Abstract
Yeast Spt16/Cdc68 and Pob3 form a heterodimer that acts in both DNA replication and transcription. This is supported by studies of new alleles of SPT16 described here. We show that Spt16–Pob3 enhances HO transcription through a mechanism that is affected by chromatin modification, since some of the defects caused by mutations can be suppressed by deleting the histone deacetylase Rpd3. While otherwise conserved among many eukaryotes, Pob3 lacks the HMG1 DNA-binding motif found in similar proteins such as the SSRP1 subunit of human FACT. SPT16 and POB3 display strong genetic interactions with NHP6A/B, which encodes an HMG1 motif, suggesting that these gene products function coordinately in vivo. While Spt16–Pob3 and Nhp6 do not appear to form stable heterotrimers, Nhp6 binds to nucleosomes and these Nhp6–nucleosomes can recruit Spt16–Pob3 to form SPN–nucleosomes. These complexes have altered electrophoretic mobility and a distinct pattern of enhanced sensitivity to DNase I. These results suggest that Spt16–Pob3 and Nhp6 cooperate to function as a novel nucleosome reorganizing factor.
Keywords: cdc68/Nhp6/nucleosome/Pob3/Spt16
Introduction
SPT16/CDC68 was identified in several genetic screens in Saccharomyces cerevisiae as a factor that affects transcription initiation globally (Prendergast et al., 1990; Malone et al., 1991; Rowley et al., 1991; Lycan et al., 1994), possibly by altering some property of chromatin (Schnell et al., 1989; Winston and Carlson, 1992; Xu et al., 1993; Wittmeyer et al., 1999). Pob3 forms a stable heterodimer with Spt16, and both proteins are localized to the nucleus (Xu et al., 1995; Wittmeyer and Formosa, 1997; Brewster et al., 1998) and partially associated with chromatin (Wittmeyer et al., 1999). pob3 mutations also cause defects in transcription, and display severe synthetic defects with spt16 mutations (Costa and Arndt, 2000; Schlesinger and Formosa, 2000). Along with a role in transcription, Spt16–Pob3 also appears to act in DNA replication since it binds to DNA polymerase α (Miles and Formosa, 1992; Wittmeyer and Formosa, 1997; Wittmeyer et al., 1999). Additionally, pob3 mutants are sensitive to the dNTP synthesis inhibitor HU, they are dependent on the Mec1 S phase checkpoint and spt16 or pob3 mutations interact genetically with DNA replication factors such as Pol1, Ctf4, Dna2 and Ctf18 (Wittmeyer and Formosa, 1997; Formosa and Nittis, 1999; Wittmeyer et al., 1999; Schlesinger and Formosa, 2000). Taken together, these results indicate that Spt16–Pob3 promotes both replication and transcription, perhaps by altering chromatin, the template for both processes.
Both SPT16 and POB3 are essential for viability in yeast, and are highly conserved among eukaryotes (Wittmeyer and Formosa, 1997; Evans et al., 1998). The formation of heterodimers is also conserved, since such complexes have been purified from both human (FACT; Orphanides et al., 1999) and frog (DUF1; Okuhara et al., 1999) cells. FACT allows RNA polymerase II to elongate past template sites incorporated into nucleosomes (Orphanides et al., 1998), suggesting a role in transcription elongation on chromatin. However, the Spt– phenotype, which arises from altered start site selection, suggests that Spt16 and Pob3 affect transcription initiation (Winston and Sudarsanam, 1998). Spt16–Pob3 associates with the histone acetyltransferase complex NuA3 (John et al., 2000) and human FACT interacts with the transcription initiation factor TFIIE (Kang et al., 2000). Spt16–Pob3/FACT therefore appears to have a complex role in transcription, which includes both initiation and elongation functions. In addition, depletion of DUF1 from frog oocyte extracts blocked DNA synthesis (Okuhara et al., 1999), indicating that the role in replication observed in yeast is also conserved. The processes affected by Spt16–Pob3/FACT/DUF1 all involve chromatin, and these diverse observations could all be explained by a single activity modulating the properties of nucleosomes.
Pob3 is a member of a conserved family, but it and the two other currently known proteins from this family that are encoded by yeasts lack a DNA-binding motif found in other homologs. This motif was first noted in the high mobility group (HMG)1/2 chromatin proteins, and confers DNA binding, unwinding and bending properties (Bustin, 1999) on proteins such as SSRP1 and DUF87 (the Pob3 homologs in FACT and DUF1). Since this feature is broadly conserved in this family outside of yeasts and seems likely to provide an important activity for a chromatin-associated factor, we considered the possibility that some other protein with an HMG1 motif acts with Spt16–Pob3. S.cerevisiae has several candidates, including Nhp6A, Nhp6B, Hmo1, Hmo2, Abf2 and Ixr1. Abf2 is localized to mitochondria, and null mutants cause phenotypes consistent with a role limited to these organelles (Diffley and Stillman, 1991). Ixr1 assists the repression of COX5b transcription under aerobic conditions and also enhances the damage caused by cis-platin (Brown et al., 1993; Lambert et al., 1994), phenotypes that do not match the profile expected for an Spt16–Pob3 partner. Our initial characterization of genetic interactions between Spt16–Pob3 and Hmo1 or Hmo2 (Lu et al., 1996) indicated only minor effects (T.Formosa, unpublished data). However, we report here that Nhp6A/B (Kolodrubetz and Burgum, 1990) displays strong genetic interactions with Spt16–Pob3, suggesting that Nhp6 could provide HMG1-motif function for Spt16–Pob3. Results consistent with this interpretation have also recently been reported by Brewster et al. (2001).
Nhp6A and Nhp6B are 92- and 98-residue proteins representing single HMG1 motifs. They are 88% identical and are functionally redundant since only deletion of both genes causes significant phenotypes, which include aberrant transcription, slow growth and temperature sensitivity (Costigan et al., 1994; Paull et al., 1996; Sidorova and Breeden, 1999; Yu et al., 2000). We find that purified Nhp6 binds nucleosomes, and this structure can then bind Spt16–Pob3. The complex of Spt16–Pob3 and Nhp6 (SPN) with nucleosomes causes changes in the electrophoretic mobility and nuclease sensitivity of the nucleosomes. We therefore propose that SPN is a novel nucleosome modifying factor.
Results
A screen for new spt16 alleles
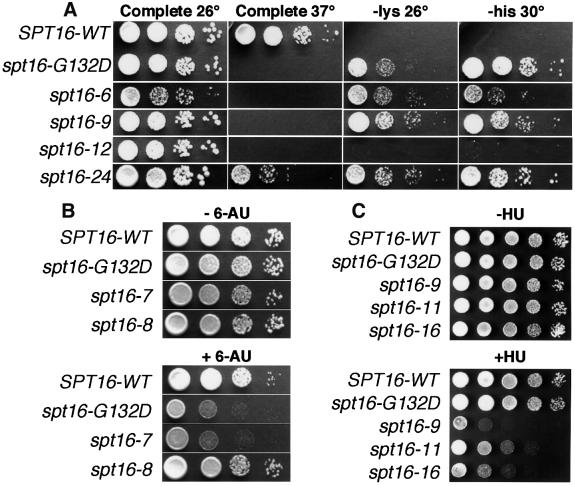
spt16 mutations were isolated previously in three independent screens (Malone et al., 1991; Rowley et al., 1991; Lycan et al., 1994). Remarkably, all three obtained the same mutation, a G132D substitution (Evans et al., 1998). To examine a broader range of alleles, we isolated additional spt16 mutants. The G132D mutation was found in four of 18 temperature-sensitive isolates (Figure 1A and Table I). While many of the remaining alleles were found to have multiple mutations, tight Ts– alleles with single substitutions were found (e.g. spt16-6).
Fig. 1. Alleles of SPT16 cause variable effects. (A) Strain 7784-1-1 (his4-912δ lys2-128δ spt16-Δ) carrying pTF128 or its derivatives with the alleles shown were grown and aliquots of 10-fold dilutions were placed on complete synthetic medium or medium lacking lysine or histidine and incubated at the temperature indicated. (B) Strains 4053-5-2 URA+ (SPT16-WT) and the isogenic 7782 URA+ set with the spt16 alleles indicated were diluted as in (A) and incubated at 30°C on synthetic medium lacking uracil and either without (– 6-AU) or with (+ 6-AU) 75 µg/ml 6-AU. (C) As in (A), except that cells were incubated at 26°C on rich medium without (–HU) or with (+HU) 90 mM HU.
