Abstract
C3G is a guanine nucleotide exchange factor (GEF) for Rap1, and is activated via Crk adaptor protein. To understand the physiological role of C3G, we generated C3G knockout mice. C3G–/– homozygous mice died before embryonic day 7.5. The lethality was rescued by the expression of the human C3G transgene, which could be excised upon the expression of Cre recombinase. From the embryo of this mouse, we prepared fibroblast cell lines, MEF-hC3G. Expression of Cre abolished the expression of C3G in MEF-hC3G and inhibited cell adhesion-induced activation of Rap1. The Cre-expressing MEF-hC3G showed impaired cell adhesion, delayed cell spreading and accelerated cell migration. The accelerated cell migration was suppressed by the expression of active Rap1, Rap2 and R-Ras. Expression of Epac and CalDAG-GEFI, GEFs for Rap1, also suppressed the accelerated migration of the C3G-deficient cells. This observation indicated that Rap1 activation was sufficient to complement the C3G deficiency. In conclusion, C3G-dependent activation of Rap1 is required for adhesion and spreading of embryonic fibroblasts and for the early embryogenesis of the mouse.
Keywords: C3G/cell adhesion/cell migration/knockout mouse/Rap1
Introduction
Ras-family G proteins function as molecular switches in growth, differentiation, survival and adhesion of eukaryotic cells (Bos, 1997; Campbell et al., 1998). They cycle between GDP-bound inactive and GTP-bound active forms. This cycling is mediated by guanine nucleotide exchange factor (GEF), the activator, and GTPase-activating proteins (GAPs), the inactivator (Bourne et al., 1990; Downward, 1992). A variety of signals, external or internal, control the on/off status of the Ras-family G proteins via the activation or inactivation of GEFs and GAPs (Overbeck et al., 1995; Bos, 1997).
C3G was identified as one of the two major proteins bound to the Src homology 3 (SH3) domain of the Crk oncogene product (Knudsen et al., 1994; Tanaka et al., 1994). The C-terminus of C3G consists of the CDC25 homology domain, which is a catalytic domain of the Ras-family GEF. C3G contains three Crk SH3-binding sequences and one p130Cas SH3-binding sequence in the central region (Kirsch et al., 1998). The N-terminal region of C3G negatively regulates its GEF activity (Ichiba et al., 1999). Many types of stimulation induce binding of the Crk–C3G complex to phosphotyrosine-containing proteins, including receptor-type tyrosine kinases, p130Cas and paxillin (reviewed by Kiyokawa et al., 1997). Following translocation from the cytosol to the cell membrane, C3G becomes phosphorylated on Tyr504, and the negative regulation by the N-terminal region is repressed to increase the GEF activity (Ichiba et al., 1999).
C3G promotes the guanine nucleotide exchange reaction of Rap1 and Rap2, and, to a lesser extent, it also stimulates R-Ras and TC21 (Gotoh et al., 1995, 1997; Ohba et al., 2000a,b). In contrast to Ras, the function of which has been studied extensively, the physiological role of the substrates of C3G remains elusive. A function of Rap1 is to antagonize Ras (Kitayama et al., 1989; Yatani et al., 1991; Sakoda et al., 1992; Boussiotis et al., 1997; Okada et al., 1998), probably by inhibiting the Ras-dependent activation of mitogen-activated protein kinase (MAPK) (Cook et al., 1993; Hu et al., 1997). However, in some cell types, Rap1 activates the MAPK cascade, as does Ras (Yoshida et al., 1992; Vossler et al., 1997; Altschuler and Ribeiro-Neto, 1998; York et al., 1998). Moreover, it is reported that growth factor-induced activation of Rap1 does not correlate with the repression of Ras-dependent MAPK activation (Zwartkruis et al., 1998). Recently, it has been shown that Rap1 contributes integrin-mediated cell adhesion (Posern et al., 1998; Tsukamoto et al., 1999; Katagiri et al., 2000; Reedquist et al., 2000), although the mechanism underlying this phenomenon is yet to be analyzed.
Rap2, the amino acid sequence of which shows 60% identity with Rap1, is regulated by the same set of GEFs and GAPs as is Rap1 (Ohba et al., 2000a). Rap2 binds to a set of effectors very similar to those of Rap1 (Janoueix-Lerosey et al., 1998; Nancy et al., 1999), and it inhibits Ras-dependent MAPK activation, as does Rap1 (Ohba et al., 2000a). A unique feature of Rap2 is its low sensitivity to GAPs, which permits more than half of Rap2 to remain in a GTP-bound active state in adherent cells.
R-Ras and TC21 are the two members of the R-Ras subfamily and are regulated by the same GEFs and GAPs (Ohba et al., 2000b). R-Ras and TC21 activate MAPK, as does the classical Ras (Cox et al., 1994; Graham et al., 1999; Movilla et al., 1999; Rosario et al., 1999); however, they may have unique functions, such as inhibition of apoptosis (Suzuki et al., 1997) and stimulation of cell adhesion (Zhang et al., 1996).
The C3G–Rap1 signaling cascade has been studied genetically in Drosophila melanogaster. Overexpression of wild-type C3G (DC3G) does not cause any detectable abnormality in the developing eye; however, expression of membrane-targeted active C3G leads to an adult rough-eye phenotype, as does the expression of active Ras or active Rap1 (Hariharan et al., 1991; Ishimaru et al., 1999). This rough-eye phenotype due to the membrane-targeted C3G is suppressed by reduction of the gene dose of Ras1, ksr, rolled (MAPK) or Rap1, indicating that the effect of DC3G overactivation is mediated by the RAS–MAPK pathway and RAP1. Overexpression of Rapgap, the inactivator, also induces a rough-eye phenotype, which is exacerbated by reduction of the Rap1 gene dosage (Chen et al., 1997). Removal of maternal Rap1 inhibits ventral furrow closure and head involution of the embryo (Hariharan et al., 1991), and it also perturbs the migration of pole cells and mesodermal cells (Asha et al., 1999). Rap1 is required for imaginal disc development, but not for adult survival (Asha et al., 1999). Thus, the C3G–Rap1 pathway in Drosophila has at least two roles: as a regulator of morphogenesis in the adult stage and as a mediator of cell proliferation and cell fate specification in the developmental stage.
Here, we demonstrate that the C3G gene is essential for mouse embryogenesis and that C3G-dependent Rap1 activation promotes cell adhesion and cell spreading, but represses cell migration.
Results
Structure of the mouse C3G gene
We sequenced the mouse C3G genome to determine the exon–intron structure (Figure 1A). We could not obtain a genomic clone that contained the initiation codon of mouse C3G. Therefore, an exon, which corresponded to cDNA from nucleotide 281 to 394 of human C3G cDNA, was temporarily designated as exon 2. The 23rd exon contained the stop codon.
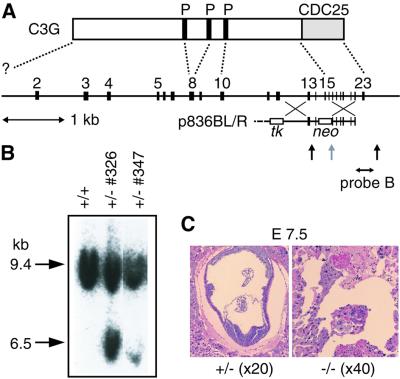
Fig. 1. Generation of C3G knockout mice. (A) Schematic structure of the mouse C3G gene and C3G protein. Coding exons 2–23 are shown at the top. A targeting vector, p836BL/R, consisted of 3.5 and 6.0 kb homologous DNA fragments, the PGK-neo cassette replacing exons 15 and 16, and the MC1-tk cassette at the 5′ end of the targeting vector. The diagnostic probe (probe B) and the EcoRV sites (vertical arrows) are indicated at the bottom. P, proline-rich Crk-binding regions; CDC25, CDC25 homology region. (B) Southern blot analysis of mouse genomic DNA. EcoRV-digested DNAs from ES cells were separated on a 0.5% agarose gel, transferred to a nylon membrane and hybridized with fluorescein isothiocyanate (FITC)-labeled probe B. The mouse genotype was identified by 9.4 and 6.5 kb fragments that were derived from the wild-type and mutant alleles, respectively. (C) Horizontal sections of uterus of a C3G+/– mouse crossed with a C3G+/– mouse and examined at E7.5. Embryos of C3G–/– mice did not conform to any histological structure, as shown on the right. A control section from a C3G+/– embryo is shown on the left.
