Abstract
Signaling by Decapentaplegic (Dpp), a member of the TGFβ superfamily of signaling molecules similar to vertebrate BMP2 and BMP4, has been implicated in many developmental processes in Drosophila melanogaster. Notably, Dpp acts as a long-range morphogen during imaginal disc growth and patterning. Genetic approaches led to the identification of a number of gene products that constitute the core signaling pathway. In addition to the ligand-activated heteromeric receptor complex and the signal-transducing intracellular Smad proteins, Dpp signaling requires two nuclear proteins, Schnurri (Shn) and Brinker (Brk), to prime cells for Dpp responsiveness. A complex interplay between the nuclear factors involved in Dpp signaling appears to control the transcriptional readout of the Dpp morphogen gradient. It remains to be seen whether similar molecular mechanisms operate in the nucleus in vertebrate systems.
Keywords: Dpp/morphogen gradient/nuclear signaling/selector proteins/TGFβ
Introduction
Cell–cell interactions are key to the development of all tissues and cell types in multicellular animals. It has become clear during the past decade that a limited number of evolutionarily conserved signaling pathways exist, and that each of them is involved in numerous, distinct cellular decisions throughout development. Using genetic, cell culture and biochemical approaches, these signaling pathways are presently being characterized at the molecular level. It appears that tissue- and stage-specific responses to signaling are partly generated via the interplay of signaling components and pre-existing nuclear proteins on cis-acting enhancer elements. In this review, we would like to summarize what is currently known about the molecules involved in Decapentaplegic (Dpp) signaling in Drosophila melanogaster with special emphasis on nuclear factors involved in signal response and interpretation. For the first time, molecular scenarios are emerging that explain how an extracellular morphogen gradient can be translated into discrete nuclear transcription patterns in a cellular field.
Dpp signaling in induction and patterning
In Drosophila, Dpp signaling has been implicated in many developmental processes. These processes include the determination of the dorsal–ventral axis in the blastoderm embryo, the dorsal closure of the embryo, the shaping of the tracheal system and the subdivision of the mesodermal anlage, the patterning of the imaginal discs and the maintenance of germline stem cells. In some situations, the action of Dpp is limited to ligand-secreting cells and their immediate neighbors. In most cases, however, Dpp acts on several cells neighboring the secreting cells. During imaginal disc development, for example, it has been shown in an elegant series of experiments that Dpp acts as a long-range morphogen and instructs cell fate in a dosage-dependent manner (Lecuit et al., 1996; Nellen et al., 1996). More recently, the Dpp gradient has been visualized in the wing imaginal disc (Entchev et al., 2000; Teleman and Cohen, 2000) and studies addressing how the gradient is formed are now underway. Bioactive Dpp has been produced in bacteria in large amounts (Groppe et al., 1998), allowing for a biochemical and biophysical characterization of its interaction with receptors and antagonists. The level to which Dpp receptors are activated critically depends on the availability of the active ligand(s), which in turn depends on the local production of the ligand, the presence of extracellular binding proteins and the distribution of antagonists, among other parameters. The distribution of the Dpp ligand in a cellular field can also be influenced by the amount of its cell surface receptors. These issues have been considered in other reviews and will not be further discussed here (Podos and Ferguson, 1999; Raftery and Sutherland, 1999).
To understand how extracellular ligand gradients can be interpreted by a responding cellular field, the cell surface receptors as well as the intracellular transduction pathway and the molecular machinery generating an appropriate response (including DNA response elements and nuclear proteins binding to them) have to be studied in detail. Much has been learned from the analysis of Dpp signaling in Drosophila and a complex picture is emerging.
Signal reception and transmission
While the gene encoding the Dpp ligand was discovered via classical genetics (and called decapentaplegic because of its requirement in 15 discs; Spencer et al., 1982), cDNAs encoding the essential Dpp receptors were cloned based on knowledge of TGFβ/activin receptor structure, and the corresponding genes were subsequently associated with previously identified mutations (Kawabata et al., 1998; Raftery and Sutherland, 1999). The heteromeric Dpp receptor complex consists of two types of transmembrane serine/threonine kinases. Based on biochemical work with the Drosophila receptors and the vertebrate homologs, it seems that the type I receptor Thick veins (Tkv) recruits the Dpp ligand and the type II receptor Punt (Put) into a heteromeric complex. In this signaling complex, the constitutive active kinase of Put phosphorylates Tkv at a type I receptor-specific, juxtamembrane GS domain. This phosphorylation activates the associated type I kinase, which in turn results in the phosphorylation of the cytoplasmic protein Mothers against Dpp (Mad). Although it has been shown that both Put and Tkv are absolutely essential for all Dpp signaling effects throughout development, it turns out that another type I receptor, Saxophone (Sax), is required for pattern ing in many tissues. Sax mediates signaling by other Drosophila members of the TGFβ superfamily (Screw and 60A) and synergizes with Tkv signaling (Neul and Ferguson, 1998; Nguyen et al., 1998). Since high levels of Tkv can bypass the requirement for Sax, it is likely that the two receptors use a similar intracellular signaling pathway, including the nuclear components discussed in more detail in this review. At what level in the signaling pathway Sax and Tkv signaling intersect remains to be investigated.