Table I. Amino acid changes in spt16 alleles.
| Allele | Amino acid changes | Phenotypes | Relative HO expression (% WT) | |||||
|---|---|---|---|---|---|---|---|---|
| 37°C | Spt | 6-AU | HU | A364a (genomic) | A364a (plasmid) | W303 (genomic) | ||
| SPT16-WT | ++ | ++ | ++ | ++ | 100 | 100 | 100 | |
| spt16-1, –2, –10 | G132D | – – | – – | – | ++ | 100 | 96 | 36 |
| spt16-4 | P565S P570L | – – | – – | – | ++ | 100 | ||
| spt16-6 | P920L | – – | – | ++ | ||||
| spt16-7 | T848I T849I D850Y | – – | – – | – | ++ | 86 | ||
| spt16-8 | G369D R373T | – – | – – | ++ | ++ | 85 | ||
| spt16-9 | G132D G836S P838S | – – | – – | – – | 40 | |||
| spt16-9a | G836S P838S | ++ | – – | |||||
| spt16-9b | G836S | ++ | ++ | |||||
| spt16-9c | P838S | ++ | ++ | |||||
| spt16-11 | T828I P859S | – – | – – | +/– | – | 114 | ||
| spt16-12 | A417T G568S R569K P599L | – – | ++ | ++/– | ++ | 107 | ||
| spt16-16 | R204W A273V C290V D318N R801Q A802T | – – | – – | – –/+ | 50 | |||
| spt16-16a | R204W A273V C290V D318N | – – | – – | – –/+ | ||||
| spt16-16b | R801Q A802T | ++ | ++ | ++ | ||||
| spt16-24 | T434I | – | – – | ++ | ++ | 63 | ||
Sequencing of the SPT16 ORF from mutants revealed the predicted amino acid changes shown. Phenotypes were tested either in strains lacking the genomic SPT16 and containing a mutant allele on a plasmid (for growth at 37°C, the Spt– phenotype and sensitivity to HU) or in strains in which the spt16 allele was integrated into the genome (for testing sensitivity to 6-AU). Effects were scored from ++ (WT) to – (mutant). HO mRNA was measured as in Figure 2.
We tested the new alleles to see whether different mutations impact different processes. spt16-G132D causes the Spt– phenotype, which is suppression of the lysine and histidine auxotrophies caused by insertion of Ty1 δ elements into LYS2 and HIS4 (Malone et al., 1991). This phenotype is associated with relaxation of the specificity of transcription start site selection (Winston and Sudarsanam, 1998). Most of the spt16 alleles cause a strong Spt– phenotype, as indicated by growth on media lacking lysine or histidine (Figure 1A and Table I). Since most mutants identified as Ts– are also Spt–, we conclude that full Spt16 activity is needed for normal transcription initiation, as was the case with Pob3 (Schlesinger and Formosa, 2000).
spt16-G132D also causes sensitivity to 6-azauracil (6-AU; Figure 1B and Orphanides et al., 1999), which alters rNTP pools in yeast (Exinger and Lacroute, 1992), and is therefore considered to reveal defects in transcription elongation (Powell and Reines, 1996). The sensitivity of spt16 alleles to 6-AU varied considerably, suggesting that different mutations affect elongation to different extents (Figure 1B and Table I; compare spt16-7 with spt16-8). Since none of the alleles of pob3 tested (pob3-L78R, -2 and -7) displayed any 6-AU sensitivity, this phenotype might reveal a unique function of Spt16.
Defects in POB3 lead to hydroxyurea (HU) sensitivity (Schlesinger and Formosa, 2000). HU inhibits ribonucleotide reductase, which is required for dNTP production (Kornberg and Baker, 1992), so this phenotype suggests a role in a process that requires dNTPs, presumably DNA synthesis. Some spt16 alleles also cause sensitivity to HU (Figure 1C and Table I; spt16-9a shows that the HUs phenotype is separable from the Ts–), suggesting that SPT16 also functions in DNA synthesis. However, while pob3 mutations were found to cause dependence on the S-phase checkpoint promoted by MEC1 (Schlesinger and Formosa, 2000), spt16 mutations did not have this effect. Strains with various spt16 alleles (including those causing HU sensitivity) died at 37°C at the same rates whether or not the MEC1 checkpoint was functional (our unpublished data). The nature or consequences of the defects caused by spt16 and pob3 mutations therefore appear to be different, at least with respect to this checkpoint.
We showed previously that spt16-G132D displays a strong synthetic defect (a decrease in the maximal temperature permissive for growth) with all pob3 alleles tested, and that spt16-4 is lethal when combined with most pob3 mutations (Schlesinger and Formosa, 2000). Table II extends these results to show that pob3-L78R is lethal when combined with all spt16 alleles tested with the notable exception of the original spt16-G132D, underscoring the importance of examining additional spt16 alleles. We also showed previously that spt16-G132D displays synthetic defects with mutations in the nuclease– helicase encoded by DNA2, the catalytic subunit of DNA polymerase α encoded by POL1 and the Pol1-binding protein encoded by CTF4 (Wittmeyer and Formosa, 1997; Formosa and Nittis, 1999; Wittmeyer et al., 1999). Table II extends these results to include other alleles of spt16. The previous results linking SPT16 to POL1, CTF4 and DNA2 were supported by this survey, with the interactions with dna2-2 being particularly extensive, including complete lethality with spt16-16. dna2-2 alters a conserved helicase motif and causes both sensitivity to DNA damage and synthetic lethality with a CTF4 mutation (Formosa and Nittis, 1999). The data also demonstrate synthetic defects with a deletion of CTF18, whose similarity to RFC subunits (Cullmann et al., 1995; Kouprina et al., 1994) implicates some process involving the DNA polymerase processivity clamp protein PCNA. These interactions provide further evidence for a complex of Spt16 and Pob3 in vivo, and support the involvement of Spt16–Pob3 in a process that includes the activities of DNA replication factors.
Table II. Properties of spt16 mutants.
| Change in MPT (°C) when combined with a second mutation | |||||
|---|---|---|---|---|---|
| spt16 allele | MPT (°C) | pob3-L78R | nhp6-Δ | dna2-2 | ctf18-Δ |
| SPT16 | >37 | 0 | 0 | 0 | 0 |
| spt16-G132D | 34 | –3 | –1 | –2 | –2 |
| spt16-6 | 33.5 | SL | –3.5 | –2.5 | –2.5 |
| spt16-7 | 34 | SL | –3 | –1 | –2 |
| spt16-8 | 35.5 | SL | –3.5 | –3.5 | –2 |
| spt16-9 | 32 | SL | –2 | –2 | –2 |
| spt16-9a | >37 | 0 | |||
| spt16-11 | 34.5 | SL | –5.5 | –2.5 | –2.5 |
| spt16-16 | 36.5 | SL | SL | SL | –2.5 |
| spt16-16a | <37 | SL | |||
| spt16-24 | 37 | SL | –6 | –3 | –3 |
7737-3-2 (spt16-Δ) with pTF128 (SPT16-WT) or derivatives with the alleles shown were used to test the MPT. Congenic strains 7810-4-3 (spt16-Δ pob3-L78R), 7847-2-4 (spt16-Δ, nhp6a-Δ, nhp6b-Δ), 7800-3-2 (spt16-Δ, dna2-2) and 7806-2-3 (spt16-Δ, ctf18-Δ) carrying the same SPT16 alleles on plasmids were also tested and the difference between the MPT for the double mutant and the MPT for the single mutant with the lower restrictive temperature is shown. SL indicates that the combination was synthetically lethal.
The new alleles of spt16 therefore suggest that Spt16 acts in transcription initiation, transcription elongation and DNA replication. The varying phenotypes of the alleles show that these processes are affected to different extents by different mutations, suggesting that Spt16 and Pob3 have multiple distinct functional domains.
Spt16–Pob3 transcription activation is affected by histone acetylation
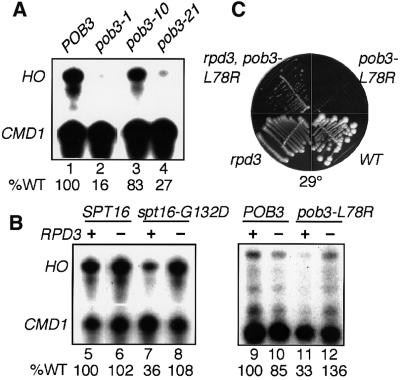
Mutating SPT16 or POB3 caused a varying decrease in expression from the HO locus (Figure 2A and Table I), ranging from no change to a 6-fold drop with pob3-1. In a W303 strain, spt16-G132D and pob3-L78R each caused reduced HO expression, but removal of the histone deacetylase encoded by RPD3 caused a return to at least normal levels (Figure 2B). This suppression extended to general growth as well, since the Ts– phenotype of a pob3 mutant was found to be partially suppressed by an rpd3 deletion (Figure 2C). Since deletion of a deacetylase restores activation lost when SPT16 or POB3 are mutated, at least part of the activation function of Spt16–Pob3 may involve changes in chromatin mediated by histone acetylation. Consistent with this, Spt16–Pob3 has been shown to be partially associated with the histone acetyltransferase complex, NuA3 (John et al., 2000). We conclude that Spt16–Pob3 is an activator of HO transcription that depends at least partly on chromatin modifications.
