Abstract
Aggregated TDP-43 is a hallmark of frontotemporal dementia (FTD), amyotrophic lateral sclerosis (ALS), and limbic-predominant age-related TDP-43 encephalopathy (LATE), and a common co-pathology in other neurodegenerative diseases. Currently, no specific biomarkers exist to assess TDP-43 pathology in vivo. We developed two small-molecule radiopharmaceuticals, [18F]ACI-19278 and [18F]ACI-19626, for visualizing TDP-43 inclusions by positron emission tomography (PET). Both ligands bind with high affinity to aggregated, but not soluble, TDP-43 in patient brain samples from diverse TDP-43 proteinopathies, including frontotemporal lobar degeneration with TDP-43 pathology (FTLD-TDP), ALS, and LATE, and in cell models. Both compounds display excellent selectivity for TDP-43 over Aβ, Tau, and α-synuclein aggregates. In non-human primates, [18F]ACI-19278 and [18F]ACI-19626 show a pharmacokinetic profile suitable for brain PET imaging (rapid brain uptake; fast and complete washout). ACI-19278 and ACI-19626 are promising first-in-class TDP-43 PET tracers with the potential to revolutionize the diagnosis and treatment of neurodegenerative proteinopathies, enabling a precision medicine approach.
Subject terms: Neurodegenerative diseases, Biomarkers, Diseases of the nervous system
High-affinity ligands were developed that specifically bind aggregated TDP-43, are selective over common co-pathologies, and possess pharmacokinetic profiles suitable for PET imaging to visualize TDP-43 pathology in the brains of patients.
Introduction
Pathological transactive response DNA-binding protein 43 kDa (TDP-43) is the main component of inclusions found in several neurodegenerative disorders, including amyotrophic lateral sclerosis (ALS), frontotemporal dementia (FTD)1,2, limbic predominant age-related TDP-43 encephalopathy (LATE)3, and found as co-pathology in Alzheimer’s disease (AD)4, dementia with Lewy bodies (DLB)5 and chronic traumatic encephalopathy (CTE)6. The mislocalization from nucleus to cytoplasm and aggregation of TDP-43 in neurons and glial cells drives neurodegeneration, resulting in progressive loss of motor and cognitive functions7,8.
TDP-43 proteinopathy is found postmortem in about 45% of FTD cases, being reported as FTLD-TDP to differentiate from FTLD characterized by accumulation of aggregated Tau (FTLD-Tau) or fused in sarcoma (FUS)–Ewing sarcoma–TAF15 family (FTLD-FUS or FTLD-FET) proteins2,9–11. Clinically, FTD represents a heterogeneous spectrum characterized by changes in behavior, personality, and language12. The three major FTD phenotypes, behavior variant (bvFTD), semantic variant primary progressive aphasia (svPPA), and the non-fluent agrammatic primary progressive aphasia (nfPPA), are each associated with the involvement of distinct parts of the frontal and temporal lobes. The availability of reliable biomarkers to support early differential diagnosis of FTD would fill a large knowledge gap.
ALS is a progressive, fatal motor neuron disease (MND) characterized by neuronal death leading to the loss of motor function, with an average life expectancy of 2–5 years after diagnosis13. In ALS, TDP-43 proteinopathy is found in approximately 97% of patients, originating in the motor cortex, brainstem, and spinal cord14. ALS and FTD represent a disease spectrum sharing clinical features with up to 50% ALS patients showing some executive function deficits15 and up to 15% of bvFTD patients showing motor dysfunction16, further highlighting the need for reliable biomarkers for accurate diagnosis.
LATE describes an aging-associated disease entity with TDP-43 proteinopathy (LATE-neuropathological changes (NC)) with or without the presence of amyloid plaques and Tau neurofibrillary tangles (AD neuropathological change, ADNC) in the aging brain, which mimics the clinical features of AD3,17. Large autopsy series indicate that LATE-NC affects ~1/3rd of individuals past 85 years of age and is strongly associated with cognitive impairment18,19. While the clinical diagnostic guidelines for LATE are being developed20,21, a definitive diagnosis is currently only possible post-mortem, and development of sensitive molecular-specific TDP-43 biomarkers has been specifically noted as a high priority in the field3.
The histopathology underlying the above-mentioned TDP-43 proteinopathies is described by four main subtypes: FTLD-TDP types A, B, C, and D9,22,23 based on morphology, localization, and association with clinical and genetic variants. Clinically, type A is associated with bvFTD, naPPA, AD23, and LATE24 and found in patients with mutations in progranulin (GRN), C9orf72 and TBK1 genes. Type B is common in FTD-MND and associated with mutations in the C9orf72 and TBK1 genes. Type C is found in svPPA, with no associated genes identified so far. Type D is rare and associated with valosin-containing protein (VCP) mutations. In addition to intracellular morphology and location, distinct folds, seeding capacity, and toxicity for FTLD-TDP type A25, type B26, and type C27 fibrils have been established28,29, confirming structural and biochemical heterogeneity. Despite a growing understanding of the pathomechanism, sensitive biomarkers for visualization, spatial distribution, and spreading patterns of pathological TDP-43 in the living brain are not available.
Positron emission tomography (PET) is a powerful imaging modality that enables visualization and quantification of molecular pathology in the brain. Significant progress has been made in the development of PET tracers targeting specific misfolded proteins such as amyloid beta (Aβ)30,31, Tau32,33, and a-synuclein34, significantly improving our understanding of neurodegenerative disorders. Aβ and Tau PET tracers provided data unraveling the temporal evolution of AD pathology accumulation in the brain. Indeed, the Aβ PET imaging allowed for the approval of Aβ immunotherapies35,36: Aβ PET positivity was applied for the inclusion of AD subjects in the studies, and a decrease in Aβ PET signal was used as a surrogate biomarker for measuring clinical effect. In addition, employing a Tau PET tracer facilitated patient classification and enrollment in anti-Aβ clinical trials and ultimately enabled the interpretation of their outcomes32,37,38. Beyond use as a diagnostic, prognostic, and staging biomarker, a TDP-43 PET tracer would be utilized for patient selection, proof of target engagement, as well as pharmacodynamic readout, enabling the assessment of efficacy in therapeutic trials in the ALS-FTD spectrum, LATE-NC, or other TDP-43 proteinopathy-related diseases. In addition, it will allow patient stratification in AD, where TDP-43 often appears as an aggravating co-pathology. Also, TDP-43 PET imaging would complement and validate the development of fluid biomarkers, while furthermore providing critical insight into the spatial distribution of pathological TDP-43 in the brain.
In this study, we report the characterization of first-in-class TDP-43 PET tracers, [18F]ACI-19278 and [18F]ACI-19626. Both compounds demonstrate high affinity to aggregated TDP-43, excellent selectivity over other amyloids found as co-pathologies, and a suitable pharmacokinetic profile for PET imaging in the human brain. ACI-19626 was prioritized for first-in-human (FIH) evaluation in healthy volunteers and patients with TDP-43 proteinopathies. Based on the data presented here, both compounds have the potential to detect TDP-43 by PET in the brains of living patients.
Results
[³H]ACI-19278 and [³H]ACI-19626 bind to patient brain-derived pathological TDP-43 aggregates
ACI-19278 and ACI-19626 (Supplementary Fig. 1a) are small molecular weight compounds identified by rational design leveraging the AC Immune Morphomer® platform. This is a large library of CNS-penetrant small molecular weight compounds designed to bind specifically to conformationally altered and beta-sheet-rich structures in protein aggregates commonly found in proteinopathies. Taking advantage of the autofluorescence properties of some compounds available in the library, initial hits were identified using simple target engagement screening assays. Identified hits were then improved through a multi-parameter medicinal chemistry optimization approach, radiolabeled with tritium, and their binding properties were characterized in orthogonal assays using patient-derived material. Using classical autoradiography (ARG), binding of [3H]ACI-19278 and [3H]ACI-19626 was assessed in brain sections obtained from the frontal cortex of FTLD-TDP type A, frontal and temporal cortex of FTLD-TDP type B, frontal and temporal cortex of FTLD-TDP type C, hippocampus of LATE-NC with ADNC (LATE-NC + ADNC), motor cortex of ALS and frontal and temporal cortex of healthy control cases (Fig. 1). Strong binding, substantially above the control tissues, was observed for the samples from FTLD-TDP type A and B, as well as LATE-NC (Fig. 1a, Total). When evaluating the signal in the presence of excess non-labeled compound, displacement was observed for the FTLD-TDP type A and B as well as LATE-NC tissues (Fig. 1a, NSB), indicating specific binding. [3H]ACI-19278 showed significant differentiation of FTLD-TDP type A tissue samples from control tissue, while the specific binding for [3H]ACI-19626 was significantly greater for both FTLD-TDP types A and B compared to that of the control tissues (Fig. 1b).
