Abstract
The >1 kb XL-exon of the rat XLαs/Gαs gene encodes the 37 kDa XL-domain, the N-terminal half of the 78 kDa neuroendocrine-specific G-protein α-subunit XLαs. Here, we describe a novel feature of the XL-exon, the presence of an alternative >1 kb open reading frame (ORF) that completely overlaps with the ORF encoding the XL-domain. The alternative ORF starts 32 nucleotides downstream of the start codon for the XL-domain and is terminated by a stop codon exactly at the end of the XL-exon. The alternative ORF encodes ALEX, a very basic (pI 11.8), proline-rich protein of 356 amino acids. Both XLαs and ALEX are translated from the same mRNA. Like XLαs, ALEX is expressed in neuroendocrine cells and tightly associated with the cytoplasmic leaflet of the plasma membrane. Remarkably, ALEX binds to the XL-domain of XLαs. Our results reveal a mechanism of gene usage that is without precedent in mammalian genomes.
Keywords: gene usage/G-protein/overlapping open reading frames/translation/XLαs
Introduction
Our laboratory previously identified a new type of G-protein α-subunit, XLαs (for ‘extra large’ αs), which is characterized by a bipartite structure (Kehlenbach et al., 1994). The C-terminal half of XLαs, referred to as the αs-domain, is encoded by exons 2–13 of the Gαs gene and hence contains the entire Gαs sequence except for the N-terminal 47 amino acids encoded by exon 1 of the Gαs gene. In XLαs, the latter residues are replaced by a novel sequence, referred to as the XL-domain, resulting in a protein with a molecular mass of 78 kDa and an electro phoretic mobility of 94 000 (rat XLαs) (Kehlenbach et al., 1994, 1995). Thus, XLαs is the largest known variant of a G-protein α-subunit. XLαs is expressed specifically in neuroendocrine tissues and cell lines (Kehlenbach et al., 1994; Pasolli et al., 2000) and is involved in signal transduction at the plasma membrane (Klemke et al., 2000; Pasolli et al., 2000) where it activates, in its GTP-bound form, adenylyl cyclase (Klemke et al., 2000).
The XL-domain of XLαs is encoded by a single exon, referred to as the XL-exon (Hayward et al., 1998a; Peters et al., 1999; Y.Wang and W.B.Huttner, in preparation), which is located ∼35 kb upstream of exon 1 of the Gαs gene (Hayward et al., 1998a; Peters et al., 1999). Expression of XLαs, in contrast to that of Gαs, is allele specific due to genomic imprinting of the XL-exon (Hayward et al., 1998a,b; Peters et al., 1999), with only the paternal allele being expressed. Here, we describe a remarkable feature of the XL-exon, the presence of an alternative, >1 kb open reading frame (ORF) that (i) completely overlaps with the ORF encoding the XL-domain of XLαs and (ii) encodes a novel protein, which binds to the XL-domain.
Results
The XL-exon of the XLαs/Gαs gene contains a second, overlapping open reading frame
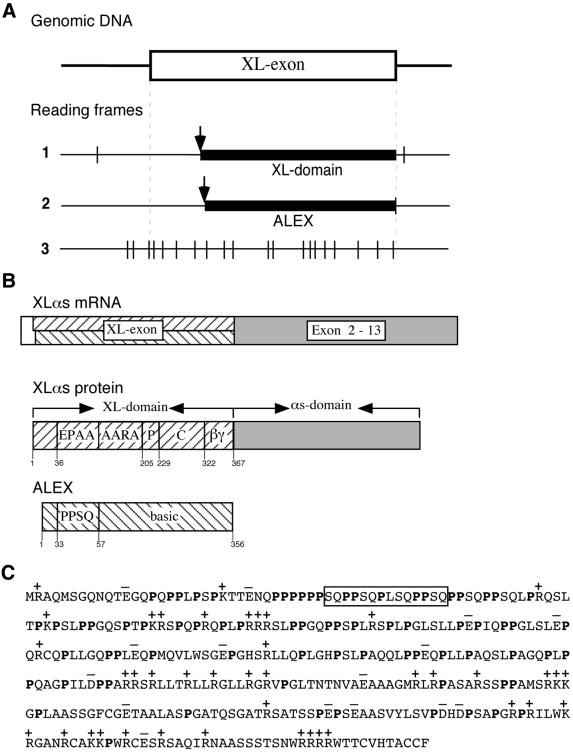
The rat XL-exon not only contains the ORF encoding the XL-domain that, in the XLαs mRNA, is continuous with the ORF encoding the αs-domain of XLαs (exons 2–13 of the XLαs/Gαs gene; Figure 1B), but, remarkably, also contains a second ORF of ∼1 kb (ORF2) that overlaps with the ORF encoding the XL-domain (ORF1, Figure 1A). ORF2 starts 32 nucleotides downstream of ORF1, is shifted by +1 in its phase compared with ORF1, extends to the 3′ end of the XL-exon and is closed by a TAG stop codon exactly at the end of the XL-exon (Figure 1A). Reading frame 3 of the XL-exon is closed by several stop codons (Figure 1A).
Fig. 1. Existence of two overlapping open reading frames in the XL-exon. (A) The XL-exon of the rat XLαs/Gαs gene (DDBJ/EMBL/GenBank accession No. AF093569) contains two ORFs. ORF1 (1) encodes the XL-domain of XLαs, ORF2 (2) the ALEX protein. The third reading frame (3) does not encode a protein due to the presence of multiple stop codons. The protein-encoding region of the XL-exon is indicated by the thick line, start codons by arrows, and stop codons by vertical lines. (B) Structure of the XLαs mRNA and the two proteins, XLαs and ALEX, derived from it. XLαs: EPAA, ARAA, alanine-rich repeats; P, proline-rich region; C, cysteine-rich region; βγ, βγ-binding region; numbers refer to the corrected translational start (Kehlenbach et al., 1995). ALEX: PPSQ, proline-rich repeats. (C) Deduced amino acid sequence of the rat ALEX protein. The N-terminal methionine corresponds to the 5′ most ATG in the second reading frame of the XL-exon, whose 5′ end was determined by primer extension analysis of PC12 cell mRNA (Y.Wang and W.B.Huttner, unpublished data). Basic amino acids are indicated by (+), acidic amino acids by (–); proline residues are shown in bold. The box indicates the peptide used as antigen to raise the anti-ALEX antibody.
Translation of ORF2 of the XLαs mRNA would give rise to a distinct protein, referred to as ALEX (for alternative gene product encoded by the XL-exon). Rat ALEX would lack any Gαs sequence (Figure 1B), consist of 356 amino acids with a calculated molecular mass of 38 kDa, and be a very proline-rich (21%) and highly basic (pI = 11.8) protein (Figure 1C).
The existence of a second, overlapping ORF is conserved between the rat, mouse and human XL-exon of the XLαs/Gαs gene
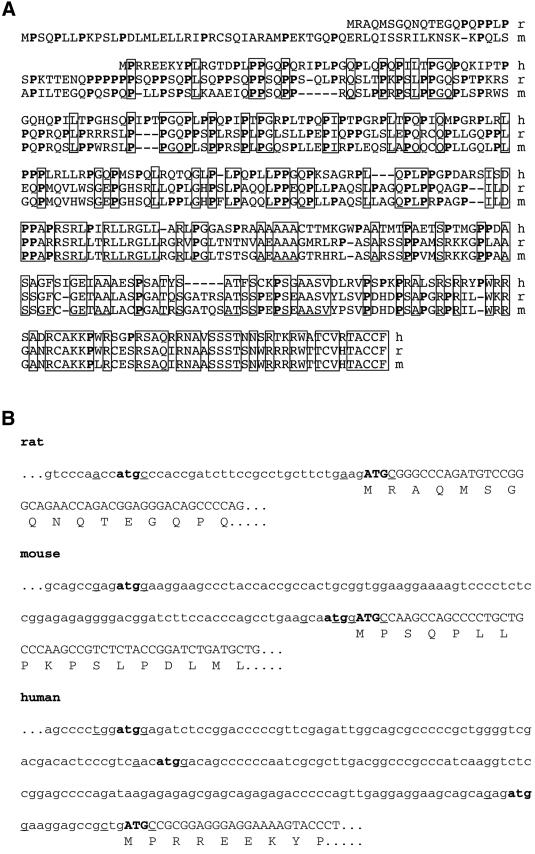
Sequence analysis of the human (Hayward et al., 1998a) and mouse (Peters et al., 1999; Klemke et al., 2000) XL- exon revealed that they, too, contain two overlapping ORFs. As in the case of the rat XL-exon, ORF2 of the human and mouse XL-exon is also shifted by +1 in its phase as compared with ORF1, starts downstream of ORF1, extends to the 3′ end of the exon and is closed by a TAG stop codon exactly at the end of the XL-exon. Translation of ORF2 of the human and the mouse XL-exon would also generate highly basic, proline-rich proteins (Figure 2A). Comparison of the nucleotide sequence of the rat, mouse and human XL-exon and alignment of the amino acid sequence of rat, mouse and human ALEX reveal that the degree of conservation is relatively low (Table I), being highest in the C-terminal region (Figure 2A).