Fig. 2. Mutations in SPT16 and POB3 decrease transcription of HO and are suppressed by loss of RPD3. (A) and (B) HO and CMD1 transcripts were measured as described in Materials and methods. The amount of HO transcript was normalized to wild type (% WT shown) assuming constant CMD1 expression (the relative efficiency of labeling of each probe causes variable ratios of HO to CMD1 in each experiment). RNA was isolated from 7697 (pob3-Δ) with pTF139 (YCp LEU2 POB3, Schlesinger and Formosa, 2000) carrying the POB3 alleles indicated (lanes 1–4), and from the congenic strains DY150, DY1539, DY5391, DY5394, DY5699, DY7380, DY7379 and DY7375 (lanes 5–12). (C) Congenic W303 strains with a deletion of RPD3, the pob3-L78R mutation, neither mutation (WT) or both mutations were placed on rich medium at 29°C.
spt16, pob3 and nhp6 mutations share phenotypes and interact genetically
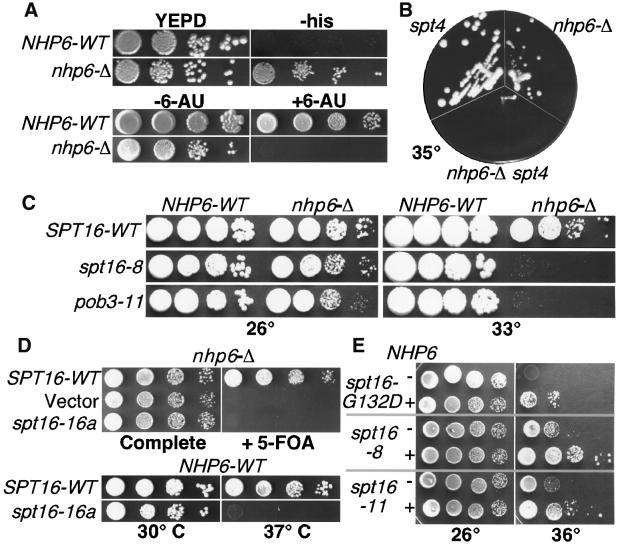
Since Pob3 lacks the HMG1 motif found in FACT and DUF1 subunits, we investigated the possibility that a protein with this feature might function with Spt16–Pob3. Nhp6A/B has this motif and, as expected for proteins that act together, Nhp6 has some functions that are similar to those of Spt16–Pob3. For example, Nhp6 is also an activator of HO transcription and this role is opposed by the deacetylase Rpd3 (Yu et al., 2000). Unlike SPT16 and POB3, NHP6A/B is non-essential, so we examined cells lacking both NHP6 genes for phenotypes associated with spt16 or pob3 defects. An nhp6a/b his4-912δ lys2-128δ strain is phenotypically His+ Lys– (Figure 3A, and our unpublished data), which indicates a weak Spt– phenotype (Prelich and Winston, 1993; Sherwood et al., 1993; Jiang and Stillman, 1996). Like some spt16 mutants, nhp6a/b strains are also sensitive to the transcription elongation inhibitor 6-AU (Figure 3A and Brewster et al., 2001), but not to HU (our unpublished data). The nhp6a/b deletion also displays a strong synthetic defect with a deletion of SPT4, such that either nhp6 or spt4 mutants are viable at 35°C, but a strain lacking all three genes is not (Figure 3B). spt4 mutants are 6-AU sensitive, and Spt4 is a member of a complex required for transcription elongation (Hartzog et al., 1998). Spt5 is also found in this complex and an spt5 mutation also displayed a synthetic defect with nhp6a/b (Brewster et al., 2001). These results suggest that Spt4–Spt5 and Nhp6 each contribute to elongation, but in different pathways. Loss of Nhp6 therefore affects both transcription initiation and elongation, as also noted for Spt16 and Pob3.
Fig. 3. Genetic effects of deleting or overexpressing NHP6. (A) DY2623 (his4-912δ) and DY6863 (his4-912δ nhp6a/b-Δ) were grown to saturation, diluted and placed on either rich medium (YEPD) or medium lacking histidine (–his). DY150 URA+ (NHP6-WT) and DY2382 (nhp6-Δ) were tested on 6-AU as in Figure 1B. (B) Congenic W303 strains lacking SPT4, both copies of NHP6 or all three genes were placed on rich medium at 35°C. (C) 7737-3-2 (spt16-Δ NHP6-WT) and 7847-2-4 (spt16-Δ, nhp6a-Δ, nhp6b-Δ) carrying pTF128 (YCp LEU2 SPT16) with the SPT16 alleles indicated, or 7697 (pob3-Δ) and 7746–5-4 (pob3-Δ, nhp6a-Δ, nhp6b-Δ) carrying pTF139-11 (YCp LEU2 pob3-11), were diluted and placed on rich medium at 26 or 33°C. (D) 7847-2-4 (spt16-Δ) with pCDC68 (YEp URA3 SPT16) was transformed with pTF128, YCplac111 (Gietz and Sugino, 1988; vector), or pTF128-16a, grown in rich medium, washed and dilutions were placed on complete synthetic medium or medium containing 5-FOA. In the bottom panel, dilutions of 7737-3-2 (spt16-Δ) with pTF128 or pTF128-16a were placed at the temperatures shown. (E) 7737-3-2 carrying pTF128 with the SPT16 alleles shown was transformed with YEplac195 (Gietz and Sugino, 1988, –NHP6) or pTF146 (+NHP6), and dilutions were placed on medium lacking uracil at the temperatures indicated.
We next tested whether removing Nhp6 exacerbates the phenotypes of spt16 or pob3 mutants, as expected if Nhp6 promotes the activity of Spt16–Pob3 in vivo. As shown in Table II and Figure 3C, deletion of NHP6A/B caused a dramatic defect when combined with some spt16 and pob3 mutations. An nhp6a/b strain grows slowly but remains viable at 33°C, as do strains with spt16-8 and pob3-11 mutations. However, combining these mutations caused as much as a 10 000-fold decrease in viability (Figure 3C and Brewster et al., 2001). The effect was allele specific, ranging from no change in the maximal permissive temperature (MPT) to synthetic lethality with spt16-16a (Table II and Figure 3D). In this case, a strain was constructed that had deletions of spt16 and nhp6a/b, and carried a plasmid with both URA3 and SPT16. Since strains with URA3 cannot grow on media containing 5-FOA (Boeke et al., 1987), and the plasmid supplies the essential SPT16 function, this strain cannot grow on 5-FOA. Introducing a LEU2 SPT16 plasmid allows loss of the URA3 plasmid and growth on 5-FOA (Figure 3D, top line). However, a plasmid with spt16-16a did not allow growth on 5-FOA, indicating that spt16-16a cannot support growth in a cell lacking Nhp6. The same plasmid supported growth in a strain with Nhp6 (Figure 3D, bottom line; the strain is Ts– as expected), demonstrating that spt16-16a is synthetically lethal with the nhp6 deletion.
The strong, allele-specific, synthetic defects caused by removal of Nhp6 from spt16 and pob3 mutants suggest that these proteins function together in vivo. In this case, some Spt16–Pob3 defects might be ameliorated by increasing the amount of Nhp6. While most alleles were not affected, the Ts– phenotypes of three spt16 alleles were partially suppressed by a high copy NHP6B plasmid (Figure 3E). Increased expression of Spt16–Pob3 did not affect the Ts– phenotype of an nhp6a/b deletion strain (our unpublished data). This pattern is consistent with formation of a complex; some spt16 or pob3 mutations cause diminished stability of this complex, but this can be suppressed by increasing the level of the binding partner Nhp6.
