Abstract
Exon enhancers are accessory pre-mRNA splicing signals that stimulate exon splicing. One class of proteins, the serine-arginine-rich (SR) proteins, have been demonstrated to bind enhancers and activate splicing. Here we report that A/C-rich exon enhancers (ACE elements) are recognized by the human YB-1 protein, a non-SR protein. Sequence-specific binding of YB-1 was observed both to an ACE derived from an in vivo iterative selection protocol and to ACE elements in an alternative exon (v4) from the human CD44 gene. The ACE element that was the predominant YB-1 binding site in CD44 exon v4 was required for maximal in vivo splicing and in vitro spliceosome assembly. Expression of wild-type YB-1 increased inclusion of exon v4, whereas a truncated form of YB-1 did not. Stimulation of exon v4 inclusion by wild-type YB-1 required the ACE necessary for YB-1 binding in vitro, suggesting that YB-1 stimulated exon inclusion in vivo by binding to an exonic ACE element. These observations identify a protein in addition to SR proteins that participates in the recognition of exon enhancers.
Keywords: alternative splicing/CD44/exonic splicing enhancer/pre-mRNA/YB-1
Introduction
Pre-mRNA splicing involves the cumulative recognition of multiple short elements by a large number of RNA and protein factors. It has become clear that sequences in addition to splice sites play an important role in discerning exons from introns during the earliest steps of precursor RNA recognition (reviewed in Black, 1995; Reed, 1996; Cooper and Mattox, 1997). Accessory elements located within the recognized exon or flanking introns can either stimulate or depress recognition. Despite the ramifications for coding capacity, a large number of exons, including both alternatively spliced exons and constitutively recognized exons, contain short enhancer sequences that activate both splicing and spliceosome formation.
The splicing factors known to recognize exon enhancers are the arginine-serine-rich (SR) proteins (Tacke and Manley, 1999). This large family of proteins is characterized by the presence of one or more RNA recognition motifs (RRMs) and a trans-activation domain characterized by SR dipeptides. When bound to exon enhancers, SR proteins are thought to interact with constitutive splicing factors bound to 3′ and 5′ splice sites, including U2AF and U1 snRNPs, thereby activating the earliest steps in exon recognition. The major members of this family of proteins bind with high affinity to the major class of characterized exon enhancer—the purine-rich enhancers characterized by internal repeats of sequence GAR.
Sequences other than purine-rich repeats have also been characterized as exon enhancers (Dominski and Kole, 1994; Staknis and Reed, 1994; van Oers et al., 1994; Wang et al., 1995; Coulter et al., 1997; Cooper, 1999; Gersappe and Pintel, 1999; Schaal and Maniatis, 1999a,b). Some of these also bind SR proteins. Interestingly, no other class of factor has been implicated as binding exon enhancer sequences other than SR proteins.
In this report we focus on a newly discovered class of exon enhancers—the A/C-rich enhancers, or ACE elements—that have been identified both in natural genes and in experiments designed to select exon enhancer elements (van Oers et al., 1994; Wang et al., 1995; Coulter et al., 1997; Schaal and Maniatis, 1999b). We show that the major nuclear protein binding the ACE element is the single-stranded nucleic acid binding protein YB-1. The element did not bind classic SR proteins nor was the binding of YB-1 stimulated by SR proteins, suggesting that YB-1-mediated exon recognition could be an alternative mechanism of exon enhancer activity.
ACE sequences were observed in a natural alternative exon from the human CD44 gene. Pre-mRNA from the human CD44 gene undergoes extensive alternative splicing within a block of at least 10 exons (Günthert, 1993; Mackay et al., 1994). Increased inclusion of some of these exons has been correlated to cancer and metastasis (Günthert et al., 1991; Fox et al., 1994). The alternative exon v4 contains three ACE elements that were required for efficient splicing in vivo and spliceosome assembly in vitro. YB-1 bound the exon v4 and stimulated exon inclusion in vivo in an ACE-dependent fashion. Our results demonstrate that YB-1 is required for ACE-dependent exon v4 inclusion and provide the first identification of a sequence-specific RNA binding protein affecting CD44 alternative splicing.
Results
A 50 kDa nuclear protein binds to ACE elements
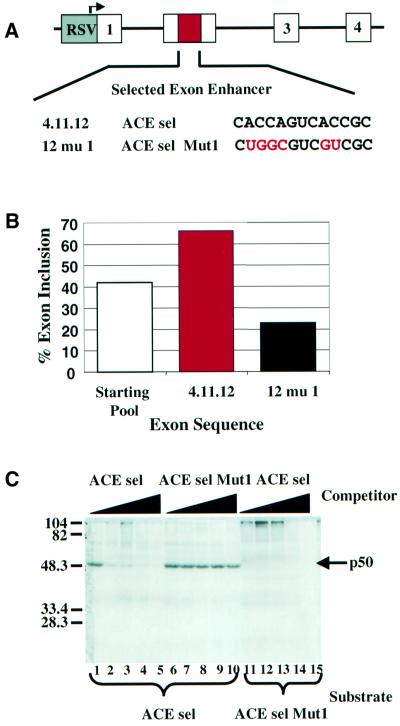
We have previously identified a class of ACE sequences via an in vivo iterative selection strategy (Coulter et al., 1997). In this strategy, a 13 nucleotide cassette of random nucleotides (6 × 107 potential sequences) was inserted into a weak alternative exon that was predominantly skipped in the absence of an enhancer (Figure 1A and B). Three rounds of transient transfection and RT–PCR were used to select for sequences that enhanced exon inclusion in vivo. Figure 1A shows the sequence of one of the A/C-rich sequences selected in this study. When placed into a natural weak exon from the cardiac troponin T gene, this sequence increased in vivo exon inclusion to 66% compared with 42% inclusion in the population of sequences used for selection (Figure 1B), demonstrating that this activity was independent of the mini-gene used for selection. Mutation of the A/C-rich portion of the sequence as diagrammed in Figure 1A eliminated enhancing activity (Coulter et al., 1997). In this paper we used wild-type and mutant ACEs (herein termed ACE sel and ACE sel Mut1) containing a duplication of the sequence 4.11.12 and the mutant 12 mu1 shown in Figure 1.
Fig. 1. A/C-rich exon enhancers bind a 50 kDa nuclear protein. (A) Sequence of one of the 13 nucleotide ACE exon enhancers identified by iterative in vivo selection from 13 random nucleotides using the diagrammed mini-gene (Coulter et al., 1997). The 4.11.12 isolate, referred to here as ACE sel, is shown along with a mutant version. (B) In vivo exon inclusion activity of the sequences shown in (A). Activity was tested in a heterologous exon context (Coulter et al., 1997). (C) In vitro UV cross-linking of a 51 nucleotide RNA including two copies of the ACE sel sequence from (A) (see Materials and methods). Radiolabeled RNA substrates as indicated beneath the gel were incubated for 10 min under standard in vitro splicing conditions using HeLa nuclear extract in the presence of increasing concentrations (0, 1, 5, 10 and 25 pmol) of competitor RNAs containing the sequences shown above the gel. Following cross-linking and RNase digestion, cross-linked proteins were resolved by 11% SDS–PAGE. A prominent 50 kDa band that was cross-linked to wild-type but not mutant RNA is indicated by an arrow.