Fig. 2. Comparison of the human, mouse and rat amino acid sequence of ALEX and the 5′ nucleotide sequence of the XL-exon. (A) Deduced amino acid sequence of ALEX. As for rat (r) ALEX, the N-terminal methionine in human (h) and mouse (m) ALEX corresponds to the 5′ most ATG in the second reading frame of the human (Hayward et al., 1998a) and mouse (AF116268) XL-exon, respectively. Proline residues are shown in bold; boxes indicate residues conserved between all three species. (B) Nucleotide sequence at the 5′ end of the XL-exon. Potential translational start codons are shown in bold; lower case letters, XLαs-ORF; upper case letters, ALEX-ORF. Nucleotides at the –3 and +4 position around the putative start codons, known to be crucial for initiation of translation (Kozak, 1991), are underlined. DDBJ/EMBL/GenBank accession Nos for the nucleotide sequence of the XL-exon are: AF093569 (rat), AJ245739 (mouse) and AJ224868 (human).
Table I. Nucleotide sequence identity of the human, mouse and rat XL-exon and amino acid sequence identity of human, mouse and rat ALEX.
| Species | Nucleotide sequence identity (%) | Amino acid sequence identity (%) |
|---|---|---|
| Rat/mouse | 85 | 79 |
| Rat/human | 59 | 55 |
| Mouse/human | 71 | 53 |
The fact that the existence of a second, overlapping ORF in the XL-exon is conserved suggests that the ORF2-encoded ALEX may be a naturally existing protein. To investigate this issue, we raised an anti-ALEX antibody directed against a peptide in the N-terminal region of ALEX (Figure 1C, box). For artificial expression of ALEX, a fragment of the XLαs cDNA, referred to as ALEX cDNA, was used. This cDNA fragment starts 23 nucleotides downstream of the ORF1 ATG at which translation of the XL-domain of XLαs is initiated, so that the first ATG is one in ORF2 (Figure 2B, rat), and ends eight nucleotides downstream of the 3′ end of the XL-exon, i.e. of the TAG stop codon of ORF2.
Characterization of cDNA-expressed ALEX in transfected PC12 cells
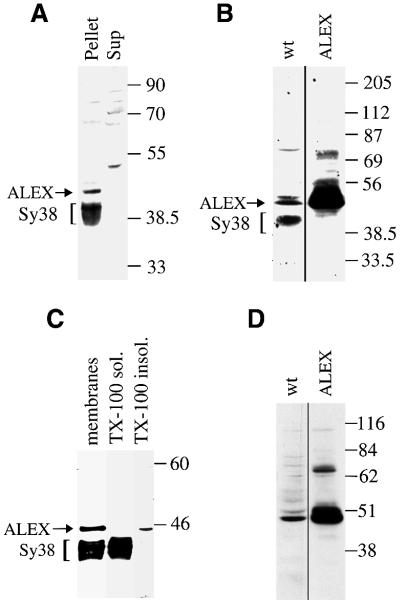
PC12 cells, which endogenously express the XLαs mRNA and contain the XLαs protein (Kehlenbach et al., 1994; Pasolli et al., 2000), were transfected with the ALEX cDNA to express ALEX. Immunoblotting using the anti-ALEX antibody of a post-nuclear supernatant (PNS) from wild-type and ALEX-transfected PC12 cells revealed the overexpression of an immunoreactive protein with an apparent electrophoretic mobility of ∼48 kDa upon transfection, i.e. ALEX (Figure 3A, lane ALEX). The discrepancy between the apparent molecular weight of ALEX on SDS–PAGE (48 kDa) and its calculated molecular mass of 38 kDa presumably reflects its unusual primary structure (see Figure 1C). Interestingly, an immunoreactive protein of the same electrophoretic mobility, perhaps endogenous ALEX (see below), was detected in wild-type PC12 cells, albeit at a much lower concentration (Figure 3A, lane wt).
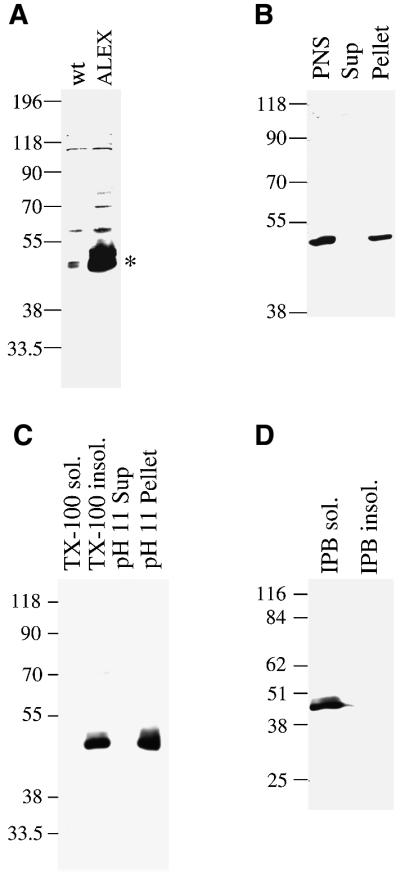
Fig. 3. Membrane association of cDNA-expressed ALEX. (A) PNS from PC12 cells either untransfected (wt) or transfected with the ALEX cDNA (ALEX) was analyzed by SDS–PAGE followed by immunoblotting using the anti-ALEX antibody. ALEX is indicated by an asterisk. (B) A PNS from PC12 cells transfected with the ALEX cDNA, as well as a supernatant (Sup) and pellet derived from it by ultracentrifugation, were analyzed by SDS–PAGE followed by immunoblotting using the anti-ALEX antibody. (C) Total membranes from PC12 cells transfected with the ALEX cDNA were treated with Triton X-100 (TX-100) or carbonate (pH 11). Soluble (TX-100 sol., pH 11 Sup) and insoluble (TX-100 insol., pH 11 Pellet) material obtained by ultracentrifugation was analyzed by SDS–PAGE followed by immunoblotting using the anti-ALEX antibody. (D) Total membranes from PC12 cells transfected with the ALEX cDNA were solubilized in immunoprecipitation buffer (IPB) containing ionic detergents, and subjected to ultracentrifugation. Supernatant (IPB sol.) and pellet (IPB insol.) were analyzed by SDS–PAGE followed by immunoblotting using the anti-ALEX antibody.
To characterize further the cDNA-expressed ALEX in the transfected cells, the PNS was subjected to high-speed centrifugation to obtain a soluble (cytosolic) and particulate (membranes and cytoskeletal elements) fraction. As shown in Figure 3B, virtually all of ALEX was recovered in the particulate fraction. To characterize the association of ALEX with the particulate fraction further, this fraction was either extracted with the non-ionic detergent Triton X-100 (1%) or stripped by incubation at pH 11.5. ALEX was recovered in the Triton X-100-insoluble fraction and not solubilized by pH 11.5 treatment (Figure 3C). In contrast, extraction of the particulate fraction with a buffer containing the ionic detergent SDS (0.3%) led to a complete solubilization of ALEX (Figure 3D). These results indicate that ALEX, whose sequence does not reveal a transmembrane segment (see Figure 1C), is tightly associated with particulate material of PC12 cells.
Identification and characterization of endogenous ALEX in wild-type PC12 cells
We exploited the particulate nature of ALEX and its resistance to Triton X-100 extraction (Figure 3) to investigate whether the ALEX-like immunoreactive band observed in untransfected PC12 cells (Figure 3A, lane wt) is endogenous ALEX. Immunoblotting using the anti-ALEX antibody revealed two immunoreactive bands in the particulate fraction of PC12 cells, one of ∼48 kDa and another of ∼38 kDa (Figure 4A); the latter was found to be synaptophysin, which cross-reacts with the anti-ALEX antibody (data not shown). PC12 cells of the subclone 27, which lack neurosecretory vesicle membrane proteins including synaptophysin but contain XLαs (Corradi et al., 1996), lacked the immunoreactive 38 kDa band but still contained the 48 kDa band (data not shown). Direct comparison of the particulate fraction from wild-type and ALEX-transfected PC12 cells showed that the 48 kDa band had the same electrophoretic mobility as cDNA-expressed ALEX (Figure 4B).