Nuclear factors implicated in Dpp signaling
The signal transducers Mad and Medea
Although the founding member of the Smad superfamily of signal transducers was Drosophila Mothers against Dpp (Mad) (Raftery et al., 1995; Sekelsky et al., 1995), much of what is currently known about Smad function has come from studies of the vertebrate homologs (for recent comprehensive reviews see Massagué, 2000; Shi, 2001). Studies mostly performed using cultured cells have provided a biochemical model of TGFβ signal transduction; activation of the type I serine/threonine kinase leads to C-terminal phosphorylation of receptor-regulated Smad proteins (Mad in Drosophila with regard to the Dpp signaling pathway), association of Phospho-Mad with a second, co-mediator Smad protein (Medea in Drosophila) and translocation of a heteromeric Smad complex (Mad–Medea) into the nucleus. Using antibodies against the phosphorylated form of Smads, support for this scenario has been obtained from in vivo Mad localization studies (Tanimoto et al., 2000; Dorfman and Shilo, 2001). In the nucleus, the Mad–Medea complex can bind to cis-acting elements in target genes, as first shown in Drosophila (Kim et al., 1997), and activate or repress transcription. Mad has been shown to interact with the transcriptional co-activator CBP/p300, suggesting that Mad may recruit CBP to effect the transcriptional activation of Dpp-responsive genes during development (Waltzer and Bienz, 1998). Indeed, Drosophila CBP mutants show defects which mimic those seen in mutants that lack Dpp or Mad (Waltzer and Bienz, 1998; Ashe et al., 2000). Growing evidence from studies in vertebrates indicates that Smad proteins achieve higher affinity (and specificity) in their interaction with cis-regulatory elements by associating with partner DNA-binding cofactors, and it is thought that the interaction of Smads with cofactors on target enhancers directs the signaling pathway to distinct response genes in different tissues (see also below).
While little is known about interaction of Mad and Medea with tissue-specific factors, genetic studies in Drosophila have led to the isolation of two additional nuclear proteins, Brinker (Brk) and Schnurri (Shn), which appear to be instrumental in Dpp signaling and that have not yet been identified or analyzed in mammalian systems.
Brinker, a repressor of Dpp target genes
The isolation of Brk as a component in the Dpp signaling pathway in Drosophila came as something of a surprise (Campbell and Tomlinson, 1999; Jaźwińska et al., 1999a,b; Minami et al., 1999). Mutations in brk produce relatively subtle phenotypes that are similar to those generated by an activation of the Dpp pathway in certain cells that normally see little or no signal; cells responding to high endogenous levels of Dpp are not affected in brk mutants. In most cells, brk expression is negatively controlled by Dpp signaling, resulting in transcript levels that are undetectable in Dpp-secreting cells and high in cells at a distance from the site of Dpp production. Ectopic expression experiments demonstrated that brk is capable of efficiently counteracting signal-dependent activation, leading to cell-autonomous repression of Dpp targets when expressed in Dpp-responding cells. Based on these observations and the fact that brk encodes a protein with features of a transcriptional repressor (with sequence similarity to homeodomains and to motifs found in transcriptional repressors), it was proposed that Brk acts as a direct transcriptional repressor of Dpp target genes.
A number of recent reports have now demonstrated that Brk is indeed a sequence-specific DNA-binding protein that interacts directly with cis-regulatory elements of a number of Dpp target genes and represses their expression via these sites (Sivasankaran et al., 2000; Kirkpatrick et al., 2001; Rushlow et al., 2001; Saller and Bienz, 2001; Zhang et al., 2001). Since brk transcription is negatively regulated by Dpp signaling, and Brk repressor levels thus increase in cells with increasing distance from the Dpp source, it has been proposed that the function of brk is most prominent in contexts in which Dpp acts as a morphogen (see below). In situations in which Dpp acts in inductive cell–cell interactions, Dpp signaling might simply counteract the Brk repressor in a first step by suppressing its transcription, thus allowing signal-controlled gene regulation to proceed.
Schnurri, a large zinc finger protein required for brk repression
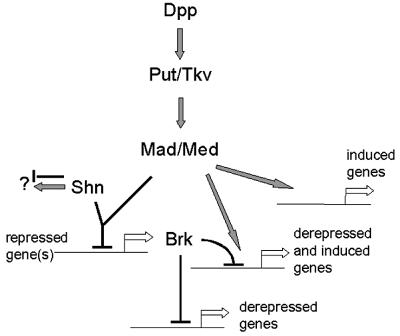
Dpp signaling has different effects on gene transcription in responding cells. Throughout development, Dpp signaling activates a large number of genes, some of which will be discussed in more detail below. However, Dpp signaling represses transcription of the brk gene in most of the responding cells. How is it that the same signaling pathway activates and represses genes in the same cellular context? It turns out that the large nuclear zinc-finger protein Schnurri (Shn) plays an important role in this process. In a large-scale screen performed by Nüsslein-Volhard et al. (1984), shn mutants have initially been isolated due to their dorsal open phenotype, a phenotype that shn shares with the two essential Dpp receptors punt and tkv. Subsequently it was shown that shn mutants mimic a large number of dpp, tkv and punt loss-of-function phenotypes and that cells lacking shn do not respond to ectopic Dpp (Arora et al., 1995; Grieder et al., 1995; Staehling-Hampton et al., 1995). Somewhat surprisingly, the early function of dpp, which is to pattern the dorsal–ventral axis of the developing embryo, is not affected in shn mutants; this finding was often taken as evidence that shn does not fulfill an essential role in Dpp signaling and might be a tissue-specific cofactor. However, recent results demonstrate that the function of shn is tightly linked to the regulation of brk expression and that both are intrinsic components of a widely targeted Dpp response system in stimulated cells (Marty et al., 2000). It turns out that most defects observed in shn mutant cells are due to their inability to repress brk transcription in response to Dpp; brk levels remain high in shn mutant cells even upon the activation of the Dpp signaling cascade and these high levels of Brk repress Dpp target genes in a dominant fashion. Indeed, the strong defects observed in shn mutants closely mimic brk gain-of-function defects (T.Marty and A.Jaźwińska, unpublished results). When brk is genetically inactivated in a shn mutant, a large number of target genes are normally activated by Dpp signaling; thus, the removal of Brk in a shn mutant re-establishes Dpp-dependent gene activation. This surprising finding indicates that Shn is required for brk repression (allowing the cells to get rid of a repressor of target genes) but is dispensable for the activation of many (maybe most) Dpp-induced genes. This is in sharp contrast to Mad, which is required for brk repression and for the activation of most target genes. Therefore, the Dpp signaling pathway appears to split into two major branches in the nucleus downstream of the activated Mad–Medea complex: a shn-dependent branch involved in brk repression and a largely shn-independent branch leading to gene activation (Figure 1).
