Abstract
To survive ionic, pH and pheromone stress, the yeast Saccharomyces cerevisiae activates signaling through the Ca2+-activated phosphatase calcineurin to the transcription factor Crz1p/Tcn1p. We show that the overexpression of SKN7, a response-regulator transcription factor, activates transcription from a calcineurin/Crz1p-dependent response element (CDRE). Ca2+-induced, calcineurin/Crz1p-dependent activation of several genes is reduced in skn7 mutants. Skn7p modulates CDRE-dependent transcription by affecting Crz1p protein levels. Specifically, the rate of Crz1p turnover is increased in skn7 mutants. Calcineurin, but not its phosphatase activity, is required for Skn7p-mediated Crz1p stabilization. Skn7p binds to both calcineurin and Crz1p in vitro, and we suggest that this interaction is required for Skn7p regulation of Crz1p. The DNA-binding and internal coiled-coil domains, but not the response- regulator phosphorylation of Skn7p, are necessary for Crz1p-dependent transcriptional activation and Crz1p stabilization by Skn7 in vivo. The DNA-binding domain of Skn7p is also required for binding to Crz1p and calcineurin in vitro. Thus, we propose that Skn7p protects Crz1p from degradation by binding to it and calcineurin through its DNA-binding domain.
Keywords: calcineurin/Crz1/response regulator/Saccharomyces cerevisiae/Skn7
Introduction
Many different physiological conditions elicit changes in intracellular Ca2+ concentration. Calcineurin (PP2B), a Ca2+- and calmodulin-dependent Ser/Thr phosphatase, is conserved from yeast to higher eukaryotes. Calcineurin regulates a large variety of processes in mammalian cells including cardiac hypertrophy (Molkentin et al., 1998), cardiac valve and skeletal muscle development (Chin et al., 1998; de la Pompa et al., 1998; Ranger et al., 1998), and neutrophil chemotaxis (Hendey and Maxfield, 1993; Lawson and Maxfield, 1995). Calcineurin-mediated regulation of the immune system has been particularly well studied in part by the use of FK506 and cyclosporin A, immunosuppressant drugs that inhibit the catalytic activity of calcineurin (J.Liu et al., 1991). Specifically, calcineurin regulates T-cell activation by dephosphorylating the transcription factor NF-AT (Clipstone and Crabtree, 1992; O’Keefe et al., 1992), which permits it to enter the nucleus and initiate transcription (Jain et al., 1993; Northrop et al., 1993).
In the budding yeast Saccharomyces cerevisiae, calcineurin allows the cell to adapt to environmental stress. Mutants lacking the genes encoding either the calcineurin catalytic subunits CNA1 and CNA2 (Cyert et al., 1991; Y.Liu et al., 1991), or the regulatory subunit CNB1 (Kuno et al., 1991; Cyert and Thorner, 1992) are viable under normal growth conditions, but die in the presence of high concentrations of different ions including Mn2+, Na+, Li+ and OH– (Nakamura et al., 1993; Mendoza et al., 1994; Farcasanu et al., 1995; Pozos et al., 1996), as well as in the presence of the cationic aminoglycoside hygromycin B (Withee et al., 1998). Calcineurin mutants also lose viability during prolonged exposure to mating pheromone (Moser et al., 1996; Withee et al., 1997). These calcineurin mutant phenotypes are rescued by overexpression of the zinc finger transcription factor Crz1p/Tcn1p (Matheos et al., 1997; Stathopoulos and Cyert, 1997). Calcineurin directly dephosphorylates Crz1p, which then translocates to the nucleus after Ca2+ stimulation. Crz1p binds directly to a 24 bp DNA element from the β1-3 glucan synthase FKS2 promoter, the calcineurin-dependent response element (CDRE) (Stathopoulos and Cyert, 1997; Stathopoulos-Gerontides et al., 1999). In addition to FKS2, calcineurin and Crz1p (Matheos et al., 1997; Stathopoulos and Cyert, 1997) activate the transcription of numerous genes including PMR2, a Na+ ATPase (Rudolph et al., 1989; Haro et al., 1991), and PMC1 and PMR1, both Ca2+ ATPases (Rudolph et al., 1989; Cunningham and Fink, 1994, 1996; Mendoza et al., 1994; Mazur et al., 1995). To identify other molecules that might regulate calcineurin signaling, we screened for genes which, when overexpressed, activate calcineurin/Crz1p-dependent transcription. In this way we isolated Skn7p, a response-regulator transcription factor.
Skn7p shows homology to bacterial response-regulator proteins (Brown et al., 1994; Morgan et al., 1995), which function in two-component systems with histidine kinases (reviewed in Parkinson, 1993; Loomis et al., 1998). In a typical prokaryotic two-component signaling pathway, a sensor histidine kinase, located at the cell membrane, auto-phosphorylates on a histidine residue upon stimulation. This phosphate is then transferred to an aspartyl residue on the receiver protein, which often, but not always, contains a DNA-binding domain and initiates transcription. Skn7p has been shown to participate in yeast two-component signaling through its canonical response-regulator aspartate residue (Asp427) with the histidine kinase Sln1p and the small phospho-transfer protein Ypd1p (Ketela et al., 1998; Li et al., 1998). Skn7p also contains a coiled-coil domain, which is necessary for several protein–protein interactions (Alberts et al., 1998; Bouquin et al., 1999).
Skn7p functions in a multiplicity of pathways, including cell wall stress, oxidative stress, heat stress and the cell cycle, although its role in this variety of processes is not completely understood. First, Skn7p helps the cell to respond to cell wall stress. Skn7p has genetic and biochemical interactions with cell wall synthesis genes (Brown et al., 1993) and members of a PKC-mediated cell-integrity pathway (Levin and Bartlett-Heubusch, 1992; Alberts et al., 1998), and transcribes at least one cell wall synthesis gene, OCH1 (S.Li and J.S.Fassler, in preparation). Secondly, Skn7p also plays a role in adaptation to oxidative stress; the mutants are sensitive to hydrogen peroxide (Krems et al., 1996), and Skn7p induces the transcription of many genes upon exposure to oxidative stress (Kuge and Jones, 1994; Morgan et al., 1997; Lee et al., 1999). Thirdly, Skn7p responds to heat stress. skn7 mutants are sensitive to acute heat stress and Skn7p contributes to the transcription of several heat shock proteins (Raitt et al., 2000). Finally, in the cell cycle, Skn7p has also been shown to activate G1 transcriptional events in the absence of normal G1–S regulatory transcription factors (Morgan et al., 1995; Bouquin et al., 1999).
Thus, Skn7p plays a part in numerous biological stress responses. In this paper we further elucidate a role for Skn7p in stress-activated signal transduction. We present biochemical and genetic evidence that Skn7p affects calcineurin-mediated signaling and physiology. Specific ally, Skn7p modulates calcineurin-dependent transcriptional output in vivo, by affecting the rate of Crz1p turnover. Furthermore, we show that Skn7p binds to both Crz1p and calcineurin in vitro, suggesting that Skn7p modulates Crz1p stability through direct protein–protein interactions. Thus, these studies identify a novel activity of Skn7p as a regulator of protein stability and establish a new mechanism for the regulation of calcineurin/Crz1p-dependent transcription. We also demonstrate for the first time a phosphatase-independent role for the calcineurin protein.
Results
Identification of SKN7 as a multicopy enhancer of calcineurin- and Crz1p-dependent transcription
A wild-type strain containing a previously characterized CDRE fused to the lacZ gene (Stathopoulos and Cyert, 1997) (KWY242) was transformed with two multicopy genomic libraries to identify genes whose overexpression caused increased calcineurin-dependent transcription (see Materials and methods). To determine whether the reporter gene activity resulted specifically from calcineurin- and Crz1p-dependent transcription, the plasmids were re-transformed into strains with a mutant version of the CDRE as well as strains that lacked either the calcineurin regulatory subunit, Cnb1p, or the transcription factor Crz1p. For each plasmid isolated, the open reading frame (ORF) responsible for reporter gene activation was identified by subcloning.
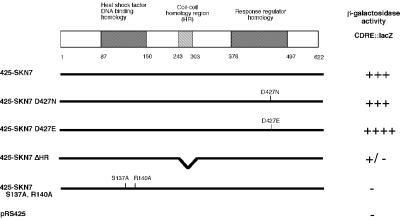
Five plasmids contained SKN7. Skn7p is a previously characterized response-regulator protein with a DNA-binding domain homologous to heat shock factor (HSF) (Brown et al., 1993) (Figure 1). Multicopy SKN7 did not activate the CDRE::lacZ reporter gene in calcineurin (cna1Δcna2Δ or cnb1Δ) or crz1Δ mutant strains, indicating that the Skn7p-mediated increase in CDRE::lacZ activity reflected signaling through the calcineurin pathway (Table I). Only one other gene, SWI5, was identified multiple times. SWI5, however, activated the CDRE::lacZ reporter gene independently of both calcineurin and Crz1p (Table I). Swi5p, like Crz1p, is a C2H2 zinc finger protein whose DNA-binding domain is almost identical to that of Crz1p. Thus, when present at high levels, Swi5p likely binds to the CDRE and activates transcription. Similarly, when overexpressed, Swi5p activates transcription of genes regulated by Ace2p, another transcription factor with a DNA-binding domain highly related to that of Swi5p (Dohrmann et al., 1992). Swi5p was not characterized further. Additional plasmids that were only isolated once will not be discussed further in this paper. These plasmids all had similar phenotypes in that they caused an increase in CDRE::lacZ activity in the wild-type strain, but not in the cnb1Δ or crz1Δ strains, or in the wild type + FK520. None of the plasmids identified in the screen activated the mutant CDRE::lacZ reporter gene. Because of its frequency of identification, Skn7p was further characterized by examining the effects of the skn7 mutation on calcineurin-dependent transcription.
