Abstract
The promyelocytic leukaemia (PML) protein localizes in the nucleus both in the nucleoplasm and in matrix-associated multiprotein complexes known as nuclear bodies (NBs). The number and the intensity of PML NBs increase in response to interferon (IFN). Overexpression of PML affects the replication of vesicular stomatitis virus and influenza virus. However, PML has a less powerful antiviral activity against these viruses than the IFN mediator MxA. Here, we show that overexpression of PML, but not that of Mx1 or MxA, leads to a drastic decrease of a complex retrovirus, the human foamy virus (HFV), gene expression. PML represses HFV transcription by complexing the HFV transactivator, Tas, preventing its direct binding to viral DNA. This physical interaction requires the N-terminal region of Tas and the RING finger of PML, but does not necessitate PML localization in NBs. Finally, we show that IFN treatment inhibits HFV replication in wild-type but not in PML–/– cells. These findings point to a role for PML in transcriptional repression and suggest that PML could play a key role in mediating an IFN-induced antiviral state against a complex retrovirus.
Keywords: HFV/Mx/NBs/Tas/transcriptional repression
Introduction
The promyelocytic leukaemia (PML) gene was originally identified through its fusion with the RARα gene in the t(15;17) translocation found in acute promyelocytic leukaemia (APL) (de Thé et al., 1991; Goddard et al., 1991; Kakizuka et al., 1991; Pandolfi et al., 1991). PML belongs to a family of proteins defined by the presence of the RBCC motif, a C3HC4 (RING finger) zinc binding motif, one or two other cysteine-rich motifs, the B boxes and a coiled-coil region. PML is expressed in the nuclear diffuse fraction of the nucleoplasm and in a discrete sub-nuclear compartment, the nuclear bodies (NBs) (Daniel et al., 1993; Dyck et al., 1994; Koken et al., 1994; Weis et al., 1994). In APL, the chimeric PML–RARα protein alters the normal localization of PML from the punctate nuclear patterns of NBs to micro-dispersed tiny dots.
n addition to PML, the NBs contain several other proteins, such as Sp100, Sp140, ISG20, CBP, pRB, Daxx, BLM and p53 (reviewed in Maul et al., 2000). However, PML was found to be responsible for the proper localization of all other NB-associated proteins since they are dispersed in PML–/– cells (Ishov et al., 1999). PML as well as Sp100 and p53 are covalently modified by the small ubiquitin-related modifier SUMO-1 (Muller et al., 1998, 2000; Sternsdorf et al., 1999). PML is modified by SUMO-1 at three sites: K65 in the RING finger, K160 in the first B box and K490 in the nuclear localization sequence (NLS) (Kamitani et al., 1998). SUMO-1 modification appears to be necessary for the localization of PML on NBs (Kamitani et al., 1998; Muller et al., 1998; Ishov et al., 1999). PML acts as a cell growth and tumour suppressor protein (Koken et al., 1995; Liu et al., 1995), is regulated during the cell cycle (Everett et al., 1999a) and is involved in apoptosis (Quignon et al., 1998; Wang et al., 1998a).
Expression of PML, ISG20, Sp140 and Sp100 is increased in response to interferon (IFN) (Guldner et al., 1992; Chelbi-Alix et al., 1995; Grötzinger et al., 1996; Gongora et al., 1997). The PML promoter contains functional IFNα-stimulated response elements and IFNγ activation sites (Stadler et al., 1995), revealing that PML is a primary target gene of IFNs. PML expression is essential for the ability of IFN to induce programmed cell death (Wang et al., 1998a).
We have shown that overexpression of PML can affect the replication of RNA viruses such as vesicular stomatitis virus (VSV) and influenza virus by interfering with viral mRNA and protein synthesis (Chelbi-Alix et al., 1998). These findings suggest a role for PML in IFN-induced antiviral defence (Chelbi-Alix et al., 1996). The well-known disorganization of PML NBs by various viruses may be part of a general viral strategy to counteract IFN action. HSV-1 infection leads to NB disruption; this effect is mediated by the viral immediate-early RING finger protein ICP0 (Everett and Maul, 1994), which first accumulates at NBs and then induces the degradation of PML and Sp100 in a proteasome-dependent manner (Chelbi-Alix and de Thé, 1999; Everett et al., 1999b). In addition to ICP0, the cytomegalovirus IE1 protein, the Epstein–Barr virus protein EBNA5 and the adenovirus E4 ORF3 protein all alter, to various extents, the localization of the NB-associated PML (reviewed in Sternsdorf et al., 1997). This alteration of PML localization and/or expression could facilitate the infection process.
Foamy viruses (FVs) are complex animal retroviruses encoding auxiliary proteins from the 3′ end of their genome in addition to the structural and enzymic gag, pol and env genes. In the case of the human foamy virus (HFV), the prototype of FVs, also called PVF-1, two additional open reading frames have been described: ORF1 [encoding transactivator of spumaviruses (Tas)] and ORF2 (for review see Lecellier and Saib, 2000). The 301 bp Tas, a 36 kDa nuclear phosphoprotein, transactivates viral gene expression by binding directly to the long terminal repeat (LTR) and an internal promoter (IP) (He et al., 1996). This latter is located upstream of the auxiliary genes, 3′ of the env gene, and directs their expression early in the replication cycle (Löchelt et al., 1993). As for most viral transactivators, Tas harbours distinct autonomous domains that are implicated in DNA binding (residues 88–200) and transactivation (residues 272–300) (He et al., 1993; Lee et al., 1995). Bet is another auxiliary protein. This 482 amino acid protein is generated by alternative splicing, which fuses the first 88 residues of Tas to 394 amino acids from ORF2. Recent works have reported that the FVs, although harbouring most of the retroviral attributes (HIV, HTLV-I), present several characteristics relating them to the hepatitis B virus (HBV), such as the formation of a specific pol mRNA and the infectivity of the viral DNA contained in extracellular virions. Moreover, as for HBV, reverse transcription of FV genomes occurs during the late phase of infection (Linial, 1999).
Although IFN treatment was shown to inhibit HFV gene expression (Sabile et al., 1996), nothing is known about the mechanisms involved and especially about the IFN effectors implicated in this inhibition. Thus, we have tested in this report the capacity of three IFN mediators, PML, MxA and Mx1, to interfere with HFV replication. Our study indicates that PML, but not MxA or Mx1, confers a high resistance to HFV infection. We evaluated the action of PML or IFN on virus production from viral entry to the expression of viral antigens. Whereas neither PML nor IFN altered HFV entry, each inhibited viral antigen expression at the transcriptional level.
Results
Overexpression of PML, but not of Mx1 or MxA, confers resistance to HFV infection
PML expression as well as its localization in NBs are altered during some viral infections; however, infection with HFV does not alter either the expression of PML or its localization to NBs (data not shown). To test a possible antiviral effect of PML against HFV infection, U373 MG control cells transfected with the empty vector and U373 MG cells stably expressing PML, U373 MG-PML, were infected with HFV at a multiplicity of infection (m.o.i.) of 0.1. Three days post-infection, viral antigen expression was monitored by immunofluorescence using anti-HFV antisera. PML expression in U373 MG-PML or IFN-treated cells was associated with a lower expression of HFV antigens, as revealed by immunofluorescence (Figure 1A and data not shown). The effect of PML overexpression on this inhibition was confirmed by western blotting. To compare the rate of viral inhibition in U373 MG-PML cells with that observed in IFN-treated cells, U373 MG cells were pretreated for 1 day with 1000 U/ml IFNα. IFN-treated, U373 MG-PML and control cells were infected with HFV at a m.o.i. of 0.1. Total protein extracts from infected cells were analysed by western blotting using anti-Gag antibodies, 3 days post-infection. The same blots were reprobed with anti-Bet and anti-Actin antibodies (Figure 1B). In infected control cells, the Gag doublet at 72 and 68 kDa (Giron et al., 1997) was clearly detected 1 day post-infection and expression of both proteins increased during the course of infection (data not shown). Conversely, in cells overexpressing PML and in IFN-treated cells, expression of both 72 and 68 kDa Gag proteins was lower than that of control cells (Figure 1B). Whereas expression of Bet (62 kDa) and Tas was detected during infection of control cells, these proteins were not detected in U373 MG-PML or IFN-treated cells (Figure 1B and data not shown). Inhibition of HFV protein expression in cells expressing PML was comparable to that observed in IFN-treated cells (Figure 1B). To determine whether this inhibition was also encountered at a higher m.o.i. and in other cell types expressing PML, GP+E-86 hygro, GP+E-86-PML, CHO hygro and CHO-PML cells were infected at a m.o.i. of 0.1 and 1. At a low or high m.o.i., expression of Gag or Bet protein was lower in cells overexpressing PML compared with controls (Figure 1C and data not shown). Yet PML induction in a tetracycline-inducible cell line did not significantly alter HFV antigen expression (data not shown), suggesting that only high levels of PML expression interfere with HFV infection.