Fig. 1. The Dpp signaling pathway in Drosophila melanogaster. The Mad–Medea complex is essential for gene activation and gene repression. Shn is essential for Mad/Medea-mediated repression of brk transcription, but is dispensable for Dpp-mediated transcriptional activation of most target genes. The question mark indicates the possibility that Shn has additional functions since the brk, shn double mutant embryos are not identical to the brk single mutant embryos (T.Marty and M.Affolter, data not shown but see Torres-Vazquez et al., 2001). Evidence suggesting that Shn also acts as an activator of transcription together with Mad has been provided (Dai et al., 2000); it remains to be investigated in vivo whether Shn is required for gene activation in certain cases. Arrows denote upregulation; bars denote downregulation.
Further experiments are required to determine the precise role of Shn in the repression of brk and the role of Mad and Medea in this process. Since it has been shown that Shn can interact directly with Mad (Dai et al., 2000; Udagawa et al., 2000), it is possible that Dpp-induced nuclear accumulation of Mad recruits the Shn repressor to target sites and that the amount of nuclear Mad determines the amount of recruited Shn repressor. Along the same lines, it is also possible that DNA-bound Shn recruits Mad and that this interaction turns the complex into a repressor. Alternatively, Shn might act indirectly and activate transcription of an unknown repressor. It is also important to determine whether direct gene repression by Dpp signaling generally requires shn or whether the function of shn is limited to brk repression.
If Shn provides such an important role in Dpp signaling why is shn function not required in the early embryo, in which Dpp patterns the dorsal ectoderm in a dosage-dependent manner? During the time period in which cell fates are specified in the early embryo, brk is expressed in a ventrolateral stripe abutting but not overlapping the dorsal dpp expression domain (Jaźwińska et al., 1999b). However, brk transcription is not repressed by Dpp signaling in the dorsal part of the embryo; rather, brk transcription is activated laterally by the Dorsal morphogen, the concentration of which is too low in the dorsal part to activate brk. Therefore, if the principle function of Shn were to repress brk transcription in a Dpp-dependent manner, shn would not be required in the early embryo. This is indeed what is observed.
Since the uncoupling of Dpp signaling and brk transcription in the early embryo appears to be the exception rather than the rule, we consider Brk and Shn as essential components of the canonical Dpp signaling system. In the following, we consider their involvement in the cellular responses of Dpp in patterning and induction processes.
A scenario for the nuclear readout of Dpp morphogen gradients
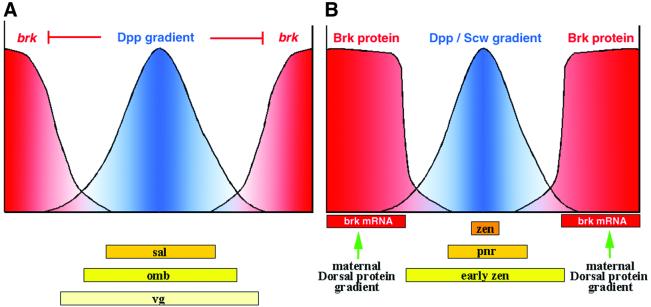
Genetic analyses of the function of Dpp in wing development have shown that Dpp functions as a morphogen and patterns the wing along its anterior–posterior (A/P) axis in a dose-dependent fashion. dpp is expressed in a narrow anterior stripe of cells at the A/P compartment boundary and is necessary both for cell proliferation and for organization of the A/P pattern across the entire imaginal disc. Dpp signaling leads to the activation of spalt (sal), optomotor blind (omb) and vestigial (vg) at successively lower thresholds across the wing blade primordium of the growing disc (Figure 2). Since Mad and Medea transmit the signal from the cell surface to the nucleus, a straightforward molecular view explaining the dose-dependent regulation of target genes would assume the presence of Mad/Med-binding sites in different numbers, local contexts and/or affinities in response genes that are differentially expressed along the A/P axis. However, genetic and biochemical data argue for a quite different, more complex scenario.
Fig. 2. Schematic model for Dpp-dependent patterning of the wing imaginal disc (A) and the embryonic dorsal–ventral axis (B). (A) In the imaginal wing disc, Dpp is secreted from a narrow stripe of cells anterior to the A/P compartment boundary and forms a long-range gradient. Dpp signaling represses the transcription of brk, which encodes a repressor of Dpp target genes. Different target genes are expressed in nested domains; sal is expressed in a less broad domain than omb, and vg is expressed in the entire wing blade primordia. A model for the regulation of expression of these genes is illustrated in Figure 3. (B) In the early Drosophila embryo, dpp is expressed in the dorsal ectoderm and amnioserosa primordium and its activity is graded due to the action of a number of components (see text). brk RNA is expressed under the control of the Dorsal morphogen in the ventral ectoderm abutting the dpp expression domain, and the Brk protein appears to diffuse dorsally during the syncytial blastoderm stages. brk-dependent repression leads to the establishment of different expression borders for early zen and pnr. zen is activated in a dorsal stripe under the control of dpp and screw.
It turns out that the expression domains of genes along the A/P axis are controlled to a large extent by the level of the Brk repressor protein, the distribution of which displays a gradient in the developing wing primordia that is inversely correlated to the amount of Dpp across the field (Campbell and Tomlinson, 1999; Jaźwińska et al., 1999a; Minami et al., 1999). Genetic experiments have demonstrated that the lateral extent of expression of both omb and sal is determined by brk; expression of both target genes expands in the absence of the Brk repressor. Since the lateral expression boundaries of omb and sal are distinct, different levels of Brk appear to be required to repress these two target genes. A third Dpp target gene, vg, which is expressed in the entire wing blade primordia, is not repressed in the wing blade primordia by extant Brk levels but is suppressed by high levels of brk in the more distant wing hinge region. Thus, it appears that a major readout of the dose of Dpp that a cell senses in the morphogenetic field translates into a distinct level of brk transcription: high amounts of Dpp signaling abolish brk transcription completely, intermediate amounts of Dpp only partially repress brk transcription, while the absence of Dpp results in high levels of brk transcription. Thus, Dpp response genes appear to read their position in the morphogen field to a large extent by measuring the amount of the Brk repressor in the nucleus.
