Abstract
Activating and inhibitory phosphorylation mechanisms play an essential role in regulating Raf kinase activity. Here we demonstrate that phosphorylation of C-Raf in the kinase activation loop (residues T491 and S494) is necessary, but not sufficient, for activation. C-Raf has additional activating phosphorylation sites at S338 and Y341. Mutating all four of these residues to acidic residues, S338D/Y341D/T491E/S494D (DDED), in C-Raf results in constitutive activity. However, acidic residue substitutions at the corresponding activation loop sites in B-Raf are sufficient to confer constitutive activity. B-Raf and C-Raf also utilize similar inhibitory phosphorylation mechanisms to regulate kinase activity. B-Raf has multiple inhibitory phosphorylation sites necessary for full kinase inhibition where C-Raf requires only one. We examined the functional significance of these inhibitory and activating phosphorylations in Caenorhabditis elegans lin-45 Raf. Eliminating the inhibitory phosphorylation or mimicking activating phosphorylation sites is sufficient to confer constitutive activity upon lin-45 Raf and induce multi-vulva phenotypes in C.elegans. Our results demonstrate that different members of the Raf family kinases have both common and distinct phosphorylation mechanisms to regulate kinase activity and biological function.
Keywords: B-Raf/C-Raf/kinase/lin-45/phosphorylation
Introduction
The Raf serine/threonine kinases are key signal transducers of diverse extracellular stimuli that activate the mitogen-activated protein kinase (MAPK) signaling pathway. Receptor-mediated activation of the small GTPase Ras recruits Raf to the plasma membrane where Raf kinase activity is regulated by a mechanism not completely understood (Morrison and Cutler, 1997). Raf propagates signals by activating the dual specificity kinase MEK (MAPK/ERK kinase), which then activates MAPK/ERK (extracellular signal-regulated kinase). Activated ERK phosphorylates a variety of different regulatory proteins including nuclear transcription factors such as Elk-1 and Myc as well as many cytoplasmic proteins (Kerkhoff and Rapp, 1998; Kerkhoff et al., 1998).
Raf kinases have three conserved regions, CR1, CR2 and CR3 (Morrison and Cutler, 1997). The CR1 region consists of two domains, a Ras-binding domain (RBD) and a cysteine-rich domain, both of which bind Ras (Vojtek et al., 1993; Nassar et al., 1995; Morrison and Cutler, 1997). The CR2 region is rich in serines and threonines and is the site of several phosphorylation events in C-Raf (Morrison et al., 1993; Yao et al., 1995; Zimmermann and Moelling, 1999). The CR3 region is the kinase domain that is regulated by phosphorylation events. Despite the high sequence and structural similarities between the three mammalian Raf isoforms, A-, B- and C-Raf are regulated differently. For example, although all Raf isoforms require activated Ras, the small GTPase Rap-1 is involved in B-Raf but not C-Raf activation in PC12 cells (Vossler et al., 1997).
Raf kinase activity is controlled by multiple pathways which utilize phosphorylation as a key mechanism of regulation. Phosphorylation of residue S621 is necessary for C-Raf and 14-3-3 protein interactions (Morrison et al., 1993; Tzivion et al., 1998; Yip-Schneider et al., 2000). Residues T268/T269 have been shown to be autophosphorylated as well as Ksr phosphorylation sites (Morrison et al., 1993; Yao et al., 1995). The inhibition of C-Raf kinase activity by cyclic AMP originally identified a protein kinase A phosphorylation site at residue S43 (Morrison et al., 1993; Wu et al., 1993). However, recent studies have shown that the phosphorylation state of S43 has no effect on Raf activity and that cyclic AMP can also inhibit B-Raf activity, which lacks a homologous site (Sidovar et al., 2000). Furthermore, S497/S499 were identified as protein kinase C (PKC) phosphorylation sites (Kolch et al., 1993; Carroll and May, 1994); however, this study as well as other laboratories (Barnard et al., 1998; Schonwasser et al., 1998) has shown that residues S497 and S499 are not necessary for C-Raf activity.
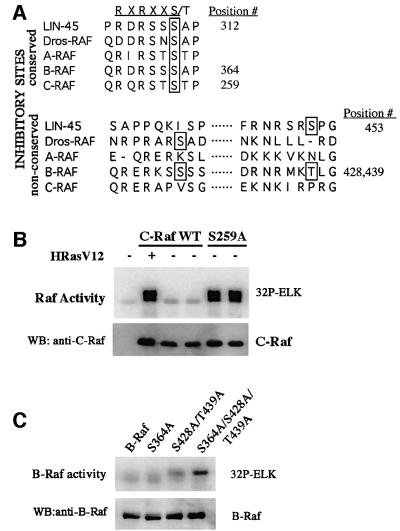
Raf kinases are also negatively regulated by phosphorylation by Akt (also known as protein kinase B). There are Akt consensus phosphorylation sites (RXRXXS/T) in all three Raf isoforms as well as in the Caenorhabditis elegans lin-45 Raf (Guan et al., 2000). There is a divergence, however, in the number of consensus sites in the different Raf kinases and where they are positioned in the protein. Substituting these phosphorylation sites to alanine results in loss of inhibition and higher basal activity (Zimmermann and Moelling, 1999; Guan et al., 2000). Recently, the serine/threonine phosphatase PP2A has been shown to complex with C-Raf and affect the phosphorylation state of S259 (Abraham et al., 2000). The balance between phosphorylation states of critical residues is an essential factor in Raf regulation.
Two important phosphorylation sites have proved to be critical for the activation of C-Raf. One phosphorylation event occurs on Y341 by Src kinase (Fabian et al., 1993; Marais et al., 1995). The tyrosine at this position is conserved in A-Raf and C-Raf, but is substituted with an aspartic acid residue in B-Raf and in C.elegans lin-45 Raf. Maximal activation of A-Raf and C-Raf requires both Ras and Src. In contrast, B-Raf, which does not have a tyrosine at this position, is maximally induced by Ras alone (Marais et al., 1997). The recruitment of Raf to the plasma membrane by activated Ras is essential for tyrosyl phosphorylation by membrane-bound Src (Marais et al., 1995). Loss of the RBD results in the loss of Src-inducible phosphorylation and Raf activity. Similarly, the substitution of Y341 to phenylalanine results in a loss of Raf activity (Mason et al., 1999). The difference between B-Raf and C-Raf in Src-stimulated phosphorylation and kinase activation demonstrates divergent mechanisms in regulating Raf proteins.
Another critical phosphorylation event on C-Raf occurs at S338 (Morrison et al., 1993; Mason et al., 1999). This serine residue is conserved among the Raf isoforms; however, unlike C-Raf, the phosphorylation state of S445 in B-Raf is constitutive (Mason et al., 1999). The constitutive phosphorylation of S445 in B-Raf may increase its basal kinase activity, which can be stimulated further by activated Ras (Mason et al., 1999). For C-Raf, phosphorylation on S338 is stimulated by Ras (Diaz et al., 1997; King et al., 1998; Mason et al., 1999). Substitution of S338 to alanine diminishes C-Raf activation by Ras as well as by Src and other mitogenic factors. p21-activated protein kinase (Pak) has been identified as a regulator of C-Raf by phosphorylating S338 (King et al., 1998). The Pak family kinases are regulated by the Rho family of small GTPases, specifically CDC42 and Rac. An alternative Ras-effector pathway involving PI3-kinase can activate Rac. Residue S338 appears to be important not only for C-Raf activity but also for integrating signals from different pathways.
The complexity of Raf regulation is further revealed by the recent identification of two phosphorylation sites in the activation loop of B-Raf (Zhang and Guan, 2000). These sites are conserved in all of the Raf isoforms as well as in the C.elegans lin-45 Raf. A previous report by Barnard et al. (1998) concluded that phosphorylation of the activation loop plays no significant role in C-Raf activation. In this study, we investigate the role of these activation loop sites in C-Raf and their cooperativity with the S338 and Y341 phosphorylation sites. Both C-Raf and B-Raf require phosphorylation of the two conserved residues in the activation loop for full activity. However, phosphorylation of the activation loop is sufficient for B-Raf activation but not for C-Raf. Furthermore, phosphorylation of residues S338 and Y341 in the N-terminus of the C-Raf kinase domain is required for C-Raf activation in addition to the activation loop phosphorylation sites. Constitutively active C-Raf can be created only when all four residues (S338, Y341, T491 and S494) are substituted by acidic residues. We have also demonstrated that similar mechanisms involving the activation loop and inhibitory phosphorylation sites are used in the C.elegans lin-45 Raf to regulate kinase activity and biological functions. This report demonstrates a common mechanism utilized to control Raf kinase activity in C.elegans and mammals, although subtle variations exist to accommodate different Raf isoforms in response to specific physiological stimulation.
