Abstract
PKB/Akt, S6K1 and SGK are related protein kinases activated in a PI 3-kinase-dependent manner in response to insulin/growth factors signalling. Activ ation entails phosphorylation of these kinases at two residues, the T-loop and the hydrophobic motif. PDK1 activates S6K, SGK and PKB isoforms by phosphorylating these kinases at their T-loop. We demonstrate that a pocket in the kinase domain of PDK1, termed the ‘PIF-binding pocket’, plays a key role in mediating the interaction and phosphorylation of S6K1 and SGK1 at their T-loop motif by PDK1. Our data indicate that prior phosphorylation of S6K1 and SGK1 at their hydrophobic motif promotes their interaction with the PIF-binding pocket of PDK1 and their T-loop phosphorylation. Thus, the hydrophobic motif phosphorylation of S6K and SGK converts them into substrates that can be activated by PDK1. In contrast, the PIF-binding pocket of PDK1 is not required for the phosphorylation of PKBα by PDK1. The PIF-binding pocket represents a substrate recognition site on a protein kinase that is only required for the phosphorylation of a subset of its physiological substrates.
Keywords: AGC kinase/docking sites/PKC/protein kinase/RSK
Introduction
Protein kinases are key mediators of diverse cellular signalling pathways. Thus, the phosphorylation of a substrate by a given protein kinase following a stimulus, must be efficient, specific and tightly regulated to avoid cross-talk between different signalling pathways and to ensure the accurate timing of the phosphorylation. However, the molecular basis for regulating the efficiency and fidelity of protein kinases has been poorly understood, until recently. There is now increasing evidence that one mechanism that enables a protein kinase to phosphorylate a specific substrate involves residues of the protein kinase that lie outside the active centre which can interact specifically with the substrate (referred to as docking sites) (Holland and Cooper, 1999; Tanoue et al., 2001). There is growing interest in characterizing the specific interactions between kinases and their substrates, as these are likely to play important roles in controlling the specificity and function of signal transduction pathways.
In the insulin and growth factors signalling pathways, phosphoinositide 3-kinase (PI 3-kinase) family members are activated, leading to the generation of the lipid second messengers phosphatidylinositol-3,4,5-triphosphate [Ptd Ins(3,4,5)P3] and PtdIns(3,4)P2 (Vanhaesebroeck and Alessi, 2000). One of the mechanisms by which these 3-phosphoinositides regulate cellular processes is by their ability to induce the phosphorylation, and hence the activation, of members of the AGC subfamily of protein kinases (Belham et al., 1999). These include isoforms of protein kinase B (PKB) (Vanhaesebroeck and Alessi, 2000), p70 ribosomal S6 kinase (S6K) (Dufner and Thomas, 1999) and serum and glucocorticoid induced kinase (SGK) (Kobayashi and Cohen, 1999; Kobayashi et al., 1999; Park et al., 1999).
PKB is activated usually within 2 min of a cell being stimulated with insulin and growth factors. It possesses an N-terminal pleckstrin homology (PH) domain that interacts with PtdIns(3,4,5)P3/PtdIns(3,4)P2, resulting in the recruitment of PKB to the plasma membrane where it becomes activated by the phosphorylation of two residues. One lies in the T-loop (also known as the activation loop) of the kinase domain (Thr308 in PKBα) and the other is located C-terminal to the catalytic domain, in a region termed the ‘hydrophobic motif’ (Ser473 in PKBα; reviewed in Vanhaesebroeck and Alessi, 2000). S6K and SGK also possess residues equivalent to Thr308 (Thr252 in S6K1 and Thr256 in SGK1) and Ser473 (Thr412 in S6K1 and Ser422 in SGK1), whose phosphorylation is required for activation of these kinases in vivo. The phosphorylation of S6K and SGK at both their T-loop and hydrophobic motifs, like that of PKB, is dependent upon PI 3-kinase activation. However, in contrast to PKB, S6K and SGK do not possess a PH domain and do not interact with PtdIns(3,4,5)P3/PtdIns(3,4)P2. S6K and SGK isoforms are also activated more slowly than PKBα following cell stimulation, with maximal activation occurring typically after 10–40 min.
PKB, S6K1 and SGK are phosphorylated at their T-loop by the 3-phosphoinositide-dependent protein kinase-1 (PDK1) (Belham et al., 1999; Toker and Newton, 2000). PDK1 is also an AGC family member, and possesses a PtdIns(3,4,5)P3/PtdIns(3,4)P2-binding PH domain C- terminal to the catalytic domain. Following PI 3-kinase activation, PDK1 and PKB are thought to co-localize at the plasma membrane through their interaction with PtdIns(3,4,5)P3/PtdIns(3,4)P2. In addition to recruiting PKB to cell membranes, the binding of PtdIns(3,4,5)P3/PtdIns(3,4)P2 to the PH domain of PKB may induce a conformational change that enables PDK1 to phosphorylate Thr308 (reviewed in Vanhaesebroeck and Alessi, 2000). As S6K and SGK do not interact with PtdIns(3,4,5) P3/PtdIns(3,4)P2, nor is the rate at which they are phosphorylated by PDK1 in vitro enhanced in the presence of PtdIns(3,4,5)P3/PtdIns(3,4)P2, the mechanism by which activation of PI 3-kinases induces activation of S6K and SGK is likely to be distinct from PKB.
The kinase domain of PDK1 was found in a yeast two-hybrid screen to interact with a region of the protein kinase C-related kinase-2 (PRK2), termed the PDK1 interacting fragment (PIF) (Balendran et al., 1999a). PRK2 is an AGC kinase that, unlike PKB, SGK and S6K isoforms, is not activated following stimulation of cells with agonists that activate PI 3-kinase, and may be regulated by the Rho family of GTPase proteins (Flynn et al., 2000). The PIF region in PRK2 is situated C-terminal to the kinase domain of PRK2, and contains a hydrophobic motif (Phe-Xaa-Xaa-Phe-Asp-Tyr), similar to that found in PKBα (Phe-Xaa-Xaa-Phe-Ser-Tyr), except that the residue equivalent to Ser473 is Asp. Mutation of the conserved aromatic residues in the hydrophobic motif of PIF or mutation of the Asp residue to either Ala or Ser inhibits the interaction of PIF with PDK1 (Balendran et al., 1999a). Subsequent work demonstrated that a 24 amino acid fragment of PIF (termed PIFtide) that encompasses the hydrophobic motif of PRK2 bound to a hydrophobic pocket on the small lobe of the kinase domain of PDK1, which was termed the ‘PIF-binding pocket’ (Biondi et al., 2000). Furthermore, some evidence was obtained that this site on PDK1 could serve as a ‘docking site’, enabling the recruitment of PDK1 by PRK2 (Balendran et al., 2000). PKCζ like PRK2 also possesses an acidic residue in its hydrophobic motif (Glu579) but, in contrast to PRK2, mutation of this residue to Ala does not affect its interaction with PDK1 (Balendran et al., 2000).
Another AGC kinase termed p90RSK, which is not activated downstream of the PI 3-kinase pathway but instead by the classical MAP kinase pathway (Frodin and Gammeltoft, 1999), is also a PDK1 substrate (Jensen et al., 1999; Richards et al., 1999). The phosphorylation of p90RSK by the ERK1/ERK2 MAP kinase members enables p90RSK to phosphorylate its own hydrophobic motif. Frodin and colleagues (Frodin et al., 2000) have recently demonstrated that PDK1 is able to interact directly with the phosphorylated hydrophobic motif of p90RSK, thus enabling PDK1 to phosphorylate the T-loop of p90RSK.
Here, we investigated the role of the PIF-binding pocket on PDK1 in enabling PDK1 to phosphorylate and activate three of its AGC kinase substrates which are activated by a PI 3-kinase-dependent mechanism, namely, S6K1, SGK1 and PKBα. Our data indicate that the PIF-binding pocket of PDK1 plays a critical role in enabling PDK1 to phosphorylate and activate S6K1 and SGK1, but not wild-type PKBα. The data suggests that the prior phosphorylation of S6K1 and SGK1 at their hydrophobic motif promotes the interaction and phosphorylation of these enzymes by PDK1. The PIF-binding pocket on PDK1 represents, to our knowledge, the first example of a substrate recognition site on a protein kinase that is only required for the phosphorylation of a subset of its physiological substrates.