Fig. 1. Characterization of binding of [³H]ACI-19278 and [³H]ACI-19626 to brain samples from TDP-43 proteinopathies.
a Autoradiographic images of [3H]ACI-19278 and [3H]ACI-19626 binding in brain tissue sections from control (temporal cortex), FTLD-TDP (type A, frontal cortex; type B, temporal cortex), and LATE-NC + ADNC (hippocampus) cases. Total: total binding; Non-specific binding (NSB): residual binding in the presence of 2 μM unlabeled ACI-19278 or ACI-19626. Scale bar, 5 mm. Seven cases were tested for FTLD-TDP type A, five ([3H]ACI-19278) or seven ([3H]ACI-19626) cases for type B, and one case from LATE-NC + ADNC in independent experiments. b Specific binding (for ACI-19278: n = 9 control; n = 7 type A; n = 5 type B; n = 5 type C; n = 1 LATE-NC; n = 5 ALS. For ACI-19626: n = 7 control; n = 7 type A; n = 6 type B; n = 4 type C; n = 1 LATE-NC; n = 3 ALS). Data is shown as mean ± SD. One-way ANOVA with Dunnett’s multiple comparisons test; for ACI-19278 **p = 0.006 for control vs type A; for ACI-19626 *p = 0.01 for control vs type A, and **p = 0.009 for control vs type B. c Representative autoradiograms of [3H]ACI-19626 with higher magnification images of selected areas in adjacent sections following labeling with anti-phospho-TDP-43 antibody. Top row (images 1, 3, and 5), high-density TDP-43 pathology; bottom row (images 2, 4, 6), mid-low density TDP-43 pathology. Scale bar, 20 μm. d Saturation binding studies with [3H]ACI-19278 and [3H]ACI-19626 on human brain tissue sections from an FTLD-TDP type A donor. Data from one experiment. e Saturation binding studies with [3H]ACI-19278 and [3H]ACI-19626 in brain homogenates from control (n = 1), FTLD-TDP type A, type B, and type C cases. Data is shown as mean ± SD for [3H]ACI-19626 on type A (six independent experiments) and for both compounds on type C (three independent experiments). In other cases, representative data from two independent experiments is shown. f Kd values from saturation binding studies in e and Supplementary Fig. 1e. Mean ± SD is reported for FTLD-TDP type A and B across two cases. n.a., no fit or R2 < 0.85. Kd values in panels d and f were assessed by different techniques and on different tissue preparations. Source data are provided as a Source Data file.
Low to no displaceable binding for both compounds was detected in brain sections from FTLD-TDP type C despite abundant phospho-TDP-43 (pTDP-43)-positive dystrophic neurites characteristic of this subtype of FTLD-TDP39 (Supplementary Fig. 1b). When tissue sections from ALS cases were incubated with [3H]ACI-19278 or [3H]ACI-19626, no displaceable signal was detected, possibly due to the very low density of pTDP-43 pathology in ALS tissue (Supplementary Fig. 1c), at least in the motor cortex region tested. Both compounds displayed an ARG signal comparable to that of the control on tissue from FTLD-TDP type C and ALS cases (Fig. 1b). Brain samples used for classical ARG included both grey and white matter (Supplementary Fig. 1e). Some non-specific white matter binding was observed for ACI-19278 whereas ACI-19626 showed no non-specific retention to white matter regions that could affect the interpretation of the tracer binding to TDP-43 inclusions present in the white matter areas.
The binding specificity for TDP-43 aggregates in FTLD-TDP type A and B as well as LATE-NC + ADNC tissue was confirmed by the association of the ARG signal intensity with the distribution and density of TDP-43 pathology across the same tissue, as visualized using anti-pS409-410 labeling (ACI-19626 ARG data and immunofluorescence in adjacent tissue sections in Fig. 1c). A stronger ARG signal was observed in regions dense in pTDP-43 aggregates (Fig. 1c, inserts 1, 3, and 5) while signal was less intense in regions with low levels of TDP-43 pathology (Fig.1c, inserts 2, 4, 6).
To measure affinity to pathological aggregated TDP-43, saturation binding studies were performed with [3H]ACI-19278 and [3H]ACI-19626 using ARG on FTLD-TDP type A frontal cortex brain sections (Fig. 1d). The equilibrium dissociation constant (Kd) and in vitro binding potential or target occupancy (ratio of target density, Bmax, over Kd) were evaluated. In brain sections from two FTLD-TDP type A donors (one sporadic case and one genetic case with mutation in the progranulin gene), [3H]ACI-19278 showed a mean Kd value of 25 ± 25 nM with Bmax/Kd of 16 ± 4 and [3H]ACI-19626 a mean Kd value of 18 ± 1 nM with Bmax/Kd of 13 ± 1 (Fig. 1d and Supplementary Fig. 1f). A Kd of 27 nM was calculated for LATE-NC + ADNC tissue for ACI-19626 (Supplementary Fig. 1g). The high binding affinity to FTLD-TDP aggregates was further confirmed in an orthogonal radiobinding assay using sarkosyl-insoluble brain extracts from samples with FTLD-TDP pathology (Fig. 1e–f and Supplementary Fig. 1h). Across two FTLD-TDP extracts tested (one from a sporadic case and one from a genetic case with mutation in the progranulin gene), [3H]ACI-19278 and [3H]ACI-19626 showed a mean Kd value of 37 ± 4 nM and 24 ± 1 nM, respectively, for FTLD-TDP type A aggregates. Furthermore, [3H]ACI-19278 and [3H]ACI-19626 displayed a mean Kd value of 38 ± 25 nM and 25 ± 18 nM, respectively, in two C9orf72 mutation cases with FTLD-TDP type B pathology. Low levels of non-saturable binding were detected for both ligands in FTLD-TDP type C tissue (Fig. 1e, f), in agreement with the autoradiography studies. Limited, non-saturable levels of binding were detected in ALS brain homogenates (Supplementary Fig. 1i) in which the levels of pathological aggregates were notably lower compared with those from FTLD-TDP type A and B tissue (Supplementary Fig. 1j). Finally, no relevant, saturable binding was observed in control brain-derived homogenates, further highlighting the tracer specificity for pathological TDP-43.
Taken together, these data demonstrate that ACI-19278 and ACI-19626 bind to pathological TDP-43 in FTLD-TDP type A and B with affinity values in the nanomolar range. In addition, both compounds display specific binding to LATE-NC + ADNC tissue.
Binding specificity of ACI-19278 and ACI-19626 for aggregated versus physiological TDP-43
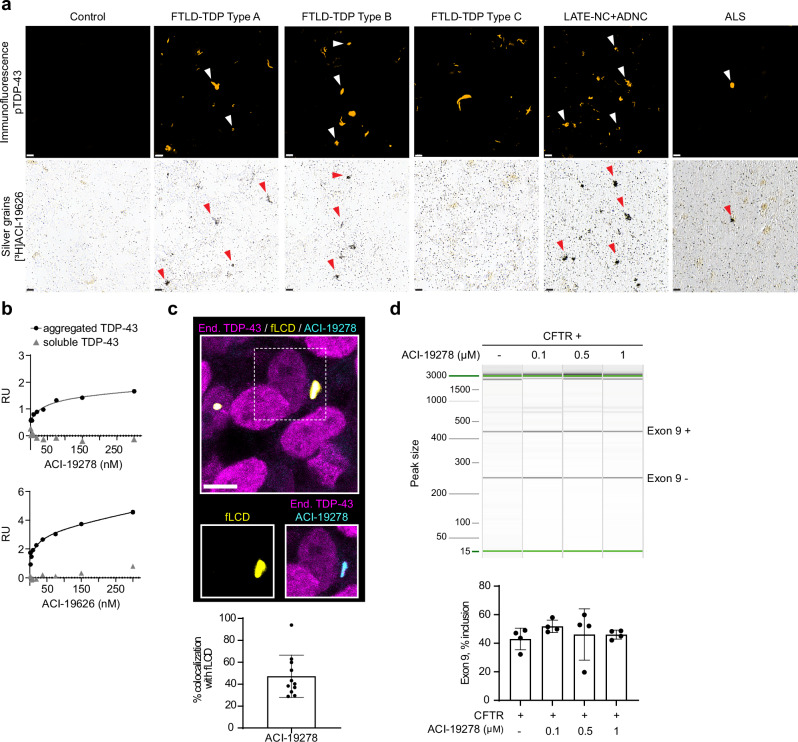
To assess target engagement of [3H]ACI-19278 and [3H]ACI-19626 to individual TDP-43 inclusions, high-resolution ARG studies were performed, allowing for a resolution of ~1 µm. No signal was observed in the absence of TDP-43 pathology in temporal cortex from control (Fig. 2a and Supplementary Fig. 2a). Immunofluorescent labeling of pTDP-43 aggregates (Fig. 2a and Supplementary Fig. 2a, top panel) and high-resolution ARG with [3H]ACI-19626 (Fig. 2b, bottom panel) or [3H]ACI-19278 (Supplementary Fig. 2a, bottom panel) performed on the same sections showed an extensive co-localization in FTLD-TDP type A and type B frontal and temporal cortex tissue, respectively, demonstrating target engagement of both compounds to TDP-43 inclusions. No ARG signal was observed on TDP-43 aggregates in temporal cortex sections from FTLD-TDP type C cases for both compounds, despite the abundance of pTDP-43 pathology. Notably, [3H]ACI-19278 (Supplementary Fig. 2a) and [3H]ACI-19626 (Fig. 2a, Supplementary Fig. 2b) displayed target engagement to pTDP-43 inclusions present in LATE-NC + ADNC tissue from multiple cases, even in case of low abundance of TDP-43 pathology. Finally, high-resolution autoradiography studies in motor cortex sections from ALS cases showed target engagement to TDP-43 inclusions in this tissue, regardless of their low density (Fig. 2a, Supplementary Fig. 2c). Altogether, these high-resolution ARG data are consistent with the classical ARG findings and radiobinding studies and further highlight binding specificity to pathological TDP-43 for ACI-19278 and ACI-19626.
