Abstract
CENP-H has recently been discovered as a constitutive component of the centromere that co-localizes with CENP-A and CENP-C throughout the cell cycle. The precise function, however, remains poorly understood. We examined the role of CENP-H in centromere function and assembly by generating a conditional loss-of-function mutant in the chicken DT40 cell line. In the absence of CENP-H, cell cycle arrest at metaphase, consistent with loss of centromere function, was observed. Immunocytochemical analysis of the CENP-H-deficient cells demonstrated that CENP-H is necessary for CENP-C, but not CENP-A, localization to the centromere. These findings indicate that centromere assembly in vertebrate cells proceeds in a hierarchical manner in which localization of the centromere-specific histone CENP-A is an early event that occurs independently of CENP-C and CENP-H.
Keywords: CENP-C/CENP-H/centromere/DT40/mitosis
Introduction
The centromere plays a fundamental role in accurate chromosome segregation during mitosis and meiosis in eukaryotes. Its functions include sister chromatid adhesion and separation, microtubule attachment, chromosome movement and mitotic checkpoint control (Choo, 1997). Although chromosome segregation errors cause genetic diseases including some cancers (Lengauer et al., 1998), the mechanism by which centromeres interact with microtubules of the spindle apparatus during cell division is not fully understood.
To understand the function of the centromere, many mutants that display errors in chromosome segregation have been characterized. This strategy has been applied in yeasts and flies, and has led to the identification of genes that encode centromere components. For example, several centromere components have been identified in Saccharomyces cerevisiae (Doheny et al., 1993; Pidoux and Allshire, 2000). Of these, CSE4p (Stoler et al., 1995; Meluh et al., 1998) and MIF2p (Brown et al., 1993) have homologs that are associated with the centromeres of other organisms. Several genes involved in the evolutionarily conserved mitotic checkpoint pathway (Amon, 1999) have also been identified through analysis of S.cerevisiae mutants. Vertebrate homologs of the checkpoint proteins localize to the centromere prior to association of the centromere with the spindle (Chen et al., 1996; Taylor and McKeon, 1997; Taylor et al., 1998). The Drosophila melanogaster mitotic checkpoint protein ZW10 has no known homolog in yeasts, but is conserved in vertebrates and Caenorhabditis elegans (Starr et al., 1997; Chan et al., 2000).
It has been difficult to apply genetic analyses to vertebrate centromeres because of the lack of suitable assays for mutations that affect the accuracy of chromosome segregation. Controversy regarding the identity of vertebrate centromeric DNA has hindered the identification of vertebrate centromere-binding proteins by biochemical methods. Centromere proteins in vertebrate cells have been identified primarily by immunological methods. Antibodies against centromere proteins have been isolated from patients with autoimmune disease (Moroi et al., 1980) and from immunized animals (Compton et al., 1991), and this has led to the identification of several centromere-associated proteins known as CENPs (Pluta et al., 1995; Craig et al., 1999; Dobie et al., 1999; Choo, 2000; Tyler-Smith and Floridia, 2000). Of the CENPs, CENP-A and CENP-C are likely to be important because their functional homologs are present in species from yeast to humans. CENP-A is a 140 amino acid centromere-specific protein in which the C-terminal 90 amino acids are 60% identical to those of histone H3 (Shelby et al., 1997). CENP-A is found only at active centromeres (du Sart et al., 1997; Warburton et al., 1997). CENP-A co-purifies with nucleosomes, suggesting that it is a component of nucleo somes, which form at active centromeres (Vafa and Sullivan, 1997). Data from analyses of CENP-A knockout mice suggest that CENP-A plays a role in early organization of the centromeric chromatin during interphase (Howman et al., 2000). The S.cerevisiae homolog of CENP-A, CSE4p, contains a C-terminal region that is 60% identical to H3 and is a constituent of a specific type of nucleosome that forms at S.cerevisiae centromeres (Meluh et al., 1998). It therefore seems likely that a centromeric nucleosome containing a specific variant of H3 is a funda mental feature of eukaryotic centromere organization.
The S.cerevisiae homolog of CENP-C, MIF2p, binds to centromeric DNA (Brown, 1995; Lanini and McKeon, 1995; Meluh and Koshland, 1995, 1997). Mutation or overexpression of MIF2 causes errors in chromosome segregation (Brown et al., 1993). CENP-C has been localized to the inner kinetochore plates adjacent to the centromeric DNA (Saitoh et al., 1992) and it is known to bind DNA directly (Yang et al., 1996). Disruption of the CENP-C gene results in cell death (Fukagawa and Brown, 1997; Kalitsis et al., 1998) due to chromosome missegregation and metaphase arrest (Fukagawa et al., 1999a). Metaphase arrest has also been observed after microinjection of anti-CENP-C antibodies into HeLa cells (Tomkiel et al., 1994). Like CENP-A, CENP-C is found only at active centromeres (Earnshaw et al., 1989; Sullivan and Schwartz, 1995) and is needed to induce formation of a functional centromere (Fukagawa et al., 1999a). The data suggest that CENP-C is an important component in kinetochore assembly.
CENP-H was initially identified as a component of the mouse centromere (Sugata et al., 1999) and recently, human CENP-H protein was isolated and its localization determined. CENP-H is localized at the inner kinetochore plate and is found in neocentromeres (Sugata et al., 2000). Although these localization data suggest that CENP-H may be a fundamental component of active centromere complexes, its precise function is unclear. To understand the role of CENP-H in centromere function, we used a conditional knockout approach in chicken B lymphocyte cell line DT40 (Buerstedde and Takeda, 1991). The high level of homologous recombination in DT40 cells allowed targeted disruption of the CENP-H gene. Furthermore, we created ΔCENP-H clones in which the chicken CENP-H gene was expressed conditionally under the control of a repressible promoter. Phenotypic analysis of these mutants revealed that CENP-H plays an important role in functional centromere formation and mitotic cell division. Immunofluorescence analysis of interphase ΔCENP-H cells revealed that CENP-H is necessary for recruiting CENP-C to form functional centromeres in higher vertebrate cells.
Results
Isolation and chromosomal mapping of the chicken CENP-H gene
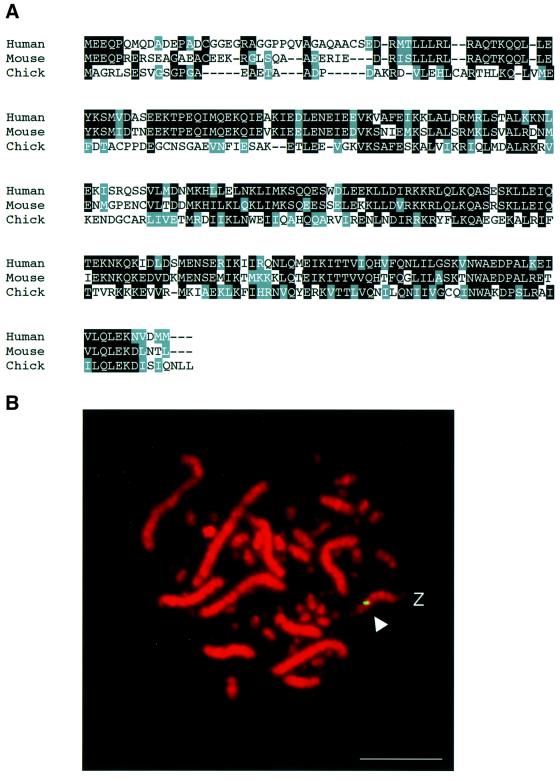
We used a region homologous in the mouse and human CENP-H genes to design degenerate primers for isolating a short stretch of the chicken CENP-H cDNA by reverse transcription–PCR (RT–PCR). Using this RT–PCR product as a probe, we screened a chicken cDNA library and isolated several cDNA clones containing the entire CENP-H gene. The full-length sequence encodes a 235 amino acid polypeptide with a predicted molecular mass of 26.8 kDa. Chicken CENP-H has 34% amino acid sequence identity and 60% amino acid similarity with human CENP-H, and 37% amino acid identity and 61% amino acid similarity with mouse CENP-H (Figure 1A). In both human and mouse CENP-H, we found predicted, long coiled-coil regions (Sugata et al., 2000). Although the predicted coiled-coil region is also present in the chicken CENP-H sequence, it is shorter than those found in the human and mouse proteins.
Fig. 1. Identification of the chicken CENP-H gene. (A) Comparison of amino acid sequences of human, mouse and chicken CENP-H proteins. Amino acid sequence is shown as a single-letter code. Amino acids that are conserved between species are shaded. Nucleotide sequence of the chicken CENP-H cDNA is available from DDBJ/EMBL/GenBank (accession No. AB055783). (B) Chromosomal localization of the CENP-H gene by FISH. DT40 metaphase spreads were hybridized with the CENP-H probe. The arrowhead indicates a probe-specific hybridization signal (green). DNA is stained with 4′,6-diamidine-2-phenylindole (DAPI) and colored red. CENP-H was mapped to chromosome Z, indicating that there is a single CENP-H allele in each DT40 cell. The scale bar corresponds to 10 µm.