Fig. 4. Membrane association of endogenous ALEX. (A) PNS from untransfected PC12 cells was subjected to ultracentrifugation, and supernatant (Sup) and pellet were analyzed by SDS–PAGE followed by immunoblotting using the anti-ALEX antibody. (B) Immunoblot analysis, using the anti-ALEX antibody, of a total membrane fraction of PC12 cells either untransfected (wt) or transfected with the ALEX cDNA (ALEX). (C) Total membranes from PC12 cells were treated with Triton X-100 (TX-100). Soluble (TX-100 sol.) and insoluble (TX-100 insol.) material obtained by ultracentrifugation, as well as an aliquot of the total membranes, were analyzed by SDS–PAGE followed by immunoblotting using the anti-ALEX antibody. (D) Immunoblot analysis, using the anti-ALEX antibody, of the Triton X-100-insoluble material of total membranes of PC12 cells either untransfected (wt) or transfected with the ALEX cDNA (ALEX). (A–C) The positions of ALEX and of synaptophysin (Sy38), which is also detected by the anti-ALEX antibody, are indicated by arrows and brackets, respectively. (B and D) Exposures of the immunoblot lanes ‘ALEX’ were shorter than those of the lanes ‘wt’.
Immunoblotting using the anti-ALEX antibody of the Triton X-100-soluble and insoluble fractions obtained from the particulate fraction of PC12 cells showed that the endogenous immunoreactive 48 kDa band, like cDNA-expressed ALEX (see Figure 3C), was recovered with the Triton X-100-insoluble material, whereas the cross- reacting synpatophysin was solubilized by Triton X-100 (Figure 4C), consistent with previous observations (Hannah et al., 1998). Direct comparison of the Triton X-100-insoluble fraction of wild-type and ALEX-transfected PC12 cells showed that the 48 kDa band had the same electrophoretic mobility as cDNA-expressed ALEX (Figure 4D).
We used peptide mapping to corroborate the identity of the endogenous immunoreactive 48 kDa band as ALEX. The Triton X-100-insoluble material of wild-type and ALEX-transfected PC12 cells was subjected to SDS– PAGE, and the 48 kDa region known to contain ALEX was subjected to limited proteolysis by Staphylococcus aureus V8 protease during SDS–PAGE, followed by immunoblotting using the anti-ALEX antibody. The molecular weight of the immunoreactive peptide fragments obtained from wild-type and ALEX-transfected PC12 cells was essentially identical (Figure 5), demonstrating that wild-type PC12 cells did contain endogenous ALEX. The relative abundance of the immunoreactive peptide fragments obtained from wild-type and ALEX-transfected PC12 cells was similar, but not identical (Figure 5), which may reflect the difference in the substrate to protease ratio, or variation in post-translational modification, between wild-type and ALEX-transfected PC12 cells.
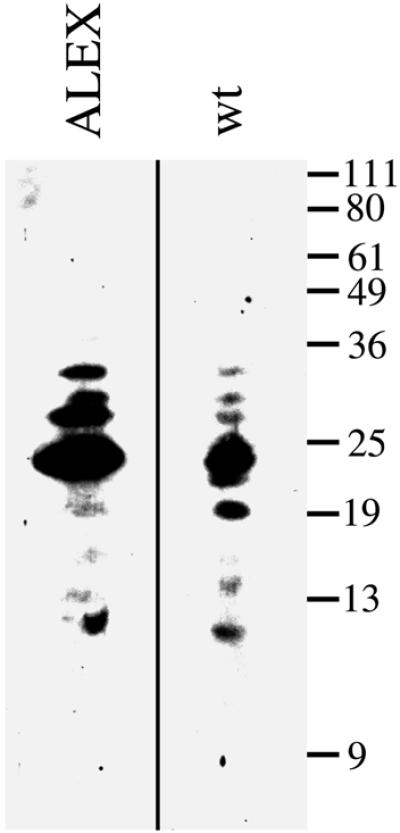
Fig. 5. Comparison of cDNA-expressed and endogenous ALEX by peptide mapping. The Triton X-100-insoluble material obtained from total membranes of PC12 cells, either transfected with the ALEX cDNA (ALEX) or untransfected (wt), was subjected to SDS–PAGE. The region of the gel containing ALEX was subjected to limited proteolysis by the S.aureus V8 protease during a second SDS–PAGE, and ALEX-derived peptides were detected by immunoblotting using the anti-ALEX antibody. Exposure of the immunoblot lane ‘ALEX’ was shorter than that of the lane ‘wt’.
Subcellular localization of ALEX
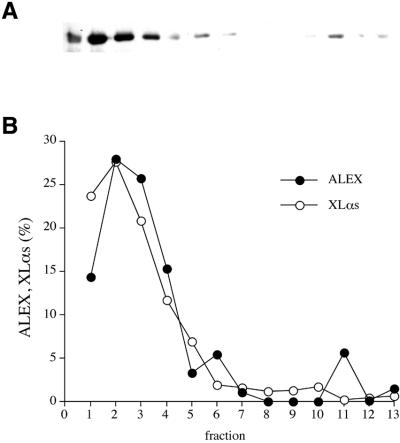
A PNS of ALEX-transfected PC12 cells was subjected to a standardized velocity sucrose density gradient centrifugation protocol (Tooze and Huttner, 1992). Immunoblotting using the anti-ALEX antibody showed that the vast majority of ALEX was detected in the top fractions of the gradient (Figure 6A and B, filled circles), which are known to contain most of the plasma membrane (Tooze and Huttner, 1992). ALEX showed a distribution across the gradient very similar to that of XLαs (Figure 6B, open circles), which is known to be associated with the plasma membrane (Pasolli et al., 2000).
Fig. 6. Subcellular fractionation of cDNA-expressed ALEX. PNS from PC12 cells either untransfected (B, XLαs) or transfected with the ALEX cDNA (A and B, ALEX) was subjected to velocity sucrose gradient centrifugation. (A) Fractions were analyzed by SDS–PAGE followed by immunoblotting with either the anti-ALEX antibody, as shown, or an antibody specific for the XL-domain of XLαs (not shown). (B) Immunoreactive ALEX (filled circles) and XLαs (open circles) in the fractions were quantitated and expressed as a percentage of total recovered per gradient (1 = top of gradient).
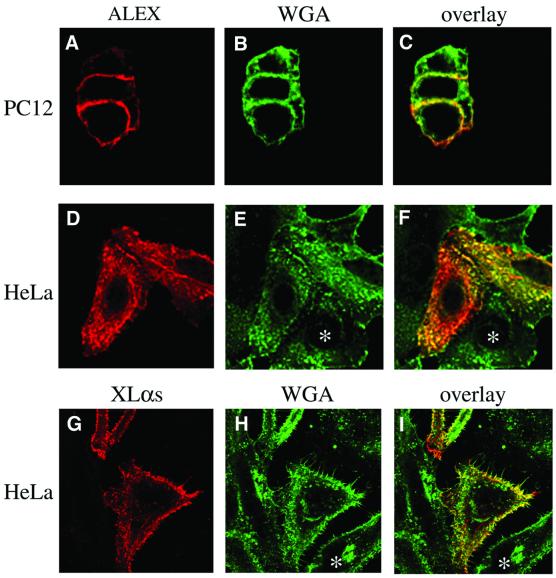
On immunofluorescence of ALEX-transfected PC12 cells using the anti-ALEX antibody, a peripheral, ring-like staining, which overlapped with wheat germ agglutinin (WGA) staining, was observed, consistent with a localization of ALEX at the plasma membrane (Figure 7A–C). It was difficult to detect specific staining with the anti-ALEX antibody in untransfected PC12 cells (data not shown), presumably because the abundance of endogenous ALEX, as compared with cDNA-expressed ALEX, is relatively low (see Figure 3A).
Fig. 7. Immunofluorescence analysis of PC12 and HeLa cells transfected with the ALEX cDNA or the XLαs cDNA. PC12 cells (A–C) transfected with the ALEX cDNA and HeLa cells (D–I) transfected with either the ALEX cDNA (D–F) or the XLαs cDNA (G–I) were double-stained with the anti-ALEX antibody (ALEX, red) or the anti-XL antibody (XLαs, red) and FITC-conjugated wheat germ agglutinin (WGA, green); (A–C) double-staining after fixation and permeabilization; (D–I) cell surface staining with WGA followed by fixation, permeabilization and immunostaining. Analysis using confocal microscopy; single optical X–Y sections through the middle of the cells (A–C) or at the level of the coverslip (D–I) are shown.
In ALEX-transfected HeLa cells, immunofluorescence staining for ALEX was largely associated with ruffles of the plasma membrane (Figure 7D and F), as revealed by counterstaining with WGA added to the intact cells at 4°C (Figure 7E and F). No staining with the anti-ALEX antibody was observed in untransfected HeLa cells (Figure 7D and F, asterisks), further documenting the specificity of the antibody. The predominant association of ALEX with plasma membrane ruffles was very similar to that observed for XLαs (Figure 7G–I) (Pasolli et al., 2000).