Results
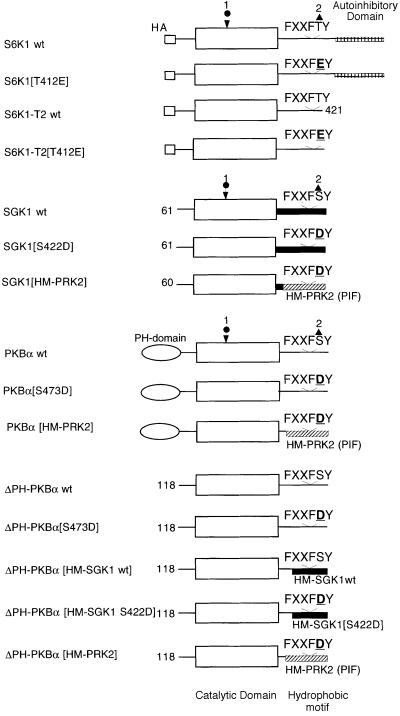
All the wild-type and mutant forms of AGC kinase substrates employed in this study are defined in Figure 1. The form of S6K that lacks the C-terminal 104 residues which encompass an autoinhibitory domain is called S6K1-T2 and is used in preference to the full-length S6K1 because it is an efficient substrate for PDK1 in vitro, in contrast to full-length S6K1, which is very poorly phosphorylated by PDK1 (Alessi et al., 1998; Pullen et al., 1998). The form of SGK1 used in this study lacks the N-terminal 60 amino acids as the full-length SGK1 protein cannot be expressed at sufficient levels to be useful as a substrate for PDK1 (Kobayashi and Cohen, 1999).
Fig. 1. The wild-type and mutant AGC kinases used in this study. The circle indicates the position of the T-loop phosphorylation site and the triangle marks the position of the hydrophobic motif phosphorylation site. wt indicates wild type. The C-terminal 104 residues of S6K1 encompass an autoinhibitory domain and this is deleted in the S6K1-T2 construct. The SGK1 protein expressed in this study lacks the 60 N-terminal amino acids. ΔPH-PKBα is a mutant of PKBα that lacks the PH domain. ΔPH-PKBα[HM-SGK1 wt] comprises residues 118–439 of PKBα fused to residues 288–431 of human SGK1. PKBα[HM-SGK1 S422D] comprises residues 118–439 of PKBα fused to residues 288–431 of human SGK1 in which the hydrophobic motif phosphorylation site Ser422 is changed to Asp. PKBα[HM-PRK2] and SGK1[HM-PRK2] comprise residues 118–425 of PKBα and residues 60–372 of SGK1, respectively, fused to residues 934–984 of human PRK2.
Role of the PDK1 ‘PIF-binding pocket’ on phosphorylation and activation of S6K1, SGK1 and PKB by PDK1
We first investigated the role of the hydrophobic PIF-binding pocket on PDK1 in enabling PDK1 to phosphorylate and activate S6K1, SGK1 and PKB. We initially tested whether a PDK1 mutant (PDK1[L155E]) in which the hydrophobic PIF-binding pocket has been disrupted (Biondi et al., 2000) could phosphorylate and activate these AGC kinase substrates. Strikingly, the phosphorylation of S6K1-T2 and SGK1 by PDK1[L155E] was drastically reduced compared with wild-type PDK1 (Figure 2A and B and Table I). We also employed mutants of S6K1 (S6K1-T2[T412E]) and SGK1 (SGK1[S422D]) in which their hydrophobic motif phosphorylation sites were changed to an acidic residue, which increased significantly the rate at which they were phosphorylated and activated by PDK1 (Table I). Although PDK1 phosphorylates S6K1-T2, this does not significantly activate this enzyme as it also requires phosphorylation of Thr412 (or mutation of this residue to a Glu) to become activated (Figures 2A and 3A). We found that S6K1-T2[T412E] and SGK1[S422D] were both phosphorylated and activated very poorly by PDK1[L155E] compared with wild-type PDK1 (Figure 2). In contrast, PKBα and its acidic hydrophobic motif mutant PKBα[S473D] were equally good substrates for wild-type PDK1 and PDK1[L155E] (Figure 2C). Interestingly, mutant forms of PKBα and PKBα[S473D] that lack the PH domain (ΔPH-PKBα and ΔPH-PKBα[S473D]) and are thus more similar in structure to S6K1-T2 and SGK1 (Figure 1) were very poor substrates for PDK1[L155E] compared with wild-type PDK1 (Figure 2D). ΔPH-PKBα was phosphorylated by PDK1 at a 50- to 100-fold lower rate than full-length PKBα (Table I), and its phosphorylation by PDK1, like that of S6K1-T2 and SGK1, was not influenced by PtdIns(3,4,5)P3 (Alessi et al., 1998; Kobayashi and Cohen, 1999). It should be noted, however, that ΔPH-PKBα was phosphorylated by PDK1 at a 10-fold lower initial rate than S6K1-T2 and SGK1, and ΔPH-PKBα[S473D] was phosphorylated at a ∼70-fold lower rate than S6K1-T2[T412E] and SGK1[S422D] (Table I).
Fig. 2. Role of the PIF-binding pocket of PDK1 in the phosphorylation of S6K1, SGK1 and PKBα. The indicated AGC substrates were incubated with wild-type GST–PDK1 or mutant GST–PDK1[L155E] in the presence of magnesium and [γ-32P]ATP as described in the Materials and methods. The activation of each AGC kinase was assessed by its ability to phosphorylate the peptide substrate Crosstide (GRPRTSSFAEG). Phosphorylation of the AGC kinase substrate was determined following electrophoresis on a 4–12% gradient polyacrylamide gel and the Coomassie Blue-staining bands corresponding to each substrate were analysed on a phosphoimager. Phosphorylation was also analysed by subjecting the AGC kinases to immuno blotting with antibodies that specifically detect the T-loop-phosphorylated form of S6K1 (Thr252), SGK1 (Thr256) and PKBα (Thr308). Under the conditions used, phosphorylation of each substrate by PDK1 was linear with time and with the amount of enzyme added to the assay. Experiments were performed in duplicate using at least two separate time points. Duplicates within one experiment did not vary >10%, and usually <5%.
Table I. Relative initial rates by which the indicated forms of S6K1, SGK1 and PKB are phosphorylated by wild type (wt) PDK1, PDK1[L155E] and PDK1[A277V].
| PDK1-wt | PDK1-L155E | PDK1-A277V | ||||
|---|---|---|---|---|---|---|
| PIFtide | – | + | – | + | – | + |
| S6K1-T2 | 10.4 | 0.58 | 0.6 | 0.27 | 6.9 | 0.6 |
| S6K1-T2[T412E] | 54 | 7.4 | 0.5 | 0.108 | 51 | 5.6 |
| SGK1 wt | 8 | 3.8 | 3.3 | 2.4 | 7 | 2 |
| SGK1[S422D] | 71 | 9.6 | 3.5 | 3.4 | 67 | 10 |
| PKBα wt | 28 | 19.3 | 35 | 35 | 30 | 29 |
| PKBα [S473D] | 100 | 85.4 | 178 | 162 | 105 | 100 |
| ΔPH-PKBα wt | 0.4 | 0.25 | 0.23 | 0.25 | 1.2 | 0.5 |
| ΔPH-PKBα[S473D] | 0.9 | 0.2 | 0.18 | 0.15 | 2.1 | 0.4 |
The phosphorylations were carried out in the absence (–) and presence (+) of 2 µM PIFtide and analysed as described in the Materials and methods section. The data shown are from a representative experiment from three separate experiments, with each determination carried out in duplicate. The error of each point ±SEM is <10% of the average value of the determination. The rate at which PDK1 phosphorylates PKBα[S473D] was given a value of 100.