Results
T491 and S494 are important for C-Raf activation
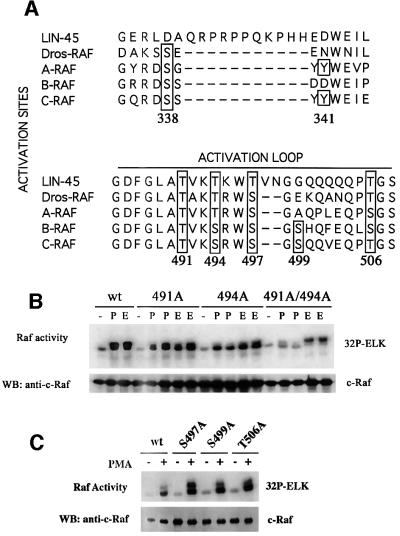
Raf is known to be activated by protein phosphorylation in response to growth factor or Ras stimulation. Phosphorylation in the activation loop between kinase subdomains VII and VIII is a mechanism utilized by many kinases including MEK and ERK (Payne et al., 1991; Rossomando et al., 1992; Alessi et al., 1994; Zheng and Guan, 1994). There are five putative phosphorylation sites that lie in the activation loop of the kinase domain of Raf. T491, S494, S497 and T506 in C-Raf are conserved among the different isoforms of Raf (Figure 1A), while S499 is present only in C-Raf and B-Raf. S508 was not analyzed because it is highly conserved in most protein kinases and is unlikely to serve as a regulatory phosphorylation site (Hanks and Quinn, 1991). Previous studies have indicated that S497/S499 is the activation phosphorylation site by PKC (Kolch et al., 1993; Carroll and May, 1994). However, Barnard et al. reported that S497 and S499 are not required for C-Raf activation by PKC and Ras (Barnard et al., 1998). These authors also concluded that phosphorylation of the activation loop is not important for C-Raf activation. Recently, we have demonstrated in B-Raf that T598 and S601, corresponding to T491 and S494 in C-Raf, are Ras-dependent phosphorylation sites essential for B-Raf activation (Zhang and Guan, 2000). To assess whether these sites have any effect on C-Raf activity, we mutated these residues in C-Raf to alanine by site-directed mutagenesis. Flag-tagged T491A and S494A were transfected in HEK293 cells, treated with phorbol 12-myristate 13-acetate (PMA) (to activate PKC) or epidermal growth factor (EGF), immunoprecipitated and subjected to a coupled Raf kinase assay. Wild-type C-Raf activity was stimulated by EGF or PMA; however, activation of the T491A or S494A mutant by PMA and EGF was decreased (Figure 1B). When these mutations were combined together, the double alanine mutation T491A/S494A showed a further decrease in inducible activity (Figure 1B). This C-Raf mutant is also compromised in activation by H-RasV12 (Figure 2A and B) and carbachol (Figure 3D), which is mediated by trimeric G-protein-coupled signaling. The loss in activity is unlikely to be due to structural changes by the amino acid substitutions. The T491A/S494A double mutant (and other mutants to be discussed later) still binds to several proteins that normally associate with wild-type C-Raf (Figure 4).
Fig. 1. (A) Alignment of sequences of C.elegans, Drosophila and human Raf proteins surrounding amino acids 338 and 341 and the activation loop of the kinase domain. The potential phosphorylation sites are boxed. The residue numbers are based on C-Raf sequence. (B) The effects of alanine substitution at residues 491 and 494 on C-Raf activation by PMA and EGF. HEK293 cells were transfected with wild-type, T491A, S494A or T491A/S494A C-Raf constructs. After 2 days, cells were treated with PMA (P) or EGF (E) and C-Raf proteins were immunoprecipitated from lysates and assayed for kinase activity in a coupled kinase assay (see Materials and methods). PMA- and EGF-stimulated alanine mutants were assayed in duplicate. C-Raf protein present in the immunoprecipitates was detected by western blotting (WB) with anti-C-Raf antibody. The results in this figure and all the other figures are a representative of at least three independent experiments. (C) Other putative phosphorylation sites in the activation loop have no effect on PMA-inducible activity. S497A, S499A and T506A constructs were assayed as mentioned in (B).
Fig. 2. (A) T491 and S494 are important for C-Raf kinase activation by RasV12. Alanine or acidic residue substitution mutants of C-Raf and B-Raf were transfected with or without RasV12. Raf proteins were immunoprecipitated and assayed for kinase activity as mentioned in Figure 1. C-Raf mutants co-transfected with Ras are shown in duplicate. (B) C-RafT491A/S494A mutant is not stimulated by RasV12. Raf kinase activity was determined by phosphoimage quantitation. The data presented are the average of duplicated assays from a representative experiment. (C) T491 in C-Raf is inducibly phosphorylated. C-Raf wild type, T491A and S494A were transfected with or without RasV12 and resolved for western blotting analysis using anti-pT598 antibody for phosphorylation and anti-Flag for Raf protein level. (D) Stimulation of T491 phosphorylation of endogenous C-Raf by EGF and Raf. HEK293 cells were stimulated with 50 ng/ml EGF and 10% fetal bovine serum for 5 min as indicated. Cell lysates were immunoblotted with anti-pT598 antibody or anti-phosphoERK antibody as indicated. The anti-pERK antibody recognizes phosphorylated forms of both ERK1 and ERK2 while the anti-ERK1 antibody recognizes ERK1 protein. (E) Phosphorylation of S494 in C-Raf is stimulated by RasV12. The experiments are similar to (C) except anti-pS601 antibody, which recognizes the phosphorylated S494 in C-Raf, was used.
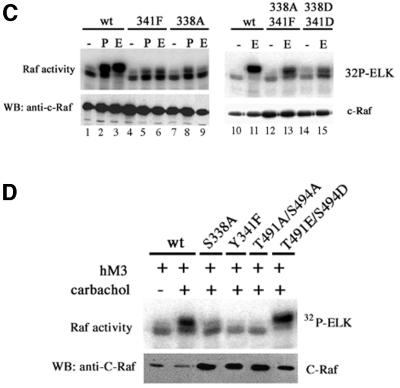
Fig. 3. (A) Effect of S338A and S338D on C-Raf kinase activation by RasV12. Cells were cotransfected with wild-type, S338A or S338D C-Raf constructs with or without RasV12 and assayed for kinase activity. (B) Y341F and Y341D diminishes C-Raf kinase activation by RasV12. Cells were transfected with wild-type, Y341F, Y341D, S338A/Y341F or S338D/Y341D C-Raf constructs with or without RasV12 and assayed for kinase activity. (C) Mutations at S338 and/or Y341 also affect PMA- and EGF-stimulated C-Raf kinase activity. Constructs noted were transfected and wild-type and mutant C-Raf proteins were assayed for kinase activity. Stimulation by PMA (P) and EGF (E) are indicated. (D) Alanine substitutions at S338, Y341 and T491/S494 also affect C-Raf kinase activation through the trimeric G-protein pathway. Cells were cotransfected with the human muscarinic receptor, hM3, and the C-Raf constructs noted. Twenty-four hours after transfection, cells were treated with carbachol and C-Raf proteins were assayed for kinase activity.
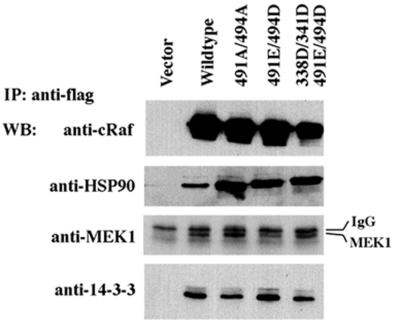
Fig. 4. C-Raf mutants can form complexes with HSP90, MEK and 14-3-3. Cells were transfected with vector, wild-type, T491A/S494A, T491E/S494E or DDED Flag-tagged C-Raf constructs. Immuno precipitates by anti-Flag antibody were resolved followed by western blotting analysis to detect associated endogenous proteins using anti-HSP90, anti-MEK and anti-14-3-3 antibodies.
