Abstract
Heat shock factor 1 (HSF1) is a serine-rich constitutively phosphorylated mediator of the stress response. Upon stress, HSF1 forms DNA-binding trimers, relocalizes to nuclear granules, undergoes inducible phosphorylation and acquires the properties of a transactivator. HSF1 is phosphorylated on multiple sites, but the sites and their function have remained an enigma. Here, we have analyzed sites of endogenous phosphorylation on human HSF1 and developed a phosphopeptide antibody to identify Ser230 as a novel in vivo phosphorylation site. Ser230 is located in the regulatory domain of HSF1, and promotes the magnitude of the inducible transcriptional activity. Ser230 lies within a consensus site for calcium/calmodulin-dependent protein kinase II (CaMKII), and CaMKII overexpression enhances both the level of in vivo Ser230 phosphorylation and transactivation of HSF1. The importance of Ser230 was further established by the S230A HSF1 mutant showing markedly reduced activity relative to wild-type HSF1 when expressed in hsf1–/– cells. Our study provides the first evidence that phosphorylation is essential for the transcriptional activity of HSF1, and hence for induction of the heat shock response.
Keywords: heat shock factor 1/heat shock response/phosphorylation/transcription
Introduction
The ability of a cell to rapidly change its gene expression pattern in response to extracellular signals usually involves modulation of the activity of pre-existing transcription factors. Protein phosphorylation has been identified as a major post-translational mechanism regulating the activity of transcription factors (Hunter and Karin, 1992; Hunter, 2000). Recent studies have revealed that phosphorylation is not simply used to switch the activity of a protein on or off, but that complex multisite phosphorylation is a common key mechanism for greatly increasing the regulatory potential of proteins (Cohen, 2000). With regard to heat shock factor 1 (HSF1), the transcription factor responsible for stress-induced expression of heat shock proteins (Hsps), it has been known since the end of the 1980s that the factor is constitutively and inducibly phosphorylated (Sorger et al., 1987; Larson et al., 1988; Sorger and Pelham, 1988; Sorger, 1990). While the inducible phosphorylation remains a diagnostic feature of the fully active DNA-binding and transcriptional state of HSF1, attempts to identify key residues and to establish the mechanistic consequences of inducible phosphorylation have not been forthcoming. A mechanistic understanding of the role of HSF1 phosphorylation has been complicated by the large number of potential sites for serine phosphorylation, and certain sites are constitutively phosphorylated whereas others are stress inducible.
Under normal growth conditions, mammalian HSF1 exists as a phosphorylated monomer in which DNA-binding and transcriptional activities are repressed. In response to heat shock and other chemical, environmental or physiological stresses, HSF1 undergoes trimerization, binds to the heat shock element in the promoter regions of target genes, and exhibits transcriptional activity. Simultaneously, HSF1 becomes inducibly phosphorylated and relocalizes to form nuclear granules (Wu, 1995; Morimoto, 1998). To date, evidence for the role of some proposed constitutively phosphorylated sites on HSF1 has emerged, whereas the inducible sites and their function have remained elusive. Studies using transactivation assays and phosphopeptide mapping of chimeric GAL4-HSF1 proteins and recombinant HSF1 have shown that serines 303 and 307 are phosphorylated, and repress the transcriptional activity of HSF1 under normal growth conditions and perhaps during attenuation (Chu et al., 1996, 1998; Knauf et al., 1996; Kline and Morimoto, 1997). Serines 303 and 307 have been suggested to be phosphorylated by glycogen synthase kinase 3β (GSK-3β) and the extracellular signal-regulated kinase (ERK) based on in vitro analyses and overexpression of these kinases (Chu et al., 1996, 1998; Knauf et al., 1996; Kim et al., 1997; He et al., 1998; Bijur and Jope, 2000; Xavier et al., 2000). Constitutive phosphorylation of Ser363 by overexpressed protein kinase C (PKC) has also been implicated in repression of HSF1 (Chu et al., 1998), whereas Dai et al. (2000) showed that Ser363 is a good substrate for overexpressed c-Jun N-terminal kinase (JNK). While these observations begin to establish a role for HSF1 regulation by phosphorylation, there is no in vivo demonstration on these phosphorylation sites or kinases.
In concert with stress-induced acquisition of transcriptional activity, HSF1 and its yeast homolog are inducibly serine phosphorylated (Sorger et al., 1987; Larson et al., 1988; Sorger and Pelham, 1988; Sorger, 1990; Baler et al., 1993; Sarge et al., 1993; Chu et al., 1996; Cotto et al., 1996; Liu and Thiele, 1996; Kline and Morimoto, 1997). In Drosophila, heat shock induces both phosphorylation and dephosphorylation of several sites, although no increase in the steady-state phosphorylation of HSF can be detected (Fritsch and Wu, 1999). In contrast to the promoting effect of inducible phosphorylation on the transcriptional capacity of Saccharomyces cerevisiae HSF, heat-inducible phosphorylation of some sites of the Kluyveromyces lactis HSF has been proposed to enhance deactivation (Høj and Jakobsen, 1994). As in yeast and fruit fly, inducible phosphorylation does not influence the DNA-binding activity of mammalian HSF1 (Jurivich et al., 1992, 1995; Cotto et al., 1996). Thus, it is likely that inducible phosphorylation influences the transcriptional competence of HSF1.
To decipher the complex phosphorylation-mediated regulation of HSF1, it is necessary to characterize each of the in vivo sites of HSF1 phosphorylation and the kinases/phosphatases involved. In this study, we have used multiple methods to identify Ser230 as a novel in vivo phosphorylation site on human HSF1. We demonstrate that Ser230, located in the regulatory domain, is constitutively and stress-inducibly phosphorylated, and contributes to the transcriptional activity of HSF1. Hence, we have identified the first phosphorylation site on HSF1 that promotes stress-induced transactivation.
Results
Heterogeneity in serine phosphorylation of endogenous human HSF1
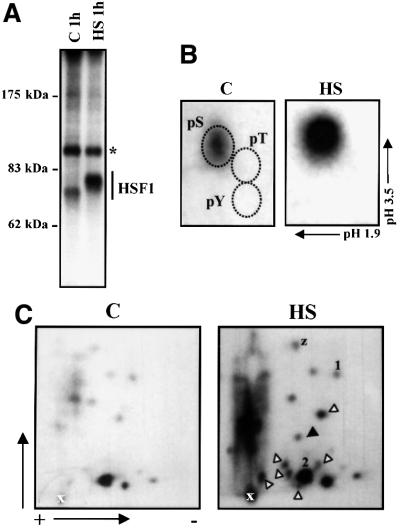
Our initial approach to identify the in vivo phosphorylation sites of human HSF1 was to map the sites by tryptic phosphopeptide analysis followed by manual Edman degradation. K562 cells were labeled with [32P]ortho phosphate for 3 h before exposure to a 1 h heat shock at 42°C and HSF1 was immunoprecipitated. HSF1 was constitutively phosphorylated, and heat shock increased phosphorylation by 2.5- to 4-fold, which was accompanied by slower migration of HSF1 on SDS–PAGE, as compared with HSF1 in untreated cells (Figure 1A). Both the constitutive and inducible phosphorylation of HSF1 occurred on serines and no trace of threonine or tyrosine phosphorylation was detected (Figure 1B).
Fig. 1. Heterogeneous in vivo phosphorylation of HSF1. K562 cells were in vivo labeled with [32P]orthophosphate for 3 h before they were subjected to heat shock (HS) or left untreated (C). HSF1 was immunoprecipitated with anti-hHSF1 antibodies and resolved on 8% SDS–PAGE. (A) Autoradiograph of the immunoprecipitated HSF1. The asterisk indicates unknown phosphoprotein. (B) Phosphoamino acid analysis of the immunoprecipitated HSF1. The relative positions of phosphoSer, phosphoThr and phosphoTyr are indicated. (C) Tryptic phosphopeptide mapping of HSF1. The black arrowhead indicates a new phosphopeptide detected upon heat shock, and the white arrowheads indicate phosphopeptides, the intensity of which is markedly enhanced upon heat stress. Phosphopeptide-1, -2 and -z are explained in Results.
