Abstract
IHF or supercoiling is required early in Tn10 transposition, but at later stages they inhibit the reaction in a classic homeostatic loop. We investigated the mechanism of transpososome assembly and regulation using hydroxyl radical DNA protection and interference. We present a three-dimensional molecular model for the IHF-bent end of Tn10 wrapped around a transposase core. Contacts span some 80 bp at the transposon end, but after assembly of an active complex containing metal ion, most contacts become dispensable. These include transposase contacts beyond the IHF site that chaperone assembly of the complex and are needed for efficient cleavage. Single and double-end breaks do not affect the complex but divalent metal ions promote large conformational changes at bp +1 and the flanking DNA.
Keywords: complex/IHF/nucleus/structure/transposase
Introduction
Tn10 is a composite transposon flanked by inverted repeats of IS10 that cooperate to mobilize the unique sequences in between (Kleckner, 1989; Kleckner et al., 1996; Chalmers et al., 2000). IS10-Right is a fully functional insertion element that encodes an active transposase which can mobilize either of the flanking IS10 elements or the whole of Tn10. IS10/Tn10 transposes by a non-replicative mechanism. The first step of the reaction is the assembly of a synaptic complex that contains a dimer of transposase and both transposon ends (Sakai et al., 1995; Bolland and Kleckner, 1996; D.Haniford, unpublished). Before the element can select a target site for integration, each monomer of transposase must cut both strands of DNA (Sakai and Kleckner, 1997). This is achieved at each end using three sequential one-step transesterification reactions, which proceed via a hairpin intermediate (Kennedy et al., 1998).
The ends of IS10 are inverted repeats, but there are some functional distinctions (Kleckner, 1989; Kleckner et al., 1996). The inside end is regulated by Dam methylation, which couples transposition to replication, whereas the outside end has an IHF binding site involved in transducing a response to changes in the superhelical density (Roberts et al., 1985; Signon and Kleckner, 1995; Chalmers et al., 1998). With a total of four transposon ends, two facing inwards and two facing outwards, Tn10 promotes rearrangements as well as transposition, at a frequency thought to reflect the distance between participating ends and the regulatory mechanisms operating on such ends.
In vivo, IHF has both positive and negative effects on Tn10 transposition and rearrangements, depending on the identity and topological arrangement of the participating ends (Signon and Kleckner, 1995). The direction of the effects measured is thought to depend on the level of supercoiling and perhaps also on the concentration of IHF. The interaction between supercoiling, IHF and its binding site, therefore, has the potential to integrate a response to factors such as replication, conjugal transfer and the general physiological state of the cell.
The Tn10 transpososome is remarkably versatile given the relatively small size. As well as responding to regulatory signals, it accommodates several different configurations of the DNA during synapsis, cleavage, flanking DNA release, target capture and strand transfer. We therefore set out to analyze how protein–DNA contacts change during the reaction and to investigate the molecular mechanism underlying regulation of transposition by IHF and supercoiling. Initially, helically phased arrays of protein contacts span ∼80 bp of DNA at the transposon end. However, most of the contacts become dispensable after activation of the complex for cleavage by a metal ion-dependent conformational change. Sig nificantly, we have identified transposase-specific contacts beyond the IHF site that are required for efficient transpososome assembly and cleavage. We also speculate on some structural and mechanistic parallels with phage λ recombination.
Results
Experimental system
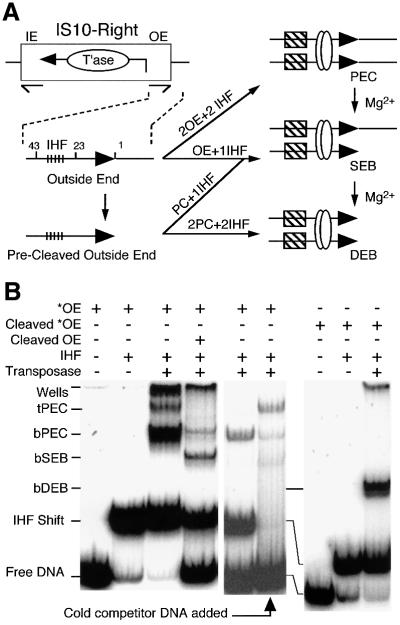
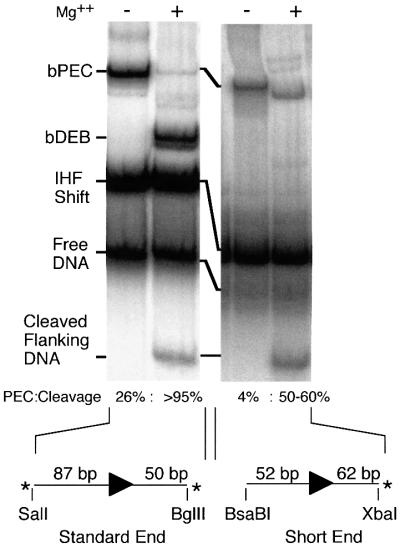
In the presence of IHF, Tn10 transposition can be reconstituted using transposase and the outside end of IS10 on a short linear fragment of DNA (Figure 1; Sakai et al., 1995). A paired end complex (PEC), comprised of a pair of transposon ends, two dimers of IHF and a dimer of transposase, assembles spontaneously in the absence of a divalent metal ion. Addition of Mg2+ initiates cleavage of the DNA, which proceeds through a single end break complex (SEB) to a double end break (DEB) (Figure 1A). The SEB and DEB complexes can be assembled directly if a pre-cleaved transposon end fragment is provided (Sakai and Kleckner, 1997). Complexes formed in this way appear to behave as true intermediates: they have the correct mobility in the gel shift assay and upon addition of Mg2+ they complete the cleavage step of the reaction and perform normal integration into a target DNA, if present.
Fig. 1. The short linear fragment assay for Tn10 transposition. (A) IS10-Right has a gene for transposase (T’ase) between the terminal inverted repeats (thin half arrows). The ends of IS10-Right are defined as ‘inside’ (IE) and ‘outside’ (OE) based on their positions in Tn10. The standard OE fragment for gel shift analysis, shown in the expanded section, contains the internal IHF binding site (tick marks), terminal inverted repeat (bp 1–23 as indicated), transposon terminus (filled horizontal arrows) and flanking sequences. Pre-cleaved OE fragment (PC) lacks flanking DNA. Combinations of OE and pre-cleaved OE fragments are synapsed by transposase (open circles) in the presence of IHF (hatched boxes) to give PEC, SEB and DEB complexes as shown. SEB and DEB complexes also arise by addition of Mg2+ to pre-formed PECs and SEB, respectively. (B) The gel shift assay for Tn10 synaptic complexes was performed as described previously (Sakai et al., 1995). 32P-labeled end fragments are indicated by *. The figure is a composite of three gels and some lanes are duplicated to help comparison. Free DNA, unbound OE fragment; IHF shift, IHF-bound OE fragment; bPEC contains two DNA frag ments, two transposase monomers and two dimers of IHF; tPEC contains two DNA fragments and two transposase monomers. Protein– DNA aggregates that barely enter the gel are visible in some lanes.
