Abstract
Telomerase uses a short stretch of its intrinsic RNA molecule as template for telomere repeat synthesis. Reverse transcription of the RNA template is catalyzed by the telomerase reverse transcriptase (TERT) protein subunit. We demonstrate that human telomerase reconstituted from recombinant TERT and telomerase RNA runs as a dimer on a gel filtration column and that it contains two telomerase RNA molecules. Significantly, a telomerase heterodimer reconstituted from wild-type and mutant telomerase RNA is barely active when compared with the wild-type homodimer. We conclude that the telomerase RNA templates in the active enzyme are interdependent and functionally cooperate with each other. We discuss models that may explain the biological and enzymatic roles of telomerase dimerization.
Keywords: dimer/reverse transcriptase/ribonucleoprotein/telomerase/telomere
Introduction
Telomere ends are replicated by a specialized machinery. In vertebrates, the G-rich strand containing the 3′ end of the chromosome consists of TTAGGG repeats and is synthesized by telomerase, a cellular reverse transcriptase (Greider and Blackburn, 1985, 1989; Yu et al., 1990; Lingner et al., 1997). The C-rich complementary 5′ end of the chromosome is replicated by the conventional DNA polymerases α and δ (Fan and Price, 1997; Diede and Gottschling, 1999; Adams Martin et al., 2000). Telomerase is a ribonucleoprotein enzyme. The telomerase RNA contains a short segment that provides the template for telomere repeat synthesis. Telomerase RNA molecules have diverged quickly in sequence and length, but their secondary structure has been conserved to a large extent during evolution (Romero and Blackburn, 1991; Lingner et al., 1994; Chen et al., 2000). In addition to providing the template, the telomerase RNA contains binding sites for telomerase protein subunits (Gilley and Blackburn, 1999; Mitchell et al., 1999a; Mitchell and Collins, 2000), several of which have been identified (Nugent and Lundblad, 1998). The protein subunit containing the active site is referred to as telomerase reverse transcriptase (TERT) since it carries the active site motifs that are conserved in retroelement-encoded reverse transcriptases (RT) (Eickbush, 1997; Nakamura and Cech, 1998). In addition to the retroviral-related RT domain, TERT contains a telomerase-specific N-terminal region, which has been implicated in RNA binding (Bryan et al., 2000b; Xia et al., 2000). Dyskerin, a protein associated with both human telomerase and small nucleolar ribonucleoprotein particles (snoRNPs), is required for the stability of telomerase RNA, and patients with a defective dyskerin gene have short telomeres (Mitchell et al., 1999b). Est1p is a telomerase protein subunit identified in Saccharomyces cerevisiae that mediates access of telomerase to the telomere (Lundblad and Szostak, 1989; Evans and Lundblad, 1999).
The TERT-related HIV-1 RT is a dimer of two related chains: a 66 kDa subunit and a 51 kDa subunit derived from p66 by proteolytic cleavage (di Marzo Veronese et al., 1986). p51 lacks the RNase H domain, but has in common with p66 the polymerase domain, which is often compared to a right hand with fingers, palm and thumb (Joyce and Steitz, 1994). The palm contains residues critical for polymerase catalytic activity. Despite their identity in amino acid sequence, the two subunits exhibit remarkably distinct folding, resulting in an asymmetric dimer structure. While p66 folds into an open structure, p51 adopts a closed conformation without catalytic function (Kohlstaedt et al., 1992).
There is evidence that yeast telomerase contains more than one active site per telomerase complex (Prescott and Blackburn, 1997a). Using the unusual property of yeast telomerase that it remains stably associated with its elongated product, differentially marked substrates that were elongated in vitro by yeast telomerase were shown to be associated with each other upon elongation, presumably via binding to different active sites in the same telomerase RNP. Additionally, it was found that a yeast telomerase RNA with a mutant non-functional template sequence became a functional template when co-expressed with the wild-type RNA, indicating interaction between separate telomerase RNA strands (Prescott and Blackburn, 1997a,b). On the other hand, size measurements by glycerol gradient centrifugation of purified telomerase from the ciliate Euplotes aediculatus suggest that this telomerase forms a monomer in solution rather than a dimer (Lingner and Cech, 1996). Size measurements of telomerase from Tetrahymena thermophila by gel filtration (Wang and Blackburn, 1997) and of human telomerase by glycerol gradient centrifugation (Schnapp et al., 1998) could not determine the state of multimerization since the polypeptide composition of the analyzed complexes remained unclear.
Here we measure in quantitative experiments the number of telomerase RNA molecules present in recombinant human telomerase complexes. We found that human telomerase contains two telomerase RNA molecules in each RNP. Furthermore, a telomerase RNA template mutant could inactivate the wild-type telomerase counterpart when co-assembled in a heterodimer, indicating an obligatory interaction between the two RNA templates. This has fundamental implications for the mechanism of telomerase and chromosome end maintenance.
Results
Reconstitution of active telomerase
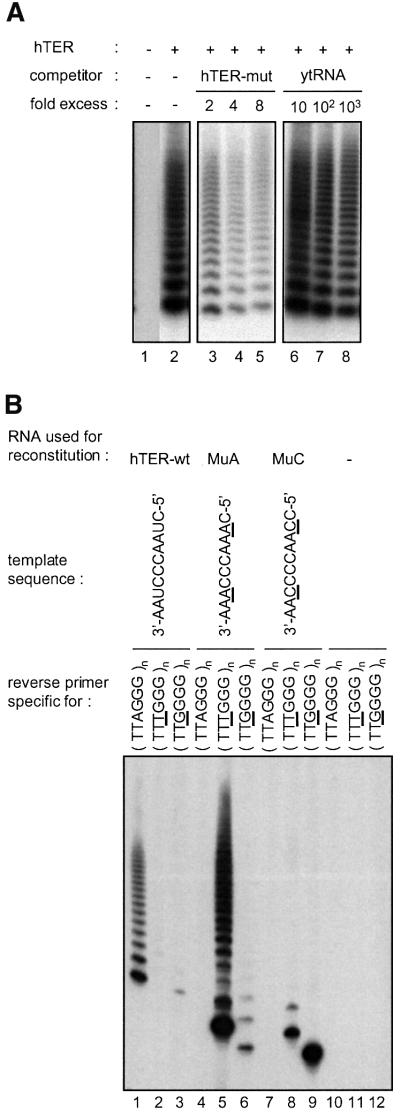
It has been shown previously that human telomerase reverse transcriptase (hTERT) translated in reticulocyte lysates assembles in the presence of telomerase RNA (hTER) into a complex that contains telomerase activity (Weinrich et al., 1997). Reconstitution was dependent on the presence of chaperone proteins in the lysate (Holt et al., 1999). More recently, hTERT expressed in insect cells was also shown to be catalytically active when combined with in vitro transcribed hTER (Masutomi et al., 2000). We independently developed a strategy to express hTERT in insect cells upon infection with baculovirus carrying the hTERT cDNA. Telomerase was reconstituted by incubating insect cell lysates containing recombinant hTERT with in vitro transcribed telomerase RNA. Telomerase activity was detected using the TRAP assay (Kim et al., 1994) in which the products of telomere repeat addition to oligonucleotide substrates are amplified by PCR (Figure 1A). Telomerase activity depended on the presence of full-length hTERT and on hTER. No activity was obtained if either telomerase RNA was omitted (Figure 1A, lane 1) or if a truncated hTERT variant lacking the first 541 amino acids was used in the reconstitution (data not shown). The reconstitution conditions were optimized for time and temperature to 90 min at 30°C, and it was found that high concentrations of ATP (10 mM) stimulated the reconstitution by more than one order of magnitude. An energy requirement for telomerase assembly is consistent with the involvement of chaperones, which have been implicated in the assembly of telomerase in rabbit reticulocyte lysates (Holt et al., 1999). Optimal TRAP activity was obtained when reconstitution was carried out with an estimated 3-fold molar excess of hTER over hTERT. The interaction between the recombinant hTERT and the telomerase RNA was specific since a large excess of an unrelated RNA in the reconstitution reaction did not interfere with the assembly of active telomerase (Figure 1A, lanes 6–8). In contrast, addition of a telomerase RNA containing a mutant template sequence resulted in a significant decrease in activity (Figure 1A, lanes 3–5). However, assembly of telomerase RNA template mutants with recombinant hTERT produced active enzyme when assayed with primers specific for the appropriate mutant telomere sequence in a modified TRAP assay (Figure 1B).