Fig. 2. Binding specificity of ACI-19278 and ACI-19626 to aggregated TDP-43.
a High-resolution autoradiography with [³H]ACI-19626 (20 nM) in tissue from FTLD-TDP type A, type B and type C, LATE-NC + ADNC, ALS cases and a healthy control. Immunofluorescence with phospho-TDP-43 pS409/410 (pTDP-43) antibody (top panels, white arrowheads). Accumulation of silver grains on pTDP-43 inclusions on the same section (bottom panels, red arrowheads), showing co-labeling of pTDP-43 aggregates. Scale bar, 10 μm. Representative images of two independent experiments and donors for each tissue type (except for LATE-ND + ADNC, 3 cases tested). b Surface plasmon resonance studies for the determination of binding affinity of ACI-19278 and ACI-19626 to aggregated and soluble TDP-43. Single-cycle kinetics analysis was performed on immobilized brain-derived TDP-43 aggregates from an FTLD-TDP type A donor and on full-length recombinant TDP-43. Response signals (RU) are plotted against ACI-19278 or ACI-19626 concentration and steady-state affinity fits to calculate Kd. Data from one out of four independent experiments are shown. c ACI-19278 efficiently binds to TDP-43 fibrils and not to nuclear physiological TDP-43. Top, representative immunofluorescence image showing SH-SY5Y cells treated with TDP-43 fibrils (fLCD, yellow) and endogenous TDP-43 (magenta) and with ACI-19278 (cyan). Scale bar, 5 μm. Bottom, quantification depicting the percentage of overlapping area of exogenous TDP-43 aggregates (fLCD) and ACI-19278. Data is shown as mean ± SD from two independent experiments (n = 11). d ACI-19278 does not interfere with the physiological function of TDP-43. RT-PCR amplification and capillary electrophoresis analysis of CFTR exon 9 inclusion in CTFR minigene transfected SH-SY5Y cells in the presence of ACI-19728 (0.1–1 µM). Results from four independent experiments) (bottom panel), data shown as mean ± SD. One-way ANOVA with Dunnett’s multiple comparisons test showed no statistically significant differences. Source data are provided as a Source Data file.
Additionally, we evaluated the binding specificity to pathological versus physiological soluble TDP-43 in an orthogonal, label-free assay using surface plasmon resonance (SPR) spectroscopy. More specifically, insoluble fractions from the frontal cortex of FTLD-TDP type A brain and recombinant soluble TDP-43 were immobilized on the biosensor surface to assess binding of ACI-19278 and ACI-19626 (Fig. 2b and Supplementary Fig. 3). Both ligands showed binding specificity to aggregated TDP-43 with mean Kd values of 60 ± 9 nM for ACI-19278 and 80 ± 26 nM for ACI-19626 and no binding to soluble TDP-43.
To further explore the specificity of binding to aggregated versus physiological TDP-43 in a cellular environment40, the inherent fluorescent properties of ACI-19278 were exploited to evaluate target engagement. In agreement with the high-resolution ARG and SPR studies, the fluorescence signal from ACI-19278 colocalized with the fluorescence signal of internalized, cytoplasmic TDP-43 fibrils (fLCD) but not with endogenous physiological nuclear TDP-43 (Fig. 2c and Supplementary Fig. 4a). Notably, ACI-19278 showed an average colocalization of approximately 50% with the fLCD signal, indicating significant overlap with TDP-43 aggregates and therefore, extensive target engagement (Fig. 2c).
Finally, to rule out potential interference with physiological function of TDP-43, we evaluated ACI-19278 in a cystic fibrosis transmembrane conductance regulator (CFTR) exon 9 splicing assay in SH-SY5Y cells, surrogate for the function of TDP-43 in vitro41 (Supplementary Fig. 4b). Incubation of CFTR minigene-transfected cells with a range of concentrations of ACI-19278 (0.1–1 µM) showed no interference with the natural TDP-43 controlled RNA splicing of exon 9 (Fig. 2d). In contrast, a positive control comprising siRNA-mediated knockdown of TDP-43 resulted in increased inclusion of exon 9, as expected (Supplementary Fig. 4c).
The findings for ACI-19278 in cellular models are expected to also apply for ACI-19626 since the two ligands have a close chemical structure (Supplementary Fig. 1a), display a similar binding profile and ACI-19278 can compete with ACI-19626 (Supplementary Fig. 4d). Collectively, these data generated using patient-derived brain material, cellular models, and multiple orthogonal experimental techniques, show that ACI-19278 and ACI-19626 bind specifically to pathological TDP-43 inclusions and neither bind to soluble TDP-43 nor alter the physiological function of TDP-43.
[³H]ACI-19278 and [³H]ACI-19626 are selective for aggregated TDP-43 over common brain amyloid co-pathologies
Having shown desirable on-target binding to TDP-43 of ACI-19278 and ACI-19626, we next evaluated selectivity for TDP-43 versus other amyloidogenic proteins by assessing binding to Aβ and Tau aggregates on frontal cortex sections from an AD brain devoid of TDP-43 pathology (Fig. 3a). Radiolabeled reference molecules for Aβ (Aβ ref cmpd which is structurally related to Pittsburg compound B and can be displaced by it (Supplementary Fig. 5a) and Tau (PI-262042) displayed strong, displaceable binding (Fig. 3a, middle and bottom panel) that correlated with the distribution of Aβ plaques and Tau aggregates, as indicated by immunofluorescence labeling on the same tissue sections (Fig. 3a, top panel). In contrast, [3H]ACI-19278 and [3H]ACI-19626 displayed minimal to no retention on this tissue (Fig. 3a), indicating their preferential binding to TDP-43 over Aβ and Tau aggregates.
Fig. 3. ACI-19278 and ACI-19626 selectivity for TDP-43 over common co-pathologies.
a Autoradiography in Alzheimer’s disease (AD) tissue containing amyloid and Tau aggregates with an Aβ reference compound ([³H]Aβ ref cmpd) and Tau reference ligand ([³H]PI-2620). NSB: non-specific binding. Immunolabeling of adjacent sections with phospho-TDP-43 (orange), phospho-Tau (green) or Aβ (red) antibody. Scale bar, 2 mm. Representative autoradiograms from one donor from 3 independent experiments. b High-resolution autoradiography in the entorhinal cortex sections of an AD patient. [3H]PI-2620 included for reference. Thioflavin S staining in an adjacent section (left, green). Scale bar, 50 µm. Representative data of two independent experiments. c High-resolution autoradiography in FTLD-Tau tissue sections. Immunolabeling with phospho-Tau (top panels, green). Scale bar, 50 µm. Representative data of two donors tested in one experiment. d High-resolution autoradiography in tissue sections from a LATE-NC + ADNC case. Immunolabeling of the same sections with phospho-TDP-43 (orange), phospho-Tau (green), or Aβ (red) antibodies. Autoradiography images from adjacent sections with [3H]PI-2620 and [3H]Aβ reference compounds are shown on the bottom. Scale bar, 20 µm. Representative data of two donors tested in one experiment. White arrowheads: pTDP-43 pathology; red arrowheads: compound binding. e Saturation binding studies with [³H]ACI-19278 and [³H]ACI-19626 in brain homogenates from: From left to right, AD brain tissue containing Aβ and Tau aggregates (AD, insoluble fraction), AD brain tissue enriched for insoluble Tau paired helical filaments (AD, Tau PHF), and Parkinson’s disease (PD) brain tissue enriched for α-synuclein aggregates (PD, α -syn enriched). Reference ligands specific for Aβ (Aβ ref cmpd), Tau (PI-2620, Tau ref cmpd), and α-synuclein (ACI-12589, α-syn ref cmpd), respectively, were assessed in each homogenate. Representative data of at least two independent experiments for [³H]ACI-19278 and [³H]ACI-19626, and mean ± SD of three independent experiments for the [³H]Aβ ref cmpd in AD brain homogenates. Data from one out of two donors tested in independent experiments are shown for PHF and PD brain homogenates. Dotted line shows the limit under which binding is considered not relevant. Source data are provided as a Source Data file.
To confirm these findings, binding to pathological Tau present in different Tauopathies was assessed by high-resolution ARG combined with immunolabeling of phosphorylated Tau (pTau, Fig. 2b–c). While the Tau tracer, [3H]PI-2620, provided the positive control for a specific signal of the Tau tangles in AD entorhinal cortex tissue, [3H]ACI-19278 and [3H]ACI-19626 displayed no binding (Fig. 3b). Furthermore, no signal was detected for [3H]ACI-19278 or [3H]ACI-19626 on the pTau inclusions in FTLD-Tau frontal cortex tissue (Fig. 3c and Supplementary Fig. 5b), further confirming their selectivity over aggregated Tau. Evaluation of binding to non-AD 3R and 4R tauopathies has also showed no binding for ACI-19626 (Supplementary Fig. 5b).
Next, selectivity was assessed for TDP-43 over Aβ and Tau aggregates in the context of mixed protein pathologies. For this, high-resolution ARG experiments were conducted in LATE-NC + ADNC hippocampal tissue. The ARG signal from [3H]ACI-19278 and [3H]ACI-19626 (Fig. 3d, middle panels) colocalized with immunolabeling of pTDP-43 (Fig. 3d, top panels, white arrows), demonstrating binding to distinct aggregates of a smaller size as compared to the larger Aβ- or Tau-immunopositive aggregates in the same section (Fig. 3d, top panels, Aβ and pTau). Minimal co-localization with Aβ-positive structures was observed for [3H]ACI-19278, while no co-localization with Aβ or Tau-positive aggregates was observed for [3H]ACI-19626. In contrast, as expected, the Aβ binding reference compound ([3H]Aβ ref cmpd) and Tau binding reference compound, [3H]PI-2620, respectively revealed the characteristic forms of Aβ plaques and Tau tangles in adjacent sections (Fig. 3d, bottom panels).