Fig. 1. Overexpression of PML confers resistance to infection by HFV. U373 MG neo, U373 MG-PML, U373 MG untreated or treated with 1000 U/ml IFNα for 24 h were infected with HFV at a m.o.i. of 0.1 for 3 days. (A) Immunofluorescence microscopy was performed with mouse anti-PML antibodies visualized with Texas red and rabbit anti-HFV antibodies followed by FITC labelling. (B) Twenty micrograms of protein extracts from infected U373 MG control, IFN-treated, U373 MG neo and U373 MG-PML cells were analysed by western blotting and revealed by polyclonal anti-Gag antibodies. The same blots were reprobed with monoclonal anti-Bet and polyclonal anti-Actin antibodies. (C) GP+E-86 hygro and GP+E-86-PML cells were infected at a m.o.i. of 0.1 or 1 for 3 days, 20 µg of protein extracts were analysed by western blotting and revealed by anti-HFV antibodies. The lower panel shows the Coomassie Blue-stained proteins. (D) Expression of Mx1 or MxA, two other IFN-induced mediators, did not confer resistance to HFV. NIH 3T3 neo, overexpressing Mx1 or MxA, were infected with HFV at a m.o.i. of 0.1 for 3 days; 20 µg of protein extracts were analysed by western blotting and revealed by anti-HFV antibodies. The lower panel shows the Coomassie Blue-stained proteins.
Since no IFN activity could be detected in the culture media from infected control or PML-overexpressing cells (data not shown), HFV resistance of PML-expressing cells could not be due to IFN induction following viral infection.
To quantify precisely the inhibitory effect of PML, we evaluated the reverse transcriptase (RT) activity in culture supernatants (reflecting the virus yield). U373 MG control cells, pretreated with 1000 U/ml IFNα, or PML-overexpressing cells, were infected with HFV at a m.o.i. of 0.1. Five days post-infection, RT activity was assayed in the supernatant of each cell culture. Compared with control infected cells (84 500 c.p.m. representing 100%), RT activity was reduced by >88% in cells expressing PML (9750 c.p.m.) or in cells pretreated with IFNα (5750 c.p.m.). Taken together, these results demonstrate that PML expression inhibits HFV replication.
Since inhibition of viral infection could involve IFN mediators other than PML, the effect of human MxA or mouse Mx1 expression was tested on HFV replication. Mx proteins are IFN-induced members of a guanosine triphosphatase superfamily. MxA or Mx1 function as mediators of the IFN-induced antiviral state (reviewed in Staeheli et al., 1993). Interestingly, we have shown previously that the nuclear organelles harbouring Mx1 are associated with or juxtaposed to PML NBs (Chelbi-Alix et al., 1995). 3T3 cells transfected with the empty vector or overexpressing MxA or Mx1 were infected with HFV at a m.o.i. of 0.1. Protein extracts from uninfected or infected cells were analysed by western blotting for HFV protein expression 3 days post-infection. Expression levels of Gag and Bet were similar in control 3T3, 3T3 MxA- and 3T3 Mx1-infected cells (Figure 1D). Moreover, MxA and Mx1 proteins did not alter HFV expression after infection at a lower m.o.i. or during a shorter period of infection (data not shown). These results demonstrate that MxA and Mx1 do not confer resistance to HFV, and therefore are unlikely to be implicated in the IFN-induced antiviral state against HFV.
Viral DNA and mRNA detection in PML-expressing cells and in IFN-treated cells
To know whether internalization of HFV was blocked by cells overexpressing PML, U373 MG control and U373 MG-PML cells were infected with HFV at a m.o.i. of 1. Three hours post-infection, unintegrated viral DNA was prepared from infected cells by the Hirt procedure (Hirt, 1967). Similar amounts of DNA from Hirt supernatant were analysed by Southern blotting and hybridization with full-length HFV DNA (Figure 2A). The unique fragment of 12 kb corresponding to HFV unintegrated DNA was detected at the same levels in cells overexpressing PML or in IFN-treated cells as in control cells (Figure 2A and data not shown). These results suggest that overexpression of PML or IFN treatment did not affect HFV entry, and that the effect of PML on the replication cycle takes place after virus adsorption and penetration.

Fig. 2. Viral DNA and mRNA in PML-expressing or in IFN-treated cells. (A) U373 MG neo and U373 MG-PML cells were infected with HFV for 3 h at a m.o.i. of 1. After extensive washing with cold PBS, cultures were treated with acetic acid (0.2 M, pH 2.5) containing 0.5 M NaCl for 6 min at 4°C in order to remove HFV on cell surface membrane. Similar amounts of DNA from Hirt supernatant were analysed by Southern blotting and hybridization with full-length HFV DNA. (B) Similar amounts of Hirt DNA from U373 MG neo, U373 MG-PML and U373 MG pretreated or not with 1000 U/ml IFNα, all infected with HFV for 30 h at a m.o.i. of 0.1, were analysed by Southern blotting using a full-length HFV probe. (C) Total cellular RNA was extracted from U373 MG controls, U373 MG-PML and U373 MG pretreated for 24 h with 1000 U/ml IFNα, all infected with HFV at a m.o.i. of 0.1 for 30 h. Twenty micrograms of RNA were analysed with HFV tas and GAPDH probes.
To assess whether the decrease in HFV expression occurred at the transcriptional level, DNA from Hirt supernatant and viral mRNA expression was analysed by Southern and northern blotting. For that purpose, Hirt supernatant DNA and total RNAs were extracted 30 h post-HFV infection (m.o.i. 0.1) from U373 MG controls, U373 MG-PML and U373 MG cells pretreated with IFNα. At the DNA level, the unique fragment of 12 kb was expressed less in cells overexpressing PML or in IFN-treated cells than in control cells (Figure 2B). At the RNA level, the 12 and 2.2 kb bands corresponding to the genomic and regulatory mRNA (tas and/or bet) transcripts, respectively, were easily detected in control infected cells, whereas no such signals were observed in PML-expressing or IFN-treated cells (Figure 2C). These results provide evidence that PML expression interferes with HFV mRNA and DNA synthesis.
Inhibition of Tas-mediated transactivation from the LTR and the internal promoter by PML: requirement of the PML RING domain
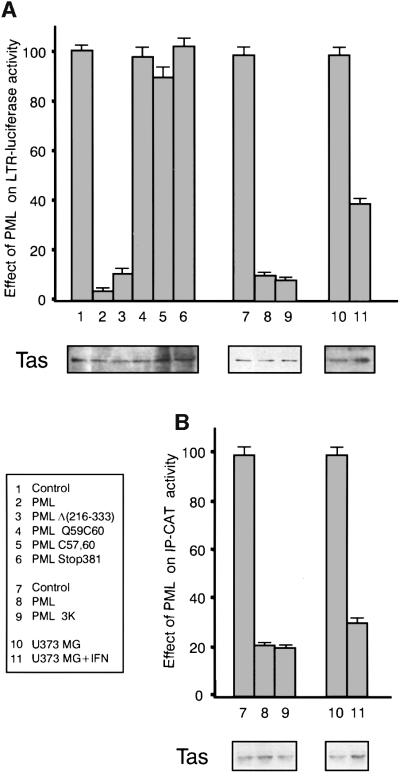
Tas activates viral gene expression from the 5′ LTR and from the internal promoter (IP) located at the 3′ end of the env gene. In order to assess whether PML expression could alter HFV primary transcription, the luciferase or the chloramphenicol acetyltransferase (CAT) reporter gene, under the transcriptional control of the HFV LTR (LTR-luc) or the IP (IP-CAT), respectively, was used in transactivation assays. CHO cells were transfected with both LTR-luc and tas or with IP-CAT and tas in the presence of the empty vector or PML. Luciferase or CAT activities were measured 48 h later (Figure 3). Compared with control cells, transactivation of the viral LTR was reduced by 90% in the presence of PML and by 60% in IFN-treated U373 MG cells (Figure 3A, lanes 1, 2, 7, 8, 10 and 11). IP transactivation was reduced by 84% in cells overexpressing PML and by 69% in IFN-treated cells (Figure 3B, lanes 7, 8, 10 and 11). In control transfections without tas, PML had no effect on the basal promoter activity from LTR-luc or IP-CAT constructs (data not shown). An equal fraction from all samples analysed by western blotting and revealed by anti-HFV antibodies shows that similar levels of Tas were detected in the transfected cells (Figure 3). These results suggest that PML acts as a repressor of Tas-mediated transactivation of HFV LTR and IP.