To initially identify proteins in HeLa nuclear extracts that bind ACE elements, we performed UV cross-linking using wild-type and mutant ACE sel RNAs (Figure 1C). We identified a prominent 50 kDa protein (denoted p50) that strongly cross-linked to wild-type ACE sel RNA but not the ACE sel Mut1 mutant RNA (Figure 1C, compare lanes 1 and 11). Furthermore, binding of p50 to ACE sel was not efficiently competed by unlabeled ACE sel Mut1 RNA despite being strongly competed by wild-type RNA (Figure 1C, compare lanes 1–5 with 6–10). This result indicated that p50 was a candidate protein for an effector of ACE activity.
p50 is distinct from the SR proteins that recognize purine-rich enhancers
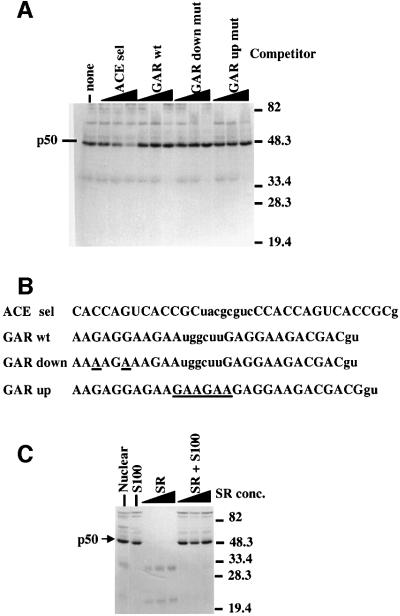
Our original iterative selection experiment isolated two classes of exon enhancers: ACE sequences and classic purine-rich enhancers (Coulter et al., 1997). The latter are known to be recognized by the SR proteins (Tacke and Manley, 1999). To assess whether SR proteins are involved in ACE function, we first asked whether RNAs containing known purine-rich enhancers were effective competitors for the UV cross-linking of p50 to ACE sel RNA (Figure 2A). Competitors containing the well characterized purine-rich enhancer from the cardiac troponin T alternative exon 5 (Figure 2B; Ramchatesingh et al., 1995) were ineffective competitors for p50 binding to ACE sel RNA (Figure 2A). Even a powerful mutant that causes higher enhancement than wild type in an exon inclusion assay (Ramchatesingh et al., 1995) was an ineffective competitor. This experiment suggests that unlike classic SR proteins, p50 does not bind strongly to purine-rich enhancers.
Fig. 2. p50 is distinct from the SR proteins that recognize purine-rich enhancers. (A) RNAs containing purine-rich enhancers do not compete UV cross-linking of p50 to ACE elements. The ACE sel RNA described in Figure 1 was subjected to UV cross-linking in the presence of competitor RNAs containing the sequences indicated above the gel. The positions of p50 and molecular weight markers are indicated. (B) Sequences of the competitor RNAs which are derived from exon 5 of the chicken cardiac troponin T gene [Ramchatesingh et al., 1995, where they are referred to as D2WT (GAR wt), D2A2 (GAR down mutant), D2EY1 (GAR up mutant)]. Not shown are the first 17 nucleotides (ACE sel; see Materials and methods) or 15 nucleotides (GAR wt, GAR down mutant and GAR up mutant) derived from the transcription vector. (C) Preparations of HeLa SR proteins do not contain p50, and SR proteins do not stimulate the binding of p50. UV cross-linking was performed using in vitro splicing conditions (Materials and methods) in either HeLa nuclear (lane 1) or S100 (lanes 2, 6, 7 and 8) extract or with increasing amounts (50, 100 or 200 ng) of SR proteins (Zahler, 1999) in the absence (lanes 3, 4 and 5) or presence (lanes 6, 7 and 8) of S100 extract. The positions of p50 and marker proteins are indicated.
Next, we asked whether semi-purified SR proteins would effectively UV cross-link to the ACE sel RNA or if they would enhance UV cross-linking of p50 to the ACE sel RNA (Figure 2C). For the latter we first determined that p50 was present in both HeLa nuclear extract and in cytoplasmic S100 extract that is deficient in nuclear SR proteins (Figure 2B, lanes 1 and 2). As shown in Figure 2C, the purified SR preparation showed only minimal UV cross-linking to the ACE sel sequence (Figure 2C, lanes 3–5) and had no effect on the ability of p50 in the S100 extract to cross-link to the ACE sel RNA (Figure 2C, lanes 6–8).
Identification of p50 as the human Y-box binding protein YB-1
The p50 protein was purified from HeLa nuclear extract using a UV cross-linking assay with the ACE sel RNA. As an initial fractionation attempt we took advantage of the affinity of p50 for poly(U)–agarose in 1 M NaCl. Proteins were eluted by repeated washing with buffer containing 2 M NaCl, and subsequently with guanidine hydrochloride. Fractions 2–4 of the 2 M NaCl wash contained a prominent 50 kDa protein (Figure 3A), and p50 UV cross-linking to the ACE sel RNA was seen in the same fractions (Figure 3B).
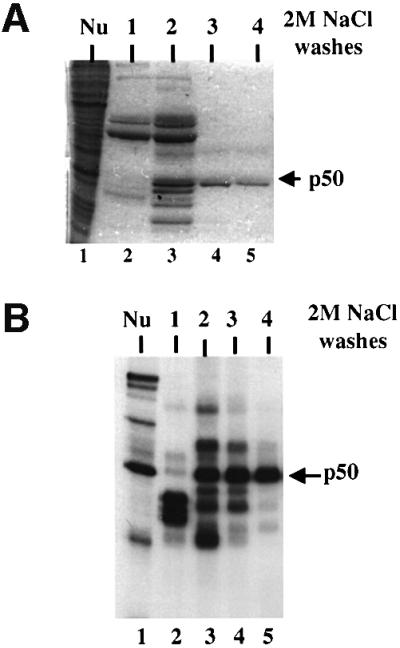
Fig. 3. Purification of p50. (A) HeLa nuclear extract was fractionated on poly(U)–agarose (Materials and methods). The starting material (lane 1) and four successive elution washes using 2 M NaCl are shown (lanes 2–5) in Coomassie Blue-stained 10% SDS–polyacrylamide gel. The position of a 50 kDa protein in fractions 2, 3 and 4 is denoted with an arrow. (B) UV cross-linking of the column fractions shown in (A). Column fractions were dialyzed against Roeder D (Dignam et al., 1983) and used in a standard UV cross-linking assay with the ACE sel RNA. The cross-linked p50 in nuclear extract (lane 1) and fractions 2–4 is indicated.
Preparative amounts of the 50 kDa protein were gel isolated. The extracted protein UV cross-linked to the ACE sel RNA in a sequence-specific manner (data not shown). The sequences of the tryptic peptides (Materials and methods) identified p50 as human YB-1 [also known as DNA binding protein B (DbpB); reviewed in Wolffe et al., 1992].
To confirm the identification of p50 as YB-1, we prepared a rabbit polyclonal antibody to a C-terminal peptide of human YB-1, AENSSAPEAEQGGAE (Anaspec, CA). The antibody immunoprecipitated a 50 kDa protein that strongly cross-linked to the ACE sel RNA, confirming that p50 was YB-1 (Figure 4A). In addition, purified N-terminal His-tagged recombinant YB-1 UV cross-linked to the ACE sel RNA (Figure 4B). Competition with ACE sel and ACE sel Mut1 RNAs demonstrated that recombinant YB-1 bound with the same sequence specificity as endogenous p50 (Figure 4B, compare lanes 1–3 with 4–6).
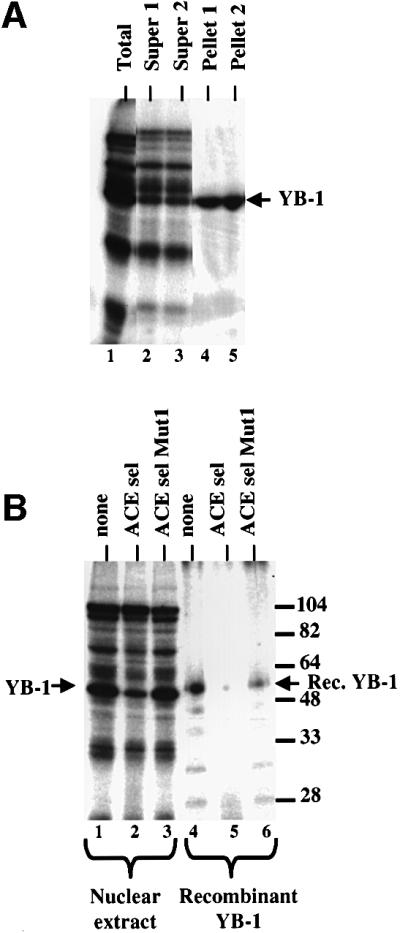
Fig. 4. Antibodies to YB-1 peptides immunoprecipitate p50. Polyclonal antibodies raised against a C-terminal peptide of human YB-1 were used to immunoprecipitate proteins cross-linked to the ACE sel RNA. (A) Immunoprecipitation of cross-linking proteins in HeLa nuclear extract. Duplicate immunoprecipitations are shown. Lane 1, total cross-linking; lanes 2 and 3, supernatants for immunoprecipitations 1 and 2; lanes 4 and 5, pellets from immunoprecipitations 1 and 2. (B) Competition of UV cross-linking of nuclear proteins (lanes 1–3) or gel-purified and renatured recombinant human YB-1 (lanes 4–6). Cross-linking reactions were performed without competitor (lanes 1 and 4) or with 10 pmol of the ACE sel competitor (lanes 2 and 5) or the ACE sel Mut 1 competitor (lanes 3 and 6). The positions of HeLa YB-1 and recombinant YB-1 are indicated; the latter is slightly larger as a result of the presence of a His tag.