We isolated genomic clones that covered the entire CENP-H locus with a chicken CENP-H cDNA. Limited sequencing revealed the genomic organization of the CENP-H gene, which spans ∼7 kb and contains nine exons (Figure 2A). Southern blot analysis of chicken genomic DNA showed a single band (data not shown), indicating that there is a single CENP-H gene in the chicken genome. To determine the chromosomal position of the chicken CENP-H gene, we used fluorescence in situ hybridization (FISH) analysis with a genomic clone as a probe. Fluorescent hybridization signals were localized to the Z chromosome (Figure 1B). The DT40 cell line has 38 autosomal chromosome pairs, with the exception of chromosome 2, which is triploid, and the Z/W sex chromosomes. FISH showed that DT40 cells contain a single CENP-H allele on the Z chromosome.
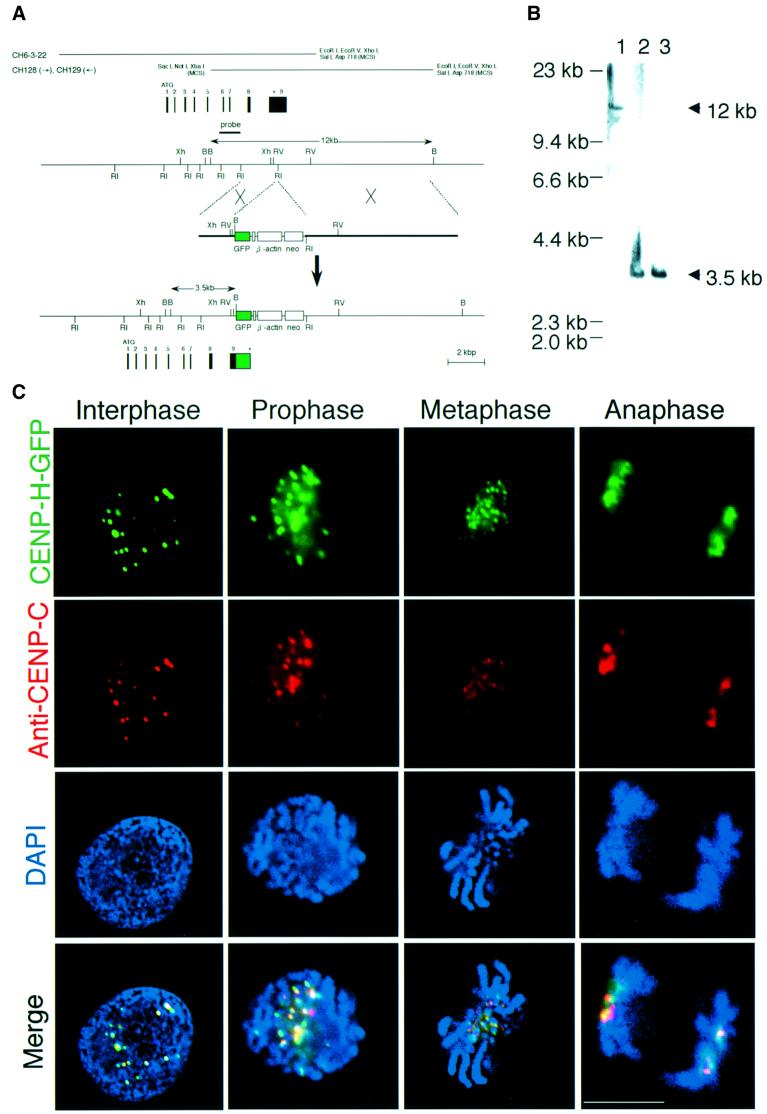
Fig. 2. Chicken CENP-H localizes to centromeres throughout the cell cycle. (A) Restriction maps of the CENP-H gene, the CENP-H–GFP replacement construct and the configuration of the targeted locus. Black boxes indicate the positions of exons. Exon 9, which contains the stop codon, was replaced with a portion of the sequence of exon 9 fused to a GFP tag followed by the neo-resistance gene (neo) and 3′ homology region. Relevant restriction enzyme sites shown are as follows: RI, EcoRI; Xh, XhoI; B, BamHI; RV, EcoRV. The position of the probe used for Southern hybridiza tion is indicated. A new 3.5 kb BamHI fragment hybridizes with the probe if targeted integration of the indicated replacement construct occurs. (B) Restriction analysis of the CENP-H–GFP allele. Genomic DNAs from wild-type DT40 cells (lane 1) and the targeted clones (lanes 2 and 3) were digested with BamHI, size fractionated by 0.7% agarose gel electrophoresis, transferred to a nylon filter and hybridized with the probe indicated in (A). (C) Localization of CENP-H–GFP at progressive stages of the cell cycle in DT40 cells. Cells were fixed and stained with anti-CENP-C antibody (red), and green signals are specific for CENP-H–GFP. Nuclei and chromosomes were visualized by counterstaining with DAPI (blue). The scale bar corresponds to 10 µm. As shown in the merged images, CENP-H–GFP signals are co-localized with CENP-C signals throughout the cell cycle.
Chicken CENP-H associates with chicken centromeres
To verify that chicken CENP-H exhibits a pattern of cell cycle-dependent centromere association similar to that of human CENP-H (Sugata et al., 2000), we generated a DT40 cell line in which the CENP-H gene was replaced with a gene encoding CENP-H–green fluorescent protein (GFP). The replacement-targeting strategy is illustrated in Figure 2A. The replacement-targeting construct was transfected into wild-type DT40 cells, and G418-resistant clones were screened by hybridizing BamHI digests with a probe located beyond the 5′-end of the targeting construct. Successful targeting was expected to replace the 12 kb cognate fragment with a 3.5 kb fragment (Figure 2A). We identified several clones in which production of CENP-H was replaced by production of the CENP-H–GFP fusion protein (Figure 2B). Because this cell line proliferates in the same way as wild-type DT40, the CENP-H–GFP fusion protein appears to be functionally identical to the CENP-H protein.
We used this cell line for immunostaining with anti-CENP-C antibody. CENP-C was used as a centromeric marker because it is localized to centromeres throughout the cell cycle (Saitoh et al., 1992; Fukagawa et al., 1999a). Figure 2C shows CENP-H–GFP signals and anti-CENP-C signals throughout the cell cycle. During interphase, CENP-H–GFP was co-localized with CENP-C as discrete signals in the nucleus (Figure 2C, panel 1). Co-localization of CENP-H–GFP with CENP-C was also observed during prophase (Figure 2C, panel 2), metaphase (Figure 2C, panel 3) and anaphase (Figure 2C, panel 4) of mitosis. We also generated a polyclonal antibody against the N-terminal region of chicken CENP-H and confirmed co-localization of CENP-H with CENP-C by double staining of the wild-type DT40 cells (data not shown). Our results indicate that, like mammalian CENP-H, chicken CENP-H is localized to centromere regions throughout the cell cycle.
Generation of CENP-H-deficient DT40 clones
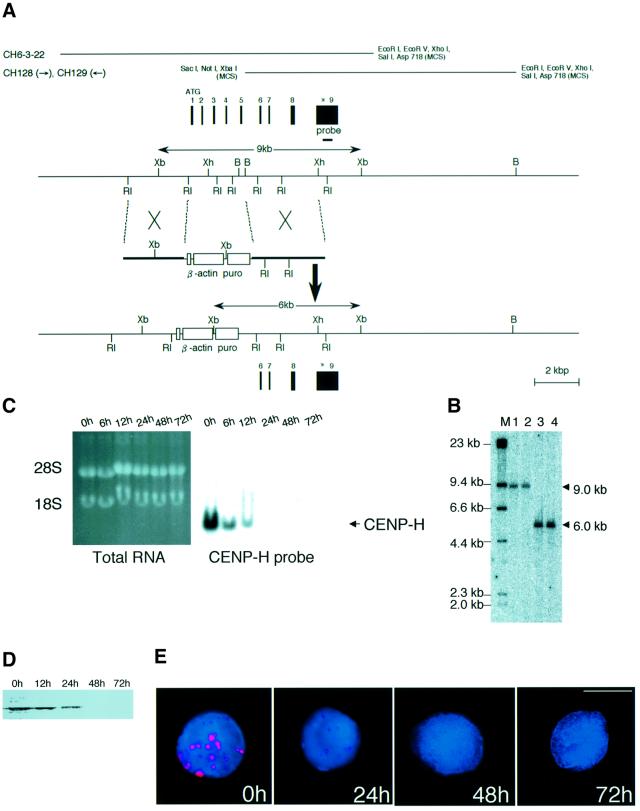
To generate conditional CENP-H mutant clones, we co-transfected a conditional expression construct with the CENP-H gene under the control of a tetracycline (tet)-repressible promoter (Gossen and Bujard, 1992) and a tet-repressible transactivator-containing zeocin (zeo)-resistance cassette into DT40 cells. We selected zeo-resistant colonies and identified several clones carrying these constructs integrated at random sites in the genome (CENP-H+/CENP-H transgene). The strategy for generation of the CENP-H deletion mutant is shown in Figure 3A. Targeted integration of the deletion construct deletes amino acids 1–112, and targeted events were identified by the presence of a 6.0 kb band upon Southern hybridization analyses of XbaI-digested genomic DNAs from puromycin (puro)-resistant clones (Figure 3A). One clone with the CENP-H+/CENP-H transgene genotype was transfected with the targeted deletion construct to generate several conditional CENP-H-deficient clones (CENP-H–/CENP-H transgene) (Figure 3B). One clone, #5-5, was chosen for further analysis. Northern blot analysis showed a steady reduction in the expression of CENP-H mRNA with time in #5-5 cells incubated with tet (Figure 3C). CENP-H mRNA was not detected 24 h after addition of tet. We also carried out western blotting and immunocytochemical analysis to investigate the level of CENP-H protein in #5-5 cells after addition of tet. We observed a steady reduction in levels of CENP-H protein with time in the presence of tet (Figure 3D and E). CENP-H protein was not detected 48 h after addition of tet. After addition of tet to the medium, we examined cell growth and viability.
