Abstract
Marine surface waters contain complex mixtures of chemicals that can adversely affect microzooplankton. There is a lack of toxicity data for this organism group, and we used two different methodologies to fill this gap. We tested the toxicity of three chemical mixtures of polar organic chemicals extracted from marine surface water, using a component-based and a whole-mixture approach. The component-based approach estimates cumulative toxic units for each mixture based on concentrations of individual compounds. The observed hazard data for zooplankton was supplemented with ECOSAR-generated QSAR daphnid LC50s when observed data was missing. ECOSAR performance was evaluated for zooplankton, where 65% of the observed hazard data for zooplankton was predicted within a factor of 10. This approach suggested that none of the mixtures should be toxic to zooplankton at their respective measured environmental concentrations. We found contrasting results using the whole-mixture approach with a reduction in ciliates and dinoflagellates, and change in microzooplankton diversity, at the measured environmental concentrations. We suggest an assessment factor of at least 1000 when using additive toxic units in a component-based risk assessment approach to cover for the extrapolation from acute to chronic toxicity data and for the range of sensitivities among microzooplankton species.
Keywords: biodiversity, ECOSAR, whole-mixture and components-based approach: risk assessment
Mixtures of contaminants extracted from seawater affect microzooplankton at measured environmental concentrations.
Introduction
Many chemicals used in human activities are emitted to the aquatic environment and finally end up in marine coastal waters. Marine surface water can contain complex mixtures of contaminants such as pesticides, pharmaceuticals, or personal care products that could adversely affect marine organisms (Gustavsson et al. 2017a, Vanryckeghem et al. 2019). Mixtures of contaminants generally cause at least additive toxicity when combined (Barata et al. 2005, Bao et al. 2013), but mixtures of certain compounds can also cause antagonistic or synergistic toxicity (Cedergreen 2014, Nys et al. 2015, Martin et al. 2021).
Mixture toxicity can be modelled using prediction models such as concentration addition (CA), which has been recommended as a first-tier assessment for mixtures (Backhaus and Faust 2012). Although CA assumes that the substances in the mixture have a similar mode of action (Löewe and Muischnek 1926), it has also been found to predict toxicity within a factor of two for chemical mixtures with different modes of action (Deneer 2000, Belden et al. 2007, Spilsbury et al. 2020). CA calculations are commonly based on toxic units (TUs), which corresponds to the ratio of a substance concentration to its effect concentration, and the individual TUs can be ranked to determine the toxicity-driving substances within a mixture. Typically, there are a few chemicals in each mixture that are driving the toxicity, with much smaller contributions of the remaining ones (Gustavsson et al. 2017b, Syberg et al. 2017, Jönander and Dahllöf 2020).
When hazard data for single substances are missing, one option is to model the toxicity of each individual substance using Quantitative Structure-Activity Relationships (QSARs). The ECOlogical Structure-Activity Relationship Model (ECOSAR) is one of the well-known QSARs that predict toxicity of substances based on their molecular structure, which can be correlated to physicochemical properties and toxicity to model organisms of different trophic levels (US EPA 2022a). For green algae and daphnids, ECOSAR has predicted the acute toxicity of 60%–64% of industrial chemicals within a factor of 10 (Reuschenbach et al. 2008), whereas toxicity of 22%–25% of the chemicals has been underestimated and 21%–28% has been overestimated (Reuschenbach et al. 2008, Golbamaki et al. 2014). Given this relatively high uncertainty, it is important to compare the modelled toxicity to observed hazard data to validate the performance of QSARs. The QSARs generally produce hazard data for a few model species within the trophic level ‘invertebrates’, but given that species within the same trophic level can have different tolerances to the same contaminant, it is also important to evaluate how representative these data are for other species in this trophic level.
Some of the organisms that are exposed to contaminants in marine surface water are microzooplankton, which are heterotrophic and mixotrophic organisms in the size range of 20–200 µm. The community consists of many types of protists, including dinoflagellates and ciliates, but also smaller metazoans like copepod nauplii or meroplankton (Sieburth et al. 1978). Microzooplankton has a key role in the pelagic food web as the link between primary producers and secondary consumers, as they are the main consumers of primary production (Calbet and Landry 2004), and an important food source for mesozooplankton like copepods (Calbet and Saiz 2005).
There is a variation in tolerance among microzooplankton towards contaminant exposure. Auto- and mixotrophic dinoflagellates have been found to increase their growth rate during exposure to crude oil, both with and without chemical dispersants, whereas oligotrich and tintinnid ciliates were negatively affected (Almeda et al. 2018). Among ciliates, larger tintinnid ciliates were less sensitive to oil and chemical dispersants than smaller ciliates (Almeda et al. 2014), and hetero- and mixotrophic ciliates were more sensitive to the pesticide rotenone than both hetero- and mixotrophic dinoflagellates (Ferreira and Calbet 2020).
Microzooplankton, particularly entire communities, are rarely used in ecotoxicological assays to evaluate the effects of contaminants on marine waters. Instead, most studies use freshwater daphnids to test toxicity of both individual substances and mixtures, and while these tests are conducted under controlled and easily repeatable conditions, they often lack information about how intra- and interspecific variation is affected by exposure. Species diversity in a community can be closely linked to its stability and function (Naeem and Li 1997, Hooper et al. 2005, Downing et al. 2014), and it is therefore important to assess whether certain microzooplankton species are more sensitive to contaminant mixtures that they encounter in their environment. By exposing entire communities of microzooplankton to contaminants, we can assess how their different sensitivities affect the diversity as well as abundance of different taxa. Both ciliates and dinoflagellates can divide daily or every 2 days (Banse 1982, Havskum and Hansen 2006, Nishitani et al. 2008), so the community diversity can change quickly over a few days if some species are more sensitive to contaminant exposure.
We used two different approaches to study how contaminants mixtures affect marine microzooplankton: the component-based approach that uses data for single substances, and the whole-mixture approach, where organisms are exposed to the entire mixture. Both approaches fall under the concept of New Approach Methods (NAMs) in ecotoxicology, reducing the use of protected taxonomic groups (Basu et al. 2025). One of the many challenges of assessing the toxicity of mixtures is the lack of hazard data for both individual chemicals and whole mixtures (Backhaus and Karlsson 2014), which can partly be overcome using modelled toxicity of chemicals. Effect-based approaches such as testing the toxicity of whole mixtures are independent of the lack of hazard data and assess the combined toxicity of the content of a mixture (Bopp et al. 2019).
The aim of this study was to evaluate toxicity of polar organic substances extracted from surface seawater on microzooplankton, as well as compare the assessed risks by the whole-mixture using naturally occurring microzooplankton with the component-based approach.
We used chemical mixture extracts collected during a sampling survey of a marine fjord in 2020, where 750 polar organic chemicals were analysed in a total of six surface water samples collected near Stenungsund, Sweden. The samples were collected in a transect from south to north, near several possible sources of contaminants such as agriculture, industry, wastewater treatment plants, and marinas, and between 62 and 80 chemicals were detected at each site (Inostroza et al. 2023). To determine how microzooplankton diversity is affected by these types of chemical mixtures, and to identify the toxicity-driving substances in the mixtures, we exposed a natural microzooplankton community to whole mixtures from three out of the six sites, selected based on difference in chemical profiles. We further proceeded to rank the toxicity-driving substances within each of the three mixtures based on their respective toxic units using the component-based approach. Missing hazard data for microzooplankton was supplemented with ECOSAR QSAR data for daphnids, and to evaluate whether this supplementation was appropriate, we compared observed and modelled hazard data for zooplankton taxa.
Materials and methods
Sampling sites, chemical mixtures, and analysis of their content
Seawater was collected from six sites along Askeröfjorden in 2020 close to various sources of contaminants such as agriculture, industry, and two wastewater treatment plants (Fig. 1, Table A.1). A total of 100 L of seawater was sampled at each site using an onsite large volume solid phase extraction (LVSPE) device (Schulze et al. 2017), and the solid phase extraction (SPE) cartridges were stored immediately at 4°C until the laboratory extractions. The content of each cartridge was extracted with methanol/ethyl acetate 1:1 (v/v, 500 mL each, neutral fraction), methanol containing 2% of 7 N ammonia in methanol (500 mL, acidic fraction), and methanol with 1% of formic acid (500 mL, basic fraction). The eluates were filtered (GF/F Whatman) to remove precipitates and reduced to dryness using a rotary evaporator (40°C water bath) and a gentle stream of nitrogen. The samples were then transferred to methanol and adjusted to a final enrichment factor of 1000 (1000x the measured environmental concentrations). The final extracts were stored at −20°C until the microzooplankton experiments. The content of each extract was analysed using LC-HRMS (Liquid Chromatography-High Resolution Mass Spectrometry), and in total 845 substances (including 100 internal standards) were analysed for. The number of substances detected at the six sites varied between 62 and 80. More detailed information about sample preparation, extraction procedure, and target screening can be found in Inostroza et al. (2023).
Figure 1.
Map displaying the six sampling sites M1-M6 and land use near the sites. The layers and information in the large map were extracted and put together by Elisabeth Fenske using GSD-Topographic Map 1:50 000, © Lantmäteriet.