DNA fragments for footprinting were 3′-end labeled with Klenow fragment using a single radioactive nucleotide to avoid double labeling. Next, complexes were treated with hydroxyl radicals and purified using the gel shift assay (Materials and methods). Individual complexes were recovered from gel slices by the crush-and-soak method, denatured and run on a DNA sequencing gel to display the protection pattern. The results were recorded on a phosphoimager and were entirely reproducible between different experiments. To provide an unbiased representation, the footprint traces are presented without manipulation, except scaling to fit inside equally sized boxes.
The paired ends complex
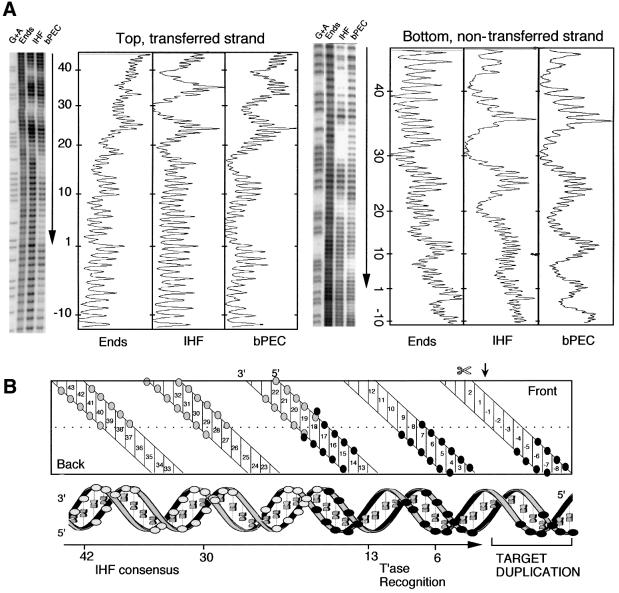
bPEC was assembled under standard conditions in the absence of divalent metal ions and footprinted with hydroxyl radicals (Figure 2A). The pattern of protection is displayed on a two-dimensional projection of the DNA helix and also on a quasi-three-dimensional representation (Figure 2B).
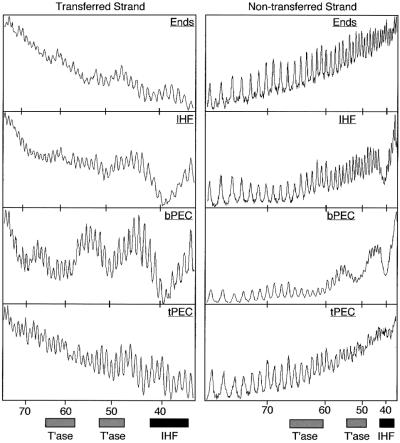
Fig. 2. Hydroxyl radical footprint analysis of the bPEC. (A) Footprints and profiles. The relative intensities of the bands are plotted to the right of each gel. The position of the transposon end and the direction in which it is facing are indicated by the vertical arrows. Nucleotide positions are indicated; +1 is defined as the terminal nucleotide of IS10; –1 is the first nucleotide in the flanking DNA. The transferred strand has a 3′-hydroxyl group at the end of the transposon and is transferred to the target site during transposition. The non-transferred strand is the opposite strand. The sequencing gel and footprint profiles for the non-transferred strand have been inverted for clarity. G + A, Maxam–Gilbert sequence ladder; Ends, free DNA fragment; IHF, IHF-bound end fragment; bPEC, bottom paired ends complex. (B) Summary of protein–DNA contacts. The double helix is represented in two dimensions (top) and three dimensions (bottom). White circles indicate IHF contacts and black circles transposase contacts. The site of first strand cleavage by transposase is indicated by a small vertical arrow and  .
.
The IHF protections are each separated by 10 bp, in almost perfect 10 bp phasing, placing them on the same face of the DNA helix (Figure 2) (Yang and Nash, 1989). Transposase protects a further three regions of DNA, also with near perfect 10 bp helical phasing (Figure 2). Together, the IHF and transposase contacts present a symmetrical array of five protected regions covering five turns of the helix. Both proteins contribute to the central region of protection, with the IHF-specific and transposase-specific protections extending outwards on either side. Note the absence of protection around bp +1 on both strands.
Hydroxyl radical interference
An interference assay with missing nucleosides was performed to investigate which regions of the inverted repeat are important for assembly of the complex. Transposon ends were treated with hydroxyl radicals to delete single nucleosides and used to form bPEC. The PEC was purified by gel shift, the DNA recovered and run on a sequencing gel to yield the interference footprint (Figure 3).
Fig. 3. Missing nucleoside interference assay for bPEC assembly. The terminal inverted repeat of IS10 is indicated by horizontal arrows. Filled boxes, positions where missing nucleosides inhibit PEC assembly; hatched boxes, missing nucleoside enhancement.
On the transferred strand, a continuous block of interference at bp +20 to +6 is directly abutted with enhancements at bp +6 to –1. On the non-transferred strand, modest interference at bp +18 to +6 is adjoined by very strong enhancements from bp +6, at least as far as bp –8 and possibly further. Enhancements probably occur at positions where the specific sequence is unimportant but where missing nucleosides promote complex assembly by reducing the energy required to bend the DNA.
Note that in the protection experiments (above) there is always an IHF signal when IHF is present. However, in the bPEC interference footprint there is little or no interference signal at the IHF binding site. This effect results from the dependence of PEC formation on IHF bound at only one of the pair of ends, and from the fact that most DNA fragments with missing nucleosides are expected to pair with an unmodified partner that is present in large excess.