Recently, it has been shown that a Brk-binding site in a wing-specific enhancer of the omb gene directly mediates the repression of omb transcription lateral to the A/P boundary in the developing wing disc (Sivasankaran et al., 2000). Somewhat surprisingly, active transcription of omb in the wing disc does not require Mad; while omb is inactive in mad mutant clones due to high levels of Brk repressor, omb transcript levels are normal in brk, mad double mutant clones (Jaźwińska et al., 1999a). These results can be taken as an argument to classify omb as an indirect Dpp target, regulated by the product of the Dpp target gene brk and by unknown transcriptional activators. However, since we consider Brk to be an integral component of the nuclear Dpp response, we favor the view that omb represents a direct target of the Dpp signaling system.
Although no detailed enhancer analysis has been published concerning the control of sal expression, genetic studies demonstrate that brk repression is crucial for setting the expression border of sal (Campbell and Tomlinson, 1999; Jaźwińska et al., 1999a). However, the level of expression of sal is also dependent on mad, since reduced expression is observed in brk, mad double mutant cells as compared with brk mutant cells. Thus, sal appears to read out gradient information based on the protein levels of both Brk and nuclear Mad. Again, the situation is somewhat different for vg, which is expressed throughout the developing wing field and requires functional Mad/Medea-binding sites to sense Dpp over this entire range (Kim et al., 1997; Certel et al., 2000).
At present, the analysis of cis-regulatory elements driving differential expression in the developing wing primordia is at an early stage and none of the elements has been analyzed carefully with regard to both Brk and Mad binding. It is therefore difficult to propose a molecular model for how the Dpp gradient is interpreted in the wing imaginal disc solely based on the few cases for which limited data are available. However, interesting insights regarding this issue have now come from studying Dpp response genes in the early embryo.
In Drosophila, a Dpp activity gradient patterns the dorsal region of the embryo during the early developmental stages. Within the dorsal field of cells patterned by Dpp, dpp transcripts are present at uniform levels and the graded activity of Dpp is generated via the differential distribution of antagonists and the modification of other TGFβ ligands (for an excellent review see Podos and Ferguson, 1999). As a result of the Dpp activity gradient, nested domains of gene expression are generated (Figure 2B). Genes [e.g. early zerknüllt (zen) and tolloid] responding to lower doses are broadly expressed, genes responding only to higher doses [e.g. pannier (pnr)] are expressed in a narrower dorsal domain, and genes requiring highest levels are expressed in a dorsal stripe (e.g. refined zen and u-shaped).
Brk is also expressed in the early embryo, and brk RNA is detected in the lateral ectoderm abutting dorsally the Dpp expression domain and ventrally the prospective mesoderm (Jaźwińska et al., 1999b). Although the expres sion of brk is not controlled by Mad/Med and Shn in the early embryo, and the Brk protein gradient probably arises via diffusion in the syncytial blastoderm (Ashe et al., 2000), Brk acts in very much the same way on Dpp target genes in early dorsal–ventral patterning as it does in the wing imaginal disc (Jaźwińska et al., 1999b). Genes (e.g. early zen) that require low levels of Dpp activity uniformly expand in brk mutants, and do not require Dpp for activation (similar to omb). Genes (e.g. pnr) dependent on higher Dpp concentrations expand at low levels in brk mutants and require Dpp signaling for normal activation levels (similar to sal). Genes (refined zen) activated at highest levels of Dpp activity require dpp and mad activity but do not expand in brk mutants (no genes regulated in this manner in the wing primordia have been identified so far).
The similarity between these two different developmental systems in the control of target genes suggests that similar molecular mechanisms might read the Dpp gradient in the nucleus. A recent careful analysis of the distribution and functional requirement of DNA-binding sites for Mad and Brk in the Dpp response gene zen suggests an elegant scenario for the nuclear readout of the Dpp morphogen gradient in the early embryo (Rushlow et al., 2001). Examination of the zen regulatory region revealed the presence of ten Mad and six Brk-binding sites. Interestingly, many of the Mad and Brk sites overlap; there is only one Brk-binding site that does not bind Mad and conversely, there are five Mad-binding sites that are not recognized by Brk. Examining the expression pattern of mutant and wild-type enhancers, two major observations were made: (i) occupancy of all Mad sites due to high concentrations of nuclear Mad appears to be the primary mechanism for zen expression in a narrow dorsal stripe (zen refinement); (ii) occupancy of Brk sites appears to be the major mechanism of repression. Since Brk competes out Mad at overlapping sites in DNA-binding assays in vitro, it has been proposed (Rushlow et al., 2001) that the competition between Mad and Brk could be a determining factor in setting up the spatially restricted expression domains of Dpp target genes; accordingly, genes would be expressed in the morphogen field according to a net balance of positive and negative inputs. Interestingly, such a scenario, combined with the observation that Mad and Brk sites do not necessarily overlap, can explain the establishment of expression domains that vary largely in size over the morphogen field. The presence of Mad sites and the concomitant absence of high affinity Brk sites in a Dpp-responsive enhancer would result in an expression domain that spans the entire cellular field (e.g. similar to vg). The presence of high affinity sites for Brk could result in a narrower domain, the width of which might be determined based on the number and/or affinity of overlapping Brk and Mad sites.
