Time zero is considered as 100%. In (C), DNA plugs from 100 and 150 min were prepared from W2057-2B (mec1Δ) and W2057-11A (+) and analyzed by pulsed-field gel electrophoresis. The DNA was blotted and probed with labeled DNA from the DUN1 gene to detect chromosome IV. α-mating factor- (G1) and HU-arrested (HU-arrested) samples were used as controls for chromosome separation. (D) Six tetrads are shown for diploid strain W1986 (MATa/α mec1-3/+ rad53-1/+ sml1Δ::HIS3/+). The genotypes of the three inviable spores are deduced from those of the sister spore clones and in each case is mec1-3 rad53-1. The arrows indicate two mec1-3 rad53-1 sml1Δ spore clones. Spore clones of the other genotypes grow equally well.
Similarly, rad53Δ cells also exhibit DNA replication defects after release from G1 arrest at the same time as Sml1 induction (Figure 7A and B). Notably, rad53Δ GAL-SML1 strains grow more slowly than wild-type in the absence of induction. This is probably due to a Mec1-independent role for RAD53 in cell growth (Zhao et al., 1998). The viability of rad53Δ GAL-SML1 cells decreases gradually as cells traverse S phase (Figure 7A). This lethality and the incomplete DNA replication of rad53Δ strains are suppressed by deletion of SML1 (data not shown).
DNA replication in mec1 cells was examined further by subjecting chromosomal DNA to pulsed-field gel electrophoresis. It has been shown that fully replicated chromosomal DNA can enter a pulsed-field gel while chromosomal DNA undergoing replication cannot (Hennessy et al., 1991). As shown in Figure 7C, 93% (100 min) and 95% (150 min) of chromosome IV in wild-type cells runs into the gel, while only 54 and 33% of the chromosome IV in mec1 cells at the same time points enters the gel. Similar results were obtained for chromosome V (data not shown). These results further demonstrate that Mec1 is required to complete DNA replication.
Hypomorphic mec1 and rad53 mutants are defective in mitochondrial DNA propagation and exhibit decreased dNTP levels
To assess further the importance of the regulation of Sml1 by Mec1 and Rad53, we examined another cellular process that is sensitive to dNTP levels. Like chromosomal DNA replication, mitochondrial DNA replication is also sensitive to dNTP levels. Lower RNR activity or dNTP levels increase the formation of mitochondrial DNA-deficient cells (petite cells), while higher dNTP levels result in reduced petite formation (Elledge and Davis, 1987; Huang and Elledge, 1997; Zhao et al., 1998). If MEC1/RAD53 play a positive role in dNTP synthesis, mutation of these genes will cause increased petite formation. This prediction was tested by measuring the frequency of petite formation in strains containing the hypomorphic alleles, mec1-3 and rad53-1 (Weinert et al., 1994). These strains grow normally; however, they exhibit a 2- to 3-fold higher frequency of petite formation than wild-type strains (Table I). Removal of Sml1 in these strains reduces the petite formation rate to that of sml1Δ strains, which is 3-fold lower than that of wild-type strains (Table I). Experiments with mec1-3 rad53-1 double mutants cannot be performed as the double mutant is inviable. However, the triple mutant mec1-3 rad53-1 sml1Δ not only grows as well as wild-type strains but also exhibits wild-type rates of petite formation (Figure 7D and Table I). The suppression of the mitochondrial defects as well as the synthetic lethality of mec1-3 and rad53-1 mutations by sml1Δ further support the notion that removal of Sml1 by Mec1 and Rad53 is important for normal cell growth.
Table I. Percentage petite formation in mec1 and rad53 mutants.
| MEC1 RAD53 | mec1-3 | rad53-1 | mec1-3 rad53-1 | |
|---|---|---|---|---|
| SML1 | 3.37 ± 0.10 | 6.90 ± 1.13 | 10.2 ± 1.35 | inviable |
| sml1Δ | 1.08 ± 0.34 | 1.42 ± 0.04 | 1.43 ± 0.02 | 2.96 ± 0.08 |
Two to four strains were measured for each genotype. The average and standard deviation for petite formation is shown as a percentage.