Fig. 3. Generation of a ΔCENP-H clone carrying a chicken CENP-H transgene under the control of a tet-repressible promoter. (A) Restriction maps of the CENP-H locus, the gene disruption construct and configuration of the targeted locus. The targeted construct is expected to disrupt exons 1–5. Relevant restriction enzyme sites shown are as follows: RI, EcoRI; Xh, XhoI; B, BamHI; Xb, XbaI. The position of the probe used for Southern hybridization is indicated. A new 6.0 kb XbaI fragment hybridizes with the probe if targeted integration of the construct occurs. (B) Restriction analysis of the targeted integration of the CENP-H disruption construct. Genomic DNAs from wild-type DT40 cells (CENP-H+, lane 1), a clone with random integration of the CENP-H transgene (CENP-H+/CENP-H transgene, lane 2) and targeted clones (CENP-H–/CENP-H transgene, lanes 3 and 4) were digested with XbaI, size fractionated by 0.7% agarose gel electrophoresis, transferred to a nylon filter and hybridized with the probe indicated in (A). (C) Suppression of CENP-H expression from the transgene. Total RNA was isolated from #5-5 cells at the times indicated following addition of tet. RNA samples (20 µg/lane) were fractionated on formaldehyde gels (left side of figure). Northern blot hybridization was carried out with the full-length CENP-H cDNA as the probe. Bands other than that for CENP-H band were not detected, indicating that the truncated form of CENP-H was not expressed. (D) Western blot analysis with anti-CENP-H antibody of the #5-5 cell extract at the times indicated following addition of tet. (E) Immunofluorescence analysis of #5-5 cells at times indicated following the addition of tet using anti-CENP-H antibody (red). DNA was counterstained with DAPI (blue). The scale bar corresponds to 10 µm.
Deletion of CENP-H results in accumulation of cells in metaphase and subsequent cell death
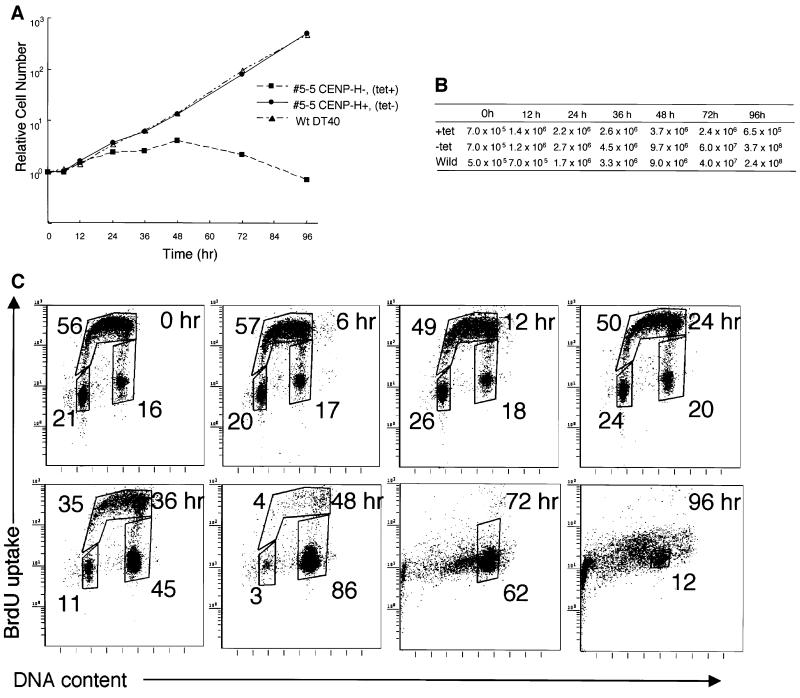
The proliferative properties of CENP-H+ #5-5 cells (tet–) were monitored by growth curves and cell cycle analysis. Viable and dead cells were assessed with Trypan blue exclusion. The growth curve of CENP-H+ #5-5 cells (tet–) was indistinguishable from that of wild-type DT40 cells (Figure 4A and B). The total cell number at each time point is shown in Figure 4B. CENP-H+ #5-5 cells (tet–) and wild-type DT40 cells divided nine times during a 96 h culture, which shows that the doubling time of these cells is once per ∼10.6 h (Figure 4A and B). For analysis of the consequence of CENP-H deletion, the proliferative properties of CENP-H-deficient #5-5 cells were monitored by growth curves and cell cycle analysis after addition of tet. Cells stopped proliferating at ∼2.5 cell cycles after the addition of tet (Figure 4A and B), and most cells had died by 96 h. Tet had no effect on the growth or viability of wild-type DT40 cells (Wang et al., 1996). These observations indicate that depletion of CENP-H results in growth arrest and subsequent cell death.
Fig. 4. CENP-H is essential for normal progression of the cell cycle. (A) Representative growth curves for the cell cultures indicated. Tet was added at time 0 to the #5-5 CENP-H– (tet+) culture. The number of cells not stained with Trypan blue was counted. Each experiment was performed twice, and each time point was examined in duplicate. (B) Total cell number plotted in (A). (C) Cell cycle distribution of #5-5 cells following inhibition of CENP-H transgene expression by adding tet at time 0. Cells were stained with FITC–anti-BrdU (y-axis, log scale) to detect BrdU incorporation and with propidum iodide to detect total DNA (x-axis, linear scale). The lower-left box represents G1-phase cells, the upper box represents S-phase cells, and the lower-right box represents G2–M-phase cells. The numbers given in the boxes indicate the percentage of gated events.
We then examined the delayed growth and cell death following CENP-H depletion in #5-5 cells. For this purpose, we measured both cellular DNA content and DNA synthesis by fluorescence-activated cell sorting (FACS) after pulse-5-bromodeoxyuridine (BrdU) labeling (Figure 4C). Asynchronous wild-type DT40 and CENP-H+ #5-5 cells (tet–) showed similar cell cycle patterns (data not shown), with S phase accounting for two-thirds of the entire cell cycle time (Figure 4C, 0 h). In contrast, when mutant cells were cultured with tet, BrdU uptake had decreased by 12 h and had disappeared completely by 48 h. At the same time, cells began to accumulate in G2–M phase, and the proportion of cells in G2–M phase reached 86% by 48 h (Figure 4C). Degradation of chromosomal DNA due to massive cell death was observed between 72 and 96 h (Figure 4C).
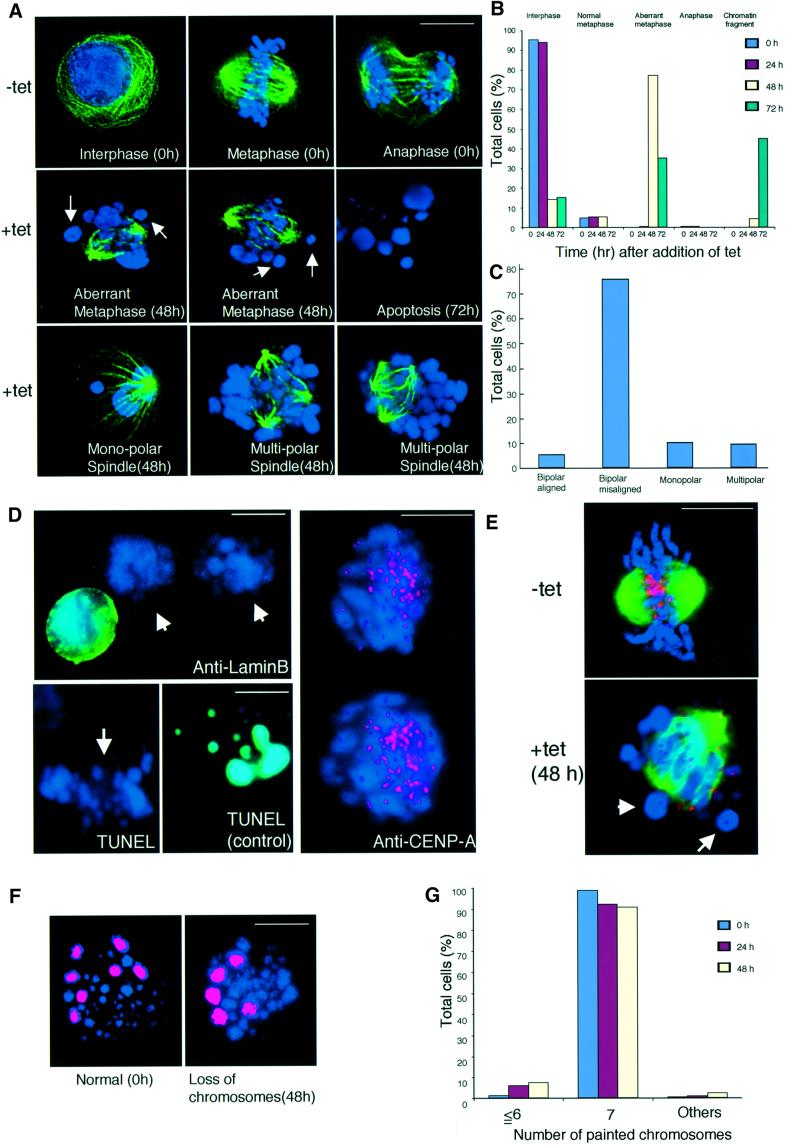
While FACS analysis showed that #5-5 cells accumulated in G2–M phase in the presence of tet (Figure 4C), we could not distinguish the exact stage of G2–M phase by FACS analysis. We then used DNA staining and immunocytochemical staining of microtubules to examine the time course of kinetics of the mitotic index (Figure 5A and B). FACS analysis showed that 16% of CENP-H+ #5-5 cells (tet–) were in G2–M phase (Figure 4C, 0 h), whereas 4.5% of cells were mitotic (Figure 5B). These percentages are similar to those of asynchronous wild-type DT40 cells (data not shown). By 48 h after the addition of tet, the percentage of #5-5 cells in G2–M phase had increased to 86%, and the percentage of metaphase cells reached 85% (Figure 5B). We did not observe any anaphase cells at 48 h, indicating that deletion of CENP-H causes metaphase arrest, failure of the cells to enter anaphase, and subsequent cell death.
