Abstract
The absolute requirement for the histone deacetylase activity of Sir2p in silencing coupled with the conservation of Sir2p-like proteins in larger eukaryotes suggests that this molecule plays an important role in gene regulation in all organisms. Here we report the purification and characterization of two Sir2p-containing protein complexes; one of which contains Sir4p and the other Net1p. The Sir4p-containing complex has an NAD-dependent histone deacetylase activity, while the Net1p-containing complex possesses deacetylase activity but only weak NAD-dependent histone deacetylase activity. Finally, we demonstrate that the Sir2p-containing complexes bind nucleosomes efficiently and partially restrict accessibility of the linker DNA to enzymatic probes.
Keywords: NET1/nucleosome/SIR2/SIR3/SIR4
Introduction
Silencing is a form of transcriptional repression that renders large chromatin domains inaccessible to the transcription machinery. Furthermore, silenced domains are refractile to recombination and replicate late in the S-phase of the cell cycle. Silenced domains are packaged into precisely positioned nucleosomes (Reimer and Buchman, 1997; Weiss and Simpson, 1998; Ravindra et al., 1999; Venditti et al., 1999) that result in a chromatin structure that is markedly altered from active chromatin. The formation of a silenced domain is manifested by its inaccessibility to various probes such as the HO endonuclease (Strathern et al., 1982), DNase I (Nasmyth, 1982), methyltransferases (Gottschling, 1992; Singh and Klar, 1992; Fritze et al., 1997) and various restriction enzymes (Loo and Rine, 1994; Donze et al., 1999).
There are four transcriptionally silenced loci in the yeast Saccharomyces cerevisiae; the cryptic mating type loci HML and HMR, the telomeres and the rDNA locus. Silencing of the mating type genes at HML and HMR requires inactivation centers termed silencers, which flank the genes. The silencers contain binding sites for ORC, Rap1p and Abf1p. In addition to the proteins that bind the silencer elements, histones and the products of the four SIR genes SIR1, SIR2, SIR3 and SIR4 (silent information regulator) are necessary for complete silencing (Rine and Herskowitz, 1987). However, none of the Sir proteins binds DNA in a sequence-specific manner. Instead the proteins bound to the silencer are believed to recruit the Sir proteins to the silenced domain (Kamakaka, 1997). An important advance in understanding the molecular mechanism of silencing was made by the observation that mutations of the histones result in derepression of the silent loci (Kayne et al., 1988; Megee et al., 1990; Park and Szostak, 1990; Johnson et al., 1992; Thompson et al., 1994; Dhillon and Kamakaka, 2000) and that Sir3p and Sir4p bind the N-terminal domains of histones H3 and H4 in vitro (Hecht et al., 1995). Additionally, it has been demonstrated that specific lysine residues in the histone tails need to be deacetylated for Sir protein binding and silencing (Braunstein et al., 1996). These results suggest that Sir3p and Sir4p are structural components of the silenced domain and that they function by binding to the histone tails in nucleosomes. The recent demonstration that Sir2p is a histone deacetylase (Imai et al., 2000; Landry et al., 2000; Smith et al., 2000) suggests that its role in silencing might be enzymatic.
Silencing at the telomeres shares many features of silencing at HML and HMR. Telomeric silencing requires Rap1p and the Sir proteins Sir2p, Sir3p and Sir4p (Grunstein, 1997). Multiple Rap1p binding sites at the telomere are believed to act as a silencer that recruits the Sir proteins, which are then thought to function by the same mechanism as they function at HML and HMR.
The rDNA consists of ∼200 copies of the 9.1 kb rDNA repeat that is repressed by Sir2p (Smith and Boeke, 1997). In vivo analysis suggests that approximately half of the genes at the rDNA are transcriptionally repressed at any given time (Dammann et al., 1995). Specific cis-acting silencers have not been defined at the rDNA, but numerous proteins have been implicated in repression of this locus, including Sir2p, which has been shown to be part of the RENT complex (regulator of nucleolar silencing and telophase exit) (Straight et al., 1999). Recruitment of Sir2p to the rDNA appears to occur by a different mechanism than at HML, HMR and telomeres. Recruitment of Sir2p to the rDNA locus requires Net1p (nucleolar silencing establishing factor and telophase regulator 1), which is part of the RENT complex (Shou et al., 1999; Straight et al., 1999).
SIR2 is unique among the SIR genes in that it is essential for transcriptional silencing of all four loci. In addition, multiple paralogs of Sir2p exist in S.cerevisiae and orthologs of Sir2p are present in numerous organisms from Caenorhabditis elegans to humans (Freeman-Cook et al., 2000). The requirement for Sir2p at all known silenced loci in yeast coupled with the conservation of Sir2p-like proteins in larger eukaryotes suggests that this molecule plays an important role in silencing (Brachmann et al., 1995). Recent evidence shows that Sir2p is an NAD-dependent histone deacetylase in vitro (Imai et al., 2000; Smith et al., 2000). These results are consistent with earlier data demonstrating that increasing Sir2p levels in the cell correlated with a decrease in histone acetylation (Braunstein et al., 1993b).
However, none of these studies demonstrated deacetylase activity of native Sir2p purified from yeast cells. Furthermore, as mentioned above, genetic and molecular studies have shown that Sir2p interacts with numerous proteins and probably exists in multiple complexes in the cell. However, there has been no systematic study aimed at purifying and characterizing native Sir2p-containing complexes from yeast cells. We, therefore, decided to perform a thorough biochemical fractionation of Sir2p-containing protein complexes. Here we report the purification and characterization of two Sir2p-containing protein complexes; one of which contains Sir4p and the other Net1p. We further demonstrate that the Sir4p-containing complex is active as an NAD-dependent histone deacetylase, whereas the Net1p-containing complex is active as a deacetylase but has very weak NAD-dependent histone deacetylase activity, even though it contains Sir2p. Finally, we demonstrate that while recombinant Sir2p is unable to bind nucleosomes, both native complexes bind nucleosomes efficiently and partially restrict accessibility of the linker DNA to enzymatic probes.
Results
Analysis of Sir2p-containing complexes
A yeast strain carrying a His6-HA3 epitope tag at the N-terminus of the SIR2 gene under its own promoter, at its endogenous locus was generated (ROY 1515). The epitope-tagged Sir2p was functionally active in our silencing assays (data not shown).
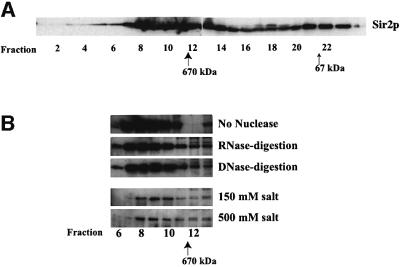
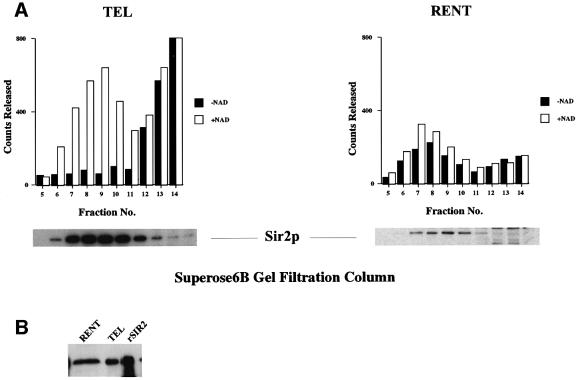
Whole cell lysates, prepared from logarithmically grow ing yeast cells, were applied directly on a Superose 6B gel-filtration column to determine the apparent molecular weight of Sir2p-containing complexes. Column fractions were analyzed by SDS–PAGE followed by immunoblotting using antiserum specific to Sir2p (Figure 1A). This analysis indicated that Sir2p eluted from the column in two distinct peaks of apparent molecular weight of ∼900 and ∼70 kDa. In all probability, the 70 kDa peak represented free Sir2p, which has a predicted molecular weight of 63 kDa, while the 900 kDa peak represented the major Sir2p-containing complexes.
Fig. 1. Sir2p is part of a large stable multi-protein complex in yeast. (A) Elution profile of Sir2p fractionated on a gel-filtration column. A whole cell extract from ROY 1515 was fractionated on a Superose 6B gel-filtration column and 15 µl aliquots of each fraction were analyzed by protein immunoblotting using antisera specific to yeast Sir2p. (B) Partially purified fractions containing Sir2p were treated with either DNase I, RNase A or 500 mM KCl and then fractionated on a Superose 6B gel-filtration column followed by protein immunoblotting using antisera specific to yeast Sir2p.
We next determined the stability of the Sir2p-containing complexes. A partially purified fraction of Sir2p was treated with DNase I, RNase A or high concentrations of KCl (500 mM) for 30 min at room temperature and was then subjected to gel-filtration analysis as in Figure 1A. The results of the immunoblotting indicated that the size of the complex was not altered by nuclease treatment, suggesting either that this complex did not contain nucleic acids or, alternatively, that removal of the nucleic acids did not affect the stability of the complex (Figure 1B). In addition, the complex was unaffected by high concentrations of monovalent cations, suggesting that the complex might be hydrophobic in nature, which was consistent with the observation that this complex had a very high affinity for phenyl-Sepharose (data not shown).