YB-1 binds to ACE sequences in a natural alternative exon
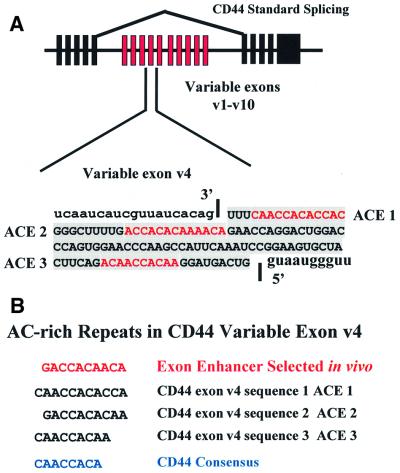
To assess the potential role of YB-1 in enhancer recognition and alternative splicing, we investigated the function of both YB-1 and ACE elements in a natural alternative exon. The CD44 variable exon 4 contains three A/C-rich sequences (denoted in red in Figure 5A) that resemble the ACE derived from iterative selection (Figure 5B). The strong similarity suggested that YB-1 might be involved in recognition of CD44 exon v4.
Fig. 5. Human CD44 alternative splicing. (A) The exon–intron architecture of the human CD44 gene is drawn at the top. The 10 alternative cassette exons are depicted in red. The exon studied in this report is the fourth variable exon v4. The sequence of this exon is shown at the bottom of the figure. Sequences within the exon are indicated with a gray background. The red sequence indicates A/C-rich elements ACE 1, ACE 2 and ACE 3 within exon v4 that are potential binding sites for YB-1. (B) A/C-rich repeats within CD44 variable exons 4 and 5. The A/C-rich sequences from the two exons are aligned and a consensus repeat sequence (blue) is derived. At the top is the derived repeat consensus ACE from the exon selection experiments.
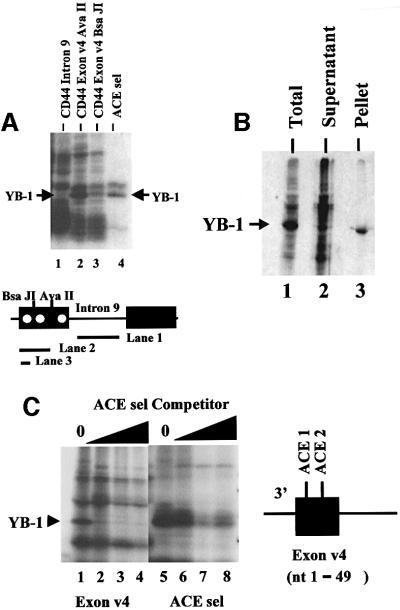
A prominent protein of 50 kDa cross-linked to an RNA containing the first 49 nucleotides of CD44 exon v4 containing ACE 1 and ACE 2 (Figure 6A, lane 2). The protein co-migrated with the band identified as YB-1, which UV cross-linked to the ACE sel RNA (compare lanes 2 and 4). An RNA containing sequences from intron 9 of CD44 did not cross-link to p50 (lane 1). The 50 kDa band that cross-linked to the exon v4 substrate RNA was effectively immunoprecipitated with the YB-1-specific antibody (Figure 6B), indicating that it was indeed YB-1. Competition studies also indicated that the 50 kDa protein UV cross-linking to sequences in CD44 exon v4 was YB-1 (Figure 6C).
Fig. 6. CD44 exon v4 can be UV cross-linked to YB-1. (A) The substrate RNAs diagrammed below the gel were UV cross-linked in a standard splicing assay using HeLa nuclear extract. A diagram of the region of the human CD44 gene containing the fourth and fifth alternative exons is indicated below the gel. Intron 9 is the naturally occurring intron separating exons v4 and v5. ACE elements within exon v4 are indicated by white circles. The partial exon v4 RNAs shown in lanes 2 and 3 were generated by transcript termination at the indicated restriction sites. The ACE sel RNA used for lane 4 is longer than the RNA used for previous figures (see Materials and methods) resulting in the cross-linking of a protein slightly larger thanYB-1 in addition to YB-1. The position of YB-1 is indicated. (B) Immunoprecipitation of cross-linked proteins with an antibody specific for YB-1. A substrate RNA containing the first half of exon 4 and including the ACE 1 and ACE 2 elements (diagrammed in C) was subjected to UV cross-linking in a standard HeLa in vitro splicing assay. Cross-linked proteins were immunoprecipitated using the anti-YB-1 antibody and displayed by SDS–PAGE. Lane 1, total proteins; lane 2, supernatant; lane 3, precipitated protein (3-fold more of the reaction was loaded in lanes 2 and 3 than 1; lanes 2 and 3 came from the same reaction). (C) Competition of YB-1 cross-linking to CD44 exon v4 with an RNA containing ACE sel. Two substrate RNAs were employed: the exon 4 substrate described in (B) (lanes 1–4) and the ACE sel RNA itself (lanes 5–8). Competitor concentrations were 0, 0.3, 3.0 and 10 pmol. The position of cross-linked YB-1 is indicated.
YB-1 activates CD44 exon v4 inclusion in vivo
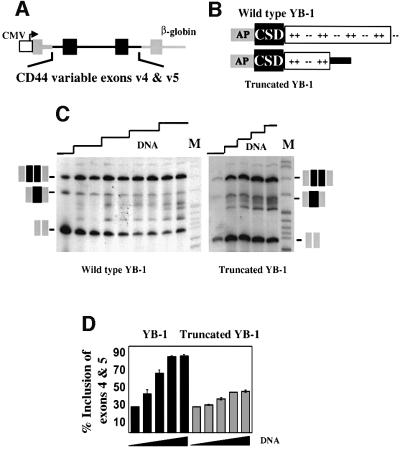
To determine the effect of YB-1 on CD44 splicing, we co-transfected expression plasmids coding for wild-type or a mutant version of YB-1 into HeLa cells along with a reporter construct containing CD44 exons v4 and v5 placed within a human β-globin gene (Figure 7A). The mutant YB-1 contains a premature termination codon resulting in a truncated YB-1 protein (Figure 7B). Western blot analysis demonstrated that both full-length and truncated proteins were expressed at equivalent levels following transfection, and both were transported to the nucleus (data not shown). RT–PCR analysis of the RNA produced from the co-transfected CD44 mini-gene demonstrated that only 28% of the expressed RNA included exons v4 and v5 in the absence of co-transfecting expression plasmid (Figure 7C, lane 1). Little RNA was produced that resulted from the inclusion of only one of the CD44 exons. Inclusion of both exons v4 and v5 rose from 28 to >80% when increasing amounts of co-transfecting YB-1 plasmid were used (Figure 7C; quantified in D). The quantitative nature of the RT–PCR assay was demonstrated using co-transfection of two constructs for CD44 cDNAs, one containing v4 + v5 and one lacking v4 + v5. The two constructs were co-transfected at varying ratios and the ratio of the RT–PCR products closely matched the ratio of the input DNA (data not shown). Co-transfection with the mutant form of YB-1 was unable to cause full activation of exon inclusion. This mutant protein contains the cold shock domain (CSD), which is thought to be the nucleic acid binding domain of the protein (Wolffe et al., 1992) but is lacking other domains. Consistent with these reports, recombinant truncated YB-1 displays sequence-specific binding to ACE elements (data not shown). In its inability to perform wild-type function, the mutant YB-1 behaves like truncated SR proteins, which also fail to function when their SR domains have been deleted (Tacke and Manley, 1999).