Generation of C3G knockout mice
To produce a targeted disruption of the mouse C3G gene, we constructed a C3G disruption vector, p836BL/R, containing 3.5 and 6.0 kb homologous DNA fragments, the PGK-neo cassette replacing exons 15 and 16, and the MC1-tk cassette being at the 5′ end of the targeting vector (Figure 1A). Embryonic stem (ES) cells were transfected with linearized p836BL/R by electroporation, and clones resistant to both G418 and 1-(2-deoxy-2-fluoro-β-d-arabinofuranosyl)-5-iodouracil were isolated. Homo logous recombinant clones were detected by Southern blot analysis with probe B, a flanking sequence on the 3′ side (Figure 1B), and the neo probe (data not shown). Of 165 clones, two were targeted for the C3G gene. Both of the targeted clones were injected into C57BL/6/blastocysts, which gave rise to chimeras. Chimeric mice were crossed with C57BL/6 mice, and mice heterozygous for the mutation in the C3G locus were identified among offspring with agouti coat color by Southern blot analysis. Homozygous mutant offspring could not be recovered from crosses between heterozygous mice, suggesting that the mutation causes lethality during embryogenesis.
To determine the stage at which mutant embryos died, we collected embryos from staged matings and genotyped them by using a PCR assay (Table I). When we examined implantation sites at embryonic days 7.5 and 8.5 (E7.5 and E8.5), nearly a quarter of the sites contained only degenerating embryonic tissue within the decidual stroma. In some cases, little DNA for the PCR assay was retrieved from the degenerating tissue and, therefore, these cases were excluded from the data shown in Table I. However, the remaining samples with sufficient amounts of DNA were identified with C3G–/– mice. Histological examination of the implantation sites with these abnormal embryos revealed the absence of typical fetal or placental structures (Figure 1C). These findings indicate that C3G was essential for embryogenesis, and the mutant embryos died shortly after implantation (around day 5).
Table I. Genotyping of embryos arising from C3G heterozygous crosses.
| Gestational age | No. of embryos or postnatal mice |
|||
|---|---|---|---|---|
| Total | +/+ | +/– | –/– | |
| E7.5 | 23 | 7 | 14 | 3 |
| E8.5 | 19 | 5 | 13 | 1 |
| Postnatal | 91 | 36 | 55 | 0 |
Complementation of mouse C3G by the human C3G gene
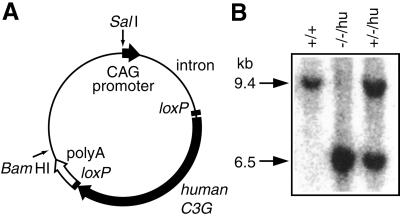
To confirm that the embryonic lethality of C3G homozygous mice resulted from the C3G deficiency, we expres sed human C3G to rescue the viability of C3G–/– mice. The human C3G cDNA was placed under the regulation of the chicken β-actin promoter and the cytomegalovirus (CMV) immediate early enhancer (Figure 2A). A 5.7 kb SalI– BamHI fragment was microinjected into the pronuclei of fertilized eggs. Of 36 offspring, seven founders (F0 mice) were identified by PCR screening at the age of 4 weeks. All founders were fertile and were crossed with C3G+/– mice. In F1 mice, we could not observe any effect of the expression of the human C3G gene. C3G+/– mice carrying the human C3G transgene, indicated hereafter as C3G+/–/hu, were then crossed. From two transgenic lines, we obtained mice deficient in mouse C3G and expressing human C3G, C3G–/–/hu (Figure 2B). However, in both mouse lines, the numbers of C3G–/–/hu mice were lower than expected (Table II), indicating that expression of human C3G did not completely rescue the mouse C3G deficiency. We used a CAG promoter, which shows potent activity in a wide range of cells (Niwa et al., 1991), to express the human C3G gene; however, in some tissues and at some developmental stages, it may not provide sufficient C3G. We never obtained C3G-deficient mice without the human C3G transgene, confirming that the C3G gene was essential for embryogenesis and that the human C3G gene complemented the C3G deficiency.
Fig. 2. Complementation of mouse C3G deficiency by the human C3G transgene. (A) Schematic illustration of an expression plasmid of the human C3G transgene. The human C3G gene, which is sandwiched with two loxP recombination sites, is placed downstream of the chicken β-actin promoter and the CMV immediate early enhancer. SalI and BamHI sites, which were used to isolate the transgene expression unit, are indicated by arrows. (B) Southern blot analysis of transgenic mice crossed with C3G knockout mice. The genotype of mouse C3G and the presence of the human C3G transgene are indicated at the top.
Table II. Genotyping of living mice from C3G+/–/hu crosses.
| Parents | No. of mice |
||||
|---|---|---|---|---|---|
| +/+/hu | +/+/– | +/–/hu | +/–/– | –/–/hu | |
| +/–/huA × +/–/– | 9 | 6 | 8 | 17 | 3 |
| –/–/huA × +/–/– | 0 | 0 | 11 | 16 | 9 |
| +/–/huB × +/–/– | 10 | 17 | 10 | 29 | 3 |
| –/–/huB × +/–/– | 0 | 0 | 14 | 15 | 2 |
A and B indicate transgenic mouse lines A and B, respectively.
Isolation of embryonic fibroblasts from C3G–/–/hu mice
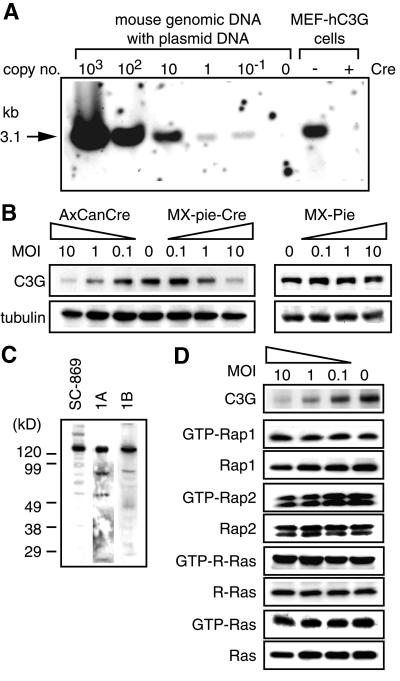
To study the role of C3G further, we obtained mouse embryonic fibroblasts (MEFs) from C3G–/–/hu embryos at E12.5. In these cell lines, which were named MEF-hC3G, the integrated human C3G transgene could be excised by infection with the Cre-expressing retrovirus, MX-pie-Cre, or adenovirus, AxCANCre. Equal amounts of genomic DNA of MEF-hC3G and irrelevant mouse genomic DNA mixed with standard C3G plasmid were digested with XhoI and analyzed by Southern blotting (Figure 3A). MEF-hC3G cells were estimated to carry 20 copies of the human C3G transgene per cell. By Cre expression, the quantity of integrated C3G was decreased to less than one copy per cell. We further confirmed by immunoblotting that removal of the human C3G transgene abolished the expression of C3G protein in MEF-hC3G cells (Figure 3B). We did not detect any truncated C3G protein that might be generated by homologous recombination by use of antibodies raised against three different regions of C3G (Figure 3C). However, contrary to our expectation, the loss of C3G in MEF-hC3G did not decrease the basal level amounts of GTP-bound forms of Rap1, Rap2 and R-Ras, which are the substrates of C3G (Figure 3D).