Previous reports regarding the functions of S497 and S499 in C-Raf activation by PKC are inconsistent with each other (Kolch et al., 1993; Carroll and May, 1994; Barnard et al., 1998; Schonwasser et al., 1998). We examined the effects of these two residues in Raf activation. Flag-tagged S497A, S499A and T506A were transfected into cells, stimulated with PMA for 5 min, and Raf kinase activity was determined. These mutants showed PMA-stimulated Raf activity similar to that of wild-type C-Raf (Figure 1C). Similar results were obtained when S497A, S499A or T506A was co-transfected with H-RasV12 or stimulated with EGF (data not shown). These data are consistent with the observation reported by Barnard et al. (1998), suggesting that S497/S499 as well as T506 are not necessary for Raf activation. However, T491 and S494 play a role in Raf activation.
Different contributions of the activation loop phosphorylation to C-Raf and B-Raf kinase activities
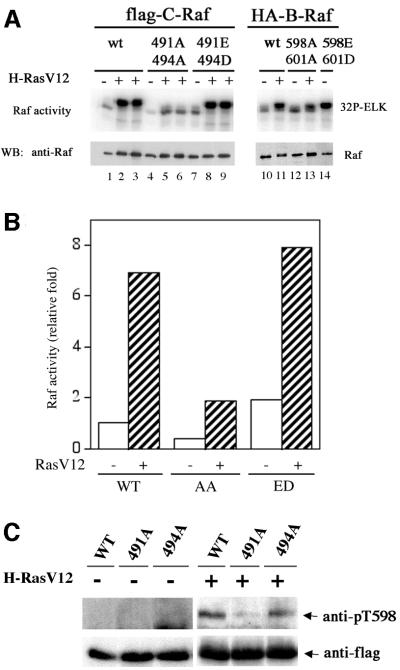
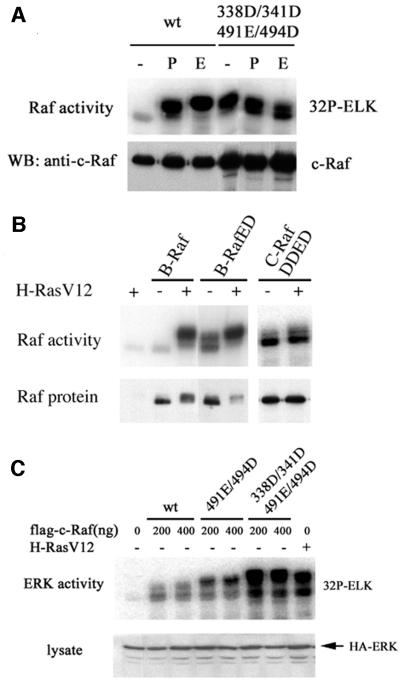
In B-Raf, substitution of T598 and S601 by acidic residues, B-Raf-ED, results in constitutively active kinase activity (Zhang and Guan, 2000). We tested the corresponding mutations in C-Raf. Acidic residue substitutions, T491E/S494D, were made and assessed for basal and H-RasV12-stimulated C-Raf kinase activity. The T491E/S494D mutation in C-Raf resulted in a slightly higher basal activity than the wild-type C-Raf and was able to be activated further by H-RasV12 (Figure 2A and B). As a control, the ED mutation in B-Raf was constitutively active (Figure 2A). The T491A/S494A mutant showed a dramatic decrease in Ras-stimulated activity. Similarly, mutating both of the corresponding residues in B-Raf to alanine also showed a decrease in activation by H-RasV12. These results suggest that residues in the activation loop, T491 and S494 in C-Raf and T598 and S601 in B-Raf, are important for the activation of both Raf isoforms but that the relative contributions to B-Raf and C-Raf activity are different. Phosphorylation of the activation loop is sufficient for B-Raf activation but insufficient for C-Raf.
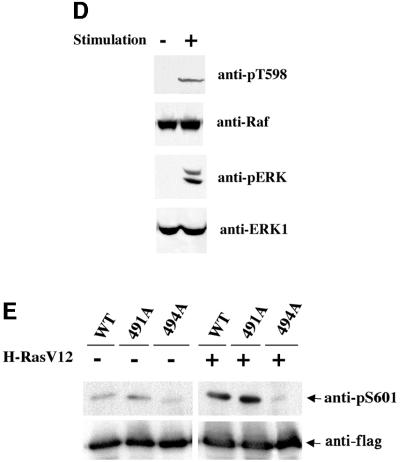
Previously, we have utilized anti-phospho-specific antibodies generated against T598 and S601 of B-Raf (Zhang and Guan, 2000). We tested the anti-phospho-specific antibodies and found that the anti-pT598 (corresponding to T491 in C-Raf) antibody recognized C-Raf (Figure 2C). The recognition of C-Raf by anti-pT598 was dependent on RasV12 stimulation, consistent with the notion that it is the phosphorylated C-Raf being recognized by anti-pT598. To confirm that anti-pT598 indeed specifically recognizes phosphorylated T491 in C-Raf, alanine substitutions of both T491 and S494 were examined. Mutation of T491 eliminated the recognition by the anti-pT598 antibody while mutation at S494 had no effect (Figure 2C). These results demonstrate the specificity of anti-pT598 antibody and further confirm that T491 in C-Raf is phosphorylated in response to RasV12 stimulation. We extended our experiments with endogenous C-Raf and found that phosphorylation of T491 was rapidly stimulated by EGF and serum (Figure 2D). Stimulation of the downstream MAPK, ERK, was included as a positive control (Figure 2D).
Phosphorylation of S494 was also examined using the anti-pS601 antibody, which recognizes the corresponding residue in B-Raf. Our results demonstrated that RasV12 stimulated S494 phosphorylation of the co-transfected C-Raf (Figure 2E). Furthermore, the specificity of the anti-pS601 antibody was confirmed by the elimination of the signal in the S494A mutant but not in the T491A mutant (Figure 2E). The above data support that T491 and S494 in C-Raf are phosphorylated in response to stimulation.
Functional cooperation between phosphorylation of S338/Y341 and T491/S494 in C-Raf activation
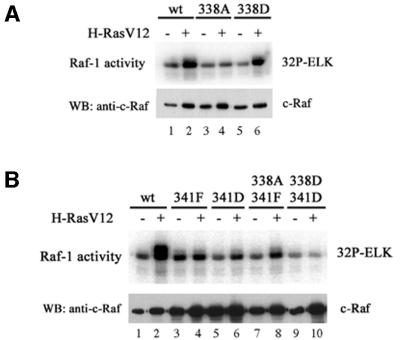
Several phosphorylation sites have been reported to be important in C-Raf regulation. One critical phosphorylation site previously identified is S338 within the N-terminus of the kinase domain (Figure 1A) (Mason et al., 1999). S338 was identified as a Pak phosphorylation site (King et al., 1998), and phosphorylation at this site was Ras inducible. Mutating S338 to alanine (S338A) diminished C-Raf activation by H-RasV12, PMA and EGF (Mason et al., 1999; Figure 3A and C). Another key phosphorylation site is Y341. Mutation of Y341 to phenylalanine inhibits Raf activation by Src (Marais et al., 1995; Barnard et al., 1998), Ras, PMA and EGF (Barnard et al., 1998; Figure 3B and C), confirming its importance in C-Raf regulation. Mutating Y341 to aspartic acid behaved similarly to the phenylalanine substitution, indicating that an acidic residue can not fully substitute the effect of tyrosine phosphorylation at this position. However, the homologous site in B-Raf is an aspartic acid residue.
We examined further the role of S338 and Y341 in Raf activation by the trimeric G-protein-coupled receptor signaling. Activation of C-Raf338A or 341F was defective in response to stimulation by carbachol (Figure 3D). When S338 was mutated to aspartic acid (S338D) to mimic phosphorylation, the mutant can be stimulated by H-RasV12, although more weakly than the wild-type C-Raf (Figure 3A). However, the basal activity of S338D was not elevated above wild-type C-Raf, supporting the concept that other phosphorylation sites, such as Y341, T491 and S494, are necessary for activation. Previous reports have demonstrated a stimulation-dependent relationship between phosphorylation at S338 and Y341 (Mason et al., 1999). Mutating both S338 and Y341 to aspartic acid residues (S338D/Y341D) did not significantly increase the basal kinase activity (Figure 3B and C), suggesting that other sites may play a role in positive activation of C-Raf. Surprisingly, S338D/Y341D can not be fully activated by Ras and EGF stimulation (Figure 3B and C). Again, this inhibition may be due to the aspartic acid substitution at Y341 being unable to fully mimic tyrosine phosphorylation at this site.