The analysis of 32P-labeled HSF1 by two-dimensional tryptic phosphopeptide mapping showed a complex pattern of phosphopeptides both in untreated and heat-shocked cells, indicating multiple phosphorylation sites (Figure 1C). A phosphopeptide, which was not detected in untreated cells, was induced upon heat shock and the intensity of several phosphopeptides was markedly enhanced upon heat stress. Furthermore, the intensity of most other phosphopeptides was moderately increased. Because some of the phosphopeptides above the loading spot were not well resolved, the trypsin-digested 32P-labeled HSF1 was also separated using a C-18 reversed-phase HPLC column. Substantial differences in the intensity of the 32P-labeled phosphopeptides from untreated and heat-shocked HSF1 were observed; however, the overall profiles of the phosphopeptide radiograms were similar upon both treatments (data not shown). The phosphopeptide analyses showed heterogeneity in HSF1 phosphorylation under normal growth conditions and a moderate or prominent increase in phosphorylation of most phosphopeptides upon stress. In addition, one phosphopeptide seemed to be specific for the stress-induced sample.
Identification of Ser230 as a novel in vivo phosphorylation site on hHSF1
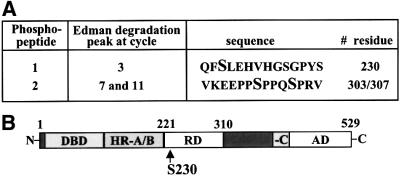
To determine the phosphorylation site of the phosphopeptides separated by TLC, manual Edman degradation was conducted. Among all possible tryptic peptides, based on sequence data, we found several serines that could match the observed phosphate-release cycles. As the transcriptional activity of HSF1 has been shown to be restricted by the regulatory domain (Green et al., 1995; Shi et al., 1995; Zuo et al., 1995; Newton et al., 1996), we searched for phosphorylation sites in this region and found from phosphopeptide-1 a phosphate release on the third cycle that would correspond to Ser230 (Figures 1C and 2). We were confident to continue analyzing this site, as our in vitro phosphorylation studies also pointed towards Ser230 (see below). The Ser230-containing phosphopeptide-1 was phosphorylated under normal growth conditions and its phosphorylation was enhanced upon heat stress (Figure 1C). In addition, we found a phosphopeptide, phosphopeptide-2, from both untreated and heat-shocked samples, in which the phosphate-release cycles revealed phosphorylation in positions 7 and 11, corresponding to serines 303 and 307 (Figures 1C and 2A). This is consistent with previous studies (Chu et al., 1996, 1998; Knauf et al., 1996; Kline and Morimoto, 1997), and provides an independent corroboration based on direct sequencing analysis of endogenous HSF1.
Fig. 2. Identification of Ser230 phosphorylation of HSF1 in vivo. (A) Manual Edman degradation was performed on the phospho peptide-1 and -2 derived from in vivo 32P-labeled endogenous HSF1 (Figure 1C). The cycle of 32P release, the corresponding phospho peptide sequence and the phosphorylated residues are shown. (B) Schematic representation of human HSF1. The position of Ser230 in the regulatory domain is indicated. DBD, DNA-binding domain; HR-A/B and -C, hydrophobic repeat A/B and C; RD, regulatory domain; AD, activation domain.
HSF1 is phosphorylated in vitro on Ser230 by CaMKII
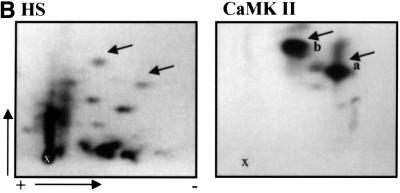
To identify the potential protein kinase that would phosphorylate HSF1 on Ser230, in vitro kinase assays were performed. Since the Ser230 is a consensus site for calcium/calmodulin-dependent protein kinase II (CaMKII), we examined whether CaMKII was able to phosphorylate recombinant human HSF1 (rhHSF1) in vitro. As shown in Figure 3A, rhHSF1 was a good substrate for CaMKII. Comparison of the tryptic phosphopeptide map of rhHSF1, phosphorylated by CaMKII, with the in vivo tryptic phosphopeptide map of HSF1 upon heat shock, indicated that CaMKII generates two phosphopeptides (phosphopeptide-a and -b) with similar migration patterns to two of the in vivo phosphopeptides (Figure 3B). Phosphopeptide-a was phosphorylated on its third amino acid, as determined by manual Edman degradation (data not shown), which is in agreement with the results from the in vivo phosphopeptide-1 (Figure 2A). No phosphate release of phosphopeptide-b was detected by 14 cycles of Edman degradation (see below).
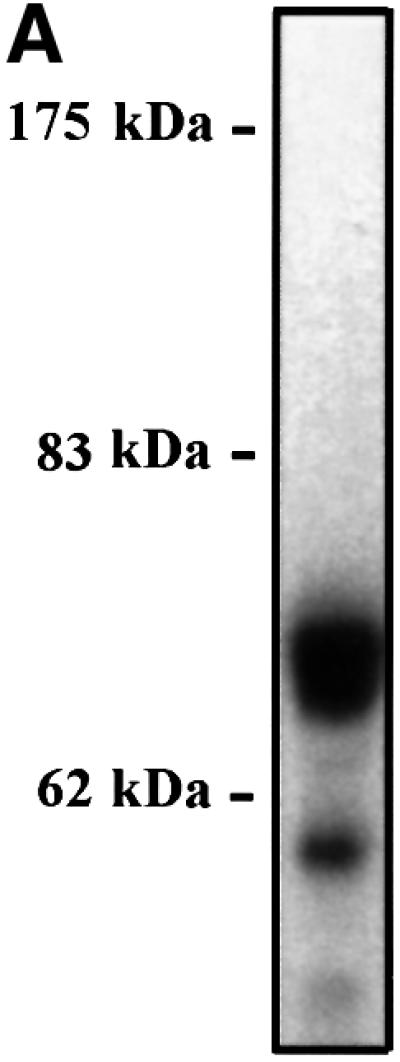
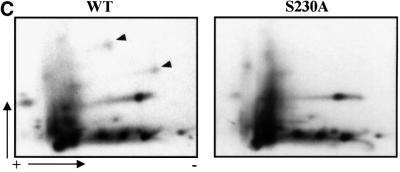
Fig. 3. CaMKII phosphorylates rhHSF1 on Ser230. (A) Autoradiograph of rhHSF1 phosphorylated in vitro by CaMKII. (B) Tryptic phosphopeptide mapping of immunoprecipitated 32P-labeled HSF1 from heat-shocked K562 cells (HS), or of rhHSF1 phosphorylated in vitro by CaMKII. The arrows mark phosphopeptides with similar migration patterns in vivo and in vitro. (C) Tryptic phosphopeptide mapping of mutant HSF1. K562 cells were transfected with wild-type or S230A HSF1-Myc-His, in vivo labeled with [32P]orthophosphate, and subjected to heat shock. HSF1 was immuno precipitated with anti-Myc antibodies followed by tryptic phosphopeptide mapping. The arrowheads represent phosphopeptides that are absent in S230A.
To directly identify the site of CaMKII phosphorylation, trypsin-digested rhHSF1 was first separated using HPLC. Interestingly, only one fraction contained a strong radioactive signal and was further used for sequencing and mass spectrometric analysis. The obtained sequence corresponding to amino acids 228–296 was phosphorylated on Ser230 by CaMKII (data not shown). The mass spectrometric analysis also detected a chemically modified pyroglutamate form of the phosphopeptide with a mass of 7021.7. As N-terminal pyroglutamate inhibits the Edman reaction, phosphopeptide-b on the TLC separation is most likely the pyroglutamate form of this peptide. These results show that CaMKII is highly specific in its action on HSF1, as it has only one phosphorylation site among the plethora of potential serine targets in the sequence.
