Abstract
The cytoskeletal protein α-catenin, which shares structural similarity with vinculin, is required for cadherin-mediated cell adhesion, and functions to modulate cell adhesive strength and to link the cadherins to the actin-based cytoskeleton. Here we describe the crystal structure of a region of α-catenin (residues 377–633) termed the M-fragment. The M-fragment is composed of a tandem repeat of two antiparallel four-helix bundles of virtually identical architectures that are related in structure to the dimerization domain of α-catenin and the tail region of vinculin. These results suggest that α-catenin is composed of repeating antiparallel helical domains. The region of α-catenin previously defined as an adhesion modulation domain corresponds to the C-terminal four-helix bundle of the M-fragment, and in the crystal lattice these domains exist as dimers. Evidence for dimerization of the M-fragment of α-catenin in solution was detected by chemical cross-linking experiments. The tendency of the adhesion modulation domain to form dimers may explain its biological activity of promoting cell–cell adhesiveness by inducing lateral dimerization of the associated cadherin molecule.
Keywords: cadherins/α-catenin/cell adhesion/protein structure/X-ray crystallography
Introduction
The maintenance and regulation of cell–cell and cell– matrix adhesion are critical to processes such as cell morphogenesis, motility, regulation of growth and differentiation, wound healing, malignant transformation and the formation of organized tissues (Takeichi, 1991; Gumbiner, 1996; Yap et al., 1997a). A family of cell surface receptors termed the cadherins (E-, N- and Xenopus C-cadherins) function to mediate cell–cell adhesion. The ectodomain of the cadherin molecule participates in Ca2+-dependent homophilic protein–protein interactions (Briecher et al., 1996), whereas numerous studies implicate the cytoplasmic tail as being necessary for generating cell adhesive strength. For example, CHO cells expressing mutant forms of Xenopus C-cadherin lacking its cytoplasmic tail maintain adhesion that is considerably weaker than that of wild-type C-cadherin (Briecher et al., 1996). Moreover, tailless mutants of E-cadherin expressed in L cell fibroblasts were unable to mediate cell adhesion in aggregation assays (Nagafuchi and Takeichi, 1988, 1989; Ozawa et al., 1990). The β- and γ-catenins associate with the cadherin cytoplasmic tail (Nathke et al., 1994) and interact with α-catenin (Aberle et al., 1994), which in turn links the cadherin–catenin complex to the actin-based cytoskeleton (Ozawa et al., 1990) via a complex network of protein interactions involving actin (Rimm et al., 1995), vinculin and α-actinin (Knudsen et al., 1995; Nieset et al., 1997; Watabe-Uchida et al., 1998; Weiss et al., 1998). Compelling evidence implicates the catenins, and in particular α-catenin, in mediating fundamental aspects of cell adhesiveness. First, mutant cadherin molecules consisting of only the cytoplasmic tail, or the catenin-binding domain, exert a dominant-negative phenotype of reduced adhesiveness when overexpressed in cultured cells and in mouse intestinal epithelia (Fujimori and Takeichi, 1993; Hermiston and Gordon, 1995), presumably by titrating β-catenin away from wild-type cadherin molecules (Kintner, 1992). Secondly, genetic evidence from non-adherent human lung carcinoma PC9 cells that lack expression of α-catenin indicated that the exogenous expression of α-catenin in these cells conferred cadherin-mediated cell adhesion and also cadherin–cytoskeletal association (Hirano et al., 1992; Watabe et al., 1994). Moreover, the expression of E-cadherin–α-catenin fusion molecules in L cell fibroblasts has revealed that the generation of strong adhesion is dependent upon the C-terminal half of α-catenin and its association with the intact actin-based cytoskeleton (Nagafuchi et al., 1994; Imamura et al., 1999). This experimental system has enabled the roles of domains of α-catenin that mediate cell adhesion to be delineated. For example, an adhesion modulation domain comprising residues 509–643 was identified as a region of α-catenin capable of conferring rapid aggregation upon L cells when exogenously expressed as a fusion with E-cadherin lacking its cytoplasmic tail. However, the adhesion induced by the adhesion modulation domain–E-cadherin fusion molecule is weak, in contrast to the strong adhesion characteristic of E-cadherin fused to full-length α-catenin, and this possibly results from the absence of actin-based cytoskeletal interactions with the adhesion modulation domain (Imamura et al., 1999).
A fundamental mechanism for the generation of cell adhesive strength involves the lateral association of cadherin molecules at the cell surface (reviewed by Yap et al., 1997a). In decisive experiments employing the ectodomain and transmembrane segment of Xenopus C-cadherin fused to tandem repeats of the FKBP, Yap et al. (1997b) demonstrated that forced clustering of this fusion protein using FK1012 significantly increased cellular adhesive strength. These studies indicate that the promotion of lateral dimers of cadherin influences adhesion independently of direct interactions between the cadherins and cytosolic proteins. Lateral dimerization of cadherins correlates with adhesive strength since purified dimers of the ectodomain of C-cadherin cross-linked to Covaspheres exhibit substantially greater homophilic binding activity, as judged by bead aggregation assays, than do monomers (Briecher et al., 1996). Moreover, the juxtamembrane region of E-cadherin, which inhibits receptor dimerization, negatively regulates cell adhesion (Ozawa and Kemler, 1998). These functional results are consistent with the crystal structures of the N-terminal domain of N-cadherin (NCD1) and of a fragment of E-cadherin encompassing the EC1 and EC2 domains, which revealed a lateral homodimeric organization (Shapiro et al., 1995; Nagar et al., 1996). In the crystal structure of NCD1, lateral dimers are organized into a self-assembling linear zipper that may represent a model for understanding how numerous weak adhesive interactions act cooperatively to generate strong adhesion at the cellular level (Shapiro et al., 1995).
Defects in cell adhesion underlie tumour metastasis, and the cadherins and α-catenin have been implicated as tumour suppressor proteins (reviewed by Birchmeier and Behrens, 1994). The down-regulation of E-cadherin in metastatic carcinomas contributes to the invasive characteristics of these tumours. In several human cancer cell lines, the expression of catenins is absent or decreased (Shimoyama et al., 1992; Morton et al., 1993; Ewing et al., 1995), and the re-expression of α-catenin in PC3 (prostate adenocarcinoma) cells suppresses the invasive phenotype by restoring E-cadherin-dependent adhesion (Ewing et al., 1995). Moreover, deletions of the α-catenin-binding region of β-catenin occur in a human gastric cell line and are associated with extremely weak cell adhesion properties and loss of α-catenin interaction (Oyama et al., 1994; Kawanishi et al., 1995).
