Abstract
Introduction
Colorectal cancer (CRC) is a molecularly diverse disorder arising from gradual accumulation of genetic and epigenetic changes giving interest in characterization of genetic alterations and microsatellite instability (MSI) for identification of new personalized therapeutic targets.
Objectives
Evaluating the incidence of somatic mutations in clinically relative signaling pathways involved in CRC tumorigenesis that include mainly Wnt and MAPK pathways together with MSI and gut microbiota.
Methods
Twenty four CRC patients were enrolled in the study. Cancer hotspot V2 panel was tested using targeted NGS on MiSeqDx device in addition to MSI identification, also gut microbiota was detected using conventional techniques. Patients received FOLFOX regimen in addition to Xeloda especially in early stages.
Results
Non-synonymous genetic variants were detected inTP53, PIK3CA, KDR, KIT, APC, FGFR3 and MET. Twelve patients (50%) had MSI-LO, 25% had MSI-HI and 20.8% were MSS. Missense mutations in PIK3CA, TP53, and KDR were identified in 2, 3, and 2 patients with MSI-HI status, respectively.
In MSI-LO, missense alterations in PIK3CA, TP53, KIT, and KDR were detected in (6, 11, 3, 2 cases, respectively). More than 50% of examined patients revealed mixed GIT flora, 18.8% of patients had E. Coli, 12.5% of patients showed Klebsiella spp. and only 6.3 % of patient revealed Proteus spp. H. pylori antigen was detected in 37.5%of patients and Blastocystis hominis cysts in only 4 patients.
Conclusion
CRC genetic mutational statuses as well as contributing environmental stress factors such as gut microbiota dysbiosis are prognostically crucial, associated with high risk potential of gene-environment interactions based on machine learning.
Keywords: Colorectal cancer, Next generation sequencing, Activating mutations, Microsatellite instability, Gut microbiota & protists infections
1. Background
Global cancer burden is increasing annually as GLOBOCAN 2018 data showed an overall increase of 18.1 million new cancer cases and 9.6 million cancer deaths in 2018 worldwide with Africa and Asia had a higher proportion of cancer mortality.1 Colorectal cancer (CRC) is the 3rd leading cause of cancer related death and its pathogenesis is highly complicated that includes both genetic and environmental factors.2 Recently in 2023, consensus molecular subtypes has been defined according to comprehensive molecular studies as well as microsatellite instability (MSI) for CRC differentiation into prognostically distinct groups identifying new treatment targets.3 Fast food & bad dietary patterns, smoking, obesity, heavy alcohol intake together with intestinal microbiota & parasitic infection play crucial roles as environmental risk factors. The human intestinal microbiota is about 1013 to 1014 microbes, and contains >100 times as many genes as in the human genome.4 A healthy human intestinal microbiota plays an important physiological role in protecting and maintaining immunity against pathogens andmicrobiota dysbiosis alters host physiological functions, favoring CRC initiation, progression, and metastasis. The link between tumor microbiota and cancer are demonstrated by 4 mechanisms: increased gene mutations directly promoting tumorigenesis, regulation of oncogenes/oncogenic pathways, modulation of the host immune system, and production of small molecules or metabolites that influence cancer development, progression, and even response to therapeutic agents. Gut microbiota can predict how immunotherapy will work and can be modulated to raise immunotherapy's effectiveness in CRC treatment.3, 4 Regarding parasitic infection, Blastocystis specie isa common parasitic protozoan worldwide, that is present in both human and many animal gastrointestinal (GI) tract. It has faecal-oral route of transmission leading toseveral non specific GIT symptoms e.g. nausea, vomiting, diarrhea and may be associated with irritable bowel syndrome (IBS). Its predominance in chronic GI illness patients was reported in Malaysia, Singapore and Egypt.5 Direct positive association between Blastocystis and CRC has been reported as it increases inflammatory cells infiltration and pro-inflammatory cytokines as tumor necrosis factor α (TNF α).6 A lot of genetic mutations that play a pivotal role in CRC pathogenesis has been identified with several targeted therapies approved regarding these genetic variations. Next Generation sequencing (NGS) technology has been globally used in clinical and translational research in CRC providing large amount of information on genetic mutations obtained. This offers the benefit of personalized medicine approaches through using targeted treatments according to the gene mutations after analyzing the tumor genome.7, 8 Our study aimed at detection of activating genetic mutations in liquid biopsy of Egyptian CRC patients to overcome the challenges posed by poor quality of FFPE samples via targeted NGS together with assessing microsatellite instability (MSI) status. Moreover, evaluating the incidence of dysbiotic intestinal microbiota and pathogenic protists among those CRC patients as well as investigating the effect of diet on human intestinal microbiota. Throughout the work, correlation with the different demographic and clinicopathological factors was done.
2. Methods
2.1. Study participants
The study included 24 newly diagnosed Egyptian colorectal cancer patients who were recruited from our Clinical Oncology department. Under strict sterile conditions, peripheral blood & stool samples were withdrawn from those patients. All clinico-pathological features were collected from patients' records. Our project was approved by Cairo University under code ≠ 39–2021.We started our study that was approved by our department IRB (unit A) by the end of2021. Those patients received chemotherapy and were followed up for a maximum period of 24 months.
2.2. Library preparation, sequencing & data analysis
Genomic DNA (gDNA) was extracted from those liquid biopsies (LB) according to the manufacturer protocol, with the QIAamp DNA Mini kit (Qiagen, Germany, Cat No./ID: 51304). Evaluation of the extracted gDNA concentration was carried out using the Qubit dsDNA High Sensitivity (HS) Assay Kit (Life Technologies, Fisher Scientific, Cat No.: Q32851), along with quantitative PCR (qPCR) to assess quality and amplifiability with ΔCq value below 5 selected for further use. Library preparation was done according to the manufacturer protocol using AmpliSeqTM for Illumina Cancer Hotspot Panel V2 (Illumina, Inc., US, Cat. No.: 20019161) which is a targeted next generation sequencing methodology detecting hotspot mutations for 50 genes with detection limits of 5 % variant allele frequency across 207 amplicons with >95 % of bases covered at ≥500×. Library quality testing which indicates successful library amplification was done by Agilent 2100 Bioanalyzer device utilizing the DNA 1000 reagents & Chips (Agilent Technologies, Santa Clara, California, Cat. Code: 5067–1504) with expected PCR product 186–277 bp. Patients' libraries as well as PhiX control library were normalized and equal volumes were pooled to form the terminal sequencing library. Sequencing was done using Cancer Hotspot Panel V2 Nano kit on MiSeqDx device (llumina) with a 2 × 150 bp read length and total time of ∼17 h.
