Introduction
For a long time it was believed that the nucleotide sequence of every RNA would represent a simple copy of its coding DNA. This axiom of molecular biology, however, has suffered several jolts during the last 25 years. The discovery of intervening or intronic sequences (splicing) and the finding that individual nucleotides can be inserted into and deleted from RNAs (RNA editing) demonstrated that genetic information can be changed at the RNA level. Post-transcriptional modification of individual ribonucleotides, namely deamination of adenosines and cytidines, can also change the readout of mRNAs (reviewed by Maas and Rich, 2000). Stable cellular RNAs, such as tRNAs, rRNAs and snRNAs, have long been known to contain a large number of post-transcriptionally synthesized irregular ribonucleotides (Limbach et al., 1994). In tRNAs, the modified nucleotides can facilitate the formation of correct anticodon–codon interaction and thereby increase the efficiency and fidelity of translation (reviewed by Agris, 1996). In rRNAs and spliceosomal snRNAs, methylation of the ribose moiety at the 2′-hydroxyl group and conversion of uridines into pseudouridine are the most prevalent nucleotide modifications. Since 2′-O-methylated nucleotides and pseudouridines are restricted to the functionally essential regions of rRNAs and snRNAs (Reddy et al., 1988; Maden, 1990), they are expected to contribute to the faithful function of the ribosome and the spliceosome. Consistent with this view, lack of ribosomal pseudouridines can reduce the growth rate or confer a selective disadvantage when it is competed against wild-type ribosomes (Raychaudhuri et al., 1999; Wrzesinski et al., 2000). More tellingly, nucleotide modifications in the 5′-terminal region of the U2 spliceosomal snRNA are absolutely essential for its function in pre-mRNA splicing (Yu et al., 1998).
Ribosomal 2′-O-methylated nucleotides and pseudouridines are synthesized by small nucleolar RNPs
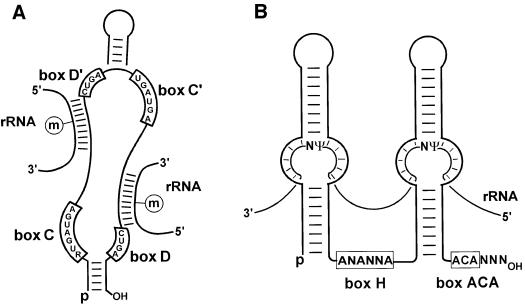
The human 18S, 5.8S and 28S rRNAs together carry ∼110 2′-O-methyl groups and almost 100 pseudouridines (Maden, 1990). Synthesis of this large number of modified nucleotides in rRNAs is directed by guide snoRNAs. The 2′-O-methylation and pseudouridylation guide snoRNAs possess distinct sequence and structural elements (Figure 1). The methylation guide snoRNAs carry the conserved box C (RUGAUGA, where R stands for any purine) and D (CUGA) motifs near their 5′ and 3′ ends, respectively (Figure 1A). The C and D boxes are frequently folded together by a short (4–5 bp) terminal helix. Additional, often imperfect copies of the C and D boxes, called C′ and D′ boxes, are located internally (Tycowski et al., 1996a; Kiss-László et al., 1998). The distance between the D′ and C′ boxes is restricted to 3–9 nucleotides, and frequently an internal stem brings these elements closer to each other. The 2′-O-methylation guide snoRNAs possess one or sometimes two 10–21 nucleotide antisense elements, which can form perfect double helices with rRNA sequences (Bachellerie et al., 1995). The ribosomal nucleotide positioned 5 bp upstream of the D or D′ box of the snoRNA is selected for 2′-O-methylation (Cavaillé et al., 1996; Kiss-László et al., 1996; Tycowski et al., 1996b).
Fig. 1. Structure and function of (A) 2′-O-methylation and (B) pseudouridylation guide snoRNAs. The consensus sequences of boxes C, C′, D, D′, H and ACA are indicated (R is a purine and N stands for any nucleotide). Models for molecular selection of 2′-O-methylated nucleotides and pseudouridine were adopted from Kiss-László et al. (1998) and Ganot et al. (1997a), respectively.
The pseudouridylation guide snoRNAs consist of two hairpins and two short single-stranded regions, which contain the conserved H (ANANNA, where N stands for any nucleotide) and ACA boxes (Balakin et al., 1996; Ganot et al., 1997b) (Figure 1B). An internal loop in the 5′- and/or 3′-terminal hairpin of the snoRNA forms a complex pseudoknot structure with rRNA sequences. The substrate ribosomal uridine is positioned at the base of the upper stem closing the recognition loop of the snoRNA (Ganot et al., 1997a). The distance between the substrate uridine and the H or ACA box of the snoRNA (∼14–16 nucleotides) is an important structural determinant for selection of the correct pseudouridylation site (Ni et al., 1997; Bortolin et al., 1999).