Fig. 3. Effect of PIFtide on the ability of PDK1 to phosphorylate S6K1, SGK1 and PKBα. The indicated AGC kinase substrates were phosphorylated with wild-type PDK1 in the absence (–) or presence (+) of 2 µM PIFtide in the presence of magnesium and [γ-32P]ATP as described in the legend to Figure 2.
In Figure 3, we demonstrate that PDK1’s ability to phosphorylate and activate S6K1-T2, SGK1 and ΔPH-PKBα, as well as the hydrophobic motif mutants S6K1-T2[T412E], SGK1[S422D] and ΔPH-PKBα[S473D], was markedly inhibited in the presence of an excess of the peptide PIFtide, which interacts with the PIF-binding pocket of PDK1 (see Introduction). In contrast, the PDK1-catalysed phosphorylation of PKBα and PKBα[S473D] was unaffected by the same concentration of PIFtide. These results confirm that PDK1 uses its hydrophobic PIF-binding pocket in order to interact with, phosphorylate and activate S6K1-T2, SGK1 and ΔPH-PKBα, but that this pocket is not required for PDK1 to phosphorylate PKBα.
Role of Lys115, Ile119, Gln150 and Ala277 in the PDK1 ‘PIF-binding pocket’ in the activation of S6K1 and SGK1
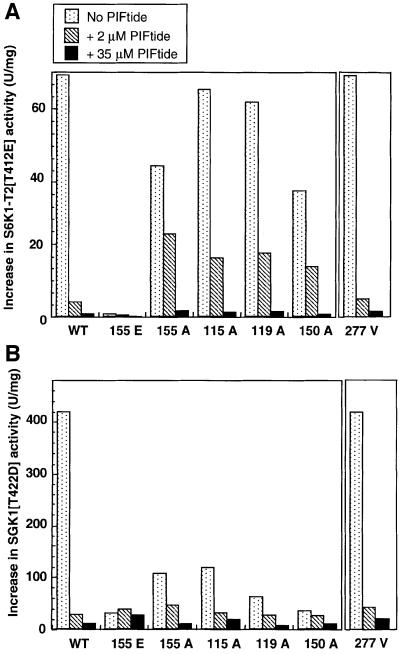
In addition to Leu155, other residues were predicted to form part of the PIF-binding pocket on PDK1. These include Lys115, Ile119 and Gln150, and mutation of these residues to Ala was shown to reduce ∼10-fold the affinity of PDK1 for PIFtide (Biondi et al., 2000). These PDK1 mutants still retained normal activity towards peptide substrates that do not interact with the PIF-binding pocket [e.g. T308tide (Biondi et al., 2000)], indicating that they were not impaired catalytically. Although mutation of Leu155 on PDK1 to Glu abolishes its interaction with PIFtide (Biondi et al., 2000), we also found that a mutant of PDK1 in which Leu155 was changed to Ala possessed 10-fold reduced affinity for PIFtide, similar to PDK1 [K115A], PDK1[I119A] and PDK1[Q150A] mutants of PDK1 (R.M.Biondi, unpublished data). We therefore decided to compare the rate at which all of these PIF-binding pocket mutants of PDK1 phosphorylated S6K1-T2[T412E] (Figure 4A) and SGK1[S422D] (Figure 4B) in the absence or presence of a low (2 µM) or high (35 µM) concentration of PIFtide. We found that the rate at which PDK1[K115A], PDK1[I119A], PDK1[Q150A] and PDK1[L155A] activated S6K1-T2[T412E] was only marginally lower than wild-type PDK1 (Figure 4A). In contrast, the rate at which these mutants of PDK1 activated SGK1[S422D] was reduced more markedly (Figure 4B), indicating that Lys115, Ile119 and Gln150 may be more critical in permitting PDK1 to phosphorylate SGK1 [S422D] than S6K1-T2[T412E]. PIFtide (2 µM) inhibited the activation of S6K1-T2[T412E] and SGK1[S422D] by wild-type PDK1 over ∼20-fold but only reduced by ∼2- to 3-fold the rate at which S6K1-T2[T412E] and SGK1[S422D] were phosphorylated by PDK1[K115A], PDK1[I119A], PDK1[Q150A] and PDK1[L155A]. This is consistent with the reduced affinity of these PDK1 mutants for PIFtide. In contrast, 35 µM PIFtide was required to achieve ∼20-fold inhibition of the phosphorylation of S6K1-T2[T412E] and SGK1[S422D] by PDK1[K115A], PDK1[I119A], PDK1[Q150A] and PDK1[L155A]. These findings provide further evidence that the inability of PDK1[L155E] to phosphorylate S6K1 and SGK1 (Figure 2) and the inhibition by PIFtide of the PDK1-catalysed phosphorylation of S6K1 and SGK1 (Figure 3) are directly related to their destruction and blockade, respectively, of the PIF-binding pocket of PDK1.
Fig. 4. Further evidence that the PIF-binding pocket of PDK1 is required for PDK1 to activate S6K1 and SGK1. S6K1-T2[T412E] (A) and SGK1[S422D] (B) were phosphorylated with the indicated mutants of PDK1 in the absence (dotted bars) or presence of 2 µM PIFtide (hatched bars) or 35 µM PIFtide (filled bars) in the presence of magnesium and [γ-32P]ATP as described in the Materials and methods. The activation of each substrate was assessed by its ability to phosphorylate the peptide substrate Crosstide (GRPRTSSFAEG). The results are presented ±SEM for two separate experiments; each determination was carried out in duplicate.
A point mutation in the kinase catalytic domain that converts Ala277 to a Val has been reported to enhance the rate at which PDK1 can activate PKB in vivo (Paradis et al., 1999; Wick et al., 2000). To investigate whether this mutant had any relevance to the PIF-binding pocket we expressed and purified mutant PDK1[A277V] as a glutathione S-transferase (GST) fusion protein. The specific activity of this mutant towards a PDK1 peptide substrate [T308tide (Biondi et al., 2000)] and its activation by PIFtide were identical to that of wild-type PDK1 (data not shown). As previously described, the mutant PDK1[A277V] was 2.3-fold more active towards ΔPH-PKBα than wild-type PDK1 (Table I and Paradis et al., 1999). In marked contrast, PDK1[A277V] phosphorylated full-length PKBα in the presence of PtdIns(3,4,5)P3 and S6K1 or SGK1 in the absence of PtdIns(3,4,5)P3 at the same rate as wild-type PDK1 (Table I). The phosphorylation (Table I) and activation (Figure 4) of S6K1 and SGK1 by PDK1[A277V] was inhibited by PIFtide similarly to wild-type PDK1. Therefore, the mechanism by which the Ala277 to Val mutation might activate PKB in vivo appears unrelated to the PIF-binding pocket.
Interaction of S6K1 and SGK1 with PDK1
Full-length S6K1 is a very poor substrate for PDK1 compared with S6K1 lacking the C-terminal 104 amino acids (termed S6K1-T2) in its regulatory domain (Alessi et al., 1998; Pullen et al., 1998). We therefore tested whether this was explained by the inability of full-length S6K1 to interact with PDK1 compared with S6K1-T2. We co-expressed in 293 cells GST–PDK1 together with HA-epitope tagged full-length S6K1, full-length S6K1 [T412E], S6K1-T2 and S6K1-T2[T412E]. Glutathione– Sepharose ‘pull-downs’ of GST–PDK1 from cell lysates prepared from these transfections were then immunoblotted for the presence of HA-epitope tagged S6K1. Although GST–PDK1 and wild-type and mutant forms of S6K1 were expressed to a similar level, full-length S6K1 and full-length S6K1[T412E] failed to interact with GST–PDK1, while the S6K1-T2 and S6K1-T2[T412E] readily interacted with GST–PDK1. As expected, we were unable to detect an interaction between GST–PDK1
Wild-type SGK1 is phosphorylated at a 10-fold lower rate than SGK1[S422D] by PDK1 (Table I). We therefore tested whether this could be accounted for by differences in the affinity of wild-type SGK1 and SGK1[S422D] for PDK1. To investigate this we co-expressed GST–SGK1 and GST–SGK1[S422D] with Myc-PDK1 in 293 cells. Glutathione–Sepharose ‘pull-downs’ of GST–SGK1 were immunoblotted for the presence of Myc-PDK1. As shown in Figure 5B, Myc-PDK1 only interacted with SGK1 [S422D], but did not interact with the wild-type SGK1. A weak interaction was still observed between SGK1 [S422D] and Myc-PDK1[L155E], indicating that SGK1 may significantly interact with an additional site on PDK1 other than the PIF-binding pocket (data not shown).