Fig. 1. Schematic diagram of Skn7p with alleles used to identify regions of Skn7p important for CDRE::lacZ activation. The HR region (see text) and the DNA-binding domain are essential for activation of the CDRE::lacZ by the overexpression of Skn7p. Multicopy plasmids were transformed into KWY242, containing the 2×CDRE::lacZ reporter gene. β-galactosidase activity was assessed by plate assay. All plasmids are derivatives of pRS425. 425-SKN7ΔHR is pKW47; 425-SKN7 S137A, R140A is pKW37. Western blot analysis revealed approximately equal levels of expression from these plasmids (data not shown).
Table I. Multicopy SKN7 activates the CDRE::lacZ reporter gene.
| CDRE::lacZ β-galactosidase activitya |
||||
|---|---|---|---|---|
| Wild typeb | crz1Δc | cnb1Δd | Wild typee + FK520 | |
| Vectorf | – | – | – | – |
| 2µ CRZ1 | ++++ | ++++ | ++++ | ++++ |
| 2µ SKN7 | +++ | – | – | +++ |
| 2µ SWI5 | +++ | +++ | +++ | +++ |
aβ-galactosidase activity was determined by plate assay.
bKWY242.
cASY834.
dASY461 and KWY246.
eASY459 and KWY242; plates contain 2 µg/ml FK520.
fVector, YEP351; 2µ CRZ1, pAMS435; 2µ SKN7, pRS425-SKN7 and library plasmids; 2µ SWI5, M1485 (D.Stillman) and library plasmids.
None of the plasmids identified in the screen activated the mutant CDRE::lacZ reporter.
skn7Δ reduces Ca2+-, calcineurin- and Crz1p-dependent transcription
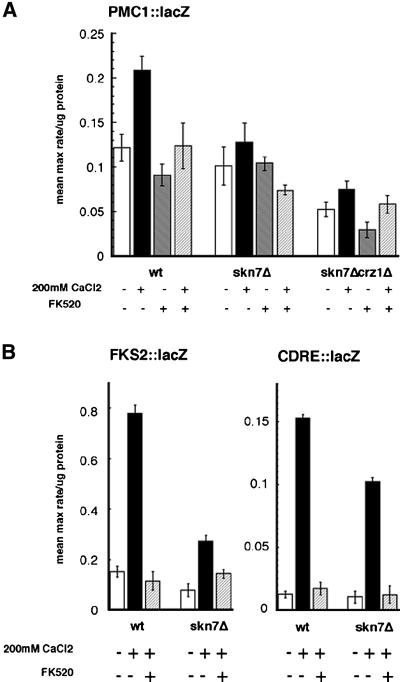
Ca2+ stimulates the expression of many genes in a calcineurin- and Crz1p-dependent fashion, including the P-type ATPases PMC1 and PMR2 (Rudolph et al., 1989; Haro et al., 1991; Cunningham and Fink, 1994, 1996), as well as the β1-3 glucan synthase FKS2 (Mazur et al., 1995; Matheos et al., 1997; Stathopoulos and Cyert, 1997). The effect of Skn7p on calcineurin signaling was measured by examining pmc1::lacZ and fks2::lacZ reporter gene activity in different strain backgrounds. FK520, a calcineurin phosphatase inhibitor (Liu et al., 1992), was used to examine the calcineurin dependence of this transcription. Skn7p was required for the full Ca2+ activation of these genes in a calcineurin- and Crz1p-dependent fashion (Figure 2). The reduced expression of pmc1::lacZ observed in the skn7Δcrz1Δ strain was equivalent to its expression in the crz1Δ strain (data not shown). Similar effects were observed with a pmr2::lacZ reporter gene (data not shown). In addition, the CDRE/Crz1p binding region of the FKS2 promoter was sufficient for this effect, as a decrease in Ca2+-activated transcription of a CDRE(24 bp)::lacZ reporter gene was seen in the skn7Δ strain (Figure 2B).
Fig. 2. skn7Δ strains can not fully activate Crz1p- and calcineurin-dependent transcription in response to Ca2+. Strains contain plasmid-based reporter genes (A) PMC1::lacZ (pAMS381), (B) FKS2::lacZ (pAMS317) and CDRE::lacZ (pAMS342) (Stathopoulos and Cyert, 1997). β-galactosidase activities are shown for wild-type (YPH499), skn7Δ (KWY266) and skn7Δcrz1Δ (KWY289) cell extracts, in either the presence or absence of 200 mM CaCl2 and 2 µg/ml FK520. For each condition, three cell extracts were assayed three times each; the SD is the error between the samples.
Skn7p and calcineurin interact genetically
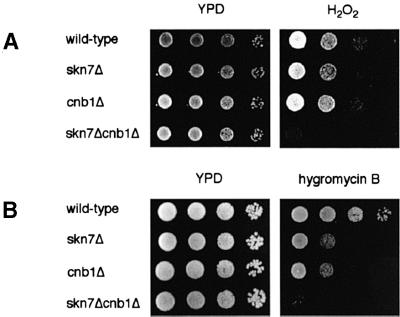
We further explored the relationship between Skn7p and calcineurin by examining the phenotypes of skn7Δcnb1Δ double mutants. Skn7p is required for the response to oxidative stress, and skn7 mutants are sensitive to several oxidizing agents including hydrogen peroxide (Krems et al., 1996) (Figure 3A). In contrast, cnb1Δ mutants exhibit no sensitivity to hydrogen peroxide (Figure 3A). However, the skn7Δcnb1Δ strain was dramatically more sensitive to hydrogen peroxide than the skn7Δ strain, suggesting that calcineurin does participate in the oxidative stress response in vivo in combination with Skn7p.
Fig. 3. Skn7p and calcineurin interact genetically. (A) Deletion of CNB1 exacerbates the hydrogen peroxide sensitivity of the skn7Δ strain. (B) skn7Δcnb1Δ mutants are more hygromycin B sensitive than skn7Δ and cnb1Δ strains. Wild-type (YPH499), skn7Δ (KWY266), cnb1Δ (DD12) and skn7Δcnb1Δ (KWY267) strains were grown to saturation, then diluted to an OD600 of 1. Ten-fold serial dilutions were spotted onto YPD plates with either no added chemicals, 1.8 mM hydrogen peroxide (A), or 70 µg/ml hygromycin B (B), and grown at 30°C for 1–3 days.
Interestingly, previous studies have established that both calcineurin and Skn7p exhibit genetic interactions with the PKC pathway, which mediates the response to cell wall stress (Heinisch et al., 1999). Calcineurin mutants are synthetically lethal with pkc1 mutants and other downstream members of the Pkc1p-regulated MAP kinase cascade (Garrett-Engele et al., 1995). In addition, overexpressing a constitutively active calcineurin allele ameliorates the pkc1Δ growth defect on low osmolarity media (Garrett-Engele et al., 1995). skn7 mutations are similarly synthetically lethal with pkc1Δ, and overexpressing Skn7p rescues a pkc1Δ cell lysis defect (Brown et al., 1994). To identify possible interactions between calcineurin and Skn7p with respect to cell wall stress, we tested their effects on hygromycin B sensitivity. Several mutants that have defects in cell wall synthesis or structure exhibit sensitivity to this drug (Dean, 1995; Lussier et al., 1997), and calcineurin and skn7 mutants are both sensitive to hygromycin B (Withee et al., 1998; S.Li and J.S.Fassler, in preparation) (Figure 3B). Notably, the skn7Δcnb1Δ strain is significantly more sensitive than either single mutant (Figure 3B). Thus, Skn7p and calcineurin act synergistically to provide hygromycin B resistance. Together, these phenotypic observations indicate that calcineurin and Skn7p participate in similar physiological functions and work together to regulate the yeast response to several different types of stress.
Skn7p stabilizes Crz1p
These genetic observations, as well as the effect of Skn7p on calcineurin-dependent transcription, suggested that Skn7p and calcineurin could be acting together in a signal transduction pathway. Several approaches were used to determine the mechanism of the Skn7p effect on calcineurin signaling. First, electrophoretic mobility shift assays (EMSA) were performed to determine whether Skn7p could bind to the CDRE DNA element. As previously reported (Stathopoulos and Cyert, 1997), a DNA–protein complex containing Crz1p was observed when extracts were incubated with oligonucleotides containing the CDRE sequence. However, the mobility of this complex was identical in extracts of wild-type and skn7Δ cells and was unaffected by addition of Skn7p-specific antiserum (data not shown). Thus, Skn7p does not appear to bind to the CDRE. Secondly, green fluorescent protein (GFP) microscopy was also used to test the effect of Skn7p on GFP–Crz1p localization. As reported previously, adding Ca2+ causes GFP–Crz1p to translocate to the nucleus in a calcineurin-dependent manner (Stathopoulos-Gerontides, 1999). The deletion or overexpression of Skn7p had no discernible effect on Crz1 protein localization, either in the absence of presence of Ca2+ (data not shown).