A Ca2+-dependent conformational change
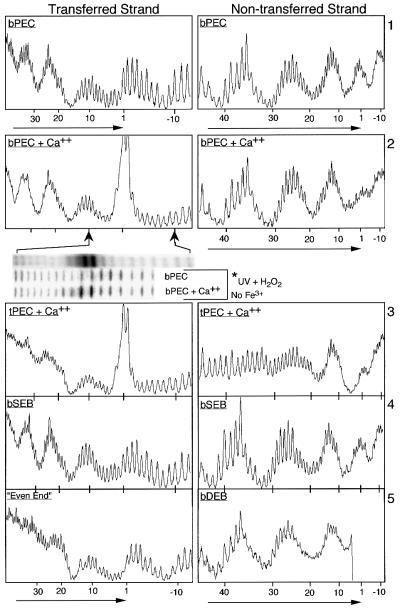
Previously, calcium ions were often included in the standard buffers for complex formation because they promote the assembly and stability of the PEC and facilitate target interactions for the DEB complex (Sakai et al., 1995; Junop and Haniford, 1996; Sakai, 1996). In the present work Ca2+ is omitted from the buffers unless stated otherwise. When the hydroxyl radical protection pattern for the bPEC is determined in the presence of 4 mM Ca2+ there are substantial changes at bp +1 and in the flanking DNA (Figure 4, panel 2). There are no changes whatsoever in the five regions of protection in the arm of the transposon contributed by transposase and IHF (Figure 4, compare panels 1 and 2).
Fig. 4. Hydroxyl radical footprints of transposition intermediates. Nucleotide positions are as defined in Figure 2. The location and direction of the transposon end is indicated by horizontal arrows. The first insert below panel 2 shows an image of the actual gel from which the trace is taken. The second and third insert show the same region of the transposon end in an experiment where hydroxyl radicals are generated without the presence of Fe3+. Hyperreactivity is clearly present at bp +1 and –1, although the result is somewhat less spectacular due to the non-specific destruction of the DNA by UV treatment.
In the presence of Ca2+, bp +1 and bp –1 on the transferred strand are hyperreactive to hydroxyl radicals. This is not due to a low level of first strand nicking by transposase as Ca2+ does not support any of the chemical steps of the transposition reaction (Bolland and Kleckner, 1996). Also, hyperreactivity is reminiscent of the Holliday junction-resolving enzyme RuvC, in which Fe3+ from the footprinting reaction appears to enter the active site, greatly increasing the local concentration of radicals around the recombination site (Hagan et al., 1998). We ruled out the possibility that any of the footprinting reagents was responsible for hyperreactivity by repeating the result with hydroxyl radicals generated directly by the action of UV light on hydrogen peroxide (Macgregor, 1992; insert under panel 2 in Figure 4).
On the non-transferred strand of the PEC, addition of Ca2+ eliminates the peak of reactivity centered on bp +1. Careful inspection of the traces (and gel images) shows that this is due to a combination of a small increase in protection at bp +1 and a greater decrease in protection in the flanking DNA between bp –4 and bp –7 (Figure 4, panel 2). This suggests that flanking interactions become dispensable after the active site has become fully structured around the metal ion, possibly as a prelude to release of the flanking DNA following cleavage.
The top-PEC
Although IHF is an absolute requirement for complex assembly, it becomes dispensable at later stages and can be stripped from the bPEC by the addition of cold competitor DNA or heparin (Junop and Haniford, 1997; Sakai et al., 2000). The new complex is known as the top-PEC because of a reduced mobility in the gel shift assay (Figure 1B). The bPEC and tPEC have very similar kinetics during the cleavage step of the reaction (Sakai et al., 2000). After cleavage, however, the bPEC is unable to interact with target DNA until it has been converted to a tPEC by the loss of IHF. Previously, DNase footprint analysis of the bPEC revealed an essentially continuous block of protection from bp –10 to ∼bp +43 (Sakai et al., 2000). Upon conversion of bPEC to tPEC, protection was reduced to a region extending from bp –10 at least as far as bp +24 and possibly as far as bp +30.
PEC was prepared under standard conditions, IHF was stripped from the complex with heparin to yield tPEC and the hydroxyl radical footprint was determined for both strands (Figure 4, panel 3). As expected the tPEC appears to have lost all IHF at the specific binding sites. There are no significant changes in the transposase-dependent pattern of protection when bPEC is converted to tPEC. Careful inspection of the central region of protection shared by IHF and transposase shows that overlapping positions are protected by IHF and transposase on both strands of DNA (compare IHF in Figure 2A with tPEC in Figure 4, panel 3).
DEB and SEB complexes
The Tn10 transpososome accommodates several configurations of the DNA at different stages of the transposition reaction. To investigate whether conformational changes occur during the cleavage steps of the reaction, footprints were determined for the SEB and DEB complexes.
SEB complex was assembled by mixing the standard transposon end fragment with a precleaved transposon end (see Figure 1). In this experiment the full-length fragment carried the radioactive label and the pre-cleaved transposon end was unlabeled; DEB complex formed in this mixture was therefore not detected. The protection pattern was determined for both strands of the uncleaved transposon end but they were identical to the bPEC (Figure 4, panel 4).
DEB complex was prepared under standard conditions and the hydroxyl radical footprint was determined for the non-transferred strand (Figure 4, panel 5, right). The footprint extended to within 5 bp of the cleaved transposon end before it was obscured by the large amount of DNA that had not reacted with the hydroxyl radicals. However, this was sufficient to show that the transposase and IHF footprints in the DEB are virtually identical to the bPEC.
IHF-independent interaction of transposase with the inverted repeat
The inside end of IS10 lacks an IHF consensus site. A requirement for IHF binding at only one end of the transposon can be inferred from rescue of the activity of a mini-IS10 on nicked plasmid DNA (R.Chalmers and N.Kleckner, unpublished). However, interpretation of this result is complicated by the degenerate IHF consensus sequence and an affinity for distorted DNA such as is present at both ends of IS10. Indeed, distortion of the outside end is demonstrated here by the ghost image of the PEC footprint on the free DNA (Figure 2). Furthermore, in mixed PECs with a single outside end, the footprints reveal a weak interaction between IHF and the partner inside end or ‘IHF-down’ mutant end (not shown). This raised the possibility that IHF must interact with both transposon ends when they are recruited into the developing synaptic complex.
To circumvent the problem of weak IHF interactions a new transposon end was constructed in which the DNA between bp +19 and +47 was replaced by a tandem repeat of the bases 5′-CTGA. This is referred to as an ‘even-end’ because of the evenness of the hydroxyl radical footprint in the presence of a high concentration of IHF. Labeled even-end fails to form PEC on its own but is recruited into mixed PEC when an outside end partner is added (not shown). The footprint of the even-end in the mixed PEC in the presence of a saturating amount of IHF shows no evidence of IHF binding and is identical to the tPEC in the three regions protected by transposase (Figure 4, panel 5, left). This confirms that one of the two transposon ends recruited into a developing transpososome is recognized by transposase independently of a transient interaction with IHF, and that IHF is strictly required at only one end of the transposon. Also, the transposase contacts in the tPEC, which has been stripped of IHF, are identical to the even-end, which was always free of IHF.