Fig. 3. Inhibition by PML of Tas-mediated transactivation of the HFV LTR and IP. CHO cells transiently co-transfected with LTR-luc and tas or with IP-CAT and tas and a plasmid expressing the empty vector (hygro), wild-type PML, PML mutant PML Δ(216–333), the RING finger PML mutants Q59C60 and C57,60, the C-terminal PML mutant (Stop381), PML 3K mutant or U373 MG. In all cases, pCMV βgal was used to monitor transfection efficiency. Twelve hours later, U373 MG cells were untreated or treated with 1000 U/ml IFNα. All cells were harvested 48 h post-transfection. Luciferase (A) and CAT (B) activities were measured. The graphs show the fold luciferase or CAT activity exhibited by the reporter constructs upon Tas transactivation in the absence or presence of PML. The data represent the average calculated from triplicates of two independent experiments and are expressed as a percentage of control cells (transfected with tas and LTR or IP) with the standard deviation indicated. Equivalent aliquots from all samples were analysed by western blotting for Tas expression.
To determine precisely the PML domains implicated in this phenomenon, similar experiments were performed with four PML mutants: the PML coiled-coil mutant (PMLΔ216–333), the C-terminal PML mutant (PML Stop381) and two PML RING finger mutants, PML Q59C60 and PML Cys57,60 (Figure 3A, lanes 3–6). While deletion of the coiled-coil domain leads to an altered nuclear localization characterized by a fine intranuclear network without speckles, the two RING finger PML mutants are nuclear diffuse, and PML Stop381 is mainly cytoplasmic (Chelbi-Alix et al., 1998). In the presence of Tas, the PML Stop381 mutant and both PML RING finger mutants did not alter either luciferase or CAT activities from the HFV LTR and IP, respectively, whereas the PMLΔ(216–333) coiled-coil mutant was as efficient as wild-type PML in inhibiting viral gene expression (Figure 3A, lanes 3–6 and data not shown). The N- and C-terminal regions of PML are not implicated in this repression since PML ΔN1–15 and PML Stop504 mutants still diminished Tas-induced LTR and IP transactivations (data not shown). As K490 is essential for the NLS function (Duprez et al., 1999), the SV40 NLS was fused to the N-terminal end of the PML mutant to generate PML 3K bearing a K to R mutation on lysines 65, 160 and 490. We showed that despite the absence of SUMO-1 conjugation sites in the PML 3K mutant, Tas-induced LTR and IP transactivations were still inhibited (Figure 3A and B, lanes 9). Similar results were obtained when we examined the effects of wild-type PML and PML mutants in a stable expression system on Tas-induced HFV LTR and IP transactivation (data not shown). Thus, the nuclear localization of PML and its RING finger motif, but not its modification by SUMO-1, its coiled-coil domain or its N- or C-terminal regions, are required for its transcriptional repression activity.
Tas co-localizes with PML in the NBs and they interact physically
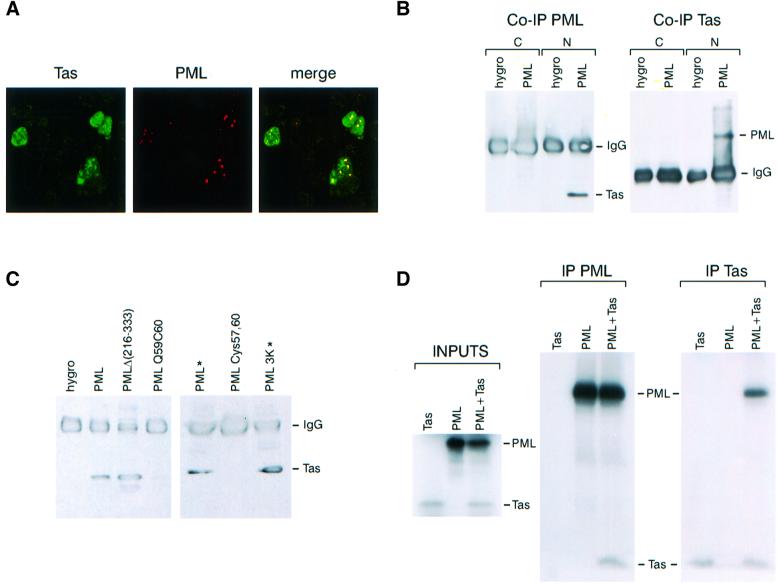
Given PML’s specific inhibition of Tas-mediated activity on HFV promoters, a possible interaction between PML and Tas was investigated first by immunofluorescence. To test whether Tas co-localized with PML on NBs, tas and PML were transiently co-transfected into CHO cells. PML was detected using a monoclonal anti-PML antibody and visualized with Texas red labelling, while Tas was detected with rabbit anti-HFV antibodies, followed by anti-rabbit fluorescein isothiocyanate (FITC) staining. As revealed in Figure 4A, confocal immunofluorescence microscopic analysis demonstrated colocalization of PML and Tas within speckles in the NBs. This suggested that in the presence of PML, the diffuse Tas protein was recruited to NBs.
Fig. 4. (A) Tas co-localizes with PML on to NBs. Tas and PML were transiently transfected in CHO cells. After fixation, PML was detected using monoclonal anti-PML antibodies and visualized with Texas red labelling. Tas was detected with rabbit anti-HFV antibodies, followed by anti-rabbit FITC. The right panel shows two-colour overlay results, demonstrating co-localization of Tas and PML proteins (yellow). (B) Tas interacts with PML. CHO cells expressing the empty vector (hygro) or stably expressing PML (CHO-PML) were transfected with tas, lysed and fractionated into cytoplasmic (C) and nuclear (N) components. One portion of these fractions was immunoprecipitated with polyclonal anti-PML antibodies and subsequent western blot probed with anti-HFV antibodies (left panel). The second portion was immunoprecipitated with anti-HFV antibodies and subsequent westerns probed with serum anti-PML (right panel). (C) Implication of the PML RING domain in Tas–PML interaction. CHO cells stably expressing the empty vector (hygro), PML, the PML coiled-coil mutant Δ(216–333), the RING finger PML mutants Q59C60 and Cys57,60 were transfected with tas. CHO cells were co-transfected with tas and PML or the PML 3K mutant (*). Two days later, cells were lysed and nuclear fractions were immunoprecipitated with anti-PML antibodies and subsequent western blot probed with anti-HFV antibodies. (D) Immunoprecipitation of PML and Tas from transcribed–translated proteins. Expression vectors encoding Tas, PML or a mixture of both were in vitro translated in the presence of [35S]methionine using the TnT rabbit-coupled reticulocyte lysate system (Promega) and analysed by 10% SDS–PAGE (INPUTS). Equimolar amounts of labelled proteins were immunoprecipitated by anti-HFV antibodies or anti-PML antibodies. Bound proteins were analysed by 13% SDS–PAGE followed by autoradiography.