To clarify whether phosphorylated Ser230 was present in several generated phosphopeptides from HSF1 in the cell, wild-type and a serine to alanine (S230A) mutant human HSF1 were introduced to K562 cells. Tryptic phosphopeptide mapping of exogenous HSF1 revealed that two phosphopeptides were not detected in the S230A as compared with the wild-type HSF1 (Figure 3C). The migration of these Ser230-containing phosphopeptides corresponded to the a- and b-phosphopeptide of rhHSF1 (Figure 3B) and the phosphopeptide-1 and -z of endogenous HSF1 (Figure 1C). In further support for phosphopeptide-z being the pyroglutamate form of phosphopeptide-1, no phosphate release was observed upon manual Edman degradation of this phosphopeptide (data not shown). Therefore, the relative intensity of phosphorylated Ser230 on in vivo phosphopeptide maps is divided on two spots (phosphopeptide-1 and -z). In addition, our results showing the presence of pyroglutamate phosphopeptides could explain why several phosphopeptides on a tryptic map have been shown to disappear upon mutation of a single amino acid residue (Kline and Morimoto, 1997).
Analyses with phosphopeptide-specific antibody reveal that Ser230 is phosphorylated in vivo and is present in the HSF1 DNA-binding complex as well as in HSF1 granules
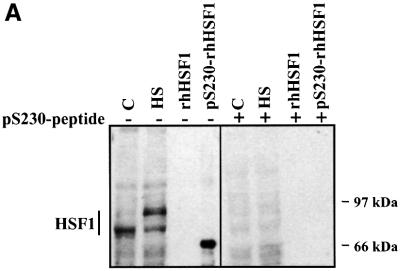
As a novel approach for HSF1 phosphorylation studies, a phosphopeptide antibody was generated to demonstrate that Ser230 is indeed phosphorylated in cells, as well as to enable characterization of phosphoSer230 on activated HSF1. The antibody raised against phosphorylated Ser230 (anti-pS230-hHSF1) recognized rhHSF1 phosphorylated in vitro on Ser230, but not the unphosphorylated rhHSF1 (Figure 4A). The anti-pS230-hHSF1 antibody also detected HSF1 from whole-cell extracts of untreated and heat-shocked K562 cells (Figure 4A). Heat shock induced a slower migration of HSF1 on the SDS–PAGE compared with HSF1 from untreated cells, which is consistent with earlier results using antibodies against total HSF1 (Sarge et al., 1993; Holmberg et al., 2000). The pS230-hHSF1 antibody also recognized a faster migrating form of HSF1 in the heat-shocked cells (Figure 4A). The sum of these two bands contributes to a prominent elevation in the amount of phosphorylated Ser230 upon heat shock as compared with untreated cells, which is in agreement with our phosphorylation results (Figures 1C and 3C). To confirm the specificity of the anti-pS230-hHSF1 antibody, the antibody was pre-incubated with an excess of free phosphopeptide, which blocked the appearance of the HSF1 band (Figure 4A). The specificity of the anti-pS230-hHSF1 antibody was further confirmed, as S230A was not recognized by the antibody (Figure 6C).
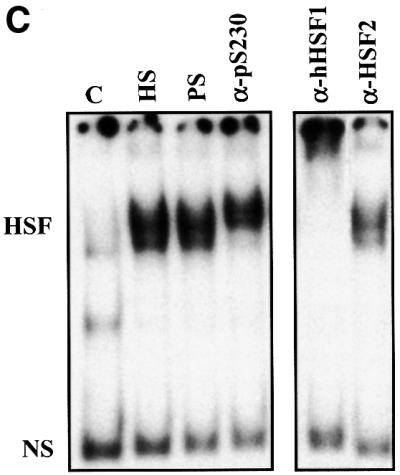
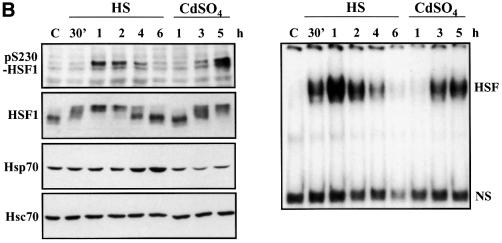
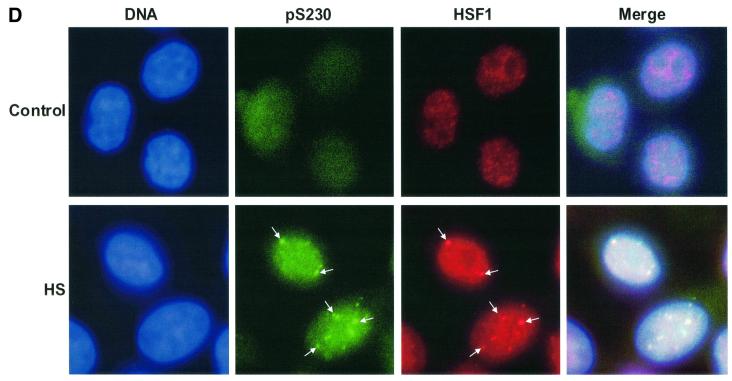
Fig. 4. Characterization of in vivo phosphorylated Ser230 on hHSF1 with phosphopeptide-specific antibodies. (A) Whole-cell extracts (60 µg) of untreated (C) and heat-shocked (HS) K562 cells, as well as rhHSF1 (5 ng) and rhHSF1 phosphorylated in vitro by CaMKII (pS230-rhHSF1, 5 ng) were resolved on 8% SDS–PAGE and immunoblotted with the anti-pS230-hHSF1 antibody (–). The antibody was pre-incubated with free phosphopeptide for competitive inhibition of the immunoreaction (+). (B) HeLa cells were exposed to heat shock (HS) and CdSO4 (100 µM) for the indicated time periods or were left untreated (C). Whole-cell extracts were analyzed by western blotting using antibodies to phosphorylated Ser230 (pS230-HSF1), HSF1, Hsp70 and Hsc70 (left panel). The HSF1 DNA-binding activity in the whole-cell extracts was examined by gel mobility shift assay (right panel). HSF, HSF1–HSE complex; NS, non-specific DNA-binding activity. (C) Whole-cell extracts from HeLa cells exposed to heat shock (HS) were incubated with anti-pS230-hHSF1 (α-pS230), anti-hHSF1 (α-hHSF1), anti-HSF2 (α-HSF2) antibodies or with pre-immune serum (PS) prior to the gel mobility shift assay. (D) HeLa cells were heat shocked (HS) or kept at 37°C (control) and the immunofluorescence analysis was performed using an anti-pS230-hHSF1 (pS230, green) and anti-hHSF1 (HSF1, red) antibody. The DNA was stained with DAPI. The arrows indicate HSF1 nuclear granules.
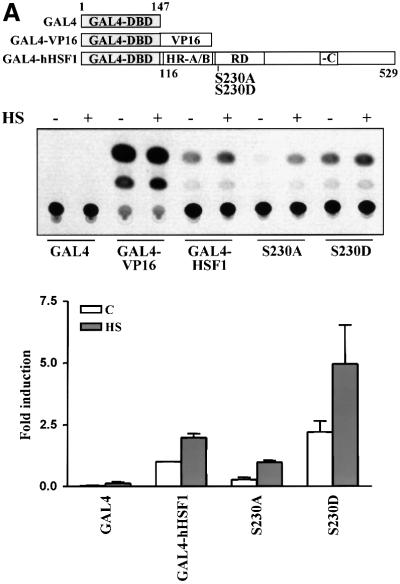
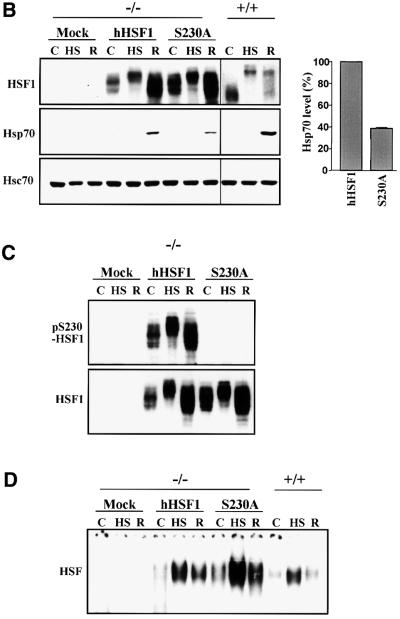
Fig. 6. S230A has diminished transcriptional activity. (A) K562 cells were transfected with wild-type or mutant GAL4-HSF1, the G5BCAT reporter plasmid and the internal control RSVβ-gal. The transfectants were exposed to heat shock followed by 3 h recovery (+). Untreated transfectants are denoted by (–). The CAT activity in untreated wild-type GAL4-HSF1 transfectants was assigned a fold induction of 1. The data represent mean ± SEM from two independent experiments performed in duplicate. (B–D) hsf1–/– MEFs (–/–) were transfected with wild-type (hHSF1) or S230A (S230A) hHSF1-Myc-His plasmids. +/+, MEFs containing endogenous HSF1. The cells were exposed to heat shock (HS), heat shock followed by 3 h recovery (R) or were left untreated (C). (B) Western blot analysis using antibodies against HSF1, Hsp70 and Hsc70. Note that the HSF1 blot of +/+ was exposed for a longer time than the HSF1 blot of –/– cells. For quantification, the Hsp70 level in wild-type hHSF1 transfectants was assigned to 100%. The data represent mean ± SEM from three independent experiments. (C) Western blotting using the anti-pS230-hHSF1 (upper panel) and anti-hHSF1 (lower panel) antibody. (D) Analysis of the HSF1 DNA-binding activity by gel mobility shift assay.