The 2′-O-methylation and pseudouridylation guide snoRNAs function in the form of small ribonucleoprotein particles (snoRNPs). Each snoRNP consists of a specific snoRNA and a set of associated proteins common to all box C/D or H/ACA snoRNPs. The conserved box C/D and H/ACA motifs are believed to function as protein binding signals. The box C/D 2′-O-methylation guide snoRNPs contain at least four evolutionarily conserved, essential proteins: fibrillarins Nop56p, Nop58p/Nop5p and Snu13p (Schimmang et al., 1989; Tyc and Steitz, 1989; Lafontaine and Tollervey, 1999, 2000; Lyman et al., 1999; Newman et al., 2000; Watkins et al., 2000). Fibrillarin shares a conserved domain with known S-adenosylmethionine-dependent methyltransferases (Niewmierzycka and Clarke, 1999; Wang et al., 2000). A point mutation in the putative methyltransferase domain of the yeast fibrillarin, Nop1p, inhibits the overall ribose methylation of rRNAs (Tollervey et al., 1993), suggesting that fibrillarin is the methyltransferase in box C/D snoRNPs.
Thus far, four snoRNP proteins, Cbf5p, Gar1p, Nhp2p and Nop10p, have been identified in box H/ACA pseudouridylation guide snoRNPs (Balakin et al., 1996; Ganot et al., 1997b; Henras et al., 1998; Watkins et al., 1998). Cbf5p (Nap57p in mammals), which is highly homologous to the Escherichia coli tRNA:Ψ55 pseudouridine synthase, provides the pseudouridine synthase activity for the snoRNA-directed pseudouridylation reaction (Lafontaine et al., 1998; Zebarjadian et al., 1999). Gar1p, Nhp2p and Nop10p are also essential for the pseudouridylation reaction, but their function remains unknown (Bousquet-Antonelli et al., 1997; Henras et al., 1998).
snoRNPs function in modification of various classes of cellular RNAs
Besides synthesis of >200 modified nucleotides in rRNAs, snoRNPs also function in 2′-O-methylation and pseudouridylation of spliceosomal snRNAs. The five major mammalian spliceosomal snRNAs, U1, U2, U4, U5 and U6, carry 30 2′-O-methyl groups and 24 pseudouridines (Reddy and Busch, 1988). Involvement of snoRNPs in modification of spliceosomal snRNAs was first demonstrated by identification of two box C/D snoRNAs, mgU6-47 and mgU6-77, which function in 2′-O-methylation of the RNA polymerase (pol) III-transcribed U6 snRNA (Tycowski et al., 1998). Later, it was shown that all trans-acting factors directing the synthesis of the eight 2′-O-methylated nucleotides and three pseudouridines in the U6 snRNA are present and are functionally active in the nucleolus (Ganot et al., 1999). This observation, coupled with the finding that each U6 modification factor recognizes short sequences around the target nucleotide, led to the proposal that 2′-O-methylation and pseudouridylation of the U6 snRNA are mediated exclusively by snoRNPs.
An unusual ‘hybrid’ snoRNA, called U85, that contains both box C/D and H/ACA domains has been identified in human and Drosophila cells (Jády and Kiss, 2001). The U85 snoRNA is associated with both box C/D- and H/ACA-specific snoRNP proteins. In vitro and in vivo RNA modification experiments demonstrated that the U85 snoRNA functions in 2′-O-methylation and pseudouridylation of the RNA pol II-transcribed U5 snRNA. Current observations suggest that U85 is not the only snoRNA that directs modification of a pol II-specific spliceosomal snRNA. Several putative 2′-O-methylation and pseudouridylation guide snoRNAs with significant sequence complementarities to the U1, U2, U4 and U5 snRNAs have been identified (Hüttenhofer et al., 2001; B.E.Jády and T.Kiss, unpublished data). The new snoRNAs possess the potential to select known 2′-O-methylated nucleotides and pseudouridines in U1, U2, U4 and U5 snRNAs, further supporting the conclusion that snoRNAs function in post-transcriptional modification of both pol II- and pol III-transcribed spliceosomal snRNAs. To what extent snoRNPs participate in 2′-O-methylation and pseudouridylation of pol II-specific snRNAs remains unclear. However, it seems that synthesis of at least some pseudouridines is achieved by protein enzymes (Massenet et al., 1999; Jády and Kiss, 2001).