ALEX is translated from the XLαs mRNA
To determine whether ALEX can be translated from the full-length XLαs mRNA, i.e. that still contains the first (ORF1) ATG (see Figure 2B, rat), we used two approaches: (i) in vitro translation of the XLαs mRNA, obtained by in vitro transcription from the XLαs cDNA, followed by immunoprecipitation using the anti-ALEX antibody; and (ii) transient transfection of PC12 cells with the XLαs cDNA, followed by immunoblotting using the anti-ALEX antibody.
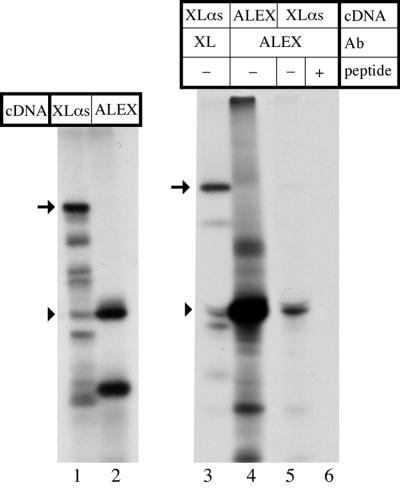
In vitro translation of the XLαs mRNA. The XLαs cDNA, as well as the ALEX cDNA fragment derived from it, were in vitro transcribed and translated in the rabbit recticulocyte lysate in the presence of [35S]methionine/cysteine. Translation products were analyzed by SDS–PAGE and autoradiography, with or without immunoprecipitation using either the Gαs/XLαs C-terminal antibody (Kehlenbach et al., 1994) or the anti-ALEX antibody. Translation of the XLαs cDNA generated a major product with an apparent mol. wt of 94 kDa, i.e. full-length XLαs (Figure 8, lane 1, arrow), and several minor bands of lower apparent molecular weight, most of which are known to be C-terminally truncated forms of XLαs (Klemke et al., 2000). Translation of the ALEX cDNA generated two translation products, one with an apparent mol. wt of 48 kDa, i.e. ALEX (Figure 8, lane 2, arrowhead), and another of lower molecular weight. (The identity of the latter product is unknown; it may be an N-terminally truncated form of ALEX, given the existence of downstream potential start codons in the cDNA.) Comparison of the ALEX mRNA-derived translation products (Figure 8, lane 2) with the XLαs mRNA-derived translation products (Figure 8, lane 1) revealed that the apparent molecular weight of one of the minor XLαs mRNA-derived translation products was identical to that of ALEX (Figure 8, lanes 1 and 2, arrowhead). Immunoprecipitation using the anti-ALEX antibody showed that the 48 kDa translation product obtained from the XLαs mRNA was indeed ALEX (Figure 8, lane 5), as shown by comparison with authentic ALEX immunoprecipitated after translation of the ALEX mRNA (Figure 8, lane 4). Immunoprecipitation by the anti-ALEX antibody of the 48 kDa translation product obtained from the XLαs mRNA was specific because (i) the more abundant XLαs protein, which was precipitated by the Gαs/XLαs C-terminal antibody (Figure 8, lane 3, arrow), was not immunoprecipitated; and (ii) the 48 kDa protein was not immunoprecipitated in the presence of the peptide used to raise the anti-ALEX antibody (Figure 8, lane 6). We conclude that in vitro translation of the XLαs mRNA generates both XLαs and ALEX.
Fig. 8. Generation of ALEX upon in vitro translation of the XLαs cDNA. The original PC12 cell full-length XLαs cDNA (Kehlenbach et al., 1994) or the ALEX cDNA fragment derived from it were subjected to in vitro transcription/translation in the presence of [35S]methionine/cysteine. The 35S-labeled translation products were analyzed by SDS–PAGE, either before (lanes 1 and 2) or after (lanes 3–6) immunoprecipitation, and visualized by phosphoimaging. Immunoprecipitation was performed with antibodies (Ab) directed against the C-terminus of XLαs (lane 3) or against ALEX (lanes 4–6), in the absence (–) or presence (+) of 10 µg/ml of the peptide used as antigen to raise the anti-ALEX antibody. Arrows, full-length XLαs; arrowheads, ALEX.
Interestingly, the immunoprecipitate obtained with the Gαs/XLαs C-terminal antibody from the XLαs mRNA translation products contained not only XLαs (Figure 8, lane 3, arrow), but also a 48 kDa band, most probably ALEX (Figure 8, lane 3, arrowhead). This raises the possibility of a physical interaction between XLαs and ALEX. Consistent with this possibility, the Gαs/XLαs C-terminal antibody did not immunoprecipitate a 48 kDa band from the ALEX mRNA translation products (data not shown), which lack XLαs. This issue is investigated further below.
Transfection of PC12 cells with the XLαs cDNA. Total membranes from PC12 cells transiently transfected with the XLαs cDNA were analyzed by immunoblotting using either the anti-XL antibody or the anti-ALEX antibody. As controls, total membranes from either wild-type PC12 cells or cells transfected with the ALEX cDNA were analyzed. One half of the membrane preparation was used directly for immunoblotting with the anti-XL antibody (Figure 9A), the other half was extracted with Triton X-100, and the insoluble material, which contains all of ALEX (see Figures 3C and 4C), was then analyzed by immunoblotting using the anti-ALEX antibody (Figure 9B). XLαs was clearly overexpressed (∼3-fold, Figure 9C) after transfection with the XLαs cDNA (Figure 9A, lane 2), as shown by comparison with untransfected, wild-type PC12 cells (Figure 9A, lane 1). As expected, PC12 cells transfected with the ALEX cDNA contained the same level of XLαs (Figure 9A, lane 3) as untransfected cells. Immunoblotting using the anti-ALEX antibody revealed that ALEX was massively overexpressed (∼10-fold, Figure 9C) after transfection with the ALEX cDNA (Figure 9B, lane 3). Interestingly, ALEX was also overexpressed in the XLαs-transfected PC12 cells (Figure 9B, lane 2). The fold increase in the ALEX level in the XLαs-transfected PC12 cells was the same (∼3-fold, Figure 9C) as that observed for XLαs. These results are consistent with both XLαs and ALEX being translated from the XLαs mRNA in vivo.
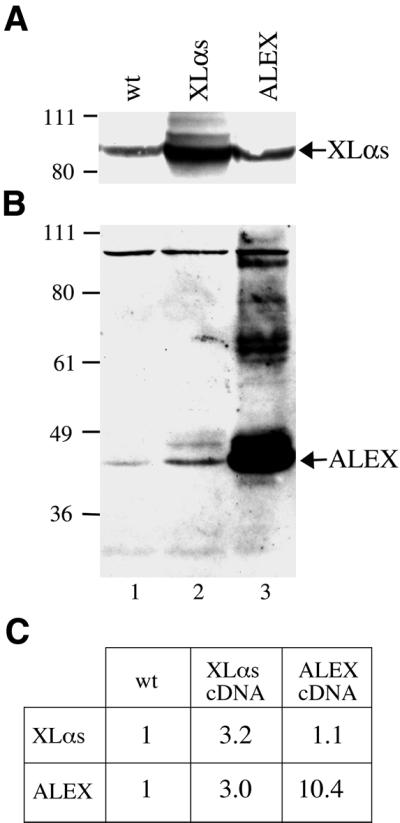
Fig. 9. Generation of ALEX upon transfection of the XLαs cDNA in PC12 cells. PC12 cells were either not transfected (wt) or transfected with the full-length XLαs cDNA (Kehlenbach et al., 1994) (XLαs) or the ALEX cDNA fragment derived from it (ALEX). (A) Total cell membranes (280 µg protein) were analyzed by immunoblotting using the anti-XL antibody. (B) The Triton X-100-insoluble material obtained from these membranes was analyzed by immunoblotting using the anti-ALEX antibody. (C) Quantification of the XLαs band of (A) and the ALEX band of (B) by densitometric scanning. The values obtained for the untransfected cells were set arbitrarily to 1 and the other values expressed relative to this.
ALEX binds to the XL-domain of XLαs
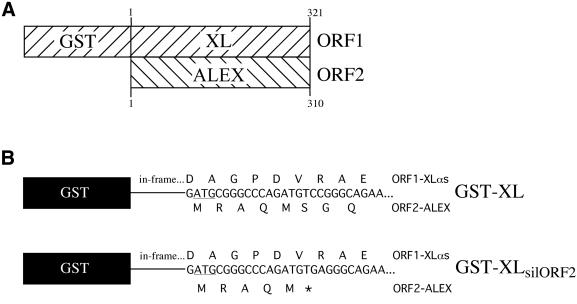
Given the presence of ALEX in immunoprecipitates of in vitro translated XLαs (Figure 8, lane 3, arrowhead), we investigated the possibility that ALEX binds to the XL-domain of XLαs. For this purpose, we constructed a fusion protein in which amino acids 1–321 of XLαs (Kehlenbach et al., 1994; see correction of translational start, Kehlenbach et al., 1995) are fused to GST (Figure 10A), referred to as GST–XL. This fusion protein (62 kDa apparent molecular weight on SDS–PAGE) contains the entire XL-domain of XLαs except for the βγ-binding region (see Figure 1B; Kehlenbach et al., 1994).
