TO THE EDITOR:
The SH2B3 gene encodes the cytoplasmic adaptor protein LNK that acts as a negative regulator of hematopoietic progenitor cell expansion and self-renewal, particularly through its interaction with JAK2 and regulation of the JAK-STAT pathway [1]. Loss of LNK function leads to enhanced sensitivity to cytokines such as thrombopoietin and to a lesser extent erythropoietin, contributing to enhanced cell proliferation [1]. Structurally, the LNK protein consists of three major conserved domains: an N-terminal dimerization domain (DD) implied in protein-protein interactions and oligomerization, a central pleckstrin homology (PH) domain involved in membrane co-localization through lipid binding, and a C-terminal Src homology 2 (SH2) domain that is critical for binding to phosphorylated tyrosine residues thereby enabling LNK to inhibit downstream signaling events [1].
In humans, SH2B3 alterations have been identified across a spectrum of hematologic malignancies, especially myeloproliferative neoplasms (MPN) [2, 3], myelodysplastic syndromes (MDS) [4], MDS/MPN overlap syndromes including chronic myelomonocytic leukemia (CMML) [5], and acute lymphoblastic leukemia (ALL) [6]. In the germline setting, SH2B3 alterations have also been observed in myeloid proliferations, in either monoallelic or biallelic configurations [7–14]. This heterogeneity, as well as their relative rarity, raises important questions regarding clonal hierarchy, functional impact, and diagnostic interpretation of SH2B3 variants in hematologic diseases.
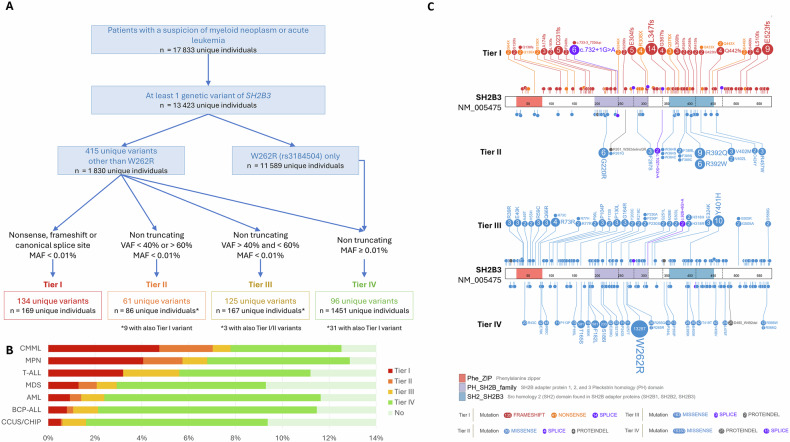
Here, we retrospectively investigated a cohort of 17,833 patients referred for suspicion of a myeloid neoplasm or acute leukemia. All patients were screened using a next-generation sequencing (NGS) panel. SH2B3 variants were classified into four tiers based on mutation type, variant allele frequency and description in normal population databases as described in the Fig. 1A (Supplementary methods, Supplementary Table 1).
Fig. 1. Landscape and classification of SH2B3 variants.
A Flowchart of the study showing selection and classification of SH2B3 variants. B Lollipop plot showing the distribution and classification of SH2B3 variants along the LNK protein structure (https://pecan.stjude.cloud/). Top panel: Tier I/II. Bottom panel: Tier III/IV. Functional domains are indicated: DD (red, aa 24-81), PH (purple, aa 194-307), and SH2 domain (blue, aa 364-462). C Frequency of SH2B3 variants by diagnostic category and tier classification in cases with clonality evidence.
Overall, Tier I and/or Tier II variants, hereafter referred as likely pathogenic SH2B3 (SH2B3LP) variants, were identified in 246 unique individuals (~1.4% of the total 17,833). Tier I (null) variants were observed throughout the entire protein, without restriction to specific domains while Tier II variants (mostly missense) exhibited a non-random distribution, with an enrichment in the PH and SH2 domains (Fig. 1B). Since the cohort included individuals referred for suspicion of hematologic malignancy without systematic diagnostic confirmation, it is likely that a proportion of cases presented with non-malignant hematologic abnormalities. Therefore, to refine the interpretation of SH2B3 variant frequencies, we analyzed their distribution according to the clinical indication for NGS, but restricted this analysis to individuals with evidence of clonality, defined by the presence of a SH2B3LP variant or in another gene from the panel. Accordingly, the prevalence of SH2B3LP variants was 2.2% (246 out of 11,113 individuals), with notable enrichment in CMML (7.2%) and MPN (5.6%) (Fig. 1C). By contrast, Tier III and IV variants were more uniformly distributed and frequently located outside of functional domains.