Further assessment of the selectivity towards TDP-43 versus other aggregation-prone proteins was made by evaluating the binding affinity of [3H]ACI-19278 and [3H]ACI-19626 in brain homogenates derived from: a) AD brain tissue with confirmed burden of Aβ pathology (AD insoluble fraction, Fig. 3e, b) AD brain tissue enriched for insoluble Tau paired helical filaments (AD Tau PHF, Fig. 3e, and c) Parkinson’s disease (PD) brain tissue enriched for α-synuclein aggregates (PD α-syn enriched, Fig. 3e). Neither compound showed any relevant, saturable binding in any of these conditions, suggesting that both [3H]ACI-19278 and [3H]ACI-19626 are indeed selective over co-pathologies such as Aβ, Tau and α-syn aggregates. In contrast, the reference molecules for Aβ (Aβ ref cmpd), Tau (PI-2620), and α-syn (ACI-1258934, α-syn ref cmpd) displayed binding with high affinity to their respective targets with Kd values of 26 nM, 21 nM, and 66 nM, respectively.
Finally, we evaluated the potential off-target binding of ACI-19278 and ACI-19626 against a panel of more than 100 receptors, enzymes, ion channels, and transporters (Supplementary Table 1) at a concentration of 1 µM. No relevant binding to any of the tested proteins was observed, indicating excellent selectivity. Since binding to monoamine oxidase (MAO) A and B has been reported as a frequent off-target liability for Tau PET tracers43, displacement assays with the radiolabeled MAO-A inhibitor, [3H]Harmine, and the MAO-B inhibitor, [3H]deprenyl, were performed using microsomes expressing recombinant human MAO-A or MAO-B enzyme. No significant competition was measured for ACI-19278 or ACI-19626 (Supplementary Table 2), indicating no binding to MAO-A or B. Taken together, ACI-19278 and ACI-19626 display excellent selectivity for TDP-43 versus other aggregation-prone proteins, including pathological Aβ, Tau, and α-syn aggregates, and have clean off-target profiles.
Evaluation of the pharmacokinetic profile of [18F]ACI-19278 and [18F]ACI-19626 in rhesus macaques
Prior to the evaluation of the radiotracer pharmacokinetic (PK) profile in rhesus macaques, the PK of ACI-19278 and ACI-19626 was assessed in mice (Supplementary Fig. 6a). Both molecules showed brain uptake and fast washout. Next, to evaluate the suitability of the pharmacokinetic (PK) profile for use as a PET tracer, ACI-19278 and ACI-19626 were radiolabeled with fluorine-18 ([18F], Supplementary Fig. 6b) and administered intravenously in separate sessions to the same rhesus macaque. Dynamic PET scans (0–180 min) were conducted to assess brain uptake, distribution, and washout parameters in vivo. Both ligands entered the brain quickly, showed good brain permeability and exhibited a fast washout (Fig. 4). Specifically, [18F]ACI-19278 demonstrated a peak whole brain uptake at 5.5 min with standardized uptake value (SUV) of 2.9 (2.7% of injected dose, ID) while [18F]ACI-19626 reached the peak uptake at 4.5 min with SUV of 1.1 (1% ID) (Fig. 4b, c and Supplementary Fig. 6c, d). [18F]ACI-19626 showed lower brain uptake compared to [18F]ACI-19278 (Fig. 4b and Supplementary Fig. 6c), which is consistent with its lower lipophilicity as compared to ACI-19278. Conversely, [18F]ACI-19278 demonstrated slightly higher retention across various brain regions, including the dentate nucleus, cortical white matter and thalamus. [18F]ACI-19626 displayed a more homogeneous distribution with no obvious regional retention (Fig. 4a, b). Nonetheless, the washout of both ligands occurred rapidly, with a brain concentration ratio between peak and 60 min at 5.4 for [18F]ACI-19278 and 15.5 for [18F]ACI-19626 (Fig. 4b, c). Finally, the two ligands were quickly metabolized in a similar manner, with ~30% parent remaining at 30 min post tracer administration (Supplementary Fig. 6d).
Fig. 4. Assessment of the pharmacokinetic profile of [18F]ACI-19278 and [18F]ACI-19626 as brain PET tracers in rhesus macaques.
The same rhesus macaque, receiving either [18F]ACI-19278 or [18F]ACI-19626 intravenously at different points in time, was imaged by PET. A magnetic resonance imaging (MRI) of the brain was acquired on a separate day for anatomical reference and image analysis. a Transverse, sagittal and coronal views for MRI and PET images are provided to illustrate the determination of standardized uptake values (SUV) averaged over 0–30 min (middle row) and 30–90 min post-injection (bottom row). b Time activity curves (TACs) generated from the data obtained by quantification of the SUV for either [18F]ACI-19278 or [18F]ACI-19626 from different brain regions. Data from one experiment. c Comparison of the TACs for [18F]ACI-19278 and [18F]ACI-19626 averaged across the whole brain and reported as a percentage of the highest uptake (SUVmax) for each molecule. Source data are provided as a Source Data file.
Finally, metabolite identification for ACI-19626 was performed in vitro in rat, monkey, and human hepatocytes (Supplementary Table 3). Sixteen metabolites were detected in human hepatocytes, with oxidation and dehydrogenation to acid in methylpiperidine (M7) being the most abundant. The carboxylic acid moiety in M7 makes it highly unlikely to be brain penetrant and thus mitigates the risk of having a radioactive metabolite interfering with the parent’s signal.
Discussion
In vivo imaging of aggregated TDP-43 would open a new era for the development of disease-modifying therapies for TDP-43 proteinopathies. Here, we report for the first time the characterization of two compounds, [18F]ACI-19278 and [18F]ACI-19626, which exhibit all the desired characteristics for the successful visualization of TDP-43 pathology in the human brain by PET.
In the absence of available TDP-43 binding reference compounds, the development of TDP-43 PET tracers has proven challenging. This is at least in part due to the low density of TDP-43 inclusions as compared to the relatively higher Aβ and Tau pathology burden44–46. The data provided in this paper support the potential successful translation of the in vitro/ex vivo binding signal to in vivo PET, as the in vitro techniques/assays that were used were optimally selected based on their translational value. First, ACI-19278 and ACI-19626 show strong displaceable binding to brain sections with FTLD-TDP pathology by ARG, a technique with a resolution comparable to PET. Second, the intensity of the ARG signal correlates with the abundance and distribution of TDP-43 aggregates, reflecting a faithful quantification of pathology. Third, ACI-19278 and ACI-19626 display Kd values below 20 nM, and the in vitro binding potentials (Bmax/Kd ratios) estimated using ARG on FTLD-TDP brain sections are above 10, which is higher than the threshold considered optimal for the successful translation of PET tracers to in vivo studies47 (>5). These parameters compare favorably to those of the first a-syn PET tracer ACI-12589 (with an estimated Bmax/Kd ratio of 12.5 in MSA), able to visualize a-syn pathology in MSA patient brain34 where the pathology is less dense than Aβ and Tau pathology and more comparable to the density of pathological TDP-43.
Another key challenge in brain PET tracer development is achieving selectivity over other aggregation-prone proteins found in the aging brain and across the spectrum of neurodegenerative diseases48,49. Our data show selectivity of ACI-19278 and ACI-19626 over Aβ and Tau in AD brain samples, aggregated Tau in FTLD-Tau, Pick’s disease, and PSP brain samples, and aggregated a-syn in PD brain samples. Importantly, selectivity for aggregated TDP-43 over Aβ and Tau was further confirmed in LATE-NC brain samples where TDP-43 pathology was present together with abundant Aβ and Tau pathology (i.e., comorbid ADNC). In addition, ACI-19278 and ACI-19626 bind selectively to aggregated TDP-43 and not to physiological soluble TDP-43 and do not interfere with the physiological function of TDP-43. These data are also aligned with the absence of binding of these ligands in age-matched control brain sections and homogenates. Finally, in vitro pharmacology binding assays showed no significant interaction with any of the tested CNS receptors and enzymes, including MAO A and MAO B, further supporting the safety and selectivity of both compounds.
Although ACI-19278 and ACI-19626 are similar in terms of chemical structure and binding characteristics, ACI-19626 has a more advantageous profile as a potential TDP-43 PET tracer. First, ACI-19626 is devoid of non-specific retention to white matter and Aβ plaques on LATE-NC/ADNC tissue, which is sometimes seen with ACI-19278. Second, ACI-19626 showed a more homogenous distribution across different brain regions in PET PK profiling in NHPs. Indeed, ACI-19626 was designed to be less lipophilic than ACI-19278 to decrease non-specific retention. In line with its lower lipophilicity, brain uptake was lower for ACI-19626 compared to ACI-19278, but to its advantage, ACI-19626 washout was faster and more complete. This may result in a higher signal-to-noise ratio for ACI-19626, which is particularly critical when imaging a low-abundance target such as aggregated TDP-43. Based on this rationale, ACI-19626 was prioritized for first-in-human (FIH) evaluation in healthy volunteers and patients with TDP-43 proteinopathies.
The characterization of the binding and selectivity profile of ACI-19278 and ACI-19626 using orthogonal assays and a diverse set of postmortem patient brain samples indicates their capacity to detect Type A and Type B pathology, present in patients with FTD, FTD-MND, ALS, and LATE, with no binding to other amyloids, such as Abeta, aggregated Tau, and a-synuclein. The potential of the tracers to be used broadly across heterogeneous TDP-43 proteinopathies will depend on the abundance of TDP-43 pathology in living patient brains and has to be tested in the clinic. Binding to FTLD-TDP type A and B together with selectivity over aggregated Tau holds promise for differential diagnostics in FTD which is currently unavailable. Target engagement on FTLD-TDP type A-like aggregates on LATE-NC + ADNC brain samples, together with selectivity over Aβ and Ta,u might enable differential diagnosis for LATE-NC + ADNC subjects, which is currently challenging based purely on clinical features21,50. Patients with LATE-NC with comorbid ADNC show a faster rate of hippocampal atrophy, memory impairment, and overall cognitive decline than patients with either ADNC alone or LATE-NC alone. Data from multiple longitudinal cohorts demonstrate that LATE-NC + ADNC has a large impact on public health, while the presence of LATE-NC might influence outcomes for disease-modifying therapies targeting ADNC, with a potentially lower efficacy in mixed LATE-NC + ADNC for therapies targeting amyloid beta. Therefore, a TDP-43 PET tracer could significantly improve the design and efficiency of clinical trials in these populations.