We also examined the dNTP levels in mec1-3 and rad53-1 strains. As shown in Table II, both strains exhibit a 25–56% reduction of all four types of dNTPs. This result confirms the genetic evidence and directly demonstrates that the Mec1 and Rad53 proteins play roles in the maintenance of normal dNTP levels.
Table II. The dNTP concentrations of mec1-3 and rad53-1 cells relative to that of wild-type cells.
| dCTP | dTTP | dATP | dGTP | |
|---|---|---|---|---|
| mec1-3 | 66 ± 0% | 58 ± 4% | 44 ± 1% | 48 ± 5% |
| rad53-1 | 70 ± 3% | 75 ± 10% | 58 ± 12% | 58 ± 8% |
The percentages of dNTP levels in mec1-3 or rad53-1 strains relative to that of wild-type strains are the averages from two trials. The standard deviations of the percentages are also shown.
Discussion
The essential function of Mec1 and Rad53
MEC1 and RAD53 not only play important roles in DNA damage checkpoint pathways, they are also essential for normal growth. While extensive studies have demonstrated that they act as signal transducers in the checkpoint response, their functions during cell growth have not been clear (reviewed in Elledge, 1996; Weinert, 1998). Sup pressors of mec1 and rad53 lethality, including deletion of the RNR inhibitor Sml1 or transcriptional repressor Crt1, and overexpression of the RNR genes, can increase RNR activity (Desany et al., 1998; Huang et al., 1998; Zhao et al., 1998). These observations suggest a role for Mec1/Rad53 in the up-regulation of RNR activity and, consequently, dNTP levels. However, these genetic data do not exclude alternative interpretations.
To delineate the essential function of Mec1/Rad53, it is crucial to demonstrate the molecular connections between these kinases and their target proteins during normal cell growth. Here, we show that Mec1 and Rad53 regulate the phosphorylation and the levels of the Sml1 protein at S phase. This evidence strongly supports a direct role for Mec1 and Rad53 in dNTP regulation. Further examination of the terminal phenotype of mec1Δ and rad53Δ cells using a regulatable Sml1 construct revealed that the absence of Mec1 and Rad53 does not affect the initiation of DNA replication but rather results in incomplete DNA replication. Therefore, Mec1 and Rad53 are needed to remove Sml1 so that sufficient dNTPs are available to complete DNA replication. Consistent with this notion, Merrill and Holm (1999) reported that certain alleles of mec1 (mec1-srf) accumulate short DNA replication intermediates and that such impaired DNA replication is suppressed by sml1Δ (Merrill and Holm, 1999).
The regulation of Sml1 by Mec1/Rad53 may complement the transcriptional induction of the RNR genes via the Mbp1/Swi6 pathway at S phase (Koch et al., 1993; Figure 8). The existence of this dual-layered control of RNR activity underscores the importance of dNTP regulation. In fact, dNTPs are required in virtually every chromosomal and mitochondrial DNA metabolic process, including replication, recombination and repair. There fore, a defect in dNTP regulation due to mec1 and rad53 mutations could lead to a wide spectrum of deficiencies during normal growth. Here we show that although partial defects in Mec1 and Rad53 function (e.g. mec1-3 and rad53-1 mutations) do not affect cell growth on their own, each mutation results in poor mitochondrial DNA propagation (Table I). In addition, cells are inviable in the presence of both mutations (Figure 7D). The fact that all of these defects are suppressed by sml1Δ suggests that they are caused by lower dNTP levels. This conclusion is supported further by the finding that mec1-3 and rad53-1 cells exhibit a 25–56% decrease in dNTP levels. Other defects seen in mec1 mutants include shorter telomeres and loss of telomere position effect, and again these defects are suppressed by removal of Sml1 (Ritchie et al., 1999; Craven and Petes, 2000).