Fig. 5. Interaction of S6K1 and SGK1 with PDK1. (A) 293 cells were transiently transfected with DNA constructs expressing GST, wild type (wt) GST-PDK1 or mutant GST–PDK1[L155E] together with the indicated wild type and mutant forms of HA-epitope-tagged S6K1. Thirty-six hours post-transfection the cells were lysed and the GST fusion proteins were purified by affinity chromatography on glutathione–Sepharose beads. Aliquots of the purified protein were electrophoresed on a 10% SDS–polyacrylamide gel, and immunoblotted using an anti-HA antibody to detect HA-S6K1 or Coomassie to ensure similar expression of GST fusion proteins. To establish that the wild-type and mutant S6K1 forms were expressed at similar levels, 10 µg of total 293 cell lysate (termed Crude extract) for each condition were electrophoresed on a 10% SDS–polyacrylamide gel and immunoblotted with anti-HA antibodies. (B) As above except that 293 cells were transiently transfected with DNA constructs expressing Myc-PDK1 and GST–SGK1 or GST–SGK1[S422D]. Detection of PDK1 was performed by immunoblotting with anti-Myc-antibodies. Duplicates of each condition are shown. Similar results were obtained in two different experiments.
Role of the hydrophobic motif of AGC kinases in regulating interaction and phosphorylation by PDK1
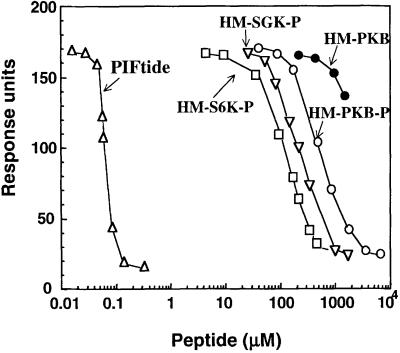
To investigate further the role of the hydrophobic motifs of AGC kinases, we synthesized peptides encompassing the phosphorylated hydrophobic motifs of PKBα, SGK1 and S6K1. We estimated the relative affinities of each peptide for the PIF-binding pocket of PDK1 by the ability of these peptides to compete for the interaction of PDK1 to biotinylated PIFtide using a surface plasmon resonance-based assay (Figure 6). The peptide corresponding to the hydrophobic motif of PKBα phosphorylated at Ser473 interacted with PDK1 with 4- and 8-fold lower relative affinity than the equivalent peptides derived from SGK1 and S6K1, respectively. As expected, the peptide corresponding to the hydrophobic motif of PKBα unphosphorylated at Ser473 did not interact significantly with PDK1. Remarkably, the peptide derived from the hydrophobic motif of PRK2 (PIFtide) interacted with the PIF-binding pocket of PDK1 with >1000-fold higher affinity than the S6K1, SGK1 and PKB hydrophobic motif peptides.
Fig. 6. Relative affinities of peptides derived from the hydrophobic motif of S6K1, SGK1, PKBα and PRK2. Surface plasmon resonance measurements were carried out on a BIAcore instrument as described in Materials and methods. Biotinylated PIFtide was immobilized on a strepavidin-coated sensor chip and the specific interaction with GST–PDK1 was recorded in the absence or presence of the indicated concentrations of peptides that encompass the hydrophobic motif (HM) of PRK2, S6K1, SGK1 and PKBα. The peptide PIFtide (open triangles) corresponds to residues 957–980 of PRK2; the peptide HM-S6K1-P (open squares) corresponds to residues 401–418 of S6K1 in which Thr412 is phosphorylated; the peptide HM-SGK1-P (inverted triangles) corresponds to residues 411–428 of SGK1 in which Ser422 is phosphorylated; the peptide HM-PKBα-P (open circles) corresponds to residues 461–480 of PKBα in which Ser473 is phosphorylated; the peptide HM-PKBα (closed circles) corresponds to the non-phosphorylated peptide comprising residues 461–480 of PKBα. The PRK2-derived peptide PIFtide was previously characterized to specifically interact with the PIF-binding pocket on PDK1 (Biondi et al., 2000). Data are single determinations from a representative experiment that was repeated at least twice with similar results.
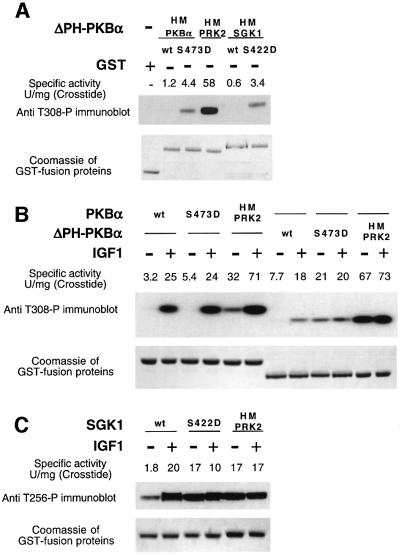
The observation that ΔPH-PKBα[S473D] is a far poorer substrate for PDK1 than SGK1[S422D] (Table I) might be explained by the difference in affinity of the hydrophobic motifs of these enzymes for PDK1. To study this we exchanged the hydrophobic motif of ΔPH-PKBα, with that of SGK1(ΔPH-PKBα[HM-SGK1]) and PRK2 (ΔPH-PKBα[HM-PRK2]) (the structure of these fusion proteins are described in Figure 1). These mutants were expressed as GST fusion proteins in serum-starved 293 cells and glutathione–Sepharose pull-downs were immunoblotted with a phospho-specific Thr308 antibody to measure the extent of phosphorylation of the T-loop of these fusion proteins. The activity of these proteins towards the peptide Crosstide was also assessed (Figure 7A). ΔPH-PKBα and ΔPH-PKBα[HM-SGK1 wt] were not phosphorylated at their T-loop and, consistent with this, were essentially inactive. However, ΔPH-PKBα[S473D] and ΔPH-PKBα [HM-SGK1 S422D] were detectably phosphorylated to a similar extent and possessed a low specific activity of ∼3 U/mg towards Crosstide. In contrast, ΔPH-PKBα[HM-PRK2] was highly phosphorylated at its T-loop residue and possessed a specific activity of ∼60 U/mg (Figure 7A), which is equivalent to that of PKBα fully phosphorylated at its T-loop residue (Balendran et al., 1999a). The finding that the T-loop phosphorylation of ΔPH-PKBα[HM-SGK1 S422D] was similar to ΔPH-PKBα[S473D], together with the data shown in Figure 6, indicate that the residues encompassing the hydrophobic motif of SGK1 alone cannot account for the markedly higher rate at which SGK1[S422D] is phosphorylated by PDK1 in vitro compared with ΔPH-PKBα[S473D] (Table I). These results may indicate that SGK1[S422D] (and S6K1-T2[T412E]) may possess additional PDK1-binding determinant(s) distinct from the hydrophobic motif which could be part of a cooperative binding mechanism.