Fig. 5. CENP-H-deficient cells show metaphase arrest associated with aberrant chromosomes and spindles that lead to chromosome missegregation. (A) Chromosome morphology and α-tubulin staining (green) in the absence or presence of tet. DNA was counterstained with DAPI (blue). In the absence of tet, cells show the normal staining pattern for α-tubulin (upper three panels). In the presence of tet, cells in which chromosomes were not aligned at the metaphase plate were observed. Arrows indicate the misaligned chromosomes (middle panels) 48 h after addition of tet. Cells with apoptosis were observed 72 h after addition of tet (middle right panel). We also detected cells with monopolar and multipolar spindles (lower panels). The scale bar corresponds to 10 µm. (B) Quantitation of aberrant #5-5 cells following inhibition of CENP-H transgene expression after addition of tet at time 0. We scored the number of interphase cells, normal metaphase cells, aberrant metaphase cells described in (A), anaphase cells and cells with micronuclei. We scored ∼2000 cells at each time point. (C) Quantitation of cells with misaligned chromosomes 48 h after addition of tet. Misaligned chromosomes were scored as bipolar spindle with misaligned chromosomes, monopolar spindle or multispindle. (D) Immunofluorescence analysis of #5-5 cells 48 h after addition of tet with anti-LaminB antibody (upper left). Aberrant chromosomes (arrow) are not stained. Apoptotic cells can be detected by TUNEL assay (control). Aberrant chromosomes (arrow) were not stained (lower left). Immunofluorescence analysis of #5-5 cells 72 h after addition of tet with anti-CENP-A antibody (right). Aberrant chromosomes are stained with CENP-A antibody (red). (E) Double staining #5-5 cells 0 (–tet) or 48 h (+tet) after addition of tet with anti-α-tubulin (green) and anti-CENP-A (red) antibodies. Arrows indicate misaligned chromo somes that do not appear to attach to microtubules, although many other chromosomes appear to have formed microtubule attachments. (F) To examine chromosome loss, we used FISH analysis with chromosome-specific painting probes. We used painting probes for chicken chromosomes 1, 2 and 3. Because DT40 cells have three copies of chromosome 2, we detected seven painted chromosomes in normal cells (left panel). #5-5 cells with loss of chromosomes (right panel) were detected after addition of tet. The scale bars correspond to 10 µm. (G) Distribution of the number of painted chromosomes per cell. #5-5 cells were cultured after addition of tet. At the indicated time, cells were treated with colcemid for 3 h. The number of painted chromosomes was scored in ∼200 metaphase cells.
Deletion of CENP-H causes chromosome aberrations and leads to chromosome missegregation
In the course of cytological analysis of CENP-H-deficient cells, we observed that 78% of cells displayed abnormal mitotic cells 48 h after addition of tet (Figure 5A and B). In large populations of metaphase-arrested cells, some chromosomes were highly condensed and appeared misaligned on the metaphase plate (Figure 5A, 48 h), whereas chromosomes appeared ordered and aligned on the metaphase plate in control cells (Figure 5A, 0 h, metaphase). Mitotic cells with highly condensed chromosomes appeared to condense the DNA further, leading to apoptosis (Figure 5A, 72 h).
Control cells cultured in the absence of tet showed well ordered bipolar spindle during metaphase and anaphase, respectively (Figure 5A, 0 h). At 48 h, cells cultured in the presence of tet appeared to have normal microtubule spindle assemblies, but some contained multiple poles or monopoles (Figure 5A and C, lower panel). This might account for the abnormal chromosome alignments described above; however, even in 75% of the mutant cells with proper bipolar spindle poles, some chromosomes were not aligned on the metaphase plate (Figure 5A and C), and in others, the alignment was completely disordered. These results indicate that CENP-H is required either directly or indirectly for congression and/or maintenance of stable chromosome alignment and for progression from metaphase to anaphase.
The images in Figure 5A show that the chromosomes of CENP-H-deficient cells are shaped differently from those of normal chromosomes. These are similar to micronuclei or apoptotic bodies. We then carried out LaminB staining and TUNEL assays to rule out these possibilities. At 48 h after addition of tet, most chromosomes of #5-5 cells were not stained by anti-LaminB antibody and by TUNEL assays (Figure 5D). We also stained these aberrant chromosomes with an anti-CENP-A antibody (V.Regnier and W.Brown, unpublished). These chromosomes showed clear double dots (Figure 5D), indicating that they are indeed chromosomes.
We observed that several chromosomes had not congressed, whereas many have aligned at the midzone in CENP-H-deficient cells (Figure 5A). It was, therefore, of interest to investigate attachment between microtubules and chromosomes in CENP-H-deficient cells. We performed double staining of cells with anti-tubulin and anti-CENP-A antibodies. At 48 h after addition of tet, congressed chromosomes appeared to attach to microtubules, while misaligned chromosomes were not attached (Figure 5E).
We then examined whether mitotic defects caused by CENP-H depletion were associated with chromosome missegregation. We performed FISH analysis of metaphase spreads with chromosome painting probes. The probe used detects chicken chromosomes 1, 2 and 3. Because DT40 cells contain three copies of chromosome 2, we can observe seven fluorescent chromosomes for each wild-type cell (Figure 5F, left panel). As shown in Figure 5F (right panel) and G, CENP-H-deficient cells with chromosome loss were observed in the presence of tet, and the proportion of these cells gradually increased after 24 h. At 48 h, 5.9% of the cells had lost at least one copy of chromosome 1, 2 or 3, which is a significant increase compared with the proportion observed in wild-type DT40 cells (Figure 5G). These results suggest that CENP-H deficiency induces chromosome missegregation due to a mitotic defect.
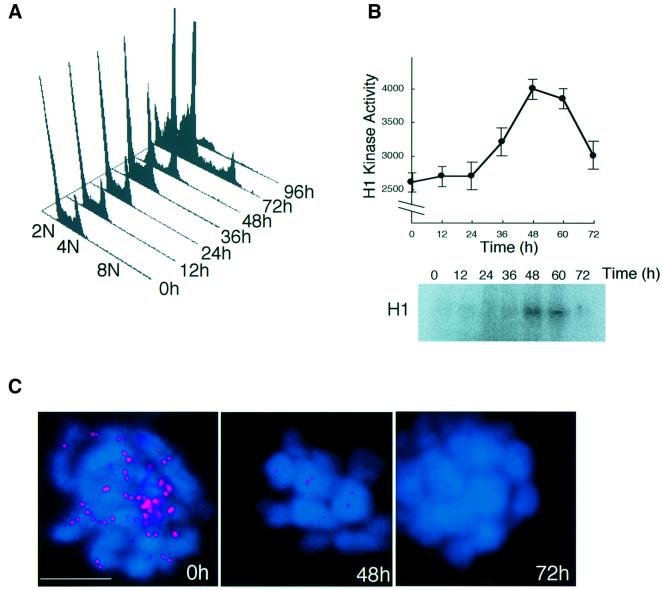
CENP-H-deficient cells proceed through the cell cycle without normal chromosome segregation
In cytological analysis of CENP-H-deficient cells, we often observed tetraploid cells at 72 h after addition of tet. We then performed FACS analysis of #5-5 cells to investigate whether the cell cycle progressed to further rounds without cell division. As mentioned above, cells accumulated at 4N DNA content 48 h after addition of tet (Figures 4C and 6A). After arrest at metaphase, cells progressed to the next cell cycle without completing cell division, and these cells completed further DNA replication with 8N DNA content 72 h after the addition of tet (Figure 6A) This observation suggested that the mitotic checkpoint was disrupted by 72 h after the addition of tet. To investigate cell cycle stage at these time points, histone H1 kinase activities in p34CDC2 immunoprecipitates were measured (Figure 6B). High activity of H1 kinase at 48 h is consistent with high accumulation of metaphase cells, and the low activity at 72 h corresponds to progression of the cell to the next cell cycle. We also performed immunostaining with an antibody against an outer centromere protein ZW10 (Fukagawa et al., 1999a; Chan et al., 2000). We detected ZW10 signals on some metaphase cells 48 h after addition of tet, though the signal intensity was a little weak. However, almost no signals were detected 72 h after addition of tet (Figure 6C), suggesting that the kinetochore structure is completely disrupted by 72 h and that kinetochore function including mitotic checkpoint function is lost.