Fig. 3. Cre-dependent disruption of the C3G gene. (A) MEF-hC3G cells were infected with a recombinant retrovirus, MX-pie (indicated as Cre –) or MX-pie-Cre (Cre +), and selected in DMEM containing 2 µg/ml puromycin for 48 h. A 5 µg aliquot of DNA from the cells and from an irrelevant mouse tail biopsy sample containing the indicated copy numbers of the human C3G genes were digested with XhoI and analyzed by Southern blotting. (B) MEF-hC3G cells were infected with Cre-carrying adenovirus (AxCanCre), Cre-carrying retrovirus (MX-pie-Cre) and control virus (MX-pie) at the multiplicity of infection (MOI) indicated at the top of panels. Forty-eight hours after infection, cells were lysed in lysis buffer and separated by SDS–PAGE, followed by immunoblotting by use of anti-C3G antibody. The filter was reprobed with anti-tubulin monoclonal antibody to confirm that similar amounts of lysates were analyzed (lower panels). (C) Cell lysates of MEF-hC3G were separated by SDS–PAGE, followed by western blotting by use of anti-C3G polyclonal antibody, sc-869, anti-C3G serum 1A or 1B. (D) MEF-hC3G cells were infected with Cre-carrying retrovirus. After 48 h, cells were lysed in lysis buffer and GTP-bound G proteins were collected by use of either GST–Raf-RBD (for Ras and R-Ras) or GST–RalGDS-RBD (for Rap1 and Rap2). The resulting complexes were precipitated by glutathione–Sepharose beads and analyzed by SDS–PAGE and western blotting by use of antibodies. Small aliquots of lysates were analyzed by immunoblotting to confirm a similar level of expression of G proteins. For the detection of GTP-bound R-Ras, MEF-hC3G cells were infected with MSCV-R-Ras retrovirus and maintained in DMEM containing 2 µg/ml puromycin for 48 h. The cells were analyzed as described, except that anti-FLAG antibody was used to detect the expressed R-Ras.
Impaired cell attachment and cell adhesion of C3G-deficient cells
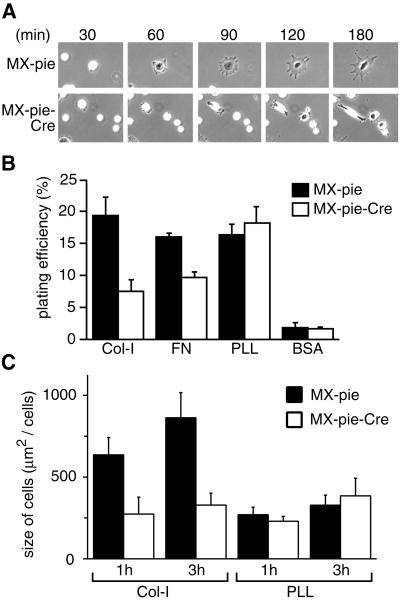
Although the morphology of Cre-expressing MEF-hC3G cells was indistinguishable from that of the parent MEF- hC3G cells, cell attachment and cell spreading after replating were significantly impaired by C3G deficiency (Figure 4A). The replating efficiency was quantitated by labeling of cells with the fluorescent dye 2′,7′-bis- (2-carboxyethyl)-5-(and-6)-carboxyfluorescein acetoxymethyl ester (BCECF, AM). As shown in Figure 4B, C3G enhanced cell attachment to dishes coated with collagen type I or fibronectin (FN), but not those coated with poly-l-lysine. Cell spreading was quantitated by measurement of cell sizes 1 and 3 h after replating. Again, C3G induced cell spreading on the dishes coated with collagen type I, but not with poly-l-lysine (Figure 4C). These results suggested that C3G was required for the cell attachment and cell spreading that were mediated by specific interaction between the extracellular matrices and integrin.
Fig. 4. Requirement for C3G in cell adhesion and cell spreading. (A) MEF-hC3G cells were infected with MX-pie-Cre or MX-pie. After 48 h, cells were trypsinized, kept in suspension for 1 h and plated on dishes coated with collagen type I. The cells were observed by time-lapse microscopy. We show representative photographs at the indicated time points. (B) MEF-hC3G cells were infected with MX-pie or MX-pie-Cre. After 48 h, cells were trypsinized and labeled with BCECF, AM. The cells were plated on 96-well black-colored plates coated with the reagents indicated at the bottom and incubated for 1 h at 37°C in a CO2 incubator. Cells were washed three times with HBSS, and the fluorescence intensity was measured at excitation and emission wavelengths of 488 and 530 nm, respectively. Plating efficiency is shown as the average for three wells, with the SE. (C) MEF-hC3G cells infected with MX-pie or MX-pie-Cre were plated to dishes coated with collagen (Col-I) or poly-l-lysine (PLL). Twenty EGFP-positive cells were photographed after 1 and 3 h and measured for size. Average and SE are shown.
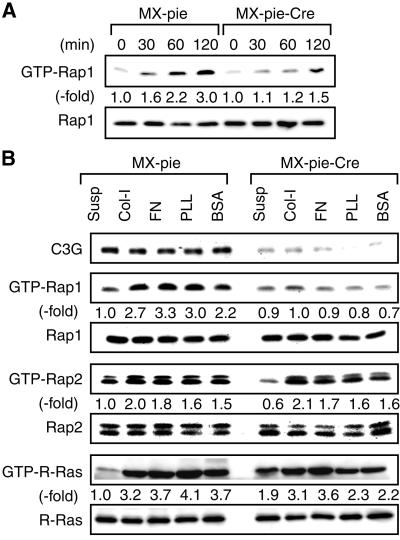
Cell attachment-induced Rap1 activation in C3G-deficient cells
To understand the mechanism by which C3G modulated cell attachment and cell spreading, we examined the activation of Rap1 after replating (Figure 5A). Rap1 was activated rapidly upon cell attachment in the untreated MEF-hC3G cells, whereas Rap1 activation was significantly attenuated in the MEF-hC3G cells infected with MX-pie-Cre. C3G activates the Rap and R-Ras subfamilies in vitro and in 293T cells (Ohba et al., 2000a,b). Therefore, we next proceeded to examine the effect of C3G deficiency on Rap2 and R-Ras (Figure 5B). The results are summarized as follows: (i) Rap1, Rap2 and R-Ras were all activated upon attachment of MEF-hC3G cells to the culture dishes; (ii) C3G deficiency inhibited the activation of Rap1 upon cell attachment, but not that of Rap2: neither of these phenomena was affected by the type of extracellular matrix; (iii) C3G deficiency inhibited R-Ras activation partially, suggesting the involvement of other GEF(s) in cell attachment-induced R-Ras activation. These results indicated that lack of Rap1 activation caused a decrease in cell attachment and cell spreading of C3G-deficient cells.
Fig. 5. Rap1 activation induced by cell adhesion. (A) MEF-hC3G cells were infected with MX-pie-Cre or MX-pie. After 48 h, cells were trypsinized, kept in suspension for 1 h and plated on dishes coated with collagen type I. Cells were lysed at the indicated times. After normalizing on the protein quantity, the level of GTP-Rap1 was analyzed by Bos’s method. (B) MEF-hC3G cells were infected with MX-pie or MX-pie-Cre, maintained for 48 h, trypsinized, kept in suspension for 1 h and plated on dishes pre-coated as noted at the top. After 1 h, we harvested cells and examined the levels of GTP-bound Rap1, Rap2 and R-Ras. For the detection of GTP-R-Ras, we used MEF-hC3G cells infected with MSCV-R-Ras retrovirus as described in Figure 3C.