Current observations also suggest that the snoRNA-based guide mechanism is not limited to the modification of ribosomal and spliceosomal RNAs. Homologs of eukaryotic box C/D snoRNAs present in archaebacteria, besides directing ribose methylation of rRNAs, also function in 2′-O-methylation of tRNAs (Omer et al., 2000; B.Clouet-D’Orval and J.-P.Bachellerie, personal communication; C.Daniels, personal communication). In human and rodents, several novel putative 2′-O-methylation and pseudouridylation snoRNAs lacking significant complementarities to rRNAs, snRNAs or other known stable cellular RNAs have been identified (Cavaillé et al., 2000; Jády and Kiss, 2000; Hüttenhofer et al., 2001; B.E.Jády and T.Kiss, unpublished data). Most probably, these ‘orphan’ guide snoRNAs function in 2′-O-methylation and pseudouridylation of thus far unidentified target RNAs. What types of cellular RNAs might be the substrates of the new guide snoRNAs? During the last decade, several non-coding RNAs with important catalytic and regulatory functions have been identified (Eddy, 1999; Filipowicz, 2000). We can anticipate that numerous additional non-coding RNAs have remained still undiscovered in eukaryotic cells. These RNAs, of course, might carry 2′-O-methylated nucleotides and pseudouridines that are synthesized by snoRNPs.
According to a current, even more fascinating concept, snoRNAs might also function in modification of mRNAs. Seven novel box C/D snoRNAs and a box H/ACA snoRNA with predominant expression in brain tissues have been identified in human and mouse (Cavaillé et al., 2000; de Los Santos et al., 2000; J.Cavaillé, P.Vitali, Z.Basyuk, J.-P.Bachellerie, J.Brosius and A.Hüttenhofer, personal communication). The function of the new brain-specific snoRNAs remains elusive. One of the novel box C/D snoRNAs carries an 18 nucleotide phylogenetically conserved target recognition element that is perfectly complementary to the serotonin receptor 5-HT2C mRNA. Intriguingly, the putative 2′-O-methylation target nucleotide in the serotonin receptor mRNA is known to undergo an adenosine-to-inosine editing reaction, leading to the intriguing possibility that snoRNA-guided 2′-O-methylation might have a regulatory function in the expression of this important brain-specific protein (Cavaillé et al., 2000).
Cellular locations for snoRNA-directed RNA modification reactions
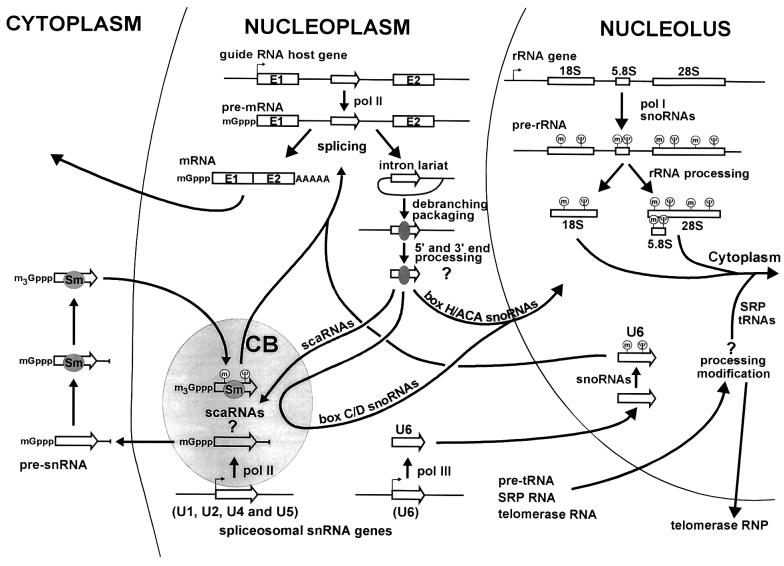
In vertebrates, all 2′-O-methylation and pseudouridylation guide snoRNAs are processed from pre-mRNA introns (reviewed in Tollervey and Kiss, 1997; Weinstein and Steitz, 1999) (Figure 2). The conserved box C/D and H/ACA motifs, through binding snoRNP proteins, direct the correct processing and nucleolar transportation of snoRNAs (Lange et al., 1998, 1999; Samarsky et al., 1998; Narayanan et al., 1999a, b). Since mature 2′-O-methylation and pseudouridylation snoRNPs are localized in the nucleolus, the most immediate issue that comes to mind is how they can function in post-transcriptional modification of various classes of cellular RNAs that accumulate in the cytoplasm or nucleoplasm. Recent attempts at understanding this apparent enigma shed new light on the intracellular trafficking of RNAs.