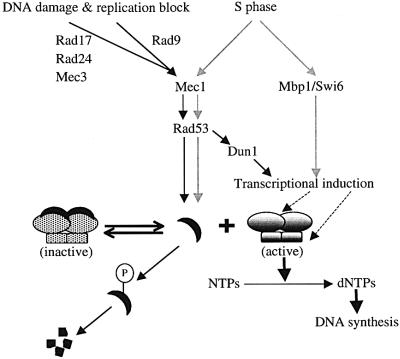
Fig. 8. A model for the regulation of RNR activity by the Mec1/Rad53 kinase cascade. As cells enter S phase (gray lines) or after they are challenged by DNA-damaging agents or by replication blocks (black lines), the Mec1/Rad53 kinase cascade leads to phosphorylation of the unbound form of Sml1. Note that the phosphorylation depicted here may be direct or indirect. Subsequently, phosphorylated Sml1 is targeted for protein degradation. This degradation drives the equilibrium from the inactive Sml1–RNR complex to the active form of RNR. The transcripts of the RNR genes are also induced at S phase by the Mbp1–Swi6 complex (gray lines) and by the Mec1/Rad53/Dun1 kinase cascade after DNA damage (black lines). Transcriptional up-regulation (dashed lines) in both situations probably increases the amount of the RNR enzyme.
Although they share similar functions during cell growth, Mec1 and Rad53 appear to differ in Sml1 regulation. The fact that the removal of Sml1 at S phase is only partially defective in rad53Δ strains but is completely defective in mec1Δ strains suggests that an additional pathway, which is probably parallel to the RAD53 pathway, may lie downstream of MEC1. Further more, the difference between the cell growth function of Mec1 and Rad53 is also indicated by the fact that sml1Δ rad53Δ double mutants grow more slowly than mec1Δ sml1Δ. It is likely that RAD53 has a Mec1-independent function(s) during the normal cell cycle and that the lack of both functions in rad53Δ strains leads to cell death, and suppression of dNTP regulation can only partially rescue cell growth. This Mec1-independent function of Rad53 may, in fact, be the control of late DNA replication origin firing as suggested by Shirahige et al (1998) and Santocanale and Diffley (1998).
The removal of Sml1 is a new facet of the checkpoint response
dNTP levels are also crucial for successful DNA repair. Indeed, dNTP levels rapidly increase in yeast cells in response to γ-irradiation (Eckstein et al., 1974). One mechanism used to achieve this outcome is the transcriptional induction of the RNR genes via the Mec1, Rad53 and Dun1 checkpoint pathways (Zhou and Elledge, 1993; Sanchez et al., 1996; Sun et al., 1996) (Figure 8). We show here that Sml1 protein levels diminish rapidly after DNA damage in a checkpoint-dependent manner. For example, 97% of Sml1 protein disappears 10 min after irradiation. We suggest that this rapid response is another important feature of the DNA repair process. This notion is supported by the fact that sml1Δ strains are more resistant to DNA damage than wild-type strains (Zhao et al., 1998). Additionally, sml1Δ rescues the DNA damage sensitivity of dun1Δ strains in the absence of increased RNR transcription (Zhao et al., 1998). However, the fact that deletion of SML1 does not suppress the DNA damage sensitivity of mec1 and rad53 strains demonstrates that additional roles of Mec1 and Rad53 in the checkpoint response (e.g. cell cycle arrest) are critical for cell survival after damage.
The up-regulation of RNR activity during S phase and after DNA damage shares similar features: transcriptional induction of the RNR genes and a precipitous decrease in Sml1 levels. However, during S phase, Mec1 and Rad53 only regulate Sml1 levels, while after DNA damage they control both features (Figure 8). In addition, the regulation of Sml1 after DNA damage requires other checkpoint genes including RAD9, RAD17, RAD24 and MEC3 (Figure 8). Furthermore, our studies with double mutants support a previous notion that RAD9 and the RAD17 group (Rad17, Rad24 and Mec3) function in a bifurcated pathway (de la Torre-Ruiz et al., 1998) (Figures 4D and 8).