Selection of mixtures for exposure experiments
The chemical content of each extract was retrieved from Inostroza et al. (2023). Differences in chemical composition between sites were visualized through principal component analysis (PCA), using normalized concentrations. The individual chemicals that contributed most to the explained variance in PC1 and PC2 were plotted as vectors pointing in the directions of increasing normalized concentration. The three sites (M1, M2, and M5) differed the most from each other in terms of composition and were selected to be used in the microzooplankton community experiment.
Whole-mixture approach
The whole-mixture approach involved exposing microzooplankton communities to the various chemical mixtures in a laboratory experiment. The microzooplankton community experiment was conducted at Kristineberg Centre during March 8th–13th 2022. Seawater with plankton was collected from approximately 2 m depth near the monitoring station Släggö (N 58° 15.5′, E 11° 26.0′) (Fig. A.1) using a 1.7 L Ruttner water sampler (Hydro-X, Swedaq). The water was reverse filtrated with a 200 µm filter to remove larger organisms and stored in a plastic container at experimental conditions for approximately 8 h until the start of the exposure.
A dilution series of stocks 10 X the aimed concentrations of each chemical extract from three sites M1, M2, and M5 were prepared to ensure that the same volume of methanol extract was added to each bottle. Stock aliquots corresponding to the three aimed measured environmental concentrations (MECs) for each site (0.1x, 1x, and 10x) were added to the respective fluorinated 295 mL HDPE bottle (n = 5, 45 bottles in all) and left in a fume hood until the methanol had evaporated. Pure methanol without extract was added to each control bottle to ensure equal conditions. Once all methanol had evaporated, 10 mL of filtered seawater (0.2 µm) was added to each bottle to resuspend the chemical mixtures, together with an Isochrysis galbana culture from Gothenburg University marine algae culture collection (GUMACC, strain no 108, source ccmp11323) that was used as a food source for the ciliates and dinoflagellates (Strom and Morello 1998). Concentrated culture of I. galbana corresponding to 80 000 cells/mL with cells sizes between 6 and 8 µm (estimated from measurements on a Beckman Coulter Multisizer 3 Particle Counter) was added to each bottle, and the bottles were filled to the rim with the reverse-filtered seawater containing microzooplankton. Each bottle was covered with Teflon sheet to avoid air bubble formation before the lids were screwed on, and the bottles were mounted on a plankton wheel rotating at 0.2 r/m. Five bottles with MilliQTM water for sequencing blanks were incubated and treated equally to the rest of the experimental bottles throughout the experiment. Start bottles without chemicals (n = 5) were taken to measure the initial diversity and abundance of ciliates and dinoflagellates prior to the exposure, and these were sampled at the start of the exposure according to the procedure described below.
The microzooplankton (0–200 µm) was exposed for five days in a thermoconstant room at 7.8°C ± 0.4 with a 11:13 h light:dark cycle of 42.9 lux ± 18 (SD) to resemble the natural ambient conditions at the time of the exposure.
Each experimental bottle with the chemical treatments or control water, as well as the blanks, was sampled at the end of the exposure for RNA extraction and microscopy counts. Eighty millilitre samples were collected from each bottle for the RNA extractions and diversity assessment. The samples were filtered onto Millipore filters (25 mm, 8 µm pore size) and frozen immediately after filtration by placing them into 1.5 mL RNase-free Eppendorf tubes submerged in a dry ice and ethanol bath. The samples were then transferred to a −80°C freezer, where they were stored until further use. The samples were filtered to ensure that most of the biological material in the samples should be within the microzooplankton size range of 20–200 µm. Some phytoplankton taxa also fit in this size span.
One hundred twenty millilitre samples were then collected from each bottle for the microscopy counts of ciliates and dinoflagellates. The samples were collected in brown glass bottles, pre-filled with 1.5% acidified Lugol’s solution to ensure a quick fixation, and stored in the dark at 5°C until further analysis. The microscopy counts of ciliates and dinoflagellates in the samples (n = 3 randomly selected out of the n=5) were performed by allowing 50-mL subsample to settle for at least 15 h in 50-mL Utermöhl chambers (Utermöhl 1958), and the whole subsamples were later counted using a Carl Zeiss™ Axio Vert.A1 Inverted Microscope. Species were identified to species level when possible, otherwise genus or unknown. It is not possible to distinguish ciliate species after the Lugol staining, and identification was therefore made by size and shape of body and cilia.
RNA extraction, cDNA synthesis, PCR amplification, and sequencing
RNA was isolated from the filters using the RNeasy PowerWater kit (Qiagen) following the manufacturers guidelines with some minor modifications and stored at −80°C until further use (see extended materials and methods in Appendix A). cDNA was synthesized from the RNA through reverse transcription using the iScript™ Select cDNA Synthesis Kit (Bio-Rad) together with the reverse primer TAReukREV3 (5´-ACTTTCGTTCTTGAT(C ⁄T) (A ⁄G)A-3´) (Stoeck et al. 2010) and stored at −20°C until further use (see extended materials and methods in Appendix A).
The cDNA samples were used as templates for amplification of the V4 region of 18S rRNA with primers TAReuk454FWD1(5´-CCAGCA(G ⁄C)C(C ⁄T)GCGGTAATTCC-3´) and TAReukREV3 (5´-ACTTTCGTTCTTGAT(C ⁄T) (A ⁄G)A-3´) (Stoeck et al. 2010). The PCR was run using KAPA HiFi Hotstart ReadyMix (Kapa Biosystems) (see extended materials and methods in Appendix A) with an activation step of 95°C at 180 s, followed by 10 three-step cycles of 98°C for 20 s, 65.5°C for 15 s, and 72°C for 30 s, and 15 three-step cycles of 98°C for 20 s, 56.8°C for 15 s, and 72°C for 30 s. The PCR ended with a final extension step of 72°C for 60 s, and the samples were then cooled to 4°C and stored at −20°C until further use.
The PCR products were quantified on a TapeStation (Agilent Technologies) using the high-sensitivity D1000 ScreenTape and reagents (Agilent Technologies), and the samples were normalized to 4 ng/µL (see extended materials and methods in Appendix A). The remaining library preparation steps and sequencing were performed at SciLifeLab National Genomics Infrastructure (Solna, Sweden) and paired-end sequenced (2×300 bp) using the Illumina MiSeq system.
Sequence data processing and taxonomic assignment
The analysis of the Illumina paired-end reads was conducted using the pipeline nf-core/ampliseq (version 2.4.0) (Straub and Peltzer 2019) using Nextflow (version 22.04.5) and Singularity (version 3.5.3) with the optional parameter ‘–double_primer’ to remove potential double primers. The nf-core/ampliseq pipeline used FastQC (version 0.11.9) (Andrews 2010) for quality control of sequences, and Cutadapt (version 3.4) (Martin 2011) to trim the reads of leftover primers. DADA2 (version 1.22.0) (Callahan et al. 2016) was used to infer Amplicon Sequence Variants (ASVs) and to taxonomically classify them using the PR2 reference database version 4.14.0 (Guillou et al. 2012). Alpha rarefaction curves were produced with QIIME2 (version 2021.8.0) (Bolyen et al. 2019) using an input metadata sheet with the different treatments. The ASVs were aligned using MAFFT (version 7.407) and visualized in Seaview (version 5.0.5) to check that there were no leftover primers that could inflate the number of ASVs.
Filtering of the unique ASVs was done using RStudio (R version 4.2.2). The ASVs annotated to the domain Bacteria or to the family Isochrysidaceae were removed. Isochrysidaceae was removed since this includes sequences for I. galbana, that was used as a food source for the microzooplankton in the experiment. The remaining ASVs with confidence >0.8 were kept and sequence reads for each ASV were merged based on genus. 38 ASVs lacked genus annotation and were therefore removed. Twenty eight out of these 38 had less than 100 reads in total among all samples, and the remaining ten occurred randomly in all samples. In total, 1977 ASVs were produced by the nf-core/ampliseq pipeline, and after filtering 209 were left on which the downstream analyses were run. Sequences are available at NCBI GenBank (www.ncbi.nlm.nih.gov) with accession numbers: PV221747-PV221784, PV232159-PV232220, PV278679-PV278752, PV262617-PV262641, PV273691-PV273715, and PV364189-PV364205.
Component-based approach
Acquisition of observed zooplankton hazard data, modelling of hazard data using ECOSAR, and calculation of cumulative toxic units
Observed hazard data for zooplankton was supplemented with QSAR data generated by ECOSAR (US EPA 2022) to fill in gaps of missing data. The complete datasets were then used to estimate cumulative toxic units for the three chemical mixtures used in the microzooplankton community experiment. Observed hazard data for the substances detected in any of the six sites were acquired from US EPA ECOTOX Knowledgebase (US EPA 2023) in February 2023, and from the European Food Safety Authority (EFSA) (Kovarich et al. 2020) and the respective REACH dossiers in August 2020. The data from REACH was accessed in August 2020 from substance-specific individual dossiers from a database hosted by the European Chemicals Agency (ECHA) (ECHA 2020). The EFSA and REACH data was provided to us in an assembled and curated form in 2022.