Fig. 7. Effect of swapping the hydrophobic motif of AGC kinases. GST fusion proteins coding for indicated wild-type and mutant forms of PKBα (A and B) and SGK1 (C), whose structure is described in Figure 1, were expressed in 293 cells. The cells were deprived of serum overnight and either left unstimulated or stimulated with 100 ng/ml IGF1 for either 10 min (B) or 15 min (C). The cells were lysed and the GST fusion proteins affinity purified on glutathione–Sepharose. A 0.5 µg sample of each protein was electrophoresed on a 4–12% SDS–polyacrylamide gel, and immunoblotted using a phospho-specific antibody recognizing PKBα (A and B) or SGK1 (C) phosphorylated at their T-loop residue, namely Thr308 and Thr256, respectively. The gels were also stained with Coomassie to ensure similar loading of the GST-ΔPH-PKBα fusion proteins. The specific activity of each GST fusion protein kinase (50 ng) was assessed by its ability to phosphorylate the peptide substrate Crosstide (GRPRTSSFAEG) as described in Materials and methods. The results of a single experiment are shown, similar results were obtained in three separate experiments for (A) and two experiments for (B and C).
Additionally, we investigated the effect of exchanging the hydrophobic motif of PKBα and SGK1 with that of PRK2 in serum-starved and IGF1-stimulated 293 cells (Figure 7B and C). In unstimulated cells, as expected, ΔPH-PKBα, as well as full-length PKBα, were not phosphorylated at their T-loop and were essentially inactive. ΔPH-PKBα[S473D] and PKBα[S473D] were phosphorylated at their T-loop to a moderate extent in unstimulated cells and possessed a low specific activity towards Crosstide. As expected, stimulation with IGF1 promoted T-loop phosphorylation and activation of PKBα, PKBα[S473D] and ΔPHPKBα, but did not further stimulate phosphorylation/activation of ΔPHPKBα[S473D]. The activity/phosphorylation of full-length PKBα[HM-PRK2] was also markedly increased by IGF1, in contrast to ΔPH-PKBα[HM-PRK2] whose activity and T-loop phosphorylation was maximal in unstimulated cells.
In Figure 7C we compared T-loop phosphorylation and activity of SGK1, SGK1[S422D] and SGK1[HM-PRK2]. In unstimulated cells, SGK1 was phosphorylated to a low extent at its T-loop site and possessed low activity. As expected, IGF1 stimulated the activity and T-loop phosphorylation of wild-type SGK1. In contrast, SGK1 [S422D] and SGK1[HM-PRK2] possessed high activity in unstimulated cells and were phosphorylated at their T-loop to the same extent as SGK1 derived from IGF1-stimulated cells. The activity and T-loop phosphorylation of SGK1[S422D] and SGK1[HM-PRK2] was not further increased by IGF1.
Discussion
The results presented in this paper highlight the importance of the PIF-binding pocket of PDK1 in directly mediating the interaction with the hydrophobic motif of S6K1 and SGK1. This functions to recruit PDK1 to S6K1 and SGK1, enabling PDK1 to phosphorylate these enzymes at their T-loop site. This conclusion is supported by the finding that a mutant of PDK1 in which the PIF-binding pocket has been disrupted (PDK1[L155E]) cannot phosphorylate and activate S6K1 or SGK1 (Figure 2). Furthermore, PIFtide inhibits the phosphorylation of S6K1 and SGK1 (Figure 3), as it interacts with the PIF-binding pocket of PDK1, preventing it from binding to S6K1 and SGK1. We further provide evidence that when these substrates are designed to have higher affinities for the PIF-binding pocket of PDK1, they are more efficiently phosphorylated by PDK1. We thus provide a model by which phosphorylation of some PDK1 substrates are regulated by their ability to interact with PDK1. Interestingly, we show that PKB is activated by PDK1 through a different mechanism, which does not require the interaction of PKB with the PIF-binding pocket of PDK1.
S6K1 requires phosphorylation of both the T-loop and hydrophobic motif for it to be activated, and thus phosphorylation of S6K1 at its T-loop site by PDK1 alone does not induce significant activation. Full-length S6K1 is a very poor substrate for PDK1 compared with a mutant of S6K1 that lacks its C-terminal 104 residues (S6K1-T2), encompassing the four Ser-Pro/Thr-Pro sequences that become phosphorylated in vivo (Alessi et al., 1998; Pullen et al., 1998) and are thought to comprise an autoinhibitory domain. Phosphorylation of the Ser-Pro/Thr-Pro sequences in the autoinhibitory domain does not activate S6K1, but plays a role in enabling S6K1 to become activated (Weng et al., 1995; Dennis et al., 1998). Removal of the autoinhibitory domain of S6K1 bypasses the need of S6K1 to be phosphorylated at the Ser-Pro/Thr-Pro sites to become activated. Here, we demonstrate that S6K1-T2 and S6K1-T2[T412E] can interact with PDK1 but not PDK1[L155E] (Figure 5). S6K1-T2[T412E] is phosphorylated by PDK1 at a higher initial rate than S6K1-T2 (Alessi et al., 1998; Pullen et al., 1998), and this could be explained by the observation that S6K1-T2[T412E] displayed an increased interaction with PDK1 than S6K1-T2. Our results also indicate that phosphorylation and activation of S6K requires the integrity and availability of the PIF-binding pocket of PDK1 (Figures 2A, 3A and 4A). These findings suggest a model for the activation of S6K1 in which the first step would involve the phosphorylation of the C-terminal Ser-Pro/Thr-Pro sequences by a proline- directed kinase that has yet to be identified. This would not activate S6K1 directly, but would induce a conformational change exposing the hydrophobic motif for interaction with the PIF-binding pocket of PDK1, enabling PDK1 to phosphorylate the T-loop residue of S6K1. This is consistent with the finding that, in response to insulin, the C-terminal Ser-Pro/Thr-Pro residues of S6K1 become phosphorylated before phosphorylation of the T-loop and the hydrophobic motif (Weng et al., 1998). The interaction of PDK1 with S6K1 would be enhanced further if S6K1 was phosphorylated at its hydrophobic motif. However, this may not be an absolute prerequisite for T-loop phosphorylation as a mutant of S6K1 in which Thr412 is mutated to Ala is still phosphorylated at its T-loop residue, albeit to a significantly lower extent than wild-type S6K1 (Weng et al., 1998).
SGK1, like S6K1, requires phosphorylation of both its T-loop and hydrophobic motif to be activated in cells, but does not possess a C-terminal tail following the hydrophobic motif. Wild-type SGK1 that has not been phosphorylated at its hydrophobic motif (Ser422) is a poor substrate for PDK1, and mutation of Ser422 to an acidic residue increases ∼5-fold the rate at which it becomes phosphorylated by PDK1 (Table I and Kobayashi and Cohen, 1999). Similar results have been obtained with the related kinases SGK2 and SGK3 (Kobayashi et al., 1999). Furthermore, when SGK1[S422D] is expressed in unstimulated 293 cells it becomes significantly phosphorylated at its T-loop residue (Thr256), whereas wild-type SGK1 does not (Kobayashi and Cohen, 1999; Park et al., 1999) and stimulation of cells with IGF1 does not further increase the T-loop phosphorylation or activity of SGK1[S422D] (Figure 7C). The finding that wild-type SGK1 did not interact with PDK1, while SGK1[S422D] did (Figure 5), also suggests that the phosphorylation of SGK1 at its hydrophobic motif plays a major role in regulating phosphorylation of SGK1 at its T-loop residue. Our observations also suggest that phosphorylation and activation of SGK1 requires the interaction with the PDK1 PIF-binding pocket.
Although the activation of S6K1 and SGK1 is dependent upon PI 3-kinase in vivo, it is not clear how PtdIns(3,4,5)P3/PtdIns(3,4)P2 regulates this process. First, the intrinsic activity of PDK1 is not stimulated by agonists that activate PI 3-kinase (Alessi et al., 1997b; Pullen et al., 1998). Secondly, the phosphorylation of S6K1 and SGK1 in vitro by PDK1 is not enhanced by the presence of PtdIns(3,4,5)P3/PtdIns(3,4)P2, which interacts with the PH domain of PDK1. This indicates that the binding of PDK1 to PtdIns(3,4,5)P3/PtdIns(3,4)P2 may not activate these enzymes directly. Instead, PtdIns(3,4,5)P3/PtdIns(3,4)P2 may induce activation of the kinase(s) that phosphorylates the hydrophobic motif of S6K1 and SGK1 and/or the proline-directed kinase(s) that phosphorylates S6K1 at its C-terminal tail. If this mechanism operated in vivo, PDK1 would not phosphorylate S6K1 or SGK1 until these enzymes became phosphorylated at their hydrophobic motif/C-terminal tail. This would be analogous to the mechanism by which PDK1 phosphorylates p90RSK (Frodin et al., 2000). Another protein kinase termed glycogen synthase kinase-3 has also recently been shown to possess a phosphorylation-dependent interaction with some of its substrates (Frame et al., 2001).