Fig. 10. Fusion protein constructs between GST and the XL-domain of XLαs that allow or prevent translation of ALEX. (A) Structure of the GST–XL cDNA construct, which allows translation of (i) a fusion protein comprising GST linked to amino acid residues 1–321 of the XL-domain of XLαs (ORF1) and (ii) a C-terminally truncated ALEX, ALEX1–310 (ORF2, see also Figure 1B). (B) The GST–XL cDNA construct (top) was mutated to the GST–XLsilORF2 construct (bottom) by altering a nucleotide triplet [TCC (top) to TGA (bottom)] such that the ORF1-encoded protein sequence, which is in-frame with GST, remains unaffected but a stop codon (asterisk) is introduced into ORF2.
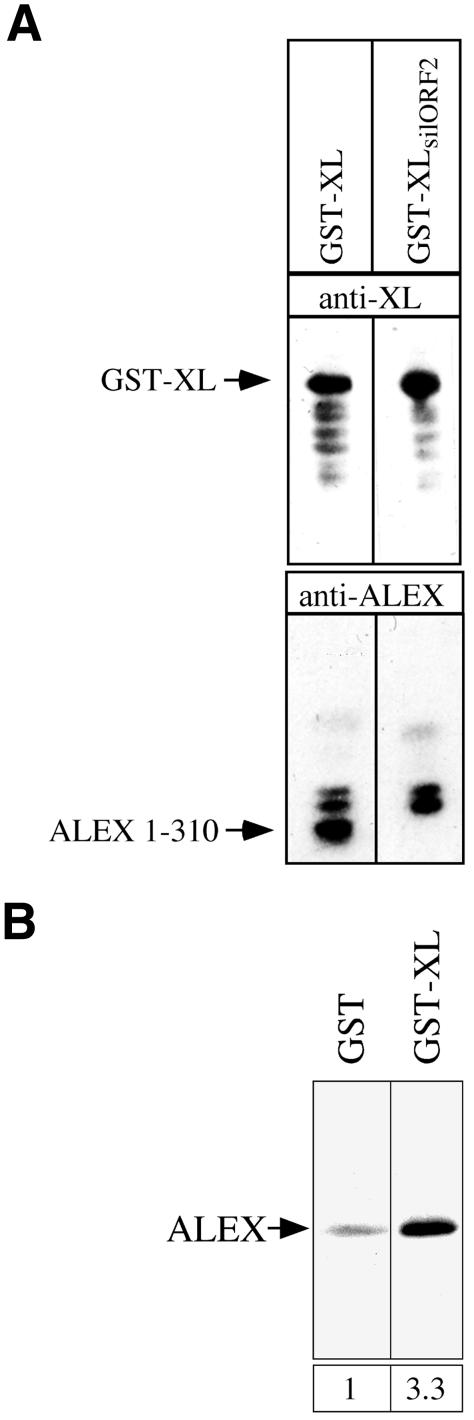
In light of the synthesis of both XLαs and ALEX in PC12 cells transfected with the XLαs cDNA (Figure 9), we first determined whether bacteria transformed with the GST–XL construct would synthesize both the GST–XL fusion protein and the corresponding C-terminally truncated ALEX1–310 (Figure 10A). Immunoblotting using the anti-ALEX antibody of a lysate obtained from bacteria transformed with the GST–XL construct showed the presence of ALEX1–310, in contrast to a lysate of bacteria transformed with GST only (data not shown). We then purified the GST–XL fusion protein from the lysate of bacteria transformed with the GST–XL construct by glutathione–Sepharose affinity chromatography and analyzed the eluate, obtained by an excess of reduced glutathione, by immunoblotting using either the anti-XL antibody (Figure 11A, top) or the anti-ALEX antibody (Figure 11A, bottom). This revealed the presence not only of the GST–XL fusion protein (Figure 11A, top left panel), but also of ALEX1–310 (Figure 11A, bottom left panel).
Fig. 11. ALEX binds specifically to the XL-domain of XLαs. (A) Generation of ALEX, and its binding to the XL-domain of XLαs, upon transformation of bacteria with the GST–XL cDNA construct. Escherichia coli were transformed with either the GST–XL or the GST–XLsilORF2 cDNA construct. The GST–XL fusion protein was purified via glutathione–Sepharose 4B and eluted by an excess of reduced glutathione. Eluates (15 µg of protein) were analyzed by SDS–PAGE and immunoblotting using either the anti-XL antibody (top) or the anti-ALEX antibody (bottom). (B) In vitro translated ALEX binds specifically to the XL-domain of a GST–XL fusion protein. Escherichia coli were transformed with pGEX-2T or the GST–XLsilORF2 cDNA construct, and GST and the GST–XL fusion protein, respectively, were purified by binding to glutathione– Sepharose 4B. 35S-Labeled ALEX, obtained by in vitro transcription/translation of the ALEX cDNA, was incubated with 50 µg of either GST or GST–XL bound to glutathione–Sepharose 4B. GST and GST–XL, as well as the 35S-labeled ALEX bound to either protein, were eluted by an excess of reduced glutathione, and eluates analyzed by SDS–PAGE and fluorography. The ALEX band was quantified by densitometric scanning and the amount of ALEX bound to the GST–XL fusion protein is expressed relative to that bound to GST only, which was set arbitrarily to 1. Both lanes of the fluorogram are the same exposure.
To prevent, upon transformation of bacteria with the GST–XL construct, the synthesis of ALEX1–310 and hence its binding to, and co-elution with, the GST–XL fusion protein upon glutathione–Sepharose affinity chromatography, we generated the GST–XLsilORF2 construct in which ORF2 encoding ALEX1–310 is closed by an in-frame stop codon shortly after the ORF2 ATG start codon (Figure 10B). Introduction of this stop codon did not change the amino acid sequence of the XL-part of the GST–XL fusion protein encoded by ORF1 (Figure 10B). Immunoblotting of the glutathione–Sepharose eluate obtained from bacteria transformed with the GST– XLsilORF2 construct showed the presence of the GST–XL fusion protein (Figure 11A, top right panel), but no longer that of ALEX1–310 (Figure 11A, bottom right panel). We conclude that (i) bacteria transformed with the GST–XL construct translate both ORF1 (encoding the GST–XL fusion protein) and ORF2 (encoding ALEX1–310), and that (ii) ALEX1–310 binds to the GST–XL fusion protein.
To determine whether full-length ALEX also binds to the GST–XL fusion protein, and to exclude the possibility that ALEX binds to the GST part rather than the XL part of the GST–XL fusion protein, we incubated in vitro translated 35S-labeled ALEX (see Figure 8, lane 2) with Sepharose beads containing either GST or the GST–XL fusion protein (synthesized from the GST–XLsilORF2 construct). Analysis of the eluates obtained by an excess of reduced glutathione showed that at least 3-fold more ALEX bound to the GST–XL fusion protein than to GST alone (Figure 11B). (The low level binding of ALEX to GST alone presumably reflects an unspecific sticking of the highly basic ALEX.) We conclude that ALEX specifically interacts with the XL-domain of XLαs and that this interaction does not require the βγ-binding region of the latter domain (as this region is lacking in the GST–XL fusion protein).
Discussion
In vivo occurrence of ALEX
Several lines of evidence indicate that ALEX indeed exists in vivo. First, upon immunoblotting of PC12 cell membranes, an anti-ALEX antibody detected a 48 kDa protein whose electrophoretic mobility was indistinguishable from that of cDNA-expressed ALEX. Detection of the 48 kDa protein by the anti-ALEX antibody reflected a specific recognition event because the 48 kDa immunoreactive band was increased selectively upon expression of the XLαs cDNA, along with XLαs. Secondly, upon limited proteolysis using V8 protease, the peptide fragments derived from the 48 kDa protein that were detected by the anti-ALEX antibody had the same molecular weight as those derived from cDNA-expressed ALEX. This indicates that V8 protease cleavage sites in the 48 kDa protein are located in the same sequence position as in cDNA-expressed ALEX. Thirdly, the 48 kDa protein, like cDNA-expressed ALEX, was recovered in the Triton X-100-insoluble fraction of PC12 cell membranes.