Fig. 1. Reconstitution of active telomerase. (A) The assembly of active telomerase is specific for hTER. TRAP assay for telomerase activity reconstituted with recombinant hTERT expressed in insect cells and in vitro transcribed hTER. An excess of either a telomerase RNA with a mutant template (hTER-mut with the template sequence: 5′-GAUUGGGAUU-3′; lanes 3-5) or unspecific competitor RNA (yeast tRNA; lanes 6-8) was added to the reconstitution mixture. (B) Telomerase was reconstituted with wild-type hTER, one of two different mutant hTER molecules, or without RNA. The telomerase activity of all four samples was measured using either the standard TRAP assay that detects synthesis of TTAGGG repeats (lanes 1, 4, 7, 10) or modified TRAP assays (Feng et al., 1995) that are specific for TTTGGG (lanes 2, 5, 8, 11) and TTGGGG repeats (lanes 3, 6, 9, 12), respectively.
Salt-dependent processivity of recombinant and native telomerase
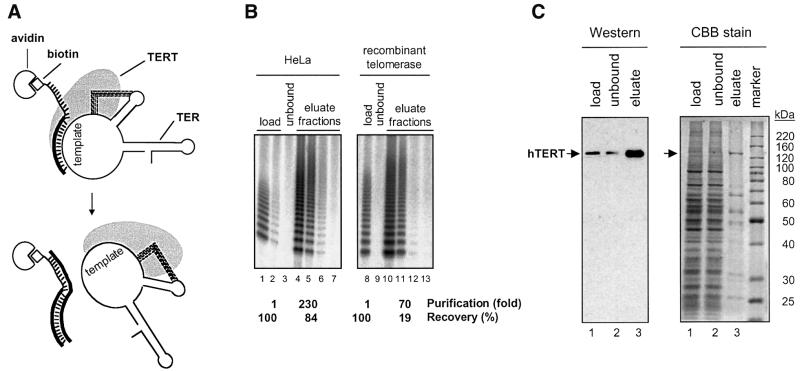
In order to characterize recombinant telomerase and compare it with native human telomerase isolated from HeLa cells, we purified both enzymes on an affinity column with an antisense oligonucleotide complementary to the template sequence (Figure 2A). This method was initially developed for telomerase from Euplotes (Lingner and Cech, 1996) and then adapted to the human system (Schnapp et al., 1998). It resulted in a 230-fold enrichment of native telomerase activity from HeLa cell nuclear extracts and in a 70-fold enrichment of the recombinant activity from insect cell lysates (Figure 2B). Enrichment of telomerase activity correlated with enrichment of the hTERT polypeptide (Figure 2C).
Fig. 2. Purification of telomerase. (A) Affinity purification of telomerase. After binding of telomerase to avidin beads via a biotinylated antisense oligonucleotide and extensive washing, telomerase is eluted with a displacement oligonucleotide. Because the base pairing potential of the displacement oligonucleotide with the affinity oligonucleotide is greater than with hTER, it forms a thermodynamically more stable duplex thus displacing telomerase from the beads. (B) Enrichment of telomerase activity by affinity purification. Telomerase from HeLa cell nuclear extracts and recombinant telomerase were affinity purified and the activity of the load, the unbound and four eluted fractions was determined using the TRAP assay. The fraction volumes used in the TRAP assay were 0.05 µl (lanes 1, 3, 8, 9), 0.02 µl (lane 2), 0.1 µl (lanes 4–7) and 1 µl (lanes 10–13). The recovery refers to the total amount of activity that was recovered after purification. The purification factor was calculated as the ratio of TRAP activity per amount of total protein present in the purified fraction divided by the TRAP activity per amount of total protein present in the load. Titration experiments and mixing experiments between eluate fractions and extract fractions indicated that measured activities were not perturbed by the presence of diffusible inhibitors (compare lane 1 with lane 2; and data not shown). (C) PAGE of recombinant telomerase fractions before and after affinity purification. Analysis by western blotting with an anti-hTERT antibody (left panel) and by staining with Coomassie Brilliant Blue (right panel). Lanes 1 and 2 contain 0.01% and lanes 3 contain 5% of the total fraction. In the eluate (lanes 3) hTERT (arrow) accounts for ∼7% of the total protein.