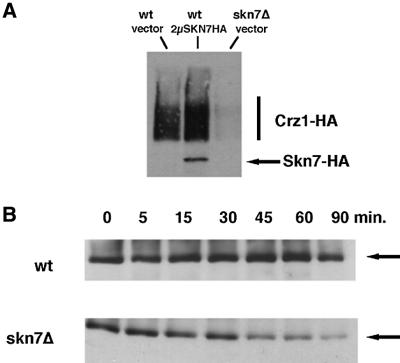
Finally, we did observe an effect of Skn7p on Crz1 protein levels. In whole-cell extracts, multicopy SKN7 increased and skn7Δ decreased Crz1 protein levels, respectively (Figure 4A). In contrast, calcineurin protein levels were unaffected by Skn7p (data not shown). Surprisingly, Skn7p had no effect on CRZ1 mRNA levels (data not shown), thus we investigated the effect of Skn7p on Crz1 protein stability. Crz1 protein levels were examined at a series of time intervals after incubation of wild-type and skn7Δ cells with cycloheximide. Crz1p in the wild-type strain was stable even 90 min after cycloheximide treatment, but Crz1 protein declined more rapidly in the skn7Δ strain, and exhibited significantly reduced levels after 45 min (Figure 4B).
Fig. 4. Skn7p stabilizes the Crz1 protein. (A) The overexpression of Skn7p results in more Crz1 protein; the deletion of Skn7p results in less Crz1 protein. Whole-cell extracts from strains either overexpressing or deleting Skn7 [YPH499 with YEP352, YPH499 with YEP352-SKN7-HA, skn7Δ::TRP1 (KWY266)] were subjected to SDS–PAGE and anti-HA western analysis. All strains contain pRS315-CRZ1-HA (pAMS450). These results were reproduced using anti-Crz1p antibodies to examine endogenous Crz1p levels (data not shown). (B) Crz1p is less stable in skn7Δ. Samples were taken at the indicated times after the addition of 100 µg/ml cycloheximide from both wild type (YPH499) and skn7Δ (KWY266), and subjected to anti-Crz1p western analysis.
Calcineurin, but not its phosphatase activity, is required for Skn7p-mediated Crz1p stabilization
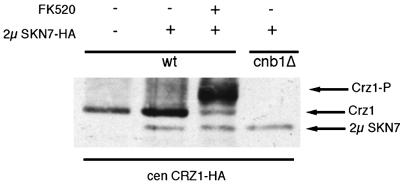
The finding that Skn7p regulates Crz1p at the level of protein turnover led us to analyze further potential regulators of Skn7p-mediated Crz1p stability. As phosphorylation often affects transcription factor protein stability (reviewed in Hochstrasser and Kornitzer, 1998), we analyzed the effect of calcineurin on Skn7p-mediated Crz1p stabilization. The overexpression of Skn7p caused an increase in Crz1 protein level in a wild-type background. However, when this experiment was performed in a cnb1Δ strain, the Crz1p levels not only failed to increase, but actually decreased (Figure 5). This observation is consistent with the fact that multicopy SKN7 failed to activate the CDRE::lacZ in a calcineurin mutant strain (Table I). To understand better the role of calcineurin in Skn7p-mediated Crz1p stability, the effects of a calcineurin inhibitor were analyzed. FK520 is a pharmaceutical closely related to FK506 that inhibits the phosphatase activity of calcineurin (Liu et al., 1992). In all previously reported cases, the addition of FK506 or FK520 causes wild-type cells to acquire calcineurin mutant phenotypes (Foor et al., 1992; Breuder et al., 1994). However, in this case, multicopy SKN7 did activate the expression of CDRE::lacZ in strains grown with FK520, in contrast to its failure to activate the same reporter construct in the cnb1Δ mutant strain (Table I). This difference was not caused by a dosage- or drug-specific effect as either increasing the concentration of FK520 or using cyclosporin A, another calcineurin inhibitor, produced similar results (data not shown). In addition, these treatments were effective in reducing the activation of CDRE::lacZ by other plasmids isolated from this genetic screen (data not shown). Furthermore, extracts of FK520-treated cells expressing multicopy SKN7 displayed increased Crz1 protein levels (Figure 5), whereas adding FK520 in the absence of multicopy SKN7 did not cause increased Crz1p levels (data not shown). These findings are consistent with the observation that multicopy SKN7 increased CDRE::lacZ activity in the presence of FK520 (Table I). Notably, Crz1p displayed the reduced mobility that represents its phosphorylated form in extracts of FK520-treated cells (Figure 5) (Stathopoulos-Gerontides et al., 1999). This confirms that FK520 treatment effectively inhibits calcineurin phosphatase activity. Thus, Skn7p stabilized Crz1p in the absence of calcineurin phosphatase activity caused by the inhibitor FK520, but not in the absence of the calcineurin B subunit gene product (cnblΔ). This suggests that the physical presence of the calcineurin polypeptides, as opposed to their phosphatase activity, is essential for Skn7p-mediated Crz1p stabilization.
Fig. 5. Calcineurin, but not its phosphatase activity, is essential for Skn7p to stabilize Crz1p. Anti-HA western blot analysis of either wild-type (YPH499) or cnb1Δ (DD12) strains containing the plasmids pAMS450, YEP352 or YEP352-SKN7-HA, with or without FK520 (2 µg/ml).
Skn7p, Crz1p and calcineurin interact physically
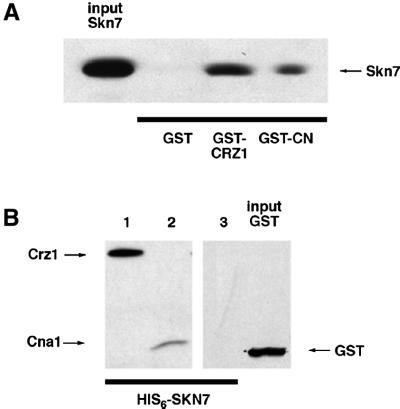
The fact that the presence of calcineurin was required for maintenance of Crz1 protein levels suggested that protein– protein interactions might be required for Skn7p- dependent Crz1p stabilization. Several experiments were performed to test for interactions between calcineurin, Crz1p and Skn7p. We were unable to detect such interactions by co-immunoprecipitation, but Skn7p from yeast extracts bound weakly to recombinant glutathione S-transferase (GST)–calcineurin immobilized on glutathione-Sepharose beads (data not shown). To develop a more sensitive assay for Skn7p, Crz1p and calcineurin protein–protein interactions, two additional experiments were performed. First, to determine whether Skn7p binds to Crz1p and/or calcineurin, recombinant GST–Crz1p, GST–calcineurin or GST purified from Escherichia coli was immobilized on glutathione-Sepharose beads and incubated with recombinant His6-tagged Skn7p (see Materials and methods). Western analysis indicated that Crz1p and calcineurin, but not GST, bound to Skn7p (Figure 6A). Secondly, in the reverse experiment, His6-tagged Skn7p was immobilized on Ni+-NTA-agarose beads, and incubated with either purified GST, Crz1p or calcineurin. Immunoblotting revealed that again calcineurin and Crz1p, but not the non-specific protein GST, interact with Skn7p (Figure 6B). These results indicate that in vitro Skn7p forms a specific interaction with both calcineurin and Crz1p.
Fig. 6. Skn7p binds to calcineurin and Crz1p in vitro. (A) Immobilized GST–Crz1p and GST–calcineurin, but not GST, can bind Skn7p. GST, GST–Crz1p and GST–calcineurin were purified from E.coli on glutathione-Sepharose, and then incubated with recombinant His6-Skn7p. Following washing, bound proteins were eluted and fractionated by SDS–PAGE. Skn7p was detected using anti-His4 antibody. (B) Immobilized His6-Skn7p binds calcineurin and Crz1p, but not GST. His6-Skn7p was purified on Ni+-NTA resin and then incubated with recombinant GST–Crz1p (lane 1), GST–calcineurin (lane 2), or GST (lane 3). Samples were processed as in (A), except that proteins were detected using anti-GST antibody.