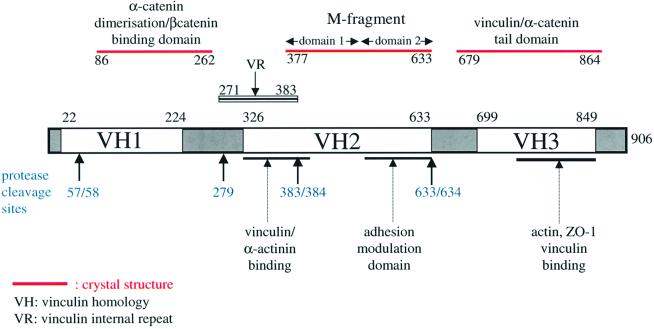
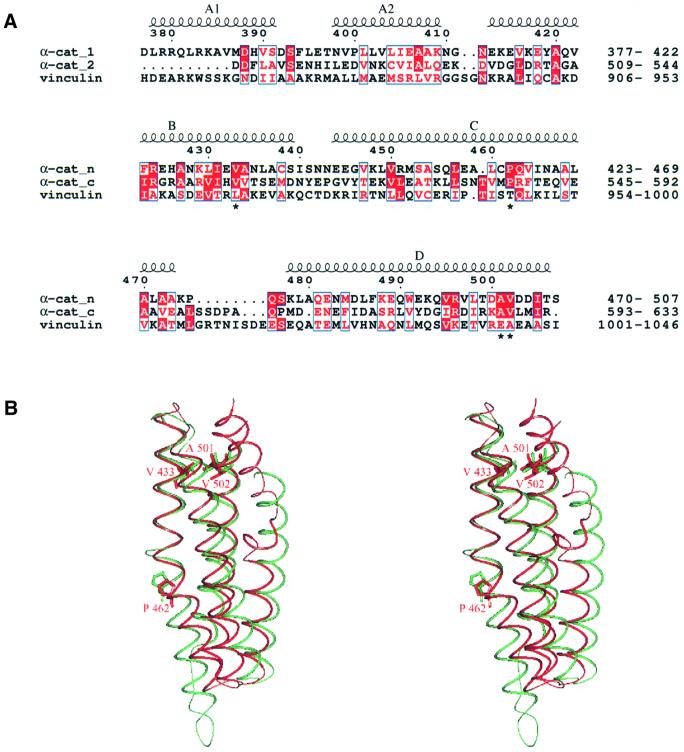
The α-catenin protein, which links the cadherin– β-catenin complex with the actin cytoskeleton, functions as a structural and scaffolding molecule. The protein shares structural and functional similarity with vinculin (Price et al., 1989; Herrenknecht et al., 1991; Nagafuchi et al., 1991), a cytoskeletal protein present within cell adherens junctions as well as the focal adhesion junctions that link integrins, the extracellular matrix receptors, to the actin cytoskeleton. Three regions of sequence similarity shared by α-catenin and vinculin are termed vinculin homology domain 1 (VH1), VH2 and VH3 (reviewed by Rudiger, 1998) (Figure 1). The crystal structure of the homodimerization and β-catenin-binding region of α-catenin established the molecular mechanisms defining α- and β-catenin interactions (Pokutta and Weiss, 2000), whereas domains of α-catenin that interact with other components of adherens junctions have been identified by means of biochemical and genetic approaches (reviewed by Rudiger, 1998; Provost and Rimm, 1999). An overlapping region of α-catenin (residues 325–402) interacts with both α-actinin and vinculin (Nieset et al., 1997; Watabe-Uchida et al., 1998; Imamura et al., 1999), whereas the C-terminal 280 residues of α-catenin interact with ZO-1 (Imamura et al., 1999), vinculin (Weiss et al., 1998) and actin (Rimm et al., 1995) (Figure 1).
Fig. 1. Schematic of α-catenin indicating that the protein is composed of tandem repeats of antiparallel α-helical bundles with defined biological functions. The positions of the VH1, VH2, VH3 and the α-catenin VR regions, together with sites of proteolysis are indicated. The domains of α-catenin for which structural data are available (β-catenin/dimerization domain; M-fragment of α-catenin; α-catenin tail) and the position of the predicted VR are shown.
To understand the molecular basis underlying the function of α-catenin in mediating adhesion, we have undertaken a crystallographic analysis of the protein. We report the crystal structure of a fragment of α-catenin (residues 377–633), referred to here as the M-fragment, delineated by a combination of limited proteolysis and sequence conservation. Our results indicate that the M-fragment is composed of a tandem repeat of a four-α-helix bundle. The bundles are structurally related to the α-catenin dimerization domain and the vinculin tail, suggesting that α-catenin is composed of a series of repeating antiparallel α-helical domains. The region of α-catenin previously defined as an adhesion modulation domain (Imamura et al., 1999) corresponds to the C-terminal four-helix bundle of the M-fragment, and in the crystal lattice these domains exist as dimers, related by a non-crystallographic symmetry operation. Evidence for dimerization of the M-fragment of α-catenin in solution was detected by chemical cross-linking experiments. The tendency of the adhesion modulation domain to form dimers may explain its biological activity of promoting cell–cell adhesiveness by inducing lateral dimerization of the associated cadherin molecule.
Results and discussion
Protein purification and crystallization
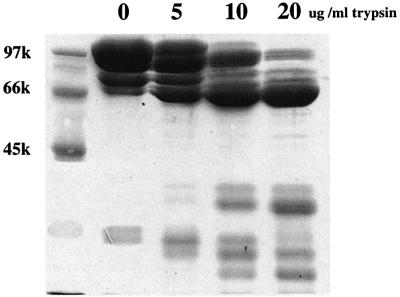
To understand the three-dimensional structure of α-catenin, we expressed full-length human α(E)-catenin (906 amino acids) using an Escherichia coli expression system in order to obtain protein suitable for crystallization studies. Although α-catenin was expressed at high levels, the protein was susceptible to proteolysis and was not suitable for crystallization. However, this expressed protein provided starting material for the delineation of stable domains within the molecule by means of defined proteolysis. Limited trypsinolysis of the partially degraded 102 kDa full-length protein yielded a stable 63 kDa intermediate (Figure 2). A similar sized fragment was obtained using limited subtilisin digestion (data not shown). N-terminal sequencing and mass spectrometric analysis of the trypsin-generated 63 kDa product indicated that this fragment corresponded to residues Ala58– Arg633, encompassing almost a complete VH1 and the entire VH2 region (Figure 1). The C-terminal domain boundary defined as Arg633 by limited proteolysis corresponds to the C-terminus of VH2 (residues 326– 633), established by sequence comparisons of α-catenin and vinculin. However, the N-terminus of VH1, corresponding to Thr22 of α-catenin, extends beyond the domain boundary defined by limited proteolysis. The 63 kDa fragment of residues 58–633 of α-catenin was expressed at high levels in soluble form in E.coli and the protein was purified to homogeneity. The purified protein readily crystallized using a number of different conditions, forming large hexagonal-shaped crystals. Unfortunately, none of these crystals diffracted to better than 7 Å resolution, which prevented their structural analysis. The low limiting resolution attainable from the 63 kDa α-catenin fragment crystals suggested additional flexible regions within the protein separating more rigid stable domains. We therefore attempted to define these domains by subjecting the 63 kDa α-catenin fragment to further limited proteolysis. Partial trypsinolysis yielded a 29 kDa fragment corresponding to residues Lys384–Arg633, somewhat shorter than VH2 defined as residues 326– 633. These regions of α-catenin include the adhesion modulation domain (residues 509–643) (Imamura et al., 1999) and a portion of the vinculin–α-actinin interaction domain (residues 325–402) (Knudsen et al., 1995; Nieset et al., 1997; Imamura et al., 1999). Significantly, residues 384–633 of the α-catenin fragment defined by limited trypsinolysis correspond almost exactly to exons 7–11 (residues 381–633) of the human α(E)-catenin gene (Furukawa et al., 1994). Although this protein fragment was produced in soluble form at high levels in E.coli, the purified protein failed to crystallize. Lys384 occurs within a highly conserved region of α-catenin that also shares sequence similarity with vinculin. Moreover, this region corresponds to a predicted α-helix as defined by the PHD protein secondary structure prediction server (Rost et al., 1994; http://www.ebi.ac.uk/∼rost/predictprotein). Previous α-catenin and vinculin sequence alignments (Herrenknecht et al., 1991; Nagafuchi et al., 1991) and examination of α-catenin and vinculin sequence similarities in this region guided us to Asp377 as the start of the sequence similarity between α-catenin and vinculin. A fragment of α-catenin corresponding to residues Asp377–Arg633, which we define as the α-catenin M-fragment, was overproduced and purified from an E.coli expression system, and crystals were obtained that diffracted to 2.1 Å when exposed to synchrotron radiation. The crystals are monoclinic and belong to space group P21 with two molecules per asymmetric unit. The structure was solved using the MAD method from SeMet-incorporated protein crystals using data collected at the Elettra synchrotron (Table I) with the programs SOLVE (Terwilliger and Berendzen, 1999) and SHARP (De la Fortelle and Bricogne, 1997), and the atomic coordinates were refined using CNS (Brünger et al., 1998). Model building was performed using O (Jones et al., 1991). The two copies of the α-catenin M-fragment present per asymmetric unit were refined independently and adopt slightly different protein conformations.