Weak physical interactions suggest that Nhp6 is not usually in stable complexes with Spt16–Pob3
Nhp6 does not copurify with Spt16–Pob3 (J.Wittmeyer, unpublished data), so these proteins do not appear to form a stable heterotrimer fully analogous to FACT or DUF1. To see whether Spt16–Pob3 and Nhp6 form less stable complexes, we fused the myc epitope to the C-terminus of Spt16 or Pob3 (Longtine et al., 1998; expression is from the native promoters) and performed immunoprecipitations from lysates. Spt16–Pob3 did not coprecipitate with Nhp6 using standard conditions (P.Eriksson, unpublished data; Brewster et al., 2001 have also recently reported that these proteins coprecipitate only under conditions of relaxed stringency). To test for even weak interactions, we next added purified His10–Nhp6A to lysates and recovered the Nhp6 using antisera against the histidine tag. With this approach, Spt16 and Pob3 were enriched in the precipitated material only when His10–Nhp6 was added (P.Eriksson, unpublished data). No similar enrichment was observed when a protein that was not expected to interact with Spt16–Pob3 (His10–Swi5) was used, so the enhanced recovery of Spt16–Pob3 with His10–Nhp6 is not due to a spurious association with other components of the assay. We also tested various combinations of fusions in a two-hybrid assay (Bartel and Fields, 1995), and obtained a weak but reproducible signal with Spt16-DBD–Nhp6-AD fusions relative to controls (P.Eriksson, unpublished data). We conclude that most of the Spt16–Pob3 and Nhp6 molecules in a cell are not stably associated with one another, but some complexes containing these three proteins do form. Such complexes could include additional proteins that mediate the interaction; for example, Spt16–Pob3 and Nhp6 could interact indirectly with one another by associating with chromatin. The genetic data therefore indicate that Spt16–Pob3 acts with Nhp6, but the physical data suggest that these proteins are not usually together in a free, stable heterotrimeric complex.
The migration of Nhp6–nucleosome complexes is altered by Spt16–Pob3
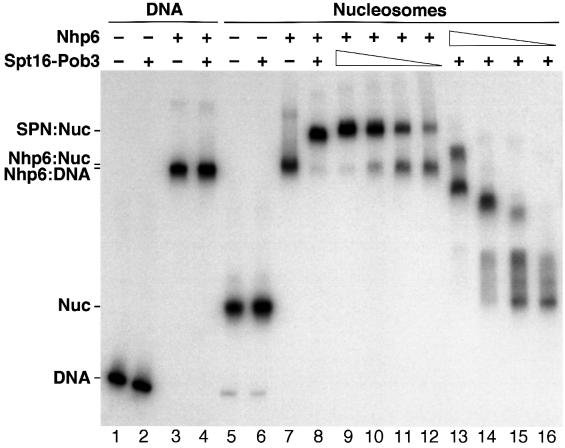
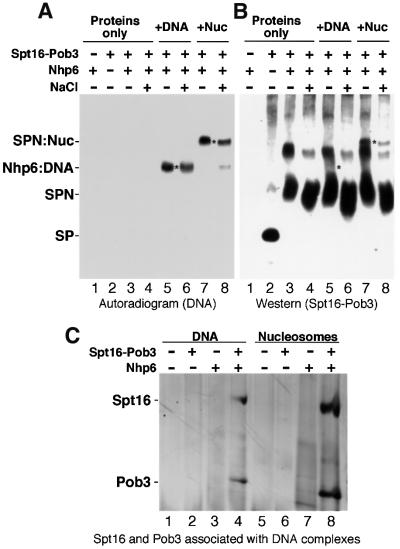
Our results lead to the hypothesis that the DNA-binding protein Nhp6 (Yen et al., 1998) assists binding of Spt16–Pob3 to chromatin without first forming a free heterotrimeric complex. We tested this by examining the migration of DNA or nucleosomes on native polyacrylamide gels in the presence of purified His10–Nhp6 and Spt16–Pob3. Spt16–Pob3 alone had no effect on DNA or nucleosomes (Figure 4, lanes 2 and 6), whereas addition of Nhp6 significantly decreased the mobility of both (Figure 4, lanes 3 and 7). Addition of both Spt16–Pob3 and Nhp6 to DNA had about the same effect as Nhp6 alone (Figure 4, lane 4). In contrast, when both Spt16–Pob3 and Nhp6 were added to nucleosomes, a complex was formed with mobility different from that of the Nhp6–Nuc complex (Figure 4, compare lanes 7 and 8). Therefore, Spt16–Pob3 binds to Nhp6–Nuc complexes to form SPN–Nuc complexes with properties distinct from those of the Nhp6–Nuc form. Optimal complex formation required a ∼4-fold molar excess of Spt16–Pob3 over nucleosomes but a ∼200-fold excess of Nhp6 (Figure 4, lanes 9–16). Similar concentrations of Nhp6 were needed whether or not the protein was affinity tagged, purified from yeast or bacteria, or purified using trichloroacetic acid (TCA) precipitation or standard chromatographic methods (our unpublished data). Most of the Nhp6 is not associated with complexes in these experiments (see below), so we conclude that high concentrations of Nhp6 are needed because Nhp6 interacts weakly with nucleosomes (perhaps due to suboptimal reaction conditions, but we note that the concentrations of Spt16–Pob3 and Nhp6 in these assays are lower than in vivo).
Fig. 4. Nhp6 interacts with nucleosomes to form a binding site for Spt16–Pob3. Purified His10–Nhp6 (+ = 30 pmol, lanes 13–16 are 15, 7.5, 3.8 and 1.9 pmol) and Spt16–Pob3 (+ = 1.5 pmol, lanes 9–12 are 0.75, 0.38, 0.19 and 0.09 pmol) were mixed with 0.1 pmol of a 200 bp DNA fragment (lanes 1–4) or the same fragment incorporated into nucleosomes (lanes 5–16). After native PAGE, the DNA was detected by autoradiography. The positions of the free DNA (DNA), nucleosomes (Nuc) and complexes described in the text are indicated.
Spt16 has an acidic C-terminal domain and Nhp6 is basic. Increasing the NaCl concentration in the binding mixture from 30 to 380 mM (Figure 5A, lanes 7 and 8) or 600 mM (our unpublished data) caused minimal loss of the SPN–Nuc form. This stability at high ionic strengths suggests that SPN–Nuc complexes are not simply aggregates.
Fig. 5. Spt16–Pob3 binds specifically to Nhp6–nucleosome complexes. (A) and (B) Nhp6 (lane 1), Spt16–Pob3 (lane 2) or both (lanes 3–8) were incubated alone (lanes 1–4) or with either DNA (lanes 5 and 6) or nucleosomes (lanes 7 and 8) at 30 (–) or 350 mM (+) NaCl, and electrophoresed as in Figure 4. DNA (A) or Spt16–Pob3 (B) were then detected in duplicate gels by autoradiography or with antisera. Asterisks mark equivalent positions in the two panels, and complexes are labeled as described in the text. (C) Samples in 30 mM NaCl were prepared as in Figure 4 except that 0.6 pmol of DNA or nucleosomes, 2 pmol of Spt16–Pob3 and 40 pmol of Nhp6 were mixed in 11 µl. After electrophoresis, regions containing DNA were excised and subjected to SDS–PAGE, then stained with silver (Ausubel et al., 1994). Lanes 1–8 are as in Figure 4, and the forms excised were those labeled in that figure as DNA (lanes 1 and 2), Nhp6:DNA (lanes 3 and 4), Nuc (lanes 5 and 6), Nhp6:Nuc (lane 7) and SPN:Nuc (lane 8).
Nhp6 loads Spt16–Pob3 specifically to nucleosomes
Spt16–Pob3 migrates in a discrete band on a native gel in the absence of Nhp6 (Figure 5B, lane 2, ‘SP’), and in two slower-migrating bands when Nhp6 is added (Figure 5B, lane 3), with only the faster form being stable in high salt (Figure 5B, lane 4). This pattern was also observed when either DNA or nucleosomes were added (Figure 5B, lanes 5–8), and suggests that Spt16–Pob3 can interact with Nhp6 during native gel electrophoresis. Since the results above indicate that Spt16–Pob3 and Nhp6 do not form stable complexes in cell extracts, this interaction is likely to be non-physiological. Very little Spt16–Pob3 comigrates with the Nhp6–DNA complexes, especially under high salt conditions (compare the regions near the asterisks in lanes 5 and 6 in Figure 5A and B), suggesting that this form contains only Nhp6 and DNA. In contrast, the SPN–Nuc form is associated with a salt-stable band of Spt16–Pob3 protein (see lanes 7 and 8 in Figure 5A and B). Therefore, Spt16–Pob3 does not stably interact with Nhp6–DNA complexes, but it does bind to Nhp6–Nuc complexes. We conclude that Spt16–Pob3 does not simply bind to Nhp6 or even to Nhp6 bound to DNA, but instead specifically recognizes Nhp6–nucleosome complexes.