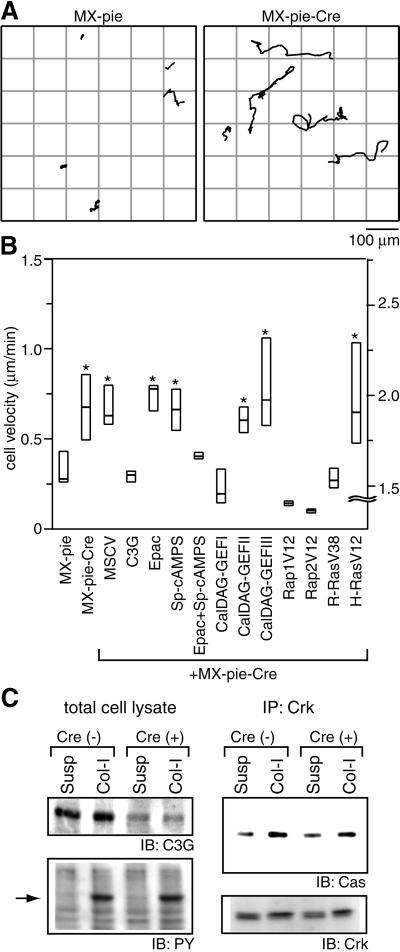
Increase in cell motility by C3G deficiency
Another phenotype of the C3G-deficient cells that we noticed was an increase in cell motility. To quantitate this observation, we recorded phase-contrast and fluorescence images of MEF-hC3G cells infected with MX-pie or MX-pie-Cre for 8 h with a time-lapse fluorescence microscope and obtained the cell paths with a cell-tracking program (Figure 6A). The velocity of C3G-deficient MEF-hC3G cells was higher than that of the control MEF-hC3G cells on collagen-coated dishes (Figure 6B). The difference was statistically significant by t-test and Welch’s test (P <0.01). When we used poly-l-lysine-coated dishes, the velocity of MEF-hC3G cells was not affected by the expression of Cre (data not shown). Similar results were obtained by using different isolates of MEF-hC3G cells. In another cell line derived from C3G+/–/– embryos, we did not find any effect of Cre on the cell motility (data not shown).
Fig. 6. Increased cell motility of C3G-deficient cells. (A) MEF-hC3G cells infected with MX-pie or MX-pie-Cre were trypsinized, kept in suspension for 1 h and plated on collagen-coated dishes. Starting after 1 h, cell images were collected every 3 min under time-lapse fluorescence microscopy equipped with a cooled CCD camera. Paths of the center of EGFP-positive cells during 8 h recording time were traced with MetaMorph2 software. (B) MEF-hC3G cells or MEF-hC3G cells expressing the proteins listed at the bottom were infected with MX-pie-Cre as indicated. We analyzed the cells as in (A) and obtained the mean velocities of 20 cells for each sample. Mid-line, top and bottom of each box indicate median, 75th quartile and 25th quartile, respectively. Cells that show a significant difference from the control, MX-pie infected cells, by t-test and Welch test (P <0.01) are marked with an asterisk at the top of the box. Note that scales shown on the right are for the cells expressing H-RasV12 used. (C) MEF-hC3G cells prepared as described in (A) were lysed and immunoprecipitated with anti-Crk monoclonal antibody and a mixture of protein G– and protein A–Sepharose. Total cell lysates and immunoprecipitated proteins were separated by SDS–PAGE, followed by immunoblotting with anti-C3G anbibody, anti-phosphotyrosine antibody (PY), anti-p130Cas antibody (Cas) or anti-Crk antibody.
We next examined whether the expression of other GEFs for Ras-family G proteins could reduce the cell motility of C3G-deficient MEF-hC3G cells (Figure 6B). As expected, re-introduction of C3G into C3G-deficient MEF-hC3G cells reduced the cell motility to the level of the parent MEF-hC3G cells. Expression of Epac, a cAMP-responsive GEF for Rap1 and Rap2, reduced the cell motility to the level of the parent MEF-hC3G cells only in the presence of a cAMP analog, Sp-cAMPS. CalDAG-GEFI, which is another GEF for Rap1, Rap2 and R-Ras and is constitutively active in many cell types (Yamashita et al., 2000), also reduced the cell motility. In contrast, CalDAG-GEFII, a GEF for the Ras and R-Ras subfamilies, and CalDAG-GEFIII, a pan-Ras GEF, did not reduce the cell motility of C3G-deficient MEF-hC3G cells. We further tested the effect of GTPase-deficient mutants of Ras-family G proteins. Rap1 and Rap2 reduced the cell motility most strongly, and R-Ras did so moderately. GTPase-deficient H-Ras, in contrast, remarkably increased the cell motility. Thus, the effect of C3G deficiency in cell migration was antagonized by the activation of its substrates, Rap1, Rap2 and R-Ras. Finally, because Crk and p130Cas are postulated to function upstream to C3G and to increase cell motility (Ohashi et al., 1999; Uemura and Griffin, 1999; Cho and Klemke, 2000; Yano et al., 2000), we confirmed that the C3G deficiency did not affect cell adhesion-induced phosphorylation of p130Cas or Crk binding to p130Cas (Figure 6C).
Discussion
Requirement for C3G in early embryogenesis
At least eight GEFs have been reported to activate Rap1 in mammalian cells (reviewed by Zwartkruis and Bos, 1999). The mortality of C3G-deficient embryos showed that none of these GEFs could complement the loss of C3G during development. This is not surprising because, except for C3G, Rap1 GEFs are expressed in a more or less tissue-specific manner. Moreover, it should be noted that C3G is the only Rap1 GEF that has been linked definitively to the tyrosine kinase signaling pathway. The defect in the very early stage of embryogenesis also suggested that C3G functions in a signal transduction cascade that is universal and essential to most animal cells. In concordance with this view, C3G is expressed ubiquitously in mammalian cells, and the orthologs of C3G have been identified in organisms from D.melanogaster to man (Ishimaru et al., 1999).
Substrates of C3G
We have shown previously that C3G promotes the guanine nucleotide exchange reaction of Rap1, Rap2 and R-Ras in vitro and in 293T cells (Gotoh et al., 1995, 1997; Ohba et al., 2000a). Although Rap1, Rap2 and R-Ras were all activated upon cell attachment to the substratum, only Rap1 activation was abolished by C3G deficiency. This observation suggests the activation of other GEFs, which stimulate Rap2 and/or R-Ras upon cell adhesion. A plausible candidate for such a molecule is PDZ-GEF1, which may interact with cell adhesion molecules via its PDZ domain and which stimulates Rap2 more efficiently than Rap1 (de Rooij et al., 1999; Liao et al., 1999; Ohtsuka et al., 1999). Alternatively, Rap2 may not require any GEF to restore its basal GTP level after cell attachment because of its low sensitivity to GAP (Ohba et al., 2000a). It is of note that our finding does not negate the possibility that C3G activates Rap2 or R-Ras under other conditions. For example, we have found that the v-crk oncogene product activates R-Ras in a manner dependent on C3G (Mochizuki et al., 2000). Compared with Crk-II, v-Crk protein possesses viral Gag at its N-terminus and lacks the SH3 domain at the C-terminus. This difference may be responsible for the recruitment of the v-Crk–C3G complex to a different subcellular localization, where R-Ras is a preferable substrate for C3G.
Requirement for C3G for cell spreading
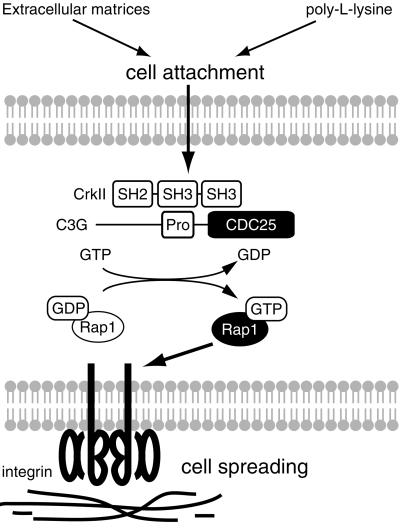
Accumulating evidence has shown that the Crk–C3G– Rap1 pathway plays a pivotal role in integrin-mediated signaling; however, the precise function of this pathway in cell adhesion remains elusive (Zwartkruis and Bos, 1999; Caron et al., 2000; Katagiri et al., 2000; Reedquist et al., 2000). Rap1 is activated not only by integrin-mediated cell attachment, but also by non-specific attachment to the positively charged substratum (Figure 5B; and Posern et al., 1998). By use of conditional knockout of C3G, we found that C3G was required for Rap1 activation in both cases. However, Rap1 activation by C3G was not sufficient for cell spreading, which required specific extracellular matrices for integrin (Figure 4C). This view is supported by several reports that Rap1 activates some types of integrin to increase cell adhesion (Caron et al., 2000; Katagiri et al., 2000; Reedquist et al., 2000). Thus, Rap1 activation by cell adhesion is dependent on C3G, but it is not necessarily mediated by a specific interaction between integrin and the substratum. In contrast, Rap1-dependent cell spreading requires a specific interaction between integrin and the substratum (Figure 7).