One exon with two almost completely overlapping ORFs, one mRNA, two proteins
In animal genomes, the existence of a >1 kb ORF (encoding ALEX) that completely overlaps with another slightly longer ORF (encoding the XL-domain of XLαs) present in the same exon has, as far as we are aware, no precedence. In contrast, the usage of overlapping reading frames is a common phenomenon in genomes of bacteriophages (e.g. Barrell et al., 1976, 1978; Fiddes and Godson, 1978), bacteria (e.g. Smith and Parkinson, 1980; Dong et al., 1993) and certain viruses (e.g. Lamb and Horvath, 1991). However, even in the latter cases, the overlap is usually rather small, i.e. at most a few hundred nucleotides, as in the paradigmatic example of the bacteriophage φX174 (Barrell et al., 1976). Here, as with ALEX, the ORF of gene E completely overlaps with that of gene D, being shifted in its phase by +1 compared with that of gene D; however, gene E encodes a protein of 91 amino acids and hence the reading frame overlap is only <300 bp rather than >1000 bp as in the present case.
Usage of overlapping reading frames to generate alternative protein products from the same gene has been demonstrated in the case of the mouse INK4a tumor supressor gene (Mao et al., 1995; Quelle et al., 1995) and the Aplysia CREB1 gene (Bartsch et al., 1998). However, only a portion of the two alternative proteins (p16INK4a and p19ARF in the case of the mouse INK4a gene; CREB1a and CREB1c in the case of the Aplysia CREB1 gene) is encoded by exons containing the overlapping reading frames, and each of the two proteins is translated from its own mRNA (Quelle et al., 1995; Bartsch et al., 1998). In contrast, in the present case, a single, large exon encodes the entire XL-domain of XLαs and all of ALEX, and both XLαs and ALEX appear to be translated from the same mRNA, i.e. the XLαs mRNA. First, northern blot analysis of total RNA from various rat tissues (Pasolli et al., 2000) does not reveal the existence of a shorter transcript that would start downstream of the ORF1 (XLαs) ATG start codon. Secondly, in vitro translation of the XLαs mRNA gave rise to both XLαs and ALEX. Thirdly, transfection of PC12 cells with the XLαs cDNA results in the expression of not only XLαs but also ALEX.
The translation of two proteins from the same mRNA raises the issue of the control mechanism that determines which of the two proteins is translated. For viral mRNAs and in yeast, several mechanisms for the translation of two proteins from one mRNA are known, for example programmed translational frameshifting (Farabaugh, 1996) or ribosome shunting (Fütterer et al., 1993). Another mechanism, which has also been reported for higher eukaryotes, is leaky scanning. If the first ATG of an mRNA does not display an optimal consensus sequence for translation initiation (Kozak, 1991), the 40S ribosomal subunit may continue scanning the mRNA and initiate translation at the next ATG (Kozak, 1999). In the case of the rat XLαs mRNA, this would be the ALEX (ORF2) ATG start codon, which is located 32 nucleotides downstream of the XLαs (ORF1) ATG start codon (Figure 2B, rat). Indeed, the rat XLαs ATG start codon does not display an optimal Kozak sequence (Figure 2B, rat), although it fits better with the Kozak consensus sequence than the mouse and human ATG start codon. However, in the case of the mouse (Figure 2B, mouse) and human (Figure 2B, human) XLαs mRNA, two and three ATG start codons, respectively, all located in ORF1, would have to be skipped by the scanning ribosome before reaching the first ATG start codon in ORF2: this is unlikely (Kozak, 1999). Hence, translation of XLαs and ALEX from the XLαs mRNA may, perhaps, be regulated by a novel mechanism whose elucidation would be interesting.
Subcellular localization and membrane attachment of ALEX
Both cDNA-expressed and endogenous ALEX were found to be membrane associated. Subcellular fractionation as well as immunofluorescence showed that cDNA-expressed ALEX, like XLαs, is localized at the plasma membrane, presumably (given the absence of a signal peptide and an obvious transmembrane segment) at its cytoplasmic surface. The association of ALEX with the plasma membrane is very tight, being resistant to carbonate extraction at pH 11.5. One may therefore speculate that ALEX is bound to the plasma membrane via a lipid anchor attached to, for example, the conserved cysteine residues in its C-terminal region. In addition, membrane binding of ALEX may be mediated by the interaction of the clusters of basic amino acids, the largest one of which is also located in the C-terminal region, with negatively charged headgroups of membrane phospholipids such as PIP2. In the case of MARKS (myristoylated alanine-rich C kinase substrate), the presence of a lipid anchor together with a polybasic region was shown to serve as a myristoyl–electrostatic switch that regulates the membrane affinity of this protein by phosphorylation of serine residues in the polybasic region (McLaughlin and Aderem, 1995). Interestingly, some of the polybasic clusters in ALEX constitute a part of the consensus sites for phosphorylation by protein kinase A (Figure 1C).
Putative function of ALEX
The function of ALEX is unknown. However, (i) the occurrence of ALEX in cells containing XLαs, (ii) its localization at the plasma membrane, as is the case for XLαs, and (iii) its binding to the XL-domain of XLαs suggest possible explanations for two previous, unresolved observations on XLαs. First, the majority of XLαs was found to undergo in vitro ADP-ribosylation by cholera toxin only poorly, in contrast to Gαs (Pasolli et al., 2000). Perhaps the interaction of XLαs with ALEX, which (extrapolating from the present data) was present in the membrane preparation used for ADP-ribosylation, renders the former a poor substrate for cholera toxin.
Secondly, XLαs, in contrast to Gαs, was found to be unable to undergo receptor-mediated GDP–GTP exchange (Klemke et al., 2000). Perhaps ALEX, which (extrapolating from the present data) was present in the membrane preparation used for investigating the activation of XLαs, did not allow GDP–GTP exchange on XLαs under the conditions used. The considerations also suggest that the physiological role of ALEX is in regulating signal transduction via XLαs.
Evolutionary implications
The nucleotide sequence of the XL-exon shows a remarkably low level of conservation between species (Table I) [Hayward et al., 1998a, AJ224868 (human); AF093569 (rat); AJ245739 (mouse)], which is much less than that of the exons encoding Gαs (Kozasa et al., 1988, human; NM_010309, mouse; NM_019132, rat). None theless, the unusual feature of the ORF encoding ALEX completely overlapping with the slightly longer ORF encoding the XL-domain of XLαs is conserved. This strongly suggests an important reason for this kind of genomic organization. The latter has interesting evolutionary implications, in particular in light of our finding that ALEX and the XL-domain bind to each other. The vast majority of mutations in the ORF overlap will result in alterations in the amino acid sequence of both ALEX and the XL-domain of XLαs. Natural selection would then favor those mutations that still allow the interaction of the two proteins (which in itself has interesting implications for the type of interaction between the two proteins). This would result in the co-evolution of ALEX and the XL-domain of XLαs.
Materials and methods
Antibodies
The anti-XL antibody, directed against the XL-domain of XLαs, was the same as described previously (Pasolli et al., 2000). The antibody against the common C-terminal decapeptide of Gαs and XLαs was the same as described previously (Kehlenbach et al., 1994) and is referred to as Gαs/XLαs C-terminal antibody. The peptide NH2-SQPPSQPLSQPPSQ-CONH2, corresponding to amino acid residues 35–48 of the rat ALEX sequence (Figure 1, box), was coupled to keyhole limpet hemocyanin via glutaraldehyde and used to raise a rabbit antiserum, referred to as anti-ALEX antibody.
cDNAs and mutagenesis
The plasmid CDM8-XLαs, originally called CDM8-XL (Kehlenbach et al., 1994), contains an ∼2.6 kb insert starting at nucleotide position 380 of the originally published sequence (Kehlenbach et al., 1994) and encodes the entire rat XLαs protein sequence (see correction of translational start in Kehlenbach et al., 1995) under the control of the cytomegalovirus (CMV) promoter.
For construction of the pCS2+-ALEX plasmid, the fragment of CDM8-XLαs corresponding to nucleotides 85–1170 of the 2.6 kb XLαs cDNA was amplified by PCR introducing BglII restriction sites on both ends. The PCR product was inserted into the BamHI restricition site of the eukaryotic expression vector pCS2+ (kindly provided by M.Brand, Heidelberg) and completely sequenced. The resulting pCS2+-ALEX plasmid encodes the entire rat ALEX protein sequence under the control of the CMV promoter.