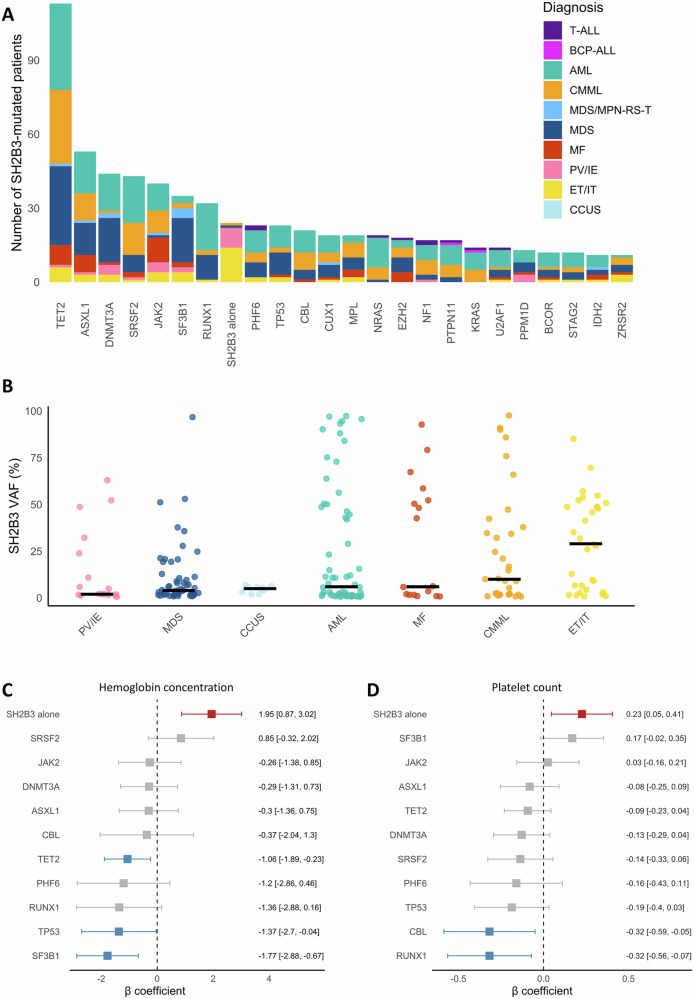
Among patients with SH2B3LP variants, the retained diagnosis was: AML (n = 61), MDS (n = 56), CMML (n = 33), essential thrombocythemia or idiopathic thrombocytosis (ET/IT, n = 31), MF (n = 17), polycythemia vera or idiopathic erythrocytosis (PV/IE, n = 17), BCP-ALL (n = 4), MDS/MPN with ring sideroblasts and thrombocytosis (MDS/MPN-RS-T, n = 4), T-cell ALL (n = 4), and clonal cytopenia of undetermined significance (CCUS, n = 9). The term “idiopathic” was used here to designate cases showing sustained erythrocytosis or thrombocytosis that did not fulfill the full WHO diagnostic criteria for PV or ET, respectively, and in which no alternative cause could be identified. Diagnosis remained undetermined in 10 cases. The overall median age of this cohort was 72 years (IQR 61–78). Additional mutations were found 212 patients (86.2%), with a median of 3 co-mutations per patient (Fig. 2A). The most frequent co-mutations were observed in TET2 (47%), ASXL1 (22%), DNMT3A (20%), SRSF2 (17%), JAK2 (16%), SF3B1 (14%), and RUNX1 (13%). Notably, a subset of patients (n = 34, 13.8%) harbored isolated SH2B3LP variants (SH2B3 alone).
Fig. 2. Molecular and hematological features of patients carrying SH2B3LP variants.