Each run quality was checked through the specifications of Illumina PhiX control library that show cluster densities between 865 and 965 k/mm2 clusters passing filter for v2 chemistry. The prediction of the probability of an error in base calling through the quality score (Q-score) was assessed with the percentage of bases >Q30 is averaged across the entire run and >80 % bases higher than Q30 at 2 × 150 bp was accepted for v2 chemistry. Reads assembly was done according to Genome Reference Consortium Human Build 37 (GRCh37) which is the human reference genome (version hg19). Processing of image generating Variant Call Format (VCF) file format was further annotated and analyzed using BaseSpace variant interpreter (Illumina).Numerical identifier in Catalogue of Somatic Mutations in Cancer (COSMIC) database was illustrated for each variant. Amino acid changesʹ impact was assessed with In Silico Predictions (Sift & PolyPhen), Functional Analysis through Hidden Markov Models (v2.3) (FATHMM) prediction, and finally ClinVar database categorize the variants as benign or pathogenic. Mutations with low depth, which indicate ≤50x depths, mutations with ≤5 % variant allele frequency and variants quality if <80 % were filtered out.9, 10
2.3. Microsatellite instability (MSI) analysis
Three primer sequences (BAT25, BAT26, and NR27) were used to examine MSI status in our CRC patients and the PCR products were analyzed on Agilent 2100 Bioanalyzer system as previously described.11 Tumors and healthy controls DNA were compared to each other with peaks detected in the tumor and were not in the normal subjects indicating marker instability. Patients with no varied, only one varied and those with ≥2 varied markers were considered as microsatellite stable (MSS), microsatellite instability-low (MSI-L) and microsatellite instability-high (MSI-HI), respectively.
2.4. Gut microbiota identification
Nowadays, improvements have been achieved by the use of a reproducible and modular short-read library 16S rRNA gene sequencing pipeline ‘sl1p’. These techniques produce high diversity and functional complexity of the tumor microbiota that nessacitates computational modeling and robust databases. In our study, microbiological conventional detection methods were followed due to budget constraints.
2.4.1. Stool analysis &culture
Fresh random stool in clean plastic leak proof container or swab transport system was collected. Stool analysis was done through checking for color with microscopic examination for pus, RBCs, and the presence of mucus.12 Stool culture was done on Mackoncy, Blood and XLD agar to detect salmonella, shigella and differentiate between enteric pathogens and normal GIT flora. Biochemical reactions in form of TSI, MIO, Citrate, LIA and urea, catalase, coagulase were further used to detect the isolated pathogen.13
2.4.2. Helicobacter pylori antigen & fecal occult blood examination
The stool was examined within 1 h of reaching the laboratory for Helicobacter pylori (H. pylori) antigen &fecal occult blood (FOB) as described by the manufacturer. Test was interpreted as follows; Positive: Two distinct red lines appear with one line should be in the control region (C) and another line should be in the test region (T). Negative: One red line appears in the control region (C). No apparent colored line appears in the test region (T).14, 15
2.5. Detection of Blastocystis hominis
Each fresh stool sample was further subdivided into three aliquots to perform the following techniques:
A-Direct microscopy of stool samples: A small portion of each stool sample was microscopically examined immediately after collection using saline and iodine wet preparations (Direct wet smear method)16 and then after formalin-ethyl acetate concentration technique for detection of Blastocystis parasites (Fecal Concentration method) and Blastocystis diagnosis was based on morphology of parasites observed.
B-in vitro cultivation on modified Jones’ medium
The second portion of each fresh stool sample was inoculated into sterile screw-capped tubes containing 5 ml Jones’ medium enhanced with 10 %horse serum. Culture tubes were incubated at 37 °C, and the sediment was examined by the × 40 objective lens after 24, 48, 72, and 96 h using a sterile pipette. The cultures were considered negative if no Blastocystis cysts were seen up to 96 h later.17
2.6. Treatment protocol
Over the last several decades, combinations of several chemotherapeutic agents have been incorporated in routine clinical practice. Also, treatment of metastatic CRC has been considered palliative for many years; aiming to increase the quality of life of the patients highlighting the need of targeted therapies in CRC treatment. According our department treatment protocol, our patients received according to their clinical stage, FOLFOX regimen (infusional 5-fluorouracil [5-FU], leucovorin calcium, and oxaliplatin) every 2 weeks for 6 months. Early stage patients received Xeloda for 6 cycles.
3. Results
3.1. Patients demographic and clinico-pathological characteristics
The clinical and pathological features of our study group were described in Table 1. All patients live in the cities of Cairo and Giza governorates, except for only three patients from Fayoum and Beni Suef. All patients are non-vegetarian except for one male patient is vegetarian and lives in Boulaq. Regarding our studied group gender, 13 patients (54.2 %) were males and 11 patients (45.8 %) were females. Their ages range from 24 to 71 years having median age of 52.5 years with 13 patients (54.2 %) above 40 years considered as late onset CRC patients and 11 patients (45.8 %) less than or equal 40 years considered as early onset CRC patients. Fourteen patients (58.4 %) had a primary tumor in colon, 5 patients (20.8 %) had rectal tumor and5 patients (20.8 %) had colorectal mass. Most of our patients had tumor stage III (41.7 %), followed by stage IV (25 %) and stage II(20.8 %).Patients were mostly T3, N1b, M1 with metastasis mainly in liver. Only 4 patients (16.7 %) had lymphovascular invasion (LVI) at diagnosis. Out of the 24 CRC cases, 18 patients (75 %) were non metastatic while 6 patients (25 %) having metastasis. After the conventional treatment, 10/18 (55.5 %) cases are still alive without progression, while the rest progressed into visceral metastasis mainly liver. Those patients could probably benefit from targeted therapy e.g. Anti angiogenic drug as Bevacizumab & anti-EGFR therapy.