For construction of the GST–XL fusion proteins, two distinct cDNA fragments encoding most of the XL-domain of XLαs (amino acids 1–321; see correction of translational start in Kehlenbach et al., 1995) were amplified from CDM8-XLαs by PCR and inserted into the pGEX-2T vector (Pharmacia) at the BamHI site downstream of, and in-frame with (regarding ORF1, see Figure 10A), the cDNA encoding GST. One cDNA fragment, referred to as GST–XL, contained a stop codon terminating ORF1 after amino acid residue 321, with ORF2 being terminated by a stop codon following the multiple cloning site of the pGEX-2T vector. The other, referred to as GST–XLsilORF2, contained the same stop codon for ORF1 and an additional stop codon at the nucleotide triplet corresponding to Ser6 of ALEX (see Figures 1C and 10B), resulting in termination of ORF2 after five amino acid residues.
Preparation of GST fusion proteins
Escherichia coli HB101 were transformed with pGEX-2T alone, or with GST–XL or GST–XLsilORF2 in pGEX-2T. Twenty milliliter cultures in LB containing ampicillin were grown overnight at 37°C, diluted into 200 ml of fresh LB containing ampicillin and grown to an optical density of 1.0–1.5. Expression of the fusion protein was induced by the addition of 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) followed by further culture for 5 h at 37°C. Bacterial cell pellets were resuspended in 10 ml of cold phosphate-buffered saline (PBS), the cells broken by repeated freezing and thawing, and proteins solubilized by addition of 1% (final concentration) Triton X-100 followed by incubation for 30 min at 4°C. Insoluble material was removed by centrifugation at 12 000 g for 10 min at 4°C, and the supernatant was incubated with 200 µl of glutathione–Sepharose (50% v/v, Pharmacia) in PBS for 2 h at 4°C. Sepharose beads were washed with PBS and either used for the binding of in vitro translated [35S]ALEX (see below), or the fusion protein was eluted with 50 mM Tris–HCl pH 8.0, containing 10 mM reduced glutathione, followed by SDS–PAGE and immunoblotting.
In vitro transcription/translation
After linearization by NotI, 1 µg of CDM8-XLαs or pCS2+-ALEX was in vitro transcribed for 2 h at 37°C in a final volume of 100 µl containing 20 µl of 5× transcription buffer (MBI Fermentas), 10 µl of rNTP mix (2.5 mM each final), 2 µl of RNase inhibitor (40 U/µl, MBI Fermentas), and either 2 µl of T7 RNA polymerase (40 U/µl) in the case of CDM8-XLαs or 2 µl of SP6 RNA polymerase (40 U/µl) in the case of pCS2+-ALEX.
Cell-free translation of in vitro transcribed RNAs was carried out at 30°C for 1.5 h using the Promega nuclease-treated reticulocyte lysate following the manufacturer’s instructions. Briefly, a typical translation mixture contained 35 µl of the reticulocyte lysate, 7 µl of nuclease-free dH2O, 1 µl of RNase inhibitor (40 U/µl), 1 µl of the amino acid mixture without methionine, 4 µl of the [35S]l-Met/Cys ProMix™ (Amersham, 1000 Ci/mmol) and 2 µl of the total in vitro transcription mixture containing the RNA template.
Immunoprecipitation
For immunoprecipitation of in vitro translated proteins, 25–75 µl of the in vitro translation mixture containing the 35S-labeled proteins and 20 µl of either the Gαs/XLαs C-terminal antibody or the anti-ALEX antibody were incubated in 600 µl of immunoprecipitation buffer [IPB; 100 mM Tris–HCl pH 7.2, 100 mM KCl, 5 mM MgCl2, 1% Triton X-100, 1% Na-deoxycholate, 0.3% SDS and 0.5 mM phenylmethylsulfonyl fluoride (PMSF)] overnight at 4°C end-over-end. After incubation with 50 µl of protein A–Sepharose (50% slurry in IPB) for 2 h at 4°C, beads were washed three times with IPB and immune complexes analyzed by SDS–PAGE and autoradiography. To test the specificiy of the anti-ALEX antibody, immunoprecipitation was carried out with the antibody solution containing 10 µg/ml of the SQPPSQPLSQPPSQ peptide.
Binding of ALEX to the GST–XL fusion protein
A 20 µl aliquot of the translation mixture containing 35S-labeled ALEX was incubated for 2 h at room temperature in 500 µl of PBS containing 50–100 µl of glutathione–Sepharose with an equal amount of either GST or GST–XL fusion protein bound to it. The Sepharose beads were washed three times with 500 µl of cold PBS containing 0.3% Triton X-100. Proteins bound to the GST fusion protein on the Sepharose beads were eluted by dissociation of the fusion protein from the beads using 100 µl of 50 mM Tris–HCl pH 8.0 containing 10 mM reduced glutathione, and analyzed by SDS–PAGE followed by autoradiography.
Cell culture and transfection
PC12 cells were grown as described (Tooze and Huttner, 1992). For transient transfection, PC12 cells harvested from a subconfluent 15 cm dish after trypsinization were subjected to electroporation (Bio-Rad Gene Pulser, 960 µF, 300 V) in 0.8 ml of PBS containing 45 µg of circular plasmid DNA.
HeLa cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal calf serum. HeLa cells on 15 cm dishes were transiently transfected using the calcium phosphate protocol (Ausubel et al., 1997) using 40 µg per dish of circular plasmid DNA.
Wild-type and transfected cells were plated on coverslips (polylysine-coated in the case of PC12 cells) for immunofluorescence, or on 15 cm dishes for subcellular fractionation. Cells were used 2 days after transfection, with 10 mM sodium butyrate being added in the case of XLαs- and ALEX-transfected PC12 cells during the last 16 h to increase the expression of the transgene (Gorman et al., 1983).
Subcellular fractionation
PNS from untransfected or transfected PC12 cells was prepared as described (Tooze and Huttner, 1992). Total membrane and soluble fractions from the PNS were obtained by centrifugation at 100 000 g for 1 h at 4°C. Velocity sucrose gradient centrifugation of the PNS was performed as described (Tooze and Huttner, 1992).
Carbonate extraction was done as described (Kehlenbach et al., 1994). Briefly, total membranes (100 µg of protein) were resuspended in 50 µl of H2O, mixed with 50 µl of extraction buffer (0.2 M Na2CO3 pH 11.5, 4 mM EDTA, 0.05% saponin, 0.5 mM PMSF), incubated under agitation for 30 min at 4°C, centrifuged at 100 000 g for 30 min at 4°C, and supernatant and pellet were collected.
For Triton X-100 extraction, total membranes (100 µg of protein) were resuspended in 100 µl of 10 mM Tris–HCl pH 8.0 containing 1 mM dithiothreitol (DTT), mixed with 100 µl of 2% Triton X-100 in the same buffer, incubated on ice for 30 min, centrifuged at 100 000 g for 30 min at 4°C and supernatant and pellet were collected.
All fractions were analyzed by SDS–PAGE and immunoblotting.
SDS–PAGE and immunoblotting
SDS–PAGE and immunoblotting were carried out according to standard procedures. The nitrocellulose membranes were blocked with 5% low-fat milk powder in PBS and incubated with either the anti-ALEX antibody (1:3000–1:5000 dilution) or the anti-XL antibody (1:400–1:800 dilution) followed by horseradish peroxidase-conjugated secondary antibody and the enhanced chemiluminescence (ECL) system (Amersham Corp.).
Peptide mapping of ALEX by limited proteolysis
Peptide mapping of ALEX by limited proteolysis during SDS–PAGE was performed using a modification (Lee and Huttner, 1983) of the procedure of Cleveland et al. (1977). Gel pieces containing the 48 kDa region were excised from the wet gel and equilibrated for 15 min at room temperature in 125 mM Tris–HCl pH 6.8, 0.1% SDS with two changes of the buffer. The gel pieces were then placed into the slots of a 4.5% stacking gel and each overlaid with 100 µl of a solution containing 125 mM Tris–HCl pH 6.8, 0.1% SDS, 15% glycerol, a trace of pyronin Y and 10 µg of S.aureus V8 protease. Electrophoresis was performed at 50 V until the dye had reached the end of the 15% resolving gel. The resulting ALEX fragments were transferred to nitrocellulose and detected using the anti-ALEX antibody.
Immunofluorescence
Indirect immunofluorescence of paraformaldehyde-fixed PC12 and HeLa cells was performed as described (Rosa et al., 1989), using either the anti-ALEX antibody at 1:1000 dilution or the anti-XL antibody at 1:400 dilution. As indicated in the legend to Figure 7, immunofluorescence was combined with double labeling using 50 µg/ml of fluorescein isothiocyanate (FITC)-conjugated WGA. WGA was either added to intact HeLa cells in PBS followed by incubation for 30 min at 4°C prior to fixation and permeabilization, or added together with secondary antibody to fixed, permeabilized and primary antibody-labeled PC12 cells. The secondary antibody was tetramethylrhodamine isothiocyanate (TRITC)-conjugated goat anti-rabbit IgG. Samples were examined by confocal laser scanning microscopy (Leica TCS4D).