A functional Skn7p DNA-binding domain is required for transcriptional, stability and protein–protein interaction effects
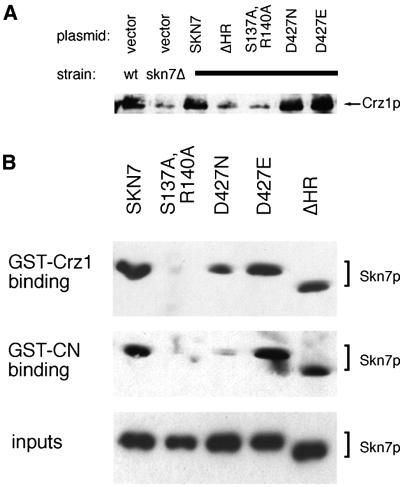
A functional Skn7p DNA-binding domain is essential for its ability to stimulate transcription. Two residues that are essential for DNA binding have been structurally determined in the DNA-binding domain of HSF (Hubl et al., 1994), which is highly homologous to the DNA-binding domain of Skn7p. Mutations in the analogous Skn7p residues (S137A, R140A) eliminate activation of the Skn7p-dependent promoter elements in OCH1 (S.Li and J.S.Fassler, in preparation). Interestingly, these two residues in the Skn7p DNA-binding domain proved essential for all aspects of Skn7p-mediated effects on calcineurin/Crz1p signaling. First, the multicopy SKN7 S137A,R140A plasmid completely failed to activate the CDRE in vivo (Figure 1). Secondly, SKN7 S137A,R140A completely failed to complement the low levels of Crz1p in the skn7Δ strain (Figure 7A). Finally, recombinant Skn7p with DNA-binding domain mutations (S137A, R140A) consistently failed to bind to either GST–Crz1p or GST–calcineurin (Figure 7B). Therefore, these residues are essential for Skn7p function in Crz1p- and calcineurin-dependent transcription, Skn7p stabilization of Crz1 protein, and Skn7p binding to calcineurin and Crz1p.
Fig. 7. The Skn7p HR and DNA-binding domain are required for Crz1p stabilization; the DNA-binding domain is essential for binding to Crz1p and calcineurin. (A) Anti-Crz1p western blot analysis of YPH499 or KWY266 with Skn7p plasmids as in Figure 1. All plasmids are derivatives of pRS425. SKN7ΔHR is pKW47; S137A,R140A is pKW37. Each Skn7p allele was expressed at comparable levels (data not shown). (B) GST–Crz1p and GST–calcineurin were purified on glutathione-Sepharose, and then incubated with 0.2 µg of His-tagged Skn7p, Skn7p S137A,R140A, Skn7p D427N, Skn7p D427E, or Skn7pΔHR (from pKW25, 39, 40, 42 and 44, respectively). The Sepharose was washed, and the bound proteins eluted and analyzed by SDS–PAGE and anti-His4 western blotting (top two panels). Purified Skn7 protein (0.1 µg) (expressed from pKW25, 39, 40, 42 and 44) was similarly run on a gel (bottom panel).
The Skn7p homology region (HR) is required for CDRE activation and Crz1p stabilization
The coiled-coil (HR) region in the center of the Skn7 protein is required for its binding to both the Mbp1p transcription factor (Bouquin et al., 1999) and to Rho1p (Alberts et al., 1998), a GTPase that activates Pkc1p as well as other targets (Nonoka et al., 1995). This part of the protein is essential for most Skn7p functions as a protein lacking this region fails to complement most skn7Δ phenotypes (Alberts et al., 1998). We consequently tested the SKN7ΔHR allele for its ability to affect CDRE::lacZ transcription, Crz1p stabilization, and binding to Crz1p and calcineurin. When multicopy SKN7ΔHR was transformed into the CDRE::lacZ-expressing strain, it failed to activate transcription (Figure 1). In addition, this allele was tested for its effects on Crz1 protein stability. SKN7ΔHR also failed to complement the low Crz1 protein levels observed in extracts of a skn7Δ strain (Figure 7A). These two observations indicated that the coiled-coil HR domain of Skn7p is essential for its ability both to activate calcineurin-dependent transcription and to stabilize Crz1p in vivo. Surprisingly, when this allele was tested for in vitro biochemical interaction with both calcineurin and Crz1p, purified recombinant Skn7ΔHRp bound as well as the wild-type Skn7p allele (Figure 7B) (see Discussion).
Skn7p response-regulator phosphorylation is not required for CDRE activation, Crz1p stabilization, or binding to Crz1p
In response-regulator proteins, a canonical aspartate residue is phosphorylated by a histidine kinase. The phosphorylation of this aspartate residue in Skn7p (Asp427) is not essential for all Skn7p functions in yeast. Several experiments were performed to determine whether Skn7p response-regulator aspartate phosphorylation is required to mediate Crz1p stabilization. First, to test the effects of this residue on CDRE::lacZ reporter gene activation, multicopy SKN7 D427N, an unphosphorylatable allele, and SKN7 D427E, an allele that mimics aspartate phosphorylation, were transformed into the KWY242 wild-type CDRE::lacZ reporter strain to compare their activities with wild-type SKN7. The activated D427E allele showed greater reporter activity than the wild-type allele. However, the D427N construct resulted in reporter gene activity indistinguishable from wild type, indicating that Asp427 is not required for increased CDRE::lacZ expression (Figure 1).
Secondly, these two alleles were tested for their effects on Crz1p stabilization. Plasmids containing either the SKN7 D427N or the SKN7 D427E allele were transformed into the skn7Δ strain. Western blot analysis was performed to determine whether either allele would complement the reduced levels of Crz1 protein displayed by the skn7 mutant. The levels of Crz1p in the strains expressing either SKN7 D427N or wild-type SKN7 were equivalent, indicating that for Crz1 protein stability, as for CDRE::lacZ reporter activation, response-regulator aspartate phosphorylation is not required (Figure 7A). Interestingly, slightly higher Crz1 protein levels were consistently observed in the D427E-expressing strain, indicating that this residue may play some functional role in Crz1p stability, although it is not required to complement the skn7Δ strain (Figure 7A).
Finally, the Skn7p D427N and D427E alleles were tested for their ability to bind in vitro to recombinant calcineurin and Crz1p. Binding of Skn7p D427E to both GST–Crz1p and GST–calcineurin was equivalent to that of wild-type Skn7p (Figure 7B). Skn7p D427N also bound to GST–Crz1p comparably to wild-type Skn7p. In contrast, Skn7p D427N showed reduced binding to GST–calcineurin (Figure 7B).
Discussion
We have demonstrated that Skn7p, a previously identified response-regulator protein, modulates Crz1p/calcineurin-dependent signaling. First, overexpression of Skn7p increases Crz1p- and calcineurin-dependent reporter gene activity, and skn7Δ strains show reduced Ca2+-, calcineurin- and Crz1p-dependent transcription of multiple genes. Secondly, overexpression of Skn7p increases Crz1 protein levels, and skn7Δ cells show greatly reduced Crz1 protein levels due to increased Crz1 protein turnover. Thirdly, the elimination of calcineurin subunits, but not the addition of a calcineurin phosphatase inhibitor, blocks both the Skn7p-mediated activation of Crz1p-dependent transcription and its effect on Crz1p stability. Finally, Skn7p binds specifically to both calcineurin and Crz1p in vitro. A functional Skn7p DNA-binding domain, but not Asp427 phosphorylation, was required for all of these functions. The Skn7p coiled-coil region was required for both Skn7p-mediated in vivo CDRE activation and Crz1p stabilization, but not for in vitro protein–protein interactions.
These observations show that Skn7p affects Crz1p-dependent transcription by modifying Crz1 protein stability. The in vitro binding of Skn7p, Crz1p and calcineurin suggests that Skn7p may modulate Crz1p stability through direct interactions with these proteins. This role for Skn7p is different from previous work on the protein in one key regard: we implicate Skn7p in the regulation of protein stability. Skn7p has three well characterized domains: a response-regulator region, which resembles two-component signaling proteins; a coiled-coil domain, which mediates protein–protein interactions; and an HSF-like DNA-binding domain, which interacts with specific promoter regions. It is particularly interesting to consider the different motifs present in Skn7p in light of this novel role for the protein.
Skn7p is a response regulator implicated in protein stability
Skn7p is the first eukaryotic response regulator to be implicated in protein stability. However, a prokaryotic equivalent exists that shares several key similarities with our model. Like Skn7p, the prokaryotic response regulator RssB/SprE binds to and affects the stability of a transcriptional regulator. RssB/SprE destabilizes the RNA polymerase subunit σs under specific environmental conditions, and RssB/SprE can bind directly to σs to modulate its degradation by the ClpXP protease (Muffler et al., 1996; Pratt and Silhavy, 1996; Zhou and Gottesman, 1998). In contrast though, many details of the two systems are different. First, while Skn7p has an HSF-like DNA-binding domain and can bind to DNA, RssB/SprE does not have any type of previously characterized DNA-binding domain (Muffler et al., 1996; Pratt and Silhavy, 1996; Zhou and Gottesman, 1998).
Secondly, phosphorylation of the conserved response-regulator aspartate plays different roles in RssB/SprE and Skn7p. For RssB/SprE, aspartyl phosphorylation greatly enhances binding to σs, the protein whose stability it modulates (Becker et al., 1999). On the other hand, Skn7p is an unusual response-regulator protein in that many of its functions operate independently of receiver phosphorylation. Although aspartyl phosphorylation of Skn7p is required for its suppression of kre9Δ (Brown et al., 1994) and its activation of OCH1 transcription (S.Li and J.S.Fassler, in preparation), Skn7p D427N, an allele that can not be phosphorylated by two-component proteins, is fully functional in many situations. For example, Skn7p D427N complements the sensitivity of skn7Δ strains to oxidative stress (Morgan et al., 1997) and, when overexpressed, rescues a pkc1Δ growth defect (Brown et al., 1994).