Fig. 7. A model for the role of C3G–Rap1 signaling pathway in cell attachment and cell spreading. In mouse embryonic fibroblasts, Rap1 is activated by C3G upon cell attachment, irrespective of the substratum. The activated Rap1 triggers an inside-out signal of integrin to induce cell adhesion, which requires specific interaction between integrin and extracellular matrices. SH2, SH3, Pro and CDC25 indicate Src homology 2, Src homology 3, proline-rich and yeast CDC25 homology domains, respectively.
Increased cell migration in C3G-deficient fibroblasts
Crk and its related protein, CrkL, are known to enhance migration in various cell types, probably through binding to p130Cas (Ohashi et al., 1999; Uemura and Griffin, 1999; Cho and Klemke, 2000; Yano et al., 2000). In Ba/F3 hematopoietic cells, expression of C3G enhances CrkL-dependent cell migration, suggesting a positive role for C3G in cell migration. In clear contrast to these previous reports, we found that C3G deficiency increased cell migration in MEFs. This increase in cell migration was suppressed by the expression of Epac or CalDAG-GEFI, as well as C3G, indicating that Rap1 activation inhibited cell migration. The discrepancy between the role of C3G and Crk in cell migration may be explained by the fact that previous reports relied mostly on the overexpression of Crk, which elevates tyrosine phosphorylation of p130Cas and paxillin (Birge et al., 1993; Sakai et al., 1994). It has been reported that tyrosine phosphorylation of p130Cas increases cell migration, whereas tyrosine phosphorylation of paxillin suppresses cell migration (Yano et al., 2000). Thus, under physiological conditions, the Crk–C3G complex may function downstream of paxillin, whereas, in cells overexpressing Crk, the complex may function downstream of p130Cas to increase cell migration. Alternatively, the difference in the cell types used in each study may explain the discrepancy. For example, upon cell adhesion, Cbl is the major phosphotyrosine-containing protein in Ba/F3 hematopoietic cells (Uemura and Griffin, 1999), whereas p130Cas and paxillin are phosphorylated predominantly on tyrosine in fibroblasts or epithelial cells (Birge et al., 1993; Sakai et al., 1994).
Downstream of Rap1 and Ras in cell migration
Rap1 inhibits Ras-induced transformation and MAPK activation in some cell types (Kitayama et al., 1989; Yatani et al., 1991; Sakoda et al., 1992; Boussiotis et al., 1997; Okada et al., 1998), which is, at least partially, due to the inhibition of Ras-induced activation of Raf serine/threonine kinase (Cook et al., 1993; Hu et al., 1997). We showed that expression of active Ras transformed the sluggish C3G-deficient cells into rapidly migrating cells (Figure 6), providing another example that Rap1 and Ras antagonize each other. However, between the C3G-deficient cells and the parental cells, we did not find any difference in MAPK activation induced by cell adhesion or epidermal growth factor stimulation (data not shown). Thus, a target or targets other than Raf appear to respond to Rap1 and Ras in opposite ways and control the velocity of cell migration. Such a candidate molecule is AF-6/canoe, which binds to both Ras and Rap1 and serves as a peripheral component of tight junctions in epithelial cells (Kuriyama et al., 1996; Yamamoto et al., 1997; Linnemann et al., 1999; Boettner et al., 2000).
In conclusion, we have shown that C3G-dependent Rap1 activation promotes cell adhesion and cell spreading, but suppresses cell migration. These functions of the C3G–Rap1 pathway appear to underlie the lethality of C3G-deficient mice in the very early stage of development.
Materials and methods
Generation of C3G–/– mice, and creation and identification of transgenic mice
The generation of the knockout mice and transgenic mice is described in the Supplementary data (available at The EMBO Journal Online).
Establishment of embryonic fibroblast cell lines
C3G+/– mice expressing the human C3G transgene were intercrossed, and E12.5 embryonic fibroblasts were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum (FCS) as described previously (Brugarolas et al., 1998). The genotype of each cell line was determined by Southern blotting and PCR as described in the Supplementary data. The cell lines that lacked the mouse C3G and carried the human C3G were named MEF-hC3G.
Virus packaging and infection
Coding sequences of C3G, Epac (de Rooij et al., 1998; Kawasaki et al., 1998a), CalDAG-GEFI, II and III (Kawasaki et al., 1998b; Yamashita et al., 2000), Rap1V12 (Kitayama et al., 1989), Rap2V12 (Ohba et al., 2000a), R-RasWT and R-RasV38 (Rey et al., 1994) were subcloned into pMSCV-pac-Flag, which was a derivative of pMSCV-pac (Hawley et al., 1994) with a Flag tag sequence at the 5′ end of the cloning site. The coding sequence of Cre recombinase was subcloned into another retroviral plasmid, pMX-pie (Onishi et al., 1996), which carries enhanced green fluorescent protein (EGFP) downstream of the internal ribosomal entry site (IRES). The resulting plasmid was named pMX-pie-Cre.
Recombinant retroviruses were produced as described previously (Pear et al., 1993). Bosc23 cells were transfected with retroviral vectors and a replication-incompetent helper vector, pCL-Eco (Naviaux et al., 1996), by the calcium phosphate co-precipitation method. After 48 h, the culture medium was cleared by centrifugation and used as virus stock. For virus titration, we developed an NIH-3T3-derived cell line that was transfected stably with loxP-LacZ (Kanegae et al., 1995). These NIH3T3-loxP-LacZ cells were infected with serially diluted virus stocks. At day 2, puromycin (Sigma, St Louis, MO) was added to 2 µg/ml. At day 4, the number of cell colonies was counted under a microscope for determination of the virus titer. Expression of EGFP or LacZ in the remaining colonies was confirmed by fluorescence microscopy or by staining with X-gal. By this procedure, we obtained virus stocks with at least 2 × 106 colony-forming units/ml. Cre-expressing adenovirus, AxCANCre (RIKEN Gene Bank, 1748), was propagated in HEK293 cells (Japan Cell Resource Bank) as described previously (Kanegae et al., 1995). Cells were infected with AxCANCre at multiplicities of infection of 0.1–10 and were assayed at least 48 h after infection.
Antibodies and reagents
Anti-C3G polyclonal antibody, sc-869 and anti-Rap1 polyclonal antibody, sc-065, were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Anti-C3G antisera 1A and 1B have been reported previously (Tanaka et al., 1994). Anti-pan-Ras and anti-tubulin monoclonal antibodies were from Calbiochem (San Diego, CA). Anti-Rap2 and anti-FLAG monoclonal antibodies were from BD Transduction Laboratories (Bluegrass Lexington, KY) and Sigma, respectively. Sp-cAMPS triethylamine (Sp-cAMPS) was purchased from Research Biochemical International (Natick, MA).
Detection of GTP-bound Ras-family G proteins
Escherichia coli expression vectors for the GST-fused Ras/Rap1-binding domain (RBD), pGEX-RalGDS-RBD and pGEX-Raf-RBD, were obtained from J.L.Bos (Utrecht University, The Netherlands) and S.Hattori (National Center for Neurology and Psychiatry, Tokyo, Japan), respectively. Detection of GTP-bound Ras-family G proteins and purification of GST fusion proteins were performed by Bos’s method with slight modifications (Franke et al., 1997; Ohba et al., 2000a). Briefly, cells were lysed in lysis buffer (50 mM Tris pH 7.5, 150 mM NaCl, 5 mM MgCl2, 1% NP-40, 0.5% deoxycholic acid, 0.1% SDS, 1 mM Na3VO4), clarified by centrifugation, and incubated with GST–RBD fusion proteins. We used GST–Raf-RBD for the detection of GTP-Ras and R-Ras, and GST–RalGDS-RBD for Rap1 and Rap2, respectively. The resulting complexes of GTP-bound G proteins and GST–RBD were precipitated by use of glutathione–Sepharose beads, and proteins bound to the beads were separated by SDS–PAGE, followed by immunoblotting with specific antibodies for each G protein. Bound antibodies were detected by an ECL chemiluminescence system (Amersham Pharmacia) and analyzed with an LAS-1000 image analyzer (Fuji-Film). We could not detect endogenous GTP-R-Ras in MEF-hC3G cells; therefore, cells were infected with an R-Ras encoding retrovirus, MSCV-R-RasWT, selected with puromycin and analyzed as described above.