Table 1.
Characteristics of the studied 24 Egyptian CRC patients.
| Patient’s characteristics | Total No. = 24 |
|---|---|
| No. (%) | |
| Demographic and Clinico-pathological Characteristics(baseline) | |
| Gender: | |
| Males | 13 (54.2 %) |
| Females | 11 (45.8 %) |
| Age (years): | |
| Range | 24–71 |
| Mean ± SD | 49.1 ± 13.3 |
| Median | 52.5 |
| > 40 | 13(54.2 %) |
| ≤ 40 | 11 (45.8 %) |
| Residence: | |
| Cairo | 3 (12.5 %) |
| Giza | 13 (54.2 %) |
| Beni Suef | 1 (4.2 %) |
| Fayoum | 1 (4.2 %) |
| Unknown | 6 (25 %) |
| Diet history | |
| Non-vegetarian | 23(95.8 %) |
| Vegetarian | 1(4.2 %) |
| Body Mass Index (BMI) | |
| Range | 20.2–36.2 |
| Mean ± SD | 27.5 ± 5.4 |
| Median | 42.3 |
| Average | 5 (20.8 %) |
| Overweight | 5 (20.8 %) |
| Obese | 6 (25 %) |
| Missing data | 8(33.3 %) |
| Smoking | |
| Yes | 4(16.7 %) |
| No | 15(62.5 %) |
| Missing data | 5(20.8 %) |
| Comorbidities | |
| DM | 5(20.8 %) |
| HTN | 5(20.8 %) |
| No | 14 (58.33 %) |
| Intestinal obstruction: | |
| Yes | 4(16.7 %) |
| No | 17(70.8 %) |
| Missing data | 3(12.5 %) |
| Perforation: | |
| Yes | 2(8.3 %) |
| No | 19(79.2 %) |
| Missing data | 3(12.5 %) |
| Stage at diagnosis | |
| II | 5(20.8 %) |
| III | 10(41.7 %) |
| IV | 6(25 %) |
| Missing data | 3(12.5 %) |
| Tumor site: | |
| Colon | 14(58.4 %) |
| Right | 6 (25 %) |
| Left | 5 (20.9 %) |
| Transverse | 3 (12.5 %) |
| Rectum | |
| Colorectal | 5(20.8 %) |
| 5(20.8 %) | |
| Grade | |
| II | 19(79.2 %) |
| III | 2(8.3 %) |
| Missing data | 3(12.5 %) |
| Lymphovascular invasion (LVI) | |
| Yes | 4(16.7 %) |
| No | 17(70.8 %) |
| Missing data | 3(12.5 %) |
| Routine Lab. | |
| Hemoglobin(g/dl) | |
| Range | 7.7–13.7 |
| Mean ± SD | 10.8 ± 1.8 |
| Median | 8.3 |
| Mild anemia | 7(29.2 %) |
| Moderate anemia | 2(8.3 %) |
| Moderate anemia | 1 (4.1 %) |
| No anemia | 14(58.4 %) |
| Total leucocytic count(x103/ cmm) | |
| Range | 04-Oct |
| Mean ± SD | 7.3 ± 2.1 |
| Median | 5.6 |
| Platelets(x106) | |
| Range | 255–575 |
| Mean ± SD | 369.7 ± 104.4 |
| Median | 329 |
| ALT(U/L) | |
| Range | 15–35 |
| Mean ± SD | 22.7 ± 6.9 |
| Median | 17.3 |
| Total bilirubin(mg/dl) | |
| Range | 0.25–0.97 |
| Mean ± SD | 0.5 ± 0.2 |
| Median | 0.32 |
| Creatinin(mg/dl) | |
| Range | 0.46–1 |
| Mean ± SD | 0.7 ± 0.2 |
| Median | 0.5 |
| Micobiological culture | |
| Klebsiella spp. | 2(8.3 %) |
| E. Coli | 3(12.5 %) |
| Proteus spp. | 1(4.2 %) |
| Mixed GIT flora | 10(41.7 %) |
| Missing data | 8(33.3 %) |
| Fecal occult blood (FOB) | |
| Yes | 10(41.7 %) |
| No | 6(25 %) |
| Missing data | 8(33.3 %) |
| H. Pylori Ag | |
| Yes | 6(25 %) |
| No | 10(41.7 %) |
| Missing data | 8(33.3 %) |
| Pathogenic protists infections | |
| Yes | 4(16.7 %) |
| No | 7(29.2 %) |
| Missing data | 13(54.2 %) |
| Microsatellite instability (MSI) | |
| MSI-HI | 6(25 %) |
| MSI-L | 12(50 %) |
| MSS | 5(20.8 %) |
| Missing data | 1(4.2 %) |
3.2. Gut microbiota patients results
As it‘s not always easy to get stool samples from patients; we succeeded to get only 16 stool samples. Regarding microbiological culture, only 10samplesout of these 16(62.5 %) revealed mixed GIT flora, 3 (18.75 %) had E. Coli, 2 (12.5 %) showed Klebsiella spp. and only 1 (6.25 %) revealed Proteus spp. Fecal occult blood was detected in 10/16 patients (62.5 %) and H. pylori was found in 6/16 patients (37.5 %).Blastocystis hominis cysts were detected in only 4 patients.
3.3. MSI status
Among the examined 24 CRC patients, 12 patients (50 %) had MSI-low (MSI-L) CRC, 6 patients (25 %) had MSI-high (MSI-HI) CRC and 5 patients (20.8 %) were MSS CRC. PIK3CA, TP53, KIT & KDR missense genetic mutations were found in 6, 11, 3 & 2 MSI-L CRC patients, respectively and in 2, 3, 0 & 2 MSI-HI CRC patients, respectively.
3.4. NGS results
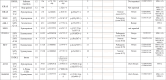
Twenty four CRC patients were enrolled in our study and their mutational analysis were described in details in Table 2. Substitution – Missense mutations were detected in 7 genes as follows: Tumor protein TP53 (TP53) drug responsive genetic mutation (c.215C > G) was found in 18 patients. Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA) benign c.1173A > G variant was detected in 10 patients. Kinase Insert Domain Receptor (KDR) gene is a Vascular Endothelial Growth Factor Receptor 2 (VEGFR2) and KDR c.1416A > T genetic variant was found in 6 patients. Five patients showed likely benign missense genetic substitution in KIT Proto-Oncogene, Receptor Tyrosine Kinase (KIT)gene at codon 1621 with A > Csubstitution. Each APC Regulator of WNT Signaling Pathway (APC), fibroblast growth factor receptor 3 (FGFR3) and MET Proto-Oncogene, Receptor Tyrosine Kinase (MET)missense genetic substitutions was detected in only one patient. As we are working on cell free DNA (cfDNA),KRAS mutations were detected in only 3 patients (12.5 %), one had inframe insertion (homozygous),one had stop gained (heterozygous) mutation and the other had both inframe insertion & stop gained mutation.
Table 2.
List of activating mutations observed in the studied 24 Egyptian CRC patients:
 |
 |
 |
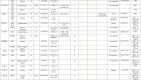 |
Comparison between different genetic mutations, clinical and laboratory analyses was described in Table 3. Incidence ofPIK3CAmissense mutations was found to be higher in patients more than 40 years old, males, average weight, stage III, and patients who had FOB. In those patients, MSI-L was 50 % while MSI-HI was 33.3 %.Regarding, TP53 missense mutation was higher in patients less than 40 years old (69.2 %), males (83.3 %), stage II (100 %), FOB positive (70 %), H. pylori positive (66.7 %) and MSI-L (91.7 %).
Table 3.
Comparison of most common genetic mutations in the studied 24 Egyptian CRC patients.