Fig. 7. Transfection of human YB-1 increased the inclusion of CD44 exons v4 and v5. (A) The reporter mini-gene containing CD44 variable exons v4 and v5. CD44 sequences are in black, human β-globin sequences are in gray, and the CMV promoter is in white. The CD44 insert is a contiguous genomic fragment mapping from 792 nucleotides upstream of exon v4 to 515 nucleotides downstream of exon v5. (B) Diagram of the wild-type and mutant YB-1 proteins made from co-expressing plasmids. The three indicated domains include an N-terminal alanine- and proline-rich region (AP), the single-stranded nucleic acid binding CSD and the C-terminal highly charged region. (C) RT–PCR analysis of RNAs produced upon transfection of HeLa cells with the reporter shown in (A) and increasing amounts (0, 1, 2, 3 or 4 µg) of either wild-type or mutant YB-1 expression plasmids. Duplicate lanes are shown for each concentration of the wild-type YB-1. PCR oligonucleotides were complementary to sequences in the flanking β-globin exons. Product RNAs resulting from inclusion of neither CD44 exon, either exon v4 or v5, or both exons v4 and v5 are indicated. Note that exons v4 and v5 have such similar sizes that inclusion bands resulting from either of the exons will be in the same region of the gel. Increasing amounts of the YB-1 expression plasmid resulted in a linear increase in the amount of YB-1 mRNA (data not shown). (D) Quantification of the effect of YB-1 on inclusion of CD44 exons v4 and v5. Results of four independent experiments are shown with 1 SD indicated.
YB-1 was unable to affect the inclusion of a heterologous weak exon inserted into the same mini-gene backbone (data not shown). The tested exon lacked notable A/C-rich sequences and would not be expected to respond to ACE binding proteins. YB-1 also had minimal effect on the ratio of mRNAs expressed from co-transfected CD44 cDNAs containing and lacking v4 + v5 (data not shown), indicating that the effect in Figure 7C was not a result of mRNA stability. We cannot rule out the possibility of an indirect effect of YB-1 on CD44 splicing caused by the transcriptional up-regulation of an unknown splicing protein. However, as YB-1 binds to CD44 exon sequences, the most straightforward interpretation of the data in Figure 7 is that YB-1 participates directly in the splicing of CD44 exons v4 and v5.
The CD44 exon v4 ACE 1 and ACE 3 elements are required for normal splicing and binding of YB-1
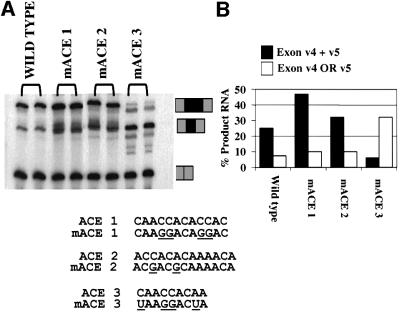
If YB-1 is important for exon v4 recognition, ACE sequences within exon v4 should be required for maximal activity. To test this hypothesis, each of the three ACE elements within exon v4 was mutated (Figure 8). These mutants were transfected into HeLa cells in the context of the mini-gene described in Figure 7. The ACE 3 mutation had the strongest phenotype (Figure 8B). It depressed the percentage of RNA that included both exons v4 and v5 from 25 to 6%. A major new PCR product in which only v4 or v5 was included appeared. This product was sequenced and found to be RNA that included only exon v5. Thus, the mutation had no effect on inclusion of v5 as the fraction of mRNAs that include v5 was comparable in wild type and mACE3, but it significantly depressed inclusion of exon v4.
Fig. 8. Mutation of CD44 exon v4 ACE 1 or 3 alters in vivo recognition of exon v4. (A) The diagrammed mutations were introduced into exon v4 of the CD44 mini-gene described in Figure 7. Wild-type and ACE 1, 2 or 3 mini-genes were transfected in duplicate into HeLa cells and the splicing phenotype was determined by RT–PCR as described in Figure 7. Note that because exons v4 and v5 are almost the same size, products containing exons v4 or v5 alone are indistinguishable in size. Sequencing of the band produced with the ACE 3 mutant indicates that this RNA product contains exon v5 but not v4. Product RNAs including no, one or two CD44 alternative exons are indicated. (B) Quantification of the results in (A).
The ACE 2 mutation had little effect, suggesting that it, alone, is not a major sequence regulating exon v4 inclusion. The ACE 1 sequence had an unusual effect. When mutated, exon v4 and v5 inclusion was increased such that 47% of the product RNA included v4 and v5 as compared with 25% for the wild type, suggesting that this sequence represents a modest exon silencer. A silencer with a different sequence has been detected experimentally in the beginning of the murine CD44 exon v5 as well (König et al., 1998).
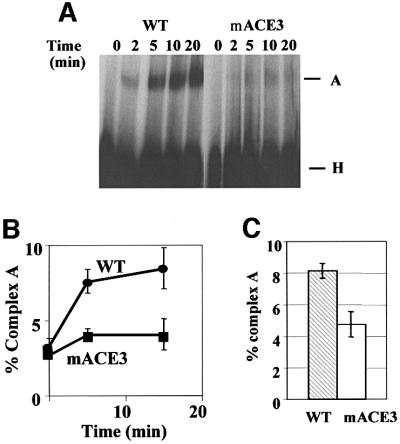
Figure 9 displays the ability of the ACE 1 and 3 mutants to direct in vitro spliceosome assembly using a substrate RNA containing intact exon v4 with its surrounding splice sites. Such substrates have the ability to assemble the first ATP-dependent spliceosome complex, complex A. Wild-type RNA forms complex A (Figure 9A), as does the ACE 1 mutant (data not shown). The ACE 3 mutant is depressed for complex A formation, in agreement with the in vivo depression of production of spliced RNA containing exon 4 (Figure 9). This result suggests that at least a portion of the phenotype of an ACE 3 mutation in vivo is a result of a reduced ability to form an early spliceosome complex.
Fig. 9. Mutation of ACE 3 depressed spliceosome assembly on CD44 exon v4. The wild-type or ACE3 mutant substrate RNAs including all of variable exon v4 and its flanking 3′ and 5′ splicing signals were incubated under in vitro splicing conditions for 0, 2, 5, 10 or 20 min. Mutations are identical to those described in Figure 8. (A) Formed complexes were analyzed by native gel electrophoresis (Carlo et al., 2000). The non-specific H complex and initial ATP-dependent spliceosomal A complex are indicated. (B) Quantification of the reaction in (A). The gel was scanned in the phosphoimager and the percentage of RNA in complex A was plotted. (C) Quantification of the amount of complex A formed at 20 min from multiple experiments using mutant and wild-type substrate RNA.
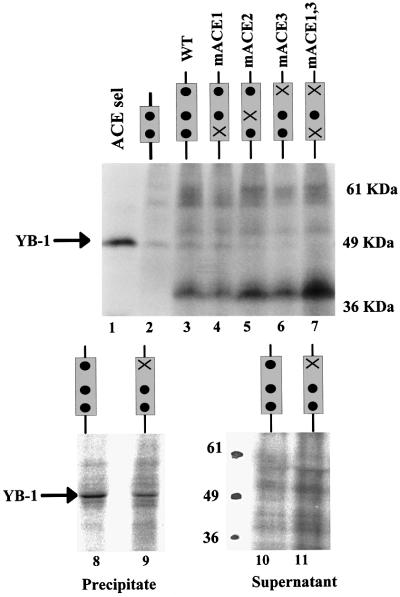
The ACE 1–3 mutations also depressed the UV cross-linking of YB-1 to exon v4 sequences (Figure 10). Some cross-linking was detected with the ACE 1 and ACE 2 mutants (Figure 10, top), consistent with the inability of these mutants to adversely affect either in vivo splicing or in vitro assembly. Reduced cross-linking was observed with the ACE 3 mutant, as detected by both the cross-linking of total proteins (Figure 10, top) and immunoprecipitation of cross-linked proteins with the YB-1-specific antibody (Figure 10, bottom). This absence is consistent with reduced inclusion of exon v4 in vivo and the reduction of in vitro assembly of exon 4 with the ACE 3 mutant.