Fig. 2. Biogenesis and function of 2′-O-methylation and pseudouridylation guide snoRNAs. In mammalian cells, all guide RNAs are synthesized within introns of pre-mRNAs in the nucleoplasm. Most intronic snoRNAs are processed from the removed and debranched host intron by exonucleolytic activities. It remains unclear whether 5′ and 3′ end processing of snoRNAs occurs already in the nucleoplasm, or later in the nucleolus. Guide snoRNAs accumulating in the nucleolus direct 2′-O-methylation and pseudouridylation of the 18S, 5.8S and 28S rRNAs, the U6 snRNA and perhaps other cellular RNAs, including tRNAs, the signal recognition particle (SRP) and telomerase RNAs. It seems that box C/D, but not box H/ACA snoRNAs, transiently appear in the Cajal body before accumulating in the nucleolus. Some guide RNAs (scaRNAs) directing modification of the pol II-transcribed spliceosomal snRNAs accumulate in the Cajal body (CB). For other details, see the text.
Certainly, the snoRNA-directed 2′-O-methylation and pseudouridylation of rRNAs take place within the nucleolus where the 18S, 5.8S and 28S rRNAs are synthes ized as long precursor rRNAs (pre-rRNAs) (Figure 2). Modification of the 18S, 5.8S and 28S rRNAs occurs during or immediately after transcription, but before nucleolytic processing of the pre-rRNA (Maden, 1990). This implies that each pre-rRNA transcript transiently interacts with ∼200 modification guide snoRNPs.
The earlier finding that the U6-specific modification factors reside within the nucleolus raised the possibility that the U6 snRNA might cycle through the nucleolus to undergo snoRNP-mediated 2′-O-methylation and pseudouridylation (Tycowski et al., 1998; Ganot et al., 1999). Indeed, upon injection into the Xenopus oocyte nucleus, fluorescent U6 snRNA transiently localizes to the nucleolus before its accumulation in the nucleoplasm (Lange and Gerbi, 2000), supporting the idea that post-transcriptional modification of the U6 snRNA takes place in the nucleolus. Besides U6, several small stable RNAs, including RNase P RNA, signal recognition particle (SRP) RNA and telomerase RNA, have been detected in the nucleolus (reviewed by Pederson, 1998). In yeast cells, the RNase P-catalyzed endonucleolytic processing of the 5′ terminus of a major portion of tRNAs occurs within the nucleolus (Bertrand et al., 1998). Last, but not least, some mRNAs have also been reported to localize transiently to the nucleolus (Bond and Wold, 1993). These observations raised the fascinating possibility that various cellular RNAs can transit through the nucleolus to undergo nucleolar processing including snoRNA-directed nucleotide modifications.
In contrast to the pol III-transcribed U6 snRNA, maturation of the pol II-specific U1, U2, U4 and U5 snRNPs includes a cytoplasmic phase (Figure 2). The newly synthesized precursor snRNAs (pre-snRNAs) are exported to the cytoplasm, where the seven common Sm proteins bind to the pre-snRNAs before processing of their 3′ ends and hypermethylation of their primary monomethylguanosine cap to trimethylguanosine. After re-importation from cytoplasm, the newly assembled snRNPs transiently appear in Cajal (coiled) bodies before accumulating in the nucleoplasm (Carvalho et al., 1999; Sleeman and Lamond, 1999). Earlier, it was found that the actively transcribed human U1 and U2 genes frequently co-localize with Cajal bodies (Frey and Mateira, 1995; Smith et al., 1995; Frey et al., 1999). Although it seems that transcription does not occur within the Cajal body, the newly synthesized pre-U2 snRNA was also detected in this nucleoplasmic organelle (Smith and Lawrence, 2000). The Cajal body has long been considered as a possible site for assembly and/or modification of spliceosomal snRNPs (reviewed in Bohmann et al., 1995; Matera and Frey, 1998). We have recently investigated the subcellular distribution of the human U85 box C/D/H/ACA snoRNA that functions in both 2′-O-methylation and pseudouridylation of the U5 snRNA (Jády and Kiss, 2001). To our surprise, in situ hybridization experiments demonstrated that the U85 snoRNAs co-localize with Cajal bodies in human HeLa cells (X.Darzacq, B.E.Jády, C.Verheggen, E.Bertrand and T.Kiss, in preparation). Likewise, three other currently identified snoRNAs that are predicted to function in pseudouridylation of the U2 snRNA and 2′-O-methylation of the U1, U4 and U5 snRNAs were also found to accumulate within Cajal bodies. These observations raised the possibility that the novel small Cajal body-specific RNAs (scaRNAs) may direct modification of the U1, U2, U4 and U5 snRNAs within Cajal bodies either after the synthesis of nascent pre-snRNAs or after re-importation of the newly assembled snRNPs from the cytoplasm. A recent study, however, indicates that nucleolar factors may also contribute to the internal modification of the U2 snRNA (Yu et al., 2001). Therefore, several exciting questions remain to be answered in the future. Do all 2′-O-methylation and pseudouridylation guide RNAs that function in modification of pol II-specific snRNAs reside within Cajal bodies? Can bona fide nucleolus-localized RNAs also function in snRNA modification? What are the cis- and trans-acting factors that direct scaRNAs into the Cajal body?