Data for all freshwater and marine taxa of microzooplankton, mesozooplankton, and invertebrates with planktonic egg or larval stages (meroplankton) with the endpoints EC50, IC50, and LC50, and the effects ‘Intoxication’, ‘Mortality’, and ‘Population’, from experiments with up to 5 days exposure time was included. The data requirements were set to ensure that the conditions in the experimental data were similar to the microzooplankton exposure scenario. Freshwater zooplankton data was included, as data for marine zooplankton was very scarce, and mesozooplankton was included, as they occur in the microzooplankton community during certain life stages. If more than one EC50 was present for any individual chemical, the arithmetic mean for each substance was used.
QSAR predictions for acute LC50 hazard data for daphnids were obtained with ECOSAR (US EPA, 2022) using the simplified molecular-input line-entry system (SMILES) for each chemical detected above its respective limit of quantitation (LOQ) in any of the six sites. The SMILES data were obtained via PubChem and ChemSpider online retrieval tools using the webchem R package (Szöcs et al. 2020). Where more than one prediction was available, the geometric mean of LC50 values for daphnids was used.
Cumulative toxic units (cumulative TUs) were calculated for each of the three chemical mixtures selected to be used in the microzooplankton community experiment. When observed hazard data was missing, data was supplemented with acute daphnid LC50s generated from ECOSAR for the remaining detected chemicals. The cumulative TU of each mixture was defined using the principles of CA (Equation 1) and was derived by calculating the sum of the individual TUs for each substance (i) in the mixture, i.e. its ratio of concentration (c) to effect concentration (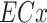 ). The cumulative TU indicates whether a mixture is near the concentration that causes toxicity at the level of chosen effect concentration (
). The cumulative TU indicates whether a mixture is near the concentration that causes toxicity at the level of chosen effect concentration ( ), i.e. when the cumulative TU ≥ 1, and is defined as:
), i.e. when the cumulative TU ≥ 1, and is defined as:
 |
(1) |
Comparison of observed and QSAR (ECOSAR) hazard data for zooplankton
To evaluate the performance of the QSAR predictions, the sensitivity of each type of zooplankton was compared to the QSAR-modelled daphnid LC50s. The observed hazard data and the QSAR data were plotted for each detected chemical in any of the six sites and ranked based on the QSAR-generated toxicity. Observed hazard data was plotted for each of the groups of zooplankton, meroplankton, and holoplanktonic crustaceans, and freshwater and marine organisms were separated. The arithmetic means for each chemical and species was used.
To compare how well the QSAR data could be used as a supplement for missing zooplankton data, the observed hazard arithmetic mean was correlated to the QSAR hazard geometric mean for each detected chemical. Furthermore, the ratio of observed to QSAR-modelled toxicity was also calculated for each detected substance to evaluate for how many substances the model could predict toxicity within a factor of 10.
Data analysis and visualization
All data were visualized and analysed using RStudio (R version 4.2.2) (© 2009–2022 RStudio, PBC) with the exception of the PERMANOVA and PERMDISP test that were run in PRIMER (version 7.0.21) (Clarke and Gorley 2015).
The PCA used to visualize the compositions of the chemical mixtures, and the non-metric multidimensional scaling (NMDS) used to visualize the beta diversity from the metabarcoding of the microzooplankton communities were both generated with R packages vegan (version 2.6–4) (Oksanen et al. 2013) and ggplot2 (version 3.4.0) (Wickham 2011). The PCA was based on normalized concentrations of each chemical, and the NMDS was based on Bray–Curtis dissimilarity generated from square root transformed proportions of each detected genus. The boxplots, barplots, and scatterplots were all generated using the R package ggplot2 (version 3.4.0). The alpha rarefaction curves were generated by QIIME2 using the nf-core/ampliseq pipeline.
Differences in alpha diversity (Shannon diversity and species richness), as well as ciliate and dinoflagellate abundances between control and chemical treatments, were tested with one-way ANOVAs and Dunnett’s post hoc tests using R packages stats (version 4.4.2) and multcomp (version 1.4–20) (Hothorn et al. 2016). The assumptions of normally distributed residuals and homogeneous variances were checked visually from residual and Q-Q plots. If assumptions were violated, the data was transformed accordingly to meet them.
Differences in species composition between all treatments were tested using a PERMANOVA, and differences in variation of composition between treatments (dispersion) were tested using a PERMDISP. Both tests were based on Bray–Curtis dissimilarity generated from square root transformed proportions of each detected genus. To test whether QSAR daphnid LC50 data was a good supplement for missing observed hazard data, a linear model was fitted using the R package stats (version 4.4.2).
Results
Characterization of sites and chemical mixtures used in experiments
Sites M1, M2, and M5 were the most separated in the PCA when comparing composition of the chemical mixtures based on normalized concentrations (Fig. 2). Separation of site M1 from the other ones was caused by relatively higher concentrations of benzoylecgonine, cotinine, and cyclohexyl phenyl ketone. Benzoylecgonine and cotinine are metabolites of cocaine and nicotine (NCBI 2023a, Benowitz et al. 2009), whereas cyclohexyl phenyl ketone is a chemical used in many industries, such as the processing of textiles and metals, and as a component in flooring, furniture, and toys (ECHA 2023a).
Figure 2.
PCA based on normalized concentrations of each detected chemical in the mixtures. The vector arrows with chemical names display the substances that most explain the data variance of PC1 and PC2.
Site M2 had higher concentrations of chloridazon, 2-hydroxyquinoline, tetraglyme, and norfloxacin relative to the other sites. Chloridazon is a pyridazinone herbicide that is used on, for example, sugar beets (May 2001), and 2-hydroxyquinoline (also sometimes named 2-quinolone) can be used as a component in drugs and fluorescent materials (Tashima 2015). Tetraglyme is a solvent that can be found in several products such as flooring, furniture, and toys, as well as in paper-based products (ECHA 2023b), and norfloxacin is a fluoroquinolone antibiotic used in both humans and animals (Song et al. 2004).
M5 was separated from the other sites by relatively higher concentrations of Tris(1,3-dichloro-2-propyl)phosphate (TDCPP) and 1,3-diphenylguanidine. TDCPP is an organophosphate that is used in processing of many materials such as textile and metals and can be used as a component in, for example, flooring, furniture, and toys (ECHA 2023c). 1,3-diphenylguanidine is used in a variety of materials and products such as construction and building materials, tyres, and brake pads in cars, as well as in ships (ECHA 2023d).
Sequencing data from experimental samples
The Illumina MiSeq sequencing of the samples produced a total of 22.9 million read pairs, and after processing the data with the nf-core ampliseq pipeline a total of 1977 unique ASVs were produced, each with between 1 and 1 287 721 reads, and the rarefaction curves show that all these samples have reached their respective saturations of observed ASVs (Fig. A.3). Eukaryotic ASVs were fewer in the x10 samples, and these treatments also held a larger amount of bacterial ASV (Fig. A.2.a). Bacterial and phytoplankton ASVs were detected in all the samples but these were not included in the diversity analysis, as the focus was on microzooplankton.
Whole-mixture approach: microzooplankton diversity
Based on the proportions of sequence reads, all ciliate and most dinoflagellate taxa were more abundant in the control, x0.1 and x1 treatments than in the x10 treatment, except for the dinoflagellate genus Dinophysis that was more abundant in the x10 treatments (Fig. 3).
Figure 3.
Alpha and beta diversity of microplankton exposed to concentrations of 0.1x, 1x, and 10x measured environmental concentrations from sites M1, M2, and the control (n = 5). The graph also includes start samples collected before exposure, and blanks that were incubated together with the experimental bottles (n = 5). The diversity metrics are based on proportions of 18S sequence reads of each taxa in a sample. (A) Alpha diversity in the form of Shannon index in each sample. The boxes display median and the 25th and 75th percentiles. The whiskers extend from the box to the highest and smallest datapoints (at most 1.5 * the interquartile range from the box), and any point outside this range is plotted individually as an outlier. (B) Beta diversity in the form of Bray–Curtis index on standardized and square root transformed data, visualized in an NMDS. The direction of the vector arrows display the direction of increased proportions of the respective ciliate and dinoflagellate taxa that most explain the variance of NMDS1 and NMDS2.
Dinoflagellate abundances increased in all treatments throughout the exposure, except for in the x10 treatments, as assessed by microscopy counts (Fig. 4), and the abundances were lower on average in all chemical treatments compared to the controls (Table A.5). The dinoflagellate abundances were significantly reduced for site M1 by 86% at x10 MEC. Sites M2 and M5 had reduced abundances of 69% and 43%, respectively, already at x1 MEC, and at 87% and 82% at x10 MEC (Table A.5). Unlike the dinoflagellates, the ciliates decreased in numbers over time in all treatments compared to the start samples, and the treatments were on average lower than control starting at 0.1x MEC from all sites (Fig. 4, Fig. A.5). The ciliate abundance for site M2 was significantly reduced by 51% at 1x MEC, and 99% at 10x MEC (Table A5), with a near-significant reduction of 48% in 0.1x MEC. There were also significant reductions in abundance for M1 and M5 at x10 MEC with 77% and 99% reduction, respectively, and near-significant reductions of 51 and 48% at 1x MEC (Table A5).
Figure 4.
Ciliate and dinoflagellate abundances in experimental exposed to extracts of concentration 0.1x, 1x, and 10x measured environmental concentrations from sites M1, M2, M5, and control (n=3). The graph also includes start samples collected before exposure (n = 3). The abundances are based on microscopy counts. The boxes display median and the 25th and 75th percentiles. The whiskers extend from the box to the highest and smallest datapoints (at most 1.5 * the interquartile range from the box).