In addition to K562 cells, a heat-induced elevation of the phosphorylation state of Ser230 occurred in HeLa cells (Figure 4B, left panel). A robust increase in the phosphoSer230 amount was detected by 1 h of heat stress, after which the amount returned to the basal level. The Ser230 phosphorylation and the inducible phosphorylation of total HSF1 occurred with identical kinetics and preceded the accumulation of Hsp70 (Figure 4B, left panel). HSF1 DNA-binding activity was detected prior to a clear increase in Ser230 phosphorylation upon heat shock (Figure 4B, right panel). In addition to heat shock, another stressor, cadmium, increased phosphorylation on Ser230 with kinetics resembling the inducible HSF1 phosphorylation (Figure 4B, left panel), indicating that inducible phosphorylation on Ser230 could be a common feature in response to various stressors.
As the increase in Ser230 phosphorylation agreed well with the stress-induced activation of HSF1, we investigated by antibody perturbation assay whether phos phoSer230 was present in the HSF1 molecules that acquired DNA-binding activity upon heat shock. The anti-pS230-hHSF1 antibody induced a shift in the HSF1–HSE complex, indicating the presence of phosphoSer230 in active HSF1 (Figure 4C). The migration shift induced by the anti-pS230-hHSF1 antibody was not as prominent as the one induced by the anti-hHSF1 antibody, which could be due to the lower reactivity against the restricted epitope of the phosphopeptide antibody. The presence of phosphoSer230 was not detected by the antibody perturbation assay in samples exposed to heat shock for <0.5 h (data not shown). An antibody against the non-heat-inducible HSF2 did not induce a shift in migration of the heat-induced HSF1–HSE complex (Figure 4C). Another parameter of HSF1 activation is its relocalization in nuclear granules, in concert with acquisition of transcriptional activity upon stress (Sarge et al., 1993; Cotto et al., 1997; Jolly et al., 1997, 1999; He et al., 1998; Holmberg et al., 2000). To study directly the phosphorylation state of HSF1 granules in intact cells, untreated or heat-shocked HeLa cells were double labeled with the anti-hHSF1 and anti-pS230-hHSF1 antibodies. Brightly staining nuclear HSF1 granules were detected upon heat shock and the localization of these granules was identical with both antibodies (Figure 4D).
Active CaMKII enhances heat-induced transactivation and Ser230 phosphorylation of endogenous HSF1
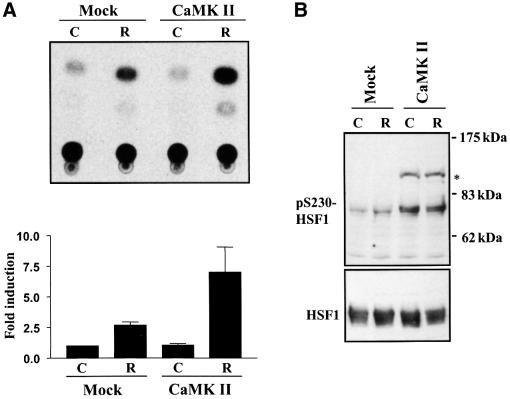
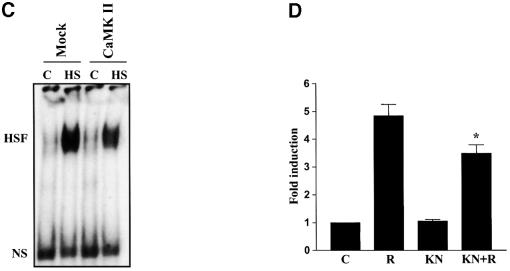
Prompted by the finding that Ser230 phosphorylation correlated with HSF1 activity (Figures 1, 2 and 4) and that CaMKII phosphorylated Ser230 in vitro (Figure 3), we examined whether CaMKII affected the phosphorylation and transcriptional activity of HSF1 in vivo. K562 cells were transfected with active CaMKII together with a CAT reporter plasmid containing the proximal HSE of human hsp70 promoter, and were exposed to heat shock followed by a 6 h recovery. A 2.5-fold increase in CAT activity was consistently observed upon heat shock, whereas overexpression of CaMKII did not affect the basal activity (Figure 5A). Interestingly, the heat-induced CAT activity was enhanced by 2- to 3-fold in cells over expressing CaMKII compared with the mock transfectants (Figure 5A). The overexpression of CaMKII increased the phosphorylation of Ser230 on endogenous HSF1 (Figure 5B), but it did not alone induce HSF1 DNA-binding activity (Figure 5C), showing that increased Ser230 phosphorylation is not sufficient to activate HSF1. To further investigate whether endogenous CaMKII modified the transactivating capacity of HSF1, the effect of the CaMKII inhibitor KN-62 was analyzed by the CAT reporter assay. Exposure to KN-62 alone did not affect the transactivating capacity of HSF1, whereas the heat-induced transcription was decreased in the presence of KN-62 (Figure 5D).
Fig. 5. CaMKII increases heat-induced transactivation and phosphorylation of Ser230 on HSF1. K562 cells were cotransfected with a plasmid coding for constitutively active CaMKII and an LSN reporter construct. The transfectants were exposed to heat shock followed by 6 h recovery at 37°C (R) or were left untreated (C). Mock, cells cotransfected with an empty vector and the reporter plasmid. (A) The CAT assay was quantified and the CAT activity in untreated mock transfectants was assigned a fold induction of 1. The data represent mean ± SEM from two independent experiments performed in duplicate. (B) Whole-cell extracts were prepared from the cells described above and analyzed by western blotting using the anti-pS230-hHSF1 (upper panel) and anti-hHSF1 (lower panel) antibody. The asterisk indicates an unknown protein band. (C) The HSF1 DNA-binding activity in the transfectants exposed to heat shock (HS) was examined by gel mobility shift assay. (D) HeLa cells were transfected with the LSN plasmid and exposed to heat shock followed by 6 h recovery at 37°C in the absence (R) or presence of KN-62 (KN+R, 10 µM KN-62 was added 15 min prior to heat shock) or KN-62 alone (KN). The data represent mean ± SEM from three independent experiments performed in duplicate. *p <0.05 when compared with the heat-shocked cells (R).