| No. of patients (%) | PIK3CA |
P value | TP53 |
P value | KIT |
P value | KDR |
P value | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Missense mutation | Others | Missense mutation | Others | Missense mutation | Others | Missense mutation | Others | |||||
| Age: | ||||||||||||
| ≤40 yrs | 2/6 (33.3 %) | 4/6 (66.7 %) | 0.63 | 6/6 (100 %) |
0/6 | 0.26 | 1/6 (16.7 %) | 5/6 (83.3 %) | 1 | 3/6 (50 %) |
3/6 (50 %) | 0.32 |
| >40 yrs | 7/13 (53.8 %) |
6/13 (46.2 %) | 9/13 (69.2 %) |
4/13 (30.8 %) | 4/13 (30.8 %) | 9/13 (69.2 %) |
3/13 (23.1 %) | 10/13 (76.9 %) |
||||
| Gender: | ||||||||||||
| Female | 3/9 (33.3 %) | 6/9 (66.7 %) | 0.39 | 7/9 (77.8 %) | 2/9 (22.2 %) | 1 | 3/9 (33.3 %) | 6/9 (66.7 %) | 0.61 | 2/9 (22.2 %) | 7/9 (77.8 %) | 0.66 |
| Male | 7/12 (58.3 %) | 5/12 (41.7 %) | 10/12 (83.3 %) | 2/12 (16.7 %) | 2/12 (16.7 %) | 10/12 (83.3 %) | 4/12 (33.3 %) | 8/12 (66.7 %) | ||||
| BMI: | ||||||||||||
| Average | 4/5 (80 %) |
1/5 (20 %) |
0.26 | 4/5 (80 %) |
1/5 (20 %) |
0.84 | 1/5 (20 %) |
4/5 (80 %) |
0.79 | 1/5 (20 %) |
4/5 (80 %) |
0.25 |
| Overweight | 2/5 (40 %) |
3/5 (60 %) |
4/5 (80 %) |
1/5 (20 %) |
2/5 (40 %) |
3/5 (60 %) |
3/5 (60 %) |
2/5 (40 %) |
||||
| Obese | 2/6 (33.3 %) | 4/6 (66.7 %) | 4/6 (66.7 %) | 2/6 (33.3 %) | 2/6 (33.3 %) | 4/6 (66.7 %) | 1/6 (16.7 %) | 5/6 (83.3 %) | ||||
| Stage: | ||||||||||||
| II | 2/5 (40 %) |
3/5 (60 %) |
0.54 | 5/5 (100 %) | 0/5 | 0.37 | 1/5 (20 %) |
4/5 (80 %) |
0.81 | 1/5 (20 %) |
4/5 (80 %) |
0.88 |
| III | 6/10 (60 %) |
4/10 (40 %) |
7/10 (70 %) | 3/10 (30 %) |
3/10 (30 %) |
7/10 (70 %) | 3/10 (30 %) |
7/10 (70 %) | ||||
| IV | 2/6 (33.3 %) | 4/6 (66.7 %) | 5/6 (83.3 % |
1/6 (16.7 %) | 1/6 (16.7 %) | 5/6 (83.3 % |
2/6 (33.3 %) | 4/6 (66.7 %) |
||||
| Site: | ||||||||||||
| Colon | 5/14 (35.7 %) |
9/14 (64.3 %) |
0.19 | 11/14 (78.6 %) |
3/14 (21.4 %) | 0.77 | 3/14 (21.4 %) | 11/14 (78.6 %) |
0.49 | 6/14 (42.9 %) | 8/14 (57.1 %) | 0.12 |
| Colorectal | 2/2 (100 %) | 0/2 | 2/2 (100 %) | 0/2 | 0/2 | 2/2 (100 %) | 0/2 | 2/2 (100 %) | ||||
| Rectal | 3/5 (60 %) |
2/5 (40 %) |
4/5 (80 %) |
1/5 (20 %) |
2/5 (40 %) |
3/5 (60 %) |
0/5 | 5/5 (100 %) |
||||
| FOB: | ||||||||||||
| Negative | 0/6 | 6/6 (100 %) |
0.09 | 5/6 (83.3 %) | 1/6 (16.7 %) | 1 | 1/6 (16.7 %) | 5/6 (83.3 %) | 1 | 3/6 (50 %) |
3/6 (50 %) |
0.29 |
| Positive | 5/10 (50 %) |
5/10 (50 %) |
7/10 (70 %) | 3/10 (30 %) | 3/10 (30 %) | 7/10 (70 %) | 2/10 (20 %) |
8/10 (80 %) | ||||
| H. pylori: | ||||||||||||
| Negative | 2/10 (20 %) | 8/10 (80 %) |
0.29 | 8/10 (80 %) |
2/10 (20 %) |
0.6 | 2/10 (20 %) |
8/10 (80 %) |
0.6 | 4/10 (40 %) | 6/10 (60 %) |
0.59 |
| Positive | 3/6 (30 %) |
3/6 (30 %) |
4/6 (66.7 %) |
2/6 (33.3 %) |
2/6 (33.3 %) |
4/6 (66.7 %) |
||||||
| Blastocyst: | ||||||||||||
| Negative | 2/7 (28.6 %) |
5/7 (71.4 %) |
1 | 5/7 (71.4 %) |
2/7 (28.6 %) |
0.49 | 2/7 (28.6 %) |
5/7 (71.4 %) |
0.49 | 3/7 (42.9 %) | 4/7 (57.1 %) | 1 |
| Positive | 1/4 (25 %) |
3/4 (75 %) |
4/4 (100 %) |
0/4 | 0/4 | 4/4 (100 %) |
||||||
| MSI status: | ||||||||||||
| MSI-L | 6/12 (50 %) |
6/12 (50 %) |
0.49 | 11/12 (91.7 %) |
1/12 (8.3 %) |
0.12 | 3/12 (25 %) |
9/12 (75 %) |
0.4 | 2/12 (16.7 %) |
10/12 (83.3 %) |
0.54 |
| MSI-HI | 2/6 (33.3 %) |
4/6 (66.7 %) |
3/6 (30 %) |
3/6 (30 %) |
0/6 | 6/6 (100 %) |
2/6 (33.3 %) | 4/6 (66.7 %) |
||||
| MSS | 1/5 (20 %) |
4/5 (80 %) |
3/5 (60 %) |
2/5 (40 %) |
1/5 (20 %) |
4/5 (80 %) |
2/5 (40 %) |
3/5 (60 %) |
||||
Regarding characteristics of the 5 patients with rectal mass, 4 patients (80 %) were males. Four patients (80 %) were late onset CRC (above 40 years) and only one was 24 years old harboring MSI-HI, PIK3CA c.1173A > G & TP53 c.215C > G mutations. Details described in Table 4, Table 5.
Table 4.
Characteristics of rectal cancer patients.
| Patient’s characteristics | Total No. = 5 |
|---|---|
| No. (%) | |
| Gender: | |
| Males | 4(80 %) |
| Females | 1 (20 %) |
| Age (years): | |
| Range | 24–71 |
| Mean ± SD | 47 ± 19.4 |
| Median | 46.5 |
| >40 | 4(80 %) |
| ≤40 | 1 (20 %) |
| Intestinal obstruction: | |
| Yes | 1 (20 %) |
| No | 4(80 %) |
| Stage at diagnosis | |
| II | 2(40 %) |
| III | 2(40 %) |
| IV | 1(20 %) |
| Lymphovascular invasion (LVI) | |
| Yes | 1 (20 %) |
| No | 4(80 %) |
| Micobiological culture | |
| Klebsiella spp | 2(40 %) |
| E. Coli | 1(20 %) |
| Mixed GIT flora | 2(40 %) |
| Fecal occult blood (FOB) | |
| Yes | 3(60 %) |
| No | 2(40 %) |
| H. Pylori Ag | |
| Yes | 3(60 %) |
| No | 2(40 %) |
| Microsatellite instability (MSI) | |
| MSI-HI | 1(20 %) |
| MSI-L | 4(80 %) |
Table 5.
Detailed mutational analysis of rectal cancer patients.
| Patient | Gender | Age (years) | Intestinal obstruction | Stage at diagnosis | Lymphovascular invasion (LVI) | Micobiological culture | Fecal occult blood (FOB) | H. Pylori Ag | Blastocystis hominis | MSI status | Missense genetic mutations |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Male | 50 | No | II | No | E. Coli | Positive | Positive | Negative | MSI-L | PIK3CA c.1173A > G TP53 c.215C > G KIT c.1621A > C |
| 2 | Female | 71 | No | III | yes | Klebsiella spp | Positive | Positive | – | MSI-L | APC c.2608C > T FGFR3 c.1156 T > C |
| 3 | Male | 55 | No | IV | No | Klebsiella spp | Positive | Positive | Positive | MSI-L | PIK3CA c.1173A > G TP53 c.215C > G |
| 4 | Male | 43 | Yes | II | No | Negative | MSI-L | TP53 c.215C > G | |||
| 5 | Male | 24 | No | III | No | – | – | – | – | MSI-HI | PIK3CA c.1173A > G TP53 c.215C > G |
Six patients had metastatic CRC, 3 patients (50 %) were males and 3 patients (50 %) were females. Their ages ranging from 35 to 57 years with median age of 49.5 years and 5 patients (83.3 %) were above 40 years. Regarding their anatomical site, 5 patients (83.3 %) had colon cancer and only one had stage rectal tumor. Regarding the gut microbiota examination, 4 patients revealed mixed GIT flora, one patient had Klebsiella spp. and one revealed E. Coli. Only one out of 6 metastatic cancer patients had H. pylori & fecal occult blood. Detailed mutational analysis of the metastatic cancer patients described in Table 6.