In our in vivo studies, Skn7p D427N resembles the wild-type allele in its ability to stabilize Crz1p and activate CDRE::lacZ transcription. However, expression of the D427E allele did lead to slightly increased Crz1p levels and CDRE::lacZ activity, indicating that regulation of Skn7p through this residue, while not required, may still play some role in this response. In contrast, Skn7p D427N bound less well than wild-type Skn7p to calcineurin in vitro. This binding, though weaker, may still be sufficient for Skn7p-mediated Crz1p stabilization in vivo. Alternatively, the presence of other proteins may improve the association of Skn7p D427N with Crz1p/calcinerin in vivo.
Finally, the mechanism of Skn7p-regulated Crz1p stabilization is distinct from RssB/SprE-mediated turnover in requiring the physical presence of the eukaryotic phosphatase calcineurin, as well as the DNA-binding domain of Skn7p. These features outline a unique mechanism of transcription factor stabilization. Future studies may reveal whether other Skn7p binding proteins are regulated by Skn7p-mediated stabilization, and whether other response-regulator proteins are implicated in protein stability.
The Skn7p coiled-coil domain is essential for in vivo function
Interestingly, Skn7p has been shown to bind to several other signaling proteins, including several transcription factors, Mbp1p, Hsf1p and itself (Bouquin et al., 1999; Raitt et al., 2000). Coiled-coil domains typically mediate protein–protein interaction, and the coiled-coil homology region (HR) is essential for Skn7p binding to both Rho1p and Mbp1p (Alberts et al., 1998; Bouquin et al., 1999). Hence, we were surprised to observe that while the coiled-coil region is essential for the effect of Skn7p on both Crz1p- and calcineurin-dependent transcription and Crz1p stabilization in vivo, it was not necessary for Skn7p binding to either Crz1p or calcineurin in vitro.
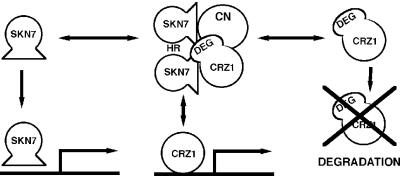
Several models may explain this result. First, Raitt et al. (2000) observed that while the Skn7p coiled-coil domain is not homologous to the HSF multimerization domain at the amino acid level, both regions are helical in nature and are at the same proximity from their HSF DNA-binding domains. Thus, the HR domain may mediate Skn7p– Skn7p dimerization. Skn7p alleles interact similarly with both Crz1p and calcineurin, so it is possible that Skn7p dimerization allows one molecule of Skn7p to interact with calcineurin and another to interact with Crz1p, forming a tetramolecular complex that could be required for Crz1p stabilization in vivo (Figure 8). Secondly, the coiled-coil domain may mediate protein–protein interaction with other proteins in vivo that are required for Crz1p stabilization but are not present in our in vitro binding reaction. Future experiments will attempt to identify in vivo protein complexes containing Skn7p and Crz1p to address this possibility. In addition to mediating in vivo effects on Crz1p stability, the HR domain is essential for binding to several additional signaling proteins including Rho1p, a GTPase that activates Pkc1p as well as other targets, and Mbp1p, a G1–S transcription factor (Alberts et al., 1998; Bouquin et al., 1999). Hence, Skn7p may integrate responses between different signaling pathways through the HR domain.
Fig. 8. Model describing the Skn7p interaction with calcineurin-mediated signal transduction (see Discussion).
A multipurpose Skn7p DNA-binding domain
Skn7p is a transcription factor and a DNA-binding protein, and although the CDRE was sufficient for the effect of Skn7p on Crz1p-dependent transcription, we were unable to detect Skn7p or Skn7p complex binding to the CDRE. In addition, Li et al. have observed Skn7p binding to an OCH1 promoter element which was unaffected in a crz1Δ mutant (S.Li and J.S.Fassler, personal communication). Interestingly, a functional Skn7p DNA-binding domain was completely essential for every Skn7p-mediated effect on calcineurin signaling that we examined. The Skn7p DNA-binding domain was required for activation of Crz1p-dependent transcription through the CDRE, Crz1p stabilization, and binding to both Crz1p and calcineurin in vitro.
Thus, the Skn7p DNA-binding domain is required for binding to both Crz1p and calcineurin as well as binding to promoters involved in stress responses. This suggests that Skn7p may exist in an equilibrium between protein– protein and protein–DNA complexes in vivo. Regulation of the relative abundance of these complexes may modulate the amount of Skn7p-dependent transcription available to the many different signal-transduction pathways in which it participates (Figure 8).
A phosphatase-independent role for calcineurin
We found that Skn7p was able both to activate Crz1p- and calcineurin-dependent transcription and to stabilize Crz1p in the presence of the calcineurin phosphatase inhibitor FK520, but not in the cnb1Δ calcineurin mutant. Consequently, we conclude that Skn7p requires calcineurin, but not its phosphatase activity, for these functions. This is the first example of a phosphatase-independent role for calcineurin. Catalysis-independent roles for signaling molecules have been uncovered in other pathways, such as the scaffolding function of the yeast MAPKK Pbs2p (Posas and Saito, 1997) and the repressive function of the inactive MAPK Kss1p (Cook et al., 1997). The fact that the presence of the complete calcineurin holoenzyme is required for Skn7p-dependent stabilization of Crz1p suggests that protein–protein interactions may be present. We found that Skn7p binds to both calcineurin and Crz1p; however, Skn7p can bind to Crz1p in the absence of calcineurin in our in vitro binding assay. Our data are consistent with the possibility that calcineurin, Crz1p and Skn7p form a complex that stabilizes Crz1p. Without the presence of all of the calcineurin subunits, Skn7p may not completely mask a degradation signal on Crz1p. However, we can not exclude the possibility that calcineurin may have some other, uncharacterized phosphatase-independent function that may affect Crz1p in vivo stability.
A novel mechanism of Crz1p regulation
The Skn7p-mediated control of Crz1p stability represents a new mechanism of regulating calcineurin signaling. Calcineurin also stabilizes Crz1p in a phosphorylation-dependent manner, independently of Skn7p (our unpublished observations). In addition, Crz1p has been previously shown to be regulated at the levels of nuclear localization (Stathopoulos-Gerontides et al., 1999) and transcription, since Crz1p initiates its own transcription upon calcineurin-responsive stress (Matheos et al., 1997). We show that the control of turnover represents a new mechanism of Crz1p regulation that affects Crz1p- and calcineurin-dependent transcription. We postulate that the complex regulation of Crz1p may enable the cell to ‘fine-tune’ the stress response.
The physiology of Skn7p
We have described a novel function for Skn7p as a regulator of protein stability, and have shown that Skn7p modulates stress-activated calcineurin signaling. Pre viously, the role of Skn7p in several stress responses, including cell integrity, oxidative stress and heat shock, has been documented (Brown et al., 1993; Kuge and Jones, 1994; Krems et al., 1996; Morgan et al., 1997; Lee et al., 1999; Raitt et al., 2000) as well as its role in the cell cycle (Morgan et al., 1995; Bouquin et al., 1999). While the precise connection between these responses remains unclear, the fact that Skn7p participates in this variety of functions suggests that they are physiologically related. In particular, Skn7p and calcineurin both affect aspects of cell wall function. Skn7p and calcineurin each activate transcription of cell wall synthesis genes and display extensive genetic interactions with PKC1, a major regulator of cell wall structure and function (Levin and Bartlett-Heubusch, 1992; Brown et al., 1993; Garrett-Engele et al., 1995; S.Li and J.S.Fassler, in preparation). In this report we show that cells lacking both Skn7p and calcineurin are synergistically sensitive to hygromycin B and hydrogen peroxide (Figure 3). These observations indicate that Skn7p and calcineurin function together in response to oxidative stress and cell wall damage, and thus regulate similar physiological processes in vivo.
The major role of Skn7p in vivo may be to integrate different stress signaling events. Some domains of Skn7p are clearly required for more than one of its functions. This may indicate that regulation of these functions is coordinated, or instead, that Skn7p signaling is modulated by direct competition for Skn7p by its different effectors. The discovery that Skn7p affects both protein stability and calcineurin signaling will contribute to future studies that elucidate mechanisms of global stress-response coordination.