The finding in this study that the PIF-binding pocket of PDK1 was not required for PDK1 to activate PKB was perhaps unexpected since PKB, like S6K and SGK, possesses a hydrophobic motif that becomes phosphorylated by agonists that activate PI 3-kinase. These observations explain our previous findings that overexpression of GST–PIF in cells did not affect PKBα activation but inhibited the activation/T-loop phosphorylation of S6K1, PRK2 and PKCζ (Balendran et al., 1999a,b, 2000). This indicates that the binding of PDK1 and PKB to PtdIns(3,4,5)P3/PtdIns(3,4)P2 could be a primary determinant for enabling PDK1 to phosphorylate PKB, rather than the interaction of PKB with the PIF-binding pocket of PDK1. This is also supported by the finding that the activation of PKB, like the activation of PI 3-kinase, occurs very rapidly in cells, indicating that the activation of PKB occurs shortly after the formation of PtdIns(3,4,5)P3/PtdIns(3,4)P2. Furthermore, it had been previously suggested that PtdIns(3,4,5)P3 could also produce a conformational change in PKB, permitting it to become phosphorylated by PDK1(Alessi et al., 1997a; Stokoe et al., 1997). This model is supported by our finding that full-length PKB possessing the hydrophobic motif of PRK2 still required IGF1 stimulation for complete phosphorylation at the T-loop site (Figure 7B). The activation of S6K1 (and SGK1) occurs more slowly than PKB. This delay could be accounted for by the time it takes for PtdIns(3,4,5)P3/PtdIns(3,4)P2 to activate the S6K1-autoinhibitory terminal kinase(s) and or the S6K1/SGK1 hydrophobic motif kinase(s), which are likely be rate limiting for the activation of S6K1/SGK1 in cells.
When ΔPH-PKBα is expressed in 293 cells it is phosphorylated at Thr308 and Ser473 in response to insulin, and this is prevented by inhibitors of PI 3-kinase (Kohn et al., 1996; Andjelkovic et al., 1997) (also see Figure 7B). This observation was originally interpreted as evidence that PDK1 was activated in vivo by PtdIns(3,4,5)P3/PtdIns(3,4)P2. However, in the present study, we demonstrate that ΔPH-PKBα is not phosphorylated by PDK1 by the same mechanism as wild-type PKBα, because ΔPH-PKBα is not phosphorylated by PDK1[L155E] and the phosphorylation of ΔPH-PKBα by PDK1 was not inhibited by PIFtide (Figures 2 and 3). Thus, the mechanism by which PDK1 phosphorylates ΔPH-PKBα resembles the activation of S6K1 and SGK1, rather than PKBα. The mutation of Ser473 to Asp in ΔPH-PKBα increases the rate at which it is phosphorylated by PDK1 in vitro (Table I). Consistent with this, ΔPH-PKBα[S473D] when expressed in unstimulated 293 cells is partially phosphorylated at its T-loop site, and this phosphorylation is not increased following stimulation of cells with IGF1 (Figure 7B). These data support the view that when ΔPH-PKBα is expressed in cells, PtdIns(3,4,5)P3/PtdIns(3,4)P2 does not activate PDK1 but instead induces phosphorylation of Ser473 through the same hydrophobic motif kinase(s) that phosphorylates S6K1 and SGK1, thereby converting ΔPH-PKBα into an efficient PDK1 substrate.
When PRK2 is overexpressed in unstimulated 293 cells, unlike S6K1, SGK1 and PKB, it becomes phosphorylated at its T-loop residue to a high stoichiometry (Balendran et al., 2000). In Figure 6 we demonstrate that the hydrophobic motif of PRK2 (PIF) interacts with >1000-fold higher relative affinity with PDK1 than the hydrophobic motifs of S6K1, SGK1 and PKBα. Furthermore, when the hydrophobic motif of ΔPH-PKBα, full-length PKBα or SGK1 is replaced with PIF, it becomes phosphorylated at the T-loop in unstimulated cells. These observations demonstrate that, in contrast to the hydrophobic motifs of PKB and SGK1, the hydrophobic motif of PRK2 is able to interact with PDK1, resulting in PRK2, ΔPH-PKBα[HM-PRK2] and SGK1[HM-PRK2] becoming phosphorylated at their T-loop residues in the absence of stimulus. The finding that artificially promoting the interaction of AGC-kinases with PDK1 stimulates their phosphorylation at the T-loop supports our model that phosphorylation of PDK1 substrates can be regulated through the interaction of the PIF-binding pocket of PDK1 with the hydrophobic motif of these enzymes. This high affinity of PIF for PDK1 compared with the equivalent motif from PKB, S6K1 and SGK suggests that there are residues encompassing the hydrophobic motif of PRK2 that are not present in PKB, S6K and SGK which contribute to the very high affinity of PIF for PDK1.
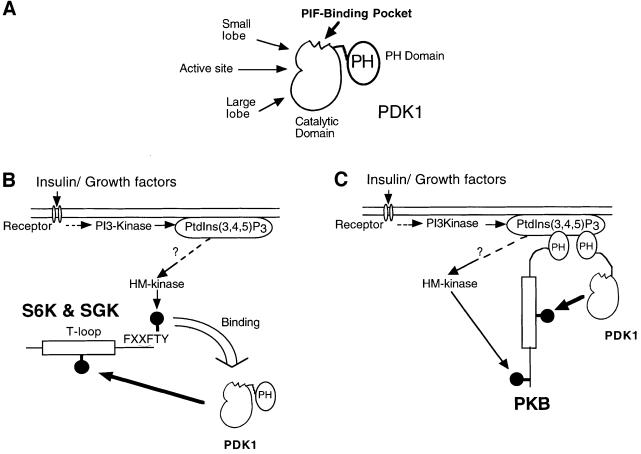
The model we propose in Figure 8 highlights our conclusion that phosphorylation of the PDK1 substrates S6K and SGK is regulated by the direct interaction of their hydrophobic motifs with the PIF-binding pocket of PDK1. In contrast, PKB does not rely upon the PIF-binding pocket of PDK1 for its T-loop phosphorylation. Another important feature of our model is that PDK1 activity does not need to be regulated and is considered to be constitutively active. Instead, it is the substrates of PDK1 that are subjected to regulation and are converted into forms that can interact with PDK1, thereby enabling them to be phosphorylated at their T-loop residue. Although PRK2 and PKCζ can interact directly with PDK1 when overexpressed in 293 cells (Balendran et al., 2000), the interaction of S6K1 and SGK1 is regulated by the phosphorylation of these enzymes at their C-terminal residue(s). In contrast, our results support the model that PKB activation entails first the interaction of PKB with PtdIns(3,4,5)P3 resulting in a conformational change that is required for PDK1-mediated phosphorylation, and secondly, the co-localization of PKB and PDK1 through their mutual interaction with PtdIns(3,4,5)P3. This model could account for the differences in the time course of activation of S6K1, SGK1 and PKB in insulin/growth factor-stimulated cells. The interaction of the hydrophobic motifs of PRK2 (Biondi et al., 2000) and p90RSK (Frodin et al., 2000) with PDK1 may not only bring these enzymes in proximity, but also significantly increase the activity of PDK1. This may also occur when S6K1 and SGK1 interact with PDK1.