Purification of Sir2p-containing complexes
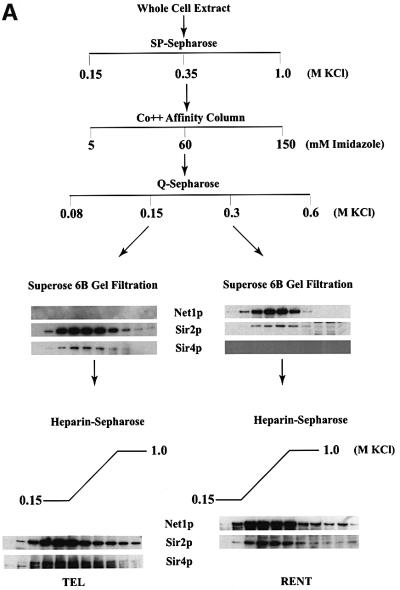
Having ascertained the stability and native size of the complex, we proceeded to purify this complex from whole cell extracts using a combination of affinity and conventional chromatography. The purification scheme for the Sir2p complex is outlined in Figure 2A. The first step in the purification entailed loading the extract onto an SP-Sepharose cation exchange column, and the vast majority of Sir2p eluted from this column in a single peak. The eluate from the 0.35 M peak was next loaded onto an affinity chromatography column using a cobalt-TALON resin to which Sir2p bound via its His6-epitope tag. The Sir2p complex was found by immunoblot analysis to elute from the column in 60 mM imidazole–HCl. This fraction was next loaded onto an anion exchange column (Q-Sepharose). Sir2p fractionated into two distinct peaks: one peak of Sir2p eluting in the 0.15 M KCl step and the second peak eluting from the Q-Sepharose column in 0.3 M KCl. The two peak fractions were further analyzed for the presence of Sir2p, Sir3p, Sir4p or Net1p using antiserum specific to these proteins. This analysis (data not shown) demonstrated that the 0.15 M fraction contained Sir2p as well as Sir4p but did not contain any Sir3p or Net1p. The presence of Sir4p in this complex suggested that this complex might be involved in transcriptional repression at the telomeres and will be referred to as the TEL complex in this manuscript. The 0.3 M fractions contained Net1p along with Sir2p, but not the other Sir proteins, and is probably the previously identified RENT complex (Straight et al., 1999), and will, therefore, be referred to as the RENT complex in this study.
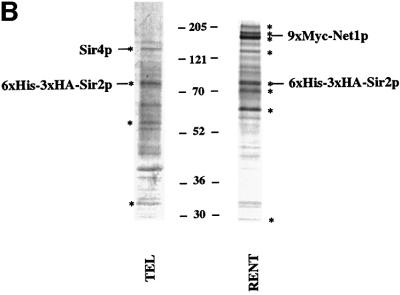
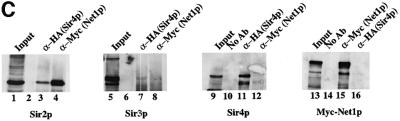
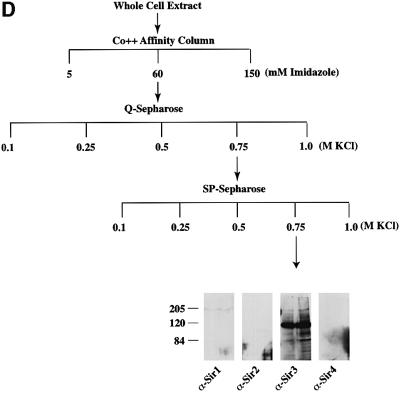
Fig. 2. Purification of Sir protein complexes from yeast whole cell extracts. (A) The fractionation scheme of Sir2p containing complexes from yeast whole cell extracts is summarized in the flow chart. The immunoblots depict selected fractions from the columns probed with antisera against Sir2p, Sir4p and Net1p. (B) Polypeptide composition of the TEL and RENT complexes. Aliquots of the peak fractions from the heparin-Sepharose column were resolved on a 3–18% SDS–polyacrylamide gel and stained with silver. The asterisks denote bands that co-fractionate with Sir2p. Identities of Sir2p, Sir4p and Net1p were ascertained by protein immunoblotting with antibodies specific to these proteins. (C) Co-immunoprecipitation analysis of Sir4p and Net1p. Whole cell extracts (2 mg) generated from ROY 1914 were subjected to immunoprecipitation with commercial antibodies against the Myc and HA epitopes. The input extract as well as aliquots of the immunoprecipitated proteins were resolved on an SDS–polyacrylamide gel and analyzed by protein immunoblotting with antibodies that recognize Sir2p (lanes 1–4), Sir3p (lanes 5–8), Sir4p (lanes 9–12) and Net1p (lanes 13–16). (D) The fractionation scheme of Sir3p containing complexes from yeast whole cell extracts is summarized in the flow chart. The immunoblots depict selected fractions from the SP-Sepharose column probed with antisera against Sir1p, Sir2p Sir3p and Sir4p.
The TEL and RENT complexes were further purified by gel-filtration chromatography on a Superose 6B column. We monitored the elution of various proteins from the gel-filtration column using immunoblots with specific antiserum and the results are shown in Figure 2A. The TEL complex appeared to be slightly smaller than the RENT complex and the complexes were ∼800 kDa and ∼1 MDa, respectively, in size. Sir4p co-fractionated with Sir2p, but not Sir1p, Sir3p or Net1p, which is consistent with it being part of the TEL complex, while Net1p co-fractionated with Sir2p in the RENT complex and did not contain Sir1p, Sir3p or Sir4p.
The final step in the purification involved fractionation over a heparin-Sepharose column using a linear salt gradient from 0.15 to 1.0 M KCl. In both the TEL and the RENT complexes, a single Sir2p-containing peak eluted from this column. Furthermore, in the TEL complex Sir4p co-fractionated with Sir2p fraction for fraction, and similarly Net1p co-fractionated with Sir2p in the RENT complex. Thus, these proteins precisely co-fractionated with Sir2p across four different columns and are most likely integral components of the two complexes.
Figure 2B shows a silver-stained protein profile of the purified TEL and RENT complexes. The asterisks mark the specific polypeptides that reproducibly co-purify with Sir2p and are most likely integral to these complexes. There are probably four proteins in the TEL complex and eight proteins in the RENT complex. The greater number of polypeptides in the RENT complex is consistent with the observation that this complex is larger than the TEL complex.
We confirmed the existence of two distinct Sir2p-containing complexes by co-immunoprecipitation experiments (Figure 2C). We initially generated a strain (ROY 1914) containing Myc9-epitope-tagged Net1p and HA3-epitope-tagged Sir4p at the N-terminus. Both modified genes were under the control of their own promoters and were located at their endogenous loci in the yeast cell. Commercially available monoclonal antibodies against the Myc and HA epitopes were used to immunoprecipitate the tagged proteins from whole cell extracts. The immunoprecipitated proteins were analyzed by SDS–PAGE followed by immunoblotting using antibodies specific to Sir2p, Sir3p, HA-Sir4p and Myc-Net1p. Immunopre cipitation of Net1p using antibodies against the Myc epitope resulted in the co-immunoprecipitation of Sir2p but not Sir3p or Sir4p (compare lanes 4, 8, 12 and 15). Similarly, immunoprecipitation of Sir4p with anti-HA antibodies resulted in the co-immunoprecipitation of Sir2p but not Net1p nor Sir3p (compare lanes 3, 7, 11 and 16). These results are consistent with the data from Figure 2A. Mock experiments without any antibodies (lanes 10 and 14) confirmed the specificity of the immunoprecipitation reactions.
Sir3p is not present in the Sir2p-containing complexes
Surprisingly neither the purified nor the immunoprecipitated TEL or RENT complexes contained Sir3p while previous studies have revealed interactions between Sir3p and Sir4p (Ivy et al., 1986; Marshall et al., 1987; Moretti et al., 1994; Hecht et al., 1996; Moazed and Johnson, 1996). We, therefore, addressed this issue by partially purifying Sir3p from yeast cells and analyzing the co-purified proteins. A yeast strain carrying a functionally active Sir3p with a His6-HA-epitope tag at the C-terminus was generated (ROY 1501) under the control of its own promoter at its native locus. We partially purified Sir3p-containing complexes using affinity and ion exchange chromatography (Figure 2D). Yeast whole cell extracts were initially loaded onto an affinity chromatography column using a cobalt-TALON resin to which Sir3p bound via its His6 epitope tag. The Sir3p complex was found by immunoblot analysis to elute off the column in 60 mM imidazole–HCl. This fraction was next loaded onto an anion exchange column (Q-Sepharose) where Sir3p eluted in one peak at 0.75 M KCl. The peak fraction was next loaded onto a cation exchange column (SP-Sepharose) and eluted from this column at 0.75 M KCl. The peak fractions from the SP-Sepharose column were analyzed for the presence of Sir1p, Sir2p, Sir3p and Sir4p by immunoblotting. These results (Figure 2D) indicated that Sir3p-containing complexes were devoid of the other Sir proteins. Further purification of this protein should help to identify the other proteins, if any, that may be present in this complex.
Analysis of the deacetylation activity of recombinant Sir2p
Recombinant Sir2p has been shown to be an NAD-dependent deacetylase that can deacetylate peptides and core histones in vitro (Imai et al., 2000; Landry et al., 2000; Smith et al., 2000). We wanted to test whether native Sir2p, purified from yeast cells, also behaved as an NAD-dependent histone deacetylase. Substrates for the deacetylase assay were initially generated using purified chicken core histones acetylated with recombinant catalytic fragment of Xenopus PCAF and [3H]acetyl-CoA as described (Wade et al., 1999). Radioactively labeled histones were purified away from the unincorporated label and were subsequently used as substrates for Sir2p-mediated deacetylation.