Given that Tas and PML proteins colocalize in the nucleus, we investigated whether PML and Tas could interact by co-immunoprecipitation. CHO cells stably expressing PML as well as control cells, both transfected with a Tas-expressing vector, were lysed and separated into cytoplasmic and nuclear fractions. One part of these fractions was immunoprecipitated with polyclonal anti-PML antibodies and analysed by western blotting using anti-HFV antibodies (Figure 4B, left panel). The second part was immunoprecipitated with anti-HFV antibodies and analysed by western blotting using anti-PML antiserum (Figure 4B, right panel). PML antiserum precipitated Tas and, reciprocally, HFV antiserum precipitated PML only from nuclear fractions derived from cells overexpressing PML and transfected with tas. Tas was not detected in samples from control cells transfected with tas and immunoprecipitated with anti-PML antibodies. As a negative control, blots were probed with anti-Actin antibodies, which was not immunoprecipitated with either anti-PML or anti-HFV antisera (data not shown).
We next analysed the PML domains implicated in these protein interactions. As expected from the HFV LTR and IP inhibition studies (Figure 3), the PML RING finger mutants Q59C60 and Cys57,60 failed to interact with Tas (Figure 4C). Conversely, the PMLΔ(216–333) coiled-coil mutant still interacted with the viral transactivator. Finally, in agreement with our previous observations, the PML 3K mutant still interacted with Tas (Figure 4C), demonstrating that SUMO-1 modification is not necessary for Tas–PML interaction.
The direct interaction between PML and Tas was confirmed by immunoprecipitation of PML and Tas from transcribed–translated proteins (Figure 4D). Equimolar amounts of labelled proteins were immunoprecipitated by anti-HFV or anti-PML antibodies. It was possible to immunoprecipitate both proteins from a mixture of PML and Tas, using either anti-HFV or anti-PML antibodies (Figure 4D), strongly suggesting that Tas and PML interact directly.
LTR-induced transactivation by Tas mutants in the absence or presence of PML
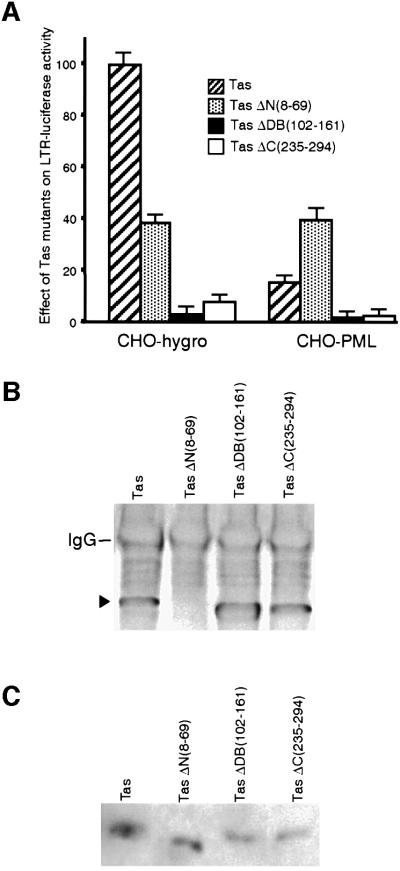
To delineate the PML binding domain in Tas, three Tas mutants were generated: Tas N-teminal mutant TasΔN (8–69), Tas DNA binding mutant TasΔDB(102–161) and Tas C-terminal mutant TasΔC(235–294). All these mutants are nuclear since they still contain the NLS spanning residues 209–226. The properties of these Tas mutants were evaluated in the absence or presence of PML. Cells stably expressing either the empty vector or PML were transfected with LTR-luc and either Tas, TasΔN(8–69), TasΔDB(102–161) or TasΔC(235–294) and harvested 48 h post-transfection to quantify the luciferase activity (Figure 5A). The Tas mutants deleted in the DNA binding domain, TasΔDB(102–161), or in the transactivation region located in the C-terminal region, TasΔC(235–294), were unable to induce LTR-derived expression, whereas the Tas mutant deleted in the N-terminal region, TasΔN(8–69), still transactivates HFV LTR by 40%. Interestingly, in this case, expression of PML did not alter this transactivation (Figure 5A), suggesting that PML could interact with the N-terminal region of Tas. Similar results were obtained when we examined the effect of wild-type PML and Tas mutants on HFV LTR in a cotransfection system (data not shown). To assess this possibility, nuclear extracts from CHO-PML cells transfected with Tas, TasΔN(8–69), TasΔDB(102– 161) or TasΔC(235–294) were immunoprecipitated with polyclonal human anti-PML antibodies and analysed by western blotting using anti-HFV antibodies. PML interaction was observed with Tas, TasΔDB(102–161) or TasΔC(235–294) but not with TasΔΝ(8–69) (Figure 5B). As observed in Figure 5C, all mutants are correctly expressed and migrated at the expected sizes.
Fig. 5. (A) LTR transactivation by Tas mutants in the absence or presence of PML. CHO cells expressing the empty vector (hygro) or CHO-PML were transfected with LTR and Tas, TasΔN(8–69), TasΔDB(102–161) or TasΔC(235–294). In each case, pCMV βgal was used to monitor transfection efficiency. All cells were harvested 48 h post-transfection and luciferase activity was measured. The data represent averages calculated from triplicates of three independent experiments and are expressed as a percentage of control cells (transfected with tas and LTR) with the standard deviation indicated. (B) The N-terminal region of Tas is necessary for Tas–PML interaction. CHO-PML were transfected with Tas, TasΔN(8–69), TasΔDB(102–161) or TasΔC(235–294). Two days later, cells were lysed and nuclear fractions were immunoprecipitated with polyclonal anti-PML antibodies and subsequent western blot probed with anti-HFV antibodies. (C) Expression of Tas mutants corresponding to the INPUTS of (B).
These results suggest that PML disturbs Tas-induced HFV transactivation by forming a complex between its RING finger domain and the N-terminal region of Tas. Since Tas and Bet share their N-terminal region (88 common N-terminal amino acids) (Lecellier and Saib, 2000), we examined the interaction between Bet and PML. We also observed by co-immunoprecipitation an interaction between these two proteins (data not shown).
PML inhibits the binding of Tas to the LTR and IP
It was previously shown by electrophoretic mobility shift assay (EMSA) that Tas binds directly to the HFV LTR and IP (He et al., 1996). Therefore, DNA binding studies were carried out with Tas binding sequences in extracts from cells stably expressing the empty vector, PML and PML RING finger mutants Q59C60 and Cys57,60. The complexes formed in the extracts from cells transfected with tas, which bound to the LTR (Figure 6A, lane 2) or IP sequences (Figure 6B, lane 10), were supershifted with mouse polyclonal anti-Tas antibodies (Figure 6A, lane 4 and B, lane 12). These complexes could be removed by competing with unlabelled LTR (Figure 6A, lane 3) or IP probes (Figure 6B, lane 11). Expression of PML diminished the complex formation between LTR and Tas (Figure 6A, lane 5) and IP and Tas (Figure 6B, lane 13), whereas both PML RING mutants Q59C60 and Cys57,60 had no effect on their formation (Figure 6A, lanes 7 and 8, and B, lanes 14 and 15). An equal fraction from EMSA samples, analysed by western blotting with anti-HFV antibodies, revealed that all cells expressed similar quantities of Tas (Figure 6C).
Fig. 6. (A) Binding of Tas to the LTR or IP sequences in the absence or presence of PML. EMSA was performed with extracts from CHO-hygro, CHO-PML, CHO-PML Q59C60 and CHO-PML Cys57,60 transfected with tas. Radiolabelled sequences derived from HFV LTR or IP were used and super-shifting was performed with mouse polyclonal anti-Tas antibodies. (B) Tas expression in EMSA samples. Equivalent aliquots from EMSA samples were analysed by western blotting and revealed by anti-HFV antibodies. (C) Expression of Tas corresponding to the inputs of (A) and (B).
Taken together, these studies indicate that interaction with PML leads to a direct inhibition of the DNA binding property of Tas to the LTR and to the IP.