Fig. 8. Model by which PDK1 specifically recognizes S6K, SGK and PKB. (A) Domain structure of PDK1 indicating where the PIF-binding pocket is located on the small lobe of the kinase domain. (B) Summary of the model by which PDK1 can recognize, interact and then phosphorylate S6K and SGK. In this model PtdIns(3,4,5)P3 regulates the activity of an unidentified hydrophobic motif (HM) kinase that phosphorylates S6K1 and SGK1, thus triggering the docking to the PIF-binding pocket of PDK1. (C) In contrast, the PIF-binding pocket of PDK1 is not involved in the binding of PDK1 to PKB. Instead it is the interaction of the PH domains of PKB and PDK1 with PtdIns(3,4,5)P3 that brings PKB and PDK1 together.
Recent work, especially on the MAP kinase signalling pathways, has highlighted the importance of a single docking region on the MAP kinases that permits their specific interaction with MAP kinase kinase activators and substrates, as well with the dual-specificity MAP kinase phosphatases that inactivate them (Tanoue et al., 2000, 2001). The docking sites on MAP kinase family members appear to be required for interaction with all substrates tested so far. This contrasts with the PIF-binding pocket of PDK1, which is not required for the phosphorylation of PKB. Furthermore, it is not yet known how the specific interactions of MAP kinases with MAP kinase kinases and MAP kinase phosphatases are regulated. In the case of PDK1 interaction with S6K, SGK and p90RSK, the phosphorylation of these enzymes at their hydrophobic motif is a key determinant in regulating this interaction. It is not yet clear how many protein kinases will turn out to possess a docking site that only interacts with a specific subset of substrates. If this were a general phenomenon, it would raise the possibility of developing compounds directed towards such sites that would inhibit the phosphorylation of a group of substrates without affecting the phosphorylation of others. Such drugs should have improved specificity compared with compounds that target the ATP binding site, and hence produce fewer side effects when used for the treatment of disease.
Materials and methods
The description of material, antibodies, buffers, cDNA constructs, purification of GST fusion proteins and immunoblotting protocols are described in the Supplementary data, available at The EMBO Journal Online.
Phosphorylation of AGC kinase substrates by PDK1
The phosphorylation of PDK1 substrates was performed in a final volume of 20 µl in a buffer containing 50 mM Tris–HCl pH 7.5, 0.1% (by vol.) 2-mercaptoethanol, 10 mM magnesium chloride, 100 µM [γ-32P]ATP (∼1000 c.p.m./pmol), 0.5 µM microcystin-LR, 0.6 µM AGC kinase substrate and 0.6–30 nM wild-type PDK1 or the indicated mutant of PDK1. After 10 min the reactions were stopped by addition of Laemmli sample buffer [100 mM Tris–HCl pH 6.8, 4% (by mass) SDS, 20% (by vol.) glycerol and 200 mM dithiothreitol (DTT)], boiled, and the samples subjected to separation by SDS–PAGE. The gels were exposed and analysed with a Fuji PhosphoImager with known amounts of [γ-32P]ATP spotted onto blank gels to permit quantification of the data. The experiments were performed so that the amount of PDK1 did not phosphorylate >20% of the substrate. A control in which PDK1 was omitted from the reaction was taken as the blank value.
Activation of AGC kinase PDK1 substrates
Phosphorylation reactions were carried out as above except that non-radioactive ATP replaced [γ-32P]ATP. Following 10 min at 30°C, cocktail was added (30 µl) containing 50 mM Tris–HCl pH 7.5, 0.1% (by vol.) 2-mercaptoethanol, 10 mM magnesium chloride, 100 µM [γ-32P]ATP (∼1000 c.p.m./pmol), 0.5 µM microcystin-LR and 100 µM peptide substrate Crosstide (GRPRTSSFAEG) and incubated for a further 30 min. Reactions were stopped by the addition of 25 µl of 0.2 M EDTA pH 8.0, spotted onto P81 phosphocellulose paper, washed and analysed as described for the assay of MAP kinase (Alessi et al., 1995). The amount of PDK1 in the assay was varied so that the assay was in the linear range. One unit of activity is defined as phosphorylation of 1 nmol of substrate in 1 min.
Binding of PDK1 to SGK1 and S6K1
For the data presented in Figure 5, 293 cells were co-transfected with 10 µg of the wild-type or mutant PDK1 plasmid and 10 µg of either the wild-type or mutant S6K1 or SGK1. Thirty-six hours post-transfection the cells were lysed in 0.6 ml of buffer A, the lysates were cleared by centrifugation at 13 000 g for 10 min at 2°C, and 0.5 ml of supernatant were incubated for 2 h at 4°C with 30 µl of glutathione–Sepharose. The beads were washed twice in buffer A containing 0.5 M NaCl, followed by two further washes in buffer A. The beads were resuspended in 30 µl of Laemmli sample buffer and subjected to SDS–PAGE. The gels were either stained with Coomassie Blue, or analysed by immunoblotting with either anti-Flag or anti-Myc antibodies (described below).
Relative affinities of peptides corresponding to the hydrophobic motifs of S6K1, SGK1, PKBa and PRK2 for PDK1
Binding was analysed by surface plasmon resonance in a BIACore 3000 system. Biotinylated PIFtide (Biotin-C12-REPRILSEEEQEMFRDFAYIADWC) was bound to streptavidin-coated Sensor chip (SA) (BIACore AB, Stevenage, UK) (20 response units, RU). Wild-type GST–PDK1 (60 nM), injected at 20 µl/min, bound specifically giving a response of 165 RU in a buffer containing 10 mM HEPES pH 7.4, 0.15 M NaCl, 0.005% (by vol.) polysorbate-20 and 1 mM DTT. To study the relative affinities of the peptides derived from the C-terminus of S6K1 [HM-S6K1-P, SESANQVFLGFT(P)YVAPSV], SGK1 [HM-SGK1-P, VKEAAEAFLGFS(P)YAPPTP], PKBα [HM-PKBα-P, VDSERRPHFPQFS(P)YSASSTA; HM-PKBα, VDSERRPHFPQFSYSASSTA] and PRK2 (PIFtide, REPRILSEEEQEMFRDFAYIADWC), each peptide was incubated at the specified concentrations with GST–PDK1 prior to injection. Steady state binding was determined at each concentration. HM-PKBα peptide produced unspecific effects on GST–PDK1 binding at concentrations >2 mM.
Supplementary data
Supplementary data for this paper are availale at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Joe Avruch for providing us with S6K1 expression constructs, Chris Armstrong for preparation of baculovirus expressed S6K1-T2 and S6K1-T2[412E], Jane Leitch for preparation of all the antibodies and the Sequencing Service (School of Life Sciences, University of Dundee, Scotland for DNA sequencing). This work was supported by the UK Medical Research Council and Diabetes UK (D.R.A.) as well as the pharmaceutical companies that support the Division of Signal Transduction Therapy (AstraZeneca, Boehringer-Ingelheim, GlaxoSmithKline, Novo-Nordisk and Pfizer).