Alpha diversity (within-treatment diversity) was reduced on average in the chemical treatments compared to the control, starting at either 0.1x the measured environmental concentration (MEC) or 1x MEC depending on the site and specific alpha diversity index used. Shannon diversity was on average reduced starting at x1 MEC in all sites (Fig. 3A), and species richness was reduced on average starting at 0.1x MEC in all sites (Fig. A.4). For Shannon diversity, significant differences between control and treatments were only found in the x10 treatments, with a 35% reduction in the x10 MEC exposure at site M1, and by 23% and 20% at sites M2 and M5, respectively (Table A.2).
Species richness for site M1, M2, and M5 at x10 MEC was significantly reduced compared to the control with effect sizes of 42%, 36%, and 31%, respectively (Table A2). Site M5 species richness was also significantly reduced by 14% at 1 X MEC, and at M2 with near-significance reduction by 14%. (Table A2). Beta diversity (community composition between treatments) was significantly altered for all sites at x1 and x10 MEC, and there was also a clear separation between all sites, the start samples, the blanks, and the rest of the samples within the ordination space for the x10 MEC (Fig. 3B, Table A.3). The direction of change in species composition was the same for the three different mixtures as they did no differ significantly (Table A.3). The PERMDISP analysis showed that the dispersion within the x1 treatments was similar, as within the control but that dispersion within the x10 treatments was significantly larger than within the controls and lower treatments (Table A.3).
Component-based approach: ranking of toxicity-driving substances in the mixtures and their cumulative toxicity
The cumulative toxic unit (TU) of the substances in site M1 was estimated to 0.0039 (Fig. 5A), and the estimation was built on observed zooplankton toxicity data for 18 substances and QSAR daphnid LC50s for another 56 substances out of the 74 detected at the site. Ursolic acid, telmisartan, and formetanate were the main drivers of toxicity in the order listed and contributed to 92% of the toxicity of the mixture. Ursolic acid is a substance found in many plants and has primarily been used in traditional medicine (Liu 1995, Wen and Xiong 2011). Telmisartan is an angiotensin II receptor antagonist used to reduce blood pressure (McClellan and Markham 1998), and formetanate is a carbamate insecticide and acaricide (NCBI 2023b). Neither of the three chemicals had a large role in distinguishing site M1 from the other sites based on their concentrations alone (Fig. 2).
Figure 5.

Cumulative toxic units and toxicity-driving substances from the three sites used in the experiments. (A) Site M1, (B) site M2, and (C) site M5. Individual toxic units are based on geometric mean of QSAR (ECOSAR) toxicity or arithmetic mean of experimental toxicity for plankton. The dashed lines correspond to the cumulative toxic units and numbers on top of bars represent % contribution to total toxicity of the mixture.
The cumulative TU of the substances in site M2 was estimated to 0.0037 (Fig. 5B). The estimated toxic unit was generated from observed zooplankton toxicity data for 19 individual substances and QSAR daphnid LC50s for another 59 substances out of the 78 substances detected at the site. In this site, seven individual substances contributed to most of the toxicity (Fig. 5B). Ursolic acid, 2-hydroxyquinoline, tramadol, 5-methyl-1H-benzotriazole, raloxifene, formetanate, and dicyclohexylurea were the main toxicity drivers in the order listed and contributed with 91% of the toxicity of the entire mixture. 2-hydroxyquinoline is a component in drugs and fluorescent materials, tramadol is an opioid analgesic (Raffa et al. 1992), and 5-methyl-1H-benzotriazole is used as a component in, for example, lubricants, cleaning products, and antifreeze agents (ECHA 2023e). Raloxifene is a selective oestrogen receptor modulator (Quintanilla Rodriguez and Correa 2022), formetanate is a carbamate insecticide and acaricide (NCBI 2023b), and dicyclohexylurea has been found to lower blood pressure in rats (Ghosh et al. 2008). The only substance out of the seven that contributed to compositional differences in M2 based on concentration relative to other sites was 2-hydroxyquinoline (Fig. 2).
The cumulative TU of the substances in site M5 was estimated to 0.0043 (Fig. 5C), and the estimate was generated from observed zooplankton toxicity data for 21 individual substances and QSAR daphnid LC50s for another 59 substances out of the 80 substances detected at the site. The two substances ursolic acid and lauramidopropylbetaine were the main drivers of toxicity and contributed to 92% of the total toxicity of the mixture. Lauramidopropylbetaine is used in many consumer products such as soaps and skin care products (Clendennen and Boaz 2019). Neither of the two toxicity-driving substances were the main explanatory variables distinguishing site M5 from the other sites based on their concentrations alone (Fig. 2).
Component-based approach: comparison of observed and QSAR toxicity to zooplankton
Observed toxicity data for zooplankton was available for 31 out of the 114 individual substances detected at sites M1-M6. This data comprised 18 individual taxa belonging to ten freshwater and eight marine microzooplankton and mesozooplankton taxa.
Comparison of the observed toxicity data for these microplankton groups to QSAR toxicity for daphnids showed that the QSAR-generated daphnid LC50s correspond to approximately the average of the observed EC50s for the microzooplankton community (Fig. 6, Fig. A.5a). Overall, there is no general difference in sensitivity to this set of contaminants for freshwater and marine taxa; however, the sensitivity to some individual chemicals differed by several orders of magnitude between the species. The largest difference was found for acetaminophen, for which sensitivity differed by a factor of 100 000 between the species.
Figure 6.
Observed and modelled zooplankton EC50s of each detected substance in sites M1-M6. The graph includes ECOSAR LC50s for daphnids (triangles) observed toxicity data (EC50s, IC50s, and LC50s) for different types of zooplankton (circles). (A) Comparison of geometric means of ECOSAR LC50s for daphnids (grey triangles) to observed toxicity data (EC50s, IC50s, and LC50s) for holoplanktonic crustacean, meroplankton, and microzooplankton . Data for freshwater taxa are displayed in small triangles and circles, and marine taxa are displayed in large ones. The substances on the y-axis are ranked based on high-to-low QSAR (ECOSAR) acute toxicity data (LC50s) for daphnids. (B) Comparison of geometric means of ECOSAR LC50s for daphnids to means of observed toxicity data (EC50s, IC50s, and LC50s) . The substances on the y-axis are ranked based on high to low QSAR (ECOSAR) acute toxicity data (LC50s) for daphnids.
The average ratio of observed to modelled toxicity was 1.2% and 65% of the observed datapoints were within a factor of 10 of the modelled toxicity (Fig. A.6a). However, the overall correlation between QSAR and observed hazard data was poor (R2=0.36) (Fig. A.6b).
Discussion
The overall aim of this study was to assess toxicity and effects of chemical mixtures from three sites on marine microzooplankton communities. The secondary aim was to compare the risk assessed between the whole-mixture and component-based approach.
Whole-mixture approach: effects on microzooplankton diversity and abundance
The chemical mixtures from all three sites affected the microzooplankton community at the measured environmental concentrations (MEC), but effect concentrations varied depending on endpoint used. Univariate estimates such as the Shannon index and species richness used in this study, were less sensitive than the multivariate Bray–Curtis dissimilarity index in detecting significant effects. Both univariate indices gave significant effects at x10 MEC, whereas the PERMANOVA analysis of Bray–Curtis dissimilarity index revealed significant compositional changes already at x1 MEC, as well as increased dispersion at x10 MEC. Although pairwise tests of groups have shown to be sensitive to dispersion heterogeneity, PERMANOVA has been shown to be one of the more robust tests and is less affected by this than other ones (Anderson and Walsh 2013). Furthermore, we observed both separation between treatments in the ordination plot, as well as significant differences from the PERMANOVA pairwise test. We can therefore conclude that the chemical mixtures at x1 MEC did change species composition as well as increased the dispersion in the microzooplankton community, and that multivariate measures of diversity are more powerful than univariate ones.
All effects on diversity were generally similar across all three chemical mixtures. The alpha diversity effect sizes were similar across all three sites, but with the largest average reduction in diversity for site M1 compared to the other ones. As for the beta diversity, no compositional changes could be detected in the community between the three sites, which indicates that the chemical mixtures, although being different in composition, have a similar effect on microzooplankton diversity. The opposite has been found in studies with individual chemicals, where the identity and mode of action of individual chemicals altered species composition differently, which has been shown for both mesozooplankton (Jönander et al. 2022) and periphyton communities (Backhaus et al. 2011).
Even though the mixtures had a similar effect on diversity, there were differences in their effects on ciliate and dinoflagellate abundances based on the microscopy counts. Survival of both organism groups was lower for site M2 than for other sites, most prominent at x1 MEC. This indicates that the chemical mixture from site M2 is more toxic, but not to the extent that it altered diversity more than in other sites.