S230A mutation impairs the transcriptional activity of HSF1
To examine whether phosphorylation of Ser230 has a functional role in regulating the transcriptional activity of HSF1, we performed site-directed mutagenesis on chimeric HSF1. A chimeric construct coding for the DNA-binding domain of the yeast transcription factor GAL4 fused to human HSF1 lacking its DNA-binding domain was prepared, and Ser230 was substituted with Ala or Asp (S230A, S230D; Figure 6A). The wild-type GAL4-hHSF1 showed some basal CAT activity, which was enhanced by 2.5-fold upon heat shock (Figure 6A). Although heat-inducible transactivity of the S230A was detected, the basal and maximal transactivation were clearly less than in the wild type (Figure 6A). When phosphorylation of Ser230 was mimicked by an S230D mutation, the basal CAT activity was similar to the heat-induced activity of the wild type, and the activity was increased to 5-fold upon heat shock (Figure 6A). Both the wild-type GAL4-hHSF1 and the mutants showed similar DNA-binding activities (data not shown). In comparison with a phosphorylation site that usually undergoes phosphorylation–dephosphorylation cycles, introduction of a constitutive negatively charged Asp enhanced the function conferred by this site, whereas introduction of an uncharged Ala reduced the activity of the protein. Thus, our results suggest that phosphorylation is used to modulate the magnitude of the transcriptional response.
To further address the physiological role of Ser230 phosphorylation in the stress response, wild-type or S230A human HSF1 was introduced into mouse embryonic fibroblasts deficient in endogenous HSF1 (hsf1–/– MEFs; McMillan et al., 1998). Both the wild-type and S230A HSF1 were expressed at similar levels and were inducibly phosphorylated upon heat shock (Figure 6B). In accordance with the study by McMillan et al. (1998), no heat-inducible expression of Hsp70 was detected in the mock-transfected hsf1–/– MEFs, whereas a prominent induction was detected in the cells transfected with wild-type HSF1 (Figure 6B). In comparison, the hsf1–/– MEFs transfected with the S230A HSF1 also expressed Hsp70 after heat stress, but the amount of Hsp70 was ∼60% lower than in the wild-type HSF1 transfectants (Figure 6B). Similarly to HSF1 in human cells, HSF1 expressed in the hsf1–/– MEFs was phosphorylated on Ser230 and the anti-pS230-hHSF1 antibody did not recognize the S230A HSF1 (Figure 6C). Both the wild-type and S230A HSF1 exhibited inducible DNA-binding activity upon heat shock in the hsf1–/– MEFs (Figure 6D), showing that the diminished transcriptional activity of S230A was not due to impaired ability to bind DNA. No heat-induced HSF1 granules were detected in hsf1–/– MEFs expressing wild-type or mutant HSF1 (data not shown), confirming that endogenous HSF1 does not form stress-induced nuclear granules in mouse cells (Sarge et al., 1993). Phosphorylation on Ser230 was not required for formation of HSF1 granules, as demonstrated by introducing wild-type and S230A HSF1 to HeLa cells (data not shown). Taken together, these results reveal that phosphorylation on Ser230 is not needed for either the heat-induced DNA-binding activity or the granule formation, but this phosphorylation event is essential for the transcriptional activity of HSF1 and, therefore, for the potency of the stress response.
Discussion
To date, some evidence for a negative function of constitutive phosphorylation in regulating HSF1 transactivity has been provided, but only correlative data on the promoting effect of phosphorylation on transactivation are available. Here, we demonstrate that phosphorylation indeed contributes to the stress-induced transcriptional activity of HSF1. Specifically, we have identified Ser230 as the first phosphorylation site involved in determining the magnitude of HSF1 transactivating capacity. While the previous studies have focused on in vitro phosphorylated or overexpressed HSF1, we took a novel approach by using phosphoprotein analyses of endogenous HSF1, phosphopeptide antibodies and mutagenesis to characterize the role of specific in vivo phosphorylation sites of human HSF1.
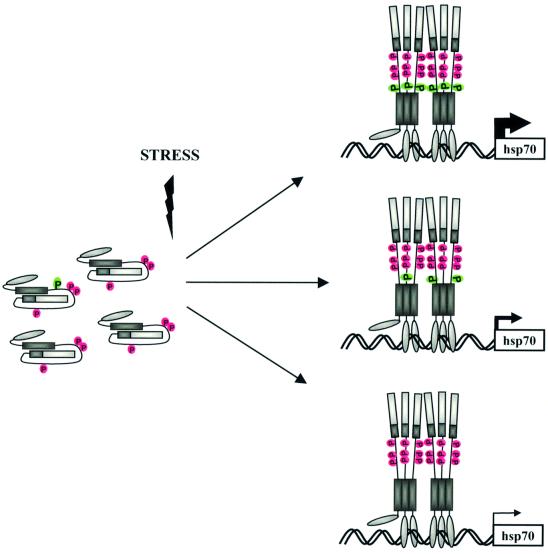
Based on our results and those of others identifying the repressive phosphorylation sites, we propose a model describing how phosphorylation controls the magnitude of HSF1-mediated transcription (Figure 7). Ser230 is phosphorylated under normal growth conditions, but its phosphorylation is clearly elevated upon heat shock, showing a stoichiometric shift towards phosphoSer230 in a given population of HSF1 molecules upon stress. Moreover, phosphorylation of Ser230 is required to generate adequate stress-inducible transactivation. In addition to phosphoSer230, the repressing function of phosphoserines 303, 307 and 363 (Chu et al., 1996, 1998; Knauf et al., 1996; Kline and Morimoto, 1997; Dai et al., 2000) affects the HSF1 activity, and our results suggest that the molar ratio between the activating and repressing phosphorylation sites determines the magnitude of transcriptional activity. In support of a control mechanism, which would be based on a stoichiometric relationship, introduction of a constitutive negative charge at residue 230 in all chimeric HSF1 molecules enhanced the transactivating capacity of HSF1 when compared with the net negative charge obtained by the normal phosphorylation–dephosphorylation cycles of Ser230. In further agreement with this hypothesis, our tryptic phosphopeptide analysis revealed heterogeneity among the HSF1 molecules phosphorylated in vivo. A modest stress-induced transcriptional activity occurs in the presence of the S230A HSF1, which is most likely mediated by other as yet uncharacterized inducible and/or constitutive phosphorylation sites on HSF1. Our results also show that the DNA-binding properties of HSF1 are not dependent on Ser230 phosphorylation, reinforcing the assumption that HSF1 DNA-binding and transcriptional activity are governed by distinct regulatory mechanisms. Our results suggest that HSF1 phosphorylation is a dynamic process, which allows for fine tuning of the transactivating potential of the factor in response to various stress conditions, rather than being a mechanism to only turn on and off the activity.
Fig. 7. Model of phosphorylation-mediated modulation of the magnitude of hHSF1 transcriptional activity. Based on the phosphorylation sites identified to date, HSF1 contains an activating site S230 (green) and three repressing sites, S303, S307 and S363 (red). Among these sites, the serines 230, 303 and 307 are localized in the regulatory domain of the factor. Upon stress, the Ser230 phosphorylation is enhanced, and the molar ratio between the activating and the repressing sites determines the magnitude of the transcriptional activity. Thickness of the arrow indicates degree of transcriptional activity. The transactivating potency is conferred by a heterogeneous population of phosphorylated HSF1. A modest transcriptional activity is acquired upon stress in the absence of Ser230 phosphorylation, which is likely to be mediated by the as yet uncharacterized inducible and constitutive phosphorylation sites. Ser230 phosphorylation is not needed for the DNA-binding activity of HSF1.
The transcriptional activity of HSF1 has been shown to be controlled by the regulatory domain composed of amino acids 221–310 (Green et al., 1995; Shi et al., 1995; Zuo et al., 1995; Newton et al., 1996). The study by Green et al. (1995) on GAL4-HSF1 chimeras indicated that the region containing amino acids 221–240 would be important for sensing heat stress. Ser230 is located in this part of the regulatory domain, a location that matches the suggested role for the novel phosphorylation site in regulating HSF1 activity. The hypothesis of Ser230 as a transactivating site is supported by our results showing that the heat-induced increase in Ser230 phosphorylation correlates with the transcriptional activity of HSF1, and that phosphoSer230 is present in the HSF1 DNA-binding complex as well as in the stress-induced nuclear granules. Furthermore, overexpression of CaMKII increases phosphorylation on Ser230 of endogenous HSF1 and results in an enhanced heat-induced transactivation of HSF1. Finally, compelling evidence for the involvement of Ser230 phosphorylation in regulating the transcriptional activity of HSF1 was obtained with the S230A mutant exhibiting impaired transactivating capacity in heat-shocked hsf1–/– MEFs. Hence, our study provides new evidence for the importance of the regulatory domain in controlling the transcriptional activity of HSF1.