Subterminal transposase contacts beyond the IHF site
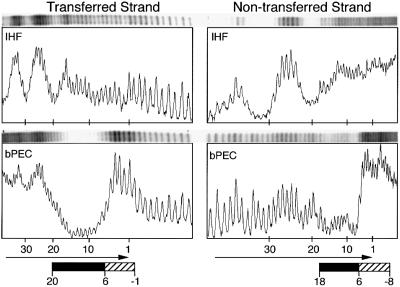
A mixed bPEC containing an even-end and a labeled outside end was assembled under standard conditions and the hydroxyl radical footprint was determined for the outside end partner (Figure 5, but see fifth section of Discussion for the reason why an even-end partner was required). Under these conditions, an additional region of protection becomes apparent beyond the IHF site, centered on bp +50 and bp +60 of the transferred strand. We can infer that this is due to transposase contacting the DNA because it is absent from the free and the IHF-shifted DNA (also see molecular modeling below). In principle, these contacts could be the result of transposase-dependent recruitment of an additional IHF protomer. However, this is not likely as there was no evidence of an additional IHF-dependent gel shift for this complex. Finally, we note that the subterminal contacts, together with those at the transposon end, have the potential to define a microdomain encompassing the IHF binding site, to constrain the torsion proposed in the ‘molecular spring’ model, to eject IHF from the developing transpososome (Chalmers et al., 1998; Sakai et al., 2000).
Fig. 5. Hydroxyl radical footprint of the subterminal regions beyond the IHF site. Regions judged to be protected by IHF or transposase are indicated by filled and shaded boxes, respectively. Nucleotide positions are as defined in Figure 2. Ends, uncomplexed transposon end fragment; IHF, IHF-bound transposon end fragment.
Elimination of the subterminal contacts
To investigate the function of the subterminal transposase contacts (Figure 5), a transposition reaction was performed with a 52 bp transposon end in order to deprive the complex of most, although not all, of the subterminal contacts at bp 48–65 (Figure 6). PEC assembly was reduced to ∼15% of that with the standard 87 bp transposon end, consistent with earlier experiments with 50 and 56 bp transposon ends (Sakai et al., 2000).
Fig. 6. Disruption of the subterminal contacts using a short transposon end fragment. The standard gel shift assay was performed using the standard BglII–SalI transposon end fragment or a short fragment with only 52 bp of transposon DNA. Assignment of the bands is as in Figure 1B. With the standard fragment, the quantity of bDEB appears greater than the cleaved flanking DNA because the SalI site is labeled more efficiently than the BglII site. The short end is only labeled on the flanking DNA because the opposite end is blunt.
Previously, the cleavage step of the reaction has always proven to be extremely robust. However, with the short transposon end, 35% of the PEC remained uncleaved even after 3 h incubation with Mg2+. Also, the remaining uncleaved PEC appeared to have undergone a conformational change as evidenced by the increase in mobility through the gel (Figure 6). Presumably, the conformational change is an aberrant version of the metal ion-dependent conformational changes at bp 1 and in the flanking DNA (above and Discussion), resulting from the defect caused by the absence of the subterminal contacts.
Discussion
General transpososome architecture
The other transposons that have been examined all have helically phased regions of protection, but most are much less extensive than for Tn10. One exception, γδ, a replicative transposon unrelated to Tn10, also has an IHF binding site adjacent to the inverted repeat, and helically phased contacts cover ∼80 bp of DNA. In this case, the transposase and IHF contacts are not on opposite faces of the helix but are rotated by ∼70° (Wiater and Grindley, 1991).
After assembly of the bPEC, many protein binding sites are not needed for the rest of the reaction. When IHF exits from the complex, the tPEC retains only two regions of protection and is very similar to the related Tn5 (Table I). Protein binding sites that have a role only during the assembly of an active complex feature in many reactions, including phage Mu, DNA inversion, and the initiation of transcription and replication (for example, Baker and Mizuuchi, 1992; Watson and Chaconas, 1996; Travers and Muskhelishvili, 1998 and references therein). In many cases the purpose of these extra sites is thought to be a combination of regulation and as chaperones to ensure the fidelity of the reaction.
Table I. Summary of interference and protection for Tn10 and Tn5.
| HO interference |
HO protection |
|||
|---|---|---|---|---|
| TS | NTS | TS | NTS | |
| bPEC | ||||
| T’ase | 63 to 58a | 65 to 61a,b | ||
| T’ase | 51 to 48a | 54 to 50a | ||
| IHF | 42 to 36 | 43 to 38 | 41 to 37a | 44 to 40a |
| IHF | 32 to 28 | 34 to 29 | 32 to 27a | 35 to 29a |
| IHF | 23 to 17 | 23 to 19 | 22 to 18a | 23 to 19a |
| T’ase | 20 to 7 | 18 to 7 | 18 to 14 | 19 to 15 |
| T’ase | 7 to 2 | 9 to 4 | ||
| T’ase | 5 to –1 enh. | 6 to –8 enh. | –6 to –9b | –4 to –7c |
| Tn5 | 20 to 7 | 20 to 7 | 17 to 13 | 19 to 6 |
| Mono. | 9 to 8 | 4 to 1 enh. | ||
| 5 to –1 enh. | 1 to –15 enh. | –6 to –10 enh. | ||
| Tn5 | 30 to 21 enh. | |||
| PEC | 20 to 5 | 20 to 4 | 17 to 13 | 20 to 16 |
| 8 to 5 | 11 to 2 | |||
| 3 to –2 enh. | 1 to –3 enh. | 3 to –2 enh. | ||
Hydroxyl radical interference and protection for the Tn10 bPEC, Tn5 monomer and Tn5 PEC. Tn5 data was abstracted from the figures in Bhasin et al. (2000).
aLost in conversion from bPEC to tPEC.
bRegion of very weak protection.
cLost in the presence of Ca2+.
enh., regions of enhancement.
Mono., monomer.
TS, transferred strand.
NTS, non-transferred strand.