While in the ALS brain, TDP-43 pathology typically appears as FTLD-TDP type B23, ACI-19278, and ACI-19626 displayed limited binding in ALS tissues tested here. A likely explanation for the weak signal is the typically low level of TDP-43 pathology found in post-mortem ALS tissue compared to FTLD-TDP51, consistent with immunolabeling experiments performed in our study. Nonetheless, when using high-resolution ARG or ALS brain fractions enriched in insoluble TDP-43, both compounds displayed binding that was distinguishable from controls. Variability in the levels of pathology across donors and brain regions used, as well as tissue quality, can greatly impact the strength of binding. Another potential explanation could be due to differences in ultrastructure between FTLD-TDP and ALS TDP-43 aggregates; however, this is less likely since the same filament fold has been recently reported in type B aggregates from ALS or FTLD-TDP donors by electron cryo-microscopy (cryo-EM) studies26. An additional consideration is that the abundance of TDP-43 pathology in ALS might decline in terminal stages of disease following the overt motor neuron death and pathology clearance as reported for FTLD-TDP52,53. Longitudinal PET imaging of TDP-43 pathology in the ALS patients' brains will help address these possibilities. Exploration of ACI-19278 and ACI-19626 binding to non-motor brain regions from ALS patients54 and tissues with FTLD-TDP type E pathology23,39 should be performed in the future to further understand the potential of these TDP-43 PET tracers across the heterogeneous ALS pathology spectrum.
In contrast to FTLD-TDP type A and B, binding of ACI-19278 and ACI-19626 to FTLD-TDP type C aggregates was minimal to absent across the different assays tested, despite the relatively abundant levels of cortical TDP-43 pathology in the characteristic form of dystrophic neurites. This might be attributed to structural and biochemical differences between FTLD-TDP type C and type A or type B aggregates. Indeed, a recent discovery showed that FTLD-TDP type C fibrils are formed as heteromeric amyloid filaments with annexin A1127,55 whereas the presence of ANXA11 was limited to only ~3 to 6% in other types of FTLD-TDP55. It appears that the region that co-assembles with ANXA11 in FTLD-TDP Type C filaments is hidden in the TDP-43 folds of ALS and FTLD-TDP type A and B. Therefore, the co-assembly with ANXA11 might hide the binding pockets of ACI-19278 and ACI-19626 on TDP-43 fibrils. These differences in the structure of the fibrillar core challenge the feasibility of the development of a pan-TDP-43 PET tracer. On the other hand, additional binding regions remain to be explored.
In conclusion, the unique molecules described in this study have the potential to open new opportunities for more timely and efficient differential diagnosis and therapeutic interventions in diseases with high unmet medical need, such as ALS, FTD, LATE, and AD. Specifically, they may improve the design and interpretation of clinical trials, enabling patient stratification, selection of optimal timing for therapeutic intervention, as well as target engagement and pharmacodynamic effect evaluation. [18F]ACI-19626 and [18F]ACI-19278, potential first-in-class TDP-43 PET tracers, might lead to substantial advances in the field of neurodegenerative proteinopathies by enabling a unique precision medicine approach. In the near future, ACI-19626 will be evaluated in a first-in-human (FIH) study for its ability to detect pathological TDP-43 in the brains of patients with TDP-43 proteinopathies and healthy volunteers.
Methods
Human brain samples
All tissues were collected from donors from whom a written informed consent for a brain autopsy and the use of the material and clinical information for research purposes had been obtained by the respective institution. All samples were anonymized and coded. Post-mortem frozen tissue blocks from different brain regions of control donors and donors with confirmed TDP-43 or other (Aβ, Tau, and α-syn) pathology were acquired from the Netherlands Brain Bank (NBB; Netherlands Institute for Neuroscience, Amsterdam (open access www.brainbank.nl)), Queen Square Brain Bank (QSBB), UCSF, LNDBB, University of Kentucky – Alzheimer’s Disease Research Center (UK ADRC NIA P30-AG-072946), Barrow Neurological Institute, and commercial providers (Tissue Solutions Ltd.) and kept at −80 °C until use. In some cases, frozen brain tissue blocks were processed using a cryotome to generate sections of 10 μm thickness and mounted on glass slides. Sections were kept at -80 °C until use. The presence or absence of pathological TDP-43 (and/or Aβ, Tau, and α-syn) aggregates in tissue sections from diseased or control donors, respectively, was confirmed by immunostaining. The available demographic information, genetic status, clinical phenotype, and presence of co-pathologies are summarized in Supplementary Tables 4 and 5.
Immunofluorescence staining
Sections were fixed for 15 min at 4 °C with 4% formaldehyde (Sigma, 252549) and washed three times 5 min with 1× PBS (Dulbecco’s phosphate-buffered saline, Sigma D1408) at room temperature (RT). Next, sections were blocked and permeabilized in blocking buffer (PBS, 10% NGS, 0.25% Triton X-100) for 1 h at RT in a moist chamber. Subsequently, slides were drained off on paper and incubated overnight at 4 °C with an antibody specific for phosphorylated serine at amino acids 409/410 TDP-43 (anti-pTDP-43 pS409/410, 1:500, Biolegend, 829901), or the phosphorylated anti-Tau antibody (AT8, 1:500, Pierce, MN1020), or an anti-Aβ antibody (4G8, Covance, SIG-39240), or the phosphorylated (Ser129) anti-a-syn antibody (S129, Abcam, ab51253). In some cases, AD sections were stained using Thioflavin S (Sigma T1892, diluted at 0.05% w/v in PBS) at RT for 8 min. The following day, sections were washed three times 8 min with 1× PBS before incubation with a secondary, AlexaFluor647-labeled goat-anti-rabbit antibody (Abcam, ab150079, 1:500) or AlexaFluor488-labeled goat-anti-mouse antibody (Invitrogen, A11001, 1:500) or AlexaFluor633-labeled goat-anti-rat antibody (Invitrogen, A-21094, 1:500) for 45 min at RT in a moist chamber. Following incubation with secondary antibody, the sections were washed three times in PBS.
For image acquisition, sections were mounted using ProLong Gold Antifade reagent (Invitrogen P36930) and imaged on a Panoramic Scan II (3DHistech) with a ×20 objective. For white matter visualization, brain sections were incubated with Luxol fast blue (ab150675) following the manufacturer’s instructions.
Autoradiography
Frozen brain sections were fixed for 15 min at 4 °C with 4% formaldehyde (Sigma, 252549) on ice and washed three times for 8 min with 1X PBS at RT. Then, sections were equilibrated in assay buffer (50 mM Tris-HCl, 0.9% NaCl, 0.1% BSA) for 30 min. Sections were subsequently incubated with 10 nM [3H]ACI-19278 (specific activity 45.3 Ci/mmol) or [3H]ACI-19626 (specific activity 55.4 Ci/mmol) or [3H]Aβ-ref (specific activity 83.9 Ci/mmol) or [3H]PI-2620 (specific activity 61.4 Ci/mmol) in assay buffer for 120 min at RT or increasing concentrations of [3H]ACI-19278 from 0.01 to 40.7 nM or [3H]ACI-19626 from 0.28 to 50.0 nM. To determine non-specific binding (NSB), adjacent brain sections were incubated with [3H]ACI-19278 or [3H]ACI-19626 or [3H]Aβ-ref or [3H]PI-2620 mixed with 2 µM of non-radiolabeled ACI-19278 or ACI-19626 or Aβ-ref or PI-2620, respectively. Sections were then washed as follows: once in ice-cold 50 mM Tris-HCl pH 7.4 buffer for 1 min, twice in ice-cold PBS for 1 min, once in ice-cold 50 mM Tris-HCl pH 7.4 buffer for 1 min, and finally rinsed briefly in ice-cold distilled water.
Sections were allowed to air-dry before being exposed and scanned in a real-time autoradiography system (BeaQuant instrument, ai4R) for two hours. Beavacq software v1.14.2 (AI4R) was used to acquire the images. ROI delimitation and quantification of signal were performed by using the image analysis software Beamage (ai4R). Specific binding was determined by subtracting the non-specific signal (NSB) from the total signal. For Kd (dissociation constant) and Bmax (maximum number of binding sites) determination, the specific binding values were first transformed into nM using a standard curve for [3H]ACI-19626, taking into account the tissue thickness. After transformation, Kd and Bmax values were calculated in GraphPad Prism by applying a nonlinear regression curve fit using a one-site, specific binding model.
Preparation of human brain homogenates from FTLD-TDP, ALS, and control donors
Sarkosyl-insoluble human brain extracts were prepared as described in Laferriere et al., 201929. Brain tissue was homogenized at 1:4 (w/v) ratio in the homogenization-solubilization (HS) buffer at 4 °C. Homogenized samples were aliquoted and stored at ‒80 °C in low protein-binding tubes. Brain homogenates were thawed on ice and resuspended in HS buffer to obtain a final concentration of 2% sarkosyl, 1 unit/µL Benzonase, and 1 mM MgCl2. The samples were then incubated at 37 °C under constant shaking at 600 rpm on a thermomixer for 45 min. The supernatants were collected in a new tube (sarkosyl-soluble fraction, S1). The pellet was resuspended in 1000 µL of myelin floatation buffer and centrifuged at 20,000 g for 1 h at 4 °C. The supernatant was carefully removed to remove all the floating lipids. This step was repeated if all the lipids could not be removed in a single step. The pellet was subsequently washed with Phosphate-Buffered Saline (PBS) and centrifuged for 30 min at 20,000 g at 4 °C. The final pellet was resuspended in 200 µL PBS and stored at −80 °C (sarkosyl-insoluble fraction). The samples were analyzed by immunoblotting in denaturing conditions as described in Afroz et al.56.