Response to HFV infection after IFN treatment in PML–/– and parental embryonic fibroblasts
PML knockout mice are viable, but are susceptible to tumour development and to some infections (Wang et al., 1998a,b). To test further the role of PML in the antiviral effect of IFN against HFV, wild-type mouse embryonic fibroblasts (wild-type MEF) and PML–/– MEF (Wang et al., 1998a), immortalized by the SV40 large T antigen, were analysed for their resistance to viral infection upon IFN treatment. Untreated cells or cells pretreated with IFNα were infected with HFV at a m.o.i. of 0.1. Viral proteins were analysed by western blotting 72 h post-infection. In wild-type MEF, IFNα inhibited viral protein synthesis. This effect was almost completely abrogated in PML–/– MEF (Figure 7A). Mouse PKR was similarly induced by IFNα in wild-type and PML–/– MEF (Figure 7A), indicating no deficiency in the IFNα pathway. In addition, Tas-mediated LTR and IP transactivation were repressed by IFNα treatment in different cell lines, including wild-type MEF, but not in PML–/– MEF (Figures 4 and 7B). However, introducing PML into PML–/– MEF by transient expression repressed these transactivations in a dose-dependent manner (Figure 7B).
Fig. 7. (A) Response to HFV infection after IFN treatment in wild-type and PML–/– MEF. Wild-type and PML–/– MEF untreated or treated with 1000 U/ml mouse IFNα for 24 h were infected with HFV at a m.o.i. of 0.1. Two days post-infection, western blots were performed using polyclonal anti-HFV, anti-Actin or anti-PKR antibodies. (B) Effect of IFN or PML on Tas-mediated transactivation of LTR in MEF cells. Left panel: wild-type and PML–/– MEF were transfected with LTR–luc and tas. Twelve hours later, cells were untreated or treated with 1000 U/ml IFNα. Right panel: PML–/– MEF were transfected with LTR–luc and tas, and increasing concentrations of PML. In all cases, pCMV βgal was used to monitor transfection efficiency. Cells were harvested 48 h post-transfection and luciferase activity was measured. The data represent averages calculated from triplicates of three independent experiments and are expressed as a percentage of control cells (transfected with tas and LTR) with the standard deviation indicated. Equivalent aliquots from all samples were analysed by western blotting for Tas expression.
Taken together, these findings indicate that PML is efficient in inhibiting the replication of this virus and is an essential mediator of HFV inhibition by IFN.
Discussion
Any stage in virus replication may be a target for inhibition by IFNs, including entry, transcription, RNA stability, initiation of translation, maturation, assembly and release (reviewed in Stark et al., 1998). All activities of IFNs are believed to be mediated by IFN-regulated cellular proteins. Although the physiological functions of the majority of IFN-induced proteins remain unclear, several of them, namely the double-stranded protein kinase (PKR), the 2′5′ oligoadenylate (2′5′A) synthetase, certain Mx proteins and PML, have been shown to display intrinsic antiviral activities (reviewed in Staeheli et al., 1993; Stark et al., 1998). Both 2′5′A synthetase and PKR pathways, the best characterized, lead to inhibition of protein synthesis and require dsRNA for their activation. Overexpression of 2′5′A synthetase or PKR confers resistance to encephalomyocarditis virus (EMCV). The Mx proteins have an intrinsic GTPase activity necessary for their intracellular antiviral actions, and interfere with virus replication at the level of virus transcription and other steps in the virus life cycle. The human cytoplasmic MxA protein is active against several RNA virus families, including Orthomyxoviridae, Paramyxoviridae, Rhabdo viridae, Bunyaviridae and Togaviridae. The murine nuclear Mx1 inhibits the replication of members of the Orthomyxoviridae. We have shown previously that PML overexpression induces resistance to infections by VSV and influenza virus (Chelbi-Alix et al., 1998). However, PML has a less powerful antiviral activity against these viruses than the IFN mediator MxA.
IFNs play a protective role against lytic replication of HFV by inhibiting viral protein and RNA synthesis (Sabile et al., 1996). Nevertheless, their effects on primary transcription and the IFN mediators implicated in this antiviral state were unknown. HFV infection does not alter either PML localization in NBs or its expression. Moreover, we provide, in this study, evidence that overexpression of PML, but not that of Mx1 or MxA, confers resistance to HFV, suggesting that PML participates in the IFN-induced antiviral state. The degree of inhibition of HFV replication by PML was similar to that obtained in control cells treated with 1000 U/ml IFNα. It should be noted that HFV infection did not induce IFN secretion in control or PML-expressing cells (Sabile et al., 1996; data not shown). Upon HFV infection, the characteristic syncytial effect was observed in control cells, but not in cells expressing PML or in cells pretreated with IFN (data not shown). This effect reflected a decrease in viral mRNA, DNA, protein synthesis and RT activity. PML expression in transfected and in IFN-treated cells inhibited Tas-mediated transactivation of the HFV LTR and the internal promoter. The role of PML in IFN-induced antiviral action against HFV was further demonstrated by the inability of IFN to inhibit HFV replication and to diminish Tas-mediated LTR transactivation in MEF PML–/–. Introducing PML into this cell line by transient expression repressed this transactivation in a dose-dependent manner.
Note that the expression of the human PKR or the catalytically inactive form of PKR (PKR K-R296) (Meurs et al., 1995) did not affect Tas-mediated transactivation of the HFV LTR (data not shown). Mouse PKR was similarly induced by IFN in wild-type and PML–/– MEF; however, IFNα inhibited viral protein synthesis much more in wild-type MEF. Taken together, these results suggest that PKR could not be implicated in the antiviral state against HFV in response to IFN.
The RING domain of PML and its nuclear localization, but not its modification by SUMO-1, are necessary for its transcriptional repression activity. PML, from the nuclear diffuse compartment or from the NBs, exerts its repression via its interaction with the N-terminal region of Tas. This interaction has been observed with Bet. This is not surprising since Tas and Bet share their N-terminal region (88 common N-terminal amino acids) (Lecellier and Saib, 2000). The RING finger of PML is implicated in protein–protein interaction. Neither a RING finger nor a coiled-coil motif has been detected by protein sequence analysis in the N-terminal region of Tas.
There is evidence demonstrating that PML regulates either negatively or positively the expression of target genes at the transcription level (reviewed in Ruggero et al., 2000; Zhong et al., 2000). The RBCC motif, shared by PML and other RING proteins, seems to prevent Tas from having direct contact with DNA. However, it is becoming apparent that PML can control gene expression through regulation of transcriptional activity of transcription factors. For instance, PML interacts with SP1 and interferes with its ability to bind DNA, leading to negative regulation of the SP1 target gene EGFR (Vallian et al., 1998). PML also interacts directly with non-phosphorylated pRb, exerting a negative effect on pRb-mediated transactivation (Alcalay et al., 1998).
In this report, we show that PML interacts directly with Tas, and interferes with its ability to bind HFV LTR and IP, leading to the repression of Tas-induced transactivation and inhibition of HFV replication in PML-expressing and IFN-treated cells. As far as we know, PML is the first example of an IFN-induced protein that inhibits the func tion of a viral transactivator through a physical interaction. The RING domain of PML, its nuclear localization, but not its coiled-coil domain or its SUMOlation, are required for the repression of the transcription, suggesting that both forms of PML, the nucleoplasmic and the NB associated, are active. This is in contrast to a recently proposed mechanism involving SUMO-1 modification of PML for efficient inhibition of Daxx-mediated transcriptional repression (Li et al., 2000). It could be interesting to determine whether other functions attributed to PML (i.e. apoptosis, tumour suppression, senescence) are maintained when PML is no longer localized on NBs. These results indicate that PML plays a key role in mediating the IFN-induced antiviral state against HFV. PML did not decrease the transactivation of HIV-1 or HTLV-1 expression by Tat or Tax, respectively, whereas transactivation of HIV-1 by Tas was reduced by 64% (data not shown). However, it remains to be studied whether PML could interfere with the replication of HIV-1 or HTLV-I through other mechanisms.
Materials and methods
Cell cultures and antibodies
Human glioblastoma astrocytoma U373 MG and WISH cells, CHO cells, mouse GP+E-86 and L929 cells, wild-type MEF and PML–/– MEF (Wang et al., 1998a), immortalized by the SV40 large T antigen, were grown at 37°C in Dulbecco’s modified Eagle’s medium. All cells were supplemented with 10% fetal calf serum. CHO and GP+E-86 cells (transfected with the empty vector, PML or PML mutants encoding vectors) were kept in medium supplemented with 0.5 mg/ml hygromycin (Gibco). U373 MG control (transfected with the empty vector) or overexpressing PML, were kept in medium supplemented with 0.5 mg/ml G418. Swiss 3T3 mouse cells transfected by the empty vector or vectors expressing MxA or Mx1(Pavlovic et al., 1990) were a kind gift from J.Pavlovic. They were grown in the same medium supplemented with 0.5 mg/ml G418. Mouse and rabbit anti-human PML antibodies were as described (Daniel et al., 1993). Polyclonal anti-HFV antibodies produced in rabbit after HFV infection, polyclonal anti-Gag, mouse polyclonal anti-Tas and monoclonal anti-Bet antibodies were described elsewhere (Giron et al., 1997). Rabbit anti-mouse PKR antibodies were from Santa Cruz.