Ciliates and dinoflagellates appeared to have different sensitivity to the chemical mixtures. The dinoflagellates were able to divide and increase in abundance throughout the exposure at concentrations up to x1 MEC from all sites, which indicates that they may be more tolerant than the ciliates. The same trend was observed in the results of the metabarcoding, where the x10 MEC treatments had a larger proportion of reads of the dinoflagellate species Dinophysis acuminata, relative to other microzooplankton species. D. acuminata is a bloom-forming species that can produce toxins that can accumulate in filter-feeders and cause diarrhetic shellfish poisoning (DSP) (Hattenrath-Lehmann et al. 2013, Mafra Jr et al. 2019). Dinoflagellates have also been found more tolerant than ciliates to other contaminants, such as those in crude oil (Almeda et al. 2014). Furthermore, Almeda et al. (2018) found that other species of bloom-forming dinoflagellates increased in abundance after exposure to oil and dispersants, whereas ciliate abundances decreased. Dinoflagellates, including those in the genus Dinophysis, have been shown to feed on ciliates such as Mesodinium (Hernández-Urcera et al. 2018, Smith et al. 2018). This could to some extent explain the general higher abundance and growth rate of the dinoflagellates compared to the ciliates, although the main changes in diversity should be attributed to effects from the chemical mixtures, as we compare with untreated controls. The control communities changed in composition themselves during the exposure time, in relation to the sampled start community. This suggests that the laboratory conditions favoured some species over other ones and can be regarded as a source of uncertainty when interpreting the results.
We can conclude with certainty that the dinoflagellates were more abundant than the ciliates in the x10 MEC treatments based on the microscopy counts, revealing that the results of the metabarcoding have some weaknesses regarding estimation of relative species abundance. Microzooplankton species, like many other organisms, can have different numbers of 18S gene copies that can affect the total number of sequencing reads, and ciliates and dinoflagellates both have more copies than other groups in the community (Yarimizu et al. 2021, Martin et al. 2022). In comparisons of microscopy cell counts and raw sequence reads, dinoflagellates had the most inflated abundance when based on raw sequence reads compared to other taxa, which could be improved with a correction factor (Martin et al. 2022). However, the relative abundances of taxa in our samples are based on sequence reads that originate from cDNA reversely transcribed from rRNA, hence, the gene copy bias should be reduced (Not et al. 2009).
Other difficulties of quantifying species using metabarcoding include the divergence in priming sites across species that can affect primer binding and amplification of certain species during PCR (Stadhouders et al. 2010, Elbrecht and Leese 2015). Furthermore, amplification efficiency can also be affected by individual taxa biomass, 18S sequence length, as well as GC content of the sequence (Elbrecht et al. 2017, Nichols et al. 2018, Saad et al. 2020). All of these could potentially favour amplification of D. acuminata over other dinoflagellates, but as we infer effects only from relative changes between treatments, any of these biases should have limited effects on the outcome of our results.
Another uncertainty of the laboratory microzooplankton exposure is the lack of chemical validation data. The concentrations of polar organic contaminants in the water from sites M1-M6 were quantified in the methanol extracts that were added to the test medium, but analysis on the actual test medium during or after the exposure could not be performed. There is therefore an uncertainty of whether it was a pulse or continuous exposure, which should be regarded when interpreting the results.
Component-based approach: supplementing missing observed zooplankton hazard data with QSAR-generated data
Observed hazard data for zooplankton was only found for 27% of the detected substances in the six sites, and thus most of the data used to calculate cumulative TUs for the sites was QSAR-generated LC50s for daphnids. When comparing the observed hazard EC50s to the QSAR-modelled daphnid LC50s, we found that the average estimate for each individual substance was similar. However, the poor correlation between QSAR data and observed hazard data suggests that QSAR daphnid LC50s may not be ideal supplements for missing zooplankton hazard data. We also found that the sensitivity among zooplankton species to the same substance could differ by several orders of magnitude and that only 65% of the observed data was within a factor of 10 of the modelled alternatives. Hence, if the most sensitive species are to be considered in the toxicity estimation, the QSAR daphnid LC50 would not be suitable as data to substitute with. Melnikov et al. (2016) used ECOSAR (v 1.11) to evaluate acute toxicity of 83 different chemicals to freshwater fish and found that 63% of the substances predicted toxicity within a factor of 10. Zhou et al. (2021) used ECOSAR (v 2) to model toxicity of 37 different chemicals and found that 85% of the modelled LC50s were in a factor of 10 for daphnids, but that 68% were in a factor of 10 for freshwater fish. Reuschenbach et al. (2008) used ECOSAR (v0.99 g) to model toxicity of 493 substances to daphnids and 279 substances to algae and found that toxicity of 64% and 60% were predicted within a factor of 10, respectively. The accuracy of these predictions is in line with our predictions, and it is possible that the differences depend on the specific subset of chemicals for which the predictions are modelled, as well as to what observed data they are compared. Standard test species were used in the comparisons made by Melnikov et al. (2016). Zhou et al. (2021), and Reicenbach et al. (2008), whereas we compared the modelled daphnid LC50s to a range of zooplanktonic species with different sensitivities to individual chemicals. This suggests that ECOSAR modelling is less suitable for predicting effects when the biodiversity is high in the system of interest.
Toxicity drivers in the chemical mixtures
Ranking of the toxicity-driving chemicals in the three mixtures showed that the toxicity drivers differed both in quantity and in composition between each site, with the exception that ursolic acid was the main driver of toxicity in all of them. We were not able to retrieve observed zooplankton hazard data for ursolic acid, and thus the individual TU for this substance is based on the QSAR daphnid LC50. When comparing observed toxicity of ursolic acid to other organisms than zooplankton, is becomes clear that it is relatively non-toxic. An in vitro assay with a parasitic protozoan found an activity IC50 for ursolic acid at 12 000 µg/L (da Silva Ferreira et al. 2010), and a motility inhibition IC50 for nematodes has been estimated to 4000 µg/L (Kalani et al. 2014), whereas the QSAR-generated daphnid LC50 was estimated to 18 µg/L. Hence, we suspect that the QSAR prediction for ursolic acid is a very conservative overestimation of toxicity, which inflates the role of ursolic acid as a toxicity driver in the chemical mixture used in this study. We did not attempt to find the cause for this overestimation but accepted that the cumulated TUs would be conservative.
Examining the general patterns of cumulative TUs and toxicity drivers, we can conclude that the identity, as well as the quantity, of toxicity-driving substances appear to have a limited role in explaining the toxicity of these mixtures. Site M2, the site with the largest quantity of toxicity drivers, did reduce ciliate and dinoflagellate survival more than the other sites at one of the tested concentrations, but in general all three mixtures caused similar toxicities to the microzooplankton.
Comparison of component-based approach and whole-mixture approach
The component-based approach indicated that neither of the chemical mixtures should be toxic to the microzooplankton at x1 MEC, given that they were based on EC50s and LC50s, and that the cumulative TU was so low, even with the overestimated toxicity of ursolic acid. However, this was not the outcome of our whole-mixture exposures, as we observed significant effects on both diversity and reduced ciliate survival at 1x MEC. This suggests that modelling effects based on single species of standard organisms, like Daphnia, does not capture effects when a higher biodiversity of microzooplankton is targeted.
The cumulative TUs generated with the component-based approach were nearly identical between all three sites at approximately 0.004, which indicate that they should be equally toxic to the microplankton community. The same conclusion can be drawn from the diversity analysis using the whole-mixture approach, but for the effects on ciliate and dinoflagellate abundances, were there were lower abundances for the M2 site extract compared to the other sites.
Environmental and risk assessment implications
The observed effects using the whole-mixture approach in this study suggests that the present-day contamination in coastal waters has the potential to disrupt the pelagic food web, in contrast to the component-based approach. Reduction in biomass, as well as loss of species in the microzooplankton community could increase the prevalence of algae blooms, as microzooplankton are the main grazers of phytoplankton (Calbet and Landry 2004). The potential for effects is even higher considering that we only studied the effects of polar organic contaminants, and not the total mixture also including non-polar organic compounds and heavy metals. Such consideration was taken by Simmons et al. (2004), who complemented the component-based approach with a whole-mixture approach to include chemicals that could not be chemically identified, and hence not included in the component-based assessment.
The difference between the approaches emphasizes the importance of caution when using only component-based approaches for assessing effects of mixtures in the environment. This is emphasized by Health Canada in their approach for using NAMs in risk assessment (Health Canada 2025). Here NAMs can support a high-throughput approach to prioritize substances and classify potential ecological risk, when deemed to provide a scientifically valid measure of the endpoint under investigation. Another approach was used by Maloney et al. (2023) combining multiple whole-mixture bioassays with the component-based approach and random forest analyses to successfully identify 14 mixtures and 16 chemicals that contributed to cumulative effects.
A more common method is to use assessment factors that vary depending on availability of ecotoxicological data, as in the EU Water Framework Directive (EC 2018). Assessment factors account for the range of sensitivities that natural communities can have compared to standard test species such as daphnids, as well as differences in length of exposure. The discrepancy between the two approaches in this study can be explained both by difference in species sensitivity and length of exposure, as the majority of available toxicity data for zooplankton are daphnids, both observed and QSAR-generated, and are mainly based on acute toxicity scenarios. The whole-mixture approach, in contrast, contained a wide range of species, and was a chronic exposure scenario as 5 days exceed the generation time for most microzooplankton. It is therefore appropriate to add an assessment factor to the results of the component-based approach that takes both differences in species sensitivity and exposure duration into account. Based on our results, an assessment factor of 1000 aligns the two approaches for these three sites. An assessment factor of 1000 for each EC50 would generate cumulative TUs in the range of 4 that better reflects the effect of the exposures, such as reductions in dinoflagellate and ciliate abundances up to approximately 70%, as well as reduced species diversity.