Preparation of human brain homogenates from an AD donor
The procedure used was adapted from Bagchi et al.57, describing extraction of an insoluble fraction containing protein aggregates from human brain tissue for in vitro binding and competition studies. Frozen brain tissue from an AD donor with confirmed burden of Aβ and Tau aggregates was used. The tissue was homogenized in high salt buffer (50 mM Tris-HCl pH 7.5, 0.75 M NaCl, 5 mM EDTA) supplemented with protease inhibitors (Complete; Roche 11697498001) at 4 °C using a glass Dounce homogenizer. The homogenate was transferred into polycarbonate centrifuge bottles (16 ×76 mm; Beckman 355603) and centrifuged at 100,000 x g (38,000 RPM) in an ultracentrifuge (Beckman, XL100K) for 60 min at 4 °C using a pre-cooled 70.1 rotor (Beckman, 342184). Pellets were resuspended in high salt buffer supplemented with 1% Triton X-100 and homogenized at 4 °C. The homogenates were centrifuged again at 100,000 x g (38,000 RPM, 70.1 Ti rotor) for 60 min at 4 °C. Pellets were resuspended in high salt buffer supplemented with 1% Triton X-100 and 1 M sucrose at 4 °C. The homogenates were centrifuged at 100,000 x g (38,000 RPM, 70.1 Ti rotor) for 60 min at 4 °C. The resulting pellets containing Aβ and Tau aggregates were resuspended in PBS, aliquoted, and stored at −80 °C until use.
Preparation of human brain PHF-Tau homogenates from an AD donor
The procedure used was adapted from Jicha et al.58 and Rostagno and Ghiso et al.59 for the enrichment of Tau paired-helical filaments (PHF). Briefly, frozen brain tissue from an AD donor was thawed on ice and homogenized with homogenization buffer (0.75 M NaCl in RAB buffer (100 mM 2-(N-morpholino) ethanesulfonic acid (MES), 1 mM EGTA, 0.5 mM MgSO4, 2 mM DTT, pH 6.8) supplemented with protease inhibitors (Roche, 693124001) using a glass Dounce homogenizer. The homogenate was then incubated at 4 °C for 20 min to let depolymerize any residual microtubules, before being transferred into polycarbonate centrifuge bottles (16 ×76 mm; Beckman 355603) and centrifuged at 11,000 g (12,700 RPM) in an ultracentrifuge (Beckman, XL100K) for 20 min at 4 °C using the pre-cooled 70.1 rotor (Beckman, 342184). Pellets were kept on ice. Supernatants were pooled into polycarbonate bottles and centrifuged again at 100,000 g (38,000 RPM) for one hour at 4 °C in the 70.1 Ti rotor to isolate PHF-rich pellets, whereas soluble Tau remained in the supernatant. The pellets from the first and second centrifugations were resuspended in extraction buffer (10 mM Tris-HCl, pH 7.4, 10% sucrose, 0.85 M NaCl, 1 tablet/50 ml protease inhibitor (Roche, 4693124001), 1 mM EGTA, 1% phosphatase inhibitor (Sigma, P5726 and P0044)). The solution was then transferred into polycarbonate centrifuge bottles (16 ×76 mm; Beckman 355603) and centrifuged at 15,000 g (14,800 RPM) in an ultracentrifuge (Beckman, XL100K) for 20 min at 4 °C using the 70.1 Ti rotor. In the presence of 10% sucrose and at low-speed centrifugation, most PHF remained in the supernatant, whereas intact or fragmented neurofibrillary tangles (NFTs) and larger PHF aggregates were pelleted. The pellets were stored at −80 °C. 30% sarkosyl (Fluka analytical, 61747) was added to the supernatants to a final concentration of 1% and stirred at RT for one hour. This solution was then centrifuged in polycarbonate bottles at 100,000 g (38,000 RPM) for one hour at 4 °C in the 70.1 Ti rotor, and the pellets containing PHF-rich material were resuspended in PBS. Aliquots were snap frozen and stored at −80 °C.
Preparation of human brain homogenates from a PD donor
The procedure used was adapted from Spillantini et al., 199860, which described the extraction of dispersed a-syn filaments from brain of PD cases. Frozen brain tissue from a PD donor was thawed on ice and homogenized in homogenization buffer using a glass Dounce homogenizer. The homogenization buffer consisted of RAB buffer (100 mM 2-(N-morpholino) ethanesulfonic acid (MES), 1 mM EGTA, 0.5 mM MgSO4, 2 mM DTT, pH 6.8) containing 0.75 M NaCl and 1X protease inhibitors (Complete; Roche 11697498001). The homogenate was then incubated at 4 °C for 20 min to allow depolymerization of any residual microtubules, before being transferred into polycarbonate centrifuge bottles (16 ×76 mm; Beckman 355603) and centrifuged at 11,000 g (12,700 RPM) in an ultracentrifuge (Beckman, XL100K) for 20 min at 4 °C using a pre-cooled 70.1 rotor (Beckman, 342184). Pellets were kept on ice while supernatants were pooled into polycarbonate bottles and centrifuged again at 100,000 g (38,000 RPM) for one hour at 4 °C at the 70.1 Ti rotor. The pellets from the first and second centrifugations were re-suspended in extraction buffer (10 mM Tris-HCl pH 7.4, 10% sucrose, 0.85 M NaCl, 1% protease inhibitor (Calbiochem 539131), 1 mM EGTA, 1% phosphatase inhibitor (Sigma P5726 and P0044)). The solution was then transferred into polycarbonate centrifuge bottles (16 × 76 mm; Beckman 355603) and centrifuged at 15,000 g (14,800 RPM, 70.1 Ti rotor) for 20 min at 4 °C. Pellets were discarded, and 20% sarkosyl (20% stock solution, Sigma L7414) was added to the supernatants to a final concentration of 1% and stirred at RT for one hour. This solution was then transferred to polycarbonate bottles at 100,000 g (38,000 RPM, 70.1 Ti rotor) for one hour at 4 °C. Pellets containing enriched a-syn aggregates were resuspended in PBS, aliquoted and stored at −80 °C until use.
Radiobinding assay for determination of the dissociation constant (Kd) on control, FTLD-TDP, Tau PHF, and PD brain homogenates
Human brain extracts were spotted onto microarray slides. The slides were incubated with [3H]ACI-19278, [3H]ACI-19626, [3H]PI-2620, or [3H]ACI-12589 at concentrations ranging from 1.3 nM to 200 nM in 50 mM Tris pH 7.4 supplemented with 0.1% BSA and 0.9% NaCl. Non-specific binding was determined with an excess of non-radiolabeled reference compound (2 µM). After two hours of incubation at RT, slides were washed and scanned by a real-time autoradiography system (BeaQuant, ai4R). Beavacq software (AI4R) was used to acquire the data, and quantification of the signal was performed by using the Beamage image analysis software (ai4R). Specific binding was calculated by subtracting the non-specific signal from the total signal. Kd values were obtained by fitting the specific binding data with non-linear regression analysis, using a one-site specific binding model in GraphPad Prism.
Filter radiobinding assay for Kd and Ki determination on AD brain homogenates
AD brain homogenates were incubated with [3H]ACI-19278 or [3H]ACI-19626 at different concentrations, ranging from 2 nM to 250 nM, or with [3H]ACI-Aβ ref at different concentrations, ranging from 2 nM to 50 nM, respectively. The reaction was performed in assay buffer (50 mM Tris, pH 7.5 in 0.9% NaCl, 0.1% BSA) and incubated for two hours at RT. Samples in duplicate were then filtered under vacuum in GF/C filter plates (PerkinElmer) to trap the aggregates with the bound radioligand and washed five times with ice-cold 50 mM Tris pH 7.5. The GF/C filters were then dried, and scintillation liquid (UltimateGold, PerkinElmer) was added to each well. The filters were analyzed on a Microbeta2 scintillation counter (PerkinElmer). Co-incubation with a non-radiolabeled compound at 1 µM was used to determine non-specific binding. Specific binding was calculated by subtracting the non-specific signal from the total signal. Kd values were calculated by nonlinear regression, using a one-site specific binding model using GraphPad Prism. Same conditions were applied for the evaluation of competition between Pittsburg compound B (2-(4-(Methylamino)phenyl)benzo[d]thiazol-6-ol) and [3H]ACI-Aβ ref. Serial dilution of PiB, ranging from 2000 nM to 0.14 nM, was tested against [3H]ACI-Aβ ref at 15 nM.