A model of the IHF-bent end of Tn10 from the IHF cocrystal structure
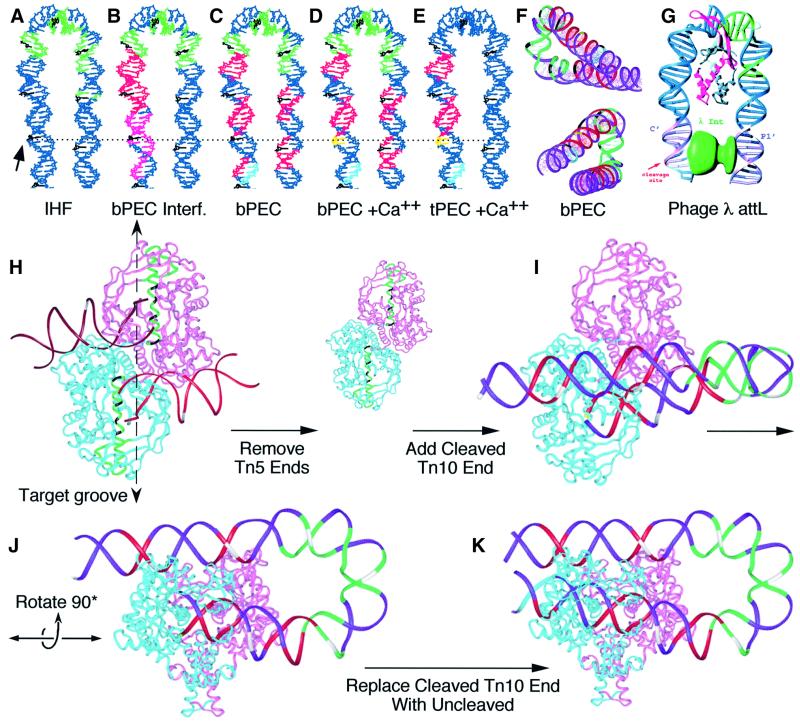
The crystal structure of IHF bound to attL of phage λ was used to model the shape of the IHF-bound outside end of Tn10 (Figure 7A–F; Rice et al., 1996). To build the model, the transposon end was aligned with attL using the IHF consensus present in both sequences. B DNA extensions were added to the DNA in the IHF cocrystal structure to represent bp –10 to +70 of Tn10. The model was colored to display the changing protein–DNA interactions as summarized in Table I.
Fig. 7. Model for the IHF-bent outside end of IS10 and summary of footprint data. A model for the IHF-bent outside end of IS10 was built by adding B DNA extensions to the structure of the IHF–attL cocrystal, as described in the Discussion. The solid arrow on the left and horizontal dashed line indicate the position of bp +1. Every tenth base on the transferred strand is colored black counting from bp +1 (A–F). (A) Hydroxyl radical protection by IHF is colored green. (B) Missing nucleotide interference for bPEC. Green, IHF interference; red, transposase interference; magenta, transposase enhancements. (C–E) IHF and transposase hydroxyl radical protection. Green, IHF; red, transposase; light blue, region of faint transposase protection; yellow, hyperreactivity induced by Ca2+. (F) The model from (C) was rendered with a ribbon backbone and displayed from two angles to convey the shape of the bend. (G) Model for λ integrase interaction with attL, based on the cocrystal structure of IHF and taken from Rice et al. (1996), with permission from Elsevier Science. (H) Ribbon model of the Tn5 cocrystal. On the transposon ends, bp +10 of the transferred strand is highlighted in white. The most highly conserved region of the protein between Tn5 and Tn10 is rendered in green with the ‘YREK’ residues highlighted in black. (I and J) The IHF-bent end of Tn10 from (C) was superimposed on the Tn5 structure by minimizing the RMS difference in the position of atoms at bp 6–12 of the non-transferred strand of both transposon ends. Some regions of the DNA in the Tn5 structure are distorted and the region at bp 6–12 most nearly matched the structure of canonical B DNA. The flanking DNA has been truncated to match the Tn5 ends. Colors summarize the protein–DNA contacts as defined in (C) and every tenth bp on the transferred strand is highlighted in white. (K) Same view as (J), but flanking DNA has been rendered.
Beyond the IHF site, there are two regions of protection, between bp +48 and +65, that are dependent on both IHF and transposase (Figure 5; Table I). The present model (Figure 7) gives several very clear indications that these are due to transposase contacts with the DNA: (i) the sub terminal protections are located exactly opposite the end of the transposon where transposase binds; (ii) the sub terminal protections are phased 10 bp apart, on the same face of the helix, oriented toward the middle of the folded structure where they could present a surface to contact a protein; (iii) the distance between the inverted repeat and the subterminal protections in the model is <20Å, a distance easily spanned by a dimer of transposase estimated to measure ∼100 × 70 × 50Å based on the crystal structure of Tn5 transposase (Davies et al., 2000).
Congruent structures at the Tn10 and phage λ recombination sites
Previously, the DNA in the IHF cocrystal structure was extended (see above) to model the juxtaposition of the C′ and P′1 sites in nucleoprotein array at phage λ attL (Figure 7G; Rice et al., 1996). The model showed that IHF would facilitate binding of the large and small domains of a single λ integrase protomer to sites C′ and P′1, respectively. The model of the Tn10 outside end revealed an interesting congruence with phage λ attL. In both systems the site of DNA cleavage is on the same strand 30 bases 3′ to the start of the IHF consensus. Also, the center of the Tn10 subterminal protection at bp +58 is at exactly the same position as the center of the λ P′1 site, with respect to the cleavage site.
Conservation of the relative locations of the three subsites in the λ and Tn10 recombinosomes is not likely to be due to descent from a common ancestor. More probably, IHF per se imposes a powerful constraint on the location of two sites that have to be brought into close juxtaposition. The molecular models (Figure 7) point to one potentially powerful constraint. The major grooves of the DNA at each of the juxtaposed sites face directly towards each other across the middle of the folded structure. This arrangement allows a protein that binds in the major groove of the DNA to span the distance between the pair of sites. To fulfill this criterion at an alternative location, both sites would have to be moved either 10 bp towards or away from the center of the IHF bend. Moving the sites further apart would tend to degrade the specificity inherent in the system, either by allowing greater flexibility in the spatial arrangement of components or by allowing for the redistribution of mechanical stress in the DNA between untwisting and writhe.
A model of the Tn10 bPEC from the Tn5 cocrystal structure
The coordinates of the cocrystal structure of Tn5 transposase were used to model the geometric relationship between the IHF-bent arm of Tn10 and the synaptic complex (Figure 7H–K). Members of the DDE family of transposons are likely to all share a common structural core (Rice and Mizuuchi, 1995; Mahillon and Chandler, 1998; Davies et al., 2000), but further evidence for the similarity between Tn10 and Tn5 transposases comes from the identical target site duplications and the identity of the three terminal nucleotides, which are important to allow the chemistry of the reaction to take place efficiently (Huisman et al., 1989; Haniford and Kleckner, 1994). To support further the relationship between Tn10 and Tn5, the BLAST algorithm was used to identify the most similar regions of the proteins. The most highly significant region of the alignment contains the ‘YREK’ motif and forms a very long α-helix that spans the full width of the dimer and appears to define the lip and bottom of the target binding groove (amino acids 298–334 of the Tn5 sequence, highlighted in green in Figure 7H). Taken together with the conserved target site duplication, it seems likely that the spatial relationship between the transposon ends and active site will be similar for Tn5 and Tn10.