Materials and methods
Yeast strains and culture conditions
Yeast strains KWY242 and KWY246 were created from YPH499, DD12 (cnb1Δ) (Cyert and Thorner, 1992), respectively, by using the pKW11 integration vector to introduce two tandem copies of the CDRE upstream of lacZ at the URA3 locus. skn7Δ::TRP1 strains, as described (Brown et al., 1993), were generated in the YPH499 background using standard techniques (Sherman et al., 1986). Yeast cells were grown on standard YPD or SCD media (Sherman et al., 1986), except that amino acids were added at approximately twice the recommended level to SCD. FK520 (Merck), an FK506 analog and calcineurin inhibitor (Liu et al., 1992), in 90% ethanol, 10% Tween-20, was added to a final concentration of 2 µg/ml where noted. In protein stability studies, cycloheximide was added to log phase cells to a final concentration of 100 µg/ml. Recombinant DNA procedures were performed according to standard techniques (Ausubel et al., 1987). Sequencing was performed either using the Sequenase system (US Biochemical Corporation) with [α-35S]dATP (Amersham) or at the Stanford University PAN facility.II
Table II. Yeast strains used in this work.
| Straina | Genotype | Reference |
|---|---|---|
| DD12 | cnb1::hisG | Cyert and Thorner (1992) |
| ASY472 | crz1Δ::loxP-kanMX-loxP | Stathopoulos and Cyert (1997) |
| KWY242 | ura3-52::2×CDRE::lacZ | this study |
| KWY246 | DD12, except ura3-52::2×CDRE::lacZ | this study |
| ASY459 | ura3-52::4×CDRE::lacZ | Stathopoulos and Cyert (1997) |
| ASY460 | ura3-52::mutant 4×CDRE::lacZ | Stathopoulos and Cyert (1997) |
| ASY461 | DD12, except 4×CDRE::lacZ | Stathopoulos and Cyert (1997) |
| ASY834 | ASY472, except 4×CDRE::lacZ | Stathopoulos and Cyert (1997) |
| KWY266 | skn7Δ::TRP1 | this study |
| KWY267 | DD12, except skn7Δ::TRP1 | this study |
| KWY289 | ASY472, except skn7Δ::TRP1 | this study |
aAll strains are in the YPH499 (MATa, ura3-52, lys2-801, ade2-101, trp1-Δ63, his3-Δ200, leu2-Δ1) background (Sikorski and Hieter, 1989).
Growth assays
Saturated cultures were diluted to an OD600 of 1. Ten-fold serial dilutions were spotted onto YPD plates that contained either no added chemicals, 1.8 mM hydrogen peroxide (Fischer) or 70 µg/ml hygromycin B (Roche Diagnostics). Cells were grown at 30°C for 1–3 days.
Plasmids
pKW11 was constructed from pAMS363 (Stathopoulos and Cyert, 1997), a 2 micron URA3 vector containing two copies of the CDRE upstream of the minimal promoter CYC1 fused to lacZ. The 2 micron sequence was removed by HindIII digestion and subsequent re-ligation to generate an integration vector. pKW25, pKW39, 40, 42 and 44 are His6-tagged Skn7p bacterial expression vectors. SKN7 coding sequence was amplified from pRS425-SKN7 (Ketela et al., 1998), pSL1108 (S.Li and J.S.Fassler, in preparation), pRS425-SKN7D427N, pRS425-SKN7D427E (Ketela et al., 1998), pAB93 (Bouquin et al., 1999), respectively, introducing an NdeI site before the ATG and a BamHI site after the TAA, with the exception of pKW44, which introduces a XhoI site at the TAA. These fragments were introduced into the NdeI, BamHI or XhoI sites of pET15b (Novagen). pKW37 was created by inserting a 3.5 kb SalI–HindIII fragment from pSL1108 (S.Li and J.S.Fassler, in preparation) into pRS425. pKW47 was created by amplifying the SKN7ΔHR allele and promoter region from pAB93 (Bouquin et al., 1999), introducing XbaI and HindIII sites outside the YEPlac195 polylinker. This piece was introduced into the same sites of pRS425. BJP3003 was created by introducing BamHI sites at the ATG and after an introduced stop codon after Ser417 in CNA1; this fragment was inserted into the BamHI site of pGEX-5X-1 (Amersham Pharmacia), a GST bacterial expression vector (B.Jiang and M.Cyert, unpublished data). BJP3003 expresses a truncated version of CNA1 that removes the autoinhibitory domain, producing a constitutively active calcineurin allele. This is referred to in the text as GST–CNA1. pLMB117 was created by using PCR mutagenesis to introduce an in-frame HindIII site at the ATG and a SalI site after the stop codon of CRZ1. This fragment was inserted on the C-terminal side of GST into the respective sites in pRD56, a pRS316 GAL1–GST vector (L.Boustany and M.Cyert, unpublished data). A BamHI–SalI GST–CRZ1 fragment from pLMB117 was cloned into the BamHI–XhoI sites of pGEX4T-3 (Amersham Pharmacia) to form pGEX-CRZ1, a bacterial expression vector (K.Saltsman and M.Cyert, unpublished data). pAMS435 is YEP351-CRZ1 (Stathopoulos and Cyert, 1997); M1485 is YEPlac181-SWI5 (D.Stillman, personal communication); YEP352-SKN7HA contains the SKN7 ORF in-frame with an HA epitope tag (H.Bussey, personal communication).
β-galactosidase assays
Quantitative assays. To examine the effect of various stimuli on reporter gene activity, cells were grown to log phase in synthetic media, then spun down and diluted to an OD of 0.2–0.3 in YPD pH 5.5, 50 mM succinic acid. CaCl2 (200 mM) and FK520 (2 µg/ml) or control solvent (90% ethanol, 10% Tween-20) were added, and the cells were grown for 4 h. Protein extracts were made from the pelleted cells as described previously (Withee et al., 1997), breaking the cells with glass beads in 0.1 M Tris pH 8.0, 10% glycerol. Protein concentration was determined by BCA assay (Pierce). Activity was determined at room temperature in a microplate reader (Bio-Rad) using 10 µl of 4 mg/ml ONPG (O-nitrophenyl-β-d-galactopyranoside; Sigma) and 90 µl of Z buffer (100 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 0.027% β-mercaptoethanol). Maximum rates are given in rate of OD415 change per minute per µg protein.
Qualitative assays. Colonies were scored for β-galactosidase activity as described (Stathopoulos and Cyert, 1997), except that blue colonies were selected at a time point before the negative control (KWY242 with the empty vector YEP351) turned blue—usually at ∼7 h. Xgal (Sigma) was used at a concentration of 50 µg/ml.
Genetic screening
KWY242 was transformed with two multicopy genomic libraries. Cells were grown at room temperature. Approximately 160 000 colonies transformed with the 2J351 library (Hill et al., 1986; Engebrecht et al., 1990) and 79 000 colonies transformed with the YEP13 library (Nasmyth and Reed, 1980) were screened. Plasmids from blue colonies were retransformed into ASY461 (4×CDRE::lacZ) and ASY462 (4× mutant CDRE::lacZ) to eliminate enhancers of general transcription. Plasmids were plated on media containing 2 µg/ml FK520 (Merck) to assess calcineurin dependence. The plasmids were also re-transformed into the original strain (KWY242), a cnb1Δ strain (KWY246), and a crz1Δ strain (ASY834), to ensure that the β-galactosidase activity was plasmid-, calcineurin- and Crz1p-dependent. Inserts were identified by sequencing the ends of the genomic DNA and matching this sequence to regions of the genome with BLAST (Altschul et al., 1990) and the Saccharomyces Genome Database (Cherry et al., http://genome-www.stanford.edu/Saccharomyces).
Protein purification
Recombinant proteins were prepared from French press E.coli extracts (BL21pLysS with His-SKN7 constructs, BLR with GST–CRZ1 and GST–CNA1) according to the manufacturers’ directions (GST tags, Amersham Pharmacia; His tags, Qiagen). After elution, proteins were dialyzed into Buffer 88 (20 mM HEPES pH 6.8, 150 mM potassium acetate, 250 mM sorbitol, 2 mM magnesium acetate) with a 10 000 MWCO DispoDialyzer (Amersham Pharmacia). Recombinant yeast calcineurin was purified by first mixing two bacterial strains containing either BJP3003 (GST–CNA1) or pET-CNB1 (Okano et al., 1998), followed by lysis and glutathione chromatography (as above).
Binding assays
Recombinant protein binding assays were performed as in Lai et al. (1998). Approximately 0.2 µg of recombinant protein were added to protein immobilized on beads in 500 µl binding buffer [50 mM Tris–HCl pH 7.4, 100 mM NaCl, 2 mM CaCl2, 2 mM MgCl2, 0.2% Triton-X, 0.5 mg/ml bovine serum albumin, aprotinin (5 µg/ml), leupeptin (5 µg/ml), pepstatin A (5 µg/ml), benzamidine (1.25 mM) and PMSF (0.5 mM) (Sigma) as well as 1 mM dithiothreitol where glutathione-Sepharose was included] and rotated at 4°C for 2–3 h. Twenty microliters of 50% (w/v) Sepharose were used in all binding assays, except that 5 µl of beads with GST–CRZ1 were used to pull down His6-tagged Skn7 alleles. Beads were washed three times with 800 µl binding buffer, before adding loading buffer, boiling, and performing SDS–PAGE.
Western blotting
SDS–PAGE and western blotting were performed by standard procedures. BCA assays (Pierce) were used to normalize protein loading (15–40 µg) for westerns of whole-cell extracts. Immunoblots were performed with either anti-HA 12CA5 antibodies (Roche Molecular Diagnostics), anti-Crz1 (H-X.Li and M.Cyert, unpublished data), anti-GST (Berkeley Antibody Co.), or anti-tetra-His antibodies (Qiagen), and anti-mouse-HRP or anti-rabbit-HRP (Amersham Pharmacia) and ECL detection reagents (Amersham Pharmacia).