Several protein kinases have been suggested to phosphorylate and, thereby, repress the transcriptional activity of HSF1 (Chu et al., 1996, 1998; Knauf et al., 1996; Kim et al., 1997; He et al., 1998; Dai et al., 2000). Currently, it is not known how ERK, PKC, GSK-3 and JNK interact to affect HSF1 regulation. We propose that CaMKII signaling is involved in the positive regulation of HSF1-mediated transactivation, which is supported by three major lines of evidence. First, HSF1 is a good substrate for CaMKII in vitro and in vivo. Secondly, overexpression of active CaMKII enhances both the level of endogenous Ser230 phosphorylation and transactivation of HSF1. Thirdly, inhibition of CaMKII decreases the transactivating capacity of HSF1. Since CaMKII is abundantly expressed and regulated by a common calcium–calmodulin mechanism (Schulman, 1993), it is a good candidate kinase that could regulate the activity of HSF1 in many cell types. Our data show that CaMKII-directed phosphorylation of Ser230 enhances the transcriptional capacity of DNA-bound HSF1, yet it is by itself not sufficient to induce HSF1 DNA binding. Further studies are, however, required to provide conclusive evidence on the mechanism of HSF1 regulation by CaMKII. Furthermore, we cannot exclude the possibility that other protein kinases might also phosphorylate Ser230 in vivo. In fact, the increasing number of reported protein kinases capable of phosphorylating HSF1 indicates that several kinases might target the same amino acid residue in vivo.
Why is HSF1 regulated in such a complex manner, involving multiple phosphorylation sites that potentially allow interaction with a multitude of signaling pathways? The heat shock response is a fast responding adaptive process, in which input signals from the ever-changing environment must be integrated appropriately, to generate an HSF1 activation adequately adjusted to any given environmental stress. As HSF1 is activated in response to a myriad of stress conditions, distinct protein kinases/phosphatases are likely to be required to orchestrate the responses to these stimuli. The presence of both inhibitory and stimulatory phosphorylation sites that are constitutively and inducibly regulated by protein kinases and phosphatases provides a precise and elegant mechanism of tuning proteins whose regulation requires both high stringency and accurate kinetics. Regulation of HSF1 by multisite phosphorylation adds this mediator to the growing group of transcription factors, including the stress-activated tumor repressor p53, whose activity is strictly controlled by phosphorylation on multiple sites (Appella and Anderson, 2000). Another example of the complex multisite phosphorylation-mediated regulation of a transcription factor is provided by Okamura et al. (2000), showing that NFAT1 is phosphorylated on at least 21 serines. The mechanism of this multisite phosphorylation-based regulation of NFAT1 is not clear, but apparently an interplay between multisite dephosphorylation, inducible phosphorylation and constitutive phosphorylation mediates activation of the factor. In the case of HSF1, the precise regulation is most likely a necessity to avoid HSF1-mediated disorders in whole organisms. Although HSF1 has an essential survival promoting function, an excessive and prolonged HSF1 activation is not likely to be beneficial for the cell. A recent report by Nakai et al. (2000) showed that male mice overexpressing an active HSF1 suffered from infertility due to apoptosis of developing germ cells.
In summary, our results reveal that HSF1 phosphorylation is a dynamic process where the stoichiometric ratios between the phosphorylation sites determine the magnitude of the transcriptional activity of the factor. Our study establishes a direct role of phosphorylation in acquisition of the transcriptional activity of HSF1 under stressful conditions. Future studies are directed to characterize all in vivo phosphorylation sites and the kinases/phosphatases involved to decipher the phosphorylation-mediated regulation of HSF1.
Materials and methods
Cell culture, treatments and preparation of cell extracts
Human K562 erythroleukemia and HeLa cervical carcinoma cells were maintained in RPMI 1640 and Dulbecco’s modified Eagle’s medium (DMEM), respectively, supplemented with 10% fetal calf serum (FCS), 2 mM l-glutamine, 100 U/ml penicillin and 100 µg/ml streptomycin. MEFs from wild-type and HSF1-deficient mice were cultured in supplemented DMEM (McMillan et al., 1998). Heat shock treatments were performed for 1 h at 42°C if not otherwise indicated. Whole-cell extracts were prepared as described previously (Mosser et al., 1988). KN-62 was purchased from Sigma.
Plasmid construction and mutagenesis
The nucleotide sequence coding for amino acids 116–529 of hHSF1 was amplified using PCR with primers that incorporated the EcoRI and XbaI restriction sites, and subcloned into pSG424 (Sadowski and Ptashne, 1989). The nucleotide sequence coding for full-length hHSF1 was amplified using PCR with primers that incorporated the EcoRI and HindIII restriction sites, and the PCR product was subcloned in-frame with the C-terminal Myc-His tag in pcDNA3.1A (Invitrogen). The point mutations were made using the QuikChange™ site-directed mutagenesis kit (Stratagene). The constructs were confirmed by sequencing.
Transfections and reporter systems
The activities of the GAL4-hHSF1 fusion proteins were measured by cotransfection with a reporter plasmid G5BCAT (Shi et al., 1995). RSVβ-gal was used as an internal control for transfection efficiency. pSG424 and pSGVP, which is a GAL4-VP16 chimera (Shi et al., 1995), were used as a negative and positive control, respectively, of transactivation capacity. For transfection, K562 cells (5 × 106) or subconfluent HeLa cells were electroporated (200 V, 975 µF; Bio-Rad Gene Pulser) with 25 µg of DNA (e.g. 10 µg of GAL4-hHSF1, 10 µg of G5BCAT and 5 µg of RSVβ-gal) and incubated for 40 h prior to use. For transfection with CaMKII, 25 µg of the plasmid coding for the constitutively active truncated form of CaMKII-α (amino acids 1–290) were cotransfected with 15 µg of LSN and 10 µg of RSVβ-gal. The LSN plasmid contains the proximal HSE of the human hsp70 promoter fused to the CAT gene (Williams et al., 1989). The CAT assay was performed with 50–100 µg of total cell extract and analyzed by chromatography as previously described (Holmberg et al., 1998). The β-galactosidase activity was measured spectrophotometrically at 420 nm to normalize transfection efficiency before the CAT assay. Student’s t-test was used for statistical analysis. The wild-type and hsf1–/– MEFs were electroporated using 280 V (975 µF) and 30 µg of plasmid and incubated 24 h before use.
In vivo phosphoprotein analyses
See Supplementary data, available at The EMBO Journal Online. For 32P metabolic in vivo labeling, K562 cells were incubated in phosphate-free MEME supplemented with 10% dialyzed FCS and 0.3 mCi/ml of [32P]orthophosphate (Amersham Pharmacia Biotech) for 3 h at 37°C before the treatments. HSF1 was immunoprecipitated, run on 8% SDS–PAGE and visualized by autoradiography. Quantification of phosphorylated HSF1 was performed using a FujiFilm Bas-1800 II PhosphorImager. The values for the fold induction of HSF1 phosphorylation were calculated relative to the control, which was assigned a value of 1. The phosphoamino acid analysis of immunoprecipitated HSF1 was performed as described in Holmberg et al. (2000). For phosphopeptide mapping, the phosphorylated HSF1 band was cut out from the dried gel and digested with TPCK trypsin (Sigma). Phosphopeptide mapping was performed in two dimensions on cellulose TLC plates (Merck), as described previously (van der Geer et al., 1993). The electrophoresis was performed at pH 1.9 using the HTLE-7000 apparatus (CBS Scientific Co.) followed by chromatography. For separation of phosphopeptides by HPLC (Hewlett Packard, Series 1050), the trypsin-digested 32P-labeled HSF1 was loaded on a C-18 reversed-phase column (Aquapore RP-300; Applied Biosystems) and eluted using a linear gradient of up to 80% acetonitrile. Fractions were collected during the gradient run, aliquots were spotted on Whatman paper and the radioactivity was detected with a PhosphorImager. Manual Edman degradation was performed as described by Eriksson et al. (1998).