To assess whether the amino acid substitutions were interfering with the ability of C-Raf to bind substrates and other proteins necessary for function, a co-immunoprecipitation was performed. Flag-tagged C-Raf wild-type, T491A/S494A, T491E/S494D and S338D/Y341D/T491E/S494D (DDED) constructs were expressed in HEK293 cells. Cell lysates were immunoprecipitated with an anti-Flag antibody and interacting proteins were detected by western blot analysis. Blots were probed for endogenous MEK, HSP90 and 14-3-3 proteins (Figure 4). Mutations at residues S338, Y341, T491 and S494 do not interfere with the ability of C-Raf to complex with these proteins, indicating that the mutations may not alter the global three-dimensional structure of C-Raf. However, our data can not exclude possible conformational changes and alteration of binding to other proteins of the Raf mutants.
We have shown that double acidic residue substitutions at S338/Y341 or T491/S494 were insufficient to confer constitutive activity. We then combined these mutations to create a quadruple mutant, S338D/Y341D/T491E/S494D (DDED), and assessed whether there is a functional relationship between these two sets of phosphorylation sites. This quadruple mutant was transfected into cells, treated with PMA or EGF, and C-Raf activity was determined. The DDED mutant showed high basal activity comparable to that of stimulated wild-type C-Raf (Figure 55A). These results suggest that all four phosphorylation sites are important for the positive regulation of C-Raf. The effect of RasV12 on C-Raf-DDED was tested. RasV12 induced a small but reproducible activation of B-Raf-ED (Figure 5B). RasV12 had an even smaller but reproducible activation on C-Raf-DDED (Figure 5B). The stronger effect of RasV12 on B-Raf-ED than C-Raf-DDED could be due to an increased phosphorylation of S445 in B-Raf, which corresponds to S338 in C-Raf.
Fig. 5. (A) C-Raf DDED has constitutive activity. Cells were transfected with wild-type or DDED C-Raf constructs, treated with PMA (P) or EGF (E), and assessed for kinase activity. (B) B-Raf-ED and C-Raf DDED display residual activation by RasV12. The constitutively active B-Raf and C-Raf mutants were co-transfected with RasV12. Kinase activity was determined. (C) C-Raf DDED displays high activity to stimulate ERK activation. Cells were cotransfected with HA-ERK and wild-type, T491E/S494D or DDED C-Raf constructs at two different concentrations or RasV12. ERK was immunoprecipitated from lysates and assayed for kinase activity.
To determine whether the DDED mutant was active in vivo and sufficient to stimulate ERK activity, cells were co-transfected with hemagglutinin (HA)-ERK and C-Raf constructs or H-RasV12. HA-ERK was immunoprecipitated from cell lysates and incubated with glutathione S-transferase (GST)–Elk-1 in an ERK kinase assay. The T491E/S494D mutant was able to stimulate ERK activity slightly higher than the wild-type C-Raf. However, the DDED mutant was able to induce ERK kinase activity similar to H-RasV12-induced levels (Figure 5C). These data are consistent with our in vitro biochemical assays and demonstrate that DDED is constitutively active both in vitro and in transfected cells.
To test the relationship between different phosphorylation sites, we examined the phosphorylation of S338 and T491 with phospho-specific antibodies. Phosphorylation of S338 was not elevated in the T491E/S494D mutant but is stimulated by Ras. Similarly, phosphorylation of T491 was not increased in the S338D background (data not shown). These results indicate that phosphorylation of these two residues, S338 and T491, is independent of each other, although both are required for full C-Raf activation. The above observations are also consistent with the kinase activity data of different C-Raf mutants.
Biological function and conservation of activating phosphorylations in Raf
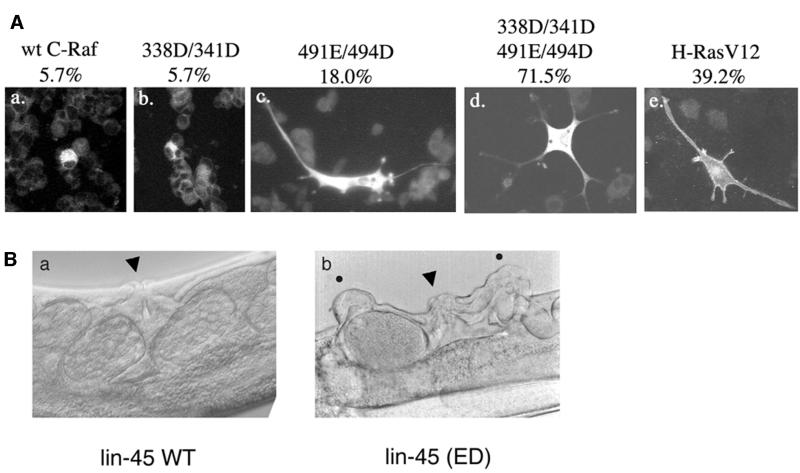
Raf kinases have been shown to participate in a variety of biological functions such as cell growth and differentiation. PC12 cells can be induced by activated Raf to differentiate and display neurite outgrowths (Wood et al., 1993). We have previously shown that B-Raf-ED is sufficient to induce neurite outgrowths, suggesting that the sites T598 and S601 have a physiological relevance. To address the role of C-Raf phosphorylation sites in PC12 differentiation, PC12 cells were transfected with wild-type, S338D/Y341D, T491E/S494D or DDED C-Raf constructs or H-RasV12. Three days after transfection, immunofluorescence assays were performed to detect cells expressing the C-Raf or Ras constructs. PC12 cells positive for transfection were scored for neurite outgrowths (Figure 6A). Vector transfected cells displayed a basal differentiation of 5%. Wild-type and S338D/Y341D C-Raf constructs (panels a and b) were unable to stimulate neurite outgrowths. However, the T491E/S494D (panel c) mutant did induce more outgrowths than wild-type C-Raf. As predicted, the quadruple mutant, DDED, which was the most active in the biochemical assays, was able to induce a nearly 2-fold higher percentage of cells to differentiate relative to H-RasV12-induced neurite outgrowths (panels d and e). Consistent with the in vitro biochemical assays (Figure 5A and C), DDED is constitutively active in vivo. The differentiation data also suggest that in C-Raf, S338, Y341, T491 and S494 all cooperate in vivo and contribute to its physiological regulation.
Fig. 6. (A) C-Raf DDED is sufficient to induce neurite outgrowths in PC12 cells. PC12 cells were transfected with (a) wild-type, (b) S338D/Y341D, (c) T491E/S494D or (d) DDED Flag-tagged C-Raf constructs or (e) HA-tagged H-RasV12. After 48 h in differentiation media, cells were fixed and incubated with anti-Flag or anti-HA antibody, washed and incubated with an anti-mouse IgG–Texas Red secondary antibody. Cells positive for expression were marked for neurite outgrowths. The percent of immunofluorescence-positive cells with neurites are indicated on top of each panel. (B) Effect of lin-45-ED transgene on C.elegans vulval induction. Normal vulva structures are indicated by an arrowhead in (a) a wild-type lin-45 transgenic line and (b) a lin-45-ED transgenic line where ectopic vulva structures are indicated by a black dot.
In C.elegans, lin-45 Raf is involved in vulval development of hermaphrodite animals and is also activated by Ras (Han et al., 1993). Interestingly, residues corresponding to C-Raf S338 and Y341 are aspartic acid resides in lin-45 Raf (Figure 1A), suggesting that these sites in lin-45 Raf are not subject to regulation by phosphorylation. In contrast, residues corresponding to C-Raf T491 and S494 in the kinase domain are conserved in lin-45 Raf (Figure 1A). To address the role of these activation loop phosphorylation sites in C.elegans, the T626/T629 sites in lin-45 were mutated to glutamic acid and aspartic acid, respectively (lin-45-ED). Both wild-type lin-45 and lin-45-ED mutant were injected into animals and stable lines were obtained. Worms expressing the lin-45-ED mutant had a multi-vulva phenotype (Figure 6B, panel b) similar to activated Ras transgenic animals (data not shown, Beitel et al., 1990; Han and Sternberg, 1990) while the wild-type lin-45 stable lines showed normal vulval induction (Figure 6B, panel a). We also obtained stable lines expressing lin-45 T626A/T629A mutant. These worms showed normal vulval induction (data not shown), indicating that the alanine substitution did not activate lin-45 Raf nor did the mutant display a dominant-negative effect on the endogenous wild-type lin-45 Raf. These observations suggest that T626 and T629 are important positive phosphorylation sites in lin-45 Raf, and substitution of these two residues by acidic amino acids resulted in elevation of lin-45 Raf kinase activity. These data also suggest that the mechanisms regulating phosphorylation at the activation loop sites are evolutionarily conserved and have physiological importance.