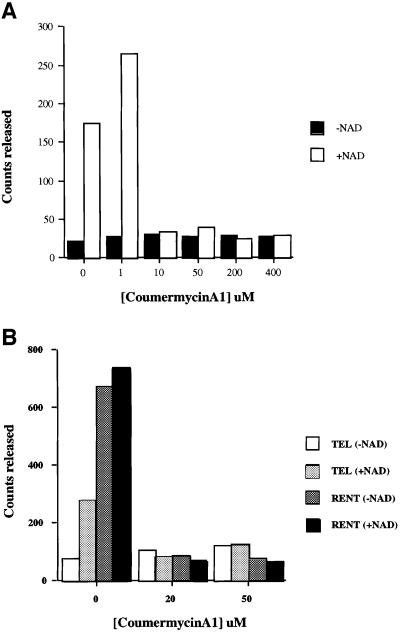
We first tested for deacetylation using recombinant yeast Sir2p purified from Escherichia coli. Consistent with previously published results, the recombinant Sir2p was capable of deacetylating the core histones and this deacetylation was dependent on NAD (data not shown). Recombinant Sir2p has also been shown to possess weak ADP-ribosylation activity and was capable of ADP-ribosylating histones and bovine serum albumin (BSA) (Tanny et al., 1999). However, we were unable to observe any ADP-ribosylation activity with our recombinant Sir2p or the purified TEL and RENT complexes (data not shown). It has also been reported that the ADP-ribosylation of substrates and NAD hydrolysis by Sir2p was sensitive to coumerycin A1 (Imai et al., 2000). We, therefore, examined the coumermycin A1 resistance of the Sir2p deacetylase activity and observed a striking inhibition of deacetylation activity in the presence of as little as 10 µM coumermycin A1 (Figure 3A). This inhibitor should be a useful tool in future studies of Sir2p.
Fig. 3. Analysis of the NAD-dependent histone deacetylase activity. (A) Coumermycin A1 sensitivity of recombinant Sir2p. Deacetylase assays were performed as described using 1 µg of acetylated histones and 0.5 µg of recombinant Sir2p for 30 min at 30°C with varying concentrations of coumermycin A1. The amount of acetate released is depicted in the graph in the presence or absence of added NAD. (B) Coumermycin A1 sensitivity of the TEL and RENT complexes. Deacetylase assays were performed as described using 1 µg of acetylated histones and approximately equal amounts of Sir2p present in the TEL and RENT complexes for 30 min at 30°C with varying concentrations of coumermycin A1. The amount of acetate released is depicted in the graph in the presence or absence of added NAD.
Deacetylation activity of the Sir2p-containing complexes
Recombinant Sir2p and its homologs are NAD-dependent histone deacetylases; however, this enzymatic activity has never been demonstrated for native Sir2p protein purified from yeast cells. We, therefore, assayed every fraction across the gel-filtration column for NAD-dependent deacetylase activity (Figure 4A).
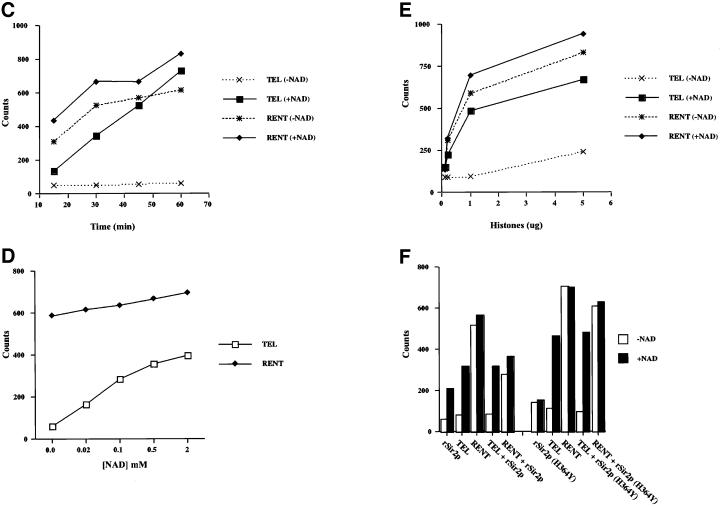
Fig. 4. Analysis of the deacetylase activity of purified Sir2p containing complexes. (A) Co-purification of Sir2p and NAD-dependent deacetylase activity from yeast extracts. Aliquots of the indicated fractions across the Superose 6B gel-filtration column were assayed for NAD-dependent deacetylase activity. A fixed volume of the same fractions was also assayed by protein immunoblotting with antisera against Sir2p. The immunoblot panels are the same as in Figure 2A. (B) Protein immunoblot analysis of Sir2p present in the TEL and RENT complexes. Protein immunoblot analysis with anti-Sir2p antibodies was used to determine approximately equal amounts of Sir2p in the TEL and RENT complexes. Ten microliters (10 µg) of the TEL complex and 40 µl (40 µg) of the RENT complex gave approximately equivalent amounts of signal (equal to ∼0.1 µg of rSir2p). (C) Time course of deacetylation with purified TEL and RENT complexes. Approximately equal amounts of Sir2p present in the TEL and RENT complexes were assayed for deacetylase activity with 1 µg of acetylated histones for varying lengths of time. The amount of acetate released is indicated in the graph. (D) Effect of NAD concentration on the deacetylase activity of the TEL and RENT complexes. The peak fractions containing the TEL and RENT complexes were assayed for deacetylase activity with approximately equal amounts of Sir2p present in the TEL and RENT complexes and 1 µg of histones for 30 min at 30°C with varying concentrations of NAD in the reaction. The amount of acetate released is shown in the graph. (E) Effect of substrate concentration on the deacetylase activity of the TEL and RENT complexes. The peak fractions containing the TEL and RENT complexes were assayed for deacetylase activity for 30 min at 30°C and varying amounts of the histones. The amount of acetate released is shown in the graph. (F) Addition of rSir2p to the TEL and RENT complexes. The deacetylase activity of 0.5 µg of recombinant wild-type or the H364Y mutant Sir2p was measured following addition of these proteins to fractions containing the TEL and RENT complexes. The amount of acetate released is shown in the graph in the presence (black bars) or absence (white bars) of NAD.
Robust NAD-dependent deacetylase activity was observed in the TEL complex that peaked precisely with Sir2p across both the gel-filtration (Figure 4A) and the heparin-Sepharose columns (data not shown). This activity was not sensitive to trichostatin A (data not shown).
Surprisingly, the NAD-dependent deacetylation activity for the RENT complex was very weak. The lack of NAD-dependent histone deacetylation activity in the RENT complex was particularly intriguing as its enzymatic activity is essential for nucleolar silencing (Imai et al., 2000). The apparent lack of NAD-dependent deacetylase activity in the RENT complex was probably a result of the presence of another deacetylase activity that was NAD-independent. We do not believe that the absence of NAD-dependent activity was a result of inhibitors in the buffers used, as both the TEL and RENT complexes were present in the same buffer. The NAD-dependent and -independent activities of the RENT complexes were not significantly sensitive to trichostatin A (data not shown).
We next compared the enzymatic properties of the purified TEL and RENT complexes. In order for these experiments to be comparable, we used an approximately equal amount of Sir2p in each reaction. The amount of Sir2p in the TEL and RENT complexes was quantitated by immunoblot analysis (Figure 4B and data not shown). Approximately 40 µg of the RENT complex gave a signal equivalent to 10 µg of the TEL complex. As the stoichiometry of Sir2 in each of these complexes was not known we are currently unable to determine the number of molecules of the TEL and RENT complexes added in each reaction.
The first experiment was a time course of deacetylation activity with both purified complexes (Figure 4C). With a fixed amount of Sir2p in each complex, there was a linear increase in the amount of acetate released from the histones as a function of time. The TEL complex was more efficient than the RENT complex with respect to NAD-dependent deacetylation. However, the overall extent of deacetylation was approximately the same for both complexes.
Recent data have shown that NAD functions as an acceptor molecule for the acetate hydrolyzed from the histones (Tanner et al., 2000). We, therefore, analyzed the NAD utilization by the two complexes (Figure 4D). Results from this analysis suggested that for a fixed amount of substrate (1 µg of histones) and length of time (30 min), increasing concentrations of NAD gave a concomitant increase in deacetylation by both the TEL and RENT complexes, although the fold increase was significantly higher for the TEL complex.
As the RENT complex appeared to be far less active in NAD-dependent deacetylation compared with the TEL complex, we reasoned that this might be because of occlusion of the substrate by other proteins. If this were the case, then increasing the substrate concentration should ultimately lead to increased availability of the substrate for NAD-dependent deacetylation by the Sir2p present in the RENT complex. However, even a 20-fold increase in substrate concentration did not dramatically stimulate the NAD-dependent deacetylase activity of the RENT complex (Figure 4E), suggesting that occlusion or sequestration of the substrate was not the reason for the lack of NAD-dependent activity observed.
We also wished to determine whether the NAD-dependent enzymatic activity of recombinant Sir2p would be affected when this protein was mixed with either the TEL or the RENT complex. To perform this analysis a fixed amount of wild-type or mutant rSir2p was first added to either the RENT or the TEL complex and incubated for 15 min in the presence of NAD. Following this incubation, acetylated histones were added and the reaction was allowed to proceed for an additional 30 min. The results from this analysis suggest that the NAD-dependent deacetylase activity of rSir2p was not dramatically affected by the presence of either the TEL or the RENT complex (Figure 4F). The addition of wild-type rSir2p to the RENT complex led to a diminution of the NAD-independent deacetylation relative to the NAD-dependent activity.
As the activity mediated by the recombinant Sir2p was sensitive to coumermycin A1, we wished to determine whether the same was true for the TEL and RENT complexes. We analyzed the NAD-dependent histone deacetylation activity of both complexes at 20 and 50 µM coumermycin A1 (Figure 3B). Both complexes were highly sensitive to the inhibitor, and furthermore, even the NAD-independent deacetylase activity of the RENT complex was sensitive to coumermycin A1 but not dimethylsulfoxide (DMSO). Identification of the polypeptides in these complexes should aid in the identity of the various activities present in these complexes and their relationship to each other.