Concluding remarks
Eubacterial rRNAs contain only a handful of 2′-O-methylated nucleotides and pseudouridines, which are synthesized by specific protein enzymes. To synthesize the numerous 2′-O-methylated nucleotides and pseudouridines in eukaryotic rRNAs, eukaryotes probably adopted the snoRNA-guided modification mechanism before the split between archaebacteria and eukaryotes (Gaspin et al., 2000; Omer et al., 2000; Watanabe and Gray, 2000). The primordial 2′-O-methylation and pseudouridylation guide snoRNAs likely derived from cis-acting rRNA or perhaps tRNA sequences which acquired the ability to function as trans-acting cofactors. The complex world of present-day guide snoRNAs probably evolved through duplications and random mutations of the snoRNA genes (Lafontaine and Tollervey, 1998). The guide snoRNA-based RNA modification mechanism provides several advantages for eukaryotic cells. Selection of modification sites by guide snoRNAs is independent of local structures of the substrate RNA, and guide RNAs can evolve much more rapidly than protein enzymes that are highly dependent on pre-existing modification sites. During evolution, therefore, the flexible snoRNA-based modification systems can continuously test novel 2′-O-methyl groups and pseudouridines in eukaryotic rRNAs, snRNAs, tRNAs and probably other cellular RNAs.
Acknowledgments
Acknowledgements
This work was supported by grants from la Ligue Nationale Contre le Cancer, l’Association pour la Recherche sur le Cancer and the Hungarian Research Foundation (OTKA, T29042 and T31738).
References
- Agris P.F. (1996) The importance of being modified: roles of modified nucleosides and Mg2+ in RNA structure and function. Prog. Nucleic Acid Res. Mol. Biol., 53, 79–129. [DOI] [PubMed] [Google Scholar]
- Bachellerie J.-P., Michot,B., Nicoloso,M., Balakin,A., Ni,J. and Fournier, M.J. (1995) Antisense snoRNAs: a family of nucleolar RNAs with long complementarity to rRNA. Trends Biochem. Sci., 20, 261–264. [DOI] [PubMed] [Google Scholar]
- Balakin A.G., Smith,L. and Fournier,M.J. (1996) The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell, 86, 823–834. [DOI] [PubMed] [Google Scholar]
- Bertrand E., Houser-Scott,F., Kendall,A., Singer,R.H. and Engelke,D.R. (1998) Nucleolar localization of early tRNA processing. Genes Dev., 12, 2463–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohmann K., Ferreira,J., Santama,N., Weis,K. and Lamond,A.I. (1995) Molecular analysis of coiled body. J. Cell Sci., 19, 107–113. [DOI] [PubMed] [Google Scholar]
- Bond V.C. and Wold,B. (1993) Nucleolar localization of myc transcripts. Mol. Cell. Biol., 13, 3221–3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortolin M.-L., Ganot,P. and Kiss,T. (1999) Elements essential for accumulation and function of small nucleolar RNAs directing site-specific pseudouridylation of ribosomal RNAs. EMBO J., 18, 457–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousquet-Antonelli C., Henry,Y., Gélugne,J.-P., Caizergues-Ferrer,M. and Kiss,T. (1997) A small nucleolar RNP protein is required for pseudouridylation of eukaryotic ribosomal RNAs. EMBO J., 16, 4770–4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho T., Almeida,F., Calapez,A., Lafarga,M., Berciano,M.T. and Carmo-Fonseca,M. (1999) The spinal muscular atrophy disease gene product, SMN: a link between snRNP biogenesis and the Cajal (coiled) body. J. Cell Biol., 147, 715–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaillé J., Nicoloso,M. and Bachellerie,J.-P. (1996) Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature, 383, 732–735. [DOI] [PubMed] [Google Scholar]