The inhibitory phosphorylation mechanisms are conserved in the Raf family
Raf kinases are also regulated by inhibitory phosphorylation events. Akt can suppress the Ras–Raf pathway (Rommel et al., 1999) by phosphorylating B-Raf (Guan et al., 2000) and C-Raf (Zimmermann and Moelling, 1999). B-Raf has three consensus Akt sites, one (S364) of which is conserved among all Raf kinases (S259 in C-Raf and S312 in lin-45; Figure 7A). Phosphorylation at this site in C-Raf, the only Akt consensus site, has been suggested to decrease kinase activity by promoting complex formation with 14-3-3 (Rommel et al., 1996; Morrison and Cutler, 1997; Tzivion et al., 1998). It was reported that mutation of S259 to alanine causes an increase in C-Raf basal kinase activity (Michaud et al., 1995). We also found a significant increase of basal activity in C-Raf S259A mutant (Figure 7B). The same mutation in B-Raf (S364A) has a minor effect on the basal B-Raf activity (Figure 7C). However, B-Raf has two additional Akt consensus sites at S428 and T439. When these Akt sites are mutated to alanine (S428A/T439A), basal activity is further increased. When all three Akt sites in B-Raf are mutated to alanine, B-Raf basal kinase activity is very high relative to wild-type B-Raf (Figure 7C). While S259 in C-Raf and S364 in B-Raf are consensus sites for Akt phosphorylation and 14-3-3 binding, the two additional Akt phosphorylation sites in B-Raf are not 14-3-3 consensus binding sites. Overall, these results suggest that C-Raf and B-Raf both utilize Akt phosphorylation as a mechanism for inhibitory regulation.
Fig. 7. (A) Alignment of sequences of C.elegans, Drosophila and human Raf proteins surrounding the inhibitory phosphorylation sites. Akt consensus recognition sequences are indicated in the top line. The putative inhibitory phosphorylation sites are boxed. (B) C-RafS259A has high basal kinase activity. Cells were transfected with wild-type C-Raf with or without RasV12 or with S259A C-Raf. C-Raf proteins were assayed for kinase activity. (C) Mutation of three inhibitory phosphorylation sites in B-Raf produces high basal kinase activity. Cells were transfected with wild-type, S364A, S428A/T439A or S364A/S428A/T439A B-Raf constructs and assayed for kinase activity.
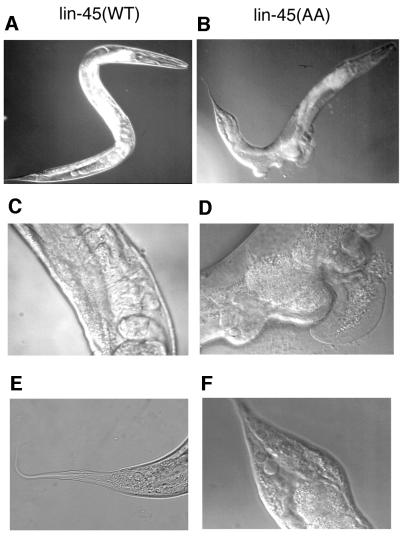
We again addressed the biological relevance of the inhibitory sites in C.elegans. The lin-45 Raf has two Akt consensus phosphorylation sites. S312 is common to all Rafs (S259 in C-Raf and S364 in B-Raf), and the other, S453, is homologous to the third Akt site of B-Raf, T439 (Figure 7A). To determine the physiological functions of these putative inhibitory phosphorylation sites, either S312 or S453 was singly mutated to alanine and the mutant lin-45 was injected into worms. Animals expressing lin-45RafS312A or lin-45RafS453A were analyzed. Vulval induction of the transgenic animals is apparently normal (data not shown). We also examined the effect of the double mutant, lin-45Raf-AA. Animals expressing lin-45-AA displayed a multi-vulva phenotype (Figure 8B and D) as well as having defects in tail development (Figure 8B and F). These phenotypes show that the mutant lin-45-AA is constitutively active and mimics activated Ras phenotypes in C.elegans (Beitel et al., 1990; Han and Sternberg, 1990). Our results indicate further that both S312 and S453 play inhibitory roles in regulation of lin-45 Raf in vivo. We also combined these alanine mutations with the acidic residue substitutions in the activation loop of lin-45 Raf. We were unable to obtain a stable C.elegans line despite our extensive efforts (data not shown). The mutant lin-45 Raf containing mutations of both inhibitory and activating sites may be too active and toxic to the worm. Our observations clearly demonstrated that the inhibitory mechanism by phosphorylation is conserved during evolution and plays an important role in regulation of the physiological functions of Raf.
Fig. 8. Effect of lin-45-AA transgene on vulva induction. (A, C and E) A wild-type lin-45 transgenic line. (B, D and F) A lin-45-AA transgenic line shows a multi-vulva phenotype (D) and abnormal tail morphology (F).
Discussion
We have demonstrated in vitro and in vivo that the activation loop plays a key regulatory role in regulation of C-Raf as well as lin-45. The biochemical data show that the single mutants T491A and S494A display partially impaired Raf activation when stimulated with EGF and PMA. Inducible activity of C-Raf is further decreased when the mutations are combined. These data are similar to what is seen in B-Raf (Zhang and Guan, 2000). When these residues in C-Raf are substituted with acidic residues (T491E/S494D) to mimic phosphorylation, there is a modest increase in basal kinase activity that can be further induced by RasV12. This result is unlike B-Raf where acidic residue substitutions lead to high kinase activity. The difference between the basal activities of the C-Raf-ED mutant and the B-Raf-ED mutant suggests that there are other regulatory mechanisms that play a role in C-Raf activation.
It has previously been established that phosphorylation sites S338 and Y341 are critical for C-Raf activation. The requirement for phosphorylation at S338 and Y341 in C-Raf is likely to be the reason that C-Raf T491E/S494D is insufficient to confer high basal activity. Consistent with this hypothesis, when the ED mutation was combined with mutations of S338D/Y341D in C-Raf, high activity was seen in in vitro kinase assays. Furthermore, the C-Raf-DDED mutant displayed constitutive biological activity in the PC12 differentiation assay and a high potency to stimulate ERK activation in HEK293 cells. In B-Raf, the site homologous to C-Raf S338 is constitutively phosphorylated (Mason et al., 1999) and the site corresponding to C-Raf Y341 is an aspartic residue. Therefore, the ED mutation in the activation loop of B-Raf is sufficient for high kinase activity. The lin-45 Raf contains aspartic residues at both positions corresponding to S338 and Y341 of C-Raf. Similarly, the ED mutation in the activation loop of lin-45 Raf is sufficient to activate lin-45 in vulval induction. While other sites may be involved in regulating the activation of C-Raf, our data support that S338, Y341, T491 and S494 are the main phosphorylation sites for activating C-Raf.
Fabian et al. (1993) have shown that C-RafY341D had an increase of basal kinase activity. However, we found that C-RafS338D/Y341D mutant did not show an elevated basal kinase activity. These results indicated that mutation of phosphorylation sites may not be simply additive. Consistent with these observations, Fabian et al. showed that C-RafY340D/Y341D double mutant is less active than either Y340D or Y341D single mutant. Our data show that C-RafS338D/Y341D is compromised in activation by EGF and Ras. Similarly, C-RafS338A/Y341F displays residual activation by Ras and EGF. These results are consistent with a model that other modifications (likely phosphorylations) are also involved in Raf activation by Ras and EGF, and acidic residue substitution may not entirely mimic the effects of phosphorylation.