Fig. 2. Detection of protease-resistant domains of α-catenin using limited proteolysis. Incubation of α-catenin with increasing concentra tions of trypsin for 20 min reveals the formation of a partially resistant 63 kDa fragment.
Table I. Crystallographic data statistics for α-catenin.
| Data collection and MAD phasing statistics | ||||
|---|---|---|---|---|
| Data set | Native | MAD1 | MAD2 | MAD3 |
| X-ray source | SRS PX14.1 | Elettra synchrotron | Elettra synchrotron | Elettra synchrotron |
| Wavelength (Å) | 1.488 | 0.9806 | 0.9804 | 0.935 |
| Resolution (Å) | 2.2 | 2.4 | 2.4 | 2.4 |
| Observations | 171 946 | 116 023 | 119 220 | 121 370 |
| Unique reflections | 27 803 | 21 233 | 21 248 | 21 246 |
| Completeness (%) | 99.1 (98.4) | 99.4 (97.1) | 99.1 (94.5) | 99.7 (99.6) |
| Anomalous completeness (%) | 98.8 (94.6) | 98.4 (90.6) | 98.8 (93.3) | |
| Rmergea (%) | 4.9 (20.5) | 6.3 (24.7) | 6.9 (24.0) | 5.4 (25.8) |
| Ranom (%) | 4.3 (15.8) | 5.6 (16.2) | 4.0 (17.3) | |
| I/σ(I) | 13.3 | 14.0 | 14.6 | 14.5 |
| Phasing powerb acentric/centric | 0.68/0.48 | 2.45/0.18 | ||
| Anomalous phasing power | 1.05 | 2.43 | 1.28 | |
| RCullisc acentric/centric | 0.59/0.61 | 0.53/0.54 | ||
| Anomalous RCullis | 0.92 | 0.68 | 2.45 | |
| Figure of merit |
0.828 |
|
|
|
| Refinement summary |
|
|
|
|
| Resolution range (Å) | 25.0–2.2 | |||
| Reflections | 27 080 | |||
| Space group | P21 (two molecules per asymmetric unit) | |||
| Unit cell | a = 42.863 Å, b = 85.703 Å, c = 75.481 Å, β = 100.47° | |||
| Protein atoms | 3792 | |||
| Solvent atoms | 410 (399 waters, two Ca2+, one Cl–, one MPDd) | |||
| R-valuee (%) | 19.3 (21.0) | |||
| Free R-valuef (%) | 25.7 (29.0) | |||
| Deviation from idealityg | ||||
| bond lengths (Å) | 0.0145 | |||
| bond angles (°) | 1.51 | |||
| Average B-factor (Å2) | 49.15 | |||
| Ramachandran ploth (%) | 93.8/6.3/0/0 | |||
Values in parentheses are for the highest resolution shell (2.25–2.20 Å for the native data set, 2.46–2.40 Å for MAD data sets).
aRmerge = ΣhΣj|<I(h)> – I(h)j|/ΣhΣj <I(h)>, where <I(h)> is the mean intensity of symmetry-equivalent reflections.
bPhasing power = r.m.s. (<FH>/E), where FH is the heavy atom structure factor amplitude and E is the residual lack of closure error.
cRCullis = Σ|(FPH – FP) – FH|/Σ|FPH – FP|
dMPD, 2-methyl-2,4-pentanediol.
eR-value = Σ||Fobs| – |Fcalc||/Σ|Fobs|, where Fobs and Fcalc are the observed and calculated structure factors, respectively.
fThe free R-value was calculated using 5% of the data.
gRoot-mean-square deviations relate to the Engh and Huber parameters.
hDistribution of the residues in the most favoured/additionally allowed/generously allowed/disallowed regions of the Ramachandran plot, according to PROCHECK (Laskowski et al., 1993).
Overall structure of the α-catenin M-fragment
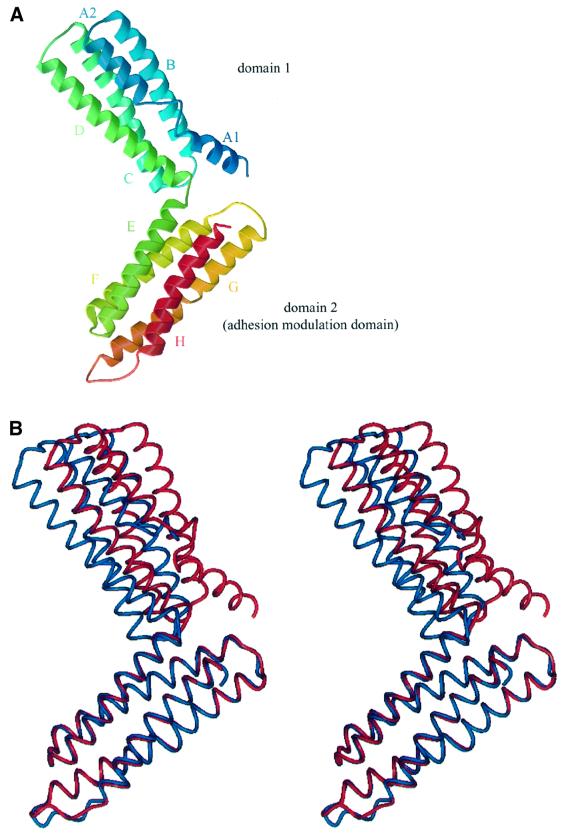
The 29 kDa α-catenin M-fragment adopts a bi-lobal structure formed from two domains with nearly identical overall architectures (Figure 3). Both domains comprise an antiparallel four-α-helix bundle with overall dimensions of 25 Å × 25 Å × 50 Å. Domain 1 is defined by residues 377–507 and domain 2 by residues 509–633. Within the α-catenin M-fragment, the two domains are arranged approximately perpendicular to one another and are connected by a short turn of one residue linking helix D of domain 1 with helix E of domain 2. Relatively few contacts are formed between the two domains of the M-fragment and there is a small buried solvent-accessible surface area at the domain interface of 400 Å2. This small interface permits relative inter-domain motion, resulting in different conformations of the two α-catenin M-fragment molecules within the asymmetric unit (referred to as molecules A and B; Figure 3B). Comparison of molecules A and B indicated that the disposition of domain 1 relative to domain 2 differs by 10° between the two molecules. The short linker between domains 1 and 2 may play a significant role in restricting the variety of possible conformations of the α-catenin M-fragment in solution, although it is possible that larger relative motions of domain 1 and domain 2 occur than that observed in the crystal lattice.
Fig. 3. Structure of α-catenin M-fragment. (A) Overall view of the structure with two four-helix domains, coloured from the N-terminus (blue) to the C-terminus (red). (B) Stereo-view showing the two crystallographically independent molecules (A and B) superimposed using equivalent Cα atoms of domain 2. Red denotes molecule A and blue denotes molecule B. The figure was prepared using BOBSCRIPT (Esnouf, 1997), Raster3D (Merrit and Murphy, 1994) and PREPI (S.Islam and M.Sternberg).