- Cavaillé J., Buiting,K., Kiefmann,M., Lalande,M., Brannan,C.I., Horsthemke,B., Bachellerie,J.-P., Brosius,J. and Hüttenhofer,A. (2000) Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl Acad. Sci. USA, 97, 14311–14316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Los Santos T., Schweitzer,J., Rees,C.A. and Francke,U. (2000) Small evolutionarily conserved RNA, resembling C/D box small nucleolar RNA, is transcribed from PWCR1, a novel imprinted gene in the Prader–Willi region, which is highly expressed in brain. Am. J. Hum. Genet., 67, 1067–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy S.R. (1999) Noncoding RNA genes. Curr. Opin. Genet. Dev., 9, 695–699. [DOI] [PubMed] [Google Scholar]
- Filipowicz W. (2000) Imprinted expression of small nucleolar RNAs in brain: time for RNomics. Proc. Natl Acad. Sci. USA, 97, 14035–14037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey M.R., Bailey,A.D., Weiner,A.M. and Matera,A.G. (1999) Association of snRNA genes with coiled bodies is mediated by nascent snRNA transcripts. Curr. Biol., 9, 126–135. [DOI] [PubMed] [Google Scholar]
- Frey M.R. and Mateira,A.G. (1995) Coiled bodies contain U7 small nuclear RNA and associate with specific DNA sequences in interphase cells. Proc. Natl Acad. Sci. USA, 92, 5915–5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganot P., Bortolin,M.-L. and Kiss,T. (1997a) Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell, 89, 799–809. [DOI] [PubMed] [Google Scholar]
- Ganot P., Caizergues-Ferrer,M. and Kiss,T. (1997b) The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev., 11, 941–956. [DOI] [PubMed] [Google Scholar]
- Ganot P., Jády.B.E., Bortolin,M.-L., Darzacq,X. and Kiss,T. (1999) Nucleolar factors direct the 2′-O-ribose methylation and pseudo uridylation of U6 spliceosomal RNA. Mol. Cell. Biol., 19, 6906–6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspin C., Cavaillé,J., Erauso,G. and Bachellerie,J.-P. (2000) Archaeal homologs of eukaryotic methylation guide small nuclear RNAs: lessons from the Pyrococcus genome. J. Mol. Biol., 297, 895–906. [DOI] [PubMed] [Google Scholar]
- Henras A., Henry,Y., Bousquet-Antonelli,C., Noaillac-Depeyre,J., Gélugne,J.-P. and Caizergues-Ferrer,M. (1998) Nhp2p and Nop10p are essential for the function of H/ACA snoRNPs. EMBO J., 17, 7078–7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüttenhofer A., Kiefmann,M., Meier-Ewert,S., O’Brien,J., Lehrach,H., Bachellerie,J.-P. and Brosius,J. (2001) RNomics: an experimental approach that identifies 201 candidates for novel, small non-messenger RNAs in mouse. EMBO J., 20, 2943–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jády,B.E. and Kiss T. (2000) Characterisation of the U83 and U84 small nucleolar RNAs: two novel 2′-O-ribose methylation guide RNAs that lack complementarities to ribosomal RNAs. Nucleic Acids Res., 28, 1348–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jády,B.E. and Kiss T. (2001) A small nucleolar guide RNA functions both in 2′-O-ribose methylation and pseudouridylation of the U5 spliceosomal RNA. EMBO J., 20, 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss-László Z., Henry,Y., Bachellerie,J.-P., Caizergues-Ferrer,M. and Kiss,T. (1996) Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell, 85, 1077–1088. [DOI] [PubMed] [Google Scholar]
- Kiss-László Z., Henry,Y. and Kiss,T. (1998) Sequence and structural elements of methylation guide snoRNAs essential for site-specific ribose methylation of pre-rRNA. EMBO J., 17, 797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine D.L.J. and Tollervey,D. (1998) Birth of the snoRNPs: the evolution of the modification-guide snoRNAs. Trends Biochem. Sci., 23, 383–388. [DOI] [PubMed] [Google Scholar]