Fig. 10. Mutation of CD44 ACE sequences depresses UV cross-linking of YB-1. Top: the binding of YB-1 to wild-type and mutant CD44 exon 4 sequences was compared by UV cross-linking. The substrates are diagrammed above the gel. The position of YB-1 is indicated. Bottom: immunoprecipitation of YB-1 UV cross-linked to wild-type or mutant substrates. Cross-linked reactions were immunoprecipitated using the YB-1-specific antibody. Proteins from both the precipitate and the supernatant were displayed by SDS–PAGE.
The ACE 3 mutation was less responsive to increasing concentrations of YB-1 than wild type in vivo (Figure 11A, compare lanes 1–4 with 5–8). YB-1 increased the percentage of wild-type product RNA including both exons v4 and v5 by 40% (Figure 11B). In the mutant, however, little RNA was produced that contained both v4 and v5. In the presence of YB-1, the amount of mutant RNA including both exons increased by only 17% and the amount of RNA including exon v5 alone increased 12%. Thus, increasing the concentration of YB-1 was unable to reverse the effect of the ACE 3 mutant in exon v4 significantly. Given the ability of YB-1 to bind to both ACE 1 and ACE 2 elements, it is not surprising that the ACE 3 mutant retained some ability to respond to YB-1.
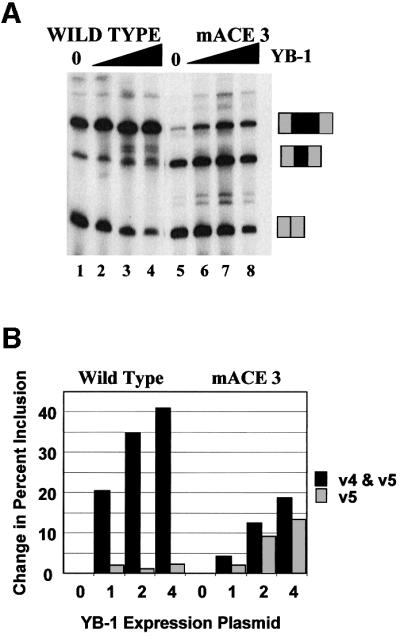
Fig. 11. Mutation of CD44 ACE sequences depresses the ability to respond to YB-1 in vivo. (A) The mini-gene containing CD44 variable exons v4 and v5 was co-transfected with an expression vector coding for YB-1 as in Figure 9. The mini-gene contained a wild-type exon v4 (lanes 1–4) or an exon v4 containing the ACE 3 mutation (lanes 5–8). Increasing amounts of YB-1 vector were used (0, 1, 2 or 4 µg). The positions of RNA species resulting from the inclusion of no, one or two CD44 exons are indicated. (B) Quantification of the results in (A).
Discussion
Exon enhancers are abundant accessory elements that facilitate the recognition of exons during the earliest steps of pre-mRNA splicing. Although most of the experimental attention to the mechanism of enhancer recognition has concentrated on the purine-rich enhancers and the SR proteins that recognize them, a number of other exon sequences are potential enhancers (Dominski and Kole, 1994; Staknis and Reed, 1994; van Oers et al., 1994; Wang et al., 1995; Coulter et al., 1997; Gersappe and Pintel, 1999; Schaal and Maniatis, 1999a,b). One common exon enhancer is the A/C-rich ACE element (van Oers et al., 1994; Wang et al., 1995; Coulter et al., 1997; Schaal and Maniatis, 1999b). Here we identify the human Y-box binding protein YB-1 as a nuclear protein that binds to ACE elements and increases in vivo inclusion of exons containing ACE elements in a fashion that is dependent upon both the presence of exonic ACE elements and full-length YB-1 protein.
YB-1 was originally identified as a transcription factor that binds single-stranded DNA within the reverse CCAAT promoter element (Dorn et al., 1987; Didier et al., 1988), which is similar to the RNA ACE elements. Recently, considerable attention has been focused on YB-1 as an RNA binding protein (Tafuri and Wolffe, 1990; Deschamps et al., 1992; Murray et al., 1992; Minich et al., 1993; Evdokimova et al., 1995). YB-1 is an abundant protein in both the nucleus and the cytoplasm of many cell types (Tafuri and Wolffe, 1990; Ranjan et al., 1993). YB-1 binds to cytoplasmic mRNA and participates in translation regulation. In Xenopus, YB-1 is the major protein within storage mRNA particles in oocytes (Darnbrough and Ford, 1981; Kick et al., 1987; Tafuri and Wolffe, 1990; Murray et al., 1991, 1992; Marello et al., 1992). In this case, multiple copies of the protein bind to RNA with a density of one protein molecule per 50 nucleotides (Darnbrough and Ford, 1981; Marello et al., 1992). In vitro SELEX revealed that the Xenopus protein (FRGY2) binds best to the sequence AACUAC (Bouvet et al., 1995), similar to the ACE elements described in this report. No RNA binding role for YB-1 within the nucleus had been reported prior to this study.
The structure of the YB-1 protein is ideal for a splicing regulatory protein (reviewed in Wolffe et al., 1992). Like other RNA binding proteins, the sequence-specific binding domain of the protein contains β-sheets. In contrast to the four-stranded β-sheet associated with the SR and hnRNP proteins or the three-stranded β-sheet found in the KH proteins, YB-1 contains a five-stranded β-barrel known as the CSD (Wistow, 1990). YB-1 also contains accessory domains that are highly enriched in both basic amino acids and aromatic groups, referred to as existing in basic/aromatic (B/A) islands (Murray et al., 1992). Separating the B/A islands are acidic regions predicted to form α-helices. It has been proposed that the accessory domains are responsible for sequence-independent RNA binding and that the CSD provides sequence specificity (Bouvet et al., 1995). This arrangement of a specificity domain containing β-sheets and a basic domain providing generic RNA binding is highly reminiscent of the organization and functionality of the SR proteins. The mutant YB-1 used in this study is truncated shortly after the CSD, thereby eliminating many of the B/A islands. Its lowered ability to stimulate CD44 splicing suggests a role for the accessory domain in splicing.
Also, like the SR proteins, YB-1 is highly phosphorylated. In Xenopus, phosphorylation is required for binding to RNA and specific dephosphorylation is associated with mRNA release from storage particles (Kick et al., 1987; Cummings and Summerville, 1988). A similar phosphorylation–dephosphorylation cycle occurs with SR proteins (Tacke and Manley, 1999). Bacterially produced YB-1 was able to bind to ACE elements with the same sequence specificity as the HeLa protein. We were, however, unable to use bacterially produced protein to activate splicing activity in vitro, suggesting that correct phosphorylation might be required.
The ACE elements and the YB-1 preferred binding sequence are similar to the A/C-rich element required for exon recognition in the Drosophila melanogaster double-sex gene. The double-sex enhancer binds the sex-regulating proteins Tra and Tra 2 (Hoshijima et al., 1991; Tian and Maniatis, 1992; Lynch and Maniatis, 1995). The latter is an SR protein, although it does not purify with the other ‘classic’ SR proteins. Human cells have no Tra orthologs, but do have two closely related Tra 2 proteins, Tra 2α and Tra 2β (Dauwalder et al., 1996; Nayler et al., 1998). It might be predicted that these proteins will also bind to and regulate ACE elements in human cells, although it should be noted that an attempt to define a binding site for human Tra 2 using iterative selection detected purine-rich binding sites, not ACE elements (Tacke et al., 1998). Preliminary experiments indicate that human Tra 2 proteins do interact with the CD44 exon v4 sequences and may also interact with YB-1 (E.Stickeler, A.Honig, W.Mattox and S.M.Berget, manuscript in preparation). In contrast, ASF/SF2 and SC35 had no effect on CD44 exon v4 or v5 alternative splicing (data not shown). Thus, a slightly different member of the SR protein family, Tra 2, may be involved in ACE recognition.