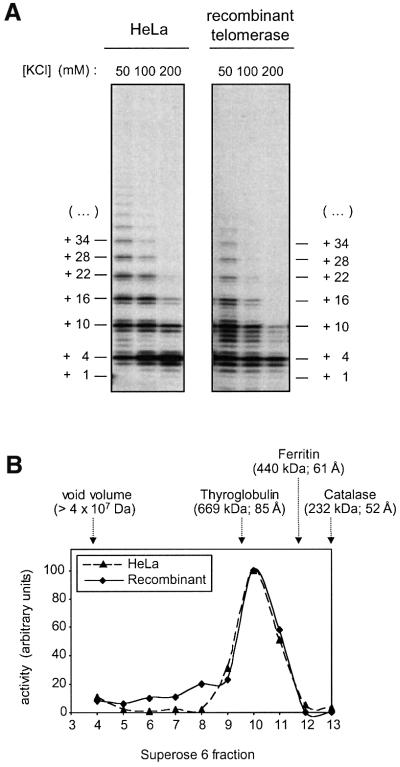
Recombinant and native telomerase were compared in their ability to extend (TTAGGG)3 oligonucleotide in the presence of [α-32P]dGTP, dATP and dTTP using a direct telomerase assay (Figure 3A). Analysis on a sequencing gel showed a similar pattern of products generated by recombinant telomerase and enzyme from HeLa cells, with the characteristic six nucleotide spacing between major bands, which corresponds to the periodicity of telomeric repeats (Figure 3A). The most abundant products correspond to the 5′ boundary of the RNA template, the position at which the substrate is predicted either to dissociate from the enzyme or to translocate to the other end of the template for the processive addition of multiple telomeric repeats. Since these experiments were carried out with a >100-fold molar excess of primer over telomerase, rebinding of the enzyme to an extended substrate after dissociation is very unlikely. Therefore, both HeLa cell telomerase and, to a slightly lesser extent, recombinant telomerase were partially processive. Processivity decreased with increasing salt concentration, consistent with published reports (Maine et al., 1999; Sun et al., 1999).
Fig. 3. Comparison of native and recombinant telomerase. (A) Direct telomerase assay. Affinity-purified telomerase was incubated with (TTAGGG)3, dATP, dTTP and [α-32P]dGTP in presence of different concentrations of KCl. Telomerase products were separated on a sequencing gel. (B) Analysis of the native molecular mass of the telomerase RNP. Affinity-purified telomerase was fractionated by gel filtration on a Superose 6 column in running buffer containing 500 mM KCl and the activity of the fractions was determined using the TRAP assay. The void volume of the column and the elution volume of three marker proteins are indicated, together with the respective molecular masses and Stokes radii (in brackets).
Native molecular mass
The sizes of affinity-purified recombinant and HeLa telomerase were analyzed by gel filtration on a Superose 6 column, which was run in 500 mM KCl (Figure 3B). The fractions were assayed for telomerase activity and the elution profile was compared with that of marker proteins of known molecular mass and Stokes radius. HeLa cell-derived telomerase activity peaked in fraction 10, which corresponds to an approximate molecular mass of 600 kDa. A very similar elution profile was obtained for the recombinant enzyme. The peak activity eluted at a position corresponding to a complex of more than twice the size of a complex composed of one molecule of hTERT (127 kDa) and one molecule of hTER (150 kDa). Thus, these data suggested that the eluted activity might be due to telomerase dimers consisting of two hTERT and two hTER moieties. Alternatively, the shape of the eluted RNP may deviate extensively from a globular form. To test the hypothesis that active telomerase forms a dimer, we developed a new method for measuring the number of telomerase RNA molecules present in the telomerase RNP.
The telomerase RNP contains two telomerase RNA molecules
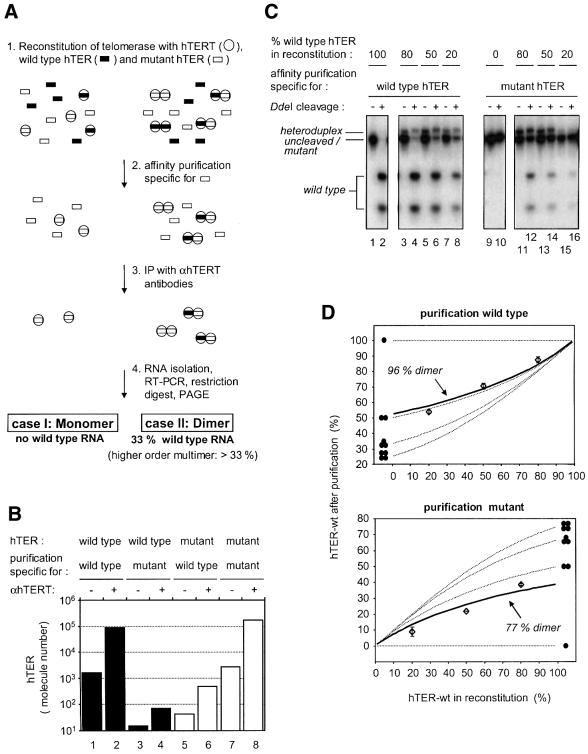
A schematic outline of the experiment that allowed determination of the number of telomerase RNA molecules in the telomerase RNP, along with the expected results for monomeric and dimeric complexes, is shown in Figure 4A. Telomerase was assembled by incubating recombinant hTERT with a mixture of wild-type and a variant hTER harboring a mutant template sequence (Figure 4A, step 1). Reconstituted telomerase was affinity purified using oligonucleotides specific for either wild-type or mutant hTER (Figure 4A, step 2; see also Figure 2A). In order to remove protein-free RNA, hTERT complexes were immunoprecipitated with anti-hTERT antibodies (Figure 4A, step 3). Telomerase RNA was isolated from the purified complexes and analyzed by RT–PCR followed by restriction digestion of the PCR product, allowing discrimination between the wild-type and mutant RNA (Figure 4A, step 4). We tested the specificity of the affinity purification by measuring the amount of wild-type hTER that purified with the mutant-specific oligonucleotide from a reconstitution containing only wild-type hTER and vice versa (Figure 4B). In both cases the purification was highly specific; purification of telomerase containing wild-type RNA with mutant oligonucleotide yielded <0.1% of the amount of RNA purified with wild-type oligonucleotide (Figure 4B, compare columns 2 and 4), while < 1% of RNA was purified using the wild-type-specific oligonucleotide from complexes containing mutant RNA (compare columns 6 and 8).
Fig. 4. Telomerase forms a dimer. (A) Scheme for the method used to determine the number of RNA molecules in the telomerase RNP (see text). (B) Affinity purification allows sequence-specific selection of telomerase complexes containing different template sequences. Telomerase was reconstituted with either wild-type or mutant hTER (template sequence: 5′-GAUUGGGAUU-3′), respectively. One half of the mixture was subjected to a purification specific for wild-type hTER and the other half to a purification specific for mutant hTER. The amount of hTER RNA was measured using quantitative RT–PCR following immunoprecipitation with αhTERT antibodies (+) or rabbit IgG (–). (C) Telomerase contains more than one hTER subunit. The experiment depicted schematically in Figure 3A was performed with different mixtures of wild-type and mutant RNA (lanes 3–8, lanes 11–16). RT–PCR products derived from wild-type hTER were cleavable with DdeI (lane 1 and 2) whereas RT–PCR products derived from mutant hTER were lacking this site (lanes 9 and 10). The band that has a slightly lower mobility than the uncleaved/mutant PCR products corresponds to heteroduplex DNA consisting of one wild-type and one mutant strand. (D) Quantitation of the experiment shown in Figure 4C. Data points (open diamonds) with standard deviations and a best-fitted solid line are shown together with theoretical curves (dotted lines) for telomerase being a monomer (labeled with one filled circle), dimer (two filled circles), trimer (three filled circles) or tetramer (four filled circles).