Using mutated Sml1 proteins that do not bind to Rnr1, we further demonstrated that the Rnr1-bound and -unbound forms of Sml1 differ in their stability. First, when Rnr1 is overexpressed, wild-type but not mutated, unbound Sml1 proteins levels increase (Figure 5B). These results suggest that Sml1 protein is stabilized upon binding to Rnr1. This conclusion is supported by the longer half-life of Sml1 proteins when Rnr1 is overexpressed (Figure 5C). Secondly, after DNA damage, mutated Sml1 proteins diminish with wild-type kinetics, suggesting that the majority of Sml1 protein turnover is of the unbound form. Thus, in wild-type cells, the rapid turnover of the unbound form of Sml1 may actually drive the equilibrium from the Sml1–RNR complex to the free form of RNR, thereby increasing RNR activity (Figure 8), especially since the KD for Sml1–Rnr1 binding is only 0.4 µM (Chabes et al., 1999). However, this does not preclude the possibility that the Rnr1-bound form of Sml1 may also be subjected to other forms of regulation.
Finally, we show that the drop in Sml1 levels controlled by the Mec1/Rad53 pathway during growth and after DNA damage is the result of post-transcriptional regulation and occurs concomitantly with the phosphorylation of Sml1. Mec1 and Rad53 are upstream protein kinases in a cascade(s) that targets multiple substrates involved in cell cycle arrest and DNA repair (reviewed in Weinert, 1998). Therefore, Sml1 is a new substrate of this kinase cascade(s) and its phosphorylation probably leads to its rapid turnover. It will be of interest to identify additional proteins involved in this regulatory circuit and to determine whether other targets of this cascade are regulated similarly.
Materials and methods
Strains, plasmids and media
The S.cerevisiae strains used in this study are listed in Table III. To construct the GAL-SML1 strain, the SML1 open reading frame was PCR-amplified and cloned behind the GAL1 promoter on vector pYX423. Next, the GAL-SML1 region of this construct was PCR-amplified and used to replace the chromosomal copy of SML1 by the cloning-free PCR-based allele replacement method (Erdeniz et al., 1997). The sequence of the GAL-SML1 region in the resulting strain was confirmed by DNA sequence analysis. The construction of sml1-S87P and sml1-I76T strains has been described (Zhao et al., 2000b). Plasmid pWJ841 was constructed by inserting a SacI fragment of the RNR1 gene into the SacI site of pRS425 (Christianson et al., 1992). Yeast media were prepared essentially as described by Adams et al. (1997). YPGL medium contains 3% glycerol, 3% lactic acid, 2% peptone and 1% yeast extract.
Table III. Yeast strains used in this study.
| Strains | Genotype | Reference/source |
|---|---|---|
| W1588-4C | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | Zhao et al. (1998) |
| W1588-4A | MATα ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | Zhao et al. (1998) |
| W1379-3C | MATa bar1Δ::LEU2 | Rothstein lab collection |
| W2057-11A | MATa bar1Δ::LEU2 GAL-SML1 | this study |
| W2079-11B | MATa bar1Δ::LEU2 GAL-SML1 rad53Δ::HIS3 | this study |
| U1195 | MATa mec1Δ::TRP1 {pWJ841} | this study |
| U1198-10C | MATa rad53Δ::HIS3 {pWJ841} | this study |
| U1475 | MATa {pRS425} | this study |
| U1476 | MATa {pWJ841} | this study |
| W2057-2B | MATa bar1Δ::LEU2 mec1Δ::TRP1 GAL-SML1 | this study |
| W1518-10B | MATα rad9Δ::HIS3 | this study |
| W1522-11B | MATα rad17Δ::LEU2 | this study |
| W1519-17B | MATα rad24Δ::LEU2 | this study |
| W1520-10B | MATα mec3Δ::URA3 | this study |
| W2097-63B | MATa sml1-S87P | Zhao et al. (2000b) |
| W2099-63D | MATa sml1-I76T | Zhao et al. (2000b) |
| W1986 | MATa/α mec1-3/+ rad53-1/+ sml1Δ::HIS3/+ | this study |
| W1729-2A | MATα HIS3 lys2Δ rad53-1 | this study |
| W1745-11C | MATa HIS3 lys2Δ mec1-3 | this study |
| W2617-4A | MATα rad9Δ::HIS3 mec3Δ::URA3 | this study |
All strains are isogenic or congenic (>6 backcrosses) to W303 (Thomas and Rothstein, 1989). The W303 derivatives W1588-4C and W1588-4A are RAD5 and their genotypes are shown. For other strains, only alleles that differ from the W1588 strains are listed.