CaMKII in vitro kinase assay
Forty-two nanograms of recombinant CaMKII-α, 25 µg of rhHSF1 (Holmberg et al., 2000) and 60 U of calmodulin (Sigma) were incubated in 100 µl of CaMK reaction buffer {50 mM PIPES pH 7.0, 20 mM MgCl2, 0.5 mM CaCl2, 0.4 mM EGTA, 40 µM ATP (Sigma), 5 µCi [γ-32P]ATP (ICN Pharmaceuticals Inc.)} for 15 min at 30°C. The reaction was terminated by boiling in Laemmli buffer. The sample was resolved on 8% SDS–PAGE.
Peptide sequencing
The HPLC-separated phosphopeptide of rhHSF1 phosphorylated in vitro by CaMKII was subjected to sequence analysis and mass spectral fingerprinting. N-terminal amino acid sequence analysis was performed with an Applied Biosystems model 477A protein sequencer equipped with an on-line Applied Biosystems model 120A phenylthiohydantoin (PTH) amino acid analyzer. For mass spectrometric analysis, the in vitro phosphorylated rhHSF1 was alkylated with iodoacetamide (100 mM; Sigma) before it was resolved on SDS–PAGE. Phosphopeptide fractions were analyzed by MALDI-TOF mass spectrometry on a PerSeptive Biosystems Elite STR mass spectrometer using α-cyano-cinnamic acid (10 mg/ml in 50% acetonitrile/0.1% trifluoroacetic acid) as the matrix. Spectra were acquired in the reflector mode and the mass spectra were internally mass calibrated.
Preparation of phosphopeptide antibody and immunoblotting
To prepare an antibody that specifically recognizes hHSF1 phosphorylated on Ser230, the phosphopeptide KYSRQFpSLEHV was conjugated to haemocyanin (Sigma) and injected into a goat. The antiserum against pS230-hHSF1 was positively and negatively affinity purified using KYSRQFpSLEHV and KYSRQFSLEHV, respectively, immobilized on a HiTrap-NHS column (Pharmacia Biotech). Whole-cell extracts were subjected to 8% SDS–PAGE, transferred to a nitrocellulose membrane (Schleicher & Schuell), and equal loading was checked using Ponceau S staining (Sigma). See Supplementary data for immunoblotting with the anti-pS230-hHSF1 antibody. For competitive inhibition of the immunoreaction, the antibody was pre-incubated for 30 min with a 500 molar excess of free phosphopeptide. Rabbit pAb to total hHSF1 (Holmberg et al., 2000), rabbit pAb to mHSF1 (Sarge et al., 1993), mouse mAb to Hsp70 (4g4; Affinity Bioreagents, Inc.) and rat mAb to Hsc70 (SPA-815; StressGen) were used as primary antibodies. Peroxidase-conjugated protein G (Pierce), horseradish peroxidase-conjugated anti-rabbit (Promega), anti-mouse and anti-rat antibodies (Amersham Life Science) were used for the blotting. The detection was performed using ECL (Amersham Life Science). The proteins were quantitated with an MCID Image Analyzer (Imaging Research MCID M4).
Indirect immunofluorescence
HeLa cells growing on coverslips were simultaneously fixed and permeabilized in 0.5% Triton X-100 in phosphate-buffered saline containing 3% paraformaldehyde. The bound primary antibodies, rabbit anti-hHSF1 and goat anti-pS230-hHSF1 antibody were detected using FITC-conjugated donkey anti-goat Ab and Cy3-conjugated donkey anti-rabbit Ab (Jackson ImmunoResearch Laboratories Inc.). The DNA was stained with DAPI (Sigma). For visualization, a Leica DMR fluorescence microscope equipped with a digital Hamamatsu ORCA CCD camera was used.
Gel mobility shift assays and antibody perturbation assay
Whole-cell extracts (15–30 µg) were incubated with a 32P-labeled oligonucleotide representing the proximal HSE of the human hsp70 promoter (Mosser et al., 1988) or with a 32P-labeled GAL4 probe (Shi et al., 1995). The protein–DNA complexes were analyzed on a native 4% polyacrylamide gel as described previously (Mosser et al., 1988). For the perturbation assay, the antibody or pre-immune serum was added in a 1:10 dilution to the whole-cell extracts and incubated at room temperature for 15 min prior to the gel mobility shift assay.
Supplementary data
Supplementary data to this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Ivor I.Benjamin (Southwestern Medical Center) for the wild-type and hsf1–/– MEFs, and Howard Schulman (Stanford University School of Medicine) for the CaMKII construct and recombinant CaMKII-α. We are grateful to Helena Saarento and Kaija-Liisa Laine for excellent technical assistance. This work was supported by the Academy of Finland, the Finnish Cancer Organizations, Sigrid Jusélius, Borg and Wihuri Foundations (L.S. and J.E.E.) and the Erna and Victor Hasselblad Foundation (J.E.E.). C.I.H. and V.H. were supported by the Turku Graduate School of Biomedical Sciences, and C.I.H. was supported by the Victoria Foundation, the Research Institute of the Åbo Akademi Foundation and the Finnish Cultural Foundation.
References
- Appella E. and Anderson,C.W. (2000) Signaling to p53: breaking the posttranslational modification code. Pathol. Biol., 48, 227–245. [PubMed] [Google Scholar]
- Baler R., Dahl,G. and Voellmy,R. (1993) Activation of human heat shock genes is accompanied by oligomerization, modification, and rapid translocation of heat shock transcription factor HSF1. Mol. Cell. Biol., 13, 2486–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijur G.N. and Jope,R.S. (2000) Opposing actions of phosphatidyl inositol 3-kinase and glycogen synthase kinase-3β in the regulation of HSF-1 activity. J. Neurochem., 75, 2401–2408. [DOI] [PubMed] [Google Scholar]
- Chu B., Soncin,F., Price,B.D., Stevenson,M.A. and Calderwood,S.K. (1996) Sequential phosphorylation by mitogen-activated protein kinase and glycogen synthase kinase 3 represses transcriptional activation by heat shock factor 1. J. Biol. Chem., 271, 30847–30857. [DOI] [PubMed] [Google Scholar]
- Chu B., Zhong,R., Soncin,F., Stevenson,M.A. and Calderwood,S.K. (1998) Transcriptional activity of heat shock factor 1 at 37°C is repressed through phosphorylation on two distinct serine residues by glycogen synthase kinase 3α and protein kinases Cα and Cζ. J. Biol. Chem., 273, 18640–18646. [DOI] [PubMed] [Google Scholar]
- Cohen P. (2000) The regulation of protein function by multisite phosphorylation—a 25 year update. Trends Biochem. Sci., 25, 596–601. [DOI] [PubMed] [Google Scholar]
- Cotto J.J., Kline,M. and Morimoto,R.I. (1996) Activation of heat shock factor 1 DNA binding precedes stress-induced serine phosphorylation. J. Biol. Chem., 271, 3355–3358. [DOI] [PubMed] [Google Scholar]
- Cotto J.J., Fox,S.G. and Morimoto,R.I. (1997) HSF1 granules: a novel stress-induced nuclear compartment of human cells. J. Cell Sci., 110, 2925–2934. [DOI] [PubMed] [Google Scholar]
- Dai R., Frejtag,W., He,B., Zhang,Y. and Mivechi,N.F. (2000) JNK targeting and phosphorylation of heat shock factor-1 suppress its transcriptional activity. J. Biol. Chem., 275, 18210–18218. [DOI] [PubMed] [Google Scholar]
- Eriksson J.E., Toivola,D.M., Sahlgren,C., Mikhailov,A. and Härmälä-Braskén,A.-S. (1998) Strategies to assess phosphoprotein phosphatase and protein kinase-mediated regulation of the cytoskeleton. Methods Enzymol., 298, 542–569. [DOI] [PubMed] [Google Scholar]