Cell adhesion assay
The cell adhesion assay was performed essentially as described previously (Newton et al., 1997). Briefly, 96-well microplates (Greiner Labortechnik GmbH, Frickenhausen, Germany) were coated for 2 h at room temperature with type-I collagen (Nitta Gelatin, Osaka, Japan), fibronectin or poly-l-lysine (Sigma). Cells were treated with 0.125% trypsin and 2 mM EDTA in phosphate-buffered saline (PBS), resuspended in minimal essential medium (MEM) containing 100 µg/ml trypsin inhibitor (Sigma), labeled with the fluorescent dye BCECF, AM (Molecular Probes, Leiden, The Netherlands) for 30 min at 37°C in Hanks’ balanced salt solution (HBSS; 140 mM NaCl, 5 mM KCl, 1 mM CaCl2, 2 mM MgSO4, 0.5 mM MgCl2, 0.3 mM Na2HPO4, 0.4 mM KH2PO4, 0.1% glucose, 0.035% NaHCO3), washed twice in HBSS and resuspended in MEM. The labeled cells were seeded to coated 96-well plates at a density of 5 × 104 cells/well in a volume of 100 µl and allowed to adhere for 1 h at 37°C. Plates were washed three times with pre-warmed HBSS. The fluorescence intensity was measured by FluoroscanII (Labsystems, Helsinki, Finland). The efficiency of cell adhesion was expressed as (fluorescence intensity after wash)/(input fluorescence intensity). In another set of experiments, MEF-hC3G cells were harvested similarly, kept in suspension for 1 h at 37°C, plated to 6-cm-diameter dishes and allowed to adhere for 1 h. The GTP-bound forms of Rap1, Rap2 and R-Ras were quantitated as described.
Time-lapse microscopy and cell-tracking analysis
MEF-hC3G cells infected with MX-pie or MX-pie-Cre were seeded on a collagen-coated 35-mm-diameter glass base dish (Asahi Techno Glass Co., Tokyo). Cells were imaged on a Zeiss Axiovert microscope (Carl Zeiss, Jena, Germany) with a cooled CCD camera (Roper Scientific, Trenton, NJ), controlled by MetaMorph2 software (Universal Image, West Chester, PA) as described previously (Miyawaki et al., 1997). Phase-contrast and fluorescence images were recorded every 3 min. We obtained a series of time-lapse images and analyzed the size, paths and velocities of EGFP-positive cells by use of a cell-tracking application handled with MetaMorph2. In other experiments, MEF-hC3G cells were inoculated with MSCV-pac-derived retroviruses and selected in the presence of puromycin for 2 days. The puromycin-resistant cells were inoculated with MX-pie-Cre and analyzed as described.
Supplementary data
Supplementary data for this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank J.L.Bos, A.Wittinghofer, A.Hall, M.R.Gold, S.Hattori, I.Saitoh, H.Kitayama, K.Kaibuchi, K.Harder, T.Akagi and M.Noda for materials, and F.Tashiro, Y.Komagata, K.Kimura, F.Ohba, K.Okuda and N.Otsuka for technical assistance. This work was supported by grants from the Ministry of Health and Welfare, from the Ministry of Education, Science, Sports, and Culture, from the Princess Takamatsu Cancer Research Fund and from The Japan Health Science Foundation.
References
- Altschuler D.L. and Ribeiro-Neto,F. (1998) Mitogenic and oncogenic properties of the small G protein Rap1b. Proc. Natl Acad. Sci. USA, 95, 7475–7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asha H., de Ruiter,N.D., Wang,M.G. and Hariharan,I.K. (1999) The Rap1 GTPase functions as a regulator of morphogenesis in vivo. EMBO J., 18, 605–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birge R.B., Fajardo,J.E., Reichman,C., Shoelson,S.E., Songyang,Z., Cantley,L.C. and Hanafusa,H. (1993) Identification and characteriz ation of a high-affinity interaction between v-Crk and tyrosine-phosphorylated paxillin in CT10-transformed fibroblasts. Mol. Cell. Biol., 13, 4648–4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettner B., Govek,E.E., Cross,J. and Van Aelst,L. (2000) The junctional multidomain protein AF-6 is a binding partner of the Rap1A GTPase and associates with the actin cytoskeletal regulator profilin. Proc. Natl Acad. Sci. USA, 97, 9064–9069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos J.L. (1997) Ras-like GTPases. Biochim. Biophys. Acta, 1333, M19–31. [DOI] [PubMed] [Google Scholar]
- Bourne H.R., Sanders,D.A. and McCormick,F. (1990) The GTPase superfamily: a conserved switch for diverse cell functions. Nature, 348, 125–132. [DOI] [PubMed] [Google Scholar]
- Boussiotis V.A., Freeman,G.J., Berezovskaya,A., Barber,D.L. and Nadler,L.M. (1997) Maintenance of human T cell anergy: blocking of IL-2 gene transcription by activated Rap1. Science, 278, 124–128. [DOI] [PubMed] [Google Scholar]
- Brugarolas J., Bronson,R.T. and Jacks,T. (1998) p21 is a critical CDK2 regulator essential for proliferation control in Rb-deficient cells. J. Cell Biol., 141, 503–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell S.L., Khosravi-Far,R., Rossman,K.L., Clark,G.J. and Der,C.J. (1998) Increasing complexity of Ras signaling. Oncogene, 17, 1395–1413. [DOI] [PubMed] [Google Scholar]
- Caron E., Self,A.J. and Hall,A. (2000) The GTPase Rap1 controls functional activation of macrophage integrin αMβ2 by LPS and other inflammatory mediators. Curr. Biol., 10, 974–978. [DOI] [PubMed] [Google Scholar]
- Chen F., Barkett,M., Ram,K.T., Quintanilla,A. and Hariharan,I.K. (1997) Biological characterization of Drosophila Rapgap1, a GTPase activating protein for Rap1. Proc. Natl Acad. Sci. USA, 94, 12485–12490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S.Y. and Klemke,R.L. (2000) Extracellular-regulated kinase activation and CAS/Crk coupling regulate cell migration and suppress apoptosis during invasion of the extracellular matrix. J. Cell Biol., 149, 223–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook S.J., Rubinfeld,B., Albert,I. and McCormick,F. (1993) RapV12 antagonizes Ras-dependent activation of ERK1 and ERK2 by LPA and EGF in Rat-1 fibroblasts. EMBO J., 12, 3475–3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox A.D., Brtva,T.R., Lowe,D.G. and Der,C.J. (1994) R-Ras induces malignant, but not morphologic, transformation of NIH3T3 cells. Oncogene, 9, 3281–3288. [PubMed] [Google Scholar]
- de Rooij J., Zwartkruis,F.J., Verheijen,M.H., Cool,R.H., Nijman,S.M., Wittinghofer,A. and Bos,J.L. (1998) Epac is a Rap1 guanine-nucleotide-exchange factor directly activated by cyclic AMP. Nature, 396, 474–477. [DOI] [PubMed] [Google Scholar]
- de Rooij J., Boenink,N.M., van Triest,M., Cool,R.H., Wittinghofer,A. and Bos,J.L. (1999) PDZ-GEF1, a guanine nucleotide exchange factor specific for Rap1 and Rap2. J. Biol. Chem., 274, 38125–38130. [DOI] [PubMed] [Google Scholar]