In addition to inter-domain conformational differences, the two crystallographically independent molecules of the α-catenin M-fragment differ at the N-terminus of domain 1. In molecule A, residues 377–392 form a short 16 residue α-helix (A1), connected to helix A2 by a segment of six residues (392–397). No density corresponding to the A1 helix is visible in molecule B, where it is assumed that the A1 helix is disordered. In molecule A, the A1 helix is poorly ordered, and some amino acid side chains are weakly defined within the electron density map. The tendency of this helix to become disordered explains the susceptibility to protease cleavage at Arg383 and, as described below, we found that Arg383 corresponds to the C-terminus of a 105 amino acid region of α-catenin that shares sequence similarity with the vinculin internal repeat sequence (Price et al., 1989). Superimposition of domains 1 and 2 of molecule A onto their counterparts in molecule B reveals the extent of their structural similarity. Excluding residues 377–392, Cα atoms of domains 1 and 2 in molecule A superimpose with equivalent Cα atoms in molecule B within an r.m.s. deviation of 0.4 Å (Figure 3B), indicating that individual domains adopt rigid conformations. An exception is the linker region (residues 602–608) connecting helix G with helix H. In domain 2 of molecule A, the linker is well defined in the electron density maps, but the equivalent region in molecule B is disordered. In molecule A, the conformation of residues 602–608 is stabilized by a crystal lattice contact.
Domain architecture of the α-catenin M-fragment
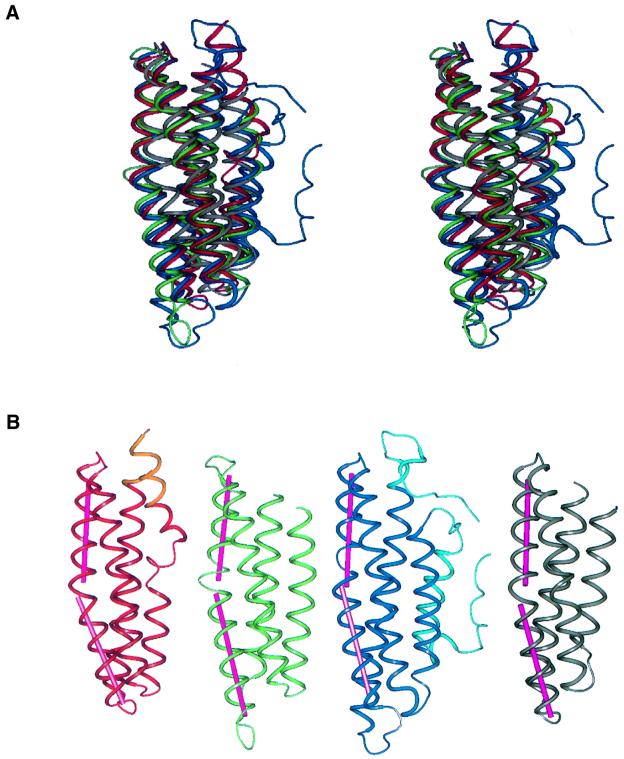
The antiparallel α-helix bundle architectures of domains 1 and 2 of the α-catenin M-fragment are strikingly similar (Figures 3 and 4). Within each domain, the first helix is the shortest, with the second and third helices being longer (25–32 residues). Using the DALI server (Holm and Sander, 1996; http://www.ebi.ac.uk/dali), structurally equivalent regions between the two domains were assigned quantitatively and the degree of structural similarity assessed. Some 116 residues of a total of 124 and 131 residues in domains 1 and 2, respectively, are structurally equivalent, such that their Cα atoms superimpose within an r.m.s. deviation of 2.1 Å. The DALI Z-score, a measure of the structural similarity in standard deviations above that expected between two molecules, is 14.2. Searching for structural similarities between domain 2 of the α-catenin M-fragment and the protein coordinate entries of the protein data bank (PDB; Berman et al., 2000) reveals matches with proteins characterized by an antiparallel α-helical bundle architecture (Table II). Strikingly, domain 2 of the α-catenin M-fragment is most closely related in architecture to other structural domains present in α-catenin and vinculin, namely the C-terminal tail domain of vinculin (DALI Z-score of 13.4) (Bakolitsa et al., 1999) and the N-terminal dimerization domain of α-catenin (DALI Z-score of 12.4) (Pokutta and Weis, 2000). Other structurally related proteins include the C-sub unit of the cytochrome oxidase complex, the aspartate receptor and apolipoprotein E (Table II). The tail domain of vinculin (residues 879–1066) formed from an antiparallel five-helix bundle shares 27% sequence identity with residues 679–864 of α-catenin (defined as the VH3 region). A structure-based sequence alignment of α-catenin and vinculin suggested that the tail domain of α-catenin will adopt a structure similar to that of helices 2–5 of vinculin (Bakolitsa et al., 1999). Thus, the protein structures with the highest structural similarity to domain 2 of the α-catenin M-fragment are other domains within α-catenin, namely domain 1 of the α-catenin M-fragment, the α-catenin tail and the α-catenin dimerization domain.
Fig. 4. Similarities between α-catenin M-fragment domains 1 and 2 and domains of other structurally related proteins. (A) Superimposition of α-catenin M-fragment domains 1 (red) and 2 (green), the vinculin tail domain (blue) and the α-catenin dimerization domain (grey). (B) A comparison of the α-catenin M-fragment domains 1 (red/orange) and 2 (green) with the vinculin tail domain (dark/light blue) and α-catenin dimerization domain (grey). Darker colours denote the structures used for alignment onto domain 2 of the α-catenin M-fragment. The figures were prepared using PREPI (S.Islam and M.Sternberg).
Table II. Highest structural relationship between domain 2 (adhesion modulation domain) of the α-catenin M-fragment and protein structures deposited at the PDB as defined by DALI.
| Residue numbers | Residues in domain 2 | R.m.s.d. of Cα (Å) | Z-score | No. of aligned residues |
|---|---|---|---|---|
| Domain 1 of the α-catenin M-fragment | ||||
| 396–459 | 518–581 | 2.1 | 14.2 | 116 |
| 460–475 | 583–598 | |||
| 476–479 | 604–607 | |||
| 481–504 | 608–631 | |||
| C-terminal domain of vinculin | ||||
| 917–940 | 510–533 | 2.2 | 13.4 | 121 |
| 943–989 | 534–580 | |||
| 990–1011 | 582–603 | |||
| 1015–1018 | 604–607 | |||
| 1020–1043 | 608–631 | |||
| N-terminal dimerization domain of α-catenin | ||||
| 150–167 | 508–525 | 2.8 | 12.4 | 115 |
| 170–216 | 535–581 | |||
| 217–231 | 583–597 | |||
| 233–258 | 606–631 | |||
| Cytochrome C oxidase C-subunit | ||||
| 2.1 | 11.9 | 116 | ||
| Aspartate receptor | ||||
| 2.7 | 9.8 | 110 | ||
| Apolipoprotein E3 fragment | ||||
| 3.2 | 8.2 | 112 | ||
Superimposing domains 1 and 2 of the α-catenin M-fragment with helices 2–5 of the vinculin tail reveals the nature of their structural relatedness (Figure 5A). The three domains share similarities in helix length, inter-helix packing geometries and helix conformation. For example, in each domain, three of the four helices are linear, the exception being the third helix. In domains 1 and 2 of the α-catenin M-fragment, the third helix is bent by 20–25°, caused by a kink at the helix centre and disruption of the helix hydrogen-bonding network. This bend is facilitated and stabilized by a conserved proline residue located at a structurally equivalent position in the two domains (Figures 4B and 5). A proline is also responsible for inducing a bend in the equivalent helix of vinculin, although it is positioned four residues N-terminal to the proline situated in helix 3 of the two α-catenin M-fragment domains (Figure 5A).