Fig. 6. CENP-H-deficient cells proceed through the cell cycle without normal chromosome segregation. (A) FACS analysis of the cell cycle pattern of #5-5 cells after addition of tet. The positions of cell populations with 2N, 4N or 8N DNA content are illustrated. At 72 h, a peak of 8N DNA content is observed. (B) Histone H1 kinase activity in chicken p34CDC2 immunoprecipitates was measured in the cells isolated in (A). Kinase activity was measured using purified histone H1 as a substrate. Total radioactivity of histone H1 was measured with a STORM imager. (C) Immunofluorescence analysis of metaphase #5-5 cells 0, 48 or 72 h after addition of tet with antibody against an outer centromere protein ZW10 (red). At 0 h (control), cells were treated with colcemid for 3 h. DNA was counterstained with DAPI (blue). The scale bar corresponds to 10 µm.
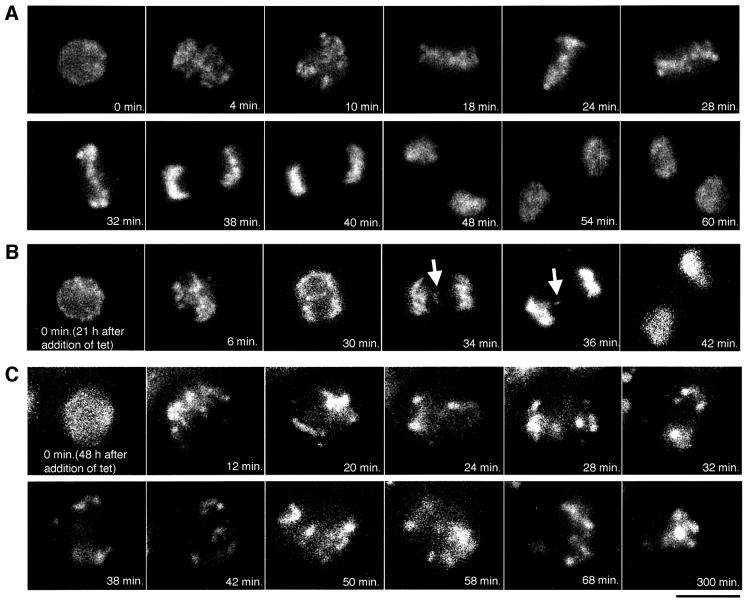
Time-lapse observation of chromosomes in CENP-H-deficient cells
We analyzed the terminal phenotype of CENP-H-deficient cells. In addition to these analyses, it is important to observe individual living cells after suppression of CENP-H. For this purpose, we stained chromosomes with Hoechst 33342 in living cells and observed the stained cells under microscopy at 37°C; microscope images were typically obtained at intervals of 2 min. Chromosomes were observed in individual cells throughout an entire mitotic cell cycle. An example of the time-lapse data of #5-5 cells in the absence of tet (control) is shown in Figure 7A. Selected frames of the dynamics of chromosomes from prophase to telophase are shown. We were able to observe well ordered metaphase plate (24–32 min) and normal cell division. It took control cells ∼40 min (n = 10) to progress from prophase to telophase. After addition of tet, we observed chromosome missegregation or long mitotic arrest. For example, we could observe lagging chromosomes 21 h after addition of tet (Figure 7B). At this stage, we detected CENP-H, but the level was reduced. These findings suggest that the low level of CENP-H affected normal centromere function. We also observed prolonged mitotic arrest from 48 h after addition of tet (Figure 7C). At this stage, cells could not exit smoothly from mitosis. A cell shown in Figure 7C was arrested in mitosis for >300 min.
Fig. 7. Dynamics of chromosomes by time-lapse observation. (A) Selected frames of chromosomes from prophase to telophase in #5-5 cells (–tet). The numbers at the bottom of each image represent the time in minutes. The scale bar corresponds to 10 µm. (B) From 21 h after addition of tet, one prophase cell was followed for time-lapse observations at intervals of 2 min. Selected frames are shown. A lagging chromosome can be observed (arrow). (C) From 48 h after addition of tet, one prophase cell was followed for time-lapse observation. Once this cell entered mitosis, the cell was arrested at metaphase for >300 min.
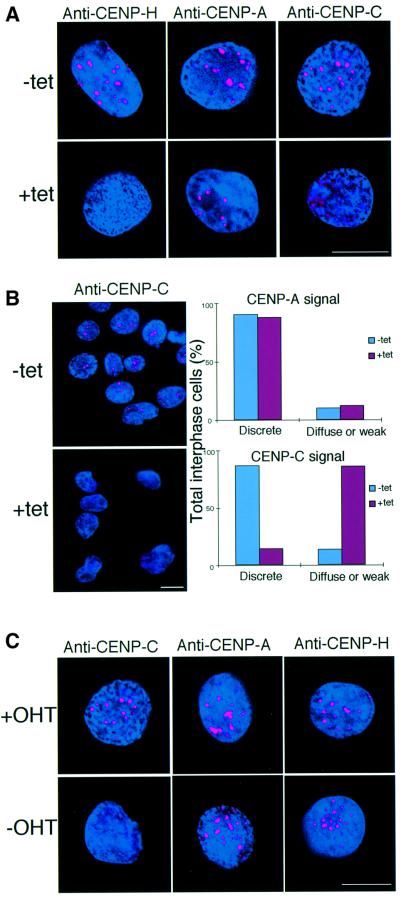
CENP-H is required for localization of CENP-C but not CENP-A to the centromere
We showed previously that human CENP-H, like CENP-A and -C, is localized to the inner kinetochore plate at the light microscopy level (Saitoh et al., 1992; Warburton et al., 1997; Sugata et al., 2000). We also showed perfect co-localization of chicken CENP-H and -C in the present study (Figure 2C). These proteins are thought to form a ‘prekinetochore structure’ that exists throughout the cell cycle and marks the location of the centromere on the chromosome. To understand the role of CENP-H in assembly of the prekinetochore structure, we performed immunofluorescence studies of a CENP-H-deficient mutant. Interphase nuclei of #5-5 cells were analyzed with antibodies against chicken CENP-A (V.Regnier and W.Brown, unpublished), CENP-C (Fukagawa et al., 1999a) and CENP-H in the presence or absence of tet. Typical results are shown in Figure 8A. In the absence of tet, all three antibodies gave strong, discrete signals on interphase nuclei of #5-5 cells (Figure 8A, upper panel). At 48 h after the addition of tet, we could not detect a CENP-H signal in interphase nuclei of #5-5 cells, suggesting complete depletion of CENP-H (Figure 8A, lower left panel). CENP-A antibody gave a positive signal on the interphase nuclei of CENP-H-deficient cells (Figure 8A, lower middle panel); however, the normal, prominent, discrete CENP-C signal (Figure 8A, upper right panel) was absent and instead a diffuse signal was observed (Figure 8A, lower right panel). We counted interphase cells that showed diffuse CENP-C signals (Figure 8B). Approximately 88% of interphase cells displayed diffuse CENP-C signals after addition of tet (Figure 8B).
Fig. 8. CENP-H is required for centromere targeting of CENP-C but not CENP-A. (A) Immunofluorescence analysis of #5-5 cells 0 (–tet) or 48 h (+tet) after addition of tet. Interphase cells were stained with anti-CENP-H, anti-CENP-A and anti-CENP-C antibodies. Antibody signals were detected with Cy3-conjugated secondary antibodies (red). DNA was counterstained with DAPI (blue). The scale bars correspond to 10 µm. (B) Quantitation of interphase cells with diffuse signals of CENP-A and CENP-C. The left panel provides an example of the diffuse CENP-C signals in CENP-H-deficient cells. As shown in the left panel, many #5-5 interphase cells displayed diffuse CENP-C signals at 48 h after addition of tet, while discrete CENP-C signals were observed in the absence of tet. The right panel shows quantitative data. We scored ∼200 interphase cells. (C) Immunofluorescence analysis of Δ/CENP-C-ER cells grown in the presence (+OHT) or absence (–OHT, restrictive conditions) of OHT for 72 h. Rabbit antibodies to anti-CENP-C, anti-CENP-A or anti-CENP-H were applied to interphase cells and detected with Cy3-conjugated second antibodies (red). DNA was counterstained with DAPI (blue).
Immunocytochemical analysis of CENP-H-deficient cells demonstrated CENP-H to be necessary for localization of CENP-C to the centromere. It was, therefore, of interest to determine whether CENP-C was necessary for CENP-H localization to the centromere. To address this question, we performed immunofluorescence analysis of the Δ/CENP-C-ER DT40 cell line, which is a conditional loss-of-function mutant of CENP-C (Fukagawa and Brown, 1997). In the presence of 4-hydroxytamoxifen (OHT) these cells proliferate similarly to wild-type cells but lose CENP-C function when OHT is removed from the culture medium. We found that the strong, discrete CENP-C signals present in nuclei in the presence of OHT (Figure 8C, upper left) were lost and that the CENP-C signal was spread throughout the cell in a diffuse manner 72 h after removal of OHT from the culture medium (Figure 8C, lower left; Fukagawa and Brown, 1997). We then analyzed localization of CENP-A and -H. As shown in Figure 8C, we detected typical centromeric signals for CENP-A and CENP-H in CENP-C-deficient cells. These results indicate that CENP-C is not necessary for localization of CENP-A and CENP-H to centromeres but that CENP-H is necessary for CENP-C localization to centromeres.