Individual mutations of T491 and S494 have been previously examined by Barnard et al. (1998). These authors concluded that the phosphorylation at the activation loop plays no significant role in C-Raf regulation. The published results are similar to ours in that mutations of T491A or S494A partially diminished C-Raf activation by RasV12 and PMA stimulation. Furthermore, mutation of all putative phosphorylation sites in the activation loop (T491, S494, S497, S499, T506 and S508) almost completely eliminated C-Raf activation by RasV12 and PMA (Barnard et al., 1998). This multi-loop mutant behaved similarly to our T491A/S494A double mutant, although these authors did not examine the T491A/S494A double mutant. Therefore, we conclude that phosphorylation of the activation loop does play an important role in Raf regulation based on the following observations: (i) T491 and S494 are conserved in all members of the Raf family including C.elegans lin-45 Raf and Drosophila D-Raf (Figure 1A), consistent with a common mechanism utilized in Raf activation by Ras. (ii) Phosphorylation of the activation loop for positive regulation is a mechanism commonly employed by protein kinases, including MEK and ERK in the same signaling pathway. (iii) We have previously provided unequivocal data that T598 and S601 are phosphorylated in B-Raf and that phosphorylation of these two residues is necessary and sufficient for B-Raf activation (Zhang and Guan, 2000). (iv) In this report, we have shown that T491 and S494 are required for C-Raf activation (Figure 1B). We have also demonstrated that RasV12 stimulated phosphorylation at both T491 and S494 of C-Raf (Figure 2C and E). Moreover, phosphorylation of T491 of endogenous C-Raf is stimulated by EGF and serum. (v) The C-Raf mutant T491E/S494D showed a measurable increase of kinase activity both in vitro and in vivo. The C-Raf-ED mutant also showed higher activity in PC12 differentiation than the wild type (Figure 6A). Furthermore, C-Raf DDED showed constitutive kinase activity and biological function while the double mutant C-Raf S338D/Y341D is not constitutively active. (vi) Mutations of the putative phosphorylation sites in the activation loop to acidic residues in lin-45 Raf resulted in a constitutively active lin-45 Raf in C.elegans vulval induction (Figure 6B), while the alanine substitution of these residues did not induce a multi-vulval phenotype (data not shown). These results further support the evolutional conservation and functional significance of the phosphorylation in the activation loop of Raf.
All Rafs, including C.elegans lin-45 Raf, Drosophila D-Raf and mammalian Rafs, are activated by Ras. It is logical to hypothesize that a common biochemical mechanism is utilized in Raf activation. Indeed, we show that phosphorylation of the two conserved residues in the activation loop, which are conserved in all Raf family members, is a common mechanism for Raf activation by Ras. However, this model does not exclude that other phosphorylation sites also play an important role in Raf activation by Ras. For example, S338 and Y341 are important for C-Raf activation but are not conserved in C.elegans lin-45 Raf. Y341 is not conserved in Drosophila D-Raf or mammalian B-Raf. Interestingly, naturally occurring residues corresponding to positions of C-Raf S338 and Y341 are always acidic residues in lin-45 Raf, D-Raf and B-Raf. Overall, our data suggest that regulation of the Raf kinase family is mediated by a common mechanism (phosphorylation of the activation loop) supported by an additional level of regulation specific to the individual Raf members.
There are also inhibitory phosphorylation sites that regulate Raf kinases. Elimination of these sites causes high basal activity. These sites in B-Raf and C-Raf may be phosphorylated by Akt, which suppresses activity possibly by keeping Raf complexed with 14-3-3 proteins. While both isoforms use Akt phosphorylation for inhibition, B-Raf contains three Akt sites whereas C-Raf has one. Mutation of the single site in C-Raf (S259A) causes a significant elevation of kinase activity. The same high level of activation of B-Raf occurs only when all three sites are eliminated. In unstimulated cells, B-Raf normally displays much higher basal activity than C-Raf, possibly due to the constitutive phosphorylation of S445 and the presence of acidic residue D448 in B-Raf. The higher basal activity of B-Raf may require a stronger inhibitory signal.
The C.elegans lin-45 Raf is apparently regulated by biochemical mechanisms more similar to mammalian B-Raf. The activation loop mutant and inhibitory site mutant in lin-45 Raf, lin-45-ED and lin-45-AA. respectively, both cause a multi-vulva phenotype in C.elegans. This phenotype is similar to transgenic animals expressing an activating let-60 Ras mutant. Previous genetic data have implicated possible negative regulation of lin-45 Raf by phosphorylation. Sur-6 was isolated as a positive regulator acting between let-60 Ras and lin-45 Raf (Sieburth et al., 1999). Sur-6 encodes a B-subunit of phosphoprotein phosphatase 2A. An attractive model is that Sur-6 is responsible for dephosphorylation of the inhibitory sites in lin-45, and, therefore, plays a positive role in lin-45 Raf regulation. Biochemical data with mammalian Raf are consistent with this model (Abraham et al., 2000). Our studies in C.elegans demonstrate further that the phosphorylation sites (described in this report) are physiologically relevant and evolutionarily conserved.
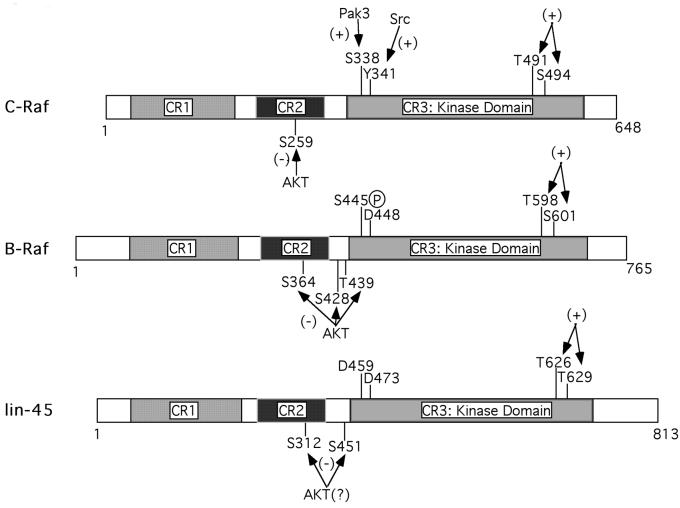
Results from this report and previous publications present a unified model for Raf regulation by phosphorylation (Figure 9). For simplicity, some Raf phosphorylation sites, of which the functional significance is not fully established, are not included in the model. Raf activation requires phosphorylation of two conserved residues (T491/S494 in C-Raf) in the activation loop and phosphorylation of residues (S338/Y341 in C-Raf) in the N-terminal region of the kinase domain. The activation loop phosphorylation sites are common to all Raf family members and represent the conserved nature of Raf activation. The N-terminal phosphorylation sites could be functionally substituted partially (B-Raf and D-Raf) or completely (lin-45 Raf) by naturally occurring acidic residues, therefore representing a divergence between Raf family members. In addition, every Raf member contains single or multiple inhibitory phosphorylation sites in the CR2 region. Dephos phorylation of the inhibitory sites is also sufficient to elevate Raf kinase activity and regulate physiological functions of Raf. The subtle differences in regulation between Raf isoforms may allow individual members to be activated by distinct extracellular signals. For example, activation of kinase(s) responsible for phosphorylation of the activation loop alone should be sufficient to activate B-Raf, but not C-Raf. C-Raf activation requires additional phosphorylations. Further studies are needed to identify upstream kinase(s) and to fully elucidate the specific molecular mechanisms of Raf regulation.
Fig. 9. Model showing similarities and differences in B-Raf, C-Raf and lin-45 kinase regulation.
Materials and methods
Plasmid constructs
All HA-tagged B-Raf constructs were previously described (Guan et al., 2000; Zhang and Guan, 2000). Flag-tagged full-length C-Raf was generously provided by Dr Kevin Pumiglia. Mutant constructs of C-Raf and lin-45 were created by PCR mutagenesis and verified by DNA sequencing. S338, Y341, T491, S494, S497, S499, T506 and S259 were individually mutated to alanine or phenylalanine (T491A, S494A, S497A, S499A, T506A, S338A and Y341F). Double alanine (phenylalanine substitution for tyrosine) substitutions were also generated in C-Raf (T491A/S494A and S338A/Y341F) and lin-45 (S312A/S453A). Individual and multiple aspartic acid and/or glutamic acid substitutions were also made for S338, Y341, S338/Y341, T491/S494 and S338/Y341/T491/S494 in C-Raf (S338D, Y341D, S338D/Y341D, T491E/S494D and DDED, respectively) and T626/T629 in lin-45 (lin-45-ED). All C-Raf constructs were Flag-tagged. Human subtype 3 muscarinic receptor (hM3) construct was generously provided by Dr Craig Lodgston (University of Michigan).
Cell culture and transfection
HEK293 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS). PC12 cells were cultured in DMEM containing 10% horse serum (HS) and 5% FBS. All cells were transfected in serum-free media using LipofectAMINE (Life Technologies, Inc.) following the manufacturer’s instructions.