SPN–Nuc complexes contain about one molecule of Spt16–Pob3 per nucleosome
To test the stoichiometry of Spt16–Pob3 in SPN–Nuc complexes, we excised the regions of the native gels containing labeled DNA and subjected them to SDS– PAGE (Figure 5C). A small amount of Spt16–Pob3 was recovered from bands containing Nhp6–DNA complexes (Figure 5C, lane 4). We attribute this to contamination by protein that coincidentally migrates with the Nhp6–DNA complexes (see Figure 5B, lane 5; Nhp6–DNA partially comigrates with the trailing edge of the Spt16–Pob3 band). Excision of SPN–Nuc forms consistently yielded larger amounts of Spt16–Pob3 (Figure 5C, lane 8). Quantitation from several experiments revealed ∼0.93 molecules of Spt16 and 1.2 molecules of Pob3 per molecule of DNA at the SPN–Nuc position. While some of this material could also be due to spurious comigration, we conclude that each nucleosome is associated with no more than one heterodimer of Spt16–Pob3. This low stoichiometry is further evidence that Spt16–Pob3 does not simply aggregate with Nhp6 in this experiment. It was not possible to estimate the Nhp6 content of complexes using a similar approach since free Nhp6 was found to migrate throughout the region containing complexes both by examination of gel slices and by western blots (our unpublished data).
SPN causes changes in the organization of nucleosomes
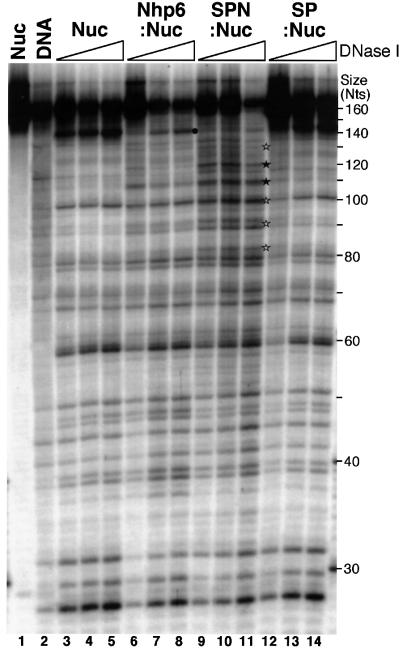
The differing electrophoretic migration rates of Nhp6–Nuc and SPN–Nuc complexes suggest that these forms have altered properties compared with nucleosomes. We treated complexes with the nuclease DNase I to see if these alterations result in a different presentation of the DNA component. The 167 bp fragment of 5S DNA used for the nucleosome assembly is able to position histone octamers uniformly such that the unique label is near the entry point of the nucleosome, with a short duplex tail extending from the distal end. DNase I preferentially digested nucleosomes alone to produce a ∼143 nucleotide fragment, indicating removal of the unprotected tail (marked with a spot in Figure 6). Addition of Spt16–Pob3 alone did not affect the pattern significantly, but digestion at this exit point was diminished when Nhp6 was added. Since Nhp6 binds to DNA, it could simply inhibit digestion by limiting overall access. However, most sites were not affected by Nhp6 addition, so the entire nucleosome is not protected. Nhp6 alone also enhanced digestion somewhat at several sites (for example, at ∼110 nucleotides in Figure 6).
Fig. 6. Sensitivity of nucleosome complexes to DNase I. Nucleosomes (Nuc) were mixed with Nhp6 (Nhp6:Nuc), SPN (SPN:Nuc) or Spt16–Pob3 (SP:Nuc) as in Figure 4, except the DNA was a restriction fragment with 167- (labeled) and 163-nucleotide strands (see Materials and methods), and the Nhp6 was purified from yeast cells. Complexes were treated with DNase I (none in lane 1, 1 arbitrary unit in lane 2, then sets of 8, 16 and 32 units), then separated by denaturing PAGE. The symbols indicate digestion sites; a black spot denotes the strong cut site corresponding to the exit of the DNA from the nucleosome, and stars indicate sites where digestion is enhanced by addition of Nhp6 and SPN, with filled symbols corresponding to more pronounced effects. Size is shown in nucleotides (Nts).
Addition of both Nhp6 and Spt16–Pob3 caused a more dramatic enhancement of DNase I digestion, particularly in the region proximal to the double-stranded tail of these nucleosomes (marked with stars in Figure 6). Use of nucleosomes reconstituted with different DNA molecules (our unpublished data) revealed that the enhanced sites appear at a constant distance from the cut marking the exit of the DNA from the nucleosome (if the black spot is considered to be 0, the enhanced cuts are at about –13, –23, –33 and –42 nucleotides, with the central sites being the most pronounced). The ∼10 nucleotide periodicity of the enhanced sites indicates that the DNA is still associated with the nucleosome (this pattern is not observed with free DNA; see Figure 6), but in a more accessible form. Since many sites remain unaffected in the SPN–Nuc form, including sites an equivalent distance from the other end of the nucleosome, the alteration appears to be localized to the DNA near the extended tail. We conclude that Nhp6 binds to nucleosomes, probably through the duplex tail, causing a large change in electrophoretic mobility and a small change in accessibility of the nucleosomal DNA. This allows binding of Spt16–Pob3, which then causes a further reorganization of the nucleosome, changing the electrophoretic migration and altering the organization of the adjacent DNA within the nucleosome.
Discussion
We present genetic and physical evidence that yeast Spt16–Pob3 functions together with the HMG1-motif protein Nhp6 to bind nucleosomes and change both their electrophoretic mobility and the accessibility of their DNA. This suggests that these highly conserved factors participate in both replication and transcription by reorganizing nucleosomes in a way that changes the presentation of the DNA. Since accessibility of chromatin is also modulated by acetylation of histones, this also explains the functional overlap between Spt16–Pob3 and acetylation. Spt16–Pob3 could be associated with factors such as the acetyltransferase Nua3 (John et al., 2000) because both promote DNA accessibility in separate ways and would therefore be expected to act coordinately. Their combined function would be opposed by deacetylases like Rpd3, explaining the genetic suppression of spt16, pob3 and nhp6 mutants by the deletion of Rpd3 (Figure 2 and Yu et al., 2000).
New spt16 alleles support roles in replication and transcription
Spt16–Pob3 or its homologs have been associated with initiation of transcription (Malone et al., 1991; Rowley et al., 1991; Lycan et al., 1994; Costa and Arndt, 2000; John et al., 2000; Kang et al., 2000; Schlesinger and Formosa, 2000), elongation of transcription (Orphanides et al., 1998, 1999) and DNA replication (Wittmeyer and Formosa, 1997; Formosa and Nittis, 1999; Okuhara et al., 1999; Wittmeyer et al., 1999; Schlesinger and Formosa, 2000). This could indicate that Spt16–Pob3 has many functions, or that it has a single activity needed during many processes. We report here that multiple alleles of SPT16 have differential effects on the Spt– phenotype (transcription initiation), sensitivity to 6-AU (transcription elongation), HU sensitivity (DNA replication) and synthetic defects with replication factors (DNA replication). If Spt16 had a single function and different mutations caused different degrees of impairment of that function, the same alleles should have the strongest effects in all tests. Since the allele-specific effects among the phenotypes do not correlate in this way, we suggest that Spt16–Pob3 has a single central activity that is needed in multiple stages of transcription and replication, and that different mutations disturb the ability of Spt16–Pob3 to participate in discrete reactions. The simplest interpretation is that Spt16–Pob3 alters nucleosomes, and this is important first during the establishment of transcription and replication initiation complexes, and then again during elongation as both RNA and DNA polymerases encounter nucleosomes on their templates. Different mutations then disturb the ability of Spt16–Pob3 to function coordinately with factors that act in these different processes.
Spt16–Pob3 functions with Nhp6
Pob3 lacks the HMG1 DNA-binding motif found in homologs from higher eukaryotes (Wittmeyer and Formosa, 1997), but we find in genetic tests that Spt16–Pob3 functions with a protein with this feature, Nhp6. Cells lacking both copies of NHP6 are viable (Costigan et al., 1994), so the essential Spt16 and Pob3 proteins cannot be entirely dependent on Nhp6 for their function. However, the robust genetic interactions we have observed demonstrate that Nhp6 supports Spt16–Pob3 function in some physiologically important way. Several approaches indicate that Nhp6 and Spt16–Pob3 are not typically associated with one another to form a free factor equivalent to FACT or DUF1. Instead, Nhp6 alone appears to interact with a nucleosome, and this forms a binding site for Spt16–Pob3. It has been suggested that HMG proteins deliver transcription factors to sites within chromatin (Bustin, 1999); our results indicate that they can also support the recruitment or action of other chromatin modulators.