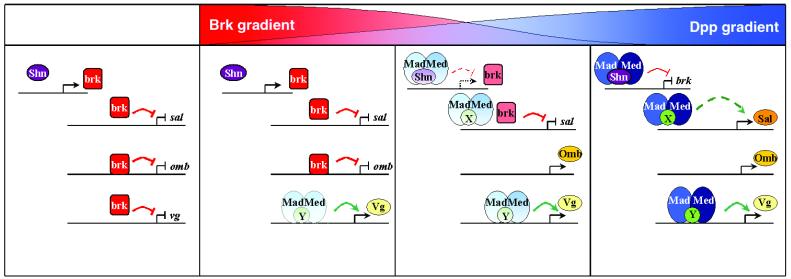
However, Brk does not repress target genes simply by competing with the Mad and Medea (or other) activators, but recruits co-repressors (e.g. Groucho) to the target sites (Zhang et al., 2001). It is very likely that it is the recruitment of co-repressors that makes Brk such a dominant repressor. Clearly, ectopic Brk can dominantly suppress the activating effect of Mad in regions of high Dpp signaling (regions of high levels of nuclear Mad); whether increasing Mad levels can indeed compete with moderate Brk levels, as would be predicted by the competition model, remains to be shown. The work on omb demonstrates that nested expression domains can be generated by Brk repression without direct competition with Mad activation. Therefore, it appears that there are many ways to generate nested expression domains using combinations of overlapping and/or non-overlapping sites for Brk, Mad/Medea and other transcriptional regulators (Figure 3). It is likely that it is the overall architecture of all transacting factor complexes of a responding enhancer element that contributes to the readout of the concentration of Mad/Medea and Brk in a responding nucleus, and protein–protein interactions might be as important as binding affinities in the net readout. Direct competition of Brk and Mad at shared sites might be important in vivo, but experiments providing direct evidence for such a mechanism occurring in vivo remain to be designed and performed.
Fig. 3. Schematic model for the patterning of a field of cells by a Dpp morphogen gradient. In the absence of Dpp signaling (a situation that might not occur in the primordia of the wing blade, but in the primordia of the wing hinge in the imaginal disc), high levels of Brk protein repress Dpp target genes (far-left panel). At the periphery of the Dpp gradient, high levels of Brk are present; the low levels of phosphorylated Mad/Med in the nucleus are sufficient to activate the vgQ enhancer, but too low to repress brk transcription. In the medial region of the gradient, brk is partially repressed under the control of Mad/Med and Shn; the reduced levels of Brk protein are insufficient to repress omb, but still sufficient to repress sal. Towards the center of the gradient, high levels of Dpp signaling strongly repress brk transcription. sal is derepressed and activated to high levels under the control of Mad/Med. Proteins for which evidence for a direct regulation of the corresponding enhancer has been provided experimentally are drawn on the DNA line; other proteins are drawn above the DNA line. For more details on the possible architecture of the enhancers controlling expression of vg, omb and sal, please refer to the text.
Although it is interesting and instructive to look at how target genes respond to the early Dpp morphogen gradient as it patterns the dorsal ectoderm, it is important to note that part of the events controlling the Dpp response (i.e. Brk diffussion) occur in a syncytium and not in a cellular field. In the wing imaginal disc, a very important sensor of the Dpp gradient appears to be the brk gene itself. To understand how Dpp patterns a cellular field, it will thus be crucial to isolate the enhancer(s) that controls the Dpp-dependent transcription of brk and to study how Mad and Medea, possibly in collaboration with Shn, control its expression. It is likely that this readout of the gradient by the brk enhancer is the same in all the imaginal discs in which Dpp establishes cellular pattern at long range, and is an instrumental component that allows the Dpp signaling system to work as a morphogen in a developmental field.
Selector genes and the interpretation of Dpp/BMP signaling
Although there are still many unanswered questions, molecular scenarios are now emerging to explain how the interaction of Dpp with its cell surface receptors leads to activation, repression and/or derepression of transcription via the essential core components Mad, Medea, Shn and Brk. But why does Dpp control target genes in such a tissue-specific manner? How does the Mad–Medea complex find the appropriate target sequences in each cell type considering the incredible complexity of the genome? A number of observations made in vertebrate systems provide first molecular explanations for this type of selectivity. Although Smads do bind DNA weakly and with moderate sequence specificity, their recruitment to selected target sites is greatly facilitated by DNA-binding cofactors (Massagué and Wotton, 2000). The most prominent of these cofactors (and the first to be identified) is the FAST-1 protein (Chen et al., 1996, 1998). FAST-1 binds a Smad2/4 or Smad3/4 complex, and all components of this complex are required for efficient DNA binding and transcription activation of certain TGFβ signaling targets. Other cofactors have been characterized recently and the differential association of Smad proteins with a wide spectrum of transcription factors of different DNA-binding specificities appears to be a characteristic of gene regulation via Smad proteins (Massagué and Wotton, 2000). The local context of Smad-binding sites in defined cis-regulatory elements combined with the availability of (positively and negatively acting) partner proteins in responding cells appears to be the molecular signature underlying tissue- and stage-specific selection of target genes.
As mentioned before, no tissue-specific cofactors that interact with Mad or Medea have been isolated so far in Drosophila. However, a few Dpp-dependent enhancers have been analyzed in some detail and in these studies a number of interesting observations have been made. Analysis of several enhancers showed that binding of a so-called selector protein was required for the enhancer to be signal inducible. In an enhancer derived from the labial (lab) gene, a direct interaction of the homeotic selector protein Lab and its cofactors Extradenticle and Homothorax is required for the enhancer’s response to Dpp signaling (Grieder et al., 1997; Ryoo et al., 1999). The sub-element to which the homeotic complex binds is distinct from the sub-element to which signaling mediators bind, suggesting that in addition to local sequence context of Mad/Medea-binding sites, distant elements can influence signaling outcome in responding cells. Similar findings were reported for an enhancer of the muscle-selector gene tinman (tin; Xu et al., 1998) and the gene even-skipped (Halfon et al., 2000), where direct binding of Tin in close proximity to Mad sites was shown to be required for Dpp inducibility. In another case, the Dpp-dependent activation of a wing blade-specific response element was proposed to require binding of the wing selector protein complex Scalloped/Vestigial (Halder et al., 1998). It appears from these cases that signal specificity is conferred in part by selector gene products. Viewed from the other side, these studies also suggest that one of the important functions of selector proteins is to assist in interpreting signaling cascades in a selector-specific manner by helping nuclear signaling mediators to provoke appropriate responses through defined enhancer elements (Mann and Affolter, 1998). It will be interesting to see whether direct interactions between selector proteins and signal transducers exist, or whether their function is indirectly linked on these enhancer elements.