In vitro kinase assay
For C-Raf kinase assays, HEK293 cells grown in 6-well plates were transfected with 250 ng of the C-Raf constructs mentioned above with or without 10 ng HRasV12 or 100 ng hM3. Twenty-four hours after transfection, some cells were treated with 50 ng/ml EGF for 3 min, PMA for 5 min or cells transfected with hM3 were treated with carbachol for 5 min. Cells were washed in ice cold phosphate-buffered saline (PBS) and lysed in RIPA buffer [50 mM Tris–HCl pH 7.5, 150 mM NaCl, 1% Triton X-100, 0.5% deoxycholate, 0.1% SDS, 1 mM dithiothreitol (DTT), 5 mM EDTA, 25 mM NaF, 1 mM phenylmethylsulfonyl fluoride, 10 µg/ml leupeptin, 10 µg/ml aprotinin]. After centrifugation, Flag-tagged kinase was immunoprecipitated with anti-M2 antibody (Sigma). Immunocomplexes were collected by incubation with protein G–Sepharose beads, washed with RIPA buffer and a final wash was carried out in HEPES buffer. Immunoprecipitated C-Raf was then incubated with 0.1 µg of GST–MEK in 20 µl of kinase buffer (10 mM HEPES pH 8.0, 10 mM MgCl2, 1 mM DTT, 50 µM ATP) for 20 min at 30°C. Fourteen microliters of supernatant from the reaction were transferred to a new tube and incubated for 10 min at 30°C with 6 µl of kinase buffer containing 0.1 µg of GST–ERK. Ten microliters of kinase buffer containing 4 µg of GST–Elk-1 and 10 µCi of [γ-32P]ATP (ICN) were added to the reaction and incubated for an additional 20 min at 30°C. Reactions were stopped upon the addition of SDS–sample buffer and boiled for 5 min. Reactions were resolved by SDS–PAGE and visualized by autoradiography. Recombinant GST–MEK1, GST–ERK1 and GST–Elk-1 were expressed and purified by glutathione agarose affinity chromatography.
For B-Raf assays, cells were transfected with 50 ng HA-B-Raf constructs with or without 10 ng of H-RasV12. Twenty-four hours after transfection, cells were lysed and assayed as mentioned for C-Raf assays, with the exceptions of 50 µl of a 250 µl total volume of lysate, which were used for the immunoprecipitation with anti-HA antibody (Babco).
For ERK assays, cells were co-transfected with 100 ng of HA-ERK and 200 and 400 ng of wild-type, T491E/S494D or DDED C-Raf constructs or 10 ng of H-RasV12. HA-tagged ERK was immunoprecipitated using anti-HA antibody and protein G–Sepharose beads. Immunocomplexes were incubated in kinase buffer containing 4 µg of GST–Elk-1 and 10 µCi of [γ-32P]ATP for 20 min at 30°C. Reactions were stopped and resolved as mentioned above.
Co-immunoprecipitation analysis
HEK293 cells grown in 6-well plates were transfected with 1 µg of empty vector, wild-type, T491A/S494A, T491E/S494D or DDED C-Raf constructs. Twenty-four hours after transfection, cells were lysed in NP-40 buffer (10 mM Tris–HCl pH 7.5, 100 mM NaCl, 1% NP-40, 50 mM NaF, 2 mM EDTA, 1 mM PMSF, 10 µg/ml leupeptin, 10 µg/ml aprotinin) and immunoprecipitated with anti-M2 antibody and protein G–Sepharose beads. Immunocomplexes were resolved by SDS–PAGE and transferred onto polyvinylene difluoride (PVDF) membranes. Co-immunoprecipitated endogenous proteins were detected by incubating membranes with anti-HSP90 (Transduction Laboratories), anti-14-3-3 (Santa Cruz Biotechnology), anti-MEK (Zheng and Guan, 1994) and anti-C-Raf (Transduction Laboratories) antibodies.
Immunoblotting analysis with anti-pT598 and anti-pS601 antibodies
Cells were transfected with wild-type, T491A and S494A C-Raf constructs with or without H-RasV12. Twenty-four hours after transfection, cells were lysed and C-Raf kinases were immunoprecipitated as mentioned above. C-Raf proteins were resolved by SDS–PAGE and transferred to PVDF membranes. Membranes were incubated in anti-pT598 or anti-pS601 antibody as previously described (Zhang and Guan, 2000). To determine the phosphorylation of endogenous C-Raf, serum-starved HEK293 cells were stimulated with 10% FBS plus EGF, 50 ng/ml, for 5 min. Cells were lysed and treated with SDS sample treatment buffer immediately. Cell lysates were subjected to western blotting without prior immunoprecipitation. Western blots with anti-ERK1 and pERK were also performed as a positive control.
PC12 cell differentiation
PC12 cells were cultured on glass coverslips and transfected with wild-type, S338D/Y341D, T491E/S494D or DDED C-Raf constructs or HA-HRasV12. After transfection for 5 h, cells were cultured in differentiation medium (DMEM containing 2% HS and 1% FBS) for 48 h. Cells were washed in PBS, fixed with formaldehyde and incubated with anti-M2 or anti-HA antibody followed by incubation with an anti-mouse IgG–Texas Red conjugate (Jackson ImmunoResearch Laboratories, Inc.). Fluorescent cells with one or more neurite outgrowths greater than two cell body lengths were counted and imaged using a confocal fluorescent microscope (Noran OZ Confocal Laser Scanning Imaging System). Percent cell differentiation was calculated by the number of differentiated cells divided by the total number of fluorescent cells.
Caenorhabditis elegans transgenics
A 3.5 kb ClaI fragment from the lin-45 genomic clone pKS(+)raf-4 containing the 5′ upstream regulatory region was fused with the lin-45 cDNA clone pSK(+)raf107a (Han et al., 1993). Subsequently, this construct was purified and injected into worms at 100 ng/µl with pRF4 at 100 ng/µl as a co-injection marker as described by Mello et al. (1991). The constructs of lin-45-ED (T626E/T629D) and lin-45-AA (S312A/S453A) were also injected in the same manner. Once stable transgenic lines were established in the wild-type strain (N2), well-fed young adult animals were picked and used for taking pictures.
Acknowledgments
Acknowledgements
We would like to thank Dr Ron Ellis and Suk-won Jin for assistance with the C.elegans microinjections; T.Zhu for technical assistance; Dr Kevin Pumiglia for the Flag-C-Raf plasmid; Drs W.Li, B.H.Zhang and A.Vojtek for Raf plasmids; Dr T.M.Lanigan and H.G.Vikis for critical review of the manuscript; Dr M.Han for suggestions and C.elegans strains; and T.E.Komorowski and the Michigan Diabetes Research Training Center for assistance with confocal microscopy. H.C. is a recipient of a pre-doctoral fellowship by the N.I.H. National Research Service Award #5-T32-GM07544 from the National Institute of General Medicine Sciences. This work is supported by grants from N.I.H., the Walther Cancer Institute, American Cancer Society and a John D. & Katherine T.MacArthur fellowship (K.-L.G.).