A Frequency of co-mutated genes (present in >10 patients) among individuals with SH2B3LP variants (n = 236). Individuals with undetermined diagnosis were removed from the figure (n = 10). B Variant allele frequencies (VAF) of SH2B3LP variants by diagnosis (in categories with >5 patients). Horizontal bars indicate the median VAF for each disease category. C, D Forest plots showing the association between mutation status and hemoglobin concentration and platelet count. Platelet counts are log-transformed. Only genes mutated in ≥10 patients were included. Error bars represent 95% confidence intervals for the β coefficients.
The median VAF of SH2B3LP variants was 7% (IQR 2–42). Higher VAFs were observed in ET/IT (median 29% [IQR 6-49%]) compared to CMML (median 10% [IQR 2–38%]), MF (median 6% [IQR 2–52%]), AML (median 6% [IQR 1–46%]), CCUS (median 5% [IQR 3–6%]), MDS (median 4% [IQR 2–11%]) and PV/IE (median 2% [IQR 2-24%]) (Fig. 2B, Supplementary Fig. 1). To infer the position of SH2B3LP variants in clonal architecture, their VAFs were compared with those of co-occurring variants in other genes. Assuming a linear model of leukemogenesis (which may not apply to all cases), SH2B3LP variants were suggested to occur as early events in ET/IT, PV/IE, and BCP-ALL, whereas they more often appeared as later events in AML, MDS, MDS/MPN-RS-T, CMML, and MF (Supplementary Fig. 2).
Considering that SH2B3 alterations are expected to potentiate JAK2-driven myeloproliferation in experimental models [2, 3, 15], we specifically compared SH2B3LP VAFs in JAK2/CALR co-mutated patients (Supplementary Fig. 3). Among JAK2-mutated cases (n = 40), SH2B3LP variants showed lower adjusted-VAFs than JAK2-V617F in 19 patients (48%), higher adjusted-VAFs in 5 patients (12%), and similar adjusted-VAFs in 16 patients (40%). In CALR-mutated patients (n = 10), SH2B3LP variants had lower adjusted-VAFs in 7 patients and comparable adjusted VAFs in 3 patients. These data suggest that SH2B3LP variants act preferentially as cooperating events rather than initiating events in JAK2 or CALR-positive MPN. In the triple-negative MPN cohort (n = 36), defined as wild-type for CALR, JAK2-V617F, and MPL-W515, SH2B3 was the sole mutated gene in 22 cases, with VAFs ranging from 1% to 57%. Eight of these harbored a VAF > 40%. The remaining 14 patients carried co-occurring mutations, most frequently affecting DNMT3A (n = 5), SF3B1 (n = 4), and TET2 (n = 4). In this latter group, SH2B3LP VAFs ranged from 1% to 70%, and seven cases had VAF > 40%. Altogether, 15/36 (42%) triple-negative MPN patients (13 ET/IT, 2 PV/IE) had SH2B3LP VAF > 40% (Supplementary Table 2).
To assess the impact of molecular alterations in SH2B3LP patients, we compared hematologic parameters across different mutational contexts. Mutations in TET2, TP53, and SF3B1 were associated with significantly lower hemoglobin levels. RUNX1 and CBL mutations were associated with decreased platelet counts. TET2 mutations were associated with higher monocyte counts while DNMT3A were associated with lower monocyte counts. Strikingly, patients harboring only SH2B3LP variants (without other molecular finding) showed significantly higher hemoglobin concentrations (β = 1.95; 95% CI: 0.87–3.02) and platelet counts (β = 0.23; 95% CI: 0.05–0.41) compared to those with other mutations (Fig. 2C, D). No consistent correlation was observed with SH2B3LP VAFs in the whole cohort or within the distinct diagnostic categories (Supplementary Fig. 4).