Fig. 5. Similarities between α-catenin M-fragment domains 1 and 2. (A) Sequence alignment of α-catenin M-fragment domains 1 and 2 and the vinculin tail domain. (B) Superimposition of α-catenin M-fragment domains 1 and 2 showing residues Val433, Pro462, Ala501 and Val502 of domain 1 and the structurally equivalent residues of domain 2 [indicated with an asterisk in (A)].
Because the individual domains of α-catenin and vinculin are more closely related to each other at the three-dimensional level than to other proteins deposited in the protein data bank (Table II), it is tempting to speculate that the different domains of α-catenin/vinculin originated as a result of gene duplication events. A sequence align ment of domains 1 and 2 of the α-catenin M-fragment, directed by their tertiary structural similarities, indicates that only 12% of residues are invariant between the two domains, a value that is not significantly higher than that expected from an alignment of two unrelated sequences (Figure 5A). Interestingly, a number of these identical residues perform structurally equivalent roles. For example, the side chains of residues Val433, Ala501 and Val502 of domain 1 pack together forming a hydrophobic cluster that is structurally identical to the hydrophobic cluster formed from residues Val555, Ala628 and Val629 of domain 2 (Figure 5B). The finding that this triad of conserved residues performs equivalent structural roles, and that conserved prolines at residues 462 and 585 are responsible for an equivalent bend in the third helix of their respective domains, suggests that the two domains may have diverged from a common ancestor.
A structure-based multiple sequence alignment of α-catenin and vinculin from diverse species suggests that the structure of the M-fragment as defined in α-catenin will be represented in vinculin. Some 18 of the 259 residues of the α-catenin M-fragment are invariant amongst all known sequences of α-catenin and vinculin, and analysis of the structure reveals that these residues play predominantly structural roles in defining the hydrophobic core and inter-helix packing interactions. The proline residues responsible for bending helix 3 in both the four-helix bundles are invariant in domain 1 and are highly conserved in domain 2 in all α-catenin and vinculin sequences analysed. Moreover, sequence insertions and deletions correspond to the loops connecting α-helices, and analysis of the profile of hydrophobic residues suggests that equivalent α-helices will have similar lengths in α-catenin and vinculin. It is also likely that the structure of the inter-domain linker of the α-catenin M-fragment will be similar in vinculin since the one-residue linker connecting helices D and E of domains 1 and 2, respectively, is conserved in the vinculin sequences.
The adhesion modulation domain of α-catenin forms dimers
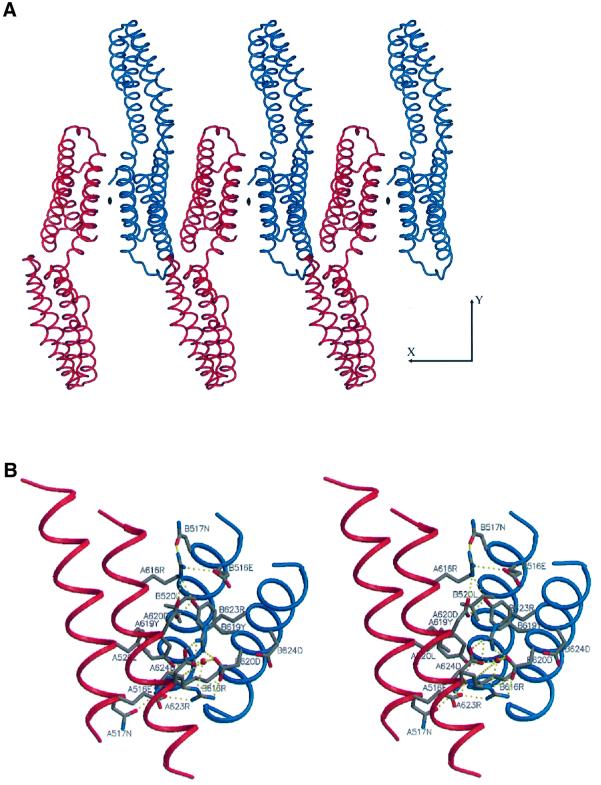
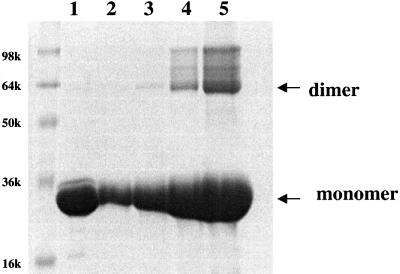
Studies using E-cadherin–α-catenin fusion proteins in L cell transfectants established that residues 509–643 of α-catenin, referred to as the adhesion modulation domain, are necessary and sufficient to mediate the cell adhesion activity of E-cadherin–α-catenin chimeras (Imamura et al., 1999). The crystal structure of the α-catenin M-fragment described here reveals that the adhesion modulation region of α-catenin corresponds to a structural domain, namely domain 2 of the α-catenin M-fragment (residues 509–633). Moreover, the nature of the interaction of α-catenin M-fragment molecules within the crystal lattice suggests a molecular mechanism by which the adhesion modulation domain may function to promote cell adhesion. The adhesion modulation domains of the two molecules of the α-catenin M-fragment in the crystallographic asymmetric unit are associated by a molecular 2-fold symmetry axis (Figure 6A). The interface between the two domains is formed by the perpendicular packing of helices E and H against their counterparts in the symmetry-related molecule and buries 1280 Å2 of solvent-accessible surface area, a value that is typical of many protein– protein interfaces and significantly higher than the surface area buried between non-specific interfaces of ∼500 Å2 generated by crystal packing contacts (Janin and Rodier, 1995). Protein–protein interactions at this interface are formed from a hydrophobic core of Leu520 and Tyr619, which pack against their symmetry-related equivalents, and a network of hydrogen bonds between the side chains of Glu516 and Asn517 of helix E, and Arg616, Tyr619 and Asp620 of helix H. Finally, Arg623 and Asp624 form a bidentate salt bridge interaction (Figure 6B). We detect the presence of dimers of the α-catenin M-fragment in solution as judged by chemical cross-linking analysis at protein concentrations of >3 µM (Figure 7), although not by size exclusion and dynamic light scattering approaches, suggesting that the interactions between molecules determined by this interface are of relatively low affinity. Since cell adhesion mediated by E-cadherin requires formation of lateral dimers of E-cadherin molecules within the plasma membrane (Yap et al., 1997b, 1998), our finding that the adhesion modulation domain has the capacity to form dimers provides a plausible molecular explanation for the results of Imamura et al. (1999). We propose that by forming homodimeric interactions, the adhesion modulation domain of α-catenin stabilizes and/or facilitates dimerization of cadherin cytoplasmic tails and hence functions to promote lateral interactions of the E-cadherin ectodomain, with a concomitant increase in cell adhesive strength.
Fig. 6. View of the dimer interface of the α-catenin M-fragment domain 2. (A) Crystal packing of dimers of molecules A and B of the α-catenin M-fragment viewed perpendicular to the 2-fold non-crystallographic symmetry axis. (B) Stereo-view showing the interactions between the monomers at the dimer interface formed from domain 2. Colouring scheme as in Figure 3B.
Fig. 7. Evidence that the M-fragment of α-catenin exists as a dimer in solution. Lane 1: 0.25 mg/ml M-fragment of α-catenin in the absence of the cross-linking reagent dimethyl suberimidate (DMS). Lanes 2–5: 0.05, 0.1, 0.25 and 0.5 mg/ml of the α-catenin M-fragment incubated with a 30-fold molar access of DMS for 1.5 h at 20°C. The formation of dimers of the α-catenin M-fragment at concentrations of >0.1 mg/ml (3 µM) (lanes 3–5) is detected by the formation of dimers cross-linked with DMS visualized on an SDS–polyacrylamide gel.