Conclusions
Chemical mixtures at concentrations present in marine coastal waters affected microzooplankton as assessed by the whole-mixture approach. The effects were similar with respect to diversity, despite having different chemical composition and toxicity drivers. However, the site with the largest quantity of toxicity drivers, did reduce ciliate and dinoflagellate abundances more than the other sites at one of the tested concentrations. Effects on ciliate and dinoflagellate abundance started at x0.1 MEC (significantly at x1-10 MEC depending on site), and significant effects on diversity started at x1 MEC. The mixture of compounds sampled from marine coastal surface water near Stenungsund, Sweden affected organisms that have a central role in the pelagic food web. This is of environmental concern, especially considering that the effects were only from polar organic compounds, excluding effects of non-polar organic compounds and heavy metals that also would have been present.
The component-based approach gave contrasting results with cumulative toxic units (TUs) for each mixture approximately 0.004, which suggests little to no toxicity to microzooplankton. We therefore conclude that the component-based risk assessment cannot stand alone but should be complemented with whole-mixture approaches using natural communities for verification of risk, and improvement of the risk models used.
We further conclude that the ECOSAR-generated QSAR data for daphnids worked well to predict the average toxicity to zooplankton for most individual chemicals in this study, but that the toxicity of many substances was under- or overestimated. Furthermore, even when the average toxicity was predicted well, we found that zooplankton species can have sensitivities that differ by a factor of 100 000, which is not considered when using the mean observed EC50, or the QSAR-generated daphnid LC50s in cumulative TU estimations. This was reflected in the whole-mixture exposures, where we detected effects at exposure levels ∼1000 times lower that the TU estimation indicated.
Supplementary Material
Acknowledgements
The authors would like to thank Mikael Gustavsson for supplying hazard data from ECHA and EFSA, and Elisabeth Fenske for providing the map of the sampling sites and assisting during the microzooplankton experiment. The authors acknowledge support from the National Genomics Infrastructure in Stockholm funded by Science for Life Laboratory, the Knut and Alice Wallenberg Foundation, and the Swedish Research Council, and SNIC/Uppsala Multidisciplinary Centre for Advanced Computational Science for assistance with massively parallel sequencing and access to the UPPMAX computational infrastructure. The bioinformatic analysis was performed on the computer cluster Albiorix at the Department of Biological and Environmental Sciences, University of Gothenburg.
Contributor Information
Christina Jönander, Department of Biological and Environmental Sciences, University of Gothenburg, Medicinaregatan 7B , Göteborg 413 90, Sweden.
Jenny Egardt, Department of Biological and Environmental Sciences, University of Gothenburg, Medicinaregatan 7B , Göteborg 413 90, Sweden.
Mats Töpel, Department of Marine Sciences, University of Gothenburg, Medicinaregatan 7B , 413 90, Sweden; IVL—Swedish Environmental Research Institute, Aschebergsgatan 44, Gothenburg 411 33, Sweden.
Francis Spilsbury, Department of Biological and Environmental Sciences, University of Gothenburg, Medicinaregatan 7B , Göteborg 413 90, Sweden.
Eric Carmona, Atmospheric Chemistry Department, Leibniz Institute for Tropospheric Research, Permoserstraße 15, Leipzig 04318, Germany.
Pedro A Inostroza, Department of Biological and Environmental Sciences, University of Gothenburg, Medicinaregatan 7B , Göteborg 413 90, Sweden; Institute for Environmental Research, RWTH Aachen University, Worringerweg 1, Aachen 52074, Germany.
Werner Brack, Atmospheric Chemistry Department, Leibniz Institute for Tropospheric Research, Permoserstraße 15, Leipzig 04318, Germany; Department of Evolutionary Ecology and Environmental Toxicology, Goethe University Frankfurt, Max-von-Laue-Straße.13, Frankfurt am Main 60438, Germany.
Ingela Dahllöf, Department of Biological and Environmental Sciences, University of Gothenburg, Medicinaregatan 7B , Göteborg 413 90, Sweden.
Author contributions
Christina Jönander (Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing), Jenny Egardt (Conceptualization, Investigation, Methodology, Writing – review & editing), Mats Töpel (Data curation, Software, Supervision, Writing – review & editing), Francis Spilsbury (Data curation, Methodology, Software, Writing – review & editing), Eric Carmona (Methodology, Writing – review & editing), Pedro A Inostroza (Data curation, Methodology, Writing – review & editing), Werner Brack (Resources, Writing – review & editing), Ingela Dahllöf (Conceptualization, Formal Analysis, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Writing – review & editing).
Conflict of interest
None declared.
Funding
This project was supported by the Swedish Research Council FORMAS [grant number FR-2017/0009], and the University of Gothenburg through the FRAM Centre.
Data availability
The data underlying this article are available in Zenodo, at https://zenodo.org/records/13988647.
References
- Almeda R, Cosgrove S, Buskey EJ. Oil spills and dispersants can cause the initiation of potentially harmful dinoflagellate blooms (“Red Tides”). Environ Sci Technol. 2018;52:5718–24. 10.1021/acs.est.8b00335. [DOI] [PubMed] [Google Scholar]
- Almeda R, Hyatt C, Buskey EJ. Toxicity of dispersant Corexit 9500A and crude oil to marine microzooplankton. Ecotoxicol Environ Saf. 2014;106:76–85. 10.1016/j.ecoenv.2014.04.028. [DOI] [PubMed] [Google Scholar]
- Anderson MJ, Walsh DC. PERMANOVA, ANOSIM, and the Mantel test in the face of heterogeneous dispersions: what null hypothesis are you testing?. Ecol Monogr. 2013;83:557–74. 10.1890/12-2010.1. [DOI] [Google Scholar]
- Andrews S. FastQC: a Quality Control Tool for High Throughput Sequence Data. Cambridge: Babraham Bioinformatics, Babraham Institute, 2010. [Google Scholar]
- Backhaus T, Faust M. Predictive environmental risk assessment of chemical mixtures: a conceptual framework. Environ Sci Technol. 2012;46:2564–73. 10.1021/es2034125. [DOI] [PubMed] [Google Scholar]
- Backhaus T, Karlsson M. Screening level mixture risk assessment of pharmaceuticals in STP effluents. Water Res. 2014;49:157–65. 10.1016/j.watres.2013.11.005. [DOI] [PubMed] [Google Scholar]
- Backhaus T, Porsbring T, Arrhenius A et al. Single-substance and mixture toxicity of five pharmaceuticals and personal care products to marine periphyton communities. Environ Toxicol Chem. 2011;30:2030–40. 10.1002/etc.586. [DOI] [PubMed] [Google Scholar]
- Banse K. Cell volumes, maximal growth rates of unicellular algae and ciliates, and the role of ciliates in the marine pelagial 1, 2. Limnol Oceanogr. 1982;27:1059–71. 10.4319/lo.1982.27.6.1059. [DOI] [Google Scholar]
- Bao VW, Leung KM, Lui GC et al. Acute and chronic toxicities of Irgarol alone and in combination with copper to the marine copepod Tigriopus japonicus. Chemosphere. 2013;90:1140–8. 10.1016/j.chemosphere.2012.09.022. [DOI] [PubMed] [Google Scholar]
- Barata C, Calbet A, Saiz E et al. Predicting single and mixture toxicity of petrogenic polycyclic aromatic hydrocarbons to the copepod Oithona davisae. Environ Toxicol Chem. 2005;24:2992–9. 10.1897/05-189R.1. [DOI] [PubMed] [Google Scholar]
- Basu N, Knapen D, LaLone C et al. Progress on new approach methods (NAMs) in ecotoxicology. Environ Toxicol Chem, 2025;44:2389–94. 10.1093/etojnl/vgaf096. [DOI] [PubMed] [Google Scholar]
- Belden JB, Gilliom RJ, Lydy MJ. How well can we predict the toxicity of pesticide mixtures to aquatic life?. Integr Environ Assess Manag. 2007;3:364–72. 10.1002/ieam.5630030307. [DOI] [PubMed] [Google Scholar]
- Benowitz NL, Hukkanen J, Jacob IIIP. Nicotine chemistry, metabolism, kinetics and biomarkers. Nicotine Psychopharmacology. 2009:29–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolyen E, Rideout JR, Dillon MR et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37:852–7. 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bopp SK, Kienzler A, Richarz A-N et al. Regulatory assessment and risk management of chemical mixtures: challenges and ways forward. Crit Rev Toxicol. 2019;49:174–89. 10.1080/10408444.2019.1579169. [DOI] [PubMed] [Google Scholar]
- Calbet A, Landry MR. Phytoplankton growth, microzooplankton grazing, and carbon cycling in marine systems. Limnol Oceanogr. 2004;49:51–7. 10.4319/lo.2004.49.1.0051. [DOI] [Google Scholar]
- Calbet A, Saiz E. The ciliate-copepod link in marine ecosystems. Aquat Microb Ecol. 2005;38:157–67. 10.3354/ame038157. [DOI] [Google Scholar]
- Callahan BJ, McMurdie PJ, Rosen MJ et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–3. 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedergreen N. Quantifying synergy: a systematic review of mixture toxicity studies within environmental toxicology. PLoS One. 2014;9:e96580. 10.1371/journal.pone.0096580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke K, Gorley R. PRIMER v7: User Manual/Tutorial. Plymouth: Primer-E Ltd, 2015. [Google Scholar]
- Clendennen SK, Boaz NW. Betaine Amphoteric Surfactants—Synthesis, Properties, and Applications. Biobased Surfactants. Amsterdam: Elsevier, 2019, 447–69. [Google Scholar]
- da Silva Ferreira D, Esperandim VR, Toldo MPA et al. Trypanocidal activity and acute toxicity assessment of triterpene acids. Parasitol Res. 2010;106:985–9. 10.1007/s00436-010-1740-2. [DOI] [PubMed] [Google Scholar]
- Deneer JW. Toxicity of mixtures of pesticides in aquatic systems. Pest Manag Sci. 2000;56:516–20. . [DOI] [Google Scholar]
- Downing AL, Brown BL, Leibold MA. Multiple diversity–stability mechanisms enhance population and community stability in aquatic food webs. Ecology. 2014;95:173–84. 10.1890/12-1406.1. [DOI] [PubMed] [Google Scholar]
- ECHA . ECHA Database. Search for Chemicals. 2020. Accessed August 2020. Available at: https://echa.europa.eu/search-for-chemicals. [Google Scholar]
- ECHA . Substance Infocard for Hydroxycyclohexyl Phenyl Ketone. 2023a. Last updated 25-01-2023. Available at: https://echa.europa.eu/substance-information/-/substanceinfo/100.012.206.(6 March 2023, date last accessed). [Google Scholar]
- ECHA . Substance Infocard for Bis(2-(2-methoxyethoxy)ethyl) ether. Last updated 03-08-2022. 2023b. Available at: https://echa.europa.eu/substance-information/-/substanceinfo/100.005.086.(6 March 2023, date last accessed).