Assessment of target engagement by high-resolution autoradiography
The protocol was adapted from Marquie et al.61. Sections were incubated with 20 nM of [3H]ACI-19278, [3H]ACI-19626, [3H]PI-2620, or [3H]Aβ Ref for one hour at room temperature (RT). Sections were then washed as follows: one time in ice-cold 50 mM Tris-HCl pH 7.4 buffer for 1 min, two times in 70% ice-cold ethanol for 1 min, one time in ice-cold 50 mM Tris-HCl pH 7.4 buffer for 1 min and finally rinsed briefly in ice-cold distilled water. Sections were subsequently dried and then exposed to Ilford Nuclear Emulsion Type K5 (Agar Scientific, AGP9281) in a light-proof slide storage box. The sections were developed by immersing them successively in the following solutions: (1) Ilford Phenisol Developer (Agar Scientific, AGP9106), (2) Ilfostop solution (Agar Scientific, AGP9104), (3) Ilford Hypam Fixer (Agar Scientific, AGP9183), and finally rinsed with distilled H2O. Immunostaining was also performed on the same section using an antibody specific for phosphorylated serine at amino acids 409/410 TDP-43 (anti-pTDP-43 pS409/410, 1:500, Biolegend, 829901), or the phosphorylated anti-Tau antibody (AT8, 1:500, Pierce, MN1020), or an anti-Aβ antibody (4G8, Covance, SIG-39240). For image acquisition, sections were mounted using ProLong Gold Antifade reagent (Invitrogen P36930) and imaged on a Panoramic Scan II (3DHistech) with a 20x objective.
PET imaging studies with [18F]ACI-19278 and [18F]ACI-19626 in non-human primates
A brain-focused PET imaging study was performed in a wild-type female rhesus monkey (Macaca mulatta) to assess the brain uptake, distribution, and washout of [18F]ACI-19278 and [18F]ACI-19626. This study was conducted in full compliance with Yale University’s Institutional Animal Care and Use Committee (IACUC) policies and procedures, which follow the recommendations of The Guide for the Care and Use of Laboratory Animals (“The Guide,” Institute of Laboratory Animal Resources, National Academy Press, Washington, D.C., 8th Edition, 2011). Yale’s animal program is accredited by AAALAC International and has an Animal Welfare Assurance statement from the National Institutes of Health (NIH) Office of Laboratory Animal Welfare (OLAW Assurance). Veterinary medicine for all research animals is provided by a board-certified veterinarian. To ensure compliance with all regulations, the research protocol governing all animal procedures was reviewed and approved by the IACUC before any investigational work began (protocol number 2021–11037 approved by Yale University’s Institutional Animal Care and Use Committee). Animals were housed in stainless-steel condominium-style enclosures cleaned daily. Each primate had access to an upper and lower level of a condominium unit measuring 32 × 36 x 83 inches with no more than two animals housed in double that space. Animal housing rooms were maintained at 22 °C ± 2 and relative humidity at 50% ± 10% with 12-hour alternating light and dark cycles. Food, Certified Primate Diet (LabDiet® 5048), Teklad 15% Monkey Diet (Envigo 8714), and Teklad Hi-Fiber Primate Diet (Envigo 7195), in addition to treats, was provided by YARC husbandry staff based on feeding instructions determined by the veterinarians. A variety of enrichment devices (e.g., foraging boards, challenger balls, puzzle feeders, etc.) were provided. City water was provided ad libitum via an automatic watering system. [18F]ACI-19278 or [18F]ACI-19626 was administered intravenously as a bolus over 3 min using a syringe pump. The injected dose was 4.2 mCi (156 MBq) or 5.3 mCi (196 MBq), respectively. Brain emission data were collected post-injection of [18F]ACI-19278 or [18F]ACI-19626 over 180 min in a microPET Focus-220 PET scanner focused over the head. Images were generated as dynamic scans. A single 3.0 T T1 MRI of the brain was acquired on a separate day for anatomical reference and image analysis. Volumes of interest (VOI) were defined by a brain atlas associated with the brain template62. The atlas was applied to each dynamic image to extract the average activity concentration (kBq/cc) within each VOI and generate time activity curves (TAC) representing the regional brain activity concentration over time. TACs were expressed in standardized uptake value (SUV) units (g/mL) by normalizing by the weight of the animal and the injected dose. Two intravenous lines were placed for the administration of fluids and radiotracer, and for the collection of blood samples. Arterial samples were collected post-injection to assess metabolism (parent fraction), blood activity, and for the determination of the input function (plasma activity x parent fraction) used for tracer kinetic modeling. Venous samples were collected following [18F]ACI-19626 injection due to failure to establish an arterial line. Metabolite analysis, to measure parent fraction, was performed using an HPLC system. For [18F]ACI-19626, metabolism could not be assessed at 60 min or later.
Statistics & Reproducibility
One-way ANOVA with Dunnett’s multiple comparisons test was used for group comparisons. Statistical method and experimental reproducibility are detailed in the figure legends. All analyses were performed using the GrpahPad Prism software (v 10.5.0).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
Work at AC Immune was partially supported by the Michael J Fox Foundation (MJFF-022232). Work at the University of Zurich, at the International Centre for Genetic Engineering and Biotechnology in Trieste, and at the Department of Neurology, Erasmus University Medical Centre, Rotterdam was supported by JPND (JPND2020-568-078). Human tissue was kindly provided by the Netherlands Brain Bank (NBB), the Queen Square Brain Bank, the University of Kentucky – Alzheimer’s Disease Center (UK ADC NIA P30-AG0-28383), and Barrow Neurological Institute. The Queen Square Brain Bank is supported by the Reta Lila Weston Institute of Neurological Studies, UCL Queen Square Institute of Neurology. We thank Prof. Tammaryn Lashley at Queen Square Brain Bank for Neurological Disorders, Prof. William Seeley at Neurodegenerative Disease Brain Bank UCSF (funding support from NIH grants P01AG019724 and P50AG023501, the Consortium for Frontotemporal Dementia Research, and the Tau Consortium), and Prof. Robert Bowser and Target ALS for guidance in tissue selection. We also thank Dr. Stella Lombardi at AC Immune for advice and guidance in radiobinding experiments and off-target binding evaluation, respectively, Dr. Jerome Molette for advice and guidance in tracer development, as well as Mathieu Clavel for his contribution in autoradiography studies, and Dr. Patrick Rodriguez for providing brain tissues from cases with tauopathies. Finally, we greatly thank Dr. Somanath Jagannath at the University of Zurich for fLCD fibril preparation and Dr. Patrizia Longone for data discussion.
Author contributions
E.V., E.C., N.D., T.J., C.S., M.P., S.C., E.B., P.T.N., H.K., and T.S. contributed to the design of experiments. E.C., D.C., T.M., J.K., M.R., M.Y.R., A.M.S., C.D., M.R., C.S., S.C., and C.C. contributed to the design and implementation of experiments and to the analysis of the results. N.D. and H.K. contributed to the invention of ACI-19278 and ACI-19626. P.T.N., R.L.C., and H.S. contributed to the selection of patient-derived material. E.V., E.C., T.J., R.L.C., T.A., H.K., F.C., P.T.N., H.S., E.B., M.P., M.D,. and C.C. contributed to the interpretation of experimental findings. E.V., M.K.V., F.C., A.P., and T.S. drafted the manuscript. All authors revised the manuscript.
Peer review
Peer review information
Nature Communications thanks Robert Mach and the other anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
The data that support the findings of this study are provided in the source data file. Additional information is available from the corresponding author upon reasonable request. Data are located in controlled access data storage at AC Immune. Source data are provided with this paper.
Competing interests
EV is a consultant of AC Immune SA. EC, ND, DC, TM, JKB, MR, AMS, TJA, CD, MYR, HK, FCA, RLC, TA, AP, MKV and TS are or were employees of AC Immune SA at the time of their contribution to this manuscript. The remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-025-64540-6.