Simultaneous signaling via different pathways might also influence the effect of Dpp signaling mediators on target enhancers (Massagué, 2000). Situations have been described in which the outcome of TGFβ signaling was changed by concomitant signaling via other pathways (Nishita et al., 2000). Also, a number of Drosophila Dpp-responsive enhancers have been described in which target elements for a second, different signaling pathway are present. The interaction between these pathways can either be positive as described for an enhancer of the even-skipped gene [inputs from Ras, Wingless (Wg) and Dpp signaling activate the enhancer synergistically; Halfon et al., 2000] and the lab gene (EGF receptor signaling synergizing with Dpp signaling; Szüts et al., 1998). Such interactions could also be negative but no such case involving Dpp signaling has been reported so far. Further work will certainly increase knowledge of the network of interactions that occur between signaling pathways and their intersection with selector protein function. It remains to be seen whether the increasing gene regulatory complexity will ultimately resolve into more simple molecular paradigms.
Dpp signaling in Drosophila and signaling via TGFβ superfamily members in vertebrates
The existence of Shn-like proteins in vertebrates (Fan and Maniatis, 1990) and the finding that Drosophila Brk functions as a repressor of BMP signaling in Xenopus (Minami et al., 1999) indicate that a regulatory cascade similar to the one identified in D.melanogaster and discussed here might be conserved in higher vertebrates. If this is the case, it will be interesting to see to what extent the morphogen function of vertebrate TGFβ superfamily members relies on an interplay between these nuclear factors. In the vertebrate system, a number of proteins have been proposed to act as repressors of TGFβ or BMP signaling (TGIF, Evi, Sno/Ski; Massagué and Wotton, 2000). In most cases, however, these factors antagonize signal-induced activation via their interaction with Smad proteins; whether any of these factors are involved in the transcriptional repression of genes upon signal transduction (similar to Dpp-induced brk repression in Drosophila) remains open. It will be interesting to find out whether any of these proteins contribute to the morphogen function of vertebrate members of the TGFβ superfamily. Along the same lines, it will be important to analyze the Drosophila homologs of genes implicated in signaling by TGFβ members and find out to what extent they are involved in signaling interpretation. It is anticipated that numerous similarities will be uncovered but also that each system uses the repertoire of available factors in different and novel ways.
Repression in signaling
At first sight, the Brk repressor plays a somewhat peculiar role in the Dpp signaling pathway: in most cells that respond to Dpp signaling, Brk is removed and does not have an important function. This mode of regulation does not allow Dpp-induced gene activation without an intervening step of transcription regulation: the repression of brk. The existence of repressors that downregulate inducible genes in the absence of the inducing signal has been documented for many other signaling systems. In the Wingless (Wg) pathway, Lef1/TCF/Pan acts as a repressor of target genes prior to Wg signal reception/transduction. Similarly, a proteolytic cleavage product of Cubitus interruptus (Ci) acts as a repressor of Hh target genes prior to Hh signal reception/transduction. However, in both of these cases, the gene encoding the repressor also encodes the activator and regulation occurs post-transcriptionally. Therefore, the repressor and the activator carry the same DNA-binding domain and have the same intrinsic DNA-binding specificity in vitro. This is not the case in the Dpp signaling pathway since Brk and Mad have different DNA-binding domains and recognize different (albeit similar) DNA sites. Conceptually, this allows for more variation in the architecture of cis-regulatory elements integrating Dpp signaling levels, and might contribute to the readout of the Dpp morphogen gradients, as outlined above. However, the repressor activity of Ci and Pan can be influenced by local sequence context and/or by distant elements (Müller and Basler, 2000; Piepenburg et al., 2000), possibly allowing as much variation in the architecture of cis-acting elements as is the case for response elements of the Dpp signaling pathway. More detailed studies are required to see whether there are basic differences between Dpp signaling and other signaling cascades. Many more surprises might surface, hopefully increasing insight into the molecular mechanisms by which signals are integrated during development and can be read in a dosage-dependent manner by cellular fields in order to control patterning and growth.
Acknowledgments
Acknowledgements
We thank Konrad Basler, Bruno Müller and the members of the Affolter laboratory for comments and discussion. We apologize to colleagues whose work has not been cited in full due to space limitations. Our own studies in this field were supported by grants from the Swiss National Science Foundation and by the Kantons Basel-Stadt and Basel-Land. A.J. acknowledges an EMBO postdoctoral fellowship. M.A.V. was supported in part by the Roche Research Foundation and the Ciba-Geigy-Jubiläums-Stiftung.