However, when telomerase was reconstituted with a mixture of wild-type and mutant RNA, significant amounts of wild-type hTER co-purified with the mutant RNA (Figure 4C, lanes 11-16), and mutant hTER co-purified with wild type in the reciprocal experiment (Figure 4C, lanes 3–8). The ratio of wild-type and mutant hTER used in the reconstitution step was varied, and the relative amounts of the two RNAs in the purified RNPs were quantified (Figure 4C and D). Comparison of the measured values with the theoretical values expected for higher order complexes indicated that telomerase forms predominantly a dimer under the experimental conditions, which included incubation steps at 30°C in the presence of 300 mM KCl (see Materials and methods). At salt concentrations below 100 mM KCl, telomerase formed even higher order complexes, whereas in the presence of 500 mM KCl some of the telomerase dimers started to dissociate (data not shown). This dependence on the ionic strength indicates that interactions between charged residues mediate the multimerization of telomerase; it is not known whether this involves the hTERT protein subunit or also the RNA moiety. Nevertheless, from these data and the native molecular mass determined by gel filtration, we conclude that telomerase forms a dimer under physiological salt concentrations.
Functional interaction between subunits of the telomerase dimer
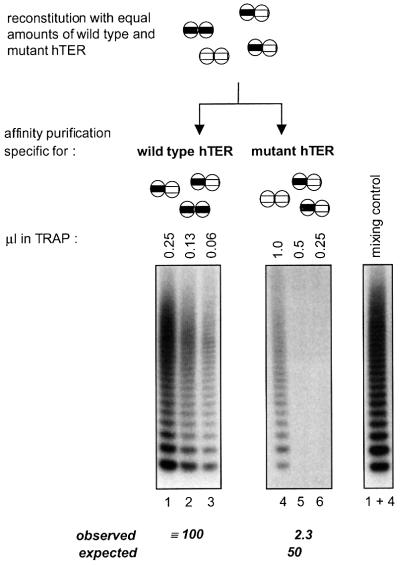
To determine whether both telomerase RNA templates in the RNP dimer would function independently of each other, we compared the specific activity of telomerase homodimers containing two wild-type active sites with that of heterodimers having one wild-type and one mutant active site (Figure 5). As described above, telomerase was reconstituted with a mixture of wild-type and mutant hTER molecules, and homo- and heterodimers were purified either with an oligonucleotide specific for wild-type hTER or an oligonucleotide specific for the mutant. The fraction purified with the wild-type oligonucleotide included wild-type homodimers and wild-type/mutant heterodimers, but no mutant homodimers, whereas the fraction purified with the mutant oligonucleotide only contained mutant homodimers and wild-type/mutant heterodimers but no wild-type homodimers. We compared the activity of the two fractions with a wild-type substrate that is not extended by telomerase containing only mutant hTER, the latter of which was also inactive with other primers that were tested (data not shown). If the activity of each reconstituted telomerase subunit were independent of that of the other subunit in the dimer, we would have expected the activity of enzyme purified with the mutant oligonucleotide to be 50% of that purified with the wild-type oligonucleotide. However, the telomerase activity present in the latter fraction was ∼40 times higher than that in the fraction purified with the mutant oligonucleotide (Figure 5). As expected, this fraction did not contain a diffusible inhibitor as it did not reduce the activity of the fraction that was purified via the wild-type template (Figure 5, compare lanes 1 and 4 with lane 1 + 4). Based on these data we conclude that the two telomerase RNA templates present in the RNP cooperate during extension of an oligonucleotide substrate.
Fig. 5. Functional interaction of telomerase RNA templates. (Upper panel) Schematic outline of the distribution of homo- and heterodimers after reconstitution of telomerase with equal amounts of wild-type and mutant hTER and affinity purification. The fraction obtained with the antisense oligonucleotide specific for the mutant template lacks wild-type homodimers and vice versa. The number of wild-type templates after purification with the antisense oligonucleotide specific for the wild type is twice the number of wild-type templates present in the fraction that was purified via the mutant template. (Lower panel) The activity of eluates was measured using the TRAP assay. Below the gel, measured activities are indicated together with calculated values for non-cooperating subunits (expected), assuming that a heterodimer containing one wild-type active site would be half as active as a wild-type homodimer. The right panel shows a control experiment in which the fractions obtained with both antisense oligonucleotides were mixed after the purification (0.25 µl wild type + 1 µl mutant).
Discussion
We have reconstituted telomerase from recombinant hTERT expressed in insect cells and from telomerase RNA transcribed in vitro to gain an understanding of its quaternary structure. We find that telomerase functions as a dimer with two telomerase RNA moieties per RNP complex. A telomerase dimer of two hTERT polypeptides each associated with an hTER moiety can explain the native molecular mass of the enzyme corresponding to ∼600 kDa. This does not exclude the possibility that other proteins present in the insect cell lysate may be present in the recombinant complex or may be involved in mediating the assembly of the active complex. In fact, an interaction between telomerase and chaperones in the extract appears likely since reconstitution was strongly stimulated by ATP, and the molecular chaperone proteins p23 and Hsp90 have been found to associate with telomerase (Holt et al., 1999). On the other hand, affinity-purified telomerase from HeLa cell nuclear extracts gave a similar profile on the gel filtration column as did the recombinant enzyme. This result is surprising given the number of proteins that have been reported to be associated with telomerase (Bryan and Cech, 1999). However, we find that the molecular mass of the enzyme is considerably larger in crude extracts and at lower salt concentrations (data not shown). Therefore, we suspect that weakly interacting telomerase-associated proteins were stripped from the telomerase complex during purification.