The model for the bPEC was created by substituting the IHF-bent arm of Tn10 from Figure 7C in place of one of the Tn5 ends. Superimposition of the Tn10 end was achieved by minimizing the root mean square (RMS) difference in the position of the equivalent atoms in the Tn5 DNA. The hydroxyl radical protection and interference patterns for Tn10 and Tn5 are very similar in the terminal 20 bp and the flanking DNA (Table I) and correspond well with the protein–DNA contacts in the model (Figure 7K). However, Tn5 lacks an IHF binding site and the subterminal transposase contacts of Tn10. Remarkably, the subterminal hydroxyl radical protections in the folded arm of Tn10 are perfectly located to interact with symmetrical positions on either side of the protein dimer interface (Figure 7J). In the model, the location of the folded arm relative to the target binding groove and the active site explains most of the regulatory features of the Tn10 transpososome as discussed below.
Subterminal contacts function during transpososome assembly and cleavage
A transposition reaction was performed with a short transposon end to test the effect of abolishing most of the subterminal contacts (Figure 6). PEC formation was reduced by 85% compared with the standard end, ∼35% of the complex remained uncleaved after 3 h incubation with Mg2+ and the remaining uncleaved bPEC had increased mobility in the gel. The subterminal contacts thus appear to be important during assembly of the transpososome and also during a subsequent step in which the complex is activated for cleavage, possibly during a metal ion-dependent conformational change (below).
The importance of the subterminal contacts for the cleavage step of the reaction raises a paradox about the behavior of the tPEC. The tPEC arises by loss of IHF from the bPEC, yet despite the complete absence of protein contacts beyond bp +20, the kinetics of cleavage are indistinguishable from the bPEC (Figure 4; Sakai et al., 2000). This means that the subterminal contacts must go through at least one cycle of engagement and disengagement to activate the PEC for the chemical steps of the reaction. Disengagement of the subterminal contacts must therefore leave the transpososome in a different state than if the subterminal contacts are never engaged.
In the model of the bPEC (Figure 7I–K), the single IHF-bent arm of the transposon passes across the middle of the dimer interface. A second folded transposon arm can not be accommodated in the structure because the second set of subterminal contacts would occupy almost the same space as those on the first transposon end. This explains why the subterminal contacts were indistinct when footprinted on an OE × OE complex, but became clear when the complex contained an unlabeled even-end and a single labeled outside end. The transpososome, therefore, appears to accommodate only one folded transposon end, which is analogous to the transposition of the IS10 element in which the inside end lacks an IHF binding site.
Regulation by IHF
Evidence that the DNA in the bPEC was tightly wrapped led to the development of a model for PEC assembly on supercoiled substrates in which DNA bending extrudes a single toroidial node that provides a plectonemic branch point mechanistically required for synapsis of the ends (Chalmers et al., 1998). Transposase itself is assumed to lack the binding energy to extrude the node unless aided by negative supercoiling or IHF. Our finding, that IHF binding is strictly required at one transposon end only (Figure 4), fits well with synapsis in a branched plectosome. However, one inconsistency in this model is that Tn10 recombination is equally efficient with ends configured either as direct or inverted repeats (Chalmers and Kleckner, 1996). If synapsis occurred at a branched plectosome, one of the two configurations would be likely to introduce an energetically unfavorable DNA tangle, depending on whether the synapsis of Tn10 ends is largely in parallel or antiparallel.
In vitro, with oligonucleotide substrates, IHF is essential for PEC assembly and inhibits target interaction after the cleavage step (Junop and Haniford, 1997; Sakai et al., 2000). On plasmid substrates, Tn10 transposition is dependent on the presence of either IHF or supercoiling. However, when both are present, at moderate to high levels, intermolecular target interactions are inhibited and the reaction is channeled into intramolecular events (Chalmers et al., 1998). The molecular model of the bPEC (Figure 7I–K) suggests a physical basis for all of these observations, except for channeling of the reaction by IHF to target sites extremely close to the transposon end (130 bp).
First, transpososome assembly depends on IHF because of the subterminal contacts and their location and orientation with respect to the end of the transposon. The subterminal contacts are located close to the transposon end, oriented towards the middle of the complex, forcing the DNA to adopt a 180° bend. From the model it is easy to visualize why a contribution from the energy of IHF binding or negative supercoiling is needed to establish the contacts. Secondly, the inhibition of target interactions is the result of the IHF-bent arm of the transposon passing directly across the top of the positively charged target binding groove. The subterminal regions of protection even appear to engage the rim of the target groove and clearly preclude target entry. Overall, the activity of IHF in promoting transpososome assembly and inhibiting target interactions can be explained as consequences of DNA bending and twisting, rather than an allosteric protein– protein interaction.
Interference enhancements indicate distorted flanking DNA
In the missing nucleotide interference assay for bPEC assembly (Figure 3), the strong enhancements in the terminal and flanking nucleotides probably reflect a distortion of the DNA. Presumably, these positions are over represented in the bPEC because the increased flexibility overcomes a limitation on PEC assembly imposed by the energy required to bend the DNA around the complex. The idea that distortion of the flanking DNA is one factor limiting assembly and stability of the PEC is supported by the observation that the DEB assembles more efficiently and is more stable than the PEC (Sakai, 1996; P.Crellin and R.Chalmers, unpublished). In vivo, perhaps supercoiling in the flanking DNA would normally help to overcome any such limitation. Although at a different stage of the reaction, we note that the superhelical density in the flanking DNA appears to affect the kinetics of Mu transposition following synapsis of the ends (Wang and Harshey, 1994).
The bPEC is an intermediate, trapped in the absence of divalent metal ions
Ca2+ induces substantial changes in the protection pattern at bp +1 and in the flanking DNA of the bPEC (Figure 4). Three lines of evidence suggest that this is a reflection of a conformational change that normally precedes the first chemical step of the transposition reaction: (i) hyper reactivity at bp +1 is restricted to the transferred strand, which is the site of the first nick at the transposon end. The non-transferred strand is not hyperreactive and is always cleaved last because of the involvement of a hairpin intermediate (Bolland and Kleckner, 1995; Kennedy et al., 1998). (ii) Ca2+ has similar electrochemical properties to Mg2+ and is likely to be properly liganded in the active site, although it does not support catalysis. (iii) Mutations in the DDE residues abolish hyperreactivity (P.Crellin, S.Sewitz and R.Chalmers, to be presented elsewhere), suggesting that the effect of Ca2+ is specific and not due to ionic interactions altering the properties of the DNA. Together, these three lines of evidence suggest that the hyperreactivity and loss of protection in the flanking DNA are a direct result of a conformational change triggered by Ca2+ entering the active site.