Analysis of mutant Sir2p complexes
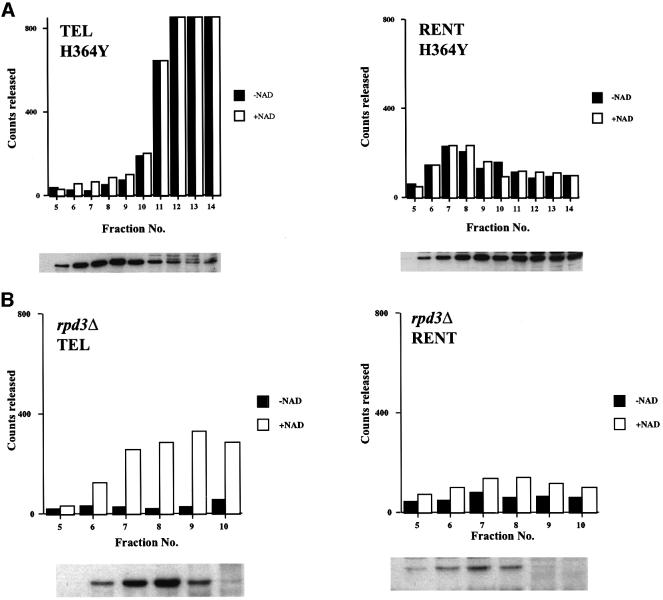
A single point mutation in Sir2p (H364Y) has been shown to abolish silencing in vivo. While this mutation does not affect its association with either Sir4p or Net1p, a recombinant version of this mutant is enzymatically inactive (Tanny et al., 1999). However, it was not known whether the entire complex also remained intact in this mutant. A strain (ROY 1840), bearing a genomic copy of His6-HA3-tagged Sir2p containing the H364Y mutation under the control of its own promoter was constructed. Sir2p-containing complexes were purified from this strain using the fractionation scheme outlined in Figure 2A and protein immunoblotting was used to monitor the purification. The size (Figure 5A) and chromatographic profiles of the mutant complexes were very similar to the wild-type complexes (data not shown), indicating that loss of silencing in the mutant was because of, primarily, the loss of deacetylation activity and not because of dissociation of the complexes. We also monitored deacetylation activity and found that the TEL complex did not possess any NAD-dependent deacetylation activity (Figure 5A). The RENT complex was similarly inactive for the NAD-dependent but not for the NAD-independent activity in the mutant strain. These results are similar to the Swi/Snf complex where mutations in ATPase function caused loss of nucleosome remodeling but did not affect complex assembly (Cote et al., 1994).
Fig. 5. Purification and analysis of the deacetylase activity of Sir2p-containing complexes purified from mutant cells. Aliquots of the indicated fractions across the gel-filtration column were assayed for NAD-dependent deacetylase activity. (A) The amount of deacetylation mediated by Sir2p containing complexes purified from cells with a mutant Sir2p (H364Y) are shown in graphical form. The immunoblot profile under the graphs is for Sir2p. (B) The amount of deacetylation mediated by Sir2p-containing complexes purified from cells lacking Rpd3p is shown. The immunoblot profile under the graphs is for Sir2p.
Rpd3 is a class I trichostatin A-sensitive histone deacetylase that negatively regulates silencing at the telomeres and the rDNA. As the RENT complex had low NAD-dependent deacetylase activity, we wondered whether this complex might be under negative control of Rpd3p; if this were the case, cells lacking Rpd3p should show a concomitant increase in NAD-dependent deacetylase activity. We purified Sir2p-containing complexes from a rpd3Δ::LEU2 strain (ROY 1834) using the fractionation scheme outlined in Figure 2A. The size and chromatographic profiles of the Sir2p complexes were identical to those isolated from wild-type strains. Deacetylation activity across the gel-filtration column revealed that the RENT complex was still inactive for NAD-dependent deacetylation activity (Figure 5B), while the TEL complex exhibited robust activity.
Sir protein interactions with nucleosomes
The Sir proteins are required for maintaining genes in a silenced state and it has been suggested that these proteins silence genes by creating higher-order chromatin structures that prevent the transcription machinery from gaining access. Previously published data have shown that Sir proteins, primarily Sir3p and Sir4p, interact with the core histone tails when these tails are presented as glutathione S-transferase (GST) fusion proteins (Hecht et al., 1995). As free histone tails are unlikely targets for the Sir proteins in vivo, we decided to examine the interactions of the Sir proteins with reconstituted rotationally positioned nucleosomes.
Either a 200 or a 400 bp fragment of DNA containing one or two repeated 5S rRNA genes were used in the reconstitution experiments. These DNA fragments have been shown to rotationally position nucleosomes (Ura et al., 1996). Mono- and dinucleosomes were reconstituted onto these fragments using previously described methods (Ura et al., 1996). Appropriately assembled nucleosomes (see Figure 6B and D) were then utilized for all of the binding studies described below.
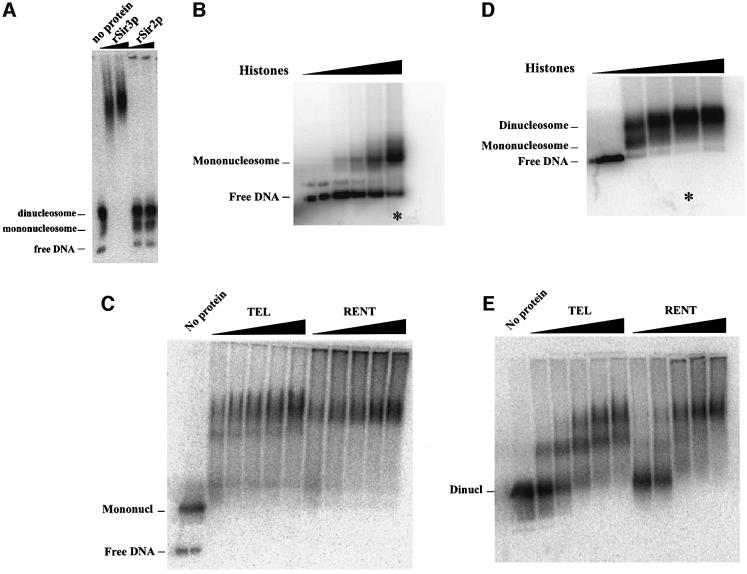
Fig. 6. Sir protein binding to dinucleosomes. (A) Recombinant Sir protein binding to dinucleosomes. Dinucleosomes were reconstituted as described and then bound with varying amounts of recombinant Sir3p or Sir2p. Following binding, the complexes were resolved on a 0.7% agarose gel, the gels were dried and the bands visualized by autoradiography. (B) Reconstitution of mononucleosomes. Mononucleosomes were reconstituted onto a 200 bp fragment of DNA containing a 5S rRNA gene using the octamer exchange method. Increasing amounts of unlabeled mononucleosomes were added to a fixed amount of end-labeled DNA at high salt and the salt concentration was gradually reduced by dilution resulting in the transfer of octamers from unlabeled to labeled DNA fragments. The reconstituted nucleosomes were analyzed by gel mobility shift analysis on a 1% agarose gel in 0.5× TBE. The reaction marked by the asterisk was used for the gel mobility shift data presented in (C). (C) Binding of purified TEL and RENT complexes to mononucleosomes. Reconstituted mononucleosomes were bound with increasing amounts of the purified TEL and RENT complexes and the tertiary complexes were resolved on a 1% agarose gel in 0.4× TBE. (D) Reconstitution of dinucleosomes. Dinucleosomes were reconstituted onto a 400 bp fragment of DNA containing two tandemly repeated 5S rRNA genes using the octamer exchange method as described in (B). The reaction marked by the asterisk was used for the gel mobility shift data presented in (E). (E) Binding of purified TEL and RENT complexes to dinucleosomes. Reconstituted dinucleosomes were bound with increasing amounts of the purified TEL and RENT complexes and the tertiary complexes were resolved on a 0.7% agarose gel in 0.4× TBE.
Binding assays were initially performed using recombinant Sir2p isolated from E.coli and recombinant Sir3p purified from baculovirus. Recombinant Sir3p bound to the dinucleosome and reduced the mobility of the nucleosomal band (Figure 6A). Furthermore, Sir3p showed a marked preference for binding to naked DNA over dinucleosomes and a preference for binding dinucleosomes over mononucleosomes (data not shown). Recombinant Sir2p, however, was unable to bind to nucleosomes as observed by changes in gel mobility. The addition of NAD did not affect the gel mobility profile with recombinant Sir2p (data not shown). It is not clear if this is a function of E.coli-expressed Sir2p or an intrinsic property of the Sir2 protein.
Having established conditions that allowed Sir3p binding to nucleosomes, we next tested the ability of the purified RENT and TEL complexes to bind nucleosomes. Increasing amounts of the two complexes were added to reconstituted dinucleosomes and the bound complexes were resolved on a 0.7% agarose gel. Both the RENT and the TEL complex were able to alter the mobility of the dinucleosomes (Figure 6E). The appearance of gel-shifted bands increased with increasing amounts of the complexes added. Inspection of the gel-shifted complexes indicated that the TEL complex initially gave rise to an intermediate migrating band that appeared to shift to a slower migrating band on addition of greater amounts of the complex. The RENT complex also gave two bands but the transition from the faster to the slower migrating band occurred at lower amounts of Sir2p added to the reaction. However, in both cases the observed pattern suggested that two molecules of the complex bound per dinucleosome fragment.
To confirm this conclusion, we reconstituted core mononucleosomes onto a 200 bp fragment of DNA (Figure 6B). The purified complexes were allowed to bind to the core mononucleosomes and resolved on a 1% agarose gel. As observed in Figure 6C the Sir2p-containing complexes gave rise to a single gel-shifted band, indicating that one molecule of each complex bound to a single core mononucleosome.
Mock reconstitution experiments were also performed with fractions purified from a sir2Δ strain (JRY 4568). Gel mobility shifts with these fractions did not result in any of the bands observed with the RENT or TEL complexes, indicating the specificity of the gel shifts (data not shown).
Accessibility analysis of Sir protein-bound dinucleosomes
Biochemical analyses of silenced chromatin have relied primarily on measuring the accessibility of various enzymatic probes to their cognate binding sites when those sites are packaged into silenced chromatin (Nasmyth, 1982; Strathern et al., 1982; Gottschling, 1992; Loo and Rine, 1994; Fritze et al., 1997; Donze et al., 1999).