Overall, our study confirms that SH2B3LP variants are relatively rare but enriched in MPN (∼5-6%) and CMML (∼7.2%). The complex interplay with other mutations, including their clonal hierarchy, allelic configuration, and clonal burden, likely contributes to the pleomorphic clinical manifestations observed in hematologic neoplasms with SH2B3LP variants. Interestingly, our analysis delineated a subgroup of patients harboring isolated SH2B3LP variants (more frequently truncating variants), with frequent isolated thrombocytosis or erythrocytosis, suggesting that SH2B3 variants alone may drive early, or indolent myeloproliferative phenotypes. This profile was distinct from other SH2B3LP individuals who presented with more complex mutational landscapes and more advanced malignancies. Additionally, younger patients (<60 y) had more frequently VAFs close to 50% (median 49% [IQR 47-51]) compatible with a germline origin, compared to older patients (median 4% [IQR 2–7%]). Supporting this hypothesis, we identified three independent families in which SH2B3LP variants co-segregated with thrombocytosis or erythrocytosis (Supplementary Table 3).
The scope of our study regarding variant interpretation has three main limitations. First, the absence of systematic germline sampling precluded definitive classification of constitutional versus somatic origin, which is relevant for SH2B3 given its role in both acquired and inherited hematopoietic disorders. Second, the present study used stringent criteria for variant categorization, prioritizing variant type, allele frequency in the general population, and somatic VAF thresholds. This strategy, while enhancing confidence in Tier I/II assignments, may have excluded some rare, non-truncating but potentially pathogenic variants, and may therefore have underestimated the full spectrum of clinically relevant SH2B3 alterations. Also, some variants with a minor allele frequency ≥0.1% previously reported in the literature as potentially pathogenic, such as E208Q, E395K and E400K were intentionally excluded. While our data suggest an enrichment of the E395K and E400K variants in patients referred for suspected MPN, their relatively high frequency in the general population challenges their pathogenicity (Supplementary Fig. 5). Further functional studies and appropriately matched control cohorts are warranted to clarify their clinical relevance. Third, our classification of variants as likely pathogenic is based solely on molecular features, population frequencies and in silico analyses, without direct functional validation.
In conclusion, our study provides a comprehensive overview of SH2B3 variants in a large cohort of patients referred for suspected myeloid neoplasms or acute leukemia. By applying a standardized and conservative classification strategy, we identified a subset of clinically significant SH2B3 variants enriched in CMML and MPN, including triple-negative cases. We further highlighted the existence of a distinct subgroup of patients, often younger, with isolated thrombocytosis or erythrocytosis and high SH2B3 VAFs, some of whom presented familial co-segregation. These findings underscore the biological and clinical relevance of SH2B3 in myeloid disorders and support its inclusion in diagnostic screening panels. Future prospective and functional studies are needed to better define the pathogenicity, germline contribution, and therapeutic implications of SH2B3 alterations.
Supplementary information
Acknowledgements
The authors thank the patients and their families, as well as all the physicians, nurses, and healthcare professionals who contribute to patient care. The authors thank Christophe Roumier and the Tumor Bank of the Lille University Hospital (certification NF 96900-2014/65453-1) for handling, conditioning, and storing patient samples.
Author contributions
ON and ND designed the study and supervised the project. LF, AM-R, EF, CP, ON and ND performed molecular analyses. MW, ST, SD, CB, MDu, EP, AC, DL, JH, SD, AD, LP, JBri, BC, AW, VC and LG collected clinical data. BD, MDe, PH, IB, EM, TB, ZM and JBru collected biological data. LBD, IB, ON and ND collected and curated data; prepared the figures and tables and wrote the manuscript. All authors critically reviewed the manuscript and approved the final version.
Data availability
The data that support the findings of this study are available from the corresponding author (N.D.), upon reasonable request.
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
In accordance with local regulations, informed consent was obtained by the referring physicians from all patients, who were informed that their clinical and molecular data could be used for research purposes. No additional samples or analyses were performed for the purpose of this study; all data were derived from routine clinical care and were anonymized prior to analysis to ensure patient confidentiality. This study was approved by an Institutional Review Board (Number approval CSTMT359) at Lille University Hospital and conducted in accordance with the Declaration of Helsinki.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Olivier Nibourel, Nicolas Duployez.
Supplementary information
The online version contains supplementary material available at 10.1038/s41408-025-01392-9.