References
- Abraham D. et al. (2000) Raf-1-associated protein phosphatase 2A as a positive regulator of kinase activation. J. Biol. Chem., 275, 22300–22304. [DOI] [PubMed] [Google Scholar]
- Alessi D.R., Saito,Y., Campbell,D.G., Cohen,P., Sithanandam,G., Rapp,U., Ashworth,A., Marshall,C.J. and Cowley,S. (1994) Identification of the sites in MAP kinase kinase-1 phosphorylated by p74 raf-1. EMBO J., 13, 1610–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard D., Diaz,B., Clawson,D. and Marshall,M. (1998) Oncogenes, growth factors and phorbol esters regulate Raf-1 through common mechanisms. Oncogene, 17, 1539–1547. [DOI] [PubMed] [Google Scholar]
- Beitel G.J., Clark,S.G. and Horvitz,H.R. (1990) Caenorhabditis elegans ras gene let-60 acts as a switch in the pathway of vulval induction. Nature, 348, 503–509. [DOI] [PubMed] [Google Scholar]
- Carroll M.P. and May,W.S. (1994) Protein kinase C-mediated serine phosphorylation directly activates Raf-1 in murine hematopoietic cells. J. Biol. Chem., 269, 1249–1256. [PubMed] [Google Scholar]
- Diaz B., Barnard,D., Filson,A., MacDonald,S., King,A. and Marshall,M. (1997) Phosphorylation of Raf-1 serine 338-serine 339 is an essential regulatory event for Ras-dependent activation and biological signaling. Mol. Cell. Biol., 17, 4509–4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian J.R., Daar,I.O. and Morrison,D.K. (1993) Critical tyrosine residues regulate the enzymatic and biological activity of Raf-1 kinase. Mol. Cell. Biol., 13, 7170–7179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan K.L., Figueroa,C., Brtva,T.R., Zhu,T., Taylor,J., Barber,T.D. and Vojtek,A.B. (2000) Negative regulation of the serine/threonine kinase B-Raf by Akt. J. Biol. Chem., 275, 27354–27359. [DOI] [PubMed] [Google Scholar]
- Han M. and Sternberg,P.W. (1990) let-60, a gene that specifies cell fates during C.elegans vulval induction, encodes a ras protein. Cell, 63, 921–931. [DOI] [PubMed] [Google Scholar]
- Han M., Golden,A., Han,Y. and Sternberg,P.W. (1993) C.elegans lin-45 raf gene participates in let-60 ras-stimulated vulval differentiation. Nature, 363, 133–140. [DOI] [PubMed] [Google Scholar]
- Hanks S.K. and Quinn,A.M. (1991) Protein kinase catalytic domain sequence database: identification of conserved features of primary structure and classification of family members. Methods Enzymol., 200, 38–62. [DOI] [PubMed] [Google Scholar]
- Kerkhoff E. and Rapp,U.R. (1998) Cell cycle targets of Ras/Raf signalling. Oncogene, 17, 1457–1462. [DOI] [PubMed] [Google Scholar]
- Kerkhoff E., Houben,R., Loffler,S., Troppmair,J., Lee,J.E. and Rapp,U.R. (1998) Regulation of c-myc expression by Ras/Raf signalling. Oncogene, 16, 211–216. [DOI] [PubMed] [Google Scholar]
- King A.J., Sun,H., Diaz,B., Barnard,D., Miao,W., Bagrodia,S. and Marshall,M.S. (1998) The protein kinase Pak3 positively regulates Raf-1 activity through phosphorylation of serine 338. Nature, 396, 180–183. [DOI] [PubMed] [Google Scholar]
- Kolch W., Heidecker,G., Kochs,G., Hummel,R., Vahidi,H., Mischak,H., Finkenzeller,G., Marme,D. and Rapp,U.R. (1993) Protein kinase Cα activates RAF-1 by direct phosphorylation. Nature, 364, 249–252. [DOI] [PubMed] [Google Scholar]
- Marais R., Light,Y., Paterson,H.F. and Marshall,C.J. (1995) Ras recruits Raf-1 to the plasma membrane for activation by tyrosine phosphorylation. EMBO J., 14, 3136–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marais R., Light,Y., Paterson,H.F., Mason,C.S. and Marshall,C.J. (1997) Differential regulation of Raf-1, A-Raf and B-Raf by oncogenic ras and tyrosine kinases. J. Biol. Chem., 272, 4378–4383. [DOI] [PubMed] [Google Scholar]
- Mason C.S., Springer,C.J., Cooper,R.G., Superti-Furga,G., Marshall,C.J. and Marais,R. (1999) Serine and tyrosine phosphorylations cooperate in Raf-1, but not B-Raf activation. EMBO J., 18, 2137–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C.C., Kramer,J.M., Stinchcomb,D. and Ambros,V. (1991) Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J., 10, 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud N.R., Fabian,J.R., Mathes,K.D. and Morrison,D.K. (1995) 14-3-3 is not essential for Raf-1 function: identification of Raf-1 proteins that are biologically activated in a 14-3-3- and Ras-independent manner. Mol. Cell. Biol., 15, 3390–3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison D.K. and Cutler,R.E. (1997) The complexity of Raf-1 regulation. Curr. Opin. Cell. Biol., 9, 174–179. [DOI] [PubMed] [Google Scholar]
- Morrison D.K., Heidecker,G., Rapp,U.R. and Copeland,T.D. (1993) Identification of the major phosphorylation sites of the Raf-1 kinase. J. Biol. Chem., 268, 17309–17316. [PubMed] [Google Scholar]
- Nassar N., Horn,G., Herrmann,C., Scherer,A., McCormick,F. and Wittinghofer,A. (1995) The 2.2 Å crystal structure of the Ras-binding domain of the serine/threonine kinase c-Raf1 in complex with Rap1A and a GTP analogue. Nature, 375, 554–560. [DOI] [PubMed] [Google Scholar]
- Payne D.M., Rossomando,A.J., Martino,P., Erickson,A.K., Her,J.H., Shabanowitz,J., Hunt,D.F., Weber,M.J. and Sturgill,T.W. (1991) Identification of the regulatory phosphorylation sites in pp42/mitogen-activated protein kinase (MAP kinase). EMBO J., 10, 885–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rommel C., Radziwill,G., Lovric,J., Noeldeke,J., Heinicke,T., Jones,D., Aitken,A. and Moelling,K. (1996) Activated Ras displaces 14-3-3 protein from the amino terminus of c-Raf-1. Oncogene, 12, 609–619. [PubMed] [Google Scholar]
- Rommel C., Clarke,B.A., Zimmermann,S., Nunez,L., Rossman,R., Reid,K., Moelling,K., Yancopoulos,G.D. and Glass,D.J. (1999) Differentiation stage-specific inhibition of the Raf–MEK–ERK pathway by Akt. Science, 286, 1738–1741. [DOI] [PubMed] [Google Scholar]
- Rossomando A.J., Wu,J., Michel,H., Shabanowitz,J., Hunt,D.F., Weber,M.J. and Sturgill,T.W. (1992) Identification of Tyr-185 as the site of tyrosine autophosphorylation of recombinant mitogen-activated protein kinase p42mapk. Proc. Natl Acad. Sci. USA, 89, 5779–5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonwasser D.C., Marais,R.M., Marshall,C.J. and Parker,P.J. (1998) Activation of the mitogen-activated protein kinase/extracellular signal-regulated kinase pathway by conventional, novel and atypical protein kinase C isotypes. Mol. Cell. Biol., 18, 790–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidovar M.F., Kozlowski,P., Lee,J.W., Collins,M.A., He,Y. and Graves,L.M. (2000) Phosphorylation of serine 43 is not required for inhibition of c-Raf kinase by the cAMP-dependent protein kinase. J. Biol. Chem., 275, 28688–28694. [DOI] [PubMed] [Google Scholar]
- Sieburth D.S., Sundaram,M., Howard,R.M. and Han,M. (1999) A PP2A regulatory subunit positively regulates Ras-mediated signaling during Caenorhabditis elegans vulval induction. Genes Dev., 13, 2562–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzivion G., Luo,Z. and Avruch,J. (1998) A dimeric 14-3-3 protein is an essential cofactor for Raf kinase activity. Nature, 394, 88–92. [DOI] [PubMed] [Google Scholar]
- Vojtek A.B., Hollenberg,S.M. and Cooper,J.A. (1993) Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell, 74, 205–214. [DOI] [PubMed] [Google Scholar]
- Vossler M.R., Yao,H., York,R.D., Pan,M.G., Rim,C.S. and Stork,P.J. (1997) cAMP activates MAP kinase and Elk-1 through a B-Raf- and Rap1-dependent pathway. Cell, 89, 73–82. [DOI] [PubMed] [Google Scholar]
- Wood K.W., Qi,H., D’Arcangelo,G., Armstrong,R.C., Roberts,T.M. and Halegoua,S. (1993) The cytoplasmic raf oncogene induces a neuronal phenotype in PC12 cells: a potential role for cellular raf kinases in neuronal growth factor signal transduction. Proc. Natl Acad. Sci. USA, 90, 5016–5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Dent,P., Jelinek,T., Wolfman,A., Weber,M.J. and Sturgill,T.W. (1993) Inhibition of the EGF-activated MAP kinase signaling pathway by adenosine 3′,5′-monophosphate. Science, 262, 1065–1069. [DOI] [PubMed] [Google Scholar]
- Yao B., Zhang,Y., Delikat,S., Mathias,S., Basu,S. and Kolesnick,R. (1995) Phosphorylation of Raf by ceramide-activated protein kinase. Nature, 378, 307–310. [DOI] [PubMed] [Google Scholar]
- Yip-Schneider M.T., Miao,W., Lin,A., Barnard,D.S., Tzivion,G. and Marshall,M.S. (2000) Regulation of the Raf-1 kinase domain by phosphorylation and 14-3-3 association. Biochem. J., 351, 151–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B.H. and Guan,K.L. (2000) Activation of B-Raf kinase requires phosphorylation of the conserved residues thr598 and ser601. EMBO J., 19, 5429–5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C.F. and Guan,K.L. (1994) Activation of MEK family kinases requires phosphorylation of two conserved Ser/Thr residues. EMBO J., 13, 1123–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann S. and Moelling,K. (1999) Phosphorylation and regulation of Raf by Akt (protein kinase B). Science, 286, 1741–1744. [DOI] [PubMed] [Google Scholar]