Synchronization and genotoxin treatment
Cultures were always grown to early- or mid-log phase before arrest and application of DNA-damaging agents. Cells were arrested in G1 phase by α-mating factor (3.4 µg/ml; from Sigma) in YPD, SC-LEU or YPGL for one doubling time (Figures 1, 2 and 3). To measure the half-life of Sml1 (Figure 1E), strain W2057-11A was first grown in YPRaffinose medium and next arrested at G2/M phase by nocodazole (5 µg/ml). Cells were then transferred to YPGal medium containing 5 µg/ml nocodazole to induce Sml1 expression and maintain the G2/M arrest. After 45 min, the expression of Sml1 was turned off by addition of 2% glucose.
HU was added to YPD or SC-LEU medium to a final concentration of 200 mM from a 2 M stock solution. MMS was added to these media at a final concentration of 0.03% (Figure 6) or 0.05% (Figures 4, 5 and 7). Before UV irradiation, cells were washed once with 0.9% NaCl and resuspended in this solution. Next, a thin suspension of cells was spread on empty Petri dishes and irradiated with UV light. After UV treatment, cells were collected and transferred to liquid YPD or SC-LEU media and grown for 20–40 min before harvesting. Cell cultures were irradiated with doses corresponding to 10, 30 or 80 krads of γ-ray using a Gammacell-220 60Co irradiator (Atomic Energy of Canada).
Protein extraction, immunoblot analysis and phosphatase treatment
Two protein extraction methods were used. To detect steady-state levels of Sml1 protein, 1.5 ml of a mid-log phase culture was spun down and washed once with phosphate-buffered saline (PBS). 1× SDS– polyacrylamide gel loading buffer [also called boiling buffer, 62.5 mM Tris pH 6.8, 2% SDS, 10% glycerol, 5% β-mercaptoethanol, 0.025% bromophenol blue] was added to the cell pellet and the samples were boiled for 5 min before loading onto 15% SDS–polyacrylamide gels. In Figure 6A and B, mixtures of phosphatase inhibitors (10 mM NaF, 2 mM Na4P2O7, 5 mM EDTA, 0.5 mM Na3VO4) were included in the SDS–polyacrylamide gel loading buffer. For better detection of phospho-Sml1 and for phosphatase treatment, the protein extracts were made employing the trichloroacetic acid (TCA) method described by Sambrook et al. (1989), except that NP-40 buffer (150 mM NaCl, 1% NP-40, 50 mM Tris pH 8.0) containing 1× proteinase inhibitor cocktail (Boehringer Mannheim) was used instead of RIPA buffer. Protein extracts were incubated with 2 U of calf intestinal phosphatase (CIP; Boehringer Mannheim) at 37°C for 15 min. β-glycerophosphate was added at a concentration of 60 mM. Proteins were transferred to polyvinylidene difluoride (PVDF) membranes and the blots were probed with anti-Sml1 antibody. Sml1 bands were detected by ECL (+) or ECF (Amersham).