Table 6.
Characteristics of metastatic CRC patients.
| Patient | Gender | Age (years) | Site | Intestinal obstruction | Site of metastasis | Lymphovascular invasion (LVI) | Micobiological culture | Fecal occult blood (FOB) | H. Pylori Ag | Blastocystis hominis | MSI status | Missense genetic mutations |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Male | 55 | Rectum | No | Liver | Yes | Klebsiella spp | Positive | Positive | Positive | MSI-L | PIK3CA c.1173A > G TP53 c.215C > G |
| 2 | Female | 57 | Colon | No | Liver | No | Mixed GIT flora | Negative | Negative | Negative | MSS | KIT c.1621A > C KDR c.1416A > T |
| 3 | Female | 49 | Colon | No | Liver | No | E. Coli | Negative | Negative | – | MSS | TP53 c.215C > G |
| 4 | Male | 35 | Colon | No | Peritoneum& LN | No | Mixed GIT flora | Negative | Negative | Positive | MSI-HI | TP53 c.215C > G KDR c.1416A > T |
| 5 | Male | 48 | Colon | No | Liver, pleural & LN | No | Mixed GIT flora | Negative | Negative | – | MSI-HI | PIK3CA c.1173A > G TP53 c.215C > G |
| 6 | Female | 50 | Colon | No | – | No | Mixed GIT flora | Negative | Negative | Negative | MSI-L | TP53 c.215C > G |
3.5. H. pylori positive CRC patients in relation to NGS results
Six patients were H. pylori positive with5/6 patients (83.3 %) were males and their median ages of 52.5 years. Three patients (50 %) had a primary tumor in rectum, 2 patients (33.3 %) had tumor in colon tumor and only one patient (16.7 %) had rectosigmoid mass. Most of those patients were MSI-L (83.3 %) with the most common mutations were TP53 c.215C > G: 4 patients, PIK3CAc.1173A > G: 3 patients & KIT c.1621A > C: 2 patients. Detailed characteristics of H. pylori positive CRC patients described in Table 7.
Table 7.
Characteristics of H. pylori positive CRC patients.
| Patient | Gender | Age (years) | Site | Intestinal obstruction | Site of metastasis | Lymphovascular invasion (LVI) | Stage | Micobiological culture | Fecal occult blood (FOB) | Blastocystis hominis | MSI status | Missense genetic mutations |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Male | 50 | Rectum | No | – | No | II | E. Coli | Positive | Negative | MSI-L | PIK3CA c.1173A > G TP53 c.215C > G KIT c.1621A > C |
| 2 | Female | 71 | Rectum | No | – | Yes | III | Klebsiella spp | Positive | – | MSI-L | KIT c.1621A > C APC c.2608C > T FGFR3 c.1156 T > C |
| 3 | Male | 55 | Rectum | No | Liver | Yes | IV | Klebsiella spp | Positive | Positive | MSI-L | PIK3CA c.1173A > G TP53 c.215C > G |
| 4 | Male | 40 | Colon | No | – | No | III | E. Coli | Positive | – | MSI-L | TP53 c.215C > G KDR c.1416A > T |
| 5 | Male | 69 | rectosigmoid | No | – | No | III | Mixed GIT flora | Positive | Negative | MSI-L | PIK3CA c.1173A > G TP53 c.215C > G |
| 6 | Male | 49 | Colon | No | – | No | II | Mixed GIT flora | Positive | – | MSI-HI | No missense mutations |
3.6. Late versus early onset CRC patients
Thirteen patients (54.2 %) above 40 years considered as late onset CRCs with 53.8 %of those patients were females. The vast majority of metastatic patients were late onset CRC. Regarding MSI status, 5/12 patients (41.7 %) were MSS, 5/12 patients (41.7 %) were MSI-L and only 2 Patients (16.6 %) were MSI-HI. The most common missense substitutions were found in TP53, PIK3CA, KDR, KIT, APC, FGFR3&MET, respectively. Eleven patients (45.8 %) less than or equal 40 years considered as early onset CRC patients with 54.5 % were males. The most common clinical stages were stage III which was found in 4 patients (36.4 %) & stage IV which was found in 3 patients (27.3 %). Regarding MSI status, 7 patients (63.6 %) were MSI-L and 4 Patients (36.4 %) were MSI-HI. The most common missense substitutions were found in TP53,PIK3CA, KDR, KIT. Detailed characteristics of late and early onset CRC patients were described in Table 8. According to our study, 2 male patients below 40 years, one of them was 35 years old with lymph node and peritoneum metastasis expressing TP53 c.215C > G& KDR c.1416A > T with MSI-HI favoring new treatment approaches. The other one was 37 years with no metastasis expressing PIK3CA c.1173A > G in addition to TP53 c.215C > G & KDR c.1416A > T and MSI-L considered target for PI3K inhibitors. Another male patient aged 48 years old that could have good prognosis although with metastasis to liver and lymph nodes expressing PIK3CA c.1173A > G and MSI-HI.
Table 8.
Late and early onset CRCs.