Detection of a vinculin internal repeat sequence within α-catenin
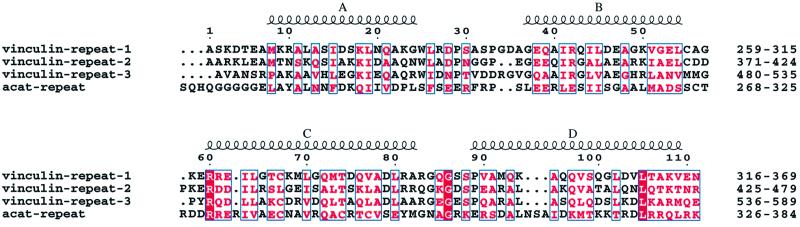
Previous sequence analysis of vinculin identified three tandem internal sequence repeats (here termed VR) of ∼110 amino acid residues located between the VH1 and VH2 regions of the molecule (Price et al., 1989). Such internal repeats have not been noted to date within α-catenin sequences. By incorporating data from α-catenin and vinculin sequence alignments, the position of protease-sensitive cleavage sites and crystal structure information, we propose that an entire VR is present between residues 271 and 383 of α-catenin (Figure 1). Moreover, secondary structure predictions and fold recognition analysis indicate that VRs adopt a four-α-helix bundle architecture. The C-terminus of the VR of α-catenin is positioned at Arg383, since the previously described VH2 regions of vinculin and α-catenin include the C-terminal 59 residues of the third VR (VR3) of vinculin, corresponding to residues 326–383 of α-catenin. Significantly, the C-terminus of this region of sequence similarity between α-catenin and the VR3 of vinculin corresponds to the site of trypsin cleavage in α-catenin between Arg383 and Lys384. The C-terminus of the N-terminal dimerization domain of α-catenin corresponds to Thr262 (Pokutta and Weis, 2000). These authors also reported a protease cleavage site C-terminal to residue 279 of α-catenin, suggesting conformational flexibility within the region defined by residues 262–279. By submitting residues 270–383 of α-catenin and the three internal repeats of vinculin to a multiple sequence alignment and secondary structure prediction using CLUSTAL W (Thompson et al., 1994) and the JPRED server (http://barton.ebi.ac.uk; Cuff and Barton, 2000), we observed the following. First, residues 270–383 of α-catenin share significant sequence similarity with the three internal repeats of vinculin (Figure 8). Secondly, the secondary structure prediction, complemented by analysis of the pattern of hydrophobic and hydrophilic residues, predicts that VRs will consist of four α-helices. Interrogating the protein database by means of a fold recognition program (3D-PSSM; Kelley et al., 2000; http://www.bmm.icnet.uk/∼3dpssm) suggested that these four helices are likely to adopt an antiparallel configuration typical of the domains of α-catenin and vinculin for which there is X-ray crystallographic information.
Fig. 8. Sequence alignment of three internal repeats of human meta-vinculin (vinculin repeats) and the related sequence in human α(E)-catenin. The mean sequence identity between the α-catenin VR and the three VRs of vinculin is ∼10%. The positions of the four predicted α-helices are shown. The figure was produced using ESPript (Gouet et al., 1999).
Concluding remarks
Together with previous studies of the N-terminus of α-catenin (Pokutta and Weis, 2000) and the C-terminal tail of vinculin (Bakolitsa et al., 1999), our structural studies of the α-catenin M-fragment provide a more fundamental understanding of the architecture of α-catenin and vinculin, molecules that maintain cell–cell and cell–matrix adhesion. A striking finding of these crystallographic studies, coupled to multiple sequence alignments and limited proteolysis analysis, is the discovery of the modular organization of α-catenin into structurally similar domains. The domains are characterized by an antiparallel α-helical organization composed of antiparallel α-helix bundles of between four and five α-helices. Secondary structure predictions indicate that the vinculin internal repeat sequences will adopt a similar antiparallel α-helix bundle. The linker regions connecting the domains vary in length, and in the degree of flexibility and susceptibility to protease cleavage.
The principle roles of α-catenin and vinculin are to mediate protein–protein and protein–phospholipid interactions. Protein molecules composed of tandemly repeated α-helical bundles provide a structural framework for the assembly of multiprotein complexes. Each domain may play a specific role and, therefore, novel biological functions within a protein may be generated as a result of the combinatorial manner in which the domains are arranged. For example, biological studies previously had identified residues 509–643 of α-catenin as an adhesion modulation domain, a region of α-catenin that corresponds to a structural domain of the α-catenin M-fragment. The vinculin tail mediates intramolecular interactions, as well as vinculin–phospholipid interactions, whereas the N-terminal domain of α-catenin is responsible for interactions with β-catenin and also mediates homodimerization. The domain of α-catenin defined by residues 273–383, which we show here to be related to the vinculin internal repeat sequences and that is predicted to form a four-α-helix bundle, encompasses the binding site for vinculin and α-actinin (residues 325–383). Helical bundles are structurally stable modules with extended surfaces suited for protein–protein interfaces. Flexible connections linking adjacent domains allow these structural and functional domains to behave independently of one another. On the basis of structural and functional studies, a functional and structural assignment of α-catenin is shown in Figure 1.
Finally, our crystal structure of the α-catenin M-fragment suggests a molecular mechanism for the promotion of cell–cell adhesion mediated by the adhesion modulation domain. The tendency of this domain to form dimers, as observed in the crystal and by chemical cross-linking in solution, would facilitate the formation of lateral dimers of the cadherin receptor to which α-catenin is attached via β-catenin.
Materials and methods
Protein expression and purification
The full-length human α-catenin cDNA was amplified by PCR and cloned into the NdeI and EcoRI sites of the pET28M vector (Hanlon and Barford, 1998), which places a His6 tag at the N-terminus of the protein. Expression of this protein was performed for 3 h at 37°C in E.coli strain B834. The protein was purified using a combination of Ni-NTA agarose chromatography (Qiagen), mono-S cation exchange chromatography (Amersham-Pharmacia Biotech), hydrophobic interaction chromatography using phenyl-TSK (Tosohoas) and finally S-200 size exclusion chromatography (Amersham-Pharmacia Biotech). Limited proteolysis was performed using trypsin and subtilisin using the approach described by Huber et al. (1997). α-catenin (2 mg/ml) in a buffer of 90 mM Tris–HCl pH 8.5, 4 mM dithiothreitol (DTT), 2 mM CaCl2 was digested at 20°C for 20 min with the following titrations of either trypsin or subtilisin: 0.1, 0.5, 1.0, 5.0, 10 and 20 µg/ml. The resultant major digest product was characterized by means of mass spectroscopic analysis and N-terminal sequencing, indicating an N-terminus at residue Ala58 and a C-terminus at Arg633. A pET28M-based expression vector corresponding to this fragment of α-catenin was constructed and the protein was expressed in E.coli as for full-length α-catenin and purified using Ni-NTA agarose chromatography (Qiagen), mono-Q anion exchange chromatography (Amersham-Pharmacia Biotech), hydrophobic interaction chromatography using phenyl-TSK (Tosohoas) and finally S-200 size exclusion chromatography. Crystals were obtained using a variety of conditions; however, none of these diffracted to better than 7 Å resolution. Limited digestion of this domain was performed using trypsin, yielding a 29 kDa fragment that by N-terminal sequencing was found to have an N-terminus at Lys384. Expression and purification of this domain using similar conditions to those described for the 63 kDa α-catenin fragment yielded pure protein that did not crystallize. By placing the N-terminus at Asp377, we obtained a protein that crystallized at 20°C in a buffer containing 30% (v/v) 2-methyl-2,4-pentanediol (MPD), 0.1 M sodium acetate pH 4.6 and 20 mM CaCl2 using the hanging drop vapour diffusion procedure. Needle-like crystals were obtained that were improved by micro-seeding using a final precipitant concentration of 20–22% (v/v) MPD, 0.1 M sodium acetate pH 5.0 and 80–100 mM CaCl2. For data collection at 100 K, crystals were transferred into a cryoprotectant buffer containing 15% (v/v) glycerol. SeMet protein was prepared using the method of Neidhardt et al. (1974).