- Downward J. (1992) Regulation of p21ras by GTPase activating proteins and guanine nucleotide exchange proteins. Curr. Opin. Genet. Dev., 2, 13–18. [DOI] [PubMed] [Google Scholar]
- Franke B., Akkerman,J.W. and Bos,J.L. (1997) Rapid Ca2+-mediated activation of Rap1 in human platelets. EMBO J., 16, 252–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotoh T. et al. (1995) Identification of Rap1 as a target for the Crk SH3 domain-binding guanine nucleotide-releasing factor C3G. Mol. Cell. Biol., 15, 6746–6753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotoh T., Niino,Y., Tokuda,M., Hatase,O., Nakamura,S., Matsuda,M. and Hattori,S. (1997) Activation of R-Ras by Ras-guanine nucleotide-releasing factor. J. Biol. Chem., 272, 18602–18607. [DOI] [PubMed] [Google Scholar]
- Graham S.M., Oldham,S.M., Martin,C.B., Drugan,J.K., Zohn,I.E., Campbell,S. and Der,C.J. (1999) TC21 and Ras share indistinguishable transforming and differentiating activities. Oncogene, 18, 2107–2116. [DOI] [PubMed] [Google Scholar]
- Hariharan I.K., Carthew,R.W. and Rubin,G.M. (1991) The Drosophila roughened mutation: activation of a rap homolog disrupts eye development and interferes with cell determination. Cell, 67, 717–722. [DOI] [PubMed] [Google Scholar]
- Hawley R.G., Lieu,F.H., Fong,A.Z. and Hawley,T.S. (1994) Versatile retroviral vectors for potential use in gene therapy. Gene Ther., 1, 136–138. [PubMed] [Google Scholar]
- Hu C.D., Kariya,K., Kotani,G., Shirouzu,M., Yokoyama,S. and Kataoka,T. (1997) Coassociation of Rap1A and Ha-Ras with Raf-1 N-terminal region interferes with ras-dependent activation of Raf-1. J. Biol. Chem., 272, 11702–11705. [DOI] [PubMed] [Google Scholar]
- Ichiba T., Hashimoto,Y., Nakaya,M., Kuraishi,Y., Tanaka,S., Kurata,T., Mochizuki,N. and Matsuda,M. (1999) Activation of C3G guanine nucleotide exchange factor for Rap1 by phosphorylation of tyrosine 504. J. Biol. Chem., 274, 14376–14381. [DOI] [PubMed] [Google Scholar]
- Ishimaru S., Williams,R., Clark,E., Hanafusa,H. and Gaul,U. (1999) Activation of the Drosophila C3G leads to cell fate changes and overproliferation during development, mediated by the RAS–MAPK pathway and RAP1. EMBO J., 18, 145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janoueix-Lerosey I., Pasheva,E., de Tand,M.F., Tavitian,A. and de Gunzburg,J. (1998) Identification of a specific effector of the small GTP-binding protein Rap2. Eur. J. Biochem., 252, 290–298. [DOI] [PubMed] [Google Scholar]
- Kanegae Y., Lee,G., Sato,Y., Tanaka,M., Nakai,M., Sakaki,T., Sugano,S. and Saito,I. (1995) Efficient gene activation in mammalian cells by using recombinant adenovirus expressing site-specific Cre recombinase. Nucleic Acids Res., 23, 3816–3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri K., Hattori,M., Minato,N., Irie,S., Takatsu,K. and Kinashi,T. (2000) Rap1 is a potent activation signal for leukocyte function-associated antigen 1 distinct from protein kinase C and phosphatidylinositol-3-OH kinase. Mol. Cell. Biol., 20, 1956–1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki H., Springett,G.M., Mochizuki,N., Toki,S., Nakaya,M., Matsuda,M., Housman,D.E. and Graybiel,A.M. (1998a) A family of cAMP-binding proteins that directly activate Rap1. Science, 282, 2275–2279. [DOI] [PubMed] [Google Scholar]
- Kawasaki H. et al. (1998b) A Rap guanine nucleotide exchange factor enriched highly in the basal ganglia. Proc. Natl Acad. Sci. USA, 95, 13278–13283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsch K.H., Georgescu,M.M. and Hanafusa,H. (1998) Direct binding of p130(Cas) to the guanine nucleotide exchange factor C3G. J. Biol. Chem., 273, 25673–25679. [DOI] [PubMed] [Google Scholar]
- Kitayama H., Sugimoto,Y., Matsuzaki,T., Ikawa,Y. and Noda,M. (1989) A ras-related gene with transformation suppressor activity. Cell, 56, 77–84. [DOI] [PubMed] [Google Scholar]
- Kiyokawa E., Mochizuki,N., Kurata,T. and Matsuda,M. (1997) Role of Crk oncogene product in physiologic signaling. Crit. Rev. Oncog., 8, 329–342. [DOI] [PubMed] [Google Scholar]
- Knudsen B.S., Feller,S.M. and Hanafusa,H. (1994) Four proline-rich sequences of the guanine-nucleotide exchange factor C3G bind with unique specificity to the first Src homology 3 domain of Crk. J. Biol. Chem., 269, 32781–32787. [PubMed] [Google Scholar]
- Kuriyama M. et al. (1996) Identification of AF-6 and canoe as putative targets for Ras. J. Biol. Chem., 271, 607–610. [DOI] [PubMed] [Google Scholar]
- Liao Y. et al. (1999) RA-GEF, a novel Rap1A guanine nucleotide exchange factor containing a Ras/Rap1A-associating domain, is conserved between nematode and humans. J. Biol. Chem., 274, 37815–37820. [DOI] [PubMed] [Google Scholar]
- Linnemann T., Geyer,M., Jaitner,B.K., Block,C., Kalbitzer,H.R., Wittinghofer,A. and Herrmann,C. (1999) Thermodynamic and kinetic characterization of the interaction between the Ras binding domain of AF6 and members of the Ras subfamily. J. Biol. Chem., 274, 13556–13562. [DOI] [PubMed] [Google Scholar]
- Miyawaki A., Llopis,J., Heim,R., McCaffery,J.M., Adams,J.A., Ikura,M. and Tsien,R.Y. (1997) Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature, 388, 882–887. [DOI] [PubMed] [Google Scholar]
- Mochizuki N., Ohba,Y., Kobayashi,S., Otsuka,N., Graybiel,A.M., Tanaka,S. and Matsuda,M. (2000) Crk Activation of JNK via C3G and R-Ras. J. Biol. Chem., 275, 12667–12671. [DOI] [PubMed] [Google Scholar]
- Movilla N., Crespo,P. and Bustelo,X.R. (1999) Signal transduction elements of TC21, an oncogenic member of the R-Ras subfamily of GTP-binding proteins. Oncogene, 18, 5860–5869. [DOI] [PubMed] [Google Scholar]
- Nancy V., Wolthuis,R.M., de Tand,M.F., Janoueix-Lerosey,I., Bos,J.L. and de Gunzburg,J. (1999) Identification and characterization of potential effector molecules of the Ras-related GTPase Rap2. J. Biol. Chem., 274, 8737–8745. [DOI] [PubMed] [Google Scholar]
- Naviaux R.K., Costanzi,E., Haas,M. and Verma,I.M. (1996) The pCL vector system: rapid production of helper-free, high-titer, recombinant retroviruses. J. Virol., 70, 5701–5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton J.P., Buckley,C.D., Jones,E.Y. and Simmons,D.L. (1997) Residues on both faces of the first immunoglobulin fold contribute to homophilic binding sites of PECAM-1/CD31. J. Biol. Chem., 272, 20555–20563. [DOI] [PubMed] [Google Scholar]
- Niwa H., Yamamura,K. and Miyazaki,J. (1991) Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene, 108, 193–199. [DOI] [PubMed] [Google Scholar]