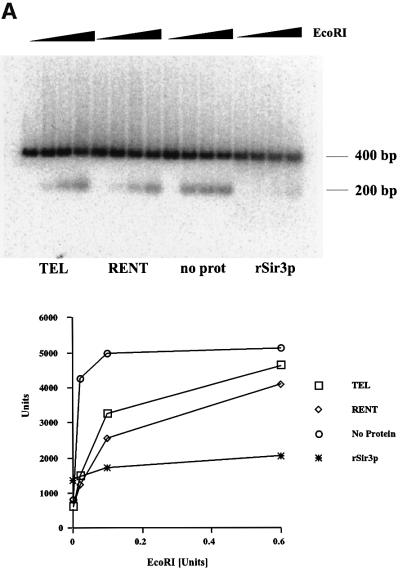
Having reconstituted Sir protein binding to dinucleosomes, we analyzed the binding of these proteins by measuring the accessibility of restriction enzymes. The dinucleosome fragment used in this study has a unique EcoRI site located between the two positioned nucleosomes. We measured the accessibility of this site to digestion by the EcoRI enzyme. Reactions containing nucleosomes alone or bound with various Sir protein complexes were digested for a fixed length of time with increasing concentrations of the enzyme. The release of a 200 bp fragment upon digestion with EcoRI was quantitated as a function of enzyme concentration and the results are presented as a graph in Figure 7A. Each experiment was performed at least three times and the data demonstrated that the EcoRI site was most accessible in samples lacking any Sir proteins. The presence of Sir3p significantly inhibited accessibility of the restriction enzyme while binding of the TEL or RENT complex reduced enzyme accessibility but not to the same extent as with Sir3p alone.
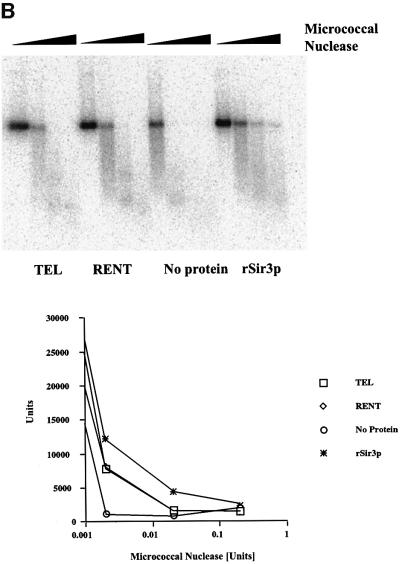
Fig. 7. Nuclease accessibility analysis of the reconstituted dinucleosomes. (A) Restriction enzyme accessibility analysis of the reconstituted dinucleosomes. Dinucleosomes bound with various Sir protein complexes (lanes 1–4 contained the TEL complex; lanes 5–8 contained the RENT complex; lanes 9–12 had no protein complex added; and lanes 13–16 contained recombinant Sir3p) were digested with increasing concentrations of EcoRI (0–0.6 U) for 15 min. The digestion was stopped, DNA was isolated and analyzed on a 1.5% agarose gel followed by autoradiography. The amount of radioactivity in the 200 bp DNA fragment was quantitated and is presented in graphical format. (B) Micrococcal nuclease accessibility analysis of the reconstituted dinucleosomes. Dinucleosomes bound with various Sir protein complexes (lanes 1–4 contained the TEL complex; lanes 5–8 contained the RENT complex; lanes 9–12 had no protein complex added; and lanes 13–16 contained recombinant Sir3p) were digested with increasing concentrations of micrococcal nuclease (0–0.25 U) for 15 min. The samples were then analyzed as described in (A).
These results were independently confirmed using a second endonuclease micrococcal nuclease that preferentially digests linker DNA present between nucleosomes. Dinucleosomes with or without added Sir proteins were digested for a fixed length of time with increasing concentrations of micrococcal nuclease and the resulting products were resolved on an agarose gel. The results of this analysis are presented in Figure 7B along with a graphical representation of the disappearance of the dinucleosome band as a function of enzyme concentration. Similar to the results obtained with EcoRI, the dinucleosome in the absence of Sir proteins was readily digested with the enzyme, while Sir3p-containing nucleosomes were highly resistant to digestion and the TEL- and RENT-associated nucleosomes were intermediate in their accessibility to micrococcal nuclease.
Discussion
The Sir proteins are domain-specific repressors that function through interactions with nucleosomes to create a specialized chromatin structure that inhibits transcription and recombination. Sir2p is the only Sir protein required for transcriptional repression of all four silenced loci in S.cerevisiae (Rine and Herskowitz, 1987; Aparicio et al., 1991; Smith and Boeke, 1997). This observation, coupled with the conservation of Sir2-like genes in prokaryotes and all eukaryotes, suggests that Sir2p plays a pivotal role in silencing (Brachmann et al., 1995). Previous genetic and biochemical data had indicated that Sir2p probably existed in two distinct protein complexes: one necessary for silencing HML, HMR and the telomeres, while a second distinct Sir2p-containing complex mediated silencing at the rDNA (Moazed et al., 1997; Straight et al., 1999; Cuperus et al., 2000).
We undertook a biochemical approach to isolate and characterize all of the major Sir2p-containing complexes from yeast cells. Our studies led to the isolation of two distinct complexes that contained Sir2p.
The TEL complex
One of the complexes that we purified was a large multi-protein complex with an approximate molecular weight of 800 kDa, which contains Sir2p and Sir4 and other unidentified subunits. This complex did not contain either Sir3p or Net1p. The presence of Sir4p in this complex suggests that it may be involved in silencing at HML, HMR and the telomeres. Studies have demonstrated an interaction between Sir2p and Sir4p, and co-immunoprecipitation experiments have revealed the absence of Sir3p from these complexes (Moazed and Johnson, 1996; Moazed et al., 1997; Straight et al., 1999). Our data showing that the Sir2p–Sir4p TEL complex was deficient in Sir3p are consistent with these studies.
However, studies using two-hybrid analysis (Moretti et al., 1994), over-expression suppressor analysis (Ivy et al., 1986; Marshall et al., 1987) as well as analysis of protein–protein interactions in vitro, (Hecht et al., 1996; Moazed and Johnson, 1996) suggest an interaction between Sir3p and Sir4p. As we were unable to detect any direct interaction between Sir3p and Sir4p in solution, our results can be reconciled with the published data by postulating that these interactions occur only following binding of these proteins to nucleosomes. Studies on extended telomeric heterochromatin demonstrate that Sir3p can associate with nucleosomes in the absence of Sir2p and Sir4p (Renauld et al., 1993; Hecht et al., 1996; Strahl-Bolsinger et al., 1997). Furthermore, in vitro binding studies between Sir2p, Sir3p and Sir4p suggest that the binding of Sir3p to Sir4p may occur subsequent to Sir4p–Sir2p interaction (Moazed et al., 1997). As we are unable to isolate a soluble complex containing Sir2p, Sir3p and Sir4p, we favor a model whereby Sir3p interacts with Sir4p following the recruitment of these proteins to the silenced loci. These results also imply that the recruitment of the Sir2p–Sir4p complex may be independent of the recruitment of Sir3p and are consistent with data demonstrating that over-expression of either Sir3p or Sir4p could suppress a defective sir1 allele (Dhillon and Kamakaka, 2000). Our demonstration that recombinant Sir3p isolated from baculovirus can bind stably to dinucleosomes in the absence of Sir2p and Sir4p is also consistent with these observations.
The role of Sir proteins in silencing
The role of Sir4p in silencing appears to be distinct from Sir3p. Our accessibility data showed that the Sir2p–Sir4p TEL complex was not as efficient in protecting the linker DNA as Sir3p and suggests that the region of the nucleosome that interacts with the TEL complex may be distinct from the region that binds Sir3p. Additionally, Sir4p could function as a chaperone for Sir2p, as we have shown the existence of a stable Sir2p–Sir4p complex and it is not clear how Sir2p is recruited to the silenced chromatin. A final role for Sir4p that is not addressed in this study could be to anchor the silenced loci to the nuclear periphery (Ansari and Gartenberg, 1997).
Recombinant Sir2p has an NAD-dependent deacetylase activity that is essential for silencing, indicating that its role is probably enzymatic (Imai et al., 2000; Landry et al., 2000; Smith et al., 2000). Additionally, it has been shown that over-expression of Sir2p promotes a global deacetylation of histones in vivo (Braunstein et al., 1993a, 1996). Our demonstration of an NAD-dependent histone deacetylase activity in the Sir2p–Sir4p containing TEL complex further supports the enzymatic role of Sir2p in silencing. While the in vivo substrates of Sir2p are not known, deacetylation of specific lysine residues in histones H3 and H4 is a pre-requisite for silencing (Thompson et al., 1993). Our data showing efficient deacetylation of core histones by the Sir2p–Sir4p TEL complex is consistent with a role for this protein in deacetylating these residues. However, this does not preclude other substrates being involved in silencing (Sekinger and Gross, 2001). Sir2p may also be involved in maintaining the histones in the deacetylated state rather than de novo deacetylation. Despite being catalytically active, recombinant Sir2p could not bind nucleosomes composed of non-acetylated histones, suggesting that tethering and/or targeting of Sir2p to the nucleosomes in silenced chromatin may be a function of the other proteins within the complex such as Sir4p (Hecht et al., 1995).
Our preliminary observations that Sir3p had a preference for binding naked DNA over nucleosomes would suggest that Sir3p interacts with linker DNA between nucleosomes. Further corroboration of this model stems from our observation that Sir3p could efficiently protect the linker DNA between nucleosomes. Genetic and biochemical data also indicate that Sir3p interacts with the N-termini of histones H3 and H4 (Johnson et al., 1990; Hecht et al., 1995) and is probably involved in arranging nucleosomes into a precisely positioned array (Reimer and Buchman, 1997; Vega-Palas et al., 1998; Weiss and Simpson, 1998; Ravindra et al., 1999). Future experiments should help determine the contribution of histone tails to the stability of Sir3p binding to nucleosomes. It should also be noted that the binding of Sir3p to linker DNA does not preclude its interactions with core histone tails, as histone H1 has been shown to interact with linker DNA as well as with the core histones (Thomas and Khabaza, 1980).