IFNs and determination of IFN titres
Human IFNα was from Schering-Plough Research Institute. Mouse cells were treated with either mouse IFNα (NIH; Ga02-901-51) or human IFNα1–8 (provided by M.Tovey, Villejuif), which is active on mouse cells. Culture media from HFV-infected mouse GP+E-86 control and GP+E-86-PML cells were titrated on L929 cells, and those from U373 MG control and U373 MG-PML on WISH cells. All cells were challenged with VSV. IFN titres were determined as the amounts of IFN required to produce 50% inhibition of the cytopathic effect (CPE).
Virus stocks and virus yield assay
HFV stocks were made after infection at a m.o.i. of 1 of the U373 MG cells. About 1 week later, when specific HFV CPEs were detectable in ∼75% of the infected monolayer, cultures were frozen, thawed three times and cell debris removed by centrifugation. Supernatants were filtered, aliquoted and kept at –70°C. Stock HFV yields were determined as previously (Santillana-Hayat et al., 1993) and are expressed as tissue culture infectious dose (ID)50/ml (106).
RT activity assay
HFV RT activity in supernatant fluids from control, PML-overexpressing or IFN-treated cells was measured using template poly(A)–oligo(dT) and divalent cation Mn2+, as described previously (Périès et al., 1979). Values represent the average of results from triplicate cultures, expressed as c.p.m. × 10–3 of [3H]TMP incorporated into poly(dT) product in 60 min.
Preparation and analysis of nucleic acids
Total RNA was extracted with the total RNA extraction kit (Quantum Bioprobe). DNA was prepared from infected cells by the Hirt procedure (Hirt, 1967) and treated with pancreatic RNase. RNAs and Hirt extracts, DNA denatured, were separated on 1% agarose. After blotting on nitrocellulose membranes (Schleicher & Schuell), northern blot analysis was performed with random priming (Promega) radiolabelled tas or GAPDH as probes, and Southern blotting with full-length HFV probe.
Transient transfection
Cells (4 × 104/ml) were seeded in six-well plates and transfected with 2 µg of plasmid DNA using the Fugene 6 transfection reagent (Roche) according to the manufacturer’s protocol. To avoid discrepancy in transfection efficiency, cells were also transfected with a pCMV β-galactosidase-encoding plasmid used to normalize luciferase or CAT activity. The medium was changed 24 h later and cells were untreated or treated with 1000 U/ml IFNα. One day later, cells were harvested and luciferase assay was performed using the Promega luciferase assay system according to the supplier’s instructions. The CAT assay was performed as described previously (Gorman et al., 1982). CAT activity was quantified with a phosphoimager. All experiments were prepared in triplicate and repeated twice. The results were normalized against β-galactosidase activity and expressed as a percentage.
Construction of expression vectors and cell lines
Stable PML-expressing U373 MG clones were obtained via transfection with the pCIN-PML construct and subsequent neomycin selection (final concentration 0.5 mg/ml) (Chelbi-Alix et al., 1998). Stable PML CHO or GP+E-86 clones and stable PML mutant CHO clones: PML Stop381; the coiled-coil PML mutant, PMLΔ(216–333); and the RING finger PML mutant, Q59C60 were obtained by lipofection (Gibco-BRL) with pSG5 constructs co-transfected with DSP-hygro- or M3P-SVhygro-derived constructs and subsequent hygromycin selection (Chelbi-Alix et al., 1998). The RING finger PML mutant, PML Cys57,60 (Duprez et al., 1999), was stably expressed in CHO clones generated by co-transfection with DSP-hygro. His tag (CATGCATCACCACCATCACCATTC) and SV40-NLS (CATGGCGCTCCCAAAAAGAAAAGAAAGGT) annealed oligonucleotides were cloned into the ATG–NcoI site of PML-pSG5 plasmid. PML 3K mutant, where SUMO-1 sites (Lys 65, 160 and 490) were mutated from K to R on His(NLS)PML-pSG5, was used in transient transfection. The IP-CAT plasmid was a kind gift from R.Flügel and M.Löchelt (Löchelt et al., 1993), and LTR-luc as well as the vector expressing Tas cloned into the psvSPORT expression vector have been described elsewhere (de Célis-Kosmas et al., 1997). Note that the expression vector pSportTas harbours the AGTAAG (Ser-Arg) to TCT AGA (Ser-Lys) mutation at the Bet donor splice site, avoiding the synthesis of a truncated Bet protein.
Mutagenesis of HFV Tas
The tas gene of HFV was cloned by inserting the 1 kb SspI fragment of pHSRV13 into the SmaI site of pSV-SportI (BRL) as described previously (de Célis-Kosmas et al., 1997). The following primers were used to generate Tas deletion mutants using the QuickChange site-directed mutagenesis kit (Stratagene): direct 5′-GATTCCTACGAAAAAGAAGACAAACATCCTCAACAT-3′, reverse 5′-ATGTTGAGGATGTTTGTCTTCTTTTTCGTAGGAATC-3′ for TasΔN(8–69); direct 5′-CTTATTTTATGTGGATTGCCCCTCAGAGGAATTGTT-3′, reverse 5′-AACAATTCCTCTGAGGGGCAATCCACATAAAATAAG- 3 for TasΔDB(102–161); and direct 5′-TCATGTGCTTCCAGTAGTGGTGAACATTCAGTTTTA-3′, reverse 5′-TAAAACTGAATGTTCACCACTACTGGAAGCACATGA-3′ for TasΔC(235–294). Mutations were verified by sequence analysis and expression by western blot analysis.
Immunoprecipitation assays
Transfected cells (107) were incubated for 30 min at 4°C in buffer containing 10 mM Tris–HCl pH 7.4, 100 mM NaCl, 2 mM EDTA, 1% NP-40, 0.5% desoxycholate (DOC) and 1 mM phenylmethylsulfonyl fluoride (PMSF). Cell extracts were centrifuged; the supernatants saved constitute the cytoplasmic fractions and the pellets lysed in buffer (20 mM Tris–HCl pH 7.4, 1 M NaCl, 5 mM MgCl2, 1% Triton, 0.5% DOC and 1 mM PMSF) constitute the nuclear fractions. PML and Tas were immunoprecipitated in separate experiments. Fractions were precleared with anti-rabbit serum. Protein G beads (Sigma) were added to supernatants in IB buffer (50 mM Tris–HCl pH 7.4, 200 mM NaCl, 0.5% DOC, 1% Triton X-100, 0.1% SDS and 10 mM EDTA) and mixed for 2 h at 4°C. Rabbit anti-PML or anti-HFV antibodies were added and incubated overnight at 4°C. Beads were washed four times with modified IB buffer (1 M NaCl). Beads were subjected to SDS–PAGE.
Expression vectors encoding Tas, PML or a mixture of Tas and PML were in vitro translated in the presence of [35S]methionine using the TnT rabbit-coupled reticulocyte lysate system (Promega). Equimolar amounts of labelled proteins were immunoprecipitated by anti-HFV or anti-PML antibodies. Binding assays were carried out in buffer (50 mM Tris–HCl pH 7.4, 150 mM NaCl, 0.5% DOC, 1% Triton X-100, 0.1% SDS and 10 mM EDTA). Beads were washed in the same buffer. Bound proteins were analysed by 13% SDS–PAGE and autoradiography.