- Lafontaine D.L.J. and Tollervey,D. (1999) Nop58p is a common component of the box C+D snoRNPs that is required for snoRNP stability. RNA, 5, 455–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine D.L.J. and Tollervey,D. (2000) Synthesis and assembly of the box C+D small nucleolar RNPs. Mol. Cell. Biol., 20, 2650–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine D.L.J., Bousquet-Antonelli,C., Henry,Y., Caizergues-Ferrer, M. and Tollervey,D. (1998) The box H+ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev., 12, 527–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange T.S. and Gerbi,S.A. (2000) Transient nucleolar localization of U6 small nuclear RNA in Xenopus laevis oocytes. Mol. Biol. Cell, 11, 2419–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange T.S., Borovjagin,A., Maxwell,E.S. and Gerbi,S.A. (1998) Conserved boxes C and D are essential nucleolar localization elements of U14 and U8 snoRNAs. EMBO J., 17, 3176–3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange T.S., Ezrokhi,M., Amaldi,F. and Gerbi,S.A. (1999) Box H and box ACA are nucleolar localization elements of U17 small nucleolar RNA. Mol. Biol. Cell, 10, 3877–3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limbach P.A., Crain,P.F. and McCloskey,A. (1994) Summary: the modified nucleosides of RNA. Nucleic Acids Res., 22, 2183–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyman S.K., Gerace,L. and Baserga,S.J. (1999) Human Nop5/Nop58 is a component common to the box C/D small nucleolar ribonucleoproteins. RNA, 5, 1597–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas S. and Rich,A. (2000) Changing genetic information through RNA editing. BioEssays, 22, 790–802. [DOI] [PubMed] [Google Scholar]
- Maden B.E.H. (1990) The numerous modified nucleotides in eukaryotic ribosomal RNA. Prog. Nucleic Acid Res. Mol. Biol., 39, 241–303. [DOI] [PubMed] [Google Scholar]
- Massenet S., Motorin,Y., Lafontaine,D.L., Hurt,E.C., Grosjean,H. and Branlant,C. (1999) Pseudouridine mapping in the Saccharomyces cerevisiae spliceosomal U small nuclear RNAs (snRNAs) reveals that pseudouridine synthase Pus1p exhibits a dual substrate specificity for U2 snRNA and tRNA. Mol. Cell. Biol., 19, 2142–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera A.G. and Frey,M.R. (1998) Nuclear structure ’98. Coiled bodies and gems: Janus or gemini? Am. J. Hum. Genet., 63, 317–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan A., Lukowiak,A., Jády,B.E., Dragon,F., Kiss,T., Terns,R.M. and Terns,M.P. (1999a) Nucleolar localization signals of box H/ACA small nucleolar RNAs. EMBO J., 18, 5120–5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan A., Speckmann,W., Terns,R. and Terns,M.P. (1999b) Role of the box C/D motif in localization of small nucleolar RNAs to coiled bodies and nucleoli. Mol. Biol. Cell, 10, 2131–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman D.R., Kuhn,J.F., Shanab,G.M. and Maxwell,E.S. (2000) Box C/D snoRNA-associated proteins: two pairs of evolutionarily ancient proteins and possible links to replication and transcription. RNA, 6, 861–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J., Tien,A.L. and Fournier,M.J. (1997) Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell, 89, 565–573. [DOI] [PubMed] [Google Scholar]
- Niewmierzycka A. and Clarke,S. (1999) S-adenosylmethionine-dependent methylation in Saccharomyces cervisiae. J. Biol. Chem., 274, 814–824. [DOI] [PubMed] [Google Scholar]
- Omer A.D., Lowe,T.M., Russell,A.G., Ebhardt,H., Eddy,S.R. and Dennis,P.P. (2000) Homologs of small nucleolar RNAs in Archaea. Science, 288, 517–522. [DOI] [PubMed] [Google Scholar]
- Pederson T. (1998) The plurifunctional nucleolus. Nucleic Acids Res., 26, 3871–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S., Niu,L., Conrad,J., Lane,B.G. and Ofengand,J. (1999) Functional effect of deletion and mutation of the Escherichia coli ribosomal RNA and tRNA pseudouridine synthase RluA. J. Biol. Chem., 274, 18880–18886. [DOI] [PubMed] [Google Scholar]
- Reddy R. and Busch,H. (1988) Small nuclear RNAs: RNA sequences, structure, and modifications. In Birnstiel,M.L. (ed.), Structure and Function of Major and Minor Small Nuclear Ribonucleoprotein Particles. Springer-Verlag, Berlin, Germany, pp. 1–37.