Is the conformation of the bPEC, as determined in the absence of metal ion, a natural intermediate of the reaction that exists fleetingly before metal ions enter the active site? For example, it is possible that the hyperreactivity that develops at bp +1 is only a trivial consequence of preparing the complex in the absence of metal ion, accompanied by a change in the electrochemical environment of the active site as Ca2+ enters. We do not believe this to be the case for the reasons given above and because the hyperreactivity that develops at bp +1 is accompanied by loss of the protection centered on bp –5 of the opposite strand. This is the same location as the hydroxyl radical interference enhancements (Figure 3) that have been interpreted as a result of a transposase-dependent bend in the flanking DNA (above). Based on the concurrence of the interference enhancements and protections in the flanking DNA, it therefore appears that the flanking DNA is initially bent around the complex until released by metal ions entering the active site.
Whether or not the flanking DNA is ever bent around the complex as suggested depends on when the catalytic metal ions enter the complex. Analogously to the way in which Mg.ATP is the cosubstrate for the ATPase enzymes, transposases probably do not bind metal ion before the substrate. In the transition state of the two metal ion catalytic mechanism used by Tn10, the metal ions have as many points of coordination with the phosphate oxygens on the DNA as with the DDE residues (Beese and Steitz, 1991; Joyce and Steitz, 1995; Kennedy et al., 2000). It is therefore highly unlikely that both metal ions can be properly located in the active site before the substrate. Taken together, we believe that the conformation of the bPEC is a natural intermediate of the transposition reaction which exists transiently before entry of the catalytic metal ion triggers an extensive conformational change.
Materials and methods
Materials were generally of the best quality available from commercial suppliers. Most chemicals were from Sigma. Enzymes were from New England Biolabs. Molecular models were constructed on a Crystal Graphics work station using the program InsightII.
DNA, proteins and PEC assembly
End fragments were derived from the outside end of IS10-Right. The BglII–SalI fragment from pNK1935, containing 87 bp of transposon and 50 bp of flanking DNA (Sakai et al., 1995), was cloned into BamHI–SalI-digested pBluescript to yield pRC98. Fragments labeled on the transferred or non-transferred strand were generated from XbaI–HincII (151 bp) or XhoI–SacI (181 bp) digests, respectively. Pre-cleaved ends were generated by PvuII–BstEII digestion of pRC35 created by ligating the ETF from pNK925 (Chalmers et al., 1998) into PvuII-digested pBluescript. The even-end fragment (118 bp) contained bp 1–19 of IS10-R followed by seven tandem repeats of the sequence CTGA cloned into pBluescript to yield pRC100. Fragments were end labeled using Klenow polymerase and [α-32P]nucleotides.
Transposase was purified as described (Chalmers and Kleckner, 1994) and IHF was the gift of Howard Nash. PECs were prepared and analyzed by gel shift also as described (Sakai et al., 1995). When added, CaCl2 and heparin were 4 mM and 250 ng/ml, respectively.
Hydroxyl radical footprinting and analysis of footprints
PECs were assembled at room temperature for 3 h to overnight and drop dialyzed against 10 mM Tris–HCl pH 7.5. Footprinting caused a small fraction of the complexes to dissociate, and control footprints of the IHF-shifted and free DNA fragments were therefore prepared from binding reactions from which transposase was omitted. Hydroxyl radical footprinting was as described by Schickor and Heumann (1994) and samples were concentrated using a Microcon YM-100 filter (Millipore). Following gel shift analysis (above) the complexes were identified by autoradiography, excised from the gel and recovered by the crush-and-soak method. Acrylamide was removed using a 0.22 µm Costar Spin-X filter and the DNA was recovered by ethanol precipitation. DNA was resuspended in 90% formamide loading buffer and then analyzed on 15% acrylamide/TBE sequencing gels. The resulting footprints were displayed by autoradiography and/or a Molecular Dynamics PhosphorImager. The relative intensities of the bands within the resultant ladders were plotted using the computer program NIH Image. Maxam–Gilbert G + A sequence ladders were prepared as described (Maxam and Gilbert, 1980).
Missing nucleosides
Labeled end fragments were treated with hydroxyl radicals as described above, precipitated with ethanol and resuspended in TE buffer to ∼100 000 c.p.m./µl. Complexes were assembled from these fragments, subjected to PAGE and isolated from the gel. The DNA was precipitated with ethanol, loaded on a sequencing gel and the resultant ladders were analyzed as described above.
Acknowledgments
Acknowledgements
We thank Phoebe Rice for critical comments, providing coordinates and valuable help with modeling; David Haniford for valuable opinion on metal ions; Yuying Liu, Nicolas Buisine and Sven Sewitz for technical help. IHF was a gift from Howard Nash. This work was funded by The Wellcome Trust. R.C. is a Royal Society University Research Fellow.