Spt16–Pob3 binding might require contact with both Nhp6 and other features of the nucleosome, or it might require nucleosomes that are somehow repositioned by the binding of Nhp6. If other proteins are also capable of causing such a change in nucleosomes in vivo, subsets of these factors could be removed without blocking all Spt16–Pob3 activity. Candidates would include other known HMG1-motif proteins, such as Hmo1 and Hmo2, which have been found to display minor genetic interactions with Spt16 (our unpublished data). However, since the quadruple deletion (nhp6a/b, hmo1, hmo2) is viable (Lu et al., 1996), if HMG1-motif proteins play an essential role in yeast their redundancy must extend beyond this set of proteins. Spt16–Pob3 might also be able to function with other classes of DNA-binding protein, or even alone, with reduced efficiency. The genetic results indicate that Nhp6 is more effective than other proteins at promoting Spt16–Pob3 function, but it remains to be determined whether the formation of SPN–nucleosome complexes involves specific protein–protein interactions between Spt16–Pob3 and Nhp6.
Nhp6 and SPN alter the properties of nucleosomes
Human FACT cosediments with nucleosomes or H2A–H2B dimers, and Orphanides et al. (1999) have suggested that FACT might act by releasing H2A–H2B from nucleosomes. We find that yeast SPN also binds to nucleosomes, and due to its modular nature we have been able to detect two distinct stages of binding. Nhp6 alone altered the electrophoretic mobility of nucleosomes, and addition of SPN caused both a further change in mobility and enhanced nuclease sensitivity. Notably, the strongest effects were in a region where H2A and H2B contact the DNA (Luger et al., 1997). This enhanced digestion suggests a reorganization of the nucleosome, specifically in the region of H2A–H2B, which is consistent with the proposed weakening of the protein–protein contacts between H2A–H2B and H3–H4. Alternatively, SPN could act by changing the contacts between histones and DNA. Nucleosome remodeling factors have been proposed to act in this way by displacing a loop of DNA (Kingston and Narlikar, 1999), which can then propagate around the nucleosome. FACT does not display standard remodeling activity (Orphanides et al., 1998), and Spt16–Pob3 does not have the ATPase activity normally associated with remodeling factors (Wittmeyer et al., 1999). However, FACT/SPN might still share some mechanistic features with these factors. Our data are consistent with a model in which the HMG1-box factor associates with DNA near the entry/exit points of the nucleosome, then Spt16–Pob3 binds to this structure in such a way that the association of the DNA with the histone core is locally disturbed. Since the DNA passes the position of the H2A–H2B dimers twice, but we only see enhanced DNase I sensitivity in one of these regions, we prefer models in which the DNA–histone interactions are disrupted locally to those in which the protein core is disturbed, although these possibilities are not mutually exclusive. In either case, SPN is a novel remodeling factor that does not require NTP hydrolysis and does not reposition nucleosomes, but reorganizes them in a way that is important for replication and transcription machinery.
Altering the interactions between the components of nucleosomes could be an important step in preparing sites for initiation of DNA replication or transcription, or in allowing polymerases to progress on nucleosomal templates. This ability would therefore explain the broad range of effects caused by mutations in SPN components. The results reported here provide tools that will promote investigation of the role of Spt16–Pob3 and Nhp6 in chromatin-mediated processes.
Materials and methods
Strains and plasmids
Strains are listed in Table III. spt16-Δ(TRP1) lacks residues 8 through the stop codon. pCDC68 (Prendergast et al., 1990) and pTF125 are high copy URA3 SPT16 plasmids. pTF128 has 746 bp upstream of the SPT16 ORF and a modified but phenotypically normal SPT16 (which now terminates GSPR) in YCplac111 (Gietz and Sugino, 1988). NHP6A and NHP6B were deleted as described (Costigan et al., 1994). A construct that inserts KanMX into URA3 (Cross, 1997, D.J.Stillman, in preparation) was used to convert nhp6a-Δ(URA3) to nhp6a-Δ(KanMX). pTF146 is NHP6B with 1039 bp upstream and 786 bp downstream in YEplac195 (Gietz and Sugino, 1988).
Table III. Strains used (all are MATa).
| Name | Genotype | Background |
|---|---|---|
| 4053-5-2 | trp1 leu2 ura3 his7 | A364a |
| 4053-5-2 URA+ | trp1 leu2 ura3::YIplac211(URA3) his7 | A364a |
| 7373-4-4 | trp1 leu2 ura3 his3 | A364a |
| 7697 | trp1 leu2 ura3 his7 pob3-Δ::TRP1 | A364a |
| 7737-3-2 | trp1 leu2 ura3 his3 spt16-Δ::TRP1 | A364a |
| 7746-5-4 | trp1 leu2 ura3 his3 pob3-Δ::TRP1 nhp6b-Δ::HIS3 nhp6a-Δ::KanMX | A364a |
| 7782-x | trp1 leu2 ura3 his7 spt16-x | A364a |
| 7782-x URA+ | trp1 leu2 ura3::YIplac211(URA3) his7 spt16-x | A364a |
| 7784-1-1 | leu2-Δ1 trp1-Δ63 ura3-52 his4-912δ lys2-128δ spt16-Δ::TRP1 | S288c |
| 7800-3-2 | trp1 leu2 ura3 his7 spt16-Δ::TRP1 dna2-2 | A364a |
| 7806-2-3 | trp1 leu2 ura3 his3 spt16-Δ::TRP1 ctf18-Δ::HIS3 | A364a |
| 7810-4-3 | trp1 leu2 ura3 his3 spt16-Δ::TRP1 pob3-L78R | A364a |
| 7847-2-4 | trp1 leu2 ura3 his3 spt16-Δ::TRP1 nhp6a-Δ::KanMX nhp6b-Δ::HIS3 | A364a |
| DY150 | ura3 ade2 trp1 can1 leu2 his3 | W303 |
| DY150 URA+ | ade2 can1 his3 leu2 trp1 ura3::YIplac211(URA3) | W303 |
| DY1539 | ura3 ade2 trp1 can1 leu2 his3 rpd3-Δ::LEU2 | W303 |
| DY2382 | ade2 can1 his3 leu2 trp1 ura3 nhp6a-Δ::URA3 nhp6b-Δ::HIS3 | W303 |
| DY2623 | his4-912δ lys2-128δ ade8 leu2 ura3 | S288c |
| DY5391 | ura3 ade2 trp1 can1 leu2 his3 lys2 spt16-G132D | W303 |
| DY5394 | ura3 ade2 trp1 can1 leu2 his3 lys2 spt16-G132D rpd3-Δ::LEU2 | W303 |
| DY5699 | ade2 can1 his3 leu2 lys2 met15 trp1 ura3 | W303 |
| DY6863 | nhp6a-Δ::URA3 nhp6b-Δ::HIS3 his4-912δ ade2 ade8 leu2 lys2 trp1 | S288c |
| DY7375 | ade2 can1 his3 leu2 lys2 met15 trp1 ura3 pob3-L78R rpd3-Δ::LEU2 | W303 |
| DY7379 | ade2 can1 his3 leu2 lys2 met15 trp1 ura3 pob3-L78R | W303 |
| DY7380 | ade2 can1 his3 leu2 lys2 met15 trp1 ura3 rpd3-Δ::LEU2 | W303 |
SPT16 alleles were integrated using YIplac211 (Gietz and Sugino, 1988) derivatives digested with SnaBI. 5-FOA-resistant isolates (Boeke et al., 1987) were obtained from Ura+ transformants and then screened for the Ts– phenotype, tested by Southern hybridization and confirmed by sequencing relevant portions after PCR amplification. Other strains were as described previously (Malone et al., 1991; Formosa and Nittis, 1999; Schlesinger and Formosa, 2000; Yu et al., 2000) or derived from them using standard genetic methods.
spt16 mutant isolation and characterization
pTF128 was mutagenized with hydroxylamine as described (Schlesinger and Formosa, 2000) and used to transform strain 7737-3-2 (spt16-Δ) carrying pCDC68 (Prendergast et al., 1990). Transformants were transferred to media containing 5-FOA (Boeke et al., 1987), then replicates were incubated at 26 and 37°C. pTF128 derivatives were recovered from Ts– isolates and retested. The MPT was determined by streaking aliquots onto agar plates at 1°C increments covering a range from 26 to 37°C, with the MPT being the highest temperature producing at least 10% of the growth obtained at 26°C.