- Fritsch M. and Wu,C. (1999) Phosphorylation of Drosophila heat shock transcription factor. Cell Stress Chap., 4, 102–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M., Schuetz,T.J., Sullivan,E.K. and Kingston,R.E. (1995) A heat shock-responsive domain of human HSF1 that regulates transcription activation domain function. Mol. Cell. Biol., 15, 3354–3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B., Meng,Y.-H. and Mivechi,N.F. (1998) Glycogen synthase kinase 3β and extracellular signal-regulated kinase inactivate heat shock transcription factor 1 by facilitating the disappearance of transcriptionally active granules after heat shock. Mol. Cell. Biol., 18, 6624–6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Høj A. and Jakobsen,B.K. (1994) A short element required for turning off heat shock transcription factor: evidence that phosphorylation enhances deactivation. EMBO J., 13, 2617–2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg C.I., Roos,P.M.K., Lord,J.M., Eriksson,J.E. and Sistonen,L. (1998) Conventional and novel PKC isoenzymes modify the heat induced stress response but are not activated by heat shock. J. Cell Sci., 111, 3357–3365. [DOI] [PubMed] [Google Scholar]
- Holmberg C.I., Illman,S.A., Kallio,M., Mikhailov,A. and Sistonen,L. (2000) Formation of nuclear HSF1 granules varies depending on stress stimuli. Cell Stress Chap., 5, 219–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T. (2000) Signaling—2000 and beyond. Cell, 100, 113–127. [DOI] [PubMed] [Google Scholar]
- Hunter T. and Karin,M. (1992) The regulation of transcription by phosphorylation. Cell, 70, 375–387. [DOI] [PubMed] [Google Scholar]
- Jolly C., Morimoto,R.I., Robert-Nicoud,M. and Vourc’h,C. (1997) HSF1 transcription factor concentrates in nuclear foci during heat shock: relationship with transcription sites. J. Cell Sci., 110, 2935–2941. [DOI] [PubMed] [Google Scholar]
- Jolly C., Usson,Y. and Morimoto,R.I. (1999) Rapid and reversible relocalization of heat shock factor 1 within seconds to nuclear stress granules. Proc. Natl Acad. Sci. USA, 96, 6769–6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurivich D.A., Sistonen,L., Kroes,R.A. and Morimoto,R.I. (1992) Effect of sodium salicylate on the human heat shock response. Science, 255, 1243–1245. [DOI] [PubMed] [Google Scholar]
- Jurivich D.A., Pachetti,C., Qiu,L. and Welk,J.F. (1995) Salicylate triggers heat shock factor differently than heat. J. Biol. Chem., 270, 24489–24495. [DOI] [PubMed] [Google Scholar]
- Kim J., Nueda,A., Meng,Y.-H., Dynan,W.S. and Mivechi,N.F. (1997) Analysis of the phosphorylation of human heat shock transcription factor by MAP kinase family members. J. Cell. Biochem., 67, 43–54. [DOI] [PubMed] [Google Scholar]
- Kline M.P. and Morimoto,R.I. (1997) Repression of the heat shock factor 1 transcriptional activation domain is modulated by constitutive phosphorylation. Mol. Cell. Biol., 17, 2107–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauf U., Newton,E.M., Kyriakis,J. and Kingston,R.E. (1996) Repression of human heat shock factor 1 activity at control temperature by phosphorylation. Genes Dev., 10, 2782–2793. [DOI] [PubMed] [Google Scholar]
- Larson J.S., Schuetz,T.J. and Kingston,R.E. (1988) Activation in vitro of sequence-specific DNA binding by a human regulatory factor. Nature, 335, 372–375. [DOI] [PubMed] [Google Scholar]
- Liu X.-D. and Thiele,D.J. (1996) Oxidative stress induces heat shock factor phosphorylation and HSF-dependent activation of yeast metallothionein gene transcription. Genes Dev., 10, 592–603. [DOI] [PubMed] [Google Scholar]
- McMillan D.R., Xiao,X., Shao,L., Graves,K. and Benjamin,I.J. (1998) Targeted disruption of heat shock transcription factor 1 abolishes thermotolerance and protection against heat-inducible apoptosis. J. Biol. Chem., 273, 7523–7528. [DOI] [PubMed] [Google Scholar]
- Morimoto R.I. (1998) Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev., 12, 3788–3796. [DOI] [PubMed] [Google Scholar]
- Mosser D.D., Theodorakis,N.G. and Morimoto,R.I. (1988) Coordinate changes in heat shock element-binding activity and HSP70 gene transcription rates in human cells. Mol. Cell. Biol., 8, 4736–4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai A., Suzuki,M. and Tanabe,M. (2000) Arrest of spermatogenesis in mice expressing an active heat shock transcription factor 1. EMBO J., 19, 1545–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton E.M., Knauf,U., Green,M. and Kingston,R.E. (1996) The regulatory domain of human heat shock factor is sufficient to sense stress. Mol. Cell. Biol., 16, 839–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura H. et al. (2000) Concerted dephosphorylation of the transcription factor NFAT1 induces a conformational switch that regulates transcriptional activity. Mol. Cell, 6, 539–550. [DOI] [PubMed] [Google Scholar]
- Sadowski I. and Ptashne,M. (1989) A vector for expressing GAL4 (1–147) fusions in mammalian cells. Nucleic Acids Res., 17, 7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarge K.D., Murphy,S.P. and Morimoto,R.I. (1993) Activation of heat shock gene transcription by HSF1 involves oligomerization, acquisition of DNA binding activity, and nuclear localization and can occur in the absence of stress. Mol. Cell. Biol., 13, 1392–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman H. (1993) The multifunctional Ca2+/calmodulin-dependent protein kinases. Curr. Opin. Cell Biol., 5, 247–253. [DOI] [PubMed] [Google Scholar]
- Shi Y., Kroeger,P.E. and Morimoto,R.I. (1995) The carboxyl-terminal transactivation domain of heat shock factor 1 is negatively regulated and stress responsive. Mol. Cell. Biol., 15, 4309–4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger P.K. (1990) Yeast heat shock factor contains separable transient and sustained response transcriptional activators. Cell, 62, 793–805. [DOI] [PubMed] [Google Scholar]
- Sorger P.K. and Pelham,H.R.B. (1988) Yeast heat shock factor is an essential DNA-binding protein that exhibits temperature-dependent phosphorylation. Cell, 54, 855–864. [DOI] [PubMed] [Google Scholar]
- Sorger P.K., Lewis,J.M. and Pelham,H.R.B. (1987) Heat shock factor is regulated differently in yeast and HeLa cells. Nature, 329, 81–84. [DOI] [PubMed] [Google Scholar]
- van der Geer P., Luo,K., Sefton,B.M. and Hunter,T. (1993) Phospho peptide mapping and phosphoamino acid analysis on cellulose thin-layer plates. In Hardie,D.G. (ed.), Protein Phosphorylation, A Practical Approach. IRL Press at Oxford University Press, Oxford, UK, pp. 31–59.
- Williams G.T., McClanahan,T.K. and Morimoto,R.I. (1989) E1a transactivation of the human HSP70 promoter is mediated through the basal transcriptional complex. Mol. Cell. Biol., 9, 2574–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. (1995) The heat shock factors: structure and regulation. Annu. Rev. Cell Dev. Biol., 11, 441–469. [DOI] [PubMed] [Google Scholar]
- Xavier I.J., Mercier,P.A., McLoughlin,C.M., Ali,A., Woodgett,J.R. and Ovsenek,N. (2000) Glycogen synthase kinase 3β negatively regulates both DNA-binding and transcriptional activities of heat shock factor 1. J. Biol. Chem., 275, 29147–29152. [DOI] [PubMed] [Google Scholar]
- Zuo J., Rungger,D. and Voellmy,R. (1995) Multiple layers of regulation of human heat shock transcription factor 1. Mol. Cell. Biol., 15, 4319–4330. [DOI] [PMC free article] [PubMed] [Google Scholar]