The RENT complex
The mechanism of silencing at the rDNA locus is not as well defined as silencing at the other loci. Silencing at the rDNA requires numerous proteins and it has been shown that Sir2p at the rDNA is part of the RENT complex (Shou et al., 1999; Straight et al., 1999), which consists of Sir2p, Cdc14p, Net1p and Nan1p (Net1p-associated nucleolar protein 1). Based on the presence of Net1p in the second Sir2p-containing complex that we have purified, we propose that this complex constitutes the previously identified RENT complex.
Recent data have shown that in addition to the RENT complex, the nucleolus contains numerous other multi-protein complexes. Two such complexes are the Werner syndrome protein complex, which is comprised of Sgs1 and topoisomerase II and I (Lebel et al., 1999) and the condensin complex, which is composed of Smc2p, Smc4p and Brn1p (Freeman et al., 2000). The third large protein complex found in the nucleolus is the rRNA transcription and processing machinery (Fath et al., 2000). We have investigated whether these complexes co-purified with the RENT complex by immunoblot analysis using antibodies against topoisomerases I and II, Smc2p, Brn1p and Nop1p. While some of the proteins were found to co-purify in the early steps of purification, none of these proteins was found in the purified RENT complex (data not shown).
As the purified RENT complex bound nucleosomes, other proteins within the complex are probably required for nucleosomal binding. Net1p may mediate the targeting of Sir2p to chromatin, since Net1p cross-links preferentially with the rDNA repeat and Sir2p is not localized to the nucleolus in its absence (Straight et al., 1999). Additional nucleosomal gel-shift experiments with complexes purified from strains lacking Net1p should help answer this question.
The most intriguing result of our analysis of the Sir2p-containing complexes is the demonstration that the RENT complex had a weak NAD-dependent deacetylase activity. Several possible models can explain this observation. As the complex has a significant amount of NAD-independent deacetylase activity we believe that there is a second deacetylase present in this fraction. However, at this point we are unable to unequivocally state that this second deacetylase is an integral component of the RENT complex.
Other possibilities to explain our results include the presence of an activity in the fraction containing the RENT complex that blocked NAD-dependent deacetylation by Sir2p. Alternatively, the in vivo substrates for the RENT complex are not free histones but some other protein (Muth et al., 2001). It is not known whether the histones on the silenced rDNA genes are hypoacetylated or whether this acetylation state changes in cells deficient in Sir2p. The final possibility is that the NAD-dependent deacetylation activity of Sir2p in the RENT complex is restricted to a specific stage in the cell cycle. This is the case for Cdc14p, a component of the RENT complex that is inactive as a protein phosphatase while it is part of the RENT complex and remains inactive until its release from Net1p prior to telophase (Shou et al., 1999).
In conclusion, we have shown that there are two major Sir2p-containing complexes in yeast cells. One of these large complexes contains Sir4p while the second complex contains Net1p. We believe that these complexes represent the Sir2p complexes present at the telomeres and rDNA, respectively. The Sir2p present in the TEL complex is highly active as an NAD-dependent histone deacetylase, while the RENT complex has strong deacetylase activity but weak NAD-dependent deacetylase activity. Further more, both the RENT and TEL complexes can stably bind nucleosomes and partially protect the linker DNA from digestion.
Materials and methods
Strains
All strains used in this study are isogenic to W-303 and are shown in Table I.
Table I. Yeast strains.
| Strain | |
|---|---|
| ROY 1515 | MATa His6-HA3-SIR2 pep4Δ::TRP1 net1::myc9-NET1/LEU2 ade2-1 LYS2 |
| ROY 1914 | MATα HA3-Sir4 pep4Δ::TRP1 net1::myc9-NET1/LEU2 ade2-1 LYS2 |
| ROY 1501 | MATa sir3Δ::SIR3-HA-His6/URA3 pep4Δ::TRP1 net1::myc9-NET1/LEU2 ade2-1 LYS2 |
| ROY 1840 | MATα sir2Δ::HIS3 His6-HA3-sir2 (H364Y)::URA3 ade2-1 LYS2 |
| ROY 1834 | rpd3Δ::LEU2 His6-HA3-SIR2 pep4Δ::TRP1 net1::myc9-NET1/LEU2 ade2-1 LYS2 |
Preparation of yeast extracts
Cells were grown overnight in yeast extract/peptone/dextrose (YPD) medium at 30°C with vigorous shaking. Most extracts were prepared from cultures harvested at OD600 of 1.5. A typical extract was prepared from 10 l of cells. The cells were spun in a GS3 rotor at 4000 r.p.m. at 4°C for 5 min. The cells were washed with 1 l of 50 mM HEPES pH 7.5 followed by two washes in extrusion buffer [50 mM HEPES, 150 mM NaCl, 0.1 mM phenylmethsulfonyl fluoride (PMSF), 0.5 mM benzamidine– HCl]. The wet weight of the cell pellet was determined and the cells were resuspended in extrusion buffer at a ratio of 1 g of cells:0.1 ml of buffer. Usually 10 l of culture yielded 50 g cells. The cell slurry was loaded into a syringe and extruded into liquid nitrogen and stored frozen.
The frozen cells were broken using a coffee grinder. A Braun aromatic coffee grinder was filled with dry ice pellets and a short pulse pulverized the pellets. Enough dry ice was used to cover the blades in the machine. Approximately 20 g of frozen cells were placed in the coffee grinder along with the dry ice and ground with shaking for 5 min. Following the grinding, all procedures were performed on ice. The powder of broken cells was transferred to an ice-cold beaker and cell extraction buffer (50 mM HEPES, 150 mM NaCl, 20 µM zinc acetate, 0.1% Tween 20, 5 mM β-mercaptoethanol along with protease inhibitors) was added to the frozen cells at a ratio of 1 g of cells:2 ml of buffer. Once the cell lysates had thawed, they were homogenized in a ground glass homogenizer, transferred to tubes and spun at 12 000 r.p.m. for 10 min in an HB6 Sorvall rotor. The supernatant was removed and processed further.
Purification of Sir2p-containing complexes
The extract was loaded directly onto a SP-Sepharose column equilibrated in HEMG buffer (25 mM HEPES, 12.5 mM MgCl2, 0.1 mM EDTA, 10% glycerol, 0.1% Tween 20) with 150 mM KCl. All HEMG buffers contained 1 mM dithiothreitol (DTT) as well as protease inhibitors. The column was washed in three steps with five column volumes of buffer at each step. The three steps of HEMG buffer contained 150 mM KCl, 350 mM KCl and 1 M KCl. The majority of Sir2p-containing complexes eluted in the 0.35 M KCl step, with a small amount eluting in the flow through.
Cobalt-TALON resin was equilibrated in 50 mM HEPES, 150 mM NaCl, 0.5% Tween 20 and 10% glycerol. The equilibration buffer was aspirated off the resin and the protein fractions were added to the resin and shaken on an end-over-end stirrer for 1 h at 4°C. The slurry was then poured into an empty chromatography column and the resin allowed to settle. The flow through was collected and the column washed sequentially with buffer containing 50 mM HEPES, 150 mM NaCl, 10% glycerol, 0.1% Tween 20 and increasing concentrations of imidazole–HCl (5 mM followed by 60 mM followed by 150 mM). The Sir2p containing complexes eluted off the column in the 60 mM imidazole–HCl fraction.
The Sir2p-containing fraction was diluted with HEMG buffer to 0.08 M KCl and loaded onto a Q-Sepharose column equilibrated in HEMG buffer with 0.08 M KCl. Step elutions were once again performed with 0.08, 0.15, 0.3 and 0.6 M KCl in HEMG buffer. Sir2p was present in the 0.15 and 0.3 M KCl fractions.
These two Sir2p-containing complexes were next loaded onto a Superose 6B gel-filtration column that had previously been equilibrated in HEMG buffer with 0.15 M KCl.
The Sir2p-containing fractions were pooled, and next loaded onto a heparin-Sepharose column equilibrated in HEMG with 0.15 M KCl. A linear gradient of KCl (10 column volumes) was applied to the column from 0.15 to 1 M. The last two columns (gel-filtration and heparin-Sepharose) were interchangeable without any change in the protein profile or enzymatic characteristics of the purified complexes. The Sir2p-containing fractions were frozen in liquid nitrogen and stored at –70°C.
Bacterial expression and purification of recombinant Sir2p
The GST–Sir2 clone was a gift of Danish Moazed. Luria–Bertani medium (400 ml) was inoculated to an OD of 0.1 from a 5 ml overnight culture and grown to an OD600 of 0.4–0.5 at 37°C. The protein expression was induced with isopropyl-β-d-thiogalactopyranoside (IPTG) (final concentration of 1 mM); then the cells were grown at 30°C for 4 h. After harvesting the cells (5000 r.p.m., 10 min, GS 3-rotor), they were resuspended in 3 ml of Tris-buffered saline (TBS) with protease inhibitors. This step was repeated. After sonication for 2 min (strength 4, 50%), 55 µl of Triton X-100 were added. The cells were incubated for 1 h at 4°C, rotating. The cell debris was centrifuged in 2 ml tubes at 14 000 r.p.m. for 30 min. The supernatant was pooled and 1.1 ml of unsettled glutathione-beads, which were washed three times in TBS with protease inhibitors, were added. After rotating at 4°C for 1 h, the mixture was poured into a small empty column. The flow through was discarded. The beads in the column were washed with 4 ml of TBS with protease inhibitors, and again with 2 ml of TBS (without protease inhibitors). This was followed by a wash step with 2 ml of cleavage buffer (25 mM Tris–HCl pH 7.5, 100 mM NaCl, 2 mM CaCl2, 2 mM MgCl2, 10% glycerol, 1 mM DTT). The column was closed and 700 µl of cleavage buffer and 72 µl of thrombin protease (1 U/µl) were added at room temperature, and the column was incubated for 3 h at room temperature, rotating. The column was centrifuged shortly and the flow through was collected which contained the purified proteins.