Measurement of dNTP levels
Yeast cultures were grown exponentially at 30°C in YPD medium for >8 generations. At a density of 1.5 × 107 cells/ml, ∼1 × 109 cells were harvested by rapid filtration through 25 mm White AAWP nitrocellulose filters (0.8 µm, Millipore). The filters were immersed immediately in 700 µl of ice-cold extract solution [12% (w/v) TCA, 15 mM MgCl2]. One nmol of dITP (deoxyinosine triphosphate) was added to monitor sample loss during the experiments. All the following steps were carried out at 4°C. The Eppendorf tubes were vortexed for 30 s, incubated for 15 min and vortexed again for 30 s. A hole was then punctured in the bottom of the tube and it was inserted into a new 1.5 ml tube. The extracts and yeast debris were then collected into a new Eppendorf tube by a low speed centrifugation. The supernatants (700 µl) were collected after centrifugation at 20 000 g for 1 min and added to 800 µl of ice-cold Freon–trioctylamine mixture [10 ml of Freon (Merck, for IR spectroscopy) and 2.8 ml of trioctylamine (Fluka, >99%)]. The samples were vortexed for 20 s and centrifuged for 1 min at 20 000 g. The aqueous phase was added to 700 µl of Freon–trioctylamine mixture and centrifuged again. A 475 µl aliquot of the aqueous phase was used to determine the dNTP pools after the addition of 25 µl of 1 M NH4HCO3 pH 8.9. A 47.5 µl aliquot of the aqueous phase was evaporated to dryness in a Speedvac (Savant), dissolved in 500 µl of water and used for the quantification of NTP pools. The values obtained for NTP pools were used as additional internal controls to monitor material loss and extraction efficiency. Separation and quantitation of dNTPs and NTPs employing HPLC were carried out as described in Hofer et al. (1998).
Other techniques
Yeast chromosomal DNA agarose blocks were prepared using the protocol of Gerring et al. (1991). Pulsed-field gel electrophoresis was performed on a CHEF DRII apparatus according to the manufacture’s instructions (Biorad). Samples for fluorescence-activated cell sorting (FACS) analysis were prepared according to Paulovich et al. (1998). The frequency of petite formation was measured as described in Zhao et al. (1998).
Acknowledgments
Acknowledgements
We thank Bilyana Georgieva, Teresa Lamb and Marisa Wagner for comments on the manuscript, and Ted Weinert for strains and discussions. This work was supported by National Institutes of Health grant GM50237 (R.R.), by the Swedish Natural Sciences Research Council (L.T.) and the Alexander and Margaret Stewart Trust Pilot Project in Cancer Research (R.R.)
References
- Adams A., Gottschling,D.E., Kaiser,C.A. and Stearns,T. (1997) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Brown E.J. and Baltimore,D. (2000) ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev., 14, 397–402. [PMC free article] [PubMed] [Google Scholar]
- Caspari T. (2000) How to activate p53. Curr. Biol., 10, R315–R317. [DOI] [PubMed] [Google Scholar]
- Chabes A., Domkin,V. and Thelander,L. (1999) Yeast Sml1, a protein inhibitor of ribonucleotide reductase. J. Biol. Chem., 274, 36679–36683. [DOI] [PubMed] [Google Scholar]
- Christianson T.W., Sikorski,R.S., Dante,M., Shero,J.H. and Hieter,P. (1992) Multifunctional yeast high-copy-number shuttle vectors. Gene, 110, 119–122. [DOI] [PubMed] [Google Scholar]