We previously demonstrated that RNAs containing purine-rich exon splicing enhancers inhibited ACE-dependent splicing in vitro, suggesting a role for SR proteins in ACE-dependent splicing (Coulter et al., 1997). Here we demonstrate that SR proteins do not bind strongly to ACEs. Neither do SR proteins affect binding of YB-1 to the selected ACE. Together, these results suggest that a role for SR proteins in ACE-dependent splicing occurs subsequent to binding of YB-1, consistent with reports describing roles for SR proteins following complex A assembly (Roscigno and Garcia-Blanco, 1995; Chew et al., 1999). YB-1 also has properties reminiscent of another class of prominent RNA binding proteins that influence splicing: the hnRNP proteins (reviewed in Weighardt et al., 1996; Krecic and Swanson, 1999). Like the hnRNP proteins, YB-1 could multimerize on RNA to form structures that look like beads under the electron microscope (Tafuri and Wolffe, 1992). The functionality of such association is unclear, but could provide interactions between exons bearing ACE elements. The CD44 gene has multiple consecutive alternative exons containing putative ACE elements. The simultaneous recognition of multiple ACE exon enhancer domains by the self-association of bound YB-1 protein could explain the observation that blocks of CD44 exons are simultaneously included upon stimulation or in certain cancers (Fox et al., 1994; Mackay et al., 1994; Bell et al., 1998; Stickeler et al., 1999). Interestingly, the increased inclusion of CD44 exons in cancer correlates with increased expression levels of YB-1 (E.Stickeler and S.M.Berget, unpublished results; Bargou et al., 1997).
The protein target by which the binding of YB-1 activates exon recognition is not known. SR proteins are thought to activate splicing by interaction with constitutive splicing factors, such as U170K and U2AF. The only RNA binding protein known to interact with YB-1 is hnRNP K (Funke et al., 1996; Shnyreva et al., 2000). Although this interaction is more relevant to the cytoplasmic activities of YB-1, it does suggest that YB-1 might interact with other members of the KH family of splicing factors. There are multiple splicing regulators that are KH proteins (Siebel et al., 1995; Arning et al., 1996; Min et al., 1997). Perhaps the most intriguing candidate is splicing factor 1 (SF1), a highly conserved protein that plays a role in early spliceosome assembly by interactions with factors bound to both 5′ and 3′ splice sites (Berglund et al., 1998). Such an interaction would place interactions between YB-1 and SF1 in a role analogous and parallel to the activation of early assembly by interactions between SR proteins and U2AF/U170K.
Materials and methods
Plasmids and transfections
In vitro transcription templates for ACE sel and ACE sel Mut1 RNAs were produced by cloning duplicate copies of the 4.11.12 or 12 mu1 selected ACE sequences described in Coulter et al., (1997) downstream of the T7 promoter in Bluescript KS+. Cut with NheI, ACE sel was transcribed into a 59 nucleotide RNA of sequence GGGCGAAUUGGCUAGAGCACCAGUCACCGCUACGCGUCCACCAGUCACCGCCGCUAGC (used for Figures 1–4). The underlined regions are the selected sequences; both copies were mutated in the 12 mu1 variant (sequence in Figure 1). Cut with AccI, a 69 nucleotide RNA was produced that added the sequence GAUACCGUCG to the 3′ end of the above RNAs (used for Figure 6). A cDNA clone isolated from a human HeLa cell line and coding for YB-1 was obtained from P.Newburger (Shen et al., 1998). This clone contained a 2 bp deletion at amino acid 207 causing a frame shift and premature termination of translation 14 amino acids downstream. A wild-type version of this expression plasmid was prepared by replacing the mutated region with a cDNA prepared by PCR amplification of a commercial cDNA preparation isolated from human skeletal muscle. Both were subcloned into pET28a for bacterial production of N-terminal His-tagged YB-1 protein and pcDNA3.1/HisC for in vivo expression. The CD44 mini-gene (Figure 7) was created by inserting a genomic fragment from the human CD44 gene containing variable exons v4 and v5 into the first intron of a β-globin in vivo expression plasmid derived from Dup 33 (Dominski and Kole, 1992) obtained from R.Kole, University of North Carolina, NC. Several templates containing CD44 exon v4 sequences were used for in vitro transcription. A short version containing nucleotides –20 upstream of the exon to +30 downstream of the exon was used to generate an RNA incapable of forming complex A (because of deletion of important sequences from the 3′ splice site) in Figures 6 and 7. A longer clone including 38 nucleotides of the intron upstream of exon v4 was used for in vitro assembly and some UV cross-linking studies (Figures 9 and 10). Transcripts were produced by truncating these plasmids within or after exon v4 sequences using BsaJI (+11 nucleotides of exon v4), AvaII (+49 nucleotides of exon v4) or SfaI (56 nucleotides after the exon v4 5′ splice site). The first RNA is truncated within ACE 1, the second contains ACE 1 and ACE 2, and the third contains all three exon v4 ACE elements. All constructs were sequenced for verification.
Transfections of HeLa cells using lipofectAMINE™ (Gibco-BRL) and isolation of total cell RNA 48 h post-transfection using TRIzol™ (Gibco-BRL) were performed following the manufacturer’s instructions. Splicing patterns were determined by RT–PCR analysis using 5′-end-radiolabeled primers specific for β-globin sequences (5′-AGACACCATGCATGGTGCACC and 3′-CCTGATCAGCGAGCTCTAG). These primers amplified no RNA from untransfected HeLa cells. Amplification conditions were 40 s at 94°C, 40 s at 58°C and 1 min at 72°C for 25 cycles (Figures 7 and 8), or 30 s at 94°C, 30 s at 62°C and 1 min at 72°C for 20 cycles (Figure 11). Product DNA was denatured and displayed on 5–6% urea gels. RNA products were quantified in the phosphoimager. Identified amplification products resulting from the inclusion of one or two variable CD44 exons were sequenced to verify identity.
In vitro assays
In vitro synthesis of RNA, splicing, spliceosome assembly and UV cross-linking assays have been described previously (Ramchatesingh et al., 1995). To immunoprecipitate YB-1, a Pansorbin suspension (20 µl) (Calbiochem) was washed once with 1× gamma bind buffer (BB; 10 mM sodium phosphate, 150 mM NaCl, 10 mM EDTA). Five microliters of rabbit antiserum were bound to Pansorbin in 300 µl of BB by rocking overnight at 4°C. The antibody beads were washed three times with RIPA buffer (50 mM Tris pH 8.0, 150 mM NaCl, 1% v/v NP-40, 0.5% deoxycholate, 0.1% SDS) and then once with buffer DG (20 mM HEPES–KOH pH 7.9 at 4°C, 80 mM monopotassium glutamate, 0.2 M EDTA) plus 0.05% Triton X-100. The cross-linking reactions were used to resuspend the Pansorbin pellet following RNase digestion. Five microliters of 3 mg/ml heparin and 0.15% Triton X-100 were added, and the mixture was agitated for 30 min at 4°C with vigorous shaking. Proteins were prepared from the supernatant by acetone precipitation. The Pansorbin pellet was washed three times with DG plus 0.05% Triton X-100, then resuspended in 40 µl of protein loading buffer. Semi-purified SR proteins were recovered in the 65–90% ammonium sulfate cuts of cleared HeLa nuclear extract (Zahler, 1999) and dialyzed against Roeder D buffer without glycerol (Dignam et al., 1983).
Purification of YB-1
YB-1 was fractionated from HeLa nuclear extract using a protocol originally described to purify U2AF (Wang et al., 1995). HeLa nuclear extract was dialyzed into HeLa equilibration buffer [20 mM HEPES pH 7.6, 3 mM MgCl2, 0.2 mM EDTA, 1 mM dithiothreitol (DTT), 1.0 M KCl, 10% glycerol]. Cleared and dialyzed extract was passed over a poly(U)–Sepharose column equilibrated with buffer A1 (20 mM HEPES pH 7.9, 3 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 1.0 M KCl, 5% glycerol) at a ratio of ∼1 ml of extract per 2 ml of bed volume. After extensive washing with buffer A1, YB-1 was eluted with four successive column volumes of buffer A1 with KCl to 2.0 M and buffer A1 with NaCl lowered to 0.1 M and supplemented with 2.0 M guanidine–HCl. The fourth 2.0 M KCl fraction and the first two guanidine–HCl fractions were pooled from multiple columns and used to gel purify the 50 kDa band for tryptic peptide mapping. Resulting peptides were sequenced by the Protein Chemistry Core Facility at Baylor College of Medicine. The four peptides sequenced each had 100% identity with human YB-1 (EDVFVHQTAIK, GAEAANVTGPGGVPVQG, PGTTGSGAGSGGPGG and ENQGDETQCQQPPQR).