Baculovirus expression of full-length Sir3p and isolation and purification of the protein from Sf9 cells
Plasmid pRO297 was cleaved with the restriction enzymes NotI–PvuII. The 3.5 kb fragment containing the Sir3p-sequence with a His6-HA-tag at the C-terminus of the protein was inserted in the NotI–SmaI sites of the pVL1392 vector (Invitrogen) for baculovirus expression of Sir3p. This plasmid was used to generate baculovirus with this gene. Infections of ∼2 × 106 Sf9 cells, 1.5 × 108 p.f.u./ml of the virus stock were used (80 µl of virus stock to an 8 cm petri dish with 10 ml of cells). The cells were incubated at 27°C for 4 days and then harvested (5 min, 4°C, 2500 r.p.m., HB 4-rotor). The supernatant was discarded, and the pellet of cells from a single petri dish was incubated in 200 µl of insect lysis buffer (10 mM Tris pH 7.5, 130 mM NaCl, 1% Triton X-100, 10 mM NaF) with protease inhibitors for 45 min on ice. The solution was centrifuged for 5 min at 4°C, and the supernatant was collected. Two milliliters of unsettled cobalt-TALON resin in cobalt wash buffer (50 mM HEPES, 150 mM NaCl, 10% glycerol, 0.5% Tween 20) were added to the cell extract. After rotating for 1 h at 4°C the slurry was poured into an empty column. The flow through was discarded. The resin in the column was washed with five column volumes of cobalt wash buffer. The bound protein was eluted with 300 mM imidazole cobalt wash buffer, dialyzed against 50 mM KCl–HEMG buffer.
Preparation of mononucleosomes from chicken erythrocytes
Chicken erythrocyte nuclei were prepared as described (Kamakaka and Thomas, 1990). To isolate chromatin, 100 OD260 of nuclei were washed with 10 ml HEPES buffer (15 mM HEPES pH 7.4, 15 mM NaCl, 60 mM KCl, 2 mM MgCl2, 0.2 mM PMSF). After spinning at 3000 r.p.m. for 5 min at 4°C, the nuclei were resuspended in 5 ml of HEPES buffer. They were incubated for 10 min at 37°C and CaCl2 was added to a final concentration of 2 mM. One-hundred and fifty Worthington units of micrococcal nuclease were added and incubated at 37°C for 30 min. The reaction was stopped by the addition of EDTA to a final concentration of 5 mM, and the reaction was placed on ice. After centrifugation at 3000 r.p.m. for 5 min, the supernatant contained mononucleosomes. The mononucleosome fraction did not contain histone H1 and was used for the reconstitution of dinucleosomes on a 32P-labeled DNA fragment.
Reconstitution of nucleosomes and mobility shift-assays in agarose gels
The DNA fragments containing one or two 5SrRNA genes were either end-labeled as described (Ura et al., 1996) or PCR labeled. Three microliters of the labeled gel-eluted fragment were added to varying amounts of chicken erythrocyte mononucleosomes, 2 µl of 5 M NaCl and 1 µl of 100 mM β-mercaptoethanol in a final volume of 10 µl. After 30 min of incubation at room temperature, the reaction was diluted by the addition of 5 µl of TE. After an additional incubation of 15 min, a second aliquot of 5 µl of TE was added. This step was repeated twice more and finally 170 µl of TE were added. The reconstituted nucleosomes were analyzed on an agarose gel in 0.4× TBE.
For the mobility shift assays, 1 µl of the reconstituted nucleosomes was further diluted with 7 µl of H2O. To 1 µl of the diluted nucleosomes, 2 µl of NS-buffer (100 mM Tris–HCl pH 7.5, 10 mM DTT, 25% glycerol, 50 mM KCl), varying amounts of protein, 0.5 µl of BSA (NEB, 10 mg/ml), 0.1 µl poly(dI–dC) (100 µg/ml) were added in a final volume of 10 µl. The reaction was incubated for 15 min at room temperature and 15 min on ice and then loaded on an agarose gel in 0.4× TBE, and run at 90 V for varying times. The gel was dried on Whatman paper at 80°C, for 1 h, and analyzed on a phosphoimager or exposed to an X-ray film.
Isolation and acetylation of core histones and deacetylase assays
Chicken core histones were acetylated and purified as described (Wade et al., 1999). A typical deacetylation reaction (200 µl) contained 0.5–1 µg of acetylated histones, 1 mM NAD+, varying amounts of the enzyme in 50 mM Tris–HCl pH 9, 4 mM MgCl2, 0.2 mM DTT. The reaction was incubated at 30°C for 30–60 min. The reaction was stopped by the addition of 50 µl of stop solution (0.1 M HCl, 0.16 M acetic acid) and 600 µl of ethylacetate. After a 10 min incubation at room temperature, 500 µl of the organic phase were removed and radioactivity measured in a Scintillation counter.
Micrococcal- and EcoRI-protection assays
The protein binding for the micrococcal- and EcoRI-protection assays was performed as described for the mobility shift assays. End-labeled DNA was used for the EcoRI digestion assays, while PCR-labeled DNA was used for the micrococcal nuclease assays. The complexes were allowed to bind to the nucleosomes for 15 min at room temperature. For the EcoRI digestion, to 10 µl of the protein–DNA reaction, 2 µl of 10× EcoRI buffer (50 mM Tris–HCl pH 7.5, 10 mM MgCl2, 50 mM NaCl, 1 mM DTT), 1 µl of EcoRI (varying dilutions of a 20 000 U/ml stock) and 7 µl of H2O were added. The reactions were incubated for 15 min at room temperature.
For the micrococcal nuclease-digestion, 1.2 µl of CaCl2 (0.05 M), 1 µl of micrococcal nuclease (varying dilutions of a 17 000 U/ml stock), and 7.8 µl of 50 mM Tris–HCl pH 7.5 were added. The reactions were incubated for 15 min at room temperature.
Then 100 µl of stop solution (20 mM EDTA pH 8.0, 0.2 M NaCl, 1% SDS) and 5 µl of proteinase K (2.5 mg/ml in TE) were added and incubated at room temperature for 5 min. The samples were phenol– chloroform extracted and then applied directly on a 1.5% agarose gel in 1× TAE. The gel was dried and analyzed as described for the mobility shift assays.
Acknowledgments
Acknowledgements
We thank Danesh Moazed, Jasper Rine and Paul Wade for various plasmids and Michael Grunstein, Susan Gasser, Ilia Ousipenski, Alex Strunnikov, Judy Berman, John Aris and Kathryn McMahon for various antibodies. We also thank David Wassarman, Mary Lilly, Jasper Rine, Ann Ehrenhofer-Murray, Paul Kaufman, Alex Strunnikov, Paul Wade, Jim Kadonaga, Yoshi Azuma and Peter Jones for many helpful comments and suggestions. We would also like to thank Simon Chandler and Alan Wolffe for help and advice in reconstituting nucleosomes.
References
- Ansari A. and Gartenberg,M.R. (1997) The yeast silent information regulator Sir4p anchors and partitions plasmids. Mol. Cell. Biol., 17, 7061–7068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio O.M., Billington,B.L. and Gottschling,D.E. (1991) Modifiers of position effect are shared between telomeric and silent mating-type loci in S.cerevisiae. Cell, 66, 1279–1287. [DOI] [PubMed] [Google Scholar]
- Brachmann C.B., Sherman,J.M., Devine,S.E., Cameron,E.E., Pillus,L. and Boeke,J.D. (1995) The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression and chromosome stability. Genes Dev., 9, 2888–2902. [DOI] [PubMed] [Google Scholar]
- Braunstein M., Rose,A.B., Holmes,S.G., Allis,C.D. and Broach,J.R. (1993a) Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev., 7, 592–604. [DOI] [PubMed] [Google Scholar]
- Braunstein M., Rose,A.B., Holmes,S.G., Allis,C.D. and Broach,J.R. (1993b) Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev., 7, 592–604. [DOI] [PubMed] [Google Scholar]
- Braunstein M., Sobel,R.E., Allis,C.D., Turner,B.M. and Broach,J.R. (1996) Efficient transcriptional silencing in Saccharomyces cerevisiae requires a heterochromatin histone acetylation pattern. Mol. Cell. Biol., 16, 4349–4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote J., Quinn,J., Workman,J.L. and Peterson,C.L. (1994) Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science, 265, 53–60. [DOI] [PubMed] [Google Scholar]
- Cuperus G., Shafaatian,R. and Shore,D. (2000) Locus specificity determinants in the multifunctional yeast silencing protein Sir2. EMBO J., 19, 2641–2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammann R., Lucchini,R., Koller,T. and Sogo,J.M. (1995) Transcription in the yeast rRNA gene locus: distribution of the active gene copies and chromatin structure of their flanking regulatory sequences. Mol. Cell. Biol., 15, 5294–5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon N. and Kamakaka,T.R. (2000) A histone variant, Htz1p and a Sir1p-like protein, Esc2p, mediate silencing at HMR. Mol. Cell., 6, 769–780. [DOI] [PubMed] [Google Scholar]
- Donze D., Adams,C.R., Rine,J. and Kamakaka,R.T. (1999) The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes Dev., 13, 698–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fath S., Milkereit,P., Podtelejnikov,A.V., Bischler,N., Schultz,P., Bier,M., Mann,M. and Tschochner,H. (2000) Association of yeast RNA polymerase I with a nucleolar substructure active in rRNA synthesis and processing. J. Cell Biol., 149, 575–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman L., Aragon-Alcaide,L. and Strunnikov,A. (2000) The condensin complex governs chromosome condensation and mitotic transmission of rDNA. J. Cell Biol., 149, 811–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman-Cook L., Kamakaka,R.T. and Pillus,L. (2000) Chromatin Contributions to Epigenetic Transcriptional States in Yeast. Oxford University Press, Oxford, UK.