Structure determination
The 29 kDa α-catenin M-fragment crystallized in space group P21 with two molecules per asymmetric unit. The structure was determined by the MAD method using SeMet-labelled crystals with data collected at the PX beam-line, Elettra Synchrotron, Trieste, Italy at λ1 (inflection), λ2 (peak) and λ3 (remote). Native data to 2.2 Å were collected at beam line 14.1, the SRS, Daresbury. All crystallographic data were processed using HKL (Otwinowski and Minor, 1997) and SCALA of the CCP4 program suite (CCP4, 1994). SeMet heavy-atom coordinates were identified using SOLVE (Terwilliger and Berendzen, 1999) and their parameters were refined and protein phase calculations were performed using the program SHARP (De la Fortelle and Bricogne, 1997) by treating the data as a special case of multiple isomorphous replacement (Ramakrishnan et al., 1993). The improved phases from SOLOMON (Abrahams and Leslie, 1996) were used to calculate electron density maps that were readily interpretable. Crystallographic statistics are listed in Table I. Electron density map interpretation and model building were performed using the program O (Jones et al., 1991) and the structure was refined using CNS (Brünger et al., 1998).
Chemical cross-linking experiments
Protein for cross-linking experiments was diluted to a final concentration of 0.05–0.5 mg/ml in a buffer containing 100 mM HEPES pH 8.5. A 30-fold molar excess of the cross-linking reagent dimethyl suberimidate (DMS) was added to the protein and cross-linking was performed for 1.5 h at 20°C. After this time, 10 µl of the reaction were loaded onto an SDS–polyacrylamide gel.
Accession number
Protein coordinates have been deposited with ID accession code 1h6g.
Acknowledgments
Acknowledgements
We thank the staff at the SRS Daresbury and the PX Station, Elettra, Trieste for access to synchrotron radiation facilities and for their help during data collection. The work was supported by grants from the CRC and MRC to D.B. and an NIH grant GM55989 to N.K.T.
References
- Aberle H., Butz,S., Stappert,J., Weissig,H., Kemler,R. and Hoschuetzky,H. (1994) Assembly of the cadherin–catenin complex in vitro with recombinant proteins. J. Cell Sci., 107, 3655–3663. [DOI] [PubMed] [Google Scholar]
- Abrahams J.P. and Leslie,A.G.W. (1996) Methods used in the structure determination of the bovine mitochondrial F1 ATPase. Acta Crystallogr. D, 52, 30–42. [DOI] [PubMed] [Google Scholar]
- Bakolitsa C., de Pereda,J.M., Bagshaw,C.R., Critchley,D.R. and Liddington,R.C. (1999) Crystal structure of the vinculin tail suggests a pathway for activation. Cell, 99, 603–613. [DOI] [PubMed] [Google Scholar]
- Berman H.M., Westbrook,J., Feng,Z., Gillialnd,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchmeier W. and Behrens,J. (1994) Cadherin expression in carcinomas: role in the formation of cell junctions and the prevention of invasiveness. Biochim. Biophys. Acta, 1198, 11–26. [DOI] [PubMed] [Google Scholar]
- Briecher W.M., Yap,A.S. and Gumbiner,B.M. (1996) Lateral dimerisation is required for the homophilic binding activity of C-cadherin. J. Cell Biol., 135, 487–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Cuff J.A. and Barton,G.J. (2000) Application of multiple sequence alignment profiles to improve secondary structure predictions. Proteins, 40, 502–511. [DOI] [PubMed] [Google Scholar]
- De la Fortelle E. and Bricogne,G. (1997) Maximum-likelihood heavy-atom refinement in the MIR and MAD methods. Methods Enzymol., 276, 472–494. [DOI] [PubMed] [Google Scholar]
- Esnouf R.M. (1997) An extensively modified version of MOLSCRIPT that includes greatly enhanced colouring capabilities. J. Mol. Graph., 15, 132–134. [DOI] [PubMed] [Google Scholar]
- Ewing C.M., Ru,N., Morton,R.A., Robinson,J.C., Wheelock,M.J., Johnson,K.R., Barrett,J.C. and Isaacs,W.B. (1995) Chromosome 5 suppresses tumorigenicity of PC3 prostate cancer cells. Correlation with re-expression of α-catenin and restoration of E-cadherin function. Cancer Res., 55, 4813–4817. [PubMed] [Google Scholar]
- Fujimori T. and Takeichi,M. (1993) Disruption of epithelial cell–cell adhesion by exogenous expression of a mutated nonfunctional N-cadherin. Mol. Biol. Cell, 4, 37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa Y., Nakatsuru,S., Nagafuchi,A., Tsukita,S., Muto,T., Nakamura,Y. and Horii,A. (1994) Structure, expression and chromosome assignment of the human catenin α 1 gene (CTNNA1). Cytogenet. Cell Genet., 65, 74–78. [DOI] [PubMed] [Google Scholar]
- Gouet P., Courcelle,P., Stuart,D.I. and Metoz,F. (1999) ESPript: multiple sequence alignments in PostScript. Bioinformatics, 15, 305–308. [DOI] [PubMed] [Google Scholar]
- Gumbiner B.M. (1996) Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell, 84, 345–357. [DOI] [PubMed] [Google Scholar]
- Hanlon N. and Barford,D. (1998) Purification and crystallisation of the CDK-associated protein phosphatase, KAP expressed in Escherichia coli. Protein Sci., 7, 508–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermiston M.L. and Gordon,J.I. (1995) In vivo analysis of cadherin function in the mouse intestinal epithelium: essential roles in adhesion, maintenance of differentiation and regulation of programmed cell death. J. Cell Biol., 129, 489–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrenknecht K., Ozawa,M., Eckerskorn,C., Lottspeich,F., Lenter,M. and Kemler,R. (1991) The uvomorulin-anchorage protein α-catenin is a vinculin homologue. Proc. Natl Acad. Sci. USA, 88, 9156–9160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano S., Kimoto,Y., Shimoyama,Y., Hirohashi,S. and Takeichi,M. (1992) Identification of a neural α-catenin as a key regulator of cadherin function and multicellular organisation. Cell, 70, 293–301. [DOI] [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1996) Mapping the protein universe. Science, 273, 595–602. [DOI] [PubMed] [Google Scholar]
- Huber A.H., Nelson,W.J. and Weis,W.I. (1997) Three-dimensional structure of the armadillo region of β-catenin. Cell, 90, 871–882. [DOI] [PubMed] [Google Scholar]
- Imamura Y., Itoh,M., Maeno,Y., Tsukita,S. and Nagafuchi,A. (1999) Functional domains of α-catenin required for the strong state of cadherin-based cell adhesion. J. Cell Biol., 144, 1311–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janin J. and Rodier,F. (1995) Protein–protein interactions at crystal contacts. Proteins, 23, 580–587. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 50, 157–160. [DOI] [PubMed] [Google Scholar]
- Kawanishi J., Kato,J., Sasaki,K., Fujii,S., Watanabe,N. and Niitsu,Y. (1995) Loss of E-cadherin-dependent cell–cell adhesion due to mutation of the β-catenin gene in human cancer cell line, HSC-39. Mol. Cell Biol., 15, 1175–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley L.A., MacCullum,R.M. and Sternberg,M.J.E. (2000) Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol., 299, 501–502. [DOI] [PubMed] [Google Scholar]
- Kintner C. (1992) Regulation of embryonic cell adhesion by the cadherin cytoplasmic domain. Cell, 69, 225–236. [DOI] [PubMed] [Google Scholar]