References
- Alessi D.R., Cohen,P., Ashworth,A., Cowley,S., Leevers,S.J. and Marshall,C.J. (1995) Assay and expression of mitogen-activated protein kinase, MAP kinase kinase and Raf. Methods Enzymol., 255, 279–290. [DOI] [PubMed] [Google Scholar]
- Alessi D.R., Deak,M., Casamayor,A. et al. (1997a) 3-phosphoinositide-dependent protein kinase-1 (PDK1): structural and functional homology with the Drosophila DSTPK61 kinase. Curr. Biol., 7, 776–789. [DOI] [PubMed] [Google Scholar]
- Alessi D.R., James,S.R., Downes,C.P., Holmes,A.B., Gaffney,P.R., Reese,C.B. and Cohen,P. (1997b) Characterization of a 3-phospho inositide-dependent protein kinase which phosphorylates and activates protein kinase Bα. Curr. Biol., 7, 261–269. [DOI] [PubMed] [Google Scholar]
- Alessi D.R., Kozlowski,M.T., Weng,Q.P., Morrice,N. and Avruch,J. (1998) 3-phosphoinositide-dependent protein kinase 1 (PDK1) phosphorylates and activates the p70 S6 kinase in vivo and in vitro. Curr. Biol., 8, 69–81. [DOI] [PubMed] [Google Scholar]
- Andjelkovic M., Alessi,D.R., Meier,R. et al. (1997) Role of translocation in the activation and function of protein kinase B. J. Biol. Chem., 272, 31515–31524. [DOI] [PubMed] [Google Scholar]
- Balendran A., Casamayor,A., Deak,M., Paterson,A., Gaffney,P., Currie,R., Downes,C.P. and Alessi,D.R. (1999a) PDK1 acquires PDK2 activity in the presence of a synthetic peptide derived from the carboxyl terminus of PRK2. Curr. Biol., 9, 393–404. [DOI] [PubMed] [Google Scholar]
- Balendran A., Currie,R.A., Armstrong,C.G., Avruch,J. and Alessi,D.R. (1999b) Evidence that PDK1 mediates the phosphorylation of p70 S6 kinase in vivo at Thr412 as well as Thr252. J. Biol. Chem., 274, 37400–37406. [DOI] [PubMed] [Google Scholar]
- Balendran A., Biondi,R.M., Cheung,P.C., Casamayor,A., Deak,M. and Alessi,D.R. (2000) A 3-Phosphoinositide-dependent protein kinase-1 (PDK1) docking site is required for the phosphorylation of protein kinase cζ (PKCζ) and PKC-related kinase 2 by PDK1. J. Biol. Chem., 275, 20806–20813. [DOI] [PubMed] [Google Scholar]
- Belham C., Wu,S. and Avruch,J. (1999) Intracellular signalling: PDK1-a kinase at the hub of things. Curr. Biol., 9, R93–96. [DOI] [PubMed] [Google Scholar]
- Biondi R.M., Cheung,P.C., Casamayor,A., Deak,M., Currie,R.A. and Alessi,D.R. (2000) Identification of a pocket in the PDK1 kinase domain that interacts with PIF and the C-terminal residues of PKA. EMBO J., 19, 979–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis P.B., Pullen,N., Pearson,R.B., Kozma,S.C. and Thomas,G. (1998) Phosphorylation sites in the autoinhibitory domain participate in p70(s6k) activation loop phosphorylation. J. Biol. Chem., 273, 14845–14852. [DOI] [PubMed] [Google Scholar]
- Dufner A. and Thomas,G. (1999) Ribosomal S6 kinase signaling and the control of translation. Exp. Cell Res., 253, 100–109. [DOI] [PubMed] [Google Scholar]
- Flynn P., Mellor,H., Casamassima,A. and Parker,P.J. (2000) Rho GTPase control of protein kinase C-related protein kinase activation by 3-phosphoinositide-dependent protein kinase. J. Biol. Chem., 275, 11064–11070. [DOI] [PubMed] [Google Scholar]
- Frame S., Cohen,P. and Biondi,R.M. (2001) A common phosphate-binding site explains the unique substrate specificity of GSK3 and its inactivation by phosphorylation. Mol. Cell, 7, 1321–1327. [DOI] [PubMed] [Google Scholar]
- Frodin M. and Gammeltoft,S. (1999) Role and regulation of 90 kDa ribosomal S6 kinase (RSK) in signal transduction. Mol. Cell. Endocrinol., 151, 65–77. [DOI] [PubMed] [Google Scholar]
- Frodin M., Jensen,C.J., Merienne,K. and Gammeltoft,S. (2000) A phosphoserine-regulated docking site in the protein kinase RSK2 that recruits and activates PDK1. EMBO J., 19, 2924–2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland P.M. and Cooper,J.A. (1999) Protein modification: docking sites for kinases. Curr. Biol., 9, R329–331. [DOI] [PubMed] [Google Scholar]
- Jensen C.J., Buch,M.B., Krag,T.O., Hemmings,B.A., Gammeltoft,S. and Frodin,M. (1999) 90-kDa ribosomal S6 kinase is phosphorylated and activated by 3-phosphoinositide-dependent protein dinase-1. J. Biol. Chem., 274, 27168–27176. [DOI] [PubMed] [Google Scholar]
- Kobayashi T. and Cohen,P. (1999) Activation of serum- and glucocorticoid-regulated protein kinase by agonists that activate phosphatidylinositide 3-kinase is mediated by 3- phosphoinositide-dependent protein kinase-1 (PDK1) and PDK2. Biochem. J., 339, 319–328. [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Deak,M., Morrice,N. and Cohen,P. (1999) Characteriz ation of the structure and regulation of two novel isoforms of serum- and glucocorticoid-induced protein kinase. Biochem. J., 344, 189–197. [PMC free article] [PubMed] [Google Scholar]
- Kohn A.D., Takeuchi,F. and Roth,R.A. (1996) Akt, a pleckstrin homology domain containing kinase, is activated primarily by phosphorylation. J. Biol. Chem., 271, 21920–21926. [DOI] [PubMed] [Google Scholar]
- Paradis S., Ailion,M., Toker,A., Thomas,J.H. and Ruvkun,G. (1999) A PDK1 homolog is necessary and sufficient to transduce AGE-1 PI3 kinase signals that regulate diapause in Caenorhabditis elegans. Genes Dev., 13, 1438–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Leong,M.L., Buse,P., Maiyar,A.C., Firestone,G.L. and Hemmings,B.A. (1999) Serum and glucocorticoid-inducible kinase (SGK) is a target of the PI 3-kinase-stimulated signaling pathway. EMBO J., 18, 3024–3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pullen N., Dennis,P.B. Andjelkovic,M., Dufner,A., Kozma,S.C., Hemmings,B.A. and Thomas,G. (1998) Phosphorylation and activation of p70s6k by PDK1. Science, 279, 707–710. [DOI] [PubMed] [Google Scholar]
- Richards S.A., Fu,J., Romanelli,A., Shimamura,A. and Blenis,J. (1999) Ribosomal S6 kinase 1 (RSK1) activation requires signals dependent on and independent of the MAP kinase ERK. Curr. Biol., 9, 810–820. [DOI] [PubMed] [Google Scholar]
- Stokoe D., Stephens,L.R., Copeland,T., Gaffney,P.R., Reese,C.B., Painter,G.F., Holmes,A.B., McCormick,F. and Hawkins,P.T. (1997) Dual role of phosphatidylinositol-3,4,5-trisphosphate in the activation of protein kinase B. Science, 277, 567–570. [DOI] [PubMed] [Google Scholar]
- Tanoue T., Adachi,M., Moriguchi,T. and Nishida,E. (2000) A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nature Cell Biol., 2, 110–116. [DOI] [PubMed] [Google Scholar]
- Tanoue T., Maeda,R., Adachi,M. and Nishida,E. (2001) Identification of a docking groove on ERK and p38 MAP kinases that regulates the specificity of docking interactions. EMBO J., 20, 466–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toker A. and Newton,A.C. (2000) Cellular signaling: pivoting around PDK1. Cell, 103, 185–188. [DOI] [PubMed] [Google Scholar]
- Vanhaesebroeck B. and Alessi,D.R. (2000) The PI3K–PDK1 connection: more than just a road to PKB. Biochem. J., 346, 561–576. [PMC free article] [PubMed] [Google Scholar]
- Weng Q.P. Andrabi,K., Kozlowski,M.T., Grove,J.R. and Avruch,J. (1995) Multiple independent inputs are required for activation of the p70 S6 kinase. Mol. Cell. Biol., 15, 2333–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Q.P., Kozlowski,M., Belham,C., Zhang,A., Comb,M.J. and Avruch,J. (1998) Regulation of the p70 S6 kinase by phosphoryl ation in vivo. Analysis using site-specific anti-phosphopeptide antibodies. J. Biol. Chem., 273, 16621–16629. [DOI] [PubMed] [Google Scholar]
- Wick M.J., Dong,L.Q., Riojas,R.A., Ramos,F.J. and Liu,F. (2000) Mechanism of phosphorylation of protein kinase B/Akt by a constitutively active 3-phosphoinositide-dependent protein kinase-1. J. Biol. Chem., 275, 40400–40406. [DOI] [PubMed] [Google Scholar]