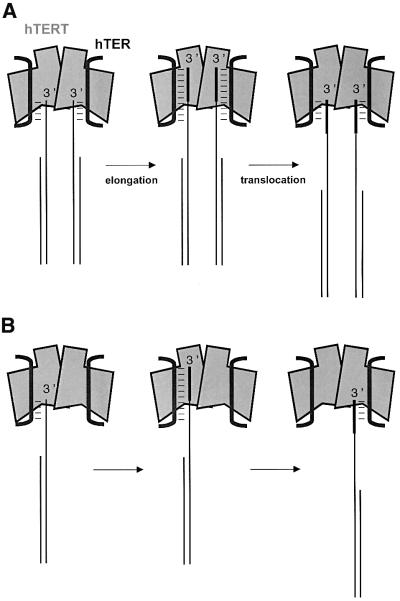
What is the role of the dimerization of human telomerase? Because a mutant template can render the wild-type counterpart nearly inactive when co-assembled in a heterodimer (Figure 5), the two telomerase RNA templates in the dimer do not function independently of each other. The HIV-1 RT dimer, in which one subunit contains the active site and the other subunit furnishes structural support, cannot provide a perfect paradigm for a telomerase dimer since, according to our knowledge, HIV-1 RT binds only one RNA template at a time. The mutant telomerase RNA template was accessible for hybridization with the telomere substrate-mimicking antisense oligonucleotide during the affinity purification, as expected if the mutant template were to occupy an active site separate from the wild-type template. Thus, it appears less likely that the mutant RNA template affected the correct folding of the wild-type template-containing active site in the heterodimer. We propose two alternative models to explain the functional interaction between the telomerase RNA templates (Figure 6). In the first model, the enzyme contains two active sites, both of which bind and extend separate substrate molecules (Figure 6A). A similar model has previously been proposed for yeast telomerase (Prescott and Blackburn, 1997a). The dominant-negative effect of the template mutant (Figure 5) we observed could be explained by a compulsory coordinated extension of the two substrate molecules. This would imply that conformational changes occurring in the two hTERT–hTER subunits during extension of the substrate or upon translocation are linked to each other in mandatory fashion. This could provide a mechanism for a coordinated extension of paired telomere 3′ ends in vivo, such as the two sister chromatids generated by semiconservative DNA replication. Telomerase-mediated extension of the telomere 3′ end may occur in parallel on the newly synthesized and the parental 3′ end of a sister chromatid pair. This is somewhat reminiscent of the dimeric DNA replication machinery associated with coordinated leading and lagging strand synthesis. However, as telomerase acts only on the chromosome 3′ (the leading strand), this model implies that extension of the replicated chromosome 3′ end would be coordinated with extension of the non-replicated parental telomeric 3′ end. This model also implies that the parental templating C-strand for the synthesized leading strand must be resected to allow both 3′ ends to be elongated by telomerase. A resection of the C-strand is supported by the presence of 3′ overhangs at both chromosomal ends, as seen in protozoa, fungi and vertebrates (Klobutcher et al., 1981; Wellinger et al., 1996; Makarov et al., 1997; Munoz-Jordan et al., 2001).
Fig. 6. Models for cooperating telomerase active sites. (A) Obligatory coordinated extension and translocation of two telomere 3′ ends (e.g. sister chromatids). (B) Template switching upon translocation during processive elongation of one telomere 3′ end.
A second model to explain the cooperation of the two template regions is shown in Figure 6B. In this case, the two active sites of telomerase would extend one substrate molecule in a cooperative manner. Substrate extension by one subunit to the 5′ end of the template would be followed by realignment of the extended substrate to the RNA of the second subunit, allowing addition of the next repeat. This template switching mechanism would provide a mechanism for translocation during processive synthesis. The substrate may contact both subunits of the heterodimer simultaneously, at least during translocation. A so-called DNA anchor site, which is separate from the catalytic site, has been proposed to exist in telomerase (Collins and Greider, 1993; Lee and Blackburn, 1993; Hammond et al., 1997; Gandhi and Collins, 1998). This site has been proposed to hold the telomere substrate and allow translocation of its 3′ end on the template without complete dissociation from telomerase. The anchor site may reside in the second hTERT subunit of the telomerase complex, rather than on the same hTERT molecule as proposed previously (Hammond et al., 1997). Thus, while one subunit extends the 3′ end of the substrate, the other subunit would still contact the substrate from the previous elongation cycle. The model in Figure 6B also provides a possible explanation for the observation that in Tetrahymena, co-expression of wild-type and mutant telomerase RNA templates in vivo leads to the synthesis of interdispersed wild-type and mutant telomeric repeats (Yu and Blackburn, 1991), although in vitro, Tetrahymena telomerase is highly processive (Greider, 1991; Lee and Blackburn, 1993; Bryan et al., 2000a). According to our model, even a processive telomerase, if assembled as a heterodimer, would catalyze the synthesis of interdispersed telomere repeats as long as template switching escorts the elongation of the substrate. Future studies will test these and other models.
Materials and methods
Expression of hTERT
The open reading frame of the hTERT cDNA was fused with an N-terminal (His)6 and XPRESS tag and cloned into the SmaI–XbaI sites of the pVL1393 baculovirus transfer vector (PharMingen). To generate recombinant baculoviruses, Sf9 cells that were grown in TNM-FH medium were transfected with the recombinant transfer plasmid and linearized Bac-N-Blue DNA according to the instructions of the supplier (Invitrogen). After 4 days at 27°C, the virus-containing supernatant was harvested and single recombinant viruses were isolated by plaque assay and amplified using High five cells (Invitrogen) that were cultivated in Insect XPRESS medium (BioWhittaker). The virus suspensions carrying the hTERT transgene were titered by end point dilution.
For hTERT production, High five cells were seeded at a density of 1.5 × 107 cells per 13.5 cm dish. After removal of the medium, recombinant virus was added in a total volume of 6 ml of medium at an m.o.i. of 10. The dishes were rocked gently (2 min–1) for 1 h and after addition of 10 ml of fresh medium they were incubated for 40 h at 27°C. Cells were harvested, washed twice with phosphate-buffered saline (PBS) and resuspended in 0.5 ml of cold lysis buffer [20 mM Tris–HCl pH 8.0, 10% v/v glycerol, 5 mM β-mercaptoethanol, 0.1 mM phenylmethylsulfonyl fluoride (PMSF)] supplemented with a protease inhibitor cocktail (complete EDTA-free; Roche) per 13.5 cm dish. All subsequent steps were performed at 4°C. Cells were lysed by sonication (three pulses of 15 s) and the non-soluble fraction was removed by centrifugation (30 min at 10 000 g). The protein concentration of the supernatant was in the range of 5–10 mg/ml as determined by a Bradford assay (Bio-Rad) and the yield of hTERT was ∼1 µg/mg protein. Approximately 50% of hTERT was soluble. hTERT protein was quantified on western blots with a polyclonal rabbit antibody raised against a C-terminal peptide of hTERT (M.Amacker and J.Lingner, in preparation). The signal strength was compared with the signal obtained with a short hTERT C-terminal peptide of known concentration that was expressed and purified from Escherichia coli (C.Wenz and J.Lingner, unpublished). For storage, the cell lysate was quick-frozen on dry ice and kept at –70°C.