Acknowledgments
Acknowledgements
We thank Lee Johnston for generously providing Skn7p antibodies; Jan Fassler for pSL1108 and for sharing data prior to publication; Howard Bussey for pRS425-SKN7 constructs and skn7Δ::TRP1 disruption construct; Nic Bouquin for pAB93; David Stillman for M1485; Yoshi Ohya for the Cnb1p expression plasmid; Angela Stathopoulos-Gerontides for numerous strains and plasmids; Kirstie Saltsman for pGEX-CRZ1; and Bo Jiang for BJP3003. We thank Valerie Denis, Victoria Heath, Kim Kafadar and Arthur Grossman for critical reading of the manuscript. This work was supported by National Institutes of Health research grant GM-48728.
References
- Alberts A.S., Bouquin,N., Johnston,L.H. and Treisman,R. (1998) Analysis of RhoA-binding proteins reveals an interaction domain conserved in heterotrimeric G protein β subunits and the yeast response regulator protein Skn7. J. Biol. Chem., 273, 8616–8622. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Gish,W., Miller,W. Myers,E.W. and Lipman,D.J. (1990) Basic Local Alignment Search Tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Ausbel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (1987) Current Protocols in Molecular Biology. John Wiley and Sons, New York, NY. [Google Scholar]
- Becker G., Klauck,E. and Hengge-Aronis,R. (1999) Regulation of RpoS proteolysis in Escherichia coli: the response regulator RssB is a recognition factor that interacts with the turnover element in RpoS. Proc. Natl Acad. Sci. USA, 96, 6439–6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouquin N., Johnson,A.L., Morgan,B.A. and Johnston,L.H. (1999) Association of the cell cycle transcription factor Mbp1 with the Skn7 response regulator in budding yeast. Mol. Biol. Cell, 10, 3389–3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuder T., Hemenway,C.S., Movva,N.R., Cardenas,M.E. and Heitman,J. (1994) Calcineurin is essential in cyclosporin A- and FK506-sensitive yeast strains. Proc. Natl Acad. Sci. USA, 91, 5372–5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J.L., North,S. and Bussey,H. (1993) SKN7, a yeast multicopy suppressor of a mutation affecting cell wall β-glucan assembly, encodes a product with domains homologous to prokaryotic two-component regulators and to heat shock transcription factors. J. Bacteriol., 175, 6908–6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J.L., Bussey,H. and Stewart,R.C. (1994) Yeast Skn7p functions in a eukaryotic two-component regulatory pathway. EMBO J., 13, 5186–5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry J.M. et al., Saccharomyces Genome Database. http://genome-www.stanford.edu/Saccharomyces
- Chin E.R. et al. (1998) A calcineurin-dependent transcriptional pathway controls skeletal muscle fiber type. Genes Dev., 12, 2499–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clipstone N.A. and Crabtree,G.R. (1992) Identification of calcineurin as a key signalling enzyme in T-lymphocyte activation. Nature, 357, 695–697. [DOI] [PubMed] [Google Scholar]
- Cook J.G., Bardwell,L. and Thorner,J. (1997) Inhibitory and activating functions for MAPK Kss1 in Saccharomyces cerevisiae filamentous-growth signalling pathway. Nature, 390, 85–88. [DOI] [PubMed] [Google Scholar]
- Cunningham K.W. and Fink,G.R. (1994) Calcineurin-dependent growth control in Saccharomyces cerevisiae mutants lacking PMC1, a homolog of plasma membrane Ca2+ ATPases. J. Cell Biol., 124, 351–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham K. and Fink,G.R. (1996) Calcineurin inhibits VCX1-dependent H+/Ca2+ exchange and induces Ca2+ ATPases in Saccharomyces cerevisiae. Mol. Cell. Biol., 16, 2226–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyert M.S. and Thorner,J. (1992) Regulatory subunit (CNB1 gene product) of yeast Ca2+/calmodulin-dependent phosphoprotein phosphatases is required for adaptation to pheromone. Mol. Cell. Biol., 12, 3460–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyert M.S., Kunisawa,R., Kaim,D. and Thorner,J. (1991) Yeast has homologs (CNA1 and CNA2 gene products) of mammalian calcineurin, a calmodulin-regulated phosphoprotein phosphatase [published erratum appears in Proc. Natl Acad. Sci. USA, 1992, 89, 4220]. Proc. Natl Acad. Sci. USA, 88, 7376–7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Pompa J.L. et al. (1998) Role of the NF-ATc transcription factor in morphogenesis of cardiac valves and septum. Nature, 392, 182–186. [DOI] [PubMed] [Google Scholar]
- Dean N. (1995) Yeast glycosylation mutants are sensitive to aminoglycosides. Proc. Natl Acad. Sci. USA, 92, 1287–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohrmann P.R., Butler,G., Tamai,K., Dorland,S., Greene,J.R., Thiele,D.J. and Stillman,D.J. (1992) Parallel pathways of gene regulation: homologous regulators SWI5 and ACE2 differentially control transcription of HO and chitinase. Genes Dev., 6, 93–104. [DOI] [PubMed] [Google Scholar]
- Engebrecht J., Hirsch,H. and Roeder,G.S. (1990) Meiotic gene conversion and crossing over: their relationship to each other and to chromosome synapsis and segregation. Cell, 62, 927–937. [DOI] [PubMed] [Google Scholar]
- Farcasanu I.C., Hirata,D., Tsuchiya,E., Nishiyama,F. and Miyakawa,T. (1995) Protein phosphatase 2B of Saccharomyces cerevisiae is required for tolerance to manganese in blocking the entry of ions into the cell. Eur. J. Biochem., 232, 712–717. [PubMed] [Google Scholar]
- Foor F., Parent,S.A., Morin,N., Dahl,A.M., Ramadan,N., Chrebet,G., Bostian,K.A. and Nielsen,J.B. (1992) Calcineurin mediates inhibition by FK506 and cyclosporin of recovery from α-factor arrest in yeast. Nature, 360, 682–684. [DOI] [PubMed] [Google Scholar]
- Garrett-Engele P., Moilanen,B. and Cyert,M.S. (1995) Calcineurin, the Ca2+/calmodulin-dependent protein phosphatase, is essential in yeast mutants with cell integrity defects and in mutants that lack a functional vacuolar H+-ATPase. Mol. Cell. Biol., 15, 4103–4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haro R., Graciadebles,B. and Rodriguez-Navarro,A. (1991) A novel P-type ATPase from yeast involved in sodium transport. FEBS Lett., 291, 189–191. [DOI] [PubMed] [Google Scholar]
- Heinisch J.J., Lorberg,A., Schmitz,H.P. and Jacoby,J.J. (1999) The protein kinase C-mediated MAP kinase pathway involved in the maintenance of cellular integrity in Saccharomyces cerevisiae. Mol. Microbiol., 32, 671–680. [DOI] [PubMed] [Google Scholar]
- Hendey B. and Maxfield,F.R. (1993) Regulation of neutrophil motility and adhesion by intracellular calcium transients. Blood Cells, 19, 143–164. [PubMed] [Google Scholar]
- Hill J.E., Myers,A.M., Koerner,J.J. and Tzagaloff,A. (1986) Yeast/E.coli shuttle vectors with multiple unique restriction sites. Yeast, 2, 163–167. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. and Kornitzer,D. (1998) Ubiquitin-dependent degradation of transcription regulators. In Peters,J.-M., Harris,J.R. and Finley,D. (eds), Ubiquitin and the Biology of the Cell. Plenum Press, New York, NY.