Acknowledgments
Acknowledgements
We thank Dr Matthew J.Hannah for help with confocal microscopy and, together with Dr Denis Corbeil and Dr Yanzhuang Wang, for helpful discussion. W.B.H. was supported by grants from the Deutsche Forschungsgemeinschaft (SPP GTPases, Hu 275/4-1, Hu 275/4-2; SFB 352, C1), the EC (ERB-FMRX-CT96-0023 and ERBBIO4CT960058), the German–Israeli Foundation for Scientific Research and Development, and the Fonds der Chemischen Industrie.
Note added in proof
Since submission of this paper, the human galectin-3 gene has been reported to contain an internal gene giving rise to transcripts with two overlapping reading frames [Guittaut,M., Charpentier,S., Normand,T., Dubois,M., Raimond,J. and Legrand,A. (2001) Identification of an internal gene to the human galectin-3 gene with two different overlapping reading frames that do not encode galectin-3. J. Biol. Chem., 276, 2652–2657]. ORF1 and ORF2 are 318 and 291 nucleotides in length, respectively, with the overlap comprising 278 nucleotides. As in the present case, ORF2 is shifted by +1 in its phase compared with ORF1. However, in contrast to the present case, it remains to be shown whether the two hypothetical proteins encocded by ORF1 and ORF2 actually exist in vivo. If so, they are unlikely to interact with each other as they are targeted to distinct compartments, i.e. the cytosol-nucleus (ORF1) and mitochondria (ORF2), upon their expression in cells by transfection.
References
- Ausubel F.M., Brent,R., Kingston,R.E., Moore,D., Seidman,J.G., Smith,J.A. and Struhl,K. (eds.) (1997) Current Protocols in Molecular Biology. John Wiley & Sons, New York, NY.
- Barrell B.G., Air,G.M. and Hutchinson,C.A. (1976) Overlapping genes in bacteriophage φX174. Nature, 264, 34–41. [DOI] [PubMed] [Google Scholar]
- Barrell B.G., Shaw,D.C., Walker,J.E., Northop,F.D., Godson,G.N. and Fiddes,J.C. (1978) Overlapping genes in bacteriophages φX174 and G4. Biochem. Soc. Trans., 6, 63–67. [DOI] [PubMed] [Google Scholar]
- Bartsch D., Casadio,A., Karl,K.A., Serodio,P. and Kandel,E.R. (1998) CREB1 encodes a nuclear activator, a repressor and a cytoplasmatic modulator that form a regulatory unit critical for long-term facilitation. Cell, 95, 211–223. [DOI] [PubMed] [Google Scholar]
- Cleveland, D.W., Fischer S.G., Kirschner M.W. and Laemmli U.K. (1977) Peptide mapping by limited proteolysis in sodium dodecyl sulfate and analysis by gel electrophoresis. J. Biol. Chem., 252, 1102–1106. [PubMed] [Google Scholar]
- Corradi N., Borgonovo,B., Clementi,E., Bassetti,M., Racchetti,G., Consalez,G.G., Huttner,W.H., Meldolesi,J. and Rosa,P. (1996) Overall lack of regulated secretion in a PC12 variant cell clone. J. Biol. Chem., 271, 27116–27124. [DOI] [PubMed] [Google Scholar]
- Dong J.M., Taylor,J.S., Latour,D.J., Iuchi,S. and Lin,E.C. (1993) Three overlapping lct genes involved in l-lactate utilization by Escherichia coli. J. Bacteriol., 175, 6671–6678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh P.J. (1996) Programmed translational frameshifting. Annu. Rev. Genet., 30, 507–528. [DOI] [PubMed] [Google Scholar]
- Fiddes J.C. and Godson,G.N. (1978) Nucleotide sequence of the J gene and surrounding untranslated regions of phage G4 DNA: comparison with phage φX174. Cell, 15, 1045–1053. [DOI] [PubMed] [Google Scholar]
- Fütterer J.Z., Kiss-Laszlo,Z. and Hohn,T. (1993) Non-linear ribosome migration on cauliflower mosaic virus 35S RNA. Cell, 73, 789–802. [DOI] [PubMed] [Google Scholar]
- Gorman C., Howard,B. and Reeves,R. (1983) Expression of recombinant plasmids in mammalian cells is enhanced by sodium butyrate. Nucleic Acids Res., 11, 7631–7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannah M.J., Weiss,U. and Huttner,W.B. (1998) Differential extraction of proteins from paraformaldehyde-fixed cells: lessons from synaptophysin and other membrane proteins. Methods, 16, 170–181. [DOI] [PubMed] [Google Scholar]
- Hayward B.E., Kamiya,M., Strain,L., Moran,V., Campbell,R., Hayashizaki,Y. and Bonthron,D.T. (1998a) The human GNAS1 gene is imprinted and encodes distinct paternally and biallelically expressed G proteins. Proc. Natl Acad. Sci. USA, 95, 10038–10043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward B.E., Moran,V., Strain,L. and Bonthron,D.T. (1998b) Bidirectional imprinting of a single gene: GNAS1 encodes maternally, paternally and biallelically derived proteins. Proc. Natl Acad. Sci. USA, 95, 15475–15480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehlenbach R.H., Matthey,J. and Huttner,W.B. (1994) XLαs is a new type of G protein. Nature, 372, 804–809. [DOI] [PubMed] [Google Scholar]
- Kehlenbach R.H., Matthey,J. and Huttner,W.B. (1995) XLαs is a new type of G-protein (Correction). Nature, 375, 253. [Google Scholar]
- Klemke M., Pasolli,A.H., Kehlenbach,R.H., Offermanns,S., Schultz,G. and Huttner,W.B. (2000) Characterization of the extra-large G-protein α-subunit XLαs. II. Signal transduction properties. J. Biol. Chem., 275, 33633–33640. [DOI] [PubMed] [Google Scholar]
- Kozak M. (1991) An analysis of vertebrate mRNA sequences: intimations of translational control. J. Cell Biol., 115, 887–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. (1999) Initiation of translation in prokaryotes and eukaryotes. Gene, 234, 187–208. [DOI] [PubMed] [Google Scholar]
- Kozasa T., Itoh,H., Tsukamoto,T. and Kaziro,Y. (1988) Isolation and characterization of the human Gsα gene. Proc. Natl Acad. Sci. USA, 85, 2081–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb R.A. and Horvath,C.M. (1991) Diversity of coding strategies in influenza viruses. Trends Genet., 7, 261–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R.W. and Huttner,W.B. (1983) Tyrosine-O-sulfated proteins of PC12 pheochromocytoma cells and their sulfation by a tyrosylprotein sulfotransferase. J. Biol. Chem., 258, 11326–11334. [PubMed] [Google Scholar]
- Mao L., Merlo,A., Bedi,G., Shapiro,G.I., Edwards,C.D., Rollins,B.J. and Sidransky,D. (1995) A novel p16INK4a transcript. Cancer Res., 55, 2995–2997. [PubMed] [Google Scholar]
- McLaughlin S. and Aderem,A. (1995) The myristoyl–electrostatic switch: a modulator of reversible protein–membrane interactions. Trends Biochem. Sci., 20, 272–276. [DOI] [PubMed] [Google Scholar]
- Pasolli A.H., Klemke,M., Kehlenbach,R.H., Wang,Y. and Huttner,W.B. (2000) Characterization of the extra-large G-protein α-subunit XLαs. I. Tissue distribution and subcellular localization. J. Biol. Chem., 275, 33622–33632. [DOI] [PubMed] [Google Scholar]
- Peters J., Wroe,S.F., Wells,C.A., Miller,H.J., Bodle,D., Beechey,C.V., Williamson,C.M. and Kelsey,G. (1999) A cluster of oppositely imprinted transcripts at the Gnαs locus in the distal imprinting region of mouse chromosome 2. Proc. Natl Acad. Sci. USA, 96, 3830–3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quelle D.E., Zindy,F., Ashmun,R.A. and Sherr,C.J. (1995) Alternative reading frames of the INK4a tumor supressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell, 83, 993–1000. [DOI] [PubMed] [Google Scholar]
- Rosa P., Weiss,U., Pepperkok,R., Ansorge,W., Niehrs,C., Stelzer,E.H.K. and Huttner,W.B. (1989) An antibody against secretogranin I (chromogranin B) is packaged into secretory granules. J. Cell Biol., 109, 17–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R.A. and Parkinson,J.S. (1980) Overlapping genes at the cheA locus of Escherichia coli. Proc. Natl Acad. Sci. USA, 77, 5370–5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooze S.A. and Huttner,W.B. (1992) Cell-free formation of immature secretory granules and constitutive secretory vesicles from the trans-Golgi network. Methods Enzymol., 219, 81–93. [DOI] [PubMed] [Google Scholar]