Immunofluorescence and western blot analysis
Cells were fixed in 4% paraformaldehyde for 15 min at 4°C. Immunofluorescence was revealed with monoclonal anti-PML antibodies, followed by Texas red-conjugated secondary antibodies and with polyclonal anti-HFV antibodies followed by FITC-conjugated secondary antibodies. For western blot analysis, cells were washed and resuspended in phosphate-buffered saline (PBS), lysed in hot Laemmli sample buffer and boiled for 5 min. Approximately 20 µg of protein were analysed on a 10% SDS–polyacrylamide gel, and transferred to nitrocellulose. Membranes were blocked with 10% skimmed milk in Tris-buffered saline for 2 h and incubated overnight at 4°C with rabbit polyclonal anti-PML (1/3000), anti-HFV (1/1000), anti-Gag (1/500), monoclonal anti-Bet (1/500) or anti-actin (1/500) antibodies. After incubation with primary antibodies, blots were washed extensively in PBS–Tween and incubated for 1 h with the appropriate peroxidase-coupled secondary antibodies (Amersham). All blots were revealed by chemoluminescence (ECL; Amersham). The membranes were reprobed if necessary; for this, they were incubated in 0.1 M glycine pH 2.9 for 30 min, washed twice in PBS–Tween and then blocked and incubated as described. To estimate the apparent molecular mass of polypeptides, prestained molecular weight standards (Bio-Rad Laboratories, Richmond, CA) were used.
EMSA
Stable CHO cell lines expressing empty vector, PML, PML Q59C60 or PML Cys57,60 were transfected with a tas expression vector. Two days later, cells were lysed in 0.3 M NaCl for 30 min at 4°C, sonicated and harvested by centrifugation at 14 000 g. The supernatants were aliquoted and kept at –20°C. HFV LTR and IP [double-stranded oligonucleotides that have been shown previously to bind to Tas (He et al., 1996)] were synthesized by oligo express. HFV LTR sense: 5′-GCAGCTTTTTATCCACTAGG GATAATGTTTTAAGGAATACTATAGTAATAGATTGATAGTTTTAACAATGATAGA-3′; HFV IP sense: 5′-AGGCCACTGGTTGCGGAAGAAAGATTG-3′. Probes were purified, radiolabelled and used in EMSA.
Acknowledgments
Acknowledgements
We acknowledge J.Pavlovic for 3T3 MxA and 3T3 Mx1 cells, E.Duprez and P.Freemont for the RING finger PML Cys57,60 mutant, and E. Meurs for the human PKR and the catalytically inactive form of PKR (PKR K-R296). The help of B.Boursin for the artwork, M.Schmid for confocal microscopy analysis and C.Chopin for technical assistance was highly appreciated. We want to thank all our colleagues for help and stimulating discussions, and R.Pine for the critical reading of the manuscript. We also thank Schering-Plough Research Institute for providing human IFNα. This work was supported by grants from the Association pour la Recherche sur le Cancer 5250.
References
- Alcalay M., Tomassoni,L., Colombo,E., Stoldt,S., Grignani,F., Fagioli,M., Szekely,L., Helin,K. and Pelicci,P.G. (1998) The promyelocytic leukemia gene product (PML) forms stable complexes with the retinoblastoma protein. Mol. Cell. Biol., 18, 1084–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelbi-Alix M. and de Thé,H. (1999) Herpes virus induces proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100. Oncogene, 18, 935–941. [DOI] [PubMed] [Google Scholar]
- Chelbi-Alix M.K., Pelicano,L., Quignon,F., Koken,M.H.M., Venturini,L., Stadler,M., Pavlovic,J., Degos,L. and de Thé,H. (1995) Induction of the PML protein by interferons in normal and APL cells. Leukemia, 9, 2027–2033. [PubMed] [Google Scholar]
- Chelbi-Alix M.K., Pelicano,L., Quignon,F., Koken,M.H.M. and de Thé,H. (1996) PML is a primary target gene of interferon and could mediate some of its biological activities. Tumor Biol., 99, 17–27. [Google Scholar]
- Chelbi-Alix M.K., Quignon,F., Pelicano,L., Koken,M.H.M. and de Thé,H. (1998) Resistance to virus infection conferred by the interferon-induced promyelocytic leukemia protein. J. Virol., 72, 1043–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel M.-T. et al. (1993) PML protein expression in hematopoietic and acute promyelocytic leukemia cells. Blood, 82, 1858–1867. [PubMed] [Google Scholar]
- de Célis-Kosmas J., Coronel,A., Grigorian,I., Emanoil-Ravier,R. and Tobaly-Tapiero,J. (1997) Non-random deletions in human foamy virus long terminal repeat during viral infection. Arch. Virol., 142, 1237–1246. [DOI] [PubMed] [Google Scholar]
- de Thé H., Lavau,C., Marchio,A., Chomienne,C., Degos,L. and Dejean,A. (1991) The PML–RARα fusion mRNA generated by the t(15;17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell, 66, 675–684. [DOI] [PubMed] [Google Scholar]
- Duprez E. et al. (1999) SUMO-1 modification of the acute promyelocytic leukaemia protein PML: implications for nuclear localisation. J. Cell Sci., 112, 381–393. [DOI] [PubMed] [Google Scholar]
- Dyck J.A., Maul,G.G., Miller,W.H., Chen,J.D., Kakizuka,A. and Evans,R.M. (1994) A novel macromolecular structure is a target of the promyelocyte–retinoic acid receptor oncoprotein. Cell, 76, 333–343. [DOI] [PubMed] [Google Scholar]
- Everett R. and Maul,G. (1994) HSV-1 protein Vmw110 causes redistribution of PML. EMBO J., 13, 5062–5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett R., Lomonte,P., Sternsdorf,T., van Driel,R. and Orr,A. (1999a) Cell cycle regulation of PML modification and ND10 composition. J. Cell Sci., 112, 4581–4588. [DOI] [PubMed] [Google Scholar]
- Everett R., Meredith,M. and Orr,A. (1999b) The ability of herpes simplex virus type 1 immediate early protein Vmw110 to bind to a ubiquitin-specific protease contributes to its roles in the activation of gene expression and stimulation of virus replication. J. Virol., 73, 417–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gäken J., Farzaneh,F., Stocking,C. and Osterlag,W. (1992) Construction of a versatile set of retroviral vectors conferring hygromycin resistance. Biotechniques, 13, 32–33. [PubMed] [Google Scholar]
- Giron M.-L., Colas,S., Wybier,J., Rozain,F. and Emanoil-Ravier,R. (1997) Expression and maturation of human foamy virus gag precursor polypeptides. J. Virol., 71, 1635–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard A.D., Borrow,J., Freemont,P.S. and Solomon,E. (1991) Characterization of a zinc finger gene disrupted by the t(15;17) in acute promyelocytic leukemia. Science, 254, 1371–1374. [DOI] [PubMed] [Google Scholar]
- Gongora C., David,G., Pintard,L., Tissot,C., Hua,T.D., Dejean,A. and Mechti,N. (1997) Molecular cloning of a new interferon-induced PML nuclear body-associated protein. J. Biol. Chem., 272, 19457–19463. [DOI] [PubMed] [Google Scholar]
- Gorman C.M., Moffat,L.F. and Howard,B.H. (1982) Recombinant genomes which express chloramphenicol acetyl-transferase in mammalian cells. Mol. Cell. Biol., 2, 1044–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grötzinger T., Jensen,K. and Will,H. (1996) The interferon (IFN)-stimulated gene Sp100 promoter contains an IFN-γ activation site and an imperfect IFN-stimulated response element which mediate type I IFN inducibility. J. Biol. Chem., 271, 25253–25260. [DOI] [PubMed] [Google Scholar]
- Guldner H., Szostecki,C., Grotzinger,T. and Will,H. (1992) IFN enhances expression of Sp100, an autoantigen in primary biliary cirrhosis. J. Immunol., 149, 4067–4073. [PubMed] [Google Scholar]