- Hubl S.T., Owens,J.C. and Nelson,H.C. (1994) Mutational analysis of the DNA-binding domain of yeast heat shock transcription factor. Nature Struct. Biol., 1, 615–620. [DOI] [PubMed] [Google Scholar]
- Jain J., McCaffrey,P.G., Miner,Z., Kerppola,T.K., Lambert,J.N., Verdine,G.L., Curran,T. and Rao,A. (1993) The T-cell transcription factor NFATp is a substrate for calcineurin and interacts with Fos and Jun. Nature, 365, 352–355. [DOI] [PubMed] [Google Scholar]
- Ketela T., Brown,J.L., Stewart,R.C. and Bussey,H. (1998) Yeast Skn7p activity is modulated by the Sln1p–Ypd1p osmosensor and contributes to regulation of the HOG pathway. Mol. Gen. Genet., 259, 372–378. [DOI] [PubMed] [Google Scholar]
- Krems B., Charizanis,C. and Entian,K.D. (1996) The response regulator-like protein Pos9/Skn7 of Saccharomyces cerevisiae is involved in oxidative stress resistance. Curr. Genet., 29, 327–334. [DOI] [PubMed] [Google Scholar]
- Kuge S. and Jones,N.J. (1994) YAP1 dependent activation of TRX2 is essential for the response of Saccharomyces cerevisiae to oxidative stress by hydrogen peroxides. EMBO J., 13, 655–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuno T., Tanaka,H., Mukai,J., Chang,C., Hiraga,K., Miyakawa,T. and Tanaka,C. (1991) cDNA cloning of a calcineurin B homolog in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun., 180, 1159–1163. [DOI] [PubMed] [Google Scholar]
- Lai M.M., Burnett,P.E., Wolosker,H. Blackshaw,S. and Snyder,S.H. (1998) Cain, a novel physiologic protein inhibitor of calcineurin. J. Biol. Chem., 273, 18325–18331. [DOI] [PubMed] [Google Scholar]
- Lawson M.A. and Maxfield,F.R. (1995) Ca2+- and calcineurin-dependent recycling of an integrin to the front of migrating neutrophils. Nature, 377, 75–79. [DOI] [PubMed] [Google Scholar]
- Lee J., Godon,C., Lagniel,G., Spector,D., Garin,J., Labarre,J. and Toledano,M.B. (1999) Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J. Biol. Chem., 274, 16040–16046. [DOI] [PubMed] [Google Scholar]
- Levin D.E. and Bartlett-Heubusch,E. (1992) Mutants in the S. cerevisiae PKC1 gene display a cell cycle-specific osmotic stability defect. J. Cell Biol., 116, 1221–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Ault,A., Malone,C.L., Raitt,D., Dean,S., Johnston,L.H., Deschenes,R.J. and Fassler,J.S. (1998) The yeast histidine protein kinase, Sln1p, mediates phosphotransfer to two response regulators, Ssk1p and Skn7p. EMBO J., 17, 6952–6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Farmer,J.D.,Jr, Lane,W.S., Friedman,J., Weissman,I. and Schreiber,S.L. (1991) Calcineurin is a common target of cyclophilin–CsA and FKBP–FK506 complexes. Cell, 66, 807–815. [DOI] [PubMed] [Google Scholar]
- Liu J. et al. (1992) Inhibition of T cell signaling by immunophilin–ligand complexes correlates with loss of calcineurin phosphatase activity. Biochemistry, 31, 3896–3901. [DOI] [PubMed] [Google Scholar]
- Liu Y. et al. (1991) The Saccharomyces cerevisiae genes (CMP1 and CMP2) encoding calmodulin-binding proteins homologous to the catalytic subunit of mammalian protein phosphatase 2B. Mol. Gen. Genet., 227, 52–59. [DOI] [PubMed] [Google Scholar]
- Loomis W.F., Kuspa,A. and Shaulsky,G. (1998) Two-component signal transduction systems in eukaryotic microorganisms. Curr. Opin. Microbiol., 1, 643–648. [DOI] [PubMed] [Google Scholar]
- Lussier M. et al. (1997) Large scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cerevisiae. Genetics, 147, 435–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheos D., Kingsbury,T., Ahsan,U. and Cunningham,K. (1997) Tcn1p/Crz1p, a calcineurin-dependent transcription factor that differentially regulates gene expression in Saccharomyces cerevisiae. Genes Dev., 11, 3445–3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazur P., Morin,N., Baginsky,W., El-Sherbeini,M., Clemas,J.A., Nielsen,J.B. and Foor,F. (1995) Differential expression and function of two homologous subunits of yeast 1,3-β-d-glucan synthase. Mol. Cell. Biol., 15, 5671–5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza I., Rubio,F., Rodriguez-Navarro,A. and Pardo,J.M. (1994) The protein phosphatase calcineurin is essential for NaCl tolerance of Saccharomyces cerevisiae. J. Biol. Chem., 269, 8792–8796. [PubMed] [Google Scholar]
- Molkentin J.D., Lu,J.R., Antos,C.L., Markham,B., Richardson,J., Robbins,J., Grant,S.R. and Olson,E.N. (1998) A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell, 93, 215–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan B.A., Bouquin,N., Merrill,G.F. and Johnston,L.H. (1995) A yeast transcription factor bypassing the requirement for SBF and DSC1/MBF in budding yeast has homology to bacterial signal transduction proteins. EMBO J., 14, 5679–5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan B.A., Banks,G.R., Toone,W.M., Raitt,D., Kuge,S. and Johnston,L.H. (1997) The Skn7 response regulator controls gene expression in the oxidative stress response of the budding yeast Saccharomyces cerevisiae. EMBO J., 16, 1035–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser M.J., Geiser,J.R. and Davis,T.N. (1996) Ca2+-calmodulin promotes survival of pheromone-induced growth arrest by activation of calcineurin and Ca2+-calmodulin-dependent protein kinase. Mol. Cell. Biol., 16, 4824–4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muffler A., Fischer,D., Altuvia,S., Storz,G. and Hengge-Aronis,R. (1996) The response regulator RssB controls stability of the σS subunit of RNA polymerase in Escherichia coli. EMBO J., 15, 1333–1339. [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., Liu,Y., Hirata,D., Namba,H., Harada,S., Hirokawa,T. and Miyakawa,T. (1993) Protein phosphatase type 2B (calcineurin)-mediated, FK506-sensitive regulation of intracellular ions in yeast is an important determinant for adaptation to high salt stress conditions. EMBO J., 12, 4063–4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K.A. and Reed,S.I. (1980) Isolation of genes by complementation in yeast: molecular cloning of a cell-cycle gene. Proc. Natl Acad. Sci. USA, 77, 2119–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonoka H., Tanaka,K., Strum,S. and Okayama,H. (1995) A downstream target of RHO1 small GTP-binding protein is PKC1, which leads to activation of the MAP kinase cascade in Saccharomyces cerevisiae. EMBO J., 14, 5931–5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northrop J.P., Ullman,K.S. and Crabtree,G.R. (1993) Characterization of the nuclear and cytoplasmic components of the lymphoid-specific nuclear factor of activated T cells (NF-AT) complex. J. Biol. Chem., 268, 2917–2923. [PubMed] [Google Scholar]
- O’Keefe S.J., Tamura,J., Kincaid,R.L., Tocci,M.J. and O’Neill,E.A. (1992) FK506- and CsA-sensitive activation of the interleukin-2 promoter by calcineurin. Nature, 357, 692–694. [DOI] [PubMed] [Google Scholar]
- Okano H., Cyert,M.S. and Ohya,Y. (1998) Importance of phenylalanine residues of yeast calmodulin for target binding and activation. J. Biol. Chem., 273, 26375–26382. [DOI] [PubMed] [Google Scholar]
- Parkinson J.S. (1993) Signal transduction schemes of bacteria. Cell, 73, 857–871. [DOI] [PubMed] [Google Scholar]
- Posas F. and Saito,H. (1997) Osmotic activation of HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2 MAPKK. Science, 276, 1702–1705. [DOI] [PubMed] [Google Scholar]
- Pozos T.C., Sekler,I. and Cyert,M.S. (1996) The product of HUM1, a novel yeast gene, is required for vacuolar Ca2+/H+ exchange and is related to mammalian Na+/Ca2+ exchangers. Mol. Cell. Biol., 16, 3730–3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt L.A. and Silhavy,T.J. (1996) The response regulator SprE controls the stability of RpoS. Proc. Natl Acad. Sci. USA, 93, 2488–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raitt D.C., Johnson,A.L., Erkine,A.M., Makino,K., Morgan,B., Gross,D.S. and Johnston,L.H. (2000) The Skn7 response regulator of Saccharomyces cerevisiae interacts with Hsf1 in vivo and is required for the induction of heat shock genes by oxidative stress. Mol. Biol. Cell, 11, 2335–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranger A.M., Grusby,M.J., Hodge,M.R., Gravallese,E.M., de la Brousse, F.C., Hoey,T., Mickanin,C., Baldwin,H.S. and Glimcher,L.H. (1998) The transcription factor NF-ATc is essential for cardiac valve formation. Nature, 392, 186–190. [DOI] [PubMed] [Google Scholar]
- Rudolph H.K., Antebi,A., Fink,G.R., Buckley,C.M., Dorman,T.E., LeVitre,J., Davidow,L.S., Mao,J.I. and Moir,D.T. (1989) The yeast secretory pathway is perturbed by mutations in PMR1, a member of a Ca2+ ATPase family. Cell, 58, 133–145. [DOI] [PubMed] [Google Scholar]
- Sherman F., Fink,G.R. and Hicks,J.B. (1986) Methods in Yeast Genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathopoulos A.M. and Cyert,M.S. (1997) Calcineurin acts through the CRZ1/TCN1-encoded transcription factor to regulate gene expression in yeast. Genes Dev., 11, 3432–3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathopoulos-Gerontides A., Guo,J.J. and Cyert,M.S. (1999) Yeast calcineurin regulates nuclear localization of the Crz1p transcription factor through dephosphorylation. Genes Dev., 13, 798–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withee J.L., Mulholland,J., Jeng,R. and Cyert,M.S. (1997) An essential role of the yeast pheromone-induced Ca2+ signal is to activate calcineurin. Mol. Biol. Cell, 8, 263–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withee J.L., Sen,R. and Cyert,M.S. (1998) Ion tolerance of Saccharomyces cerevisiae lacking the Ca2+/CaM-dependent phosphatase (calcineurin) is improved by mutations in URE2 or PMA1. Genetics, 149, 865–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y. and Gottesman,S. (1998) Regulation of proteolysis of the stationary-phase σ factor RpoS. J. Bacteriol., 180, 1154–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]