References
- 1.Maslah N, Cassinat B, Verger E, Kiladjian JJ, Velazquez L. The role of LNK/SH2B3 genetic alterations in myeloproliferative neoplasms and other hematological disorders. Leukemia. 2017;31:1661–70. [DOI] [PubMed] [Google Scholar]
- 2.Hurtado C, Erquiaga I, Aranaz P, Miguéliz I, García-Delgado M, Novo FJ, et al. LNK can also be mutated outside PH and SH2 domains in myeloproliferative neoplasms with and without V617FJAK2 mutation. Leuk Res. 2011;35:1537–9. [DOI] [PubMed] [Google Scholar]
- 3.Zhang X, Ding Y, Li P, Bourgeois K, Rangan A, Herrick JL, et al. Null-type SH2B3 mutations are potential drivers in a subset of Ph-negative myeloproliferative neoplasms. Am J Hematol. 2024;99:1841–4. [DOI] [PubMed] [Google Scholar]
- 4.Bernard E, Tuechler H, Greenberg PL, Hasserjian RP, Arango Ossa JE, Nannya Y, et al. Molecular International Prognostic Scoring System for Myelodysplastic Syndromes. NEJM Evid. 2022;1:EVIDoa2200008. [DOI] [PubMed] [Google Scholar]
- 5.Palomo L, Meggendorfer M, Hutter S, Twardziok S, Ademà V, Fuhrmann I, et al. Molecular landscape and clonal architecture of adult myelodysplastic/myeloproliferative neoplasms. Blood. 2020;136:1851–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roberts KG, Morin RD, Zhang J, Hirst M, Zhao Y, Su X, et al. Genetic alterations activating kinase and cytokine receptor signaling in high-risk acute lymphoblastic leukemia. Cancer Cell. 2012;22:153–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arfeuille C, Vial Y, Cadenet M, Caye-Eude A, Fenneteau O, Neven Q, et al. Germline bi-allelic SH2B3/LNK alteration predisposes to a neonatal juvenile myelomonocytic leukemia-like disorder. Haematologica. 2024;109:2542–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wintering A, Hecht A, Meyer J, Wong EB, Hübner J, Abelson S, et al. LNK/SH2B3 as a novel driver in juvenile myelomonocytic leukemia. Haematologica. 2024;109:2533–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coltro G, Lasho TL, Finke CM, Gangat N, Pardanani A, Tefferi A, et al. Germline SH2B3 pathogenic variant associated with myelodysplastic syndrome/myeloproliferative neoplasm with ring sideroblasts and thrombocytosis. Am J Hematol. 2019;94:E231–4. [DOI] [PubMed] [Google Scholar]
- 10.Leardini D, Flex E, Stieglitz E, Cerasi S, Bertuccio SN, Baccelli F, et al. Biallelic SH2B3 germline variants are associated with a neonatal myeloproliferative disease and multisystemic involvement. Eur J Hum Genet. 2025;33:1127–35. [DOI] [PMC free article] [PubMed]
- 11.Vermeersch G, Devos T, Devos H, Lambert F, Poppe B, Van Hecke S. Germline heterozygous SH2B3-mutations and (idiopathic) erythrocytosis: Detection of a previously undescribed mutation. EJHaem. 2023;4:1143–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rumi E, Harutyunyan AS, Pietra D, Feenstra JDM, Cavalloni C, Roncoroni E, et al. LNK mutations in familial myeloproliferative neoplasms. Blood. 2016;128:144–5. [DOI] [PubMed] [Google Scholar]
- 13.Oh ST, Simonds EF, Jones C, Hale MB, Goltsev Y, Gibbs KD Jr, et al. Novel mutations in the inhibitory adaptor protein LNK drive JAK-STAT signaling in patients with myeloproliferative neoplasms. Blood. 2010;116:988–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lasho TL, Pardanani A, Tefferi A. LNK Mutations in JAK2 Mutation–Negative Erythrocytosis. N Engl J Med. 2010;363:1189–90. [DOI] [PubMed] [Google Scholar]
- 15.Koren-Michowitz M, Gery S, Tabayashi T, Lin D, Alvarez R, Nagler A, et al. SH2B3 (LNK) mutations from myeloproliferative neoplasms patients have mild loss of function against wild type JAK2 and JAK2 V617F. Br J Haematol. 2013;161:811–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author (N.D.), upon reasonable request.