- Craven R.J. and Petes,T.D. (2000) Involvement of the checkpoint protein Mec1p in silencing of gene expression at telomeres in Saccharomyces cerevisiae. Mol. Cell. Biol., 20, 2378–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Torre-Ruiz M.A., Green,C.M. and Lowndes,N.F. (1998) RAD9 and RAD24 define two additive, interacting branches of the DNA damage checkpoint pathway in budding yeast normally required for Rad53 modification and activation. EMBO J., 17, 2687–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desany B.A., Alcasabas,A.A., Bachant,J.B. and Elledge,S.J. (1998) Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev., 12, 2956–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckstein H., Ahnefeld,S. and Albietz-Loges,K. (1974) Synthesis of deoxynucleoside tri- and monophosphates in synchronized and asynchronously growing cells. Z. Naturforsch., 29c, 272–282. [DOI] [PubMed] [Google Scholar]
- Elledge S.J. (1996) Cell cycle checkpoints: preventing an identity crisis. Science, 274, 1664–1672. [DOI] [PubMed] [Google Scholar]
- Elledge S.J. and Davis,R.W. (1987) Identification and isolation of the gene encoding the small subunit of ribonucleotide reductase from Saccharomyces cerevisiae: DNA damage-inducible gene required for mitotic viability. Mol. Cell. Biol., 7, 2783–2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge S.J. and Davis,R.W. (1989) DNA damage induction of ribonucleotide reductase. Mol. Cell. Biol., 9, 4932–4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdeniz N., Mortensen,U.H. and Rothstein,R. (1997) Cloning-free PCR-based allele replacement methods. Genome Res., 7, 1174–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerring S.L., Connelly,C. and Hieter,P. (1991) Positional mapping of genes by chromosome blotting and chromosome fragmentation. In Guthrie,C. and Fink,G.R. (eds), Guide to Yeast Genetics and Molecular Biology. Academic Press Inc., San Diego, CA, pp. 57–77. [DOI] [PubMed]
- Hennessy K.M., Lee,A., Chen,E. and Botstein,D. (1991) A group of interacting yeast DNA replication genes. Genes Dev., 5, 958–969. [DOI] [PubMed] [Google Scholar]
- Hofer A., Ekanem,J.T. and Thelander,L. (1998) Allosteric regulation of Trypanosoma brucei ribonucleotide reductase studied in vitro and in vivo. J. Biol. Chem., 273, 34098–34104. [DOI] [PubMed] [Google Scholar]
- Huang M. and Elledge,S.J. (1997) Identification of RNR4, encoding a second essential small subunit of ribonucleotide reductase in Saccharomyces cerevisiae. Mol. Cell. Biol., 17, 6105–6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M., Zhou,Z. and Elledge,S.J. (1998) The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell, 94, 595–605. [DOI] [PubMed] [Google Scholar]
- Kato R. and Ogawa,H. (1994) An essential gene, ESR1, is required for mitotic cell growth, DNA repair and meiotic recombination in Saccharomyces cerevisiae. Nucleic Acids Res., 22, 3104–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch C., Moll,T., Neuberg,M., Ahorn,H. and Nasmyth,K. (1993) A role for the transcription factors Mbp1 and Swi4 in progression from G1 to S phase. Science, 261, 1551–1557. [DOI] [PubMed] [Google Scholar]
- Merrill B.J. and Holm,C. (1999) A requirement for recombinational repair in Saccharomyces cerevisiae is caused by DNA replication defects of mec1 mutants. Genetics, 153, 595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulovich A.G., Armour,C.D. and Hartwell,L.H. (1998) The Saccharo myces cerevisiae RAD9, RAD17, RAD24 and MEC3 genes are required for tolerating irreparable, ultraviolet-induced DNA damage. Genetics, 150, 75–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichard P. (1988) Interactions between deoxyribonucleotide and DNA synthesis. Annu. Rev. Biochem., 57, 379–374. [DOI] [PubMed] [Google Scholar]
- Ritchie K.B., Mallory,J.C. and Petes,T.D. (1999) Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1 and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 6065–6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotman G. and Shiloh,Y. (1999) ATM: a mediator of multiple responses to genotoxic stress. Oncogene, 18, 6135–6144. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sanchez Y., Desany,B.A., Jones,W.J., Liu,Q., Wang,B. and Elledge,S.J. (1996) Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science, 271, 357–360. [DOI] [PubMed] [Google Scholar]