Discussion
CENP-H is required for centromere function during mitosis
Prior to the experiments reported here, it was suggested that CENP-H, a newly identified constitutive centromere component, may play an important role in centromere function (Sugata et al., 1999, 2000). Human CENP-H co-localized with inner kinetochore plate proteins CENP-A and CENP-C, and was detected at neocentromeres, but not at inactive centromeres, in stable dicentric chromosomes (Sugata et al., 2000). This observation suggested that CENP-H played a role in centromere function and in order to address this point we have constructed a conditional loss-of-function mutant in the hyper-recombinogenic chicken DT40 cell line. We describe herein the phenotype resulting from disruption of the chicken CENP-H gene in DT40 cells and present direct evidence that CENP-H is required for mitosis.
CENP-H-deficient cells arrested in metaphase possessed aberrant chromosomes and spindles, showed missegregation of chromosomes and finally died. Previously, we reported the targeted disruption of the CENP-C gene (Fukagawa and Brown, 1997; Fukagawa et al., 1999a). Although we detected an accumulation of metaphase cells among CENP-C-deficient cells, only 10% of cells were in metaphase in asynchronous cultures. One explanation for this observation is that CENP-C may serve further functions during the cell cycle in addition to its role in mitosis. In comparison, 85% of CENP-H-deficient cells were arrested in metaphase, which suggests that CENP-H is critical to centromere function during mitosis. There are many reports of knockout mice for genes encoding centromeric proteins (Kalitsis et al., 1998; Hudson et al., 1998; Cutts et al., 1999; Howman et al., 2000; Uren et al., 2000) and such studies are important for understanding the roles of centromeric proteins during early development in the mouse. All active centromere components except CENP-B, which is also localized to inactive centromeres, were found to be essential for early development. Because CENP-H is essential for proper mitotic cell division, we predict that a CENP-H-knockout mouse would be embryonic lethal.
Chromosome aberrations observed in CENP-H-deficient cells
Although analyses of knockout mice are important, our conditional knockout system is more suitable for detailed analysis of the cell cycle. In the present study, we showed quantitatively the temporal accumulation of metaphase cells by two-dimensional FACS and microscopic analysis of CENP-H-conditional mutant cells. As noted above, 85% of cells were arrested at metaphase and showed aberrant mitotic structures prior to cell death 48 h after addition of tet. This severe phenotype with metaphase arrest is similar to the phenotype of cells injected with anti-CENP-C antibody (Tomkiel et al., 1994). CENP-H is co-localized to CENP-C and is thought to be an inner kinetochore component. Because CENP-H-deficient cells show mislocalization of CENP-C, it is possible that the function of CENP-H is related to the function of CENP-C. Tomkiel et al. (1994) reported that cells injected with anti-CENP-C antibody displayed metaphase arrest and that the kinetochore structure (size) of these cells was altered. They concluded that kinetochores assembled in the absence of CENP-C are structurally weak and that a weak kinetochore can not withstand microtubule-associated forces at metaphase. In CENP-H-deficient cells with metaphase arrest, many chromosomes attach to microtubules, though several chromosomes fail to align at the metaphase plate (Figure 5). The phenotype of our cells and the reported phenotype of CENP-C-disrupted cells suggest that CENP-H-deficient cells lack proper kinetochore function and that kinetochore structure may be disrupted, as shown in cells injected with anti-CENP-C antibody. The metaphase arrest observed in CENP-H-deficient cells could be explained by the disruption of functional kinetochore structures. Even if microtubules attach to the kinetochores in CENP-H-deficient cells, the attachment is not strong to enough to ensure proper chromosome segregation, and the cells arrest at metaphase. To clarify this point, it will be important to observe directly the kinetochore structure of CENP-H-deficient cells by electron microscopy.
After a long period of metaphase arrest, CENP-H-deficient cells proceeded through the cell cycle without normal chromosome segregation. We observed cells with an 8N DNA content by FACS analysis (Figure 6A) and we also detected tetraploid cells that had multiple spindle structures (Figure 5A). This phenotype is likely to reflect the kinetochore structure and function. Although kinetochore function of #5-5 cells is weak and unstable 48 h after addition of tet, cells still have partial function. However, kinetochore function, including mitotic checkpoint function, seems to be completely disrupted by 72 h. We consistently observed immunofluorescence signals for an outer centromere protein ZW10 48 h after addition of tet, although the signal intensity was weak (Figure 6C). However, we did not observe ZW10 signal 72 h after addition of tet (Figure 6C), suggesting that the kinetochore structure is completely disrupted by 72 h. Cells arrested in metaphase finally lose mitotic checkpoint function and proceed through the cell cycle without normal chromosome segregation. The cells then form the nuclear membrane around the near-tetraploid genome. If these cells proceed through an additional cell cycle, multiple- or micro-nuclei can be present, and the cells finally die due to apoptosis.
CENP-H is required for centromere targeting of CENP-C but not CENP-A
Immunofluorescence analysis has provided insight into the formation of the prekinetochore structure and the role of CENP-H in this structure. Depletion of CENP-H does not alter the discrete, typical centromeric signals of CENP-A. CENP-A is a centromere-specific histone H3 and has an essential function in structural organization of centromeric chromatin (Shelby et al., 1997; Henikoff et al., 2000; Howman et al., 2000; Takahashi et al., 2000). Our results indicate that targeting of CENP-A to the centromere is not mediated by CENP-H. Anti-CENP-C antibody was used to investigate further whether a normal functional centromere is formed in CENP-H-deficient cells. CENP-C is a good marker for this purpose because the protein is present only on active centromeres (Earnshaw et al., 1989; Sullivan and Schwartz, 1995) and is functionally essential (Fukagawa and Brown, 1997; Kalitsis et al., 1998). In the present study, depletion of CENP-H led to an absence of discrete CENP-C signals on interphase centromeres, indicating that CENP-H is needed to recruit CENP-C to form a functional centromere. The observed failure of normal centromere formation during interphase can also explain the severe mitotic defects observed in CENP-H-deficient cells.
Considering the present data and data from recent reports, we would like to discuss functional centromere formation in cells of higher vertebrates. The first step in centromere formation is formation of CENP-A-containing nucleosomes, and CENP-H is then targeted to these structures. CENP-C then interacts with the CENP-A– CENP-H structure to form a prekinetochore. In mitosis, the components of the prekinetochore form an inner plate. Centromere proteins at the interzone, outer plate and fibrous corona associate with the inner plate to form a complete centromere structure that can ensure microtubule attachment and faithful chromosome segregation during mitosis. It is unclear whether CENP-H binds directly to CENP-A or CENP-C to form the prekinetochore. We have not detected direct binding of these proteins by in vitro biochemical analysis (Sugata et al., 2000). It is possible, however, that CENP-H binds CENP-A and/or CENP-C directly in vivo. Alternatively, CENP-H may be targeted to a CENP-A structure through the actions of unknown factors. In either case, CENP-H is required for targeting of CENP-C to form the prekinetochore.
The prekinetochore exists throughout the cell cycle and marks the locations of centromeres on chromosomes. Recent data suggest that centromere formation does not depend strictly on DNA sequences, but also requires epigenetic factors (Karpen and Allshire, 1997; du Sart et al., 1997; Williams et al., 1998). One candidate for an epigenetic factor is CENP-A. There are two models of centromere loading of CENP-A. In the replication model, CENP-A expression is coupled to replication of centromeric DNA, and CENP-A is incorporated into the centromere (Csink and Henikoff, 1998). The alternative model proposes that synthesis of CENP-A is uncoupled from replication of centromeric DNA. Centromere-specific chromatin assembly occurs through the actions of chromatin assembly factors that aid in deposition of CENP-A on the newly synthesized DNA. Shelby et al. (2000) showed recently that synthesis of CENP-A is uncoupled from replication of centromeric DNA. If the second model is correct, the CENP-A-specific assembly factors can be identified and characterized to understand formation of the prekinetochore. One candidate assembly factor is Mis6, which appears to be necessary for recruiting CENP-A to form proper centromeres in fission yeast (Takahashi et al., 2000). The DT40 system described in the present study will be useful for further investigations of the mechanism of prekinetochore formation. Creation of a collection of DT40 conditional mutants, including mutants for CENP-A and Mis6, will contribute to our understanding of mitosis in higher eukaryotes.
Materials and methods
Molecular biology
The chicken CENP-H gene was isolated by degenerate RT–PCR of mRNA extracted from DT40 cells. The sequences of the degenerate primers were 5′-GC(ACT)CA(AG)AC(ACT)AA(AG)CA(AG)CA(AG)CT and 5′-TT(CT)TG(CT)TT(AG)TT(CT)TT(CT)TC. We screened a chicken cDNA library (Stratagene) using the RT–PCR product as a probe. Several independent positive clones were sequenced and identified as full-length chicken CENP-H cDNAs. We used the chicken CENP-H cDNA as a probe to isolate genomic DNA clones specific for CENP-H from a DT40 genomic DNA library as described (Fukagawa and Brown, 1997). The genomic structure of the CENP-H clone thus obtained was determined by PCR and partial sequencing.