| Patient’s characteristics | Total No. = 24 |
|---|---|
| No. (%) | |
| Late onset CRCs (≤40 years old) | 13/24 (54.2 %) |
| Gender: | |
| Males | 6 (46.2 %) |
| Females | 7 (53.8 %) |
| Tumor site: | |
| Colon | 7 (53.8 %) |
| Rectum | 4 (30.8 %) |
| Recto-sigmoid | 2 (15.4 %) |
| Micobiological culture | |
| Klebsiella spp. | 1/8(12.5 %) |
| E. Coli | 2/8(25 %) |
| Mixed GIT flora | 5/8(62.5 %) |
| Missing data | 5 |
| Fecal occult blood (FOB) | |
| Yes | 6/8(75 %) |
| No | 2/8(25 %) |
| Missing data | 5 |
| H. Pylori Ag | |
| Yes | 3/8(37.5 %) |
| No | 5/8(62.5 %) |
| Missing data | 5 |
| Microsatellite instability (MSI) | |
| MSI-HI | 2/12 (16.6 %) |
| MSI-L | 5/12 (41.7 %) |
| MSS | 5/12 (41.7 %) |
| Missing data | 1 |
| Early onset CRCs (>40 years old). | 11/24 (45.8 %) |
| Gender. | |
| Males | 6/11 (54.5 %) |
| Females | 5/11 (45.5 %) |
| Tumor site: | |
| Colon | 7/ 11 (63.6 %) |
| Rectum | 1/11 (9.1 %) |
| Recto-sigmoid | 3/11 (27.3 %) |
| Micobiological culture | |
| Klebsiella spp. | 1/8 (12.5 %) |
| E. Coli | 1/8 (12.5 %) |
| Proteus spp. | 1/8 (12.5 %) |
| Mixed GIT flora | 5/8 (62.5 %) |
| Missing data | 3 |
| Fecal occult blood (FOB) | |
| Yes | 4/8 (50 %) |
| No | 4/8 (50 %) |
| Missing data | 3 |
| H. Pylori Ag | |
| Yes | 3/8(37.5 %) |
| No | 5/8(62.5 %) |
| Missing data | 3 |
| Microsatellite instability (MSI) | |
| MSI-HI | 4/11 (36.4 %) |
| MSI-L | 7/11 (63.6 %) |
4. Discussion
NGS mutational analysis using liquid biopsy is becoming more popular for being minimally invasive, a highly accurate, sensitive, and specific approach for cell-free DNA genomic profiling to supplement tissue testing and inform treatment decisions.18 In this research, we aimed at detection of activating genetic mutations & MSI status in cfDNA of 24 Egyptian CRC patients, in addition to evaluating the incidence of dysbiotic intestinal microbiota and pathogenic protists among those patients. Many lifestyle factors are associated with increased risk of CRC such as lack of physical activity, obesity, a diet low in fresh fruits and vegetables, a low-fiber & high-fat diet or diet high in processed meat. This was obviously clear in our study, as all patients were non-vegetarian except for one patient. This vegetarian patient was a 40 years male presenting with stage III tumor in transverse colon. He was positive for E. Coli, H. pylori and FOB harboring substitution – missense mutations in KDRc.1416A > T &TP53 c.215C > G with MSI-L status. Our patients age ranges from 24 to 71 years old with the vast majority were above 40 yearsand45.8 % were less than or equal 40 years. This is in accordance to Sanjoaquin et al., 2004 which revealed that 95 % of CRC were recorded after 17 years with risk increased in association with smoking, alcohol, and white bread consumption, and decreased with frequent consumption of fruit. The relative risk in vegetarians compared with nonvegetarians was 0.85 (95 % CI: 0.55–1.32).19
Regarding our mutational analysis, 75 % of our patients had substitution – missense mutations in TP53gene, 42 % had PIK3CA, 25 % had KDR and 21 % had KIT with mutation in APC, FGFR3 and MET genes were found in only 4 % of cases. We used AmpliSeq for Illumina Cancer Hotspot Panel v2 which is a targeted resequencing assay designed to investigate somatic mutations in the hotspot regions of 50 genes known to be associated with cancer. Its low input requirement allows for use with a variety of sample types, including formalin-fixed, paraffin-embedded (FFPE) tissues. Our study found that KRAS mutations were detected in only 3 patients (12.5 %). Of these, one patient had a homozygous inframe insertion, another had a heterozygous stop-gain mutation, and the third had both an inframe insertion and a stop-gain mutation. It is important to note that the panel specifically targets SNPs and SNVs in Exons 2 and 3 of the KRAS gene.
As 42 % of our patients harboring PIK3CA mutations; PI3K inhibitors could be considered as an effective targeted therapy. One-fourth of cases (6 cases) were metastatic, harboring variants in the following genes: TP53: 5/6 patients; PIK3CA: 2/6 patients; KIT & KDR were found in only one patient each. The analysis done by Szász et al., 2023 revealed that 7 blood samples (43.8 %) in his study had 13 mutations in the following genes: TP53: 5 samples; KRAS: 4 samples; NTRK3: 2 samples; SMAD4: 1 sample; BRAF: 1 sample, however; 9 samples (56.2 %) did not show any mutations and 1/3 of his samples were from metastatic patients with highest variant allele frequency was observed in the KRAS gene.20 Another study done by Cancer Genome Atlas (TCGA) network revealed that the most commonly altered genes were mainly APC, TP53, KRAS, PIK3CA and BRAF in their CRC patients.21 Several studies compared the mutational status of liquid biopsy and tumor tissue from the same CRC patient such as Mazouji et al., 2021 that defined the diagnostic and predictive importance of testing liquid biopsy specimen. He frequently found KRAS, BRAF, APC, and TP53 genetic alterations in the ctDNA of CRC patients having significant concordance, up to 100 % with tumor tissue of thatpatients.22 Also,Verner et al., 2023 highlighted the analytical validation of the circulating cell-free DNA genomic profiling test, including accuracy compared with conventional methods, as well as sensitivity, specificity, precision, reproducibility, and repeatability. He described that concordance with conventional methods showed percent positive agreement of 98.7 %, 89.3 %, and 96.2 % for single nucleotide variants (SNVs), insertion/deletions (indels), and copy number amplifications (CNAs), respectively, and 100.0 % for translocations and MSI. Analytical sensitivity revealed a median limit of detection of 0.7 % and 0.6 % for SNVs and indels, 1.4-fold for CNAs, 0.5 % variant allele frequency for translocations, and 0.6 % for MSI. Specificity was >99 % for SNVs/indels and 100 % for CNAs, translocations, and MSI. Average positive agreement from precision, reproducibility, and repeatability experiments was 97.5 % and 88.9 % for SNVs/indels and CNAs, and 100 % for translocations and MSI.23
MSI occurs as a result of mutation in mismatch repair (MMR) genes and its examination is considered as a prognostic marker &molecular feature used to classify CRCs into MSI-HI, MSI-L or MSS tumors. MSI-HI CRCs have the best prognosis with immune checkpoint inhibitors approved to treat those patients. Recent researches testing MSI status in CRC liquid biopsy showed that prevalence of MSI-HI is approximately 12 to 20 % among CRCs with incidence in relation to clinical stage as follows: ∼ 20 % in stage I & II, 12 % in stage III & 4–5 % in stage IV.24 While, among our examined 24 CRC patients, 12 patients (50 %) had MSI-low (MSI-L) CRC, 6 patients (25 %) had MSI-high (MSI-HI) CRC and 5 patients (20.8 %) had MSS CRC. Our previous study in 2019 on surgical specimens revealed 11/19 (57.9 %) patients had MSI-HI CRC and 8/19 (42.1 %) patients had MSI-L CRC.11 Kumar et al.,2023 revealed that the immunotherapy nivolumab when added to FOLFOX in his positive MSI-HCRC patient, the patient's metastatic disease had a complete clinical response. This case highlights the complementary role of ctDNA testing for biomarker identification. By performing simultaneous ctDNA testing at the time of diagnosis, an actionable biomarker was discovered that significantly altered this patient's prognosis and treatment options.25
As proved in a number of studies, the microbioma affects tumor initiation and progression through direct effect on the tumor cells and indirectly through manipulation of the immune system. It can even determine response to cancer therapies and predict disease progression.26 In our study, 10 patients out of 16 examined patients (62.5 %) revealed mixed GIT flora, 3 patients (18.75 %) had E. Coli, 2 patients (12.5 %) showed Klebsiella spp. and only 1 patient (6.25 %) revealed Proteus spp. Fecal occult blood was detected in 10/16 patients (62.5 %) and H. pylori antigen was found in only 6/16 patients (37.5 %). Those patients who were H. pylori positive mostly expressing microsatellite instability (MSI-L) together with TP53 &PI3KCA gene mutations in ∼50 % of them. As previously reported, the CRC microbiota include different strains such as Bacteroides fragilis, Streptococcus gallolyticus, Enterococcusfaecalis and Escherichia coli, as well as Fusobacterium nucleatum, Parvimonas, Peptostreptococcus, Porphyromonas and Prevotella.27 Also, a positive association between H. pylori infection and the risk of CRC was noticed in meta-analysis of 47 studies including 17,416 CRC cases and 55,811 healthy controls with overall, H. pylori infection was associated with an increased risk of CRC (OR = 1.70 95 % CI 1.64–1.76, I2 = 97 %).28 It has been reported that exposure to H. pylori inhibited the proliferation of CD4 + T cells isolated from the blood, and this inhibition could be blocked by anti-PD-L1 antibodies. Researchers confirmed also that H. pylori infection enhanced PD-L1 expression in human gastrointestinal epithelial cells and that co-culture experiments of H. pylori infected gastrointestinal epithelial cells with primary human T cells induced T cell apoptosis.29, 30 As well as Blastocystis species remains one of the most common intestinal parasites in humans having incidence of ∼10 % in developed countries and up to 50–60 % in developingcountries.31 Here, Blastocystis hominis cysts were detected in only 4 patients out of 11 examined patients (36.4 %) with previous studies report Blastocystis frequencies in CRC patient in various countries as follows: Saudi Arabia (17.5 %), Malaysia (25.7 %), Jordan (25 %), Egypt (31 %), 25.78 % in Venezuela and 22.9 % in Argentina.32 Several reports revealed that microbiota and intestinal parasitic infections play an essential role in CRC initiation and development. In addition, prevention of CRC development and progression together with improvement of treatment efficacy could be done through diet modulation in cancer associated microbiota, through the intake of dietary elements that avoid dysbiosis &intestinal inflammation or modulate cancer treatment response.33.