N-terminal His-tagged recombinant YB-1 was induced in Escherichia coli BL21 using 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 3–4 h. Protein was purified using Ni-NTA according to the manufacturer’s instructions (Qiagen). Recombinant protein was gel-purified from SDS–PAGE. Protein isolated from gel bands was concentrated by precipitation with four volumes of acetone at –20°C for 30 min, and centrifugation at 13 000 g for 20 min. The pellet was resuspended in 60 µl of 6 M guanidine–HCl in Roeder D buffer and incubated at room temperature to completely denature the protein. Renaturation was accomplished by dilution into 3.0 ml of Roeder D buffer and incubation at room temperature for 1 h. The resulting solution was desalted and concentrated in an ultra free centrifugal filter device (Millipore).
Acknowledgments
Acknowledgements
Special thanks to members of the Cooper and Berget laboratories for helpful suggestions and critical comments. We thank Dr P.Newburger for YB-1 clones and antibodies, R.Kole for plasmid constructs, the Baylor College of Medicine genome center for genomic human clones, Dr Richard Cook and the Baylor College of Medicine Protein Core for advice and protein sequencing, the Cell Culture Center (Endotronics Inc.) for large volume HeLa cell growth, and Gopal Singh for technical assistance. This work was supported by grants from The American Cancer Society (NP-79230) and the NIH (RO1 HL45565) to T.A.C.; the US Army (DAMD17-96-1-6084) and the NIH (GM38526) to S.M.B., the German Research Foundation (Sti. 153/1-1) to E.S., and the Foundation for Gene and Cell Therapy and the Medical Research Council of Canada (The Jesse Davidson Post-doctoral Research Fellowship) to S.D.F.
Note added in proof
While this paper was in review, YB-1 was shown to be associated with RNA polymerase II via binding to TLS or EWS, and to affect alternative splicing of an adenovirus E1A reporter construct. [Chansky,H.A., Hu,M., Hickstein,D.D. and Yang,Y. (2001) Oncogenic TLS/ERG and EWS/Fli-1 fusion proteins inhibit RNA splicing mediated by YB-1 protein. Cancer Res., 61, 3586–3590.]
References
- Arning S., Gruter,P., Bilbe,G. and Kramer,A. (1996) Mammalian splicing factor SF1 is encoded by variant cDNAs and binds to RNA. RNA, 2, 794–810. [PMC free article] [PubMed] [Google Scholar]
- Bargou R.C. et al. (1997) Nuclear localization and increased levels of transcription factor YB-1 in primary human breast cancers are associated with intrinsic MDR1 gene expression. Nature Med., 3, 447–478. [DOI] [PubMed] [Google Scholar]
- Bell M.V., Cowper,A.E., Lefranc,M.P., Bell,J.I. and Screaton,G.R. (1998) Influence of intron length on alternative splicing of CD44. Mol. Cell. Biol., 18, 5930–5941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund J.A., Abovich,N. and Rosbash,M. (1998) A cooperative interaction between U2AF65 and mBBP/SF1 facilitates branchpoint region recognition. Genes Dev., 12, 858–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black D. (1995) Finding splice sites within a wilderness of RNA. RNA, 1, 763–771. [PMC free article] [PubMed] [Google Scholar]
- Bouvet P., Matsumoto,K. and Wolffe,A.P. (1995) Sequence-specific RNA recognition by the Xenopus Y-box binding proteins. J. Biol. Chem., 270, 28297–28303. [DOI] [PubMed] [Google Scholar]
- Carlo T., Sierra,R. and Berget,S.M. (2000) A 5′ splice site-proximal enhancer binds SF1 and activates exon bridging of a microexon. Mol. Cell. Biol., 20, 3988–3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew S.L., Liu,H.X., Mayeda,A. and Krainer,A.R. (1999) Evidence for the function of an exonic splicing enhancer after the first catalytic step of pre-mRNA splicing. Proc. Natl Acad. Sci. USA, 96, 10655–10660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper T.A. (1999) Defining pre-mRNA cis elements that regulate cell-specific splicing. Methods Mol. Biol., 118, 391–403. [DOI] [PubMed] [Google Scholar]
- Cooper T.A. and Mattox,W. (1997) The regulation of splice-site selection, and its role in human disease. Am. J. Hum. Genet., 61, 259–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulter L.R., Landree,M.A. and Cooper,T.A. (1997) Identification of a new class of exonic splicing enhancers by in vivo selection. Mol. Cell. Biol., 17, 2143–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings A. and Sommerville,J. (1988) Protein kinase activity associated with stored messenger ribonucleoprotein particles of Xenopus oocytes. J. Cell Biol., 107, 45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnbrough C.H. and Ford,P.J. (1981) Identification in Xenopus laevis of a class of oocyte-specific proteins bound to messenger RNA. Eur. J. Biochem., 113, 415–424. [DOI] [PubMed] [Google Scholar]
- Dauwalder B., Amaya-Manzanares,F. and Mattox,W. (1996) A human homolog of the Drosophila sex determination factor transformer-2 has conserved splicing regulatory functions. Proc. Natl Acad. Sci. USA, 93, 9004–9009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschamps S., Viel,A., Garrigos,M., Denis,H. and LeMaire,M. (1992) mRNP4, a major RNA binding protein from Xenopus oocytes is identical to a transcription factor FRGY2. J. Biol. Chem., 267, 13799–13802. [PubMed] [Google Scholar]
- Didier D.K., Schiffenbauer,J., Woulfe,S.L., Zacheis,M. and Schwartz, B.D. (1988) Characterization of the cDNA encoding a protein binding to the major histocompatibility complex class II Y box. Proc. Natl Acad. Sci. USA, 85, 7322–7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J.D., Lebovitz,R.M. and Roeder,R.G. (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res., 11, 1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z. and Kole,R. (1992) Cooperation of pre-RNA sequence elements in splice site selection. Mol. Cell. Biol., 12, 2108–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z. and Kole,R. (1994) Identification of exon sequences involved in splice site selection. J. Biol. Chem., 269, 23590–23596. [PubMed] [Google Scholar]
- Dorn A., Durand,B., Marfling,C., Le meur,M., Benoist,C. and Mathis,D. (1987) The conserved MHC class II boxes X and Y are transcriptional control elements and specifically bind nuclear proteins. Proc. Natl Acad. Sci. USA, 84, 6249–6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evdokimova V.M., Wei,C., Sitikov,A.S., Simonenko,P.N., Lazerev, O.A., Vasilenko,K.S., Usinov,VA., Hershey,J.W.B. and Ovchinnikov, L.P. (1995) The major protein of messenger ribonucleoprotein particles in somatic cells is a member of the Y-box binding transcription factor family. J. Biol. Chem., 270, 3186–3192. [DOI] [PubMed] [Google Scholar]
- Fox S.B., Fawcett,J., Jackson,D.G., Collins,I., Gatter,K.C., Harris,A.L., Gearing,A. and Simmons,D.L. (1994) Normal human tissues, in addition to some tumors, express multiple different CD44 isoforms. Cancer Res., 54, 4539–4546. [PubMed] [Google Scholar]
- Funke B., Zuleger,B., Benavente,R., Schuster,T., Goller,M., Stevenin,J. and Horak,I. (1996) The mouse poly(C) binding protein exists in multiple isoforms and interacts with several RNA binding proteins. Nucleic Acids Res., 24, 3821–3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gersappe A. and Pintel,D.J. (1999) CA- and purine-rich elements form a novel bipartite exon enhancer which governs inclusion of the minutevirus of mice NS2-specific exon in both singly and doubly spliced mRNAs. Mol. Cell. Biol., 19, 364–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günthert U. (1993) CD44: a multitude of isoforms with diverse functions. Curr. Top. Microbiol. Immunol., 184, 47–63. [DOI] [PubMed] [Google Scholar]