Western analysis
Samples were resolved by 10% SDS–PAGE. The gel was transferred at 50 V for 2 h onto a PVDF membrane (ImmobilonTM-P, Millipore) in 25 mM Tris, 192 mM glycine and 20% v/v methanol. After transfer, the membrane was blocked for 1 h with 5% w/v milk powder and 1% w/v bovine serum albumin (BSA) in 20 mM Tris–HCl pH 7.6, 137 mM NaCl and 0.1% v/v Tween-20, and probed with 0.1 µg/ml affinity-purified polyclonal hTERT antibody and 0.02 µg/ml peroxidase-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch).
In vitro transcription of hTER
For the construction of plasmid pT7hTER, the hTER gene was fused at the 5′ end with the T7 promoter sequence and cloned into the EcoRI–BamHI sites of pUC18. This construct was linearized with BamHI prior to in vitro transcription. Five micrograms of template DNA were incubated for 2 h at 37°C in 40 mM Tris–HCl pH 8.0, 6 mM MgCl2, 10 mM dithiothreitol (DTT), 2 mM spermidine, 4 mM dNTPs, 1.2 U/µl RNasin (Roche) and 150 U of T7 RNA polymerase (Roche) in a total volume of 75 µl. The RNA was purified by gel filtration using Miniquickspin RNA columns (Roche) with a final yield of 100 µg of RNA.
Mutation of hTER
Mutations were introduced into the hTER gene by cloning double-stranded oligonucleotide cassettes into the MscI–KasI sites of pT7hTER flanking the telomerase RNA template. Prior to this step, a second KasI site located in the vector backbone was removed by digestion of pT7hTER with HindIII and NdeI, Klenow fill in and re-ligation. All mutants as well as a wild-type control harbored an additional base exchange that destroyed a DdeI site close to the template region.
Reconstitution of telomerase
In a standard reconstitution reaction 1 ml of insect cell lysate containing 0.04 nmol of recombinant hTERT was adjusted to 150 mM KCl, 15 mM MgCl2 and 10 mM ATP. After addition of 0.13 nmol of in vitro transcribed hTER, the reaction was incubated at 30°C for 90 min. For storage, reconstituted telomerase was quick-frozen on dry ice and kept at –70°C.
Affinity purification
The affinity purification of telomerase was carried out as described (Schnapp et al., 1998). To affinity purify the telomerase mutant containing the template sequence 5′-GAUUGGGAUU-3′, the oligonucleotide 5′-biotin-CTAGACCTGTCATCAGAAUCCCAAUC-3′ (underlined = 2′ O-methyl ribonucleotides) was used and bound complexes were eluted with 5′-GATTGGGATTCTGATGACAGGTCTAG-3′ as a displacement oligonucleotide.
TRAP assay
This assay was performed as described (Kim and Wu, 1997) with some modifications. Fifty microliter TRAP reactions contained TRAP buffer (20 mM Tris–HCl pH 8.3, 1.5 mM MgCl2, 63 mM KCl, 1 mM EGTA, 0.1 mg/ml BSA, 0.005% v/v Tween-20), 25 µM dNTPs, 0.1 µg of TS primer (5′-AATCCGTCGAGCAGAGTT-3′), 2 µCi of [α-32P]dCTP (3000 Ci/mmol; Amersham) and ≤1 µl of telomerase fraction. The reaction mixture was placed in a thermocycler (Biometra) and incubated at 30°C for 30 min and then heated to 94°C. After addition of 2 U of Taq DNA polymerase (Eurobio) and 0.1 µg of ACX primer (5′-GCGCGGCTTACCCTTACCCTTACCCTAACC-3′) in 1 µl TRAP buffer, the reaction was cycled 27–30 times at 94°C for 30 s and at 60°C for 30 s. The PCR products were separated on 15% non-denaturing polyacrylamide gels. Dried gels were analyzed quantitatively using a phosphoimager (Fuji) and the AIDA software (Raytest). Modified TRAP assays for the specific amplification of telomeric repeats encoded by the hTER mutants MuA and MuC were performed as described (Feng et al., 1995).
Direct telomerase assay
For the direct analysis of telomerase products, 1–7 µl of affinity-purified telomerase (containing <0.025 µM hTERT) were assayed in a final volume of 40 µl. Standard reaction conditions were 50 mM Tris–acetate pH 8.5, 50 mM KCl, 1 mM MgCl2, 1 mM spermidine, 5 mM β-mercaptoethanol, 1 mM dATP, 1 mM dTTP, 2.5 µM dGTP, 15 µCi of [α-32P]dGTP (3000 Ci/mmol; Amersham) and 2.5 µM (TTAGGG)3. The reaction mix was incubated at 37°C for 2 h. Fifty microliters of RNase stop solution [10 mM Tris–HCl pH 8.0, 20 mM EDTA, 0.1 mg/ml RNase A, 100 U/ml RNase T1 (Roche)] were added, and the mixture was incubated at 37°C for 15 min. After addition of 50 µl of proteinase K solution [10 mM Tris–HCl pH 8.0, 0.5% w/v SDS, 0.3 mg/ml proteinase K (Roche)] and a 15 min incubation at 37°C the reaction mixture was extracted with phenol/chloroform and the DNA was precipitated with ethanol and analyzed on a 12% polyacrylamide–urea gel.
Gel filtration
A SMART system equipped with a Superose 6 PC 3.2/30 column (Pharmacia) was used. The column was equilibrated with 20 mM Tris–HCl pH 8.0, 500 mM KCl, 10% v/v glycerol, 5 mM β-mercaptoethanol, 0.1 mM PMSF. Affinity-purified telomerase was adjusted to 500 mM KCl and 50 µl were injected. Eighty-microliter fractions were collected and 1 µl of each fraction was assayed for telomerase activity by TRAP. For the calibration of the column the high molecular weight calibration kit from Pharmacia was used.
Immunoprecipitation of hTERT
Twenty microliters of affinity-purified telomerase were diluted with 380 µl of IP buffer (20 mM HEPES–KOH pH 7.9, 500 mM KCl, 1 mM MgCl2, 0.1% v/v NP-40, 10% v/v glycerol, 0.5 mM PMSF, 0.5 mM DTT, 0.1 mg/ml BSA, 0.05 mg/ml yeast tRNA) and pre-cleared during a 1 h incubation at 4°C with 20 µl of protein G–Sepharose (Pharmacia). The supernatant was divided into two equal aliquots. To one aliquot, 1 µg of a polyclonal rabbit antibody raised against a C-terminal peptide of hTERT (M.Amacker and J.Lingner, in preparation) was added, whereas to the other one 1 µg of rabbit IgG was added. After 1 h at 4°C, 10 µl of protein G–Sepharose beads were added and the samples were incubated for 1 h at 4°C. Finally, the beads were washed three times with 400 µl of IP buffer.