- ECHA . Substance Infocard for Tris[2-chloro-1-(chloromethyl)ethyl] phosphate (TDCPP). 2023c. Last updated 06-05-2022. Available at: https://echa.europa.eu/substance-information/-/substanceinfo/100.033.767. (6 May 2022, date last accessed).
- ECHA . Substance Infocard for 1,3-diphenylguanidine. 2023d. Last updated 01-10-2022. Available at: https://echa.europa.eu/substance-information/-/substanceinfo/100.002.730#REGULATORY_NAMEScontainer. (6 March 2023, date last accessed).
- ECHA . Substance Infocard for Methyl-1H-benzotriazole. 2023e. Last updated 24-02-2023. Available at: https://echa.europa.eu/substance-information/-/substanceinfo/100.045.073. (14 March 2023, date last accessed). [Google Scholar]
- Elbrecht V, Leese F. Can DNA-based ecosystem assessments quantify species abundance? Testing primer bias and biomass—sequence relationships with an innovative metabarcoding protocol. PLoS One. 2015;10:e0130324. 10.1371/journal.pone.0130324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbrecht V, Peinert B, Leese F. Sorting things out: assessing effects of unequal specimen biomass on DNA metabarcoding. Ecol Evol. 2017;7:6918–26. 10.1002/ece3.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Commission (EC) . Technical guidance for deriving environmental quality standards. Common Implementation strategy for the Water Framework Directive. Guidance Document No. 2018. 27 Updated version 2018. https://rvs.rivm.nl/sites/default/files/2019-04/Guidance%20No%2027%20-%20Deriving%20Environmental%20Quality%20Standards%20-%20version%202018.pdf [Google Scholar]
- Ferreira GD, Calbet A. Caveats on the use of rotenone to estimate mixotrophic grazing in the oceans. Sci Rep. 2020;10:1–11. 10.1038/s41598-020-60764-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Chiang PC, Wahlstrom JL et al. Oral delivery of 1, 3-dicyclohexylurea nanosuspension enhances exposure and lowers blood pressure in hypertensive rats. Basic Clin Pharma Tox. 2008;102:453–8. 10.1111/j.1742-7843.2008.00213.x. [DOI] [PubMed] [Google Scholar]
- Golbamaki A, Cassano A, Lombardo A et al. Comparison of in silico models for prediction of daphnia magna acute toxicity. SAR QSAR Environ Res. 2014;25:673–94. 10.1080/1062936X.2014.923041. [DOI] [PubMed] [Google Scholar]
- Guillou L, Bachar D, Audic S et al. The Protist Ribosomal Reference database (PR2): a catalog of unicellular eukaryote small sub-unit rRNA sequences with curated taxonomy. Nucleic Acids Res. 2012;41:D597–604. 10.1093/nar/gks1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustavsson BM, Magnér J, Almroth BC et al. Chemical monitoring of Swedish coastal waters indicates common exceedances of environmental thresholds, both for individual substances as well as their mixtures. Mar Pollut Bull. 2017; 122:409–19. 10.1016/j.marpolbul.2017.06.082. [DOI] [PubMed] [Google Scholar]
- Gustavsson M, Kreuger J, Bundschuh M et al. Pesticide mixtures in the Swedish streams: environmental risks, contributions of individual compounds and consequences of single-substance oriented risk mitigation. Sci Total Environ. 2017; 598:973–83. 10.1016/j.scitotenv.2017.04.122. [DOI] [PubMed] [Google Scholar]
- Hattenrath-Lehmann TK, Marcoval MA, Berry DL et al. The emergence of dinophysis acuminata blooms and DSP toxins in shellfish in New York waters. Harmful Algae. 2013;26:33–44. 10.1016/j.hal.2013.03.005. [DOI] [Google Scholar]
- Havskum H, Hansen PJ. Net growth of the bloom-forming dinoflagellate heterocapsa triquetra and pH: why turbulence matters. Aquat Microb Ecol. 2006;42:55–62. 10.3354/ame042055. [DOI] [Google Scholar]
- Health Canada . Use of new approach methods (NAMs) in risk assessment. Fact sheet series: Topics in risk assessment of substances under the Canadian Environmental Protection Act, 1999 (CEPA). 2025. https://www. canada.ca/en/health-canada/services/chemical-substances/fact- sheets/use-new-approach-methods-risk-assessment.html. (4 August 2025, date last accessed).
- Hernández-Urcera J, Rial P, García-Portela M et al. Notes on the cultivation of two mixotrophic dinophysis species and their ciliate prey mesodinium rubrum. Toxins. 2018;10:505. 10.3390/toxins10120505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper DU, Chapin FS, Ewel JJ et al. Effects of biodiversity on ecosystem functioning: a consensus of current knowledge. Ecol Monogr. 2005;75:3–35. 10.1890/04-0922. [DOI] [Google Scholar]
- Hothorn T, Bretz F, Westfall P et al. Package ‘multcomp’. Simultaneous inference in general parametric models. 2016; Vienna, Austria: Project for Statistical Computing. 10.1002/bimj.200810425. [DOI] [PubMed] [Google Scholar]
- Inostroza PA, Carmona E, Arrhenius A et al. Target screening of chemicals of emerging concern (CECs) in surface waters of the Swedish West Coast. Data. 2023;8:93. 10.3390/data8060093 [DOI] [Google Scholar]
- Jönander C, Backhaus T, Dahllöf I. Single substance and mixture toxicity of dibutyl-phthalate and sodium dodecyl sulphate to marine zooplankton. Ecotoxicol Environ Saf. 2022;234:113406. 10.1016/j.ecoenv.2022.113406. [DOI] [PubMed] [Google Scholar]
- Jönander C, Dahllöf I. Short and long-term effects of low-sulphur fuels on marine zooplankton communities. Aquat Toxicol. 2020;227:105592. 10.1016/j.aquatox.2020.105592. [DOI] [PubMed] [Google Scholar]
- Kalani K, Kushwaha V, Sharma P et al. In vitro, in silico and in vivo studies of ursolic acid as an anti-filarial agent. PLoS One. 2014;9:e111244. 10.1371/journal.pone.0111244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovarich S, Ceriani L, Ciacci A et al. OpenFoodTox: EFSA’s chemical hazards database. 2020. 10.5281/zenodo.3693783. (1 November 2020, date last accessed). [DOI]
- Liu J. Pharmacology of oleanolic acid and ursolic acid. J Ethnopharmacol. 1995;49:57–68. 10.1016/0378-8741(95)90032-2. [DOI] [PubMed] [Google Scholar]
- Löewe S, Muischnek H. Über kombinationswirkungen mitteilung: hilfsmittel der Fragestellung. Naunyn Schmiedeberg’s Archiv Pharmacol. 1926;114:313–26. [Google Scholar]
- Mafra Jr L, Nolli P, Mota L et al. Multi-species okadaic acid contamination and human poisoning during a massive bloom of Dinophysis acuminata complex in southern Brazil. Harmful Algae. 2019;89:101662. 10.1016/j.hal.2019.101662. [DOI] [PubMed] [Google Scholar]
- Maloney EM, Villeneuve DL, Jensen KM et al. Evaluation of complex mixture toxicity in the Milwaukee Estuary (WI, USA) using whole-mixture and component-based Evaluation methods. Environ Toxicol Chem. 2023;42:1229–56. 10.1002/etc.5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JL, Santi I, Pitta P et al. Towards quantitative metabarcoding of eukaryotic plankton: an approach to improve 18S rRNA gene copy number bias. MBMG. 2022;6:e85794. 10.3897/mbmg.6.85794. [DOI] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–2. 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- Martin O, Scholze M, Ermler S et al. Ten years of research on synergisms and antagonisms in chemical mixtures: a systematic review and quantitative reappraisal of mixture studies. Environ Int. 2021;146:106206. 10.1016/j.envint.2020.106206. [DOI] [PubMed] [Google Scholar]
- May M. Crop protection in sugar beet. Pest Outlook. 2001;12:188–91. 10.1039/b108605g. [DOI] [Google Scholar]
- McClellan KJ, Markham A. Telmisartan. Drugs. 1998;56:1039–44. 10.2165/00003495-199856060-00007. [DOI] [PubMed] [Google Scholar]
- Melnikov F, Kostal J, Voutchkova-Kostal A et al. Assessment of predictive models for estimating the acute aquatic toxicity of organic chemicals. Green Chem. 2016;18:4432–45. 10.1039/C6GC00720A. [DOI] [Google Scholar]