References
- 1.Neumann, M. et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science314, 130–133 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Arai, T. et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys. Res. Commun.351, 602–611 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Nelson, P. T. et al. Limbic-predominant age-related TDP-43 encephalopathy (LATE): consensus working group report. Brain142, 1503–1527 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uryu, K. et al. Concomitant TAR-DNA-binding protein 43 pathology is present in Alzheimer disease and corticobasal degeneration but not in other tauopathies. J. Neuropathol. Exp. Neurol.67, 555–564 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arai, T. et al. Phosphorylated TDP-43 in Alzheimer’s disease and dementia with Lewy bodies. Acta Neuropathol.117, 125–136 (2009). [DOI] [PubMed] [Google Scholar]
- 6.McKee, A. C. et al. TDP-43 proteinopathy and motor neuron disease in chronic traumatic encephalopathy. J. Neuropathol. Exp. Neurol.69, 918–929 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prasad, A., Bharathi, V., Sivalingam, V., Girdhar, A. & Patel, B. K. Molecular Mechanisms of TDP-43 Misfolding and Pathology in Amyotrophic Lateral Sclerosis. Front. Mol. Neurosci.12, 25 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barmada, S. J. & Finkbeiner, S. Pathogenic TARDBP mutations in amyotrophic lateral sclerosis and frontotemporal dementia: disease-associated pathways. Rev. Neurosci.21, 251–272 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Mackenzie, I. R. & Neumann, M. Molecular neuropathology of frontotemporal dementia: insights into disease mechanisms from postmortem studies. J. Neurochem.138, 54–70 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Cairns, N. J. et al. Neuropathologic diagnostic and nosologic criteria for frontotemporal lobar degeneration: consensus of the Consortium for Frontotemporal Lobar Degeneration. Acta Neuropathol.114, 5–22 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grossman, M. et al. Frontotemporal lobar degeneration. Nat. Rev. Dis. Prim.9, 40 (2023). [DOI] [PubMed] [Google Scholar]
- 12.Kawakami, I., Arai, T. & Hasegawa, M. The basis of clinicopathological heterogeneity in TDP-43 proteinopathy. Acta Neuropathol.138, 751–770 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chio, A. et al. Prognostic factors in ALS: A critical review. Amyotroph. Lateral Scler.10, 310–323 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brettschneider, J. et al. Stages of pTDP-43 pathology in amyotrophic lateral sclerosis. Ann. Neurol.74, 20–38 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ringholz, G. M. et al. Prevalence and patterns of cognitive impairment in sporadic ALS. Neurology65, 586–590 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Burrell, J. R., Kiernan, M. C., Vucic, S. & Hodges, J. R. Motor neuron dysfunction in frontotemporal dementia. Brain134, 2582–2594 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Nelson, P. T. LATE neuropathologic changes with little or no Alzheimer disease is common and is associated with cognitive impairment but not frontotemporal dementia. J. Neuropathol. Exp. Neurol.80, 649–651 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nag, S. & Schneider, J. A. Limbic-predominant age-related TDP43 encephalopathy (LATE) neuropathological change in neurodegenerative diseases. Nat. Rev. Neurol.19, 525–541 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nelson, P. T. et al. Frequency of LATE neuropathologic change across the spectrum of Alzheimer’s disease neuropathology: combined data from 13 community-based or population-based autopsy cohorts. Acta Neuropathol.144, 27–44 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corriveau-Lecavalier, N. et al. Clinical criteria for a limbic-predominant amnestic neurodegenerative syndrome. Brain Commun.6, fcae183 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolk, D. A. et al. Clinical criteria for limbic-predominant age-related TDP-43 encephalopathy. Alzheimers Dement.21, e14202 (2025). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mackenzie, I. R. et al. A harmonized classification system for FTLD-TDP pathology. Acta Neuropathol.122, 111–113 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neumann, M., Lee, E. B. & Mackenzie, I. R. Frontotemporal Lobar Degeneration TDP-43-Immunoreactive Pathological Subtypes: Clinical and Mechanistic Significance. Adv. Exp. Med. Biol.1281, 201–217 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neumann, M. et al. Limbic-predominant age-related TDP-43 proteinopathy (LATE-NC) is associated with abundant TMEM106B pathology. Acta Neuropathol.146, 163–166 (2023). [DOI] [PubMed] [Google Scholar]
- 25.Arseni, D. et al. TDP-43 forms amyloid filaments with a distinct fold in type A FTLD-TDP. Nature620, 898–903 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arseni, D. et al. Structure of pathological TDP-43 filaments from ALS with FTLD. Nature601, 139–143 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arseni, D. et al. Heteromeric amyloid filaments of ANXA11 and TDP-43 in FTLD-TDP Type C. Nature, 10.1038/s41586-024-08024-5 (2024). [DOI] [PMC free article] [PubMed]
- 28.Porta, S. et al. Distinct brain-derived TDP-43 strains from FTLD-TDP subtypes induce diverse morphological TDP-43 aggregates and spreading patterns in vitro and in vivo. Neuropathol. Appl. Neurobiol.47, 1033–1049 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laferriere, F. et al. TDP-43 extracted from frontotemporal lobar degeneration subject brains displays distinct aggregate assemblies and neurotoxic effects reflecting disease progression rates. Nat. Neurosci.22, 65–77 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Rabinovici, G. D. & Jagust, W. J. Amyloid imaging in aging and dementia: testing the amyloid hypothesis in vivo. Behav. Neurol.21, 117–128 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chapleau, M., Iaccarino, L., Soleimani-Meigooni, D. & Rabinovici, G. D. The role of amyloid PET in imaging neurodegenerative disorders: a review. J. Nucl. Med. 63, 13S–19S (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leuzy, A. et al. Tau PET imaging in neurodegenerative tauopathies-still a challenge. Mol. Psychiatry24, 1112–1134 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Groot, C., Villeneuve, S., Smith, R., Hansson, O. & Ossenkoppele, R. Tau PET Imaging in Neurodegenerative Disorders. J. Nucl. Med.63, 20S–26S (2022). [DOI] [PubMed] [Google Scholar]
- 34.Smith, R. et al. The alpha-synuclein PET tracer [18F] ACI-12589 distinguishes multiple system atrophy from other neurodegenerative diseases. Nat. Commun.14, 6750 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Dyck, C. H. et al. Lecanemab in early Alzheimer’s disease. N. Engl. J. Med. 388, 9–21 (2023). [DOI] [PubMed] [Google Scholar]
- 36.Yadollahikhales, G. & Rojas, J. C. Anti-Amyloid Immunotherapies for Alzheimer’s disease: a 2023 clinical update. Neurotherapeutics20, 914–931 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brendel, M. et al. Assessment of 18F-PI-2620 as a biomarker in progressive Supranuclear Palsy. JAMA Neurol.77, 1408–1419 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ossenkoppele, R. et al. Discriminative accuracy of [18F]flortaucipir Positron Emission Tomography for Alzheimer disease vs other neurodegenerative disorders. Jama320, 1151–1162 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee, E. B. et al. Expansion of the classification of FTLD-TDP: distinct pathology associated with rapidly progressive frontotemporal degeneration. Acta Neuropathol.134, 65–78 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scialo, C. et al. Seeded aggregation of TDP-43 induces its loss of function and reveals early pathological signatures. Neuron, 10.1016/j.neuron.2025.03.008 (2025). [DOI] [PubMed]
- 41.Romano, V., Quadri, Z., Baralle, F. E. & Buratti, E. The structural integrity of TDP-43 N-terminus is required for efficient aggregate entrapment and consequent loss of protein function. Prion9, 1–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kroth, H. et al. Discovery and preclinical characterization of [(18)F]PI-2620, a next-generation tau PET tracer for the assessment of tau pathology in Alzheimer’s disease and other tauopathies. Eur. J. Nucl. Med. Mol. Imaging46, 2178–2189 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harada, R. et al. Imaging of reactive Astrogliosis by Positron Emission Tomography. Front. Neurosci.16, 807435 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cordts, I. et al. TDP-43 Proteinopathy specific biomarker development. Cells12, 10.3390/cells12040597 (2023). [DOI] [PMC free article] [PubMed]
- 45.Lopez-Carbonero, J. I. et al. In vivo diagnosis of TDP-43 proteinopathies: in search of biomarkers of clinical use. Transl. Neurodegener.13, 29 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCluskey, S. P., Plisson, C., Rabiner, E. A. & Howes, O. Advances in CNS PET: the state-of-the-art for new imaging targets for pathophysiology and drug development. Eur. J. Nucl. Med. Mol. Imaging47, 451–489 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pike, V. W. PET radiotracers: crossing the blood-brain barrier and surviving metabolism. Trends Pharm. Sci.30, 431–440 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spires-Jones, T. L., Attems, J. & Thal, D. R. Interactions of pathological proteins in neurodegenerative diseases. Acta Neuropathol.134, 187–205 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kulichikhin, K. Y., Malikova, O. A., Zobnina, A. E., Zalutskaya, N. M. & Rubel, A. A. Interaction of Proteins Involved in Neuronal Proteinopathies. Life13, 10.3390/life13101954 (2023). [DOI] [PMC free article] [PubMed]
- 50.Nelson, P. T., Schneider, J. A., Jicha, G. A., Duong, M. T. & Wolk, D. A. When Alzheimer’s is LATE: Why does it matter?. Ann. Neurol.94, 211–222 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Young, A. L. et al. Data-driven neuropathological staging and subtyping of TDP-43 proteinopathies. medRxiv, 10.1101/2023.01.31.23285242 (2023). [DOI] [PMC free article] [PubMed]
- 52.Yousef, A. et al. Neuron loss and degeneration in the progression of TDP-43 in frontotemporal lobar degeneration. Acta Neuropathol. Commun.5, 68 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kawles, A. et al. Cortical and subcortical pathological burden and neuronal loss in an autopsy series of FTLD-TDP-type C. Brain145, 1069–1078 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takeuchi, R. et al. Heterogeneity of cerebral TDP-43 pathology in sporadic amyotrophic lateral sclerosis: Evidence for clinico-pathologic subtypes. Acta Neuropathol. Commun.4, 61 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Robinson, J. L. et al. Annexin A11 aggregation in FTLD-TDP type C and related neurodegenerative disease proteinopathies. Acta Neuropathol.147, 104 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Afroz, T. et al. Immunotherapy targeting the C-terminal domain of TDP-43 decreases neuropathology and confers neuroprotection in mouse models of ALS/FTD. Neurobiol. Dis.179, 106050 (2023). [DOI] [PubMed] [Google Scholar]
- 57.Bagchi, D. P. et al. Binding of the radioligand SIL23 to alpha-synuclein fibrils in Parkinson disease brain tissue establishes feasibility and screening approaches for developing a Parkinson disease imaging agent. PLoS One8, e55031 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jicha, G. A. et al. A conformation- and phosphorylation-dependent antibody recognizing the paired helical filaments of Alzheimer’s disease. J. Neurochem.69, 2087–2095 (1997). [DOI] [PubMed] [Google Scholar]
- 59.Rostagno, A. & Ghiso, J. Isolation and biochemical characterization of amyloid plaques and paired helical filaments. Curr. Protoc. Cell Biol. Chapter 3, Unit 3 33 33 33 31-33, 10.1002/0471143030.cb0333s44 (2009). [DOI] [PMC free article] [PubMed]
- 60.Spillantini, M. G., Crowther, R. A., Jakes, R., Hasegawa, M. & Goedert, M. alpha-Synuclein in filamentous inclusions of Lewy bodies from Parkinson’s disease and dementia with Lewy bodies. Proc. Natl. Acad. Sci. USA95, 6469–6473 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marquie, M. et al. Validating novel tau positron emission tomography tracer [F-18]-AV-1451 (T807) on postmortem brain tissue. Ann. Neurol.78, 787–800 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rohlfing, T. et al. The INIA19 Template and NeuroMaps Atlas for primate brain image parcellation and spatial normalization. Front Neuroinform.6, 27 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are provided in the source data file. Additional information is available from the corresponding author upon reasonable request. Data are located in controlled access data storage at AC Immune. Source data are provided with this paper.