- Ohashi Y., Iwata,S., Kamiguchi,K. and Morimoto,C. (1999) Tyrosine phosphorylation of Crk-associated substrate lymphocyte-type is a critical element in TCR- and β1 integrin-induced T lymphocyte migration. J. Immunol., 163, 3727–3734. [PubMed] [Google Scholar]
- Ohba Y. et al. (2000a) Rap2 as a slowly responding molecular switch in the rap1 signaling cascade. Mol. Cell. Biol., 20, 6074–6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba Y., Mochizuki,N., Yamashita,S., Chan,A.M., Schrader,J.W., Hattori,S., Nagashima,K. and Matsuda,M. (2000b) Regulatory proteins of R-Ras, TC21/R-Ras2 and M-Ras/R-Ras3. J. Biol. Chem., 275, 20020–20026. [DOI] [PubMed] [Google Scholar]
- Ohtsuka T., Hata,Y., Ide,N., Yasuda,T., Inoue,E., Inoue,T., Mizoguchi,A. and Takai,Y. (1999) nRap GEP: a novel neural GDP/GTP exchange protein for Rap1 small G protein that interacts with synaptic scaffolding molecule (S-SCAM). Biochem. Biophys. Res. Commun., 265, 38–44. [DOI] [PubMed] [Google Scholar]
- Okada S., Matsuda,M., Anafi,M., Pawson,T. and Pessin,J.E. (1998) Insulin regulates the dynamic balance between Ras and Rap1 signaling by coordinating the assembly states of the Grb2–SOS and CrkII–C3G complexes. EMBO J., 17, 2554–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi M. et al. (1996) Applications of retrovirus-mediated expression cloning. Exp. Hematol., 24, 324–329. [PubMed] [Google Scholar]
- Overbeck A.F., Brtva,T.R., Cox,A.D., Graham,S.M., Huff,S.Y., Khosravi-Far,R., Quilliam,L.A., Solski,P.A. and Der,C.J. (1995) Guanine nucleotide exchange factors: activators of Ras superfamily proteins. Mol. Reprod. Dev., 42, 468–476. [DOI] [PubMed] [Google Scholar]
- Pear W.S., Nolan,G.P., Scott,M.L. and Baltimore,D. (1993) Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl Acad. Sci. USA, 90, 8392–8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posern G., Weber,C.K., Rapp,U.R. and Feller,S.M. (1998) Activity of Rap1 is regulated by bombesin, cell adhesion and cell density in NIH3T3 fibroblasts. J. Biol. Chem., 273, 24297–24300. [DOI] [PubMed] [Google Scholar]
- Reedquist K.A., Ross,E., Koop,E.A., Wolthuis,R.M., Zwartkruis,F.J., van Kooyk,Y., Salmon,M., Buckley,C.D. and Bos,J.L. (2000) The small GTPase, rap1, mediates CD31-induced integrin adhesion. J. Cell Biol., 148, 1151–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey I., Taylor-Harris,P., van Erp,H. and Hall,A. (1994) R-ras interacts with rasGAP, neurofibromin and c-raf but does not regulate cell growth or differentiation. Oncogene, 9, 685–692. [PubMed] [Google Scholar]
- Rosario M., Paterson,H.F. and Marshall,C.J. (1999) Activation of the Raf/MAP kinase cascade by the Ras-related protein TC21 is required for the TC21-mediated transformation of NIH 3T3 cells. EMBO J., 18, 1270–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai R., Iwamatsu,A., Hirano,N., Ogawa,S., Tanaka,T., Mano,H., Yazaki,Y. and Hirai,H. (1994) A novel signaling molecule, p130, forms stable complexes in vivo with v-Crk and v-Src in a tyrosine phosphorylation-dependent manner. EMBO J., 13, 3748–3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakoda T., Kaibuchi,K., Kishi,K., Kishida,S., Doi,K., Hoshino,M., Hattori,S. and Takai,Y. (1992) smg/rap1/Krev-1 p21s inhibit the signal pathway to the c-fos promoter/enhancer from c-Ki-ras p21 but not from c-raf-1 kinase in NIH3T3 cells. Oncogene, 7, 1705–1711. [PubMed] [Google Scholar]
- Suzuki J., Kaziro,Y. and Koide,H. (1997) An activated mutant of R-Ras inhibits cell death caused by cytokine deprivation in BaF3 cells in the presence of IGF-I. Oncogene, 15, 1689–1697. [DOI] [PubMed] [Google Scholar]
- Tanaka S. et al. (1994) C3G, a guanine nucleotide-releasing protein expressed ubiquitously, binds to the Src homology 3 domains of CRK and GRB2/ASH proteins. Proc. Natl Acad. Sci. USA, 91, 3443–3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto N., Hattori,M., Yang,H., Bos,J.L. and Minato,N. (1999) Rap1 GTPase-activating protein SPA-1 negatively regulates cell adhesion. J. Biol. Chem., 274, 18463–18469. [DOI] [PubMed] [Google Scholar]
- Uemura N. and Griffin,J.D. (1999) The adapter protein crkl links cbl to C3G after integrin ligation and enhances cell migration. J. Biol. Chem., 274, 37525–37532. [DOI] [PubMed] [Google Scholar]
- Vossler M.R., Yao,H., York,R.D., Pan,M.G., Rim,C.S. and Stork,P.J. (1997) cAMP activates MAP kinase and Elk-1 through a B-Raf- and Rap1-dependent pathway. Cell, 89, 73–82. [DOI] [PubMed] [Google Scholar]
- Yamamoto T., Harada,N., Kano,K., Taya,S., Canaani,E., Matsuura,Y., Mizoguchi,A., Ide,C. and Kaibuchi,K. (1997) The Ras target AF-6 interacts with ZO-1 and serves as a peripheral component of tight junctions in epithelial cells. J. Cell Biol., 139, 785–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita S., Mochizuki,N., Ohba,Y., Tobiume,M., Okada,Y., Sawa,H., Nagashima,K. and Matsuda,M. (2000) CalDAG-GEFIII activation of Ras, R-Ras and Rap1. J. Biol. Chem., 275, 25488–25493. [DOI] [PubMed] [Google Scholar]
- Yano H., Uchida,H., Iwasaki,T., Mukai,M., Akedo,H., Nakamura,K., Hashimoto,S. and Sabe,H. (2000) Paxillin α and Crk-associated substrate exert opposing effects on cell migration and contact inhibition of growth through tyrosine phosphorylation. Proc. Natl Acad. Sci. USA, 97, 9076–9081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatani A., Quilliam,L.A., Brown,A.M. and Bokoch,G.M. (1991) Rap1A antagonizes the ability of Ras and Ras-Gap to inhibit muscarinic K+ channels. J. Biol. Chem., 266, 22222–22226. [PubMed] [Google Scholar]
- York R.D., Yao,H., Dillon,T., Ellig,C.L., Eckert,S.P., McCleskey,E.W. and Stork,P.J. (1998) Rap1 mediates sustained MAP kinase activation induced by nerve growth factor. Nature, 392, 622–626. [DOI] [PubMed] [Google Scholar]
- Yoshida Y., Kawata,M., Miura,Y., Musha,T., Sasaki,T., Kikuchi,A. and Takai,Y. (1992) Microinjection of smg/rap1/Krev-1 p21 into Swiss 3T3 cells induces DNA synthesis and morphological changes. Mol. Cell. Biol., 12, 3407–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Vuori,K., Wang,H., Reed,J.C. and Ruoslahti,E. (1996) Integrin activation by R-ras. Cell, 85, 61–69. [DOI] [PubMed] [Google Scholar]
- Zwartkruis F.J. and Bos,J.L. (1999) Ras and rap1: two highly related small GTPases with distinct function. Exp. Cell Res., 253, 157–165. [DOI] [PubMed] [Google Scholar]
- Zwartkruis F.J., Wolthuis,R.M., Nabben,N.M., Franke,B. and Bos,J.L. (1998) Extracellular signal-regulated activation of Rap1 fails to interfere in Ras effector signalling. EMBO J., 17, 5905–5912. [DOI] [PMC free article] [PubMed] [Google Scholar]