References
- Arora K., Dai,H., Kazuko,S.G., Jamal,J., O’Connor,M.B., Letsou,A. and Warrior,R. (1995) The Drosophila schnurri gene acts in the Dpp/TGFβ signaling pathway and encodes a transcription factor homologous to the human MBP family. Cell, 81, 781–790. [DOI] [PubMed] [Google Scholar]
- Ashe H.L., Mannervik,M. and Levine,M. (2000) Dpp signaling thresholds in the dorsal ectoderm of the Drosophila embryo. Development, 127, 3305–3312. [DOI] [PubMed] [Google Scholar]
- Campbell G. and Tomlinson,A. (1999) Transducing the Dpp morphogen gradient in the wing of Drosophila: regulation of Dpp targets by brinker. Cell, 96, 553–562. [DOI] [PubMed] [Google Scholar]
- Certel K., Hudson,A., Carroll,S.B. and Johnson,W.A. (2000) Restricted patterning of vestigial expression in Drosophila wing imaginal discs requires synergistic activation by both mad and the drifter POU domain transcription factor. Development, 127, 3173–3183. [DOI] [PubMed] [Google Scholar]
- Chen X., Rubock,M.J. and Whitman,M. (1996) A transcriptional partner for MAD proteins in TGFβ signalling [published erratum appears in Nature, 384, 648]. Nature, 383, 691–696. [DOI] [PubMed] [Google Scholar]
- Chen Y.G., Hata,A., Lo,R.S., Wotton,D., Shi,Y., Pavletich,N. and Massagué,J. (1998) Determinants of specificity in TGFβ signal transduction. Genes Dev., 12, 2144–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai H., Hogan,C., Gopalakrishnan,B., Torres-Vazquez,J., Nguyen,M., Park,S., Raftery,L.A., Warrior,R. and Arora,K. (2000) The zinc finger protein schnurri acts as a Smad partner in mediating the transcriptional response to decapentaplegic. Dev. Biol., 227, 373–387. [DOI] [PubMed] [Google Scholar]
- Dorfman R. and Shilo,B.Z. (2001) Biphasic activation of the BMP pathway patterns the Drosophila embryonic dorsal region. Development, 128, 965–972. [DOI] [PubMed] [Google Scholar]
- Entchev E.V., Schwabedissen,A. and Gonzalez-Gaitan,M. (2000) Gradient formation of the TGFβ homolog Dpp. Cell, 103, 981–991. [DOI] [PubMed] [Google Scholar]
- Fan C.M. and Maniatis,T. (1990) A DNA-binding protein containing two widely separated zinc finger motifs that recognize the same DNA sequence. Genes Dev., 4, 29–42. [DOI] [PubMed] [Google Scholar]
- Grieder N.C., Nellen,D., Burke,R., Basler,K. and Affolter,M. (1995) Schnurri is required for Drosophila Dpp signaling and encodes a zinc finger protein similar to the mammalian transcription factor PRDII-BF1. Cell, 81, 791–800. [DOI] [PubMed] [Google Scholar]
- Grieder N.C., Marty,T., Ryoo,H.D., Mann,R.S. and Affolter,M. (1997) Synergistic activation of a Drosophila enhancer by HOM/EXD and DPP signaling. EMBO J., 16, 7402–7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groppe J., Rumpel,K., Economides,A.N., Stahl,N., Sebald,W. and Affolter,M. (1998) Biochemical and biophysical characterization of refolded Drosophila DPP, a homolog of bone morphogenetic proteins 2 and 4. J. Biol. Chem., 273, 29052–29065. [DOI] [PubMed] [Google Scholar]
- Halder G., Polaczyk,P., Kraus,M.E., Hudson,A., Kim,J., Laughon,A. and Carroll,S. (1998) The Vestigial and Scalloped proteins act together to directly regulate wing-specific gene expression in Drosophila. Genes Dev., 12, 3900–3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfon M.S., Carmena,A., Gisselbrecht,S., Sackerson,C.M., Jimenez,F., Baylies,M.K. and Michelson,A.M. (2000) Ras pathway specificity is determined by the integration of multiple signal-activated and tissue-restricted transcription factors. Cell, 103, 63–74. [DOI] [PubMed] [Google Scholar]
- Jaźwińska A., Kirov,N., Wieschaus,E., Roth,S. and Rushlow,C. (1999a) The Drosophila gene brinker reveals a novel mechanism of Dpp target gene regulation. Cell, 96, 563–573. [DOI] [PubMed] [Google Scholar]
- Jaźwińska A., Rushlow,C. and Roth,S. (1999b) The role of brinker in mediating the graded response to Dpp in early Drosophila embryos. Development, 126, 3323–3334. [DOI] [PubMed] [Google Scholar]
- Kawabata M., Imamura,T. and Miyazono,K. (1998) Signal transduction by bone morphogenetic proteins. Cytokine Growth Factor Rev., 9, 49–61. [DOI] [PubMed] [Google Scholar]
- Kim J., Johnson,K., Chen,H.J., Carroll,S. and Laughon,A. (1997) Drosophila Mad binds to DNA and directly mediates activation of vestigial by Decapentaplegic. Nature, 388, 304–308. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick H., Johnson,K. and Laughon,A. (2001) Repression of Dpp targets by binding of brinker to mad sites. J. Biol. Chem., 21, 18216–18222. [DOI] [PubMed] [Google Scholar]
- Lecuit T., Brook,W.J., Ng,M., Calleja,M., Sun,H. and Cohen,S.M. (1996) Two distinct mechanisms for long-range patterning by Decapentaplegic in the Drosophila wing [published erratum appears in Nature, 382, 93]. Nature, 381, 387–393. [DOI] [PubMed] [Google Scholar]
- Mann R.S. and Affolter,M. (1998) Hox proteins meet more partners. Curr. Opin. Genet. Dev., 8, 423–429. [DOI] [PubMed] [Google Scholar]
- Marty T., Müller,B., Basler,K. and Affolter,M. (2000) Schnurri mediates dpp-dependent repression of brinker transcription. Nature Cell Biol., 2, 745–749. [DOI] [PubMed] [Google Scholar]
- Massagué J. (2000) How cells read TGFβ signals. Nature Rev. Mol. Cell. Biol., 1, 169–178. [DOI] [PubMed] [Google Scholar]
- Massagué J. and Wotton,D. (2000) Transcriptional control by the TGFβ/Smad signaling system. EMBO J., 19, 1745–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami M., Kinoshita,N., Kamoshida,Y., Tanimoto,H. and Tabata,T. (1999) brinker is a target of Dpp in Drosophila that negatively regulates Dpp-dependent genes. Nature, 398, 242–246. [DOI] [PubMed] [Google Scholar]