- Günthert U. et al. (1991) A new variant of glycoprotein CD44 confers metastatic potential to rat carcinoma cells. Cell, 65, 13–24. [DOI] [PubMed] [Google Scholar]
- Hoshijima K., Inoue,K., Higuchi,I., Sakamoto,H. and Shimura,Y. (1991) Control of double-sex alternative splicing by transformer and transformer-2 in Drosophila. Science, 252, 833–836. [DOI] [PubMed] [Google Scholar]
- Kick D., Barrett,P., Cummings,A. and Sommerville,J. (1987) Phosphorylation of a 60 kDa polypeptide from Xenopus oocytes blocks messenger RNA translation. Nucleic Acids Res., 15, 4099–4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- König H., Ponta,H. and Herrlich,P. (1998) Coupling of signal transduction to alternative pre-mRNA splicing by a composite splice regulator. EMBO J., 17, 2904–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krecic A.M. and Swanson,M.S. (1999) hnRNP complexes: composition, structure, and function. Curr. Opin. Cell Biol., 11, 363–371. [DOI] [PubMed] [Google Scholar]
- Lynch K.W. and Maniatis,T. (1995) Synergistic interactions between two distinct elements of a regulated splicing enhancer. Genes Dev., 9, 284–293. [DOI] [PubMed] [Google Scholar]
- Mackay C.R., Terpe,H.-J., Stauder,R., Marston,W.L., Stark,H. and Gunthert,U. (1994) Expression and modulation of CD44 variant isoforms in humans. J. Cell Biol., 124, 71–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marello K., La Rovere,J. and Sommerville,J. (1992) Binding of Xenopus oocyte masking proteins to mRNA sequences. Nucleic Acids Res., 20, 5593–5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min H., Turck,C.W., Nikolicand,J.M. and Black,D.L. (1997) A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes Dev., 11, 1023–1036. [DOI] [PubMed] [Google Scholar]
- Minich W.B., Maidebura,I.P. and Ovchinnikov,L.P. (1993) Purification of the major 50-kDa repressor protein from cytoplasmic mRNA of rabbit reticulocytes. Eur. J. Biochem., 212, 633–638. [DOI] [PubMed] [Google Scholar]
- Murray M.T., Krohne,G. and Franke,W.W. (1991) Different forms of cytoplasmic mRNA binding proteins and particles in Xenopus laevis oocytes and embryos. J. Cell Biol., 112, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray M.T., Schiller,D.L. and Franke,W.W. (1992) Sequence analysis of cytoplasmic RNA binding proteins of Xenopus oocytes identifies a family of RNA binding proteins. Proc. Natl Acad. Sci. USA, 89, 11–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayler O., Cap,C. and Stamm,S. (1998) Human transformer-2-β gene (SFRS10): complete nucleotide sequence, chromosomal localization, and generation of a tissue-specific isoform. Genomics, 53, 191–202. [DOI] [PubMed] [Google Scholar]
- Ramchatesingh J., Zahler,A.M., Neugebauer,K.M., Roth,M.B. and Cooper,T.A. (1995) A subset of SR proteins activates splicing of the cardiac troponin T alternative exon by direct interactions with an exonic enhancer. Mol. Cell. Biol., 15, 4898–4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan M., Tafuri,S.R. and Wolffe,A.P. (1993) Masking mRNA from translation in somatic cells. Genes Dev., 7, 1725–1736. [DOI] [PubMed] [Google Scholar]
- Reed R. (1996) Initial splice site recognition and pairing during pre-mRNA splicing. Curr. Opin. Genet. Dev., 6, 215–220. [DOI] [PubMed] [Google Scholar]
- Roscigno R.F. and Garcia-Blanco,M.A. (1995) SR proteins escort the U4/U6.U5 tri-snRNP to the spliceosome. RNA, 1, 692–706. [PMC free article] [PubMed] [Google Scholar]
- Schaal T.D. and Maniatis,T. (1999a) Multiple distinct splicing enhancers in the protein-coding sequences of a constitutively spliced pre-mRNA. Mol. Cell. Biol., 19, 261–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaal T.D. and Maniatis,T. (1999b) Selection and characterization of pre-mRNA splicing enhancers: identification of novel SR protein-specific enhancer sequences. Mol. Cell. Biol., 19, 1705–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q., Wu,R., Leonard,J.L. and Newburger,P.E. (1998) Identification and molecular cloning of a human selenocysteine insertion sequence binding protein. A bifunctional role for DNA binding protein B. J. Biol. Chem., 273, 5443–5446. [DOI] [PubMed] [Google Scholar]
- Shnyreva M., Schullery,D.S., Suzuki,H., Higaki,Y. and Bomsztyk,K. (2000) Interaction of two multifunctional proteins. J. Biol. Chem., 275, 15498–15503. [DOI] [PubMed] [Google Scholar]
- Siebel C.W., Admon,A. and Rio,D.C. (1995) Soma-specific expression and cloning of PSI, a negative regulator of P element pre-mRNA splicing. Genes Dev., 9, 269–283. [DOI] [PubMed] [Google Scholar]
- Staknis D. and Reed,R. (1994) SR proteins promote the first specific recognition of pre-mRNA and are present together with the U1 small nuclear ribonucleoprotein particle in a general splicing enhancer complex. Mol. Cell. Biol., 14, 7670–7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stickeler E., Kittrell,F., Medina,D. and Berget,S.M. (1999) Stage-specific changes in SR splicing factors and alternative splicing in mammary tumorigenesis. Oncogene, 18, 3574–3582. [DOI] [PubMed] [Google Scholar]
- Tacke R. and Manley,J.L. (1999) Determinants of SR protein specificity. Curr. Opin. Cell Biol., 11, 358–362. [DOI] [PubMed] [Google Scholar]
- Tacke R., Tohyama,M., Ogawa,S. and Manley,J.L. (1998) Human Tra 2 proteins are sequence-specific activators of pre-mRNA splicing. Cell, 93, 139–148. [DOI] [PubMed] [Google Scholar]
- Tafuri S.T. and Wolffe,A.P. (1990) Xenopus Y-box binding transcription factors: molecular cloning, functional analysis and developmental regulation. Proc. Natl Acad. Sci. USA, 87, 9028–9032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tafuri S.T. and Wolffe,A.P. (1992) DNA binding, multimerization, and transcription stimulation by the Xenopus Y box proteins in vitro. New Biol., 4, 349–359 [PubMed] [Google Scholar]
- Tian M. and Maniatis,T. (1992) Positive control of pre-mRNA splicing in vitro. Science, 256, 237–240. [DOI] [PubMed] [Google Scholar]
- Van Oers C.C.M., Admea,G.J., Zandberg,H., Moen,T.C. and Baas,P.D. (1994) Two different sequence elements within exon 4 are necessary for calcitonin-specific splicing of human calcitonin/calcitonin-gene-related-peptide I pre-mRNA. Mol. Cell. Biol., 14, 951–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Hoffmann,H.M. and Grabowski,P.J. (1995) Intrinsic U2AF binding is modulated by exon enhancer signals in parallel with changes in splicing activity. RNA, 1, 21–35. [PMC free article] [PubMed] [Google Scholar]
- Weighardt F., Biamonti,G. and Riva,S. (1996) The roles of heterogeneous nuclear ribonucleoproteins (hnRNP) in RNA metabolism. BioEssays, 18, 747–756. [DOI] [PubMed] [Google Scholar]
- Wistow G. (1990) Cold shock and DNA binding. Nature, 344, 823–824. [DOI] [PubMed] [Google Scholar]
- Wolffe A.P., Tafuri,S., Ranjan,M. and Familari,M. (1992) The Y-box binding factors: a family of nucleic acid binding proteins conserved from Escherichia coli to man. New Biol., 4, 290–298. [PubMed] [Google Scholar]
- Zahler A.M. (1999) Purification of SR protein splicing factors. Methods Mol. Biol., 118, 419–432. [DOI] [PubMed] [Google Scholar]