- Knudsen K.A., Soler,A.P., Johnson,K.R. and Wheelock,M.J. (1995) Interaction of α-actin with the cadherin/catenin cell–cell adhesion complex via α-catenin. J. Cell Biol., 130, 67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur,M.W. and Thornton,J.M. (1998) Validation of protein models derived from experiment. Curr. Opin. Struct. Biol., 8, 631–639. [DOI] [PubMed] [Google Scholar]
- Merrit E.A. and Murphy,M.E.P. (1994) Raster3D version 2.0. A program for photorealistic molecular graphics. Acta Crystrallogr. D, 50, 869–873. [DOI] [PubMed] [Google Scholar]
- Morton R.A., Ewing,C.M., Nagafuchi,A., Tsukita,A. and Isaacs,W.B. (1993) Reduction of E-cadherin levels and deletion of the α-catenin gene in human prostate cancer cells. Cancer Res., 53, 3585–3590. [PubMed] [Google Scholar]
- Nagafuchi A. and Takeichi,M. (1988) Cell binding function of E-cadherin is regulated by its cytoplasmic domain. EMBO J., 7, 3679–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagafuchi A. and Takeichi,M. (1989) Transmembrane control of cadherin-mediated cell adhesion: a 94-kDa protein functionally associated with a specific region of the cytoplasmic domain of E-cadherin. Cell Regul., 1, 37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagafuchi A., Takeichi,M. and Tsukita,S. (1991) The 102-kD cadherin-associated protein: similarity to vinculin and posttranscriptional regulation of expression. Cell, 65, 849–857. [DOI] [PubMed] [Google Scholar]
- Nagafuchi A., Ishihara,S. and Tsukita,S. (1994) The roles of catenins in the cadherin-mediated cell adhesions: functional analysis of E-cadherin–α catenin fusion molecules. J. Cell Biol., 127, 235–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagar B., Overduin,M., Ikura,M. and Rini,J.M. (1996) Structural basis of calcium-induced E-cadherin rigidification and dimerisation. Nature, 380, 360–364. [DOI] [PubMed] [Google Scholar]
- Nathke I.S., Hinck,L., Swedlow,J.R., Papkoff,J. and Nelson,W.J. (1994) Defining interactions and distributions of cadherin and catenin complexes in polarised epitheial cells. J. Cell Biol., 125, 1341–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidhardt F., Bloch,P.L. and Smith D.F. (1974) Culture media for enterobacteria. J. Bacteriol., 119, 736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieset J.E., Redfield,A.R., Jin,F., Knudsen,K.A., Johnson,K.R. and Wheelock,M.J. (1997) Characterisation of the interactions of α-catenin and α-actinin and β-catenin/plakoglobulin. J. Cell Sci., 110, 1013–1022. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Oyama T. et al. (1994) A truncated β-catenin disrupts the interaction between the E-cadherin and α-catenin: a cause of loss of intercellular adhesiveness in human cancer cell lines. Cancer Res., 54, 6282–6287. [PubMed] [Google Scholar]
- Ozawa M. and Kemler,R. (1998) The membrane-proximal region of the E-cadherin cytoplasmic domain prevents dimerisation and negatively regulates adhesion activity. J. Cell Biol., 142, 1605–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa M., Ringwald,M. and Kemler,R. (1990) Uvomorulin–catenin complex formation is regulated by a specific domain in the cytoplasmic region of the cell adhesion molecule. Proc. Natl Acad. Sci. USA, 87, 4246–4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokutta S. and Weis,W.I. (2000) Structure of the dimerisation and β-catenin binding region of α-catenin. Mol. Cell, 5, 533–543. [DOI] [PubMed] [Google Scholar]
- Price G.J., Jones,P., Davison,M.D., Patel,B., Bendori,R., Geiger,B. and Critchley,D.R. (1989) Primary sequence and domain structure of chicken vinculin. Biochem. J., 259, 453–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provost E. and Rimm,D.L. (1999) Controversies at the cytoplasmic face of the cadherin-based adhesion complex. Curr. Opin. Cell Biol., 11, 567–572. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan V., Finch,C.T., Graziano,V., Lee,P.J. and Sweet,R.M. (1993) Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature, 362, 219–223. [DOI] [PubMed] [Google Scholar]
- Rimm D.L., Koslov,E.R., Kebriaei,P., Cianci,C.D. and Morrow,J.S. (1995) α1 (E)-catenin is an actin-binding and -bundling protein mediating the attachment of F-actin to the membrane adhesion complex. Proc. Natl Acad. Sci. USA, 92, 8813–8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B., Sander,C. and Scheider,R. (1994) PHD: an automatic mailserver for the prediction of protein secondary structure. Comput. Appl. Biosci., 10, 53–60. [DOI] [PubMed] [Google Scholar]
- Rudiger M. (1998) Vinculin and α-catenin: shared and unique functions in adherens junctions. BioEssays, 20, 733–740. [DOI] [PubMed] [Google Scholar]
- Shapiro L. et al. (1995) Structural basis of cell–cell adhesion by cadherins. Nature, 374, 327–337. [DOI] [PubMed] [Google Scholar]
- Shimoyama Y., Nagafuchi,A., Fujita,S., Gotch,M., Takeichi,M., Tsukita,S. and Hirochashi,S. (1992) Cadherin dysfunction in a human cancer cell line. Possible involvement of loss of α-catenin expression in reduced cell–cell adhesiveness. Cancer Res., 52, 5770–5774. [PubMed] [Google Scholar]
- Takeichi M. (1991) Cadherin cell adhesion receptors as a morphogenetic regulator. Science, 251, 1451–1455. [DOI] [PubMed] [Google Scholar]
- Terwilliger T. and Berendzen,J. (1999) Automated MAD and MIR structure solutions. Acta Crystallogr. D, 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progress multiple sequence alignment through sequence weighting position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4674–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watabe M.A., Nagafuchi,S., Tsukita,S. and Takeichi,M. (1994) Induction of polarised cell–cell association and retardation of growth by activation of the E-cadherin–catenin adhesion system in a dispersed carcinoma line. J. Cell Biol., 127, 247–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watabe-Uchida M. et al. (1998) α-Catenin–vinculin interactions function to organise the apical junctional complex in epithelial cells. J. Cell Biol., 142, 847–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss E.E., Kroemker,M., Rudiger,A.-H. and Rudiger,M. (1998) Vinculin is part of the cadherin–catenin junctional complex: complex formation between α-catenin and vinculin. J. Cell Biol., 141, 755–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap A.S., Brieher,W.M. and Gumbiner,B.M. (1997a) Molecular and functional analysis of cadherin-based adherens junctions. Annu. Rev. Cell Dev. Biol., 13, 119–146. [DOI] [PubMed] [Google Scholar]
- Yap A.S., Brieher,W.M., Pruschy,M. and Gumbiner,B.M. (1997b) Lateral clustering of the adhesive ectodomain: a fundamental determinant of cadherin function. Curr. Biol., 7, 308–315. [DOI] [PubMed] [Google Scholar]
- Yap A.S., Niessen,C.M. and Gumbiner,B.M. (1998) The juxtamembrane region of the cadherin cytoplasmic tail supports lateral clustering, adhesive strengthening and interactions with p120ctn. J. Cell Biol., 141, 779–789. [DOI] [PMC free article] [PubMed] [Google Scholar]