S1 assays
RNA levels were quantitated by S1 nuclease protection using HO and CMD1 probes followed by phosphorimager analysis as described (Bhoite and Stillman, 1998; Yu et al., 2000).
Nucleosome binding and nuclease sensitivity
DNA fragments containing the sea urchin 5S DNA gene were amplified by PCR and labeled with polynucleotide kinase and [γ-32P]ATP (Sambrook et al., 1989). DNA and chicken histone octamers (Graziano et al., 1988; a generous gift from V.Graziano and V.Ramakrishnan) were mixed in 2 M NaCl and dialyzed as described (Luger et al., 1999). Nucleosomes were isolated by velocity sedimentation in fractions with 15% maltose, 0.1 mg/ml human serum albumin (Sigma), 25 mM HEPES pH 7.6, 1 mM Na2EDTA, 0.3 µg/ml leupeptin, 1.4 µg/ml pepstatin and 0.5 mM phenylmethylsulfonyl fluoride (gradient buffer).
Spt16–Pob3 was purified as described (Wittmeyer et al., 1999). NHP6A was amplified by PCR and inserted into pET16B, and the resulting His10–Nhp6 was expressed in Escherichia coli BL21(DE3)-pLysS (Studier et al., 1990) and purified by chelated nickel chromatography as described (Brazas et al., 1995), with or without additional purification using DNA cellulose chromatography. Untagged Nhp6 was purified from E.coli carrying pRJ1228 (Paull and Johnson, 1995) or from yeast cells by differential precipitation with TCA and cation exchange chromatography essentially as described (Paull and Johnson, 1995). Binding assays were performed by mixing nucleosomes or DNA (in 5 µl of gradient buffer) with 2 µl of a solution containing 10% glycerol, 20 mM Tris–HCl pH 7.5, 100 mM NaCl, 1 mM 2-mercaptoethanol, 1 mM Na2EDTA and Nhp6 or Spt16–Pob3 as indicated in each experiment. Reactions were incubated for 10 min at 30°C, then electrophoresed at 180 V for 4–5 h at 4°C through 4% polyacrylamide, 5% (w/v) glycerol, 2 mM MgCl2 and 0.5 × TBE (Sambrook et al., 1989). Regions containing labeled DNA were excised and separated by SDS–PAGE and stained with silver (Ausubel et al., 1994) or Coomassie Blue. The amount of DNA was determined by phosphorimaging, and the ratio of protein to DNA was quantitated using NIH Image software.
For immunodetection, proteins were transferred to nitrocellulose (Schleicher and Schuell BA83), probed with antisera directed against Spt16–Pob3 (Wittmeyer and Formosa, 1997) and detected with enhanced chemiluminescence as directed (Amersham-Pharmacia Biotech).
For nuclease digestions, the 5S DNA was digested with EcoRI, labeled, then digested with ScaI, releasing a fragment with 167 and 163 nucleotide strands with the label at the 5′ overhang of the EcoRI cut. Nucleosomes and complexes were formed as above, then various amounts of DNase I (Boehringer-Mannheim) were added, the concentration of MgCl2 was adjusted to 2 mM and the samples were incubated for 15 min at 30°C. EDTA and carrier DNA were added, the DNA was extracted with CHCl3, precipitated with ethanol and electrophoresed on 8% polyacrylamide gels containing 7 M urea along with size standards.
Acknowledgments
Acknowledgements
We thank V.Graziano and V.Ramakrishnan for histone octamers, R.Singer for pCDC68, the University of Utah Bioprocessing Center and D.Kolodrubetz for antisera, R.Johnson for pRJ1228 and B.Cairns for the 5S DNA plasmid, nucleosome expertise and helpful comments on this manuscript. This work was supported by grants from the NSF to T.F., the NIH to D.S., the Swedish Foundation for International Cooperation in Research and Higher Education to P.E. and the Huntsman Cancer Institute (through B.Cairns) for J.W.
References
- Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (eds) (1994) Current Protocols in Molecular Biology. Greene Publishing Associates, Inc., and John Wiley & Sons, Inc., Boston, MA.
- Bartel P.L. and Fields,S. (1995) Analyzing protein–protein interactions using two-hybrid system. Methods Enzymol., 254, 241–263. [DOI] [PubMed] [Google Scholar]
- Bhoite L.T. and Stillman,D.J. (1998) Residues in the Swi5 zinc finger protein that mediate cooperative DNA-binding with the Pho2 homeodomain protein. Mol. Cell. Biol., 18, 6436–6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J.D., Trueheart,J., Natsoulis,G. and Fink,G.R. (1987) 5-fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol., 154, 164–175. [DOI] [PubMed] [Google Scholar]
- Brazas R.M., Bhoite,L.T., Murphy,M.D., Yu,Y., Chen,Y., Neklason,D.W. and Stillman,D.J. (1995) Determining the requirements for cooperative DNA binding by Swi5p and Pho2p (Grf10p/Bas2p) at the HO promoter. J. Biol. Chem., 270, 29151–29161. [DOI] [PubMed] [Google Scholar]
- Brewster N.K., Johnston,G.C. and Singer,R.A. (1998) Characterization of the CP complex, an abundant dimer of Cdc68 and Pob3 proteins that regulates yeast transcriptional activation and chromatin repression. J. Biol. Chem., 273, 21972–21979. [DOI] [PubMed] [Google Scholar]
- Brewster N.K., Johnston,G.C. and Singer,R.A. (2001) A bipartite yeast SSRP1 analog comprised of Pob3 and Nhp6 proteins modulates transcription. Mol. Cell. Biol., 21, 3491–3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S.J., Kellett,P.J. and Lippard,S.J. (1993) Ixr1, a yeast protein that binds to platinated DNA and confers sensitivity to cisplatin. Science, 261, 603–605. [DOI] [PubMed] [Google Scholar]
- Bustin M. (1999) Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol. Cell. Biol., 19, 5237–5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa P.J. and Arndt,K.M. (2000) Synthetic lethal interactions suggest a role for the Saccharomyces cerevisiae rtf1 protein in transcription elongation. Genetics, 156, 535–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costigan C., Kolodrubetz,D. and Snyder,M. (1994) NHP6A and NHP6B, which encode HMG1-like proteins, are candidates for downstream components of the yeast SLT2 mitogen-activated protein kinase pathway. Mol. Cell. Biol., 14, 2391–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross F.R. (1997) ‘Marker swap’ plasmids: convenient tools for budding yeast molecular genetics. Yeast, 13, 647–653. [DOI] [PubMed] [Google Scholar]
- Cullmann G., Fien,K., Kobayashi,R. and Stillman,B. (1995) Characterization of the five replication factor C genes of Saccharomyces cerevisiae. Mol. Cell. Biol., 15, 4661–4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley J.F. and Stillman,B. (1991) A close relative of the nuclear, chromosomal high-mobility group protein HMG1 in yeast mitochondria. Proc. Natl Acad. Sci. USA, 88, 7864–7868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans D.R., Brewster,N.K., Xu,Q., Rowley,A., Altheim,B.A., Johnston,G.C. and Singer,R.A. (1998) The yeast protein complex containing cdc68 and pob3 mediates core-promoter repression through the cdc68 N-terminal domain. Genetics, 150, 1393–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exinger F. and Lacroute,F. (1992) 6-Azauracil inhibition of GTP biosynthesis in Saccharomyces cerevisiae. Curr. Genet., 22, 9–11. [DOI] [PubMed] [Google Scholar]
- Formosa T. and Nittis,T. (1999) Dna2 mutants reveal interactions with DNA polymerase α and Ctf4, a Pol α accessory factor, and show that full Dna2 helicase activity is not essential for growth. Genetics, 151, 1459–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R.D. and Sugino,A. (1988) New yeast–Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene, 74, 527–534. [DOI] [PubMed] [Google Scholar]
- Graziano V., Gerchman,S.E. and Ramakrishnan,V. (1988) Reconstitution of chromatin higher-order structure from histone H5 and depleted chromatin. J. Mol. Biol., 203, 997–1007. [DOI] [PubMed] [Google Scholar]
- Hartzog G.A., Wada,T., Handa,H. and Winston,F. (1998) Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev., 12, 357–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y.W. and Stillman,D.J. (1996) Epigenetic effects on yeast transcription caused by mutations in an actin-related protein present in the nucleus. Genes Dev., 10, 604–619. [DOI] [PubMed] [Google Scholar]
- John S., Howe,L., Tafrov,S.T., Grant,P.A., Sternglanz,R. and Workman,J.L. (2000) The something about silencing protein, Sas3, is the catalytic subunit of NuA3, a yTAF(II)30-containing HAT complex that interacts with the Spt16 subunit of the yeast CP (Cdc68/Pob3)–FACT complex. Genes Dev., 14, 1196–1208. [PMC free article] [PubMed] [Google Scholar]
- Kang S.W., Kuzuhara,T. and Horikoshi,M. (2000) Functional interaction of general transcription initiation factor TFIIE with general chromatin factor SPT16/CDC68. Genes Cells, 5, 251–263. [DOI] [PubMed] [Google Scholar]
- Kingston R.E. and Narlikar,G.J. (1999) ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev., 13, 2339–2352. [DOI] [PubMed] [Google Scholar]
- Kolodrubetz D. and Burgum,A. (1990) Duplicated NHP6 genes of Saccharomyces cerevisiae encode proteins homologous to bovine high mobility group protein 1. J. Biol. Chem., 265, 3234–3239. [PubMed] [Google Scholar]
- Kornberg A. and Baker,T. (1992) DNA Replication. W.H.Freeman and Co., New York.