- Müller B. and Basler,K. (2000) The repressor and activator forms of cubitus interruptus control hedgehog target genes through common generic gli-binding sites. Development, 127, 2999–3007. [DOI] [PubMed] [Google Scholar]
- Nellen D., Burke,R., Struhl,G. and Basler,K. (1996) Direct and long-range action of a DPP morphogen gradient. Cell, 85, 357–368. [DOI] [PubMed] [Google Scholar]
- Neul J.L. and Ferguson,E.L. (1998) Spatially restricted activation of the SAX receptor by SCW modulates DPP/TKV signaling in Drosophila dorsal-ventral patterning. Cell, 95, 483–494. [DOI] [PubMed] [Google Scholar]
- Nguyen M., Park,S., Marques,G. and Arora,K. (1998) Interpretation of a BMP activity gradient in Drosophila embryos depends on synergistic signaling by two type I receptors, SAX and TKV. Cell, 95, 495–506. [DOI] [PubMed] [Google Scholar]
- Nishita M., Hashimoto,M.K., Ogata,S., Laurent,M.N., Ueno,N., Shibuya,H. and Cho,K.W. (2000) Interaction between Wnt and TGFβ signalling pathways during formation of Spemann’s organizer. Nature, 403, 781–785. [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C., Wieschaus,E. and Klundig,H. (1984) Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster. I. Zygotic loci on the second chromosome. Wilhelm Roux’s Arch. Dev. Biol., 193, 267–282. [DOI] [PubMed] [Google Scholar]
- Piepenburg O., Vorbruggen,G. and Jackle,H. (2000) Drosophila segment borders result from unilateral repression of hedgehog activity by wingless signaling. Mol. Cell, 6, 203–209. [PubMed] [Google Scholar]
- Podos S.D. and Ferguson,E.L. (1999) Morphogen gradients: new insights from DPP. Trends Genet., 15, 396–402. [DOI] [PubMed] [Google Scholar]
- Raftery L.A. and Sutherland,D.J. (1999) TGFβ family signal transduction in Drosophila development: from Mad to Smads. Dev. Biol., 210, 251–268. [DOI] [PubMed] [Google Scholar]
- Raftery L.A., Twombly,V., Wharton,K. and Gelbart,W.M. (1995) Genetic screens to identify elements of the decapentaplegic signaling pathway in Drosophila. Genetics, 139, 241–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushlow C., Colosimo,P.F., Lin,M.C., Xu,M. and Kirov,N. (2001) Transcriptional regulation of the Drosophila gene zen by competing Smad and Brinker inputs. Genes Dev., 15, 340–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoo H.D., Marty,T., Casares,F., Affolter,M. and Mann,R.S. (1999) Regulation of Hox target genes by a DNA bound Homothorax/Hox/Extradenticle complex. Development, 126, 5137–5148. [DOI] [PubMed] [Google Scholar]
- Saller E. and Bienz,M. (2001) Direct competition between Brinker and Drosophila Mad in Dpp target gene transcription. EMBO Rep., 2, 298–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekelsky J.J., Newfeld,S.J., Raftery,L.A., Chartoff,E.H. and Gelbart,W.M. (1995) Genetic characterization and cloning of mothers against dpp, a gene required for decapentaplegic function in Drosophila melanogaster. Genetics, 139, 1347–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. (2001) Structural insights on Smad function in TGFβ signaling. BioEssays, 23, 223–232. [DOI] [PubMed] [Google Scholar]
- Sivasankaran R., Vigano,M.A., Muller,B., Affolter,M. and Basler,K. (2000) Direct transcriptional control of the dpp target omb by the DNA binding protein brinker. EMBO J., 19, 6162–6172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer F.A., Hoffmann,F.M. and Gelbart,W.M. (1982) Decapentaplegic: a gene complex affecting morphogenesis in Drosophila melanogaster. Cell, 28, 451–461. [DOI] [PubMed] [Google Scholar]
- Staehling-Hampton K., Laughon,A.S. and Hoffmann,F.M. (1995) A Drosophila protein related to the human zinc finger transcription factor PRDII/MBPI/HIV-EP1 is required for dpp signaling. Development, 121, 3393–3403. [DOI] [PubMed] [Google Scholar]
- Szüts D., Eresh,S. and Bienz,M. (1998) Functional intertwining of Dpp and EGFR signaling during Drosophila endoderm induction. Genes Dev., 12, 2022–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimoto H., Itoh,S., ten Dijke,P. and Tabata,T. (2000) Hedgehog creates a gradient of DPP activity in Drosophila wing imaginal discs. Mol. Cell, 5, 59–71. [DOI] [PubMed] [Google Scholar]
- Teleman A.A. and Cohen,S.M. (2000) Dpp gradient formation in the Drosophila wing imaginal disc. Cell, 103, 971–980. [DOI] [PubMed] [Google Scholar]
- Torres-Vazquez J., Park,S., Warrior,R. and Arora,K. (2001) The transcription factor Schnurri plays a dual role in mediating Dpp signaling during embryogenesis. Development, 128, 1657–1670. [DOI] [PubMed] [Google Scholar]
- Udagawa Y., Hanai,J.I., Tada,K.I., Grieder,N.C., Momoeda,M., Taketani,Y., Affolter,M., Kawabata,M. and Miyazono,K. (2000) Schnurri interacts with mad in a dpp-dependent manner. Genes Cells, 5, 359–369. [DOI] [PubMed] [Google Scholar]
- Waltzer L. and Bienz,M. (1998) Drosophila CBP represses the transcription factor TCF to antagonize Wingless signalling. Nature, 395, 521–525. [DOI] [PubMed] [Google Scholar]
- Xu X., Yin,Z., Hudson,J.B., Ferguson,E.L. and Frasch,M. (1998) Smad proteins act in combination with synergistic and antagonistic regulators to target Dpp responses to the Drosophila mesoderm. Genes Dev., 12, 2354–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Levine,M. and Ashe,H.L. (2001) Brinker is a sequence-specific transcriptional repressor in the Drosophila embryo. Genes Dev., 15, 261–266. [DOI] [PMC free article] [PubMed] [Google Scholar]