All plasmids were constructed by standard methods. Full-length CENP-H cDNA was cloned into the EcoRI site of pUHD10-3 (Gossen and Bujard, 1992) to yield the tet-sensitive expression plasmid pUHD-CENP-H. To generate the ptTA-zeo construct, the zeo-resistance cassette driven by the β-actin promoter was inserted into pUHD15-1, which contains the tet-responsive transactivator gene under control of the CMV promoter. The left and right arms of the disruption construct were amplified as 2.0 and 3.5 kb fragments by long-range PCR with a genomic clone as the template, and were cloned into pBluescript (Stratagene). For the CENP-H-puro disruption construct, the puro-resistance cassette under control of the β-actin promoter was inserted between the two arms.
To generate the replacement construct for the CENP-H–GFP fusion gene, we cloned a 10 kb fragment containing the last exon of CENP-H into pBluescript and replaced the stop codon in the last exon with a GFP sequence followed by a neo-resistance cassette driven by the β-actin promoter. Targeted integration of this replacement construct was expected to replace the native CENP-H gene with the CENP-H–GFP fusion gene.
Cell culture and transfection
DT40 cells were cultured and transfected as described previously (Buerstedde and Takeda, 1991). G418 (Sigma) was used at a final concentration of 2 mg/ml, puromycin (Clontech) at a final concentration of 0.5 µg/ml and zeo (Invitrogen) at a final concentration of 1 mg/ml to select for stable transfectants. To suppress expression of the tetracycline-responsive CENP-H transgene, tet (Sigma) was added to the culture medium to a final concentration of 2 µg/ml. Δ/CENP-C-ER DT40 cells were cultured in the presence of OHT (Sigma) at a final concentration of 100 nM.
Cell cycle analysis
Cells were pulse labeled at the times indicated in Figure 4 with 20 µM BrdU (Roche) for 20 min. Cells were harvested and fixed at 4°C overnight with 70% ethanol. After treatment with 4 M HCl in 0.5% Triton X-100 for 30 min, cells were incubated with anti-BrdU primary antibody (Becton Dickinson) for 1 h at room temperature and then with fluorescein isothiocyanate (FITC)-conjugated anti-mouse IgG (Jackson Immuno Research Laboratories) for 30 min at room temperature. Cells were stained with 5 µg/ml propidium iodide (Sigma) in phosphate-buffered saline (PBS). Between incubations, cells were washed with PBS containing 1% bovine serum albumin (BSA). Subsequent flow cytometry was performed with an Epics Altra cytometer (Beckman–Coulter). Fluorescence data were displayed as dot plots using Altra analysis software (Beckman-Coulter).
Histone H1 kinase activity in p34CDC2 immunoprecipitates was measured as described previously (Fukagawa et al., 1999a). Labeled H1 in dried gels was visualized and quantitated with a STORM imager (Amersham Pharmacia).
Antibody production
A chicken CENP-H expression construct (amino acids 1–123) was created in vector pET28a (Novagen). The histidine-tagged recombinant protein was expressed in Escherichia coli BL21 (DE3) after 4 h induction with 0.5 mM isopropyl-β-d-thiogalactopyranoside and purified by nickel column chromatography. Purified recombinant CENP-H was used to immunize a rabbit. Serum was affinity purified against recombinant CENP-H protein on a CNBr-activated Sepharose 4B column.
Immunocytochemistry and FISH analysis
Immunofluorescent staining of whole cells was performed as described previously (Fukagawa et al., 1999a). Cells were collected onto slides with a cytocentrifuge, fixed in 3% paraformaldehyde in PBS for 15 min at room temperature, permeabilized in 0.5% NP-40 in PBS for 15 min at room temperature, rinsed three times in 0.5% BSA and incubated for 1 h at 37°C in anti-α-tubulin antibody (1:50; Sigma), anti-LaminB antibody (1:50; MartiTect), rabbit anti-CENP-A antibody (1:50; V.Regnier and W.Brown, unpublished), rabbit anti-CENP-C antibody (1:1000; Fukagawa et al., 1999a) or rabbit anti-CENP-H antibody (1:2000). Binding of primary antibodies was then detected using either FITC-conjugated rabbit anti-mouse IgG diluted 1:200 (Jackson Immuno Research Laboratories) or Cy3-conjugated goat anti-rabbit IgG diluted 1:1000 (Amersham) in PBS/0.5% BSA. Aberrant metaphase cells or mitotic spindles were observed and scored at the indicated time points. Approximately 2000 cells were scored for each time point.
TUNEL assay was performed using an apoptosis detection system (Promega). Cells were collected onto slides with a cytocentrifuge, fixed in 3% paraformaldehyde in PBS for 15 min at room temperature and permeabilized in 0.2% Triton X-100 in PBS for 15 min at room temperature. DNA breaks were detected with terminal nucleotidyl transferase and fluorescein dUTP.
FISH was performed as described previously (Fukagawa et al., 1999b). Biotin-labeled painting probes were created with degenerate-oligo-primed (DOP)-PCR as described previously (Fukagawa et al., 1999b). The probe for mapping of the CENP-H gene was labeled by nick translation. Probes were detected with Cy3-conjugated avidin (Amersham, 1:500 dilution with 4× SSC, 0.05% Tween-20, 1% BSA). Chromosomes and nuclei were counterstained with DAPI at 0.2 µg/ml in Vectashield antifade (Vector Labs). For the chromosome stability assay, the number of chromosomes 1, 2 and 3 (seven in normal cells) were scored in at least 200 metaphase cells at the time points indicated.
All immunofluorescence and FISH images were collected with a cooled CCD camera (Photometrics Image Point) mounted on a Zeiss Axioscope microscope with a 63× objective (Zeiss) together with a filter wheel. Images were manipulated with Iplab software (Signal Analytics).
Fluorescence microscopy in living cells
For fluorescence staining of chromosomes, Hoechst 33342 was added to the culture medium to a final concentration of 100 ng/ml. Cells were stained with Hoechst 33342 for 10–15 min and washed three times with culture medium. For microscopy observation, HEPES buffer pH 7.4 was added to a final concentration of 20 mM to avoid the need for CO2 gas. Fluorescently stained living cells were observed using an Olympus inverted microscope IX70 with an oil immersion objective lens (PlanApo 60×, NA = 1.40). The DeltaVision microscope system used in this study was purchased from Applied Precision Inc. For temperature control during microscope observation, the system was assembled in a custom-made, temperature-controlled room. Time-lapse images were recorded at 2 min intervals with an exposure time of 0.2–0.5 s.
Acknowledgments
Acknowledgements
The authors are very grateful to Ms Y.Miyauchi, Ms K.Suzuki and Ms K.Kita for technical assistance, Dr E.Nigg for the anti-CDC2 antibody and Dr C.Pendon for the anti-ZW10 antibody. This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas (A) ‘Genome Science’ and (C) ‘Genome Biology’ and ‘Cancer Biology’ from the Ministry of Education, Science, Sports and Culture of Japan.
References
- Amon A. (1999) The spindle checkpoint. Curr. Opin. Genet. Dev., 9, 69–75. [DOI] [PubMed] [Google Scholar]
- Brown M.T. (1995) Sequence similarities between the yeast chromosome segregation protein Mif2 and the mammalian centromere protein CENP-C. Gene, 160, 111–116. [DOI] [PubMed] [Google Scholar]
- Brown M.T., Goetsch,L. and Hartwell,L.H. (1993) MIF2 is required for mitotic integrity during anaphase spindle elongation in Saccharomyces cerevisiae. J. Cell Biol., 123, 387–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buerstedde J.M. and Takeda,S. (1991) Increased ratio of targeted to random integration after transfection of chicken B cell lines. Cell, 67, 179–188. [DOI] [PubMed] [Google Scholar]
- Chan G.K., Jablonski,S.A., Starr,D.A., Goldberg,M.L. and Yen,T.J. (2000) Human Zw10 and ROD are mitotic checkpoint proteins that bind to kinetochores. Nature Cell Biol., 2, 944–947. [DOI] [PubMed] [Google Scholar]
- Chen R.-H., Water,J.W., Salmon,E.D. and Murray,A.W. (1996) Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science, 274, 242–246. [DOI] [PubMed] [Google Scholar]
- Choo K.H.A. (1997) The Centromere. Oxford University Press, Oxford, UK.