5. Conclusion
The aim of our study was to summarize the most recent evidence on possible use of genetic biomarkers and microsatellite instability in relation to microbiota for diagnosis, prognosis and response to therapy in a group of Egyptian CRC patients. CRC is becoming a challenging problem in our society especially at younger ages; thus, it is essential to understand the disease genetic mutational status as well as contributing environmental stress factors such as gut microbiota dysbiosis and their interplay with patient genetic variations. Considering NGS as a laboratory cornerstone of precision oncology treatment, molecular characterization of CRC mutational status in liquid biopsy could be used as a non-invasive tool that will add a new perspective in clinical decision-making through providing valuable genetic information about each patient to improve diagnostic and prognostic procedures as well as response to targeted therapies for those patients. Moreover, CRC patients may benefit from MSI testing that can be helpful in guiding therapeutic decisions. Further studies in the field of precision medicine are needed to correlate variants found in liquid biopsy versus tumor tissue defining the predictive importance in clinical setting.
6. Declarations
6.2. Consent for publication
Obtained from all individual participants included in the study.
6.3. Availability of data and material
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
6.4. Authors' contributions
NK designed the study, analyzed and interpreted the clinical, microbiological & molecular data and has a valuable contribution in manuscript writing & editing. MS was responsible of molecular technical testing and correlations. MA, RA, RH & MR provided us with patients' specimens and responsible for demographic & clinical data collection. MG & AF collected the patients' stool samples & responsible of microbiological examination. ER & MA collected the patients' stool samples & responsible of parasitological examination. HK was responsible for data analysis, bioinformatics and contributor in writing the manuscript. All authors read and approved the final manuscript.
CRediT authorship contribution statement
Neemat M. Kassem: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Software, Project administration, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Mohamed G. Seadawy: Methodology, Investigation. Maha Gaafar: Methodology, Investigation. Hebatallah A. Kassem: Writing – original draft, Software, Project administration, Methodology, Investigation, Formal analysis, Data curation. Amira Farouk Ahmed Hussein: Methodology. Enas M. Ali Rizk: Methodology. Marwa A. Hassan: Investigation. Riham H. AbdelAziz: Data curation. Mentallah M. Abdelradi: Data curation. Mostafa F. El-Hosseny: Methodology. Mohamed Abdulla: Data curation. Rabab Abdel Moneim: Data curation.
6.1. Ethics approval and consent to participate
All procedures performed in the study involving human participants were in accordance with the ethical standards of the institutional research committee and with the 1964 Helsinki declaration and its later amendments (GCP guidelines) or comparable ethical standards.All samples were handled according to the rules and regulations by Cairo University.Our project was approved by Cairo University under code ≠ 39-2021. We started our study that was approved by our department IRB (unit A) bythe end of 2021
Funding
This study was approved and funded by Cairo University grant code 39-2021.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We deeply acknowledge the technical support offered by Biological prevention dep., Ministry of defense presented by Dr. Mohamed G. Seadawy. Deep sincere acknowledgment goes to the soul of Prof. Dr. Abdel Rahman Zekry for his immeasurable support given to this work.
Contributor Information
Neemat M. Kassem, Email: nkkassem@hotmail.com.
Mohamed G. Seadawy, Email: biologist202054@yahoo.com.
Maha Gaafar, Email: Gaafar.maha@yahoo.co.uk.
Hebatallah A. Kassem, Email: Heba.kasem@hotmail.com.
Amira Farouk Ahmed Hussein, Email: Amira.farouk@kasralainy.edu.eg.
Enas M. Ali Rizk, Email: enasrizk@kasralainy.edu.eg.
Marwa A. Hassan, Email: mahassan@kasralainy.edu.eg.
Riham H. AbdelAziz, Email: rihamhany@kasralainy.edu.eg.
Mentallah M. Abdelradi, Email: Dr.tma10@gmail.com.
Mostafa F. El-Hosseny, Email: mfm.memo@gmail.com.
Mohamed Abdulla, Email: mohamed.abdulla@oncology.clinic.org.
Rabab Abdel Moneim, Email: dr.rababahmed2014@hotmail.com.