- Sanchez Y., Bachant,J., Wang,H., Hu,F., Liu,D., Tetzlaff,M. and Elledge,S.J. (1999) Control of the DNA damage checkpoint by Chk1 and Rad53 protein kinases through distinct mechanisms. Science, 286, 1166–1171. [DOI] [PubMed] [Google Scholar]
- Santocanale C. and Diffley,J.F. (1998) A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature, 395, 615–618. [DOI] [PubMed] [Google Scholar]
- Shirahige K., Hori,Y., Shiraishi,K., Yamashita,M., Takahashi,K., Obuse,C., Tsurimoto,T. and Yoshikawa,H. (1998) Regulation of DNA-replication origins during cell-cycle progression. Nature, 395, 618–621. [DOI] [PubMed] [Google Scholar]
- Sidorova J.M. and Breeden,L.L. (1997) Rad53-dependent phosphoryl ation of Swi6 and down-regulation of CLN1 and CLN2 transcription occur in response to DNA damage in Saccharomyces cerevisiae. Genes Dev., 11, 3032–3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z., Fay,D.S., Marini,F., Foiani,M. and Stern,D.F. (1996) Spk1/Rad53 is regulated by Mec1-dependent protein phosphorylation in DNA replication and damage checkpoint pathways. Genes Dev., 10, 395–406. [DOI] [PubMed] [Google Scholar]
- Tanaka H., Arakawa,H., Yamaguchi,T., Shiraishi,K., Fukuda,S., Matsui,K., Takei,Y. and Nakamura,Y. (2000) A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature, 404, 42–49. [DOI] [PubMed] [Google Scholar]
- Thomas B.J. and Rothstein,R. (1989) Elevated recombination rates in transcriptionally active DNA. Cell, 56, 619–630. [DOI] [PubMed] [Google Scholar]
- Vallen E.A. and Cross,F.R. (1999) Interaction between the MEC1-dependent DNA synthesis checkpoint and G1 cyclin function in Saccharomyces cerevisiae. Genetics, 151, 459–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert T. (1998) DNA damage and checkpoint pathways: molecular anatomy and interactions with repair. Cell, 94, 555–558. [DOI] [PubMed] [Google Scholar]
- Weinert T.A., Kiser,G.L. and Hartwell,L.H. (1994) Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev., 8, 652–665. [DOI] [PubMed] [Google Scholar]
- Xu Y., Ashley,T., Brainerd,E.E., Bronson,R.T., Meyn,M.S. and Baltimore,D. (1996) Targeted disruption of ATM leads to growth retardation, chromosomal fragmentation during meiosis, immune defects and thymic lymphoma. Genes Dev., 10, 2411–2422. [DOI] [PubMed] [Google Scholar]
- Zhao X., Muller,E.G. and Rothstein,R. (1998) A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell, 2, 329–340. [DOI] [PubMed] [Google Scholar]
- Zhao R., Gish,K., Murphy,M., Yin,Y., Notterman,D., Hoffman,W.H., Tom,E., Mack,D.H. and Levine,A.J. (2000a) Analysis of p53-regulated gene expression patterns using oligonucleotide arrays. Genes Dev., 14, 981–993. [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Georgieva,B., Chabes,A., Domkin,V., Ippel,J.H., Schleucher,J., Wijmenga,S., Thelander,L. and Rothstein,R. (2000b) Mutational and structural analyses of the ribonucleotide reductase inhibitor Sml1 define its Rnr1 interaction domain whose inactivation allows suppression of mec1 and rad53 lethality. Mol. Cell. Biol., 20, 9076–9083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng P., Fay,D.S., Burton,J., Xiao,H., Pinkham,J.L. and Stern,D.F. (1993) SPK1 is an essential S-phase-specific gene of Saccharomyces cerevisiae that encodes a nuclear serine/threonine/tyrosine kinase. Mol. Cell. Biol., 13, 5829–5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z. and Elledge,S.J. (1993) DUN1 encodes a protein kinase that controls the DNA damage response in yeast. Cell, 75, 1119–1127. [DOI] [PubMed] [Google Scholar]