- He F., Sun,J.D., Garret,E.D. and Cullen,B.R. (1993) Functional organization of the Bel-1 trans activator of human foamy virus. J. Virol., 67, 1896–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F., Blair,W.S., Fukushima,J. and Cullen,B.R. (1996) The human foamy virus bel-1 transcription factor is a sequence-specific DNA binding protein. J. Virol., 70, 3902–3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt B. (1967) Selective extraction of polyoma DNA from infected mouse cells cultures. J. Mol. Biol., 26, 365–369. [DOI] [PubMed] [Google Scholar]
- Ishov A., Sotnikov,A., Negorev,D., Vladimirova,O., Neff,N., Kamitani,T., Yeh,E., Strauss,J.,III and Maul,G. (1999) PML is critical for ND10 formation and recruits the PML-interacting protein Daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol., 147, 221–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakizuka A., Miller,W.,Jr, Umesono,K., Warrell,R.,Jr, Frankel,S.R., Murty,V.V., Dmitrovsky,E. and Evans,R.M. (1991) Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RARα with a novel putative transcription factor, PML. Cell, 66, 663–674. [DOI] [PubMed] [Google Scholar]
- Kamitani T., Kito,K., Nguyen,H.P., Wada,H., Fukuda-Kamitani,T. and Yeh,E.T.H. (1998) Identification of three major sentrinization sites in PML. J. Biol. Chem., 41, 26675–26682. [DOI] [PubMed] [Google Scholar]
- Koken M.H.M. et al. (1994) The t(15;17) translocation alters a nuclear body in a RA-reversible fashion. EMBO J., 13, 1073–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koken M.H.M. et al. (1995) The PML growth-suppressor has an altered expression in human oncogenesis. Oncogene, 10, 1315–1324. [PubMed] [Google Scholar]
- Lecellier C.H. and Saib,A. (2000) Foamy viruses: between retroviruses and pararetroviruses. Virology, 271, 1–8. [DOI] [PubMed] [Google Scholar]
- Lee S.W., Chang,J., Lee,C.W., Kim,D.H., Choi,K.Y. and Sung,Y.C. (1995) Mutational analysis of human foamy virus Bel1 activation domain. Mol. Cell, 5, 467–474. [Google Scholar]
- Li H., Leo,C., Zhu,J., Wu,X., O’Neil,J., Park,E.J. and Chen,J.D. (2000) Sequestration and inhibition of Daxx-mediated transcriptional repression by PML. Mol. Cell. Biol., 20, 1784–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linial M.L. (1999) Foamy viruses are unconventional retroviruses. J. Virol., 73, 1747–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.-H., Mu,Z.-M. and Chang,K.-S. (1995) PML suppresses oncogenic transformation of NIH/3T3 cells by activated neu. J. Exp. Med., 181, 1965–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löchelt M., Muranyi,W. and Flügel,R.M. (1993) Human foamy virus genome possesses an internal, bel1-dependent and functional promoter. Proc. Natl Acad. Sci. USA, 90, 7317–7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul G., Negorev,D., Bell,P. and Ishov,A. (2000) Review: properties and assembly mechanisms of ND10, PML bodies or PODs. J. Struct. Biol., 129, 278–287. [DOI] [PubMed] [Google Scholar]
- Meurs E.F., McMillan,N., William,B.R.G., Hovanessian,A.G. and Southern,P.J. (1995) Human PKR transfected into murine cells stimulates expression of genes under control of the HIV-1 or HTLV-I LTR. Virology, 214, 653–659. [DOI] [PubMed] [Google Scholar]
- Muller S., Matunis,M.J. and Dejean,A. (1998) Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J., 17, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S., Berger,M., Lehembre,F., Seeler,J.S., Haupt,Y. and Dejean,A. (2000) c-Jun and p53 activity is modulated by SUMO-1 modification. J. Biol. Chem., 275, 13321–13329. [DOI] [PubMed] [Google Scholar]
- Pandolfi P.P., Grignani,F., Alcalay,M., Mencarelli,A., Biondi,A., LoCoco,F., Grignani,F. and Pelicci,P.G. (1991) Structure and origin of the acute promyelocytic leukemia myl/RARα cDNA and characterization of its retinol-binding and transactivation properties. Oncogene, 6, 1285–1292. [PubMed] [Google Scholar]
- Pavlovic J., Zurcher,T., Haller,O. and Staeheli,P. (1990) Resistance to influenza virus and vesicular stomatitis virus conferred by expression of human MxA protein. J. Virol., 64, 3370–3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Périès J., Canivet,M., Rhodes-Feuillette,A. and Todaro,G. (1979) Effect of interferon on chronic infection of human cells by xentropic type C viruses. Int. J. Cancer, 2, 798–802. [DOI] [PubMed] [Google Scholar]
- Quignon F., De Bels,F., Koken,M., Feunteun,J., Ameisen,J. and de Thé,H. (1998) PML induces a novel caspase-independent death process. Nature Genet., 20, 259–265. [DOI] [PubMed] [Google Scholar]
- Rees S., Coote,J., Stables,J., Goodson,S., Harris,S. and Lee,M.G. (1996) Bicistronic vector for the creation of stable mammalian cell lines that predisposes all antibiotic-resistant cells to express recombinant protein. Biotechniques, 20, 102–110. [DOI] [PubMed] [Google Scholar]
- Ruggero D., Wang,Z.G. and Pandolfi,P.P. (2000) The puzzling multiple lives of PML and its role in the genesis of cancer. BioEssays, 22, 827–835. [DOI] [PubMed] [Google Scholar]
- Sabile A., Rhodes-Feuillette,A., Jaoui,F.Z., Tobaly-Tapiero,J., Giron,M.L., Lasneret,J., Périès,J. and Canivet,M. (1996) In vitro studies on interferon-inducing capacity and sensitivity to IFN of human foamy virus. Res. Virol., 147, 29–37. [DOI] [PubMed] [Google Scholar]
- Santillana-Hayat M., Rozain,F., Bittoun,P., Chopin-Robert,C., Lasneret,J., Peries,J. and Canivet,M. (1993) Transient immunosuppressive effect induced in rabbits and mice by the human spumaretrovirus prototype HFV (human foamy virus). Res. Virol., 144, 389–396. [DOI] [PubMed] [Google Scholar]
- Stadler M. et al. (1995) Transcriptional induction of the PML growth suppressor gene by interferons is mediated through an ISRE and a GAS element. Oncogene, 11, 2565–2573. [PubMed] [Google Scholar]
- Staeheli P., Pitossi,F. and Pavlovic,J. (1993) Mx proteins: GTPases with antiviral activity. Trends Cell Biol., 3, 268–272. [DOI] [PubMed] [Google Scholar]
- Stark G.R., Kerr,I.M., William,B.R.G., Silverman,R.H. and Schreiber,R.D. (1998) How cells respond to interferons. Annu. Rev. Biochem., 67, 227–264. [DOI] [PubMed] [Google Scholar]
- Sternsdorf T., Grotzinger,T., Jensen,K. and Will,H. (1997) Nuclear dots: actors on many stages. Immunobiology, 198, 307–331. [DOI] [PubMed] [Google Scholar]
- Sternsdorf T., Jensen,K., Reich,B. and Will,H. (1999) The nuclear dot protein sp100, characterization of domains necessary for dimerization, subcellular localization, and modification by small ubiquitin-like modifiers. J. Biol. Chem., 274, 12555–12566. [DOI] [PubMed] [Google Scholar]
- Vallian S., Chin,K.V. and Chang,K.S. (1998) The promyelocytic leukemia protein interacts with Sp1 and inhibits its transactivation of the epidermal growth factor receptor promoter. Mol. Cell. Biol., 18, 7147–7156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.-G., Ruggero,D., Ronchetti,S., Zhong,S., Gaboli,M., Rivi,R. and Pandolfi,P.P. (1998a) PML is essential for multiple apoptotic pathways. Nature Genet., 20, 266–272. [DOI] [PubMed] [Google Scholar]
- Wang Z.G., Delva,L., Gaboli,M., Rivi,R., Giorgio,M., Cordon-Cardo,C., Grosveld,F. and Pandolfi,P.P. (1998b) Role of PML in cell growth and the retinoic acid pathway. Science, 279, 1547–1551. [DOI] [PubMed] [Google Scholar]
- Weis K., Rambaud,S., Lavau,C., Jansen,J., Carvalho,T., Carmo-Fonseca,M., Lamond,A. and Dejean,A. (1994) Retinoic acid regulates aberrant nuclear localization of PML/RARα in acute promyelocytic leukemia cells. Cell, 76, 345–356. [DOI] [PubMed] [Google Scholar]
- Zhong S., Salomoni,P. and Pandolfi,P. (2000) The transcriptional role of PML and the nuclear body. Nature Cell Biol., 2, E85–E90. [DOI] [PubMed] [Google Scholar]