References
- 1.Sung H., Ferlay J., Siegel R.L., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Abd El Kader Y., Emera G., Safwat E., Kassem H., Kassem N. The KRAS StripAssay for detection of KRAS mutation in Egyptian patients with colorectal cancer (CRC): a pilot study. J Egypt Natl Canc Ins. 2013;25:37–41. doi: 10.1016/j.jnci.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 3.Noack P., Langer R. Molecular pathology of colorectal cancer. Memo. 2023;16:116–121. doi: 10.1007/s12254-023-00893-2. [DOI] [Google Scholar]
- 4.Helmink B.A., Khan M.A.W., Hermann A., Gopalakrishnan V., Wargo J.A. The microbiome, cancer, and cancer therapy. Nat Med. 2019;25(3):377–388. doi: 10.1038/s41591-019-0377-7. [DOI] [PubMed] [Google Scholar]
- 5.Hussein E.M., Hussein A.M., Eida M.M., Atwa M.M. Pathophysiological variability of different genotypes of human Blastocystis hominis Egyptian isolates in experimentally infected rats. Parasitol Res. 2008;102:853–860. doi: 10.1007/s00436-007-0833-z. [DOI] [PubMed] [Google Scholar]
- 6.Kumarasamy V., Atroosh W.M., Anbazhagan D., Abdalla M.M.I., Azzani M. Association of Blastocystis hominis with colorectal cancer: a systematic review of in vitro and in vivo evidences. World J Gastrointest Oncol. 2022;14(3):734–745. doi: 10.4251/wjgo.v14.i3.734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wakai T., Prasoon P., Hirose Y., Shimada Y., Ichikawa H., Nagahashi M. Next-generation sequencing-based clinical sequencing: Toward precision medicine in solid tumors. Int J Clin Oncol. 2019;24:115–122. doi: 10.1007/s10147-018-1375-3. [DOI] [PubMed] [Google Scholar]
- 8.Normanno N., Cervantes A., Ciardiello F., De Luca A., Pinto C. The liquid biopsy in the management of colorectal cancer patients: current applications and future scenarios. Cancer Treat Rev. 2018;70:1–8. doi: 10.1016/j.ctrv.2018.07.007. [DOI] [PubMed] [Google Scholar]
- 9.Kassem N.M., Kassem H.A., Selim H., et al. Targeted next generation sequencing provides insight for the genetic alterations in liquid biopsy of Egyptian brain tumor patients. Egypt J Med Hum Genet. 2022;23:23. doi: 10.1186/s43042-022-00214-y. [DOI] [Google Scholar]
- 10.Kassem N., Kassem H., Kassem L., Hassan M. Detection of activating mutations in liquid biopsy of Egyptian breast cancer patients using targeted next-generation sequencing: a pilot study. J Egypt Natl Canc Inst. 2021;33(1):10. doi: 10.1186/s43046-021-00067-3. PMID: 33864517. [DOI] [PubMed] [Google Scholar]
- 11.Kassem N.M., Emera G., Kassem H.A., et al. Clinicopathological features of Egyptian colorectal cancer patients regarding somatic genetic mutations especially in KRAS gene and microsatellite instability status: a pilot study. Egypt J Med Hum Genet. 2019;20:20. [Google Scholar]
- 12.Al-Abri S.S., Beeching N.J., Nye F.J. Traveller’s diarrhoea. Lancet Infect Dis. 2005;5:349–360. doi: 10.1016/S1473-3099(05)70139-0. [DOI] [PubMed] [Google Scholar]
- 13.Weissfeld A., Baselski V. In: Clinical microbiology procedures handbook. Garcia L.S., editor. American Society for Microbiology; Washington DC: 2010. Specimen receipt and accessioning. section 1. Aerobic bacteriology, 1.2.1-4. [Google Scholar]
- 14.Yamamoto M., Nakama H. Cost-effectiveness analysis of immunochemical occult blood screening for colorectal cancer among three fecal sampling methods. Hepatogastroenterology. 2000;47(32):396–399. [PubMed] [Google Scholar]
- 15.Altman E, Fernanez H. et al Analysis of Helicobacter pylori isolates from Chile: Occurrence of selective type I Lewis b antigen expression in lipopolysaccharide. 2008, 57(pt 5): 585. [DOI] [PubMed]
- 16.Garcia L.S. 5th ed. ASM Press; Washington, DC., USA: 2007. Diagnostic medical parasitology; pp. 27–32. [Google Scholar]
- 17.Jones D.R. A medium for investigatingthe breakdown of pectin by bacteria. Nature. 1946;158(4018):625. doi: 10.1038/158625b0. [DOI] [PubMed] [Google Scholar]
- 18.Mazouji O., Ouhajjou A., Anouar N., et al. Mutational profiling using liquid biopsy to guide targeted therapy in patients with metastatic cancer. Sci Rep. 2025;15:11135. doi: 10.1038/s41598-025-88094-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanjoaquin M.A., Appleby P.N., Thorogood M., Mann J.I., Key T.J. Nutrition, lifestyle and colorectal cancer incidence: a prospective investigation of 10998 vegetarians and non-vegetarians in the United Kingdom. Br J Cancer. 2004;90(1):118–121. doi: 10.1038/sj.bjc.6601441. PMID: 14710217; PMCID: PMC2395312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szász I, Kiss T, Mokánszki A, et al. Identification of liquid biopsy-based mutations in colorectal cancer by targeted sequencing assays, Molecular and Cellular Probes, 67, 2023,101888, ISSN 0890-8508, https://doi.org/10.1016/j.mcp.2022.101888. [DOI] [PubMed]
- 21.Zhou H., Zhu L., Song J., et al. Liquid biopsy at the frontier of detection, prognosis and progression monitoring in colorectal cancer. Mol Cancer. 2022;21:86. doi: 10.1186/s12943-022-01556-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mazouji O., Ouhajjou A., Incitti R., Mansour H. Updates on clinical use of liquid biopsy in colorectal cancer screening, diagnosis, follow-up, and treatment guidance. Front Cell Dev Biol. 2021;24(9) doi: 10.3389/fcell.2021.660924. PMID: 34150757; PMCID: PMC8213391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verner EL, Jackson JB, Severson E, et al. Validation of the labcorp plasma focus test to facilitate precision oncology through cell-free DNA genomic profiling of solid tumors. J Mol Diagn. 2023 Jul;25(7):477-489. doi: 10.1016/j.jmoldx.2023.03.008. Epub 2023 Apr 15. Erratum in: J Mol Diagn. 2023 Nov;25(11):849. doi: 10.1016/j.jmoldx.2023.08.001. PMID: 37068734. [DOI] [PubMed]
- 24.Tieng F.Y.F., Abu N., Lee L.H., Ab Mutalib N.S. Microsatellite instability in colorectal cancer liquid biopsy-current updates on its potential in non-invasive detection, prognosis and as a predictive marker. Diagnostics (Basel). 2021;11(3):544. doi: 10.3390/diagnostics11030544. PMID: 33803882; PMCID: PMC8003257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar A., Green M., Thacker J., Jeck W.R., Strickler J.H. Circulating tumor DNA testing overcomes limitations of comprehensive genomic profiling from tumor tissue. Case Rep Oncol. 2023;16(1):210–217. doi: 10.1159/000529813. PMID: 37064498; PMCID: PMC10091230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Y., Wu F.H., Wu P.Q., Xing H.Y., Ma T. The role of the tumor microbiome in tumor development and its treatment. Front Immunol. 2022;15(13) doi: 10.3389/fimmu.2022.935846. PMID: 35911695; PMCID: PMC9334697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rebersek M. Gut microbiome and its role in colorectal cancer. BMC Cancer. 2021;21:1325. doi: 10.1186/s12885-021-09054-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zuo, Yuling PhDa; Jing, Zhao PhDa; Bie, Mingjiang PhDb; Xu, Chunyan MSc; Hao, Xinyu PhDd,∗; Wang, Baoning PhDe,∗. Association between Helicobacter pylori infection and the risk of colorectal cancer: a systematic review and meta-analysis. Medicine 99(37):p e21832, September 11, 2020. | doi: 10.1097/MD.0000000000021832. [DOI] [PMC free article] [PubMed]
- 29.Nasr R., Shamseddine A., Mukherji D., Nassar F., Temraz S. The crosstalk between microbiome and immune response in gastric cancer. Int J Mol Sci. 2020;21(18):6586. doi: 10.3390/ijms21186586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu Y.Y., Lin C.W., Cheng K.S., et al. Increased programmed death-ligand-1 expression in human gastric epithelial cells in helicobacter pylori infection. Clin Exp Immunol. 2010;161(3):551–559. doi: 10.1111/j.1365-2249.2010.04217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tan K.S. Blastocystis in humans and animals: new insights using modern methodologies. Vet Parasitol. 2004;126(1–2):121–144. doi: 10.1016/j.vetpar.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 32.Mohamed A.M., Ahmed M.A., Ahmed S.A., Al-Semany S.A., Alghamdi S.S., Zaglool D.A. Predominance and association risk of Blastocystis hominis subtype I in colorectal cancer: a case control study. Infect Agent Cancer. 2017;12(12):21. doi: 10.1186/s13027-017-0131-z. PMID: 28413436; PMCID: PMC5389010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sánchez-Alcoholado L., Ramos-Molina B., Otero A., et al. The role of the gut microbiome in colorectal cancer development and therapy response. Cancers (Basel). 2020;12(6):1406. doi: 10.3390/cancers12061406. PMID: 32486066; PMCID: PMC7352899. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.