- Samarsky D.A., Fournier,M.J., Singer,R.H. and Bertrand,E. (1998) The snoRNA box C/D motif directs nucleolar targeting and also couples snoRNA synthesis and localization. EMBO J., 17, 3747–3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmang T., Tollervey,D., Kern,H., Frank,R. and Hurt,E.C. (1989) A yeast nucleolar protein related to mammalian fibrillarin is associated with small nucleolar RNA and is essential for viability. EMBO J., 8, 4015–4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleeman J.E. and Lamond,A.I. (1999) Newly assembled snRNPs associate with coiled bodies before speckles, suggesting a nuclear snRNP maturation pathway. Curr. Biol., 9, 1065–1074. [DOI] [PubMed] [Google Scholar]
- Smith K.P. and Lawrence,J.B. (2000) Interactions of U2 gene loci and their nuclear transcripts with Cajal (coiled) bodies: evidence for preU2 within Cajal bodies. Mol. Biol. Cell, 11, 2987–2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K., Carter,K., Johnson,C. and Lawrence,J. (1995) U2 and U1 snRNA gene loci associate with coiled bodies. J. Cell. Biochem., 59, 473–485. [DOI] [PubMed] [Google Scholar]
- Tollervey D. and Kiss,T. (1997) Function and synthesis of small nucleolar RNAs. Curr. Opin. Cell Biol., 9, 337–342. [DOI] [PubMed] [Google Scholar]
- Tollervey D., Lehtonen,H., Jansen,R., Kern,H. and Hurt,E. (1993) Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell, 72, 443–457. [DOI] [PubMed] [Google Scholar]
- Tyc K. and Steitz,J.A. (1989) U3, U8 and U13 comprise a new class of mammalian snRNPs localized in the nucleolus. EMBO J., 8, 3113–3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski K.T., Shu,M.-D. and Steitz,J.A. (1996a) A mammalian gene with introns instead of exons generating stable RNA products. Nature, 379, 464–466. [DOI] [PubMed] [Google Scholar]
- Tycowski,K.T., Smith,C.M., Shu,M.-D. and Steitz,J.A. (1996b) A small nucleolar RNA requirement for site-specific ribose methylation of rRNA in Xenopus. Proc. Natl Acad. Sci. USA, 93, 14480–14485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski K.T, You,Z.-H., Graham,P.J. and Steitz,J.A. (1998) Modification of U6 spliceosomal RNA is guided by other small RNAs. Mol. Cell, 2, 629–638. [DOI] [PubMed] [Google Scholar]
- Wang H., Boisvert,D., Kim,K.K., Kim,R. and Kim,S.-H. (2000) Crystal structure of a fibrillarin homologue from Methanococcos jannaschii, a hyperthermophile, at 1.6 Å resolution. EMBO J., 19, 317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y. and Gray,M.W. (2000) Evolutionary appearance of genes encoding proteins associated with box H/ACA snoRNAs: Cbf5p in Euglena gracilis, an early diverging eukaryote, and candidate Gar1p and Nop10p homologs in archaebacteria. Nucleic Acids Res., 28, 2342–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins N.J., Gottschalk,A., Neubauer,G., Kastner,B., Fabrizio,P., Mann, M. and Lührmann,R. (1998) Cbf5p, a potential pseudouridine synthase, and Nhp2p, a putative RNA-binding protein, are present together with Gar1p in all H box/ACA-motif snoRNPs and constitute a common bipartite structure. RNA, 4, 1549–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins N.J. et al. (2000) A common core RNP structure shared between the small nucleolar box C/D RNPs and the spliceosomal U4 snRNP. Cell, 103, 457–466. [DOI] [PubMed] [Google Scholar]
- Weinstein L.B. and Steitz,J.A. (1999) Guided tours: from precursor snoRNA to functional snoRNP. Curr. Opin. Cell Biol., 11, 378–384. [DOI] [PubMed] [Google Scholar]
- Wrzesinski J., Bakin,A., Ofengand,J., and Lane,B.G. (2000) Isolation and properties of Escherichia coli 23S-RNA pseudouridine 1911, 1915, 1917 synthase. IUBMB Life, 50, 33–37. [DOI] [PubMed] [Google Scholar]
- Yu Y.-T., Shu,M.-D. and Steitz,J.A. (1998) Modifications of U2 snRNA are required for snRNP assembly and pre-mRNA splicing. EMBO J., 17, 5783–5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y.-T., Shu,M.-D., Narayanan,A., Terns,R.M., Terns,M.P. and Steitz,J.A. (2001) Internal modification of U2 small nuclear (sn)RNA occurs in nucleoli of Xenopus oocytes. J. Cell Biol., 152, 1279–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zebarjadian Y., King,T., Fournier,M.J., Clarke,L. and Carbon,J. (1999) Point mutations in yeast CBF5 can abolish in vivo pseudouridylation of rRNA. Mol. Cell. Biol., 19, 7461–7472. [DOI] [PMC free article] [PubMed] [Google Scholar]