- Naeem S, Li S. Biodiversity enhances ecosystem reliability. Nature. 1997;390:507–9. 10.1038/37348. [DOI] [Google Scholar]
- National Center for Biotechnology Information . PubChem compound summary for CID 448223. Benzoylecgonine. 2023a. https://pubchem.ncbi.nlm.nih.gov/compound/Benzoylecgonine. (21 March 2023, date last accessed). [Google Scholar]
- National Center for Biotechnology Information . PubChem compound summary for CID 31099. Formetanate. 2023b. https://pubchem.ncbi.nlm.nih.gov/compound/Formetanate. (15 March 2023, date last accessed). [Google Scholar]
- Nichols RV, Vollmers C, Newsom LA et al. Minimizing polymerase biases in metabarcoding. Mol Ecol Resour. 2018;18:927–39. 10.1111/1755-0998.12895. [DOI] [PubMed] [Google Scholar]
- Nishitani G, Nagai S, Takano Y et al. Growth characteristics and phylogenetic analysis of the marine dinoflagellate dinophysis infundibulus (Dinophyceae). Aquat Microb Ecol. 2008;52:209–21. 10.3354/ame01233. [DOI] [Google Scholar]
- Not F, del Campo J, Balagué V et al. New insights into the diversity of marine picoeukaryotes. PLoS One. 2009;4:e7143. 10.1371/journal.pone.0007143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nys C, Asselman J, Hochmuth JD et al. Mixture toxicity of nickel and zinc to Daphnia magna is noninteractive at low effect sizes but becomes synergistic at high effect sizes. Environ Toxicol Chem. 2015;34:1091–102. 10.1002/etc.2902. [DOI] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R et al. Package ‘vegan’ Community Ecology Package, Version. 2013;2:1–295. [Google Scholar]
- Quintanilla Rodriguez BS, Correa R. Raloxifene. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing, 2022. [Updated 2022 May 22]. https://www.ncbi.nlm.nih.gov/books/NBK544233/ [Google Scholar]
- Raffa RB, Friderichs E, Reimann W et al. Opioid and nonopioid components independently contribute to the mechanism of action of tramadol, an’atypical’opioid analgesic. J Pharmacol Exp Ther. 1992;260:275–85. 10.1016/S0022-3565(25)11227-5. [DOI] [PubMed] [Google Scholar]
- Reuschenbach P, Silvani M, Dammann M et al. ECOSAR model performance with a large test set of industrial chemicals. Chemosphere. 2008;71:1986–95. 10.1016/j.chemosphere.2007.12.006. [DOI] [PubMed] [Google Scholar]
- Saad OS, Lin X, Ng TY et al. Genome size, rDNA copy, and qPCR assays for Symbiodiniaceae. Front Microbiol. 2020;11:847. 10.3389/fmicb.2020.00847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze T, Ahel M, Ahlheim J et al. Assessment of a novel device for onsite integrative large-volume solid phase extraction of water samples to enable a comprehensive chemical and effect-based analysis. Sci Total Environ. 2017;581-582:350–8. 10.1016/j.scitotenv.2016.12.140. [DOI] [PubMed] [Google Scholar]
- Sieburth JM, Smetacek V, Lenz J. Pelagic ecosystem structure: heterotrophic compartments of the plankton and their relationship to plankton size fractions 1. Limnol Oceanogr. 1978;23:1256–63. 10.4319/lo.1978.23.6.1256. [DOI] [Google Scholar]
- Simmons JE, Teuschler LK, Gennings C et al. Component-based and whole-mixture techniques for addressing the toxicity of drinking-water disinfection by-product mixtures. J Toxicol Environ Health Part A. 2004;67:741–54. 10.1080/15287390490428215. [DOI] [PubMed] [Google Scholar]
- Smith JL, Tong M, Kulis D et al. Effect of ciliate strain, size, and nutritional content on the growth and toxicity of mixotrophic dinophysis acuminata. Harmful Algae. 2018;78:95–105. 10.1016/j.hal.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song G, He Y, Cai Z. A fluorescence spectroscopic study of the interaction between norfloxacin and DNA. Can J Anal Sci Spectros. 2004;49:203–9. [Google Scholar]
- Spilsbury FD, Warne MSJ, Backhaus T. Risk assessment of pesticide mixtures in Australian rivers discharging to the Great Barrier Reef. Environ Sci Technol. 2020;54:14361–71. 10.1021/acs.est.0c04066. [DOI] [PubMed] [Google Scholar]
- Stadhouders R, Pas SD, Anber J et al. The effect of primer-template mismatches on the detection and quantification of nucleic acids using the 5′ nuclease assay. J Mol Diagn. 2010;12:109–17. 10.2353/jmoldx.2010.090035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeck T, Bass D, Nebel M et al. Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol Ecol. 2010;19:21–31. 10.1111/j.1365-294X.2009.04480.x. [DOI] [PubMed] [Google Scholar]
- Straub D, Peltzer A. nf-core/Ampliseq. London: Zenodo; 2019. [Google Scholar]
- Strom LS, Morello TA. Comparative growth rates and yields of ciliates and heterotrophic dinoflagellates. J Plankton Res. 1998;20:571–84. 10.1093/plankt/20.3.571. [DOI] [Google Scholar]
- Syberg K, Backhaus T, Banta G et al. Toward a conceptual approach for assessing risks from chemical mixtures and other stressors to coastal ecosystem services. Integr Environ Assess Manag. 2016; 13:376–86. 10.1002/ieam.1849. [DOI] [PubMed] [Google Scholar]
- Szöcs E, Stirling T, Scott ER et al. webchem: an R package to retrieve chemical information from the web. J Stat Softw. 2020;93:1–17. [Google Scholar]
- Tashima T. The structural use of carbostyril in physiologically active substances. Bioorg Med Chem Lett. 2015;25:3415–9. 10.1016/j.bmcl.2015.06.027. [DOI] [PubMed] [Google Scholar]
- U.S. EPA . Operation manual for the ecological structure-activity relationship model (ECOSAR) Class Program. 2022a. Available from: https://www.epa.gov/tsca-screening-tools/ecological-structure-activity-relationships-program-ecosar-operation-manual.
- U.S. EPA . ECOTOX User Guide: eCOTOXicology Knowledgebase System. Version 5.3. 2023. Available at: http://www.epa.gov/ecotox/ (6 February 2023, date last accessed). [Google Scholar]
- Utermöhl H. Zur vervollkommnung der quantitativen phytoplankton-methodik: mit 1 tabelle und 15 abbildungen im text und auf 1 tafel. Int Ver Für Theor Und Angew Limnol Mitt. 1958;9:1–38. [Google Scholar]
- Vanryckeghem F, Huysman S, Van Langenhove H et al. Multi-residue quantification and screening of emerging organic micropollutants in the Belgian part of the North Sea by use of Speedisk extraction and Q-Orbitrap HRMS. Mar Pollut Bull. 2019;142:350–60. 10.1016/j.marpolbul.2019.03.049. [DOI] [PubMed] [Google Scholar]
- Wen J-H, Xiong Y-Q. The effect of herbal medicine danshensu and ursolic acid on pharmacokinetics of rosuvastatin in rats. Eur J Drug Metab Pharmacokinet. 2011;36:205–11. 10.1007/s13318-011-0048-7. [DOI] [PubMed] [Google Scholar]
- Wickham H. ggplot2. WIREs Comput Stats. 2011;3:180–5. 10.1002/wics.147. [DOI] [Google Scholar]
- Yarimizu K, Sildever S, Hamamoto Y et al. Development of an absolute quantification method for ribosomal RNA gene copy numbers per eukaryotic single cell by digital PCR. Harmful Algae. 2021;103:102008. 10.1016/j.hal.2021.102008. [DOI] [PubMed] [Google Scholar]
- Zhou L, Fan D, Yin W et al. Comparison of seven in silico tools for evaluating of daphnia and fish acute toxicity: case study on Chinese Priority Controlled Chemicals and new chemicals. BMC Bioinf. 2021;22:1–31. 10.1186/s12859-020-03903-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Kovarich S, Ceriani L, Ciacci A et al. OpenFoodTox: EFSA’s chemical hazards database. 2020. 10.5281/zenodo.3693783. (1 November 2020, date last accessed). [DOI]
Supplementary Materials
Data Availability Statement
The data underlying this article are available in Zenodo, at https://zenodo.org/records/13988647.