RT–PCR and quantification of hTER molecule number
After immunoprecipitation, RNA was eluted from the beads with a mixture of phenol/chloroform and precipitated with ethanol. For cDNA synthesis, the primer R3c [5′-GTTTGCTCTAGAATGAACGGTGG AAG-3′ (Nakamura et al., 1997)] and Superscript II RT (Life Technologies) were used according to the protocol of the supplier. Ten percent of the cDNA was PCR amplified in a total volume of 25 µl containing 67 mM Tris–HCl pH 8.8, 16 mM (NH4)2SO4, 0.01% v/v Tween-20, 1.5 mM MgCl2, 0.2 mM dNTPs, 0.3 µM hTER_fw (5′-GGAAGCTTGCTAGCGCACCGGGTTGCGGAGG-3′), 0.3 µM R3c, 2.5 µCi of [α-32P]dCTP (3000 Ci/mmol; Amersham) and 1.5 U of Taq DNA polymerase (Eurobio). After 1 min at 95°C, the reaction was incubated for 15 s at 95°C and for 1 min at 56°C during 27 cycles. PCR products were cleaved with DdeI and separated on non-denaturing 8% polyacrylamide gels. The dried gels were analyzed on a phosphoimager.
For Figure 4B, hTER cDNA was determined by quantitative PCR on a GeneAmp 5700 Sequence Detection System (PE Biosystems) with SYBR green as the DNA dye. Ten percent of the cDNA was PCR amplified in a total volume of 25 µl containing 1× SYBR green master mix (PE Biosystems), 0.8 µM hTER_fw and 0.8 µM R3c. The thermal cycling conditions included an initial denaturation step at 95°C for 10 min followed by 40 cycles at 95°C for 15 s and 56°C for 1 min. Comparison of the results with the Ct values obtained with known numbers of hTER cDNA (quantified spectrophotometrically) allowed calculation of the number of hTER molecules.
Theoretical curves for different multimerization states
The expected probability (P) of hTER variants A and B in telomerase complexes as a function of the degree of multimerization and the ratio of their respective concentrations in the initial reconstitution step (R) was calculated according to the following functions:
monomer: P(x):P(y) = R0/N1:R1/N1
dimer: P(xx):P(xy):P(yy) = R0/N2:2 × R1/N2:R2/N2
trimer: P(xxx):P(xxy):P(xyy):P(yyy) = R0/N3:3 × R1/N3:3 × R2/N3:R3/N3
tetramer: P(xxxx):P(xxxy):P(xxyy):P(xyyy):P(yyyy) = R0/N4:4 × R1/N4:6 × R2/N4:4 × R3/N4:R4/N4
where x = hTER variant A, y = hTER variant B and R = [hTER variant A]/[hTER variant B], B being always the hTER variant present in molar excess, and N = (R + 1).
Acknowledgments
Acknowledgements
We thank Markus Nabholz and Julie Cooper for critically reading the manuscript, and Patrick Reichenbach for help with quantitative RT–PCR. This work was supported in part by the Swiss National Science Foundation, the Human Frontier Science Program and Cancer Research Switzerland.
References
- Adams Martin A., Dionne,I., Wellinger,R.J. and Holm,C. (2000) The function of DNA polymerase α at telomeric G tails is important for telomere homeostasis. Mol. Cell. Biol., 20, 786–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan T.M. and Cech,T.R. (1999) Telomerase and the maintenance of chromosome ends. Curr. Opin. Cell Biol., 11, 318–324. [DOI] [PubMed] [Google Scholar]
- Bryan T.M., Goodrich,K.J. and Cech,T.R. (2000a) A mutant of Tetrahymena telomerase reverse transcriptase with increased processivity. J. Biol. Chem., 275, 24199–24207. [DOI] [PubMed] [Google Scholar]
- Bryan T.M., Goodrich,K.J. and Cech,T.R. (2000b) Telomerase RNA bound by protein motifs specific to telomerase reverse transcriptase. Mol. Cell, 6, 493–499. [DOI] [PubMed] [Google Scholar]
- Collins K. and Greider,C.W. (1993) Tetrahymena telomerase catalyzes nucleolytic cleavage and nonprocessive elongation. Genes Dev., 7, 1364–1376. [DOI] [PubMed] [Google Scholar]
- Chen J.L., Blasco,M.A. and Greider,C.W. (2000) Secondary structure of vertebrate telomerase RNA. Cell, 100, 503–514. [DOI] [PubMed] [Google Scholar]
- di Marzo F., Copeland,T.D., DeVico,A.L., Rahman,R., Oroszlan,S., Gallo,R.C. and Sarngadharan,M.G. (1986) Characterization of highly immunogenic p66/p51 as the reverse transcriptase of HTLV-III/LAV. Science, 231, 1289–1291. [DOI] [PubMed] [Google Scholar]
- Diede S.J. and Gottschling,D.E. (1999) Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases α and δ. Cell, 99, 723–733. [DOI] [PubMed] [Google Scholar]
- Eickbush T.H. (1997) Telomerase and retrotransposons: which came first? Science, 277, 911–912. [DOI] [PubMed] [Google Scholar]
- Evans S.K. and Lundblad,V. (1999) Est1 and Cdc13 as comediators of telomerase access. Science, 286, 117–120. [DOI] [PubMed] [Google Scholar]
- Fan X. and Price,C.M. (1997) Coordinate regulation of G- and C-strand length during new telomere synthesis. Mol. Biol. Cell., 8, 2145–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J. et al. (1995) The RNA component of human telomerase. Science, 269, 1236–1241. [DOI] [PubMed] [Google Scholar]
- Gandhi L. and Collins,K. (1998) Interaction of recombinant Tetrahymena telomerase proteins p80 and p95 with telomerase RNA and telomeric DNA substrates. Genes Dev., 12, 721–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilley D. and Blackburn,E.H. (1999) The telomerase RNA pseudoknot is critical for the stable assembly of a catalytically active ribonucleoprotein. Proc. Natl Acad. Sci. USA, 96, 6621–6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greider C.W. (1991) Telomerase is processive. Mol. Cell. Biol., 11, 4572–4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greider C.W. and Blackburn,E.H. (1985) Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell, 43, 405–413. [DOI] [PubMed] [Google Scholar]
- Greider C.W. and Blackburn,E.H. (1989) A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature, 337, 331–337. [DOI] [PubMed] [Google Scholar]
- Hammond P.W., Lively,T.N. and Cech,T.R. (1997) The anchor site of telomerase from Euplotes aediculatus revealed by photo-cross-linking to single- and double-stranded DNA primers. Mol. Cell. Biol., 17, 296–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt S.E. et al. (1999) Functional requirement of p23 and Hsp90 in telomerase complexes. Genes Dev., 13, 817–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce C.M. and Steitz,T.A. (1994) Function and structure relationships in DNA polymerases. Annu. Rev. Biochem., 63, 777–822. [DOI] [PubMed] [Google Scholar]