- Kouprina N., Kroll,E., Kirillov,A., Bannikov,V., Zakharyev,V. and Larionov,V. (1994) CHL12, a gene essential for the fidelity of chromosome transmission in the yeast Saccharomyces cerevisiae. Genetics, 138, 1067–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert J.R., Bilanchone,V.W. and Cumsky,M.G. (1994) The ORD1 gene encodes a transcription factor involved in oxygen regulation and is identical to IXR1, a gene that confers cisplatin sensitivity to Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 91, 7345–7349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie,A., 3rd, Demarini,D.J., Shah,N.G., Wach,A., Brachat,A., Philippsen,P. and Pringle,J.R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast, 14, 953–961. [DOI] [PubMed] [Google Scholar]
- Lu J., Kobayashi,R. and Brill,S.J. (1996) Characterization of a high mobility group 1/2 homolog in yeast. J. Biol. Chem., 271, 33678–33685. [DOI] [PubMed] [Google Scholar]
- Luger K., Mader,A.W., Richmond,R.K., Sargent,D.F. and Richmond,T.J. (1997) Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature, 389, 251–260. [DOI] [PubMed] [Google Scholar]
- Luger K., Rechsteiner,T.J. and Richmond,T.J. (1999) Preparation of nucleosome core particle from recombinant histones. Methods Enzymol., 304, 3–19. [DOI] [PubMed] [Google Scholar]
- Lycan D., Mikesell,G., Bunger,M. and Breeden,L. (1994) Differential effects of Cdc68 on cell cycle-regulated promoters in Saccharomyces cerevisiae. Mol. Cell. Biol., 14, 7455–7465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone E.A., Clark,C.D., Chiang,A. and Winston,F. (1991) Mutation in SPT16/CDC68 suppress cis- and trans-acting mutations that affect promoter function in Saccharomyces cerevisiae. Mol. Cell. Biol., 11, 5710–5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles J. and Formosa,T. (1992) Protein affinity chromatography with purified yeast DNA polymerase α detects proteins that bind to DNA polymerase. Proc. Natl Acad. Sci. USA, 89, 1276–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuhara K., Ohta,K., Seo,H., Shioda,M., Yamada,T., Tanaka,Y., Dohmae,N., Seyama,Y., Shibata,T. and Murofushi,H. (1999) A DNA unwinding factor involved in DNA replication in cell-free extracts of Xenopus eggs. Curr. Biol., 9, 341–350. [DOI] [PubMed] [Google Scholar]
- Orphanides G., LeRoy,G., Chang,C.-H., Luse,D.S. and Reinberg,D. (1998) FACT, a factor that facilitates transcript elongation through nucleosomes. Cell, 92, 105–116. [DOI] [PubMed] [Google Scholar]
- Orphanides G., Wu,W.H., Lane,W.S., Hampsey,M. and Reinberg,D. (1999) The chromatin-specific transcription elongation factor FACT comprises the human SPT16/CDC68 and SSRP1 proteins. Nature, 400, 284–288. [DOI] [PubMed] [Google Scholar]
- Paull T.T. and Johnson,R.C. (1995) DNA looping by Saccharomyces cerevisiae high mobility group proteins NHP6A/B. Consequences for nucleoprotein complex assembly and chromatin condensation. J. Biol. Chem., 270, 8744–8754. [DOI] [PubMed] [Google Scholar]
- Paull T.T., Carey,M. and Johnson,R.C. (1996) Yeast HMG proteins NHP6A/B potentiate promoter-specific transcriptional activation in vivo and assembly of preinitiation complexes in vitro. Genes Dev., 10, 2769–2781. [DOI] [PubMed] [Google Scholar]
- Powell W. and Reines,D. (1996) Mutations in the second largest subunit of RNA polymerase II cause 6-azauracil sensitivity in yeast and increased transcriptional arrest in vitro. J. Biol. Chem., 271, 6866–6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prelich G. and Winston,F. (1993) Mutations that suppress the deletion of an upstream activating sequence in yeast: involvement of a protein kinase and histone H3 in repressing transcription in vivo. Genetics, 135, 665–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendergast J.A., Murray,L.E., Rowley,A., Carruthers,D.R., Singer,R.A. and Johnston,G.C. (1990) Size selection identifies new genes that regulate Saccharomyces cerevisiae cell proliferation. Genetics, 124, 81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley A., Singer,R.A. and Johnston,G. (1991) CDC68, a yeast gene that affects regulation of cell proliferation and transcription, encodes a protein with a highly acidic carboxyl terminus. Mol. Cell. Biol., 11, 5718–5726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning. A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schlesinger M.B. and Formosa,T. (2000) POB3 is required for both transcription and replication in the yeast Saccharomyces cerevisiae. Genetics, 155, 1593–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell R., D’Ari,L., Foss,M., Goodman,D. and Rine,J. (1989) Genetic and molecular characterization of suppressors of SIR4 mutations in Saccharomyces cerevisiae. Genetics, 122, 29–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood P.W., Tsang,S.V. and Osley,M.A. (1993) Characterization of HIR1 and HIR2, two genes required for regulation of histone gene transcription in Saccharomyces cerevisiae. Mol. Cell. Biol., 13, 28–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorova J. and Breeden,L. (1999) The MSN1 and NHP6A genes suppress SWI6 defects in Saccharomyces cerevisiae. Genetics, 151, 45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F.W., Rosenberg,A.H., Dunn,J.J. and Dubendorff,J.W. (1990) Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol., 185, 60–89. [DOI] [PubMed] [Google Scholar]
- Winston F. and Carlson,M. (1992) Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet., 8, 387–391. [DOI] [PubMed] [Google Scholar]
- Winston F. and Sudarsanam,P. (1998) The SAGA of Spt proteins and transcriptional analysis in yeast: past, present, and future. Cold Spring Harbor Symp. Quant. Biol., 63, 553–561. [DOI] [PubMed] [Google Scholar]
- Wittmeyer J. and Formosa,T. (1997) The Saccharomyces cerevisiae DNA polymerase α catalytic subunit interacts with Cdc68/Spt16 and with Pob3, a protein similar to an HMG1-like protein. Mol. Cell. Biol., 17, 4178–4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmeyer J., Joss,L. and Formosa,T. (1999) Spt16 and Pob3 of Saccharomyces cerevisiae form an essential, abundant heterodimer that is nuclear, chromatin-associated, and copurifies with DNA polymerase α. Biochemistry, 38, 8961–8971. [DOI] [PubMed] [Google Scholar]
- Xu Q., Johnston,G.C. and Singer,R.A. (1993) The Saccharomyces cerevisiae Cdc68 transcription activator is antagonized by San1, a protein implicated in transcriptional silencing. Mol. Cell. Biol., 13, 7553–7565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q., Singer,R.A. and Johnston,G.C. (1995) Sug1 modulates yeast transcription activation by Cdc68. Mol. Cell. Biol., 15, 6025–6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen Y.M., Wong,B. and Johnson,R.C. (1998) Determinants of DNA binding and bending by the Saccharomyces cerevisiae high mobility group protein NHP6A that are important for its biological activities. Role of the unique N terminus and putative intercalating methionine. J. Biol. Chem., 273, 4424–4435. [DOI] [PubMed] [Google Scholar]
- Yu Y., Eriksson,P. and Stillman,D.J. (2000) Architectural transcription factors and the SAGA complex function in parallel pathways to activate transcription. Mol. Cell. Biol., 20, 2350–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]