References
- Baker T.A. and Mizuuchi,K. (1992) DNA-promoted assembly of the active tetramer of the Mu transposase. Genes Dev., 6, 2221–2232. [DOI] [PubMed] [Google Scholar]
- Beese L.S. and Steitz,T.A. (1991) Structural basis for the 3′–5′ exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. EMBO J., 10, 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhasin A., Goryshin,I.Y., Steiniger-White,M., York,D. and Reznikoff,W.S. (2000) Characterization of a Tn5 pre-cleavage synaptic complex. J. Mol. Biol., 302, 49–63. [DOI] [PubMed] [Google Scholar]
- Bolland S. and Kleckner,N. (1995) The two single-strand cleavages at each end of Tn10 occur in a specific order during transposition. Proc. Natl Acad. Sci. USA, 92, 7814–7818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolland S. and Kleckner,N. (1996) The three chemical steps of Tn10/IS10 transposition involve repeated utilization of a single active site. Cell, 84, 223–233. [DOI] [PubMed] [Google Scholar]
- Chalmers R.M. and Kleckner,N. (1994) Tn10/IS10 transposase purification, activation and in vitro reaction. J. Biol. Chem., 269, 8029–8035. [PubMed] [Google Scholar]
- Chalmers R.M. and Kleckner,N. (1996) IS10/Tn10 transposition efficiently accommodates diverse transposon end configurations. EMBO J., 15, 5112–5122. [PMC free article] [PubMed] [Google Scholar]
- Chalmers R., Guhathakurta,A., Benjamin,H. and Kleckner,N. (1998) IHF modulation of Tn10 transposition: sensory transduction of supercoiling status via a proposed protein–DNA molecular spring. Cell, 93, 897–908. [DOI] [PubMed] [Google Scholar]
- Chalmers R., Sewitz,S., Lipkow,K. and Crellin,P. (2000) Complete nucleotide sequence of Tn10. J. Bacteriol., 182, 2970–2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies D.R., Goryshin,I.Y., Reznikoff,W.S. and Rayment,I. (2000) Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science, 289, 77–85. [DOI] [PubMed] [Google Scholar]
- Hagan N.F., Vincent,S.D., Ingleston,S.M., Sharples,G.J., Bennett,R.J., West,S.C. and Lloyd,R.G. (1998) Sequence-specificity of Holliday junction resolution: identification of RuvC mutants defective in metal binding and target site recognition. J. Mol. Biol., 281, 17–29. [DOI] [PubMed] [Google Scholar]
- Haniford D. and Kleckner,N. (1994) Tn10 transposition in vivo: temporal separation of cleavages at the two transposon ends and roles of terminal basepairs subsequent to interaction of ends. EMBO J., 13, 3401–3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisman O., Errada,P.R., Signon,L. and Kleckner,N. (1989) Mutational analysis of IS10’s outside end. EMBO J., 8, 2101–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce C.M. and Steitz,T.A. (1995) Polymerase structures and function: variations on a theme? J. Bacteriol., 177, 6321–6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junop M.S. and Haniford,D.B. (1996) Multiple roles for divalent metal ions in DNA transposition: distinct stages of Tn10 transposition have different Mg2+ requirements. EMBO J., 15, 2547–2555. [PMC free article] [PubMed] [Google Scholar]
- Junop M.S. and Haniford,D.B. (1997) Factors responsible for target site selection in Tn10 transposition: a role for the DDE motif in target DNA capture. EMBO J., 16, 2646–2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy A.K., Guhathakurta,A., Kleckner,N. and Haniford,D.B. (1998) Tn10 transposition via a DNA hairpin intermediate. Cell, 95, 125–134. [DOI] [PubMed] [Google Scholar]
- Kennedy A.K., Haniford,D.B. and Mizuuchi,K. (2000) Single active site catalysis of the successive phosphoryl transfer steps by DNA transposases: insights from phosphorothioate stereoselectivity. Cell, 101, 295–305. [DOI] [PubMed] [Google Scholar]
- Kleckner N. (1989) Transposon Tn10. In Berg,D.E. and Howe,M.M. (eds), Mobile DNA. American Society for Microbiology, Washington, DC, pp. 227–268.
- Kleckner N., Chalmers,R.M., Kwon,D.K., Sakai,J. and Bolland,S. (1996) Tn10 and IS10 transposition and chromosome rearrangements: mechanism and regulation in vivo and in vitro. In Saedler,H. and Gierl,A. (eds), Transposable Elements. Vol. 204. Springer, Berlin, pp. 49–82. [DOI] [PubMed]
- Macgregor R.B. Jr (1992) Photogeneration of hydroxyl radicals for footprinting. Anal. Biochem., 204, 324–327. [DOI] [PubMed] [Google Scholar]
- Mahillon J. and Chandler,M. (1998) Insertion sequences. Microbiol. Mol. Biol. Rev., 62, 725–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A.M. and Gilbert,W. (1980) Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol., 65, 499–560. [DOI] [PubMed] [Google Scholar]
- Rice P. and Mizuuchi,K. (1995) Structure of the bacteriophage Mu transposase core: a common structural motif for DNA transposition and retroviral integration. Cell, 82, 209–220. [DOI] [PubMed] [Google Scholar]
- Rice P.A., Yang,S., Mizuuchi,K. and Nash,H.A. (1996) Crystal structure of an IHF–DNA complex: a protein-induced DNA U-turn. Cell, 87, 1295–1306. [DOI] [PubMed] [Google Scholar]
- Roberts D., Hoopes,B.C., McClure,W.R. and Kleckner,N. (1985) IS10 transposition is regulated by DNA adenine methylation. Cell, 43, 117–130. [DOI] [PubMed] [Google Scholar]
- Sakai J. and Kleckner,N. (1997) The Tn10 synaptic complex can capture a target DNA only after transposon excision. Cell, 89, 205–214. [DOI] [PubMed] [Google Scholar]
- Sakai J., Chalmers,R.M. and Kleckner,N. (1995) Identification and characterization of a pre-cleavage synaptic complex that is an early intermediate in Tn10 transposition. EMBO J., 14, 4374–4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai J.F. (1996) Characterization of protein:DNA complexes in Tn10 transposition. PhD thesis, Department of Molecular and Cellular Biology, Harvard University, Cambridge, MA.
- Sakai J.S., Kleckner,N., Yang,X. and Guhathakurta,A. (2000) Tn10 transpososome assembly involves a folded intermediate that must be unfolded for target capture and strand transfer. EMBO J., 19, 776–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schickor P. and Heumann,H. (1994) Hydroxyl radical footprinting. In Kneale,G.G. (ed.) DNA–Protein Interactions: Principles and Protocols. Humana Press, Totowa, NJ, pp. 21–32.
- Signon L. and Kleckner,N. (1995) Negative and positive regulation of Tn10/IS10-promoted recombination by IHF: two distinguishable processes inhibit transposition off of multicopy plasmid replicons and activate chromosomal events that favor evolution of new transposons. Genes Dev., 9, 1123–1136. [DOI] [PubMed] [Google Scholar]
- Travers A. and Muskhelishvili,G. (1998) DNA microloops and microdomains: a general mechanism for transcription activation by torsional transmission. J. Mol. Biol., 279, 1027–1043. [DOI] [PubMed] [Google Scholar]
- Wang Z. and Harshey,R.M. (1994) Crucial role for DNA supercoiling in Mu transposition: a kinetic study. Proc. Natl Acad. Sci. USA, 91, 699–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson M.A. and Chaconas,G. (1996) Three-site synapsis during Mu DNA transposition: a critical intermediate preceding engagement of the active site. Cell, 85, 435–445. [DOI] [PubMed] [Google Scholar]
- Wiater L.A. and Grindley,N.D. (1991) γδ transposase. Purification and analysis of its interaction with a transposon end. J. Biol. Chem., 266, 1841–1849. [PubMed] [Google Scholar]
- Yang C.C. and Nash,H.A. (1989) The interaction of E.coli IHF protein with its specific binding sites. Cell, 57, 869–880. [DOI] [PubMed] [Google Scholar]