- Kim N.W. and Wu,F. (1997) Advances in quantification and characterization of telomerase activity by the telomeric repeat amplification protocol (TRAP). Nucleic Acids Res., 25, 2595–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N.W. et al. (1994) Specific association of human telomerase activity with immortal cells and cancer. Science, 266, 2011–2015. [DOI] [PubMed] [Google Scholar]
- Klobutcher L.A., Swanton,M.T., Donini,P. and Prescott,D.M. (1981) All gene-sized DNA molecules in four species of hypotrichs have the same terminal sequence and an unusual 3′ terminus. Proc. Natl Acad. Sci. USA, 78, 3015–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlstaedt L.A., Wang,J., Friedman,J.M., Rice,P.A. and Steitz,T.A. (1992) Crystal structure at 3.5 Å resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science, 256, 1783–1790. [DOI] [PubMed] [Google Scholar]
- Lee M.S. and Blackburn,E.H. (1993) Sequence-specific DNA primer effects on telomerase polymerization activity. Mol. Cell. Biol., 13, 6586–6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J. and Cech,T.R. (1996) Purification of telomerase from Euplotes aediculatus: requirement of a primer 3′ overhang. Proc. Natl Acad. Sci. USA, 93, 10712–10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J., Hendrick,L.L. and Cech,T.R. (1994) Telomerase RNAs of different ciliates have a common secondary structure and a permuted template. Genes Dev., 8, 1984–1998. [DOI] [PubMed] [Google Scholar]
- Lingner J., Hughes,T.R., Shevchenko,A., Mann,M., Lundblad,V. and Cech,T.R. (1997) Reverse transcriptase motifs in the catalytic subunit of telomerase. Science, 276, 561–567. [DOI] [PubMed] [Google Scholar]
- Lundblad V. and Szostak,J.W. (1989) A mutant with a defect in telomere elongation leads to senescence in yeast. Cell, 57, 633–643. [DOI] [PubMed] [Google Scholar]
- Maine I.P., Chen,S.F. and Windle,B. (1999) Effect of dGTP concentration on human and CHO telomerase. Biochemistry, 38, 15325–15332. [DOI] [PubMed] [Google Scholar]
- Makarov V.L., Hirose,Y. and Langmore,J.P. (1997) Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell, 88, 657–666. [DOI] [PubMed] [Google Scholar]
- Masutomi K., Kaneko,S., Hayashi,N., Yamashita,T., Shirota,Y., Kobayashi,K. and Murakami,S. (2000) Telomerase activity reconstituted in vitro with purified human telomerase reverse transcriptase and human telomerase RNA component. J. Biol. Chem., 275, 22568–22573. [DOI] [PubMed] [Google Scholar]
- Mitchell J.R. and Collins,K. (2000) Human telomerase activation requires two independent interactions between telomerase RNA and telomerase reverse transcriptase. Mol. Cell, 6, 361–371. [DOI] [PubMed] [Google Scholar]
- Mitchell J.R., Cheng,J. and Collins,K. (1999a) A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol. Cell. Biol., 19, 567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell J.R., Wood,E. and Collins,K. (1999b) A telomerase component is defective in the human disease dyskeratosis congenita. Nature, 402, 551–555. [DOI] [PubMed] [Google Scholar]
- Munoz-Jordan J., Cross,G.A.M., de Lange,T. and Griffith,J.D. (2001) T-loops at trypanosoma telomeres. EMBO J., 20, 579–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T.M. and Cech,T.R. (1998) Reversing time—origin of telomerase. Cell, 92, 587–590. [DOI] [PubMed] [Google Scholar]
- Nakamura T.M., Morin,G.B., Chapman,K.B., Weinrich,S.L., Andrews,W.H., Lingner,J., Harley,C.B. and Cech,T.R. (1997) Telomerase catalytic subunit homologs from fission yeast and human. Science, 277, 955–959. [DOI] [PubMed] [Google Scholar]
- Nugent C.I. and Lundblad,V. (1998) The telomerase reverse transcriptase: components and regulation. Genes Dev., 12, 1073–1085. [DOI] [PubMed] [Google Scholar]
- Prescott J. and Blackburn,E.H. (1997a) Functionally interacting telomerase RNAs in the yeast telomerase complex. Genes Dev., 11, 2790–2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott J. and Blackburn,E.H. (1997b) Telomerase RNA mutations in Saccharomyces cerevisiae alter telomerase action and reveal nonprocessivity in vivo and in vitro. Genes Dev., 11, 528–540. [DOI] [PubMed] [Google Scholar]
- Romero D.P. and Blackburn,E.H. (1991) A conserved secondary structure for telomerase RNA. Cell, 67, 343–353. [DOI] [PubMed] [Google Scholar]
- Schnapp G., Rodi,H.P., Rettig,W.J., Schnapp,A. and Damm,K. (1998) One-step affinity purification protocol for human telomerase. Nucleic Acids Res., 26, 3311–3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D., Lopez-Guajardo,C.C., Quada,J., Hurley,L.H. and Von Hoff,D.D. (1999) Regulation of catalytic activity and processivity of human telomerase. Biochemistry, 38, 4037–4044. [DOI] [PubMed] [Google Scholar]
- Wang H. and Blackburn,E.H. (1997) De novo telomere addition by Tetrahymena telomerase in vitro. EMBO J., 16, 866–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinrich S.L. et al. (1997) Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nature Genet., 17, 498–502. [DOI] [PubMed] [Google Scholar]
- Wellinger R.J., Ethier,K., Labrecque,P. and Zakian,V.A. (1996) Evidence for a new step in telomere maintenance. Cell, 85, 423–433. [DOI] [PubMed] [Google Scholar]
- Xia J., Peng,Y., Mian,I.S. and Lue,N.F. (2000) Identification of functionally important domains in the N-terminal region of telomerase reverse transcriptase. Mol. Cell. Biol., 20, 5196–5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G.L. and Blackburn,E.H. (1991) Developmentally programmed healing of chromosomes by telomerase in Tetrahymena. Cell, 67, 823–832. [DOI] [PubMed] [Google Scholar]
- Yu G.L., Bradley,J.D., Attardi,L.D. and Blackburn,E.H. (1990) In vivo alteration of telomere sequences and senescence caused by mutated Tetrahymena telomerase RNAs. Nature, 344, 126–132. [DOI] [PubMed] [Google Scholar]