- Choo K.H.A. (2000) Centromerization. Trends Cell Biol., 10, 182–188. [DOI] [PubMed] [Google Scholar]
- Compton D.A., Yen,T.J. and Cleveland,D.W. (1991) Identification of novel centromere/kinetochore-associated proteins using monoclonal antibodies generated against human mitotic chromosome scaffolds. J. Cell Biol., 112, 1083–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig J.M., Earnshaw,W.C. and Vagnarelli,P. (1999) Mammalian centromeres: DNA sequence, protein composition and role in cell cycle progression. Exp. Cell Res., 246, 249–262. [DOI] [PubMed] [Google Scholar]
- Csink A.K. and Henikoff,S. (1998) Something from nothing: the evolution and utility of satellite repeats. Trends Genet., 14, 200–204. [DOI] [PubMed] [Google Scholar]
- Cutts S.M. et al. (1999) Defective chromosome segregation, microtubule bundling and nuclear bridging in inner centromere protein gene (Incenp)-disrupted mice. Hum. Mol. Genet., 8, 1145–1155. [DOI] [PubMed] [Google Scholar]
- Dobie K.W., Hari,K.L., Maggert,K.A. and Karpen,G.H. (1999) Centromere proteins and chromosome inheritance: a complex affair. Curr. Opin. Genet. Dev., 9, 206–217. [DOI] [PubMed] [Google Scholar]
- Doheny K.F., Soger,P.K., Hyman,A.A., Tugendreich,S., Spencer,F. and Hieter,P. (1993) Identification of essential components of the S.cerevisiae kinetochore. Cell, 73, 761–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- du Sart D. et al. (1997) A functional neo-centromere formed through activation of a latent human centromere and consisting of non-α satellite DNA. Nature Genet., 16, 144–152. [DOI] [PubMed] [Google Scholar]
- Earnshaw W.C., Ratrie,H. and Stetten,A.C.G. (1989) Visualization of centromere proteins CENP-B and CENP-C on a stable dicentric chromosome in cytological spreads. Chromosoma, 98, 1–12. [DOI] [PubMed] [Google Scholar]
- Fukagawa T. and Brown,W.R.A. (1997) Efficient conditional mutation of the vertebrate CENP-C gene. Hum. Mol. Genet., 6, 2301–2308. [DOI] [PubMed] [Google Scholar]
- Fukagawa T., Pendon,C., Morris,J. and Brown,W. (1999a) CENP-C is necessary but not sufficient to induce formation of functional centromere. EMBO J., 18, 4196–4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa T., Hayward,N., Yang,J., Azzalin,C., Griffin,D., Stewart,A.F. and Brown,W. (1999b) The chicken HPRT gene: a counter selectable marker for the DT40 cell line. Nucleic Acids Res., 27, 1966–1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen M. and Bujard,H. (1992) Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl Acad. Sci. USA, 89, 5547–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Ahmad,K., Platero,J.S. and van Steense,B. (2000) Heterochromatic deposition of centromeric histone H3-like proteins. Proc. Natl Acad. Sci. USA, 97, 716–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howman E.V., Fowler,K.J., Newson,A.J., Redward,S., MacDonald,A.C., Kalitsis,P. and Choo,K.H.A. (2000) Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc. Natl Acad. Sci. USA, 97, 1148–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson H. et al. (1998) Centromere protein B null mice are mitotically and meiotically normal but have lower body and testis weight. J. Cell Biol., 141, 309–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalitsis P., Fowler,K.J., Earle,E., Hill,J. and Choo,K.H.A. (1998) Targeted disruption of mouse centromere protein C gene leads to mitotic disarray and early embryo death. Proc. Natl Acad. Sci. USA, 95, 576–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen G.H. and Allshire,R.C. (1997) The case for epigenetic effects on centromere identity and function. Trends Genet., 13, 489–496. [DOI] [PubMed] [Google Scholar]
- Lanini L. and McKeon,F. (1995) Domains required for CENP-C assembly at the kinetochore. Mol. Biol. Cell, 6, 1049–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengauer C., Kinzler,K.W. and Vogelstein,B. (1998) Genetic instabilities in human cancers. Nature, 396, 643–649. [DOI] [PubMed] [Google Scholar]
- Meluh P.B. and Koshland,D. (1995) Evidence that the Mif2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol. Biol. Cell, 6, 793–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh P.B. and Koshland,D. (1997) Budding yeast centromere composition and assembly as revealed by in vivo cross-linking. Genes Dev., 11, 3401–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh P.B., Yang,P., Glowczewski,L., Koshland,D. and Smith,M. (1998) Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell, 94, 607–613. [DOI] [PubMed] [Google Scholar]
- Moroi Y., Peebles,C., Fritzler,M.J., Steigerwald,J. and Tan,E.M. (1980) Autoantibody to centromere (kinetochore) in scleroderma sera. Proc. Natl Acad. Sci. USA, 77, 1627–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux A.L. and Allshire,R.C. (2000) Centromeres: getting a grip of chromosomes. Curr. Opin. Cell Biol., 12, 308–319. [DOI] [PubMed] [Google Scholar]
- Pluta A.F., Mackay,A.M., Ainsztein,A.M., Goldberg,I.G. and Earnshaw,W.C. (1995) The centromere: hub of chromosomal activities. Science, 270, 1591–1594. [DOI] [PubMed] [Google Scholar]
- Saitoh H., Tomkiel,J., Cooke,C.A., Ratrie,H.,III, Maurer,M., Rothfield,N.F. and Earnshaw,W.C. (1992) CENP-C, an autoantigen in scleroderma, is a component of the human inner kinetochore plate. Cell, 70, 115–125. [DOI] [PubMed] [Google Scholar]
- Shelby R.D., Vafa,O. and Sullivan,K.F. (1997) Assembly of CENP-A into centromere chromatin requires a cooperative array of nucleosomal DNA contact sites. J. Cell Biol., 136, 501–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelby R.D., Monier,K.M. and Sullivan,K.F. (2000) Chromatin assembly at kinetochores is uncoupled from DNA replication. J. Cell Biol., 151, 1113–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr D.A., Williams,B.C., Li,Z., Etemad-Moghadam,B., Dawe,R.K. and Goldberg,M.L. (1997) Conservation of centromere/kinetochore protein ZW10. J. Cell Biol., 138, 1289–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoler S., Keith,K.C., Curnick,K.E. and Fitzgerald-Hayes,M. (1995) A mutation in CSE4, an essential gene encoding a novel chromatin associated protein in yeast, cause chromosomes nondisjunction and cell cycle arrest at mitosis. Genes Dev., 9, 573–586. [DOI] [PubMed] [Google Scholar]
- Sugata N., Munekata,E. and Todokoro,K. (1999) Characterization of a novel kinetochore protein, CENP-H. J. Biol. Chem., 274, 27343–27346. [DOI] [PubMed] [Google Scholar]
- Sugata N., Li,S., Earnshaw,W.C., Yen,T.J. Yoda,K., Masumoto,H., Munekata,E., Warburton,P.E. and Todokoro,K. (2000) Human CENP-H multimers colocalize with CENP-A and CENP-C at active centromere–kinetochore complexes. Hum. Mol. Genet., 9, 2919–2926. [DOI] [PubMed] [Google Scholar]
- Sullivan B.A. and Schwartz,S. (1995) Identification of centromeric antigens in dicentric Robertsonian translocations: CENP-C and CENP-E are necessary components of functional centromeres. Hum. Mol. Genet., 4, 2189–2197. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Chen,E.S. and Yanagida,M. (2000) Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science, 288, 2215–2219. [DOI] [PubMed] [Google Scholar]
- Taylor S.S. and McKeon,F. (1997) Kinetochore localization of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage. Cell, 89, 727–735. [DOI] [PubMed] [Google Scholar]
- Taylor S.S., Ha,E. and McKeon,F. (1998) The human homologue of Bub3 is required for kinetochore localization of Bub1 and a Mad3/Bub1 related protein kinase. J. Cell Biol., 142, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomkiel J., Cooke,C.A., Saitoh,H., Bernat,R.L. and Earnshaw,W.C. (1994) CENP-C is required for maintaining proper kinetochore size and for a timely transition to anaphase. J. Cell Biol., 125, 531–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler-Smith C. and Floridia,G. (2000) Many paths to the top of the mountain: diverse evolutionary solutions to centromere structure. Cell, 102, 5–8. [DOI] [PubMed] [Google Scholar]
- Uren A.G., Wong,L., Pakusch,M., Fowler,K.J., Burrows,F.J., Vaux,D.L. and Choo,K.H.A. (2000) Survivin and inner centromere protein INCENP show similar cell-cycle localization and gene knockout phenotype. Curr. Biol., 10, 1319–1328. [DOI] [PubMed] [Google Scholar]
- Vafa O. and Sullivan,K.F. (1997) Chromatin containing CENP-A and α-satellite DNA is a major component of the inner kinetochore plate. Curr. Biol., 7, 897–900. [DOI] [PubMed] [Google Scholar]
- Wang J., Takagaki,Y. and Manley,J.L. (1996) Targeted disruption of an essential vertebrate gene: ASF/SF2 is required for cell viability. Genes Dev., 10, 2588–2599. [DOI] [PubMed] [Google Scholar]
- Warburton P.E. et al. (1997) Immunolocalization of CENP-A suggests a novel nucleosome structure at the inner kinetochore plate of active centromeres. Curr. Biol. 7, 901–904. [DOI] [PubMed] [Google Scholar]
- Williams B.C., Murphy,T.D., Goldberg,M.L. and Karpen,G.H. (1998) Neocentromere activity of structurally acentric mini-chromosomes in Drosophila. Nature Genet., 18, 30–37. [DOI] [PubMed] [Google Scholar]
- Yang C.H., Tomkiel,J., Saitoh,H., Johnson,D.H. and Earnshaw,W.C. (1996) Identification of overlapping DNA-binding and centromere-targeting domains in the human kinetochore protein CENP-C. Mol. Cell. Biol., 16, 3576–3586. [DOI] [PMC free article] [PubMed] [Google Scholar]