- Fritze C.E., Verschueren,K., Strich,R. and Easton Esposito,R. (1997) Direct evidence for SIR2 modulation of chromatin structure in yeast rDNA. EMBO J., 16, 6495–6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschling D.E. (1992) Telomere-proximal DNA in Saccharomyces cerevisiae is refractory to methyltransferase activity in vivo. Proc. Natl Acad. Sci. USA, 89, 4062–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M. (1997) Molecular model for telomeric heterochromatin in yeast. Curr. Opin. Cell. Biol., 9, 383–387. [DOI] [PubMed] [Google Scholar]
- Hecht A., Laroche,T., Strahl-Bolsinger,S., Gasser,S.M. and Grunstein,M. (1995) Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell, 80, 583–592. [DOI] [PubMed] [Google Scholar]
- Hecht A., Strahl-Bolsinger,S. and Grunstein,M. (1996) Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature, 383, 92–96. [DOI] [PubMed] [Google Scholar]
- Imai S.-I., Armstrong,C.M., Kaeberlein,M. and Guarente,L. (2000) Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature, 403, 795–800. [DOI] [PubMed] [Google Scholar]
- Ivy J.M., Klar,A.J. and Hicks,J.B. (1986) Cloning and characterization of four SIR genes of Saccharomyces cerevisiae. Mol. Cell. Biol., 6, 688–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L.M., Kayne,P.S., Kahn,E.S. and Grunstein,M. (1990) Genetic evidence for an interaction between SIR3 and histone H4 in the repression of the silent mating loci in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 87, 6286–6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L.M., Fisher-Adams,G. and Grunstein,M. (1992) Identification of a non-basic domain in the histone H4 N-terminus required for repression of the yeast silent mating loci. EMBO J., 11, 2201–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamakaka R.T. (1997) Silencers and locus control regions: opposite sides of the same coin. Trends Biochem. Sci., 22, 124–128. [DOI] [PubMed] [Google Scholar]
- Kamakaka R.T. and Thomas,J.O. (1990) Chromatin structure of transcriptionally competent and repressed genes. EMBO J., 9, 3997–4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayne P.S., Kim,U.J., Han,M., Mullen,J.R., Yoshizaki,F. and Grunstein,M. (1988) Extremely conserved histone H4 N-terminus is dispensable for growth but essential for repressing the silent mating loci in yeast. Cell, 55, 27–39. [DOI] [PubMed] [Google Scholar]
- Landry J., Sutton,A., Tafrov,S.T., Heller,R.C., Stebbins,J., Pillus,L. and Sternglanz,R. (2000) The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl Acad. Sci. USA, 97, 5807–5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebel M., Spillare,E.A., Harris,C.C. and Leder,P. (1999) The Werner syndrome gene product co-purifies with the DNA replication complex and interacts with PCNA and topoisomerase I. J. Biol. Chem., 274, 37795–37799. [DOI] [PubMed] [Google Scholar]
- Loo S. and Rine,J. (1994) Silencers and domains of generalized repression. Science, 264, 1768–1771. [DOI] [PubMed] [Google Scholar]
- Marshall M., Mahoney,D., Rose,A., Hicks,J.B. and Broach,J.R. (1987) Functional domains of SIR4, a gene required for position effect regulation in Saccharomyces cerevisiae. Mol. Cell. Biol., 7, 4441–4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megee P.C., Morgan,B.A., Mittman,B.A. and Smith,M.M. (1990) Genetic analysis of histone H4: essential role of lysines subject to reversible acetylation. Science, 247, 841–845. [DOI] [PubMed] [Google Scholar]
- Moazed D. and Johnson,D. (1996) A deubiquitinating enzyme interacts with SIR4 and regulates silencing in S.cerevisiae. Cell, 86, 667–677. [DOI] [PubMed] [Google Scholar]
- Moazed D., Kistler,A., Axelrod,A., Rine,J. and Johnson,A.D. (1997) Silent information regulator protein complexes in Saccharomyces cerevisiae: a SIR2/SIR4 complex and evidence for a regulatory domain in SIR4 that inhibits its interaction with SIR3. Proc. Natl Acad. Sci. USA, 94, 2186–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti P., Freeman,K., Coodly,L. and Shore,D. (1994) Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev., 8, 2257–2269. [DOI] [PubMed] [Google Scholar]
- Muth V., Nadaud,S., Grummt,I. and Voit,R. (2001) Acetylation of TAF(I)68, a subunit of TIF-IB/SL1, activates RNA polymerase I transcription. EMBO J., 20, 1353–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K.A. (1982) The regulation of yeast mating-type chromatin structure by SIR: an action at a distance affecting both transcription and transposition. Cell, 30, 567–578. [DOI] [PubMed] [Google Scholar]
- Park E.C. and Szostak,J.W. (1990) Point mutations in the yeast histone H4 gene prevent silencing of the silent mating type locus HML. Mol. Cell. Biol., 10, 4932–4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindra A., Weiss,K. and Simpson,R.T. (1999) High-resolution structural analysis of chromatin at specific loci: Saccharomyces cerevisiae silent mating-type locus HMRa. Mol. Cell. Biol., 19, 7944–7950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer S.K. and Buchman,A.R. (1997) Yeast silencers create domains of nuclease resistant chromatin in an SIR4-dependent manner. Chromosoma, 106, 136–148. [DOI] [PubMed] [Google Scholar]
- Renauld H., Aparicio,O.M., Zierath,P.D., Billington,B.L., Chhablani,S.K. and Gottschling,D.E. (1993) Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength and by SIR3 dosage. Genes Dev., 7, 1133–1145. [DOI] [PubMed] [Google Scholar]
- Rine J. and Herskowitz,I. (1987) Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics, 116, 9–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekinger E.A. and Gross,D.S. (2001) Silenced chromatin is permissive to activator binding and PIC recruitment. Cell, 105, 403–414. [DOI] [PubMed] [Google Scholar]
- Shou W., Seol,J.H., Shevchenko,A., Baskerville,C., Moazed,D., Chen,Z.W., Jang,J., Charbonneau,H. and Deshaies,R.J. (1999) Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell, 97, 233–244. [DOI] [PubMed] [Google Scholar]
- Singh J. and Klar,A.J.S. (1992) Active genes in budding yeast display enhanced in vivo accessibility to foreign DNA methylases: a novel in vivo probe for chromatin structure of yeast. Genes Dev., 6, 186–196. [DOI] [PubMed] [Google Scholar]
- Smith J.S. and Boeke,J.D. (1997) An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev., 11, 241–254. [DOI] [PubMed] [Google Scholar]
- Smith J.S. et al. (2000) A phylogenetically conserved NAD+-dependent protein deacetylase activity in the sir2 protein family. Proc. Natl Acad. Sci. USA, 97, 6658–6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl-Bolsinger S., Hecht,A., Luo,K. and Grunstein,M. (1997) SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev., 11, 83–93. [DOI] [PubMed] [Google Scholar]
- Straight A.F., Shou,W., Dowd,G.J., Turck,C.W., Deshaies,R.J., Johnson,A.D. and Moazed,D. (1999) Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell, 97, 245–256. [DOI] [PubMed] [Google Scholar]
- Strathern J.N., Klar,A.J., Hicks,J.B., Abraham,J.A., Ivy,J.M., Nasmyth,K.A. and McGill,C. (1982) Homothallic switching of yeast mating type cassettes is initiated by a double-stranded cut in the MAT locus. Cell, 31, 183–192. [DOI] [PubMed] [Google Scholar]
- Tanner K.G., Landry,J., Sternglanz,R. and Denu,J.M. (2000) Silent information regulator 2 family of NAD-dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc. Natl Acad. Sci. USA, 97, 14178–14182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanny J.C., Dowd,G.J., Huang,J., Hilz,H. and Moazed,D. (1999) An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell, 99, 735–745. [DOI] [PubMed] [Google Scholar]
- Thomas J.O. and Khabaza,A.J. (1980) Cross-linking of histone H1 in chromatin. Eur. J. Biochem., 112, 501–511. [DOI] [PubMed] [Google Scholar]
- Thompson J.S., Hecht,A. and Grunstein,M. (1993) Histones and the regulation of heterochromatin in yeast. Cold Spring Harb. Symp. Quant. Biol., 58, 247–256. [DOI] [PubMed] [Google Scholar]
- Thompson J.S., Ling,X. and Grunstein,M. (1994) Histone H3 amino terminus is required for telomeric and silent mating locus repression in yeast. Nature, 369, 245–247. [DOI] [PubMed] [Google Scholar]
- Ura K., Nightingale,K. and Wolffe,A.P. (1996) Differential association of HMG1 and linker histones B4 and H1 with dinucleosomal DNA: structural transitions and transcriptional repression. EMBO J., 15, 4959–4969. [PMC free article] [PubMed] [Google Scholar]
- Vega-Palas M.A., Venditti,S. and Di Mauro,E. (1998) Heterochromatin organization of a natural yeast telomere. Changes of nucleosome distribution driven by the absence of Sir3p. J. Biol. Chem., 273, 9388–9392. [DOI] [PubMed] [Google Scholar]
- Venditti S., Vega-Palas,M.A. and Di Mauro,E. (1999) Heterochromatin organization of a natural yeast telomere. Recruitment of Sir3p through interaction with histone H4 N terminus is required for the establishment of repressive structures. J. Biol. Chem., 274, 1928–1933. [DOI] [PubMed] [Google Scholar]
- Wade P.A., Jones,P.L., Vermaak,D. and Wolffe,A.P. (1999) Purification of a histone deacetylase complex from Xenopus laevis: preparation of substrates and assay procedures. Methods Enzymol., 304, 715–725. [DOI] [PubMed] [Google Scholar]
- Weiss K. and Simpson,R.T. (1998) High-resolution structural analysis of chromatin at specific loci: Saccharomyces cerevisiae silent mating type locus HMLα. Mol. Cell. Biol., 18, 5392–5403. [DOI] [PMC free article] [PubMed] [Google Scholar]